Abstract
RNA repair has now been demonstrated to be a genuine biological process and appears to be present in all three domains of life. In this article, we consider what this might mean for the transition from an early RNA-dominated world to modern cells possessing genetically encoded proteins and DNA. There are significant gaps in our understanding of how the modern protein-DNA world could have evolved from a simpler system, and it is currently uncertain whether DNA genomes evolved once or twice. Against this backdrop, the discovery of RNA repair in modern cells is timely food for thought and brings us conceptually one step closer to understanding how RNA genomes were replaced by DNA genomes. We have examined the available literature on multisubunit RNA polymerase structure and function and conclude that a strong case can be made that the Last Universal Common Ancestor (LUCA) possessed a repair-competent RNA polymerase, which would have been capable of acting on an RNA genome. However, while this lends credibility to the proposal that the LUCA had an RNA genome, the alternative, that LUCA had a DNA genome, cannot be completely ruled out.
Keywords: RNA repair, DNA origins, evolution, RNA polymerase, proofreading, Last Universal Common Ancestor
Introduction
The term “RNA world” (Gilbert 1986) gave a name to the hypothesis that, before proteins and DNA, RNA was the main biological catalyst and genetic material (Woese 1967; Crick 1968; Orgel 1968). The hypothesis was bolstered by the discovery of catalytic RNA (Kruger et al. 1982; Guerrier-Takada et al. 1983), building on the earlier discovery that RNA is the genetic material in some viruses (Fraenkel-Conrat 1956; Gierer and Schramm 1956) and in plant viroids (Diener 1971), the latter possessing catalytic RNA but not protein (Flores et al. 2004). The theory has been extensively developed and has gained much support (White 1976; Benner, Ellington, and Tauer 1989; Jeffares, Poole, and Penny 1998; Joyce and Orgel 1999; Yarus 2002); nevertheless, there are many unresolved issues. One such issue, and the subject of this article, is the timing and nature of the evolutionary transition from RNA to DNA.
It is widely accepted that protein synthesis evolved from an RNA world (Poole, Jeffares, and Penny 1998; Noller 2004), major evidence being the central catalytic role of RNA in the ribosome (Moore and Steitz 2002). Thus, one intermediate stage in the evolution of the modern DNA-protein world was a “ribonucleoprotein” (RNP) world. Where DNA sits in the picture is less agreed upon (fig. 1). At one end of the spectrum is the proposal that DNA evolved before protein (Benner, Ellington, and Tauer 1989), possibly even before RNA (Dworkin, Lazcano, and Miller 2003)—see legend to figure 1. While feasible, there are no indications from extant organisms that DNA took on a biological role at such an early stage (Poole, Logan, and Sjöberg 2002), and we shall not discuss this further.
FIG. 1.—
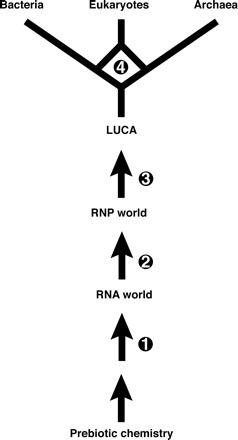
Hypotheses for the origin of DNA. There is currently great uncertainty regarding the timing of the origin of DNA. This figure summarizes the different possibilities proposed in the literature. Scenario 1: Deoxyribonucleotides could have been synthesized prebiotically, and with the discovery that DNA can be made to carry out catalysis, it is not impossible to envision DNA emerging before RNA and proteins, though this idea is not widely accepted (reviewed in Dworkin, Lazcano, and Miller [2003]). Scenario 2: It has been suggested that deoxyribonucleotide synthesis could have originally occurred via a reverse of the deoxyriboaldolase reaction. In modern cells, deoxyribose-5-phosphate is broken down to glyceraldehyde-3-phosphate and acetaldehyde during salvage. The reverse reaction is energetically favorable and has been argued to be “easier” to evolve than protein synthesis (Benner, Ellington, and Tauer 1989), although it is not a feature of modern metabolism. Scenario 3: In modern cells, deoxyribonucleotides are synthesized by ribonucleotide reductases, which are ubiquitous and share a common origin (as evidenced by structure and conservation of key catalytic residues), despite differing biochemistries. This has been taken to suggest that the LUCA possessed DNA (Pode, Penny, and Sjöberg 2000). Scenario 4: DNA replication seems to have evolved twice, as the bacterial apparatus shares no homology with the archaeal/eukaryotic apparatus. It has therefore been suggested that LUCA possessed an RNA genome or, alternatively, a composite RNA/DNA genome (Leipe, Aravind, and Koonin 1999; Forterre 2002).
Biochemical data suggest that DNA evolved after proteins, and before the three domains—archaea, bacteria, and eukaryotes—diverged. In all organisms that carry out de novo synthesis of deoxyribonucleotides, these are formed by reduction of ribonucleotides by ribonucleotide reductase (Poole, Logan, and Sjöberg 2002). Crystal structures of the catalytic subunits of the three divergent classes of this enzyme demonstrate a common origin (Uhlin and Eklund 1994; Logan et al. 1999; Sintchak et al. 2002; Larsson et al. 2004), suggesting that the Last Universal Common Ancestor (LUCA) of archaea, bacteria, and eukaryotes had a DNA genome.
In contrast, structural and phylogenomic studies of the replication apparatus demonstrate that numerous key components (notably DNA polymerases and primases, see Leipe, Aravind, and Koonin [1999] for a full list) evolved twice, the bacterial apparatus being largely unrelated to the archaeal/eukaryotic apparatus (Leipe, Aravind, and Koonin 1999; Forterre 2002; Forterre, Filée, and Myllykallio 2004). Consistent with this, a recent attempt to reconstruct the conserved core of LUCA identified only four universal components of modern DNA replication and repair (DnaN: the sliding clamp; HolB: the clamp loader; PolI-A: the 3′–5′ exonuclease subunit of DNA PolI; and RecA: involved in recombination repair). Three produced trees congruent with 16–18S ribosomal RNA (rRNA) trees, while HolB was argued to have been subject to horizontal gene transfer (Harris et al. 2003).
It is therefore unclear whether DNA appeared before LUCA (as is commonly assumed) (Penny and Poole 1999; Poole, Penny, and Sjöberg 2000) or after LUCA (suggesting LUCA contained an RNA or possibly an RNA/DNA genome [Leipe, Aravind, and Koonin 1999; Forterre 2002]). To clarify, if DNA was the genetic material in LUCA, it seems likely that one of the two forms of replication apparatus (either bacterial or archaeal/eukaryotic) was present at this stage, with the other form representing a lineage-specific replacement (nonorthologous gene displacement [NOGD]). At the same time, LUCA must have carried out ribonucleotide reduction. Alternatively, LUCA possessed an RNA genome, and DNA replication evolved twice independently (Leipe, Aravind, and Koonin 1999; Forterre 2002).
Leipe, Aravind, and Koonin (1999) suggest that LUCA possessed a hybrid DNA/RNA genome, thereby providing an explanation for the universal distribution of certain components of the DNA replication/repair apparatus. This model requires that a reverse transcriptase activity was present in some form, though no direct evidence for this function in LUCA is available. Furthermore, although the model aims to explain the presence of key components of the modern DNA apparatus in LUCA, by their own admission, the authors' model does not actually provide roles for most of these, including critical components such as the sliding clamp, clamp loader, ligase, and topoisomerase I.
