Severe acute respiratory syndrome (SARS) is a newly emerging disease that first emerged in Guangdong Province, China in November 2002 (1). The SARS coronavirus (SARS-CoV) was found to be the etiology of the disease (2)(3)(4). Subsequent surveillance studies have indicated that this virus is of animal origin and have suggested that the source of the disease is still circulating in this geographic region (5). Indeed, the potential risk of reemergence of SARS is further highlighted by a recent confirmed SARS case in January 2004 (6). Therefore, the establishment of a rapid SARS diagnostic method is a high priority for control of the disease.
Currently, there are two major diagnostic approaches for SARS. Detection of antibodies against SARS-CoV is a sensitive and specific diagnostic approach, but serconversion can be detected only around day 10 of illness (7). In contrast, PCR-based tests have been shown to be useful for early SARS diagnosis (8). Quantitative PCR approaches are a powerful tool for identifying SARS-CoV early after disease onset (4)(9)(10)(11). However, because of the requirements for sophisticated instrumentation and expensive reagents, these rapid molecular tests might not be the method of choice in basic clinical settings in developing countries or in field situations. It is therefore critical to develop simple and economical molecular tests for the above scenarios.
The invention of loop-mediated isothermal amplification (LAMP) has opened up a new horizon for molecular diagnosis (12). This method depends on autocycling strand displacement DNA synthesis performed by a Bst DNA polymerase, and a detailed amplification mechanism has been described elsewhere (12). The reaction relies on recognition of the DNA target by six independent sequences, making this kind of assay highly specific. This method is rapid and has a DNA amplification efficiency equivalent to that of PCR-based methods (12)(13)(14). More importantly, this approach is inexpensive, and all reactions can be performed in an isothermal environment. The potential clinical applications of this method have been demonstrated recently (13). Here we demonstrate the feasibility of using this technology for detection of SARS-CoV.
Thirty-one retrospective SARS samples collected between March 26, 2003, and April 9, 2003, were used in this study. All SARS patients in this study were confirmed to be seropositive for SARS-CoV by immunofluorescence assays (2). The age range for these patients was 16–74 years (median, 45 years), and the M:F ratio was 16:15. The study was approved by our local clinical research ethics committee. Nasopharyngeal aspirate (NPA) samples were collected on days 1–15 after disease onset as described previously (15). NPA samples from patients with other respiratory diseases (adenovirus, n = 8; respiratory syncytial virus, n = 10; human metapneumovirus, n = 10; influenza A virus, n = 20; influenza B virus, n = 4; rhinovirus, n = 6) and from healthy individuals (n = 30) were used as negative controls.
RNA from clinical samples was extracted, and cDNA was synthesized as described previously (9)(15). In this study, the ORF1b region of SARS-CoV (nucleotides 17741–17984; accession no. AY274119; see Fig. 1S in the Data Supplement that accompanies the online version of this Technical Brief at http://www.clinchem.org/content/vol50/issue6/) was chosen for SARS diagnosis. DNA plasmids containing the target sequences were used as positive controls. To accelerate the amplification reaction, cDNA for the SARS-CoV ORF1b sequence was amplified by a modified LAMP reaction (14) in the presence of six primers: F3 (5′-CTTAGGATTGCCTACG-3′); B3c (5′-AGTCCAGTTACATTTTCT-3′); FIP (5′-AGTGTGCTGTTTCAGTAGTGATTCATCACAGGGTT-3′); BIP (5′-TGTAATGTCAACCGCTTTGCGACGTGGTATTTC-3′); Loop B (5′-TCTTTATGACAAACTGCAAT-3′); and Loop Fc (5′-TTTGTGTGAATATGACATAGTCATA-3′; see Fig. 1S in the online Data Supplement). In a typical LAMP reaction, 0.5–1 μL of heat-denatured cDNA was amplified in a 12.5-μL reaction containing 0.4 mM each of the deoxynucleotide triphosphates, 1.6 μM each of FIP and BIP, 0.2 μM each of F3 and B3c, 0.8 μM each of Loop F and Loop Fc, 4 U of Bst DNA polymerase (New England Biolabs), and 1× Bst polymerase buffer (New England Biolabs). Reaction mixtures were incubated at 60 °C for 1 h, followed by heat inactivation at 80 °C for 5 min. Amplified products were analyzed by gel electrophoresis.
