Abstract
With the persistent threat of emerging infectious diseases and bioterrorism, it has become increasingly important that clinicians be able to identify the diseases that might signal the occurrence of these unusual events. Essential to a thoughtful diagnostic approach is understanding when to initiate a public health investigation and how to appropriately use commonly performed microbiology procedures in the sentinel laboratory to evaluate potential pathogens. Although diagnostic test development is evolving rapidly, recognizing many of these pathogens continues to challenge the capabilities of most sentinel laboratories. Therefore, effective, ongoing communication and education among clinicians, infection control personnel, sentinel laboratorians, public health authorities, and Laboratory Response Network reference laboratorians is the key to preparedness.
The emergence of West Nile fever virus (1999) and the deliberate release of anthrax (2001) in the United States were first identified by astute clinicians and laboratorians rather than by the public health surveillance systems [1, 2]. With the ongoing threat of both natural and intentional outbreaks, infectious disease specialists and primary care providers need to understand how to enhance the likelihood of diagnosing and confirming the etiology of these unusual events. The local clinical microbiology laboratory, designated as a sentinel laboratory within the Laboratory Response Network (LRN), plays an important role in facilitating the evaluation of potential bioterrorism and emerging infectious disease events [3–6]. Although testing capabilities and technology continue to rapidly expand, detection of many emerging and reemerging pathogens remains difficult for sentinel laboratories. This article is intended to help clinicians develop a thoughtful approach and communicate with the sentinel laboratory.
Communication and The LRN
Effective communication is the key to preparedness. Several of the potential bioterrorism agents require special media and growth conditions, higher biocontainment levels, and advanced molecular techniques. Many of these microorganisms also are easily overgrown by commensal flora and can be misidentified by commercial biochemical identification systems [4, 7–10]. To confirm the diagnosis in the most expeditious and appropriate manner, it is important to encourage ongoing dialogue between clinicians, sentinel laboratories, and LRN reference laboratories. When possible, it is imperative for clinicians to convey which specific pathogens are suspected and to verify periodically that certain microbiologic orders are being processed. Failure to communicate may lead to delayed detection, greater casualties (patient and laboratory), and hysteria.
Communication begins by having a preconceived outbreak preparedness plan, including a checklist of emergency contacts (e.g., infection control officer, laboratory director, hospital administrator, state public health epidemiologist, and laboratorian). In the United States, if a sentinel laboratory cannot rule out an agent of suspected bioterrorism or emerging infectious disease, then it must be referred to an LRN reference laboratory. Specimens with relatively low suspicion of being a bioterrorist agent or an unusual pathogen can be referred through routine channels. For highly suspected cases, state public health authorities should be contacted immediately. Environmental and high-risk bioterrorism specimens should be transported under the jurisdiction of law enforcement authorities. When a communicable outbreak is suspected, it is important to ensure that local county health departments are integrated early in the response effort.
The LRN [11] is an interactive partnership that organizes existing local, national, and international laboratories to provide the greatest diagnostic expertise in response to bioterrorism, chemical terrorism, and other public health emergencies [12]. Cofounded in 1999 by the Centers for Disease Control and Prevention (CDC), the Federal Bureau of Investigation (FBI), and the Association of Public Health Laboratories (APHL), the LRN is composed of >150 national, public health, hospital-based, environmental, food, chemical, agricultural, veterinary, and military testing facilities in all 50 United States, the United Kingdom, Australia, Canada, and Germany. In addition to routine diagnostic testing, the role of sentinel laboratories (local diagnostic facilities) is to recognize, rule out, and refer potential bioterrorist agents to an LRN reference laboratory using commonly performed clinical microbiology procedures. Reference laboratories in the LRN perform rapid confirmatory testing and toxin detection and maintain biosafety level (BSL)–3 facilities. National laboratories (e.g., CDC) are equipped to provide the most secure containment (BSL-4), perform definitive testing and characterization, and archive potential pathogens.
General Approach
Evaluating potential outbreaks begins with knowing whether the suspected exposure is intentional (overt or covert) or epidemiologically linked to an area where the pathogen is endemic [5, 6]. Overt events are either declared (e.g., by alleged attackers) or have a clinical presentation that is highly suggestive of a particular pathogen (e.g., smallpox). Covert events may occur insidiously and with nonspecific clinical syndromes; thus, they may go unnoticed for several days or weeks. Whereas overt outbreaks would warrant immediate notification of public health officials and LRN reference laboratories, potential covert events might be initially evaluated internally by clinicians, infection control personnel, and the sentinel laboratory. Although hospital and public health authorities should be notified as soon as a select agent is considered, an LRN-confirmed result would be needed to initiate a formal public health response. Key epidemiological clues to suspicious emergency events include sharp rises in the frequency or severity of communicable diseases (including those in animals), an unusual cluster or age distribution, occurrence of novel or rare diseases, presence or lack of an appropriate exposure history, travel to a location that has high-consequence disease transmission, unexplained deaths, or pathogens with unusual antimicrobial resistance.
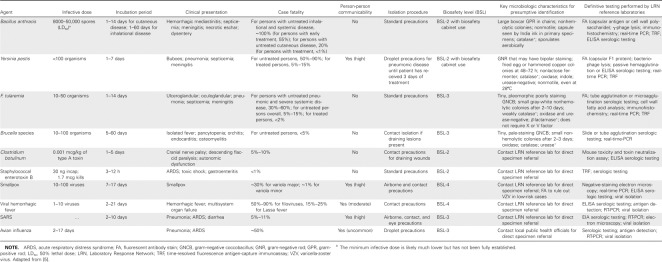
The following are several principles that should guide clinicians and laboratorians with respect to sentinel laboratory evaluation of potential bioterrorist or emerging infectious disease agents. (1) Obtain optimum specimen collection instructions from the sentinel laboratory and alert them to the possibility of an unusual, fastidious, or dangerous pathogen (table 1). (2) Limit culture manipulation by sentinel laboratories to what is required for referral to an LRN reference laboratory, to maximize speed, accuracy, and safety. (3) Do not inoculate highly suspected smallpox, hemorrhagic fever viruses, alphaviruses, or any unknown viral agents of potential bioterrorism into cell culture. Smallpox, herpes B, and Ebola viruses can replicate in human and nonhuman primate cell lines, including those used for herpesvirus culture [13]. Local public health authorities or the CDC should be contacted prior to specimen collection. (4) Do not send environmental (e.g., packages, powders, letters, soil, or water), food, animal, or plant specimens to sentinel laboratories for analysis or culture; refer them directly to an LRN reference laboratory [9, 10, 13–15]. (5) Restrict manipulation of certain potential agents (e.g., Francisella tularensis, Brucella species, Coxiella burnetii, Burkholderia mallei, and Burkholderia pseudomallei) to environments under certified class II biological safety cabinet or BSL-3 conditions, because of the high risk of laboratory-acquired infection. Plate sniffing and any technique that could generate infectious aerosols are prohibited.
