ABSTRACT
Klebsiella pneumoniae represents a growing clinical threat, given its rapid development of antibiotic resistance, necessitating new therapeutic strategies. Existing live-infection models feature high mortality rates, limiting their utility in the study of natural adaptive immune response to this pathogen. We developed a preclinical model of pneumonia with low overall mortality, in which previously exposed mice are protected from subsequent respiratory tract challenge with K. pneumoniae. Histologic analyses of infected murine lungs demonstrate lymphocytic aggregates surrounding vasculature and larger airways. Initial exposure in RAG1 knockout mice (lacking functional B and T cells) failed to confer protection against subsequent K. pneumoniae challenge. While administration of isolated K. pneumoniae capsule was sufficient to provide protection, we also found that initial inoculation with K. pneumoniae mutants lacking capsule (Δcps), O-antigen (ΔwecA) or both conferred protection from subsequent wild-type infection and elicited K. pneumoniae-specific antibody responses, indicating that non-capsular antigens may also elicit protective immunity. Experiments in this model will inform future development of multivalent vaccines to prevent invasive K. pneumoniae infections.
KEY WORDS: Klebsiella pneumoniae, Adaptive immunity, Murine model, Pneumonia, Capsule
Summary: This novel mouse model of nonlethal pulmonary Klebsiella pneumoniae infection allows for the exploration of mechanisms required to mount a protective memory response to K. pneumoniae in the lung.
INTRODUCTION
The opportunistic pathogen Klebsiella pneumoniae is widespread in the environment and can asymptomatically colonize the human gastrointestinal tract and other mucosal surfaces (Fung et al., 2012; Gorrie et al., 2017; Kock et al., 2016; Lin et al., 2012; Martin and Bachman, 2018; Podschun and Ullmann, 1998; Struve and Krogfelt, 2004). Over the past two decades, the emergence of antibiotic resistance determinants, including K. pneumoniae carbapenemases and extended-spectrum beta lactamases (Munoz-Price et al., 2013; Paterson et al., 2004; Santino et al., 2013), make this pathogen an increasingly severe clinical threat. By 2030, the global prevalence of third-generation cephalosporin and carbapenem resistance in K. pneumoniae infections is projected to exceed 50% (Alvarez-Uria et al., 2018). Owing to its carriage in the human population, pervasiveness in healthcare settings, and rise in antibiotic resistance, K. pneumoniae is responsible for a growing proportion of nosocomial infections, including pneumonia, urinary tract infection and sepsis (David et al., 2019; Podschun and Ullmann, 1992; Tsay et al., 2002). Vaccination or other immunotherapies may prove to be critical tools in the prevention or treatment of K. pneumoniae infections in the looming absence of effective antibiotics. Despite the urgency of this threat, no licensed K. pneumoniae vaccine is currently available, and vaccine development is hindered by our minimal knowledge of the immune responses to this pathogen.
Murine studies focused on the host immune response to K. pneumoniae have largely utilized model isolates (e.g. ATCC 43816) that are highly and rapidly lethal in mice (Fodah et al., 2014; Hsieh et al., 2013; Lau et al., 2007; Lavender et al., 2004; Lawlor et al., 2005; Lin et al., 2014; Wu et al., 2009), precluding their utility in illuminating natural adaptive immune responses to live K. pneumoniae. Instead, experiments have assessed adaptive immunity elicited by heat-killed organisms and a variety of specific K. pneumoniae immunogenic factors, including outer membrane vesicles, O-antigens, type 3 fimbriae and purified capsule (Chen et al., 2011; Cryz et al., 1985; Lee et al., 2015; Amezcua Vesely et al., 2019; Lavender et al., 2005; Trautmann et al., 2004; Hegerle et al., 2018). Early work in rodents and subsequently humans indicated that immunization with capsule elicits serotype-specific antibody responses (Cryz et al., 1985; Alcantar-Curiel et al., 1993; Cryz et al., 1988, 1984, 1986a,b); however, capsule is not the sole driver of protective immunity during infection (Lee et al., 2015; Lundberg et al., 2013). Moreover, it is currently unknown whether invasive infection in patients elicits durable protective immunity. If so, the host cell types imperative for this protection and the bacterial antigens responsible need to be identified.
To begin assessing these questions, we created a preclinical murine model of survivable K. pneumoniae lung infection, followed by subsequent re-challenge, in order to study the development of adaptive immune responses. We found that the majority of mice surviving initial infection with K. pneumoniae in the respiratory tract were protected from subsequent challenge in an adaptive immune-dependent manner. We further found that inoculation with live bacteria confers greater protection than inoculation with heat-killed organisms and that K. pneumoniae capsule, while an important immune stimulus, is not the sole antigen capable of eliciting protection.
RESULTS
Survivors of K. pneumoniae respiratory tract infection are protected from subsequent infection
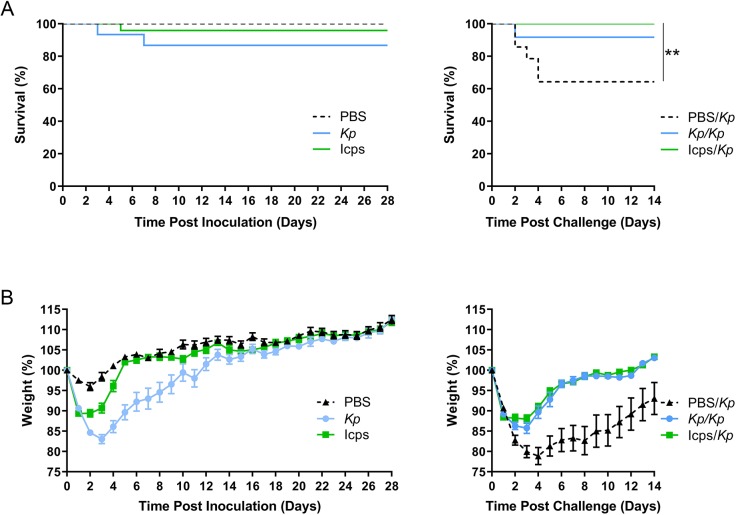
Our prior studies have introduced the K. pneumoniae strain TOP52, which causes reproducible experimental pneumonia with low overall lethality (Rosen et al., 2015). Here, we leveraged this model to elucidate whether survivors of initial inoculation would be protected from subsequent K. pneumoniae challenge. Female C57BL/6J mice were intratracheally inoculated with 107 colony-forming units (CFU) of K. pneumoniae TOP52 or sterile PBS, and weights and survival were followed for 28 days. Surviving mice were subsequently challenged intratracheally with 107 CFU of K. pneumoniae TOP52 and monitored for an additional 14 days prior to sacrifice (Fig. 1A). Following initial inoculation, all the mice that received intratracheal PBS survived for 28 days, whereas 75% of K. pneumoniae-inoculated mice survived (P=0.0170; Fig. 1B). Challenge of both groups of surviving mice with K. pneumoniae resulted in 45% mortality in mice that initially had received PBS and no deaths in the K. pneumoniae-survivor mouse group (P=0.0033; Fig. 1B). We speculate that the increase in mortality associated with TOP52 infection in the PBS/K. pneumoniae challenge group relative to the mortality of mice in the initial TOP52 inoculation group is likely related to the difference in age of the mice upon first TOP52 infection and the second invasive surgical procedure in the group with increased mortality.
