Significance
Understanding how mammalian organisms achieve the full diversity of cell types in the adult organism is a central goal of developmental cell biology. Recent work has shown that some embryonic precursor cells can self-organize into developmental structures but the mechanisms of gene regulation that contribute to this process remain unknown. Here we show embryonic stem cells self-organize into distinct gene expression states that resemble developmental gene programs. We find that microRNAs, small noncoding regulators of gene expression, play a critical role in organizing fluctuations across gene networks to help achieve this organization into distinct expression states.
Keywords: microRNAs, cell states, embryonic stem cells, heterogeneity, variation
Abstract
Pluripotent embryonic stem cells (ESCs) contain the potential to form a diverse array of cells with distinct gene expression states, namely the cells of the adult vertebrate. Classically, diversity has been attributed to cells sensing their position with respect to external morphogen gradients. However, an alternative is that diversity arises in part from cooption of fluctuations in the gene regulatory network. Here we find ESCs exhibit intrinsic heterogeneity in the absence of external gradients by forming interconverting cell states. States vary in developmental gene expression programs and display distinct activity of microRNAs (miRNAs). Notably, miRNAs act on neighborhoods of pluripotency genes to increase variation of target genes and cell states. Loss of miRNAs that vary across states reduces target variation and delays state transitions, suggesting variable miRNAs organize and propagate variation to promote state transitions. Together these findings provide insight into how a gene regulatory network can coopt variation intrinsic to cell systems to form robust gene expression states. Interactions between intrinsic heterogeneity and environmental signals may help achieve developmental outcomes.
Alan Turing first proposed the existence of chemical “morphogens” that could impart pattern formation during development (1). Later, Lewis Wolpert proposed that gradual differences in morphogen concentrations across cells’ external environment could suffice to define many different patterns (2–4). While organisms such as Drosophila and Xenopus develop by utilizing maternally deposited asymmetric morphogen gradients, mammals are unique because early stages of development take place without such obvious gradients (5). Investigations have aimed to recapitulate aspects of mammalian embryonic development through mixing cell types that signal to each other or supplying specified agonists in culture medium, often with external scaffolds to help organize cells (6–9). These studies have revealed “self-organized” body patterning axes, with formation of these axes relying to differing extents on the external signals provided by morphogens or scaffolds. Embryonic stem cells (ESCs) have proven to be particularly useful models for self-organizing processes that may shape the early embryo (6, 10, 11). Yet, the extent to which genetically identical cells can inherently generate diversity without external signals has remained less clear, as has a mechanism for such a phenomenon. This would require the gene regulatory network to give a variable output across cells that receive similar input. Prevailing views ascribe spontaneously arising cell-to-cell variation in gene expression to stochastic processes at gene loci, which include phenomena such as transcriptional bursting (12–19). However, an alternative hypothesis is that naturally arising variation within cell systems can be coordinated across multiple, specific loci by gene regulatory elements and coopted by the cell to enable diversification.
In ESCs, the core transcriptional regulatory network consists of pluripotency genes such as Pou5f1 (Oct4), Sox2, and Nanog (together “OSN”) (20–23). This regulatory network also contains microRNAs (miRNAs), small RNAs that bind and regulate genes in mammals through Argonaute (Ago) effector proteins (24), of which Ago2 is the principally active form in mammals. ESCs are known to express Nanog and Sox2 in a variable, heterogeneous fashion across cell populations (15, 25–30). The degree of cell-to-cell variation for Nanog and Sox2 is higher than that observed for Oct4 (15, 31, 32), though the molecular basis of this difference is unknown, as is the basis of Nanog and Sox2 heterogeneity. Additionally, the full extent to which the core ESC regulatory network can be subdivided based on factors driving cell-to-cell variation is unclear. In this study, we find ESCs exhibit intrinsic heterogeneity through formation of interconverting cell states in the absence of external gradients. Networks of genes and miRNAs that vary between states regulate each other, forming a circuit for variation that includes Nanog, Sox2, and Esrrb but not Pou5f1, Tcf3, or Smad1. These findings imply that the core transcriptional gene regulatory network of ESCs contains a subcircuit that coherently amplifies variation to achieve transition to new states.
Results
ESCs Exhibit Intrinsic Variation between States Expressing Distinct Developmental Gene Expression Programs.
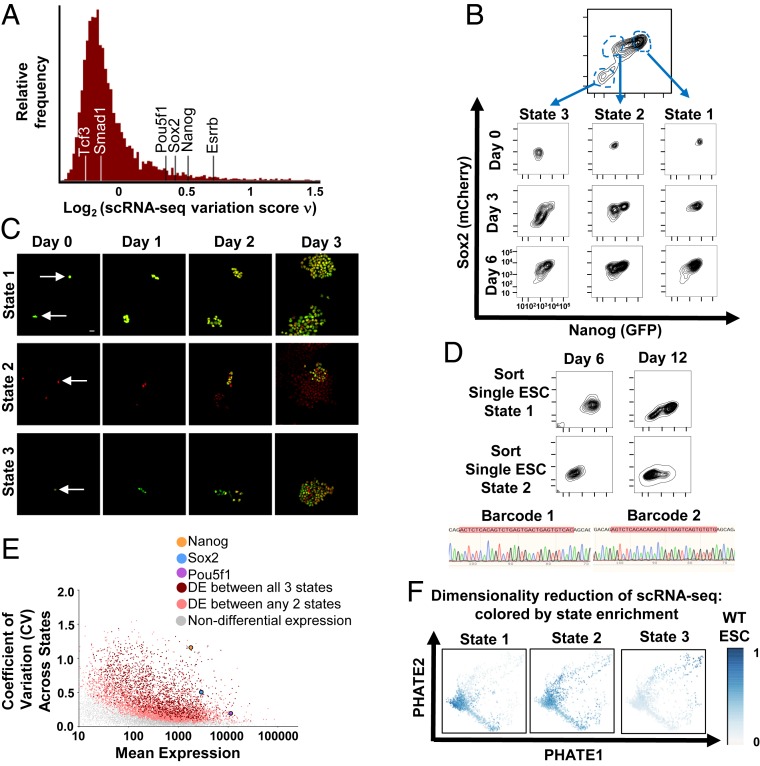
To explore ESC variation, we measured the coding transcriptomes of individual ESCs by single-cell RNA-sequencing (scRNA-seq) and identified highly variable genes by a statistic (ν-score) that corrects the coefficient of variation for technical sampling noise (32). Consistent with previous reports (15, 25, 27–30, 32, 33), we found a remarkably high degree of variation for Nanog and Sox2 transcripts across single ESCs (Fig. 1A). To further investigate variation in these pluripotency factors, we generated cells with heterozygous insertions of fluorophore tags at the endogenous loci of Nanog and Sox2 joined by posttranslational cleavage sequences (GFP-P2A-Nanog and Sox2-P2A-mCherry, respectively; SI Appendix, Fig. S1A). Identically cultured ESCs showed remarkable heterogeneity in levels of Nanog and Sox2 (Fig. 1B and SI Appendix, Fig. S1B). We analyzed the frequency distributions of Nanog and Sox2 levels, noting a dominant peak of high expression with one (Sox2) or two (Nanog) minor peaks at lower expression (SI Appendix, Fig. S1C). Additionally, Sox2-high Nanog-low ESCs have been previously identified (29). Thus, to enable a coarse grain analysis of the continuum of ESC variation, we clustered cells into three predominant states of Nanog and Sox2 expression for subsequent analysis (Fig. 1B: state 1 = high Nanog, high Sox2; state 2 = low Nanog, high Sox2; and state 3 = low Nanog, low Sox2). When cells from these states were isolated by flow cytometric sorting and cultured identically, each state recapitulated the heterogeneity of the parental population (Fig. 1 B, Bottom). To extend this analysis, we sorted single ESCs from each state and assessed their organization and evolving state distribution by fluorescence microscopy. Single ESCs from each state grew into colonies with mixed state morphology, with a high degree of intra- and intercolony variation in state distribution (Fig. 1C), consistent with a previous report tracking Nanog in single-cell–derived ESC colonies (29). We did not detect any reproducible orientation of states with respect to each other, with states 2 to 3 sometimes oriented toward the edges of colonies and sometimes located centrally with respect to state 1 (Fig. 1C and SI Appendix, Fig. S2 A and B). To further delineate whether single ESCs displayed an inherent capacity to organize into states, we introduced a unique molecular barcode into each ESC. We sorted single cells from a given state and assessed their ability to repopulate other states. Over time, single-cell–derived ESCs with unique barcodes switched into other states (Fig. 1D). Together these results establish that single ESCs contain an inherent ability to organize into a cell system containing a distribution of states. Individual cells switch between states to give rise to intrinsic heterogeneity at the population level.
