Highlights
-
•
Coronavirus nucleocapsid proteins are appealing drug targets against coronavirus-induced diseases.
-
•
A variety of compounds targeting the coronavirus nucleocapsid protein have been developed.
-
•
Many of these compounds show potential antiviral activity.
Abstract
The advent of severe acute respiratory syndrome (SARS) in the 21st century and the recent outbreak of Middle-East respiratory syndrome (MERS) highlight the importance of coronaviruses (CoVs) as human pathogens, emphasizing the need for development of novel antiviral strategies to combat acute respiratory infections caused by CoVs. Recent studies suggest that nucleocapsid (N) proteins from coronaviruses and other viruses can be useful antiviral drug targets against viral infections. This review aims to provide readers with a concise survey of the structural features of coronavirus N proteins and how these features provide insights into structure-based development of therapeutics against coronaviruses. We will also present our latest results on MERS-CoV N protein and its potential as an antiviral drug target.
Teaser
Nucleocapsid proteins are essential for coronavirus viability and constitute potential targets for the development of therapeutics against recent coronavirus outbreaks such as SARS and MERS.
Introduction
Upper and lower respiratory infections caused by viruses are very common in temperate climates 1, 2, and cause rhinitis, pharyngitis, sinusitis, bronchiolitis and pneumonia 3, 4. Coronaviruses are a large group of RNA viruses with single-stranded RNA genomes that cause 30% of upper and lower respiratory tract infections in humans. Some coronaviruses such as human coronavirus 229E (HCoV-229E), OC43 (HCoV-OC43), HKU1 (HCoV-HKU1) and NL63 (HCoV-NL63) are detected globally and only evoke mild symptoms in most individuals 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, although some individuals can have more-severe illness 18, 19. However, other human coronaviruses including the severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle-East respiratory syndrome coronavirus (MERS-CoV) can elicit severe symptoms and even death 20, 21.
Between 2003 and 2004, SARS-CoV caused a worldwide epidemic and had significant impact on the economy of the countries affected by the outbreak. SARS emerged in November 2002 and was widely prevalent in at least 27 countries with 8096 reported cases and a total of 774 deaths up until 31 July 2003 [case-fatality rate (CFR) of 9.5%; http://www.who.int/csr/sars/country/en/]. The clinical circumstances are similar among many human coronaviruses, and are particularly urgent in the case of MERS. Since the emergence of MERS in 2012, there have been 1179 laboratory-confirmed cases of infection with MERS-CoV and a total of 442 deaths as of 4 June 2015 (CFR of 37.4%; http://www.who.int/csr/don/04-june-2015-mers-korea/en/). This CFR is higher than most viruses infecting humans. Based on the sequence analysis of the RNA genome, SARS-CoV probably originated from coronaviruses infecting Chinese rhinolophid bats 22, 23. The origin of MERS-CoV is still unclear, although dromedary camels have been described as an intermediate host for the virus 24, 25, 26. These zoonotic viruses usually have lower surveillance priorities in the public health network, but once they evolve strains capable of crossing the species barrier to infect humans they can easily cause an outbreak. As is true for all coronavirus infections, there is currently no efficacious therapy available for this disease.
The CoV genome consists of positive-sense, single-stranded RNA of approximately 30 kb in length, and it contains several genes encoding several structural and nonstructural proteins that are required for progeny virion production in a conserved linear arrangement 27, 28. The CoVs have several conserved structural proteins: the matrix (M) protein, the small envelope (E) protein, the trimeric spike (S) glycoprotein and the nucleocapsid (N) protein. Some variants have a third glycoprotein, hemagglutinin-esterase (HE), which is present in lineage A betacoronaviruses. The N protein is located inside the virus particle and is one of the most abundant structural proteins in CoVs. It binds to the viral RNA genome to form a virion core comprising a ribonucleoprotein (RNP) complex that assumes a long helical structure 29, 30. The RNP is important for maintaining the RNA in an ordered conformation for replication and transcription. The CoV N protein also plays an essential part in viral RNA synthesis 31, 32, 33. In addition to its role in viral processes, the CoV N protein is also involved in the regulation of cellular processes, such as gene transcription, interferon inhibition, actin reorganization, host cell cycle progression and apoptosis 34, 35, 36, 37. Moreover, the N protein is an important diagnostic marker and immunodominant antigen in host immune responses 38, 39, 40. Recent studies suggest that N proteins of coronaviruses and other viruses could be useful antiviral drug targets against infections caused by these viruses because they serve many crucial functions during the viral lifecycle 41, 42, 43. This review aims to provide readers with (i) a concise survey of the structural features of coronavirus N proteins, (ii) an explanation of how these features correlate to ribonucleocapsid formation and (iii) insights into structure-based development of therapeutics against coronaviruses by targeting the N protein. We will also present the latest information on current efforts to develop an antiviral therapeutic against MERS-CoV using its N protein as a target.
A concise survey of the structural features of coronavirus N proteins
A combination of protein sequence alignment, secondary structure and intrinsic disorder predictions suggests that all CoV N proteins share the same modular organization [44]. Coronavirus N proteins contain three intrinsically disordered regions (IDRs): N-arm, central linker region (LKR) and C-tail, and two structural domains: N-terminal domain (NTD) and C-terminal domain (CTD), where the NTD and CTD are sandwiched between three IDRs (Fig. 1a,b). Previous studies have reported that the NTDs of the CoV N proteins are involved in RNA binding, whereas the CTDs are involved in RNA binding and oligomerization 45, 46, 47, 48, 49, 50. All three IDRs of coronaviral N protein can modulate the RNA-binding and oligomerization properties of NTD and CTD, respectively [51]. For example, N-terminal IDR has been implicated in the RNA binding of the N protein. It has also been reported that the LKR and the C-terminal IDR are involved in the oligomerization of the N protein [47]. In addition, the LKR of the N protein with Ser–Arg-rich sequences has also been shown to contain an RNA-binding region and putative phosphorylation sites that might regulate N protein functions 52, 53, 54 and N–M interaction. The RNA-binding activity of the LKR also contributes toward RNA chaperone activity, and is involved in template switching under in vitro conditions [33].