Fig. 1.
Survivors of K. pneumoniae (Kp) respiratory tract inoculation are protected upon subsequent Kp challenge. (A) Schematic of experimental course in which C57BL/6J mice are intratracheally inoculated with Kp or PBS followed by challenge with Kp 28 days later. (B) Survival of mice inoculated with Kp or PBS over 28 days (left; Kp n=40, PBS n=20) and over 14 days after challenge with Kp (right; Kp/Kp n=15, PBS/Kp n=20). (C) Daily weights of mice inoculated with Kp or PBS over 28 days (left) and over 14 days after challenge with Kp (right) are presented as a percentage of weight at day 0 inoculation (left) or day 0 challenge (right). Data are shown as mean±s.e.m. and are combined from at least three independent experiments. *P<0.05, **P<0.01; Mantel–Cox log-rank test.
We measured the weights of mice throughout the experiment as an indicator of overall morbidity after infection. Mice initially inoculated with K. pneumoniae had significantly lower weights than PBS-inoculated controls from days 1-25 post-inoculation (Fig. 1C). However, after challenging all survivors with K. pneumoniae, those mice originally exposed to K. pneumoniae had significantly less weight loss from days 2-12 post-challenge compared to control mice (Fig. 1C). Together, these data demonstrate that, compared to naïve mice, survivors of initial K. pneumoniae TOP52 infection exhibit reduced morbidity and mortality upon secondary challenge with K. pneumoniae TOP52.
The K. pneumoniae protection phenotype requires the adaptive immune system
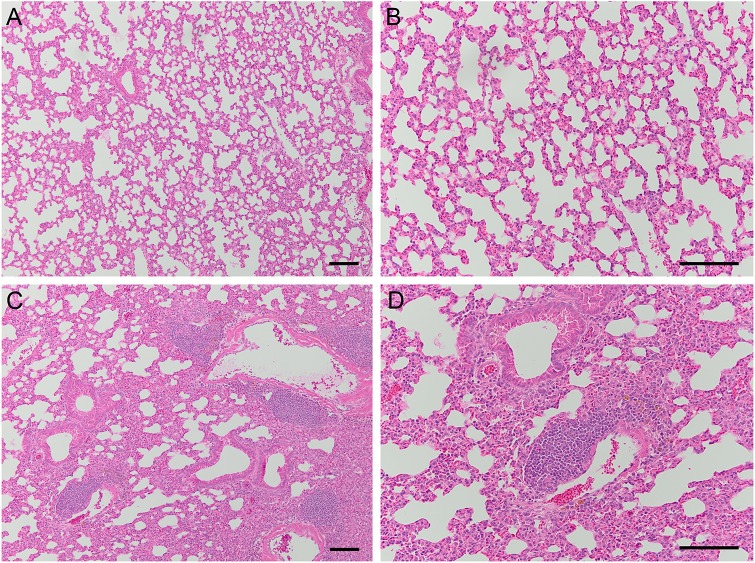
To ensure that the protective phenotype observed was not associated with persistent bacterial infection in K. pneumoniae-infected mice, we harvested spleens and lungs 28 days post-inoculation with K. pneumoniae or PBS control, prior to challenge. We found that surviving mice cleared the original inoculum, as lungs and spleen had extremely low or undetectable bacterial colonization (Fig. S1A). Histologic examination of lungs at 28 days post-inoculation with K. pneumoniae revealed a moderate inflammatory infiltrate not found in PBS-inoculated lungs (Fig. 2). Additionally, we observed perivascular collections of lymphoid cells in most K. pneumoniae-inoculated lungs (Fig. 2C,D) that were not present in PBS-inoculated lungs. Of note, mice that survived subsequent challenge with K. pneumoniae similarly harbored low or absent bacterial burden in lungs and spleens at 14 days post-challenge (Fig. S1B).
Fig. 2.
Mice inoculated with K. pneumoniae develop lymphoid aggregates in their lungs. (A-D) Representative histologic images of lungs harvested 28 days post-inoculation with PBS (A,B) or K. pneumoniae TOP52 (C,D) demonstrate a persistent inflammatory infiltrate in TOP52-inoculated organs, including the presence of perivascular lymphocytic collections. Low-power images (A,C) were taken with a 10× objective and higher-power images (B,D) were taken with a 20× objective. Scale bars: 100 µm.
To demonstrate that the observed protective phenotype upon challenge with K. pneumoniae arose from a host adaptive immune mechanism, we performed analogous experiments in RAG1−/− mice, which lack functional B and T cells. All of the RAG1−/− mice that received intratracheal inoculation with PBS survived for 28 days, whereas 68% of K. pneumoniae-inoculated RAG1−/− mice survived (P<0.0001; Fig. 3A). As expected, these rates were similar to those rates observed in initial infection of wild-type mice (Fig. 1B), indicating that the innate immune response is sufficient to control initial infection in a majority of, but not all, mice.
Fig. 3.
Survivors of Kp respiratory tract inoculation are not protected upon subsequent Kp challenge in RAG1−/− mice. (A) Survival of RAG1−/− mice inoculated with Kp or PBS over 28 days (left; Kp n=27, PBS n=31) and over 14 days after challenge with Kp (right; Kp/Kp n=25, PBS/Kp n=30). (B) Daily weights of RAG1−/− mice inoculated with Kp or PBS over 28 days (left) and over 14 days after challenge with Kp (right). Data are shown as mean±s.e.m. and are combined from at least three independent experiments. ***P<0.001; Mantel–Cox log-rank test.
Challenge of both groups of surviving RAG1−/− mice with K. pneumoniae resulted in similar mortality in the PBS and K. pneumoniae-survivor mouse groups (23% and 28% mortality, respectively; Fig. 3A). RAG1−/− mice initially inoculated with K. pneumoniae had significantly lower weights than PBS-inoculated control mice from days 1-10 post-inoculation (Fig. 3B). However, after subsequent K. pneumoniae challenge, RAG1−/− mice originally exposed to K. pneumoniae had weights similar to those of control mice, with the exception of slightly higher weights on days 8-9 (Fig. 3B). Additionally, organs collected from surviving RAG1−/− mice at 14 days post-challenge demonstrated equivalent, low bacterial burdens in both K. pneumoniae-exposed and naïve mice (Fig. S2). Together, these experiments demonstrate that the K. pneumoniae protection we observed in wild-type mice is lost in RAG1−/− mice, indicating an adaptive immune mechanism of protection.
