Significance
Human CD8+ T cells can effectively kill target cells. However, not all CD8+ T cells are equally efficient herein. We developed a method to dissect the transcriptome and proteome of FACS-sorted cytokine-producing fixed human CD8+ T cells. We show that CD29 identifies IFN-γ–producing CD8+ T cells with high expression levels of cytotoxic molecules. Preselecting for CD29+ T cells potentiated the killing capacity of CD8+ T cell products. Reanalysis of published scRNA-seq data revealed a core signature of CD29+ T cells, which had a good predictive value for survival in melanoma patients. In conclusion, our study provides fundamental insights in the heterogeneity and functionality of human CD8+ T cells, which could help potentiate the efficacy of adoptive T cell therapies.
Keywords: T cells, cytotoxicity, CD29, IFN-γ, IL-2
Abstract
Cytotoxic CD8+ T cells can effectively kill target cells by producing cytokines, chemokines, and granzymes. Expression of these effector molecules is however highly divergent, and tools that identify and preselect CD8+ T cells with a cytotoxic expression profile are lacking. Human CD8+ T cells can be divided into IFN-γ– and IL-2–producing cells. Unbiased transcriptomics and proteomics analysis on cytokine-producing fixed CD8+ T cells revealed that IL-2+ cells produce helper cytokines, and that IFN-γ+ cells produce cytotoxic molecules. IFN-γ+ T cells expressed the surface marker CD29 already prior to stimulation. CD29 also marked T cells with cytotoxic gene expression from different tissues in single-cell RNA-sequencing data. Notably, CD29+ T cells maintained the cytotoxic phenotype during cell culture, suggesting a stable phenotype. Preselecting CD29-expressing MART1 TCR-engineered T cells potentiated the killing of target cells. We therefore propose that CD29 expression can help evaluate and select for potent therapeutic T cell products.
CD8+ T cells can effectively clear cells from our body that are infected with intracellular pathogens. In addition, CD8+ T cells are critical for immunosurveillance to eliminate precancerous or cancerous cells. To exert their effector function, T cells produce proinflammatory molecules such as IFN-γ and TNF-α, granzymes and perforin, and chemokines such as CCL3 (MIP1α) and CCL4 (MIP1β) (1–3). Intriguingly, human T cells do not respond uniformly to activation, but rather show a heterogeneous production profile of effector molecules (4, 5). This may derive from stochastic and oscillating production of effector molecules, as suggested in developmental processes (6). However, it may also be due to intrinsic differences in the effector response to antigen by distinct CD8+ T cell types.
Interestingly, even though activated human CD8+ T cells efficiently produce IFN-γ and IL-2, we and others found that they generally produce only one of the two cytokines (5, 7, 8). Only a small proportion of human CD8+ T cells produces IFN-γ and IL-2 combined (5, 7, 8). Several studies proposed that human and mouse CD8+ T cells can provide T cell “help” by expressing CD154/CD40L (7, 9, 10), in addition to their well-known cytotoxic function. CD40L-expressing human CD8+ T cells were shown to produce IL-2, but not IFN-γ (7). However, whether this cytokine production profile is stochastic, or whether IFN-γ– and IL-2–producing CD8+ T cells represent two different T cell subsets and/or a divergent differentiation status is yet to be determined. It is also not known whether IFN-γ– and IL-2–producing CD8+ T cells possess a different cytotoxic potential.
In this study, we set out to dissect the properties of human CD8+ T cells that display a differential cytokine production profile. Previous studies used cytokine capture assays to isolate T cells for gene expression analysis (11). While this method is very accurate for low-abundance responses, the risk of selecting false-positive cells by capturing cytokines from the neighboring cell in high-abundance responses is substantial. We therefore developed a protocol that allowed for efficient isolation of RNA and protein from fluorescence-activated cell sorting (FACS)-sorted fixed T cells after intracellular cytokine staining. With this top-down approach, we performed an unbiased RNA-sequencing (RNA-seq) and mass spectrometry (MS) analyses on IFN-γ– and IL-2–producing primary human CD8+ T cells.
We found that IFN-γ–producing T cells exhibit a cytotoxic expression profile, which is sustained during T cell culture. The surface molecules CD29 and CD38 helped identify the cytotoxic IFN-γ–producing T cells, and the noncytotoxic IL-2–producing CD8+ T cells, respectively, in in vitro cultures, a feature that is maintained for several weeks. Using these two surface markers allowed us to distinguish T cells with a distinct cytokine production profile and cytotoxic potential already prior to T cell stimulation. Furthermore, the CD29 gene expression signature was a good prognostic marker for long-term survival in melanoma patients. In conclusion, CD29 selects for bona fide IFN-γ+ CD8+ T cells with high expression levels of cytotoxic molecules. CD29 could therefore potentially be used to evaluate the quality of therapeutic T cell products, and that could help to identify and select cytotoxic CD8+ T cells for the generation of effective T cell products.
Results
Efficient Recovery of RNA and Protein from T Cells after Intracellular Cytokine Staining.
To determine whether and how the gene expression profile of IFN-γ–producing CD8+ T cells differed from IL-2–producing cells, we needed a method to separate high percentages of cytokine producers while preventing possible contaminations from false-positive cells. We therefore developed a protocol for efficient recovery of RNA and proteins from FACS-sorted fixed cells after intracellular cytokine staining. With the standard TRIzol RNA isolation protocol, >99% of the mRNA isolated from phorbol 12-myristate 13-acetate (PMA)–ionomycin-activated and formaldehyde-fixed T cells was lost, as determined by RT-PCR of several standard housekeeping genes, and of IFNG and IL2 mRNA (SI Appendix, Fig. S1A). To conserve the RNA integrity, we performed the intracellular cytokine staining (ICCS) and FACS in an RNA-protecting high salt buffer that contained RNAse inhibitors (Fig. 1A). This alteration in the protocol preserved the quality of ICCS, and thus the distinction of different cytokine producers (5). To free RNA from protein complexes, we included a proteinase K digestion step prior to RNA isolation (12) (Fig. 1A). These adjustments yielded mRNA levels from fixed T cells comparable to “fresh” T cells that were prepared without permeabilization and formaldehyde fixation (SI Appendix, Fig. S1A). Importantly, RNA purity and integrity were compatible with RNA-seq analysis, as revealed by nanodrop analysis and RNA nano chip assay (SI Appendix, Fig. S1B).
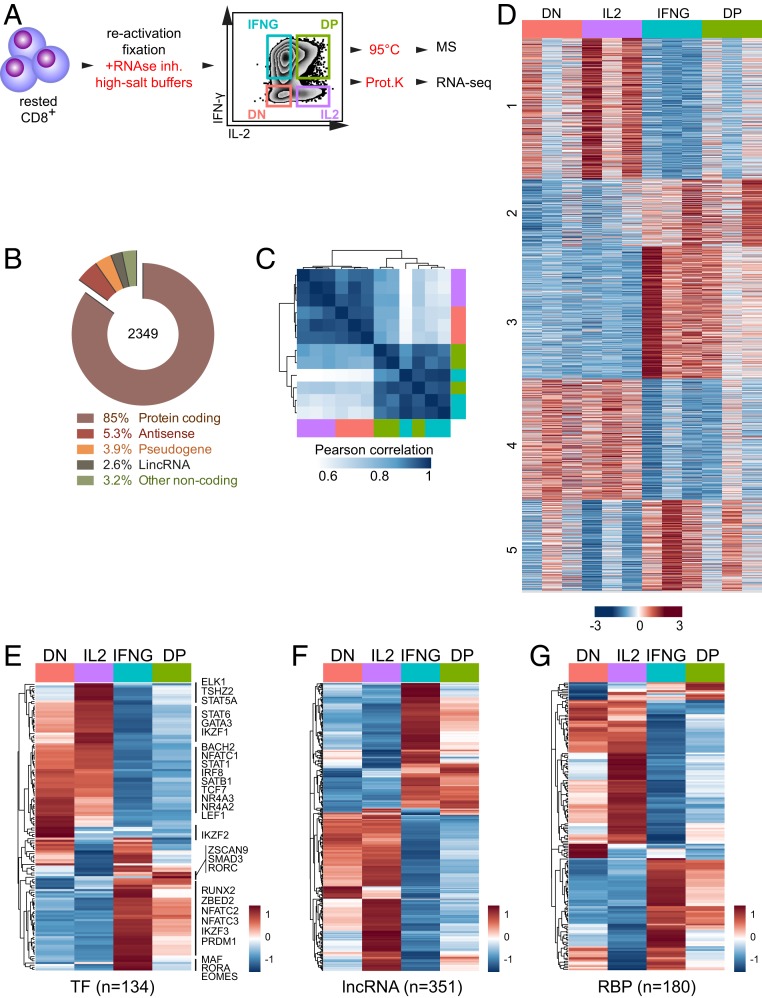
Fig. 1.
