Summary
Severe acute respiratory syndrome (SARS) is a new infectious disease that first emerged in Guangdong province, China, in November, 2002. A novel coronavirus was later identified in patients with SARS. The detection of the virus in these patients, its absence in healthy controls or other patients with atypical pneumonia, and the reproduction of a similar disease in a relevant animal model fulfilled Koch's postulates for implicating this coronavirus as the causal agent of SARS. The full genome sequence was determined within weeks of the virus's identification. The rapid progress in the aetiology, the development of laboratory diagnostic tests, and the defining of routes of viral transmission were facilitated through a unique WHO-coordinated virtual network of laboratories, which shared information on a real-time basis through daily teleconferences. Subsequent studies have indicated that the SARS coronavirus is of animal origin, that its precursor is still present in animal populations within the region, and that live-animal markets in southern China may have provided the animal-human interphase that allowed this precursor virus to adapt to human-human transmission. These findings underscore the potential for the re-emergence of SARS and the need for laboratory tests for early diagnosis. However, the low viral load in the respiratory tract makes early diagnosis of SARS a diagnostic challenge, although improvements in the sensitivity of molecular diagnostic methods continue to be made.
Severe acute respiratory syndrome (SARS), a new infectious respiratory disease typically associated with fever, shortness of breath, cough, and pneumonia, emerged in southern China in late 2002. Within months, the disease had spread globally, affecting over 8000 patients in 29 countries with 774 fatalities. Over half of these infections can be directly traced to one index patient who arrived in Hong Kong on Feb 21, 2003, after acquiring the disease in Guangdong. This is a dramatic illustration of the impact of modern air travel on the spread of an emerging human infection.1 The outbreak was brought under control through a concerted global effort coordinated by WHO, and by July 5, 2003, no further human-human transmission was taking place and the global outbreak was declared over. In many ways it was an unprecedented global experience, in the rapidity and extent of its spread, the magnitude of its impact on the health systems and economies, and in the effectiveness of its control.2, 3, 4
The first known cases of this unusual disease occurred in Foshan in Guangdong province, China, in November, 2002.5, 6, 7, 8 Similar cases were subsequently reported in Heyuan, which affected family members and healthcare workers in contact with an index case (a chef working in an exotic game restaurant in Shenzhen). Many of these early patients had epidemiological links to the live-animal market trade.5, 7 By January, 2003, the disease had spread to Guangzhou, the capital of Guangdong province, and caused major outbreaks that primarily affected healthcare workers. By the end of January, 2003, the characteristics of the disease were recognised to be a rapidly progressive atypical pneumonia without leucocytosis, which was refractory to conventional antibiotic therapy and was associated with outbreaks in family or hospital settings.9 Clinicians in Guangzhou had termed it “infectious atypical pneumonia”. Several causal agents were under consideration at that time, including chlamydia, which had been found in necropsy tissue of patients who had died from the disease.10 However, the clinical progression and the lack of a response to appropriate macrolide antibiotics argued against a role for chlamydia in this disease.
Meanwhile, the outbreak of disease in Guangzhou was causing concern and even panic among the residents, as reported in local and regional media.8 On Feb 11, 2003, WHO was informed of 305 cases from Guangdong province, over 100 of whom were healthcare workers, with five reported deaths.11 The health authorities in Hong Kong set up enhanced epidemiological and microbiological surveillance of cases of atypical pneumonia, especially for those with a history of recent travel to mainland China. Although several causal agents were identified in patients with atypical pneumonia diagnosed in Hong Kong, the first agent of note was avian influenza A subtype H5N1 from a family returning from Fujian in early February, 2003.12 This finding raised concerns that the avian influenza H5N1 virus may be responsible for the outbreak in Guangdong. A global pandemic alert was declared by WHO and preparations for vaccine development were started.13 However, further investigations revealed no further cases of H5N1 avian influenza in other patients in Guangdong or in Hong Kong.14
By the end of February, 2003, clusters of a pneumonic illness in healthcare workers were being reported in a hospital in Hanoi in association with an index patient who had recently travelled there from Hong Kong. By March 10, a similar outbreak of illness in healthcare workers was reported from the Prince of Wales Hospital in Hong Kong.15 WHO provided a preliminary case definition and a name ("severe acute respiratory syndrome”) for the disease. At this time, additional suspected cases were reported in Toronto and Singapore. WHO issued a global alert and a subsequent travel advisory warning against unnecessary travel to affected regions. The identification of the causal agent was clearly a priority in the effort to contain the spread of this novel disease.16
Aetiology
On March 18, 2003, WHO initiated a virtual network of laboratories to investigate the cause of SARS.16 Daily teleconferences updated members of the network on the progress of laboratory investigations in different parts of the world on patients with suspected SARS.16 This investigation established that influenza and other common respiratory pathogens were not the cause of this novel disease syndrome. Strategies for the detection of a novel agent included the use of direct electron microscopy on respiratory specimens, bronchoalveolar lavage, and lung tissue obtained from lung biopsy or necropsy, culture in cell lines known to support respiratory viral infection (and subsequently on other types of cells), PCR and reverse-transcription PCR (RT-PCR) with consensus primers for respiratory viral pathogens, and random-primer RT-PCR methods. Serology was used for the detection of agents antigenically related to known respiratory pathogens. One of the problems faced in this investigation was the differentiation of SARS from more routine causes of atypical pneumonia. The one feature supporting the probable diagnosis of SARS was epidemiological linkage to a cluster of similar disease cases. Substantial numbers of such cases were initially present in Guangzhou, at the hospital outbreaks at Hanoi and at the Prince of Wales Hospital, Hong Kong, and subsequently in Toronto and Singapore. Through global case notification, a suspected case from Singapore was also intercepted on a flight arriving in Frankfurt, Germany.17
Members of the network reported virus-like particles in clinical respiratory specimens by electron microscopy, and these virus particles had pleomorphic morphology compatible with paramyxoviruses.16, 17 Shortly after, human metapneumovirus was detected in the respiratory tract of patients with SARS at the Prince of Wales Hospital in Hong Kong, and in Toronto.18, 19 However, human metapneumovirus was not consistently detected in other patients with SARS in Hong Kong outside the Prince of Wales Hospital or elsewhere globally. In addition, the genetic sequence of the human metapneumovirus isolated from these patients was essentially similar to that previously known to circulate in human beings. As such, it was difficult to explain why human metapneumovirus, which was known to have been a human pathogen for many years, would cause a novel and unusual disease syndrome such as SARS. Rhinovirus and chlamydia were also detected in patients with suspected SARS.10, 17, 20