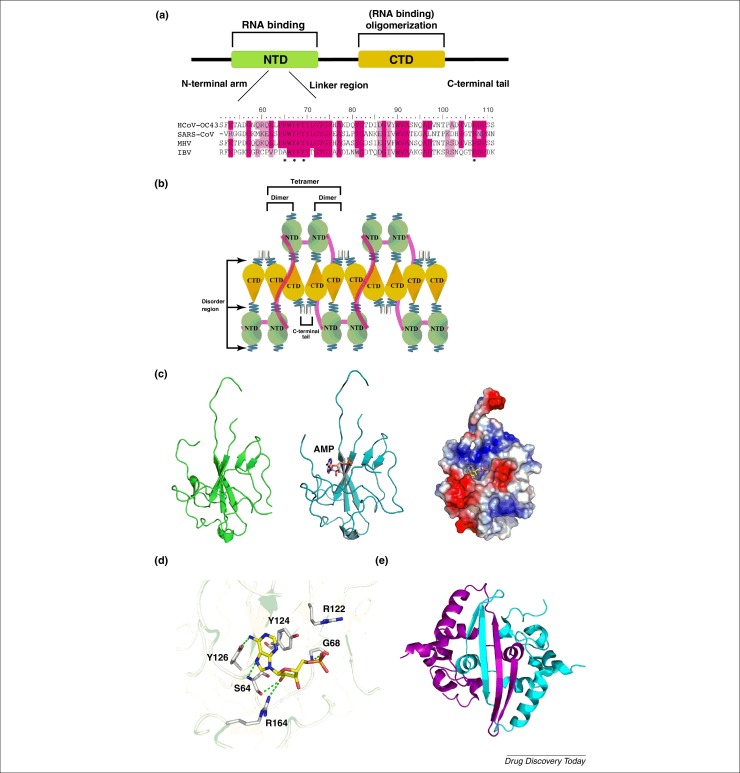
Figure 1.
(a) Domain organization of human coronavirus nucleocapsid protein. MHV: mouse hepatitis virus; IBV: avian infectious bronchitis virus. (b) A schematic mechanism of the oligomeric N protein complexed with RNA showing that the N proteins form a tetramer through the interactions between the C-terminal domains and C-terminal tails of the dimer. (c) Ribbon representation of HCoV-OC43 N-NTD structure (left). Ribbon representation of HCoV-OC43 N-NTD structure with AMP depicted as a stick structure (middle). Electrostatic surface of the HCoV-OC43 N-NTD–AMP complex (right). Blue denotes positive charge potential, red indicates negative charge potential. (d) Detailed stereoview of the interactions at the AMP-binding site. The dotted green lines represent H-bonds. (e) Ribbon representation of the SARS-CoV N-C-terminal-domain (CTD) dimer structure.
To clarify the molecular mechanism of RNP formation in CoVs, the structures of truncated fragments of the N protein, including the N-terminal and C-terminal domains, were determined. Previous studies have reported that the N terminus of the N protein provides a scaffold for RNA binding. Structural analysis revealed that the fold and the distribution of the secondary structures of the N-terminal domain of the N protein are essentially conserved across the various CoVs 41, 55. The core of the structure of CoV N-NTD adopts a unique five-stranded antiparallel β-sheet sandwiched between loops (or short 310 helix) with the topology of β4–β2–β3–β1–β5; and the whole structure presents a right-handed fist-shaped structure in which palm and finger are rich in basic residues and the flexible loops are organized around the β-sheet core of the N-terminal domain (Fig. 1c). All CoV N-NTDs possess positively charged amino acids on or near the protruding loop; however, in the SARS-CoV, mouse hepatitis virus (MHV) and avian infectious bronchitis virus (IBV) N-NTDs the protruding segment comprises a β-hairpin, whereas a flexible loop predominates in the HCoV-OC43 N-NTD. Based on the surface charge distribution, the protruding loop of the CoV N-NTD containing several positively charged residues, for example, Arg 106, Arg 107 and Arg 117 in HCoV-OC43, were identified to provide a site for binding of the phosphate backbone of RNA through electrostatic interactions [55]. In the central core of MHV N-NTDs, there are two crucial conserved Tyr residues, Tyr 127 and Tyr 129, that are involved in RNA binding and mutations of these residues to alanine can lead to loss of virus replication [56]. By contrast, alanine substitution of Tyr 94 in the IBV N-NTD, which corresponds to Tyr 126 of HCoV-OC43 and Tyr 129 of MHV, led to a significant decrease in its RNA-binding activity and a total loss of infectivity of the viral RNA to Vero cells [57]. The crystal structures of HCoV-OC43 N-NTD complexed with ribonucleoside 5′-monophosphates (AMP, UMP, GMP and CMP) have been reported to identify a distinct ribonucleotide-binding pocket for understanding the molecular interactions that govern CoV N-NTD binding to RNA [41]. The base of ribonucleoside 5′-monophosphates was inserted into a hole in the N-NTD that was almost perpendicular to the phosphate moiety and the phosphate group was bound to a basic and conserved 5′-phosphate-binding site that contained the largest positively charged region on the N-NTD surface (Fig. 1c). The detailed interactions between the AMP and HCoV-OC43 N-NTD are shown in Fig. 1d. The amino acid composition of this binding site includes Ser 64, Gly 68, Arg 122, Tyr 124, Tyr 126 and Arg 164. The Arg 122, Tyr 124, Tyr 126 and Arg 164 side-chains generate a distinct ribonucleotide-binding pocket and interact with the ribonucleoside 5′-monophosphate via H-bonding, ionic bonding and π–π stacking forces. Tyr 124 is located on the surface of the N protein in the HCoV-OC43 N-NTD and is directly involved in the interactions with the AMP base through π–π stacking. The phenolic hydroxyl group substituent on Tyr 126 forms H-bonds with the sixth amino groups present in the AMP adenine ring. These amino acids are sequentially and structurally conserved in other HCoV N proteins; therefore, they are probably essential for RNA recognition and interaction in all coronavirus N proteins. All featured three additional HCoV-OC43 N-NTD complexes [cytosine monophosphate (CMP), guanosine monophosphate (GMP) and uridine monophosphate (UMP)] are similar to the interaction of the HCoV-OC43 N-NTD–AMP complex.