Table 1.
Specimen collection guidelines listed by bacteria, toxins, and viruses.
Syndrome-Based Approach
Diagnosis of infections caused by unusual pathogens that present with nonspecific clinical syndromes (e.g., pneumonitis, rash, meningoencephalitis, and gastroenteritis) can prove to be extremely challenging. Even in the midst of a known special pathogen epidemic (e.g., severe acute respiratory syndrome [SARS]), endemic diseases that share similar manifestations (e.g., human influenza or pneumococcal pneumonia) will continue to occur. For respiratory syndromes (figure 1) that initially mimic community-acquired pneumonia or influenza-like illness, epidemiologic clues (e.g., occupational and travel histories) have been critical in identifying inhalational anthrax, SARS, and avian influenza [18]. Acute or generalized febrile vesicular (or pustular) eruptions cause great concern because of the possibility that they may be smallpox or monkeypox (figure 2).
Figure 1.
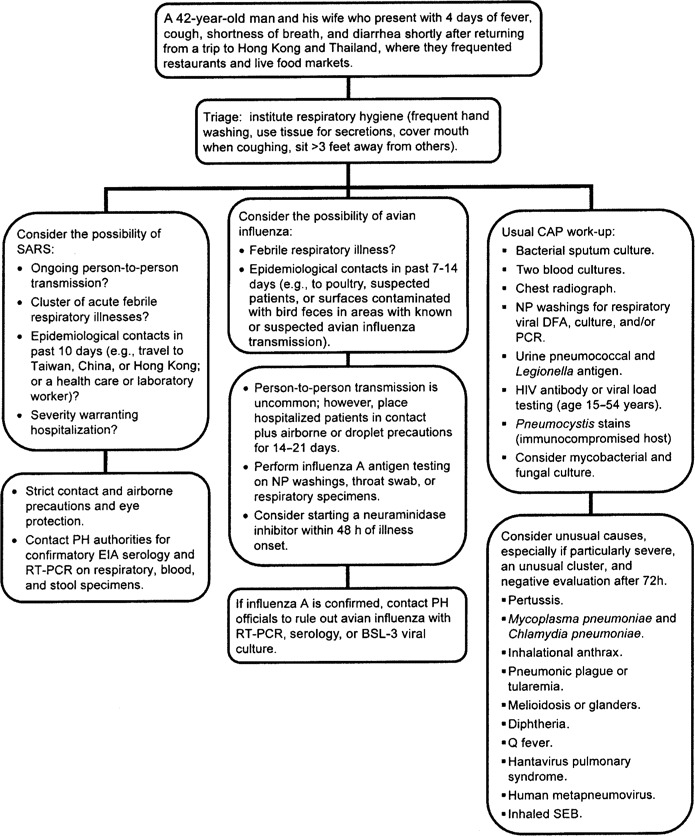
Sample algorithm illustrating the approach to a hypothetical cluster of acute febrile respiratory illnesses with epidemiological links to severe acute respiratory syndrome (SARS) or avian influenza. BSL-3, biosafety level—3; CAP, community-acquired pneumonia; DFA, direct fluorescent antibody stain; NP, nasopharyngeal; PH, public health; SEB, staphylococcal enterotoxin B. Adapted from [16, 17].
Figure 2.
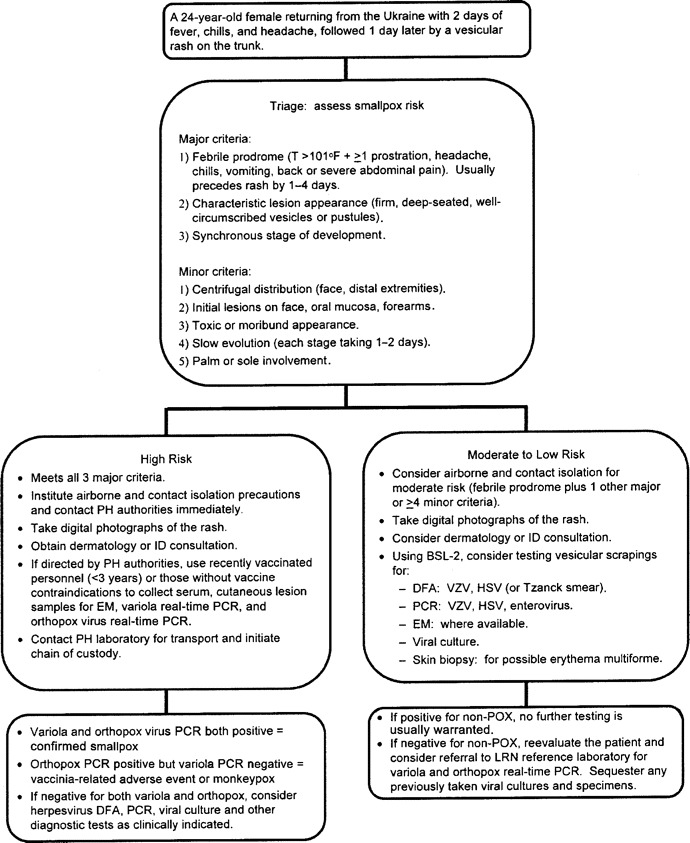
Sample algorithm illustrating the approach to a hypothetical patient with an acute generalized febrile vesicular rash. BSL-2, biosafety—level 2; DFA, direct fluorescent antibody stain; EM, electron microscopy; HSV, herpes simplex virus; ID, infectious diseases; PH, public health; VZV, varicella-zoster virus. Adapted from [50].
