Highlights
-
•
45 full FMDV genomes were analysed from farms infected in the UK during 2007.
-
•
Analyses revealed intra-herd clustering consistent with virus transmission events.
-
•
Inter-herd sequence differences were estimated to be 4.6 nucleotides.
-
•
Random selection of 1 sequence from each herd generated similar transmission trees.
-
•
These results will help to design cost-effective approaches for the control of FMD.
Keywords: Foot-and-mouth disease virus, Full genome, Within-herd genetic variation, Transmission trees
Abstract
Full-genome sequences have been used to monitor the fine-scale dynamics of epidemics caused by RNA viruses. However, the ability of this approach to confidently reconstruct transmission trees is limited by the knowledge of the genetic diversity of viruses that exist within different epidemiological units. In order to address this question, this study investigated the variability of 45 foot-and-mouth disease virus (FMDV) genome sequences (from 33 animals) that were collected during 2007 from eight premises (10 different herds) in the United Kingdom. Bayesian and statistical parsimony analysis demonstrated that these sequences exhibited clustering which was consistent with a transmission scenario describing herd-to-herd spread of the virus. As an alternative to analysing all of the available samples in future epidemics, the impact of randomly selecting one sequence from each of these herds was used to assess cost-effective methods that might be used to infer transmission trees during FMD outbreaks. Using these approaches, 85% and 91% of the resulting topologies were either identical or differed by only one edge from a reference tree comprising all of the sequences generated within the outbreak. The sequence distances that accrued during sequential transmission events between epidemiological units was estimated to be 4.6 nucleotides, although the genetic variability between viruses recovered from chronic carrier animals was higher than between viruses from animals with acute-stage infection: an observation which poses challenges for the use of simple approaches to infer transmission trees. This study helps to develop strategies for sampling during FMD outbreaks, and provides data that will guide the development of further models to support control policies in the event of virus incursions into FMD free countries.
1. Introduction
The poor fidelity and lack of proofreading activity of the polymerases of RNA viruses cause high rates of spontaneous mutation during virus replication. These rates are estimated to range from 10−5 to 2 × 10−3 mutations per nucleotide per replication event (Thebaud et al., 2010). As a consequence, these viruses evolve rapidly and have high degrees of genome variability, which is a constant challenge for molecular diagnosic tests, as well as for prophylaxis and control methods such as vaccines and antivirals. Nevertheless, these high evolution rates have been exploited to understand the transmission of human and animal RNA virus infections across fine spatial and temporal scales (Cottam et al., 2006, Baillie et al., 2011, Bataille et al., 2011, Cotten et al., 2013, Gray et al., 2011, Hughes et al., 2012, Li et al., 2010, Orton et al., 2013). These studies help to increase the knowledge on virus evolution and to identify and analyse the potential origins, patterns of transmission and spread and risks of virus infections to be ready for the prediction, early detection and/or control of the disease.
Foot-and-mouth disease virus (FMDV) is a non-enveloped, single-stranded positive-sense RNA virus from the genus Aphthovirus within the family Picornaviridae which rapidly spreads among cloven-hoofed animals. Full genome sequences of FMDV can be generated and analysed in real-time to discern the origin of outbreaks, the transmission links between infected premises, and to predict undisclosed infection to support control and eradication policies in free-without-vaccination countries (Cottam et al., 2008b, Valdazo-González et al., 2012). Furthermore, these approaches have also been used to monitor the genetic evolution of FMD viruses at the finest scales: such as within an individual animal (Wright et al., 2011) and during animal-to-animal transmission in experimental studies (Juleff et al., 2013). The interpretation of these data can be enhanced by using a range of models that have been recently developed that integrate sequence data with epidemiological information (Cottam et al., 2008a, Morelli et al., 2012). However, the practical use of these tools to confidently reconstruct transmission trees during FMD outbreaks is limited by our understanding of the genetic diversity of viruses that exist within different epidemiological units under field conditions (within-herd diversity) (Orton et al., 2013).
This study has investigated the genetic variability of viruses from field samples collected from the FMDV outbreaks that occurred in the Southeast of the United Kingdom (UK) between the 3rd of August and the 30th of September 2007 (Cottam et al., 2008b, Ryan et al., 2008). FMDV sequences from the O/EURO-SA topotype were generated and analysed from samples within each of the eight infected premises (IPs) from 10 separate locations (with individual herds/flocks of animals grazing together) confirmed in a series of FMD outbreaks that occurred in two phases (that were geographically 17 km and temporally 34 days apart). Information regarding these herds and the clinical and laboratory investigations of these outbreaks has been described previously (Cottam et al., 2008b, Reid et al., 2009, Ryan et al., 2008). A particular focus of this work has been to consider the impact of sequencing only a single sample from each epidemiological unit upon the inferred transmission trees in order to help to design rapid and cost effective sequencing approaches that can be used in the event of FMD outbreaks, when sequencing all of the infected animals within the outbreak might not be possible.
2. Material and methods
2.1. Selection of samples
In total, 34 FMD virus-positive clinical samples from 26 animals infected during the 2007 outbreak in UK (Table 1 ) were processed in this study, and were jointly analysed with a further 11 previously published full-genome sequences from these outbreaks (Cottam et al., 2008b). These samples had been selected from the samples sent to the UK National Reference Laboratory for FMD (The Pirbright Institute, United Kingdom) during the 2007 outbreak in UK on basis of the cycle threshold (CT) values (⩽27) generated by a real-time RT-PCR which targets the region encoding the FMDV non-structural protein 3D (Reid et al., 2009). These samples included vesicular epithelium (10% suspension, prepared as described (Cottam et al., 2008b), whole blood (collected in EDTA tubes), sera and oesophageal/pharyngeal scrapings (probangs).
Table 1.