Three laboratories within the WHO network independently reported the isolation of a novel coronavirus from clinical specimens of patients with SARS.14, 17, 20 In all three laboratories, the strategies that led to the detection of this novel coronavirus were broadly similar. Although culture in cell lines (including Hep-2, MRC-5, MDCK, LLC-Mk2, HeLa, RDE, NCI-H292, HUT-292, B95-8, and A549) that were usually used to culture respiratory viruses proved unproductive, the virus was isolated in Vero-E6 cells in two laboratories and FRhK-4 cells in the other. Even before the nature of the virus was identified, a link with SARS was established by showing that serum samples from patients with suspected SARS had increasing antibody titres to virus-infected cells by indirect immunofluorescence assay (IIFA). The identification of the agent as a coronavirus was made using thin-section electron microscopy of virus-infected cells and negatively stained preparations of cell-culture supernatants that were virus infected (Figure 1 ). Independent confirmation of the identity of the virus was obtained by sequencing fragments of the viral genome derived through random-primed RT-PCR in two laboratories, and the use of consensus primers for coronaviruses in the other laboratory. In all three laboratories, the region that yielded the early sequence information was the open reading frame (ORF) 1b region of the replicase gene, possibly because this is a highly conserved region of the virus. DNA array analysis using a microarray containing probes for various virus families21 was done on infected and uninfected cells. This analysis gave positive signals for a group of eight oligonucleotides whose sequence was derived from two virus families, the Coronaviridae and the Astroviridae.20 However, microarray analysis could only be done once the virus was isolated in culture and could not be done directly on the clinical specimen.
Figure 1.

Thin section electron micrograph of the surface of an infected FRhK4 cell showing SARS coronavirus with spikes. Bar=100 nm.
The independent detection by three laboratories of a coronavirus-like agent in samples from patients with SARS-like disease in Hong Kong, Vietnam, Singapore, and Germany (patient from Singapore) led particular credence to the significance of this observation. However, to conclusively establish a causal role for this novel coronavirus, the virus had to be consistently found in the relevant clinical specimens from patients with the disease and not in healthy controls. RT-PCR methods based on the initial genetic sequence of the virus replicase gene were designed and applied to detect viral RNA in patients' specimens. The virus isolate was also used in IIFAs and EIAs to detect serological responses in acute and convalescent serum specimens. With these approaches, infection with the novel coronavirus, subsequently named SARS-associated coronavirus, was detectable in 45 of 50 consecutive patients with clinically suspected SARS in Hong Kong. All 32 patients from whom paired serum samples were available had seroconverted to positive for SARS coronavirus. Viral RNA was not detectable by RT-PCR in healthy controls. Serum samples from 200 healthy blood donors in Hong Kong, even though there was an outbreak of SARS at the time, did not have antibody that was reactive to cells infected with SARS coronavirus in IIFAs.14 A second study found evidence of SARS coronavirus by RT-PCR or virus culture in 19 patients with SARS from Vietnam, Hong Kong, and Singapore, and nine patients from whom serum samples were available had increasing antibody titres to the virus.20 Furthermore, 384 randomly selected serum samples from US blood donors had no reactivity to the SARS coronavirus by EIA.20 A third study found all five patients with probable SARS and three of 13 patients with suspected SARS who originated from Singapore and Vietnam to have evidence of SARS coronavirus RNA by RT-PCR.17 Taken together, these results indicated that the novel coronavirus was associated with SARS, and given the lack of serological reactivity in human beings, suggested that it was probably recently introduced to the human population. Because, on the basis of the partial genetic information available, the virus seemed to be an agent not previously known to circulate in human populations, the likely origin of the virus was from an animal source.
Fulfilment of Koch's postulates for establishing the cause of a disease requires that the microbe should be detected at the site of the disease. SARS coronavirus was detected in the lung biopsy and bronchoalveolar lavage of a patient with SARS by virus culture, RT-PCR, and electron microscopy.10, 17, 20 In addition, virus-like particles compatible in size and morphology to coronavirus were shown in lung biopsy tissue and bronchoalveolar lavage.10, 20 The lung pathology of fatal SARS showed bronchial epithelial denudation, loss of cilia, type 2 pneumocyte hyperplasia, and in patients who died later in the course of the disease, syncytial cells were seen in the alveoli.20, 22, 23 SARS coronavirus RNA was shown in the lungs by RT-PCR23, 24 and in-situ hybridisation,25, 26, 27 and viral antigen was detected in alveolar epithelial cells and macrophages by immunohistology (our unpublished data). No evidence of human metapneumovirus was detected in the lungs by RT-PCR, by electron microscopy, or by a serological response in paired serum samples.22, 23
The final requirement for the fulfilment of Koch's postulates to establish the cause of an infectious disease is reproduction of the disease in a relevant animal model. Infection of cynomolgous macaques with an isolate of SARS coronavirus led to disease that was pathologically similar to that seen in human patients with SARS, with epithelial necrosis, serosanguinous alveolar exudates, hyaline membranes, type 2 pneumocyte hyperplasia, and the presence of syncytia.28, 29 The virus was successfully re-isolated from these lesions and an antibody response to the virus was shown in the infected animals. By contrast, infection of macaques with human metapneumovirus produced a mild suppurative rhinitis with minimal erosion in the infected airways.28 The possibility remained that co-infection with human metapneumovirus could exacerbate the disease caused by SARS coronavirus. However, there was no evidence of human metapneumovirus infection (by RT-PCR or serological methods) in patients with severe and fatal SARS. SARS coronavirus was detected in these patients, which suggested that human metapneumovirus was not necessary for development of severe or fatal SARS.23 In addition, infection of macaques with SARS coronavirus followed by human metapneumovirus did not produce enhanced disease.28 Thus, it was concluded that SARS coronavirus was both necessary and sufficient to cause the full syndrome of SARS. Subsequently, it was shown that SARS coronavirus infects and replicates in cats and ferrets, and can be transmitted to uninfected animals.30 Recently, SARS coronavirus has also been shown to infect and replicate in mice, although without overt disease.31 These experiments provide alternative animal models for the investigation of SARS. Of these, the macaque and ferret models show clinically significant disease.
