Abstract
First identified in 2001, human metapneumovirus (HMPV) is a novel pathogen and causative agent of acute respiratory tract infection. Re-infection with HMPV is common, and currently there is no available vaccine against HMPV infection. Two genotypes of HMPV have been identified, A and B, both of which can be divided further into at least two distinct sub-genotypes. Here we report the results of the first study to investigate the genetic variability of HMPV strains circulating within Cambodia. The overall incidence of HMPV infection amongst an all-ages population of patients hospitalised with ALRI in Cambodia during 3 consecutive years, between 2007 and 2009, was 1.7%. The incidence of HMPV infection was highest amongst children less than 5 years of age, with pneumonia or bronchopneumonia the most frequent clinical diagnoses across all age groups. The incidence of HMPV infection varied annually. As anticipated, genetic diversity was low amongst the conserved F gene sequences but very high amongst G gene sequences, some strains sharing as little as 56.3% and 34.2% homology at the nucleotide and amino acid levels, respectively. Simultaneous co-circulation of strains belonging to the HMPV sub-genotypes B1, B2 and lineage A2b, amongst patients recruited at 2 geographically distinct provincial hospitals, was detected. Sub-genotype B2 strains were responsible for the majority of the infections detected, and a significant (p = 0.013) association between infection with lineage A2b strains and disease severity was observed.
Keywords: Human metapneumovirus, HMPV, Genetic diversity, Acute lower respiratory tract infection, Respiratory disease, Epidemiology, Cambodia, South East Asia
1. Introduction
Human metapneumovirus (HMPV) is a negative single-stranded RNA virus, belonging to the Metapneumovirus genus of the Pneumovirinae subfamily of the Paramyxoviridae family (van den Hoogen et al., 2002). First identified by van den Hoogen and colleagues in 2001, HMPV is a causative agent of acute respiratory tract infections (van den Hoogen et al., 2001). Primarily at risk of severe HMPV-related disease are infants, immunocompromised patients and the elderly (Boivin et al., 2002, O’Gorman et al., 2006, Sivaprakasam et al., 2007, van den Hoogen et al., 2004b, van den Hoogen et al., 2003, Williams et al., 2005). Infection with HMPV can be both symptomatic and asymptomatic (Bruno et al., 2009). Clinical manifestations of symptomatic HMPV are indistinguishable from those of human respiratory syncytial virus (HRSV) and range from mild upper respiratory tract infection to severe disease requiring hospitalisation. Re-infection with HMPV is common and currently there is no vaccine available. Although reports have confirmed the global circulation of HMPV, data from tropical countries, especially Asia, remains limited. Similar to other pneumoviruses, HMPV virions are surrounded by a lipid membrane into which three transmembrane surface glycoproteins are inserted: the attachment glycoprotein (G), the fusion protein (F) and the small hydrophobic protein (SH) (Collins and Mottet, 1993, Skiadopoulos et al., 2006). HMPV strains can be classified on the basis of genetic variability within the F and G genes. Two HMPV genotypes have been identified, A and B, both of which can be divided further into the distinct sub-genotypes B1, B2, A1, A2 (van den Hoogen et al., 2004a). In addition, the sub-genotype A2 comprises two lineages, A2a and A2b (Huck et al., 2006, Ishiguro et al., 2004).
The global circulation patterns of HMPV are variable and complex. Not only can the incidence of HMPV infection vary from year to year, but it is now well understood that HMPV epidemics are highly localised and community based; that strains of both genotypes, all sub-genotypes and lineages can circulate concurrently within a given location during a given season (Kahn et al., 2008, Papenburg and Boivin, 2010, van den Hoogen et al., 2004b). Furthermore, in localised HMPV epidemics worldwide, the predominant circulating strains can vary annually. In order to generate a globally effective vaccine, the genetic heterogeneity of circulating HMPV must be well understood. Here we report the results of the first study to investigate the seasonality and genetic variability of HMPV strains circulating in Cambodia.
2. Materials and methods
2.1. Sample collection and screening
The samples analysed in this study were collected as part of the Surveillance and Investigation of Epidemic Situations in Southeast Asia (SISEA) project. The aim of the SISEA project was to investigate the prevalence of endemic and emerging pathogens in South-East Asia. Between June 2007 and December 2009, a total of 3858 samples were collected from patients admitted with symptoms of acute lower respiratory infection (ALRI) to Takeo (southern Cambodia) and Kampong Cham (central-east Cambodia) provincial hospitals. For patients aged < 5 years, ALRI was defined as an illness of < 10 days duration that consisted of cough or difficult breathing, plus tachypnea. For patients aged between 5 and 14 years, case definition included the above symptoms as well as a fever ≥ 38 °C. For patients aged 15 years or older ALRI was defined as a fever ≥ 38 °C, plus chest pain or tachypnea or auscultatory crackles (WHO, 2005). A severe case for patients < 5 years of age was defined as ALRI in addition to: respiratory rate ≥ 60/min (age < 2 months), ≥ 50/min (age < 11 months), or ≥ 40/min (ages between 1 and 5 years), oxygen saturation < 93%, cardiac frequency > 180 beats per minute (bpm) for infants < 1-year-old or > 140 bpm amongst the 1–5 years age group, clinical respiratory distress, according to the World Health Organisation (WHO) guidelines (WHO, 2005). A severe case for patients > 5 years of age was defined as the above symptoms with at least 2 of the following: systolic blood pressure < 90 mmHg, cardiac frequency > 120 bpm, respiratory rate ≥ 30/min, oxygen saturation < 93%, body temperature < 35 °C or ≥ 40 °C.
Nasopharyngeal and throat swabs were collected from each patient and immediately introduced into a sterile tube containing virus transport medium (VTM). Samples were kept in liquid nitrogen following collection and during transport to the Institut Pasteur in Phnom Penh, where samples were aliquoted and stored at −80 °C. All samples were screened using 5 multiplex reverse transcriptase-polymerase chain reaction (RT-PCR) and PCR assays at the Institut Pasteur of Cambodia for the presence of 18 common and novel respiratory viruses including HMPV, HRSV, human bocavirus (HBoV), Influenza A and B viruses, coronaviruses OC43, 229E, HKU1 and NL63, severe acute respiratory syndrome-associated coronavirus (SARS-CoV), parainfluenza viruses 1–4, adenoviruses, rhinovirus and enterovirus, as previously reported (Buecher et al., 2010). Using the RT-PCR assays described, no cross amplification or false positive results were observed (Buecher et al., 2010). Patients whose samples tested positive for HMPV were included in the study. In addition we attempted to collect sputum samples, and requested chest X-rays, from each enrolled patient.
This study was approved by the National Ethics Committee of Cambodia. All patients/parents of sick children who participated in this study provided written informed consent.
2.2. RT-PCR and sequencing
Viral RNA was extracted from 140 μL of VTM using the Qiagen Viral RNA Mini kit (Qiagen, CA, USA) as per the manufacturer's instructions. Viral RNA was eluted in 60 μL of Qiagen AVE buffer and stored at −80 °C until required.