At first glance, it would appear that the data emerging from studies of the DNA replication apparatus and that for deoxyribonucleotide synthesis are in conflict. However, while a single evolutionary origin for the catalytic subunit of the ribonucleotide reductases is established, its evolutionary history is obscured (Poole, Logan, and Sjöberg 2002). There are several reasons for this. First, the phylogenetic distribution of the paralogous catalytic subunits from the three classes is incomplete. Bacteria as a domain possess all three classes, archaea largely possess class II and III reductases, and eukaryotes thus far possess only class I reductases. Secondly, the reductases may have undergone horizontal gene transfer as well as polyphyletic losses (Torrents et al. 2002). While the three classes perform the same reaction, they do so under different oxygenic conditions: class I is strictly aerobic, class II is operational in both aerobic and anaerobic conditions and class III is strictly anaerobic. Consequently, many organisms possess more than one class, and gain and loss of reductases may be a cause/consequence of changes in habitat. Third, sequence similarity is extremely low between the classes (Tauer and Benner 1997; Torrents et al. 2002), making it unlikely that meaningful phylogenetic analyses can be carried out for the entire data set. These points, taken together, make it difficult to establish the timing of their emergence (i.e., pre- or post-LUCA).
Mirkin et al. (2003) have predicted that LUCA possessed 500–600 genes. However, a genome of this size may be too large to be maintained as RNA. Modern RNA polymerases have a fidelity of ∼10−4, and the largest RNA viruses, the single-stranded coronaviruses, are around 30 kb (Atkins 1993; Marra et al. 2003; Rota et al. 2003). While RNA virus genomes are probably under selection to be small (Crotty and Andino 2002), a genome of 30 kb causes problems both for the RNA world and for an RNA-based LUCA. Partial reconstructions of the late stages of the RNP world (Jeffares, Poole, and Penny 1998; Yarus 2002) appear to overstep this limit, and conservation of the ribosomal protein superoperon between archaea and bacteria has been interpreted as evidence for genomes in excess of 30 kb in LUCA (Leipe, Aravind, and Koonin 1999).
Several features of genomes could potentially increase information content and genome size. These include the early evolution of recombination, redundancy through polyploidy, division into chromosomes, and overlapping sequences as a means of reducing mutational targets (Reanney 1987; Jeffares, Poole, and Penny 1998; Lehman 2003; Peleg et al. 2004; Santos, Zintzaras, and Szathmáry 2004). All these mechanisms are available to modern RNA viruses; yet, the upper limit of ∼30 kb, on the same order of magnitude as predicted by RNA polymerase fidelity, implies that RNA genome size is nevertheless strongly determined by this parameter.
While any specific number (e.g., 500 genes; Mirkin et al. 2003) is open to debate, the numbers give some indication of the coding problem. For instance, if gene size in LUCA was on average half that of modern bacteria (i.e., ∼500 bp), a genome with 500 genes would be ∼250 kb, about half that of Mycoplasma genitalium. With half the number of genes, as suggested from minimal gene set studies (Mushegian and Koonin 1996; Hutchison et al. 1999), the genome would still be ∼125 kb, well above the size of the largest RNA viruses.
Santos, Zintzaras, and Szathmáry (2004) have presented simulations examining the interplay between recombination and redundancy in a simple replicator system. They show that, at modest redundancy (approximately three- to fourfold), a recombination-competent system could increase information content by at least 25%, which the authors conclude is nevertheless insufficient for emergence of a simple ribozyme replicase, let alone a complex RNA cell, in line with the consensus that limits to RNA-coding capacity is a major problem in the evolution of life (Jeffares, Poole, and Penny 1998; Joyce and Orgel 1999; Poole, Jeffares, and Penny 1999; Scheuring 2000).
Returning to the RNA to DNA transition, there are two specific problems. On one hand the late stages of the RNP world appear complex, so how could a small RNA genome have supported both protein synthesis and have evolved enzymes as chemically “sophisticated” as ribonucleotide reductases (Poole, Penny, and Sjöberg 2000)? On the other hand, how feasible is the contention that the LUCA had an RNA genome? This second question links LUCA to earlier stages, namely the RNP to DNA transition, since comparative genomics data point to the possibility of DNA replication evolving twice, post-LUCA.
Given the difficulty with RNA as genetic material, one is almost forced to accept Scenario 3 in figure 1 over Scenario 4 and conclude that there was an NOGD of the replication apparatus. However, it is now becoming apparent that RNAs are subject to both proofreading and repair (Thomas, Platas, and Hawley 1998; Reichert and Mörl 2000; Aas et al. 2003), raising the possibility that a large genome was possible in the absence of DNA. There are two forms of bona fide RNA repair now known. One is repair of methyl damage, conserved between eukaryotes and bacteria (Aas et al. 2003; Ougland et al. 2004) (Appendix 1). The second form involves proofreading and repair by RNA polymerases, analogous to that observed with DNA polymerases, and is conserved across all three domains of life (table 1). We argue from structural and functional data that the second of these, RNA polymerase-dependent proofreading and repair, is likely to have been a feature of LUCA rather than a recent innovation as part of selection for improved messenger RNA (mRNA) quality control, though this is no doubt the current function of this phenomenon. We will briefly summarize the experimental evidence for RNA repair by RNA polymerases, then consider the implications for the RNA to DNA transition and the nature of the genome of LUCA.
Table 1.
Transcript Elongation, Proofreading and Repair by RNA Polymerases
|
|
Polymerase Function |
3′→5′ Nuclease Activity |
References |
Cleavage Induction |
References |
|---|---|---|---|---|---|
| Archaeal DdRp | Transcription | ✓ | Hausner, Lange, and Musfeldt (2000) | TFS (homologous to eukaryotic TFIIS) | Hausner, Lange, and Musfeldt (2000), Lange and Hausner (2004) |
| Bacterial DdRp | Transcription | ✓ | Orlova et al. (1995) | GreA/GreB (functionally analogous to TFIIS) | Borukhov, Sagitov, and Goldfarb 1993, Erie et al. (1993) |
| Eukaryotic DdRp I | rRNA transcription | ✓ | Tschochner (1996) | TFIISa | Schnapp, Graveley, and Grummt (1996) |
| Eukaryotic DdRp II | mRNA and snRNA transcription | ✓ | Wang and Hawley (1993), Thomas, Platas, and Hawley (1998) | TFIIS | Jeon and Agarwal (1996) |
| Eukaryotic DdRp III | Transcription of tRNA, 5S rRNA, U6 snRNA, snoRNA | ✓ | Whitehall, Bardeleben, and Kassavetis (1994) | TFIIS | Chédin et al. (1998) |
| Vaccinia virus DdRp | Transcription and replication | ✓ | Hagler and Shuman (1993) | TFIIS role in proofreading unclear | Rosales et al. (1994) |
| T7 DNA phage DdRpb | Transcription | ✓ | Sastry and Ross (1997) | — | — |
| Influenza virus RdRpb |
Transcription and replication |
✓ |
Ishihama et al. (1986), Ishihama (1996) |
— |
— |
NOTE.—Abbreviations: DdRp, DNA-dependent RNA polymerase; RdRp, RNA-dependent RNA polymerase; GreA/GreB, growth regulator A/B; TFIIS, transcription factor IIS; TFS, transcription factor S.
Also known as SII.
Single subunit RNA polymerases, evolutionarily unrelated to multisubunit RNA polymerases.
Proofreading and Repair by RNA Polymerases
As with DNA, RNA can be damaged as well as incorrectly copied. How the cell checks that an mRNA is correctly synthesized and deals with incorrectly copied RNAs is a subject of intense investigation, falling under the broad term of quality control. In both eukaryotes and bacteria, there are mechanisms for eliminating transcripts that fail to pass quality control (Gillet and Felden 2001; Wilusz, Wang, and Peltz 2001; Vasudevan, Peltz, and Wilusz 2002). The general expectation has been that, following recognition of a damaged or incorrectly synthesized mRNA, the damaged molecule would be destroyed rather than repaired; repair would not pay its way, since thousands of RNAs are produced in the cell, and therefore the best way to deal with the odd damaged RNA is to break it down and start afresh.
