Abstract
Middle East respiratory syndrome (MERS) is an emerging zoonotic viral respiratory disease that was first identified in Saudi Arabia in 2012. In 2015, the largest MERS outbreak outside of the Middle East region occurred in the Republic of Korea. The rapid nosocomial transmission of MERS-coronavirus (MERS-CoV) in Korean health care settings highlighted the importance and urgent need for a rapid and reliable on-site diagnostic assay to implement effective control and preventive measures. Here, the evaluation and validation of two newly developed reverse transcription–insulated isothermal PCR (RT-iiPCR) methods targeting the ORF1a and upE genes of MERS-CoV are described. Compared with World Health Organization-recommended singleplex real-time quantitative RT-PCR (RT-qPCR) assays, both RT-iiPCR assays had comparable analytical sensitivity for the detection of MERS-CoV RNA in tissue culture fluid and in sputum samples spiked with infectious virus. Furthermore, clinical evaluation was performed with sputum samples collected from subjects with acute and chronic respiratory illnesses, including patients infected with MERS-CoV. The overall agreement values between the two RT-iiPCR assays and the reference RT-qPCR assays were 98.06% (95% CI, 94.43%–100%; κ = 0.96) and 99.03% (95% CI, 95.88%–100%; κ = 0.99) for ORF1a and upE assays, respectively. The ORF1a and upE MERS-CoV RT-iiPCR assays coupled with a field-deployable system provide a platform for a highly sensitive and specific on-site tool for diagnosis of MERS-CoV infections.
The Middle East respiratory syndrome–coronavirus (MERS-CoV), first identified in Saudi Arabia in September 2012, is an emerging zoonotic pathogen that causes severe acute respiratory illness in humans.1 To date, more than 1900 laboratory-confirmed MERS-CoV infections and 684 human deaths in 27 countries have been reported, with a mortality rate of approximately 36% [World Health Organization (WHO), http://www.who.int/emergencies/mers-cov/en, accessed March 24, 2017]. MERS-CoV is a zoonotic virus that has repeatedly moved into the human population via contact with the infected dromedary camels in the Arabian Peninsula (WHO, MERS-CoV Global Summary and risk assessment, http://www.who.int/emergencies/mers-cov/mers-summary-2016.pdf, accessed January 12, 2016). Recent phylogenetic analysis of viral isolates from humans, camels, and bats revealed that bats may have been the original primary reservoir of the virus, and they may have initially transmitted the virus to camels.2 Thus, transmission of MERS-CoV to humans is suspected to occur by direct or indirect contact with infected camels or camel-related products (eg, raw camel milk, camel urine).3, 4 Human-to-human transmission of MERS-CoV requires close contact and can occur among relatives in households and among patients and health care workers in health care settings (nosocomial infection).5
Since its emergence, most of the MERS-CoV infections have occurred in the Arabian Peninsula (Kuwait, Bahrain, Qatar, the United Arab Emirates, Oman, Yemen, and Saudi Arabia), but additional cases have been reported from countries in North Africa, Europe, North America, and Asia due to movement of infected individuals. The outbreak in the Republic of Korea in May 2015 was the largest MERS-CoV outbreak ever recorded outside of Saudi Arabia and resulted in 185 laboratory-confirmed human infections in Korea and one in China, with 36 deaths.6 The index case was traced back to an individual with a travel history to the Middle East. The MERS outbreaks have been attributed to failures of preventive and control measures in health care settings.5 Therefore, early diagnosis, prompt isolation of suspected cases, and timely tracing of case contacts are key strategies to prevent further transmission.
Following the emergence of MERS-CoV, several molecular detection methods and serological assays were developed and deployed internationally through an international collaborative laboratory response.7, 8, 9, 10 Currently, real-time quantitative RT-PCR (RT-qPCR) is the primary method for laboratory diagnosis of MERS-CoV infection, and it requires at least two different genomic targets for a positive diagnosis according to the case definition announced by WHO as of July 26, 2017 (WHO, http://www.who.int/csr/disease/coronavirus_infections/case_definition/en/index.html). The two RT-qPCR assays developed by Corman et al7, 8 shortly after the first report of the disease were designated as recommended MERS-CoV molecular diagnostics by WHO. Both assays proved to be highly sensitive and were successfully used for the diagnosis of the majority of the MERS-CoV cases. These assays target genomic regions upstream of the envelope gene (upE) and the viral open reading frame 1a (ORF1a). The RealStar MERS-CoV RT-qPCR Kit (Alotona Diagnostics, Hamburg, Germany) has been developed using these WHO-recommended assays.11 However, these assays are costly, demand expensive instrumentation, and require a dedicated laboratory environment with technically skilled personnel. Consequently, simple and rapid methods are required to meet the needs of point-of-need MERS-CoV detection. For this purpose, many isothermal RNA amplification methods were developed for exponential amplification of RNA at low and constant temperatures such as rapid one-step RNA amplification/detection assay12 and reverse transcriptional loop-mediated isothermal amplification (RT-LAMP).13, 14 The RT-LAMP assay can be performed in a simple heating block.
Recently, fluorescent probe hydrolysis–based insulated isothermal PCR (iiPCR) for amplification and detection of nucleic acid has been described.15 The iiPCR is highly sensitive and specific for the detection of both DNA and RNA and can be performed with a single heating source; thus, it does not require an expensive thermocycler.16, 17 The PCR mix in a capillary tube (R-tube; GeneReach USA, Lexington, MA) is heated at the bottom. Rayleigh-Bénard convection drives fluid cycling through temperature gradients and the three PCR steps, namely denaturation, annealing, and extension, can be completed sequentially at different zones within the capillary tube. Subsequent integration of hydrolysis probe technology and an optical detection module into the device allow automatic detection and interpretation of iiPCR results.17 Performance of iiPCR assays on a commercially available, field-deployable, and user-friendly iiPCR system, the POCKIT Nucleic Acid Analyzer (GeneReach USA), has been demonstrated to be comparable to that of real-time PCR, nested PCR, and/or virus isolation for the detection of various pathogens in different hosts, including dengue virus and malaria in human samples.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 31 Taking advantage of this system, in this study, two singleplex RT-iiPCR assays for the detection of MERS-CoV upE and ORF1a genes separately were developed and the ability of the assays for viral nucleic acid detection was determined. The analytical sensitivity, analytical specificity, and reproducibility of the two MERS-CoV-specific RT-iiPCR assays were assessed using viral tissue culture fluid (TCF) and human sputum samples spiked with known amounts of MERS-CoV. The clinical performance of these two assays was further evaluated and validated using RNA extracted from sputum samples of MERS-CoV–infected patients obtained from the recent Korean outbreak and compared with the corresponding reference singleplex real-time RT-qPCR assays recommended by WHO.