RT-PCR was used to amplify a fragment of the F and G gene open reading frames (ORFs). RT-PCR was performed using the Qiagen One-Step RT-PCR kit (Qiagen) as per the manufacturer's instructions, and the following published primer sequences: HMPVF Fwd 5′-CAATGCAGGTATAACACCAGCAATATC-3′ and HMPVF Rev 5′-GCAACAATTGAACTGATCTTCAGGAAAC-3′ (F gene) (van den Hoogen et al., 2004a); HMPVG Fwd 5′-GAGAACATTCGRRCRATAGAYATG-3′ and HMPVG Rev 5′-AGATAGACATTRACAGTGGATTCA-3′ (G ORF) (Ludewick et al., 2005). Thermocycling was performed under the following conditions: reverse transcription at 50 °C for 30 min, PCR activation at 94 °C for 15 min, 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 1 min, extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. All PCR products were visualised using ethidium bromide under UV light on a 1.5% agarose gel. When required, PCR products were purified using the QIAquick Gel Extraction protocol of the QIAquick PCR Purification kit (Qiagen) prior to sequence analysis. Single-pass sequencing reactions were performed at a contract sequencing facility using the ABI BigDye Terminator Cycle Sequencing kit on an ABI 3730XL automatic DNA Analyser (Macrogen, Seoul, Korea).
2.3. Phylogenetic analysis
Consensus sequences were generated using CLC Main Workbench software, version 5.6.1 (http://www.clcbio.com). Nucleotide sequences of reference strains from the two main HMPV genotypes and sub-lineages were obtained from Genbank, and used to construct alignments and phylogeny (Table 1 ). Cambodian and reference HMPV nucleotide sequences were aligned using the Clustal W alignment program of Mega software, version 4.0 (Tamura et al., 2007). The average pairwise Jukes–Cantor distance was found to be 0.08 and 0.6 for the HMPV F and G alignments, respectively, indicating that the data were suitable to construct Neighbour-Joining trees (Nei and Kumar, 2000). Trees were constructed using the p-distance nucleotide substitution model, with 1000 bootstrap replicates, using Mega 4.0 software. An avian metapneumovirus (AMPV) type C strain (DQ009484) was used to root both HMPV F and G phylogenetic trees. The nucleotide sequences of HMPV strains obtained in this study were deposited in GenBank under the accession numbers HQ864230–HQ864312.
Table 1.
Reference HMPV strains used in this study to construct phylogeny. Shown are the name of the strain, country/place of isolation, Genbank accession number and lineage. Whether strains were included in Fig. 2 (HMPV F phylogeny), Fig. 3 (HMPV G phylogeny) or Fig. 2, Fig. 3 (both HMPV F and G phylogenies) is indicated.
| Strain | Origin | Genbank accession number | Lineage | Figure(s) |
|---|---|---|---|---|
| NL/1/94 | Netherlands | AY355335 | B2 | Fig. 2, Fig. 3 |
| NL/1/99 | Netherlands | AY304361 | B1 | Fig. 2, Fig. 3 |
| NL/00/1 | Netherlands | AF371337 | A1 | Fig. 2 |
| NL/00/17 | Netherlands | AY355324 | A2 | Fig. 2, Fig. 3 |
| NL/12/00 | Netherlands | AY296096 | B1 | Fig. 3 |
| NL/3/93 | Netherlands | AY296025.1 | A2b | Fig. 3 |
| JPS03-240 | Japan | AY530095 | A2b | Fig. 2, Fig. 3 |
| JPS05-21 | Japan | EU131160 | B2 | Fig. 3 |
| JPS03-180 | Japan | AY530092.1 | A1 | Fig. 3 |
| JP-O0601 | Japan | EF589610.1 | A2b | Fig. 2 |
| Can98-75 | Canada | AY297748.1 | B2 | Fig. 2, Fig. 3 |
| Can40-02 | Canada | AY574226.1 | A1 | Fig. 3 |
| Can00-16 | Canada | AY145301.1 | A2a | Fig. 2 |
| Can534-02 | Canada | AY574245.1 | A1 | Fig. 3 |
| Can99-81 | Canada | AY574224.1 | A1 | Fig. 2, Fig. 3 |
| Can182-02 | Canada | AY574230.1 | A2a | Fig. 3 |
| Can215-02 | Canada | AY574236.1 | A2a | Fig. 3 |
| Can97-83 | Canada | AY297749.1 | A2a | Fig. 2, Fig. 3 |
| CHN05-06 | China | EF571504.1 | A2b | Fig. 3 |
| CHN06-06 | China | EF571505.1 | A2b | Fig. 3 |
| CHN09-06 | China | EF571508.1 | A2b | Fig. 3 |
| BJ1887 | China | DQ843659.1 | A2b | Fig. 2, Fig. 3 |
| CHN12-06 | China | EF571511.1 | B1 | Fig. 3 |
| CHN_N09 | China | GU048716.1 | B1 | Fig. 2 |
| CHN_N36 | China | GU048743.1 | B1 | Fig. 2 |
| BJ4944 | China | DQ270222.1 | B2 | Fig. 3 |
| Arg-1-98 | Argentina | DQ362948 | A1 | Fig. 3 |
| Arg-1-00 | Argentina | DQ362947.1 | B1 | Fig. 2 |
| Arg-2-00 | Argentina | DQ362954.1 | B1 | Fig. 3 |
| Arg-3-00 | Argentina |
DQ362952.1 (G) DQ362941.1 (F) |
A2a | Fig. 2, Fig. 3 |
| Arg-4-00 | Argentina |
DQ362953.1 (G) DQ362942.1 (F) |
A2a | Fig. 2, Fig. 3 |
| Arg-5-00 | Argentina |
DQ362957.1 (G), DQ362945.1 (F) |
B1 | Fig. 2, Fig. 3 |
| RSA30-01 | Republic of South Africa (RSA) | AY848906.1 | A1 | Fig. 3 |
| RSA19-01 | Republic of South Africa (RSA) | AY848892.1 | A1 | Fig. 3 |
| RSA_58_00 | Republic of South Africa (RSA) | AY848869.1 | B2 | Fig. 3 |
| KR-108-04 | South Korea | DQ061254.1 | B1 | Fig. 2 |
| KR-95-04 | South Korea | DQ061253.1 | B1 | Fig. 2 |
| SIN06-NTU84 | Singapore | EF397621.1 | A2b | Fig. 2 |
| TWN_05-00125 | Taiwan | EF535506.1 | B2 | Fig. 2, Fig. 3 |
| Peru6_2003 | Peru | DQ393720.1 | B2 | Fig. 3 |
| Peru1_2002 | Peru | DQ393715.1 | B2 | Fig. 3 |
| UY_1_07_1 | Uruguay | GQ888743.1 | B2 | Fig. 3 |
| Q01-7182 | Australia | AY327802.1 | A1 | Fig. 3 |
| Q01-4199 | Australia | AY327803.1 | A1 | Fig. 3 |
| IND06-14 | India | EU259867.1 | A2b | Fig. 3 |
| IND06-10 | India | EU259870.1 | A2b | Fig. 3 |
| FL-4-01 | Finland | AY296015.1 | A1 | Fig. 3 |
| UK-5-01 | United Kingdom (UK) | AY296047.1 | B2 | Fig. 3 |
| TN93-3-2 | Tennessee, USA | EU857589.1 | B2 | Fig. 2 |
| TN93-6-16 | Tennessee, USA | EU857590.1 | B2 | Fig. 2 |
| TN89-7-13 | Tennessee, USA | EU857569.1 | B1 | Fig. 2 |
2.4. Statistical analysis
Significant pairwise associations were assessed using Fisher exact test. A p value < 0.05 was considered significant. Statistical analyses were performed using STATA/SE 11.1 (Statacorp., College Station, TX, USA).