Agent-Specific Consideration
This section will describe the clinical presentation of category A agents, selected category B agents, SARS, and avian influenza and will discuss potential practical problems with laboratory identification and personnel exposure (table 2). All diagnostic laboratories should have access to a clinical microbiologist who has been certified by the American Board of Medical Microbiology, American Board of Pathology, or an equivalent certifying body, to strengthen their ability to assess difficult issues in laboratory operation and to support infectious disease practice. The CDC, American Society for Microbiology, and Association of Public Health Laboratories regularly update testing protocols for sentinel laboratories [19–21].
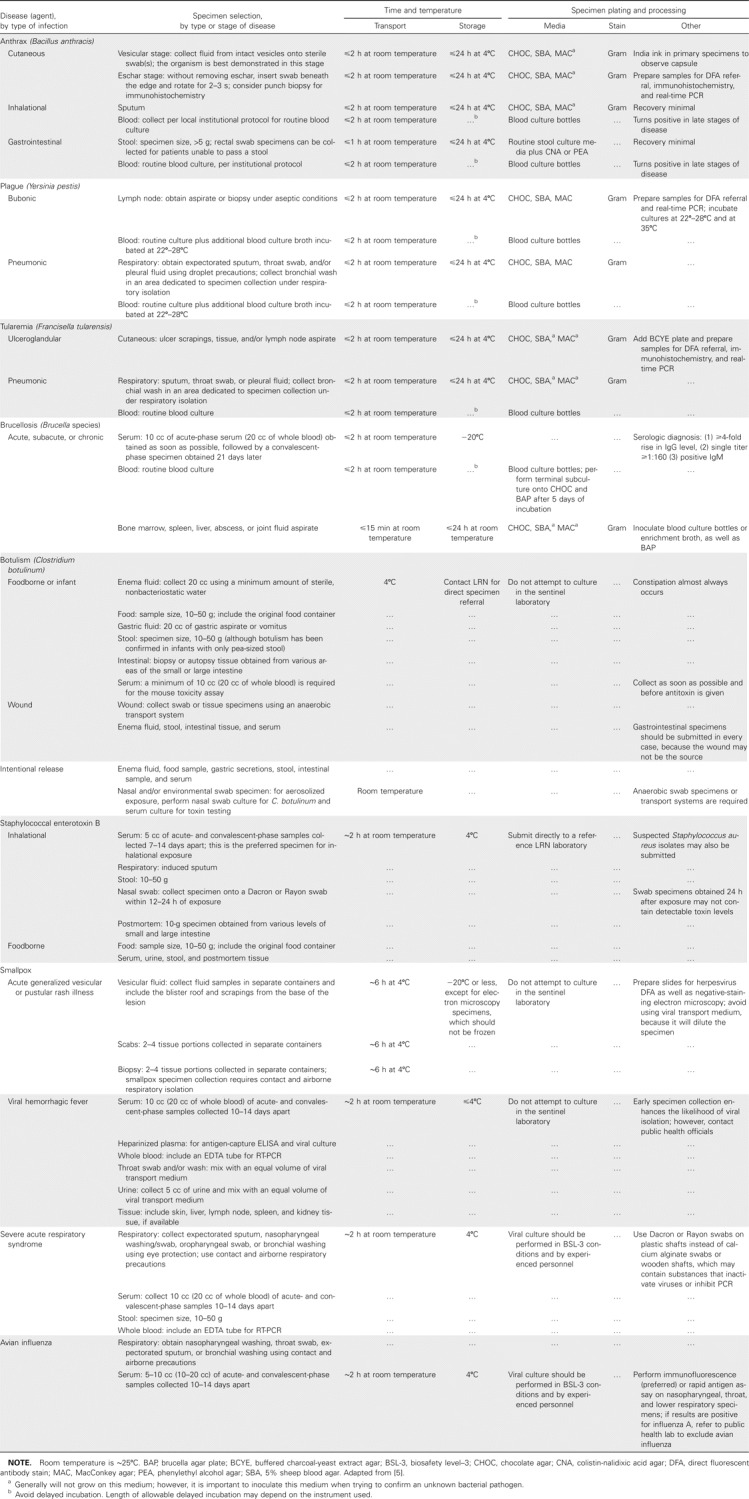
Table 2.
Clinical and microbiological characteristics listed by bacteria, toxins, and viruses.
Several unusual and infrequently encountered pathogens have been misidentified by the commercial identification systems heavily relied on by most sentinel labs [4, 7–10]; however, the biochemical profiles of each system are frequently revised to improve accuracy [22]. Multiple biothreat detection platforms, including nucleic acid amplification and antibody-based technologies, have been developed for real-time and field-condition use and are discussed elsewhere [23, 24].