Details of the sequences obtained during the FMDV outbreak in 2007 in United Kingdom.
| Viruses | Infected Premise (IP) | Specimen1 | Animal | Species | Oldest lesion age2 |
Collection date | References | GenBank | |
|---|---|---|---|---|---|---|---|---|---|
| Animal | Infected premise | ||||||||
| IAH2 | IP0 | CC | – | – | – | – | – | Cottam et al. (2008b) | EU448369 |
| MAH | IP0 | CC | – | – | – | – | – | EU448370 | |
| UKG/7/2007 | IP1b | ES | IP1b_1 | Cattle | 8 | 10 | 03/08/2007 | Cottam et al. (2008b) | EU448371 |
| UKG/7B/2007 | IP1b | ES | IP1b_1 | Cattle | 8 | 10 | 04/08/2007 | Cottam et al. (2008b) | EU448372 |
| UKG/9/2007 | IP1b | ES | IP1b_2 | Cattle | 7 | 10 | 03/08/2007 | This work | KJ560276 |
| UKG/11/2007 | IP1b | ES | IP1b_3 | Cattle | 8 | 10 | 03/08/2007 | This work | KJ560277 |
| UKG/13/2007 | IP1b | Blood | IP1b_4 | Cattle | 3 | 10 | 03/08/2007 | This work | KJ560278 |
| UKG/32/2007 | IP1b | Blood | IP1b_5 | Cattle | 5 | 10 | 03/08/2007 | This work | KJ560279 |
| UKG/91/2007 | IP2b | ES | IP2b_1 | Cattle | 6 | 7 | 06/08/2007 | This work | KJ560280 |
| UKG/92/2007 | IP2b | ES | IP2b_2 | Cattle | 6 | 7 | 06/08/2007 | This work | KJ560281 |
| UKG/93/2007 | IP2b | ES | IP2b_3 | Cattle | 6 | 7 | 06/08/2007 | Cottam et al. (2008b) | EU448373 |
| UKG/94/2007 | IP2b | ES | IP2b_4 | Cattle | 5 | 7 | 06/08/2007 | This work | KJ560282 |
| UKG/95/2007 | IP2b | ES | IP2b_5 | Cattle | 6 | 7 | 06/08/2007 | This work | KJ560283 |
| UKG/96/2007 | IP2b | Blood | IP2b_1 | Cattle | 6 | 7 | 06/08/2007 | This work | KJ560284 |
| UKG/97/2007 | IP2b | Blood | IP2b_2 | Cattle | 6 | 7 | 06/08/2007 | This work | KJ560285 |
| UKG/132/2007 | IP2c | Blood | IP2c_1 | Cattle | None | None | 07/08/2007 | This work | KJ560286 |
| UKG/150/2007 | IP2c | Blood | IP2c_2 | Cattle | None | None | 07/08/2007 | Cottam et al. (2008b) | EU448374 |
| UKG/158/2007 | IP2c | Blood | IP2c_3 | Cattle | None | None | 07/08/2007 | This work | KJ560287 |
| UKG/642/2007 | IP3b | ES | IP3b_1 | Cattle | 2 | 5 | 12/09/2007 | This work | KJ560288 |
| UKG/643/2007 | IP3b | ES | IP3b_2 | Cattle | 4–5 | 5 | 12/09/2007 | Cottam et al. (2008b) | EU448375 |
| UKG/644/2007 | IP3b | ES | IP3b_3 | Cattle | 4–5 | 5 | 12/09/2007 | This work | KJ560289 |
| UKG/645/2007 | IP3b | ES | IP3b_4 | Cattle | 2–3 | 5 | 12/09/2007 | This work | KJ560290 |
| UKG/647/2007 | IP3b | Blood/serum | IP3b_1 | Cattle | 2 | 5 | 12/09/2007 | This work | KJ560291 |
| UKG/648/2007 | IP3b | Blood/serum | IP3b_2 | Cattle | 4–5 | 5 | 12/09/2007 | This work | KJ560292 |
| UKG/649/2007 | IP3b | Blood/serum | IP3b_3 | 2–3 | 5 | 12/09/2007 | This work | KJ560293 | |
| UKG/650/2007 | IP3b | Blood/serum | IP3b_4 | Cattle | 2–3 | 5 | 12/09/2007 | This work | KJ560294 |
| UKG/1153/2007 | IP3c | ES | IP3c_1 | Cattle | ND | 5 | 15/09/2007 | Cottam et al. (2008b) | EU448376 |
| UKG/1170/2007 | IP3c | Blood | IP3c_2 | Cattle | ND | 5 | 15/09/2007 | This work | KJ560295 |
| UKG/800/2007 | IP4b | ES | IP4b_1 | Cattle | 7 | 10 | 13/09/2007 | Cottam et al. (2008b) | EU448377 |
| UKG/805/2007 | IP4b | ES | IP4b_2 | Cattle | 8 | 10 | 13/09/2007 | This work | KJ560296 |
| UKG/1418/2007 | IP5 | O/PS | IP5_1 | Sheep | ND | 21 | 17/09/2007 | This work | KJ560297 |
| UKG/1419/2007 | IP5 | O/PS | IP5_2 | Sheep | ND | 21 | 17/09/2007 | This work | KJ560298 |
| UKG/1421/2007 | IP5 | O/PS | IP5_3 | Sheep | ND | 21 | 17/09/2007 | Cottam et al. (2008b) | EU448378 |
| UKG/1425/2007 | IP5 | O/PS | IP5_4 | Sheep | ND | 21 | 17/09/2007 | This work | KJ560290 |
| UKG/1426/2007 | IP5 | O/PS | IP5_5 | Sheep | ND | 21 | 17/09/2007 | This work | KJ560300 |
| UKG/1484/2007 | IP6b | ES | IP6b_1 | Cattle | 4 | 4 | 21/09/2007 | Cottam et al. (2008b) | EU448379 |
| UKG/1485/2007 | IP6b | ES | IP6b_1 | Cattle | 4 | 4 | 21/09/2007 | This work | KJ560301 |
| UKG/1488/2007 | IP6b | Serum | IP6b_1 | Cattle | 4 | 4 | 21/09/2007 | This work | KJ560302 |
| UKG/1679/2007 | IP7 | ES | IP7_1 | Cattle | 2 | 5 | 24/09/2007 | Cottam et al. (2008b) | EU448380 |
| UKG/1684/2007 | IP7 | ES | IP7_2 | Cattle | 3 | 5 | 24/09/2007 | This work | KJ560303 |
| UKG/1693/2007 | IP7 | Blood | IP7_1 | Cattle | 2 | 5 | 24/09/2007 | This work | KJ560304 |
| UKG/1694/2007 | IP7 | Blood | IP7_3 | Cattle | 2 | 5 | 24/09/2007 | This work | KJ560305 |
| UKG/1698/2007 | IP7 | Blood | IP7_2 | Cattle | 3 | 5 | 24/09/2007 | This work | KJ560306 |
| UKG/1701/2007 | IP7 | Blood | IP7_4 | Cattle | 3 | 5 | 24/09/2007 | This work | KJ560307 |
| UKG/1704/2007 | IP7 | Blood | IP7_5 | Cattle | 1 | 5 | 24/09/2007 | This work | KJ560308 |
| UKG/1709/2007 | IP7 | Blood | IP7_3 | Cattle | 2 | 5 | 24/09/2007 | This work | KJ560309 |
| UKG/2366/2007 | IP8 | ES | IP8_1 | Cattle | 2 | 2 | 29/09/2007 | Cottam et al. (2008b) | EU448381 |
CC = cell culture; ES = epithelium suspension; O/PS = oesophageal/pharyngeal scrapings (probangs).