Materials and Methods
Viruses and Cells
A patient-derived MERS-CoV isolate (MERS-CoV/KOR/KNIH/002_05_2015; https://www.ncbi.nlm.nih.gov/nuccore; accession number KR029139.1) was kindly provided by Dr. Sung Soon Kim [Korea Centers for Disease Control and Prevention (KCDC), Osong, Republic of Korea]. A working virus stock was prepared by passaging MERS-CoV in a human hepatoma cell line, Huh7 cells (Japanese Collection of Research Bioresources Cell Bank, Osaka, Japan). The infectious viral titer of the TCF supernatant, expressed as plaque forming units per mL (PFU/mL) was determined by plaque assay using Vero cells (ATCC CCL-81; ATCC, Manassas, VA) according to a standard laboratory protocol. All procedures using live MERS-CoV were performed in the biosafety level-3 facility at Center for Virus Research and Testing, Korea Research Institute of Chemical Technology, Daejeon, Republic of Korea.
Human coronaviruses, hCoV-229E (ATCC VR-740) and hCoV-OC43 (ATCC VR-1558) were purchased from ATCC and amplified in human fetal lung fibroblast MRC-5 cells (ATCC CCL-171). Feline infectious peritonitis coronavirus (FIPV) (ATCC VR-990) and its host cell line Crandall feline kidney were obtained from ATCC and Korean Cell Line Bank (Seoul, Republic of Korea), respectively. Other human viral pathogens included in this study were influenza virus type A (H1N1, A/Puerto Rico/8/34; ATCC VR-1469), and influenza virus type B (B/Panama/45/1990; KCDC). Influenza viruses (types A and B) were propagated by infection of Madin Darby canine kidney cells.
Ethics Statement
Clinical data and specimens obtained from the patients infected with MERS-CoV were used in this study following ethical approval granted by the Institutional Review Board of Chungnam National University Hospital, Daejeon, Republic of Korea. All surviving patients provided written informed consent before participating in the study. In fatal cases, an exemption to the patients' consent was obtained from the institutional review board for the retrospective analysis of clinical samples. All experiments were performed according to the approved guidelines.
Clinical Specimens
A total of 55 sequential sputum samples collected from 12 patients infected with MERS-CoV were obtained from the Chungnam National University Hospital. These patients were diagnosed positive for MERS-CoV infection by real-time RT-qPCR assays targeting the ORF1a and upE sequences at the KCDC laboratory between May and June of 2015. Sputum samples collected from patients suffering from other acute and chronic respiratory illnesses (n = 48) were included in the study as negative samples. Sputum samples from nine healthy individuals were randomly selected and spiked with serially diluted MERS-CoV TCF working stocks.
Nucleic Acid Extraction
For analytical sensitivity analysis, TCF containing MERS-CoV (3.7 × 106 PFU/mL) was subjected to 10-fold serial dilutions (100 to 107) in Dulbecco's modified Eagle's medium (HyClone, Logan, UT) containing 10% fetal bovine serum (HyClone). Viral RNA was extracted from serial dilutions of MERS-CoV TCF and MERS-CoV–spiked sputum samples (100 μL/sample) by using the taco DNA/RNA Extraction Kit (GeneReach USA) on a taco Nucleic Acid Automatic Extraction System (GeneReach USA), according to the manufacturer’s instructions. Viral RNA was eluted in 100 μL of elution buffer. Total RNA from sputum samples collected from patients infected with MERS-CoV and from patients suffering from other acute and chronic respiratory illnesses (controls) was extracted using TRIzol LS reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instruction in a biosafety level-3 facility. The final volume of each extracted sample was 50 μL. All nucleic acid samples were placed at −80°C until further use.
In Vitro Transcribed RNA Preparation
The analytical sensitivity of the singleplex MERS-CoV RT-iiPCR assays was determined by using in vitro transcribed (IVT) RNA. Briefly, the sequences of the ORF1a and upE regions of the MERS-CoV (nucleotides 27361 to 27596 and nucleotides 11137 to 11339, respectively; https://www.ncbi.nlm.nih.gov/nuccore; accession number NC_019843) were synthesized, cloned into the pGEM-3Z vector (Promega, Madison, WI) downstream of the T7 promoter sequence, and subsequently used as the template in in vitro transcription using the mMESSAGE mMACHINE T7 Transcription Kit (Ambion/Life Technologies, Grand Island, NY). Residual DNA was removed using the Ambion Turbo DNA-free kit (Applied Biosystems, Grand Island, NY). Concentration of RNA was measured by a NanoDrop1000 Spectrophotometer (NanoDrop Technologies, Houston, TX). Single-use IVT RNA aliquots were stored at −80°C. The analytical sensitivity of the RT-iiPCR was determined using a dilution series (100 to 108 molecules/reaction) of the IVT RNA. The concentration of the IVT RNA/μL was calculated as described previously.32
Reference MERS-CoV ORF1a and upE Real-Time RT-qPCR Tests
The singleplex ORF1a and upE real-time RT-qPCR assays were performed according to the WHO-recommended protocol33 using the SuperScript III One-Step RT-PCR system with Platinum Taq polymerase (Invitrogen, Carlsbad, CA). Primers and probes targeting ORF1a and upE genes were synthesized according to the previously published sequences by GenoTech Corp. (Daejeon, Republic of Korea).7, 8 Thermocycling program was set up as follows: an reverse transcription step at 50°C for 30 minutes, followed by 95°C for 2 minutes and 40 cycles of 15 seconds at 95°C, and 1 minute at 55°C, as described previously.34 Duplicate samples with CT values >38 were considered negative. Each run included negative controls spiked with water and positive controls with the IVT RNA–containing target sequences. All clinical specimens were tested for the human RNase P gene by RT-qPCR to monitor nucleic acid extraction efficiency and the presence of PCR inhibitors as described elsewhere.34
Establishment of RT-iiPCR
The MERS-CoV–specific RT-iiPCR was designed on the basis of the probe hydrolysis-based POCKIT method as described previously.17 The primers and probe targeted the ORF1a or upE region of MERS-CoV (https://www.ncbi.nlm.nih.gov/nuccore; accession number NC_019843). The conserved regions were identified by aligning 253 sequences available in the GenBank database. The RT-iiPCR reaction conditions, such as concentrations of primers and probe, Taq DNA polymerase, and reverse transcriptase, were tested systematically to obtain the highest sensitivity and specificity. Following optimization of the RT-iiPCR assay conditions, the reagents including primers and probe were lyophilized (proprietary) and used in this study. Briefly, the lyophilized premix was reconstituted in 50 μL of Premix Buffer B (GeneReach USA), and then 5 μL of the test nucleic acid extract was added. A 50-μL volume of the premix/sample mixture was transferred into a labeled R-tube, which was subsequently sealed with a cap, spun briefly in a microcentrifuge (Cubee; GeneReach USA), and placed into the POCKIT Nucleic Acid Analyzer. The default program, including a reverse transcription step at 50°C for 10 minutes and an iiPCR step at 95°C for about 30 minutes, was completed in less than 1 hour. Signal-to-noise ratios, ie, light signals collected after iiPCR/fluorescent signals collected before iiPCR,17 were converted automatically to “+”, “−”, or “?” according to the default signal-to-noise thresholds by the built-in algorithm. The results were shown on the display screen at the end of the program. A “?” indicated that the results were ambiguous and the sample should be tested again.