Inoculation with heat-killed K. pneumoniae provides an intermediate level of protection
Past studies have used respiratory tract inoculation of heat-killed K. pneumoniae or extracellular vesicles to study adaptive protection, likely because infection with live organisms resulted in lethality (Chen et al., 2011; Lee et al., 2015). We next asked whether different levels of protection resulted from exposure to heat-killed versus live K. pneumoniae. We inoculated mice with 107 CFU of K. pneumoniae TOP52, the equivalent amount of heat-killed K. pneumoniae or sterile PBS control. Weights and survival were tracked for 28 days prior to challenge with live K. pneumoniae. All of the mice that were inoculated with PBS and 98% of the mice that received heat-killed K. pneumoniae survived 28 days post-inoculation, compared to 68% of those initially inoculated with live K. pneumoniae (P<0.0001 for both comparisons; Fig. 4A). After subsequent challenge with live K. pneumoniae, mice previously exposed to heat-killed bacteria demonstrated an intermediate level of protection between live K. pneumoniae-exposed mice and K. pneumoniae-naïve mice (Fig. 4A).
Fig. 4.
Heat-killed K. pneumoniae (HK Kp) respiratory tract inoculation provides an intermediate level of protection upon subsequent Kp challenge. (A) Survival of mice inoculated with Kp, HK Kp or PBS over 28 days (left; Kp n=65, HK Kp n=50, PBS n=48) and over 14 days after challenge with Kp (right; Kp/Kp n=35, HK Kp/Kp n=39, PBS/Kp n=38). (B) Daily weights of mice inoculated with Kp, HK Kp or PBS over 28 days (left) and over 14 days after challenge with Kp (right). Weight data are shown as mean±s.e.m. (C) Lung bacterial titers 48 h post-challenge with Kp. (D) Lung bacterial titers 96 h post-challenge. All data are combined from at least three independent experiments. For titers, short bars represent geometric means, and full dotted horizontal lines represent limits of detection. ns, not significant; *P<0.05, **P<0.01, ***P<0.001.
Mice initially inoculated with heat-killed K. pneumoniae had significantly lower weights than PBS control mice (days 1-9 post-inoculation) but significantly higher weights than mice inoculated with live K. pneumoniae (on days 2-14 post-inoculation) (Fig. 4B). After challenge with live K. pneumoniae, mice originally exposed to heat-killed K. pneumoniae had significantly lower weights than live K. pneumoniae survivors (on days 2-12 post-challenge) but significantly higher weights than PBS control mice (on days 6-9 post-challenge) (Fig. 4B).
Subsets of mice from this experiment were sacrificed at 48 h or 96 h post-K. pneumoniae challenge. Lungs from mice originally inoculated with heat-killed organisms or PBS control demonstrated higher bacterial titers at both 48 h (Fig. 4C) and 96 h (Fig. 4D) post-challenge, relative to mice that originally survived inoculation with live K. pneumoniae. Taken together, these data suggest that exposure to heat-killed bacteria provides modest protection from subsequent live challenge but does not confer the full protection evident in mice that survive live K. pneumoniae infection.
Isolated capsule provides protection from subsequent K. pneumoniae challenge
Capsule has been the best-studied and most important antigen in protection from K. pneumoniae for decades (Bachman et al., 2015; Cryz et al., 1985; Lawlor et al., 2005, 2006; Schembri et al., 2005; Wu et al., 2009). To determine if K. pneumoniae capsule plays a critical role in adaptive immunity in our model, we isolated capsule from K. pneumoniae TOP52. Mice initially inoculated intratracheally with this isolated capsule (Icps), live K. pneumoniae or PBS were subsequently challenged with live K. pneumoniae. Most mice initially inoculated with PBS or Icps survived, and mortality in K. pneumoniae-inoculated mice (Fig. 5A) was consistent with previous experiments. Mice inoculated with Icps had significantly lower weights than PBS control mice (on days 1-4 post-inoculation) and significantly higher weights than mice inoculated with live K. pneumoniae (on days 2-9 post-inoculation) (Fig. 5B).
Fig. 5.
Isolated capsule provides protection upon subsequent Kp challenge. (A) Survival of mice inoculated with Kp, PBS or isolated capsule (Icps) over 28 days (left; Kp n=15, PBS n=15, Icps n=24) and over 14 days after challenge with Kp (right; Kp/Kp n=12, PBS/Kp n=14, Icps/Kp n=21). (B) Daily weights of mice inoculated with Kp, PBS or Icps over 28 days (left) and over 14 days after challenge with Kp (right). Data are shown as mean±s.e.m. and are combined from at least three independent experiments. **P<0.01; Mantel–Cox log-rank test.
After challenge with K. pneumoniae, mice initially inoculated with Icps were protected from mortality, compared to mice initially inoculated with PBS (P=0.0031; Fig. 5A). Mice originally exposed to Icps also displayed significantly higher weights (on days 2-14 post-challenge), similar to those observed in K. pneumoniae-inoculated mice (Fig. 5B). These data demonstrate that a preparation of isolated capsule is sufficient to protect mice from subsequent K. pneumoniae challenge.