Self-association of the N protein is a crucial step in virus particle assembly for CoVs. Previous studies have shown that full-length CoV N proteins can form high-order oligomers, and the C-terminal domains of the CoV N proteins mediate the self-association of the protein to form high-order oligomers 58, 59. The crystal structures of the C-terminal domains of SARS-CoV, MHV and IBV N proteins show a similar stable fold in which the four-stranded β-sheet forms one face and the α-helices form the opposite face, suggesting that the dimerized N protein is the building block for all groups of coronaviruses 45, 46, 49, 50, 60. The crystal structure of the C-terminal domain shows a tightly intertwined twofold symmetric C-terminal domain dimer, with a β-hairpin from one subunit extending into the cavity of the opposite subunit, which forms an antiparallel β-sheet stabilized through extensive H-bonding across the dimer interface (Fig. 1e). The opposite dimerization interface is composed of several helices, where strong hydrophobic interactions involving hydrophobic residues including tryptophan, phenylalanine, leucine and proline were observed. The hydrophobic interactions between the longest helix and the intermolecular β-sheet were also observed to stabilize the dimer conformation further.
The C-tail region of N protein also has an important role in oligomerization [58]. However, owing to its highly disordered sequence, no NMR or crystal structures have been solved to date. Luo et al. report that residues 363–382 of the C-tail region from the SARS-CoV N protein are responsible for RNA binding and the C-terminal half of the protein is associated with oligomerization [48]. In the case of HCoV-229E N protein, CTD with an extended C-tail region enhanced higher-order oligomer formation [58]. The C-tail has also been found to participate in the oligomerization of the SARS-CoV N protein [48], because the removal of 40 amino acid residues from the C terminus apparently decreased the ability of the protein to oligomerize. Previous studies proposed that the H-bonds across the tetramer interface formed by the main chain atoms of the C-tail region stabilize the oligomerization of the CTD of the N protein through domain swapping.
The CTD of the N protein has been shown to lack RNA-binding activity in some cases [61]; however, the presence of the CTD can increase the RNA-binding affinity of the NTD and assist nucleocapsid formation. In SARS-CoV N protein, the CTD has been shown to bind to RNA if it includes an N-terminal charged region which is part of the CTD structure as shown by NMR spectroscopy 44, 51. These results led us to propose a helical packaging model of CoV RNP, which will be explained in more detail in the following sections.
Packaging of the CoV ribonucleocapsid
Coronavirus genomes are generally very large (>20 kb) and their accommodation into virions <100 nm in diameter requires extensive supercoiling of the nucleic acid into a well-packed RNP. Structural studies of the CoV RNP can be traced back to the late 1970s when electron microscopy (EM) studies of coronaviruses found helical entities within the virus particle 62, 63, 64. The observed helical RNPs had coil diameters of 9–16 nm and a hollow interior of 3–4 nm in diameter. Unfortunately, technical limits at the time only yielded a blurry picture of the RNP, and more-detailed information would not be available until 2006. Advances in 2D electron cryomicroscopy (cryo-EM) and single particle image analysis finally enabled Neuman et al. to investigate the structural organization of the SARS-CoV at 4 nm resolution [65]. Interestingly, viral RNP is maintained in a spherically packaged form within SARS-CoV, whereas RNP released from disrupted viral particles took the appearance of strands with a diameter of 15 nm. Later studies employing 3D cryoelectron tomography on MHV and transmissible gastroenteritis virus (TGEV) also observed coiled structures and tubular shapes within the virus particle consistent with the formation of a helical RNP [66]. The tomography results showed quasi-circular density profiles of ∼11 nm in diameter enclosing an empty space of ∼4 nm in diameter, in agreement with early EM studies. Neither the 2D cryo-EM nor the 3D cryo-EM tomography studies found consistently ordered structures within the virion, suggesting that the helical RNP is structurally flexible, a characteristic that might be important in the lifecycle of the virus. This flexibility is not unexpected because RNA is known to be dynamic and exists in multiple folded and unfolded states [67]. The interaction between RNA and CoV N protein could involve the structural and intrinsically disordered regions of the CoV N protein through an induced fit process 44, 51. Because currently there are no data supporting the existence of long-lived N protein oligomers in the absence of nucleic acid, packaging of the CoV RNP most probably employs an RNA-binding coupled-packaging mechanism such as that proposed for MHV [68].
Although the structure of any CoV RNP at atomic resolution is still lacking, current 3D structural information and biochemical data allowed Chang et al. to propose a putative series of molecular events that led to RNP formation [47]. In the model, RNP formation can be initiated by binding of genomic RNA to either the NTD or the CTD, which in turn facilitates binding to other domains through coupled allostery [69]. The model enables multiple initiation sites in parallel. This initial N-protein–RNA binary complex can then grow by either recruiting additional free N protein or by combining with other binary complexes. In the larger oligomer complex the CTD forms a helical core, such as that observed for the SARS-CoV N protein CTD crystal structure, with the RNA wrapping and twisting along a positively charged groove on the outside of the helix [46]. Presumably, the RNA binds to the CTD helix through its phosphate backbone. In addition to the RNA–CTD interaction, the CTD helix is held together by a number of weak protein–protein interactions [59]. The NTD then forms a cap on the outside of the helical CTD–RNA complex through electrostatic interactions between the NTD and the free phosphate groups on the RNA molecule. RNA bases sticking out of the groove intercalate with a set of conserved aromatic residues, further stabilizing the RNP. Two major features are underscored in this model. First, the intrinsically disordered region between the NTD and CTD plays a crucial part in the process by allowing the two structural domains to adapt a wide range of conformations and positions for optimal packing of the RNP complex. Second, RNP formation is driven by electrostatic interactions and a multitude of weak CTD–CTD interactions, consistent with the idea that local weak interactions can assist the formation of globally stable structures in viral capsids [70]. The use of multiple weak interactions to stabilize the RNP would also minimize the formation of kinetic traps and allow greater control over CoV RNP assembly. Interestingly, the RNP structure proposed for SARS-CoV based on the above process would form a helical structure with a diameter of 16 nm and containing a 4 nm diameter hole, consistent with the cryo-EM results.
It should be stated that, although the model proposed by Chang et al. generally recapitulates most of the in vitro findings in the literature, there is one minor shortcoming: the model does not attempt to explain the role of the RNA packaging signal, which is crucial for in vivo initiation of RNP packaging 34, 71. Rather, according to the model, all RNA sequences should be more or less equivalent, which might be true after RNP formation is initiated in vivo. Despite this minor flaw, this model is currently the only one that provides a biophysical working framework for CoV RNP formation.
