Abstract
To predict lung nodule malignancy with a high sensitivity and specificity for low dose CT (LDCT) lung cancer screening, we propose a fusion algorithm that combines handcrafted features (HF) into the features learned at the output layer of a 3D deep convolutional neural network (CNN). First, we extracted twenty-nine handcrafted features, including nine intensity features, eight geometric features, and twelve texture features based on grey-level co-occurrence matrix (GLCM) averaged from five grey levels, four distances and thirteen directions. We then trained 3D CNNs modified from three 2D CNN architectures (AlexNet, VGG-16 Net and Multi-crop Net) to extract the CNN features learned at the output layer. For each 3D CNN, the CNN features combined with the 29 handcrafted features were used as the input for the support vector machine (SVM) coupled with the sequential forward feature selection (SFS) method to select the optimal feature subset and construct the classifiers. The fusion algorithm takes full advantage of the handcrafted features and the highest level CNN features learned at the output layer. It can overcome the disadvantage of the handcrafted features that may not fully reflect the unique characteristics of a particular lesion by combining the intrinsic CNN features. Meanwhile, it also alleviates the requirement of a large scale annotated dataset for the CNNs based on the complementary of handcrafted features. The patient cohort includes 431 malignant nodules and 795 benign nodules extracted from the LIDC/IDRI database. For each investigated CNN architecture, the proposed fusion algorithm achieved the highest AUC, accuracy, sensitivity, and specificity scores among all competitive classification models.
Keywords: Lung Nodule Malignancy, convolutional neural network, handcrafted feature, fusion algorithm, Radiomics
I. INTRODUCTION
Lung cancer is the leading cause of cancer-related death in the United States and China. Early detection and diagnosis improve the prognosis for patients with early stage lung cancer treated with surgical resection. The landmark national lung screening trial (NLST) has shown that low-dose computed tomography (LDCT) screening reduces lung cancer mortality by 20% compared to chest radiography (National Lung Screening Trial Research et al., 2011). As more evidence on the benefits of LDCT screening emerges, the U.S. Preventive Services Task Force has recommended “annual screening for lung cancer with LDCT in adults aged 55 to 80 years who have a 30 pack-year smoking history and currently smoke or have quit within the past 15 years” (Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010)(Cho et al., 2010). The Centers for Medicare & Medicaid Services (CMS) have also determined that the evidence is sufficient to add annual screening for lung cancer with LDCT for appropriate beneficiaries. While LDCT screening has demonstrated a 20% survival benefit over chest radiography, the overall false positive rate in NLST was high (26.6%), and the positive predictive value was low (3.8%) (National Lung Screening Trial Research et al., 2011; Pinsky et al., 2015). False positive tests may lead to anxiety, unnecessary and potentially harmful additional follow-up diagnostic procedures, and associated healthcare costs. A reliable strategy is needed to reduce false-positive rates, unnecessary biopsies, and ultimately, patient morbidity and healthcare costs.
To reduce the high false-positive rate in LDCT lung cancer screening, the American College of Radiology developed a new classification scheme named Lung CT Screening Reporting and Data System (Lung-RADS) that 1) increases the size threshold to classify a nodule as positive from 4 mm to 6 mm and 2) requires growth for pre-existing nodules. While applying the Lung-RADS criteria to the NLST data greatly reduced the false-positive rate, it also reduced the baseline sensitivity by nearly 9%, adversely affecting the benefit of LDCT screening to reduce mortality (Pinsky et al., 2015). With the aim of obtaining high true-positive rate and true-negative rate by the computer-aided techniques, a number of algorithms were proposed to automatically characterize the lung nodules (Chan et al., 2008; Armato et al., 2003). In the recent years, the radiomics-based approaches were proposed to analyze various diseases based on medical imaging, presenting a promising way for lesion malignancy classification (Zhang et al., 2017; Sutton et al., 2016; Gillies et al., 2016; Hao et al., 2018). By extracting and analyzing large amounts of quantitative features from medical images, radiomics can build a predictive model by machine learning algorithms to support clinical decisions. However, even the model that achieved the highest accuracy (80.12%) using a 10-folder cross validation still had a relatively low sensitivity at 0.58. These findings indicate the difficulty of predefining quantitative features that fully reflect the unique characteristics of a particular lesion (Audrey G. Chung, 2015). Thus, developing an effective model based on other input is needed so that only patients with a high probability of developing malignancies undergo additional imaging and invasive testing.
Deep learning has achieved great success in various applications in computer vision (Ding and Tao, 2018) and medical imaging processing and analysis (Fu et al., 2018; Huang et al., 2018b; Zhu et al., 2018a; Parisot et al., 2018). In a recent lung cancer detection challenge organized by Kaggle, most top-scored models were based on a deep convolutional neural network (CNN). Unlike handcrafted feature-based classifiers (Zhou et al., 2017; Liu et al., 2017; Wimmer et al., 2016; Lian et al., 2016; Vallières et al., 2015; Parmar et al., 2015; Namburete et al., 2015), CNN-based classifiers (Kooi et al., 2017; Shin et al., 2016; Roth et al., 2016; Zhu et al., 2018b) use the original images as input and learn features automatically to classify, eliminating the need to extract predefined features. In general, to obtain a good classification performance, CNN requires a large scale annotated dataset to learn the representative nature of a lesion by training a large number of parameters (Miotto et al., 2018). Many successful applications of CNN have used more than 100,000 samples, such as ImageNet with millions of images (Krizhevsky et al., 2017), the skin cancer dataset with 129,450 images (Esteva et al., 2017), and the retinal fundus photographs dataset with 128,175 images (Gulshan et al., 2016). For many other medical problems, obtaining such a large annotated dataset is still challenging. This challenge is commonly surmounted through transfer learning, which fine-tunes a CNN model pre-trained on a large scale dataset (Kermany et al., 2018; Shin et al., 2016; Akcay et al., 2018).
However, for medical imaging with the three dimensional (3D) tensor form, such as CT, transfer learning is not optimal, as most of large datasets are 2D, and there lacks a large scale 3D dataset with pre-trained 3D CNN architecture. Using a conventional classifier (such as SVM) with the CNN features as input is another common technique to improve the performance of CNNs (Shen et al., 2015; Shen et al., 2017). This technique is available for medical imaging with the 3D tensor form, however it can’t solve the challenge intrinsically because the natural representation of a lesion can’t be learned well by CNNs without a large scale annotated training dataset.
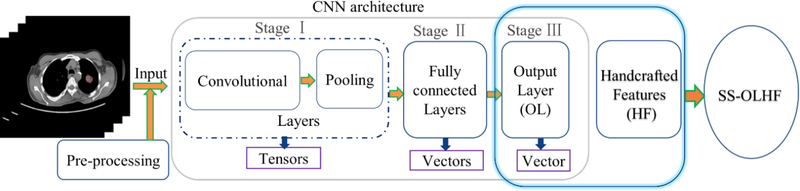
Currently, most models use either handcrafted features or features learned based on CNN alone. Model performance can be improved by incorporating visual CT findings or domain-knowledge based features into construction of predictive models (Shi et al., 2008; Causey et al., 2018). On one hand, a combining algorithm could overcome the disadvantage of the handcrafted features’ inability to fully reflect the unique characteristics of a particular lesion by combining the intrinsic CNN features. On the other hand, it could alleviate the requirement of a large scale annotated dataset for the CNN because of the complement of handcrafted features for the CNN features. In general, the architecture of a typical CNN (Fig. 1) is structured as a series of stages: 1) convolutional layers and pooling layers with tensor output; 2) hidden fully connected layers with vector output; and 3) an output layer with vector output (LeCun et al., 2015). In most existing combination methods, the representation learned at the final hidden fully connected layer is combined into the handcrafted features, improving the performance of both the handcrafted features and the CNN (Antropova et al., 2017). Causey et al. also proposed a fusion algorithm based on the random forest classifier by combining the CNN presentation learned at the last hidden fully connected layer before the output layer into the handcrafted features, achieving high accuracy for lung nodule malignancy prediction (Causey et al., 2018). However, as pointed out by Y. LeCun and G. Hinton, “deep-learning methods are representation-learning methods with multiple levels of representation, obtained by composing simple but non-linear modules that each transform the representation at one level (starting with the raw input) into a representation at a higher, slightly more abstract level” (LeCun et al., 2015). This indicates that the representation learned at the output layer is at a higher level and more abstract than the representation learned at the final hidden fully connected layer. Thus, a fusion algorithm could achieve better performance by combining handcrafted features into the CNN representation learned at the output layer, instead of the final hidden fully connected layer. To the best of our knowledge, this is the first attempt to explore a fusion algorithm between the CNN features at the output layer and the handcrafted features.
Fig. 1.