However, errors in transcription may be more common than the above reasoning might lead us to suppose. The average length of proteins in prokaryotes is between 300–400 amino acids (aa) (Skovgaard et al. 2001), equating to transcripts somewhere in excess of 1 kb. The genome of Escherichia coli K-12 (Blattner et al. 1997) revealed the longest open reading frame to code for a 2,383 aa protein. The corresponding transcript should be over 7,200 nucleotides. With errors on the order of every 10,000 bases, RNA polymerases would make one error in every 10 transcripts in bacteria, and for the largest E. coli ORF errors might turn up in 2 of 3 transcripts. The human genome encodes a muscle protein called titin that, at 26,926 aa, is the largest protein known (Labeit and Kolmerer 1995). At around 80 kb just for the coding region (i.e., minus introns), it would be expected to pick up around eight errors per mRNA during transcription. Even considering code redundancy, a lot of mRNAs would be expected to possess coding errors, and the known pathways of mRNA surveillance only deal with transcripts with premature termination signals (nonsense-mediated decay; Wilusz, Wang, and Peltz [2001]) or transcripts lacking a stop codon (nonstop decay; Vasudevan, Peltz, and Wilusz [2002]). Without some form of proofreading and repair, the number of mRNAs carrying coding errors could be significant.
Though we all learn that proofreading and repair is restricted to DNA, the potential benefits for RNA are clear for exactly the same reason that the nonsense-mediated and nonstop-decay pathways are beneficial. Significantly, both RNA- and DNA-dependent RNA polymerases possess a 3′–5′ nuclease activity, which is augmented by cleavage-stimulatory factors (table 1; Fish and Kane 2002). This enables RNA polymerases to negotiate barriers to elongation. Briefly, it seems that upon arrest of the RNA polymerase at some barrier to elongation, the cleavage-stimulatory factors interact with the polymerase, causing it to backtrack and hydrolyze the nascent transcript. Elongation is then reinitiated.
The ability to negotiate blocks to elongation (which includes misincorporation) has been interpreted as improving the fidelity of the process and therefore playing a part in quality control of transcripts (Erie et al. 1993; Jeon and Agarwal 1996). This might well be interpreted as a form of proofreading, and indeed, initial identification of 3′–5′ nuclease activity in the single subunit RNA-dependent RNA polymerase from influenza virus (Ishihama et al. 1986) led to speculation that RNA polymerases, like many of their DNA counterparts, possessed proofreading (Shirai and Go 1991).
Experiments on misincorporations suggested that mRNA transcription was subject to a high degree of fidelity in both bacteria and eukaryotes (Erie et al. 1993; Jeon and Agarwal 1996) though a direct role in proofreading was not demonstrated. Thomas, Platas, and Hawley (1998) subsequently presented in vitro evidence for proofreading by RNA polymerase II (pol II). Upon misincorporation of a nucleotide in the growing RNA, the addition of the next nucleotide is slowed, with extension occurring between 5- and 20-fold slower than for a correct incorporation. This delay provides time for the cleavage-stimulatory factor, transcription factor IIS (TFIIS) to bind arrested pol II, stimulating removal of the mismatched nucleotide via the intrinsic PolII 3′–5′ exonuclease activity (Wind and Reines 2000). Removal of the misincorporated nucleotide was observed at low levels in the absence of TFIIS, and this is known to be the case for a number of RNA polymerases.
The work on human RNA pol II provides clear evidence for RNA polymerase-dependent proofreading and error correction, but other polymerases also possess this capacity (table 1). Recently, several important papers shed further light on this process. Shaevitz et al. (2003) presented an optical trapping study of the RNA polymerase (RNAP) from E. coli, showing backtracking and recovery from pausing by single molecules of RNAP. Pausing was significantly reduced by growth regulator A (GreA) or growth regulator B (GreB)-induced cleavage. Sosunov et al. (2003) demonstrated by an elegant exploitation of the intrinsic exonuclease activity of E. coli RNAP, combined with mutagenesis and modeling, that a common two-metal mechanism is used for both elongation and repair (fig. 2), in a manner similar to that proposed for the 3′–5′ exonuclease activity of DNA polymerases (Joyce and Steitz 1994). Two further papers showed surprising similarities in the roles of the prokaryotic and eukaryotic transcription factors in stimulation of the endonuclease activity (fig. 3a–d). In a medium-resolution (3.8 Å) study of the binding of TFIIS to yeast pol II (Kettenberger, Armache, and Cramer 2003, 2004), the highly extended TFIIS is seen to insert its β-ribbon domain III via a pore into the active site. Two acidic residues at the tip of a loop in domain III are presented to the active site adjacent to the tightly bound metal ion involved in elongation (fig. 3, panels b and d). In a parallel study using electron microscopy (EM) at 15 Å resolution (Opalka et al. 2003), the prokaryotic elongation factor GreB was bound to E. coli RNAP. GreB inserts a completely α-helical domain into the RNAP active site. Despite having no structural homology whatsoever to TFIIS, it nevertheless also appears to present two completely conserved acidic residues to the active site near the metal ion (fig. 3, panels a and c). In both cases, the acidic residues are proposed to position a second metal ion and water molecule for hydrolytic RNA cleavage (fig. 2) as proposed by Sosunov et al. (2003). However, the resolution of the EM study did not allow the acidic residues to be accurately placed in the active site. Sosunova et al. (2003) nicely complemented this work using mutation, crosslinking, and modeling, locating the acidic residues unambiguously in the active site and confirming that GreB does not work allosterically but by direct recruitment of a metal ion.
FIG. 2.—

Schematic diagram of the transcription factor-catalyzed two-metal ion mechanism for the 3′–5′ exonuclease activity as proposed by Sosunova et al. (2003), extended to both the prokaryotic and eukaryotic polymerases.
FIG. 3.—
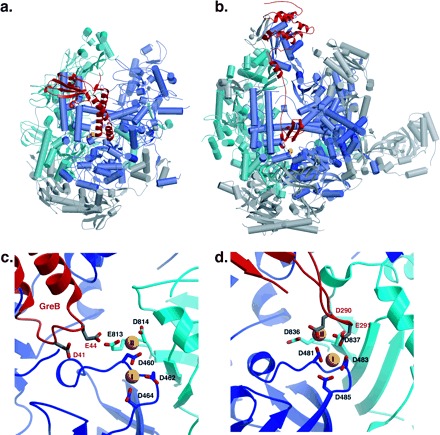
A structural comparison of the bacterial and eukaryotic RNA polymerase systems. (a) Overall view of the bacterial RNA polymerase from Escherichia coli in complex with GreB. The coordinates are of the Thermus aquaticus RNAP and a homology model for E. coli GreB, fitted to the 15-Å EM reconstruction of the E. coli complex (Opalka et al. 2003). The β and β′ subunits are colored cyan and blue, respectively, while the other subunits are colored gray for clarity. The α-helices are drawn as cylinders. GreB is colored red—α-helices are drawn as ribbons for contrast with RNAP. (b) The eukaryotic system as represented by the 3.8-Å crystal structure of yeast PolII in complex with TFIIS, PDB (Protein Data Bank) code 1Y1V (Kettenberger, Armache, and Cramer 2003, 2004). The Rpb2 and Rpb1 subunits are colored cyan and blue, respectively. TFIIS is colored red. The orientations in panels (a) and (b) are almost identical. (c) Detailed view of the active site of the E. coli complex. The side-chain positions (from coordinates kindly provided by Seth Darst) should not be interpreted literally due to the low resolution of the EM structure, which is nevertheless the best available experimentally derived model. (d) Detailed view of the yeast PolII active site. In panels (c) and (d) the two Mg2+ ions proposed to be part of the two-metal ion mechanism for RNA synthesis and editing are shown as gold spheres. Since Mg2+ ion II is not deposited in any PDB file, it was modeled by hand into the coordinates of RNAP/GreB and generated in the yeast PolII structure by superposition of the Cα atoms of the coordinating residues of RNAP and PolII. Its position in this figure is for illustrative purposes only.
