Abstract
The severe acute respiratory syndrome-coronavirus (SARS-CoV) caused an outbreak of atypical pneumonia in 2003. The SARS-CoV viral genome encodes several proteins which have no homology to proteins in any other coronaviruses, and a number of these proteins have been implicated in viral cytopathies. One such protein is 3a, which is also known as X1, ORF3 and U274. 3a expression is detected in both SARS-CoV infected cultured cells and patients. Among the different functions identified, 3a is a capable of inducing apoptosis. We previously showed that caspase pathways are involved in 3a-induced apoptosis. In this study, we attempted to find out protein domains on 3a that are essential for its pro-apoptotic function. Protein sequence analysis reveals that 3a possesses three major protein signatures, the cysteine-rich, Yxxϕ and diacidic domains. We showed that 3a proteins carrying respective mutations in these protein domains exhibit reduced pro-apoptotic activities, indicating the importance of these domains on 3a's pro-apoptotic function. It was previously reported that 3a possesses potassium ion channel activity. We further demonstrated that the blockade of 3a's potassium channel activity abolished caspase-dependent apoptosis. This report provides the first evidence that ion channel activity of 3a is required for its pro-apoptotic function. As ion channel activity has been reported to regulate apoptosis in different pathologic conditions, finding ways to modulate the ion channel activity may offer a new direction toward the inhibition of apoptosis triggered by SARS-CoV.
Abbreviations: a.a., amino acid; AO, acridine orange; ER, endoplasmic reticulum; ORF, opening reading frame; SARS-CoV, severe acute respiratory syndrome-coronavirus
Keywords: Drosophila, Ion channel, Severe acute respiratory syndrome, Site-directed mutagenesis, Vero E6
1. Introduction
Severe acute respiratory syndrome-coronavirus (SARS-CoV) is the etiological agent causing the global outbreak of atypical pneumonia in 2003. The SARS-CoV genome carries a minimum of 14 open reading frames (ORFs; Marra et al., 2003, Rota et al., 2003, Ksiazek et al., 2003, Thiel et al., 2003), encoding replicases, various structural proteins (including spike, envelope, membrane, nucleocapsid), and a number of accessory proteins (Thiel et al., 2003, Rota et al., 2003, Marra et al., 2003). A subset of these viral proteins, including 3a, are only found in SARS-CoV but not other coronaviruses (Rota et al., 2003). Understanding the molecular functions of these SARS-CoV-specific proteins would shed light on the life cycle of the virus.
The SARS-CoV 3a locus, also known as X1 (Rota et al., 2003), ORF3 (Marra et al., 2003) and U274 (Tan et al., 2004b), encodes a 274 a.a. protein (Marra et al., 2003). The 3a protein shows cytoplasmic (Oostra et al., 2006, Tan et al., 2004b, Yu et al., 2004, Yuan et al., 2005a, Zhong et al., 2006) and plasma membrane (Ito et al., 2005, Lu et al., 2006, Tan et al., 2004b) localization in both transfected and viral-infected cells. 3a possesses three transmembrane regions (a.a. 34–56; 77–99; 103–125) in its N-terminus, and an intracellular C-terminal region (Marra et al., 2003, Rota et al., 2003). The central region of 3a carries several conserved sequences which includes a cysteine-rich domain (a.a. 127–133), a Yxxϕ domain (a.a. 160–163) and a diacidic domain (a.a. 171–173), which is then followed by the C-terminal domain (a.a. 209–264) (Oostra et al., 2006, Marra et al., 2003, Zeng et al., 2004, Tan et al., 2004b, Yu et al., 2004). The cysteine-rich domain is known to be responsible for homo- and hetero-dimerization of 3a, which is crucial for its ion channel activity (Lu et al., 2006). To date, the functional significance of 3a's ion channel activity on its function is still not well defined. The Yxxϕ domain is a protein internalization signal which is involved in clathrin-mediated endocytosis (Sorkin, 2004); while the diacidic domain is a trafficking signal responsible for efficient endoplasmic reticulum (ER) protein export (Nishimura and Balch, 1997). Both the Yxxϕ and diacidic domains play important roles in intracellular protein trafficking of 3a, and a deletion mutant (a.a. 160–173) which uncovers these domains abolishes the localization of 3a to the cell surface (Tan et al., 2004b). Further, the 3a C-terminal domain (a.a. 125–200) has been demonstrated to possess RNA-binding activity (Sharma et al., 2007).
Expression of 3a is detected in patients’ intestinal surface enterocytes and pneumocytes (Zeng et al., 2004, Yu et al., 2004, Chan et al., 2005). Both its physical interactions with other viral structural proteins including Spike, Envelope and Membrane (Tan et al., 2004b) and its incorporation into newly packaged matured SARS-CoV virions (Shen et al., 2005, Ito et al., 2005) suggest that 3a plays a structural role in the SARS-CoV life cycle. Apart from being a viral structural protein, 3a has been shown to regulate various cellular responses of host cells, including the up-regulation of fibrinogen gene expression (Tan et al., 2005), and augmentation of IL-8 and NF-κB promoter activities (Kanzawa et al., 2006), possibly through its RNA-binding activity (Sharma et al., 2007).