Statistical Analysis
Limit of detection with 95% confidence was determined by statistical probit analysis (a nonlinear regression model) using SPSS software version 17.0 (SPSS Inc., Chicago, IL). The clinical performance of the assays was calculated based on the analysis of the 55 sputum samples from patients infected with MERS-CoV and 48 donors not suspected of MERS-CoV infection. The singleplex ORF1a or upE RT-qPCR assay recommended by WHO for MERS-CoV diagnosis was used as a reference test.34 The degree of agreement between the two assays was assessed by calculating Cohen's Kappa (k) values. Sensitivity was calculated as:
and specificity was calculated as:
Results
Evaluation of Analytical Sensitivity and Specificity of MERS-CoV ORF1a and upE RT-iiPCR Assays
Analytical Sensitivity
The analytical sensitivities of the singleplex MERS-CoV ORF1a and upE RT-iiPCR assays were evaluated separately by using RNA extracted from serial 10-fold dilutions of TCF containing MERS-CoV. The detection limit for both ORF1a and upE RT-iiPCR methods were determined to be approximately 3.7 × 10−1 PFU/mL of MERS-CoV in TCF, which is equivalent to a technical limit of detection with 95% confidence of <10 copies of synthetic RNA (Table 1 ). To obtain an estimate of detection limit in a more clinically relevant setting, viral RNA extracted from sputum samples of healthy individuals spiked with 10-fold serial dilutions of MERS-CoV were used. The limit of detection values for both assays for MERS-CoV in sputum were determined to be approximately 3.7 × 10−1 PFU/mL (Table 2 ), suggesting that the effect of PCR inhibition is minimal.
Table 1.
Analytical Sensitivity Analysis of MERS-CoV ORF1a and upE RT-iiPCR Using TCF Containing MERS-CoV
| Assay | PFU/mL | MERS-CoV RT-iiPCR |
MERS-CoV RT-qPCR |
|---|---|---|---|
| No. positive/total | No. positive/total | ||
| MERS-CoV ORF1a | 3.7 × 101 | 3/3 | 3/3 |
| 3.7 × 100 | 3/3 | 3/3 | |
| 3.7 × 10−1 | 3/3 | 3/3 | |
| 3.7 × 10−2 | 1/3 | 1/3 | |
| 3.7 × 10−3 | 0/3 | 0/3 | |
| 3.7 × 10−4 | 0/3 | 0/3 | |
| MERS-CoV upE | 3.7 × 101 | 3/3 | 3/3 |
| 3.7 × 100 | 3/3 | 3/3 | |
| 3.7 × 10−1 | 3/3 | 3/3 | |
| 3.7 × 10−2 | 1/3 | 1/3 | |
| 3.7 × 10−3 | 0/3 | 0/3 | |
| 3.7 × 10−4 | 0/3 | 0/3 |
MERS-CoV, Middle East respiratory syndrome–coronavirus; PFU, plaque forming units; RT-iiPCR, reverse transcription-insulated isothermal PCR; TCF, tissue culture fluid.
Table 2.
Analytical Sensitivity Analysis of MERS-CoV ORF1a and upE RT-iiPCR Assays Using Virus-Spiked Sputum Samples
| Sample no. | Virus amount (PFU/mL) |
ORF1a |
upE |
||
|---|---|---|---|---|---|
| RT-iiPCR | RT-qPCR (CT) | RT-iiPCR | RT-qPCR (CT) | ||
| SP1 | 3.7 × 101 | + | + (28.60) | + | + (28.28) |
| 3.7 × 100 | + | + (32.31) | + | + (31.92) | |
| 3.7 × 10−1 | + | + (35.58) | + | + (35.31) | |
| No virus | − | − | − | − | |
| SP2 | 3.7 × 101 | + | + (32.34) | + | + (31.35) |
| 3.7 × 100 | + | + (36.60) | + | + (35.30) | |
| 3.7 × 10−1 | + | − (38.93) | + | + (37.84) | |
| No virus | − | − | − | − | |
| SP7 | 3.7 × 101 | + | + (31.18) | + | + (30.85) |
| 3.7 × 100 | + | + (34.87) | + | + (34.75) | |
| 3.7 × 10−1 | + | + (36.92) | + | + (36.49) | |
| No virus | − | − | − | − | |
| SP8 | 3.7 × 101 | + | + (29.01) | + | + (28.65) |
| 3.7 × 100 | + | + (32.48) | + | + (32.14) | |
| 3.7 × 10−1 | + | + (36.34) | + | + (35.60) | |
| No virus | − | − | − | − | |
| SP9 | 3.7 × 101 | + | + (29.54) | + | + (29.12) |
| 3.7 × 100 | + | + (33.44) | + | + (32.78) | |
| 3.7 × 10−1 | + | + (34.61) | + | + (34.18) | |
| No virus | − | − | − | − | |
| SP10 | 3.7 × 101 | + | + (31.48) | + | + (30.94) |
| 3.7 × 100 | + | + (32.80) | + | + (32.33) | |
| 3.7 × 10−1 | + | + (36.35) | + | + (35.77) | |
| No virus | − | − | − | − | |
| SP11 | 3.7 × 101 | + | + (28.35) | + | + (28.39) |
| 3.7 × 100 | + | + (31.04) | + | + (30.80) | |
| 3.7 × 10−1 | + | + (32.87) | + | + (32.30) | |
| No virus | − | − | − | − | |
| SP13 | 3.7 × 101 | + | + (28.04) | + | + (28.23) |
| 3.7 × 100 | + | + (31.50) | + | + (31.96) | |
| 3.7 × 10−1 | + | + (32.62) | + | + (33.33) | |
| No virus | − | − | − | − | |
| SP14 | 3.7 × 101 | + | + (28.19) | + | + (28.50) |
| 3.7 × 100 | + | + (30.93) | + | + (30.86) | |
| 3.7 × 10−1 | + | + (32.30) | + | + (32.94) | |
| No virus | − | − | − | − | |
+, positive; −, negative; MERS-CoV, Middle East respiratory syndrome–coronavirus; PFU, plaque forming units; RT-iiPCR, reverse transcription-insulated isothermal PCR.
Analytical Specificity
The specificities of the singleplex ORF1a and upE MERS-CoV RT-iiPCR assays were evaluated with viral nucleic acids extracted from infectious TCF containing human coronavirus 229E (hCoV-229E), hCoV-OC43, FIPV, and influenza virus type A and B strains. All reactions yielded negative results, indicating high analytical specificity with no false-positive test results with either assay.