Insights into development of therapeutics against coronaviruses by targeting N protein
Viral nucleocapsid proteins often are multifunctional and have crucial roles in the viral lifecycle. Previous studies have suggested that the N protein could be used as a target for antiviral drug development against viral infections 41, 42, 58, 72. For example, the low sequence variation between strains of influenza viruses suggest that N proteins are genetically stable and less prone to develop resistance against putative inhibitors 42, 72. Although CoV N protein sequences from different strains of the same species also are highly conserved, the same cannot be said when comparing the N protein sequences between different CoV species. This is particularly true when comparing the degree of sequence conservation among CoV N proteins with that of CoV replicase proteins, which include more-traditional drug targets such as proteases, polymerases and helicases. However, targeting the CoV N protein has some advantages. First, there is a wealth of high-resolution structural information about N proteins from various CoV species, which provides researchers with a good starting point for structure-based drug discovery, whereas the structures of most replicase proteins are still unknown. The availability of high-resolution structures is especially important for drug discovery targeting enzymatic activities, because crucial elements for enzyme function might not be detected from primary sequence analyses [73]. Moreover, CoV N proteins share a common modular organization with highly conserved protein structures, which would allow the development of broad-spectrum anticoronavirus drugs. Second, antiviral drug targets that participate in essential oligomeric interactions are less likely to develop drug-resistant variants [74]. Compounds targeting CoV N proteins could also be used in conjunction with compounds that target the CoV replicase proteins to develop ‘cocktail’ therapies.
Two strategies to inhibit oligomeric N protein function have been reported [75]. The first strategy is to develop antiviral agents that target the RNA-binding site, which contains a number of conserved residues, and specifically block the formation of RNP during genome replication. For example, naproxen and its derivatives have been proposed to intercalate into the hydrophobic pocket on the RNA-binding groove of influenza virus N protein and inhibit its RNA-binding ability, resulting in reduced viral replication 76, 77. Inhibition of RNP formation by several families of compounds targeting the zinc fingers of HIV N protein has also been demonstrated [43]. A second strategy is to hamper normal N protein oligomerization by enhancing or inhibiting the interaction between N protein molecules. Nucleozin and its derivatives obtained by high-throughput methods have been shown to prevent RNP formation in influenza virus by interfering with N protein oligomerization 78, 79. Similar stratagems can be applied toward development of anticoronaviral agents. In the following sections, we summarize the CoV N-protein-related actions of existing ligands, newly developed chemicals, peptides and polyphenol compounds (Table 1 ). We will also briefly describe developments in coronaviral N protein vaccines.
Table 1.
Listing of compounds acting on CoV N proteins.
| Ligand | Classification | Target site | Function | Refs |
|---|---|---|---|---|
| PJ34 | Chemical | NTD | Inhibition of RNA binding | [49] |
| H3 | Chemical | NTD | Inhibition of RNA binding | [81] |
| N377–389 | Peptide | C terminus | Inhibition of oligomerization | [57] |
| (−)-Catechin gallate | Polyphenol compound | ND | Inhibition of RNA binding | [82] |
| (−)-Gallocatechingallate | Polyphenol compound | ND | Inhibition of RNA binding | [82] |
Abbreviations: CoV, coronavirus; H3, 6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5, 8-dione; ND, not determined; NTD, N-terminal domain; PJ34, N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino)acetamide hydrochloride.
The N-terminal domain of coronavirus N protein as an antiviral target
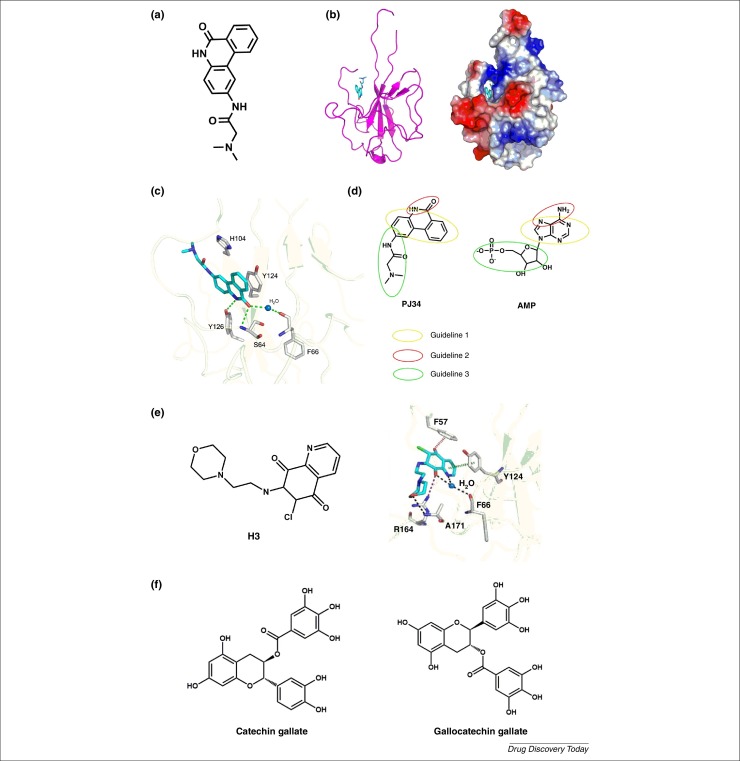
A detailed high-resolution structure of HCoV-OC43 N-NTD in complex with ribonucleotide monophosphate has been reported by Lin et al. [80]. They identified a unique ribonucleotide-binding pocket in the center of the N-NTD comprising conserved RNA-binding residues including Arg 122, Tyr 124, Tyr 126 and Arg 164. Mutation of these residues resulted in significant decrease of the RNA-binding affinity of the protein and subsequent decrease in viral replication. This led the authors to postulate that the N-terminal RNA-binding domain of a CoV N protein could serve as a target for development of RNA-binding inhibitors that could act as broad-spectrum antiviral drugs. Compounds binding to this site that act as competitive N protein inhibitors could be employed to combat highly pathogenic CoVs. One such compound is N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino)acetamide hydrochloride (PJ34) (Fig. 2a), which we found to inhibit coronavirus replication and have a potent inhibitory effect on the RNA-binding activity of HCoV-OC43N protein. PJ34 has been reported to protect mice against brain ischemia, splanchnic ischemia, reperfusion and lipopolysaccharide (LPS) toxicity in various models of local inflammation. The crystal structure of PJ34 in complex with HCoV-OC43 N-NTD shows that its aromatic core stacks onto Tyr 124 of N-NTD (Fig. 2b,c). The aromatic core also contains a number of H-bond-forming moieties that mediate specific interactions with the protein. It also has an attached branching moiety that fits into the ribonucleotide-binding pocket of N-NTD. Interestingly, binding of PJ34 to N-NTD mimics that of AMP to N-NTD, but PJ34 is buried deeper into the hydrophobic pocket of the protein. Compared with AMP, PJ34 binds more closely to the N-terminal loop of N-NTD, and the branched moiety of PJ34 inserts into an interior core of N-NTD. By contrast, PJ34 and AMP interact with Ser 64, Tyr 124 and Tyr 26, indicating that these three residues have important roles in RNA binding. Because the three residues are structurally conserved they are suitable interaction targets for drug screening endeavors against the N protein. Based on careful study of the chemical features and mechanisms of action of AMP and PJ34, we formulated three general guidelines for development of agents targeting CoV N-NTD. First, a polycyclic aromatic core is required to enable π–π stacking with the tyrosine residues in the N-NTD. Second, the aromatic core requires the presence of moieties capable of forming specific H-bond interactions with the N-NTD. Finally, attaching a branched moiety (or moieties) that fits into the ribonucleotide-binding pocket can enhance the affinity and specificity of the compound (Fig. 2d). Using these guidelines as a selection tool for virtual screening, a second compound, 6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5, 8-dione (H3), was recently identified as a potential antiviral compound (Fig. 2e) [81]. H3 was shown to inhibit the RNA-binding activity of HCoV-OC43 N-NTD with higher efficacy than PJ34 in vitro. Structural studies showed that, compared with PJ34, H3 adopted a bound conformation more similar to that of bound AMP, which could suggest that the similarity of a compound's bound conformation to that of AMP might influence its inhibitory activity toward CoV N-NTDs.