It has been reported that apoptosis initiates viral cytopathic effect in SARS-CoV-infected cells (Ren et al., 2005, Bordi et al., 2006, Yan et al., 2004). Consistent with the viral cytopathological studies, a number of SARS-CoV proteins, including 3a (Wong et al., 2005, Law et al., 2005), are found to be pro-apoptotic (Tan et al., 2004a, Tan et al., 2007a, Tan et al., 2007b, Surjit et al., 2004, Chow et al., 2005, Lin et al., 2006, Yu et al., 2004, Yuan et al., 2005b, Yang et al., 2005, Zhao et al., 2006, Khan et al., 2006, Kopecky-Bromberg et al., 2006, Chan et al., 2007). Previously, we and others showed that both caspase-8 (Law et al., 2005, Padhan et al., 2008) and cytochrome c (Padhan et al., 2008, Wong et al., 2005) are involved in 3a-induced apoptosis. More recently, Padhan et al. (2008) reported that Bax, p53 and p38 MAP kinase also play roles in 3a-induced apoptosis.
The SARS-CoV 3a protein appears to have multiple functions, including apoptosis induction. Although the cysteine-rich, Yxxϕ and diacidic domains are well-known protein motifs on 3a (Fig. 1A), it still remains unclear how these domains are involved in 3a functions. In the present study, we performed a structure–function study of 3a with an aim to investigate the roles of these domains in its pro-apoptotic function in vitro and in vivo. In addition the cysteine-rich domain had previously been shown to be critical for 3a's ion channel activity (Lu et al., 2006), our data further illustrate that the ion channel activity is indispensible for caspase-dependent apoptosis of 3a.
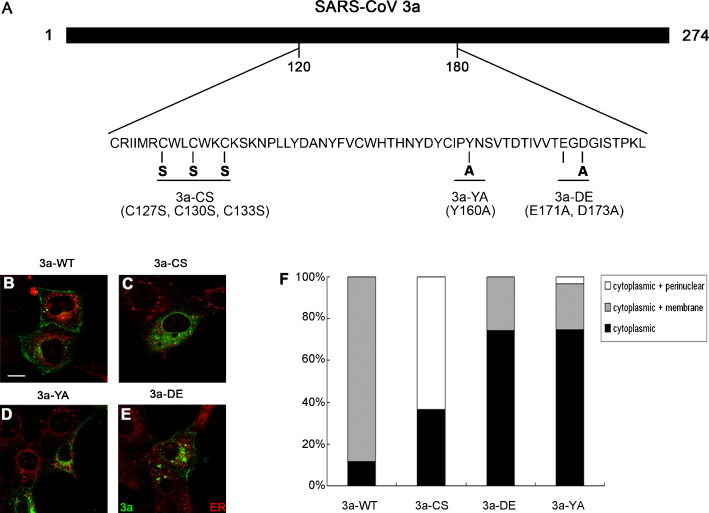
Fig. 1.
Subcellular localization of wild type and mutant SARS-CoV 3a proteins in Vero E6 cells. (A) Mutagenesis scheme of this study. (B–E) Subcellular localization of wild type and mutant 3a proteins in Vero E6 cells. Wild type 3a protein (3a-WT) displayed plasma membrane and punctate cytoplasmic staining pattern (B). The cysteine-rich domain mutant protein 3a-CS lost the plasma membrane localization, and concentrated in the cytoplasm and the perinuclear region (C). Both 3a-YA (D) and 3a-DE (E) mutant proteins showed cytoplasmic localization and fractional plasma membrane localization was retained. Different degrees of protein aggregation were also observed in 3a-YA and 3a-DE mutant proteins (D and E). ER-Tracker™ Red was used as a counter-stain to label the endoplasmic reticulum (shown in red). Scale bar represents 16 μm. (F) Quantification of subcellular distribution of 3a-WT and mutant 3a proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
2. Materials and methods
2.1. Generation of 3a mutant constructs
The wild type pUAST-3a-WT construct (Wong et al., 2005) was used as template to generate three mutant 3a constructs pUAST-3a-CS, -3a-YA and -3a-DE (Fig. 1A). Primers used were CS-F: 5′-TTA TGA GAT CTT GGC TTT CTT GGA AGT CCA AAT CCA A-3′; CS-R: 5′-TTG GAT TTG GAC TTC CAA GAA AGC CAA GAT CTC ATA A-3′; Y160A-F: 5′-CTG TAT ACC AGC TAA CAG TGT CAC-3′; Y160A-R: 5′-GTG ACA CTG TTA GCT GGT ATA CAG-3′; DE-F: 5′-CGT TAC TGC AGG TGC CGG CAT TTC A-3′; DE-R: 5′-TGA AAT GCC GGC ACC TGC AGT AAC G-3′. All mutations were confirmed by DNA sequencing. For mammalian cell expression, both wild type and mutant 3a genes were subcloned into pcDNA vectors to generate pcDNA3.1-3a or pcDNA6-3a-EGFP constructs. Standard overlapping PCR strategy was used to generate pcDNA3.1-3a-CS-YA-DE triple mutant construct.
2.2. Mammalian cell culture and transient transfection
The African green monkey kidney cell line Vero E6 was maintained at 37 °C in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 10% heat-inactivated fetal bovine serum (Gibco-BRL), streptomycin (100 g/ml) and penicillin (100 U/ml). Cells were seeded onto 24-well plates 24 h prior to transfection, and 0.3 μg of DNA was used for transient transfection. Lipofectamine (Invitrogen) and PLUS transfection reagents (Invitrogen) were used according to manufacturer's instructions.