Inoculation of mice with K. pneumoniae deficient in capsule, O-antigen or both confers protection from subsequent wild-type K. pneumoniae challenge
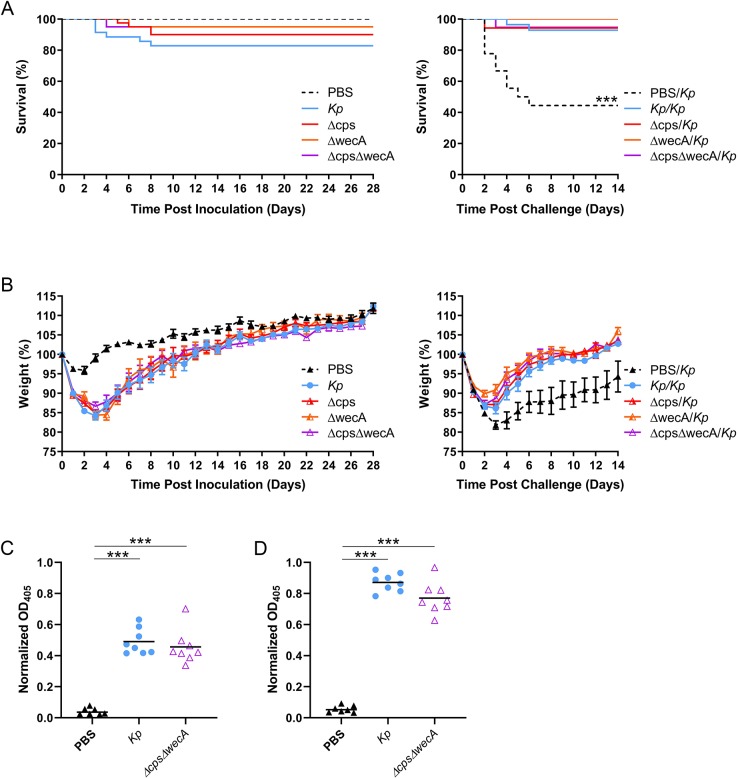
To determine if capsule was required for the protective phenotype, we constructed a capsular mutant in which a 16-kb region of the capsule synthesis operon from wzi onward was deleted in K. pneumoniae TOP52 (termed Δcps). Additionally, O-antigen may play an important role in immunity to K. pneumoniae (Trautmann et al., 2004; Hsieh et al., 2014; Pennini et al., 2017; Rollenske et al., 2018). Therefore, we also constructed a mutant deficient in the wecA gene required for O-antigen production (ΔwecA), as well as a strain deficient in both capsule and O-antigen (ΔcpsΔwecA). Upon initial inoculation of mice with 107 CFU of Δcps, ΔwecA or ΔcpsΔwecA, we observed survival rates of 90-95% (Fig. 6A). After challenge of all survivors with wild-type K. pneumoniae, we observed survival rates of 94-100% among mice initially inoculated with these mutants (Fig. 6A). This is similar to survival of mice initially inoculated with wild-type K. pneumoniae and significantly higher than survival of PBS control mice upon K. pneumoniae challenge (P<0.0001 for each of the three mutant comparisons to wild type).
Fig. 6.
Inoculation of mice with Kp deficient in capsule, O-antigen or both provides protection upon subsequent Kp challenge. (A) Survival of mice inoculated with Kp, PBS, Kp lacking capsule (Δcps), O-antigen (ΔwecA) or both (ΔcpsΔwecA) over 28 days (left; Kp n=35, PBS n=24, Δcps n=40, ΔwecA n=20, ΔcpsΔwecA n=20) and over 14 days after challenge with Kp (right; Kp/Kp n=28, PBS/Kp n=18, Δcps/Kp n=36, ΔwecA/Kp n=19, ΔcpsΔwecA/Kp n=19). (B) Daily weights of mice inoculated with Kp, PBS, Δcps, ΔwecA or ΔcpsΔwecA over 28 days (left) and over 14 days after challenge with Kp (right). Weight data are shown as mean±s.e.m. and are combined from at least three independent experiments. (C) IgG ELISAs against whole Kp TOP52 using 1:50 diluted sera at 28 days post-inoculation. (D) IgG ELISAs against whole ΔcpsΔwecA. For ELISAs, each point represents the average of a single mouse serum run in triplicate. ***P<0.001 (for survival comparing PBS/Kp to all other groups, Mantel–Cox log-rank test; for ELISAs, Mann–Whitney U-test).
Mice initially inoculated with Δcps, ΔwecA or ΔcpsΔwecA had weights similar to those of mice inoculated with wild-type K. pneumoniae and lower than those of PBS control mice (Fig. 6B). After challenge with wild-type K. pneumoniae, mice initially exposed to Δcps, ΔwecA or ΔcpsΔwecA were protected from the weight loss observed in PBS control mice (on days 3-14 post-challenge), exhibiting weights similar to those in mice initially inoculated with wild-type K. pneumoniae (Fig. 6B).
Additionally, we analyzed sera of mice 28 days after infection with K. pneumoniae TOP52, ΔcpsΔwecA or PBS (mock infection). Enzyme-linked immunosorbent assays (ELISAs), performed using plates coated with whole-cell K. pneumoniae TOP52, demonstrated high levels of reactive IgG from mice infected with either TOP52 or ΔcpsΔwecA compared to mock-infected mice (P=0.0003 for both comparisons; Fig. 6C). Moreover, we tested sera in plates coated with whole-cell ΔcpsΔwecA and similarly found high levels of reactive IgG from mice infected with either TOP52 or ΔcpsΔwecA, compared to mock-infected mice (P=0.0003 for both comparisons; Fig. 6D). Together, these data suggest that antigens beyond capsular polysaccharide and O-antigen can contribute to protective immunity against K. pneumoniae.
DISCUSSION
The emergence of hypervirulent K. pneumoniae strains and the development of extensive antimicrobial resistance make K. pneumoniae infections a worrisome and imminent threat to human health. Given this urgency, the development of vaccines or other immunotherapies to protect against K. pneumoniae infection is paramount, but is challenged by our incomplete understanding of adaptive immune responses to this pathogen. Here, we present a murine model of survivable pneumonia with live K. pneumoniae that enables exploration of natural adaptive immune response to K. pneumoniae infection of the respiratory tract. Mice that survive K. pneumoniae lung infection in our model were protected from morbidity and mortality upon subsequent challenge. While several fundamental observations regarding the nature of host response to K. pneumoniae have been made in models using heat-killed organisms (Chen et al., 2011; Amezcua Vesely et al., 2019), the present model permits the study of adaptive responses stemming from persistent bacterial-host interactions during live infection. Additionally, while most work in the field has relied on 24 h titer data as primary endpoints, this model also allows the use of morbidity and mortality to meaningfully evaluate the efficacy of potential therapies.
This murine model of immunity to K. pneumoniae can now be employed to further elucidate protective correlates of immunity. Recent work has demonstrated the contributions of T-helper 17 and tissue-resident memory T cells in the clearance of K. pneumoniae from the lung (Chen et al., 2011; Amezcua Vesely et al., 2019). Further experiments are underway in our model to identify specific host cell subsets required for protection. On the pathogen side, our most critical finding is that non-capsular antigens not only can participate in eliciting an adaptive response, but can also confer complete protection in the absence of capsule exposure. We have demonstrated serologic IgG responses to K. pneumoniae lacking both capsule and O-antigen, and although reactive IgGs have not been definitively correlated to protective immunity, they could conceivably be used to identify additional K. pneumoniae antigens. If appropriate non-capsular antigens can be identified, their inclusion in a potential vaccine might help to circumvent the limitation of serotype specificity that would accompany a capsule-based K. pneumoniae vaccine. Thus, we are currently examining the K. pneumoniae proteome in a reverse-vaccinology approach to identify bacterial antigens that could prove useful for incorporation into a multivalent vaccine to provide protection across multiple capsular serotypes.