Figure 2.
(a) Chemical structure of N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino)acetamide hydrochloride (PJ34). (b) Structural overview of human coronavirus OC43 (HCoV-OC43) N-N-terminal-domain(NTD)–PJ34 complex. A ribbon representation of the HCoV-OC43 N-NTD with PJ34 depicted as a stick model is shown on the left, whereas the electrostatic surface of the HCoV-OC43 N-NTD–PJ34 complex is shown on the right. Blue denotes positive charge potential, red indicates negative charge potential. (c) Detailed view of the interactions between PJ34 and HCoV-OC43 N-NTD, with the H-bonds depicted as green dotted lines. (d) Three general guidelines for the design of compounds that bind to the nucleotide-binding pocket of CoV N-NTD, deduced from the molecular structures of PJ34 and AMP. (e) Chemical structure of 6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5, 8-dione (H3) (left) and a detailed view of the interactions between H3 and HCoV-OC43 N-NTD. H-bonds mediated by the side- and main-chain atoms are marked as red and black dotted lines, respectively. Anion-π and stacking interactions are denoted in blue and green dashed lines, respectively. (f) Chemical structure of (−)-catechin gallate and (−)-gallocatechin gallate.
Inhibition of coronaviral N protein oligomerization by developing competing peptides
The C-terminal tail has been found to participate in the oligomerization of the SARS-CoV N protein because removal of 40 amino acids from the C terminus apparently decreased the ability of the protein to oligomerize [48]. In HCoV-229E, the HCoV-229E N protein CTD lacking 13 amino acids (residues 377–389) from the C-tail impaired higher-order oligomerization [58]. Based on computer-assisted prediction of its secondary structure and experimental CD results, the end of the C-terminal tail of HCoV-229E N protein CTD forms a short β-strand. H-bonds across the tetramer interface formed by the main chain atoms of the short β-strand can stabilize oligomerization of the C-terminal domain of the N protein through domain swapping. Inhibition of viral N protein oligomerization by developing competing peptides and small organic compounds is an attractive therapeutic strategy against viral infection 82, 83, 84. We have shown that an excess of a peptide based on the sequence of the C-terminal tail interfered with oligomerization of the C-terminal domain of HCoV-229E N protein and had an inhibitory effect on the viral titer [58]. These results suggest that small molecules or peptides designed against the oligomer interface could be potential inhibitors of CoV.
The inhibition of coronaviral N protein RNA-binding activity by gallate derivatives
Roh reported a novel method for the discovery of inhibitors targeting the CoV N protein by quantum dot (QD)-conjugated oligonucleotides [85]. This novel method provides a facile and sensitive way of screening compounds against RNA/DNA-binding virus proteins, particularly when incorporated into a biochip system. Roh immobilized SARS-CoV N protein onto a glass chip and screened chemical compounds that inhibited RNA-binding activity of the protein. Two polyphenolic compounds, (−)-catechin gallate and (−)-gallocatechin gallate (Fig. 2e), showed remarkable inhibitory activity in a concentration-dependent fashion in vitro. At a concentration of ∼110 μM (0.05 μg/ml), (−)-catechin gallate and (−)-gallocatechin gallate showed more than 40% inhibition activity on a nanoparticle-based RNA oligonucleotide biochip system. However, their binding mechanism to the CoV N protein is still unclear. Interestingly, the structures of these compounds generally conform to the guidelines presented above. First, one of the benzene rings and the dihydropyran cycle forms a bulky aromatic core that can participate in stacking interactions. Second, the multiple hydroxyl groups on the aromatic core also have the potential to form H-bonds with the residues on N-NTD. Finally, the remaining benzene rings branching out of the aromatic core can interact with the ribonucleotide-binding pocket of N-NTD. The striking agreement between the catechin gallate compounds and our proposed guidelines raises the possibility that their inhibitory activity arises from the interaction between the compounds and N-NTD. More studies will be required to test this possibility. In addition, the efficacy of these compounds in cells or tissue cultures is unknown and remains to be assessed.