Illustration of the proposed SS-OLHF algorithm by fusing the highest level CNN representation learned at the output layer (OL) of a 3D CNN into the handcrafted features (HF). In the CNN architecture, Stage I mainly includes convolutional layers and pooling layers with tensor output; Stage Ⅱ mainly includes hidden fully connected layers with vector output; Stage Ⅲ is the output layer with vector output.
Specifically, we propose a fusion algorithm (SS-OLHF) (Fig. 1) that combines the highest level CNN representation learned at the output layer (OL) of a 3D CNN into the domain knowledge, i.e. handcrafted features (HF), using the support vector machine (SVM) coupled with the sequential forward feature selection method (SFS) to select the optimal feature subset and construct the final classifier. The proposed fusion algorithm could lead to better performance in differentiating malignant and benign lung nodules for LDCT lung cancer screening.
II. Materials
We downloaded the Lung Image Database Consortium and Image Database Resource Initiative (LIDC/IDRI) (Armato et al., 2011) (http://www.via.cornell.edu/lidc) to evaluate the proposed fusion classifiers. This dataset includes 1,010 cases, each of which includes images from a clinical thoracic CT scan and an associated XML file that records the annotations from four radiologists. 7,371 lesions were marked “nodule” by at least one of the four radiologists, and 2,669 of those nodules had sizes equal to or larger than 3 mm and were rated with malignancy suspiciousness from 1 to 5 (1 indicates the lowest malignancy suspiciousness, and 5 indicates the highest malignancy suspiciousness).
We considered all nodules with sizes equal to or larger than 3 mm. In total, 2,340 nodules were considered. For each nodule, the malignancy suspiciousness rate was the average value of all rates given by all radiologists who outlined the nodule. By removing ambiguous nodules with malignancy suspiciousness rated at 3, we obtained a total of 431 malignant nodules (average rating>3) and 795 benign nodules (average rating <3) to evaluate our models’ performance. Finally, for each nodule, the 3D nodule region of interest (ROI) was extracted based on the contour given by the radiologist who gave the malignancy suspiciousness rate closest to the average. For all nodules considered in this study, the measurement conditions including pixel size in transaxial image, slice thickness, reconstruction interval and nodule volume are summarized in appendix Table A1. The difference between the reconstruction interval and the slice thickness (DIT) are presented in appendix Table A2, where DIT<0 implies that there is an overlap between neighboring slices of reconstructed CT.
III. Methods
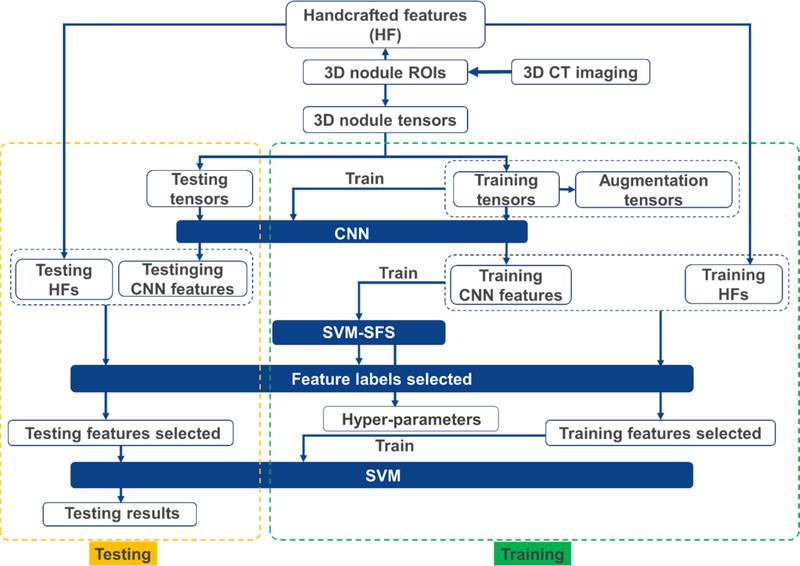
3.1. The workflow of the fusion algorithm SS-OLHF
The overall workflow of the fusion algorithm SS-OLHF is illustrated in Fig. 2. A 5-fold cross validation method was employed in this work, where 3 folds were used for training, one fold was used for validation and the remaining fold was set aside for testing. For each nodule, the handcrafted features were extracted and the nodule tensor was constructed based on the segmented 3D nodule ROI. The augmentation processing was performed on the training nodule tensors by rotating and flipping. The 3D tensor, including training tensors and augmentation tensors with the same size, were used as the input for training the 3D CNN to train CNN. Then, the CNN features learned at the output layers before softmax operation were extracted based on the trained CNN. We obtained the fusion features by combining the CNN features into the handcrafted features. We used SVM with radial basis function kernel coupled with SFS to obtain the optimal fusion feature labels selected and hyper-parameters of SVM, based on the training samples only. Once the SVM model was fixed with the hyper-parameters based on validation fold, it was applied on the test cases using optimal fusion feature subset from the testing samples.
Fig. 2.
The overall workflow of the fusion algorithm SS-OLHF.
3.2. Handcrafted feature extraction
In this study, we combined the handcrafted features into the highest level CNN features learned at the output layer with high abstract. Imaging features, including intensity, geometric and texture features, extracted from contoured nodules (Zhou et al., 2017) were used to construct the fusion algorithm. The following intensity features were extracted based on the intensity histogram: minimum, maximum, mean, standard deviation, sum, median, skewness, kurtosis, and variance. Geometric features associated with a nodule were volume, major diameter, minor diameter, eccentricity, elongation, orientation, bounding box volume, and perimeter. To obtain high level texture features, all 3D nodule ROIs were first interpolated to a fixed resolution with 0.5 mm/voxel along three axes using a 3D spline interpolation method. Thirteen 3D gray-level co-occurrence matrices (GLCMs) (Davis L.S., 1979) were constructed based on these interpolated ROIs at 13 standard 3D directions in 3D linear space, respectively, denoted as [0 1 0], [−1 1 0], [−1 0 0], [−1 −1 0], [0 1 −1], [0 0 −1], [0 −1 −1], [−1 0 −1], [1 0 −1], [−1 1 −1], [1 −1 −1], [−1 −1 −1], [1 1 −1], for a particular grey levels and displacement distance. The following twelve texture features were obtained from the GLCM by averaging the above thirteen GLCMs: energy, entropy, correlation, contrast, texture variance, sum-mean, inertia, cluster shade, cluster prominence, homogeneity, max-probability, and inverse variance. Four displacement distances 1, 2, 3, 4 and five grey levels 8, 16, 32, 64, 128 were used construct different groups to extract 20 groups of texture features. The averaging texture features over such 20 groups of texture features were used to construct the fusion algorithm SS-OLHF.
3.3. Nodule tensor construction for CNNs
After all 3D nodule ROIs were interpolated to a fixed resolution with 0.5 mm/voxel along three axes using a 3D spline interpolation method, training data were augmented by 2D rotation of [90°, 180°, 270°] along all three axes and flipping along all three coordinate planes. Subsequently, for each interpolated nodule ROI and augmentation data, we constructed the 3D nodule tensor whose center the tumors were located in and whose periphery zeros were filled in. Each nodule tensor had the same size of 105×97×129, determined by the biggest nodule size among all interpolated nodule ROIs and augmentation data. We used these nodule tensors from all interpolated nodule ROIs and augmentation data as the input for all CNNs to train the CNN architectures and extract the CNN features.