Genetic sequence of the SARS coronavirus
Within weeks of the isolation of the novel coronavirus associated with SARS, the full genome of the virus was sequenced.32, 33 Analysis of the genome of the SARS coronavirus confirmed that it was completely novel across the whole of its length, and was not derived by recombination between previously known animal or human coronaviruses. This finding also confirmed that SARS coronavirus could not have been artificially generated in the laboratory as a bioterrorist agent.
Coronaviruses are enveloped viruses with a single, positive-stranded RNA genome. All coronaviruses have five major ORFs that encode the replicase, spike, envelope, membrane, and nucleocapsid proteins.34, 35 These viruses can be subdivided into three groups. On the basis of antigenic and genetic studies, all previously known human coronaviruses can be classified into group 1 (eg, 229E) and group 2 (eg, OC43). SARS coronavirus showed some serological cross-reactivity with antisera raised to human coronavirus 229E (group 1).20 Partial initial viral sequences of the viral replicase had indicated that SARS coronavirus was genetically distinct from all previously known coronaviruses.14, 17, 20 Further characterisation of the full genome confirmed this deduction and suggested that the virus is distantly related to coronaviruses from groups 2 and 3.32, 33, 36 These findings prompted the conclusion that either the SARS coronavirus represents a new fourth group within the coronavirus genus,32, 33 or that it represented a group 2 virus that had diverged from others in the group very early in evolution.37 However, unlike other group 2 viruses, SARS coronavirus does not contain a haemagglutinin-esterase protein encoding sequence. Further work is required to confirm the taxonomic classification of this virus.
The availability of the full genetic sequence has allowed refinement of diagnostic assays (see below), rational antiviral drug design, and vaccine development.38, 39
Origins and molecular epidemiology of SARS
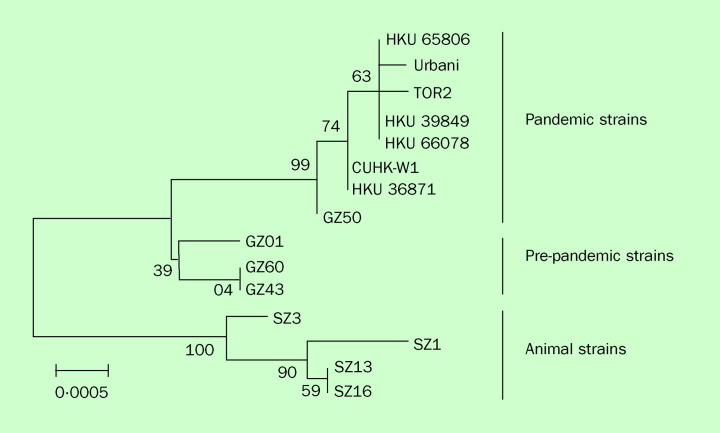
The recent introduction of a novel coronavirus in human beings without prior serological evidence of infection in the human population prompted much speculation about the origin of this virus. Whereas bioengineered or even extraterrestrial agents were adduced by some to explain its origin,40 the most probable explanation was that it was an animal virus that had recently acquired the ability for human-human transmission. This hypothesis was strengthened by reports that early patients with SARS in Guangdong were exposed to live wild game animals held in markets serving the restaurant trade for freshly cooked game meat.5, 7 A search for the animal source of the precursor virus was prompted (Figure 2 ). Sampling of several animal species found in one of the live-game markets in Guangdong revealed that some of them carried a virus genetically and antigenically related to the human SARS coronavirus.41 In particular, a SARS-like coronavirus was detected by RT-PCR in the nasal and faecal swabs of civet cats (Paguma larvata) and a raccoon dog (Nyctereutes procyonoides). Serological evidence of infection was found in these species and also in a Chinese ferret-badger (Melogale moschata).41 Genetic sequencing of the virus revealed that the animal coronavirus had more than 99% homology with human SARS coronavirus (Figure 3 ). However, when compared with the animal viruses, the human SARS coronavirus seemed to have a 29-nucleotide deletion in ORF8 (Figure 4 ). This additional sequence predicts that the animal virus will express a putative protein of 122 aminoacids that is not seen in the human SARS coronavirus. Interestingly, a subsequent study revealed other genotypes with similar deletions in the same ORF.42 In that study, viruses with a 82-nucleotide deletion were isolated from the early phase of the epidemic. In addition, viruses with a 415-nucleotide deletion resulting in the loss of the whole ORF8 region were found in May, 2003 (ie, at the late phase of the outbreak). These results indicated that the expression of ORF8 is dispensable for the SARS coronavirus in human beings. Alternatively, it is also tempting to speculate that deletions in ORF8 might give the virus an advantage to adaptation in human beings.