3. Results
3.1. HMPV circulation in Cambodia
Of the 3858 patients tested between 2007 and 2009, 65 (1.7%) were positive for HMPV by multiplex real time RT-PCR. Of these, 40 (61.5%) originated from Takeo hospital and 25 (38.5%) from Kampong Cham hospital. Between June and December 2007, two (3%) HMPV positive samples were identified. In 2008 and 2009, 58 (89%) and 5 (8%) samples were detected, respectively (Fig. 1A). Amongst the study population, the proportion of HMPV infection was 0.2% in 2007, 4.3% in 2008 and 0.3% in 2009. Cambodia is a tropical country, and as such, experiences dry and rainy seasons of approximately equal duration. Dry seasons lie typically from November to April, and wet seasons from May/June to October/November (Mardy et al., 2009, Vance et al., 2004) (Fig. 1B). In 2008, the incidence of HMPV infections peaked during the wet season from April to October, with 84.6% (n = 55) of all positive specimens collected during this period (Fig. 1A and B). Between July and September 2008, positivity rates for HMPV reached 10% of the collected samples (Fig. 1A and B). In 2007 and 2009, the few cases identified occurred during November and December. The low number of HMPV-positive specimens observed during 2007 and 2009 was not due to a particular reduction in the number of clinical samples collected during coincident time periods.
Fig. 1.

(A) Seasonal distribution of HMPV positive samples. Data represent the incidence of HMPV infections amongst the total number of specimens tested from patients hospitalised for acute lower respiratory infections between June 2007 and December 2009. (B) Average monthly rainfall in Kampong Cham and Takeo provinces. Data represent the combined average monthly rainfall (mm) in Kampong Cham and Takeo provinces, for 2001 and 2002.
3.2. Patients’ characteristics and co-infections
The patients infected with HMPV were aged between 1 month and 78 years (median: 1.2 years). Thirty (46%) patients were female. The incidence of HMPV infection was highest amongst the 0–5 years age group (Table 2 ). The most common diagnosis was pneumonia or bronchopneumonia (58.5%) (Table 2).
Table 2.
Clinical diagnosis for HMPV-positive patients according to age.
| Age (years) | Clinical diagnosis |
||||
|---|---|---|---|---|---|
| Total (%) | Bronchiolitis (%) | Bronchitis (%) | Pneumonia/Bronchopneumonia (%) | Other respiratory syndromes (%) | |
| 0– ≤ 5 | 52 (80) | 6 (12) | 9 (17) | 28 (54) | 9 (17) |
| 6–10 | 4 (6) | 0 | 1 (25) | 3 (75) | 0 |
| 11–19 | 0 | 0 | 0 | 0 | 0 |
| 20–50 | 4 (6) | 0 | 0 | 3 (75) | 1 (25) |
| ≥ 50 | 5(8) | 0 | 0 | 3 (60) | 2 (40) |
| Total (%) | 65 | 6 (9.2) | 10 (15.4) | 37 (56.9) | 12 (18.5) |
Adequate sputum samples were obtained from only 8 of the 65 patients since sputum collection is difficult in young children, who represented the majority of the HMPV-infected patients in this study. In addition, many sputum samples were discarded as they were contaminated and therefore unsuitable for bacteriological testing. As testing for co-infection with bacteria could not be performed for the majority of patients investigated in this study, the limited results obtained were not included in this report.
Of the 65 HMPV infected patients, six (9.2%) were co-infected with another respiratory virus: four with an adenovirus (6%), one with a rhinovirus (1.5%) and one with a human bocavirus (1.5%). The age of the patients co-infected with HMPV and an adenovirus ranged from 6 months to 2 years (median: 0.9 months).
Of the remaining 59 patients, 37 presented with hyperleukocytosis which is suggestive of a potential bacterial co-infection (neutrophil counts were unfortunately not performed for the majority of patients). Of these, three patients aged between 1 and 4 years were defined as severe cases. Amongst the 22 patients with a normal leukocyte count, five were defined as severe cases. All five patients were between 1 and 9 years of age.
Overall, of the 65 HMPV positive patients, F and/or G gene sequences were successfully obtained from 54 (83%) isolates. Sequences were obtained from 5 of 6 patients with virus co-infection, 31 of 37 patients with hyperleukocytosis and 18 of 22 patients with a normal leukocyte count. Of the 3 patients with hyperleukocytosis who were classified as a severe case, two children were infected by an HMPV strain belonging to sub-genotype B2 and one patient was infected with a lineage A2b strain (Section 3.3). However as it was probable that the patients with hyperleukocytosis were also co-infected with a bacteria, disease severity in these patients could not be attributed to HMPV infection alone.
Of the patients with a normal leukocyte count, sub-genotype B2, B1 and sub-lineage A2b strains were identified in 14, 1 and 3 cases, respectively. Two (14%) of the 14 patients infected with B2 strains were classified as having severe illness compared with all three (100%) of the patients infected with A2b strains (Section 3.3). Overall, lineage A2b strains accounted for only seven (13%) of all HMPV infections, but were identified in three (60%) out of 5 severe cases in patients with normal leukocyte counts compared with only two (40%) severe cases in patients with normal leukocyte counts and sub-genotype B2 infection (p = 0.013); sub-genotype B2 strains accounting for 46 (86%) of all strains successfully sequenced. In this study, infection with sub-genotype B1 HMPV did not appear to result in particularly severe illness.
3.3. Molecular epidemiology of HMPV
3.3.1. Distribution of HMPV genotypes
Twenty (37%) and 34 (63%) samples were collected at Kampong Cham and Takeo hospitals, respectively. The two specimens collected in 2007 were collected at Takeo hospital and belonged to sub-genotype B2 (Fig. 2, Fig. 4). In 2008, sub-genotype B2 (n = 43) and lineage A2b (n = 5) strains circulated concurrently (Fig. 2, Fig. 3, Fig. 4 ). Of the sub-genotype B2 samples, 27 (63%) were collected at Takeo hospital and 16 (37%) at Kampong Cham hospital. One (20%) and four (80%) sub-genotype A2b samples were collected at Takeo and Kampong Cham hospitals, respectively. In 2009, in addition to sub-genotype B2 (n = 1) and lineage A2b (n = 2), sub-genotype B1 was also detected (n = 1) (Fig. 2, Fig. 4). All HMPV positive samples collected in 2009 originated from Takeo hospital. In total, 85% of all strains analysed belonged to sub-genotype B2 (Fig. 4). Only sub-genotype B1 was not detected at both locations; a solitary sub-genotype B1 strain was collected at Takeo hospital.
Fig. 2.

Phylogenetic analysis of partial F gene sequences from Cambodian and reference HMPV strains. Phylogeny was constructed using the neighbour-joining method with 1000 bootstrap replicates. Only bootstrap values > 70% are shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Cambodian strains are indicated by ‘Cam’ followed by the year of collection. Cambodian strains isolated from Takeo province are indicated by ●; strains collected in Kampong Cham province by ▴. Lineages are indicated. An avian metapneumovirus (AMPV) strain was used as an outgroup.
Fig. 4.
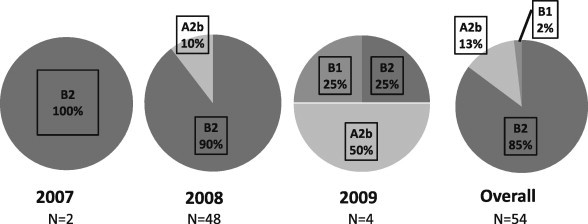
Circulation of HMPV lineages in Cambodia from 2007 to 2009. Relative percentage of HMPV lineages detected following analysis of F and G gene sequences obtained during the study period by year and overall.
Fig. 3.

Phylogenetic analysis of partial G ORF sequences from Cambodian and reference HMPV strains. Phylogeny was constructed using the neighbour-joining method with 1000 bootstrap replicates. Only bootstrap values > 70% are shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated. Cambodian strains are indicated by ‘Cam’ followed by the year of collection. Cambodian strains isolated from Takeo province are indicated by ●; strains collected in Kampong Cham province by ▴. Lineages are indicated. An avian metapneumovirus (AMPV) strain was used as an outgroup.