ND = not determined.
2.2. Full genome (FG) amplification and sequencing strategies
Samples were processed individually on separate days to prevent potential cross-contamination. Viral RNA was extracted using either the RNeasy Mini Kit (Qiagen, Crawley, West Sussex, UK), or TRIzol Reagent (Invitrogen, Paisley, UK) for those samples such as oesophageal-pharyngeal scrapings with high CT values. Reverse transcription (RT) and complete FMDV genome amplification [except for the poly(C) region] were performed with one oligo-dT reverse RT primer and 23 tagged PCR primers pairs as previously described (Cottam et al., 2008b), but using a cDNA purification step (Illustra GFX™ PCR DNA and Gel Band Purification Kit, GE Healthcare UK Limited, Buckinghamshire, UK) prior to PCR amplification. Additional PCR reactions were carried out using oligo dT reverse primers to amplify the 3′ terminus of the virus, as described (Valdazo-González et al., 2012). Negative control reactions were performed in parallel and were included in all steps and for each of the amplification reactions.
Amplified PCR products were separated by gel electrophoresis (1.8% agarose gels), stained with ethidium bromide (0.2–0.5 μg/mL) and visualized under ultraviolet light. After purification (Illustra GFX™ PCR DNA and Gel Band Purification Kit, GE Healthcare UK Limited, Buckinghamshire, UK), cycle sequencing was carried out using M13 universal forward and reverse primers (Cottam et al., 2008b) or the corresponding specific forward and reverse primers for each PCR product. One of the two following Sanger sequencing reagents and sequencers were used: the Beckman DTCS Kit (Beckman Coulter, USA) on a Beckman Coulter CEQ 8000 sequencer and the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) on an ABI PRISM®-3730 analyzer. Sequences were assembled, proof-read and edited using Lasergene® v11.0 package (DNASTAR Inc., Madison, WI). These sequences have been submitted to GenBank and have been assigned the following accession numbers: KJ560276–KJ560309.
2.3. Complete genome sequences of foot-and-mouth disease virus
The sequences generated in this study were aligned (BioEdit, Version 7.0.5.3 (Hall, 1999)) together with 11 previously published sequences from this outbreak (Cottam et al., 2008b). In total, 45 complete genome sequences from samples from 33 animals (28 cattle and 5 sheep) from eight infected premises (IPs) (ten separate herds) were analysed. This analysis comprised one to five animals and up to eight sequences per herd. Eleven out of these 33 animals were represented by two or three sequences obtained from different clinical samples within the animal (see Table 1).
2.4. Positive selection and recombination analysis
Detection of potential selection pressures at amino acid sites was calculated using the CODEML programme in the PAML 4.1 software package (Yang, 2007). Briefly, the dN/dS ratio (ω value) was obtained using eight different models (M0 to M8). Comparison of likelihood values for nested models by likelihood ratio tests (LRTs) determined if models of positive selection (M2a, M3 and M8) were significantly more likely than models of neutral evolution (M1a and M7). Bayesian methods were used to locate specific sites that have ω > 1 with high posterior probabilities. Preliminary data for the analysis (transition/transversion ratio and phylogenetic relationship between taxa) were carried out using TREE-PUZZLE version 5.2. (Schmidt et al., 2002). Detection of potential recombination between sequences was carried out using low linkage disequilibrium (observed data versus corresponding null distributions from 500 simulated datasets) as implemented in a test statistic, as described (Haydon et al., 2004).
2.5. Bayesian Markov chain Monte Carlo (MCMC) analysis (BEAST) analysis
Bayesian evolutionary analysis using Markov chain Monte Carlo (MCMC) sampling (30,000 trees from 30 million generations), as implemented using BEAST software, Version 1.6.1 (Drummond and Rambaut, 2007), was carried out to infer the phylogenetic relationships between the 45 complete sequences, to estimate the age of their most recent common ancestor (MRCA) and their rate of molecular evolution. Sampling collection dates were used to calibrate the molecular clock. The HKY model of base substitution with the gamma model of site heterogeneity was selected as described (Orton et al., 2013) and used with different combinations of molecular clocks, demographic and phylogeographic diffusion models to check the robustness of the parameters. The resulting output was checked in Tracer, Version 1.5 and visualized with FigTree (Rambaut, 2010), Version 1.3.1 (Lemey et al., 2010).
2.6. Statistical parsimony (TCS) analysis
Maximum parsimony analyses of the 45 complete FMDV sequences recovered during the outbreak and two additional FMDV sequences from isolates used in the laboratories at the time of the outbreak (IP0) were implemented in the TCS freeware, Version 1.21 (Clement et al., 2000). The tree with the 47 sequences was edited so that the main horizontal axis accommodated the two extremes of these cases: the potential sources of these outbreaks (IP0) and the sequence of the last premises infected (IP8). This tree was considered the reference tree for the following analysis.
2.7. Foot-and-mouth disease virus genetic variability and its effect on the reconstruction of transmission trees in single random sequencing strategies
For this analysis, those nucleotide sites containing International Union of Pure and Applied Chemistry (IUPAC) ambiguity codes (15 sites) were removed. One thousand datasets comprising of one randomly-selected sequence per herd were generated. Each dataset was processed with a pipeline for computing (a) a statistical parsimony tree, as implemented in TCS (TCS tree); and (b) a minimum spanning tree (MST). For each TCS tree and MST an adjacency matrix was computed. A cumulative adjacency matrix was computed as the sum of the adjacency matrices of all one thousand trees of each type. A consensus tree was constructed based on the aggregated matrix using the MST algorithm (consensus trees available in Supplementary data 1).