3.4. Convolutional neural network
3.4.1. 3D CNN architectures
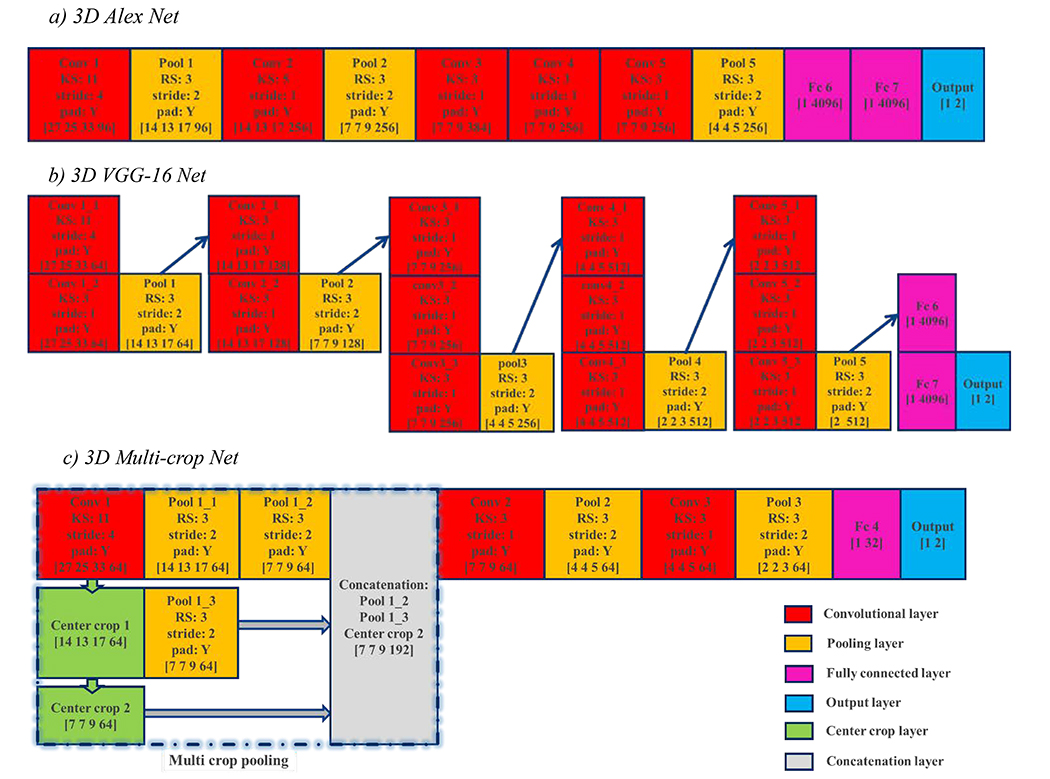
Because nodule imaging has an intrinsic 3D tensor structure, the 3D CNN architectures with the 3D nodule tensors as input were trained to extract the CNN features for the proposed fusion algorithm. This study used three 3D CNN architectures, which were modified from two 2D CNN architectures (AlexNet (Krizhevsky et al., 2012) and VGG-16 Net (Simonyan and Zisserman, 2014)) and one recently developed 2D CNN architecture dedicated to classifying lung malignancy (Multi-crop Net (Shen et al., 2017)). Unlike the original 2D CNN architectures, which use 2D convolutional kernels and 2D pooling, our CNN architectures with input in the form of 3D nodule tensors use 3D convolutional kernels to perform 3D convolution in all convolutional layers and 3D max-pooling in all pooling layers. Meanwhile, the 3D CNNs preserve structures, such as layer number at every stage and unit number at every layer (except the output layer with two units), the stride size in most convolutional and pooling layers, and padding processing. The three 3D CNN architectures with corresponding details are shown in Fig. 3 and explained below.
Fig. 3.
Three 3D CNN architectures modified from two widely used 2D architectures and one 3D CNN dedicated to lung nodule malignancy classification. The vector at bottom of each rectangle indicates the size of the output tensor at this layer for a patient case, with the last number being the unit number. The workflow is from top to bottom and left to right on adjacent rectangles, and along the arrow direction on non-adjacent rectangles. KS: kernel size; RS: region size.
a). 3D AlexNet
The first 3D CNN architecture is modified from the AlexNet, which achieved significant improvement over other non-deep learning methods for ImageNet Large Scale Visual Recognition (ILSVRC) 2012 (Krizhevsky et al., 2012). This 3D AlexNet architecture includes 5 convolutional layers, three max-pooling layers, two hidden fully connected layers, and one output layer with approximately 113 million free parameters, whose details are shown in Fig. 3(a). The features learned at all convolutional layers and max-pooling layers are in the form of 4D tensors with the size [7 7 9 256] at the final convolutional layer and [4 4 5 256] at the final max-pooling layer. All features learned at the hidden fully connected layers and the output layer are in the form of vectors with the size [1 4096] at the final hidden fully connected layer and [1 2] at the output layer.
b). 3D VGG-16 Net
The second 3D CNN architecture is modified from the 16-layer 2D CNN developed by the visual geometry group (VGG-16) at the University of Oxford for ImageNet Large Scale Visual Recognition (ILSVRC) 2014 (Simonyan and Zisserman, 2014). This architecture consists of five stacks of convolution-pooling operation, in which each max-pooling layer is tailed by a few convolutional layers (Fig. 3(b)). Overall, it includes thirteen convolutional layers, five max-pooling layers, two hidden fully connected layers, and one output layer with approximately 65 million free parameters. Unlike the original 2D CNN architecture that used all 2D kernels with size 3 and stride 1 in convolutional layers, the first convolutional layer uses 3D kernels with size 11 and stride 4, so that the large input size (105×97×129) works with the available memory of our computer with the modified 3D VGG-16. The features learned at the final convolutional layer, final max-pooling layer, final hidden fully connected layer, and output layer are in the forms of a 4D tensor with size [2 2 3 512], a 2D tensor with size [2 512], a vector with size [1 4096], and a vector with size [1 2], respectively.
c). 3D Multi-crop Net
The third 3D CNN architecture (Fig. 3(c)) is based on a recently developed multi-crop CNN dedicated to lung malignancy classification (Shen et al., 2017). The multi-crop CNN extracts salient nodule information using a multi-crop pooling strategy that crops center regions from convolutional feature maps and then applies max-pooling at different times. The multi-crop CNN outperforms other state-of-the-art models for classifying lung nodule malignancies, so we chose it as an improved 3D CNN architecture to evaluate our fusion algorithm. In this 3D CNN architecture, which has approximately 0.5 million free parameters, the multi-crop pooling strategy is used after the first convolutional layer, and there are two additional convolutional layers, two max-pooling layers, one hidden fully connected layer, and one output layer. Like the 3D CNN modified from VGG-16 Net, the kernel size is 11 and the stride is 4 at the first convolutional layer. The features learned at the final convolutional layer, final pooling layer, final hidden fully connected layer, and the output layer are in the form of a 4D tensor with size [4 4 5 64], a 4D tensor with size [2 2 3 64], a vector with size [1 32], and a vector with size [1 2], respectively.
3.4.2. CNN training procedures
For binary classification, the output of the CNN is a two dimensional vector (y0, y1) for each sample with label, where q equals either 0 or 1. The softmax of (y0, y1), defined as
| (1) |
indicates the probability distribution over the two classes. The networks were trained by minimizing the loss function defined by averaging the cross entropy along each batch with size, as follows:
| (2) |
We used the Adam optimization algorithm, based on a first-order gradient (Kingma and Ba, 2014), to optimize the objective function (2). We used minibatch Stochastic Gradient Descent (SGD) to compute the gradient in small batches for the available memory of our computer. The batch size was set to 70. Then, we used the standard backpropagation to adjust weights in all layers. We used Layer (Channels) normalization, introduced by Ba et al. (Jimmy Lei Ba, 2016), to normalize the input to one nonlinear output based on the mean and standard deviation over all channels (units) in a layer, as follows,
| (3) |
where g was the standard rectified linear unit (ReLU), W was the learned weight in this layer, and the extra parameters γ and β need to be trained during training the CNN. Dropout regularization was used for all hidden fully connected layers with the dropout ratio set to 0.9. The changing learning rate was initially set to 0.005 for the first epoch, 0.001 from the second epoch to the fourth epoch, 0.0005 from the fifth epoch to the eighth epoch, and after the ninth epoch. The training processing stopped automatically when the loss function value achieved 0.01 and iterative steps achieved 100 epochs for each CNN. To deal with the data imbalance problem in training CNNs, we selected part of the majority augmentation data rather than all majority augmentation data to obtain the balanced binary training samples.
3.5. Feature selection and classification
The fusion features (Table 1) comprise 29 handcrafted features, as described in section 3.2, and 2 CNN features learned at the output layer before softmax operating, which are the highest level representation learned by CNN. For each testing sample, one of these 2 CNN features is directly connected with the prediction probability belonging to positive class, which is denoted as CNN featureP, and the other one is directly connected with the prediction probability belonging to negative class, which is denoted as CNN featureN. Because of the redundancy and similarity among these fusion features, feature selection is needed to improve the model performance. As a traditional feature selection method, the sequential forward feature selection (SFS) method (Kohavi and John, 1997) coupled with the SVM classifier (SS) was used to select the feature subset. We used the area under the receive operating characteristic (ROC) curve (AUC) by a 5-fold cross validation method in the training dataset as the criterion to select the optimal feature subset. Finally, the SVM based classifier with RBF kernel was further trained on the training dataset and classified the testing dataset using the selected optimal feature subset as input.
Table 1.