Anthrax. Bacillus anthracis is an aerobic, spore-forming, gram-positive rod that causes cutaneous and systemic disease, usually in areas where animal vaccination is not performed [25]. Cutaneous infection usually presents as a small, painless, pruritic papule that develops into a vesicle, which then develops into a necrotic eschar with regional lymphadenopathy and significant edema. In the first cluster of anthrax attacks, several cases of cutaneous disease were not recognized until other confirmed cases were reported [26, 27]. Similarly, early symptoms of inhalational disease (fever, malaise, and myalgias) are hard to distinguish from influenza-like illness and community-acquired pneumonia [18, 28]. However, within a few days, patients' conditions rapidly deteriorate, with development of severe dyspnea, hypoxemia, widened mediastinum, and hemorrhagic pleural effusions evident on chest radiographs. At this point, the disease is frequently fatal, although early initiation of antimicrobial therapy during the 2001 anthrax attacks improved the survival rate to 55% [29, 30]. Gastrointestinal anthrax presents with pharyngitis, severe neck swelling, abdominal pain, or hemorrhagic ascites. Depending on the organ system involved, anthrax is diagnosed by culturing vesicular fluid, pleural fluid, blood, sputum, peritoneal fluid, CSF, or stool specimens. Gram staining reveals large, “boxcar” gram-positive rods, and in primary specimens, a capsule can be observed with India ink preparation. Colonies are characteristically nonhemolytic, ground-glass opaque with irregular edges, and tenaciously adherent. A presumptive identification can be made by sentinel laboratories on the basis of Gram stain appearance, rapid aerobic growth, colony morphology, catalase production, encapsulation, lack of motility, and aerobic spore formation [15]. Due to laboratory safety concerns, motility testing should be performed using motility medium tubes, rather than wet preparation. For similar reasons, India ink preparation and spore staining may not be necessary for LRN referral if the other aforementioned criteria are consistent with B. anthracis infection. Communication between the laboratory and clinicians is vital to compare whether the clinical presentation is consistent with anthrax and to help ensure that clinicians are aware of the preliminary laboratory findings, so that patients can be appropriately cared for. Definitive confirmation is conducted at reference LRN laboratories by direct fluorescent antibody staining, time-resolved fluorescence antigen-capture immunoassay, phage lysis, and real-time PCR [3, 15, 24, 25, 31]. Antimicrobial susceptibility testing may be performed to exclude natural or genetically-engineered resistance using Clinical and Laboratory Standards Institute (CLSI) criteria [32]. Screening of nasal swab specimens is reserved for formal epidemiological investigations.
Plague. Yersinia pestis is a small, gram-negative rod transmitted by bites of infected rodent fleas. Foci of plague are endemic to many parts of the world and attempted bioterrorist attacks using Y. pestis date back to the 14th century [33]. After an incubation period of 1–7 days, naturally-acquired disease often begins with febrile lymphadenitis (buboes), whereas an aerosolized attack would likely manifest as an outbreak of pneumonia. Septicemia and meningitis often result from advanced illness. The diagnosis is made by culturing lymph node aspirate, sputum, pleural fluid, blood, or CSF. Although bipolar staining is more consistently observed by Wright or Giemsa staining, it can be observed by Gram staining. It should be noted that other Enterobacteriaceae and Pasteurella species may also exhibit bipolar staining; thus, this is not exclusive for Y. pestis. Y. pestis grows on routine bacteriologic media, with optimal growth at 25–28°C. Pinpoint gray-white colonies appear after 24 h of incubation, and after 48–72 h of growth, the typical fried egg or hammered-copper colony morphologies become evident. Y. pestis is catalase positive, ferments glucose but not lactose, and is oxidase-negative, indole-negative, and urease-negative. Unlike other Yersinia species that are motile at 22–30°C, Y. pestis is nonmotile. Although older commercial systems (e.g., BBL Crystal Enteric—Nonfermenter System, version 3.0 [BD Microbiology Systems], and API, version 10.0 [bioMérieux]) have misidentified Y. pestis as Yersinia pseudotuberculosis, Shigella species, H2S-negative Salmonella species, or Acinetobacter species [10, 22, 34], newer panels (e.g., API 20E, version 4.0 [bioMérieux], and the MicroScan Rapid Gram-Negative Identification Type 4 database [Dade Behring]), appear to provide more accurate identification [35]. Y. pestis is confirmed by LRN reference laboratories using fluorescent antibody techniques and bacteriophage lysis [10]. Although it has not been adequately developed for definitive confirmatory testing, a real-time PCR assay exists in the LRN [24]. A rapid dipstick test that detects the capsular F1 antigen has demonstrated a high degree of sensitivity, specificity, and reproducibility, but it is not widely available [36]. CLSI antimicrobial susceptibility testing criteria are available for Y. pestis [32].
Tularemia. Francisella tularensis is a small, pleomorphic, gram-negative coccobacillus carried by a variety of insect vectors that feed on wild lagomorpha, deer, and rodents [37]. Laboratory-acquired infections have also occurred; thus, culture manipulation in a biological safety cabinet or BSL-3 laboratory is recommended. After 2–10 days of incubation, patients develop fever, chills, headache, and malaise, followed by ulceroglandular, glandular, oculoglandular, oropharyngeal, pneumonic, or typhoidal syndromes. In naturally acquired infection, ulceroglandular disease most frequently occurs and presents with ⩾1 papuloulcerative lesion with central eschar and tender regional lymphadenopathy. The diagnosis may be confirmed by 4-fold increases in the tube agglutination serologic titer, or the organism can be cultured from lymph node aspirate, ulcer scrapings, sputum, pleural fluid, and blood. By Gram staining, the organism appears as tiny, pleomorphic, poorly-staining, gram-negative coccobacilli. F. tularensis primarily grows on cysteine-supplemented media, including chocolate, Thayer-Martin, and buffered charcoal yeast extract agars, and >48 h must pass before the small, gray-white colonies become visible. Because of its fastidious nature, colonies may be mistaken for Haemophilus influenzae [38]; however, F. tularensis does not require X (hemin) factor, V (NAD) factor, or form satellite growth around β-hemolytic colonies. F. tularensis is weakly catalase positive but oxidase negative and urease negative. Commercial biochemical systems (e.g., Vitek NHI panel; bioMérieux) [39] may incorrectly identify it as Actinobacillus actinomycetemcomitans with 99% certainty; however, F. tularensis is β-lactamase—positive [9]. F. tularensis often fails to grow on automated biochemical panels (e.g., MicroScan NEG Combo Type 25; Dade Behring) [40]; however, reliable identification has been reported using the Vitek 2 GN card (bioMérieux) [16]. Once received by a reference LRN laboratory, the identity is confirmed with direct fluorescent antibody staining, typical growth characteristics, and real-time PCR [9, 24, 42]. As for B. anthracis and Y. pestis, CLSI antimicrobial susceptibility testing criteria is available for F. tularensis [32].
