Abstract
Ever since the discovery of SARS-CoV in the year 2003, numerous researchers around the world have been working relentlessly to understand the biology of this virus. As in other coronaviruses, nucleocapsid (N) is one of the most crucial structural components of the SARS-CoV. Hence major attention has been focused on characterization of this protein. Independent studies conducted by several laboratories have elucidated significant insight into the primary function of this protein, which is to encapsidate the viral genome. In addition, many reports also suggest that this protein interferes with different cellular pathways, thus implying it to be a key regulatory component of the virus too. In the first part of this review, we will discuss these different properties of the N-protein in a consolidated manner. Further, this protein has also been proposed to be an efficient diagnostic tool and a candidate vaccine against the SARS-CoV. Hence, towards the end of this article, we will discuss some recent progress regarding the possible clinically relevant use of the N-protein.
Keywords: SARS, Nucleocapsid protein, Coronavirus, SARS diagnosis, SARS-CoV assembly, RNA virus
1. N-protein: structure and composition
The nucleocapsid (N) protein is encoded by the 9th ORF of SARS-CoV. The same ORF also codes for another unique accessory protein called ORF9b, though in a different reading frame, whose function is yet to be defined. The N-protein is a 46 kDa protein composed of 422 amino acids (Rota et al., 2003). Its N-terminal region consists mostly of positively charged amino acids, which is responsible for RNA binding. A lysine rich region is present between 373 and 390 amino acids at the C-terminus, which is predicted to be the nuclear localization signal. Besides that, a SR-rich motif is present in the middle region encompassing 177–207 amino acids. Biophysical studies done by Chang et al. (2006) have suggested that this protein is composed of two independent structural domains and a linker region. The first domain is present at the N-terminus, inside the putative RNA binding domain. The second domain consists of the C-terminal region that is capable of self-association. Between these two structural domains, there lies a highly disordered region, which serves as a linker. This region has been reported to interact with the membrane (M) protein and human cellular hnRNPA1 protein (Fang et al., 2006, Luo et al., 2005a, Luo et al., 2005b). Besides, this region is also predicted to be a hot spot for phosphorylation. Hence, in summary, the N-protein can be classified into three distinct regions (Fig. 1 ), which may serve completely different functions during different stages of the viral life cycle. A similar mode of organization has been reported for other coronavirus nucleocapsid proteins.
Fig. 1.
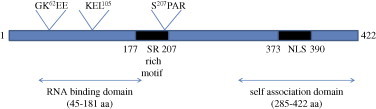
Structure of the SARS-CoV nucleocapsid protein. A schematic diagram showing various different domains identified to-date. The numbers 1–422 correspond to the length in amino acids of the N gene. GKEE represents the sumoylation motif (lysine residue). KEL is the RXL motif, responsible for binding with cyclin D and SPAR is the motif that gets phosphorylated by cyclin–CDK complex (serine residue).
2. Stability of the N-protein
In vitro thermodynamic studies done by C. Luo et al. (2004) and H. Luo et al. (2004) using purified recombinant N-protein have shown it to be stable between pH 7 and 10, with maximum conformational stability near pH 9. Further, it was observed to undergo irreversible thermal-induced denaturation. It starts to unfold at 35 °C and is completely denatured at 55 °C (Wang et al., 2004). However, chemicals such as urea or guanidium chloride-induced denaturation of the N-protein is a reversible process.
3. Post-translational modification
As in other coronavirus N-proteins, SARS-CoV N-protein has been predicted and later experimentally proven to undergo various post-translational modifications such as acetylation, phosphorylation, and sumoylation.
Acetylation is the first modification of the N-protein to be experimentally proven. By mass spectrometric analysis of convalescent sera from several SARS patients, it has been shown that the N-terminal methionine of N is removed as well as all other methionines are oxidised and the resulting N-terminal serine is acetylated. However, the functional relevance of this modification, if any, remains to be elucidated (Krokhin et al., 2003).
Another unique modification of the N-protein is its ability to get sumoylated. Studies done by Y.H. Li et al. (2005) and F.Q. Li et al. (2005) have clearly established that heterologously expressed N in mammalian cells is sumoylated. Using a site-directed mutagenesis approach, the sumoylation motif has been mapped to the lysine residue 62, which is present in a putative sumo-modification domain (GK62EE). Their data further suggests that sumoylation may play a key role in modulating homo-oligomerization, nucleolar translocation and cell-cycle deregulatory property of the N-protein. Further experimental support regarding sumoylation of N-protein came from another independent study carried out by Fan et al. (2006) wherein they have demonstrated an association between the N-protein and Hubc9, which is a ubiquitin conjugating enzyme of the sumoylation system. They have also mapped the interaction domain to the SR-rich motif, which is in agreement with the earlier report. However, they failed to detect the involvement of the GKEE motif in mediating this interaction (Fan et al., 2006).