2.3. Immunofluorescence staining of Vero E6 cells
Cells were seeded onto coverslips at a density of 1.2 × 105 cells/coverslip. After transient transfection, cells were fixed with 3.7% formaldehyde in 1× PBS for 15 min and then permeabilized by 1% Triton X-100 in 1× PBS for 5 min. After blocking with 1% goat serum in 1× PBS for 30 min, cells were incubated with mouse anti-SARS-3a antibody X98 (1:40; Wong et al., 2005) at 4 °C overnight. Alexa Fluor 488 goat anti-rabbit IgG (H + L) (1:400; Invitrogen) was used as secondary antibody. Endoplasmic reticulum was labeled by ER-Tracker™ Red (5 μM, BODIPY® TR glibenclamide, Invitrogen) and cell nuclei were stained with Hoechst 33342 (5 μM, trihydrochloride trihydrate, Molecular Probes) at room temperature for 10 min. Fluorescent images were captured using an Olympus BX51 upright fluorescence microscope or a Bio-Rad confocal microscope.
2.4. Caspase assays and potassium channel blockers treatment
Caspase activity assays were measured using the Caspase-Glo®8 and Caspase-Glo®9 assay systems (Promega) according to manufacturer's instructions. Cell permeable synthetic caspase inhibitors (Merck) z-VAD-fmk, z-IETD-fmk and z-LEHD-fmk were dissolved in DMSO. Potassium channel blockers barium chloride (Ba) and 4-aminopyridine (AP) were dissolved in sterile distilled water. Vero E6 cells transiently transfected with 3a constructs were treated with caspase inhibitors (50 μM in 1% DMSO) at 48 h post-transfection, and cells were further incubated for another 24 h. For potassium channel blockers treatment, 3a-transfected Vero E6 cells were treated with Ba or AP at 24 h post-transfection, and cells were then further cultured for another 48 h. After treatments, cells were subjected to immunofluorescence staining as described above.
2.5. Electrophysiology
The ion conducting property of 3a was assessed by whole-cell patch clamp. Human embryonic kidney (HEK) 293 cells were transfected with GFP-tagged 3a-WT-, 3a-CS, 3a-YA or 3a-DE constructs. Twenty-four hours after transfection, cells were trypsinized and seeded on poly-l-lysine-coated coverslips. Single cells with GFP fluorescence were selected for patch clamp recording. Whole cell currents were recorded by an EPC9 patch clamp amplifier (HEKA) controlled by Pulse software (HEKA). The intracellular solution contained in mM: 140 KCl, 5 NaCl, 2 MgCl2, 10 Hepes, at pH 7.2. Bath solution contained in mM: 140 NaCl, 5 KCl, 2 MgCl2, 1 CaCl2, 10 glucose, 10 Hepes, at pH 7.4. The voltage clamp protocol consisted of rectangular voltage steps from +100 to −100 mV in 20 mV increments applied from a holding potential of −60 mV. Whole cell currents were recorded before and 5 min after 10 mM Ba application (Lu et al., 2006). The experiments were performed at room temperature. The data was analyzed with PulseFit software (HEKA).
2.6. Western blotting
Western blotting was performed as described previously (Wong et al., 2008), and subcellular fractionation was performed according to (Frezza et al., 2007). Primary antibodies used were anti-Bid (1:2000; BD Biosciences), anti-cytochrome C (1:2000; Abcam), anti-glutamate dehydrogenase (GDH; 1:1000; US Biological) and anti-β-tubulin (1:10,000; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, under the auspices of the National Institute for Child Health and Human Development), and secondary antibodies used were goat anti-mouse IgG (H+L) peroxidase conjugate (1:2,500; Abcam) and goat anti-rabbit IgG (H + L) peroxidase conjugate (1:4000; Cell Signaling).
2.7. Drosophila genetics
Fly strains were grown at 29 °C on standard cornmeal medium supplemented with dry yeast. The gmr-GAL4 driver line was obtained from Bloomington Drosophila Stock Center and the UAS-3a-WT line was previously described in (Wong et al., 2005). Standard microinjection procedure was employed to generate mutant 3a transgenic lines (UAS-3a-CS, UAS-3a-YA and UAS-3a-DE). Using RT-PCR, expression level of all 3a transgenes was found to be comparable (data not shown).
2.8. Scanning electron microscopy of adult fly eyes
In brief, 2–3-day-old adult fly heads were fixed in 2.5% glutaraldehyde (EM grade, Electron Microscopy Sciences) in phosphate buffer (pH 7.4) for 4 h, then post-fixed with 1% osmium tetroxide (Electron Microscopy Sciences), dehydrated to 100% ethanol and critical-point dried with liquid CO2. Gold–palladium-coated specimens were examined with a JEOL JSM-6301FE microscope operated at 5 kV (Chau et al., 2006).
2.9. Acridine orange staining of Drosophila larval eye discs
Acridine orange (AO) staining of third-instar larval eye discs was performed as previously described (Hay et al., 1995). Larvae of corresponding genotypes were fed on either unmodified standard cornmeal medium as described above or medium supplemented with Ba since first instar larval stage. Both 1 and 10 μM Ba gave similar results. Eye disc images were captured using an Olympus BX51 upright fluorescence microscope or a Leica NT confocal microscope.
2.10. Statistical analyses
Statistical analyses were performed using Student's t-test. Data were presented as means + S.E.M. and p-values < 0.05 were considered statistically significant.