RNA-seq analysis on CD8+ T cells after intracellular cytokine staining. (A) Scheme to isolate RNA and protein from cytokine-producing, fixed T cells. Primary human CD8+ T cells were activated for 48 h with α-CD3/α-CD28, and rested for 4 d in the presence of 10 ng/mL rhIL-15. T cells were reactivated for 4 h with 10 ng/mL PMA and 1 µM ionomycin. Intracellular cytokine staining was performed in RNA-protecting buffers. Cells were FACS-sorted under RNA-protecting conditions. Total RNA and protein was recovered for RNA-seq and mass spectrometry analysis (Fig. 2) by reversing formaldehyde cross-linking. IL2: IL-2 single producers; IFNG: IFN-γ single producers; DP: double positive: IL-2 and IFN-γ coproducers; DN: double negative for IL-2 and IFN-γ production. (B–G) RNA-seq analysis of CD8+ T cells with a differential IL-2 and/or IFN-γ production profile. (B) Gene biotypes and (C) samplewise Pearson correlation coefficient of genes (n = 2,349) that are differentially expressed between all four T cell populations. (D) Heatmap of differentially expressed protein-coding genes (n = 1,984), numbers indicate k-means clusters (k = 5). (E) Heatmap of differentially expressed TFs with >100 DESeq2 normalized counts, (F) lncRNAs, and (G) RBPs with >10 DESeq2 normalized counts. (E–G) Expression levels of biological replicates were averaged (n = 3 per population). Color scale of heatmaps represents Z-score.
To achieve MS-grade protein recovery, we used the filter aided sample preparation (FASP) protocol (13). Because this protocol contains a 95 °C heating step to reverse the formaldehyde cross-linking, it is compatible with MS analysis of cells prepared with the ICCS procedure (14, 15). Indeed, the coefficient of determination of MS analysis from fixed and fresh samples was R2 = 0.973 (SI Appendix, Fig. S1C). Thus, RNA and protein could be efficiently recovered from cytokine-producing, formaldehyde-fixed T cells, which allowed us to study their expression profile in depth.
Differential Gene Expression Profile of IFN-γ– and IL-2–Producing CD8+ T Cells.
To determine the gene expression profile of IFN-γ–producing cells with that of IL-2–producing cells, human CD8+ T cells from three biological replicates, each containing a pool of 40 healthy donors, were activated for 2 d with α-CD3/α-CD28. T cells were removed from the stimulus and cultured for 4 d in the presence of human recombinant IL-15. ICCS after 4 h of activation with PMA–ionomycin identified four T cell populations: IFN-γ producers (IFNG), IL-2 producers (IL2), IFN-γ/IL-2-coproducing T cells (double positive [DP]), and T cells that did not produce detectable levels of either cytokine (double negative [DN]; Fig. 1A).
Of the RNA-sequencing data, on average 19.8 million reads (∼92%) of the total 21.4 million reads could be mapped to the genome of each T cell population. Of these, 2,349 genes were differentially expressed in at least one population, with a cutoff of log2 fold change (LFC) >0.5, p-adjusted <0.01 (Dataset S1). According to Ensembl-BioMart gene annotation, 85% (n = 1,984) of the differentially expressed genes (DEGs) were protein coding, 5.3% were antisense, 3.9% were pseudogenes, 2.6% were long-intergenic noncoding RNA (lincRNA), and 3.2% comprised other classes of noncoding RNA (ncRNA) (Fig. 1B).
Pearson’s correlation coefficient of differentially expressed genes revealed that each biological replicate clustered according to its cytokine production profile (Fig. 1C). Interestingly, IFNG cells clustered with DP cells, and IL2 cells with DN cells (Fig. 1C). This close relationship of IFNG and DP cells and of IL2 with DN cells was also revealed by gene cluster analysis using k-means (k = 5), in particular in clusters 2 and 4 (241 and 434 genes, respectively) (Fig. 1D and Dataset S1). Yet, cluster 1 (507 genes) was more specific for IL2 T cells, and for IFNG T cells, clusters 3 and 5 (474 and 326 genes, respectively; Fig. 1D and Dataset S1).
We next interrogated whether specific gene classes were differentially expressed. Gene set enrichment analysis revealed several pathways differing between IFNG and IL2 producers, which included the calcium signaling and transport for IFNG producers, and WNT signaling for IL2 producers (SI Appendix, Fig. S1 D and E). Also gene expression modulators like transcription factors (TFs), long noncoding RNAs (lncRNAs), and RNA binding proteins (RBPs) were differentially expressed in IFNG and IL2 producers. Most of the 134 differentially expressed TFs were shared between DN and IL2 producers, and between IFNG and DP producers (Fig. 1E and Dataset S2). This included GATA3 and IRF8 for IL2/DN producers, and PRDM1 (Blimp-1), NFATc2, and NFATc3 for IFNG/DP producers (SI Appendix, Fig. S1F and Dataset S2). Specific gene expression was detected for EOMES and RORA in IFNG producers, and for RORC (ROR-γt) in DP producers (Dataset S2). A small cluster of TFs including IKZF2 (Helios) was specifically up-regulated in DN cells (SI Appendix, Fig. S1F). Also the 351 differentially expressed lncRNAs revealed the close kinship between DN/IL2 producers and IFNG/DP producers (Fig. 1F and Dataset S2). For instance, the IFN-γ-promoting IFNG-AS1 (NEST) (16) was highly expressed in IFNG/DP producers (SI Appendix, Fig. S1G). Conversely, GATA3-AS1 known to reenforce the Th2 phenotype in CD4+ T cells (17) showed highest gene expression in IL2 producers (SI Appendix, Fig. S1G).
RBPs are critical regulators of RNA splicing, stability, and translational control (18). Again, most of the 180 differentially expressed RBPs were similarly expressed in IFNG/DP producers and in IL2/DN producers (Fig. 1G and Dataset S2). Combined, these data revealed a differential gene expression profile in IFNG and IL2 producers, which correlates with distinct expression of gene regulators.
Differential Protein Signature of IL-2- and IFN-γ-Producing CD8+ T Cells.
We next determined the protein expression profile of the four FACS-sorted CD8+ T cell populations by MS. We identified in total 5,995 proteins from the three biological replicates. After removing possible contaminants and after filtering for proteins that were present in each biological replicate in at least one of the four populations, we identified a total of 3,833 proteins. Each sample contained >3,500 identified proteins (Fig. 2A and Dataset S1). Pearson’s correlation and k-means clustering of the 81 differentially expressed proteins (nine clusters) also revealed the kinship in the proteome between IFNG and DP producers, and between IL2 and DN producers (Fig. 2 B and C). A total of 42 (52%) of the differentially expressed proteins overlapped with the differential gene expression profile, including IL-2 and IFN-γ (Fig. 2D). In conclusion, IL2 and IFNG producers display a distinct gene and protein expression signature.
Fig. 2.
Mass spectrometry analysis on cytokine producing, fixed CD8+ T cells. (A) Number of proteins identified by MS analysis of FACS-sorted T cell populations per sample (bars) and of all 12 samples combined after filtering (red line; n = 3,833). (B) Samplewise Pearson correlation coefficient of differentially expressed proteins (n = 81). (C) Heatmap of differentially expressed proteins, numbers indicate k-means clusters (k = 9). Proteins showing differential expression in both mRNA and protein are indicated in blue. Color scale represents log2 centered intensity. (D) Venn diagram of differentially expressed proteins and genes (Fig. 1).
IFN-γ-Producing CD8+ T Cells Display a Cytotoxic Profile.
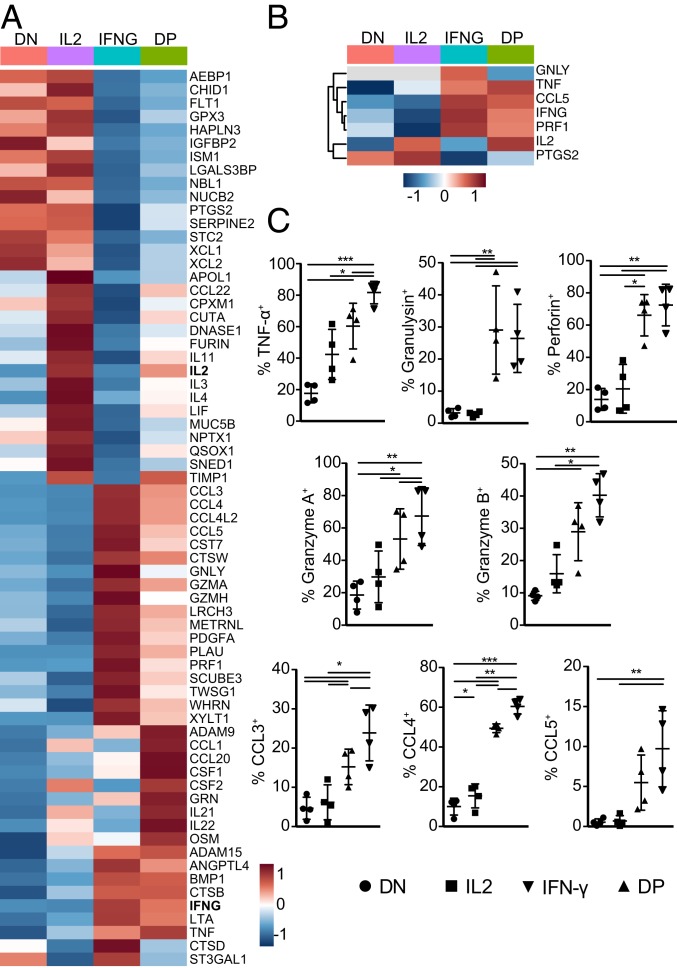
To determine whether IFNG and IL2 producers also showed a distinct gene expression of other secreted effector molecules, we selected differentially expressed genes from Fig. 1 that 1) had expression levels >100 DESeq2 normalized counts and that 2) were annotated as “secreted” in the human protein atlas (19). After removing nuclear components and mitochondrial and ribosomal proteins (Materials and Methods), we identified 67 differentially expressed genes encoding secreted proteins (Fig. 3A). For IL2 producers, these included the prototypic Th2 helper effector molecules IL-4, IL-3, leukemia inhibitory factor (LIF) (20, 21), and the Th2-inducing cytokine IL-11 and prostaglandin-endoperoxide synthase 2 (PTGS2, or COX-2) (22, 23) (Fig. 3A). IL-2 and PTGS2 were also found differentially expressed in the MS analysis (Fig. 3B). Conversely, IFNG producers—and in most cases also DP producers—contained substantially higher gene expression levels of the prototypic cytotoxic molecules TNF-α, Granulysin, Granzyme A, Granzyme H, and Perforin (Fig. 3A). Also the chemokines CCL5 (RANTES), CCL3, CCL4, and its alternative splicing variant CCL4L2, all of which are associated with antiviral and antibacterial activity of cytotoxic CD8+ T cells (24–26), were highly expressed in IFNG and DP producers (Fig. 3A). IFN-γ, TNF-α, Granulysin, Perforin, and CCL5 were also identified by MS analysis (Fig. 3B).