Initially, the SARS-CoV N-protein was predicted to be heavily phosphorylated. Later on, from results obtained in our laboratory as well as by other researchers, it is now clear that the N-protein is a substrate of multiple cellular kinases. First experimental evidence for the phosphorylation status of the N-protein came from the study done by Zakhartchouk et al. (2005) wherein by 32P orthophosphate labelling, they were able to observe phosphorylation of adenovirus vector expressed N-protein in 293T cells. Further studies done in our laboratory clearly confirmed this observation. Majority of the N-protein was found to be phosphorylated at its serine residues (although the involvement of threonine and tyrosine residues could not be detected, they may be occurring in vivo). In addition, using a variety of biochemical assays, it was proved that, at least in vitro, the N-protein could get phosphorylated by mitogen activated protein kinase (MAP kinase), cyclin-dependent kinase (CDK), glycogen synthase kinase 3 (GSK3), and casein kinase 2 (CK2). Also, this data provides preliminary indication regarding phosphorylation-dependent nucleo-cytoplasmic shuttling of the N-protein (Surjit et al., 2005). Whether these events actually bear any functional significance in vivo, remains yet to be established.
4. Localization of the N-protein
In contrast to the N-protein of many other coronaviruses, the SARS-CoV N-protein is predominantly distributed in the cytoplasm, when expressed heterologously or in infected cells (Surjit et al., 2005, You et al., 2005, Rowland et al., 2005). In infected cells, a few cells exhibited nucleolar localization (You et al., 2005). As reported by You et al. (2005) the N-protein contains pat4, pat7, and bipartite-type nuclear localization signals. It has also been predicted to possess a potential CRM-1 dependent nuclear export signal. However, no clear experimental evidence could be obtained regarding the involvement of these signature sequences in regulating the localization of the N-protein. Interestingly, studies done in our laboratory revealed that the majority of N-protein localized to the nucleus in serum starved cells. This phenomenon could be reproducibly observed both in biochemical fractionation as well as immunofluorescence studies. In addition, treatment of cells with specific inhibitors of different cellular kinases such as CK2 inhibitor and CDK inhibitor, resulted in retention of a fraction of the N-protein in the nucleus, whereas GSK3 and MAPK inhibitor had very little effect. Further, N was found to be efficiently phosphorylated by the cyclin–CDK complex, which is known to be active only in the nucleus. The N-protein was also found to associate with 14-3-3 protein in a phospho-specific manner and inhibition of the 14-3-3θ protein level by siRNA resulted in nuclear accumulation of the N-protein. Although these experiments are too preliminary to conclusively provide any answer regarding the intra-cellular localization of N-protein, nevertheless they do provide substantial clues regarding the physical presence of the N-protein in the nucleus, under certain circumstances, which may be a very dynamic phenomenon. Another study done by Timani et al. (2005) using different deletion mutants of the N-protein fused to EGFP, showed that the N-terminal of N, which contains the NLS 1 (aa 38–44) localizes to the nucleus, whereas the C-terminal region containing both NLS 2 (aa 257–265) and NLS 3 (aa 369–390) localizes to the cytoplasm and nucleolus. Using a combination of different deletion mutants, they concluded that the N-protein may act as a shuttle protein between cytoplasm–nucleus and nucleolus. Taken together, all these results further suggest that the N-protein per se has the physical ability to localize to the nucleus. Whether this localization is regulated through phosphorylation mediated activation of a potential NLS or piggy-backing by association with another cellular nuclear protein or through any other mechanism remains to be established.
5. Genome encapsidation: primary function of a viral capsid protein
Being the capsid protein, the primary function of the N-protein is to package the genomic RNA in a protective covering. In order to achieve this structure, the N-protein must be equipped with two different characteristic properties such as: (i) being able to recognize the genomic RNA and associate with it; (ii) self-associate into an oligomer to form the capsid. The N-protein of SARS-CoV has been experimentally proven to possess these properties in vitro, as discussed below.
5.1. Recognition and binding with the genomic RNA
First experimental evidence regarding the RNA binding property of the N-protein came from the work of Huang et al. (2004), wherein by NMR studies, they proved the ability of N-terminal domain to associate with several viral 3′ untranslated RNA sequence. Additionally, a recent report by Chen et al. (2007) proved the presence of another RNA binding domain at the C-terminal region (residues 248–365) of the N-protein, which is proposed to be a stronger interaction than that at the N-terminus. Based on the structural analysis of the RNA–protein interaction, they have further suggested that the genomic RNA is packaged in a helical manner by the N-protein. The RNA binding motif of N-protein has been mapped to 363–382 amino acid residues by Luo et al. (2006).