3.3.2. Variation within the HMPV F gene
Variation within the HMPV F gene was examined by amplifying a fragment of approximately 556 bp within the F ORF (nucleotide [nt]: 726–1281), corresponding to a region within the HMPV fusion protein ectodomain (van den Hoogen et al., 2004a). These partial F gene sequences were obtained from 52 (80%) clinical isolates (Genbank accession numbers HQ864261–HQ864312). Overall, 86.5% F gene sequences obtained belonged to sub-genotype B2.
The pairwise distance analysis revealed that there was between 82% and 100% nucleotide (nt) identity amongst Cambodian HMPV sequences. Homology at the amino acid (aa) sequence level was higher, ranging from 94.4% to 100%. Amongst the genotype B specimens only (n = 46), nt identity was between 92.6% and 100%, aa identity ranged from 97.2% to 100%. Nucleotide identity was 97.3% to 100%, and aa similarity was between 97.8% and 100% amongst the sub-genotype B2 strains. Genetic diversity was low between the lineage A2b strains (n = 6, nt: 99–100%, aa: 99–100%). Five previously described, conserved aa substitutions that distinguished genotype A from genotype B strains were also observed in our study (van den Hoogen et al., 2004a). Genotype A strains could be distinguished by aa substitutions Val286, Lys296, Gln312, Lys348 and Asn404; genotype B strains indeed retained aa substitutions Ile286, Asn296 (genotype B1), Asp296 (genotype B2), Lys312, Arg348 and Pro404 (numbers indicate the position of residues relative to the HMPV reference strain NL/00/1, Genbank accession number AF371337).
3.3.3. Variation within the HMPV G gene
Variation within the HMPV G gene was investigated by amplifying the G gene ORF, a region of approximately 900 bp (nt: 6285–7181) (Ludewick et al., 2005). Sequences of the G ORF were obtained from 31 (47.6%) of the 65 HMPV positive patient samples (Genbank accession numbers HQ864230–HQ864260). Of the 54 HMPV positive samples from which either F or G gene sequences were successfully amplified, both F and G gene sequences were obtained from 29 (54%) samples.
HMPV G gene sequences were obtained for, and enabled characterization of, 2 isolates that were not successfully sequenced for the F gene: strains Cam2008-P0044 and Cam2008-P113 (Fig. 3). Phylogenetic analysis of G gene sequences revealed the same clustering of HMPV strains into the predominant sub-genotype B2 and lineage A2b (Fig. 2, Fig. 3). As observed from the HMPV F gene phylogeny, there was a predominance of sub-genotype B2 strains (Fig. 2, Fig. 3). Of the 31 samples from which G gene sequences were obtained, 29 were collected in 2008, and two in 2009. Twenty-six viruses belonged to sub-genotype B2 and five strains, including the two strains collected in 2009, were classified as lineage A2b (Fig. 3). The Cambodian HMPV G proteins varied in length, ranging from 229 to 294 aa residues, as a result of multiple insertions and deletions. When sub-genotype B2 and lineage A2b strains were compared, homology at the nt and aa levels was variable (nt: 53.6% to 100%, aa: 34.2% to 100%). Nucleotide and aa sequence identity amongst sub-genotype B2 strains was between 88.2% and 100%, and 81% to 100%, respectively. Similar to corresponding results obtained following analysis of HMPV F gene sequences, diversity amongst lineage A2b strains was low (nt: 95.8% to 100%, aa: 99.5% to 100%). Variation at the aa level was greater than that at the nt for both sub-genotype B2 and lineage A2b strains.
4. Discussion
To our knowledge, the results of this study are the first to report the prevalence and also the genetic diversity of HMPV infections in an all-ages population in a South-East Asian country. Prior to this investigation, the only studies that were carried out regionally, in Thailand and Singapore, examined the prevalence and genetic diversity of HMPV amongst children (Loo et al., 2007, Pavlin et al., 2008, Samransamruajkit et al., 2006).
The overall incidence of HMPV infection amongst a population of patients hospitalised with ALRI in Cambodia between 2007 and 2009 was 1.7%, however the incidence of HMPV infection varied annually. This overall prevalence of HMPV in Cambodia was lower than reported elsewhere (Gaunt et al., 2009, Gioula et al., 2010, Loo et al., 2007, Mullins et al., 2004, Samransamruajkit et al., 2006, Sivaprakasam et al., 2007, van den Hoogen et al., 2003, Wang et al., 2008, Williams et al., 2010). Bastien and colleagues observed the incidence of HMPV infection amongst a population of Canadian patients of all age groups to be 14.8% (Bastien et al., 2003). However, Falsey et al. (2003) reported an annual incidence of 1.5%, amongst an adult population of outpatients and hospitalised patients in the USA, speculating simply that the annual prevalence of HMPV infection is highly variable.
The populations investigated by Bastein and Falsey consisted of hospitalised patients and outpatients (Bastien et al., 2003, Falsey et al., 2003). As all of the Cambodian patients investigated in this study required hospitalisation, the low incidence of HMPV infection could reflect a reduced capacity of HMPV to cause severe disease in the study population. Indeed, as nearly all children have been infected with HMPV by 2 years of age with only a minority requiring hospitalisation, a large proportion of HMPV infections in older children and adults are mild or asymptomatic (Falsey et al., 2003). The incidence of HMPV infection amongst Cambodian outpatients was unfortunately not investigated. Therefore, that we observed low incidence of HMPV amongst the population studied may also have resulted from a bias in recruitment since the study only investigated patients with severe illness and did not include outpatients with mild disease. It must also be noted that sampling frequency may have influenced detection of HMPV incidence amongst the Cambodian population studied. Sampling increased sharply during 2009 due to the H1N1 influenza pandemic, hence the increased number of samples collected and tested may have resulted in an underestimation of HMPV incidence.
However, although estimates of prevalence and seasonality can vary according to many factors, including the population studied and the methodology used, it is now accepted that circulation of HMPV between and within given populations is highly variable (Aberle et al., 2008, Falsey et al., 2003, Ludewick et al., 2005, Oliveira et al., 2009). Indeed, in this study, the annual prevalence of HMPV infection varied from 4.3% in 2008 to as low as 0.2% and 0.3% in 2007 and 2009, respectively. During the seasonal peak in 2008, between July and September, infection with HMPV accounted for 10% of all respiratory infections. Hence changes in the prevalence of HMPV between consecutive seasons, as observed in this study, are consistent with observations made by others, specifically, high and low incidence of infection alternating between consecutive years (Carneiro et al., 2009, Falsey et al., 2003, Hopkins et al., 2008, Huck et al., 2006, Ludewick et al., 2005, van den Hoogen et al., 2004a). In addition, timing of the annual seasonal peak of HMPV infection has been observed to vary. In a retrospective study conducted in Austria, Aberle et al. reported a yearly shift in the peak occurrence of HMPV infections, alternating biennially between spring and winter, and that this pattern was sustained over the 20-year period investigated (Aberle et al., 2008). We were unable to comment on the seasonality of HMPV infection in Cambodia, on account of the very low number of HMPV positive samples detected during 2007 and 2009 (Fig. 1). Despite this, in 2008, the peak of infection did occur during the rainy season, between May and September (Fig. 1). Similar to HRSV, the peak incidence of HMPV infection is thought to occur during the rainy season in tropical countries, as HMPV and HRSV typically circulate concurrently resulting in seasonal overlap (Garcia-Garcia et al., 2006, Gerna et al., 2005, McAdam et al., 2004, Noyola et al., 2005, Olsen et al., 2010). Furthermore it was observed, consistent with the findings of Oliveira et al. (2009), that HMPV circulated throughout the year, resulting in a small number of sporadic infections during the 3 consecutive years investigated (Fig. 1A).