Figure 2.
Investigation of the animal precursor of the SARS coronavirus in a live-animal market in China.
Figure 3.
Phylogeny of human and animal SARS coronaviruses isolated in 2003. The scale bar represents the genetic distance estimated by Kimura's two parameter substitution model (adapted from reference 41, with permission).
Figure 4.
Genetic organisation of SARS coronavirus-like viruses found in human beings and animals (modified from reference 41). Open reading frames (ORFs) 1a and 1b, encoding the non-structural polyproteins, and those encoding the spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins are indicated (green boxes). ORFs for putative proteins 7a, 8a, 8b and 9b are indicated by brown boxes. An extra 29-nucleotide sequence is ORF8 of the animal SARS coronavirus (based on AY278554 numbering). The presence of this 29-nucleotide sequence in animal isolates generates a new protein-encoding sequence in ORF8 (blue box). The labelling system is based on reference 37.
Animal traders working with live animals in these markets had high seroprevalence for both the human and animal SARS coronavirus, although they did not have a history of SARS-like disease.41, 43 The animal precursor of SARS coronavirus is thus likely to be inefficient at infecting human beings, and repeated exposure to the precursor animal virus probably leads to the abortive infection or antigenic stimulation that results in this observed serological response in the animal handlers. These findings, taken with the history of exposure to these same live-animal markets provided by some of the early patients with SARS, suggests that the live-animal markets were probably the site where the inter-species transfer of the animal virus to human beings occurred.
It seems that once the virus is introduced into such live-animal markets with a diverse fauna of small mammals, the precursor SARS coronavirus efficiently establishes itself in several small mammalian species. Studies with avian influenza viruses in live-poultry markets has shown that such viruses amplify within the setting of a market trading in live birds,44 and it is likely that the precursor SARS coronavirus does likewise in markets in which there is a diversity of mammalian species, with animals being removed and added to the ecosystem on a regular basis.45 However, the reservoir of this virus in nature remains to be conclusively identified. The palm civet or the raccoon dog are current candidates. Equally, another species could be the reservoir of the virus in the wild, while the civet cats and similar species serve as the amplifier of the disease in the setting of live-animal markets. Further work is required to identify the natural reservoir of this virus.
Detailed molecular epidemiological investigations indicate that the viruses detected in human beings in the early stages of the SARS outbreak in Guangdong are closely related to the animal SARS coronavirus.41, 42, 46 Some viruses had the additional 29-nucleotide sequence that was seen in the animal viruses. The early viruses were found to be genetically more diverse and more prone to mutation than viruses isolated later in the course of the outbreak, which suggests that the SARS coronavirus was in the process of host adaptation and was subjected to a high selection pressure during the early phase of the outbreak.42 Later in the outbreak, the virus had presumably become more adapted to human-human transmission and one lineage of the virus had become dominant. For example, in Feb-March, 2003, there were at least four introductions of SARS coronavirus from China to Hong Kong.46 These were genetically diverse. However, both classic and molecular epidemiology suggests that only one of these viruses seems to have given rise to the major outbreak in Hong Kong that subsequently spread globally. Thus, most of the SARS coronaviruses isolated from patients in Canada, Vietnam, and Singapore are genetically closely related to the virus isolated from a single SARS patient in Hong Kong (HKU-33), whereas other genetic lineages seem to be less transmissible.42, 46, 47, 48, 49, 50
Laboratory diagnosis
Isolation of the causal agent and determination of its partial genome sequence provided the basis for the first generation of laboratory tests for SARS.14, 17, 20 RT-PCR tests were based on the ORF1b sequence of SARS coronavirus replicase gene, the first part of the genome to be sequenced. Serological tests were based on the IIFA of virus-infected cells and EIAs were based on viral cell extracts. Virus isolation was not very sensitive, was labour intensive, and took too long to be clinically relevant. Although these tests used in combination proved robust and useful for the retrospective diagnosis of SARS, they proved less satisfactory for providing a diagnosis in the first few days after disease onset.51, 52
The first generation of RT-PCR assays were more sensitive at the end of the first week of illness, with only 35–65% of specimens testing positive in the first few days of the disease.51, 53, 54, 55 However, these assays provided important insights into the pathogenesis and transmission of the disease. Virus was detected by RT-PCR and by culture not only in the respiratory tract but also in the faeces and urine.51 Replication of the virus in the gastrointestinal tract was confirmed by electron microscopic studies of biopsies of the upper and lower intestinal mucosa in patients with SARS.56 These findings suggested that infection with SARS coronavirus was disseminated and not simply confined to the respiratory tract. They also raised the possibility that the infection could be spread by the faecal route. This route proved to be important in at least one major community outbreak, Amoy Gardens in Hong Kong, in which over 300 patients were infected within the period of a few days, presumably through contaminated faeces.57 Quantitative RT-PCR assays on the nasopharyngeal aspirates as well as rates of positivity of first-generation RT-PCR assays at different stages of the illness showed that, unlike other respiratory viral infections, viral load and rates of positivity of SARS coronavirus in the upper respiratory tract increased progressively to peak at around day 10 after disease onset.51, 53, 54, 55, 58 These findings provided the reason why RT-PCR assays were of poor sensitivity early in the course of the disease. They also predicted that virus transmission would be lower in the first few days of illness, a finding supported by epidemiological observations.59 Whereas viral RNA could be detected by RT-PCR for many weeks or even months after the onset of disease, virus culture was only possible during the first 3 weeks of illness.51 It is possible that even though virus replication continues after the first 3 weeks of disease, the virus may be complexed with antibody and therefore no longer infective or transmissible. This finding was also in agreement with epidemiological findings.