Perhaps, the most convincing proof till date regarding the ability of the N-protein to package the genomic RNA came from the work of Hsieh et al. (2005). They have established a system to produce SARS-CoV VLPs by cotransfection of spike, membrane, envelope, and nucleocapsid cDNAs into Vero E6 cells. While testing the packaging of an RNA bearing GFP fused to SARS-CoV packaging signal into this particle, they observed that the presence of the N-protein is an absolute requirement. However, the N-protein was not essential for the assembly of the empty particle per se. Further, by performing a filter binding assay using recombinant N-protein, they were able to identify two independent RNA binding domains in the N-protein; one at the N-terminus (1–235 aa) and the other at the C-terminus (236–384 aa). These results are in agreement with previous findings and further suggest that these two regions may be functional in vivo. Future experiments using a model infection system will confirm these observations.
5.2. Formation of the capsid
One of the most crucial properties required by the N-protein for genome encapsidation is its ability to self-associate. Therefore, many laboratories have focused on characterizing this phenomenon, with an eye to develop possible interference strategies that may help in limiting virus propagation.
Initial studies done in our laboratory using a yeast two-hybrid assay revealed that N-protein is able to self-associate through its C-terminal 209 amino acid residues (Surjit et al., 2004a, Surjit et al., 2004b, Surjit et al., 2004c). A parallel study done by He et al. (2004) using the mammalian two-hybrid system and sucrose gradient fractionation also proved the ability of the N-protein to self-associate to form an oligomer. They further mapped the interaction region to 184–196 amino acid residues, encompassing the SR-rich motif. However, there were some discrepancies regarding the interaction domain mapped in these two studies. Later on, extensive biophysical and biochemical analysis done by Chen lab (Yu et al., 2005, Yu et al., 2006) and Jiang lab (Luo et al., 2005a, Luo et al., 2005b, Luo et al., 2006) have enriched our understanding of the oligomerisation process of the N-protein. In summary, the SR-rich motif does possess binding affinity, but this is specific for the central region (211–290 aa) of another molecule of N-protein, instead of the SR-rich motif itself. The C-terminal region (283–422 aa) possess binding affinity for itself and to associate into a dimer, trimer, tetramer or hexamer, in a concentration-dependent manner. The essential sequence for oligomerisation of the N-protein was identified to be residues 343–402. Interestingly, this region also encompasses the RNA binding motif of the N-protein, which prompts us to speculate that there might be mutual interplay between RNA binding and oligomerisation activities of the N-protein. Further, the oligomerisation was observed to be independent of electrostatic interactions and addition of single strand DNA to the reaction mixture containing tetramers of the N-protein promoted oligomerisation. Thus, it has been proposed that once the tetramer is formed by protein–protein interaction between nucleocapsid molecules, binding with genomic RNA prompts further assembly of the complete nucleocapsid structure.
6. Perturbation of host cellular process by the N-protein
Besides being the capsid protein of the virus, the N-protein of many coronaviruses is known to double up as a regulatory protein. The N-protein of the SARS-CoV too has been shown to modulate the host cellular machinery in vitro, thereby indicating its possible regulatory role during its viral life cycle. Some of the major cellular processes perturbed by heterologous expression of the N-protein are discussed below.
6.1. Deregulation of host cell cycle
Two groups have reported the ability of the N-protein to interfere with the host cell cycle in vitro. Work done by Y.H. Li et al. (2005) and F.Q. Li et al. (2005) proved that mutation of the sumoylation motif in the N-protein leads to cell cycle arrest. Work done in our laboratory has shown the inhibition of S phase progress in cells expressing the N-protein (Surjit et al., 2006). Further, we have observed down-regulation of S phase specific gene products like cyclin E and CDK2 in SARS-CoV infected cell lysate, which suggest that the observed phenomenon may be relevant in vivo. In an attempt to further characterize the mechanism of cell cycle blockage induced by the N-protein, several biochemical and mutational analysis were carried out. The results thus obtained demonstrated that the N-protein directly inhibits the activity of the cyclin–CDK complex, resulting in hypophosphorylation of retinoblastoma protein with a concomitant down-regulation of E2F1-mediated transactivation. Analysis of RXL and CDK phosphorylation mutant N-protein identified the mechanism of inhibition of CDK4 and CDK2 activity to be different. Whereas the N-protein could directly bind to cyclin D and inhibit the activity of CDK4–cyclin D complex; inhibition of CDK2 activity appeared to be achieved in two different ways: indirectly by down-regulation of protein levels of CDK2, cyclin E, and cyclin A, and by direct binding of N-protein to CDK2–cyclin complex. Nevertheless, the mechanism of cell cycle deregulation in vivo, if any, remains to be understood.