To investigate the genetic variability of HMPV strains in Cambodia, regions of the fusion (F) and attachment (G) glycoproteins were analysed. Both F and G proteins are expressed on the surface of virus particles and infected cells. However due to its essential function, resulting in a high level of conservation and antigenic relatedness between regions of the protein amongst genotype A and B strains, only the F protein is able to elicit potent neutralising antibody responses that although typically homologous, can also be cross-protective (Ryder et al., 2010, Skiadopoulos et al., 2006, Skiadopoulos et al., 2004, Ulbrandt et al., 2008). Analogous to the findings of similar studies, a high degree of similarity at both the nt and aa levels was observed for the F gene sequences analysed in this study (Boivin et al., 2004, Endo et al., 2008, van den Hoogen et al., 2004a, Yang et al., 2009). In contrast, the G protein is highly genetically and antigenically diverse, and the least conserved region amongst HMPV strains (Endo et al., 2008, Galiano et al., 2006, Skiadopoulos et al., 2006, van den Hoogen et al., 2004a). Despite functioning as an attachment glycoprotein, Biacchesi and colleagues demonstrated that following deletion of the G glycoprotein, attenuated HMPV isolates retained the ability to replicate in vivo albeit at a reduced rate (Biacchesi et al., 2004). Hence the high rate of nucleic acid substitutions, insertions and deletions observed within this region occur potentially as the attachment glycoprotein is non-essential for HMPV replication. Potentially as a result of the high level of HMPV G gene sequence diversity between strains, successful amplification of fewer HMPV G sequences, compared with the number of HMPV F sequences obtained, was achieved in this and other studies (Pizzorno et al., 2010, van den Hoogen et al., 2004a).
In this study, when comparing genotype A and B strains, nt identity between G protein sequences was as low as 34.2%. Van den Hoogen and colleagues reported not only a similarly low rate of G protein sequence homology, but, analogous to the findings of this study, that variation was higher at the aa level compared to the nucleic acid level (van den Hoogen et al., 2004a). The level of nucleotide and amino acid diversity observed amongst G sequences of Cambodian sub-genotype B2 and lineage A2b strains was similar to published reports (Ishiguro et al., 2004, Ludewick et al., 2005). It is widely accepted that the high level of aa sequence variation occurs as a result of targeted immune pressure due to the prominent location of the G protein on the surface of the virus particle (Endo et al., 2008, van den Hoogen et al., 2004a).
Only very few studies have investigated the distribution of HMPV genotypes within South-East Asia (Loo et al., 2007, Pavlin et al., 2008, Samransamruajkit et al., 2006). Similar to the results obtained previously in Thailand, we observed that sub-genotype B2 strains clearly predominated and that lineage A2b and sub-genotype B1 strains co-circulated in Cambodia between 2007 and 2009 (Fig. 2). Similar to (Loo et al., 2007), no HMPV sub-genotype A1 strains were detected in this study (Loo et al., 2007). Indeed, globally, circulation of HMPV genotypes has been observed to vary annually, according to geographical location. Predominant genotypes are replaced every 1–3 years within a given population, which is thought to occur upon development of adaptive immunity to infection with strains of the predominating circulating genotype (Aberle et al., 2010, Carneiro et al., 2009, Chung et al., 2008, Hopkins et al., 2008, Huck et al., 2006, Mackay et al., 2006, Oliveira et al., 2009, Williams et al., 2010). As the majority of strains analysed in this study were collected in 2008 (89%), we need to exercise caution when commenting on subgroup changes of HMPV in Cambodia. Indeed, in 2009 although specimens positive for sub-genotypes B1 and B2, and lineage A2b were detected, only four samples were collected, hence it is not possible to comment on whether this represented a shift in predominant circulating HMPV genotypes.
A number of studies have reported a high incidence of co-infection with HMPV and an additional viral respiratory pathogen, frequently HRSV (Cuevas et al., 2003, Oliveira et al., 2009, Pizzorno et al., 2010, Semple et al., 2005). It is thought that the high incidence of HMPV/HRSV co-infection occurs as the annual epidemics of both viruses partially overlap, especially in temperate climates (Papenburg and Boivin, 2010). In this study, 9.2% of HMPV positive specimens were co-infected with an additional virus, with HMPV/adenovirus co-infections the most frequently detected. A similar incidence of HMPV/adenovirus co-infection was observed in children with respiratory tract infection in Japan (Kaida et al., 2007). All of the HMPV/adenovirus co-infected patients identified in this study were aged 2 years or less, which is not surprising as primary adenovirus infections typically occur in very young children (Lenaerts et al., 2008). One patient identified in this study was co-infected with HMPV and HBoV. The co-infected patient was not classified as having severe illness. The role of HBoV as a causative agent of respiratory tract disease remains contentious (Blessing et al., 2009, Chow and Esper, 2009, Mackay, 2007, Schildgen et al., 2008). A recent study by (Blessing et al., 2009) demonstrated that qualitative detection of HBoV DNA within a patient sample was not sufficient to establish HBoV as the sole cause of respiratory illness. This was due to prolonged shedding and persistence of HBoV DNA over an extended period (Blessing et al., 2009). Hence, the clinical relevance of detection of HBoV DNAin this co-infected patient identified in this study remains unclear. Interestingly, despite concurrent high circulation of HRSV during each rainy season within the same population (Buchy, Personal communication), co-infection with HMPV and HRSV was not detected in this study. The incidence of HRSV infection amongst the population investigated was similar to that reported both regionally, and similar to that reported for studies conducted worldwide (data not shown).
Pneumonia and broncho-pneumonia were the most frequent diagnoses amongst all of the age groups studied. These syndromes are commonly used in other studies as indicators of severe illness (Falsey, 2008, Ulloa-Gutierrez et al., 2004, van den Hoogen et al., 2003, Vicente et al., 2006). In a 25-year retrospective study of HMPV infection amongst young children treated at an outpatient clinic in the USA, bronchiolitis was the most frequent clinical expression of HMPV infection, and pneumonia was observed in only 8% of children (Williams et al., 2004). However, of 12 HMPV cases detected during 2 years in Thailand, 11 (91%) patients required hospitalisation. Amongst the hospitalised patients, the primary manifestation (50%) of HMPV infection was also pneumonia (Samransamruajkit et al., 2006). Similarly, amongst a population of children aged < 5 years hospitalised with ALRI symptoms in Israel, Wolf and colleagues reported a strong association between HMPV infection and pneumonia (Wolf et al., 2006). Although the pathophysiologic mechanisms underlying this association were unclear, the authors speculated that infection with HMPV might exacerbate or be involved in the pathogenesis of bacterial pneumonia (Wolf et al., 2006). Co-infection with HMPV and an additional respiratory pathogen, virus or bacteria, has been associated with more severe illness compared with HMPV mono-infection (Cuevas et al., 2003, Semple et al., 2005). In this study, comprehensive testing for co-infection with bacteria could not be performed as appropriate patient samples were largely not available or were contaminated, and therefore unsuitable for testing. Of 17 Cambodian patients with severe illness and hyperleukocytosis, 8 (47%) were diagnosed with pneumonia or bronchopneumonia. It would be interesting to further examine the association between bacterial pneumonia and HMPV infection.