Fig. 3.
IL2 and IFNG producers express unique sets of secreted proteins. (A and B) Heatmap of differentially expressed genes (A, n = 67) and proteins (B, n = 7) that are annotated as secreted proteins (see Materials and Methods). Expression levels of biological replicates were averaged (n = 3 per population). Gray: not detected. Color scale of heatmaps represents Z-score. (C) Validation of secreted effector molecules identified in A and B in four new individual donors. α-CD3/α-CD28 activated T cells were rested for 4 d prior to 4-h reactivation with PMA–ionomycin in the presence of monensin. Cytokine production was determined by intracellular cytokine staining. Plots depict the production of effector molecules in DN, IL2, IFNG, and DP producers. Repeated measurement ANOVA with Tukey posttest (*P < 0.05, **P < 0.01, ***P < 0.001).
We validated this prototypic cytotoxic gene expression profile of IFNG and DP cells in a new cohort of four individual donors. Identical to the cell preparation for RNA-seq/MS analysis, peripheral blood mononuclear cells (PBMCs) were activated for 2 d with α-CD3/α-CD28, and rested for 4 d in rhIL-15. ICCS after reactivation for 4 h with PMA–ionomycin confirmed that IFNG/DP producers produced significantly more TNF-α, Granulysin, Perforin, Granzyme A and B, and the chemokines CCL3, CCL4, and CCL5 than the DN/IL2 producers (Fig. 3C). Not only the percentage, but also the production of cytokines/chemokine per cell was higher, as determined by the geometric mean fluorescence intensity (GeoMFI; SI Appendix, Fig. S2A). We therefore conclude that IFNG and DP CD8+ T cells preferentially express cytotoxic effector molecules.
CD29 and CD38 Enrich for IFN-γ– and IL-2–Producing CD8+ T Cells.
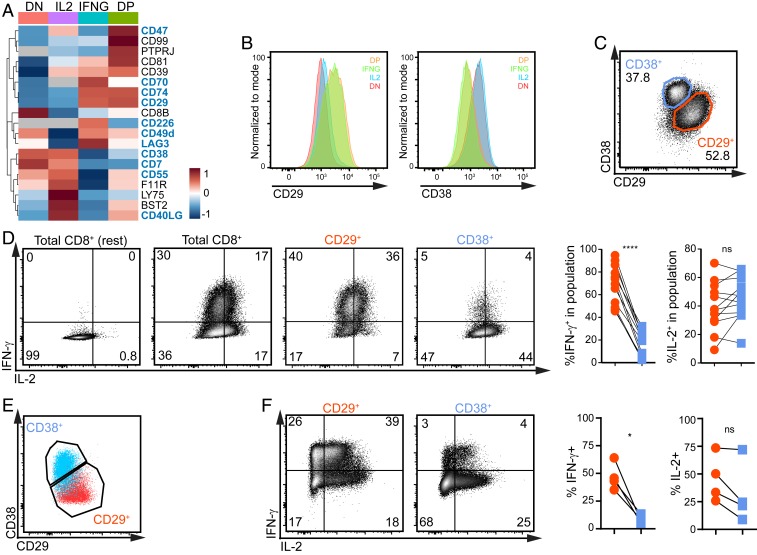
We next interrogated if surface markers could identify IFNG producers and IL2 producers. We filtered the MS data for surface molecules with an expression of ≥1.5-fold difference in one of the four T cell populations. Nineteen surface markers showed differential protein expression in at least one T cell population (Fig. 4A). RNA-seq analysis identified 72 differentially expressed annotated proteins (SI Appendix, Fig. S2B). We tested the expression pattern of 28 of these markers by flow cytometry on PMA–ionomycin-activated CD8+ T cells. None of the 16 surface markers we identified by RNA-seq analysis alone was suitable to select for IFNG and IL2 producers by flow cytometry (SI Appendix, Fig. S2C). Gene products that were detected in both MS and RNA-seq analysis, like β1-integrin (ITGB1; CD29) showed significantly higher cell surface expression in IFNG/DP producers than in IL2/DN producers (Fig. 4B and SI Appendix, Fig. S2 C and D). HLA class II histocompatibility antigen gamma chain CD74 also followed this expression pattern (SI Appendix, Fig. S2 C and E). Conversely, IL2/DN producers showed significantly higher expression levels of cyclic ADP ribose hydrolase CD38 than IFNG/DP producers (Fig. 4B and SI Appendix, Fig. S2 C and D), and CD40L and the complement decay-accelerating factor CD55 coexpressed with CD38 (SI Appendix, Fig. S2 F and G).
Fig. 4.
CD29 and CD38 identify IFNG and IL2 producers. (A) Heatmap of differentially expressed CD molecules identified by MS analysis with LFC >1.5 (gray: not detected). Expression levels of biological replicates were averaged (n = 3 per population). Color scale represents Z-score. (B) CD29 (ITGB1; Left) and CD38 (Right) expression of DN, IL2, IFNG, and DP producers in α-CD3/α-CD28 activated T cells that were rested for 4 d and then reactivated for 4 h with PMA–ionomycin (n = 6 donors). (C) Representative CD29 and CD38 expression profile of nonactivated CD8+ T cells. (D) Representative IL-2 and IFN-γ production after 4-h reactivation with PMA–ionomycin of CD8+ T cells (total), and of CD29+ and CD38+ T cells, based on gating shown in C. Resting CD8+ T cells (Left) served as control. (Right) Compiled data of 12 donors. (E and F) CD8+ T cells were FACS-sorted at day 7 of culture based on CD29 and CD38 expression. Cells were reactivated for 2 d with α-CD3/α-CD28 and rested for another 14 d with rhIL-15 (day 23). (E) Representative CD29 and CD38 expression. (F) Representative IFN-γ and IL-2 production and compiled data of four donors by CD29+ and CD38+ after 4-h reactivation with PMA–ionomycin. Paired ratio t test (*P < 0.05; ****P < 0.0001, ns: not significant).
We next sought for markers that could preidentify IFNG/DP producers and DN/IL2 producers in resting T cells. CD40L and CD74 had limited expression in nonactivated, CD8+ T cells (SI Appendix, Fig. S2G) and were thus not suitable. CD29 and CD38 however allowed for the identification of IFNG/DP producers and DN/IL2 producers prior to reactivation (Fig. 4 C and D). This included the production of TNF-α, Granzyme A, Granulysin, Perforin, CCL3, CCL4, and CCL5 by CD29+ T cells (SI Appendix, Fig. S3A). Because CD29+ T cells included DP producers (Fig. 4B), the overall IL-2 protein production was similar in CD29+ and CD38+ T cells, yet clearly distributed between CD29+ DP and CD38+ IL2 single producers (SI Appendix, Fig. S3B).
Intriguingly, even though the percentage of CD29+ T cells and CD38+ T cells was greatly variable between donors (SI Appendix, Fig. S3C), their expression pattern and cytokine production profile was maintained throughout the T cell culture (Fig. 4 E and F). Specifically, when we FACS-sorted CD29+ and CD38+ T cells on day 7 after activation, reactivated them with αCD3/αCD28 for 2 d, and cultured them for another 10 d (i.e., 12 d postsort), CD29 and CD38 expression was maintained by the vast majority (>90%) of the cells (SI Appendix, Fig. S3D). In addition, the CD29+ FACS-sorted T cells expanded slightly better than CD38+ T cells, as determined by a slightly higher loss of carboxyfluorescein succinimidyl ester (CFSE) signal, and by increased cell numbers from 8 d postsort (SI Appendix, Fig. S3 E and F). Importantly, also when T cells were cultured for 14 d after the second α-CD3/α-CD28 activation (i.e., 23 d of total T cell culture), CD29+ T cells were the prime IFNG producers, and CD38+ T cells, the IL2 producers (Fig. 4 D and E). Using different cytokines during α-CD3/α-CD28 activation, or during the resting phase did not alter the percentage of CD29+ T cells at the end of culture, or the GeoMFI expression of CD29 (SI Appendix, Fig. S3 G and H). Overall, we conclude that the differences between CD29+ and CD38+ T cells are cell intrinsic and can be used to distinguish IFNG/DP producers from IL2/DN producers.