Reproducibility
Reproducibility of the singleplex ORF1a and upE MERS-CoV RT-iiPCR assays was assessed by testing independently (three experimental runs) three replicates of the nucleic acid extract of 10−5 dilution (3.7 × 10−1 PFU/mL) of infectious TCF. All nine reactions were detected positive, suggesting excellent intra- and interassay reproducibility of the established assays (Table 3 ).
Table 3.
Intra- and Interassay Variability of MERS-CoV ORF1a and upE RT-iiPCR
| Assay run | RT-iiPCR result∗ (positive/total) | S/N (average ± SD) |
|---|---|---|
| MERS-CoV ORF1a RT-iiPCR | ||
| 1 | 3/3 | 4.95 ± 0.03 |
| 2 | 3/3 | 4.92 ± 0.02 |
| 3 | 3/3 | 4.85 ± 0.11 |
| MERS-CoV upE RT-iiPCR | ||
| 1 | 3/3 | 4.94 ± 0.05 |
| 2 | 3/3 | 4.85 ± 0.05 |
| 3 | 3/3 | 4.93 ± 0.05 |
MERS-CoV, Middle East respiratory syndrome–coronavirus; PFU, plaque forming units; RT-iiPCR, reverse transcription-insulated isothermal PCR; S/N, signal-to-noise ratio; TCF, tissue culture fluid.
Nucleic acid extract of 10−5 dilution (3.7 × 10−1 PFU/mL) of MERS-CoV infectious TCF.
Evaluation of the MERS-CoV ORF1a and upE RT-iiPCR Assays Using Clinical Samples
To determine clinical sensitivity and specificity of the singleplex MERS-CoV ORF1a and upE RT-iiPCR assays, 55 sputum samples consisting of sequential sample sets taken from 12 patients during the course of acute MERS illness were analyzed. These samples were confirmed to be positive for MERS-CoV infection by the real-time RT-qPCR assay routinely used at the KCDC laboratory during the outbreak in Korea in 2015. To estimate the diagnostic performance of the singleplex RT-iiPCR methods, the assay results were compared with the reference singleplex RT-qPCR assays that were run side-by-side (ie, the reference RT-qPCR assays were repeated with the newly extracted RNA from sputum samples).33 The results from the initial laboratory testing together with those from the RT-iiPCR and RT-qPCR assays using the clinical specimens are shown in Table 4 . The ORF1a RT-iiPCR assay was able to detect MERS-CoV RNA in 54 of 55 positively confirmed samples, whereas the reference ORF1a RT-qPCR assay positively detected 52 of 55. Meanwhile, the upE RT-iiPCR assay positively detected all 55 samples, whereas the reference upE RT-qPCR assay confirmed 54 of 55 positive samples (Table 4). Specifically, the previously positive specimen from patient ID P085 (collected 06-14-2015) was tested as negative with ORF1a RT-iiPCR and reference singleplex ORF1a and upE RT-qPCR assays. The upE RT-iiPCR was the only assay that identified the P085 specimen as positive. Two positive samples from patients ID P130 (collected 06-14-2015) and ID P148 (collected 06-21-2015) were tested as false negatives by the reference ORF1a RT-qPCR, resulting in detection of 52 of 55 positive samples (Table 4). Thus, by comparing the RT-iiPCR results to those from the initial laboratory testing at the time of the outbreak, the sensitivities of the singleplex MERS-CoV ORF1a and upE RT-iiPCR assays were 99.03% (54 of 55; 95% CI, 95.88%–100%) and 100% (55 of 55; 95% CI, 97.43%–100%), respectively, whereas those of the reference RT-qPCR were 97.09% (52 of 55; 95% CI, 87.43%–100%) for ORF1a and 99.03% (54 of 55; 95% CI, 95.88%–100%) for upE, respectively (Table 4).
Table 4.
Detection of MERS-CoV by RT-iiPCR and RT-qPCR in Sputum Samples Collected from Patients Infected with MERS-CoV
| Patient's ID | Age | Sex | Date of onset | Outcome | No. of samples | Collection date∗ | Initial laboratory testing (RT-PCR)† | MERS-CoV RT-iiPCR |
MERS-CoV RT-qPCR |
Estimated viral load‡ (log10 [copies]) |
Internal control (CT)§ |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF1a | upE | ORF1a | upE | ORF1a | upE | RNase P | ||||||||
| P016 | 40 | M | 05-19-2015 | Recovered | 8 | 06-09-2015 | + | + | + | + | + | 3.906 | 4.195 | 35.8 |
| 06-10-2015 | + | + | + | + | + | 2.681 | 3.111 | 32.8 | ||||||
| 06-10-2015′ | + | + | + | + | + | 3.507 | 3.289 | 32.2 | ||||||
| 06-11-2015 | + | + | + | + | + | 6.024 | 6.482 | 30.9 | ||||||
| 06-11-2015′ | + | + | + | + | + | 5.779 | 6.224 | 32.1 | ||||||
| 06-13-2015 | + | + | + | + | + | 5.266 | 5.837 | 31.9 | ||||||
| 06-14-2015 | + | + | + | + | + | 2.363 | 2.846 | 32.3 | ||||||
| 06-14-2015′ | + | + | + | + | + | 2.478 | 3.128 | 32.7 | ||||||
| P030 | 60 | M | 05-30-2015 | Recovered | 6 | 06-08-2015 | + | + | + | + | + | 6.527 | 7.049 | 29.7 |
| 06-09-2015 | + | + | + | + | + | 4.936 | 5.132 | 28.4 | ||||||
| 06-09-2015′ | + | + | + | + | + | 4.917 | 5.243 | 29.7 | ||||||
| 06-11-2015 | + | + | + | + | + | 6.017 | 6.406 | 27.4 | ||||||
| 06-11-2015′ | + | + | + | + | + | 5.986 | 6.360 | 28.4 | ||||||
| 06-11-2015″ | + | + | + | + | + | 6.238 | 6.521 | 24.9 | ||||||
| P031 | 69 | M | 05-30-2015 | Fatal | 5 | 06-08-2015 | + | + | + | + | + | 6.821 | 6.864 | 27.8 |
| 06-11-2015 | + | + | + | + | + | 7.878 | 8.242 | 28.2 | ||||||
| 06-12-2015 | + | + | + | + | + | 5.996 | 6.416 | 34.1 | ||||||
| 06-15-2015 | + | + | + | + | + | 6.478 | 6.125 | 25.3 | ||||||
| 06-17-2015 | + | + | + | + | + | 5.322 | 5.003 | 24.6 | ||||||
| P054 | 62 | F | 05-31-2015 | Recovered | 4 | 06-12-2015 | + | + | + | + | + | 6.306 | 6.447 | 31.4 |