Five patients, with a normal leukocyte count and severe illness, were all aged < 10 years with no detected viral co-infection. Of these, 3 patients were infected with lineage A2b strains. No difference in the severity of disease following infection with HMPV strains of the A and B genotypes has been reported (Agapov et al., 2006, Xiao et al., 2010). However conversely, and similar to the findings of this study, an association between HMPV genotype A strains and severe disease has also been observed (Esper et al., 2004, Martinello et al., 2002, Vicente et al., 2006). Although not all of the patients infected with lineage A2b strains experienced severe illness, we did observe a statistically significant (p = 0.013) association between infection with lineage A2b strains and severe illness in children with no known co-infections or underlying co-morbid conditions. To our knowledge, this is the first report of a significant association between infection with HMPV lineage A2b strains and severe illness. Similarly, an association between severe disease and infection with genotype A strains of the closely related Paramyxovirus HRSV has also been observed, relative to infection with HRSV genotype B strains (Gilca et al., 2006, Martinello et al., 2002, Sullender, 2000, Walsh et al., 1997). However, we cannot rule out the possibility of nosocomial infection amongst hospitalised patients, or prior infection with other respiratory infections as factors which may have contributed to the severity of illness amongst the patients infected with sub-lineage A2b strains identified in this study. Additional studies recruiting a larger number of patients are required to further address the question of whether infection with HMPV sub-lineage A2b strains is associated with severe disease,. In addition, the circulating genotypes amongst a Cambodian outpatient population infected with HMPV should also be investigated, to elucidate whether the same genotypes are associated with less severe infection.
In conclusion, the results of this study confirm that HMPV sub-genotypes B1 and B2, and lineage A2b strains co-circulated in Cambodia between 2007 and 2009. Sub-genotype B2 strains predominated, and demonstrated higher diversity at both the nucleotide and amino acid level compared with lineage A2b strains. The incidence of HMPV infection fluctuated annually, with the largest peak observed during the rainy season. We also report a significant association between infection with lineage A2b strains and severe illness. The results of this study further highlight the dynamic nature of localised HMPV epidemics.
Acknowledgements
This study was supported by Surveillance and Investigation of Epidemic Situations in Southeast Asia (SISEA) project, a grant from the French Agency for Development. We gratefully thank Dr Alain Viari, Dr Roger Frutos and Dr Eric Coissac for invaluable advice and help with bioinformatic and phylogenetic analysis. We would like to thank all of the technical staff in the Virology and Epidemiology units at the Institut Pasteur in Cambodia for their assistance and especially Dr. Duong Veasna for the statistical analysis. We are also grateful to the staff of Kampong Cham and Takeo Provincial Hospitals.
References
- Aberle J.H., Aberle S.W., Redlberger-Fritz M., Sandhofer M.J., Popow-Kraupp T. Human metapneumovirus subgroup changes and seasonality during epidemics. The Pediatric Infectious Disease Journal. 2010 doi: 10.1097/INF.0b013e3181e3331a. [DOI] [PubMed] [Google Scholar]
- Aberle S.W., Aberle J.H., Sandhofer M.J., Pracher E., Popow-Kraupp T. Biennial spring activity of human metapneumovirus in Austria. The Pediatric Infectious Disease Journal. 2008;27:1065–1068. doi: 10.1097/INF.0b013e31817ef4fd. [DOI] [PubMed] [Google Scholar]
- Agapov E., Sumino K.C., Gaudreault-Keener M., Storch G.A., Holtzman M.J. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. The Journal of Infectious Diseases. 2006;193:396–403. doi: 10.1086/499310. [DOI] [PubMed] [Google Scholar]
- Bastien N., Ward D., Van Caeseele P., Brandt K., Lee S.H., McNabb G., Klisko B., Chan E., Li Y. Human metapneumovirus infection in the Canadian population. Journal of Clinical Microbiology. 2003;41:4642–4646. doi: 10.1128/JCM.41.10.4642-4646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S., Skiadopoulos M.H., Yang L., Lamirande E.W., Tran K.C., Murphy B.R., Collins P.L., Buchholz U.J. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. Journal of Virology. 2004;78:12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing K., Neske F., Herre U., Kreth H.W., Weissbrich B. Prolonged detection of human bocavirus DNA in nasopharyngeal aspirates of children with respiratory tract disease. The Pediatric Infectious Disease Journal. 2009;28:1018–1019. doi: 10.1097/INF.0b013e3181a854ae. [DOI] [PubMed] [Google Scholar]
- Boivin G., Abed Y., Pelletier G., Ruel L., Moisan D., Cote S., Peret T.C., Erdman D.D., Anderson L.J. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. The Journal of Infectious Diseases. 2002;186:1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- Boivin G., Mackay I., Sloots T.P., Madhi S., Freymuth F., Wolf D., Shemer-Avni Y., Ludewick H., Gray G.C., LeBlanc E. Global genetic diversity of human metapneumovirus fusion gene. Emerging Infectious Diseases. 2004;10:1154–1157. doi: 10.3201/eid1006.031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno R., Marsico S., Minini C., Apostoli P., Fiorentini S., Caruso A. Human metapneumovirus infection in a cohort of young asymptomatic subjects. New Microbiology. 2009;32:297–301. [PubMed] [Google Scholar]
- Buchy, P. In: Arnott, A. (Ed.), Circulation of RSV in Cambodia. Personal communication.
- Buecher C., Mardy S., Wang W., Duong V., Vong S., Naughtin M., Vabret A., Freymuth F., Deubel V., Buchy P. Use of a multiplex PCR/RT-PCR approach to assess the viral causes of influenza-like illnesses in Cambodia during three consecutive dry seasons. Journal of Medical Virology. 2010;82:1762–1772. doi: 10.1002/jmv.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro B.M., Yokosawa J., Arbiza J., Costa L.F., Mirazo S., Nepomuceno L.L., Oliveira T.F., Goulart L.R., Vieira C.U., Freitas G.R., Paula N.T., Queiroz D.A. Detection of all four human metapneumovirus subtypes in nasopharyngeal specimens from children with respiratory disease in Uberlandia, Brazil. Journal of Medical Virology. 2009;81:1814–1818. doi: 10.1002/jmv.21555. [DOI] [PubMed] [Google Scholar]
- Chow B.D., Esper F.P. The human bocaviruses: a review and discussion of their role in infection. Clinics in Laboratory Medicine. 2009;29:695–713. doi: 10.1016/j.cll.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.Y., Han T.H., Kim S.W., Hwang E.S. Genotype variability of human metapneumovirus, South Korea. Journal of Medical Virology. 2008;80:902–905. doi: 10.1002/jmv.21129. [DOI] [PubMed] [Google Scholar]
- Collins P.L., Mottet G. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. Journal of General Virology. 1993;74(Pt 7):1445–1450. doi: 10.1099/0022-1317-74-7-1445. [DOI] [PubMed] [Google Scholar]
- Cuevas L.E., Nasser A.M., Dove W., Gurgel R.Q., Greensill J., Hart C.A. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerging Infectious Diseases. 2003;9:1626–1628. doi: 10.3201/eid0912.030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo R., Ebihara T., Ishiguro N., Teramoto S., Ariga T., Sakata C., Hayashi A., Ishiko H., Kikuta H. Detection of four genetic subgroup-specific antibodies to human metapneumovirus attachment (G) protein in human serum. The Journal of General Virology. 2008;89:1970–1977. doi: 10.1099/vir.0.83679-0. [DOI] [PubMed] [Google Scholar]