In summary, the available data suggest that this form of nascent transcript repair is ubiquitous, providing not only a means of reading through arrest sites, but also of removing misincorporated bases during transcription. Moreover, recent data strengthen the concept of an intrinsic unified two-metal mechanism for synthesis and repair in both DNA and RNA polymerases, greatly stimulated by external factors in the latter case. That read-through and repair is distinct from RNA surveillance demonstrates that quality control of RNA synthesis is not limited to degradative pathways. We will next consider the implications of this process for the timing of the RNA to DNA transition.
Multisubunit RNA Polymerases Date Back to LUCA
Before evaluating the possibility that LUCA possessed an RNA genome, subject to proofreading and repair, we first review the evidence that multisubunit RNA polymerases and the repair capability date back to LUCA.
There are strong lines of evidence that attest to LUCA possessing such an RNA polymerase. Firstly, archaeal, bacterial, and eukaryotic multisubunit RNA polymerases all share a common core of four subunits (corresponding to bacterial subunits α, β, β′, and ω [the α subunit is present in two copies in bacterial RNA polymerase; in Archaea there are two separate subunits with homology to α (subunits D and L), and the same is true for Eukaryotes (Rbp3 and Rbp11) (Cramer 2002a)]), as evidenced from inspection of the available structures from bacteria and eukaryotes, together with sequence similarities (Cramer 2002a). Consistent with placement in LUCA, the two largest subunits of RNA polymerase (β and β′) are routinely used as markers in phylogenetic reconstructions. There is no evidence for extensive, domain-level horizontal transfer, and the resulting trees are congruent with trees constructed using rRNA (Tourasse and Gouy 1999) (while establishing the correct topology is nontrivial, see Tourasse and Gouy [1999], topology does not affect the conclusion that these two subunits can be traced back to the LUCA). Furthermore, Harris et al. (2003) report that the α subunit also produces a tree congruent with rRNA and can thus be placed in the LUCA on phylogenetic grounds, consistent with results from other authors (Koonin 2003).
The second point regarding the antiquity of multisubunit RNA polymerase relates to catalysis. Both the polymerase and repair activities have been incontrovertibly demonstrated to lie within the conserved catalytic core; coordination of the two metal ions required for catalysis is shared between the β′ and β subunits (fig. 3c and d). Significantly, a careful sequence-based comparison has led to speculation that modern RNA polymerases evolved from a homodimeric RNA-binding domain (perhaps acting in some ancillary capacity together with a ribozyme RNA polymerase; Iyer, Koonin, and Aravind [2003]). However, this ultimate origin should be taken to represent a probable earlier intermediate stage; on evidence, LUCA would have possessed an RNA polymerase resembling a modern multisubunit RNA polymerase. That the capacity for repair appears to be a consequence of the common two-metal mechanism for elongation implies that, at the very least, the RNA polymerase in LUCA possessed the capacity for repair. Whether genomic RNA repair was a feature of the LUCA is somewhat harder to judge and will be considered next.
Does Modern RNA Repair Suggest an RNA-Based LUCA?
Owing to the nature of the catalytic site, RNA repair could most definitely have been a feature of the LUCA. However, in answering the question above, several points should be borne in mind. Firstly, the argument for genomic RNA repair in LUCA is complicated, and perhaps weakened, by the difficulty in establishing whether or not DNA was present. Secondly, this thesis extrapolates a modern function in mRNA quality control by polymerases reading off DNA templates to an ancestral function in genome replication by polymerases reading off RNA templates. The third is somewhat dependent on the first; assuming RNA repair was present, was it involved in quality control, genome replication, or both?
Given that ribonucleotide reductases cannot be unambiguously placed in LUCA, despite being ubiquitous, and given that the DNA replication apparatus has evolved twice, it is currently unclear if DNA was a feature of LUCA, with one of the two forms of DNA replication having evolved, with the ribonucleotide reductases being present (Scenario 1) or whether DNA replication evolved twice, post-LUCA (Scenario 2). Scenario 1 implies near-complete NOGD of the incumbent DNA replication apparatus. This seems hard to swallow, though this is not in itself an argument against the scenario.
The only difference in arguing for a DNA-based LUCA scenario is that it requires some unknown external source for a complex DNA replication apparatus, and Forterre (1999, 2002) and Villarreal and DeFilippis (2000) have argued for a viral origin for at least one of the modern cellular forms of replication apparatus. However, it is worth noting that even if a viral origin were unequivocally demonstrated, it is not possible to establish the timing of the origin of cellular DNA replication. DNA virus-derived DNA replication could have been co-opted concurrent with or prior to LUCA, with this original apparatus being displaced by another viral apparatus in one lineage after divergence of bacteria from archaea/eukaryotes. Likewise, this could have happened twice independently, post-LUCA (Forterre 2001).
Forterre's hypothesis, that there was “arms-race-driven coevolution of cellular and viral genomes” (Forterre 2002), where modified forms of nucleic acids evolved in stepwise fashion in viruses to evade host “immune” systems, proposes that deoxyribonucleotide synthesis evolved first in viruses, and, importantly, offers a solution to a conflicting aspect of polymerase relationships. Upon evolution of ribonucleotide reduction, the simplest explanation in terms of their use in replication of genetic material is that the incumbent polymerase incorporated deoxyribonucleotide precursors. Substrate and template selectivity, where studied, appears to be easily altered—single residues are responsible for substrate selectivity in E. coli DNA pol I (Astatke et al. 1998) and reverse transcriptase (Gao et al. 1997), and the RNA-dependent RNA polymerase from Brome Mosaic Virus will accept both DNA and RNA-DNA hybrid templates (Siegel et al. 1999). If, as per Scenario 1 above, there had been only one NOGD from a viral source, one would expect an evolutionary relationship between modern multisubunit RNA polymerases and one family of replicative DNA polymerases. However, this is not the case. DNA polymerase III (C-family polymerases), the replicative polymerase in bacteria, is unrelated to modern archaeal- and eukaryotic-replicative DNA polymerases (which are B-family polymerases) and neither are related to multisubunit RNA polymerases (Ito and Braithwaite 1991; Steitz 1999; Cramer 2002b; Grabowski and Kelman 2003). In contrast, the palm domain of A-family DNA polymerases shows structural homology to the equivalent domains of several RNA-dependent RNA polymerases, reverse transcriptases, and DNA-dependent RNA polymerases (Hansen, Long, and Schultz 1997; Steitz 1999). This speaks in favor of a viral origin for deoxyribonucleotide syntheses in so far as it is possible to envisage deoxyribonucleotide “takeover” in an RNA virus. These polymerases are nevertheless too far diverged to be useful for phylogenetic studies, and on sequence alone, it is not possible to show an evolutionary relationship between RNA-dependent RNA polymerases and reverse transcriptases (Zanotto et al. 1996). Furthermore, while the palm domains appear homologous, the finger and thumb domains are not (Hansen, Long, and Schultz 1997; Steitz 1999). The existence of a range of prereplicatively modified deoxyribonucleotides in DNA viruses does, however, provide a modern precedent for Forterre's hypothesis (examples being hydroxymethyluracil instead of T in SP01, U instead of T in PBS1 and PBS2, hydroxymethylcytosine instead of C in T-even phage, and 2-aminoadenine instead of A in cyanophage S-2L—see Kornberg and Baker (1992).
The lack of homology between multisubunit RNA polymerases and the two unrelated families of replicative DNA polymerase makes it difficult to establish the details of the RNA to DNA transition. It is possible that this transition occurred in cells, with DNA being initially replicated by the ancestor of the multisubunit RNA polymerases. However, this scenario introduces a third step, for which there is no evidence.