Coronavirus vaccination via nucleocapsid protein
Coronaviral N protein has been considered a desirable target for DNA or recombinant-protein-mediated vaccination. Because the N protein resides inside the virus particle, it does not elicit neutralizing antibodies so the goal of viral vaccine development using the N protein has focused on how to induce the generation of cytotoxic T lymphocytes (CTLs) capable of destroying infected cells. Zhu et al. and Zhao et al. introduced SARS-CoV N protein DNA in mice and were able to induce high CTL activity 86, 87. Expression of SARS-CoV N protein by other viral vectors, such as the vaccine virus Ankara (MVA-NC), measles virus (rMV-N) and baculovirus (vAc-N), were successful at generating potent SARS-CoV-specific humoral and T-cell-mediated immune responses 88, 89, 90. To enhance N-protein-induced CTLs, fusion proteins where the N protein is attached to a known antigen protein, such as lysosome-associated membrane protein and calreticulin, were also tested in mice and generated strong N-protein-specific humoral and cellular immunity 91, 92. CTL epitopes in the N protein of SARS-CoV have also been mapped and several regions within the N protein induced cellular immune response in the peptide form [93]. However, the generation of N-protein-specific T cells in different mouse strains required different peptide sequences corresponding to different regions of the N protein. Zhao et al. and Cheung et al. reported that the N protein peptides NP111, NP331, NP351 and NP220 stimulated proliferation of N-protein-specific T cells in C57BL/6, C3H, BALB/c and HLA-A2.1Kb, respectively 94, 95. A more promising approach is the development of a fusion protein vaccine containing truncated SARS-CoV S and N proteins to generate specific neutralizing antibodies and induce specific CTLs at the same time [95]. The vaccine was shown to have low cross-reactivity to HCoV-229E or HCoV-OC43. Mice immunized with the vaccine were protected from SARS-CoV when challenged with the virus. However, a major stumbling block in the development of an N-protein-based vaccine is the induction of undesired immune responses that can cause immunopathology [96]. Unfortunately, this could be a general feature of the N protein, because N proteins from other nidoviruses were also shown to induce deleterious immune responses [97]. It is debatable at this stage whether the N protein should be included in subunit vaccines, because it could cause adverse effects if the induction of neutralizing antibodies is not strong enough [98]. Provided that researchers ultimately overcome these problems, the challenge to translate laboratory results to real clinical situations still remains.
Future perspectives and concluding remarks
There is no doubt that CoVs have joined the ranks of important human pathogens in this century. This is particularly true when we consider that there have already been two major CoV outbreaks in the past 15 years showing severe clinical signs. The emergence of SARS and MERS has made the identification of potential drug targets and development of effective lead compounds against CoVs an urgent task. During the SARS outbreak, many SARS patients were treated with one of many medication combinations, including ribavirin with corticosteroids, interferon (alfacon-1) with corticosteroids or ribavirin with protease inhibitors, with some encouraging outcomes 99, 100, 101. Although a combination of ribavirin and interferon has shown synergistic effects against MERS-CoV under in vitro conditions, results from human trials have been inconclusive [102], and there is still no definitive treatment regimen established for MERS. It is worrying that there is a lack of specialized therapeutics against CoVs. Current experimental compounds developed to treat CoV infections primarily target the 3C-like (3CL) and papain-like (PLP) proteases, and helicase (Nsp13) 103, 104. However, antiviral inhibitors targeting these enzymes can act nonspecifically on homologous proteins in the cell, causing host cell toxicity and severe side effects. Therefore, new antiviral strategies and targets are needed to combat infections caused by CoV.
The N protein is an appealing target because it serves as the protein building-block of CoV RNPs involved in viral transcription and replication. In addition to the development of antiviral drugs, CoV N proteins have also been used for the development of vaccines against CoV infections. However, it is unknown whether the vaccine could successfully translate to real clinical situations, or whether it would be equally effective against other CoV diseases such as MERS. To facilitate the discovery of agents that specifically block the formation of RNP during MERS-CoV genome replication, we have solved the crystal structure of the MERS-CoV N-NTD (residues 39 to 165) to a resolution of 2.63 Å (Fig. 3a) [105]. Similar to the structures of N-NTDs from other coronaviruses, MERS-CoV N-NTD forms a single domain composed of a five-stranded β-sheet core sandwiched between loops (or short 310 helix). Owing to the flexibility of the long loop between strands β2 and β3, it was relatively difficult to locate its precise position. Based on the surface charge distribution and superimposition with HCoV-OC43 and SARS-CoV N-NTDs, we identified the ribonucleotide-binding pocket in the MERS-CoV N-NTD which also contained the conserved RNA-binding residues found in other CoV N-NTDs (Fig. 3a,b). This pocket could be a validated target for antiviral drugs that interfere with the RNA-binding activity of the MERS-CoV N protein. In addition to the ribonucleotide-binding pocket, several sites comprising conserved residues on the MERS-CoV N-NTD could be potential ligand-binding sites based on the results of drug-binding site prediction (Fig. 3c) [106]. Although these sites might not be directly involved in RNA binding, the binding of small molecule compounds to these sites could still interfere with N protein function, for example through allostery, and could be alternative targets for antiviral drug development.
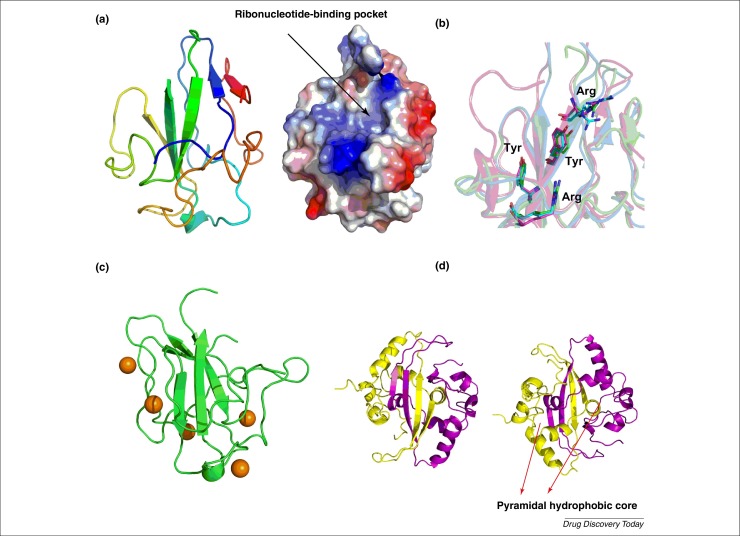
Figure 3.
(a) Ribbon representation of Middle-East respiratory syndrome coronavirus (MERS-CoV) N-N-terminal-domain (NTD) structure (left). Electrostatic surface of the MERS-CoV N-NTD structure (right). Blue denotes positive charge potential, red indicates negative charge potential. (b) Structural superimposition of the conserved residues of MERS-CoV N-NTD (green) involved in RNA binding with N-NTDs from severe acute respiratory syndrome coronavirus (SARS-CoV) (magenta) (PDB code: 2ofz) and human coronavirus OC43 (HCoV-OC43) (cyan) (PDB code: 3v3p). (c) Putative drug-binding sites predicted by metaPocket are shown as brown spheres on the MERS-CoV N-NTD structure. (d) Front (left) and back (right) views of the modeled dimer structure of MERS-CoV N-C-terminal-domain (CTD). The two red arrows indicate the position of the two pyramidal hydrophobic cores that stabilize the CTD dimer.