CD29 Is Expressed on Nonnaïve CD8+ T Cells.
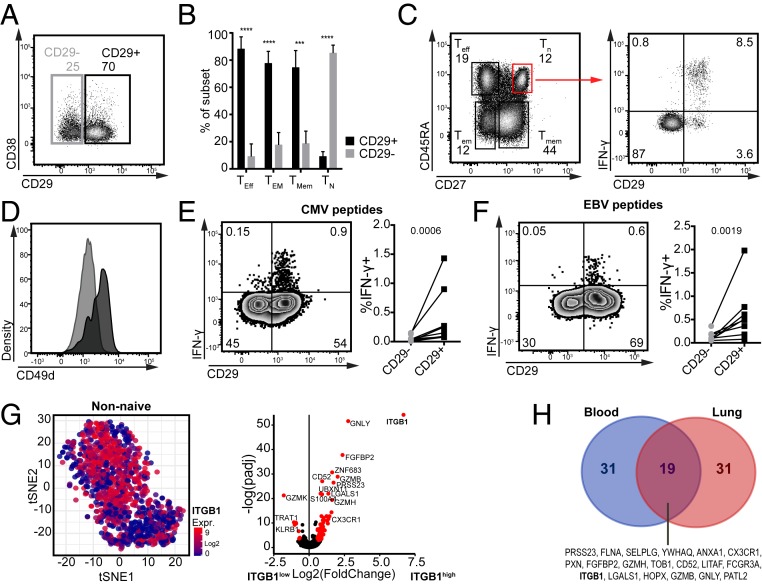
We next investigated how our findings on differential cytokine production by CD29+ and CD38+ T cells in vitro translated to CD8+ T cell reactivity ex vivo. CD38 expression on CD8+ T cells was limited (14.1 to 25.8%). CD38 expression was similarly distributed in different T cell subsets, i.e., CD45RA+CD27+ naïve T cells (TN), CD45RA+CD27− effector CD8+ T cells (TEff), CD45RA−CD27− effector memory (TEM), and CD45RA−CD27+ memory CD8+ T cells (TMem) (SI Appendix, Fig. S4A) and is described to be absent on stem cell memory cells (27). However, CD38 expression did not anticorrelate with CD29 expression ex vivo (Fig. 5A). In fact, CD38 expression was observed only after several days of T cell activation in vitro, and only then its inverse correlation with CD29 became apparent. Because CD29 expression was detected ex vivo and maintained its strong correlation with IFN-γ production (SI Appendix, Fig. S4 C and D), we further focused on CD29 expression and its relation to T cell differentiation and functionality. CD29 expression was primarily detected in TEff, TEM, and TMem CD8+ T cells with equal protein expression levels (Fig. 5B and SI Appendix, Fig. S4B). This expression profile was in concordance with that of cytotoxic molecules (28, 29) and its observed expression in effector T cells (8, 30). Of the CD45RA+CD27+ naïve T cells (TN), only 6.6 ± 4.2% expressed CD29 (Fig. 5B and SI Appendix, Fig. S4B). However, the CD29+ “naïve” T cells that rapidly produce IFN-γ upon 4 h PMA–ionomycin stimulation, and express CD49d (Fig. 5 C and D), were previously identified as memory T cells with a naïve phenotype (TMNP) (31). Irrespective of their differentiation status, the vast majority of IFN-γ–producing T cells—and less so the IL-2–producing cells—expressed CD29 (SI Appendix, Fig. S4 C and D). Of note, CD29+ TMNP were the prime IFN-γ producers in the TN subset (SI Appendix, Fig. S4C).
Fig. 5.
CD29 marks ex vivo nonnaïve CD8+ T cells with a cytotoxic signature. (A) Representative expression profile of CD29 and CD38 of peripheral blood-derived CD8+ T cells ex vivo. (B) Percentage of CD29+CD8+ T cells in naïve (TN: CD27+CD45RA+), memory (TMem: CD27+CD45RA−), effector memory (TEM: CD27−CD45RA−), and effector (TEff: CD27−CD45RA+, n = 8 donors) peripheral blood-derived CD8+ T cells. ***P < 0.001, ****P < 0.0001. (C, Left) Representative dot plot of CD45RA and CD27 expression in CD8+ T cells that were activated for 4 h with PMA–ionomycin. (C, Right) IFN-γ production and CD29 expression of TN. Representative of five donors. (D) Representative expression of CD49d by CD29+ (black) and CD29− (light gray) peripheral blood-derived CD8+ TN (n = 4 donors). (E and F) Peripheral blood-derived CD8+ T cells were activated for 6 h with MHC-I restricted peptide pools for CMV (E) or EBV (F). (Left) Representative dot plots of IFN-γ and CD29 expression and (Right) compilation of eight donors. Ratio t test. Numbers depict P value. (G) Single-cell gene expression analysis from ref. 34 of peripheral blood-derived CD8+ T cells excluding naïve T cells (for details see SI Appendix, Fig. S4 E–H). (Left) tSNE projection of ITGB1 (CD29) expression in nonnaïve CD8+ T cells [n = 1,023, log2(TPM)]. (Right) Volcano plot of differentially expressed genes in ITGB1high and ITGB1low CD8+ T cells. Cutoff for ITGB1 gene expression was determined as depicted in SI Appendix, Fig. S4I. (H) Venn diagram of the core signature of CD29+CD8+ T cells from blood and lung (the top 50 up-regulated genes in each tissue). Differential expression with absolute LFC >0.5 and p-adjusted <0.05.
To determine how CD29 expression related to antigen-specific triggering, we studied choriomeningitis virus (CMV)- and Epstein-Barr virus (EBV)-specific T cells. In healthy blood donors, CMV-specific T cells are primarily of the effector, and EBV-specific T cells of the central memory phenotype (32, 33). We activated PBMCs with CMV and EBV peptide pools and found that the IFN-γ–producing CMV- and EBV-specific T cells are primarily CD29+ (Fig. 5 E and F). In conclusion, CD29 is almost exclusively expressed by nonnaïve T cells.
scRNA-Seq Analysis Reveals the Cytotoxic Signature of CD29+ CD8+ T Cells.
To further characterize the gene expression profile of CD29+CD8+ T cells, we reanalyzed previously published single-cell RNA-seq (scRNA-seq) data of peripheral blood-derived CD8+ T cells (34). To distinguish naïve T cells from nonnaïve CD29+ T cells, we used unsupervised clustering followed by differential expression analysis. High LEF1, CCR7, and SELL gene expression and low CCL5, GZMB, and PRF1 gene expression was used to identify T cells with a TN transcriptome signature (35, 36) (SI Appendix, Fig. S4E). CCL5 gene expression highly correlated with nonnaïve T cells and was therefore used to enrich for those for downstream analysis (SI Appendix, Fig. S4 F–H).
Even though many nonnaïve CD8+ T cells express ITGB1 (CD29), its expression levels vary greatly, as revealed in the scRNA-seq analysis (Fig. 5 G, Left and SI Appendix, Fig. S4B). We therefore compared the gene expression profile of nonnaïve T cells that show high or low ITGB1 gene expression (threshold was determined as depicted in SI Appendix, Fig. S4I). High ITGB1 gene expression strongly associated with high gene expression levels of GNLY (Granulysin), GZMB (Granzyme B), and of TBX21 (T-bet) and ZNF683 (Hobit), two transcription factors associated with the cytotoxic activity of human T cells (37) (Fig. 5 G, Right and SI Appendix, Fig. S5A). We confirmed increased T-bet and Hobit protein expression in PBMC-derived CD29+CD8+ T cells (SI Appendix, Fig. S5B). Conversely, in concordance with previously described gene expression profiles (28, 38), ITGB1low CD8+ T cells associated with high gene expression levels of GZMK (Granzyme K), and the inhibitory receptor KLRB1 (CD161) (Fig. 5 G, Right and Dataset S3). The surface markers CD55 and CD127 (IL-7 receptor) that anticorrelated with IFNG/DP producers in in vitro cultured T cells (Fig. 4A and SI Appendix, Fig. S2B) were also primarily expressed by CD29− T cells ex vivo (SI Appendix, Fig. S5C).
scRNA-seq analysis of CD8+ T cells from lung tissue (34) also revealed a nonuniform expression of the ITGB1 transcript (SI Appendix, Fig. S4 J and K). Again, high ITBG1 expression associated with a cytotoxic gene expression profile (SI Appendix, Fig. S4K). When we compared the top 50 most up-regulated genes in ITGB1high T cells derived from blood and lung, and from lung and liver tissue (39), we identified a core signature gene set of ITGB1high CD8+ T cells that encompassed the cytotoxic molecules GNLY, GZMB, GZMH, and FGFBP2, and LGALS1 (Galectin-1) (Fig. 5H and SI Appendix, Fig. S4L). The fractalkine receptor CX3CR1 is also part of the ITGB1high signature (Fig. 5H). We confirmed this finding with flow cytometry analysis of blood-derived CD8+ T cells (SI Appendix, Fig. S5E). Interestingly, CX3CR1+ T cells were always CD29+ (SI Appendix, Fig. S5E). In contrast to murine CD8+ T cells (29), however, CX3CR1 expression in humans is almost exclusively found on CD45RA+CD27− effector CD8+ T cells, and only on a minority of TM or TEM cells (SI Appendix, Fig. S5F) (40). In line with the restricted expression pattern of CX3CR1, CD29 expression correlates better with production of IFN-γ and TNF-α than CX3CR1 (SI Appendix, Fig. S5 G and H). Also other surface markers such as CD49a (ITGA1), CD49b (ITGA2), CD49d (ITGA4), and CD11b (ITGAM) that we found on IFNG/DP producers (Fig. 4A and SI Appendix, Fig. S2B) were coexpressed by a subset, but not by all CD29+ blood-derived CD8+ T cells (SI Appendix, Fig. S5I). Conversely, CD74 was restricted to activated T cells and not expressed on CD8+ T cells ex vivo (SI Appendix, Fig. S5D). Thus, ITGB1 (CD29) gene expression robustly identifies human T cells with a cytotoxic gene expression profile in blood and in tissues.