Botulism. Clostridium botulinum is a neurotoxin-producing, obligate anaerobic, spore-forming, gram-positive bacillus [43]. The neurotoxin irreversibly inhibits the release of acetylcholine at presynaptic terminals, thereby causing cranial nerve palsies and afebrile descending symmetric flaccid paralysis. Other syndromes that may mimic botulism are myasthenia gravis, Lambert-Eaton syndrome, polio, tick paralysis, Guillain-Barre syndrome, brainstem stroke, and heavy metal intoxication. Botulism is usually foodborne or produced in wounds infected with C. botulinum. It is the most potent bacterial toxin and can be absorbed by mucosal surfaces or respiratory tracts. Therefore, sentinel labs should not attempt culture or toxin analysis, because skin contact may induce severe symptoms in the laboratorian. A specialized LRN reference lab should be contacted to receive suspected food sources, environmental swab specimens, stool samples, gastric secretions, and wound samples. Serum should be drawn before antitoxin is given. Mouse toxicity bioassay and toxin neutralization studies are used for confirmatory testing, making it important to ascertain whether the patient received any drugs with neuromuscular activity [14, 44]. Real-time PCR assays for neurotoxins A and B are also being developed in the LRN [24].
Smallpox. Variola virus is a large, brick-shaped, enveloped DNA Orthopoxvirus. Although no known smallpox cases have occurred since 1978, the threat of security breaches in collections previously produced by the former Union of Soviet Socialist Republics have placed it high on the list of potential bioterrorist agents [45, 46]. After 7–17 days of incubation, almost all patients experience a febrile prodrome, consisting of fever plus headache, chills, prostration, nausea, malaise, backache, or severe abdominal pain. About 1–4 days later, the typical smallpox rash emerges as papules that evolve into generalized firm, deep-seated, well-circumscribed nodules, vesicles, or pustules. Varicella-zoster virus, which causes chickenpox and shingles, is the agent most likely to be confused with smallpox, and 7%–17% of unvaccinated patients with chickenpox may meet the smallpox febrile prodrome criteria [47]. However, varicella produces crops of blisters in various stages of development, whereas smallpox lesions generally remain in a synchronous stage of development and evolve slowly, with each stage lasting 1–2 days. Furthermore, smallpox lesions first appear on the periphery (e.g., on the face, mouth, hands, and feet, including palms and soles), whereas varicella usually presents on the trunk. The early presentation of smallpox may also resemble infection with other poxviruses (e.g., monkeypox, vaccinia, orf, and molluscum contagiosum), bullous impetigo, contact dermatitis, drug eruptions, erythema multiforme, hand-foot-mouth disease, and eczema herpeticum. Only personnel who have been successfully vaccinated against smallpox within 3 years should be allowed to collect specimens. Specimens obtained from patients at moderate or low risk may undergo sentinel laboratory testing using herpesvirus-specific direct fluorescent antibody staining, PCR (for varicella-zoster virus, herpes simplex virus, and enterovirus), electron microscopy, and viral culture of vesicle scrapings. Electron microscopy is the fastest method (15–20 min) for distinguishing herpesviruses from poxviruses (figure 3), but it may not be available at all laboratories. It can, however, be performed by select LRN reference laboratories [48, 49]. Furthermore, whereas electron microscopy can differentiate between parapoxviruses (e.g., orf) and orthopoxviruses (e.g., vaccinia and variola), it cannot distinguish variola from other orthopoxviruses. Direct fluorescent antibody testing is one of the most rapid and widely available ways of confirming varicella-zoster virus, thus essentially excluding variola. A skin biopsy and bacterial culture can evaluate erythema multiforme and impetigo. If none of these tests are diagnostic, the patient's condition and the need for dermatologic and histologic testing should be reevaluated. Orthopoxvirus real-time PCR assays can be performed at LRN reference laboratories for definitive confirmation [3, 50, 51].
Figure 3.
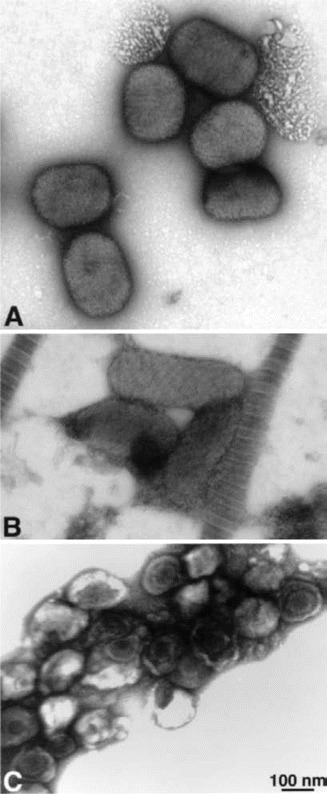
Electron microscopy of negatively stained viruses. A, Viruses in the Orthopox genus (e.g., variola, vaccinia, monkeypox) and viruses in the Molluscipox genus (e.g., molluscum contagiosum) are morphologically similar. They are large, brick-shaped particles, ∼200 nm × 250–300 nm in size. Depending on the number and configuration of membranes around the particles, the surfaces may appear somewhat rough or with deep ridges. B, Parapox viruses (e.g., orf and Milker nodule virus) are slimmer and more elongated, 140–170 nm × 220–300 nm in size. The surface of these viruses frequently appears to have criss-crossed striations, like rope wrapped diagonally around the particle. C, Herpesviruses (e.g., varicella-zoster virus, human herpes virus, or herpes simplex virus) are 120–200 nm in diameter, with icosahedral nucleocapsids of ∼100 nm in diameter. Smallpox or variola can be rapidly (in 10–20 min) and easily distinguished morphologically from herpesviruses and parapoxviruses by negatively staining vesicle fluid. Electron photomicrograph is courtesy of Dr. Sara E. Miller.