Based on SARS-CoV N-CTD crystal structures, we expect that the two β-strands of the β-hairpin in the CTD of MERS-CoV N protein would be located within residues 311–315 and 329–337, respectively (Fig. 3d). In addition to the H-bonds across the dimer interface formed by the main chain atoms, hydrophobic interactions involving several hydrophobic residues generate two pyramidal hydrophobic cores in the SARS-CoV N-CTD; these resemble a bow-tie-shaped pocket and stabilize the dimerization of the C-terminal domain of the N protein [45]. We found that the hydrophobic residues contributing to the dimer formation of the SARS-CoV N protein are conserved in MERS-CoV N-CTD. We suggest that the two pyramidal hydrophobic cores in MERS-CoV N-CTD could also participate in stabilizing the dimers of the MERS-CoV N protein. Targeting this hydrophobic pocket seems to be a promising avenue for the development of anti-MERS-CoV drugs.
In summary, finding small-molecule ligands that bind with high affinity to N protein could be a substantial step toward developing effective therapeutic agents for CoV diseases. The fact that the CoV N protein has many conserved sites that are crucial for its correct function bodes well for the future outlook of this approach. By contrast, because one is essentially trying to block physical phenomena (binding to RNA and self-association) instead of chemical reactions, intimate understanding of the biophysical aspects of the protein are essential for this approach to work. In this regard, advances in our knowledge of CoV N protein structure and physical behavior during this century has helped tremendously. We hope that this review will benefit the development of drugs that act on the assembly of CoV RNP through the N protein.
Acknowledgments
This work was supported by MOST grant 103-2113-M-005-007-MY3 (M-H.H.). We thank Professor Stanley Perlman (University of Iowa) and Professor Tai-Huang Huang (IBS, Academia Sinica) for their contribution of helpful resources that made this research possible.
Biographies

Ming-Hon Hou received his PhD in biochemical sciences from National Taiwan University. He did his postdoctoral research at the Academia Sinica and continued his research in drug design and biophysics at the National Chung Hsing University in Taiwan. He is now Professor and Director of the Institute of Genomics and Bioinformatics, National Chung Hsing University, and jointly appointed as Adjunct Faculty of Molecular and Biological Agricultural Sciences program of the International Graduate Program at Academia Sinica. His research interests cover a wide range of structural biology and drug development topics, including protein structure, unusual DNA structures, DNA–drug and protein–drug interactions.

Chung-ke Chang received his PhD from the Department of biological sciences at Carnegie Mellon University, USA. He has published a number of papers on the structural and biophysical aspects of the SARS coronavirus nucleocapsid protein. Trained as a protein NMR spectroscopist, his research interests range from protein structure elucidation, to characterization of macromolecular interactions to metabolomics. He is currently a postdoctoral fellow at the Institute of Biomedical Sciences, Academia Sinica, Taiwan.
References
- 1.Cherry D.K. National Ambulatory Medical Care Survey: 2006 summary. Natl. Health Stat. Report. 2008;3:1–39. [PubMed] [Google Scholar]
- 2.Hsiao C-J. National Ambulatory Medical Care Survey: 2007 summary. Natl. Health Stat. Report. 2010;27:1–32. [PubMed] [Google Scholar]
- 3.Canducci F. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J. Med. Virol. 2008;80:716–723. doi: 10.1002/jmv.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jevsnik M. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect. Dis. 2012;12:365. doi: 10.1186/1471-2334-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matoba Y. Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn. J. Infect. Dis. 2015;68:138–141. doi: 10.7883/yoken.JJID.2014.266. [DOI] [PubMed] [Google Scholar]
- 6.Lau S.K. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kon M. Detection of human coronavirus NL63 and OC43 in children with acute respiratory infections in Niigata, Japan, between 2010 and 2011. Jpn. J. Infect. Dis. 2012;65:270–272. doi: 10.7883/yoken.65.270. [DOI] [PubMed] [Google Scholar]
- 8.Gaunt E.R. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talbot H.K. The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr. Infect. Dis. J. 2009;28:682–687. doi: 10.1097/INF.0b013e31819d0d27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkman R. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.K. Epidemiology of respiratory viral infection using multiplex RT-PCR in Cheonan, Korea (2006–2010) J. Microbiol. Biotechnol. 2013;23:267–273. doi: 10.4014/jmb.1212.12050. [DOI] [PubMed] [Google Scholar]
- 12.Dare R.K. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J. Infect. Dis. 2007;196:1321–1328. doi: 10.1086/521308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prill M.M. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr. Infect. Dis. J. 2012;31:235–240. doi: 10.1097/INF.0b013e31823e07fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabeça T.K. Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza Other Respir. Viruses. 2013;7:1040–1047. doi: 10.1111/irv.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R. Characterization of human coronavirus etiology in Chinese adults with acute upper respiratory tract infection by real-time RT-PCR assays. PLoS One. 2012;7:e38638. doi: 10.1371/journal.pone.0038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepiller Q. High incidence but low burden of coronaviruses and preferential associations between respiratory viruses. J. Clin. Microbiol. 2013;51:3039–3046. doi: 10.1128/JCM.01078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Hoek L. Human coronaviruses: what do they cause? Antivir Ther. 2007;12:651–658. [PubMed] [Google Scholar]
- 18.Pyrc K. The novel human coronaviruses NL63 and HKU1. J. Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Hoek L. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zumla A. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perlman S. The Middle East respiratory syndrome – how worried should we be? MBio. 2013;4:e00531–e613. doi: 10.1128/mBio.00531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau S.K. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 24.Chu D.K. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haagmans B.L. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemida M.G. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St-Jean J.R. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadler K. SARS – beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masters P.S. Structure and function studies of the nucleocapsid protein of mouse hepatitis virus. Adv. Exp. Med. Biol. 1990;276:239–246. doi: 10.1007/978-1-4684-5823-7_33. [DOI] [PubMed] [Google Scholar]
- 30.Masters P.S., Sturman L.S. Background paper. Functions of the coronavirus nucleocapsid protein. Adv. Exp. Med. Biol. 1990;276:235–238. doi: 10.1007/978-1-4684-5823-7_32. [DOI] [PubMed] [Google Scholar]
- 31.Almazan F. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schelle B. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuniga S. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh P.K. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopecky-Bromberg S.A. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du L. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surjit M. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang T.K. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan K.H. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 2005;12:1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mourez T. Baculovirus expression of HCoV-OC43 nucleocapsid protein and development of a western blot assay for detection of human antibodies against HCoV-OC43. J. Virol. Methods. 2007;139:175–180. doi: 10.1016/j.jviromet.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin S.Y. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenavas S. Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target. Future Microbiol. 2013;8:1537–1545. doi: 10.2217/fmb.13.128. [DOI] [PubMed] [Google Scholar]
