Abstract
The danger posed by biological threat agents and the limitations of modern detection methods to rapidly identify them underpins the need for continued development of novel sensors. The application of nanomaterials to this problem in recent years has proven especially advantageous. By capitalizing on large surface/volume ratios, dispersability, beneficial physical and chemical properties, and unique nanoscale interactions, nanomaterial-based biosensors are being developed with sensitivity and accuracy that are starting to surpass traditional biothreat detection methods, yet do so with reduced sample volume, preparation time, and assay cost. In this review, we start with an overview of bioagents and then highlight the breadth of nanoscale sensors that have recently emerged for their detection.
Introduction
In 1346, the Mongol army that was laying siege to the Crimean city of Kaffa hurled the corpses of plague victims over the city walls, an event that likely seeded the epidemic that infected the besieged city. The disease subsequently spread across Europe as the Black Death, killing 24 million people in the next six years [1], [2]. Though by no means the first example of biological warfare, this event is an illustrative example of the power of even crude biological weapons. It was not until the 20th century, though, that advances in microbiology spurred widespread research into the production and weaponization of biological threat agents. The biological arms race that took off during World War II and escalated with Cold War tensions was thankfully tempered by the United Nation's 1972 Biological Weapons Convention (BWC), which prohibited the development, production, and stockpiling of bioweapons and their delivery systems [1], [2], [3], [4]. Since then, the threat of biowarfare has largely been supplanted by that of bioterrorism, which profits from the ‘horror factor’ and the low financial cost/casualty ratio associated with bioagents [5]. Examples such as the 2001 anthrax attacks in the United States, which led to 22 cases of confirmed anthrax, five deaths, and required prophylactic antibiotics for 10,000 people, underscore the need for vigilance and the urgency behind the development of superior sensors and monitoring techniques for weapons of mass destruction (WMD) threats [1], [2].
Bioagents include bacteria, viruses, and toxins, and they need not be intentionally disseminated in order to pose a threat to human health [1]. Moreover, the need to detect members of this diverse group with specificity, sensitivity, and rapidity augments an already substantial challenge. Traditional detection methods, which include assays based on biochemical and immunological recognition, biomolecular techniques such as polymerase chain reaction (PCR), or cell culture, often require ample time (hours to days), leaving a clinician to essentially guess as to the identity of the pathogen or toxin. For environmental monitoring purposes and the initiation of containment and prophylactic treatment of exposed individuals, these techniques leave significant room for improvement.
In recent years, efforts in the field of bioagent detection have taken advantage of the burgeoning use of nanomaterials (NMs) throughout the scientific community to create novel sensors. In addition to the properties unique to nanoscale materials, the disposability and small size of NMs make them well-suited to application in portable and multiplexed sensors. Relying on intrinsic NM properties in conjunction with immunogenic or other biological recognition mechanisms, detection schemes exploiting colorimetric, photoluminescence (PL), electrochemical, plasmon resonance, and other responses have been explored [6], [7], [8], [9], [10], [11], [12], [13], [14]. Here we examine a range of bioagents and the nanoscale sensors that have been recently developed to detect them. Building on the foundation laid by an earlier review from this group that focused on the devices under development for bio-threat agent detection, we examine the most novel and innovative ways in which NMs are being applied to this problem [15]. Many of the techniques highlighted are research-based prototypes not yet ready for integration into commercial devices, but they already offer promise when compared to the current ‘gold standards’ for detection and highlight areas in which nanosensors are able to outperform the traditional detection paradigms. Rather than an exhaustive review of these sensors, our focus is on highlighting select NMs and how they have been applied to detecting biothreat agents.
Overview of bioagents
The bioagents that have been studied over the course of the past century were selected for a variety of characteristics that made them especially suited to weaponization. The number and range of bioagents that were ultimately chosen for further development reflects how these characteristics in combination can fill different niches in an arsenal, from incapacitation to mass fatalities, from confined attack to self-spreading epidemic. Infectivity, or the ease with which a microorganism establishes itself in the host, is not necessary related to virulence, which refers to the severity of the disease induced by the pathogen. Transmissibility of the pathogen from one person to another is required to seed an epidemic, while stability is more critical in pathogens that are intended to be dispersed by environmental routes. Other attributes, including toxicity, pathogenicity, inoculation period, and lethality also come into play. A detailed description of each of these characteristics can be found in the NATO Handbook on the Medical Aspects of NBC Defensive Operations (NBC – nuclear, biological, and chemical) [16]. A sampling of historically important bioagents is shown in Table 1 . This list is by no means a comprehensive inventory of agents or of countries involved in their research but is instead meant to illustrate the variety of actors at play. In the context of this review, we also consider important biological threats that require no explicit weaponization (e.g., endemic diseases such as the 2014 Ebola outbreak in West Africa).
Table 1.
Representative bioagents
| Agent (disease) | Country or organizationa | Incidentsb(aggressor, target, year) | Transmission modec | Greatest concern |
|---|---|---|---|---|
| Bacteria | ||||
| Bacillus anthracis (anthrax) | US, UK, Canada, USSR, Iraq, Germany, Japan, Aum Shinrikyo |
Germany; Allies; WWId USSR; Sverdlosk; 1979 Aum Shinrikyo; Japan; 1993 Not determined; US; 2001 |
Inhalation, ingestion, cutaneous | Aerosol dispersion |
| Yersinia pestis (plague) | US, USSR, Japan | Japan; China; WWII | Inhalation, animal vectors | High fatality rate, secondary transmission |
| Francisella tularensis (tularemia) | US, USSR, Japan | USSR; German troops; WWIIe | Ingestion, inhalation, contact, animal vectors | Infectivity, difficult diagnosis, antibiotic resistance |
| Coxiella burnetti (Q fever) | US, USSR | USSR; German troops; WWIIe | Inhalation, animal vectors | Infectivity, stability, secondary infection (from animal vectors) |
| Brucella species (brucellosis) | US |
Germany; Allies; WWId Japan; China and Manchuria; WWII USSR; Afghanistan; 1982–4e |
Inhalation, ingestion, contact | Aerosol dispersion |
| Burkholderia mallei (glanders) | Germany, Japan, USSR, US | Inhalation, contact | Infectivity, high morbidity | |
|
Salmonella typhimurium (salmonella) |
Japan |
Japan; China; WWII Rajneeshee cult; Oregon; 1984 |
Ingestion |
Incapacitation |
| Viruses | ||||
| Variola major (smallpox) | USSR, Japan | USSR; Aralsk; 1971 | Inhalation, contact | Secondary transmission |
| Viral hemorrhagic fevers (Ebola, Marburg, etc.) |
US, USSR, Japan |
– |
Inhalation, contact |
High mortality, secondary transmission |
| Toxins | ||||
| Mycotoxins (including aflatoxin, T-2 toxins) | USSR, Iraq | USSR; Laos; 1975–81 | Contact, inhalation, ingestion | Incapacitation |
| Clostridium botulinum toxin (botulism) | US, Iraq, USSR Germany, Japan, Aum Shinrikyo | Aum Shinrikyo; Japan; 1990–95 | Inhalation, ingestion | Extreme toxicity, aerosol dispersion |
| Ricin | US, UK, USSR, Iraq, Al Queda | Soviet assassin; Georgi Markov; 1978 | Inhalation, ingestion, intramuscular | Widespread availability |
| Staphylococcal enterotoxin B (SEB) | US | – | Inhalation, ingestion | Incapacitation |
Partial list of states or groups involved in researching or weaponizing the agent from Ref. [1].
Accidental releases are in plain text. Attempted use shown in italics. Successful attacks in bold.
Common natural transmission mode shown in italics. Transmission mode posing greatest terrorist threat in bold.
Against livestock rather than human targets.
Suspected use.
Bacteria
Bacteria are unicellular organisms that reproduce by replication and cause disease either through invasion of host tissue or release of a toxin. Modern biological warfare began during World War I when German agents infected animals shipped from the United States and other neutral countries to Allied Forces with anthrax and glanders [4]. The diseases caused by bacteria, however, have long histories; Bacillus anthracis, the causative agent of anthrax, has been proposed as a possible cause of the fifth and sixth plagues mentioned in Exodus. Likewise, Plague (caused by Yersinia pestis) played a substantial role both in natural epidemics (e.g., the Black Death) and in bioweapons programs in the 20th century. Interestingly, Y. pestis still sees occasional outbreaks in the American Southwest, typically originating from small mammal and rodent vectors, although modern antibiotics have significantly reduced mortality [17]. The diversity of bacteria is reflected in the range of diseases they cause, infectious doses, etc. For example, while the mortality rate of untreated inhalation anthrax is greater than 90%, the mortality rate of brucellosis, caused by one of six Brucella species, is 2–5%. Paradoxically, as few as 10 organisms are required to cause brucellosis, however, while an infectious dose of B. anthracis is 10,000 spores.
The development of antibiotics is undoubtedly one of the most important advances in human health in the 20th century, but antibiotics may have limited effectiveness against weaponized or genetically engineered disease strains [2], [16]. Moreover, even in the absence of genetic engineering, natural antibiotic resistance seen in diseases such as tuberculosis, which still causes more than a million fatalities annually, may prove to be one of the most substantial health challenges of the next century [18].