Viral hemorrhagic fevers. The agents of viral hemorrhagic fevers include Filoviridae viruses (e.g., Ebola and Marburg viruses), Arenaviridae viruses (e.g., Lassa, Junin, Machupo, and Guanarito viruses), Bunyaviridae viruses (e.g., hantavirus, Rift Valley fever, and nairovirus viruses), and Flaviviridae viruses (e.g., yellow fever, Omsk hemorrhagic fever, and Kyasanur Forest disease viruses) [52]. Although these are all enveloped RNA viruses and many are zoonoses, they have diverse geographic distributions and host reservoirs. The recent outbreak of Marburg virus in northern Angola occurred between October 2004 and July 2005, resulting in 374 cases and a mortality rate of 88% [53]. After 5–10 days of incubation, the typical clinical presentation is acute fever, headache, and myalgias followed by prostration, maculopapular rash, sore throat, cough, chest and abdominal pain, vomiting, and diarrhea. A few days later, jaundice, delirium, severe weight loss, hemorrhagic manifestions, multisystem organ failure, and shock occur. Whereas early specimen collection gives the best chance of viral isolation, it is essential to initiate contact, eye, and respiratory precautions, to obtain a travel history, and to contact the local public health department. Paired serum samples, heparinized plasma samples, EDTA whole blood samples, throat washings, urine specimens, and tissue specimens should be submitted to a national LRN laboratory for antibody testing, ELISA antigen detection, RT-PCR, and BSL-4 viral culture [13].
SARS. Although there has been no known severe natural recurrence of SARS since July 2003, the SARS-associated coronavirus is highly contagious through aerosolized droplets, stool, and contaminated fomites [54]. After a 2–10 day incubation period, the usual presentation of SARS begins with fever, headache, and malaise followed by cough and progressive dyspnea. The use of SARS testing should be judicious, particularly in the absence of ongoing person-to-person transmission, because false-positive results are more likely to occur when the pretest probability of disease is low. Therefore, testing should be aimed toward clusters of patients with an undiagnosed febrile respiratory illness severe enough to warrant hospitalization and who have one of the following epidemiological links within 10 days of onset: travel to or close contact with an ill person from China, Hong Kong, or Taiwan; employment as a health care worker with direct patient contact; or employment as a laboratorian working near live SARS coronavirus [17]. Patients at high risk for SARS should be placed in airborne and contact isolation until public health officials are consulted. Initially, electron microscopy was invaluable in the identification of a coronavirus as the etiological agent, because molecular- and culture-based detection systems centered on respiratory viruses in other families [55]. Presently, SARS testing involves enzyme immunoassay serologic and RT-PCR on respiratory, blood, and stool specimens [56, 57]; however, because several laboratory-acquired cases have occurred, strict handling precautions must be observed [58].
Avian influenza. Highly pathogenic avian influenza is caused by certain strains of type A influenza virus that primarily cause symptomatic infection in domesticated fowl and swine [59, 60]. Until 1997, avian influenza rarely caused human disease, but beginning in December 2003, a severe H5N1 outbreak was declared in Vietnam, Thailand, Cambodia, China, Indonesia, Turkey, Azerbaijan, and Iraq, resulting in 186 confirmed human cases and 105 deaths [61]. Similar to SARS, the clinical features of avian influenza are fairly nonspecific; however, the current spread of avian influenza is likely to continue. Testing should be considered for patients with a febrile respiratory illness who, within 7–14 days, patients who have had exposure to poultry (within 1 meter), patients with influenza A, patients with a severe unexplained respiratory illness, or surfaces contaminated with bird feces in areas with confirmed or suspected avian influenza activity [59, 62]. This is particularly applicable if the illness is severe enough to warrant hospitalization, the patient has radiographically confirmed pneumonia, and an alternative diagnosis has not been established [63]. Sentinel laboratories may test for influenza A using immunofluorescence and rapid immunochromatographic antigen detection assays; however, these do not distinguish avian influenza A from human influenza A. Furthermore, unlike human influenza, rapid antigen tests may have lower sensitivity for avian influenza virus, and throat samples may provide a greater yield than nasopharyngeal specimens [59]. Confirmatory avian influenza testing using paired serum samples, BSL-3 viral culture, or a recently approved influenza A/H5 (Asian lineage) subtyping RT-PCR assay are available by contacting state public health authorities [60, 64].
Summary
Astute clinicians and laboratorians will continue to detect the emergence and reemergence of both naturally occurring and intentional infectious disease outbreaks. To be optimally prepared, effective communication among clinicians, public health authorities, sentinel laboratories, and LRN reference laboratories is critical. Knowing when to initiate a public health investigation and how to use commonly performed microbiological procedures in the sentinel laboratory to evaluate potential bioterrorism and emerging infections is essential to the accurate and rapid confirmation of the causative agent.
Acknowledgments
We thank Dr. J. Michael Miller of the Bioterrorism Preparedness and Response Program at the Centers for Disease Control and Prevention for reviewing this manuscript.
Financial support. Department of Veterans Affairs Special Fellowship Program in Health Services Research (to B.C.P.) and National Institute of Allergy and Infectious Diseases (grant UO1AI066569 to C.W.W.).
Potential conflicts of interest. C.W.W. is a consultant for Roche Diagnostics and Sanofi Pasteur and has received grants from Cubist Pharmaceuticals. All other authors: no conflicts.