Interestingly, we consistently observed an intermediate level of protection following inoculation of heat-killed organisms. This is important to note, given that others have studied adaptive responses using heat-killed inocula or extracellular vesicles in lieu of lethal live infections (Chen et al., 2011; Lee et al., 2015; Amezcua Vesely et al., 2019). We speculate that the nature and/or amplitude of immune responses to K. pneumoniae differ in hosts interacting with live versus heat-killed organisms or bacterial components, as observed in other infection systems (Bahjat et al., 2009; Moretti et al., 2017). It should be noted that we discerned full protection from isolated capsule inoculation alone compared to partial protection with heat-killed organisms, further suggesting a dependence of immune responses on the entire milieu of exposure at the host-pathogen or host-antigen interface. These observations are not trivial, as antigens found to be important for protection when delivered in isolation may not ultimately prove to be the most suitable vaccine candidates in the setting of dynamic host-pathogen interactions. Furthermore, the local environment of antigen delivery may play a large role in the nature of the subsequent immune response. While human vaccine delivery by intramuscular or subcutaneous injection offers some practical advantages (Miquel-Clopes et al., 2019; Neutra and Kozlowski, 2006), mucosal vaccines are already available for selected pathogens (e.g. influenza and polio) and are being studied in many infectious diseases. Using this murine model, additional work can compare and contrast the protection acquired via K. pneumoniae antigen exposure at the respiratory mucosa versus delivery of vaccine antigens at a distant site.
An intriguing finding of this work is the observation of perivascular and peribronchial lymphoid aggregates within the lungs of mice that survived K. pneumoniae infection. These lymphoid aggregates are morphologically consistent with previously described bronchus-associated lymphoid tissue (BALT) observed in murine lungs following several pulmonary infections (Chiavolini et al., 2010; Slight et al., 2013; Tan et al., 2019). In some models, the BALT functions as a set of tertiary lymphoid structures, enabling local T-cell priming and B-cell maturation. In K. pneumoniae-inoculated mice, the observed lymphoid aggregates could represent tertiary lymphoid structures required for the development of the robust pathogen-specific protection observed following challenge, or reflect less specific priming and activation (Halle et al., 2009). Future studies will interrogate the cellular composition and organization of these aggregates, as well as the relationship between proper formation of these structures and protection from re-infection.
This work introduces a system for studying adaptive immune responses to K. pneumoniae with the goal of informing future vaccine design. Correlates of protective immunity discovered and validated in this model system will ultimately require corroboration with human samples. While some promising K. pneumoniae vaccine candidates are already being studied (Hegerle et al., 2018; Feldman et al., 2019), these are unlikely to provide coverage across all strains capable of causing disease. By leveraging the present model, more conserved non-capsular (e.g. proteinaceous) antigens may be identified that could be combined with specific polysaccharide components to broaden K. pneumoniae vaccine coverage.
MATERIALS AND METHODS
Bacterial strains, mutant construction and culture conditions
K. pneumoniae strain TOP52 and mutants derived from this parent wild-type isolate were used for all experiments. Previous capsule K-typing by the Statens Serum Institut, using historical sera, identified this strain as capsular type K6. However, using the Institut Pasteur Klebsiella Sequence Typing Database and sequencing data, this isolate was found to be sequence type 152 and carry wzi allele 150, which corresponds to associated KL types of KL163, KL27 and KL46 (Johnson et al., 2014; Rosen et al., 2008; Wylie et al., 2019). A modified lambda Red recombinase protocol was utilized to construct Δcps (lacking a 16-kb region of cps operon starting from wzi) and ΔwecA using pKD46s (Bachman et al., 2015). Linear DNA required for recombination events was amplified from pKD4 using the following primers: cpsF, 5′-ATGATAAAAATTGCGCGCATTGCCGTGACGTTGGGTTTGCTTTCCTCACTGGGAGCCCAGGTGTAGGCTGGAGCTGCTTC-3′; cpsR, 5′-CTCTGCCAATCCTGTACTGACCTATAATGCCTAACAGGAATTATAAAATTAATTGCAAAGCATATGAATATCCTCCTTAG-3′; wecAF, 5′-GCTTGTGCTCCCGGTAATGGTTGAGTCATCACATCCCGTGTAGGCTGGAGCTGCTTC-3′; and wecAR, 5′-CGCTATACTTCCCGGATTAACTATGCTGAGAGCACATGCGCATATGAATATCCTCCTTAG-3′. Kanamycin cassettes were subsequently removed using the helper plasmid pCP20 encoding FLP recombinase (Datsenko and Wanner, 2000). To make ΔcpsΔwecA, the protocol for constructing the wecA mutant was applied to Δcps. All mutants were confirmed by sequencing of amplicons generated by PCR using the following primers: cps checkF, 5′-GGGTAAATGTACTTGCCTCGCCG-3′; cps checkR, 5′-AACACTCTGCCAATCCTGTACTGACC-3′; wecA checkF, 5′-GGTGTACACCAGCACGATGGC-3′ and wecA checkR, 5′-CCAGAGACAGAGAAAGCG-3′.
For preparation of murine inocula, bacteria were grown statically in 20-ml cultures at 37°C for 16 h in Luria-Bertani (LB) broth. Cultures were centrifuged at 8000 g for 10 min, and bacteria were subsequently resuspended in sterile PBS and diluted to the desired inoculum concentration by measuring optical density at 600 nm (OD600). Inocula were verified by serial dilution and plating. For heat-killed TOP52 experiments, inocula were incubated at 60°C for 30 min; plating of these aliquots confirmed the lack of live bacteria.
Mouse infections
All animal procedures complied with ethical regulations for animal testing and research and were approved by the Institutional Animal Care and Use Committee at Washington University School of Medicine. Female C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME, USA) or RAG1−/− mice (B6.129S7-Rag1tm1Mom/J) were 7-8 weeks old at the onset of all experiments. For initial inoculations, an intratracheal administration procedure was adapted from those previously described (Deng et al., 2004). Briefly, each mouse was anesthetized with inhaled isoflurane, and the trachea exposed through surgical dissection. Inoculum (20 μl containing either 1-2×107 CFU, sterile PBS or isolated capsule) was injected intratracheally using a 30-gauge, caudally directed needle. Overlying tissues were replaced and skin was closed using Vetbond (3 M Animal Care Products, St Paul, MN, USA). Mice received 1 mg/kg of buprenorphine SR subcutaneously for pain control. Mice were assessed for mortality and weighed daily. After 28 days, the majority of surviving mice (now aged 11-12 weeks) were challenged with 1-2×107 CFU TOP52 using the same method as above. Mortality and weight changes were assessed for an additional 14 days prior to sacrifice. In some instances, mice were sacrificed at 28 days post-inoculation or at other noted time points to perform histopathologic analyses or measure bacterial titers, as described below. For ELISAs, mice were inoculated via oropharyngeal aspiration as described below.