| 06-14-2015 | + | + | + | + | + | 6.701 | 7.025 | 28.4 | ||||||
| 06-15-2015 | + | + | + | + | + | 5.672 | 5.700 | 27.1 | ||||||
| 06-16-2015 | + | + | + | + | + | 5.983 | 5.812 | 31.9 | ||||||
| P082 | 82 | F | 06-04-2015 | Fatal | 3 | 06-11-2015 | + | + | + | + | + | 2.716 | 4.159 | 31.0 |
| 06-13-2015 | + | + | + | + | + | 6.146 | 6.719 | 26.3 | ||||||
| 06-14-2015 | + | + | + | + | + | 3.756 | 4.600 | 27.9 | ||||||
| P085 | 65 | F | 06-06-2015 | Recovered | 5 | 06-10-2015 | + | + | + | + | + | 4.347 | 4.100 | 26.6 |
| 06-13-2015 | + | + | + | + | + | 5.049 | 5.138 | 35.0 | ||||||
| 06-14-2015 | + | ND | + | ND | ND | ND | ND | 27.3 | ||||||
| 06-15-2015 | + | + | + | + | + | 4.376 | 4.590 | 29.5 | ||||||
| 06-16-2015 | + | + | + | + | + | 3.842 | 4.241 | 31.8 | ||||||
| P110 | 57 | F | 06-02-2015 | Recovered | 2 | 06-13-2015 | + | + | + | + | + | 5.538 | 5.491 | 29.7 |
| 06-13-2015′ | + | + | + | + | + | 5.040 | 5.411 | 27.5 | ||||||
| P122 | 55 | F | 06-06-2015 | Recovered | 6 | 06-10-2015 | + | + | + | + | + | 6.168 | 6.731 | 29.2 |
| 06-12-2015 | + | + | + | + | + | 5.075 | 5.221 | 34.5 | ||||||
| 06-13-2015 | + | + | + | + | + | 5.218 | 5.750 | 31.7 | ||||||
| 06-14-2015 | + | + | + | + | + | 4.866 | 5.177 | 29.5 | ||||||
| 06-15-2015 | + | + | + | + | + | 3.100 | 3.401 | 29.8 | ||||||
| 06-16-2015 | + | + | + | + | + | 3.702 | 3.789 | 33.2 | ||||||
| P130 | 65 | F | 06-12-2015 | Recovered | 2 | 06-14-2015 | + | + | + | ND | + | ND | 2.672 | 30.2 |
| 06-16-2015 | + | + | + | + | + | 2.160 | 2.604 | 30.2 | ||||||
| P148 | 38 | F | 06-06-2015 | Recovered | 4 | 06-15-2015 | + | + | + | + | + | 3.564 | 3.678 | 35.8 |
| 06-17-2015 | + | + | + | + | + | 5.781 | 6.081 | 33.1 | ||||||
| 06-18-2015 | + | + | + | + | + | 5.516 | 5.843 | 31.3 | ||||||
| 06-21-2015 | + | + | + | ND | + | ND | 2.743 | 38.5 | ||||||
| P172 | 60 | F | 06-15-2015 | Recovered | 10 | 06-22-2015 | + | + | + | + | + | 5.359 | 5.812 | 29.8 |
| 06-25-2015 | + | + | + | + | + | 3.936 | 4.074 | 28.0 | ||||||
| 06-25-2015 | + | + | + | + | + | 4.614 | 4.722 | 28.3 | ||||||
| 06-27-2015 | + | + | + | + | + | 3.133 | 3.007 | 28.0 | ||||||
| 06-28-2015 | + | + | + | + | + | 4.949 | 5.153 | 26.0 | ||||||
| 06-28-2015′ | + | + | + | + | + | 4.559 | 4.922 | 26.4 | ||||||
| 06-29-2015 | + | + | + | + | + | 3.961 | 4.157 | 28.2 | ||||||
| 06-29-2015′ | + | + | + | + | + | 2.923 | 3.328 | 26.9 | ||||||
| 06-29-2015″ | + | + | + | + | + | 2.710 | 3.308 | 25.8 | ||||||
| 06-30-2015 | + | + | + | + | + | 5.497 | 5.791 | 27.8 | ||||||
F, female; M, male; +, positive; MERS-CoV, Middle East respiratory syndrome–coronavirus; ND, not detected; RT-iiPCR, reverse transcription-insulated isothermal PCR.
Dates indicated with (′) and (′′) indicate sample replicate.
Patients were confirmed positive by real-time RT-qPCR at the Korean Center for Disease Control laboratory during the 2015 outbreak.
Viral loads (Log10 copies) were quantified by using in vitro transcribed RNA derived from the amplicon region of each assay.
All specimens were tested for the human RNase P gene to monitor nucleic acid extraction efficiency and the presence of PCR inhibitors as described in Lu et al.34
All samples from patients suffering from other respiratory illnesses were tested as negative with the corresponding assays indicating high specificity (100%). Lastly, the overall agreement values between the RT-iiPCR and reference RT-qPCR were 98.06% (95% CI, 94.43%–100%; κ = 0.96) for ORF1a signature, with two positive samples giving discrepant results; and 99.03% (95% CI, 95.88%–100%; κ = 0.99) for upE signature, with one discrepant result from a positive sample between the RT-iiPCR and reference RT-qPCR (Table 5 ).
Table 5.
Diagnostic Performance Comparison between MERS-CoV RT-iiPCR and RT-qPCR Assays
| ORF1a | MERS-CoV RT-qPCR |
||
|---|---|---|---|
| Positive | Negative | Total | |
| MERS-CoV RT-iiPCR | |||
| Positive | 52 | 2 | 54 |
| Negative | 0 | 49 | 49 |
| 52 | 51 | 103 | |
| Agreement (95% CI): 98.06% (94.43%–100%); κ = 0.96 | |||
|
upE |
MERS-CoV RT-qPCR |
||
| Positive |
Negative |
Total |
|
| MERS-CoV RT-iiPCR | |||
| Positive | 54 | 1 | 55 |
| Negative | 0 | 48 | 48 |
| 54 | 49 | 103 | |
| Agreement (95% CI): 99.03% (95.88%–100%); κ = 0.99 | |||
MERS-CoV, Middle East respiratory syndrome–coronavirus; RT-iiPCR, reverse transcription–insulated isothermal PCR; RT-qPCR, real-time quantitative RT-PCR.
Discussion
The 2015 MERS-CoV outbreak in the Republic of Korea revealed that a rapid and reliable diagnostic assay suitable for on-site detection of virus is critical and urgently needed to effectively control the spread of infection among individuals. Unfortunately, the existing RT-qPCR assays are not suitable to be used in clinical settings because they require expensive equipment and a laboratory environment staffed with skilled technicians. In this study, the development and evaluation of a rapid and highly sensitive field-deployable system is described for detection of MERS-CoV that allows mobile detection of the virus directly from clinical materials obtained from patients suspected of infection.