Under Scenarios 1 and 2 above, RNA repair could have been a feature of the LUCA but clearly in different capacities. Under Scenario 1, a DNA-based LUCA, the only possible function for RNA repair would be the modern one; mRNA quality control. Genomic RNA repair by this route may have been a feature of an earlier stage in life, prior to the RNA to DNA transition, but it would obviously not have been a feature of LUCA. Under Scenario 2, the LUCA possessed an RNA genome, and if the inherent potential for RNA repair by RNA polymerases was realized, both the genome and expressed transcripts would be subject to proofreading and repair, as both would be synthesized by RNA polymerase.
It is of course possible that the potential for repair inherent in the two-metal mechanism of catalysis was not realized in the LUCA and that proofreading and repair emerged later, twice independently (perhaps in conjunction with the appearance of GreA/B and TFIIS cleavage-stimulatory factors), as a form of mRNA quality control. However, this seems highly unlikely. The convergent evolution of cleavage-stimulatory factors suggests that the intrinsic proofreading and repair capability of RNA polymerases was already in use with these factors emerging independently as evolutionary refinements of this process. Moreover, in an RNA-based system where there were strong limits on coding capacity, increases in replication fidelity would have been selectively advantageous. We therefore think it highly unlikely that this intrinsic capability of RNA polymerases was not co-opted at a very early stage. Indirectly supporting this contention, this capacity has been made use of several times in unrelated polymerases, including both single- and multisubunit RNA polymerases (Steitz 1999; Sosunov et al. 2003; table 1), as well as in DNA polymerases, including 3′–5′ exonuclease activity of the Klenow fragment (Steitz 1999). Moreover, such a two-metal ion mechanism has been proposed to be possible with RNA alone, both for polymerization and exonuclease reactions (T. A. Steitz and J. A. Steitz 1993; Steitz 1998). Certainly, it seems somewhat difficult to contend that this catalytic capability was utilized in mRNA quality control post-LUCA but not in LUCA.
Having said that, if multisubunit RNA polymerases had a role in genome maintenance at some early stage in evolution, they must have initially recognized RNA as template, not DNA, as in modern enzymes. Two lines of evidence demonstrate this to be feasible and in fact still biologically relevant. Over 30 years ago, Biebricher and Orgel (1973) published evidence that DNA-dependent RNA polymerase from E. coli was capable of accepting an RNA template, and more recent studies confirm this (Wettich and Biebricher 2001; Pelchat, Grenier, and Perreault 2002). While the RNA-replicating capacity of E. coli RNA polymerase is not physiological, such an example is known. In plants, RNA polymerase II is co-opted into replication of viroids (Mühlbach and Sanger 1979; Schindler and Mühlbach 1992; Fels, Hu, and Riesner 2001)—pathogenic autonomously replicating single-stranded circular RNAs reaching approximately 400 bases in length (Flores et al. 2004). In both cases, it seems that there is some selectivity, with replication initiation requiring some degree of specificity, which may be some combination of sequence and structure (Fels, Hu, and Riesner 2001; Pelchat, Grenier, and Perreault 2002; Pelchat and Perreault 2004).
A second point of concern is the fidelity of modern RNA polymerases. In the absence of cleavage-stimulatory factors, proofreading and repair are reduced, and one may wonder whether it is mechanistically possible for an RNA polymerase to operate at higher fidelity than natural polymerases. In the case of DNA polymerases, it is clear that fidelity differs considerably between replicative and repair-specific polymerases (Beard and Wilson 2003), and DNA polymerase mutants with increased replication fidelity have been selected in mutagenesis screens (Fijalkowska, Dunn, and Schaaper 1993). To our knowledge, this has not been done for multisubunit RNA polymerases, but an example does exist for the single-subunit RNA-dependent RNA polymerase from poliovirus. This mutant was selected for resistance to ribavirin, which acts as a mutagen, increasing the rate of mutation. The mutant high-fidelity polymerase emerged via a single-point mutation and exhibited an elevated fidelity in the absence of the drug (Pfeiffer and Kirkegaard 2003). This is consistent with different fidelities observed for RNA polymerases from different viruses (see Pugachev et al. [2004] for a naturally occurring high-fidelity example). It thus seems reasonable to suggest that the current fidelity of RNA polymerases is a result of selection, resulting in a compromise between processivity and proofreading, and that higher fidelity RNA replication in LUCA could have been possible.
Although to our knowledge no one has examined the proofreading capability of multisubunit DNA-dependent RNA polymerases working with an RNA template, the above data demonstrate the biochemical feasibility of a scenario where LUCA possessed an RNA genome that was both replicated and subject to proofreading and repair by a multisubunit RNA polymerase.
Conclusions
A close examination of the available literature on the structure and function of DNA-dependent RNA polymerases allows us to make several statements of relevance to the nature of LUCA. First, several compelling arguments can be made that place RNA repair as a feature of the LUCA. Intrinsic proofreading appears to be a feature common to all multisubunit RNA polymerases (table 1), and available structures show that a common two-metal ion mechanism lies at the heart of both elongation and editing activities (Sosunov et al. 2003). A common evolutionary origin for this family of RNA polymerases is evident from both sequence and structural data (Cramer 2002a; Harris et al. 2003; Mirkin et al. 2003), making it relatively straightforward to argue that a polymerase resembling the modern conserved core was present in LUCA and has not been subject to extensive horizontal transfer (Tourasse and Gouy 1999; Harris et al. 2003). Furthermore, the available biochemical and structural data suggest that both proofreading and repair of genomic (and transcribed) RNA by RNA polymerases would have preceded the origins of DNA.
The main conclusion we draw is that an early origin for RNA repair is likely. RNA polymerase-mediated proofreading and repair resolves the problem of how an RNA genome could have maintained sufficient genetic information for complex processes to emerge (Poole, Penny, and Sjöberg 2000). Moreover, in providing a possible solution to the RNA-coding capacity conundrum, it lends credibility to a late-independent origin for the two forms of DNA replication apparatus, putting the RNA-based LUCA scenario on an equal footing with a DNA-based LUCA.
We believe that the simplest explanation of the data comes by invoking an RNA-based LUCA. In this scenario, cleavage-stimulatory factors evolved twice independently (GreA/GreB in bacteria and transcription factor S/TFIIS in archaea/eukaryotes) with an initial function in RNA genome repair. Subsequently, DNA replication could have evolved de novo twice independently, once in the ancestor of modern bacteria, and once in the ancestor of archaea and eukaryotes (fig. 4). Alternatively, there could have been transfer of a complete DNA replication apparatus plus deoxyribonucleotide synthetic enzymes from DNA viruses, either once or twice, being compatible with either scenario shown in figure 4 (Forterre 2001, 2002). Given that replicative DNA polymerases in cellular lineages are unrelated to multisubunit RNA polymerases, the two transfer scenarios seem likely, though we note that very few examples of C-family polymerases (bacterial replicative DNA polymerases) have been detected in viruses or plasmids (Filée et al. 2002).