Viruses
Viruses are infectious agents that require a host for propagation, a trait that has caused them to excel at finding new hosts [19], [20]. Whereas many of the bacteria that were weaponized are only poorly transmissible from person to person, viruses are the pathogens that seed pandemics. In Boston in 1752, for example, only 174 susceptible people (i.e., those who had not been previously infected or inoculated and remained in the city) escaped infection with smallpox during a significant outbreak [21]. Although the disease was eradicated in 1980 through the World Health Organization's worldwide campaign, this extraordinary infectivity, along with the corresponding virulence reflected by the high mortality rate, ensures that smallpox remains among the most high-priority bioagents.
While the public response to the 2014 West Africa outbreak of Ebola – a virus in the hemorrhagic fever family, many of whose members were weaponized – demonstrates the power that diseases with high mortality rates and dramatic presentation have on the human psyche, commonplace endemic diseases should not be underestimated. The Spanish Flu of 1918–19 caused an estimated 20–50 million deaths worldwide, and new strains of influenza are constantly emerging through mutation and genetic recombination, requiring the development of a new vaccine every year. Other viruses appear seemingly out of nowhere, as was the case with Severe Acute Respiratory Syndrome (SARS) in 2003 and Middle East Respiratory Syndrome (MERS) in 2012. Both qualify as emerging infectious diseases, defined as diseases that have increased in incidence in the past twenty years or show signs of a likely increase in the near future [22], [23], [24]. While some bacterial species fall into the category of emerging diseases (e.g., multidrug-resistant M. tuberculosis), viruses have assumed a higher public profile in recent years as outbreaks threaten to turn into pandemics. Because limited viral equivalents to antibiotics currently exist, treatment for the diseases they cause is typically palliative.
Toxins
Because toxins can originate from diverse sources, including animals, plants, and microbes, they are defined not by their source but by their common properties: they are chemicals that elicit a toxic response and are neither man-made nor volatile. Unlike bacteria and viruses, which replicate under suitable circumstances, toxins lack the ability to propagate themselves and spread to new hosts. Botulinum neurotoxin (BoNT), among the best-researched bioagents, is the most lethal toxin known. A protein toxin produced by the bacterium Clostridium botulinum, causative agent of botulism, its LD50 (lethal dose to 50% of the sample population) is a mere 0.1 μg/70 kg intravenously. A thousand-fold less toxic, ricin is a protein found in the seeds of the castor bean plant and has a history of use in assassinations and murders, such as the Bulgarian secret police's 1978 assassination of Bulgarian dissident Georgi Markov, in which a ricin pellet likely obtained from the KGB was injected into his leg by a ‘pedestrian’ agent using a modified umbrella. A third protein toxin, staphylococcal enterotoxin B (SEB), produced by the bacterium Staphylococcus aureus, is an incapacitant at low doses, while the high doses required to elicit mortality give the agent an excellent ‘safety ratio.’ Mycotoxins, produced by fungi, include trichothecenes (T-2 toxins), ochratoxin, and aflatoxin. Since these are low molecular weight compounds, these toxins result in symptoms more common to chemical agents. Some T-2 toxins are vesicants (blister agents) 400 times more potent than mustard gas, and their effects are observed in minutes rather than hours or days. Antitoxins have been developed for some toxins, although the requirement for rapid administration (e.g., before the onset of symptoms for BoNT) limits their utility in non-prophylactic situations.
Current detection methods
In the event of a biological attack or natural outbreak, identification of the bioagent is critical to both containment and diagnostic choice of appropriate prophylactic therapy. Traditional methods for the detection of bioagents include culture, immunological assays such as enzyme-linked immunosorbent assay (ELISA), PCR, and others. These methods are sensitive and specific, but they are typically used after the fact to confirm a clinician's suspected diagnosis. Pairing collection and filtration systems to these techniques lays the foundation for continuous monitoring systems that could give advanced warning of a bioattack, but many of the techniques mentioned here are cumbersome, requiring purified samples and hours or days to complete the analysis [25].
Culture
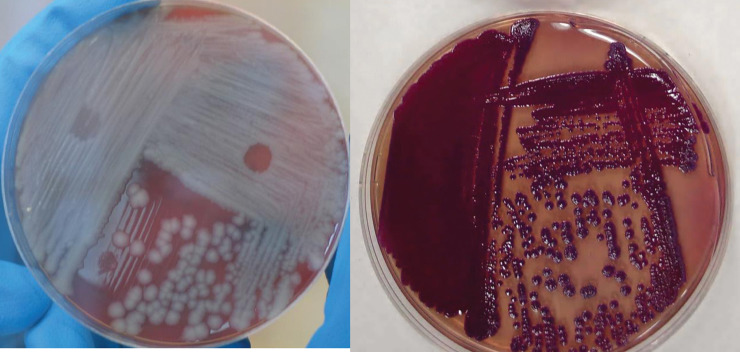
Culture is suited to the identification of both bacteria and viruses. Bacterial cultures are typically grown on a nutrient-containing substrate from clinical or environmental samples, while viral cultures must be grown in host cells. Obtaining a sample that contains the pathogen can be difficult (e.g., bacteria may not be found in the blood in the early stages of the disease and will be absent if a patient has already begun antibiotic treatment), and the difficulty in culturing some agents can generate false negatives [1], [19]. Moreover, reaching an identifiable stage of growth can be slow, typically requiring several days. Some samples are even more extreme; culture of M. tuberculosis, for example, requires nine days or more [26], [27], [28], while Brucella species can take up to three weeks and are isolated from blood with 10–90% success [1], [2], [29]. To further complicate matters, while phenotypic diagnosis via microscopy can be used to identify inclusion bodies from poxvirus infections, this method is insufficient to differentiate individual poxviridae species, meaning that smallpox and the related pox species are indistinguishable by culture. A final consideration is that many of the bioagents mentioned here are extremely pathogenic, and culturing them can invite substantial risk and may require extensive containment facilities. Examples of cultured B. anthracis and Burkholderia pseudomallei are shown in Fig. 1 .
Figure 1.
Bacterial culture of Bacillus anthracis (left), the spores of which cause anthrax, and Burkholderia pseudomallei (right), the cause of melioidosis. Reproduced from [1].
Immunoassays
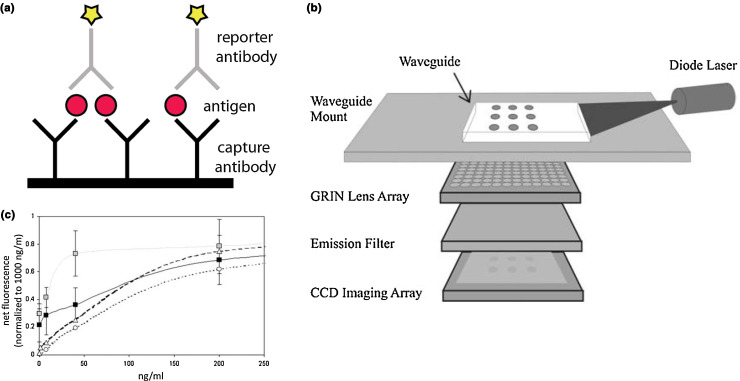
Immunodiagnostics refers collectively a range of assays that detect proteins, antigens, or host-produced antibodies [1]. These assays traditionally use antibodies produced in host animals, although aptamers, peptides, and engineered antibody fragments are also used now for similar purposes. As illustrated in Fig. 2 , for typical antigen detection, a complementary antibody, known as the capture antibody, is affixed to a substrate and captures any antigen present in the sample. Signal transduction is accomplished through the addition of a secondary reporter antibody that binds to the analyte and is labeled with a fluorophore, radioisotope, or other detectable label [30]. Because the analyte is sandwiched between the capture and reporter species, the descriptive term sandwich immunoassay is commonly used [1]. The same technique can be adapted to detect antibodies generated in an host following exposure.
Figure 2.
(a) Capture antibodies affixed to a substrate capture antigen present in the analytical sample. Reporter antibodies, which are labeled with a reporter species such as a fluorophore or radioisotope, bind the captured antigen. (b) One possible sensor array based on immunoassays, in which the fluorophores tagging the reporter antibody are excited by laser and the PL read out on a CCD imaging array. The grid setup allows for multiplexing. (c) Fluorescence response to the presence of various analytes detected by immunoassays, including two types of BoNT (black and gray squares), ricin (open circles), and cholera toxin (open triangles). Reproduced with permission from [37].
Enzyme-linked immunosorbent assays (ELISA) make use of an enzyme tethered to the reporter antibody in the sandwich assay described above. The enzyme produces a colorimetric, fluorescent, or electrochemiluminescent response in a substrate that is added in the final step of the detection sequence and serves to amplify the original signal. As one of the most well-developed detection techniques for bioagents, the U.S. Department of Defense (DoD) has ELISAs for nearly 130 antibodies and antigens, including bacteria, viruses, and protein toxins as part of its Critical Reagents Program [1], [2], [16], [31]. ELISA offers relatively fast readout times (15 min to several hours) but does so at the cost of accuracy (50–70%) and sensitivity [1]. Moreover, the environmental susceptibility of the participating species (e.g., denaturation of the antibodies) limits application for continuous monitoring and in challenging environments around the world [30], [32], [33]. This has led the DoD and especially the Defense Threat Reduction Agency (DTRA) to invest heavily in the development of more stable antibodies. These include antibodies derived from sharks and camelids (e.g., llamas), which are structurally simpler and more tolerant of heat [34], [35], [36].