- Esper F., Martinello R.A., Boucher D., Weibel C., Ferguson D., Landry M.L., Kahn J.S. A 1-year experience with human metapneumovirus in children aged < 5 years. The Journal of Infectious Diseases. 2004;189:1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R. Human metapneumovirus infection in adults. The Pediatric Infectious Disease Journal. 2008;27:S80–S83. doi: 10.1097/INF.0b013e3181684dac. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Erdman D., Anderson L.J., Walsh E.E. Human metapneumovirus infections in young and elderly adults. The Journal of Infectious Diseases. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Galiano M., Trento A., Ver L., Carballal G., Videla C. Genetic heterogeneity of G and F protein genes from Argentinean human metapneumovirus strains. Journal of Medical Virology. 2006;78:631–637. doi: 10.1002/jmv.20586. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia M.L., Calvo C., Martin F., Perez-Brena P., Acosta B., Casas I. Human metapneumovirus infections in hospitalised infants in Spain. Archives of Disease in Childhood. 2006;91:290–295. doi: 10.1136/adc.2005.082388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E., McWilliam-Leitch E.C., Templeton K., Simmonds P. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; assessment of its importance as a diagnostic screening target. Journal of Clinical Virology. 2009;46:318–324. doi: 10.1016/j.jcv.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Gerna G., Campanini G., Rovida F., Sarasini A., Lilleri D., Paolucci S., Marchi A., Baldanti F., Revello M.G. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter-spring seasons. Brief report. Archives of Virology. 2005;150:2365–2375. doi: 10.1007/s00705-005-0581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilca R., De Serres G., Tremblay M., Vachon M.L., Leblanc E., Bergeron M.G., Dery P., Boivin G. Distribution and clinical impact of human respiratory syncytial virus genotypes in hospitalized children over 2 winter seasons. The Journal of Infectious Diseases. 2006;193:54–58. doi: 10.1086/498526. [DOI] [PubMed] [Google Scholar]
- Gioula G., Chatzidimitriou D., Melidou A., Exindari M., Kyriazopoulou-Dalaina V. Contribution of human metapneumovirus to influenza-like infections in North Greece, 2005–2008. Euro Surveillance. 2010;15 doi: 10.2807/ese.15.09.19499-en. [DOI] [PubMed] [Google Scholar]
- Hopkins M.J., Redmond C., Shaw J.M., Hart I.J., Hart C.A., Smyth R.L., Semple M.G. Detection and characterisation of human metapneumovirus from children with acute respiratory symptoms in north-west England, UK. Journal of Clinical Virology. 2008;42:273–279. doi: 10.1016/j.jcv.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Huck B., Scharf G., Neumann-Haefelin D., Puppe W., Weigl J., Falcone V. Novel human metapneumovirus sublineage. Emerging Infectious Diseases. 2006;12:147–150. doi: 10.3201/eid1201.050772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro N., Ebihara T., Endo R., Ma X., Kikuta H., Ishiko H., Kobayashi K. High genetic diversity of the attachment (G) protein of human metapneumovirus. Journal of Clinical Microbiology. 2004;42:3406–3414. doi: 10.1128/JCM.42.8.3406-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J.S., Kesebir D., Cotmore S.F., D’Abramo A., Jr., Cosby C., Weibel C., Tattersall P. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. The Journal of Infectious Diseases. 2008;198:41–50. doi: 10.1086/588674. [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Goto K., Shiomi M., Kohdera U., Iritani N. Co-infection of human metapneumovirus with adenovirus or respiratory syncytial virus among children in Japan. Microbiology and Immunology. 2007;51:679–683. doi: 10.1111/j.1348-0421.2007.tb03956.x. [DOI] [PubMed] [Google Scholar]
- Lenaerts L., De Clercq E., Naesens L. Clinical features and treatment of adenovirus infections. Reviews in Medical Virology. 2008;18:357–374. doi: 10.1002/rmv.589. [DOI] [PubMed] [Google Scholar]
- Loo L.H., Tan B.H., Ng L.M., Tee N.W., Lin R.T., Sugrue R.J. Human metapneumovirus in children, Singapore. Emerging Infectious Diseases. 2007;13:1396–1398. doi: 10.3201/eid1309.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewick H.P., Abed Y., van Niekerk N., Boivin G., Klugman K.P., Madhi S.A. Human metapneumovirus genetic variability, South Africa. Emerging Infectious Diseases. 2005;11:1074–1078. doi: 10.3201/eid1107.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I.M. Human bocavirus: multisystem detection raises questions about infection. The Journal of Infectious Diseases. 2007;196:968–970. doi: 10.1086/521311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I.M., Bialasiewicz S., Jacob K.C., McQueen E., Arden K.E., Nissen M.D., Sloots T.P. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. The Journal of Infectious Diseases. 2006;193:1630–1633. doi: 10.1086/504260. [DOI] [PubMed] [Google Scholar]
- Mardy S., Ly S., Heng S., Vong S., Huch C., Nora C., Asgari N., Miller M., Bergeri I., Rehmet S., Veasna D., Zhou W., Kasai T., Touch S., Buchy P. Influenza activity in Cambodia during 2006–2008. BMC Infectious Diseases. 2009;9:168. doi: 10.1186/1471-2334-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinello R.A., Chen M.D., Weibel C., Kahn J.S. Correlation between respiratory syncytial virus genotype and severity of illness. The Journal of Infectious Diseases. 2002;186:839–842. doi: 10.1086/342414. [DOI] [PubMed] [Google Scholar]
- McAdam A.J., Hasenbein M.E., Feldman H.A., Cole S.E., Offermann J.T., Riley A.M., Lieu T.A. Human metapneumovirus in children tested at a tertiary-care hospital. The Journal of Infectious Diseases. 2004;190:20–26. doi: 10.1086/421120. [DOI] [PubMed] [Google Scholar]
- Mullins J.A., Erdman D.D., Weinberg G.A., Edwards K., Hall C.B., Walker F.J., Iwane M., Anderson L.J. Human metapneumovirus infection among children hospitalized with acute respiratory illness. Emerging Infectious Diseases. 2004;10:700–705. doi: 10.3201/eid1004.030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. Oxford University Press; Oxford/New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- Noyola D.E., Alpuche-Solis A.G., Herrera-Diaz A., Soria-Guerra R.E., Sanchez-Alvarado J., Lopez-Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. Journal of Medical Microbiology. 2005;54:969–974. doi: 10.1099/jmm.0.46052-0. [DOI] [PubMed] [Google Scholar]
- O’Gorman C., McHenry E., Coyle P.V. Human metapneumovirus in adults: a short case series. European Journal of Clinical Microbiology & Infectious Diseases. 2006;25:190–192. doi: 10.1007/s10096-006-0116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D.B., Durigon E.L., Carvalho A.C., Leal A.L., Souza T.S., Thomazelli L.M., Moraes C.T., Vieira S.E., Gilio A.E., Stewien K.E. Epidemiology and genetic variability of human metapneumovirus during a 4-year-long study in Southeastern Brazil. Journal of Medical Virology. 2009;81:915–921. doi: 10.1002/jmv.21436. [DOI] [PubMed] [Google Scholar]
- Olsen S.J., Thamthitiwat S., Chantra S., Chittaganpitch M., Fry A.M., Simmerman J.M., Baggett H.C., Peret T.C., Erdman D., Benson R., Talkington D., Thacker L., Tondella M.L., Winchell J., Fields B., Nicholson W.L., Maloney S., Peruski L.F., Ungchusak K., Sawanpanyalert P., Dowell S.F. Incidence of respiratory pathogens in persons hospitalized with pneumonia in two provinces in Thailand. Epidemiology and Infection. 2010:1–12. doi: 10.1017/S0950268810000646. [DOI] [PubMed] [Google Scholar]
- Papenburg J., Boivin G. The distinguishing features of human metapneumovirus and respiratory syncytial virus. Reviews in Medical Virology. 2010;20:245–260. doi: 10.1002/rmv.651. [DOI] [PubMed] [Google Scholar]