Apart from respiratory samples, viral RNA was also detected in stool, blood, cerebrospinal fluid, and urine.58, 60, 61, 62, 63 Of these non-respiratory samples, stool specimens had the highest proportion of positive RT-PCR results. In one study, 65 of 67 stool samples collected later in the illness (mean 14·2 days) were positive in conventional RT-PCR assays.58 This observation agreed with the report that stool samples might be a good alternative to respiratory samples for the identification of SARS patients.55 However, one study indicated that viral RNA was not detectable in stool samples within the first 4 days of illness, suggesting that stool samples might not be useful for early diagnosis of SARS.51
The low sensitivity of conventional RT-PCR assays in the first few days of disease, the period during which laboratory diagnosis is most relevant for patient care, prompted the exploration of several strategies to improve test sensitivity. A quantitative real-time RT-PCR approach was used to improve sensitivity as well as turnaround time.64, 65 These methods had the benefit of being closed systems that reduced the chance of PCR cross-contamination within the laboratory. More importantly, the technology, when combined with automated specimen extraction methods, had the potential to become high throughput assays, a significant advantage in areas with outbreaks of SARS in which large numbers of specimens had to be tested. In addition, these assays provided quantitative viral load data, which helped elucidate the pathogenesis of the disease. For example, these assays showed that viral load was highest in specimens of the lower respiratory tract (eg, bronchoalveolar lavage, sputum, endotracheal aspirates), and was higher in nasopharyngeal aspirate than in throat swabs.17 Faecal samples had very high viral loads toward the end of the first week of illness, and were the specimen of choice during the second week of disease.66
Other approaches were also used to enhance the detection rate of SARS. The sensitivity of detection could be increased by testing multiple serial samples collected from the same patient.62 However, this increased the already high workloads in clinical diagnostic laboratories. Real-time nested RT-PCR strategies were shown to be useful for detecting low copy numbers of SARS coronavirus present in the early stage of illness.67, 68 The turnaround time for this method was much shorter than that for the conventional nested RT-PCR assay. However, nested RT-PCR increases the risk of laboratory cross-contamination, leading to false-positive results.69
In infected cells, coronaviruses generate subgenomic mRNA for the synthesis of structural viral protein by discontinuous transcription. Because of the genomic organisation of SARS coronavirus, all subgenomic mRNA molecules contain the nucleocapsid gene sequence region of the SARS coronavirus.32, 33 Thus, targeting the nucleocapsid gene sequence should be expected to enhance sensitivity of diagnosis. This proved to be the case when applied to specimens collected from experimentally infected macaques.29 However, several independent studies on clinical specimens from patients with SARS proved otherwise,66, 70, 71, 72 and showed that most of the viral RNA in clinical specimens is of genomic origin.66 However, it was advisable to have RT-PCR assays that target different parts of the viral genome to serve as a confirmation of the positive result.73 This strategy provides some assurance that a positive result is not due to laboratory contamination with PCR amplicon.
Given that the analytical sensitivity of RT-PCR assays could not be increased much further, the only other option for enhancing test sensitivity was by increasing the amount of viral RNA added to the RT-PCR reaction mix.70, 74 By extraction of RNA samples from a larger volume of nasopharyngeal aspirate and by applying quantitative real-time RT-PCR technologies, the sensitivity of SARS-coronavirus detection improved to approximately 80% of nasopharyngeal aspirate specimens collected from day 1–3 of disease onset with 100% specificity.75 This method was not necessarily associated with an increase in PCR inhibitors.75 However, this strategy implies that an inhibitor control is important to assure against a false-negative result due to PCR inhibition.
In initial studies, serum or plasma were not thought to be useful for molecular diagnosis of SARS.17, 62 However, subsequent studies on plasma and serum samples from SARS patients showed that SARS coronavirus RNA was detectable in more than 50% of plasma and 78% of serum samples during the first week of illness.70 In another study, which used multiple serial plasma samples, 79% of SARS patients within the first 3 days of illness could be identified by quantitative RT-PCR.76 A recent study showed that SARS coronavirus RNA was detectable and was claimed to replicate in peripheral blood mononuclear cells from a SARS patient during early disease.61 Whether the viral RNA originated from infection of peripheral blood mononuclear cells or by phagocytosis or endocytosis of virus replicating the respiratory tract is still unclear. Nevertheless, blood leucocytes may provide yet another alternative for early SARS diagnosis. However, SARS coronavirus viraemia occurs mainly within the first week of the disease.77 In addition, outside the period of a SARS epidemic, most respiratory infections are caused by other pathogens, and a respiratory specimen still remains the most important clinical specimen for clinical investigation.
According to the results of first-generation RT-PCR assays, patients with a positive RT-PCR result on admission had worse outcomes in terms of survival, requirement for intensive care, and assisted ventilation, when compared with SARS patients with negative RT-PCR results at admission.53 This finding was presumably a surrogate for viral load early in the disease. Recently, this hypothesis has been confirmed using quantitative RT-PCR, and high viral load in the nasopharyngeal aspirate was found to be an independent predictor of mortality. Viral load from patients who died from the disease was recently shown to be about 4·5 log higher than in patients who eventually recovered from the illness.78 A high viral load is also associated with the occurrence of diarrhoea78 and the requirement for intensive care.70 Thus, quantitative PCR assays might provide critical prognostic information for clinical management. In particular, if the viral load is a major predictor of clinical outcome, effective antiviral therapy that helps to reduce viral load is likely to provide clinical benefit.