Fig. 1.
Single ESCs exhibit intrinsic variation between cell states. (A) Distribution of single-cell variation test statistic (ν) scores for 7,259 genes across 2,299 well-sampled cells measured by scRNA-seq. Nanog, Sox2, and Esrrb are indicated, as are Pou5f1, Smad1, and Tcf3. (B) ESC labeled by heterozygous insertion of fluorophore tags at the endogenous loci for Nanog and Sox2 (GFP-P2A-Nanog, Sox2-P2A-mCherry) were separated into three distinct cell states by flow cytometric sorting and cultured identically. The population is shown over time. (C) ESCs were isolated from states 1 to 3 by flow cytometric sorting and plated at low density. Cells were analyzed by widefield fluorescence for Nanog (GFP) and Sox2 (mCherry) at the indicated timepoints. (D) A unique barcode was introduced into each ESC (SI Appendix, Supplementary Materials and Methods). Single ESCs from states 1 and 2, respectively, were isolated and cultured. State distribution and sequencing of the barcode region (red highlight) are shown. (E) Coefficient of variation (CV)-mean plot of protein coding gene expression across three states. Genes with differential expression between all three states (red) or between any two states (peach) are highlighted. Nanog, Sox2, and Pou5f1 (Oct4) are indicated. (F) Dimensionality reduction applied to scRNA-seq data. Cells are plotted according to their low dimensional representations by potential of heat-diffusion for affinity-based trajectory embedding (PHATE). Each cell is colored for relative enrichment by gene expression signatures of states 1 to 3 (SI Appendix, Supplementary Materials and Methods).
To gain insight into the observed ESC states, we characterized their coding and noncoding transcriptomes by ribosomal RNA (rRNA)-depleted RNA-seq (GEO: GSE132708). Protein-coding genes differentially expressed between all three states were highly enriched for developmental regulators and certain pluripotency genes (including Nanog, Sox2, and Esrrb) and depleted for housekeeping, cell cycle, metabolic, and other pluripotency genes (including Pou5f1, Smad1, and Tcf3), suggesting variation across states was specific to particular developmental loci (Figs. 1E and 2 A and B and SI Appendix, Fig. S3 A–C). Next, we sought to determine the extent to which the chosen states reflected the scale of variation in ESCs by utilizing these state expression signatures. We represented ESC states in a gene-unbiased manner through dimensionality reduction of scRNA-seq data and colored cells by enrichment for states 1 to 3 expression programs. We chose methods of dimensionality reduction that emphasize progression along trajectories as opposed to clustering into groups (34, 35) to reflect the continuum of variation in ESCs. States 1 to 3, defined by Nanog and Sox2, captured a large portion of variation across single ESCs, as indicated by a gradual progression of cells enriched for each state’s expression across the major axes (Fig. 1F and SI Appendix, Fig. S3D). Further, variation across single cells was related to variation across states 1 to 3 for individual genes, as differentially expressed genes across states (including Nanog, Sox2, and Esrrb) showed relatively high ν-scores (Fig. 1A and SI Appendix, Fig. S3E).
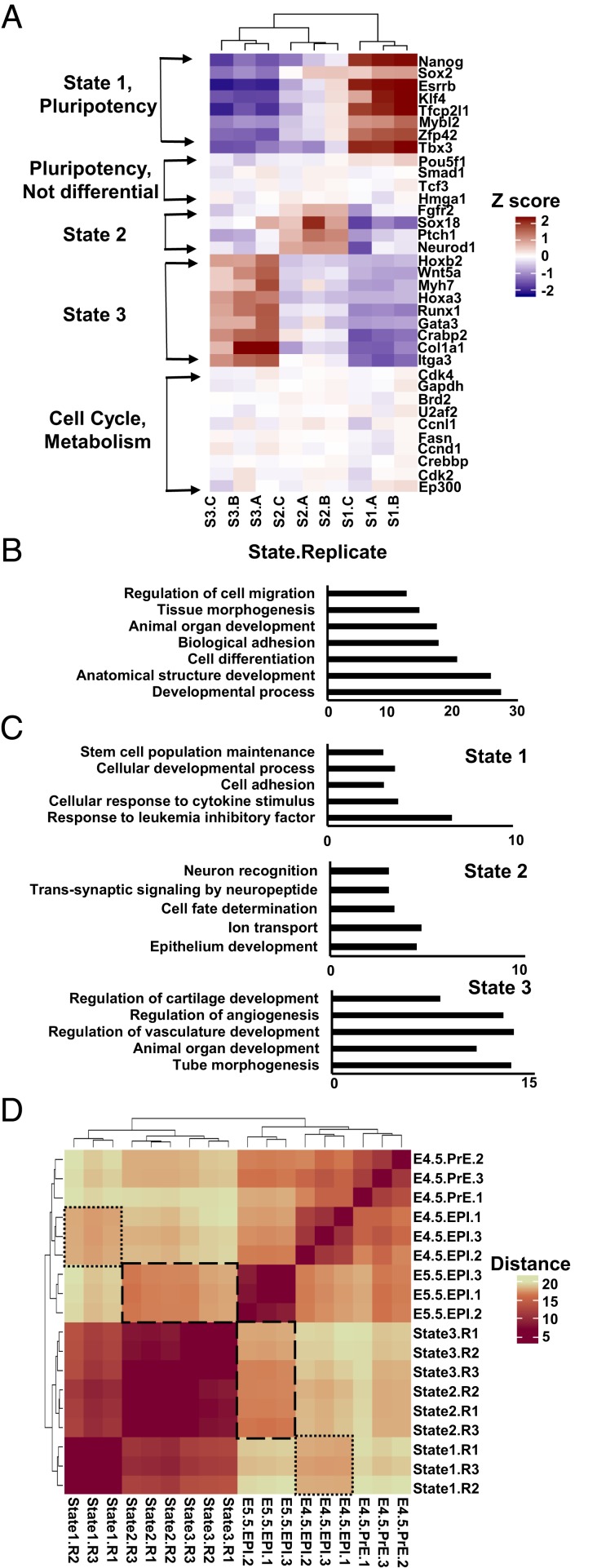
Fig. 2.