Murine organ titers and histology
Murine organs (lungs, spleens) were harvested from surviving mice at the conclusion of each experiment, or from mice at predetermined time points throughout the experiments. The right lung was prepared for bacterial titer; the left lung was processed for histology. Organs for titer were homogenized in sterile PBS via a Bullet Blender (Next Advance, Averill Park, NY, USA) for 5 min. A 200-μl aliquot was removed from the 1-ml homogenate, serially diluted and plated on LB agar. Organs for histology were washed in PBS, fixed in 10% neutral buffered formalin, dehydrated in ethanol and embedded in paraffin; 5-μm sections were stained with Hematoxylin and Eosin. Images were obtained using an Olympus DP25 camera and BX40 light microscope.
Capsule isolation
Capsular material was isolated from K. pneumoniae TOP52 as previously described (Zamze et al., 2002), with some modifications. Briefly, bacteria were cultured overnight at 37°C, shaking at 90 rpm in 3 l LB broth. The bacteria were centrifuged at 8000 g for 20 min and decanted, and the pellet was resuspended in 120 ml deionized water. The resuspended pellet was heated in a 100°C water bath for 15 min and then cooled to room temperature (RT). Then, 480 ml acetone was added to a final concentration of 80% (v/v) and stirred gently at 4°C overnight to precipitate capsular material. The precipitate was decanted, air dried for 48 h and lyophilized. Yield of isolated capsular product was quantified by uronic acid assay as previously described (Rosen et al., 2015). Mice were inoculated with 25-50 μg lyophilized capsular product resuspended in 20 μl PBS.
ELISAs
Mouse sera used for ELISAs were obtained from mice at 28 days post-oropharyngeal aspiration (Kudva et al., 2011) with 50 μl of 1-2×108 CFU of K. pneumoniae TOP52, ΔcpsΔwecA or sterile PBS. Relative IgG levels were determined for individual mice in triplicate by a method similar to one previously described (Cox et al., 2015). Briefly, 96-well flat-bottom plates were coated with ∼5×106 CFU in 100 μl per well of live K. pneumoniae TOP52 or ΔcpsΔwecA. These bacterial strains were grown statically overnight in LB broth at 37°C, centrifuged at 8000 g for 10 min, resuspended in sterile PBS (OD600=∼0.85) and diluted 1:10 in carbonate coating buffer prior to coating. Inocula were verified by serial dilution and plating. After incubating overnight at 4°C, plates were washed 3× with PBS with 0.05% Tween-20 (0.05% PBST), blocked with 5% bovine serum albumin for 1 h and washed 3× with 0.05% PBST at RT. Mouse sera were diluted 1:50 in 0.05% PBST, and 50 μl was applied per well and incubated overnight at 4°C. Plates were then washed 5× with 0.05% PBST before 100 μl of 1:5000 horseradish peroxidase-conjugated anti-mouse IgG antibodies (GE Healthcare #NA931) in 0.05% PBST were added to each well for 1 h at RT. Plates were subsequently washed 5× with 0.05% PBST, developed using ABTS peroxidase substrate (Seracare #5120-0032) for 30 min at RT and stopped with 1% sodium dodecyl sulfate. The optical density of the reaction was recorded at 405 nm using a Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA) and corrected by subtracting the negative control well values prior to analysis.
Statistical analysis
For Kaplan–Meier survival analyses, the Mantel–Cox log-rank test was used to determine differences in survival between two groups. Comparisons between two groups of normally distributed continuous variables (mouse weights) were analyzed using Student's t-tests with Holm–Sidak correction for multiple comparisons. For values not definitively normally distributed (organ titers, ELISA values), the Mann–Whitney U-test was used. All tests were two-tailed, and P-values <0.05 were considered significant. Analyses were performed using GraphPad Prism 8.02.
Supplementary Material
Acknowledgements
We thank Drs David Hunstad and Ali Ellebedy for critical feedback on this work and manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.A.R.; Methodology: J.T., C.M.S., D.A.R.; Formal analysis: D.A.R.; Investigation: J.T., J.S.N., A.A.D., D.A.R.; Data curation: J.T., J.S.N., A.A.D., D.A.R.; Writing - original draft: C.M.S., D.A.R.; Writing - review & editing: C.M.S., A.A.D., D.A.R.; Supervision: D.A.R.; Funding acquisition: D.A.R.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases [K08-AI127714], the Children's Discovery Institute of Washington University and St Louis Children's Hospital (to D.A.R.), and the National Heart, Lung, and Blood Institute [T32-HL125241 to C.M.S.].