A variety of isothermal amplification methods for nucleic acid detection similar to iiPCR, such as LAMP and recombinase polymerase amplification, have been developed for use in simple point-of-need systems.35, 36, 37 Despite their advantages, most LAMP assays still have some technical limitations, such as propensity to produce false-positive reactions and variations in visual observation of LAMP signals between different observers, in particular for weak positive samples.38 Recently, several RT-LAMP assays have been described in the literature for detection of MERS-CoV13, 14, 39 with the advantages of being rapid, simple, accurate, and cost-effective, suitable for on-site application. However, further validation of these assays is needed using specimens from infected patients to ensure their clinical performance.
The singleplex ORF1a and upE MERS-CoV RT-iiPCR assays described in this study are performed in commercially available, simple, and compact instruments, the POCKIT Nucleic Acid Analyzer series, which can process 4 to 32 samples at a time within an hour. The automated interpretation by the iiPCR machine makes the method easier because it does not require data analysis by the user. Furthermore, the ready-to-use lyophilized amplification reagents that are stable for at least 2 years at 4°C provide great ease of storage and transportation. Removal of contaminating PCR inhibitors and genomic DNA during nucleic acid extraction is critical to avoid false negative and positive results, respectively.40 Thus, optimal RNA extraction and template preparation is key for obtaining the highest sensitivity and specificity of any molecular diagnostic assay including RT-iiPCR. Automatic or semiautomatic nucleic acid extraction methods only require basic laboratory training skills and minimize the hands-on time required for template preparation before assembling the reaction. Furthermore, the clinical samples are directly added to the lysis buffer, which immediately inactivates the pathogen and, thus, reduces the risk of exposure of the personnel handling the clinical material. However, it is important to use appropriate personal protective wear and follow recommended biosecurity guidelines during handling and testing of the clinical specimens. Utilization of a light-weight, field-deployable automatic nucleic acid extraction device (taco mini Nucleic Acid Automatic Extraction System; GeneReach USA) along with the iiPCR/POCKIT system greatly reduces the hands-on time from sample collection to results (Figure 1 ). These instruments also can be connected to a car battery and are designed as a portable system to be used in clinics, hospitals, farms, and public health laboratories in remote areas without electricity. Taking advantage of this platform, the two MERS-CoV singleplex RT-iiPCR methods targeting either the ORF1a or upE gene were established to aid rapid on-site diagnosis of MERS-CoV infection. Multiple RT-qPCR assays targeting different sequences in the MERS-CoV genome have been recommended to be used as sequential primary and confirmatory tests to help reduce the risks of misidentifying MERS-CoV cases.34 Particularly, the upE assay has been recommended for screening purposes, whereas the ORF1a assay for confirmation.8 Both ORF1a- and upE-specific RT-iiPCR assays described in this study were shown to be analytically sensitive, and no cross-reactivity was observed with other respiratory viruses, including human coronaviruses such as the hCoV-229E and hCoV-OC43 strains.
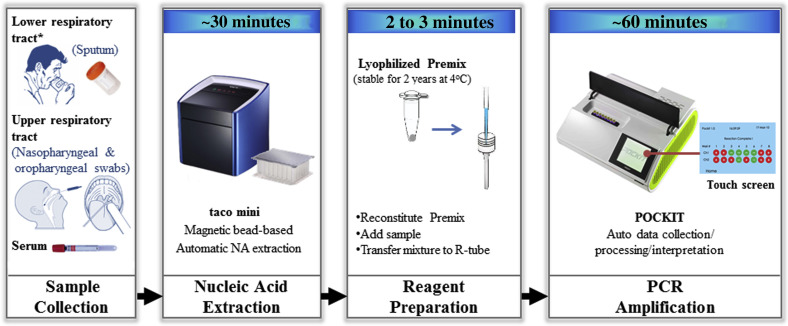
Figure 1.
POCKIT system workflow for point-of-need detection of Middle East respiratory syndrome–coronavirus (MERS-CoV) RNA. This system includes a compact automatic nucleic acid (NA) extraction device (taco mini) and a portable PCR device (POCKIT). After sample collection, nucleic acids were extracted using a preloaded extraction plate in approximately 30 minutes, and subsequently, the lyophilized reverse transcription-insulated isothermal PCR reaction was reconstituted and nucleic acids were added. The mixture was transferred to an R-tube and tested in a POCKIT device. TaqMan probe hydrolysis–based amplification signals were automatically detected, processed, and interpreted, providing qualitative results on the display screen in 60 minutes. Asterisk indicates sample collection method used in this study.
Lower respiratory tract specimens such as sputum and tracheal aspirates are the recommended sample types for accurate diagnosis because they are known to contain high viral RNA loads that persist longer compared with other sample types tested.41 Therefore, the detection limit of singleplex ORF1a and upE MERS-CoV RT-iiPCR assays were compared with the corresponding WHO-recommended reference singleplex RT-qPCR assays using RNA extracted from infectious TCF or human sputum samples spiked with serially diluted MERS-CoV and assessed the assay performance. The data showed that the singleplex ORF1a and upE RT-iiPCR and the corresponding singleplex reference RT-qPCR assays were able to detect as low as <10 copies/μL of synthetic RNA and 3.7 × 10−1 PFU/mL of infectious TCF. Similar results were obtained when analytical sensitivities of ORF1a and upE RT-iiPCR assays were evaluated using sputum samples spiked with a MERS-CoV isolate. Furthermore, the clinical sensitivity and specificity of both singleplex MERS-CoV RT-iiPCR assays were evaluated using archived sputum samples collected from confirmed cases of MERS-CoV infection during the Korean outbreak in 2015. To estimate the diagnostic performance, the singleplex RT-iiPCR assay results were compared side by side with the reference singleplex RT-qPCR assay. The data indicated that the sensitivities of the singleplex MERS-CoV ORF1a and upE RT-iiPCR assays were 99.03% (54 of 55; 95% CI, 95.88%–100%) and 100% (55 of 55; 95% CI, 97.43%–100%), respectively, whereas those of the reference RT-qPCR assays were 97.09% (52 of 55; 95% CI, 87.43%–100%) for ORF1a and 99.03% (54 of 55; 95% CI, 95.88%–100%) for upE. Possible explanation for the discrepancy with initial diagnosis seen in two specimens (P085 [06-14-2015] and P130 [06-14-2015]) could be viral RNA degradation during long-term storage. The viral RNA copy number of false-negative samples was below detection limit in both assays (assessed by reference RT-qPCR <10 copies), whereas the CT value of internal amplification control remained moderately low (RNase P CT 27.3 and 30.2, respectively), indicating the possibility of viral RNA loss during storage and/or the extraction process. Moreover, in a number of specimens tested, the overall estimated viral load detected during our study was lower compared with the viral copy number estimated a year earlier (data not shown). These observations suggest that the sensitivity of the assay can be adversely affected by the collection method and storage of clinical specimens. In the case of the specimen from patient ID P148 (collected 06-21-2015) that was tested as false negative by the reference ORF1a RT-qPCR assay, it could potentially be due to the presence of extremely low concentrations of target RNA. The RNA levels of the internal amplification control in this sample (RNase P CT 38.5) were the lowest compared with the rest of the samples, suggesting relatively low RNA extraction levels. Nevertheless, the data presented in this study demonstrate that the performance of both singleplex ORF1a and upE MERS-CoV RT-iiPCR tests is equivalent or higher compared with the reference singleplex RT-qPCR tests and provides much faster results (within an hour).