In order to assess the impact of collecting and processing only a single virus-positive sample from each of the infected herds to infer a transmission tree (single random sequencing strategy), the TCS trees were converted to putative transmission trees by merging each unlabelled node into the closest labelled node. Other approaches for merging nodes were also considered and are available from the authors on request. The MST was computed on the basis of Hamming distance trees (number of sites in which two sequences differ). The cumulative adjacency matrices record, for each possible edge (pair of premises), the number of times it occurred in the TCS transmission trees and the MSTs. These trees were further analysed by determining the frequency of all topologies obtained with the one thousand datasets and each method. For each topology, the number of edges that were not consistent with the reference tree (inferred from the TCS tree with all the 45 sequences) was determined.
In order to determine genetic distance that might be expected for individual herd-to-herd transmission events in an FMD outbreak when using a single random sequencing strategy, the Hamming distance between each source and target premise, according to the reference tree, was calculated for each of the one thousand randomly generated sequence sets. The individual distance values were aggregated by means and standard deviation for each edge in the reference tree, and by frequency of Hamming distance value.
The devised pipeline for all these analyses was driven by a command line application to enable batch processing. The software was implemented in Python 2.7.3, using Biopython 1.6.0. R 2.15.1. was used for statistical analyses and visualisation. The TCS software was modified to support non-interactive operation. Computing was carried out on Linux 3.5.0 systems (Ubuntu 12.10 “Quantal Quetzal”, 64 bit). The scripts are available from the authors on request.
3. Results
3.1. RT-PCR and sequencing strategies
The overlapping RT-PCR and sequencing strategy generated products of the expected size for all the 34 complete [except poly(C)] FMDV genomes (Table 1). The sequence coverage ranged from 3.7 to 7.8 reads/site. No amplified DNA was detected in the control reactions run in parallel. The assembled FMD virus sequences were all 8193 nucleotides (nts) in length. Of these sites, 28 nts were derived from PCR primers, 12 nts from an artificial internal poly(C) tract within the 5′ UTR and 13 nts were included to represent the 3′ terminal poly(A) tail.
3.2. Complete genome sequences of foot-and-mouth disease virus
In total, 34 complete FMDV genomes generated in the present study together with a further 11 more sequences generated in a previous study (Cottam et al., 2008b) were analysed. Nucleotide alignments of these 45 complete genome sequences revealed nucleotide changes at 64 sites distributed along the genome (Table 2 ). A total of 15 sites with 17 IUPAC ambiguities codes were also found in 11 sequences. Eight of these ambiguities were present in three out of the four sequences from probangs in sheep (IP5), whilst five and four ambiguities were present in four blood and three epithelium samples, respectively, from IP2b, IP2c and IP3b. These substitutions were broadly distributed across the FMD genome. Within the ORF, 53 nt substitutions led to 22 amino acid changes. Neither positive selection nor recombination were detected in these sequences.
Table 2.
Nucleotide and non-synonymous amino acid substitutions along the 45 FMDV sequences obtained during the 2007 outbreak in UK. Each purine and pyrimidine base has a different colour and yellow was assigned to ambiguous positions (denoted by IUPAC nucleotide code).
 |
3.3. Bayesian analysis (BEAST)
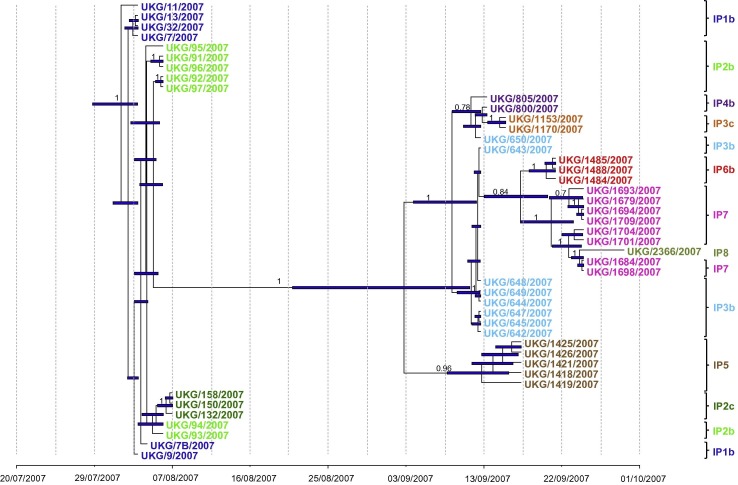
A Bayesian MCMC tree of the 45 FMDV sequences using the HKY model of base substitution (gamma model of site heterogeneity) employing a relaxed molecular clock, Bayesian skyline plot, and sampling 30,000 trees from 30 million generations (Fig. 1 ), estimated a rate of nucleotide substitution of 4.94 × 10−5 (95% highest posterior density – HPD interval: 2.92 × 10−5 – 7.02 × 10−5) per site per day. The root of the maximum clade credibility (MCC) tree or age of their most recent common ancestor (MRCA) was estimated as the 1st August 2007 (95% HPD: 29th July 2007–3rd August 2007). The estimated date for the ancestor of the second phase of the outbreak was the 3rd of September of 2007 (95% HPD: 21st August 2007–11th September 2007) whilst the estimated time for the ancestor of IPs 3 to 8 was the 9th of September 2007 (95% HPD: 4th September 2007–12th September 2007). In the context of the virus phylogeny, evolution rate and estimated rate of the MCRA, the interpretation of these results was not affected by different combinations of molecular clocks, demographic and phylogeographic diffusion models used in the analyses (data not shown).
Fig. 1.