Polymerase chain reaction
PCR provides an extremely sensitive means of detecting pathogenic bioagents (but not isolated toxins) through sequential replication/amplification of a target nucleic acid originating from the suspected bioagent [1], [30], [32]. Using target-specific oligonucleotide primers that hybridize to and flank the region of DNA to be amplified, a DNA polymerase, and a supply of nucleic acid precursors (deoxyribonucleotide triphosphates), replication of the target DNA is accomplished by subjecting the mixture to repeated, established cycles of heating, primer binding, and primer extension. The effect of this process is to render undetectably small quantities of genetic material (as few as 10 copies) sufficiently plentiful for detection. To amplify RNA from viruses, the process is modified to first convert the target RNA to complementary DNA by reverse transcription, after which the complementary DNA is amplified by PCR. Although cycling through PCR a sufficient number of times to guarantee a readout is time-consuming (6+ hours), by coupling a fluorescent reporter to the reaction, real-time monitoring is made possible [1]. PCR is the gold standard for virus detection; however, due to the nature of primer design, it may fail to detect new viral strains or mutants [1], [2], [38]. Moreover, because it is sensitive only to a region of DNA, a known pathogen could be engineered to be undetected by standard PCR through the omission of the region important for amplification [1].
Animal lethality assay
Suspected cases of foodborne botulism are confirmed using the mouse lethality assay, in which the analytical sample is administered to a mouse that has received BoNT antitoxin and a second mouse that has not. By monitoring the onset of symptoms, the presence of BoNT can be confirmed. The assay requires up to 96 hours to complete but remains the ‘gold standard’ for foodborne BoNT detection, as the complicated matrix can interfere with immunological assays. It is ineffective in detecting inhalation botulism [1].
Physical methods
Lacking the biorecognition aspect of the other bioagents, mycotoxin detection relies predominantly on physical separation technologies, including gas–liquid chromatography (GLC), high-performance liquid chromatography (HPLC), thin-layer chromatography (TLC), and capillary electrophoresis [39], [40], [41]. HPLC coupled with mass spectrometry can achieve detection limits as low as 0.1 ppb [42]. The standard method for confirming a suspected tuberculosis diagnosis is smear microscopy with a sputum sample, a diagnostic tool that can be completed within minutes but with abysmal specificity (correctly identifies 30–35% of positive cases).
Nanomaterials for bioagent detection
Developing better sensors for bioagents requires improving on the sensitivity, selectivity, or rapidity of the sensors that are currently available. While the methods described above, especially when used in combination, can achieve extraordinary sensitivity and selectivity, they are overwhelmingly slow, with shortcomings both in sample preparation (e.g., extraction from a matrix, purification) and assay time. Moreover, these techniques are often expensive and immobile, requiring laboratory conditions for successful analysis. These are precisely the shortcomings that NMs are best-suited to address [43], [44]. Their small size and disposability make NMs excellent candidates for field-based sensors. As a substrate for immunoassays, NMs provide high surface area on a platform that can be dispersed in an analytic sample and often provide feedback in less than a minute. Magnetic NMs show promise as a means of concentrating an analyte from a complex matrix and can also provide feedback even in opaque solutions. Quantum-confined semiconducting NMs possess photophysical properties that can be exploited for tagging analytes and participating in energy transfer while their physical properties make them more durable than dyes and suited to a non-laboratory environment. Table 2 presents a representative overview of some of the many NMs that have been used to detect bioagents and highlight the diversity of materials, assay formats and signal transduction mechanisms that are being exploited to create superior sensors.
Table 2.
Representative examples of biothreat agent detection with nanomaterials
| Target | Nanomaterial | Recognition/capture component | Sensing mechanism | Reference |
|---|---|---|---|---|
| Brucellaspp. | AuNPs | Complementary oligonucleotide | Colorimetry | [45] |
| Brucellaspp. | Magnetic NPs | Antibodies | Magnetic susceptibility | [50] |
| Influenza virus | Ag nanorods | Electrostatic forces | SERS | [46] |
| BoNT | QDs | Single chain variable fragment | FRET | [47] |
| B. anthracisspores | Ln-doped NPs | EDTA | PL | [48] |
| Genetic material (bacteria or virus) | Magnetic NPs | Electrostatic forces | PCR | [49] |
| Variolavirus | Heterogeneous nanowires | Antibodies | Reflectance/PL | [51] |
| M. tuberculosis | Carbon nanotubes | Complementary oligonucleotide | Impedance | [52] |
Abbreviations: BoNT, botulinum neurotoxin; AuNP, gold nanoparticles; QD, quantum dot; EDTA, ethylenediamine tetraacetic acid; SERS, surface enhanced Raman spectroscopy; FRET, Förster resonance energy transfer; PL, photoluminescence; PCR, polymerase chain reaction.
Nanomaterial bioconjugation
Among their properties, the ability of NMs to be functionalized with a wide variety of biomolecules and attachment chemistries is perhaps one of the most important criteria in their use as probes for detecting bioagents. Biofunctionalization is what allows for the capture or recognition of biological analytes on the surface of the NM, provided that suitable molecular orientation, ligand density, and binding strength exist. Standard attachment biochemistries, including electrostatic, covalent, and non-covalent bonding, have all been demonstrated in a range of nanoparticles (NPs) [53].
The diversity of bioconjugation options, combined with the many available NMs, forms an extensive and complex biochemical toolbox for tackling the challenge of biothreat detection. In combination, these options permit the design of probes that are able to respond to a variety of ‘positive’ conditions. For example, bacteria can be detected through antibody binding to specific moieties displayed on the cell surface, including proteins, carbohydrates, and sugars, while other antibodies can be used to detect secreted proteins such as toxins. An entirely different method, oligonucleotides on an NP surface can be used to capture genetic material, which can then be amplified by PCR. While either of these strategies will detect the analyte, additional factors, such as the thermal stability of the sensor, its response time and cost, analyte preparation, and assay sensitivity can inform which NM and conjugation strategy is optimally suited to address specific design constraints. As it currently stands, no technology stands out as the best in all of these areas; however, the NM toolbox provides us with many exciting paths forward [54].
Gold nanoparticles
Gold nanoparticles (AuNPs) are widespread in the detection of bacteria, viruses, and toxins [6]. At their most basic, they provide a scaffold that is compatible with a large swath of biologically and chemically active molecules without compromising their activity, and the high surface/volume ratio intrinsic to nanoscale materials further enhances this utility [6], [8]. AuNPs range from 2 to 100 nm and exhibit a localized surface plasmon resonance (LSPR) phenomenon that gives rise to excellent optical absorption properties capable of sensing changes near the AuNP surface. Below 2 nm in size, gold nanocrystals can also be fluorescent, and the term gold nanocluster is used to differentiate between materials exhibiting fluorescent and plasmonic signals. Plasmonic effects (and therefore AuNPs) dominate literature that focuses on sensing and detection using gold species. AuNPs can be coated with reactive ligands, allowing the use of covalent modification chemistries for the attachment of biomolecules (DNA, antibodies, proteins, etc.), including maleimide-thiol, NHS-ester, cycloaddition (click) chemistry, carbodiimide coupling (EDC), as well as non-covalent (e.g., biotin or NTA) or passive adsorption [55].
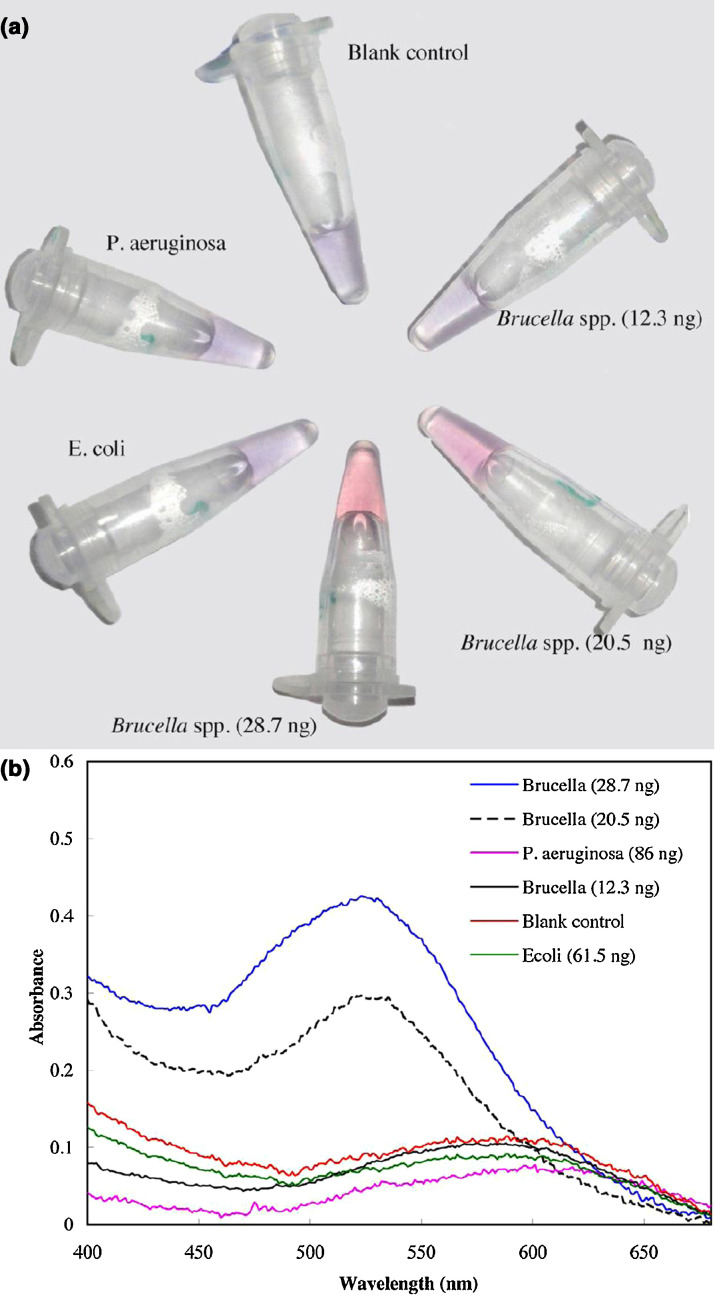
One of the most straightforward uses of AuNPs for detection involves functionalizing the AuNP surface with an oligonucleotide complementary to the target DNA, as shown in Fig. 3 , for example, Brucella species [45]. Assays were complete in 1–10 min, in contrast to hours to weeks for PCR or culture, respectively, while attaining pg/μL detection limits. In the presence of the analyte, the AuNPs aggregate in a matter of minutes, giving rise to a colorimetric change that is not observed in the presence of non-complementary DNA (Fig. 3). While the color change is visible, absorption spectroscopy can provide more quantitative feedback and lowers the detection limit. By changing the surface functionalization to other DNA sequences, antibodies, or other capture elements, this system can easily be converted to the detection of not only other examples of bacterial DNA [27], [56], [57], [58], [59] but also viruses and certain toxins [60], [61], [62]. Electrochemical transduction using AuNPs provides a means of detection that can be applied in turbid or colored solutions. Electrochemical response resulting from the aggregation in solution of antibody-functionalized AuNPs has been applied to the detection of a variety of bioagents [6], [8], [58], [63], [64]. The same principle can be applied to a solid substrate that doubles as an electrode [58], [65], [66].