ESC states differ in expression of developmental regulators. (A) Heatmap of normalized expression across ESC states for selected genes. Three biological replicates (A–C) are shown. (B) Top gene ontology (GO) analysis terms and P values for genes differentially expressed between all three states. (C) GO analysis terms and P values for top 300 genes uniquely highest expressed in each cell state. (D) Heatmap of gene expression distance between ESC states 1 to 3 compared to expression profiles of embryonic development (E4.5 preepiblasts, E4.5 epiblasts, and E5.5 epiblasts) (36). Highlighted are state 1 vs. E4.5 epiblasts (small dashes) and states 2 to 3 vs. E5.5 epiblasts (large dashes).
Next, we sought to test whether ESC states relate to specific developmental programs. Ontology analysis revealed that state 1 resembles a naïve, cytokine responsive population of cells, whereas state 2 displays increased expression of preectodermal makers such as Sox18 and Neurod1, and state 3 displays increased expression of preendodermal and premesodermal markers such as Gata3 and Hoxa3 (Fig. 2 A and C and SI Appendix, Fig. S3F). We compared states 1 to 3 to characterized gene expression profiles of the mouse blastocyst at developmental stages ranging from E4.5 to E5.5 in embryogenesis (9, 36). We calculated the distance in gene expression between conditions (SI Appendix, Supplementary Materials and Methods). While states 1 to 3 were most similar to each other, state 1 was closer in expression to E4.5 epiblasts than were states 2 and 3, whereas the latter were closer in expression to E5.0 or E5.5 (Fig. 2D and SI Appendix, Fig. S3G). Overall, we find that ESCs contain variation observable as cells transitioning between states that express distinct gene expression programs related to development.
Pluripotency Gene Neighborhoods Are Bound by miRNAs that Increase Target Variation.
Diversification of single cells into new discrete states requires coordinated expression of gene programs, raising the question of how variation is related across different gene loci. To address this question, we constructed gene interaction neighborhoods using network inference methods (32, 37). The neighborhood of a chosen “node” gene represents the set of genes most closely correlated with it and with each other (SI Appendix, Supplementary Materials and Methods). Emphasis of this method on topology helps alleviate artifacts in correlation strength that may arise from the low technical sampling of transcripts in scRNA-seq. The neighborhoods of Nanog, Sox2, and Esrrb are shown (Fig. 3A and SI Appendix, Fig. S4A). We confirmed these in silico inferred neighborhoods were meaningful by testing the covariation of Nanog with neighbors Eif2s2, Esrrb, and Hsp90ab1 by introducing a fluorophore tag at their endogenous locus in Nanog fluorophore-tagged cells. Eif2s2, Esrrb, and Hsp90ab1 all showed covariation with Nanog by this method (SI Appendix, Fig. S4B). Further, the neighborhoods of Nanog, Sox2, and Esrrb all contain each other as members and have additional mutual neighbors (SI Appendix, Fig. S4C), supporting the idea that these genes interact and form an interconnected clique.
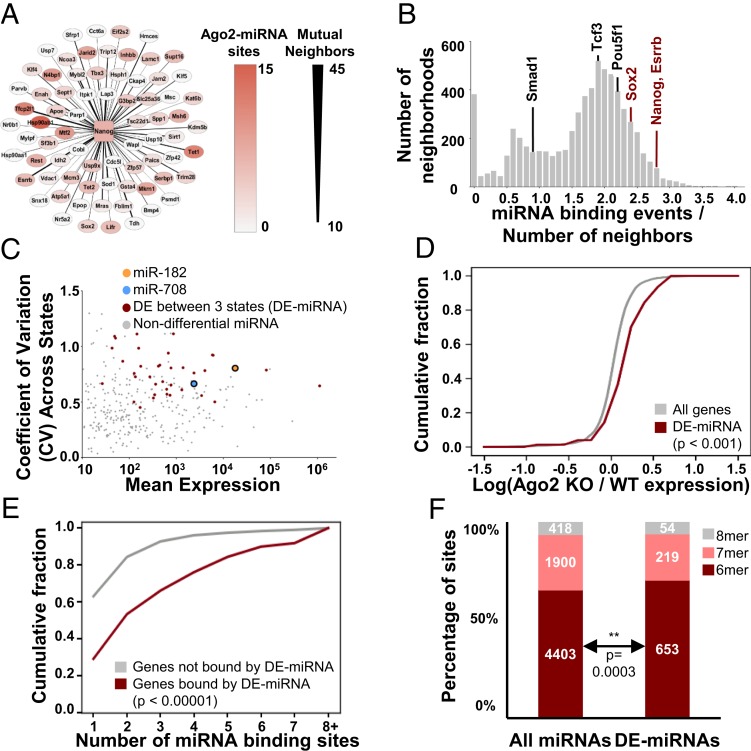
Fig. 3.
Single ESC variation is organized into neighborhoods bound by miRNAs. (A) Nanog interaction neighborhood, constructed by analyzing covariation across single cells. Thickness of Nanog connection represents number of mutual neighbors between Nanog and that gene, red shading indicates degree of Argonaute binding (miRNA activity). (B) Histogram for average miRNA binding per gene (miRNA binding events/number of neighbors) for the 6,577 nonempty neighborhoods produced for all of the genes present in WT scRNA-seq data. Values for Nanog, Sox2, and Esrrb neighborhoods are indicated in red; those for Pou5f1, Smad1, and Tcf3 neighborhoods are shown in black. (C) CV-mean plot for miRNA expression across states. DE-miRNAs are marked in red. MiR-182 and miR-708 are indicated. (D) CDF for the ratio of gene expression in Ago2-inducible ESCs with (WT) or without (KO) Ago2 induction. In these cells, Argonaute 2 is expressed from a doxycycline-inducible transgene in an endogenous Ago1−/−/Ago2−/−/Ago3−/−/Ago4−/− background, and in the absence of doxycycline for 48 h all miRNA activity is lost (43). “WT” ESCs are cultured in 1 μg/mL doxycycline (∼WT Ago2-miRNA levels) and “KO” ESCs are cultured in 0 μg/mL doxycycline for 48 h (<1% remaining miRNA activity). Expression for all genes was measured by RNA-seq. Kolmogorov–Smirnov (K–S) P values are shown. (E) CDF of number of Ago2-miRNAs binding sites for genes where at least one site is assigned to a variable DE-miRNA (red) and genes bound by Ago2-miRNA where no sites are assigned to DE-miRNAs (gray). K–S P value is shown. (F) Distribution of target site affinities (6-mer, 7-mer, or 8-mer matches for the assigned miRNA seed within the cluster of Ago2 binding) for all miRNAs or variable DE-miRNAs. Hypergeometric P value for enrichment is shown.