The fusion features used in our fusion algorithms
| Intensity features | Geometry features | Texture features | CNN features |
|---|---|---|---|
| Minimum | Volume | Energy | CNN featureP |
| Maximum | Major diameter | Entropy | CNN featureN |
| Mean | Minor diameter | Correlation | |
| Stand deviation | Eccentricity | Contrast | |
| Sum | Elongation | Texture Variance | |
| Median | Orientation | Sum-Mean | |
| Skewness | Bounding box volume | Inertia | |
| Kurtosis | Perimeter | Cluster Shade | |
| Variance | Cluster Prominence | ||
| Homogeneity | |||
| Max-Probability | |||
| Inverse Variance |
IV. Experimental Setup
4.1. Methods for comparative testing
For each CNN architecture investigated, we first compared the original 2D CNN and the modified 3D CNN with our proposed fusion algorithm SS-OLHF. We also compared our algorithm with other two conventional CNN feature based methods that utilize the features learned at the final hidden fully connected layer (FFL).
S-FFL. One common CNN feature based method first extracts the CNN features learned at FFL and then uses a third classifier to perform the classification (Shen et al., 2015; Shen et al., 2017). In this method, the 3D CNN features learned at FFL were extracted first, the variance analysis method selected the CNN feature subset to be used as input of a third classifier by removing features with a variance smaller than the mean variance, and SVM with RBF kernel was used to train the classifier and perform the classification.
S-FFLHF. Conventional fusion algorithms for combining handcrafted features (HF) and CNN extractedfeatures usually use the CNN features learned at the FFL (Antropova et al., 2017), rather than the output layer. We compared our fusion algorithm with this fusion strategy that combines the CNN features learned at FFL into 29 handcrafted features. We used ReliefF (Kononenko et al., 1997), the classical feature selection method, to obtain the optimal feature set based on the training dataset. The SVM model with an RBF kernel was used to construct the classifier.
Additionally, we compared our fusion algorithm with three other state-of-the-art methods that use different dataforms as input. Firstly, the original experimental method of Multi-crop Net that used the nodule patch without segmentation as input (oriMulti-crop) was compared with our proposed fusion algorithm using the segmented 3D nodule ROI as input. To show that the CNN features can be as the complement of handcrafted features to improve the classification accuracy, the same classifier and feature selection processing using handcrafted features (SS-HF) only as input was compared with our fusion algorithms. The third method compared with our fusion algorithms is the support tensor machine with the 3D nodule tensor as input. These three compared approaches are descripted in detail as following:
oriMulti-crop. Different from the our proposed fusion algorithm using segmented 3D nodule ROIs, the original experimental method of Multi-crop net (oriMulti-crop) used 3D nodule patches without ROI segmentation as the input (Shen et al., 2017). The experimental results show that the oriMulti-crop outperforms other state-of-art models in classifying lung nodule malignancies. We compared results from the oriMulti-crop with the 3D nodule patches covering the nodule as the input with our fusion algorithm, using the nodule patch size 32*32*32 with the same resolution and the same training protocols for both algorithms.
SS-HF. We also evaluated our fusion algorithm by comparing it with the classification using just the 29 handcrafted features with the same feature selection processing (SFS) and classifier training method (SVM).
STM. The support tensor machine (STM) (Tao et al., 2007), which uses a high order tensor as input, is a common tensor space model and has been applied successfully to pedestrian detection, face recognition, remote sensors, and medical imaging analysis (Guo et al., 2016; Biswas and Milanfar, 2017; Li et al., 2018). The 3D STM uses the 3D nodule tensors directly as input, so it doesn’t require the extraction of predefined features. Moreover, the parameters needed to train are much smaller than with CNN. When we compared STM with our proposed algorithm, we used all original 3D nodule tensors without augmentation tensors to train the STM model to perform classification.
4.2. Experimental setting
To solve the class imbalance problem, for the vector space models, we used the Synthetic Minority Over-sample Technique (SMOTE) (Chawla et al., 2002) to generate a synthetic vector sample based on minority class information to augment the decision region of the minority class using the K-nearest neighborhood (KNN) graph based on Euclidean distance. For all CNNs, we handled the imbalance problem in training processing by randomly selecting part of the augmentation tensors in the majority class and all of the augmentation tensors in the minority class to obtain a balanced training dataset. For the STM, we used the original nodule tensors to train the model, randomly selecting part of the nodule tensors in the majority class and all of the nodule tensors in the minority class to obtain a balanced training dataset.
We employed a 5-fold cross validation (CV) method to evaluate the performance of the different classifiers. All samples were randomly partitioned into 5 subsets with a size of either 245 or 246. These five subsets were fixed for each method investigated in this study, where one subset was used as the testing subset and the rest were used as training data. Within the features selection processing, we employed again the 5-fold CV method on training set to select the feature subset which consisted of all features selected for at least four times in such 5-fold CV experiments. The classification models were then trained on all samples in the training set using this selected feature subset. Finally, the trained models performed classification on the testing subset.
For each testing subset, we calculated the AUC, classification accuracy, sensitivity, and specificity. We used the average results and standard deviations from one round of 5-fold experiments which are equivalent to 5 independent experiments, as the evaluation criteria. The receiver operating characteristic curve (ROC) is also used to analyze the performance of different predicting algorithms. Additionally, the ISO-accuracy lines (Nicolas Lachiche, 2003) are used, defined as
| (4) |
where TPr and FPr are the true positive rate and false positive rate, respectively, P(N) and P(P) are the probability of the positive class and the probability of the negative class, respectively, and a is a variable. The ISO-accuracy lines are a family of lines that are parallel, i.e. that have the same slope. The point on the ROC curve where the ISO-accuracy line is tangent obtains the optimal probability threshold to obtain the optimal accuracy. Therefore, we also used the ISO-accuracy line tangent to the ROC curve to evaluate the performance of all classification approaches in this study.
V. Results
5.1. Comparison with CNN-based methods
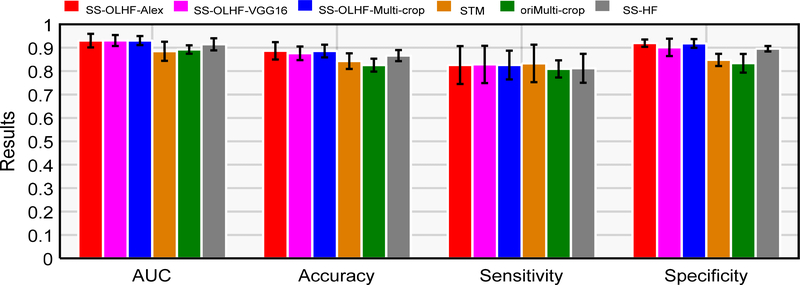
The results comparing our fusion algorithm with different CNN architectures and other methods based on CNN are summarized in Table 2. For each CNN architecture, the 3D CNN obtained better results than the 2D CNN. SVM with the CNN features learned at the final hidden fully connected layer as input (S-FFL) improved the performance of 3D CNNs in terms of AUC, classification accuracy, sensitivity, and specificity. The conventional fusion algorithm combining the CNN features learned at the final hidden fully connected layer into handcrafted features (S-FFLHF) obtained better results than S-FFL in most of cases, but not for all four evaluated metrics of the three CNN architectures. Our proposed fusion algorithm obtained the best performance for every CNN architecture. The best
Table 2.
Performance of different predictive models based on CNNs
| Methods | 2D | 3D | S-FFL | S-FFLHF | SS-OLHF | |
|---|---|---|---|---|---|---|
| AlexNet | AUC(%) | 88.87±2.87 | 90.56±1.81 | 91.13±1.70 | 91.25±1.87 | 93.03±2.92 |
| ACC(%) | 84.67±2.01 | 84.99±1.86 | 86.05±1.87 | 86.88±0.40 | 88.66±3.72 | |
| SEN(%) | 78.60±7.42 | 80.88±5.15 | 81.99±4.28 | 81.06±1.81 | 82.60±8.09 | |
| SPE(%) | 88.10±2.84 | 87.30±1.99 | 88.29±1.22 | 90.11±0.52 | 91.95±1.58 | |
| VGG16 Net | AUC(%) | 86.22±3.26 | 90.34±4.00 | 91.76±3.01 | 91.05±3.17 | 93.07±2.33 |
| ACC(%) | 85.73±2.45 | 86.13±2.84 | 87.03±2.47 | 85.94±0.45 | 87.60±2.91 | |
| SEN(%) | 80.05±8.08 | 80.29±4.36 | 80.71±7.20 | 80.84±2.95 | 82.85±7.97 | |
| SPE(%) | 88.81±3.88 | 89.30±2.20 | 90.60±2.08 | 88.81±1.21 | 90.14±3.71 | |
| Multi-crop Net | AUC(%) | 89.18±3.14 | 90.48±3.51 | 90.86±2.95 | 92.70±2.34 | 93.06±1.92 |
| ACC(%) | 85.73±2.64 | 86.46±2.53 | 86.62±1.61 | 86.05±0.80 | 88.58±2.70 | |
| SEN(%) | 80.44±6.57 | 81.91±6.28 | 82.28±5.14 | 80.37±2.08 | 82.60±6.13 | |
| SPE(%) | 88.70±1.79 | 88.93±1.88 | 89.06±0.73 | 90.37±2.71 | 91.82±1.86 |
Note. ACC is accuracy, SEN is sensitivity and SPE is specificity, which are available for following tables.