- 43.Musah R.A. The HIV-1 nucleocapsid zinc finger protein as a target of antiretroviral therapy. Curr. Top. Med. Chem. 2004;4:1605–1622. doi: 10.2174/1568026043387331. [DOI] [PubMed] [Google Scholar]
- 44.Chang C.K. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu I.M. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C.Y. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C.K. The SARS coronavirus nucleocapsid protein – forms and functions. Antiviral. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 49.Ma Y. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell. 2010;1:688–697. doi: 10.1007/s13238-010-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang C.K. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng T.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Surjit M. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C.H. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen I.J. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim. Biophys. Acta. 2013;1834:1054–1062. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grossoehme N.E. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan Y.W. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo Y.S. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang C.K. Transient oligomerization of the SARS-CoV N protein – implication for virus ribonucleoprotein packaging. PLoS One. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jayaram H. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang C.Y. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci. 2009;18:2209–2218. doi: 10.1002/pro.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caul E.O., Egglestone S.I. Coronavirus-like particles present in simian faeces. Vet. Rec. 1979;104:168–169. doi: 10.1136/vr.104.8.168. [DOI] [PubMed] [Google Scholar]
- 63.Davies H.A. Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol. 1981;53:67–74. doi: 10.1099/0022-1317-53-1-67. [DOI] [PubMed] [Google Scholar]
- 64.Macnaughton M.R., Madge M.H. The genome of human coronavirus strain 229E. J. Gen. Virol. 1978;39:497–504. doi: 10.1099/0022-1317-39-3-497. [DOI] [PubMed] [Google Scholar]
- 65.Neuman B.W. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bárcena M. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U. S. A. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dyson H.J. Roles of intrinsic disorder in protein–nucleic acid interactions. Mol. Biosyst. 2012;8:97–104. doi: 10.1039/c1mb05258f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Compton S.R. In vitro replication of mouse hepatitis virus strain A59. J. Virol. 1987;61:1814–1820. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hilser V.J., Thompson E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zlotnick A. Are weak protein–protein interactions the general rule in capsid assembly? Virology. 2003;315:269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 71.Kuo L. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monod A. Learning from structure-based drug design and new antivirals targeting the ribonucleoprotein complex for the treatment of influenza. Expert Opin. Drug Discov. 2015;10:345–371. doi: 10.1517/17460441.2015.1019859. [DOI] [PubMed] [Google Scholar]
- 73.Kwong A.D. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crowder S., Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat. Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- 75.Cianci C. Influenza nucleoprotein: promising target for antiviral chemotherapy. Antivir. Chem. Chemother. 2013;23:77–91. doi: 10.3851/IMP2235. [DOI] [PubMed] [Google Scholar]
- 76.Tarus B. Structure-based design of novel naproxen derivatives targeting monomeric nucleoprotein of influenza A virus. J. Biomol. Struct. Dyn. 2015;33:1899–1912. doi: 10.1080/07391102.2014.979230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lejal N. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob. Agents Chemother. 2013;57:2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hung H.C. Development of an anti-influenza drug screening assay targeting nucleoproteins with tryptophan fluorescence quenching. Anal. Chem. 2012;84:6391–6399. doi: 10.1021/ac2022426. [DOI] [PubMed] [Google Scholar]
- 79.Gerritz S.W. Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15366–15371. doi: 10.1073/pnas.1107906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin S.-Y. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang C-k. Structure-based virtual screening and experimental validations for discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. BioSyst. 2015 doi: 10.1039/C5MB00582E. [DOI] [PubMed] [Google Scholar]
- 82.Hayouka Z. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pujol A. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J. Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen Y.F. E339…R416 salt bridge of nucleoprotein as a feasible target for influenza virus inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16515–16520. doi: 10.1073/pnas.1113107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomedicine. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu M.S. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao P. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2005;331:128–135. doi: 10.1016/j.virol.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bai B. Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of SARS-like coronavirus generates humoral and cellular immune responses. Mol. Immunol. 2008;45:868–875. doi: 10.1016/j.molimm.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liniger M. Induction of neutralising antibodies and cellular immune responses against SARS coronavirus by recombinant measles viruses. Vaccine. 2008;26:2164–2174. doi: 10.1016/j.vaccine.2008.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schulze K. A prime-boost vaccination protocol optimizes immune responses against the nucleocapsid protein of the SARS coronavirus. Vaccine. 2008;26:6678–6684. doi: 10.1016/j.vaccine.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang K. Immune responses to T-cell epitopes of SARS CoV-N protein are enhanced by N immunization with a chimera of lysosome-associated membrane protein. Gene Ther. 2009;16:1353–1362. doi: 10.1038/gt.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim T.W. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu S.J. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheung Y.K. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25:6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deming D. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wongyanin P. Role of porcine reproductive and respiratory syndrome virus nucleocapsid protein in induction of interleukin-10 and regulatory T-lymphocytes (Treg) J. Gen. Virol. 2012;93:1236–1246. doi: 10.1099/vir.0.040287-0. [DOI] [PubMed] [Google Scholar]
- 98.Bolles >M. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adedeji A.O., Sarafianos S.G. Antiviral drugs specific for coronaviruses in preclinical development. Curr. Opin. Virol. 2014;8:45–53. doi: 10.1016/j.coviro.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Graham R.L. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Omrani A.S. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keum Y.S., Jeong Y.J. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Y.S. Crystallographic analysis of the N-terminal domain of Middle East respiratory syndrome coronavirus nucleocapsid protein. Acta Crystallogr. F: Struct. Biol. Commun. 2015;71:977–980. doi: 10.1107/S2053230X15010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Z. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]