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.043240.supplemental
References
- Alcantar-Curiel M. D., Martinez-Ramos A. and Garcia-Latorre E. (1993). [Capsular polysaccharide of Klebsiella pneumoniae. II. Immunogenic properties]. Rev. Latinoam. Microbiol. 35, 109-115. [PubMed] [Google Scholar]
- Alvarez-Uria G., Gandra S., Mandal S. and Laxminarayan R. (2018). Global forecast of antimicrobial resistance in invasive isolates of Escherichia coli and Klebsiella pneumoniae. Int. J. Infect. Dis. 68, 50-53. 10.1016/j.ijid.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezcua Vesely M. C., Pallis P., Bielecki P., Low J. S., Zhao J., Harman C. C. D., Kroehling L., Jackson R., Bailis W., Licona-Limón P. et al. (2019). Effector TH17 cells give rise to long-lived TRM cells that are essential for an immediate response against bacterial infection. Cell 178, 1176-88.e15. 10.1016/j.cell.2019.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M. A., Breen P., Deornellas V., Mu Q., Zhao L., Wu W., Cavalcoli J. D. and Gilmore M. S. (2015). Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6, e00775 10.1128/mBio.00775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahjat K. S., Meyer-Morse N., Lemmens E. E., Shugart J. A., Dubensky T. W., Brockstedt D. G. and Portnoy D. A. (2009). Suppression of cell-mediated immunity following recognition of phagosome-confined bacteria. PLoS Pathog. 5, e1000568 10.1371/journal.ppat.1000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Mcaleer J. P., Lin Y., Paterson D. L., Zheng M., Alcorn J. F., Weaver C. T. and Kolls J. K. (2011). Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity 35, 997-1009. 10.1016/j.immuni.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiavolini D., Rangel-Moreno J., Berg G., Christian K., Oliveira-Nascimento L., Weir S., Alroy J., Randall T. D. and Wetzler L. M. (2010). Bronchus-associated lymphoid tissue (BALT) and survival in a vaccine mouse model of tularemia. PLoS ONE 5, e11156 10.1371/journal.pone.0011156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B. L., Schiffer H., Dagget G. Jr, Beierschmitt A., Sithole F., Lee E., Revan F., Halliday-Simmonds I., Beeler-Marfisi J., Palmour R. et al. (2015). Resistance of Klebsiella pneumoniae to the innate immune system of African green monkeys. Vet. Microbiol. 176, 134-142. 10.1016/j.vetmic.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Cryz S. J. Jr. Fürer E. and Germanier R. (1984). Protection against fatal Klebsiella pneumoniae burn wound sepsis by passive transfer of anticapsular polysaccharide. Infect. Immun. 45, 139-142. 10.1128/IAI.45.1.139-142.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J. Jr. Fürer E. and Germanier R. (1985). Purification and vaccine potential of Klebsiella capsular polysaccharides. Infect. Immun. 50, 225-230. 10.1128/IAI.50.1.225-230.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J. Jr. Fürer E. and Germanier R. (1986a). Immunization against fatal experimental Klebsiella pneumoniae pneumonia. Infect. Immun. 54, 403-407. 10.1128/IAI.54.2.403-407.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J. Jr. Mortimer P., Cross A. S., Fürer E. and Germanier R. (1986b). Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine 4, 15-20. 10.1016/0264-410X(86)90092-7 [DOI] [PubMed] [Google Scholar]
- Cryz S. J. Jr. Cross A. S., Sadoff G. C. and Que J. U (1988). Human IgG and IgA subclass response following immunization with a polyvalent Klebsiella capsular polysaccharide vaccine. Eur. J Immun. 18, 2073-2075. 10.1002/eji.1830181230 [DOI] [PubMed] [Google Scholar]
- Datsenko K. A. and Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Nat. Acad. Sci. USA 97, 6640-6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S., Reuter S., Harris S. R., Glasner C., Feltwell T., Argimon S., Abudahab K., Goater R., Giani T., Errico G. et al. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919-1929. 10.1038/s41564-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J. C., Zeng X., Newstead M., Moore T. A., Tsai W. C., Thannickal V. J. and Standiford T. J. (2004). STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J. Immunol. 173, 4075-4083. 10.4049/jimmunol.173.6.4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. F., Mayer Bridwell A. E., Scott N. E., Vinogradov E., Mckee S. R., Chavez S. M., Twentyman J., Stallings C. L., Rosen D. A. and Harding C. M. (2019). A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc. Nat. Acad. Sci. USA 116, 18655-18663. 10.1073/pnas.1907833116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodah R. A., Scott J. B., Tam H.-H., Yan P., Pfeffer T. L., Bundschuh R. and Warawa J. M. (2014). Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS ONE 9, e107394 10.1371/journal.pone.0107394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C.-P., Lin Y.-T., Lin J. C., Chen T.-L., Yeh K.-M., Chang F.-Y., Chuang H.-C., Wu H.-S., Tseng C.-P. and Siu L. K. (2012). Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 18, 1322-1325. 10.3201/eid1808.111053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrie C. L., Mirčeta M., Wick R. R., Edwards D. J., Thomson N. R., Strugnell R. A., Pratt N. F., Garlick J. S., Watson K. M., Pilcher D. V. et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208-215. 10.1093/cid/cix270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle S., Dujardin H. C., Bakocevic N., Fleige H., Danzer H., Willenzon S., Suezer Y., Hämmerling G., Garbi N., Sutter G. et al. (2009). Induced bronchus-associated lymphoid tissue serves as a general priming site for T cells and is maintained by dendritic cells. J. Exp. Med. 206, 2593-2601. 10.1084/jem.20091472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerle N., Choi M., Sinclair J., Amin M. N., Ollivault-Shiflett M., Curtis B., Laufer R. S., Shridhar S., Brammer J., Toapanta F. R. et al. (2018). Development of a broad spectrum glycoconjugate vaccine to prevent wound and disseminated infections with Klebsiella pneumoniae and Pseudomonas aeruginosa. PLoS ONE 13, e0203143 10.1371/journal.pone.0203143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.-F., Liu J.-Y., Pan Y.-J., Wu M.-C., Lin T.-L., Huang Y.-T. and Wang J.-T. (2013). Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J. Infect. Dis. 208, 1580-1589. 10.1093/infdis/jit384 [DOI] [PubMed] [Google Scholar]
- Hsieh P.-F., Wu M.-C., Yang F.-L., Chen C.-T., Lou T.-C., Chen Y.-Y., Wu S.-H., Sheu J.-C. and Wang J.-T. (2014). D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: implication in vaccine design. Front. Microbiol. 5, 608 10.3389/fmicb.2014.00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. G., Spurbeck R. R., Sandhu S. K. and Matson J. S. (2014). Genome sequence of Klebsiella pneumoniae urinary tract isolate TOP52. Gen. Announc. 2, e00668-14 10.1128/genomeA.00668-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock R., Werner P., Friedrich A. W., Fegeler C., Becker K., Bindewald O., Bui T. T., Eckhoff C., Epping R., Kähmann L. et al. (2016). Persistence of nasal colonization with human pathogenic bacteria and associated antimicrobial resistance in the German general population. New Microb. New Infect 9, 24-34. 10.1016/j.nmni.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva A., Scheller E. V., Robinson K. M., Crowe C. R., Choi S. M., Slight S. R., Khader S. A., Dubin P. J., Enelow R. I., Kolls J. K. et al. (2011). Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J. Immunol. 186, 1666-1674. 10.4049/jimmunol.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H. Y., Clegg S. and Moore T. A. (2007). Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microbial Path 42, 148-155. 10.1016/j.micpath.2007.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender H. F., Jagnow J. R. and Clegg S. (2004). Biofilm formation in vitro and virulence in vivo of mutants of Klebsiella pneumoniae. Infect. Immun. 72, 4888-4890. 10.1128/IAI.72.8.4888-4890.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender H., Jagnow J. J. and Clegg S. (2005). Klebsiella pneumoniae type 3 fimbria-mediated immunity to infection in the murine model of respiratory disease. Int. J. Med. Microbiol. 295, 153-159. 10.1016/j.ijmm.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Lawlor M. S., Handley S. A. and Miller V. L. (2006). Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect. Immun. 74, 5402-5407. 10.1128/IAI.00244-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor M. S., Hsu J., Rick P. D. and Miller V. L. (2005). Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Micro. 58, 1054-1073. 10.1111/j.1365-2958.2005.04918.x [DOI] [PubMed] [Google Scholar]