6.2. Inhibition of interferon production
Production of interferon is one of the primary host defense mechanism. However, SARS-CoV infection does not result in IFN production. Nevertheless, pretreatment of cells with IFN blocks SARS-CoV infection (Spiegel et al., 2005, Zheng et al., 2004). Based on this observation, Palese lab has studied the IFN inhibitory property of different SARS-CoV proteins, which revealed that ORF3b, ORF6 as well as the N-protein have the ability to independently inhibit IFN production through different mechanisms. The N-protein was found to inhibit the activity of IRF3 and NFkB in host cells, resulting in inhibition of IFN synthesis. IRF3 activity was also blocked by 3b and ORF6 proteins, but inhibition of NFkB activity was a property unique to the N-protein. In addition, 3b and ORF6 proteins were able to block STAT1 activity through different mechanisms (Kopecky-Bromberg et al., 2007). All these data suggest that SARS-CoV may employ multiple factors to check the activity of host immune system and N-protein may be one of the major partners in this process. It may be possible that these different factors act independently during different stages of viral life cycle. In that case, regulatory activity of the N-protein will be as indispensible as its structural activity.
6.3. Up-regulation of COX2 production
Another major proinflammatory factor-induced during viral infection is the cyclo oxygenase-2 protein. Using 293T cells expressing the N-protein, Yan et al. (2006) have shown that expression of the N-protein leads to upregulation of COX2 protein production in a transcriptional manner. They have further demonstrated that the N-protein directly binds to the NFkB response element present in the COX2 promoter through a 68 aa residue binding domain (136–204 aa) and activates its transcription.
Although the N-protein is known to associate with stretches of nucleic acids, till date there is no other documentation or prediction of its sequence specific DNA binding activity (as a transcription factor). In such a scenario, the above observation, if reproducible in vivo, may really be a unique property of the N-protein and may further add to the established regulatory functions of the N-protein.
6.4. Up-regulation of AP1 activity
Exogenously expressed N-protein has been reported to enhance the DNA binding activity of c-fos, ATF-2, CREB-1, and fos B in an ELISA-based assay, thus suggesting an increase in AP1 activity in these cells (He et al., 2003). Mechanistic details and functional significance of this phenomenon remains to be elucidated.
6.5. Induction of apoptosis in serum starved monkey kidney cells
Earlier work done in our laboratory has shown that N-protein, when expressed in COS-1 monkey kidney cells, induces apoptosis in the absence of growth factors. Attempts to understand the mechanism of programmed cell death revealed that the N-protein down modulated the activity of pro-survival factors such as extracellular regulated kinase, Akt, and bcl 2, and upregulated the activity of pro-apoptotic factors like caspase-3 and caspase-7. This phenomenon was also associated with reorganization of the actin cytoskeleton (Surjit et al., 2004a, Surjit et al., 2004b, Surjit et al., 2004c). However, this phenomenon was not observed in another cell line of epithelial lineage (huh7). Recently, Zhang et al. (2007) have reported that serum starvation apoptosis of N expressing COS-1 cells involves activation of mitochondrial pathway. It remains to be studied whether this phenomenon is actually recapitulated in vivo.
6.6. Association with host cell proteins
C. Luo et al. (2004) and H. Luo et al. (2004) have reported the interaction between hnRNPA1 and N-protein by using a variety of biochemical and genetic assays. The interaction was found to be mediated through the middle region (161–210 aa) of N-protein. If relevant in vivo, this interaction may play a significant role in regulation of the viral RNA synthesis.
Another interesting study done by C. Luo et al. (2004) and H. Luo et al. (2004) have reported association between the N-protein and human cyclophylin A. By SPR analysis they have shown it to be a high affinity interaction. Although the significance of this interaction is not known in vivo, they have proposed that this interaction might be crucial for viral infection. Notable is the fact that HIV-1 gag also binds with human cyclophylin A and this interaction is crucial for HIV infection (Gamble et al., 1996).
In summary, although several regulatory roles have been proposed for the SARS-CoV N-protein using a variety of in vitro experimental systems, no clear evidence exists for their occurrence in vivo. In the absence of a suitable in vivo experimental system, all these functions remain speculative.
7. N-protein: an efficient diagnostic tool
One of the most essential steps to limit the outbreak of any infectious disease is the ability to diagnose the causative agent, at the earliest possible time, which can be achieved by detecting some of the markers that are specifically expressed by the pathogen or by identifying some of the host factors that are specifically produced during infection. N-protein, being one of the predominantly expressed proteins at the early stage of SARS-CoV infection, against which a strong antibody response is initiated by the host; has been proposed to be an attractive diagnostic tool.
In serum of SARS-CoV patients, the N-protein has been detected as early as day 1 of infection by ELISA using monoclonal antibodies against it (Che et al., 2004). Further, a comparative study to detect SARS-CoV specific IgG, SARS-CoV RNA, and the N-protein during early stages of infection has demonstrated that detection efficiency of the N-protein is significantly higher than the other two markers (Y.H. Li et al., 2005, F.Q. Li et al., 2005).
Researchers have been mainly focussing on two different strategies by which nucleocapsid can be used as a diagnostic tool (i) development of efficient monoclonal antibodies against the N-protein, (ii) production of recombinantly expressed, highly purified N-protein for detection of N-specific antibody in the host.