FIG. 4.—

Was the genetic material of LUCA RNA or DNA? Two possible scenarios for the nature of the LUCA are shown. (a) LUCA was RNA based. Previous studies (Leipe, Aravind, and Koonin 1999; Forterre 2002) have pointed out that the DNA replication apparatus appears to have evolved twice, once in the bacterial lineage and once in the archaeal-eukaryote lineage. This implies that the LUCA possessed an RNA genome, and in this scenario, bacterial RNA polymerase-associated cleavage-stimulatory factors (GreA/GreB) and their archaeal/eukaryotic equivalents (TFS/TFIIS) were originally involved in proofreading and repair of the genome, as indicated by [Gen]. As discussed in the text, ribonucleotide reductases have a single origin, but may have been subject to horizontal gene transfer, so a common origin is not necessarily inconsistent with a DNA genome evolving twice independently. (b) LUCA was DNA based. In this scenario, one of the extant DNA replication machineries dates back to LUCA, while the other displaced the extant (LUCA) apparatus. In the figure, the archaeoeukaryotic apparatus is depicted as ancestral, but it is equally possible that the bacterial apparatus is ancestral. The source of a second replication apparatus is unclear but has been suggested to be viral in origin. Under a DNA-LUCA, the cleavage-stimulatory factors would have evolved twice independently in mRNA quality control [QC], which is their current function. A DNA-LUCA implies that ribonucleotide reductase was present in LUCA and that it this family of enzymes have subsequently been subject to horizontal gene transfer, thereby obscuring their early origin. Note also that the cleavage-stimulatory factors could have been subject to NOGD, mirroring the events depicted for the DNA replication apparatuses, though for simplicity, this is not shown. Note that, the scenario in panel (a) is reconcilable with two independent viral transfers but requires more events than either of the depicted scenarios. For completeness, the evolutionary relationship between eukaryote nuclear genes and counterparts in archaea and bacteria, the latter being of mitochondrial endosymbiotic origin (Esser et al. 2004), is displayed. However, the eukaryotic replication apparatus is related to the archaeal apparatus, not the bacterial, excepting universal components (see text).
Even if it could be shown that the genome of LUCA was DNA based and that proofreading and repair of DNA was a constituent feature of LUCA, the data we have examined nevertheless point to RNA proofreading and repair being present in LUCA and at the very least indicate a role in transcript quality control. Under this scenario, genomic RNA repair should still be considered a likely step in the RNA to DNA transition, instead occurring pre-LUCA.
In conclusion, we maintain that RNA polymerase-dependent repair of genetic material is likely to have been a crucial step in the evolution of complex RNA- and protein-based cells. Recent data from comparative genomics have made clear the difficulty in establishing whether DNA was present in LUCA, and consequently, insights into early mechanisms of RNA repair are as important to the reconstruction of LUCA as they are to the RNA to DNA transition.
Appendix 1
As summarized in the table below, two additional forms of RNA repair have been reported; transfer RNA (tRNA) repair and repair of methyl-damaged RNA.
Table A1.
Additional Forms of RNA Repair
|
Type of Repair |
Comments |
Distribution |
References |
|---|---|---|---|
| tRNA repair | Restoration of upstream tRNA 3′ end in transcripts encoding overlapping tRNAs | Human and mouse mitochondria | Reichert and Mörl (2000) |
| Oxidative demethylation | Demethylation of 1-methyladenine and 3-methylcytosine | Conserved between Escherichia coli and human | Aas et al. (2003), Ougland et al. (2004) |
|
|
|
Proposed in plant RNA viruses |
Aravind and Koonin (2001)
|
tRNA Repair
tRNA repair is so far restricted to mammalian mitochondria. Here overlapping tRNAs are expressed as a single molecule that is processed such that the downstream tRNA is complete, leaving the upstream tRNA in a truncated form which must be “repaired” by extending the 3′ end (Reichert and Mörl 2000). This tRNA “completion” seems to occur in the absence of a template, leading to speculation that it works by trial and error—tRNAs with an incorrect 3′ sequence are degraded by exonuclease activity and re-elongated; when the correct sequence is produced, the tRNA can be aminoacylated and released from the cycle of degradation and elongation (Reichert and Mörl 2000).
This form of repair has probably emerged to tackle the peculiarities of mammalian mitochondrial genomes, where Muller's ratchet is operating (Lynch 1996). A number of features of mitochondrial genomes are likely to be attributable to this process, notably RNA editing in metazoan mitochondria (Börner et al. 1997). tRNA repair is therefore best considered a form of organellar RNA editing. While some forms of organellar editing are interesting for the RNA world insofar as they illustrate possible RNA-based processes, their narrow phylogenetic distribution suggests that they are a recent phenomenon.
Oxidative Demethylation
Recently, Aas et al. (2003) demonstrated in vivo repair of aberrantly methylated adenine (1-methyladenine) and cytosine (3-methylcytosine) in RNA by oxidative demethylases. In E. coli, AlkB is responsible for repair of these lesions in both DNA and RNA, while in human, two AlkB homologues provide a division of labor; hABH2 has a preference for double-stranded DNA, repairing lesions close to the replication fork, while hABH3 has a preference for single-stranded substrates (both single-stranded DNA and RNA), suggesting that it operates during transcription. More recently, it has been demonstrated that both E. coli AlkB and hABH3 repair both mRNA and tRNA damaged by methylation (Ougland et al. 2004). The initial lead that suggested a role for AlkB in RNA repair came via a BLAST survey for proteins with a 2-oxoglutarate-Fe(II) oxygenase fold (Aravind and Koonin 2001). Homologues of AlkB from seven plant RNA viruses were identified, suggesting a role in RNA demethylation, perhaps in countering a host-defence strategy involving viral RNA methylation. The demonstration that AlkB-family proteins can repair RNA suggests that these RNA virus homologues are involved in genome maintenance.
Oxidative demethylation is not universal in distribution (no archaeal AlkB homologues have been detected), and it is difficult to establish the antiquity of this process. Aravind and Koonin (2001) report that the 2-oxoglutarate-Fe(II) oxygenase superfamily possesses a universally conserved protein fold (the DSBH fold) but have invoked horizontal gene transfer to explain the origin of the eukaryote AlkB homologues, though no phylogenetic evidence was presented to support their claim (Aravind and Koonin 2001).
Acknowledgments
This work was supported by grants to A.M.P. from the Swedish Research Council (Vetenskapsrådet) and the Knut and Alice Wallenberg Foundation and to D.T.L. from Vetenskapsrådet. We thank Patrick Forterre for valuable comments on an early version of the manuscript and for sending preprints, Seth Darst for providing side-chain coordinates for the RNAP/GreB complex, and two anonymous reviewers for helpful comments on the manuscript.