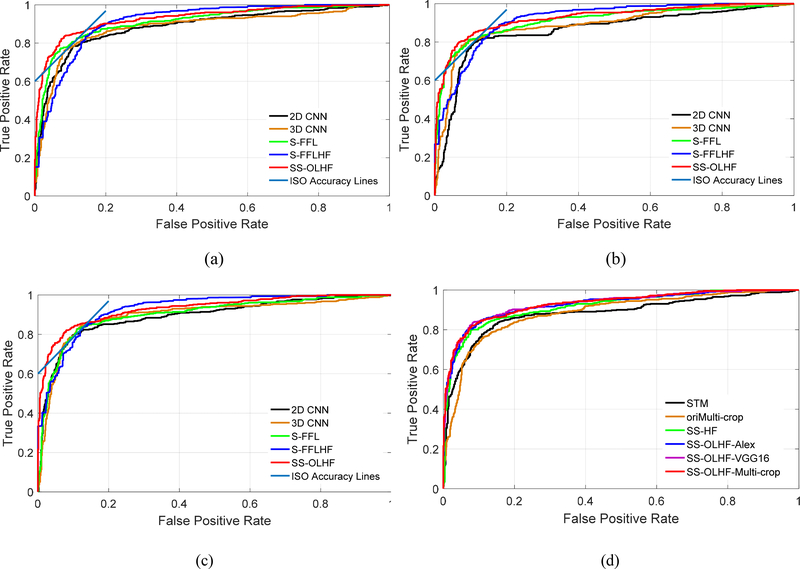
The ROC curves of our proposed algorithm always lie above the ROC curves of the 2D CNN, 3D CNN, and S-FFL, which indicates that our proposed fusion algorithm outperforms the 2D CNN, 3D CNN, and S-FFL for all three CNN architectures (Fig. 4 (a)–(c)). The ISO-accuracy line tangent to ROC for our fusion algorithm is above and to the left of the conventional fusion algorithm for all three investigated CNN architectures (Fig. 4 (a)–(c)). This indicates that our proposed fusion algorithm obtains a better TPr and smaller FPr than the conventional fusion algorithm S-FFLHF, though the ROC curve of our fusion algorithms doesn’t always lie above the ROC curve of S-FFLHF.
Fig. 4.
The ROC curves for the compared methods: (a) five methods based on AlexNet architecture; (b) five methods based on VGG-16 Net architecture; (c) five methods based on Multi-crop Net architecture; (d) six methods, including our three fusion algorithms, based on different CNN architectures and three conventional approaches with different data forms as input.
Our proposed fusion algorithms based on three CNN architectures yield results with small differences (Table 2). The fusion algorithm based on Multi-crop net has steadier results than the other algorithms, because its standard deviations (AUC: 1.92; ACC: 2.70; SEN: 6.13; SPE: 1.86) for five independent experiments are the smallest. It obtained results of 0.9306, 88.58%, 82.60%, and 91.82% for AUC, accuracy, sensitivity, and specificity, respectively.
5.2. Comparison with other state-of-the-art approaches
Table 3 shows the results obtained from the three state-of-the-art approaches with different forms of data as input for modeling and testing, as described in section 4.1. All of the fusion algorithms with three CNN architectures outperformed the other three competitive approaches (Fig. 4 (d) and Fig. 5). The corresponding p-values in T-test for AUC and accuracy and in bivariate Chi-Square test by ROCKit software for ROC are reported in Table 4 from the 5 independent experimental results compared with the fusion algorithm based on Multi-crop Net as it is steadiest. These results show that the fusion algorithm based on Multi-crop Net is significantly better than the other three methods, as shown by the p-values < 0.05.
Table 3.
Performance of three state-of-the-art approaches
| Methods | AUC(%) | ACC(%) | SEN(%) | SPE(%) |
|---|---|---|---|---|
| oriMulti-crop | 89.24±1.81 | 82.54±2.76 | 80.94±3.65 | 83.36±4.00 |
| SS-HF | 90.45±2.58 | 85.62±2.37 | 81.21±6.20 | 89.56±1.17 |
| STM | 88.47±4.09 | 84.26±3.33 | 83.29±8.02 | 84.78±2.60 |
AUC (=0.9307) was obtained based on VGG-16 Net, the best sensitivity (82.60%) was obtained based on AlexNet and Multi-crop Net, and the best specificity (91.95%) and classification accuracy (88.66%) were obtained based on AlexNet.
Fig. 5.
The comparison results among our three fusion algorithms and other three state-of-the-art approaches.
Table 4.
P-values in the unpaired T-test for AUC and ACC and in bivariate Chi-Square test by ROCKit software for ROC between our fusion algorithm based on Multi-crop net and the other three methods
| Methods | AUC | ACC | ROC |
|---|---|---|---|
| oriMulti-crop | 0.0135 | 0.0161 | <10−4 |
| SS-HF | 0.0493 | 0.0004 | 0.0058 |
| STM | 0.0462 | 0.0275 | 10−4 |
5.3. Features selected by the fusion algorithms
The features selected in each testing fold by our fusion algorithm with three CNN architectures are shown in Table 5. The fusion algorithm based on AlexNet selected 5 optimal features: 2 intensity features, one texture feature, one geometry feature, and one CNN feature. The fusion algorithm based on VGG16 selected 8 features: 4 intensity features, one texture feature, 2 geometry features, and one CNN feature. The fusion algorithm based on Multi-crop Net selected 9 features: 2 intensity features, 4 texture features, 2 geometry features, and one CNN feature. Variance as the intensity feature, contrast as the texture feature, minor diameter as geometry feature, and CNN featureP were selected by all three fusion algorithms, which indicates the complementarity among these three types of handcrafted features and CNN features.
Table 5.
The features that are selected by our fusion algorithms
| The fusion algorithms | Intensity features | Geometry features | Texture features | CNN features |
|---|---|---|---|---|
| SS-OLHF-Alex | Stand deviation Variance |
Minor diameter | Contrast | CNN featuresP |
| SS-OLHF-VGG16 | Maximum Stand deviation Sum Variance |
Major diameter Minor diameter |
Contrast | CNN featuresP |
| SS-OLHF-Multi-crop | Sum Variance |
Volume Minor diameter |
Contrast Texture Variance Cluster Shade Inverse Variance |
CNN featuresP |
5.4. The influence of CT reconstruction conditions and nodule size
The influence of nodule volume and CT reconstruction conditions including pixel size, slice thickness and the difference between reconstruction interval and slice thickness (DIT) on the predicted performance is presented Table 6 for six compared methods, including SS-OLHF-Alex, SS-OLHF-VGG16, SS-OLHF-Multi-crop, STM, oriMulti-crop and SS-HF. The nodule size measured by the volume greatly influences the predictive performance, especially for the false positive rates (FPR) on cases with the volume ≥ median and the false negative rates (FNR) on cases with the volume < median. These results suggest that that nodule size is a key factor influencing the malignancy identification. The reconstruction conditions also influence the predicted results. Specifically, the FNRs for all algorithms on cases with pixel size ≥ median are higher than the case with pixel size < median, and the FNRs for all algorithms on cases with slice thickness ≥ median are also higher than cases with slice thickness < median. Meanwhile, the FPRs for all compared algorithms on cases with slice thickness < median are lower than cases with slice thickness ≥ median. These results suggest that predictive performance of these algorithms improves with higher image resolution. Additionally, most FPRs and FNRs for our proposed three fusion algorithms are lower than the other three compared algorithms, which further shows that our proposed fusion algorithms are better than the other three compared algorithms.
Table 6.