Bats and alpacas are potential reservoirs for MERS-CoV in the wild, whereas dromedary camels may be the only animal host responsible for animal-to-human transmission of MERS-CoV.42, 43 Accordingly, regular screening and isolation of MERS-CoV–infected camels have been recommended to help control MERS-CoV spread.44 Thus, the field-deployable MERS-CoV RT-iiPCR on the POCKIT system has potential to be used for timely on-site monitoring of MERS-CoV carriers in camel and alpaca herds and in bat populations in the wild. It would be interesting to see whether these newly developed assays could be used to detect MERS-CoV in samples from dromedary camels, bats, and alpacas in epidemiological investigations.
Lastly, we conclude that the two rapid, highly sensitive and specific MERS-CoV RT-iiPCR methods coupled with the field-deployable platform described here can be effectively used as an on-site, point-of-need diagnostic tool to aid diagnosis of MERS-CoV infection in clinics, hospitals, airports, or premises where a large number of individuals congregate (eg, religious festivals) as well as in epidemiological investigations including animal reservoirs.
Acknowledgment
We thank Dr. Sung Soon Kim (Korea Centers for Disease Control and Prevention) for providing the MERS-CoV strain (MERS-CoV/KOR/KNIH/002_05_2015) used in this study.
Footnotes
Supported by Korea Research Institute of Chemical Technology (KRICT) intramural grant KK1603-C00 (Y.Y.G., S.N., K.B.K., and M.K.) and Korean Centers for Disease Control and Prevention (KCDC) grant 2015ER480800 (Y.Y.G., M.K., and H.P.).
Y.Y.G. and Y.-S.K. contributed equally to this work.
Disclosures: P.-Y.A.L., Y.-C.L., Y.-L.T., and H.-T.T.W. are employed by GeneReach USA, Lexington, MA.
Contributor Information
Yun Young Go, Email: yygo@krict.re.kr.
Udeni B.R. Balasuriya, Email: ubalasuriya@uky.edu.
References
- 1.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han H.J., Yu H., Yu X.J. Evidence for zoonotic origins of Middle East respiratory syndrome coronavirus. J Gen Virol. 2016;97:274–280. doi: 10.1099/jgv.0.000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C., Meyer B., Muller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., Alhakeem R.F., Assiri A.M., Hajomar W., Albarrak A.M., Al-Tawfiq J.A., Zumla A.I., Memish Z.A. Transmission of MERS-coronavirus in household contacts. N Engl J Med. 2014;371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- 5.Oboho I.K., Tomczyk S.M., Al-Asmari A.M., Banjar A.A., Al-Mugti H., Aloraini M.S., Alkhaldi K.Z., Almohammadi E.L., Alraddadi B.M., Gerber S.I., Swerdlow D.L., Watson J.T., Madani T.A. 2014 MERS-CoV outbreak in Jeddah–a link to health care facilities. N Engl J Med. 2015;372:846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korea Centers for Disease Control and Prevention Middle East respiratory syndrome coronavirus outbreak in the Republic of Korea, 2015. Osong Public Health Res Perspect. 2015;6:269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T.M., Muth D., Muller M.A., Drexler J.F., Zambon M., Osterhaus A.D., Fouchier R.M., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 8.Corman V.M., Muller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M., Nitsche A., Bleicker T., Landt O., Schweiger B., Drexler J.F., Osterhaus A.D., Haagmans B.L., Dittmer U., Bonin F., Wolff T., Drosten C. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 9.Palm D., Pereyaslov D., Vaz J., Broberg E., Zeller H., Gross D., Brown C.S., Struelens M.J., Joint ECDC-WHO Regional Office for Europe Novel Coronavirus Laboratory Survey participants; ECDC National Microbiology Focal Points; WHO European Region EuroFlu Network; European Network for Diagnostics of “Imported” Viral Diseases (ENIVD) Laboratory capability for molecular detection and confirmation of novel coronavirus in Europe, November 2012. Euro Surveill. 2012;17:20335. doi: 10.2807/ese.17.49.20335-en. [DOI] [PubMed] [Google Scholar]
- 10.Reusken C., Mou H., Godeke G.J., van der Hoek L., Meyer B., Muller M.A., Haagmans B., de Sousa R., Schuurman N., Dittmer U., Rottier P., Osterhaus A., Drosten C., Bosch B.J., Koopmans M. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. 2013;18:20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- 11.Corman V.M., Olschlager S., Wendtner C.M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J Clin Virol. 2014;60:168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo B., Jin C.E., Lee T.Y., Lee J.H., Park M.K., Sung H., Park S.Y., Lee H.J., Kim S.M., Kim J.Y., Kim S.H., Shin Y. An isothermal, label-free, and rapid one-step RNA amplification/detection assay for diagnosis of respiratory viral infections. Biosens Bioelectron. 2017;90:187–194. doi: 10.1016/j.bios.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.H., Baek Y.H., Kim Y.H., Choi Y.K., Song M.S., Ahn J.Y. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front Microbiol. 2016;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai S.M., Liu H.J., Shien J.H., Lee L.H., Chang P.C., Wang C.Y. Rapid and sensitive detection of infectious bursal disease virus by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J Virol Methods. 2012;181:117–124. doi: 10.1016/j.jviromet.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Chang H.F., Tsai Y.L., Tsai C.F., Lin C.K., Lee P.Y., Teng P.H., Su C., Jeng C.C. A thermally baffled device for highly stabilized convective PCR. Biotechnol J. 2012;7:662–666. doi: 10.1002/biot.201100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai Y.L., Wang H.T., Chang H.F., Tsai C.F., Lin C.K., Teng P.H., Su C., Jeng C.C., Lee P.Y. Development of TaqMan probe-based insulated isothermal PCR (iiPCR) for sensitive and specific on-site pathogen detection. PLoS One. 2012;7:e45278. doi: 10.1371/journal.pone.0045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsen H.Y., Shih C.M., Teng P.H., Chen H.Y., Lin C.W., Chiou C.S., Wang H.T., Chang H.F., Chung T.Y., Lee P.Y., Chiang Y.C. Detection of Salmonella in chicken meat by insulated isothermal PCR. J Food Prot. 2013;76:1322–1329. doi: 10.4315/0362-028X.JFP-12-553. [DOI] [PubMed] [Google Scholar]