Figure 3.
(a) Visual changes in the color of AuNPs as they aggregate in the presence of Brucella spp. are absent when the system is exposed to non-target DNA from other bacterial species. (b) The colorimetric changes can be monitored using UV–vis spectroscopy, providing a more sensitive read-out. Reproduced with permission from [45].
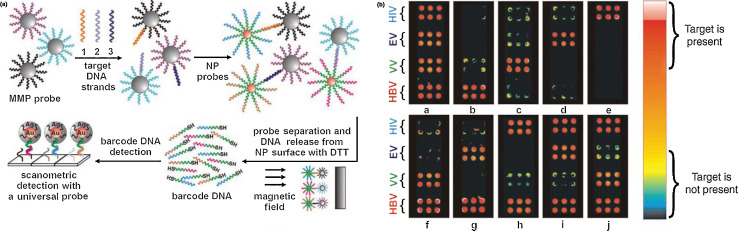
Lateral flow assays rely on colorimetric changes on a solid substrate. AuNPs loaded with antibodies (here for ricin) serve as the reporter species in a sandwich immunoassay with a banded output similar to that of a pregnancy test, the most well-known example of a lateral flow assay [67]. Alternatively, replacing the solid substrate with functionalized magnetic microparticles provides a means for pulling the probe/target complexes out of a matrix for subsequent detection using a chip-based read-out system (Fig. 4 ) [68]. Functionalizing both a magnetic microparticle probe (MMP) and a AuNP with polynucleotide strands that together are complementary to the target analyte provides a means of linking the AuNP to the MMP in the presence of the analyte. Applying a magnetic field pulls the target complexes out of the analytical matrix, and the use of ‘barcode’ DNA to isolate the complexes based on the analyte being detected allows for multiplexing using a scanometric readout.
Figure 4.
(a) Both a substrate, here magnetic NPs, and AuNP are functionalized with polynucleotide strands that together are complementary to the target analyte. Hybridization of the target results in binding of the AuNP to the magnetic microparticle probe (MMP). The application of a magnetic field pulls the target complexes out of the analyte matrix. Use of ‘barcode’ DNA to isolate the complexes based on the analyte being detected allows for multiplexing using a scanometric readout. (b) A multiplexed scanometric readout demonstrates the ability to differentiate among hepatitis B (HBV), variola (VV), Ebola, and human immunodeficiency viruses (HIV). Reproduced with permission from [68].
Surface enhanced Raman spectroscopy (SERS) has also been used in conjunction with AuNPs for bioagent detection [6], [8], [68], [69]. The presence of the target DNA results in binding of the AuNP to a surface by bridging two smaller polynucleotide strands, one anchored to the substrate and the other functionalizing the AuNP. Silver subsequently bound to the surface of the AuNP enhances the SERS signal, leading to femtomolar detection limits [69]. Another technique that relies on surface-based properties of NMs, surface plasmon resonance (SPR) has likewise been used in conjunction with AuNPs for bioagent detection [67], [70].
Finally, AuNPs are being paired with highly specialized and technical detection methods, including quartz crystal microbalance [71], [72], evanescent field-coupled waveguide-mode sensors [73], and localized SPR [74], [75]. In one example, Jin et al. employed a piezoelectric transducer to detect bovine serum albumin coated with aflatoxin using AuNPs as a ‘weight label’ in order to amplify the signal of the piezo quartz crystal [71].
NPs synthesized from alternative metals, including noble metals such as silver, platinum, and palladium, and rare earth metals such as europium, terbium, and gadolinium, have also seen interest for assay development [55]. Among these, silver nanoparticles (AgNPs) are the best characterized due to their antimicrobial properties. Although they generate a strong SPR signal, they can be challenging to work with as they are prone to oxidation; however, AgNPs show promise in theranostic applications, in which NPs play a role in both the detection and the treatment of infectious diseases.
Quantum dots
One of the most straightforward and sensitive methods of detection biological interactions is by monitoring changes in light emission properties. This can be achieved using NPs with intrinsic PL properties, most notably, quantum dots (QDs). QDs are a subset of NPs defined as quantum-confined semiconducting nanocrystals. Their unique properties, including size-dependent emission energies, small size, physical robustness, and flexible surface functionalization, have garnered significant interest for bioagent detection. QD-antibody conjugates have been highly successful in detecting protein toxins and viruses; examples include ricin [76] and S. typhi [77], as well as a multiplexed analysis of ricin, SEB, cholera, and shiga-toxin [78]. In addition to straightforward ELISAs [78], [79], antibody-labeled QDs can be used as a fluorescent tag and coupled with laboratory techniques such as flow cytometry [80], [81] or agglutination/flocculation assays [82]. The same principles can be applied to QDs that are functionalized with aptamers, synthetic DNA capture elements that have been engineered to recognize bacteria, viruses, and toxins [83], [84]. In these systems, the greatest benefit arises from the rapidity of the assay and, in some cases, the ability to multiplex by using QDs with different emission wavelengths. Furthermore, QD-antibody conjugates have been shown to amplify an SPR output signal, resulting in a 10-fold signal enhancement over analogous control single-domain antibody-based reporters used by themselves [85].
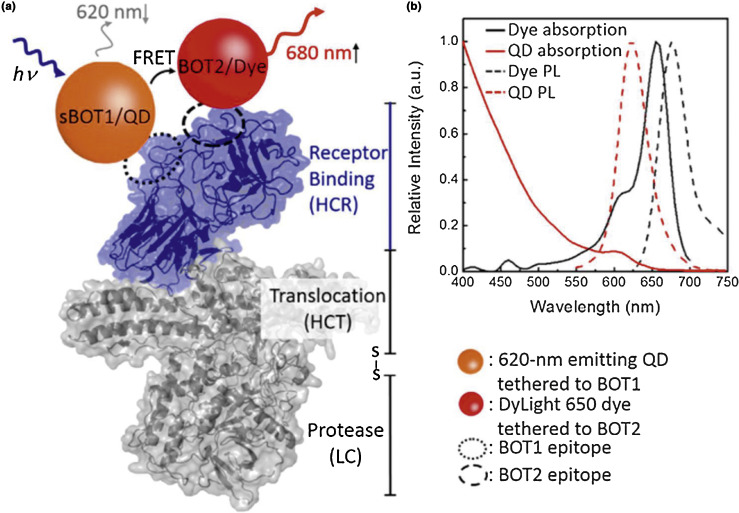
Exploiting Förster resonance energy transfer (FRET), a nonradiative energy transfer process that takes place between an excited state donor and a ground-state acceptor, between a QD and dye capitalizes on the luminescent advantages of QDs while providing an internal standard (in the form of the ratiometric donor:acceptor PL response) that ensures continued sensor integrity [86], [87]. By employing the distance-dependence of FRET between the QD-dye pair and biological recognition principles of single-chain variable fragments, Lee et al. developed a sensor for BoNT with a ratiometric PL output resulting in a limit of detection of 30 pM (Fig. 5 ) [47]. Another QD-based BoNT FRET sensor was developed by Sapsford et al., in which a Cy3-labeled peptide substrate was attached to the surface of the QD. Here the actual BoNT proteolytic activity was monitored via a loss-of-FRET between the QD and Cy3 as the peptide was cleaved and the Cy3 dissociated from the QD surface [88].
Figure 5.
A schematic of ratiometric QD-FRET of a BoNT sensor shows energy transfer between the excited state QD donor and the ground state dye emitter. The ratio of emission from the QD to emission from the dye indicates how much of the QD is participating in FRET and therefore how much toxin is present in the analytical sample. Reproduced from Ref. [47]. Copyright 2015 American Chemical Society.