False positive rates and false negative rates on different reconstruction conditions and nodule volume.
| Condition | No. | No. | % | SS-OLHF-Alex | SS-OLHF-VGG16 | SS-OLHF-Multi-crop | STM | oriMulti-crop | SS-HF |
|---|---|---|---|---|---|---|---|---|---|
| Pixel size | ≥ Median (620) | 385 | FPR | 8.31 | 8.05 | 7.53 | 11.17 | 18.44 | 8.57 |
| 235 | FNR | 13.62 | 12.34 | 13.62 | 12.34 | 15.74 | 14.47 | ||
| <Median (606) | 410 | FPR | 7.32 | 11.46 | 9.51 | 12.20 | 14.88 | 11.46 | |
| 196 | FNR | 21.94 | 21.43 | 21.94 | 29.08 | 22.96 | 23.98 | ||
| Slice thickness | ≥ Median (707) | 448 | FPR | 9.60 | 10.94 | 9.38 | 14.73 | 16.74 | 10.71 |
| 259 | FNR | 10.04 | 8.49 | 9.65 | 9.65 | 15.06 | 11.58 | ||
| <Median (519) | 347 | FPR | 5.48 | 8.36 | 7.49 | 7.78 | 16.43 | 9.22 | |
| 172 | FNR | 28.49 | 28.49 | 29.07 | 35.47 | 25.00 | 29.65 | ||
| DIT | < 0 (422) | 311 | FPR | 7.85 | 8.70 | 7.66 | 14.08 | 17.18 | 8.49 |
| 111 | FNR | 17.19 | 16.56 | 17.19 | 18.75 | 17.19 | 19.38 | ||
| =0 (804) | 484 | FPR | 7.72 | 11.54 | 9.94 | 8.01 | 15.71 | 12.50 | |
| 320 | FNR | 18.02 | 16.22 | 18.02 | 23.42 | 24.32 | 17.12 | ||
| Volume | ≥ Median (613) | 230 | FPR | 23.91 | 28.26 | 25.22 | 34.35 | 30.00 | 30.87 |
| 383 | FNR | 8.36 | 7.31 | 7.31 | 10.18 | 13.32 | 9.14 | ||
| <Median (613) | 565 | FPR | 1.24 | 2.30 | 1.77 | 2.48 | 11.15 | 1.59 | |
| 48 | FNR | 89.58 | 89.58 | 97.92 | 97.92 | 64.58 | 95.83 | ||
| All | 1226 | 795 | FPR | 7.80 | 9.81 | 8.55 | 11.70 | 16.60 | 10.06 |
| 431 | FNR | 17.40 | 16.47 | 17.40 | 19.85 | 19.03 | 18.79 | ||
Note. The number in each row in the second column presents the number of nodules satisfying the corresponding condition. The third column presents the number of benign nodules (upper) and the number of malignant nodules (bottom) in each condition.
5.5. Complementary characteristics of CNN features and handcrafted features
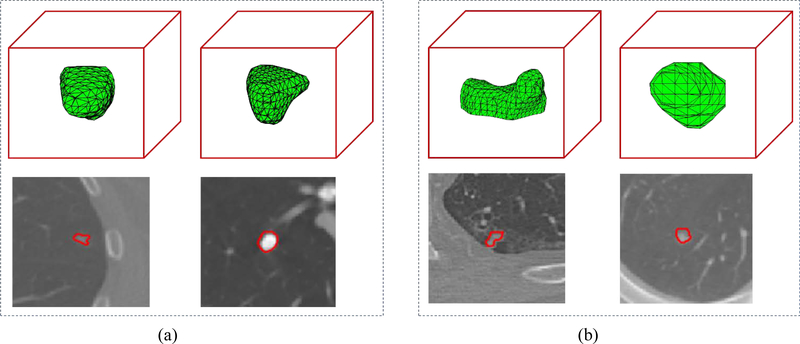
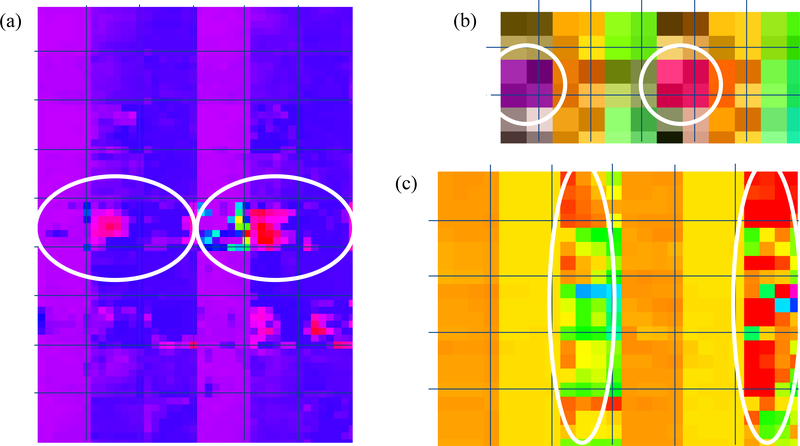
We selected a malignant nodule (Fig. 6(a) left) and a benign nodule (Fig. 6(a) right) to illustrate the complementary characteristics provided by CNN features. These two nodules were predicted correctly by the three fusion algorithms and the three corresponding CNNs but were predicted incorrectly by the SS-HF. For such two nodules and each investigated CNN, three randomly selected 3D CNN feature maps from the last convolutional layer with many channels (Fig. 3) were illustrated in Fig. 7 by unfolding into 2D feature maps in the columns, where the malignant nodule is located in the left tree columns and the benign nodule is located in the right three columns. In Fig. 7, the white circles show the major discriminative information in the CNN feature maps while they can hardly be differentiated from each other by the shapes shown in Fig. 6 (a).
Fig. 6.
Four randomly selected examples for analyzing the complementary characteristics of CNN features (a) and handcrafted features (b).
Fig. 7.
2D illustration by unfolding into the columns for three randomly selected feature maps learned in the last convolutional layers from: (a) Alex Net (a), (b) VGG16 Net and (c) Multi-crop Net. In the (a), (b) and (c), all left three columns are corresponding to the malignant nodule showed in left Fig. 6 (a) and all right three columns are corresponding to the benign nodule showed in right Fig. 6 (a). The white circles show the major discriminative information between the malignant nodule and benign nodule in the feature maps of each CNN: (a) Alex Net (a), (b) VGG16 Net and (c) Multi-crop Net.
To show the complementary characteristics provided by the handcrafted features for the CNN features, two selected nodules were presented in Fig. 6 (b), in which left column shows the malignant nodule while the right column show the benign nodule. Both nodules were predicted correctly by the three fusion algorithms and the SS-HF but predicted incorrectly by the three corresponding CNNs. This could be caused by the limited CNN features learned from the dataset with limited annotated samples, which couldn’t characterize such nodules. The handcrafted features can characterize better them since the shape of the malignant nodule (Fig. 6 (b) left) is more complex than the benign nodule (Fig. 6 (b) right).
VI. Discussions and conclusions
Lung cancer screening based on LDCT has shown to reduce the mortality of lung cancer patients in the NLST but false positive rate is very high. CNN has been a powerful tool in many fields. However, it generally requires a large scale annotated dataset to learn the natural representation of an object. The conventional radiomics method, based on handcrafted features, has performed well for many tasks with relatively small sample size. However, the handcrafted features may not fully reflect the unique characteristics of particular lesions. To solve these problems and to develop a highly sensitive and specific model that differentiates between malignant and benign lung nodules, we proposed a fusion algorithm that incorporates the CNN representation learned at the output layer of a 3D CNN into the handcrafted features, using the feature selection method SFS coupled with SVM selects the optimal feature subset and constructs the classifier. Unlike conventional fusion methods that combine the CNN representation learned at the final hidden fully connected layer into the handcrafted features, our proposed fusion algorithm combines the highest level CNN representation learned at the output layer into the handcrafted features.
We investigated three 3D CNN architectures – AlexNet, VGG-16 Net, and Multi-crop Net – modified from the 2D CNN architectures with different parameter values in our proposed fusion algorithm. The experimental results (Table 2–4) show that the fusion algorithm performed best in all competitive approaches. The fusion algorithm based on Multi-crop Net obtained steadier results than our fusion algorithms based on the other two CNN architectures. As the steadiest fusion algorithm (achieving lowest standard deviation of different evaluation criteria), the fusion algorithm based on Multi-crop Net obtained 82.60% and 91.82% for sensitivity and specificity, respectively. The false positive rate is 8.28% and false negative rate is 17.4%, which are relatively low. While both sensitivity and specificity are improved in the proposed fusion algorithm as compared to other state-of-art classifiers, the sensitivity isn’t that high (<85%), which might be induced by the imbalanced patients between the classes. One feasible solution to obtain more balanced solution is to develop a multi-objective model where both sensitivity and specificity are considered as the objectives during model optimization (Zhou et al., 2018). The three CNN investigated in this study were proposed in 2012, 2014, and 2017, respectively. The performance of the fusion algorithm could be further improved by using more advanced CNN architectures such as ResNet (He et al., 2015) and DenseNet (Huang et al., 2018a). Furthermore, a prospective study is desired to evaluate the proposed fusion algorithm.