CD29+ Gene Signature Is Prognostic for Long-Term Survival of Melanoma Patients.
We next interrogated if ITGB1 gene expression correlated with good clinical responses. We focused on skin cutaneous melanoma (SKCM), because the Cox regression analysis for survival from all The Cancer Genome Atlas (TCGA) datasets on CD8B expression identified SKCM as the only tumor type with a clear benefit of CD8+ T cell infiltration (SI Appendix, Fig. S6A, P = 0.000125). ITGB1 is expressed by many cell types, including tumor cells (41). Therefore, we used the CD29+ core signature gene set we defined in Fig. 5H as the 19 overlapping genes from the top 50 expressed genes that were coexpressed in blood- and lung-derived ITGB1high/CD29+ T cells. Intriguingly, analysis of TCGA data of melanoma patients revealed that the CD29+ core signature correlated well with a positive clinical outcome (high CD29+ signature: median = 10.08 y, low CD29+ signature: median = 4.84 y; Fig. 6A). In particular, using the patient cohort with high CD8+ T cell infiltrate levels estimated by CIBERSORT (42) further showed a clear benefit of the CD29+ core signature for long-term survival (high CD29+ signature: median = 13.5 y, low CD29+ signature: median = 5.37 y; Fig. 6B). Thus, the CD29+ core signature possesses prognostic value for melanoma patients.
Fig. 6.
CD29 gene signature is associated with better survival, and CD29+T cells are superior in killing tumor cells. (A and B) Kaplan–Meier plots of overall survival of (A) all patients suffering from SKCM or (B) of patients with high CD8+ T cell infiltrate estimates. Patients were stratified for a high or low CD29+ gene signature as defined in Fig. 5H (see Materials and Methods). Numbers indicate the numbers of patients at risk at each time point. The indicated P value was calculated using a log-rank test. (C) CD29+ or CD38+ MART1 TCR-engineered FACS-sorted CD8+ T cells were cultured for 6 h with MART1+ or MART1− tumor cells at an effector:target (E:T) ratio of 3:1. Representative dot plot of IFN-γ and IL-2 production as measured by ICCS. (D and E) MART1-TCR-expressing CD8+ T cells (total CD8+ T cells, or FACS-sorted CD29+ and CD38+) were cocultured for 20 h with CFSE-labeled MART1+ (Upper) or MART1− (Lower) tumor cells at an E:T ratio of 3:1. Dead tumor cells were determined by near-IR live-dead marker. (D) Representative dot plot, and (E) compiled data of six donors. Repeated measurement ANOVA with Tukey posttest. Numbers indicate P value. (F) Correlation of tumor killing of total CD8+ MART1-TCR–expressing T cells with the percentage of CD29+CD8+ T cells present in the T cell product (n = 34, linear regression).
MART1 TCR-Engineered CD29+ CD8+ T Cells Effectively Kill Tumor Cells.
Lastly, we examined whether CD29 expression is also indicative of high production levels of cytotoxic molecules when T cells were activated through cognate antigen recognition. We retrovirally transduced CD8+ T cells with the codon-optimized MART1-TCR that recognizes the HLA-A*0201 restricted 26 to 35 epitope of MART1 (43). FACS-sorted CD29+ and CD38+ MART1 TCR-engineered CD8+ T cells were then exposed for 6 h to a patient-derived HLA-A201+ MART1high-expressing melanoma tumor cell line (MART1+), or to the HLA-A201− MART1low-expressing cell line (MART1−) (44, 45) (Fig. 6C). MART1 TCR-engineered CD29+ T cells produced substantially higher levels of IFN-γ, TNF-α, Granulysin, and CD107α in response to MART1 antigen-expressing tumor cells than CD38+ T cells (SI Appendix, Fig. S6B). CD29+ T cells were the major source of IL-2+IFN-γ+ double producing T cells, and CD38+ T cells were enriched for IL2 single producers (Fig. 6C and SI Appendix, Fig. S6B).
We then determined whether MART1 TCR-engineered CD29+ T cells were also more potent in killing tumor cells. CFSE-labeled MART1+ and MART1− tumor cells were cocultured for 20 h with FACS-sorted MART1 TCR-engineered total CD8+ T cells or with FACS-sorted CD29+ or CD38+ T cells. We found that MART1+ tumor cells, but not MART1− cells, were effectively killed by MART1 TCR-engineered T cells (Fig. 6D). Notably, in line with their cytotoxic gene and protein expression profile, CD29+ T cells showed superior killing when compared to CD38+ T cells (Fig. 6 D and E). This augmented killing capacity by CD29+ T cells was observed at each effector:target ratio tested (SI Appendix, Fig. S6C). Intriguingly, when we tested the killing capacity of MART-1 TCR-engineered T cells from 34 individual donors, we found that the percentage of CD29-expressing T cells strongly correlated with the killing capacity of the T cell product (P = 0.0096; Fig. 6F). Thus, CD29 expression identifies T cells with the highest cytotoxic potential in response to tumor cells.
Discussion
In this study, we present how RNA-seq and mass spectrometry analysis can be performed from formaldehyde-fixed, permeabilized primary CD8+ T cells. This method uses the endogenous protein expression measured by intracellular cytokine staining for selection, resulting in a reliable isolation of different populations without the need of genetic modifications (11). We anticipate that one can readily adapt this rapid and cost-effective method to select cells with other features, such as differential chemokine or transcription factor expression.
Our study showed that the differential cytokine production of human CD8+ T cells reflects stable, T cell-intrinsic properties. Whereas IL2 producers display a gene expression profile resembling helper T cells, IFNG producers express a cytotoxic core signature. With CD29 and CD38, IFNG-DP producers can be identified and separated from DN-IL2 producers even prior to T cell stimulation, a feature that was maintained upon prolonged in vitro cultures. Importantly, scRNA-sequencing analysis on blood- and lung-derived CD8+ T cells revealed that the cytotoxic core signature of CD29+ T cells is not limited to in vitro T cell cultures but is also found ex vivo. A cytotoxic gene expression of tumor-infiltrating T cells has been associated with better patient survival (46, 47). In line with that finding, TCGA analysis in SKCM patients also indicates that the CD29 core gene signature is a good prognostic marker for long-term survival. In fact, it further stratifies long-term survival in SKCM patients with high CD8+ T cells infiltration, which is already by itself a good predictor.
We found that CD29 expression marks nonnaïve T cells. CX3CR1+ T cells are also included in this population. However, because CX3CR1 is primarily expressed in effector T cells in humans that are known to poorly expand in vitro (29), we consider CD29 superior to identify IFN-γ producers with a cytotoxic expression profile that also showed a good proliferative capacity. Selecting for CD29+ T cells resulted in the generation of TCR-engineered T cell products with improved killing of target cells at least in vitro.
How CD29 expression is induced and maintained on CD8+ T cells is yet to be determined. We found a great variation in the percentage of CD29+ T cells between blood donors. It is conceivable that this donor-individual difference in CD29 expression stems from a differential pathogenic and microbial exposure. Yet, CMV and EBV infections efficiently generated CD29+ IFN-γ–producing T cells, indicating that a variety of pathogens may yield cytotoxic CD29+ T cells. Of note, the clear correlation with CD29 expression and cytokine production was not detected in mice housed under specific pathogen-free (SPF) conditions. This may again point to the possibility that pathogen exposure contributes to the generation of nonnaïve cytotoxic CD29+ T cells. Which TF regulates ITGB1/CD29 or CD38 gene expression in IL2 or IFNG producers, however, remains to be revealed. We found that 15 TFs with increased transcript levels in IFNG producers (Fig. 1E) have putative binding sites in the IGTB1/CD29 promoter/enhancer (GeneHancerID GH10J032947): EED, RXRA, SOX13, ATF3, ARNT, ZNF395, HLF, PRDM1, RARA, CREB1, NCOA1, ZNF592, JUN, PHF21A, and SMAD5. Likewise, four TFs (STAT5A, NFATC1, TRIM22, and IKZF1) associated with IL2 producers could interact with the CD38 promoter/enhancer (GeneHancerID GH04J015778). Whether these TFs govern CD29 and/or CD38 expression is however yet to be defined.