In preliminary experiments, reactions were performed with different copy numbers of the positive control to determine the detection limit of the assay. Because the reaction products consist of stem-loop DNA structures with multiple inverted repeats of the target and cauliflower-like structures with multiple loop-stem-loops (12)(14), the reaction would produce bands of different sizes in gel electrophoresis analyses. As shown in Fig. 1 , a characteristic DNA ladder was observed in positive controls (lanes 1 and 26). The detection limit of the assay was 10 copies/reaction (see Fig. 2SA in the online Data Supplement), and positive signals were consistently observed in reactions containing ≥50 copies of the target sequence (data not shown).
Figure 1.
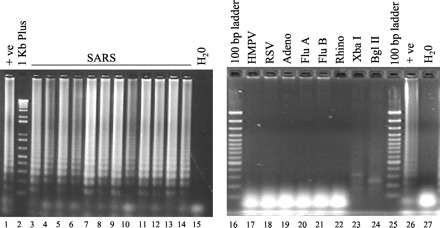
LAMP assay for SARS.
Positive signals were observed in the positive control (+ ve; lanes 1 and 26) and SARS samples (lanes 3–14), but not in the water control (lanes 15 and 27) and non-SARS samples (lanes 17–22). Lanes 23 and 24, positive LAMP products digested with XbaI and BglII, respectively. Lanes 2, 16, and 25, DNA markers as indicated. HMPV, human metapneumovirus; Adeno, adenovirus; RSV, respiratory syncytial virus; Flu A, influenza A virus; Flu B, influenza B; Rhino, rhinovirus.
Among 31 SARS samples, the SARS-CoV sequence could be detected in 20 cases (64%; Fig. 1 , lanes 3–14). The detection rate for SARS-CoV in these samples increased as the disease progressed (Table 1 ). In the early stages after disease onset, 4 of 13 (31%) were positive in the assay. For samples isolated from day 8 to day 15 after disease onset, positive signals were observed in all of the cases (n = 12). These results agreed with our previous findings that the viral load in SARS patients peaks at the second week of the disease (7). Because the targeted sequence contains BglII and XbaI restriction sites (see Fig. 1S in the online Data Supplement), we also validated the identities of these positive signals by restriction enzyme digestion. All amplified products could be digested by these restriction enzymes as expected (Fig. 2SB in the online Data Supplement and Fig 1 ). By contrast, no positive signal was observed in healthy individuals (n = 30; data not shown), non-SARS patients (n = 58; Fig. 1 , lanes 17–22), and water controls (Fig. 1 , lanes 15 and 27).
Table 1.
Detection of SARS CoV by LAMP assay.
| Day after onset | Sample size, n | Number positive, n (%) | ||
|---|---|---|---|---|
| LAMP assay | PCR1 | |||
| 1–3 | 13 | 4 (31%) | 5 (38%) | |
| 4–7 | 6 | 4 (67%) | ND2 | |
| 8–15 | 12 | 12 (100%) | 12 (100%) | |
Reverse transcription-PCR protocol adapted from Peiris et al. (2).
ND, not done.
In this study, we demonstrated the potential use of LAMP for early SARS diagnosis. Recently we also reported the use of a quantitative PCR method for SARS diagnosis (9)(16)(17). Compared with quantitative PCR assays, the LAMP assay described in this study has two main shortcomings: (a) the LAMP assay does not allow quantification of SARS-CoV RNA; and (b) the LAMP assay is less sensitive than real-time PCR assays (9). However, one should note that the detection rates for SARS in the LAMP assay (Table 1 ) are similar to those with our conventional PCR-based assays (18). To confirm this observation, we further tested some of these clinical samples with a conventional PCR assay (2). As shown in Table 1 , the detection rate of the LAMP assay was similar to that of the reverse transcription-PCR assay. These results agree with previous findings that the sensitivities of LAMP assays are equivalent to those for conventional PCR-based methods (12)(13)(14).