In this study, the following features: variance as the intensity feature, contrast as the texture feature, minor diameter as the geometry feature, and CNN featureP were selected by all our three fusion algorithms based on three CNN architectures, although different optimal feature subsets were obtained by different CNN architectures (Table 5). These selections are desirable as the first two features selected by all three fusion algorithms indicate the heterogeneity of lesions, the third one indicates the size of lesions, and the CNN featureP learned at the output layer of CNN indicates the intrinsic characteristics. On the other hand, we used the SFS feature selection method coupled with SVM classifier to select the optimal feature subset. Different optimal features were obtained by the fusion algorithms based on different CNN architectures, which implies that the feature selection results are closely correlated with the different CNN features learned by different CNNs. For example, the multi-crop Net focuses on learning information on the central region by cropping the central regions from convolutional feature maps, which could require the selected handcrafted features to focus on the global information. The outputs of the last pooling layer in VGG16 are 2D tensors, which could lose some useful information than Alex Net with the output in forms of 4D tensors (Fig. 3). Therefore, the complementary handcrafted features selected by the SS-OLHF-VGG16 are more than the SS-OLHF-Alex. Feature selection generally depends on many factors, such as classifiers, feature set, database and selection processing (Zhou et al., 2016). For CT images acquired under different acquisition conditions, one may need to identify robust features through test-retest analysis (Timmeren et al., 2016) so that only reliable features are extracted, thereby improving the repeatability of feature selection.
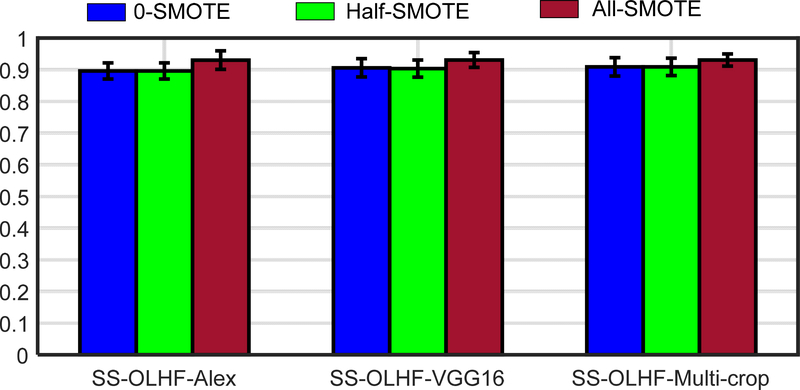
As a successful and classical machine learning algorithm, SVM was applied as the classifier for the final classification task. SVM is trained on the training set by constructing a hyper-plane in training samples space to maximize the margin between binary training samples. Thus, the imbalance database will weaken the performance of SVM. Therefore, the SMOTE technique was used to generate the synthetic vector samples to augment the decision region of the minority class for solving the imbalance problem. The influence of the size of the synthetic samples generated by SMOTE is presented on Fig. 8. The case generating the synthetic samples with the number which is the difference between the numbers of two classes (All-SMOTE) have best AUC performance compared with the case without the synthetic sample (0-SMOTE) and the case just generating the synthetic samples with half of the number which is the difference between the numbers of two classes (half-SMOTE).
Fig. 8.
The AUC performance of the different size of synthetic samples generated by SMOTE. 0-SMOTE: without the synthetic sample; Half-SMOTE: generate the synthetic samples with half of the number which is the difference between the numbers of two classes; All-SMOTE: generate the synthetic samples with the number which is the difference between the numbers of two classes.
As shown by the different predicting performance on different conditions and nodule size (Table 6), the nodules with volume smaller than median has higher FNR and the nodules with volume larger than median has higher FPR. These results indicate that the volume is a very important geometry feature but also has a negative effect on identifying large benign nodules and small malignant nodules. The lesion surrounding region may provide additional characteristics for large benign nodules and small malignant nodules. For example, the shell descriptor proposed by Hao et al. (Hao et al., 2018) descripting the different morphologic patterns in the tumor periphery could be used to improve the performance of the fusion algorithms, which will be studied in the future.
Our proposed fusion algorithms were performed on the nodule ROI with manual segmentation by four radiologists. The manual segmentation is time consuming as well as bringing difference between different radiologists. Many automatic segmentation methods were proposed (Badrinarayanan et al., 2017; Chen et al., 2018). Recently, many researchers focus on the CNN research and disease quantification without segmentation (Tong et al., 2019). These techniques can be incorporated into our fusion algorithm removing the step of manual segmentation.
Acknowledgement
This work was partly supported by the American Cancer Society (ACS-IRG-02-196), the US National Institutes of Health (5P30CA142543), and the National Natural Science Foundation of China (NSFC, 11771456 and U1708261). The authors would like to thank Dr. Jonathan Feinberg for editing the manuscript.
Appendix. The statistics on the measurement conditions and nodule characteristics
Table A1.
The statistics on the measurement conditions and nodule characteristics
| Conditions | Pixel size (mm) | Slice thickness (mm) | Reconstruction Interval (mm) | Nodule volume (mm3) |
|---|---|---|---|---|
| Mean±SD | 0.68±0.08 | 1.95±0.75 | 1.76±0.86 | 737.78±1815.3 |
| ≥Mean, no.(%) | 606 (49.43) | 707 (57.67) | 626 (51.06) | 239 (19.49) |
| <Mean, no.(%) | 620 (50.57) | 519 (42.33) | 600 (48.94) | 987 (81.51) |
| Median | 0.68 | 2.00 | 1.80 | 100.20 |
| Min | 0.46 | 0.60 | 0.50 | 4.39 |
| Max | 0.98 | 5.00 | 5.00 | 18280 |
| ≥Median, no.(%) | 620 (50.57) | 707 (57.67) | 626 (51.06) | 613 (50) |
| <Median, no.(%) | 606 (49.43) | 519 (42.33) | 600 (48.94) | 613(50) |
Note. The reconstruction interval is calculated as the difference between two adjacent slice-locations annotated in DICOM files. SD: standard deviation.
Table A2.
The difference between the reconstruction interval and the slice thickness (DIT).
| DIT | < 0, no.(%) | 423 (34.50%) |
| = 0, no.(%) | 803 (65.50%) | |
| > 0, no.(%) | 0 (0%) |
References
- Akcay S, Kundegorski ME, Willcocks CG and Breckon TP 2018. Using Deep Convolutional Neural Network Architectures for Object Classification and Detection Within X-Ray Baggage Security Imagery Ieee T Inf Foren Sec 13 2203–15 [Google Scholar]
- Antropova N, Huynh BQ and Giger ML 2017. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets Medical physics 44 5162–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armato SG, Altman MB and Wilkie J 2003. Automated lung nodule classification following automated nodule detection on CT: a serial approach Med Phys 30 1188–97 [DOI] [PubMed] [Google Scholar]
- Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI and Hoffman EA 2011. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans Medical physics 38 915–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audrey G, Chung MJS, Kumar Devinder,Khalvati Farzad,Haider Masoom A.,Wong Alexander 2015. Discovery Radiomics for Multi-Parametric MRI Prostate Cancer Detection
- Badrinarayanan V, Kendall A and Cipolla R 2017. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation Ieee T Pattern Anal 39 2481–95 [DOI] [PubMed] [Google Scholar]
- Biswas SK and Milanfar P 2017. Linear Support Tensor Machine With LSK Channels: Pedestrian Detection in Thermal Infrared Images Ieee T Image Process 26 4229–42 [DOI] [PubMed] [Google Scholar]
- Causey JL, Zhang JY, Ma SQ, Jiang B, Qualls JA, Politte DG, Prior F, Zhang SZ and Huang XZ 2018. Highly accurate model for prediction of lung nodule malignancy with CT scans Sci Rep-Uk 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HP, Hadjiiski L, Zhou C and Sahiner B 2008. Computer-aided diagnosis of lung cancer and pulmonary embolism in computed tomography - A review Acad Radiol 15 535–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla NV, Bowyer KW, Hall LO and Kegelmeyer WP 2002. SMOTE: synthetic minority over-sampling technique Journal of artificial intelligence research 16 321–57 [Google Scholar]
- Chen L, Shen C, Li S, Maquilan G, Albuquerque K, Folkert MR and Wang J 2018. Automatic PET cervical tumor segmentation by deep learning with prior information Medical Imaging 1057436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N, Kim SJ, Choi HY, Lyou CY and Moon WK 2010. Features of Prospectively Overlooked Computer-Aided Detection Marks on Prior Screening Digital Mammograms in Women With Breast Cancer Am J Roentgenol 195 1276–82 [DOI] [PubMed] [Google Scholar]
- Davis LS, J SA, Aggarwal J 1979. Texture analysis using generalized co-occurrence matrices Ieee T Pattern Anal 251–9 [DOI] [PubMed] [Google Scholar]
- Ding CX and Tao DC 2018. Trunk-Branch Ensemble Convolutional Neural Networks for Video-Based Face Recognition Ieee T Pattern Anal 40 1002–14 [DOI] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM and Thrun S 2017. Dermatologist-level classification of skin cancer with deep neural networks Nature 542 115–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HZ, Cheng J, Xu YW, Wong DWK, Liu J and Cao XC 2018. Joint Optic Disc and Cup Segmentation Based on Multi-Label Deep Network and Polar Transformation IEEE transactions on medical imaging 37 1597–605 [DOI] [PubMed] [Google Scholar]