We analyzed these neighborhoods for molecular characteristics that could account for interactions between member genes giving rise to cell states. miRNAs are intriguing candidate cell state controllers because individual miRNAs can regulate hundreds of genes, which could allow cell-to-cell fluctuations in miRNA to generate relatively large effects on cell state (38). We mapped miRNA binding to target genes in ESCs using Ago cross-linking and immunoprecipitation (CLIP) data (SI Appendix, Supplementary Materials and Methods) (39). Next, we calculated whether neighborhoods were enriched for binding by particular miRNAs by comparing them to matched control neighborhoods constructed to contain the same number of genes of similar expression distribution and total binding of miRNAs (SI Appendix, Supplementary Materials and Methods). We found high miRNA binding of Nanog transcripts and of the entire variable pluripotency gene clique (Fig. 3 A and B and SI Appendix, Fig. S4 A and D). Pluripotency gene neighborhoods were enriched for binding by particular miRNAs, such as miR-182 and miR-708 (SI Appendix, Fig. S4E). Notably, transcripts and neighborhoods of less variable pluripotency genes Pou5f1, Smad1, and Tcf3, showed lesser binding by miRNAs and contained fewer mutual neighbors than variable pluripotency genes (Fig. 3B and SI Appendix, Figs. S4 C and D and S4F). The consistent enrichment of particular miRNAs within neighborhoods suggested a role for miRNA in regulating these neighborhoods.
To test if this was the case, we first determined variably expressed miRNAs (differentially expressed [DE]-miRNA) across states 1 to 3 (Fig. 3C and SI Appendix, Fig. S5A). DE-miRNA included many miRNAs enriched for binding pluripotency gene neighborhoods, including miR-182 and miR-708. Notably, although DE-miRNAs varied across states, their activity did not vary by cell cycle phase (SI Appendix, Fig. S5B, shown for miR-182). Classically, miRNAs are thought to regulate genes by repressing targets through mRNA destabilization or translational inhibition (40). We found increased mean mRNA levels for targets of DE-miRNAs in ESCs deficient for miRNA activity (Fig. 3D and SI Appendix, Fig. S6B) (41–43). Next, we tested the effect of a single DE-miRNA by generating ESCs deficient in miR-182 using CRISPR-Cas9 targeting of the miRNA hairpin loop (Mir182indel), a strategy previously reported to generate functional miRNA knockouts (KOs) (44). We validated that Mir182indel cells had little to no detectable miR-182 activity using reporters (45) (SI Appendix, Fig. S6A). We did not detect changes for miR-182 targets on average across Mir182indel cells (SI Appendix, Fig. S6C), leading us to consider whether DE-miRNA might be acting cooperatively to regulate targets. Consistent with this idea, binding-site analysis indicated that DE-miRNA targets are more highly bound by Ago2-miRNA complexes than nonvariable miRNA targets (Fig. 3E). Interestingly, DE-miRNA binding was skewed toward lower affinity “6-mer” miRNA site type matches and therefore away from higher affinity “7/8-mer” sites (hypergeometric P value = 0.0003) when considering the degree of complementarity between the miRNA and its messenger RNA (mRNA) target (Fig. 3F).
Weak, cooperative interactions are a hallmark of molecular events prone to variation. Strikingly, targets of DE-miRNA showed significantly increased variation across single cells (ν-score) compared to all genes, with particularly high variation for miR-182 targets (Fig. 4A). We analyzed Mir182indel cells by scRNA-seq in parallel to WT cells and noted the increase in variation for miR-182 targets in wild-type (WT) ESCs was lost in Mir182indel ESCs (Fig. 4A). Therefore, while miR-182 did not have a detectable effect on average target expression across all cells, it appeared to have a significant effect increasing cell-to-cell variation of its targets (Fig. 4A).
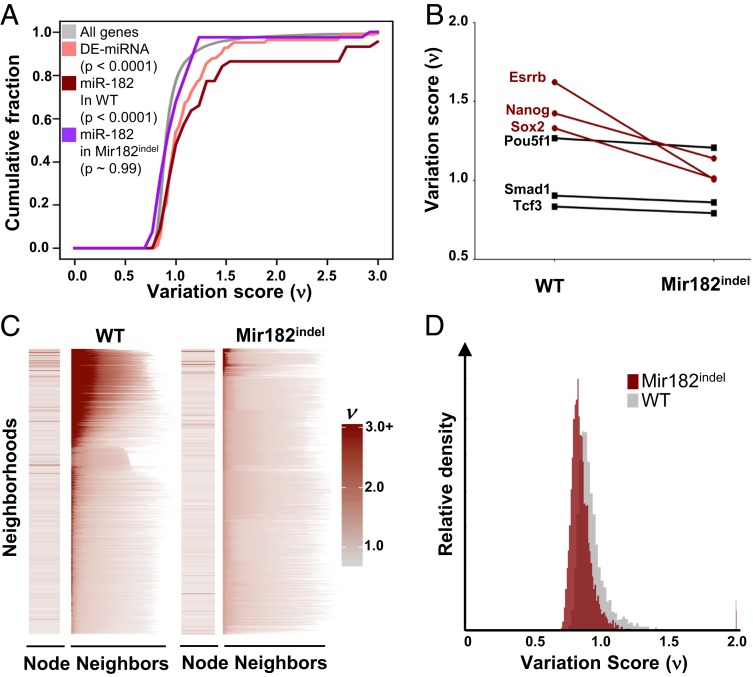
Fig. 4.
ESC variation is coordinated and propagated across neighborhoods. (A) CDF plot of variation score across single cells (ν-score) for all genes (gray), genes targeted by two or more DE-miRNAs (peach), genes targeted by miR-182 in WT ESC (red), and genes targeted by miR-182 in Mir182indel ESC (violet). Kruskal–Wallis P values are shown. (B) Variation score across single cells (ν) for variable pluripotency genes Nanog, Sox2, and Esrrb and less variable (stable) pluripotency genes Pou5f1, Smad1, and Tcf3 in WT vs. Mir182indel ESCs. (C) Variation scores (ν) for all nonempty neighborhoods in WT and Mir182indel ESCs. The variation score of the node gene is shown in the Left column. Next, in each row we plot the score of each node gene’s neighbors (arranged from highest variation score at Left to lowest at Right). Neighborhoods are plotted Top to Bottom by decreasing average variation of all neighbors for WT and Mir182indel ESCs separately. (D) Overlaid histogram of variation (ν-scores) in WT cells and Mir182indel cells for 6,107 genes well-sampled in both WT and Mir182indel cells.
Coordination and Propagation of Variation across Neighborhoods.