3. Results
3.1. Subcellular localization of SARS-CoV 3a protein and its mutants in Vero E6 cells
We performed site-directed mutagenesis on three domains of 3a, cysteine-rich (3a-CS; C127S C130S C133S), YXXϕ (3a-YA; Y160A) and diacidic (3a-DE; E171A D173A), in an attempt to investigate the functional significance of these regions on 3a function in vitro and in vivo (Fig. 1A). Immunofluorescence staining was first performed on these mutant 3a proteins in Vero E6 cells. We found that wild type 3a (3a-WT) protein localized to the plasma membrane (Fig. 1B and F), and also displayed a punctate cytoplasmic staining pattern (Fig. 1B and F). In contrast to 3a-WT, the 3a-CS mutant protein lost the plasma membrane localization and became more concentrated in the cytoplasm and the perinuclear region (Fig. 1C and F). Similarly, the plasma membrane localization of 3a-YA (Fig. 1D and F) and 3a-DE (Fig. 1E and F) mutants was partially reduced with a concomitant increase in cytoplasmic signals.
3.2. Protein domains required for inducing 3a's caspase-dependent apoptosis
We next determined whether these mutations would affect 3a's pro-apoptotic function. Previously, we demonstrated the pro-apoptotic properties of 3a-WT in Vero E6 cells using multiple methodologies, including nuclear condensation, DNA fragmentation and TUNEL assay (Law et al., 2005). Here, we used nuclear condensation as the indicator to assess the pro-apoptotic potential of 3a mutants (Fig. 2 ). Consistent with previous findings (Law et al., 2005), we showed that up to 60% of 3a-WT-expressing Vero E6 cells displayed nuclear condensation (Fig. 2A). To determine the roles of CS, YA and DE motifs in 3a-induced apoptosis, we generated a 3a-CS-YA-DE triple mutant and examined its ability to induce nuclear condensation. When compared with 3a-WT, we observed a ∼5-fold reduction of the number of condensed nuclei in cell population expressing the 3a-CS-YA-DE triple mutant protein (Fig. 2B). Unlike 3a-CS-YA-DE triple mutant, cells expressing single mutant proteins (3a-CS, 3a-DE and 3a-YA) displayed more prominent nuclear condensation (Fig. 2C). This indicates that CS, YA and DE motifs in 3a each contribute independently and significantly to apoptosis induction.
Fig. 2.
Wild type and mutant SARS-CoV 3a proteins possess caspase activities and cause nuclear condensation in Vero E6 cells. (A) Expression of wild type 3a protein (3a-WT) induced nuclear condensation (arrows) in Vero E6 cells. Scale bar represents 16 μm. (B) Vero E6 cells transfected with 3a-WT construct showed nuclear condensation while cells transfected with the 3a triple mutant (3a-CS-YA-DE) construct showed a much reduced level of nuclear condensation. At least 100 cells were counted in each experiment. *p < 0.05, 3a-CS-YA-DE triple mutant versus 3a-WT control. (C) Vero E6 cells transfected with 3a-WT and mutant 3a (3a-CS, 3a-YA and 3a-DE) constructs displayed different degrees of nuclear condensation. Nuclear condensation induced by 3a-WT, but not mutant 3a proteins, was inhibited by caspase-8 inhibitor II (z-IETD-fmk) and caspase-9 inhibitor I (z-LEHD-fmk). #p < 0.05, caspase inhibitor-treated 3a-WT cells versus 3a-WT control; *p < 0.05, 3a mutant-expressing cells versus 3a-WT-expressing cells. At least 100 cells were counted in each experiment. (D) General caspase inhibitor (z-VAD-fmk) significantly reduced nuclear condensation induced by 3a-WT protein in Vero E6 cells. At least 100 cells were counted in each experiment. *p < 0.05, caspase inhibitor-treated 3a-WT cells versus 3a-WT control. (E and F) Caspase-8 and -9 activities were detected in Vero E6 cells transfected with 3a-WT construct but these activities were significantly reduced in mutant 3a-expressing cells. *p < 0.05, 3a mutant-expressing cells versus 3a-WT-expressing cells.
Caspase-8 and -9 are enzymes that mediate the cell surface death receptor- and mitochondria-mediated apoptotic pathways, respectively (Chen and Wang, 2002). We have previously demonstrated a role of caspase-8 in 3a-induced apoptosis (Law et al., 2005). Here, we found that both caspase-8- and -9-specific inhibitors significantly suppressed nuclear condensation induced by 3a-WT in Vero E6 cells (Fig. 2C). This indicates 3a triggers apoptosis through both the death receptor- and mitochondria-mediated pathways. When we treated 3a-WT-expressing Vero E6 cells with a broad-spectrum caspase inhibitor z-VAD-fmk (Van Noorden, 2001) which blocks caspase-dependent apoptosis in general, a similar level of inhibition was again observed (Fig. 2D). Consistent with the caspase inhibition results, we further showed that the activities of caspase-8 and -9 were significantly reduced in all 3a mutant-expressing cells (Fig. 2E and F). Truncation of Bid and mitochondrial cytochrome c release are hallmark features of death receptor- and mitochondria-mediated apoptotic pathways, respectively (Strasser et al., 2009). Here, we show that 3a-WT protein induced Bid cleavage (Fig. 3A) and mitochondrial cytochrome c release (Fig. 3B). Taken together, our data demonstrate that the disruption of the CS, YA or DE motif individually does not diminish the ability of 3a to activate the death receptor and mitochondrial apoptotic pathways (Fig. 3), which is indicative of functional redundancy of these motifs on 3a-induced apoptosis.