- Gillies RJ, Kinahan PE and Hricak H 2016. Radiomics: Images Are More than Pictures, They Are Data Radiology 278 563–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL and Webster R 2016. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs Jama-J Am Med Assoc 316 2402–10 [DOI] [PubMed] [Google Scholar]
- Guo X, Huang X, Zhang L, Zhang L, Plaza A and Benediktsson JA 2016. Support Tensor Machines for Classification of Hyperspectral Remote Sensing Imagery IEEE Transactions on Geoscience and Remote Sensing 54 3248–64 [Google Scholar]
- Hao HX, Zhou ZG, Li SL, Maquilan G, Folkert MR, Iyengar P, Westover KD, Albuquerque K, Liu F, Choy H, Timmerman R, Yang L and Wang J 2018. Shell feature: a new radiomics descriptor for predicting distant failure after radiotherapy in non-small cell lung cancer and cervix cancer Phys Med Biol 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He KM, Zhang XY, Ren SQ and Sun J 2015. Deep Residual Learning for Image Recognition arXiv:1512.03385 [Google Scholar]
- Huang G, Liu Z, Maaten L v d and Weinberger KQ 2018a. Densely Connected Convolutional Networks arXiv:1608.06993 [Google Scholar]
- Huang H, Hu XT, Zhao Y, Makkie M, Dong QL, Zhao SJ, Guo L and Liu TM 2018b. Modeling Task fMRI Data Via Deep Convolutional Autoencoder IEEE transactions on medical imaging 37 1551–61 [DOI] [PubMed] [Google Scholar]
- Jimmy Lei Ba JRK, Hinton Geoffrey E. 2016. Layer Normalization arXiv:1607.06450v1 [Google Scholar]
- Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H and Zhang K 2018. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning Cell 172 1122–31 e9 [DOI] [PubMed] [Google Scholar]
- Kingma D and Ba J 2014. Adam: A method for stochastic optimization arXiv preprint arXiv:1412.6980 [Google Scholar]
- Kohavi R and John GH 1997. Wrappers for feature subset selection Artificial intelligence 97 273–324 [Google Scholar]
- Kononenko I, Šimec E and Robnik-Šikonja M 1997. Overcoming the myopia of inductive learning algorithms with RELIEFF Applied Intelligence 7 39–55 [Google Scholar]
- Kooi T, Litjens G, van Ginneken B, Gubern-Merida A, Sanchez CI, Mann R, den Heeten A and Karssemeijer N 2017. Large scale deep learning for computer aided detection of mammographic lesions Medical image analysis 35 303–12 [DOI] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I and Hinton GE Advances in neural information processing systems,2012), vol. Series) pp 1097–105 [Google Scholar]
- Krizhevsky A, Sutskever I and Hinton G E 2017. ImageNet Classification with Deep Convolutional Neural Networks Commun Acm 60 84–90 [Google Scholar]
- LeCun Y, Bengio Y and Hinton G 2015. Deep learning Nature 521 436–44 [DOI] [PubMed] [Google Scholar]
- Li SL, Yang N, Li B, Zhou ZG, Hao HX, Folkert MR, Iyengar P, Westover K, Choy H, Timmerman R, Jiang S and Wang J 2018. A pilot study using kernelled support tensor machine for distant failure prediction in lung SBRT Medical image analysis 50 106–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C, Ruan S, Denœux T, Jardin F and Vera P 2016. Selecting radiomic features from FDG-PET images for cancer treatment outcome prediction Medical image analysis 32 257–68 [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang J, Yap P-T and Shen D 2017. View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data Medical image analysis 36 123–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto R, Wang F, Wang S, Jiang X and Dudley JT 2018. Deep learning for healthcare: review, opportunities and challenges Brief Bioinform 19 1236–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namburete AI, Stebbing RV, Kemp B, Yaqub M, Papageorghiou AT and Noble J A 2015. Learning-based prediction of gestational age from ultrasound images of the fetal brain Medical image analysis 21 72–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD 2011. Reduced lung-cancer mortality with low-dose computed tomographic screening N Engl J Med 365 395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas Lachiche PF 2003. Improving accuracy and cost of two-class and multi-class probabilistic classifiers using ROC curves PRoceedings of the Twentieth International Conference on Machine Learning (ICML-2003), Washington DC. [Google Scholar]
- Parisot S, Ktena SI, Ferrante E, Lee M, Guerrero R, Glocker B and Rueckert D 2018. Disease prediction using graph convolutional networks: Application to Autism Spectrum Disorder and Alzheimer’s disease Medical image analysis 48 117–30 [DOI] [PubMed] [Google Scholar]
- Parmar C, Grossmann P, Bussink J, Lambin P and Aerts HJ 2015. Machine Learning methods for Quantitative Radiomic Biomarkers Scientific reports 5 13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky PF, Gierada DS, Black W, Munden R, Nath H, Aberle D and Kazerooni E 2015. Performance of Lung-RADS in the National Lung Screening Trial: a retrospective assessment Ann Intern Med 162 485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L and Summers RM 2016. Improving Computer-Aided Detection Using Convolutional Neural Networks and Random View Aggregation IEEE transactions on medical imaging 35 1170–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Zhou M, Yang F, Yang C and Tian J 2015. Multi-scale Convolutional Neural Networks for Lung Nodule Classification Information processing in medical imaging : proceedings of the … conference 24 588–99 [DOI] [PubMed] [Google Scholar]
- Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y and Tian J 2017. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification Pattern Recognition 61 663–73 [Google Scholar]
- Shi JZ, Sahiner B, Chan HP, Ge J, Hadjiiski L, Helvie MA, Nees A, Wu YT, Wei J, Zhou C, Zhang YH and Cui J 2008. Characterization of mammographic masses based on level set segmentation with new image features and patient information Med Phys 35 280–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D and Summers RM 2016. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning IEEE transactions on medical imaging 35 1285–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K and Zisserman A 2014. Very deep convolutional networks for large-scale image recognition arXiv preprint arXiv:1409.1556 [Google Scholar]
- Sutton EJ, Dashevsky BZ, Oh JH, Veeraraghavan H, Apte AP, Thakur SB, Morris EA and Deasy JO 2016. Breast cancer molecular subtype classifier that incorporates MRI features J Magn Reson Imaging 44 122–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao D, Li X, Wu X, Hu W and Maybank SJ 2007. Supervised tensor learning Knowledge and Information Systems 13 1–42 [Google Scholar]
- Timmeren J E v, Leijenaar RTH, Elmpt W v, Wang J, Zhang Z, Dekker A and Lambin P 2016. Test–Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2 361–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Udupaa JK, Odhner D, Wu C, Schuster SJ and Torigian DA 2019. Disease quantification on PET/CT images without explicit object delineation Medical image analysis 51 169–83 [DOI] [PubMed] [Google Scholar]
- Vallières M, Freeman C, Skamene S and El Naqa I 2015. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities Physics in medicine and biology 60 5471–96 [DOI] [PubMed] [Google Scholar]
- Wimmer G, Tamaki T, Tischendorf JJ, Häfner M, Yoshida S, Tanaka S and Uhl A 2016. Directional wavelet based features for colonic polyp classification Medical image analysis 31 16–36 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Oikonomou A, Wong A, Haider MA and Khalvati F 2017. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer Scientific reports 7 46349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Folkert M, Iyengar P, Westover K, Zhang Y, Choy H, Timmerman R, Jiang S and Wang J 2017. Multi-objective radiomics model for predicting distant failure in lung SBRT Physics in medicine and biology 62 4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZG, Folkert M, Cannon N, Iyengar P, Westover K, Zhang YY, Choy H, Timmerman R, Yan JS, Xie XJ, Jiang S and Wang J 2016. Predicting distant failure in early stage NSCLC treated with SBRT using clinical parameters Radiotherapy And Oncology 119 501–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZG, Li SL, Qin GG, Folkert M, Jiang S and Wang J 2018. Automatic multi-objective based feature selection for classification arXiv:1807.03236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu JZ, Cauley SF, Rosen BR and Rosen MS 2018a. Image reconstruction by domain-transform manifold learning Nature 555 487–+ [DOI] [PubMed] [Google Scholar]
- Zhu WT, Liu CC, Fan W and Xie XH 2018b. DeepLung: Deep 3D Dual Path Nets for Automated Pulmonary Nodule Detection and Classification IEEE Winter Conf. on Applications of Computer Vision [Google Scholar]