Coordinated regulation of gene neighborhoods by miRNAs could provide a mechanism for individual miRNAs to impact the variation of many genes by binding and regulating their interacting neighbors. First, we asked what effect loss of miR-182 activity has on the DE-miRNA–bound variable pluripotency gene clique. We compared the variation across single cells (ν-score) for Nanog, Sox2, and Esrrb in WT vs. Mir182indel ESCs. These genes had lower variation in Mir182indel than in WT ESCs, even if they were not direct targets of miR-182 (Fig. 4B). By contrast, variation for Pou5f1, Smad1, and Tcf3, three pluripotency genes not bound by DE-miRNAs, was unchanged (Fig. 4B). This suggested that miR-182 propagates variability through the neighborhoods it binds in addition to promoting variability of its direct targets. To further assess whether variation can be propagated across neighborhoods, we plotted variation of all genes and their neighbors (Fig. 4C). Remarkably, highly variable genes across single cells showed a strong tendency to group into the same interaction neighborhoods, showing synchronous covariation when measured across a population of uniformly cultured cells (Fig. 4C, note that red shading indicating higher ν-score is clustered in a subset of neighborhoods rather than being evenly distributed across all neighborhoods). This indicates that individual genes do not vary stochastically with respect to each other. Rather, variation is organized at the level of neighborhoods. The degree and concentration of variation is significantly decreased in Mir182indel ESCs (Fig. 4 C and D), consistent with the idea that miR-182 loss reduces cell-to-cell variation of bound targets, which in turn leads to less variation across Mir182indel ESC neighborhoods compared to WT ESC neighborhoods. This synchronized covariation of a group of genes within a cell could give that cell an inherent ability to diversify into a new state.
Variation in miRNA Can Drive Variation in ESC States.
Cell-to-cell variation in DE-miRNA expression and binding of these miRNAs within key neighborhoods could drive cell diversification into new states. This raised the possibility of miRNA regulation of Nanog and Sox2 contributing to the observed distribution of cell states through variation of miRNA levels. We explored qualitative models that could recapitulate the observed distribution of states. Previous work has established that bimodal distributions of target expression can emerge from the interplay between cell-to-cell miRNA variation and the threshold-like response of their targets (45, 46). Thus, we explored minimal qualitative models that could yield the observed distribution of states. We found that a model in which two distinct miRNA pools regulate Nanog and Sox2, one targeting both genes while the other controls Nanog specifically, recapitulated three cell states (SI Appendix, Fig. S7). In this configuration, cell-to-cell variation in miRNA levels results in highly variable expression of Nanog and Sox2. Loss of the shared miRNA pool, Nanog-specific pool, or of all miRNAs results in changes in the distribution of cell states (SI Appendix, Fig. S7).
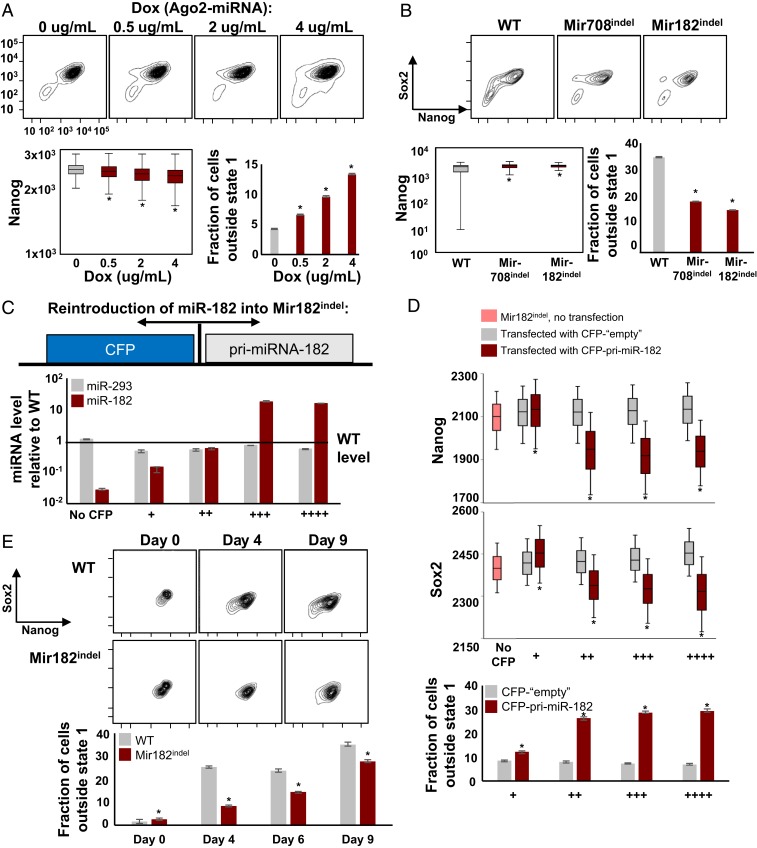
To experimentally test this idea, we measured variation in ESC states in cells with inducible Ago2 expression (and therefore inducible miRNA activity) in an endogenous Ago1−/−/2−/−/3−/−/4−/− background (43). We observed a titratable increase in Nanog variation and cells exiting state 1 with increasing Ago-miRNA activity (Fig. 5A and SI Appendix, Fig. S8A). A similar effect was observed upon loss of individual DE-miRNA, as Mir182indel and Mir708indel ESCs showed reduced state diversity compared to WT, with reduction in variation of Nanog and fewer cells exiting state 1 (Fig. 5B and SI Appendix, Fig. S8B). These results were consistent with the qualitative predictions of our model, and together they established that loss of miRNA could reduce cell state variation in ESCs.
Fig. 5.
Variation in microRNA can drive variation in ESC states. (A) Cell state distributions in Ago2-inducible ESCs (Fig. 3D) labeled at Nanog and Sox2 loci by fluorophores (SI Appendix, Fig. S1). Cells were cultured in the indicated concentrations of doxycycline (Ago2-miRNA) for 48 h. Shown are variation in Nanog (*P < 0.0001, Levene’s test) and the fraction of cells outside state 1 (*P < 0.0001, binomial test). Error is 95% CI from bootstrapping. See also SI Appendix, Fig. S8A. (B) Cell state distributions in WT, Mir182indel, and Mir708indel ESCs labeled at Nanog and Sox2 loci by fluorophores (SI Appendix, Fig. S1). Shown are variation of Nanog and the fraction of cells outside state 1. Statistics were calculated as in A. See also SI Appendix, Fig. S8B. (C) Reintroduction of miR-182 into Mir182indel cells. Mir182indel cells were transfected with a bidirectional expression plasmid expressing pri-miR-182 tightly coupled to cerulean fluorescent protein (CFP), or with a CFP-only plasmid (“empty” control). Cells were isolated by flow cytometric sorting at increasing levels of CFP. miR-182 and miR-293 levels are quantified and plotted relative to WT ESCs. Error is SD for n = 3 replicates. (D) Nanog (GFP) and Sox2 (mCherry) levels in transfected Mir182indel cells, plotted by CFP (miR-182) levels as in C; *P < 0.0001, Levene’s test, pooled maroon vs. gray. Also shown is fraction of cells outside of state 1 (*P < 0.0001, binomial test). See also SI Appendix, Fig. S8C. (E) State 1 WT and Mir182indel ESCs were isolated by flow cytometric sorting and cultured. The fraction of cells outside state 1 is shown (*P < 0.0001, binomial test). See also SI Appendix, Fig. S8D.