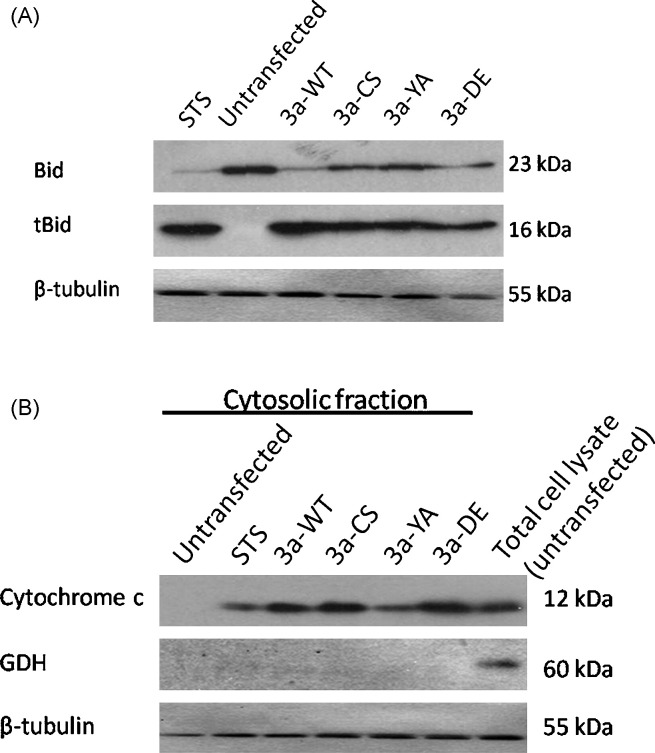
Fig. 3.
3a induces Bid truncation and mitochondrial cytochrome C release. (A) Truncation of Bid was observed when untransfected HEK293 cells were treated with staurosporine (STS). Similar to STS treatment, HEK293 cells transfected with 3a-WT, -CS, -YA, and -DE constructs all showed Bid truncation. Bid: uncleaved Bid (23 kDa); tBid: truncated Bid (16 kDa). (B) Upon STS treatment, cytochrome c was detected in the cytosolic fraction of untransfected HEK293 cells. Cytosolic cytochrome c was also detected in cells transfected with 3a-WT, -CS, -YA and -DE constructs. Glutamate dehydrogenase (GDH), a mitochondrial marker, was used to demonstrate the cytosolic fractions were free of mitochondrial contamination. Staurosporine (1 μM) was used to induce apoptosis, and β-tubulin was used as loading control.
3.3. 3a mutants are less potent in triggering apoptosis in vivo
We further investigated the pro-apoptotic property of 3a mutants in vivo. We previously reported that overexpression of 3a-WT protein caused a rough-eye phenotype, and the eye size was also reduced due to excessive cell death (Wong et al., 2005; Fig. 4B). In contrast, overexpression of 3a-CS, 3a-YA, and 3a-DE mutants showed only mild rough eye phenotype (Fig. 4C–E), and the eye size of these flies was also comparable to the gmr-GAL4 driver control (Fig. 4A). Nevertheless, we found that both 3a-WT (Fig. 4G) and 3a mutants (Fig. 4H–J) caused disruption of external eye morphology by scanning electron microscopy.
Fig. 4.
In vivo expression of wild type and mutant 3a disrupt adult eye structures in Drosophila. (A–E) Light microscopic examination of adult fly eyes. When compared to the gmr-GAL4 driver alone control (A), expression of the 3a-WT protein in eye tissues resulted in rough-eye phenotype (B) as characterized by loss of regularity of the adult external eye structure. Expression of mutant 3a proteins (3a-CS, 3a-YA and 3a-DE) showed minimal dominant external eye phenotype (C–E). (F–J) Scanning electron microscopic examination of adult fly eyes. Expression of 3a-WT caused severe loss of sensory bristles (G) when compared to the gmr-GAL4 control (F); while the expression of 3a-CS, 3a-YA and 3a-DE mutants caused less severe loss of sensory bristles (H–J). Scale bar represents 20 μm.
Acridine orange (AO) staining was performed to determine the pro-apoptotic property of 3a mutants in flies. Acridine orange is a dye which specifically stains apoptotic cells (Hay et al., 1995). As previously reported (Wong et al., 2005), 3a-WT-expressing transgenic animals displayed an increased number of AO-positive cells (Fig. 5B) when compared to the gmr-GAL4 driver control (Fig. 5A). In contrast to 3a-WT, relatively few number of AO-positive cells were detected in 3a-CS, 3a-YA and 3a-DE mutant animals (Fig. 5C–E). Consistent with the Vero E6 cells data (Fig. 2), our in vivo results further validate the importance of cysteine-rich, Yxxϕ and diacidic domains of 3a (Fig. 1A) in 3a's pro-apoptotic function.
Fig. 5.
In vivo expression of wild type and mutant 3a induce apoptosis in Drosophila. Acridine orange (AO) staining was performed on third instar larval eye imaginal discs to label apoptotic cells. (A) The gmr-GAL4 control showed low levels of AO-stained cells. (B) 3a-WT expression induced apoptosis as indicated by an increase in the number of AO-stained cells. The 3a-CS (C), 3a-YA (D) and 3a-DE (E) mutants also induced apoptosis but to a less extent when compared to 3a-WT (B) as indicated by the relatively less number of AO-stained cells detected. Arrows indicate morphogenetic furrows. Scale bar represents 50 μm.