Footnotes
William Martin, Associate Editor
References
- Aas, P. A., M. Otterlei, P. O. Falnes et al. (11 co-authors). 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421:859–863. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and E. V. Koonin. 2001. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2:RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astatke, M., K. Ng, N. D. Grindley, and C. M. Joyce. 1998. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. USA 95:3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins, J. F. 1993. Contemporary RNA genomes. Pp. 535–556 in R. F. Gesteland and J. F. Atkins, eds. The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Beard, W. A., and S. H. Wilson. 2003. Structural insights into the origins of DNA polymerase fidelity. Structure (Camb.) 11:489–496. [DOI] [PubMed] [Google Scholar]
- Benner, S. A., A. D. Ellington, and A. Tauer. 1989. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 86:7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher, C. K., and L. E. Orgel. 1973. An RNA that multiplies indefinitely with DNA-dependent RNA polymerase: selection from a random copolymer. Proc. Natl. Acad. Sci. USA 70:934–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner, F. R., G. Plunkett III, C. A. Bloch et al. (17 co-authors). 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474. [DOI] [PubMed] [Google Scholar]
- Börner, G. V., S. Yokobori, M. Mörl, M. Dörner, and S. Pääbo. 1997. RNA editing in metazoan mitochondria: staying fit without sex. FEBS Lett. 409:320–324. [DOI] [PubMed] [Google Scholar]
- Borukhov, S., V. Sagitov, and A. Goldfarb. 1993. Transcript cleavage factors from E. coli. Cell 72:459–466. [DOI] [PubMed] [Google Scholar]
- Chédin, S., M. Riva, P. Schultz, A. Sentenac, and C. Carles. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 12:3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P. 2002a Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 12:89–97. [DOI] [PubMed] [Google Scholar]
- ———. 2002b Common structural features of nucleic acid polymerases. Bioessays 24:724–729. [DOI] [PubMed] [Google Scholar]
- Crick, F. H. C. 1968. The origin of the genetic code. J. Mol. Biol. 38:367–379. [DOI] [PubMed] [Google Scholar]
- Crotty, S., and R. Andino. 2002. Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin. Microbes Infect. 4:1301–1307. [DOI] [PubMed] [Google Scholar]
- Diener, T. O. 1971. Potato spindle tuber “virus.” IV. A replicating, low molecular weight RNA. Virology 45:411–428. [DOI] [PubMed] [Google Scholar]
- Dworkin, J. P., A. Lazcano, and S. L. Miller. 2003. The roads to and from the RNA world. J. Theor. Biol. 222:127–134. [DOI] [PubMed] [Google Scholar]
- Erie, D. A., O. Hajiseyedjavadi, M. C. Young, and P. H. von Hippel. 1993. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262:867–873. [DOI] [PubMed] [Google Scholar]
- Esser, C., N. Ahmadinejad, C. Wiegand et al. (15 co-authors). 2004. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21:1643–1660. [DOI] [PubMed] [Google Scholar]
- Fels, A., K. Hu, and D. Riesner. 2001. Transcription of potato spindle tuber viroid by RNA polymerase II starts predominantly at two specific sites. Nucleic Acids Res. 29:4589–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska, I. J., R. L. Dunn, and R. M. Schaaper. 1993. Mutants of Escherichia coli with increased fidelity of DNA replication. Genetics 134:1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée, J., P. Forterre, T. Sen-Lin, and J. Laurent. 2002. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54:763–773. [DOI] [PubMed] [Google Scholar]
- Fish, R. N., and C. M. Kane. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577:287–307. [DOI] [PubMed] [Google Scholar]
- Flores, R., S. Delgado, M. E. Gas, A. Carbonell, D. Molina, S. Gago, and M. De la Pena. 2004. Viroids: the minimal non-coding RNAs with autonomous replication. FEBS Lett. 567:42–48. [DOI] [PubMed] [Google Scholar]
- Forterre, P. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33:457–465. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Genomics and early cellular evolution. The origin of the DNA world. C. R. Acad. Sci. III 324:1067–1076. [DOI] [PubMed] [Google Scholar]
- ———. 2002. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 5:525–532. [DOI] [PubMed] [Google Scholar]
- Forterre, P., J. Filée, and H. Myllykallio. 2004. Origin and evolution of DNA and DNA replication machineries. Pp. 145–168 in L. R. de Pouplana, ed. The genetic code and the origin of life. Landes Bioscience, Georgetown, Tex.
- Fraenkel-Conrat, H. 1956. The role of the nucleic acid in the reconstitution of active Tobacco Mosaic Virus. J. Am. Chem. Soc. 78:882–883. [Google Scholar]
- Gao, G., M. Orlova, M. M. Georgiadis, W. A. Hendrickson, and S. P. Goff. 1997. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. USA 94:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer, A., and G. Schramm. 1956. Infectivity of ribonucleic acid from Tobacco Mosaic Virus. Nature 177:702–703. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. 1986. The RNA world. Nature 319:618. [Google Scholar]
- Gillet, R., and B. Felden. 2001. Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol. Microbiol. 42:879–885. [DOI] [PubMed] [Google Scholar]
- Grabowski, B., and Z. Kelman. 2003. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 57:487–516. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., K. Gardiner, T. Marsh, N. Pace, and S. Altman. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857. [DOI] [PubMed] [Google Scholar]
- Hagler, J., and S. Shuman. 1993. Nascent RNA cleavage by purified ternary complexes of vaccinia RNA polymerase. J. Biol. Chem. 268:2166–2173. [PubMed] [Google Scholar]
- Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109–1122. [DOI] [PubMed] [Google Scholar]
- Harris, J. K., S. T. Kelley, G. B. Spiegelman, and N. R. Pace. 2003. The genetic core of the Universal Ancestor. Genome Res. 13:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner, W., U. Lange, and M. Musfeldt. 2000. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 275:12393–12399. [DOI] [PubMed] [Google Scholar]
- Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169. [DOI] [PubMed] [Google Scholar]
- Ishihama, A. 1996. A multi-functional enzyme with RNA polymerase and RNase activities: molecular anatomy of influenza virus RNA polymerase. Biochimie 78:1097–1102. [DOI] [PubMed] [Google Scholar]
- Ishihama, A., K. Mizumoto, K. Kawakami, A. Kato, and A. Honda. 1986. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J. Biol. Chem. 261:10417–10421. [PubMed] [Google Scholar]
- Ito, J., and D. K. Braithwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L. M., E. V. Koonin, and L. Aravind. 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares, D. C., A. M. Poole, and D. Penny. 1998. Relics from the RNA world. J. Mol. Evol. 46:18–36. [DOI] [PubMed] [Google Scholar]
- Jeon, C., and K. Agarwal. 1996. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl. Acad. Sci. USA 93:13677–13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, C. M., and T. A. Steitz. 1994. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 63:777–822. [DOI] [PubMed] [Google Scholar]
- Joyce, G. F., and L. E. Orgel. 1999. Prospects for understanding the origin of the RNA world. Pp. 49–77 in R. F. Gesteland, T. R. Cech, and J. F. Atkins, eds. The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Kettenberger, H., K. J. Armache, and P. Cramer. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347–357. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16:955–965. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V. 2003. Comparative genomics, minimal gene-sets and the Last Universal Common Ancestor. Nat. Rev. Microbiol. 1:127. [DOI] [PubMed] [Google Scholar]
- Kornberg, A., and T. A. Baker. 1992. DNA replication. Pp. 90–93. W. H. Freeman and Company, New York.
- Kruger, K., P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling, and T. R. Cech. 1982. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31:147–157. [DOI] [PubMed] [Google Scholar]
- Labeit, S., and B. Kolmerer. 1995. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 270:293–296. [DOI] [PubMed] [Google Scholar]
- Lange, U., and W. Hausner. 2004. Transcriptional fidelity and proofreading in Archaea and implications for the mechanism of TFS-induced RNA cleavage. Mol. Microbiol. 52:1133–1143. [DOI] [PubMed] [Google Scholar]
- Larsson, K. M., A. Jordan, R. Eliasson, P. Reichard, D. T. Logan, and P. Nordlund. 2004. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat. Struct. Mol. Biol. 11:1142–1149. [DOI] [PubMed] [Google Scholar]
- Lehman, N. 2003. A case for the extreme antiquity of recombination. J. Mol. Evol. 56:770–777. [DOI] [PubMed] [Google Scholar]
- Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, D. T., J. Andersson, B. M. Sjoberg, and P. Nordlund. 1999. A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science 283:1499–1504. [DOI] [PubMed] [Google Scholar]