Our LAMP reaction relies on recognition of viral sequences by six primers, potentially making this kind of assay more specific than conventional PCR assays. Indeed, none of the negative control samples (n = 88) was positive in our assay. Recently, Parida et al. (19) reported a real-time closed-tube detection method for West Nile virus in which the amounts of magnesium pyrophosphate precipitates generated in LAMP reactions are measured. This real-time approach for LAMP might further reduce the risk of cross-contamination problems.
The primary goal of this study was to develop a simple and inexpensive test for SARS diagnosis. Unlike the quantitative PCR-based detection approach, the LAMP assay does not require sophisticated instrumentation. Because reactions are performed in an isothermal environment (e.g., a water bath), there is no time loss from thermal changes during DNA amplification. The LAMP assay is rapid and does not require expensive reagents or instruments. In a SARS outbreak, a diagnostic laboratory might routinely receive hundreds of clinical samples each day for SARS diagnosis. The application of this LAMP test might help to reduce the running cost for SARS diagnosis. From a practical point of view, highly sensitive quantitative reverse transcription-PCR assays should be used to test samples collected from patients within the first week of illness. For samples collected from patients after the first week of disease onset, the LAMP assay might be an inexpensive and accurate alternative for SARS diagnosis.
In conclusion, we report a simple LAMP assay for SARS diagnosis. We believe the inexpensive running costs of the assay make this technology very applicable to laboratories for SARS diagnosis in developing countries. The technique might have great potential to be used in field situations or at the bedside as a preliminary screening test. Regardless of the method used, testing in a suitably accredited laboratory is important, especially during an outbreak, when quality-assured diagnoses are essential. We expect that, with this rapid diagnostic method, prompt identification of this pathogen will facilitate control of the disease and provision of prompt treatment of patients.
Supplementary Material
Acknowledgments
This work was funded in part by the US National Institute of Allergy and Infectious Diseases (Public Health Research Grant A195357), the Research Grant Council of Hong Kong (HKU 7543/03M), The University of Hong Kong (HKU SARS Research Fund), and Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003;362:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 5.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003;302:276-278. [DOI] [PubMed] [Google Scholar]
- 6.Cyranoski D. Swift response greets return of SARS in China. Nature 2004;427:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peiris JSM, Chu CM, Cheng VCC, Chan KS, Hung IF, Poon LLM, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003;361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger A, Drosten C, Doerr HW, Sturmer M, Preiser W. Severe acute respiratory syndrome (SARS)-paradigm of an emerging viral infection. J Clin Virol 2004;29:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon LLM, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol 2003;28:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng EK, Ng PC, Hon KL, Cheng WT, Hung EC, Chan KC, et al. Serial analysis of the plasma concentration of SARS coronavirus RNA in pediatric patients with severe acute respiratory syndrome. Clin Chem 2003;49:2085-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant PR, Garson JA, Tedder RS, Chan PK, Tam JS, Sung JJ. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med 2003;349:2468-2469. [DOI] [PubMed] [Google Scholar]
- 12.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000;28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enosawa M, Kageyama S, Sawai K, Watanabe K, Notomi T, Onoe S, et al. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis J Clin Microbiol 2003;41:4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 2002;16:223-229. [DOI] [PubMed] [Google Scholar]
- 15.Poon LLM, Wong OK, Luk W, Yuen KY, Peiris JS, Guan Y. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin Chem 2003;49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poon LLM, Chan KH, Wong OK, Cheung TK, Ng I, Zheng B, et al. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem 2004;50:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poon LLM, Wong BW, Chan KH, Leung CS, Yuen KY, Guan Y, et al. A one step quantitative RT-PCR for detection of SARS coronavirus with an internal control for PCR inhibitors. J Clin Virol (published online Feb 3, 2004; DOI accession number: doi:10.1016/j.jcv.2003.12.007; available at: http://dx.doi.org).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KH, Poon LLM, Cheng VCC, Guan Y, Hung IFN, Kong J, et al. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis 2004 (published online December 24, 2003; available at: http://www.cdc.gov/ncidod/EID/vol10no2/03-0610.htm).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol 2004;42:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