3.4. Ion channel activity of 3a is involved in caspase-dependent apoptosis
It was reported that 3a possesses ion channel property (Lu et al., 2006), here we performed electrophysiological measurements to validate the ion channel activity of 3a-WT in mammalian cells (Fig. 6A and B). Isolated HEK293 cells expressing 3a-WT-EGFP protein were picked according to the GFP signal. Cells expressing 3a-WT-EGFP displayed a larger membrane current over the potential range (Fig. 6B) when compared to the mock transfected cells (Fig. 6A). We also detected channel activities in 3a-YA-EGFP and 3a-DE-EGFP-expressing cells (Fig. 6C and D), however, the activities were found to be different from that observed in 3a-WT-EGFP-expressing cells. Notably, 3a-CS-EGFP-expressing cells displayed whole cell currents that were similar to that of mock transfected cells (Fig. 6E), indicating the 3a-CS mutant protein lacks ion channel activity. We further found that addition of ion channel blocker barium chloride (Ba) altered the cell membrane current mediated by 3a-WT-EGFP, 3a-YA-EGFP and 3a-DE-EGFP proteins (Fig. 6B–D). However, Ba displayed no observable effect on 3a-CS-EGFP-expressing cells (Fig. 6E).
Fig. 6.
Potassium ion channel blockers suppress 3a-WT-induced apoptosis. (A–E) Ion channel properties of 3a. The 3a-WT (B), -YA (C), -DE (D) but not 3a-CS (E) proteins display ion channel activity. The I–V relationship of mock transfected HEK293 cells (A), and cells transfected with cDNA encoding for 3a-WT-EGFP (B), 3a-YA-EGFP (C) and 3a-DE-EGFP (D) and 3a-CS-EGFP (E) were measured by whole cell patch clamping. Whole cell currents were recorded under voltage steps before (diamond) and 5 min after (square) 10 mM barium chloride (Ba) application. (F) Vero E6 cells transfected with 3a-WT and 3a-CS constructs were treated with potassium ion channel blockers 4-aminopyridine (AP) or barium chloride (Ba). AP or Ba treatment alone only induced minimal nuclear condensation on untransfected cells. Nuclear condensation induced by 3a-WT expression was significantly suppressed by potassium channel blockers (AP and Ba), while such treatments did not result in any significant inhibitory effect on 3a-CS-expressing cells. Untransfected Vero E6 cells treated with 1 μM staurosporine (STS) for 8 h were used as a control because neither AP nor Ba exerted any suppressive effect on STS-induced cell death. Results were plotted as percentage of cells showed nuclear condensation and expressed as means + S.E.M. of three independent experiments. At least 100 cells were counted in each experiment. *p < 0.05, AP- or Ba-treated 3a-WT-expressing cells versus 3a-WT control.
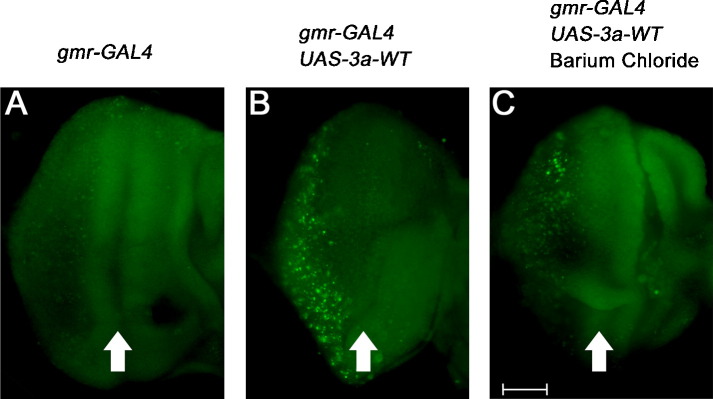
It has been reported that the Cys133 residue of the 3a-WT protein is responsible for homo-dimer/-tetramer formation, and is also essential for 3a's ion channel activity (Lu et al., 2006). As ion channel activity has been implicated in apoptosis (Burg et al., 2006), we investigated whether disrupting the ion channel property of 3a would affect its pro-apoptotic function. To intervene the ion channel function we treated 3a-WT-transfected Vero E6 cells with ion channel blockers, 4-aminopyridine (AP) or Ba, and found that both inhibitors were able to suppress 3a-WT-induced nuclear condensation (Fig. 6F). It has previously been shown that the C133S point mutation (Lu et al., 2006) compromised 3a's ion channel activity. As expected, both AP and Ba treatment exerted no further suppressive effect on 3a-CS-induced nuclear condensation (Fig. 6F). We next fed 3a-WT flies with Ba and determined the extent of 3a-induced apoptosis by AO staining in vivo. We found that the number of AO-positive cells was largely reduced in 3a-WT-expressing larvae that were treated with Ba (Fig. 7C) when compared to the untreated control (Fig. 7B). As Ba is an inhibitor of the 3a ion channel conductance (Lu et al., 2006; Fig. 6), both our in vitro and in vivo data clearly demonstrate a linkage between the pro-apoptotic property and ion channel activity of 3a.
Fig. 7.
Barium chloride treatment suppresses 3a-WT-induced apoptosis in Drosophila. Acridine orange (AO) staining in third instar larval eye imaginal discs was performed to identify apoptotic cells in flies. (A) The gmr-GAL4 control showed low levels of AO-stained cells in third instar imaginal eye discs. (B) 3a-WT expression induced apoptosis as indicated by the increase in the number of AO-stained cells. (C) Feeding 3a-WT-expressing transgenic animals with barium chloride (Ba) reduced the number of AO-stained cells. Arrows indicate morphogenetic furrows. Scale bar represents 50 μm.