Silica nanoparticles
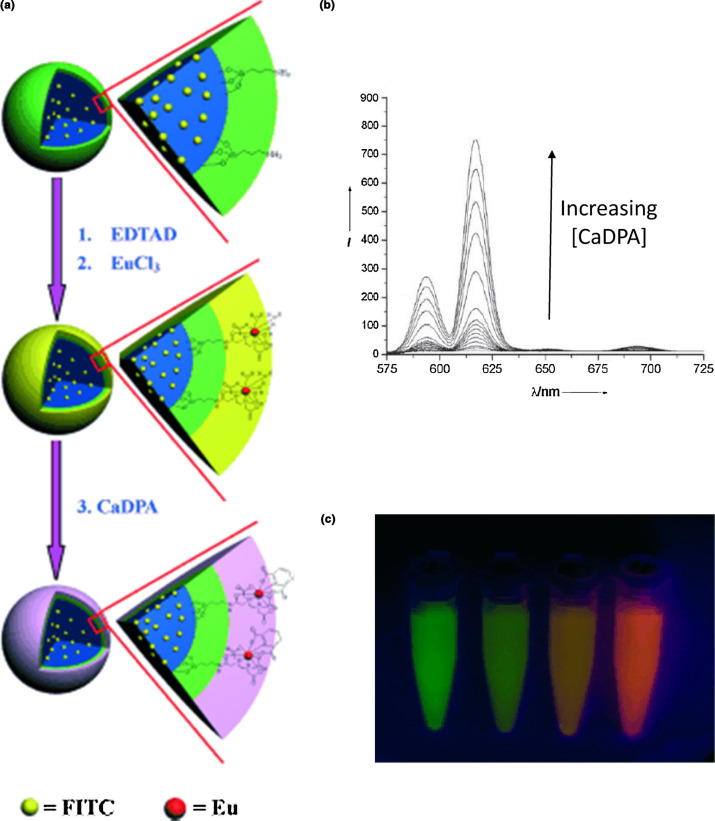
Dye-doped porous silica NPs produce bright signals with low dye leakage and less aggregation compared to polymer-based NPs [89]. Like QDs, silica NPs can accommodate a variety of attachment chemistries, and the large number of encapsulated fluorophores ensures that they are highly photostable and resistant to photobleaching. In one notable example, Ai et al. have created a detector for calcium dipicolinate (CaDPA) – a biomarker for B. anthracis spores that comprises about 10% of the spore's dry weight – using silica NPs doped with fluorescein isothiocyanate (FITC) [48]. An organic complex, ethylenediaminetetraacetic acid dianhydride (EDTAD), is covalently coordinated to the surface of the NP, and europium(III) introduced into the synthetic reaction is chelated by the EDTA (Fig. 6 ). Detection of CaDPA is indicated by an increase in luminescence from the Eu3+ by capitalizing on its water sensitivity. While coordinated by the EDTA, two water molecules remain in the inner coordination sphere of the Eu3+, reducing emission from what would be expected in an anhydrous environment by providing a pathway to nonradiative quenching for the Eu3+; thus green fluorescence from the FITC dominates. The addition of CaDPA, however, displaces the coordinated water molecules, resulting in an improvement in the emission intensity from the Eu3+ complex, while the FITC emission intensity remains unchanged. The assay is able to detect concentrations of CaDPA six orders of magnitude lower than the infectious dose of spores in a mere two minutes. The same EDTA/lanthanide/CaDPA chemistry has been used on other substrates, including polyacrylonitrile and carbon NPs, both of which are inherently luminescent and eliminate the need for a doped NP at the core of the device [90], [91].
Figure 6.
(a) The core of this sensor for calcium dipicolinate (CaDPA) consists of silica doped with fluorescein isothiocyanate (FITC). The surface is functionalized with ethylenediaminetetraacetic acid dianhydride (EDTAD), which chelates Eu3+. While the FITC emission dominates in the absence of CaDPA, the red Eu3+ emission becomes visible upon addition of the analyte. (b) Increase in the intensity of the spectral features associated with Eu3+ emission results from the increase in CaDPA concentrations. (c) The addition of CaDPA to the NPs changes emission from green, arising from the FITC, to red, originating with the Eu3+. Vials shown contain concentrations of CaDPA, from left to right, of 0 μM, 25 μM, 50 μM, and 100 μM. Reproduced from [48].
Magnetic nanoparticles
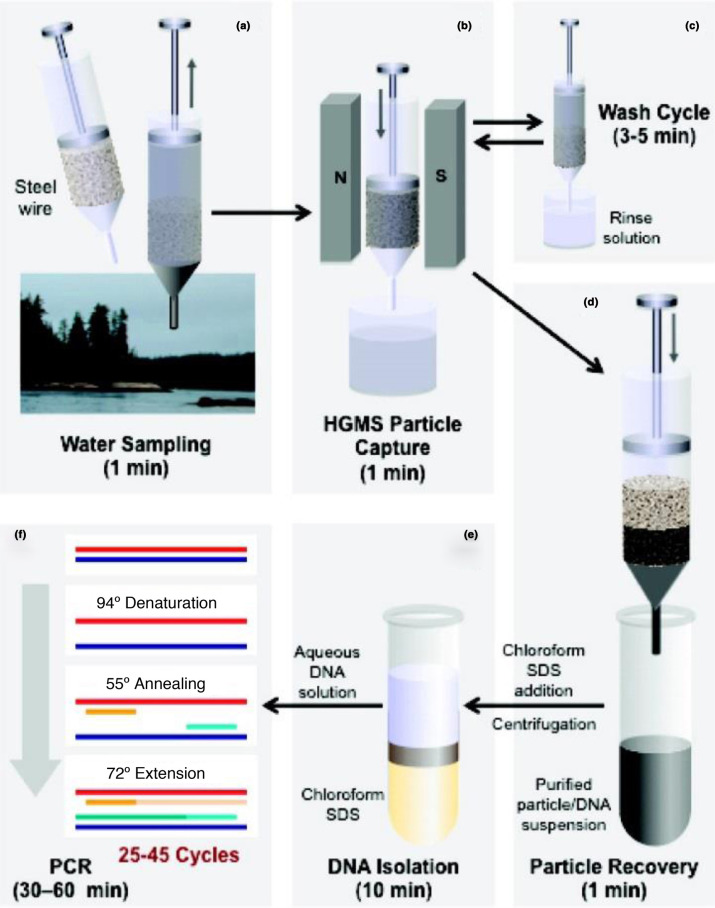
Although magnetic NPs do not have the same photophysical properties as other NPs, their ability to be manipulated in a magnetic field provides a straightforward method for both purifying and concentrating analytes, thereby improving detection limits and significantly simplifying sample preparation [49], [92]. For example, using polycationic magnetic NPs, Bromberg et al. were able to nonspecifically capture polyanionic DNA from solutions, including environmental samples, and the now-sequestered genetic material could be identified by PCR, as shown in Fig. 7 [49]. While employing a traditional technique for bioagent identification, this method facilitates analyte isolation from environmental samples. Biological material is scavenged from the analytical solution by the magnetic NPs, which are concentrated using high-gradient magnetic separation and then washed before being released from the magnetic field and processed for PCR. Decorating the surface of a magnetic NP with antibodies as a capture element can simplify the detection process and eliminate the need for pairing with PCR, although that convenience comes at the cost of the ability to do general monitoring. Magnetic capture and concentration has been demonstrated with B. anthracis spores using antibody-functionalized magnetic NPs followed by detection through charge-transfer mechanisms [93]. Pairing a second antibody immobilized on a column surface to the antibody-driven capture of analyte in solution results in a specialized sandwich assay with signal transduction through a magnetic detection system, as has been demonstrated with Y. pestis [94]. In a solution-based analysis, changes in magnetic susceptibility with increasing hydrodynamic radius (arising from then capture of the analyte) can be used, a phenomenon demonstrated with Brucella antibodies [50]. Magnetic NPs have also been employed in bioagent detection schemes that use SPR [95] and nuclear magnetic resonance (NMR) [96] for signal transduction.
Figure 7.
(a) Magnetic NPs scavenge DNA, bacteria, and viruses from an environmental sample. (b) Particles are captured and concentrated using high-gradient magnetic separation (HGMS). (c) Washing removes other suspended matter. (d) Removing the magnetic field and flushing the system with buffer washes out the magnetic NPs and associated biological material. (e) DNA is separated from NPs using a chloroform/SDS solution and centrifugation. (f) Real-time PCR identifies nucleic acids. Reprinted with permission from [49]. Copyright 2009 American Chemical Society.