Using a phage display approach, Flego et al. (2005) have identified human antibody fragments that recognise distinct epitopes of the N-protein. These may help to develop efficient reagents to detect N-protein in the infected host. Further, several laboratories have been trying to develop efficient monoclonal antibodies against the major immunodominant epitopes of the N-protein, that can be used in ELISA to detect SARS-CoV at an early stage of infection (Shang et al., 2005, Liu et al., 2003, He et al., 2005, Woo et al., 2005). In another interesting study, Liu et al. (2005) have developed an immunofluorescence assay using antirabbit N-antibody that can specifically detect N-protein from throat wash sample of SARS-CoV patients at day 2 of illness.
Several other workers have focused on economical production of highly purified recombinant N-protein using a variety of heterologous expression systems that can be used in ELISA to detect N-specific antibody in the patient sample. N-protein has been produced in abundant quantity using codon optimised gene in Escherichia coli (Das and Suresh, 2006). Saijo et al. (2005) have successfully expressed recombinant N-protein using a baculovirus expression system, which was found to be 92% efficient in neutralizing antibody assay. In another study, Liu et al. (2004), have expressed full length N-protein using yeast expression system. However diagnostic use of recombinant N-protein has been a problematic issue because of several reasons as discussed below.
Bacterially expressed N-protein has been reported to produce false sero-positivity owing to interference of bacterially derived antigens (Leung et al., 2006, Yip et al., 2007). In addition, several studies have shown cross-reactivity between full-length N-protein of SARS and polyclonal antisera of group 1 animal coronaviruses, which may lead to faulty detection (Sun and Meng, 2004). Another study done by Woo et al., have also reported crossreactivity of full-length recombinant N-protein with antisera of H CoV-OC43 and H CoV-229E infected patients, thus giving false positive results. They were able to minimise this false positivity by further verifying the ELISA results with Western blot assay using recombinant N and spike protein of SARS-CoV (Woo et al., 2004).
Later on, studies done by Qiu et al. (2005) and Bussmann et al. (2006), showed that recombinantly expressed C-terminal of the N-protein acts more specifically in detecting SARS-CoV specific antisera in comparision to full-length N-protein. It is noteworthy that this region is predicted to encompass major antigenic sites of the N-protein.
Also, several reports have been published dealing with the detection of N specific IgM by ELISA or indirect immunofluorescent assay (Chang et al., 2004, Hsueh et al., 2004, Woo et al., 2004). However, in these studies, IgM antibodies became detectable later than IgG antibodies, which is in contrast to the phenomena observed in most other pathogens.
A recent report published by Yu et al. (2007), have attempted to solve this problem by using a truncated N-protein (122–422 aa) as an antigen in IgM ELISA. They found the IgM response to appear 3 days before detection of IgG response, which is in agreement with the results obtained from other known pathogens. Further, their results showed 100% specificity and sensitivity of the truncated protein in detecting N-specific IgM from patients with laboratory confirmed SARS cases in comparison to healthy volunteers. The authors have suggested that the IgM capture ELISA using this truncated N-protein may be more effective in serodiagnosis of SARS-CoV at an earlier time.
In another interesting report, Woo et al. (2005) have carried out comparative studies to evaluate the relative diagnostic efficacy of recombinantly expressed N- and S-proteins. They observed sensitivity of recombinant N-IgG ELISA to be significantly higher than that of recombinant S-IgG ELISA. The reverse was true in case of IgM ELISA using recombinant N- and S-proteins. Based on this data, they have suggested the practice of an ELISA for detection of IgM against both S- and N-protein instead of N alone (Woo et al., 2005).
Taken together, all these data does support the notion that the N-protein may be used as an efficient diagnostic tool for detection of SARS-CoV infection. Nevertheless, production scale-up and further validation of specificity using patient samples will determine the possible clinical use of these reagents.
8. N-protein: a suitable vaccine candidate
One of the most clinically relevant uses of the N-protein can be its use as a protective vaccine against SARS-CoV infection. N-protein is one of the major antigens of the SARS-CoV. Also, N-protein analysed from different patient samples shows least variation in the gene sequence (Tong et al., 2004), therefore indicating it to be a stable protein, which is a primary requirement for an efficient vaccine candidate.
Earlier studies carried in Collins lab, Rao lab, and Li lab have clearly shown that anti-serum to the N-protein does not contain neutralizing antibodies against SARS-CoV (Buchholz et al., 2004, Pang et al., 2004, and Liang et al., 2005). This may be attributed to the localization of N-protein inside the viral envelope, which will not be accessible to the antibody during infection. It is noteworthy that the most effective SARS-CoV structural protein that can induce neutralizing antibody production is the S-protein (Buchholz et al., 2004). The S antibody could block viral infection with 100% efficiency. On the other hand, although unable to induce humoral immunity, expression of N-protein-induced significant cytotoxic T lymphocyte (CTL) response (Buchholz et al., 2004, Gao et al., 2003, Zhu et al., 2004). Induction of N specific CTLs will help limit the infection by lysing virus infected cells. This will also limit the spread of virus. Thus, N-based vaccines may further augment the protection efficiency when co-administered with S-based vaccine. Several laboratories have been exploring various strategies to evaluate the potential of N-protein as a vaccine candidate.