To determine if reintroduction of miRNA could restore diversity, we constructed an inducible bidirectional plasmid expressing miR-182 tightly coupled to a fluorophore (CFP) and introduced it into Mir182indel ESCs. This allows single-cell measurement of miR-182, Nanog, and Sox2 levels through their respective fluorophores (CFP, GFP, and mCherry, respectively). CFP levels tracked miR-182 expression and the latter was restored to wild-type levels (“++”) or overexpressed (“+++,” Fig. 5C). As a control, we transfected Mir182indel cells with a CFP-only “empty” plasmid in parallel to reexpression of miR-182. Mir182indel cells with miR-182 reexpressed showed an increase in cell state variation, with an increased fraction of cells outside state 1 and increased variation in Nanog and Sox2 levels (Fig. 5D and SI Appendix, Fig. S8C, note that levels of Nanog and Sox2 change across red “miR-182” bins but stay similar across gray empty bins). This indicates that adding exogenous variation in cell-to-cell miRNA levels by taking advantage of natural variation in plasmid transfection and expression efficiency increases variation in cell states across the population. Finally, we isolated state 1 ESCs from WT and Mir182indel ESCs by flow cytometric sorting and assessed their ability to diversify into other states over time. Mir182indel ESCs were delayed in diversifying out of state 1 compared to WT ESCs (Fig. 5E and SI Appendix, Fig. S8D). This supports the idea that miR-182 fluctuations impact transition of cells out of state 1, which is expected when miRNAs act as feedback repressors of genes subject to transient fluctuations (47). We conclude that cell-to-cell variation in DE-miRNA levels can drive variation in bound pluripotency gene neighborhoods in a subset of cells, enabling their diversification into new states.
Discussion
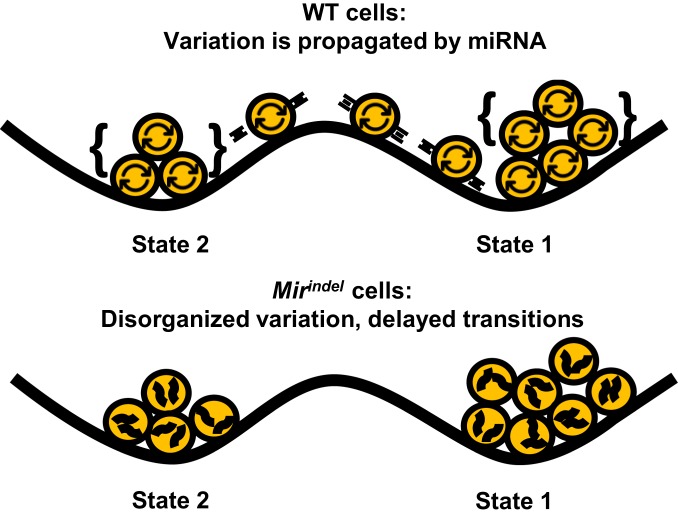
We find naturally arising variation in ESC gene expression can be described by three cell states with distinct expression programs related to embryonic development. Genes that vary across these three states encode specific developmental factors such as Nanog, Sox2, and Esrrb, but not others such as Pou5f1, Tcf3, or Smad1 that are equally well expressed. When isolated, single ESCs form a cell system that recapitulates the heterogeneity of the parental distribution despite the absence of an externally supplied morphogen gradient or scaffold. Variation within the cell system is concentrated at specific genes and miRNAs and excluded from others, forming an integrated genetic subcircuit that organizes variation into three cell states. Deletion of variable miRNAs reduces variation of target genes and reduces propagation of variation across the gene network, resulting in delayed ability for states to repopulate one another (Fig. 6). Reexpression of variable miRNA restores variation. These results define variation within cell systems as a fundamentally regulated process subject to modulation by noncoding elements such as miRNAs.
Fig. 6.
MicroRNAs organize intrinsic variation into cell states. In WT cells, the presence of microRNAs allows coordination of cell-intrinsic fluctuations across loci (curved arrows). Concentration of fluctuations at particular gene neighborhood cliques can lead to a state transition due to changes in expression across co-related genes. In contrast, cells lacking variation-prone miRNAs do not coordinate fluctuations across loci (squiggles) as effectively and are delayed in transitioning between cell states.
These results are consistent with previous reports describing heterogeneity in the expression of pluripotency factors in ESCs (15, 25–30). The molecular basis of this heterogeneity is still under debate, and the results here suggest miRNAs are important to ensure ESC heterogeneity by accelerating the dynamics of interconversion between states. A previous report utilized continuous long-term single-cell tracking to examine Nanog expression dynamics (29), similarly identifying that Nanog-low/negative ESCs can revert to high Nanog expression under standard culture conditions. Interestingly, this study found that the context of cells surrounding Nanog-low cells influenced their propensity to differentiate into precursor cells expressing Foxa2 and Sox1. Future work aimed at understanding how the intrinsic state within a cell interacts with the milieu of states surrounding it to achieve appropriate diversity will be of great interest.
Our results show variable miRNAs such as miR-182 act on a clique of variable transcription factors (Nanog, Sox2, and Esrrb). More broadly, we show that variation across ESCs is organized into a specific subset of interacting gene neighborhoods. This contrasts with stochastic models in which variation at different loci is unrelated to one another or all loci are equally variable. Further, we find genes that vary between states encode developmental factors such as Nanog and Sox2. Since Nanog and Sox2 are themselves involved in promoting expression of many miRNAs (24), this provides opportunity for higher-order organization of intrinsic fluctuations within cells. Even within a genetically identical population of cells cultured under uniform conditions, fluctuations will inevitably arise in a subset of cells. If these fluctuations are always focused to a particular set of interacting genes in a highly ordered way, as identified here, and those genes encode developmental factors, variation of these factors within this subset of cells will inevitably result in their transition to another developmental cell state. This causes intrinsic heterogeneity or the ability of cells to repopulate specific cell states when separated.
Previous descriptions of cell-to-cell variation in gene expression have attributed it to noise or stochastic processes (12–19). Yet we find manipulating the level of miR-182 in ESCs can directly change cell-to-cell variation in Nanog levels. The presence of a genetic element (in this case, miRNA) whose manipulation explicitly impacts the distribution of a gene’s expression across cells indicates distribution of expression across cells for a gene is in fact highly regulated.
We identified a role for miRNA to increase variation, in contrast to their previously emphasized role reducing cell-to-cell variation in gene expression (48). Many additional gene regulatory components beyond miRNAs, such as enhancers, RNA binding proteins, or splicing factors may also contribute to cell state variation by virtue of their ability to regulate many genes. Although challenging, it will be important to further define the structural and regulatory features of molecular classes that vary in activity cell to cell.