References
- 1.Gerberding JL, Hughes JM, Koplan JP. Bioterrorism preparedness and response: clinicians and public health agencies as essential partners. JAMA. 2002;287:898–900. doi: 10.1001/jama.287.7.898. [DOI] [PubMed] [Google Scholar]
- 2.Nash D, Mostashari F, Fine A, et al. The outbreak of West Nile virus infection in the New York City area in 1999. N Engl J Med. 2001;344:1807–14. doi: 10.1056/NEJM200106143442401. [DOI] [PubMed] [Google Scholar]
- 3.Cockerill FR, III, Smith TF. Response of the clinical microbiology laboratory to emerging (new) and reemerging infectious diseases. J Clin Microbiol. 2004;42:2359–65. doi: 10.1128/JCM.42.6.2359-2365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klietmann WF, Ruoff KL. Bioterrorism: implications for the clinical microbiologist. Clin Microbiol Rev. 2001;14:364–81. doi: 10.1128/CMR.14.2.364-381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JM. Agents of bioterrorism. Preparing for bioterrorism at the community health care level. Infect Dis Clin North Am. 2001;15:1127–56. doi: 10.1016/s0891-5520(05)70189-6. [DOI] [PubMed] [Google Scholar]
- 6.Snyder JW. Role of the hospital-based microbiology laboratory in preparation for and response to a bioterrorism event. J Clin Microbiol. 2003;41:1–4. doi: 10.1128/JCM.41.1.1-4.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Society for Microbiology . Sentinel laboratory guidelines for suspected agents of bioterrorism. Burkholderia mallei and B. pseudomallei. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 8.American Society for Microbiology . Sentinel laboratory guidelines for suspected agents of bioterrorism: Brucella species. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 9.Centers for Disease Control and Prevention, American Society for Microbiology, Association of Public Health Laboratories . Basic protocols for level A laboratories for the presumptive identification of Francisella tularensis. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 10.American Society for Microbiology . Sentinel laboratory guidelines for suspected agents of bioterrorism: Yersinia pestis. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 11.The Laboratory Response Network . Available at: http://www.bt.cdc.gov/lrn. Accessed 24 March 2006. [Google Scholar]
- 12.Morse S, Kellog R, Perry S, et al. In: The Laboratory Response Network Preparedness against bioterrorism and re-emerging infectious diseases. Kocik J, Janiak M, Negut M, editors. Amsterdam, the Netherlands: IOS Press; 2004. [Google Scholar]
- 13.American Society for Microbiology . Sentinel laboratory guidelines for suspected agents of bioterrorism: unknown viruses. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 14.American Society for Microbiology . Sentinel laboratory guidelines for suspected agents of bioterrorism: botulinum toxin. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 15.Centers for Disease Control and Prevention, American Society for Microbiology, Association of Public Health Laboratories . Basic diagnostic testing protocols for level A laboratories for the presumptive identification of Bacillus anthracis. Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 21 September 2005. [Google Scholar]
- 16.Centers for Disease Control and Prevention . Interim recommendations for persons with possible exposure to avian influenza during outbreaks among poultry in the United States. Available at: http://www.cdc.gov/flu/avian/professional/possible-exposure.htm. Accessed 24 March 2006. [Google Scholar]
- 17.Centers for Disease Control and Prevention . In the absence of SARS-CoV transmission worldwide: guidance for surveillance, clinical and laboratory evaluation, and reporting, version 2. Available at: http://www.cdc.gov/ncidod/sars/absenceofsars.htm. Accessed 24 March 2006. [Google Scholar]
- 18.Howell JM, Mayer TA, Hanfling D, et al. Screening for inhalational anthrax due to bioterrorism: evaluating proposed screening protocols. Clin Infect Dis. 2004;39:1842–7. doi: 10.1086/426080. [DOI] [PubMed] [Google Scholar]
- 19.Sentinel level clinical microbiology laboratory guidelines . Available at: http://www.asm.org/Policy/index.asp?bid=6342. Accessed 24 March 2006. [Google Scholar]
- 20.Emergency preparedness and response: laboratory information. Available at: http://www.bt.cdc.gov/labissues/. Accessed 24 March 2006.
- 21. Available at: http://www.aphl.org/programs/emergency_preparedness/. Accessed 24 March 2006.
- 22.O'Hara CM. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev. 2005;0:147–62. doi: 10.1128/CMR.18.1.147-162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim DV, Simpson JM, Kearns EA, Kramer MF. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin Microbiol Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen DR, Hartman LJ, Loveless BM, et al. Detection of biological threat agents by real-time PCR: comparison of assay performance on the R.A.P.I.D., the LightCycler, and the Smart Cycler Platforms. Clin Chem. 2006;52:141–5. doi: 10.1373/clinchem.2005.052522. [DOI] [PubMed] [Google Scholar]
- 25.Inglesby TV, O'Toole T, Henderson DA, et al. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA. 2002;287:2236–52. doi: 10.1001/jama.287.17.2236. [DOI] [PubMed] [Google Scholar]
- 26.Freedman A, Afonja O, Chang MW, et al. Cutaneous anthrax associated with microangiopathic hemolytic anemia and coagulopathy in a 7-month-old infant. JAMA. 2002;287:869–74. doi: 10.1001/jama.287.7.869. [DOI] [PubMed] [Google Scholar]
- 27.Jernigan DB, Raghunathan PL, Bell BP, et al. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg Infect Dis. 2002;8:1019–28. doi: 10.3201/eid0810.020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehnert MJ, Doyle TJ, Hill HA, et al. Clinical features that discriminate inhalational anthrax from other acute respiratory illnesses. Clin Infect Dis. 2003;36:328–36. doi: 10.1086/346035. [DOI] [PubMed] [Google Scholar]
- 29.Jernigan JA, Stephens DS, Ashford DA, et al. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis. 2001;7:933–44. doi: 10.3201/eid0706.010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sejvar JJ, Tenover FC, Stephens DS. Management of anthrax meningitis. Lancet Infect Dis. 2005;5:287–95. doi: 10.1016/S1473-3099(05)70113-4. [DOI] [PubMed] [Google Scholar]
- 31.Bell CA, Uhl JR, Hadfield TL, et al. Detection of Bacillus anthracis DNA by LightCycler PCR. J Clin Microbiol. 2002;40:2897–902. doi: 10.1128/JCM.40.8.2897-2902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. CLSI document M100-S15. Wayne, PA: CLSI; 2005. pp. 136–7. [Google Scholar]