- 19.Balasuriya U.B., Lee P.Y., Tiwari A., Skillman A., Nam B., Chambers T.M., Tsai Y.L., Ma L.J., Yang P.C., Chang H.F., Wang H.T. Rapid detection of equine influenza virus H3N8 subtype by insulated isothermal RT-PCR (iiRT-PCR) assay using the POCKIT Nucleic Acid Analyzer. J Virol Methods. 2014;207:66–72. doi: 10.1016/j.jviromet.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Tsai Y.L., Wang H.C., Lo C.F., Tang-Nelson K., Lightner D., Ou B.R., Hour A.L., Tsai C.F., Yen C.C., Chang H.F., Teng P.H., Lee P.Y. Validation of a commercial insulated isothermal PCR-based POCKIT test for rapid and easy detection of white spot syndrome virus infection in Litopenaeus vannamei. PLoS One. 2014;9:e90545. doi: 10.1371/journal.pone.0090545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkes R.P., Tsai Y.L., Lee P.Y., Lee F.C., Chang H.F., Wang H.T. Rapid and sensitive detection of canine distemper virus by one-tube reverse transcription-insulated isothermal polymerase chain reaction. BMC Vet Res. 2014;10:213. doi: 10.1186/s12917-014-0213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambagala A., Pahari S., Fisher M., Lee P.A., Pasick J., Ostlund E.N., Johnson D.J., Lung O. A rapid field-deployable reverse transcription-insulated isothermal polymerase chain reaction assay for sensitive and specific detection of bluetongue virus. Transbound Emerg Dis. 2017;64:476–486. doi: 10.1111/tbed.12388. [DOI] [PubMed] [Google Scholar]
- 23.Lung O., Pasick J., Fisher M., Buchanan C., Erickson A., Ambagala A. Insulated isothermal reverse transcriptase PCR (iiRT-PCR) for rapid and sensitive detection of classical swine fever virus. Transbound Emerg Dis. 2016;63:e395–e402. doi: 10.1111/tbed.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkes R.P., Kania S., Tsai Y.L., Lee P.Y., Chang H.H., Ma L.J., Chang H.F., Wang H.T. Rapid and sensitive detection of feline immunodeficiency virus using an insulated isothermal polymerase chain reaction-based assay with a point-of-need PCR detection platform. J Virol Methods. 2015;27:510–515. doi: 10.1177/1040638715593597. [DOI] [PubMed] [Google Scholar]
- 25.Wilkes R.P., Lee P.Y., Tsai Y.L., Tsai C.F., Chang H.H., Chang H.F., Wang H.T. An insulated isothermal PCR method on a field-deployable device for rapid and sensitive detection of canine parvovirus type 2 at points of need. J Virol Methods. 2015;220:35–38. doi: 10.1016/j.jviromet.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chua K.H., Lee P.C., Chai H.C. Development of insulated isothermal PCR for rapid on-site malaria detection. Malar J. 2016;15:134. doi: 10.1186/s12936-016-1183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Go Y.Y., Rajapakse R.P., Kularatne S.A., Lee P.Y., Ku K.B., Nam S., Chou P.H., Tsai Y.L., Liu Y.L., Chang H.F., Wang H.T., Balasuriya U.B. A pan-dengue virus reverse transcription-insulated isothermal PCR assay intended for point-of-need diagnosis of dengue virus infection by use of the POCKIT nucleic acid analyzer. J Clin Microbiol. 2016;54:1528–1535. doi: 10.1128/JCM.00225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo H.C., Lo D.Y., Chen C.L., Tsai Y.L., Ping J.F., Lee C.H., Lee P.A., Chang H.G. Rapid and sensitive detection of Mycoplasma synoviae by an insulated isothermal polymerase chain reaction-based assay on a field-deployable device. Poult Sci. 2017;96:35–41. doi: 10.3382/ps/pew228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soltan M.A., Tsai Y.L., Lee P.A., Tsai C.F., Chang H.G., Wang H.T., Wilkes R.P. Comparison of electron microscopy, ELISA, real time RT-PCR and insulated isothermal RT-PCR for the detection of Rotavirus group A (RVA) in feces of different animal species. J Virol Methods. 2016;235:99–104. doi: 10.1016/j.jviromet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Tsai Y.L., Lee P.A., Chen Q., Zhang Y., Chiang C.J., Shen Y.H., Li F.C., Chang H.G., Gauger P.C., Harmon K.M., Wang H.T. Evaluation of two singleplex reverse transcription-Insulated isothermal PCR tests and a duplex real-time RT-PCR test for the detection of porcine epidemic diarrhea virus and porcine deltacoronavirus. J Virol Methods. 2016;234:34–42. doi: 10.1016/j.jviromet.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carossino M., Lee P.Y., Nam B., Skillman A., Shuck K.M., Timoney P.J., Tsai Y.L., Ma L.J., Chang H.F., Wang H.T., Balasuriya U.B. Development and evaluation of a reverse transcription-insulated isothermal polymerase chain reaction (RT-iiPCR) assay for detection of equine arteritis virus in equine semen and tissue samples using the POCKIT system. J Virol Methods. 2016;234:7–15. doi: 10.1016/j.jviromet.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Lu Z., Branscum A.J., Shuck K.M., Zhang J., Dubovi E.J., Timoney P.J., Balasuriya U.B. Comparison of two real-time reverse transcription polymerase chain reaction assays for the detection of Equine arteritis virus nucleic acid in equine semen and tissue culture fluid. J Vet Diagn Invest. 2008;20:147–155. doi: 10.1177/104063870802000202. [DOI] [PubMed] [Google Scholar]
- 33.WHO Laboratory Testing for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) June 2015. Updated June 2015. Available at http://apps.who.int/iris/bitstream/10665/176982/1/WHO_MERS_LAB_15.1_eng.pdf?ua=1. (accessed January 12, 2017)
- 34.Lu X., Whitaker B., Sakthivel S.K., Kamili S., Rose L.E., Lowe L., Mohareb E., Elassal E.M., Al-sanouri T., Haddadin A., Erdman D.D. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James A., Macdonald J. Recombinase polymerase amplification: emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev Mol Diagn. 2015;15:1475–1489. doi: 10.1586/14737159.2015.1090877. [DOI] [PubMed] [Google Scholar]
- 36.Parida M., Sannarangaiah S., Dash P.K., Rao P.V., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 39.Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS One. 2015;10:e0123126. doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 41.Oh M.D., Park W.B., Choe P.G., Choi S.J., Kim J.I., Chae J., Park S.S., Kim E.C., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim D.K., Lee S.M., Song K.H., Bang J.H., Kim E.S., Kim H.B., Park S.W., Kim N.J. Viral Load Kinetics of MERS Coronavirus Infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 42.Mohd H.A., Al-Tawfiq J.A., Memish Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virol J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omrani A.S., Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction. Pathog Glob Health. 2015;109:354–362. doi: 10.1080/20477724.2015.1122852. [DOI] [PMC free article] [PubMed] [Google Scholar]