Previous studies showed that the cytokine production profile of CAR T cells was highly diverse (2). As culturing T cells with different cytokines did not alter the relative distribution of CD29+ and CD29− T cells, we propose to select for donors with high CD29 expression, or to select for CD29+ T cells in case of low percentages of CD29+ T cells to generate effective cytotoxic T cell products. Similarly, it could help identify vaccines with a high potential to induce cytotoxic T cells. Selecting for CD29+ cytotoxic T cells to generate T cell products may also come with a positive side effect. IL2 producers displayed a Th2 gene expression profile, which includes IL-3, IL-4, IL-11, LIF, and PTGS2 (COX-2). A Th2 cytokine profile may impede antitumoral responses (48). In fact, IL2 and DN populations produce PTGS2 (COX-2) that was recently shown to suppress antitumoral immunity (49, 50). It is therefore conceivable that removing IL2/DN producers from T cell products may already support the antitumoral responses of CD29+ T cells.
Another factor that impedes effective antitumoral T cell responses is the failure to migrate into the tumor tissue (47, 48). Whereas β1 (CD29) and β2 integrins appear not to be required for T cell migration into at least some tumor types (51), the expression of metalloproteinases is critical for this process (52). Intriguingly, we found that IFNG/DP producers also express higher transcript levels of the metalloproteinase ADAM15 and ADAM9, which could potentially support their intratumoral migratory capacity. An additional critical feature of an effective T cell response is their capacity to expand. We show here that FACS-sorted CD29+ T cells divide at least as effectively, if not slightly better than CD38+ T cells. It is therefore conceivable that CD29+ T cells also accumulate in vivo, a critical feature for, e.g., the killing of B cell lymphomas by CAR T cells (53).
In summary, our data show that RNA-seq and mass spectrometry analysis after intracellular cytokine staining is a powerful tool to define distinct T cell populations. This allowed us to uncover the differential make-up of CD8+ T cell responses that is conserved and sustained, marked by CD29. Our findings may not only help improve T cell products for therapeutic purposes in the future, but they also provide a robust marker to study IFN-γ+ CD8+ T cells with cytotoxic properties.
Materials and Methods
T Cell Activation.
PBMCs from deidentified healthy volunteers were isolated by Lymphoprep (density gradient separation; StemCell) and stored in liquid nitrogen until further use. The study was performed according to the Declaration of Helsinki (seventh revision, 2013). Written informed consent was obtained (Sanquin Research, Amsterdam, The Netherlands).
For RNA-seq and MS analysis, cryopreserved blood from three independently obtained pools of 40 blood donors depleted for monocytes was used. CD8+ T cells were enriched to a purity of ±80% with the CD8 magnetic-activated cell sorting (MACS) positive isolation kit (Miltenyi). Validation essays were performed with cryopreserved blood from individual donors without MACS isolation.
T cells were activated as previously described (5). Briefly, nontissue culture-treated 24-well plates (Corning) were precoated overnight at 4 °C with 4 μg/mL rat α-mouse IgG2a (MW1483, Sanquin) in phosphate-buffered saline (PBS). Plates were washed and coated for >2 h with 1 μg/mL α-CD3 (clone Hit3a, eBioscience) at 37 °C. The 0.8 × 106 CD8+ enriched PBMCs/well were seeded with 1 µg/mL soluble α-CD28 (clone CD28.2, eBioscience) in 1 mL Iscove’s Modified Dulbecco’s Media (IMDM) (LONZA) supplemented with 8% fetal calf serum (FCS), 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM l-glutamine. After 48 h of incubation at 37 °C, 5% CO2, cells were harvested, washed, and further cultured in standing T25/75 tissue culture flasks (Thermo Scientific) at a density of 0.8 × 106/mL, supplemented with 10 ng/mL human recombinant IL-15 (Peprotech). Medium was refreshed every 5 to 7 d. Alternatively, cells were cultured without cytokine or, with human recombinant IL-2 (50 IU/mL), IL-6 (10 ng/mL), IL-9 (10 ng/mL), IL-11 (10 ng/mL), and IL-21 (50 ng/mL).
Preparation of Activated and Fixed T Cells for RNA-Seq and MS Analysis.
T cells were activated with 10 ng/mL PMA and 1 µM ionomycin (Sigma-Aldrich) for 4 h in the presence of 1:1,000 monensin (eBioscience). Cells were stained in sterile PBS for 30 min at 4 °C with anti-CD8α (RPA-T8, BD) and near-infrared (near-IR) live-dead marker (Thermo Fisher). ICCS was performed with CytoFix-CytoPerm kit (BD) according to the manufacturer’s protocol. Prior to use, CytoFIX was preincubated on ice for 2 min with 40 IU/mL murine RNase inhibitors (M0314, NEB). After 15 min of incubation at 4 °C, cells were washed once with 1xCytoPerm that was preincubated with 40 IU/mL RNase inhibitors. Cells were then incubated in 1xCytoPerm (preincubated with RNase inhibitors) for 15 min at 4 °C. Cells were then washed and resuspended in sorting buffer (2× SSC: 300 mM NaCl, 30 mM sodium citrate, 80 IU/mL RNase inhibitor). Antibodies directed against IL-2 (MQ1-17H12) and IFN-γ (4S.B3) were incubated overnight in sorting buffer at 4 °C. Cells were washed once and resuspended in sorting buffer prior to FACS sorting.
To evaluate the protein recovery of fixed samples compared to fresh samples, we used 5 × 106 PBMCs treated with CytoFix-CytoPerm kit (BD) according to the manufacturer’s protocol or left in PBS. The samples were washed once with PBS, snap frozen in liquid nitrogen, and used for MS analysis.
FACS Sorting.
The precooled FACSAria III (BD) sorter was washed once with 70% ethanol, followed by a 5-min wash with cleaning buffer (20× SSC: 3 M NaCl, 300 mM sodium citrate, 400 IU/mL RNase inhibitors). Sorted cells (excluding cell doublets and dead cells) were collected in 500 µL sorting buffer. Cells were spun for 20 min at 4 °C at 4,000 rpm (2,820 relative centrifugal force [RCF]). The cell pellet was transferred in 1 mL sorting buffer in lo-Bind tubes (Eppendorf) and spun at 10,000 × g for 5 min in a table-top centrifuge (Eppendorf). Supernatant was removed, and pellet was frozen at −80 °C until further use.
Live cells were FACS-sorted according to standard procedures. Antibody staining was performed in PBS + 1% FCS for 30 min at 4 °C for live-dead marker (Invitrogen), α-CD8 (RPA-T8, SK1), α-CD29 (Mar4, BD), α-CD38 (HIT2, Biolegend), and for murine α-TCRβ when selecting for MART1-TCR-expressing T cells (H57-597). Cells were washed once with culture medium and sorted on a precooled FACSAria III sorter. Sorted cells were collected in culture medium.
Flow-Cytometry Analysis.
Cells were stained in PBS + 1% FCS for live-dead marker and antibodies against: CD3 (UCHT1), CD8 (RPA-T8, SK1), CD29 (Mar4), CD38 (HIT2), CD55 (JS11), CD11b (ICRF44), CD27 (CLB-27), CD49a (TS2/7), CD49b (P1E6-C5), CD49d (9F10), CD39 (EbioA1), CD40L (24-31), CD45RA (HI100), CD5 (M1649), CD70 (Ki-24), CD74 (M-B741), CD80 (L307.4), CD81 (JS-81), CD103 (B-ly7), CD122 (Mik-B3), CD137 (4B4-1), CD226 (11-A8), CD360 (17A12), CCR4 (1G1), CX3CR1 (2A9-1), GITR (108-17), HLA-DR (LN3), ICOS (C398.4A), IL18R1 (H44), IL7R (HIL-7R-M21), KLRG1 (HP-3D9), lag3 (3DS223H), Ox40 (ACT35), PD-L1 (MIH1), for MART1-TCR selection murine TCR-B (H57-597). The cytokine production profile was determined by ICCS after activation with PMA–ionomycin for 4 h. Cells were prepared with a CytoFIX-CytoPerm kit following manufacturer’s protocol. Cells were stained with antibodies against IFN-γ (4S.B3), IL-2 (MQ1-17H12), TNF-α (MAb11), Granulysin (DH2), Granzyme A (CB9), Granzyme B (GB11), CCL3 (CR3M), CCL4 (FL34Z3L), CCL5 (21445), and Perforin (dG49). Staining with antibodies against T-bet (4B10) and Hobbit (Sanquin-Hobit/1) was performed with the Foxp3/Transcription Factor Staining kit (eBioscience) according to the manufacturer’s protocol. To measure cell proliferation, CD29+ and CD38+ FACS-sorted T cells were labeled with 3 μM CFSE (Cayman Chemicals) according to standard protocols. To measure CMV or EBV reactivity of CD8+ T cells, MHC Class I-restricted peptide pools (Promix, ProImmune) supplemented with α-CD28 (1 μg/mL) were used to stimulate cells for 6 h. Cells were acquired on LSR II, Fortessa, or Symphony (all BD) using FACS Diva v8.0.1 (BD). Data were analyzed with FlowJo VX (TreeStar).
Sample Preparation for RNA-Sequencing.
For validation assays, cell pellets were defrosted on ice and resuspended in proteinase K digestion buffer (20 mM Tris⋅HCl [pH 8.0], 1 mM CaCl2, 0.5% SDS) and 75 µg molecular grade proteinase K (Life Technologies). After a 1-h incubation at 55 °C, TRIzol-chloroform extraction was performed with 500 μL according to the manufacturer’s protocol (Invitrogen).