- Pavlin J.A., Hickey A.C., Ulbrandt N., Chan Y.P., Endy T.P., Boukhvalova M.S., Chunsuttiwat S., Nisalak A., Libraty D.H., Green S., Rothman A.L., Ennis F.A., Jarman R., Gibbons R.V., Broder C.C. Human metapneumovirus reinfection among children in Thailand determined by ELISA using purified soluble fusion protein. The Journal of Infectious Diseases. 2008;198:836–842. doi: 10.1086/591186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Masner M., Medici C., Sarachaga M.J., Rubio I., Mirazo S., Frabasile S., Arbiza J. Molecular detection and genetic variability of human metapneumovirus in Uruguay. Journal of Medical Virology. 2010;82:861–865. doi: 10.1002/jmv.21752. [DOI] [PubMed] [Google Scholar]
- Ryder A.B., Tollefson S.J., Podsiad A.B., Johnson J.E., Williams J.V. Soluble recombinant human metapneumovirus G protein is immunogenic but not protective. Vaccine. 2010;28:4145–4152. doi: 10.1016/j.vaccine.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samransamruajkit R., Thanasugarn W., Prapphal N., Theamboonlers A., Poovorawan Y. Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations. The Journal of Infection. 2006;52:254–263. doi: 10.1016/j.jinf.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Schildgen O., Muller A., Allander T., Mackay I.M., Volz S., Kupfer B., Simon A. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clinical Microbiology Reviews. 2008;21:291–304. doi: 10.1128/CMR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple M.G., Cowell A., Dove W., Greensill J., McNamara P.S., Halfhide C., Shears P., Smyth R.L., Hart C.A. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. The Journal of Infectious Diseases. 2005;191:382–386. doi: 10.1086/426457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprakasam V., Collins T.C., Aitken C., Carman W.F. Life-threatening human metapneumovirus infections in West of Scotland. Journal of Clinical Virology. 2007;39:234–237. doi: 10.1016/j.jcv.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos M.H., Biacchesi S., Buchholz U.J., Amaro-Carambot E., Surman S.R., Collins P.L., Murphy B.R. Individual contributions of the human metapneumovirus F G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology. 2006;345:492–501. doi: 10.1016/j.virol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Skiadopoulos M.H., Biacchesi S., Buchholz U.J., Riggs J.M., Surman S.R., Amaro-Carambot E., McAuliffe J.M., Elkins W.R., St Claire M., Collins P.L., Murphy B.R. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. Journal of Virology. 2004;78:6927–6937. doi: 10.1128/JVI.78.13.6927-6937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullender W.M. Respiratory syncytial virus genetic and antigenic diversity. Clinical Microbiology Reviews. 2000;13:1–15. doi: 10.1128/cmr.13.1.1-15.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Ulbrandt N.D., Ji H., Patel N.K., Barnes A.S., Wilson S., Kiener P.A., Suzich J., McCarthy M.P. Identification of antibody neutralization epitopes on the fusion protein of human metapneumovirus. The Journal of General Virology. 2008;89:3113–3118. doi: 10.1099/vir.0.2008/005199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa-Gutierrez R., Skippen P., Synnes A., Seear M., Bastien N., Li Y., Forbes J.C. Life-threatening human metapneumovirus pneumonia requiring extracorporeal membrane oxygenation in a preterm infant. Pediatrics. 2004;114:e517–e519. doi: 10.1542/peds.2004-0345. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B.G., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295:119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A., Osterhaus A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nature Medicine. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., Herfst S., Sprong L., Cane P.A., Forleo-Neto E., de Swart R.L., Osterhaus A.D., Fouchier R.A. Antigenic and genetic variability of human metapneumoviruses. Emerging Infectious Diseases. 2004;10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., Osterhaus D.M., Fouchier R.A. Clinical impact and diagnosis of human metapneumovirus infection. The Pediatric Infectious Disease Journal. 2004;23:S25–S32. doi: 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B.G., van Doornum G.J., Fockens J.C., Cornelissen J.J., Beyer W.E., de Groot R., Osterhaus A.D., Fouchier R.A. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. The Journal of Infectious Diseases. 2003;188:1571–1577. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- Vance W.H., Bell R.W., Seng V. School of Environmental Science, Murdoch University; Western Australia, Australia: 2004. Rainfall Analysis for the Provinces of Battambang, Kampong Cham and Takeo, The Kingdom of Cambodia. http://www.environment.murdoch.edu.au/groups/aciar/images/RainfallAnalysis190704.pdf. [Google Scholar]
- Vicente D., Montes M., Cilla G., Perez-Yarza E.G., Perez-Trallero E. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clinical Infectious Diseases. 2006;42:e111–e113. doi: 10.1086/504378. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., McConnochie K.M., Long C.E., Hall C.B. Severity of respiratory syncytial virus infection is related to virus strain. The Journal of Infectious Diseases. 1997;175:814–820. doi: 10.1086/513976. [DOI] [PubMed] [Google Scholar]
- Wang H.C., Huang S.W., Wang S.W., Tsai H.P., Kiang D., Wang S.M., Liu C.C., Su I.J., Wang J.R. Co-circulating genetically divergent A2 human metapneumovirus strains among children in southern Taiwan. Archives of Virology. 2008;153:2207–2213. doi: 10.1007/s00705-008-0242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . In: Pocket Book of Hospital Care for Children. WHO, editor. WHO Press; 2005. [Google Scholar]
- Williams J.V., Edwards K.M., Weinberg G.A., Griffin M.R., Hall C.B., Zhu Y., Szilagyi P.G., Wang C.K., Yang C.F., Silva D., Ye D., Spaete R.R., Crowe J.E., Jr. Population-based incidence of human metapneumovirus infection among hospitalized children. The Journal of Infectious Diseases. 2010;201:1890–1898. doi: 10.1086/652782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.V., Harris P.A., Tollefson S.J., Halburnt-Rush L.L., Pingsterhaus J.M., Edwards K.M., Wright P.F., Crowe J.E., Jr. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. The New England Journal of Medicine. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.V., Martino R., Rabella N., Otegui M., Parody R., Heck J.M., Crowe J.E., Jr. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. The Journal of Infectious Diseases. 2005;192:1061–1065. doi: 10.1086/432732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D.G., Greenberg D., Kalkstein D., Shemer-Avni Y., Givon-Lavi N., Saleh N., Goldberg M.D., Dagan R. Comparison of human metapneumovirus, respiratory syncytial virus and influenza A virus lower respiratory tract infections in hospitalized young children. The Pediatric Infectious Disease Journal. 2006;25:320–324. doi: 10.1097/01.inf.0000207395.80657.cf. [DOI] [PubMed] [Google Scholar]
- Xiao N.G., Xie Z.P., Zhang B., Yuan X.H., Song J.R., Gao H.C., Zhang R.F., Hou Y.D., Duan Z.J. Prevalence and clinical and molecular characterization of human metapneumovirus in children with acute respiratory infection in China. The Pediatric Infectious Disease Journal. 2010;29:131–134. doi: 10.1097/inf.0b013e3181b56009. [DOI] [PubMed] [Google Scholar]
- Yang C.F., Wang C.K., Tollefson S.J., Piyaratna R., Lintao L.D., Chu M., Liem A., Mark M., Spaete R.R., Crowe J.E., Jr., Williams J.V. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virology Journal. 2009;6:138. doi: 10.1186/1743-422X-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]