Nanowires
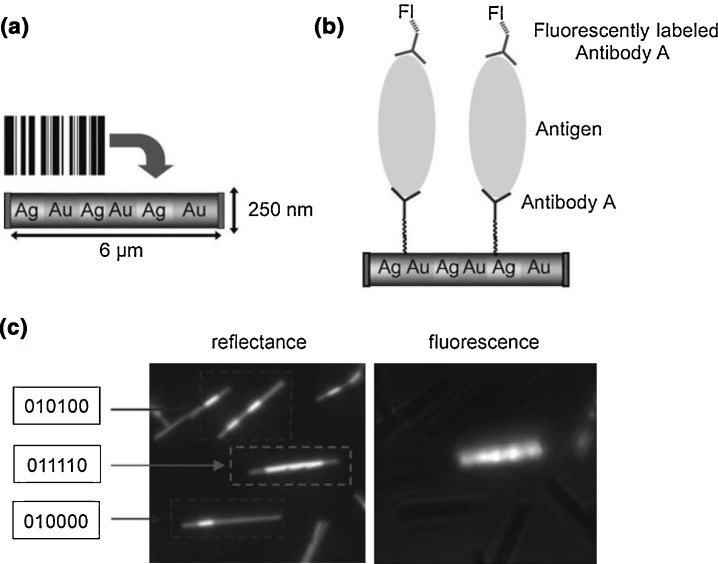
Unlike NPs, which have capture or analyte biomolecules evenly dispersed on the surface, nanowires provide a somewhat heterogeneous substrate. Metallic nanowires synthesized through the sequential deposition of different metals can be used to create a barcode-like system in which the order of metals is characteristic to the target analyte, and different barcodes used together allow for multiplexing [51]. Using the barcode as a substrate for a sandwich assay with a fluorophore tag, the presence of an analyte can be ascertained through fluorimetry, and the identity of the analyte by ‘reading the barcode,’ as shown in Fig. 8 . Other nanowire- or nanorod-based sensors rely less on the morphology of the NM and more on the intrinsic properties of the constituent elements. For example, a silicon nanowire was functionalized with a peptide nucleic acid to which was attached an oligonucleotide complementary to the analyte (here the dengue virus), and hybridization led to a change in the resistance of the wire [97]. Antibody-functionalized nanowires or nanorods of a variety of compositions, including indium oxide [98], zinc oxide [99], silver [46], and silicon [100], have been used for the detection of bioagents using various detection paradigms. For example, Mishra et al. report a silicon nanowire transistor exploiting antibody binding that uses charge transfer resistance to detect SEB [100], while Shanmukh et al. relied on silver nanorods as a SERS substrate [46].
Figure 8.
(a) A metal nanowire formed through sequential deposition of different metals serves as a substrate for a sandwich assay. (b) By labeling the wire with antibodies, a standard sandwich immunoassay can be set up. A fluorescent label acts as the indicator for detection of the antigen. (c) Fluorimetry indicates the presence of an analyte, while using reflectance allows for ‘reading the barcode’ to determine which analyte is present in a multiplexed system. Reproduced with permission from [51].
Carbon-based nanomaterials
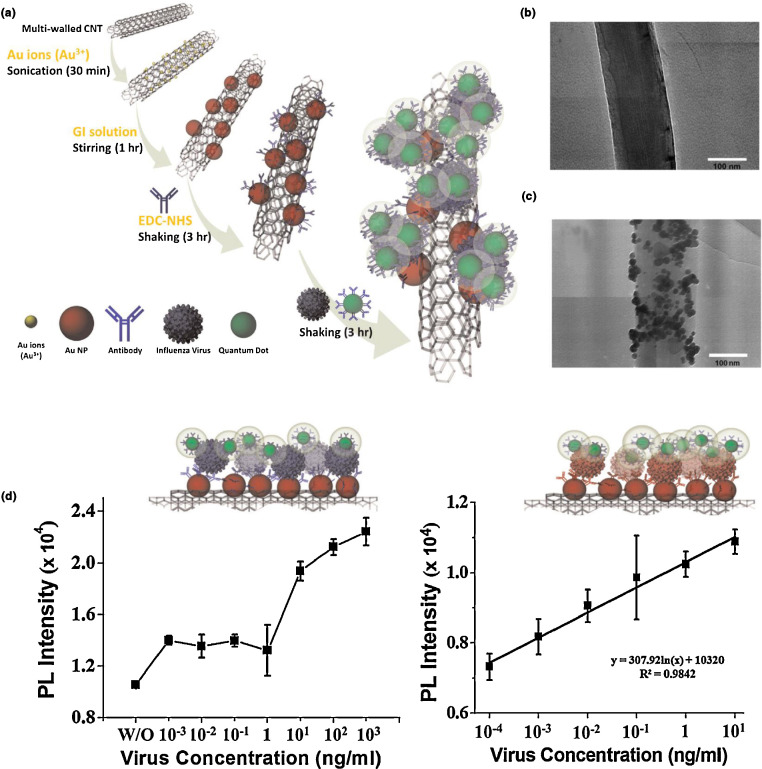
Carbon NMs are valued in biosensors for their high electrical conductivity and biocompatibility. Simple aggregation and aggregate visualization of carbon nanotubes can be used to indicate the presence of an analyte, as demonstrated with B. anthracis [101], while other nanoscale carbon-based sensors exploit a wide range of properties. Changes in the emission wavelength of nanoscale carbon arising from hybridization of surface-decorating single-stranded DNA complementary to an analyte can indicate the presence of an analyte in solution [90], [102]. In addition to exploiting their own intrinsic photophysical properties, carbon nanotubes have been used as substrates for fluorescence-based assays, as demonstrated by Lee et al. in the development of a carbon nanotube/AuNP/QD NM composite for the detection of influenza virus [103]. As shown in Fig. 9 , carbon nanotubes (CNTs) are seeded with AuNPs in situ, and these are subsequently functionalized with influenza antibodies. Antibody-functionalized QDs added to the CNT/AuNP assemblies adhere to any viral analyte that has been captured. The detection ensemble gives PL feedback that can be used to detect the virus at concentrations spanning six orders of magnitude. Carbon NMs can also serve as quenching species; coordinating single-stranded DNA labeled with a fluorophore to the surface of a C60 cluster results in quenched PL from the fluorophore, while hybridization with complementary (analyte) DNA results in the release of the DNA/fluorophore complex from the C60 and initiation of PL [104]. Carbon NMs have also been incorporated into sensors that rely on electrochemical feedback for analyte detection [105], [106], [107]. For example, employing a composite of carbon nanotubes and nanoscale zirconia on an indium-tin-oxide electrode, Das et al. were able to demonstrate changes in electrochemical impedance arising from modifications of the surface [52]. For a more comprehensive examination of the role of carbon NMs in the detection of bio- and chemical agents, the reader may wish to refer to the review by Kumar et al. [10].
Figure 9.
(a) Schematic showing the synthesis and detection strategy for CNT/AuNP/QD influenza probes. (b) TEM of CNT. (c) TEM of AuNPs on CNT. (d) Photoluminescence intensity from the probe ensemble at various concentrations of two different influenza strains (A/Beijing/262/95 on the left; New Caledonia/20/991vR116 on the right). Reproduced from Ref. [103] with permission.
Nanobarcodes
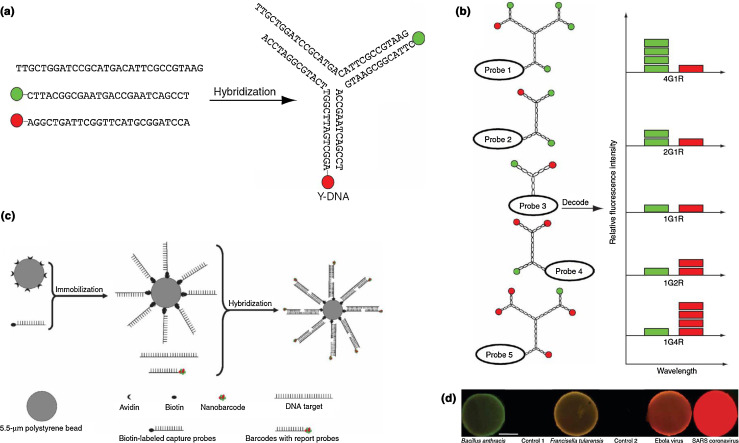
While the sensors described above are derived from predominantly inorganic NMs, the use of biological molecules on a nano-length scale also falls within the purview of this review. Among these are nanobarcodes, which consist of three single-stranded oligonucleotides that hybridize to form Y-DNA [108]. Two of these ssDNA molecules are tagged with a fluorophore, one red and one green, while the third possesses a region complementary to the target analyte, as shown in Fig. 10 . This basic design can be expanded to create a library of nanobarcodes with two important properties: the un-tagged oligonucleotide is complementary to different target analytes (region labeled as Probe 1–5), and the ratio of green to red fluorophores changes, differentiating each probe (and therefore each analyte) through the ratiometric measurement of red and green channels. By immobilizing capture DNA complementary to a different region of the target analyte on a substrate, here polystyrene microbeads, the analyte can be scavenged from a solution and then labeled through the introduction of the nanobarcodes. The method has been shown to detect attomole concentrations of multiple synthetic bioagent signature sequences, including those from anthrax, Ebola, and SARS, in 30 seconds under multiplexing conditions.
Food- and water-borne illnesses.
Food- and water-borne pathogens naturally have low infective doses and high virulence; they are widespread and therefore available; and they are inherently stable in food or water [1]. Despite practical impediments (e.g., dilution, food and water treatment), an attack on food or water supplies could be quite powerful as a means of inciting mass casualty and fear [109]. For example, a 2005 study identified vulnerable points in the processing of milk at which the introduction of a gram of BoNT would sicken 100,000 consumers [110]. Other food- and water-borne pathogens identified on the CDC's list of bioterrorism agents include Salmonella and Shigella species, Escherichia coli, Vibrio cholera, Campylobacter jejuni, Listeria monocytogenes, the protozoa Cryptosporidium parvum, and the hepatitis A virus. In addition to the general requirements for bioagent detection, detecting these pathogens require sensors that function in unusual matrices (e.g., milk) [49], [111], [112], [113].
Figure 10.