Serodiagnosis is currently the gold standard for confirmation of a diagnosis of SARS. IIFA using virus-infected cells spotted onto Teflon-coated microwell slides,51 or EIA based on extracts of virus-infected cells coated on microwell EIA plates have proved reliable methods for the diagnosis of SARS.79 However, seroconversion usually occurred in weeks 2 or 3 of illness, and a few patients seroconverted as late as 28 days after disease onset.58 Thus, serology is not an option for early diagnosis of the disease. Detection of IgM or IgA subclass-specific antibody did not allow earlier diagnosis.58, 80 These tests require culture of live SARS coronavirus to provide the relevant antigen used in the assays, and require reliable inactivation of virus infectivity before they can be used outside biohazard level 3 containment. Recombinant viral nucleoprotein and spike protein antigens expressed in Escherichia coli and other substrates have been assessed as alternative serological test platforms.79, 81, 82 These have the potential for providing low cost, reproducible, and non-hazardous serological tests in an EIA format that is amenable to high throughput testing.79, 83, 84 However, it should be borne in mind that each antigen and assay format must be individually tested to assess the possibility of cross-reaction with other related human coronaviruses. Whereas such serological cross-reactions did not prove to be a major problem with the whole native viral antigen assays mentioned above, this should not be assumed to mean that recombinant antigens are equally specific until they have been shown to be so. Furthermore, although serological cross-reactivity proved not be a problem during the period of the SARS outbreak, in the post-SARS era when the prior probability of SARS is extremely low, a positive serological result must be treated with even greater caution. In addition to the two known human coronaviruses 229E and OC43, a new human coronavirus has been discovered.85 The possibility of cross-reaction with other human coronaviruses, although rare, is real and must be taken seriously. Furthermore, individuals vary in the antigenic epitopes to which they produce antibodies. Hence, cross-reactivity may well be a rare event, although such a result, if not interpreted with care, may have major implications for public health. We therefore suggest that all positive serological results should be confirmed by neutralisation tests in reference laboratories.
In periods in which there are no outbreaks of SARS, clinical specimens for investigation of SARS should be tested with particular care. Positive RT-PCR results should be confirmed by an independent molecular test on a different specimen. RT-PCR tests for different genome targets (eg, replicase gene and nucleoprotein gene) should be available in reference laboratories to confirm positive results, and for selective use in confirming negative results on patients on whom there is a high degree of suspicion. Serial specimens from many sites (eg, respiratory, faeces, and serum) should be tested. In addition, to avoid obtaining false-negative results, assays with internal PCR inhibitor control should be encouraged. Positive results should be confirmed in an independent reference laboratory. Serological assays should show a significant rise in antibody titres to native viral antigens (eg, IIFA) and be confirmed by virus neutralisation. Human infection with an animal SARS-like coronavirus, which is what we are most likely to encounter, may behave atypically compared with SARS. For example, the clinical disease may be milder and the viral load may be even lower and more transient than with SARS. This means even greater vigilance may be required in making the diagnosis of a infection with the SARS-related animal coronavirus.
Re-emergence of SARS?
Possible options for the re-emergence of SARS includes the undetected continued transmission of the virus from the outbreak in the summer of 2003, escape of the virus from laboratories that contain live SARS coronavirus, or the re-emergence of the virus from an animal reservoir. There is at present no evidence for the virus persisting in the human population. Although the virus may be detected by RT-PCR in the faeces for months after the onset of disease,51 virus isolation is rarely possible after the third week of illness, and there is no evidence for disease transmission by patients in late convalescence. The possibility that immunocompromised patients, such as those with AIDS, may harbour the virus long term has to be considered. However, if epidemiologically relevant transmission from such cases occurred, SARS should have been manifest by now, given the intense global surveillance and awareness that now exists. Escape of the virus from a laboratory has indeed occurred on three occasions86 and highlights the importance of biosafety. WHO has provided strict biohazard guidelines for the handling of this virus and its secure storage.87 At least one of these events involved the handling of virus that was thought to be chemically inactivated.86, 88 Thus, researchers should use appropriate and internationally accepted methods for the inactivation of live viruses.89 A detailed study on SARS-coronavirus inactivation has been recently reported.90 To prevent any inadvertent laboratory-acquired infection leading to secondary transmission in the community, as happened after the laboratory infections in Beijing,88 a high level of alert for febrile and respiratory illness in all laboratory workers who handle infectious SARS coronavirus needs to be maintained.
The re-emergence of the virus from its animal reservoir remains possible, given that the virus is detectable in the faeces and respiratory secretions of small mammals within live-animal markets in southern China. Re-emergence in four patients with SARS in Guangdong in December, 2003, and January, 2004, and epidemiological genetic linkage to live-game markets and viruses isolated from them, prompted the wholesale cull of civet cats and other small mammals in these markets in January, 2004.91 No further human cases of SARS acquired from animals have since been detected.
Search strategy and selection criteria
PubMed searches and references from relevant articles were used for this review. Search terms used were “SARS”, “severe acute respiratory syndrome”, “coronavirus”, “etiology”, “diagnosis”, “PCR”, and “serology”. Only papers published in English (except reference 10) were reviewed.
Acknowledgments
Acknowledgments
This work is supported by National Institute of Allergy and Infectious Diseases (Public Health Research Grant A195357), Research Grant Council of Hong Kong (grants HKU 7543/03M and HKU 7542/03M), and SARS research funds from the University of Hong Kong.
Conflicts of interest
We declare that we have no conflicts of interest.