Bayesian maximum-clade-credibility time-scaled phylogenetic tree (BEAST) generated using 45 sequenced FMDV full genomes obtained from infected animals during the 2007 outbreak in UK. Sequences from the same holdings are coloured with the same colour as follows: in dark blue, sequences from IP1b; in light and dark green, sequences from IP2b and c, respectively; in brown, sequences from IP5; in light blue and orange, sequences from IP3b and c, respectively; in purple, sequences from IP4b; in red, sequences from IP6; in pink, sequences from IP7; and in green pistachio, sequences from IP8. The analysis was undertaken using the HKY model of base substitution (gamma model of site heterogeneity), exponential relaxed molecular clock, Bayesian skyline plot, sampling 30,000 trees from 30 million generations. Uncertainty for the date of each node (95% highest posterior density – HPD -–intervals) is displayed in bars. Only node labels with posterior over 0.7 are indicated. Overall, a rate of nucleotide substitution of 4.94 × 10−5 (95% HPD: 2.92 × 10−5–7.02 × 10−5) per site per day was estimated. The ancestor is estimated to be on the 01/08/2007 (95% HPD = 29/07/2007–03/08/2007). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Statistical parsimony analysis
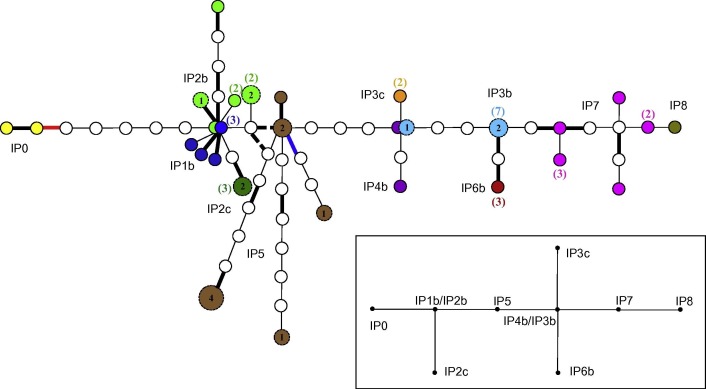
The tree edited from the statistical parsimony analysis of the 45 FMDV sequences recovered in the outbreak and including two more sequences from the putative outbreak sources suggested a chain of transmission events (Fig. 2 ). Two nodes of the tree were represented by identical sequences from two different IPs, whilst the number of nucleotide differences between all of the sequences differed by up to four nucleotides (nts) between herds. The phylogenetic relationship between sequences from different IPs was compatible with the Bayesian MCMC tree.
Fig. 2.
Statistical parsimony tree as implemented by TCS using 45 full FMDV genomes obtained from infected animals during the 2007 outbreak in UK. Sequences from the same holdings are coloured with the same colour as per Fig. 1. The sequences in yellow belonged to isolates used at the Pirbright campus during July 2007. When two or more samples within the same premise provided identical sequences, the number of samples represented is shown in brackets in the same colour than the premise to which the sequences belong to. Samples that contained sequence ambiguities are shown as larger labelled circles with the actual number of sites in which ambiguities were present. Lines in bold correspond to non-synonymous nucleotide substitutions. The two intermittent lines represent the two options corresponding to one ambiguity in one site of a virus sequence from IP5. The lines in red correspond to a nucleotide substitution causing an amino acid change (His to Arg) important for heparan sulphate binding (cell culture adaptation). Finally, the lines in blue correspond to nucleotide substitutions causing an amino acid change (Asp to Gly) associated with, but not critical for, heparan sulphate binding. In the right down square a simplified view of the tree (reference tree) was drawn. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The TCS tree revealed strong evidence for within-herd clustering of the sequences (Fig. 2) and within these herds the Hamming distances (i.e. number of nt differences) between sequences obtained from cattle with acute infections ranged from zero to six. The sequences obtained from sheep with healed lesions (IP5) had one to 13 nt differences between each other. These sequences obtained from probangs had evolved independently from a putative common ancestor with branch lengths of up to seven nt substitutions.
Consensus sequences from different specimens within the same animal were obtained from 11 out of 33 infected animals. In eight out of 11 animals, the sequences from different specimens were found to be identical (five animals), or had one (one animal) or two ambiguity (two animals) differences. Sequences obtained from the remaining three animals differed at one nt (in case of one animal in IP1b and another in IP7) or four sites (three nts and a further site with an ambiguity, in case of the virus collected from the sera of one animal in IP3b).
3.5. Reconstruction of transmission trees with single randomly sampled sequences
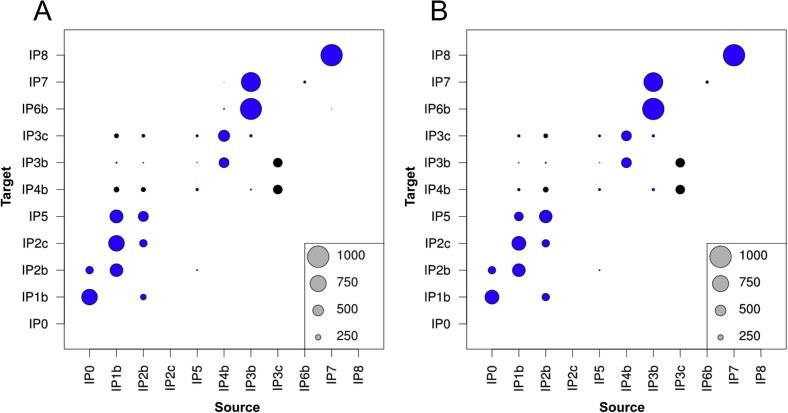
The cumulative adjacency matrices generated out of one thousand TCS transmission trees (i.e. after merging each unlabelled node of the TCS trees to the closest labelled node) and the one thousand MSTs (Fig. 3 ) were similar between each other. However, the transmission route from IP1b to IP2b was more frequently inferred when using the MST approach rather than the TCS method (see consensus tree for each of these methods in Supplementary data 1). Most of the trees placed IP5 as a leaf node (cul-de-sac) between the two phases of the outbreaks, and a number of trees located IP3c as the source of infection of herds IP3b and IP4.
Fig. 3.