No matter how cell-to-cell variation originates, gene regulatory networks are poised to organize and direct it to achieve diversification of states in a robust manner without requiring varied input to each cell from an external gradient. Thus, organization of variation intrinsic to cell systems is a sufficient principle to allow for single cells to diversify into distinct states, providing a mechanism for cell-intrinsic symmetry breaking. This feature of gene regulatory networks may in part explain how self-organization occurs, and its description here builds upon previously observed links between gene expression variation and differentiation (49–51). We show that single ESCs can both self-organize into three states and that these states can repopulate one another. In adult tissues such as intestine and lung, stem-like cell types have shown the ability to repopulate each other (52–54). Development of tissues is also known to be intimately tied to miRNA function (47). While cells in complex tissues receive external signals in addition to experiencing intrinsic fluctuations, these may cooperate in homeostatic ratios of cell states. Future studies will be necessary to determine the extent to which phenomena in mammalian development and tissue homeostasis are related to the intrinsic heterogeneity observed in ESCs. In addition to development, other biological systems demonstrate spontaneous organization of seemingly equivalent cells into states, such as bacteria in forming persister states or cancer cells in forming therapy resistant subpopulations (55–59). Determining the configurations of gene regulatory networks within such systems will be of great interest. Nevertheless, the results presented here suggest that naturally arising cell-to-cell variation, sometimes described as stochastic fluctuation, is in fact coherently organized biology.
Materials and Methods
For additional information, please see SI Appendix, Supplementary Materials and Methods.
Cell Lines.
Ten mouse ESC lines were used in this study: 1) DGCR8−/− ESC (42); 2) Ago2-inducible ESCs (43), Nanog-GFP/Sox2-mCherry; 3) V6.5 ESC (Jaenisch Laboratory, Whitehead Institute, Massachusetts Institute of Technology [MIT]), Nanog-GFP/Sox2-mCherry; 4) V6.5 ESC, Nanog-GFP/Sox2-mCherry (nuclear localization sequence [NLS]-tagged fluorophores); 5) V6.5 ESC, Mir182indel, Nanog-GFP/Sox2-mCherry; 6) V6.5 ESC, Mir708indel, Nanog-GFP/Sox2-mCherry; 7) V6.5 ESC, Nanog-GFP/Sox2-mCerulean3; 8) V6.5 ESC, Nanog-GFP/Esrrb-E2-Crimson; 9) V6.5 ESC, Nanog-GFP/Eif2s2-mCherry; and 10) V6.5 ESC, Nanog-GFP/Hsp90ab1-mCherry.
All fluorophore tags, NLSs, and microRNA gene indels were added by us (see relevant sections in SI Appendix).
Flow Cytometry and Fluorescence Activated Cell Sorting (FACS).
A total of 50 to 90% confluent ESCs were analyzed on BD LSRII or LSRFortessa with FACSDiva v8.0 acquisition software. Flow cytometry standard files were analyzed with FlowJo V9.9. Samples were gated first for live cells based on forward scatter area vs. side scatter area and then for single cells (forward scatter width vs. forward scatter height). Cells singly transfected with transient fluorophore expression constructs or singly tagged with either GFP or mCherry were used as fluorescence compensation controls. Channel gains were adjusted based on native V6.5 ESC.
BD FACSARIA was used to sort Nanog-GFP/Sox2-mCherry ESCs into states (as defined by GFP/mCherry protein levels), and to sort single cells. States were sorted into fresh culture medium in 5-mL collection tubes, then immediately spun at 1,000 rpm for 5 min and resuspended for either plating or RNA isolation. Single cells were sorted into 200 μL fresh culture medium in one well of a 96-well flat bottom plate (VWR catalog no. 29442-054).
RNA-Sequencing Sample Preparation.
Standard TRIzol protocol was used to isolate total RNA from sorted states 1/2/3 or the total unseparated ESC population passed through the sorter. Following DNase I (NEB M0303) treatment and ethanol precipitation, samples were analyzed by Agilent BioAnalyzer and accepted for sample RNA integrity number > 7.0. RNA-sequencing represents three biological replicates isolated by flow sorting on 3 separate days. rRNA-depleted RNA-sequencing libraries (∼100 ng RNA/sample) were prepared using Kapa RNA HyperPrep Kit with RiboErase (HMR) KK8561 using 11 rounds of PCR amplification after addition of External RNA Controls Consortium spike-in controls at the recommended concentration. The final libraries were quality control (QC) checked by fragment electrophoresis and qPCR for colony-forming units prior to pooling and loading on an Illumina FlowCell (NextSEq. 500, 150 bp paired-end reads). Each sample library was sequenced to 30- to 45-M read depth.
Single-Cell RNA-Seq.
scRNA-seq was performed by the Koch Institute Nanowell Cytometry Core using SeqWell (60) technology. In brief, single-cell suspensions of fluorophore-tagged V6.5 ESCs were made by trypsinization followed by serial passage through 50-μm cell strainer meshes. Approximately 10,000 cells were loaded onto a SeqWell array, lysed, and prepared as single-cell cDNA libraries as described (60). Libraries were sequenced using a NextSEq. 500 and aligned to the mm10 genome using the SeqWell analysis pipeline (Love Lab, MIT). The entire process was repeated on a different day to generate two data tables representing reads per cell across mm10 annotated transcripts (Gencode M15). These two tables were merged together and analyzed, prior to gene neighborhood construction (SI Appendix, Supplementary Methods and Materials).
miRNA-Seq and Data Analysis Pipeline.
Total RNA samples were prepared from ESC states identically to RNA-seq above for two biological replicates isolated by flow cytometric sorting. Small RNA libraries were then prepared using the NEB small RNA-sequencing kit (E7300S) according to the manufacturer’s instructions using 13 cycles of PCR amplification. QC assessment was done by electrophoresis and colony forming units prior to loading pooled samples onto an Illumina FlowCell (HiSeq2000, one lane for eight samples, 40-bp SE reads) with 10- to 15-M reads/sample. For complete details of the data analysis pipeline, including commands used, please see the document “MiRNA-sequencing data analysis pipeline” Zenodo (DOI: 10.5281/zenodo.3694341).
Statistical Analyses.
For all analyses with P values, significance was determined at P ≤ 0.05. P values are shown in the figure, or in the figure legend, wherever they are used. The Kolmogorov–Smirnov (K–S) test was used to assess whether paired cumulative distribution functions (CDFs) were significantly different (Fig. 3 D and E and SI Appendix, Fig. S6 B and C). The Kruskal–Wallis one-way analysis of variance test was used to assess differences between 3+ CDFs (Fig. 4A). Both the K–S and Kruskal–Wallis test statistics were calculated in GraphPad Prism. In all cases, a CDF based on the population was compared to CDF(s) based on sample(s). The cumulative hypergeometric statistical test for enrichment was used in Fig. 3F. This test detects enrichment for a property in a population sample, compared to what would have been expected based on the prevalence of that property in the whole population. Four numbers are required: population size (N), number of population successes (n), sample size (K), and number of sample successes (k). The test statistic was calculated using Python v3.6, using scipy.stats.hypergeom.pmf and summing from k to min(n,K) to calculate the cumulative value. Box-whisker plots in Fig. 5 A and B were generated using matplotlib (Python v3.6), and associated P values reflect the results of Levene’s test for equality of variances (Python v3.6, scipy.stats.levene), an alternative to Bartlett’s test which is robust to deviations from nonnormality. In Fig. 5A, Levene’s test confirmed significance of the higher Nanog variances for “+Dox” conditions, compared to “no Dox.” In Fig. 5B, Levene’s test confirmed significance of the lower Nanog variances for Mirindel cell populations, compared to WT. Box-whisker plots in Fig. 5D were generated using plot.ly for Python, with whiskers from 25th to 75th percentiles. Here, Levene’s test confirmed significance of the higher Nanog and Sox2 variances for cells transfected with CFP-pri-miR-182 (pooled across CFP levels), compared to those transfected with the CFP-empty plasmid. Error bars for “fraction of cells outside state 1” plots in Fig. 5 A, B, D, and E represent 95% CIs, and were generated by bootstrapping using 10 bootstrap samples. Associated P values reflect the results of a binomial test (scipy.stats.binom_test, Python v3.6), with expected number of successes calculated from the fraction represented by the relevant gray/control bar. Error bars for all plots in SI Appendix, Fig. S8 represent 95% CIs generated by bootstrapping using 10 bootstrap samples. Exact cell numbers for all plots Fig. 5 and SI Appendix, Fig. S8 are recorded in SI Appendix, Supplementary Materials and Methods.