- Lynch, M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller′s ratchet in mitochondrial genomes. Mol. Biol. Evol. 13:209–220. [DOI] [PubMed] [Google Scholar]
- Marra, M. A., S. J. Jones, C. R. Astell et al. (59 co-authors). 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404. [DOI] [PubMed] [Google Scholar]
- Mirkin, B. G., T. I. Fenner, M. Y. Galperin, and E. V. Koonin. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, P. B., and T. A. Steitz. 2002. The involvement of RNA in ribosome function. Nature 418:229–235. [DOI] [PubMed] [Google Scholar]
- Mühlbach, H. P., and H. L. Sanger. 1979. Viroid replication is inhibited by alpha-amanitin. Nature 278:185–188. [DOI] [PubMed] [Google Scholar]
- Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H. F. 2004. The driving force for molecular evolution of translation. RNA 10:1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka, N., M. Chlenov, P. Chacon, W. J. Rice, W. Wriggers, and S. A. Darst. 2003. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114:335–345. [DOI] [PubMed] [Google Scholar]
- Orgel, L. E. 1968. Evolution of the genetic apparatus. J. Mol. Biol. 38:381–393. [DOI] [PubMed] [Google Scholar]
- Orlova, M., J. Newlands, A. Das, A. Goldfarb, and S. Borukhov. 1995. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. USA 92:4596–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland, R., C. M. Zhang, A. Liiv, R. F. Johansen, E. Seeberg, Y. M. Hou, J. Remme, and P. O. Falnes. 2004. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell 16:107–116. [DOI] [PubMed] [Google Scholar]
- Pelchat, M., C. Grenier, and J. P. Perreault. 2002. Characterization of a viroid-derived RNA promoter for the DNA-dependent RNA polymerase from Escherichia coli. Biochemistry 41:6561–6571. [DOI] [PubMed] [Google Scholar]
- Pelchat, M., and J. P. Perreault. 2004. Binding site of Escherichia coli RNA polymerase to an RNA promoter. Biochem. Biophys. Res. Commun. 319:636–642. [DOI] [PubMed] [Google Scholar]
- Peleg, O., V. Kirzhner, E. Trifonov, and A. Bolshoy. 2004. Overlapping messages and survivability. J. Mol. Evol. 59:520–527. [DOI] [PubMed] [Google Scholar]
- Penny, D., and A. Poole. 1999. The nature of the last universal common ancestor. Curr. Opin. Genet. Dev. 9:672–677. [DOI] [PubMed] [Google Scholar]
- Pfeiffer, J. K., and K. Kirkegaard. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 100:7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, A., D. Jeffares, and D. Penny. 1999. Early evolution: prokaryotes, the new kids on the block. Bioessays 21:880–889. [DOI] [PubMed] [Google Scholar]
- Poole, A., D. Penny, and B.-M. Sjöberg. 2000. Methyl-RNA: an evolutionary bridge between RNA and DNA? Chem. Biol. 7:R207–R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, A. M., D. C. Jeffares, and D. Penny. 1998. The path from the RNA world. J. Mol. Evol. 46:1–17. [DOI] [PubMed] [Google Scholar]
- Poole, A. M., D. T. Logan, and B.-M. Sjöberg. 2002. The evolution of the ribonucleotide reductases: much ado about oxygen. J. Mol. Evol. 55:180–196. [DOI] [PubMed] [Google Scholar]
- Pugachev, K. V., F. Guirakhoo, S. W. Ocran et al. (11 co-authors). 2004. High fidelity of yellow fever virus RNA polymerase. J. Virol. 78:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reanney, D. C. 1987. Genetic error and genome design. Cold Spring Harb. Symp. Quant. Biol. 52:751–757. [DOI] [PubMed] [Google Scholar]
- Reichert, A. S., and M. Mörl. 2000. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 28:2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales, R., N. Harris, B. Y. Ahn, and B. Moss. 1994. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J. Biol. Chem. 269:14260–14267. [PubMed] [Google Scholar]
- Rota, P. A., M. S. Oberste, S. S. Monroe et al. (35 co-authors). 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. [DOI] [PubMed] [Google Scholar]
- Santos, M., E. Zintzaras, and E. Szathmáry. 2004. Recombination in primeval genomes: a step forward but still a long leap from maintaining a sizable genome. J. Mol. Evol. 59:507–519. [DOI] [PubMed] [Google Scholar]
- Sastry, S. S., and B. M. Ross. 1997. Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J. Biol. Chem. 272:8644–8652. [DOI] [PubMed] [Google Scholar]
- Scheuring, I. 2000. Avoiding catch-22 of early evolution by stepwise increase in copying fidelity. Selection 1:135–145. [Google Scholar]
- Schindler, I.-M., and H.-P. Mühlbach. 1992. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84:221–229. [Google Scholar]
- Schnapp, G., B. R. Graveley, and I. Grummt. 1996. TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA. Mol. Gen. Genet. 252:412–419. [DOI] [PubMed] [Google Scholar]
- Shaevitz, J. W., E. A. Abbondanzieri, R. Landick, and S. M. Block. 2003. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426:684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai, T., and M. Go. 1991. RNase-like domain in DNA-directed RNA polymerase II. Proc. Natl. Acad. Sci. USA 88:9056–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R. W., L. Bellon, L. Beigelman, and C. C. Kao. 1999. Use of DNA, RNA, and chimeric templates by a viral RNA-dependent RNA polymerase: evolutionary implications for the transition from the RNA to the DNA world. J. Virol. 73:6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintchak, M. D., G. Arjara, B. A. Kellogg, J. Stubbe, and C. L. Drennan. 2002. The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat. Struct. Biol. 9:293–300. [DOI] [PubMed] [Google Scholar]
- Skovgaard, M., L. J. Jensen, S. Brunak, D. Ussery, and A. Krogh. 2001. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 17:425–428. [DOI] [PubMed] [Google Scholar]
- Sosunov, V., E. Sosunova, A. Mustaev, I. Bass, V. Nikiforov, and A. Goldfarb. 2003. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 22:2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova, E., V. Sosunov, M. Kozlov, V. Nikiforov, A. Goldfarb, and A. Mustaev. 2003. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl. Acad. Sci. USA 100:15469–15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391:231–232. [DOI] [PubMed] [Google Scholar]
- ———. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395–17398. [DOI] [PubMed] [Google Scholar]
- Steitz, T. A., and J. A. Steitz. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 90:6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauer, A., and S. A. Benner. 1997. The B12-dependent ribonucleotide reductase from the archaebacterium Thermoplasma acidophila: an evolutionary solution to the ribonucleotide reductase conundrum. Proc. Natl. Acad. Sci. USA 94:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. J., A. A. Platas, and D. K. Hawley. 1998. Transcriptional fidelity and proofreading by RNA polymerase II. Cell 93:627–637. [DOI] [PubMed] [Google Scholar]
- Torrents, E., P. Aloy, I. Gibert, and F. Rodriguez-Trelles. 2002. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55:138–152. [DOI] [PubMed] [Google Scholar]
- Tourasse, N. J., and M. Gouy. 1999. Accounting for evolutionary rate variation among sequence sites consistently changes universal phylogenies deduced from rRNA and protein-coding genes. Mol. Phylogenet. Evol. 13:159–168. [DOI] [PubMed] [Google Scholar]
- Tschochner, H. 1996. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3′ end. Proc. Natl. Acad. Sci. USA 93:12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin, U., and H. Eklund. 1994. Structure of ribonucleotide reductase protein R1. Nature 370:533–539. [DOI] [PubMed] [Google Scholar]
- Vasudevan, S., S. W. Peltz, and C. J. Wilusz. 2002. Non-stop decay—a new mRNA surveillance pathway. Bioessays 24:785–788. [DOI] [PubMed] [Google Scholar]
- Villarreal, L. P., and V. R. DeFilippis. 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 74:7079–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., and D. K. Hawley. 1993. Identification of a 3′→5′ exonuclease activity associated with human RNA polymerase II. Proc. Natl. Acad. Sci. USA 90:843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettich, A., and C. K. Biebricher. 2001. RNA species that replicate with DNA-dependent RNA polymerase from Escherichia coli. Biochemistry 40:3308–3315. [DOI] [PubMed] [Google Scholar]
- White, H. B. III. 1976. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7:101–104. [DOI] [PubMed] [Google Scholar]
- Whitehall, S. K., C. Bardeleben, and G. A. Kassavetis. 1994. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J. Biol. Chem. 269:2299–2306. [PubMed] [Google Scholar]
- Wilusz, C. J., W. Wang, and S. W. Peltz. 2001. Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev. 15:2781–2785. [DOI] [PubMed] [Google Scholar]
- Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C. R. 1967. The genetic code: The molecular basis of genetic expression. Harper and Row, New York.
- Yarus, M. 2002. Primordial genetics: phenotype of the ribocyte. Annu. Rev. Genet. 36:125–151. [DOI] [PubMed] [Google Scholar]
- Zanotto, P. M., M. J. Gibbs, E. A. Gould, and E. C. Holmes. 1996. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 70:6083–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]