4. Discussion
We previously showed that the SARS-CoV 3a protein induces caspase-dependent apoptosis both in vitro and in vitro (Wong et al., 2005, Law et al., 2005). Various structural/functional domains have been identified in 3a, which include the cysteine-rich, Yxxϕ and diacidic domains (Fig. 1A; Tan et al., 2006). The cysteine-rich domain is crucial for homo-/hetero-dimerization and ion channel activity of 3a (Lu et al., 2006); while both the Yxxϕ and diacidic domains are well-characterized protein intracellular trafficking signals (Nishimura and Balch, 1997, Sorkin, 2004). Although the properties of these protein domains have been well characterized, their roles in 3a's pro-apoptotic function had not been studied. Although these three domains are located in close proximity, it is possible that each of them is independently responsible for eliciting 3a's cellular activities, such as apoptosis induction. In this study, we generated mutant 3a constructs, 3a-CS, 3a-YA and 3a-DE (Fig. 1A), and investigated the functional significance of the cysteine-rich, Yxxϕ and diacidic domains on 3a's pro-apoptotic activity. We previously demonstrated that 3a-WT induces nuclear condensation and DNA fragmentation in Vero E6 cells; 3a-expressing cells are also found to be TUNEL-positive (Law et al., 2005). Here, we used nuclear condensation as readout to measure the pro-apoptotic potential of 3a mutants. We found that all 3a-CS, 3a-YA and 3a-DE mutant protein-expressing cells possessed reduced caspase activities, and these proteins were also less potent in inducing cell death (Fig. 2). Consistent with the in vitro data, all the 3a mutants displayed diminished pro-apoptotic activities in vivo. We observed elevated number of apoptotic cells in flies overexpressed with 3a-WT (Fig. 5B; Wong et al., 2005), whereas the number of apoptotic cells was much reduced in mutant 3a-expressing flies (Fig. 5C–E). Altogether, our data clearly show that all cysteine-rich, Yxxϕ and diacidic domains are required for 3a's pro-apoptotic function.
We previously reported caspase-8 activation in 3a-WT-expressing Vero E6 cells (Law et al., 2005), and our in vivo data also showed that cytochrome c can modulate 3a-WT-induced apoptosis (Wong et al., 2005). As the cytochrome c-mediated apoptotic pathway links tightly with caspase-9 activation, in this study we confirmed the involvement of caspase-9 in 3a-WT-induced cell death (Fig. 2). Indeed, the involvement of caspase-9 in 3a-induced apoptosis has also been reported in a recent study performed by Padhan et al. (2008). Both our and others’ data indicate that 3a-WT utilizes more than one caspase pathway to trigger cell death. We further showed that both Bid truncation and mitochondrial cytochrome c release are involved in 3a-induced apoptosis (Fig. 3).
As ion channels are known to regulate different phases of apoptosis (Burg et al., 2006) and that 3a possesses potassium-sensitive ion channel activity (Lu et al., 2006), we investigated the relationship between ion channel function and pro-apoptotic potential of 3a. Barium chloride (Ba; Lu et al., 2006, Nietsch et al., 2000) and 4-aminopyridine (AP; Grishin et al., 2005) are two commonly used potassium channel blockers. We found that both Ba- and AP-treatment were able to rescue 3a-WT-induced nuclear condensation (Fig. 6), and a similar suppressive effect was also observed in vivo (Fig. 7). Besides, we further found that the ion channel activity of 3a is linked to caspase-dependent apoptosis. When we co-treated 3a-WT-expressing cells simultaneously with ion channel blocker and broad-range caspase inhibitor, no further inhibition of nuclear condensation was observed (Fig. 8 ). This indicates that the ion channel activity and caspase-induced nuclear condensation of 3a-WT are linked. To conclude, this is the first report which describes the functional significance of 3a's ion channel activity in apoptosis induction. Apart from itself being an ion channel, the influence of 3a on the activity of other endogenous ion transporters in viral-infected cells also warrants further investigation.
Fig. 8.
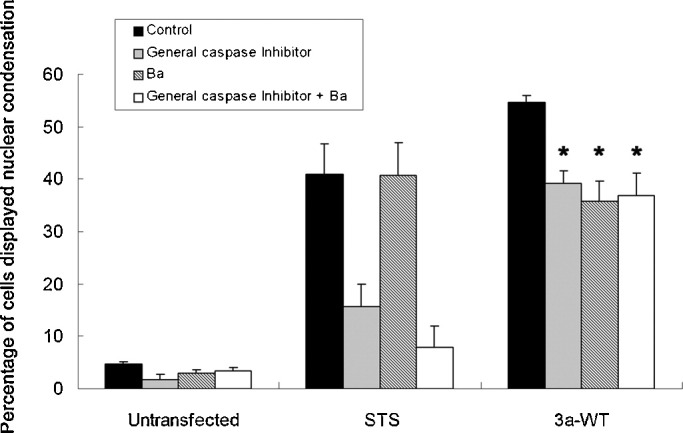
Pro-apoptotic and ion channel activities of 3a are linked. Vero E6 cells, either untransfected or transfected with 3a-WT construct, were treated with general caspase inhibitor (z-VAD-fmk) and/or Ba. z-VAD-fmk and/or barium chloride (Ba) treatment alone only induced minimal nuclear condensation on untransfected cells. Cells transfected with 3a-WT construct displayed nuclear condensation. z-VAD-fmk and/or Ba treatment significantly reduced nuclear condensation induced by 3a-WT expression in Vero E6 cells. Untransfected Vero E6 cells treated with 1 μM staurosporine (STS) for 8 h were used as control, only z-VAD-fmk but not Ba was able to suppress STS-induced nuclear condensation. All results were plotted as percentage of cells that showed nuclear condensation, and were expressed as means + S.E.M. of three independent experiments. At least 100 cells were counted in each experiment. *p < 0.05, Ba and/or caspase inhibitor-treated 3a-WT-expressing cells versus 3a-WT control.