In an elegant work done by Kim et al. (2004), calreticulin fused N-protein expressing vaccinia virus has been shown to generate potent N-specific humoral and T-cell immune responses in mice. As reported by the authors, fusion with calreticulin specifically enhanced the efficiency and significantly reduced the titre of challenging vector (vaccinia virus). The authors have proposed that N-protein may be the logical choice as a target antigen in the event of S antibody dependent enhancement (ADE) of infection. However, ADE phenomenon has not been observed during spike-mediated vaccination (Buchholz et al., 2004). Another study done by Wang et al. (2005) has attempted to use plasmid DNA expressing S-, M-, and N-proteins as an efficient vaccine candidate. Although they report the production of some B-cell and T-cell responses against N-protein, however stringent immune response was obtained for the S- and M-proteins, thus scaling down the choice of N-protein as a suitable candidate vaccine (Wang et al., 2005). A similar plasmid mediated vaccination approach has also been reported by Zhao et al. (2004) wherein they have immunised mice with the DNA construct (pCI vector) expressing the N-protein. They too have reported the generation of a robust B-cell and T-cell immune response in animals. Another group of workers have also reported successful use of the N-protein as a DNA vaccine. They immunised mice by intra-mucosal injection of the N-protein expressing plasmid vector and were able to obtain specific humoral and T-cell responses (Zhu et al., 2004).
The N-protein has also been reported to be of potential interest as a peptide-based vaccine. A systematic study done by Liu et al. (2006) has revealed the immunodominant epitopes of the N-protein which could efficiently stimulate immune response. They have also deduced some conserved immunodominant epitopes in mouse, monkey, and humans, which may help in design of the vaccine.
A recent report published by Gao's laboratory provides further evidence regarding the efficiency of an N-based vaccine (Zhao et al., 2007). By using overlapping synthetic peptides spanning the N-protein, they have identified dominant helper T-cell epitopes in the nucleocapsid protein of SARS-CoV. Immunization of mice with peptides emcompassing these dominant TH-cell epitopes resulted in strong cellular immunity in vivo. Priming with the helper peptides significantly accelerated the immune response induced by the N-protein. Further, by fusing with a conserved neutralizing epitope from the spike protein of SARS-CoV, two of the TH-cell epitope bearing peptides assisted in the production of higher titre neutralizing antibodies in vivo, in comparision to spike epitope alone or its mixture with TH epitope of N. Thus, it is practically possible to generate a better immune response by using a fusion of N- and S-protein. However, the TH epitopes identified in their report is specific to mouse. Therefore, TH epitopes identified in that study will not be useful for human. Nevertheless, their data provides useful information for the design of peptide-based anti-SARS-CoV vaccines.
Another interesting study conducted by Pei et al. (2005) reports the possible use of the N-protein as a mucosal vaccine candidate. They expressed the N-protein in Lactobacillus lactis, which is a food grade bacteria, and challenged the mice either orally or intra-mucosally. As preliminary evidence, they were able to observe significant N-specific IgG in the sera of orally challenged animals.
9. Future perspective
It is a significant achievement for the research community that within a short span of time, we have been able to obtain more or less a clear understanding regarding the structural and functional properties of the N-protein. However, it is a fact worth mentioning that all the studies done here were performed with in vitro experiments, using recombinantly expressed N-protein, in isolation. So at present, all we can conclude is that, the N-protein per se has the physical ability to perform the above described functions, in other words N-protein does bear the necessary signature sequence or motifs or conformation to perform these functions under suitable circumstances. Whether a similar event is recapitulated in vivo during viral infection, will be dependent on several criteria: (i) net effect of other viral factors on the activity of N-protein, (ii) net translation and turn over rate of N-protein, (iii) a conducive intra-cellular milieu, and (iv) net modulation of an already skewed cellular pathway by other viral factors. Hence, it will be interesting to re-evaluate the properties of N-protein in a SARS-CoV infection model. However, owing to the limited user friendliness and accessibility of an infection system, probably we still have to resort to in vitro systems for further analysis of the characteristics of N-protein. One of the better experimental system has already been established by the Chang lab (Hsieh et al., 2005), wherein all the structural proteins were co-expressed to form VLP in 293T cells. If this system can be further improved to optimise the rate of synthesis of these different proteins to a near in vivo level, it will at least enable us to study the net effect of the N-protein with respect to other viral proteins. Further establishment of a replicon system may also be helpful. In addition, some of the interesting preliminary observations reported by several laboratories need to be elaborately analysed. To begin with, the reported interaction of the N-protein with genomic RNA packaging signal needs to be further characterised and mapped. Since the oligomerisation domain and the RNA binding regions of the N-protein overlap with each other, the suggested possibility of regulated genome incorporation and capsid assembly should be further characterised with the aid of a replicon system or a particle assembly system. In addition, the reported ability of the N-protein to modulate different cellular pathways should be further characterised in the particle assembly system or at least in the presence of other viral accessory proteins.