For RNA-seq analysis, RNA was isolated with the RNAeasy formalin-fixed paraffin embedded (FFPE) kit (Qiagen) according to the manufacturer’s protocol, omitting the steps directed at deparaffinization. RNA was resuspended in RNase-free water. RNA concentration was measured with nanodrop (Thermo Fisher), and RNA integrity was determined with the RNA 6000 Nano assay on the Bioanalyser 2100 (Agilent). Sequencing was performed with stranded “FFPE-library preparation” (GenomeScan), including ribosomal RNA depletion with the Next rRNA Depletion kit (NEB E6310). Sequencing was performed on Illumina HiSEq. 4000, with an average depth of 21 million paired-end 150-nt (PE150) reads.
Quantitative PCR Analysis.
For validation experiments, cDNA preparation was performed with random hexamers using Super Script III reverse transcription (Invitrogen) according to the manufacturer’s protocol. RT-qPCR primers were designed with the National Center for Biotechnology Information (NCBI) primer blast tool (54), and manufactured by Sigma. qPCR primer pairs were used when standard curves showed an r2 >0.98. RT-qPCR was performed with Power SYBR-green (Thermo Fisher) with the standard protocol (Tm = 60 °C for 1 min) on a 7500 Real-Time qPCR system (Applied Biosystems). RT-qPCR data were analyzed with 7500 Software v2.3 (Applied Biosystems), and plotted using GraphPad PRISM. Primers are as follows: β-Actin (Fwd AGAGCTACGAGCTGCCTGAC, Rev AGCACTGTGTTGGCGTACAG); RPS18 (Fwd CAGAAGTGACGCAGCCCTCTA, Rev AGACAACAAGCTCCGTGAAGA); and GAPDH (Fwd GAGACTCGTGCAATGGAGATTCT, Rev ACCCTGTTGCTGTAGCCA). IL2 and IFNG primers were previously described (5).
RNA-Seq Data Analysis.
Reads quality was inspected using fastQC version 0.11.7 (55). Reads were aligned with STAR version 2.5.0a (56) on the human genome hg38-release 92 from ENSEMBL (57). STAR was used to count reads per genes (option–quantMode GeneCounts) and DESeq2 (58) using Wald test with p-adjusted <0.01 and LFC >0.5 to isolate differentially expressed genes. DESeq2 normalized counts were used for heatmaps and plotting. Further annotation was obtained from Ensembl BioMart (57, 59) to discriminate between different types of coding and noncoding genes. Gene set enrichment analysis was performed using GSEA (60). Identification of transcription factors between CD29, CD38, and IFNG/IL2 populations was done using GeneHancer (61) annotation.
Mass Spectrometry Analysis.
Sample preparation was performed as previously described (62). Tryptic peptides were separated by nanoscale C18 reverse-phase chromatography coupled on line to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) via a nanoelectrospray ion source (Nanospray Flex Ion Source, Thermo Scientific). The MS2 ion count target was set to 1.5 × 104. Only precursors with charge state 2 to 7 were sampled for MS2. The instrument was run in top speed mode with 3-s cycles. All data were acquired with Xcalibur software.
The RAW mass spectrometry files were processed with the MaxQuant computational platform, 1.5.2.8. Proteins and peptides were identified using the Andromeda search engine by querying the human Uniprot database (downloaded February 2015). Standard settings with the additional options match between runs, label-free quantification (LFQ), and razor + unique peptides for quantification were selected. The generated “proteingroups.txt” table was filtered for potential contaminants, reverse hits and “only identified by site” using Perseus 1.5.1.6. The LFQ values were normalized using a log2 transformation. Data were imported in R and processed with the Differential Enrichment analysis of Proteomics data (DEP) R package (63) and filtered for proteins that were found in all biological replicates of at least one population. Filtered data were normalized and imputed using random draws from a Gaussian distribution centered around a minimal value (q = 0.01). Differential protein expression was determined with DEP (which uses Limma) with p-adjusted <0.05 and log2 fold change >0.5.
Single-Cell RNA-Seq Analysis.
Single-cell RNA-seq datasets were reanalysed from Guo et al. (34) and Zheng et al. (39). Count matrix was filtered for “PTC” for peripheral blood CD8+ T cells, “NTC” for normal lung or liver CD8+ T cells. Single-cell toolkit (SCTK) or ASAP (for clustering of naïve-like cells) was used for scRNA-seq data analysis (64, 65). First, the batch effect from the different patients was corrected using ComBaT in SCTK (66). Cells were filtered for >1,000 expressed genes per cell, keeping the 50% most-expressed genes after zero removal. We then log transformed and performed differential expression (DE) analysis (absolute LFC ≥0.5 and p-adjusted <0.05). CD29 grouping was determined based on the “double peak” expression distribution of ITGB1 (CD29), PTC from blood CD8+ (cutoff: 10 reads/cell), NTC from lung or liver CD8+ cells (cutoff: 32 reads/cell [lung] and 10 reads/cell [liver]) (SI Appendix, Fig. S7). Unsupervised clustering was performed in ASAP, using k-means (set to k = 4) on t-Distributed Stochastic Neighbor Embedding (t-SNE). For validation, naïve-like cells were compared to the remaining “nonnaïve-like” cells with ASAP (using Limma, LFC >1 and p-adjusted <0.05). All DE genes were used for volcano plots, and Venn diagrams show the top 50 most-upregulated DE genes.
Filtering of Differentially Expressed Genes for Functional Annotation.
Genes encoding secreted proteins were extracted from ref. 67, and DEGs, or proteins with an absolute LFC >1.5, were identified. Of note, we manually curated the list for secreted genes for obvious misannotations (histones, membrane, TF, RNA binding, CD molecules, protein without protein evidence, collagen, nuclear, nucleus, ribosomal, and mitochondrial). Similarly, genes and proteins encoding CD molecules were extracted with the corresponding gene and protein names (68), and DEGs or proteins with an absolute LFC >1.5 were identified.
Genes encoding for transcription factor were obtained from ref. 69 and filtered from the DEG. LncRNAs were filtered from DEG, excluding protein-coding genes and TCR/IG pseudogenes. Genes of experimentally validated RNA binding proteins were obtained from refs. 70–72, and filtered from the DEG.
Survival Analysis.
Oncolnc (73) was used to assess the effect of CD8B expression on different types of cancer. A Cox regression analysis for CD8B was perfomed and the −log(false discovery rate [FDR]) was used to identify responsive cancer types (FDR < 0.01).
Datasets for SKCM RNA-seq and patient survival were obtained from TCGA (https://www.cancer.gov/tcga) using University of California, Santa Cruz Xena (ref. 74; https://xenabrowser.net/datapages/). Gene signature of CD29+ CD8+ T cells (Fig. 5G) was used to isolate the gene expression from the RNA-seq SKMC TCGA dataset (in TPM+1, where TPM is transcripts per kilobase per million). TPM+1 counts were transformed into a genewise Z-score. The average of all Z-scores was calculated per patient. Patients were then stratified in two groups (high or low CD29 signature expression) using the median of the Z-score average (as described in ref. 34). Patients were also stratified for CD8+ T cell infiltrates using estimates obtained by CIBERSORT (42) on SKCM TPM+1 counts. Survminer R package was used to prepare and visualize the patient survival with Kaplan–Meier plot. Log-rank test was used to determine statistical differences in survival between group.
Generation of MART1-TCR-Expressing T Cells.
PBMCs from individual donors were activated for 48 h with α-CD3/α-CD28 as described above. Cells were harvested and retrovirally transduced with the MART1-TCR, as previously described (43). Briefly, nontissue cultured-treated 24-well plates were coated with 50 μg/mL Retronectin (Takara) overnight and washed once with 1 mL/well PBS. A total of 300 µL/well viral supernatant was added to the plate, and plates were centrifuged for 1 h at 4 °C at 4,000 rpm (2,820 RCF). The 0.5 × 106 T cells were added per well, spun for 10 min at 1,000 rpm, and incubated overnight at 37 °C. The following day, cells were harvested and cultured in T25 flasks at a concentration of 0.8 × 106 cells/mL for 6 to 8 d in presence of 10 ng/mL rhIL-15.
Functional Assays with MART1-TCR-Expressing T Cells.
MART1-TCR-transduced CD8+ T cells were FACS-sorted based on TCR expression and rested overnight in medium at 37 °C. Cytokine production was determined, as described above, by ICCS after 6 h of coculture with HLA-A2+ MART1hi Mel 526 (MART1+), or HLA-A2− MART1lo Mel 888 (MART1−) tumor cell lines (44), in a 3 to 1 effector to target (E:T) ratio. For CD107α staining, cocultured cells were supplemented with anti-CD107a, Brefeldin A, and Monensin, in the culture medium for 6 h. Cells were subsequently measured by flow cytometry. For killing assays, total CD8+ MART1 TCR-engineered T cells were FACS-sorted or sorted based on their CD29 and CD38 expression profile (CD29+ and CD38+). Tumor cells were labeled with 1.5 μM CFSE for 10 min at 37 °C in FCS-free medium and washed three times with warm culture medium. The 15 × 103 tumor cells were cocultured with MART1-TCR+ FACS-sorted T cells for 20 h, in a 3:1, 1:1, and 0.3:1 E:T ratio. Dead cells were quantified by flow cytometry with near-IR live-dead marker on CFSE+ tumor cells.
Statistical Analysis and Data Visualization.