References
- 1.Fleck F. How SARS changed the world in less than six months. Bull WHO. 2003;81:625–626. [PMC free article] [PubMed] [Google Scholar]
- 2.Caulford P. SARS: aftermath of an outbreak. Lancet. 2003;362(suppl):S2–S3. doi: 10.1016/S0140-6736(03)15052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enserink M. SARS: a pandemic prevented. Science. 2003;302:2045. doi: 10.1126/science.302.5653.2045. [DOI] [PubMed] [Google Scholar]
- 4.Heymann DL, Rodier G. Global surveillance, national surveillance, and SARS. Emerg Infect Dis. 2004;10:173–175. doi: 10.3201/eid1002.031038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiman RF, Evans MR, Priser W. Role of China in the quest to define and control severe acute respiratory syndrome. Emerg Infect Dis. 2003;9:1037–1041. doi: 10.3201/eid0909.030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z, Zhang F, Xu M. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52:715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhong NS, Zheng BJ, Li YM. Epidemiological and aetiological studies of patients with severe acute respiratory syndrome (SARS) from Guangdong in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosling L, Rosling M. Pneumonia causes panic in Guangdong province. BMJ. 2003;326:416. doi: 10.1136/bmj.326.7386.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong NS, Zeng GQ. Our strategies for fighting severe acute respiratory syndrome (SARS) Am J Respir Crit Care Med. 2003;168:7–9. doi: 10.1164/rccm.200305-707OE. [DOI] [PubMed] [Google Scholar]
- 10.Hong T, Wang JW, Sun YL. Chlamydia-like and coronavirus-like agents found in dead cases of atypical pneumonia by electron microscopy [in Chinese] Zhonghua Yi Xue Za Zhi. 2003;83:632–636. [PubMed] [Google Scholar]
- 11.WHO Acute respiratory syndrome, China. Wkly Epidemiol Rec. 2003;78:41. [Google Scholar]
- 12.Peiris JSM, Yu WC, Leung CW. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO Influenza A (H5N1), Hong Kong, Special Administrative Region of China. Wkly Epidemiol Rec. 2003;78:49–50. [PubMed] [Google Scholar]
- 14.Peiris JS, Lai ST, Poon LL. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee N, Hui D, Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 16.WHO Multicentre Collaborative Network for SARS Diagnosis A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet. 2003;361:1730–1733. doi: 10.1016/S0140-6736(03)13376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drosten C, Gunther S, Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 18.Chan PKS, Tam JS, Lam CW. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis. 2003;9:1058–1063. doi: 10.3201/eid0909.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poutanen SM, Low DE, Henry B. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 20.Ksiazek TG, Erdman D, Goldsmith C. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Coscoy L, Zylberg M. Microarray based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franks TJ, Chong PY, Chui P. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol. 2003;34:43–48. doi: 10.1016/S0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholls JM, Poon LL, Lee KC. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzulli T, Farcas GA, Poutanen SM. Severe acute respiratory syndrome-associated coronavirus in lung tissue. Emerg Infect Dis. 2003;10:20–24. doi: 10.3201/eid1001.030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima N, Asahi-Ozaki Y, Nagata N. SARS coronavirus-infected cells in lung detected by new in situ hybridization technique. Jpn J Infect Dis. 2003;56:139–141. [PubMed] [Google Scholar]
- 26.Chong PY, Chui P, Ling AE. Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore. Arch Pathol Lab Med. 2004;128:195–204. doi: 10.5858/2004-128-195-AODDTS. [DOI] [PubMed] [Google Scholar]
- 27.To KF, Tong JH, Chan PK. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouchier RAM, Kuiken T, Schutten M. Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken T, Fouchier RAM, Schutten M. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martina BE, Haagmans BL, Kuiken T. SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subbarao K, McAuliffe J, Vogel L. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra MA, Jones SJ, Astell CR. The genome sequence of the SARS associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 33.Rota PA, Oberste MS, Monroe SS. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 34.Holmes KV, Enjuanes L. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 35.Lai MMC. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng FY, Chan CW, Chan MN. The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39) Exp Biol Med (Maywood) 2003;228:866–873. doi: 10.1177/15353702-0322807-13. [DOI] [PubMed] [Google Scholar]
- 37.Snijder EJ, Bredenbeek PJ, Dobbe JC. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SRS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 40.Wickramasinghe C, Wainwright M, Narlikar J. SARS: a clue to its origins? Lancet. 2003;361:1832. doi: 10.1016/S0140-6736(03)13440-X. [DOI] [PubMed] [Google Scholar]
- 41.Guan Y, Zheng BJ, He YQ. Isolation and characterization of viruses related to SARS cornavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 42.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 43.Centers for Disease Control and Prevention Prevalence of IgG antibody to SARS-associated coronavirus in animal traders: Guangdong Province, China, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:986–987. [PubMed] [Google Scholar]
- 44.Kung NY, Guan Y, Perkins NR. The impact of a monthly rest day on avian influenza virus isolation rates in retail live poultry markets in Hong Kong. Avian Dis. 2003;47:1037–1041. doi: 10.1637/0005-2086-47.s3.1037. [DOI] [PubMed] [Google Scholar]
- 45.Webster RG. Wet markets: a continuing source of severe acute respiratory syndrome and influenza. Lancet. 2004;363:234–236. doi: 10.1016/S0140-6736(03)15329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Y, Peiris JSM, Poon LLM. Molecular epidemiology of SARS coronavirus in Hong Kong. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chim SS, Tsui SK, Chan KC. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362:1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan Y, Wei CL, Ee LA. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsui SKW, Chim SSC, Lo YMD. Coronavirus genomic sequence variations and the epidemiology of the severe acute respiratory syndrome. N Engl J Med. 2003;349:187–188. doi: 10.1056/NEJM200307103490216. [DOI] [PubMed] [Google Scholar]
- 50.Yeh SH, Wang HY, Tsai CY. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc Natl Acad Sci USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan KH, Poon LLM, Cheng VCC. Detection of SARS coronavirus (SCoV) by RT-PCR, culture, and serology in patients with acute respiratory syndrome (SARS) Emerg Infect Dis. 2003;10:294–299. [Google Scholar]