The most unique and significant property of the N-protein revealed by preliminary studies is its ability to act as sequence specific DNA binding factor. It has been shown to bind NFkB response element of COX2 promoter and enhance COX2 gene expression. This activity may be further empowering the N-protein to manipulate the entire gene expression programme of the infected cell. Therefore, studies should be initiated to elaborately analyse this phenomenon. It seems to deserve so much attention because another study done by Palese lab has proved the ability of the N-protein to inhibit NFkB activity, which results in inhibition of IFN synthesis. Further, Liao et al. (2005) have reported the activation of NFkB by N-protein in Vero E6 cells and He et al. (2005) failed to detect any change in NFkB activity in the same cells. Therefore it needs to be clarified whether N enhances NFkB activity and if yes; whether upregulation of COX2 transcription by direct DNA binding is a property specific to that promoter or it is a global phenomenon. In such a scenario, there may be complicated crosstalk between the ability of N-protein to deregulate the expression of COX2 and IFN in infected cells.
Lastly, the N-protein is known to be the most abundantly expressed protein of the SARS-CoV. Therefore, any information generated from the analysis of this protein, whether in vivo or ex vivo, will definitely help to increase our understanding of the biology of SARS-CoV and may someday help design better protective tools against it.
Acknowledgements
The authors wish to thank Ms. Alisha Lal for helping out in typing and formatting this review. We apologize to all those colleagues whose work we might have missed to cite in this article.
References
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann B.M., Reiche S., Jacob L.H., Braun J.M., Jassoy C. Antigenic and cellular localisation analysis of the severe acute respiratory syndrome coronavirus nucleocapsid protein using monoclonal antibodies. Virus Res. 2006;122:119–126. doi: 10.1016/j.virusres.2006.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.T., Kao C.L., Chung M.Y., Chen S.C. SARS exposure and emergency department workers. Emerg. Infect. Dis. 2004;10:1117–1119. doi: 10.3201/eid1006.030972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Hao W., Wang Y. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Suresh M.R. Copious production of SARS-CoV nucleocapsid protein employing codon optimized synthetic gene. J. Virol. Methods. 2006;137:343–346. doi: 10.1016/j.jviromet.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Zhuo Y., Tan X. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J. Med. Virol. 2006;78:1365–1373. doi: 10.1002/jmv.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Ye L.B., Zhang Y. Nucleocapsid amino acids 211 to 254, in particular, tetrad glutamines, are essential for the interaction between the nucleocapsid and membrane proteins of SARS-associated coronavirus. J. Microbiol. 2006;44:577–580. [PubMed] [Google Scholar]
- Flego M., Di Bonito P., Ascione Generation of human antibody fragments recognizing distinct epitopes of the nucleocapsid (N) SARS-CoV protein using a phage display approach. BMC Infect. Dis. 2005;5:73. doi: 10.1186/1471-2334-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T.R., Vajdos F.F., Yoo S. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Andonov A. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Du Q., Lau S. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods. 2005;127:46–53. doi: 10.1016/j.jviromet.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.K., Chang S.C., Huang C.C. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.R., Huang L.M., Chen P.J. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., Hung C.F. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A. Mass spectrometric characterization of proteins from the sars virus: a preliminary report. Mol. Cell Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T., van Maren W.W., Chan F.K., Chan W.S., Lo A.W., Ma C.H., Tam F.C., To K.F., Chan P.K., Sung J.J., Lim P.L. Extremely low exposure of a community to severe acute respiratory syndrome coronavirus: false seropositivity due to use of bacterially derived antigens. J. Virol. 2006;80:8920–8928. doi: 10.1128/JVI.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Li J., Liu X.E. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J. Virol. Methods. 2005;130:45–50. doi: 10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M.F., Du R.L., Liu J.Z. SARS patients-derived human recombinant antibodies to S and M proteins efficiently neutralize SARS-coronavirus infectivity. Biomed. Environ. Sci. 2005;18:363–374. [PubMed] [Google Scholar]
- Liao Q.J., Ye L.B., Timani K.A. Activation of NF-kappaB by the full-length nucleocapsid protein of the SARS coronavirus. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:607–612. doi: 10.1111/j.1745-7270.2005.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Hu S., Hu Y. The C-terminal portion of the nucleocapsid protein demonstrates SARS-CoV antigenicity. Genom. Proteom. Bioinform. 2003;1:193–197. doi: 10.1016/S1672-0229(03)01024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.S., Yang K.Y., Lin J. High-yield expression of recombinant SARS coronavirus nucleocapsid protein in methylotrophic yeast Pichia pastoris. World J. Gastroenterol. 2004;10:3602–3607. doi: 10.3748/wjg.v10.i24.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I.J., Chen P.J., Yeh S.H. Immunofluorescence assay for detection of the nucleocapsid antigen of the severe acute respiratory syndrome (SARS)-associated coronavirus in cells derived from throat wash samples of patients with SARS. J. Clin. Microbiol. 