Acknowledgements
We thank members of the Laboratory of Drosophila Research for critical reading of the manuscript. This work was supported by a research grant from the Health, Welfare and Food Bureau of Hong Kong (Research Fund for the Control of Infectious Diseases; 02040302).
References
- Bordi L., Castilletti C., Falasca L., Ciccosanti F., Calcaterra S., Rozera G. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch Virol. 2006;151:369–377. doi: 10.1007/s00705-005-0632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg E.D., Remillard C.V., Yuan J.X. K+ channels in apoptosis. J Membr Biol. 2006;209:3–20. doi: 10.1007/s00232-005-0838-4. [DOI] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Ma C.W., Chan W.Y., Chan H.Y. The SARS-coronavirus membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophys. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau K.W., Chan W.Y., Shaw P.C., Chan H.Y. Biochemical investigation of Tau protein phosphorylation status and its solubility properties in Drosophila. Biochem Biophys Res Commun. 2006;346:150–159. doi: 10.1016/j.bbrc.2006.05.112. [DOI] [PubMed] [Google Scholar]
- Chen M., Wang J. Initiator caspases in apoptosis signaling pathways. Apoptosis. 2002;7:313–319. doi: 10.1023/a:1016167228059. [DOI] [PubMed] [Google Scholar]
- Chow K.Y., Yeung Y.S., Hon C.C., Zeng F., Law K.M., Leung F.C. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 2005;579:6699–6704. doi: 10.1016/j.febslet.2005.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Grishin A., Ford H., Wang J., Li H., Salvador-Recatala V., Levitan E.S. Attenuation of apoptosis in enterocytes by blockade of potassium channels. Am J Physiol Gastrointest Liver Physiol. 2005;289:G815–821. doi: 10.1152/ajpgi.00001.2005. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wassarman D.A., Rubin G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Fielding B.C., Tan T.H., Chou C.F., Shen S., Lim S.G. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Nietsch H.H., Roe M.W., Fiekers J.F., Moore A.L., Lidofsky S.D. Activation of potassium and chloride channels by tumor necrosis factor alpha. Role in liver cell death. J Biol Chem. 2000;275:20556–20561. doi: 10.1074/jbc.M002535200. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Balch W.E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., de Groot R.J., Rottier P.J. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J Virol. 2006;80:2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K., Minakshi R., Towheed M.A., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Ren L., Yang R., Guo L., Qu J., Wang J., Hung T. Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol. 2005;24:496–502. doi: 10.1089/dna.2005.24.496. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sharma K., Surjit M., Satija N., Liu B., Chow V.T., Lal S.K. The 3a accessory protein of SARS coronavirus specifically interacts with the 5’UTR of its genomic RNA, using a unique 75 amino acid interaction domain. Biochemistry. 2007;46:6488–6499. doi: 10.1021/bi062057p. [DOI] [PubMed] [Google Scholar]
- Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem Biophys Res Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A. Cargo recognition during clathrin-mediated endocytosis: a team effort. Curr Opin Cell Biol. 2004;16:392–399. doi: 10.1016/j.ceb.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Strasser A., Jost P.J., Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Tham P.Y., Chan D.Z., Chou C.F., Shen S., Fielding B.C. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J Virol. 2005;79:10083–10087. doi: 10.1128/JVI.79.15.10083-10087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Regulation of cell death during infection by the severe acute respiratory syndrome coronavirus and other coronaviruses. Cell Microbiol. 2007 doi: 10.1111/j.1462-5822.2007.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.X., Tan T.H., Lee M.J., Tham P.Y., Gunalan V., Druce J. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Van Noorden C.J. The history of Z-VAD-FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem. 2001;103:241–251. doi: 10.1078/0065-1281-00601. [DOI] [PubMed] [Google Scholar]
- Wong S.L., Chen Y., Chan C.M., Chan C.S., Chan P.K., Chui Y.L. In vivo functional characterization of the SARS-coronavirus 3a protein in Drosophila. Biochem Biophys Res Commun. 2005;337:720–729. doi: 10.1016/j.bbrc.2005.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.L., Chan W.M., Chan H.Y. Sodium dodecyl sulfate-insoluble oligomers are involved in polyglutamine degeneration. FASEB J. 2008;22:3348–3357. doi: 10.1096/fj.07-103887. [DOI] [PubMed] [Google Scholar]
- Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K. SARS coronavirus induces apoptosis in Vero E6 cells. J Med Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.J., Chen Y.C., Hsiao C.H., Kuo T.C., Chang S.C., Lu C.Y. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q. Characterization of the 3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J Mol Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Shi S.Q., Yang Y., Peng J.P. M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol Toxicol. 2006;22:313–322. doi: 10.1007/s10565-006-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Guo Z., Yang H., Peng L., Xie Y., Wong T.Y. Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients. J Gen Virol. 2006;87:369–373. doi: 10.1099/vir.0.81078-0. [DOI] [PubMed] [Google Scholar]