Data generated with flow cytometry were analyzed with paired t test, repeated measurement ANOVA using GraphPad PRISM version 7. Differences were considered significant with a P value <0.05.
Plots were generated with ggplot2 version 3.0, DEP version 1.2.0, and with GraphPad. Heatmaps were generated with pHeatmap version 1.0.10 and DEP. Venn diagrams were generated with http://bioinformatics.psb.ugent.be/webtools/Venn/.
Data Availability.
All sequencing data are deposited on NCBI GEO under the accession no. GSE125497, and all MS data were deposited on PRIDE PXD012874.
Supplementary Material
Acknowledgments
We thank the blood donors for donation and E. Mul, M. Hoogenboezem, and S. Tol for FACS sorting. We are grateful to B. Popovic and D. Amsen for critical reading of this manuscript. This research was supported by the Landsteiner Foundation of Blood Transfusion Research (LSBR; Fellowship 1373), by the Dutch Science Foundation (VIDI grant 917.14.214), and by the Dutch Cancer Society (KWF Kankerbestrijding; grant 10132) all to M.C.W.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing dataset of this paper has been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/ (accession no. GSE125497), and all the mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the PRoteomics IDEntifications Database (PRIDE) partner repository with the dataset identifier no. PXD012874.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913940117/-/DCSupplemental.
References
- 1.Donia M., et al. , PD-1+ polyfunctional T cells dominate the periphery after tumor-infiltrating lymphocyte therapy for cancer. Clin. Cancer Res. 23, 5779–5788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue Q., et al. , Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J. Immunother. Cancer 5, 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma C., et al. , Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 3, 418–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Q., et al. , Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl. Acad. Sci. U.S.A. 109, 1607–1612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolet B. P., Guislain A., Wolkers M. C., Combined single-cell measurement of cytokine mRNA and protein identifies T cells with persistent effector function. J. Immunol. 198, 962–970 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Huang S., Non-genetic heterogeneity of cells in development: More than just noise. Development 136, 3853–3862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frentsch M., et al. , CD40L expression permits CD8+ T cells to execute immunologic helper functions. Blood 122, 405–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamann D., et al. , Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186, 1407–1418 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong K. L., Lew F. C., MacAry P. A., Kemeny D. M., CD40L-expressing CD8 T cells prime CD8α(+) DC for IL-12p70 production. Eur. J. Immunol. 38, 2251–2262 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Hernandez M. G. H., Shen L., Rock K. L., CD40-CD40 ligand interaction between dendritic cells and CD8+ T cells is needed to stimulate maximal T cell responses in the absence of CD4+ T cell help. J. Immunol. 178, 2844–2852 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Okoye I. S., et al. , Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. U.S.A. 111, E3081–E3090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson H., Krawczyk K. M., Johansson M. E., High salt buffer improves integrity of RNA after fluorescence-activated cell sorting of intracellular labeled cells. J. Biotechnol. 192, 62–65 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Wiśniewski J. R., Zougman A., Nagaraj N., Mann M., Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Shi S. R., Liu C., Balgley B. M., Lee C., Taylor C. R., Protein extraction from formalin-fixed, paraffin-embedded tissue sections: Quality evaluation by mass spectrometry. J. Histochem. Cytochem. 54, 739–743 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Magdeldin S., Yamamoto T., Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics 12, 1045–1058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez J. A., et al. , The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152, 743–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbons H. R., et al. , Divergent lncRNA GATA3-AS1 regulates GATA3 transcription in T-helper 2 cells. Front. Immunol. 9, 2512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman R., McHugh J., Turner M., RNA binding proteins as regulators of immune cell biology. Clin. Exp. Immunol. 183, 37–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlén M., et al. , Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Mosmann T. R., Cherwinski H., Bond M. W., Giedlin M. A., Coffman R. L., Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136, 2348–2357 (1986). [PubMed] [Google Scholar]
- 21.Piccinni M. P., et al. , Defective production of both leukemia inhibitory factor and type 2 T-helper cytokines by decidual T cells in unexplained recurrent abortions. Nat. Med. 4, 1020–1024 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Iñiguez M. A., Punzón C., Fresno M., Induction of cyclooxygenase-2 on activated T lymphocytes: regulation of T cell activation by cyclooxygenase-2 inhibitors. J. Immunol. 163, 111–119 (1999). [PubMed] [Google Scholar]
- 23.Curti A., et al. , Interleukin-11 induces Th2 polarization of human CD4(+) T cells. Blood 97, 2758–2763 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Cocchi F., et al. Identification the major HIV-suppressive factors produced by CD8+ T cells. Science 270, 1811–1815 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Auerbach D. J., et al. , Identification of the platelet-derived chemokine CXCL4/PF-4 as a broad-spectrum HIV-1 inhibitor. Proc. Natl. Acad. Sci. U.S.A. 109, 9569–9574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narni-Mancinelli E., et al. , Memory CD8+ T cells mediate antibacterial immunity via CCL3 activation of TNF/ROI+ phagocytes. J. Exp. Med. 204, 2075–2087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gattinoni L., et al. , A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengsch B., et al. , Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J. Immunol. Methods 453, 3–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Böttcher J. P., et al. , Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat. Commun. 6, 8306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohen S., et al. , The functional heterogeneity of CD8+ cells defined by anti-CD45RA (2H4) and anti-CD29 (4B4) antibodies. Cell. Immunol. 128, 314–328 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Pulko V., et al. , Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 17, 966–975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appay V., et al. , Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8, 379–385 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Hertoghs K. M. L., et al. , Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J. Clin. Invest. 120, 4077–4090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo X., et al. , Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Willinger T., et al. , Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J. Immunol. 176, 1439–1446 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Yang C., et al. , Transcriptome signatures reveal rapid induction of immune-responsive genes in human memory CD8(+) T cells. Sci. Rep. 6, 27005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira Braga F. A., et al. , Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 45, 2945–2958 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Kragten N. A. M., et al. , Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur. J. Immunol. 48, 1644–1662 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Zheng C., et al. , Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Nishimura M., et al. , Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J. Immunol. 168, 6173–6180 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Sun Q., et al. , Prognostic value of increased integrin-beta 1 expression in solid cancers: A meta-analysis. OncoTargets Ther. 11, 1787–1799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Eerland R., et al. , Manufacture of gene-modified human T-cells with a memory stem/central memory phenotype. Hum. Gene Ther. Methods 25, 277–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marincola F. M., et al. , Locus-specific analysis of human leukocyte antigen class I expression in melanoma cell lines. J. Immunother. Emphasis Tumor Immunol. 16, 13–23 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topalian S. L., Solomon D., Rosenberg S. A., Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J. Immunol. 142, 3714–3725 (1989). [PubMed] [Google Scholar]
- 46.Becht E., et al. , Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mlecnik B., et al. , Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44, 698–711 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Chen D. S., Mellman I., Oncology meets immunology: The cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Zelenay S., et al. , Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böttcher J. P., et al. , NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salmon H., et al. , Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J. Clin. Invest. 122, 899–910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahrends T., et al. , CD4+ T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity 47, 848–861.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Cazaux M., et al. , Single-cell imaging of CAR T cell activity in vivo reveals extensive functional and anatomical heterogeneity. J. Exp. Med. 216, 1038–1049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J., et al. , Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews S., FASTQC: A quality control tool for high throughput sequence data (Version 0.11.9, Bioinformatics Group, Babraham Institute, 2019).
- 56.Dobin A., Gingeras T. R., “Mapping RNA-seq reads with STAR” Current Protocols in Bioinformatics (John Wiley & Sons, Inc., Hoboken, NJ, 2015), pp. 11.14.1–11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerbino D. R., et al. , Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinsella R. J., et al. , Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, bar030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fishilevich S., et al. , GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Aalderen M. C., et al. , Label-free analysis of CD8+ T cell subset proteomes supports a progressive differentiation model of human-virus-specific T cells. Cell Rep. 19, 1068–1079 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Zhang X., et al. , Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Gardeux V., David F. P. A., Shajkofci A., Schwalie P. C., Deplancke B., ASAP: A web-based platform for the analysis and interactive visualization of single-cell RNA-seq data. Bioinformatics 33, 3123–3125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenkins D. F., et al. , Interactive single cell RNA-seq analysis with the single cell toolkit (SCTK). bioRxiv:329755 (24 May 2018).
- 66.Johnson W. E., Li C., Rabinovic A., Li C., Rabinovic A., Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Human Protein Atlas , The human secretome and membrane proteome. https://www.proteinatlas.org/humanproteome/tissue/secretome. Accessed 15 January 2018.
- 68.SIB Swiss Institute of Bioinformatics, European Bioinformatics Institute (EBI), Protein Information Resource (PIR) , UniProt - Swiss-Prot Protein Knowledgebase. https://www.uniprot.org/docs/cdlist.txt. Accessed 14 July 2017.
- 69.Human Protein Atlas , Protein classes. https://www.proteinatlas.org/humanproteome/proteinclasses. Accessed 17 January 2018.
- 70.Castello A., et al. , Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Perri J. I., et al. , Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat. Commun. 9, 4408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gerstberger S., Hafner M., Tuschl T., A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anaya J., OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2, e67 (2016). [Google Scholar]
- 74.Goldman M., et al. , The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. 10.1101/326470 (26 September 2019). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are deposited on NCBI GEO under the accession no. GSE125497, and all MS data were deposited on PRIDE PXD012874.