2005;43:2444–2448. doi: 10.1128/JCM.43.5.2444-2448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Leng C.H., Lien S.P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Luo H., Zheng S. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Ye F., Sun T.Y. In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys. Chem. 2004;112:15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Ye F., Chen K. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- Luo H., Chen Q., Chen J. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Chen J., Chen K. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., Kong X., Bartlam M., Rao Z. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85:3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- Pei H., Liu J., Cheng Y. Expression of SARS-coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Appl. Microbiol. Biotechnol. 2005;68:220–227. doi: 10.1007/s00253-004-1869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Wang J., Wang H., Chen Z., Dai E., Guo Z., Wang X., Pang X., Fan B., Wen J., Wang J., Yang R. Use of the COOH portion of the nucleocapsid protein in an antigen-capturing enzyme-linked immunosorbent assay for specific and sensitive detection of severe acute respiratory syndrome coronavirus. Clin. Diagn. Lab. Immunol. 2005;12:474–476. doi: 10.1128/CDLI.12.3.474-476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Rowland R.R., Chauhan V., Fang Y. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells. J. Virol. 2005;79:11507–11512. doi: 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M., Ogino T., Taguchi F. Recombinant nucleocapsid protein-based IgG enzyme-linked immunosorbent assay for the serological diagnosis of SARS. J. Virol. Methods. 2005;125:181–186. doi: 10.1016/j.jviromet.2005.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Wang X.Y., Yuan J.W. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Martinez-Sobrido L. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.F., Meng X.J. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: implication for SARS diagnosis. J. Clin. Microbiol. 2004;42(5):2351–2352. doi: 10.1128/JCM.42.5.2351-2352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Chow V.T. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Kumar P. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Kumar R., Mishra R.N. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Chow V.T. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani K.A., Liao Q., Ye L. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Lingappa J.R., Chen Q. Direct sequencing of SARS-coronavirus S and N genes from clinical specimens shows limited variation. J. Infect. Dis. 2004;190:1127–1131. doi: 10.1086/422849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu X., Wang Y. Low stability of nucleocapsid protein in SARS virus. Biochemistry. 2004;43:11103–11108. doi: 10.1021/bi049194b. [DOI] [PubMed] [Google Scholar]
- Wang Z., Yuan Z., Matsumoto M. Immune responses with DNA vaccines encoded different gene fragments of severe acute respiratory syndrome coronavirus in BALB/c mice. Biochem. Biophys. Res. Commun. 2005;327:130–135. doi: 10.1016/j.bbrc.2004.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Wong B.H.L., Chan K.H., Chu C.M., Tsoi H.W., Huang Y., Peiris J.S.M., Yuen K.Y. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 2004;11:665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2005;43:3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Hao Q., Mu Y. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006;38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yip C.W., Hon C.C., Zeng F., Chow K.Y., Chan K.H., Peiris J.S., Leung F.C. Naturally occurring anti-Escherichia coli protein antibodies in the sera of healthy humans cause analytical interference in a recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay for serodiagnosis of severe acute respiratory syndrome. Clin. Vaccine Immunol. 2007;14:99–101. doi: 10.1128/CVI.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Dove B.K., Enjuanes L. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- Yu I.M., Gustafson C.L., Diao J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Oldham M.L., Zhang J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Le M.Q., Inoue S. Recombinant truncated nucleocapsid protein as antigen in a novel immunoglobulin M capture enzyme-linked immunosorbent assay for diagnosis of severe acute respiratory syndrome coronavirus infection. Clin. Vaccine Immunol. 2007;14:146–149. doi: 10.1128/CVI.00360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Viswanathan S., Mahony J.B. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J. Gen. Virol. 2005;86:211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wei L., Jiang D., Wang J., Cong X., Fei R. SARS-CoV nucleocapsid protein induced apoptosis of COS-1 mediated by the mitochondrial pathway. Artif. Cells Blood Substit. Immobil. Biotechnol. 2007;35:237–253. doi: 10.1080/10731190601188422. [DOI] [PubMed] [Google Scholar]
- Zhao P., Cao J., Zhao L.J. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2004;331:128–135. doi: 10.1016/j.virol.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.-M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(11):6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., He M.L., Wong K.L. Potent inhibition of SARS-associated coronavirus (SCOV) infection and replication by type I interferons (IFNalpha/beta) but not by type II interferon (IFN-gamma) J. Interferon Cytokine Res. 2004;24:388–390. doi: 10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- Zhu M.S., Pan Y., Chen H.Q. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