(a) Hybridization of three single-stranded DNA molecules forms a unit of Y-DNA, a nanobarcode. The probe consists of two fluorophore-tagged ssDNA molecules and an un-tagged ssDNA molecule that has a free end complementary to the target analyte. (b) Using the design principle for making a basic nanobarcode, multiple fluorophores can be incorporated into a single barcode. By using only two fluorophores, which can be read off a two-channel fluorimeter, at different ratios, the probes and therefore their target analytes can be ratiometrically differentiated. (c) Capture DNA loaded onto a polystyrene bead provides a substrate for collection of the analyte. Addition of the nanobarcode then identifies the captured species in multiplexed systems. (d) Demonstrated success of capturing and labeling several bioagents, including B. anthracis, from a mixture. Reproduced with permission from [108].
Alternate and promising nanomaterials
While many of the NMs mentioned above are now well-developed in the laboratory and perhaps on their way to commercial application, more newly emerging NMs also show promise for eventual application in bioagent detection and perhaps even mitigation or deactivation of bioagents. For example, the use of a virus as a biological nanoscaffold hosting antibodies for bioagent recognition and fluorescent dyes for labeling has been demonstrated in the detection of SEB and ricin [114], [115]. More recently, growing research on nanomotors, electrochemically propelled nanoscale materials, has shown promise in both capturing and transporting targeted species, with recent demonstrations having shown nanomotors sequestering and deactivating anthrax spore simulants from environmental matrices [14], [116].
Device integration and commercialization
Most of the NMs described in the context of this review are still several steps removed from commercial application. While their success has been demonstrated in the laboratory, factors such as scale-up, device fabrication, systems integration, and interfacing, all critical on a traditional path to commercialization, have yet to be tackled or feature less prominently in the scientific literature. These factors contribute to the cost of the sensors and require performance trade-offs (e.g., sensitivity vs. shelf stability) that the sensors highlighted here have not yet addressed. Exploring these requirements to bring a promising sensor to market are beyond the scope of this review; instead we refer the reader to several resources with that process as their focus [25], [117] and to the limited literature that has done side-by-side testing of some commercially available sensors [118], [119]. It is worth noting that the proliferation of sophisticated smartphones provides an exciting alternate route to use that dramatically simplifies the laboratory-to-consumer pathway by exploiting an already ubiquitous technology [120], [121].
Outlook
While traditional methods of detecting bioagents can achieve excellent sensitivity and specificity, these techniques are often slow and expensive and require ample sample preparation and laboratory conditions for their execution. There are certainly many circumstances under which these methods are not only appropriate but warranted and thus worth the effort, but a wide range of conditions also exist in which these techniques are insufficient. Whether continuously monitoring environmental samples, screening thousands of samples from a long-term monitoring program, or rapidly responding to a bioterrorism attack or virulent outbreak, devoting even a few hours to the preparation and analysis of one sample may prove too costly. Moreover, detection of these biothreats is most needed in the field at the point of release, where access to laboratory-grade instrumentation is not feasible and funding is often scarse. It is in these areas that the nanoscale sensors highlighted in this review truly excel. Many build on immunogenic assays that have already been established, exploiting the scale and consequent dispersability of NMs to improve the assay time. Others use NMs as a means of processing environmental samples to achieve lower detection limits than could be reached in a similar timeframe by traditional means. Although conveying only a small cross-section of what has been actually applied or even considered, the diversity of the NMs and conjugate biochemistries highlighted here offers a compelling case for their continued development and expanded use in accommodating the specific demands of bioagent detection in the laboratory, in the field, or in complex matrices.
Further information
For the curious reader, several outstanding resources provide substantially more information on historical and clinical aspects of bioagents [1], [2], [122]. We also note several excellent reviews on current and emerging detection methods for bioagents [2], [20], [25], [41], [119], [123], [124], [125], as well as methods specific to the detection of pathogens in food [112], [113], [126].
Acknowledgements
The authors acknowledge the Office of Naval Research, the Naval Research Laboratory Nanosciences Institute, and DTRA JSTO MIPR #B112582M. CER acknowledges a National Research Council Research Associateship Fellowship through NRL.
References
- 1.Dembek Z.F. Office of the Surgeon General; 2007. Medical Aspects of Biological Warfare. [Google Scholar]
- 2.Dembek Z.F. USAMRIID; 2011. Medical Management of Biological Casualties Handbook. [Google Scholar]
- 3.Jin Z., Hildebrandt N. Trends Biotechnol. 2012;30:394. doi: 10.1016/j.tibtech.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Georgiev V. National Institute of Allergy and Infectious Diseases, NIH, Humana Press; 2009. Defense Against Biological Weapons (Biodefense) [Google Scholar]
- 5.Primmerman C.A. Lincoln Lab. J. 2000;12:3. [Google Scholar]
- 6.Upadhyayula V.K.K. Anal. Chim. Acta. 2012;715:1. doi: 10.1016/j.aca.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Syed M.A. Biosens. Bioelectron. 2014;51:391. doi: 10.1016/j.bios.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Saha K. Chem. Rev. 2012;112:2739. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y. Chem. Soc. Rev. 2012;41:2283. [Google Scholar]
- 10.Kumar O. Def. Sci. J. 2008;58:617. [Google Scholar]
- 11.Kaittanis C. Adv. Drug Deliv. Rev. 2010;62:408. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier-Wirth C., Coste J. Biologicals. 2010;38:9. doi: 10.1016/j.biologicals.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Driskell J.D., Tripp R.A. Clin. Microbiol. Newslett. 2009;31:137. [Google Scholar]
- 14.Campuzano S. Analyst. 2011;136:4621. doi: 10.1039/c1an15599g. [DOI] [PubMed] [Google Scholar]
- 15.Sapsford K.E. Mater. Today. 2008;11:49. [Google Scholar]
- 16.NATO; 1996. NATO Handbook on the Medical Aspects of NBC Defensive Operations AMedP-6(B) [Google Scholar]
- 17.Kwit N. Morb. Mortal. Wkly. Rep. 2015;64:918. doi: 10.15585/mmwr.mm6433a6. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization; 2014. Global Tuberculosis Report. [Google Scholar]
- 19.Knipe D.M., Howley P.M. Lippincott Williams & Wilkins; 2001. Fields Virology. [Google Scholar]
- 20.Cheng X. Anal. Bioanal. Chem. 2009;393:487. doi: 10.1007/s00216-008-2514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenner F. World Health Organization; 1988. Smallpox and Its Eradication. [Google Scholar]
- 22.Jones K.E. Nature. 2008;451:990. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morens D.M. Nature. 2004;430:242. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederberg J. National Academies Press; 1992. Emerging Infections: Microbial Threats to Health in the United States. [PubMed] [Google Scholar]
- 25.Lim D.V. Clin. Microbiol. Rev. 2005;18:583. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L. Clin. Dev. Immunol. 2011;2011 doi: 10.1155/2011/193963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain M.M. Clin. Biochem. 2013;46:633. doi: 10.1016/j.clinbiochem.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Wang S. Biotechnol. Adv. 2013;31:438. doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pappas G. N. Engl. J. Med. 2005;352:2325. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 30.Bogomolova A. Sens. Chem. Biol. Appl. 2010:333. [Google Scholar]
- 31.Ler S.G. J. Chromatogr. A. 2006;1133:1. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 32.Iqbal S.S. Biosens. Bioelectron. 2000;15:549. doi: 10.1016/s0956-5663(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 33.Byrne B. Sensors. 2009;9:4407. doi: 10.3390/s90604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldman E.R. Anal. Chem. 2008;80:8583. doi: 10.1021/ac8014774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson G.P. Anal. Chem. 2010;82:7202. doi: 10.1021/ac100961x. [DOI] [PubMed] [Google Scholar]
- 36.Walper S.A. PLoS ONE. 2012;7:e32801. doi: 10.1371/journal.pone.0032801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ligler F.S. Anal. Bioanal. Chem. 2003;377:469. doi: 10.1007/s00216-003-1992-0. [DOI] [PubMed] [Google Scholar]
- 38.Yanik A.A. Nano Lett. 2010;10:4962. doi: 10.1021/nl103025u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer J., Gareis M. J. Vet. Med. B. 1987;34:613. doi: 10.1111/j.1439-0450.1987.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 40.Young J.C., Games D.E. J. Chromatogr. 1992;627:247. doi: 10.1016/0021-9673(92)87204-l. [DOI] [PubMed] [Google Scholar]