Cumulative adjacency matrices of one thousand (A) TCS transmission trees, after merging the empty nodes of the TCS tree to the closest premise; and (B) MSTs. The common edges with the reference TCS tree with all 47 sequences (Fig. 2) were coloured in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
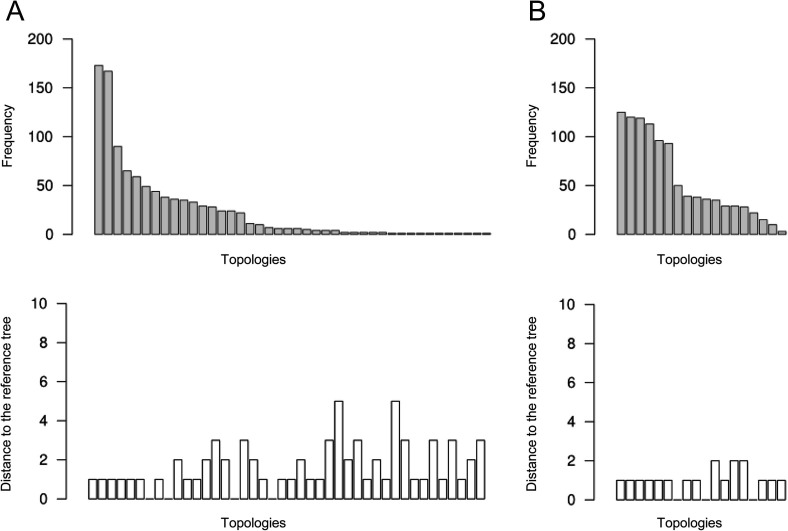
Only 10.8% of the trees were identical to the TCS tree. However, up to 73.7% of the generated TCS transmission trees and 80.0% of the trees generated using the MST approach differed by only one edge with the reference TCS tree (Fig. 4 and Supplementary data 2). The frequency of the different tree topologies generated with the one thousand trees using each of the TCS and MST approaches as well as the distance for each of these tree topologies with the reference TCS tree are shown in Fig. 4. The MST approach produced fewer tree topologies (18) than the TCS approach (42). The TCS trees had up to five edges not included in the reference tree while there were only up to two such edges in the MSTs. However, the mean number of different edges was similar (1.1 for TCS trees, 0.98 for MSTs).
Fig. 4.
Frequencies of tree topologies (up) and distances of topologies (down) to the reference tree (TCS tree using 45 sequences) obtained from the one thousand TCS transmission trees (A) and the MSTs (B). Topologically identical trees (i.e. those with a topological distance of 0) were “binned”. Bars show cardinalities (“sizes”) of bins. They are ordered by descending cardinality.
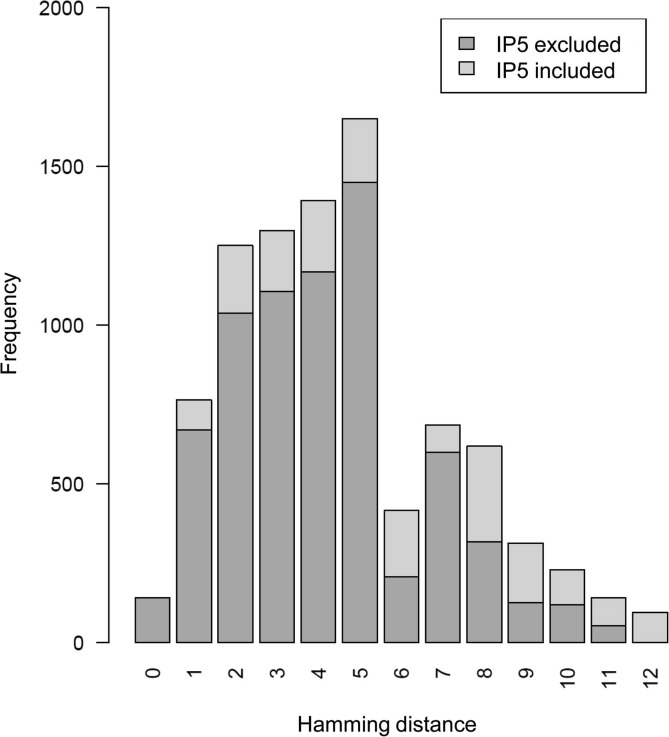
The Hamming distance between sequence pairs representing a source and recipient herd according to the reference tree for each of the one thousand MSTs presented a mean of 4.6 nts. The removal of sequences from IP5, where some sequences derived from chronic animals, reduced the distance to 4.1 nts. A histogram with the distribution of the Hamming distances including and excluding the sequences from IP5 is shown in Fig. 5 .
Fig. 5.
Cumulative herd-to-herd Hamming distance distribution within the one thousand MSTs. Bars were plotted including (light grey colour) and excluding (dark grey colour) the Hamming distances between the sequences of IP5 and the rest of the sequences.
4. Discussion
This study describes for the first time the within-herd genetic diversity of the FMD viruses collected during a field outbreak, and the impact of this sequence variability on trees that are reconstructed to describe the relationship between infected herds. The TCS and Bayesian analysis of all the generated 45 sequences yielded trees that exhibited clustering that corresponded to each of the infected herds, and were consistent with genetic conclusions generated in real-time during the outbreaks (Cottam et al., 2008b). However, further integration of epidemiological data (provided by the Department for Environment, Food and Rural Affairs (Defra) was required to resolve two nodes in the TCS reference tree which comprised identical sequences (Fig. 2: IP1b and IP2b, and IP3b and IP4b, respectively). Epidemiological evidence used to discriminate these herds included field observations where mixing of animals had been observed prior to the clinical cases (for the IP3b/IP4b node: FMD 2007 Epidemiology report, 21 September 2007, Defra) and the date of earliest infection estimated from FMDV lesion ages (for both the IP1b/IP2b and the IP3b/IP4b nodes, already compiled elsewhere: (Cottam et al., 2008b, Ryan et al., 2008)). Accordingly, accounting for sequence and epidemiological data, the most parsimonious order in which the premises and herds were infected was described as: IP0-IP1b-(IP2b(-IP2c))-IP5-IP4-(IP3b-(IP3c)(IP6))-IP7-IP8.
These data support a single source of virus for these cases, as reported previously (Cottam et al., 2008b, Ellis-Iversen et al., 2011, Schley et al., 2008). A particular epidemiological feature of these cases was the role played by IP5 to link the two distinct phases of outbreaks that occurred during August and September. All the sequences generated from convalescent animals on this premise had phylogenetic ancestors between the two phases of the outbreaks, close to the main line of infection. An alternative hypothesis has suggested that the different phases of these outbreaks were seeded by two separate releases from the Pirbright complex (Schley et al., 2008). However, this scenario is only possible if two unlikely events had occurred: firstly un-sampled cases on IP5 would need to have identical sequences to the node shared by IP1b and IP2b (at sites different to the closest root of IP5 sequences); and secondly, and most importantly, the six nt substitutions separating the closest source to IP1b would need to have arisen independently in the proposed second release to IP5, findings that are not supported by data from experimentally infected cattle with a FMD virus from the same lineage (Juleff et al., 2013).