Data and Code Availability.
RNA-sequencing and small-RNA sequencing data are available at the Gene Expression Omnibus (GEO) with accession number GSE132708. SI Appendix, Tables S2 and S3 show RNA and miRNA expression in states. Other data, scripts, and detailed descriptions of sequencing analysis pipelines are available in the SI Appendix or on Zenodo (DOI: 10.5281/zenodo.3694341).
Supplementary Material
Acknowledgments
We would like to thank Paige Coles for technical assistance. Additionally, we thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Flow Cytometry Core and Shanu Metha, Stuart Levine, and Vincent Butty for assistance with single-cell RNA-seq. S.G. acknowledges funding from a Charles W. and Jennifer C. Johnson Clinical Investigator Award and NIH NCI T32 CA009216; P.A.S. acknowledges funding from NIH P01 CA042063; and C.B., M.D.G., and A.D.M. acknowledge funding from Horizon 2020 Marie Skłodowska-Curie Action-Research and Innovation Staff Exchange (MSCA-RISE) 2016 grant agreement 734439 (INFERNET, new algorithms for inference and optimization from large-scale biological data). This work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute.
Footnotes
The authors declare no competing interest.
Data deposition: RNA-sequencing and small-RNA sequencing data are available on the Gene Expression Omnibus (GEO) with accession number GSE132708. SI Appendix, Tables S2 and S3 show RNA and miRNA expression in states. Other scripts, data, and detailed descriptions of sequencing pipelines are available through Zenodo (DOI: 10.5281/zenodo.3694341).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920695117/-/DCSupplemental.
References
- 1.Turing A. M., The chemical basis of morphogenesis. Philos T Roy Soc B 237, 37–72 (1952). [Google Scholar]
- 2.Wolpert L., Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). [DOI] [PubMed] [Google Scholar]
- 3.Wolpert L., Positional information and pattern formation. Curr. Top. Dev. Biol. 6, 183–224 (1971). [DOI] [PubMed] [Google Scholar]
- 4.Green J. B., Sharpe J., Positional information and reaction-diffusion: Two big ideas in developmental biology combine. Development 142, 1203–1211 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Chen Q., Shi J., Tao Y., Zernicka-Goetz M., Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat. Commun. 9, 1819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beccari L., et al. , Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Harrison S. E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M., Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Rivron N. C., et al. , Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Shahbazi M. N., et al. , Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature 552, 239–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siggia E. D., Warmflash A., Modeling mammalian gastrulation with embryonic stem cells. Curr. Top. Dev. Biol. 129, 1–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spagnoli F. M., Hemmati-Brivanlou A., Guiding embryonic stem cells towards differentiation: Lessons from molecular embryology. Curr. Opin. Genet. Dev. 16, 469–475 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Chang H. H., Hemberg M., Barahona M., Ingber D. E., Huang S., Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Q., Ramsköld D., Reinius B., Sandberg R., Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Eldar A., Elowitz M. B., Functional roles for noise in genetic circuits. Nature 467, 167–173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R. M., et al. , Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516, 56–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj A., van Oudenaarden A., Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serra D., et al. , Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A., Razooky B., Cox C. D., Simpson M. L., Weinberger L. S., Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys. J. 98, L32–L34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarro P., et al. , OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 31, 4547–4562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowen J. M., et al. , Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hnisz D., et al. , Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagey M. H., et al. , Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whyte W. A., et al. , Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marson A., et al. , Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers I., et al. , Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Ying Q. L., et al. , The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abranches E., et al. , Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development 141, 2770–2779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochiai H., Sugawara T., Sakuma T., Yamamoto T., Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci. Rep. 4, 7125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filipczyk A., et al. , Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 17, 1235–1246 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kalmar T., et al. , Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karwacki-Neisius V., et al. , Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell 12, 531–545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein A. M., et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer Z. S., et al. , Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon K. R., et al. , Visualizing structure and transitions for biological data exploration. bioRxiv:10.1101/120378 (04 April 2019). [Google Scholar]
- 35.Paul F., et al. , Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Boroviak T., et al. , Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell 35, 366–382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li A., Horvath S., Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics 23, 222–231 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Garg S., Sharp P. A., GENE EXPRESSION. Single-cell variability guided by microRNAs. Science 352, 1390–1391 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Bosson A. D., Zamudio J. R., Sharp P. A., Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 56, 347–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartel D. P., Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory R. I., et al. , The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R., DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamudio J. R., Kelly T. J., Sharp P. A., Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 156, 920–934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S., et al. , Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukherji S., et al. , MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 43, 854–859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Giudice M., Bo S., Grigolon S., Bosia C., On the role of extrinsic noise in microRNA-mediated bimodal gene expression. PLOS Comput. Biol. 14, e1006063 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cassidy J. J., et al. , Repressive gene regulation synchronizes development with cellular metabolism. Cell 178, 980–992 e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmiedel J. M., et al. , Gene expression. MicroRNA control of protein expression noise. Science 348, 128–132 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Moris N., et al. , Histone acetyltransferase KAT2A stabilizes pluripotency with control of transcriptional heterogeneity. Stem Cells 36, 1828–1838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahrends R., et al. , Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science 344, 1384–1389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peláez N., et al. , Dynamics and heterogeneity of a fate determinant during transition towards cell differentiation. eLife 4, e08924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tata P. R., et al. , Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hogan B. L., et al. , Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beumer J., Clevers H., Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143, 3639–3649 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S., Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Sharma S. V., et al. , A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaffer S. M., et al. , Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jordan N. V., et al. , HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tammela T., et al. , A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gierahn T. M., et al. , Seq-well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing and small-RNA sequencing data are available at the Gene Expression Omnibus (GEO) with accession number GSE132708. SI Appendix, Tables S2 and S3 show RNA and miRNA expression in states. Other data, scripts, and detailed descriptions of sequencing analysis pipelines are available in the SI Appendix or on Zenodo (DOI: 10.5281/zenodo.3694341).