- 33.Inglesby TV, Dennis DT, Henderson DA, et al. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 2000;283:2281–90. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- 34.Wilmoth BA, Chu MC, Quan TJ. Identification of Yersinia pestis by BBL crystal enteric/nonfermenter identification system. J Clin Microbiol. 1996;34:2829–30. doi: 10.1128/jcm.34.11.2829-2830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roe-Carpenter D, Nothaft DM, Bobolis J, et al. Program and abstracts of the 105th General Meeting of the American Society for Microbiology (Atlanta) Washington, DC: American Society for Microbiology; 2005. Performance of the MicroScan rapid gram-negative identification type 4 (RNID4) database [abstract C-200] [Google Scholar]
- 36.Chanteau S, Rahalison L, Ralafiarisoa L, et al. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet. 2003;361:211–6. doi: 10.1016/S0140-6736(03)12270-2. [DOI] [PubMed] [Google Scholar]
- 37.Dennis DT, Inglesby TV, Henderson DA, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–73. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 38.Ross JJ. A young man from Nantucket. Clin Infect Dis. 2004;38:1139–7. doi: 10.1086/383061. [DOI] [PubMed] [Google Scholar]
- 39.Clarridge JE, III, Raich TJ, Sjosted A, et al. Characterization of two unusual clinically significant Francisella strains. J Clin Microbiol. 1996;34:1995–2000. doi: 10.1128/jcm.34.8.1995-2000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roe DE, Janda JM, Lindquist D, et al. Program and abstracts of the 101st General Meeting of the American Society for Microbiology (Orlando) Washington, DC: American Society for Microbiology; 2001. Evaluation of the MicroScan panels for the growth and detection of selected group 2 agents [abstract C-340] p. 230. [Google Scholar]
- 41.Garin-Bastuji B, Vaissaire J, Albert D, et al. Program and abstracts of the 104th General Meeting of the American Society for Microbiology (New Orleans) Washington, DC: American Society for Microbiology; 2004. Evaluation of the VITEK 2 system for the identification of highly pathogenic bacteria: Brucella spp., Burkholderia mallei, and B. pseudomallei, Francisella tularensis subsp. holarctica, Yersinia pestis, and Bacillus anthracis [abstract C-177] [Google Scholar]
- 42.Versage JL, Severin DD, Chu MC, Petersen JM. Development of a multitarget real-time TaqMan PCR assay for enhanced detection of Francisella tularensis in complex specimens. J Clin Microbiol. 2003;41:5492–9. doi: 10.1128/JCM.41.12.5492-5499.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnon SS, Schechter R, Inglesby TV, et al. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–70. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 44.Sobel J. Botulism. Clin Infect Dis. 2005;41:1167–73. doi: 10.1086/444507. [DOI] [PubMed] [Google Scholar]
- 45.Breman JG, Henderson DA. Diagnosis and management of smallpox. N Engl J Med. 2002;346:1300–8. doi: 10.1056/NEJMra020025. [DOI] [PubMed] [Google Scholar]
- 46.Henderson DA, Inglesby TV, Bartlett JG, et al. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 1999;281:2127–37. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 47.Moore ZS, Seward JF, Watson BM, Maupin TJ, Jumaan AO. Chickenpox or smallpox: the use of the febrile prodrome as a distinguishing characteristic. Clin Infect Dis. 2004;39:1810–7. doi: 10.1086/426026. [DOI] [PubMed] [Google Scholar]
- 48.Hazelton PR, Gelderblom HR. Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg Infect Dis. 2003;9:294–303. doi: 10.3201/eid0903.020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller SE. Bioterrorism and electron microscopic differentiation of poxviruses from herpesviruses: dos and don'ts. Ultrastruct Pathol. 2003;27:133–40. doi: 10.1080/01913120309932. [DOI] [PubMed] [Google Scholar]
- 50.Centers for Disease Control and Prevention . Acute, generalized vesicular or pustular rash illness testing protocol in the United States. Available at: http://www.bt.cdc.gov/agent/smallpox/diagnosis/rashtestingprotocol.asp. Accessed 21 September 2005. [Google Scholar]
- 51.Olson VA, Laue T, Laker MT, et al. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J Clin Microbiol. 2004;42:1940–6. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borio L, Inglesby T, Peters CJ, et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 53.World Health Organization . Marburg haemorrhagic fever in Angola—update 25. Available at: http://www.who.int/csr/don/2005_08_24/en/index.html. Accessed 21 September 2005. [Google Scholar]
- 54.Christian MD, Poutanen SM, Loutfy MR, Muller MP, Low DE. Severe acute respiratory syndrome. Clin Infect Dis. 2004;38:1420–7. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 56.Emery SL, Erdman DD, Bowen MD, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–6. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lau SK, Che XY, Woo PC, et al. SARS coronavirus detection methods. Emerg Infect Dis. 2005;11:1108–11. doi: 10.3201/eid1107.041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang G, Chen Q, Xu J, et al. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg Infect Dis. 2004;10:1774–81. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beigel JH, Farrar J, Han AM, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–85. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 60.World Health Organization (WHO) WHO manual on animal influenza: diagnosis and surveillance. Available at: http://www.wpro.who.int/NR/rdonlyres/EFD2B9A7-2265-4AD0-BC98-97937B4FA83C/0/manualonanimalaidiagnosisandsurveillance.pdf. Accessed 21 September 2005. [Google Scholar]
- 61.World Health Organization (WHO) Cumulative number of confirmed human cases of avian influenza A/ (H5N1) reported to the WHO. Available at: http://www.who.int/csr/disease/avian_influenza/country/cases_table_2006_03_24/en/index.html. Accessed 24 March 2006. [Google Scholar]
- 62.Centers for Disease Control and Prevention . Interim recommendations for persons with possible exposure to avian influenza during outbreaks among poultry in the United States. Available at: http://www.cdc.gov/flu/avian/professional/possible-exposure.htm. Accessed 21 September 2005. [Google Scholar]
- 63.United States Department of Health and Human Services . Appendix 2. Interim recommendations: enhanced US surveillance and diagnostic evaluation to identify cases of human infection with avian influenza A (H5N1) Available at: http://www.hhs.gov/pandemicflu/plan/sup2.html#app2. Accessed 24 March 2006. [Google Scholar]
- 64.Centers for Disease Control Prevention New laboratory assay for diagnostic testing of avian influenza A/H5 (Asian lineage) Morb Mortal Wkly Rep. 2006;55:127–0. [PubMed] [Google Scholar]