When individual sequences were randomly selected from each of the 10 herds to generate one thousand datasets, only 10.8% of the resulting trees were identical to the reference tree. The majority (73.7% of the generated TCS transmission trees and 80.0% of the trees generated using the MST approach) of the remaining trees differed by only one single edge to the reference tree. These results provide confidence in the use of single samples and corresponding sequences to represent each epidemiological unit as a cost-effective approach in FMD outbreaks. In line with general limitations of sequence analysis, these approaches and alternative analysis of these datasets using a Bayesian inference framework (modified from (Morelli et al., 2012): data not shown) cannot resolve transmission events where genome sequences are too similar (and they may also be confounded by accumulation of nucleotide differences during chronic infection).
In this current study, sequences were obtained from samples collected from animals with both acute and chronic stages of infection, which have been considered for the analysis regardless of the origin of the samples. Only samples from chronically infected animals were available in IP5 and acutely infected animals from the other herds were slaughtered as soon as the disease was suspected. A previous study showed that sequences obtained from probang samples were difficult to interpret since they had a high number of sites containing ambiguities, and that the phylogenetic relationship of these sequences were divergent from the main animal-to-animal transmission line (Juleff et al., 2013). This was not the case for all the probangs processed in this study, as only one of the five sequences obtained from these probangs (with similar CT values by real-time RT-PCR (Reid et al., 2009) between each other) had more than two sites with ambiguities, and although sequences with ambiguities diverged in long-length branches, they shared the same ancestor within the main line of transmission together with other sequences from probangs within IP5.
This study demonstrates that simple computational methods, applied to full genome sequences, can produce trees that accurately reflect transmission events in an outbreak. These methods are applicable in real-time during an outbreak, as they are only limited by the speed of sequencing. The computational analyses (alignment, TCS and MST) are highly efficient and well scalable, and they require only FG sequences as their input. Further support for using sequences to reconstruct transmission trees at high resolution has been generated for other RNA viruses infecting humans, such as influenza virus (Baillie et al., 2011), hepatitis C virus (Gray et al., 2011), HIV (Li et al., 2010) or Middle East respiratory syndrome coronavirus (MERS-CoV) (Cotten et al., 2013) which have focussed on sequencing samples from individuals (all those available infected individuals or a representative subset of them) to determine host-to-host transmission events, or even reveal intra host infection pathways. However, the ability to resolve these events is dependent upon two factors: the sequence length (i.e. number of available substitution sites) adopted for the study, and the evolutionary rate of the pathogen. Previous transmission studies of equine influenza virus have indicated that more than one sequence per individual was considered to be required to reliably reconstruct the transmission dynamics of the outbreak (Hughes et al., 2012). However, this study was based on comparison of only 6% of the full genome of the virus, and in other work, transmission studies based on full genome analysis of avian influenza provided data to confidently define inter-herd transmission events (Bataille et al., 2011). This present study was based on consensus data from Sanger sequencing protocols. Future work in this area will need to accommodate next-generation sequencing data (Logan et al., 2014) using new protocols that may increase the number of sequences that can be handled and processed, as well as providing deep-sequencing data with high resolution of polymorphisms at individual sites (Wright et al., 2011).
5. Conclusion
For the first time, this study describes the within-herd genetic variability of FMDV within an outbreak, and how this genetic variability affects the herd-to-herd transmission tree when sequencing one virus per epidemiological unit. These data indicate that inferred transmission trees generated using single viral sequences from epidemiological units are robust, although attention should be paid when using the sequences of samples from chronically infected animals. This study is useful to design cost-effective sampling approaches in case of FMDV epidemics and will help to develop further models to support control policies in case of exotic incursions of FMDV in FMDV free countries.
Acknowledgements
This work was supported by the Department for Environment, Food and Rural Affairs (Defra projects SE2938 and SE2940). The authors would like to express their gratitude to field teams for providing samples, colleagues within the Vesicular Disease Reference Laboratory at The Pirbright Institute for associated laboratory analysis, and Kate Sharpe and her team at the Animal Health and Veterinary Laboratory Agency for providing further epidemiological data.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2015.03.032.
Appendix A. Supplementary data
Consensus trees ((A) TCS transmission tree, and (B) MST) approach computed as MTs from the cumulative adjacency matrices shown in Fig. 3. The thickness of the edges is proportional to the number of the trees containing a corresponding edge.
Histograms of topological divergences to the reference tree (TCS tree with 45 sequences) for (A) the TCS transmission tree (merging nodes to the closest premises); and (B) for the MSTs. Divergence is quantified by the asymmetric difference of the reference tree edge set and the edge of the topology considered.