- 41.Turner N.W. Anal. Chim. Acta. 2009;632:168. doi: 10.1016/j.aca.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Kostiainen R. J. Chromatogr. 1991;538:323. doi: 10.1016/0378-4347(91)80607-e. [DOI] [PubMed] [Google Scholar]
- 43.Denkbas E.B. Springer; 2012. Technological Innovations in Sensing and Detection of Chemical, Biological, Radiological, Nuclear Threats and Ecological Terrorism. [Google Scholar]
- 44.Vaseashta A. Springer; 2012. Technological Innovations in Sensing and Detection of Chemical, Biological, Radiological, Nuclear Threats and Ecological Terrorism. [Google Scholar]
- 45.Sattarahmady N. Biochem. Eng. J. 2015;97:1. [Google Scholar]
- 46.Shanmukh S. Nano Lett. 2006;6:2630. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 47.Lee J. Nano Lett. 2015;15:7161. doi: 10.1021/acs.nanolett.5b03442. [DOI] [PubMed] [Google Scholar]
- 48.Ai K. Angew. Chem. Int. Ed. 2009;48:304. doi: 10.1002/anie.200804231. [DOI] [PubMed] [Google Scholar]
- 49.Bromberg L. Anal. Chem. 2009;81:5637. doi: 10.1021/ac9003437. [DOI] [PubMed] [Google Scholar]
- 50.Fornara A. Nano Lett. 2008;8:3423. doi: 10.1021/nl8022498. [DOI] [PubMed] [Google Scholar]
- 51.Tok J.B.H. Angew. Chem. Int. Ed. 2006;45:6900. doi: 10.1002/anie.200601104. [DOI] [PubMed] [Google Scholar]
- 52.Das M. Appl. Phys. Lett. 2011;99:143702. [Google Scholar]
- 53.Field L.D. Acc. Chem. Res. 2015;48:1380. doi: 10.1021/ar500449v. [DOI] [PubMed] [Google Scholar]
- 54.Medintz I. Nat. Mater. 2006;5:842. doi: 10.1038/nmat1776. [DOI] [PubMed] [Google Scholar]
- 55.Algar W.R. Bioconjug. Chem. 2011;22:825. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- 56.Veigas B. Nanotechnology. 2010;21:415101. doi: 10.1088/0957-4484/21/41/415101. [DOI] [PubMed] [Google Scholar]
- 57.Gill P. Nanobiotechnology. 2008;4:28. [Google Scholar]
- 58.Thiruppathiraja C. Anal. Biochem. 2011;417:73. doi: 10.1016/j.ab.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 59.Guarise C. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3978. doi: 10.1073/pnas.0509372103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jannetto P.J. J. Clin. Microbiol. 2010;48:3997. doi: 10.1128/JCM.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shyu R.H. Toxicon. 2002;40:255. doi: 10.1016/s0041-0101(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 62.Uzawa H. Biosens. Bioelectron. 2008;24:923. doi: 10.1016/j.bios.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 63.Jarocka U. Sensors. 2014;14:15714. doi: 10.3390/s140915714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Owino J. Sensors. 2008;8:8262. doi: 10.3390/s8128262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labib M. Analyst. 2013;138:1865. doi: 10.1039/c3an36771a. [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Paredes G. Electroanalysis. 2009;21:379. [Google Scholar]
- 67.Shyu R.-H. Toxicon. 2002;40:255. doi: 10.1016/s0041-0101(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 68.Stoeva S.I. Angew. Chem. Int. Ed. 2006;45:3303. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 69.Cao Y.C. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 70.Uzawa H. Biosens. Bioelectron. 2008;24 doi: 10.1016/j.bios.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 71.Jin X. Biosens. Bioelectron. 2009;24:2580. doi: 10.1016/j.bios.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 72.Kleo K. Anal. Biochem. 2011;418:260. doi: 10.1016/j.ab.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Gopinath S.C. PLOS ONE. 2013;8:e69121. doi: 10.1371/journal.pone.0069121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu S. Opt. Mater. 2009;31:1608. [Google Scholar]
- 75.Shen W.Z. J. Mater. Chem. 2012;22:8127. [Google Scholar]
- 76.Boeneman Gemmill K. Bioconjug. Chem. 2013;24:269. doi: 10.1021/bc300644p. [DOI] [PubMed] [Google Scholar]
- 77.Yang L.J., Li Y.B. J. Food Prot. 2005;68:1241. doi: 10.4315/0362-028x-68.6.1241. [DOI] [PubMed] [Google Scholar]
- 78.Goldman E.R. Anal. Chem. 2004;76:684. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 79.Li X.P. Talanta. 2012;100:1. doi: 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 80.Wang L. Bioconjug. Chem. 2005;16:194. doi: 10.1021/bc0498020. [DOI] [PubMed] [Google Scholar]
- 81.Zahavy E. J. Fluoresc. 2010;20:389. doi: 10.1007/s10895-009-0546-z. [DOI] [PubMed] [Google Scholar]
- 82.Generalova A.N. Colloids Surf. A. 2009;342:59. [Google Scholar]
- 83.Roh C., Jo S.K. J. Chem. Technol. Biotechnol. 2011;86:1475. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kiel J.L. Proc. SPIE. 2004;5617:382. [Google Scholar]
- 85.Anderson G.P. Anal. Chim. Acta. 2013;786:132. doi: 10.1016/j.aca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 86.Medintz I., Hildebrandt N. Wiley-VCH; Weinheim, Germany: 2013. FRET – Förster Resonance Energy Transfer: From Theory to Applications. [Google Scholar]
- 87.Algar W.R. Coord. Chem. Rev. 2014;263:65. [Google Scholar]
- 88.Sapsford K.E. ACS Nano. 2011;5:2687. doi: 10.1021/nn102997b. [DOI] [PubMed] [Google Scholar]
- 89.Yan J. Nano Today. 2007;2:44. [Google Scholar]
- 90.Chen H. Chem. Commun. 2015;51:5036. doi: 10.1039/c5cc00757g. [DOI] [PubMed] [Google Scholar]
- 91.Oh W.-K. Biosens. Bioelectron. 2011;29:172. doi: 10.1016/j.bios.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 92.Ashtari P. Talanta. 2005;67:248. doi: 10.1016/j.talanta.2005.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pal S. IEEE Sens. J. 2008;8:647. [Google Scholar]
- 94.Meyer M.H. J. Microbiol. Methods. 2007;68:218. doi: 10.1016/j.mimet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 95.Kamikawa T.L. Nanotechnology. 2012;11:88. doi: 10.1109/TNANO.2011.2157936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koh I. Angew. Chem. Int. Ed. 2008;47:4119. doi: 10.1002/anie.200800069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang G.-J. Sens. Actuators B: Chem. 2010;146:138. [Google Scholar]
- 98.Ishikawa F.N. ACS Nano. 2009;3:1219. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jang Y. J. Nanosci. Nanotechnol. 2012;12:5173. doi: 10.1166/jnn.2012.6361. [DOI] [PubMed] [Google Scholar]
- 100.Mishra N.N. Lab Chip. 2008;8:868. doi: 10.1039/b802036a. [DOI] [PubMed] [Google Scholar]
- 101.Wang H. J. Am. Chem. Soc. 2006;128:13364. doi: 10.1021/ja065455o. [DOI] [PubMed] [Google Scholar]
- 102.Jeng E.S. Nano Lett. 2006;6:371. doi: 10.1021/nl051829k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee J. Biosens. Bioelectron. 2015;64:311. doi: 10.1016/j.bios.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 104.Li H. Small. 2011;7:1562. doi: 10.1002/smll.201100068. [DOI] [PubMed] [Google Scholar]
- 105.Singh C. Sens. Actuators B: Chem. 2013;185:258. [Google Scholar]
- 106.Singh R. Analyst. 2014;139:5415. doi: 10.1039/c4an01335b. [DOI] [PubMed] [Google Scholar]
- 107.Das M. Biomacromolecules. 2011;12:540. doi: 10.1021/bm1013074. [DOI] [PubMed] [Google Scholar]
- 108.Li Y. Nat. Biotechnol. 2005;23:885. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.LeClaire R., Pitt M.L. Humana Press; 2005. Biological Weapons Defense. [Google Scholar]
- 110.Wein L.M., Liu Y. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9984. doi: 10.1073/pnas.0408526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Velusamy V. Springer; 2012. Technological Innovations in Sensing and Detection of Chemical, Biological, Radiological, Nuclear Threats and Ecological Terrorism. [Google Scholar]
- 112.Vinayaka A.C., Thakur M.S. Anal. Bioanal. Chem. 2010;397:1445. doi: 10.1007/s00216-010-3683-y. [DOI] [PubMed] [Google Scholar]
- 113.Koppen R. Appl. Microbiol. Biotechnol. 2010;86:1595. doi: 10.1007/s00253-010-2535-1. [DOI] [PubMed] [Google Scholar]
- 114.Sapsford K.E. Biosens. Bioelectron. 2006;21:1668. doi: 10.1016/j.bios.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 115.Goldman E.R. Sensors. 2009;9:542. doi: 10.3390/s90100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orozco J. Analyst. 2015;140:1421. doi: 10.1039/c4an02169j. [DOI] [PubMed] [Google Scholar]
- 117.Fatah A.A. U.S. Department of Justice; Washington, DC: 2001. An Introduction to Biological Agent Detection Equipment for Emergency First Responders. [Google Scholar]
- 118.King D. J. Clin. Microbiol. 2003;41:3454. doi: 10.1128/JCM.41.7.3454-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zasada A.A. Lett. Appl. Microbiol. 2015;60:409. doi: 10.1111/lam.12392. [DOI] [PubMed] [Google Scholar]
- 120.Petryayeva E., Algar W.R. Anal. Chem. 2014;86:3195. doi: 10.1021/ac500131r. [DOI] [PubMed] [Google Scholar]
- 121.Contreras-Naranjo J.C. IEEE J. Sel. Top. Quantum Electron. 2016;22:1. [Google Scholar]
- 122.Carus W.S. Health Secur. 2015;13:219. doi: 10.1089/hs.2014.0092. [DOI] [PubMed] [Google Scholar]
- 123.Ahmed A. Clin. Microbiol. Rev. 2014;27:631. doi: 10.1128/CMR.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Čapek P., Dickerson T. Toxins. 2010;2:24. doi: 10.3390/toxins2010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mirski T. Ann. Agric. Environ. Med. 2014;21:224. doi: 10.5604/1232-1966.1108581. [DOI] [PubMed] [Google Scholar]
- 126.Alocilja E.C., Pal S. 2014. Wiley Handbook of Science and Technology for Homeland Security. [Google Scholar]