- Lee W.-H., Choi H.-I., Hong S.-W., Kim K.-S., Gho Y. S. and Jeon S. G. (2015). Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 47, e183 10.1038/emm.2015.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T., Siu L. K., Lin J.-C., Chen T.-L., Tseng C.-P., Yeh K.-M., Chang F.-Y. and Fung C.-P. (2012). Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 12, 13 10.1186/1471-2180-12-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-L., Chuang Y.-P., Huang Y.-T., Hsieh P.-F., Lin Y.-T. and Wang J.-T. (2014). Identification of an immuno-dominant protein from Klebsiella pneumoniae strains causing pyogenic liver abscess: implication in serodiagnosis. BMC Microbiol. 14, 321 10.1186/s12866-014-0321-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg U., Senn B. M., Schüler W., Meinke A. and Hanner M. (2013). Identification and characterization of antigens as vaccine candidates against Klebsiella pneumoniae. Hum. Vacc. Immunother. 9, 497-505. 10.4161/hv.23225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. M. and Bachman M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell Infect. Microb. 8, 4 10.3389/fcimb.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel-Clopes A., Bentley E. G., Stewart J. P. and Carding S. R. (2019). Mucosal vaccines and technology. Clin. Exp. Immunol. 196, 205-214. 10.1111/cei.13285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J., Roy S., Bozec D., Martinez J., Chapman J. R., Ueberheide B., Lamming D. W., Chen Z. J., Horng T., Yeretssian G. et al. (2017). STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171, 809-23.e13. 10.1016/j.cell.2017.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Poirel L., Bonomo R. A., Schwaber M. J., Daikos G. L., Cormican M., Cornaglia G., Garau J., Gniadkowski M., Hayden M. K. et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785-796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutra M. R. and Kozlowski P. A. (2006). Mucosal vaccines: the promise and the challenge. Nat. Rev. Immunol. 6, 148-158. 10.1038/nri1777 [DOI] [PubMed] [Google Scholar]
- Paterson D. L., Ko W. C., Von Gottberg A., Mohapatra S., Casellas J. M., Goossens H., Mulazimoglu L., Trenholme G., Klugman K. P. and Bonomo R. A. (2004). International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial Infections. Ann. Intern. Med. 140, 26-32. 10.7326/0003-4819-140-1-200401060-00008 [DOI] [PubMed] [Google Scholar]
- Pennini M. E., De Marco A., Pelletier M., Bonnell J., Cvitkovic R., Beltramello M., Cameroni E., Bianchi S., Zatta F., Zhao W. et al. (2017). Immune stealth-driven O2 serotype prevalence and potential for therapeutic antibodies against multidrug resistant Klebsiella pneumoniae. Nat. Commun. 8, 1991 10.1038/s41467-017-02223-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podschun R. and Ullmann U. (1992). Klebsiella capsular type K7 in relation to toxicity, susceptibility to phagocytosis and resistance to serum. J. Med. Microbiol. 36, 250-254. 10.1099/00222615-36-4-250 [DOI] [PubMed] [Google Scholar]
- Podschun R. and Ullmann U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589-603. 10.1128/CMR.11.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenske T., Szijarto V., Lukasiewicz J., Guachalla L. M., Stojkovic K., Hartl K., Stulik L., Kocher S., Lasitschka F., Al-Saeedi M. et al. (2018). Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immun. 19, 617-624. 10.1038/s41590-018-0106-2 [DOI] [PubMed] [Google Scholar]
- Rosen D. A., Pinkner J. S., Jones J. M., Walker J. N., Clegg S. and Hultgren S. J. (2008). Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76, 3337-3345. 10.1128/IAI.00090-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D. A., Hilliard J. K., Tiemann K. M., Todd E. M., Morley S. C. and Hunstad D. A. (2015). Klebsiella pneumoniae FimK promotes virulence in murine pneumonia. J. Infect. Dis. 213, 649-658. 10.1093/infdis/jiv440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santino I., Bono S., Nuccitelli A., Martinelli D., Petrucci C. and Alari A. (2013). Microbiological and molecular characterization of extreme drug-resistant carbapenemase-producing Klebsiella pneumoniae isolates. Int. J. Immunopath. Ph. 26, 785-790. 10.1177/039463201302600325 [DOI] [PubMed] [Google Scholar]
- Schembri M. A., Blom J., Krogfelt K. A. and Klemm P. (2005). Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 73, 4626-4633. 10.1128/IAI.73.8.4626-4633.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slight S. R., Rangel-Moreno J., Gopal R., Lin Y., Fallert Junecko B. A., Mehra S., Selman M., Becerril-Villanueva E., Baquera-Heredia J., Pavon L. et al. (2013). CXCR5(+) T helper cells mediate protective immunity against tuberculosis. J. Clin. Invest. 123, 712-726. 10.1172/JCI65728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C. and Krogfelt K. A. (2004). Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 6, 584-590. 10.1111/j.1462-2920.2004.00590.x [DOI] [PubMed] [Google Scholar]
- Tan H. X., Esterbauer R., Vanderven H. A., Juno J. A., Kent S. J. and Wheatley A. K. (2019). Inducible bronchus-associated lymphoid tissues (iBALT) serve as sites of B cell selection and maturation following influenza infection in mice. Front. Immun. 10, 611 10.3389/fimmu.2019.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann M., Held T. K. and Cross A. S. (2004). O antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Vaccine 22, 818-821. 10.1016/j.vaccine.2003.11.026 [DOI] [PubMed] [Google Scholar]
- Tsay R.-W., Siu L. K., Fung C.-P. and Chang F.-Y. (2002). Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch. Int. Med. 162, 1021-1027. 10.1001/archinte.162.9.1021 [DOI] [PubMed] [Google Scholar]
- Wu M.-F., Yang C.-Y., Lin T.-L., Wang J.-T., Yang F.-L., Wu S.-H., Hu B.-S., Chou T.-Y., Tsai M.-D., Lin C.-H. et al. (2009). Humoral immunity against capsule polysaccharide protects the host from magA+ Klebsiella pneumoniae-induced lethal disease by evading Toll-like receptor 4 signaling. Infect. Immun. 77, 615-621. 10.1128/IAI.00931-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie K. M., Wylie T. N., Minx P. J. and Rosen D. A. (2019). Whole-genome sequencing of Klebsiella pneumoniae isolates to track strain progression in a single patient with recurrent urinary tract infection. Front. Cell Infect. Microbiol. 9, 14 10.3389/fcimb.2019.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamze S., Martinez-Pomares L., Jones H., Taylor P. R., Stillion R. J., Gordon S. and Wong S. Y. C. (2002). Recognition of bacterial capsular polysaccharides and lipopolysaccharides by the macrophage mannose receptor. J. Biol. Chem. 277, 41613-41623. 10.1074/jbc.M207057200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.