- 52.WHO WHO SARS international reference and verification laboratory network: policy and procedures in the inter-epidemic period (published online Jan 23, 2004) http://www.who.int/csr/resources/publications/en/SARSReferenceLab.pdf (accessed Sept 22, 2004)
- 53.Tsang OT, Chau TN, Choi KW. Coronavirus-positive nasopharyngeal aspirate as predictor for severe acute respiratory syndrome mortality. Emerg Infect Dis. 2003;9:1381–1387. doi: 10.3201/eid0911.030400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang P, Louie M, Richardson SE. Interpretation of diagnostic laboratory tests for severe acute respiratory syndrome: the Toronto experience. Can Med Assoc J. 2004;170:47–54. [PMC free article] [PubMed] [Google Scholar]
- 55.Wu HS, Chiu SC, Tseng TC. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg Infect Dis. 2004;10:304–310. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung WK, To KF, Chan PK. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Department of Health, Hong Kong Outbreak of severe acute respiratory syndrome (SARS) at Amoy Gardens, Kowloon Bay, Hong Kong. Main findings of the investigation (published online April, 2003) http://www.info.gov.hk/info/ap/pdf/amoy_e.pdf (accessed Sept 22, 2004)
- 58.Peiris JS, Chu CM, Cheng VC. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipsitch M, Cohen T, Cooper B. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung EC, Chim SS, Chan PK. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin Chem. 2003;49:2108–2109. doi: 10.1373/clinchem.2003.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Wo J, Shao J. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J Clin Virol. 2003;28:239–244. doi: 10.1016/S1386-6532(03)00195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yam WC, Chan KH, Poon LL. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J Clin Microbiol. 2003;41:4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lau KK, Yu WC, Chu CM, Lau ST, Sheng B, Yuen KY. Possible central nervous system infection by SARS coronavirus. Emerg Infect Dis. 2004;10:342–344. doi: 10.3201/eid1002.030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McIntosh K. The SARS coronavirus: rapid diagnostics in the limelight. Clin Chem. 2003;49:845–846. doi: 10.1373/49.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poon LL, Wong OK, Chan KH. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin Chem. 2003;49:953–955. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poon LL, Chan KH, Wong OK. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lau LT, Fung YW, Wong FP. A real-time PCR for SARS-coronavirus incorporating target gene pre-amplification. Biochem Biophys Res Commun. 2003;312:1290–1296. doi: 10.1016/j.bbrc.2003.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang SS, Chen TC, Yang JY. Sensitive and quantitative detection of severe acute respiratory syndrome coronavirus infection by real-time nested polymerase chain reaction. Clin Infect Dis. 2004;38:293–296. doi: 10.1086/380841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu AC. The difficulties of testing for SARS. Science. 2004;303:469–471. doi: 10.1126/science.303.5657.469. [DOI] [PubMed] [Google Scholar]
- 70.Ng EK, Ng PC, Hon KL. Serial analysis of the plasma concentration of SARS coronavirus RNA in pediatric patients with severe acute respiratory syndrome. Clin Chem. 2003;49:2085–2088. doi: 10.1373/clinchem.2003.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bressler AM, Nolte FS. Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:987–991. doi: 10.1128/JCM.42.3.987-991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drosten C, Chiu LL, Panning M. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol. 2004;42:2043–2047. doi: 10.1128/JCM.42.5.2043-2047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emery SL, Erdman DD, Bowen MD. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–316. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poon LL, Chan KH, Wong OK. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poon LLM, Wong BW, Chan KH. A one step quantitative RT-PCR for detection of SARS coronavirus with an internal control for PCR inhibitors. J Clin Virol. 2004;30:214–217. doi: 10.1016/j.jcv.2003.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grant PR, Garson JA, Tedder RS, Chan PK, Tam JS, Sung JJ. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med. 2003;349:2468–2469. doi: 10.1056/NEJM200312183492522. [DOI] [PubMed] [Google Scholar]
- 77.Ng LF, Wong M, Koh S. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol. 2004;42:347–350. doi: 10.1128/JCM.42.1.347-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng VC, Hung IF, Tang BS. Viral replication in the nasopharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;38:467–475. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu HS, Chiu SC, Tseng TC. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg Infect Dis. 2004;10:304–310. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 81.Ho TY, Wu SL, Cheng SE, Wei YC, Huang SP, Hsiang CY. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem Biophys Res Commun. 2004;313:938–947. doi: 10.1016/j.bbrc.2003.11.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, Wen J, Li J. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin Chem. 2003;49:1989–1996. doi: 10.1373/clinchem.2003.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi Y, Yi Y, Li P. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J Clin Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woo PC, Lau SK, Tsoi HW. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Hoek L, Pyrc K, Jebbink MF. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orellana C. Laboratory-acquired SARS raises worries on biosafety. Lancet Infect Dis. 2004;4:64. doi: 10.1016/S1473-3099(04)00911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.WHO WHO post-outbreak biosafety guidelines for handling of SARS-CoV specimens and cultures (published Dec 18, 2003) http://www.who.int/csr/sars/biosafety2003_12_18/en/ (accessed Sept 22, 2004)
- 88.WHO Investigation into China's recent SARS outbreak yields important lessons for global public health (published July 2, 2004) http://www.wpro.who.int/sars/docs/update/update_07022004.asp (accessed 22 Sept, 2004)
- 89.WHO First data on stability and resistance of SARS coronavirus compiled by members of WHO laboratory network (published May 4, 2003) http://www.who.int/csr/sars/survival_2003_05_04/en/index.html (accessed Sept 22, 2004)
- 90.Rabenau HF, Cinatl J, Morgenstern B, Bauer G, Preiser W, Doerr HW. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol (Berl) April 29, 2004 doi: 10.1007/s00430-004-0219-0. Published online; doi:10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Normile D. Viral DNA match spurs China's civet roundup. Science. 2004;303:292. doi: 10.1126/science.303.5656.292. [DOI] [PubMed] [Google Scholar]