References
- Baillie G.J., Galiano M., Agapow P.M., Myers R., Chiam R., Gall A., Palser A.L., Watson S.J., Hedge J., Underwood A., Platt S., McLean E., Pebody R.G., Rambaut A., Green J., Daniels R., Pybus O.G., Kellam P., Zambon M. Evolutionary dynamics of local pandemic H1N1/09 influenza lineages revealed by whole genome analysis. J. Virol. 2011;86:11–18. doi: 10.1128/JVI.05347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille A., van der Meer F., Stegeman A., Koch G. Evolutionary analysis of inter-farm transmission dynamics in a highly pathogenic avian influenza epidemic. PLoS Pathog. 2011;7:e1002094. doi: 10.1371/journal.ppat.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cottam E.M., Haydon D.T., Paton D.J., Gloster J., Wilesmith J.W., Ferris N.P., Hutchings G.H., King D.P. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J. Virol. 2006;80:11274–11282. doi: 10.1128/JVI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Thebaud G., Wadsworth J., Gloster J., Mansley L., Paton D.J., King D.P., Haydon D.T. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc. Biol. Sci. 2008;275:887–895. doi: 10.1098/rspb.2007.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Wadsworth J., Shaw A.E., Rowlands R.J., Goatley L., Maan S., Maan N.S., Mertens P.P.C., Ebert K., Li Y., Ryan E.D., Juleff N., Ferris N.P., Wilesmith J.W., Haydon D.T., King D.P., Paton D.J., Knowles N.J. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog. 2008;4:e1000050. doi: 10.1371/journal.ppat.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A., Al-Tawfiq J.A., Alhakeem R.F., Madani H., AlRabiah F.A., Al Hajjar S., Al-nassir W.N., Albarrak A., Flemban H., Balkhy H.H., Alsubaie S., Palser A.L., Gall A., Bashford-Rogers R., Rambaut A., Zumla A.I., Memish Z.A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis-Iversen J., Smith R.P., Gibbens J.C., Sharpe C.E., Dominguez M., Cook A.J. Risk factors for transmission of foot-and-mouth disease during an outbreak in southern England in 2007. Vet. Rec. 2011;168:128. doi: 10.1136/vr.c6364. [DOI] [PubMed] [Google Scholar]
- Gray R.R., Parker J., Lemey P., Salemi M., Katzourakis A., Pybus O.G. The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol. Biol. 2011;11:131. doi: 10.1186/1471-2148-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Haydon D.T., Bastos A.D.S., Awadalla P. Low linkage disequilibrium indicative of recombination in foot-and-mouth disease virus gene sequence alignments. J. Gen. Virol. 2004;85:1095–1100. doi: 10.1099/vir.0.19588-0. [DOI] [PubMed] [Google Scholar]
- Hughes J., Allen R.C., Baguelin M., Hampson K., Baillie G.J., Elton D., Newton J.R., Kellam P., Wood J.L., Holmes E.C., Murcia P.R. Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks. PLoS Pathog. 2012;8:e1003081. doi: 10.1371/journal.ppat.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juleff N., Valdazo-González B., Wadsworth J., Wright C.F., Charleston B., Paton D.J., King D.P., Knowles N.J. Accumulation of nucleotide substitutions occurring during experimental transmission of foot-and-mouth disease virus. J. Gen. Virol. 2013;94:108–119. doi: 10.1099/vir.0.046029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Welch J.J., Suchard M.A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bar K.J., Wang S., Decker J.M., Chen Y., Sun C., Salazar-Gonzalez J.F., Salazar M.G., Learn G.H., Morgan C.J., Schumacher J.E., Hraber P., Giorgi E.E., Bhattacharya T., Korber B.T., Perelson A.S., Eron J.J., Cohen M.S., Hicks C.B., Haynes B.F., Markowitz M., Keele B.F., Hahn B.H., Shaw G.M. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan G., Freimanis G.L., King D.J., Valdazo-González B., Bachanek-Bankowska K., Sanderson N.D., Knowles N.J., King D.P., Cottam E.M. A universal protocol to generate consensus level genome sequences for foot-and-mouth disease virus and other positive-sense polyadenylated RNA viruses using the Illumina MiSeq. BMC Genomics. 2014;15:828. doi: 10.1186/1471-2164-15-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli M.J., Thebaud G., Chadoeuf J., King D.P., Haydon D.T., Soubeyrand S. A bayesian inference framework to reconstruct transmission trees using epidemiological and genetic data. PLoS Comput. Biol. 2012;8:e1002768. doi: 10.1371/journal.pcbi.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton R.J., Wright C.F., Morelli M.J., Juleff N., Thebaud G., Knowles N.J., Valdazo-González B., Paton D.J., King D.P., Haydon D.T. Observing micro-evolutionary processes of viral populations at multiple scales. Philos. Trans. R. Soc. London B-Biol. Sci.2013;368:20120203. doi: 10.1098/rstb.2012.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. 2010. FigTree 1.3.1, available at http://tree.bio.ed.ac.uk/software/figtree.
- Reid S.M., Ebert K., Bachanek-Bankowska K., Batten C., Sanders A., Wright C., Shaw A.E., Ryan E.D., Hutchings G.H., Ferris N.P., Paton D.J., King D.P. Performance of real-time reverse transcription polymerase chain reaction for the detection of foot-and-mouth disease virus during field outbreaks in the United Kingdom in 2007. J. Vet. Diagn. Invest. 2009;21:321–330. doi: 10.1177/104063870902100303. [DOI] [PubMed] [Google Scholar]
- Ryan E., Gloster J., Reid S.M., Li Y., Ferris N.P., Waters R., Juleff N., Charleston B., Bankowski B., Gubbins S., Wilesmith J.W., King D.P., Paton D.J. Clinical and laboratory investigations of the outbreaks of foot-and-mouth disease in southern England in 2007. Vet. Rec. 2008;163:139–147. doi: 10.1136/vr.163.5.139. [DOI] [PubMed] [Google Scholar]
- Schley D., Knowles N.J., Gubbins S., Gloster J., Burgin L., Paton D.J. Probable route of infection for the second UK 2007 foot-and-mouth disease cluster. Vet. Rec. 2008;163:270–271. doi: 10.1136/vr.163.9.270. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Thebaud G., Chadoeuf J., Morelli M.J., McCauley J.W., Haydon D.T. The relationship between mutation frequency and replication strategy in positive-sense single-stranded RNA viruses. Proc. Biol. Sci. 2010;277:809–817. doi: 10.1098/rspb.2009.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdazo-González B., Polihronova L., Alexandrov T., Normann P., Knowles N.J., Hammond J.M., Georgiev G.K., Ozyoruk F., Sumption K.J., Belsham G.J., King D.P. Reconstruction of the transmission history of RNA virus outbreaks using full genome sequences: foot-and-mouth disease virus in Bulgaria in 2011. PLoS ONE. 2012;7:e49650. doi: 10.1371/journal.pone.0049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C.F., Morelli M.J., Thebaud G., Knowles N.J., Herzyk P., Paton D.J., Haydon D.T., King D.P. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J. Virol. 2011;85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consensus trees ((A) TCS transmission tree, and (B) MST) approach computed as MTs from the cumulative adjacency matrices shown in Fig. 3. The thickness of the edges is proportional to the number of the trees containing a corresponding edge.
Histograms of topological divergences to the reference tree (TCS tree with 45 sequences) for (A) the TCS transmission tree (merging nodes to the closest premises); and (B) for the MSTs. Divergence is quantified by the asymmetric difference of the reference tree edge set and the edge of the topology considered.