Abstract
Background and Aims
Previous twin research suggests relationship status can moderate underlying genetic liability towards alcohol misuse. This paper examined: (1) whether genome-wide polygenic scores (GPS) for alcohol consumption are associated with alcohol misuse; (2) whether these GPS are moderated by romantic relationships (gene–environment interaction; G × E)and (3)whether G × E results are consistent across sex.
Design
Linear mixed-effects models were used to test associations between genome-wide polygenic scores, relationship status and alcohol use/misuse.
Setting
Finnish twins born between 1983 and 1987 identified through Finland’s central population registry.
Participants
An intensively studied subset of Finnish Twin Study (FinnTwin12) during the young adult phase (aged 20–26 years). The analytical sample includes those with complete interview and genetic data (n = 1201).
Measurements
Key measurements included involvement in a romantic partnership, drinking frequency, intoxication frequency and DSM-IV alcohol dependence (AD) symptoms. Genome-wide polygenic scores (GPS) were created from available summary statistics from a large genome-wide association study (GWAS) of drinks per week.
Results
GPS predicted drinking frequency [b = 0.109; 95% confidence interval (CI) = 0.050, 0.168], intoxication frequency (b = 0.111; 95% CI = 0.054, 0.168) and AD symptoms (b = 0.123; 95% CI = 0.064, 0.182). Having a romantic relationship negatively influenced the association between GPS and drinking frequency(b = −0.105;95% CI= −0.211,0.001), intoxication frequency(b = −0.118;95%CI = −0.220, −0.016) and AD symptoms (b = −0.119; 95% CI = −0.229, −0.009). There was a three-way interaction between sex, relationship status and GPS for intoxication frequency (b = 0.223; 95% CI = 0.013, 0.433), such that the reduced association between GPS and intoxication frequency for those in a relationship was only apparent in males. We found no evidence of three-way interactions for drinking frequency or AD symptoms.
Conclusions
Being in a romantic relationship reduced the association between genetic predisposition and drinking, high-risk drinking and alcohol problems. However, for high-risk drinking the protective effect was limited to males, mapping onto earlier findings suggesting that males benefit more from romantic partnerships.
Keywords: Alcohol misuse, gene–environment interaction, polygenic risk scores, romantic partnerships, sex differences, young adulthood
INTRODUCTION
Alcohol use is one of the leading contributors to preventable mortality and morbidity world-wide [1–3]. Twin and family studies indicate that genetic influences account for approximately 50% of the variation in the population [4]; however, there is strong evidence that the importance of genetic influences changes across environmental contexts, otherwise referred to as gene–environment interaction, or G × E [5,6]. Environments that allow greater access to alcohol, or acceptance of alcohol use, may create opportunity for increased manifestation of individual predispositions toward alcohol misuse and consequently the development of problems [7–11]. Conversely, environments that exert more social control, such as greater parental monitoring in adolescence, appear to reduce the importance of genetic predispositions [7,12]. Mapping which environments reduce alcohol misuse among those at greater genetic risk will be critical for developing tailored prevention intervention strategies as we move into an era of precision medicine.
Much of the foundational work on G × E in alcohol outcomes has been conducted in twin studies [6–9,12]. Most G × E studies to date using measured genotypes on alcohol use outcomes have focused on candidate genes or single nucleotide polymorphisms (SNPs), where the effect of a specific candidate gene or single SNP varies as a function of the environment [6]. However, candidate gene research has generated inconsistent results, probably a reflection of being underpowered to robustly detect moderations, false positives and publication bias [13,14]. Furthermore, the use of single genes in G × E studies does not align with our current molecular genetic understanding that complex behaviors, including alcohol use [15], problems [16] and dependence [17], have a polygenic architecture, driven by many genetic variants of very small effect [18,19]. Large sample sizes are needed to detect robust genetic associations for complex behavioral outcomes in genome-wide association studies (GWAS), which use data from the entire genome rather than relying on pre-defined SNPs [20,21].
To characterize individual risk across hundreds or thousands of alleles associated with an outcome in a GWAS, genome-wide polygenic risk scores (GPS) have emerged as a way to aggregate this information into a single score. As we begin to identify GPS robustly associated with substance use and dependence, one of the critical next steps toward precision medicine will be to characterize the pathways by which risk unfolds [22]. For alcohol related outcomes, this will necessitate characterizing how specific environments moderate the likelihood that individuals carrying risky genetic predispositions will develop excessive use, problems and dependence, providing important information about targeted areas for intervention.
In this study, we focused on romantic relationships, as epidemiological research has consistently shown that being in committed relationships is associated with health benefits [23]. Alcohol use patterns vary as a function of relationship/marital status. Those in committed relationships (especially marriage) engage in less problem drinking [24,25] and have a lower risk for alcohol use disorder [26,27] than those who are not married, and these findings are generally consistent across males and females. This reduction in risky behaviors is due in part to increased social control and monitoring associated with being in a relationship [23], as well as individuals’ motivation to align their behavior with the social expectations typically associated with the spousal role [28,29]. Although marriage-like relationships are linked with health benefits for both married men and women [30], men, in general, benefit more from marriage than women through positive life-styles with fewer health-deteriorating behaviors [31,32]. Theoretical reasoning of sex differences in the potential protective effects of marriage is complex, but marriage appears to provide more social control for men, with empirical evidence demonstrating that women engage in greater monitoring of their partners’ health-promoting behaviors than do men [33]. Finally, twin studies have found that the heritability of alcohol consumption is decreased among individuals in committed relationships [34,35], suggesting that being with a partner may act as a ‘social control’ that limits expression of genetic predispositions toward alcohol problems.
Here, we test this hypothesis using molecular genetic data in a population-based sample of young adults [36]. We focused on young adulthood because it is a critical period for the development of alcohol use patterns and problems [34], with heavy alcohol use at its highest point [37] and the peak age of onset for alcohol related disorders falling during this period [38]. Young adulthood is also a period when romantic partnerships become increasing salient, as young adults in committed relationships consume less alcohol than their single peers [39]. We used results from the largest mega-analysis to date on alcohol consumption [15], which used drinks per week in ~1 million individuals, to calculate genome-wide polygenic scores in our independent, population-based sample. We tested: (1) whether these polygenic risk scores were associated with alcohol use, heavy consumption and alcohol problems; (2) whether being in a romantic relationship changed the association between genetic risk and alcohol outcomes; and (3) because there are sex differences in patterns of alcohol use and in the prevalence of alcohol use disorders [38] and heavy consumption [37] and the fact that social control processes may operate differently for men and women in the context of relationships [31–33], we examined whether there were sex differences in G × E [40].
METHODS
Design
We used data from the youngest cohort of the Finnish Twin Cohort Study (FinnTwin12) when twins were in young adulthood (aged 20–26 years). We fitted a series of linear mixed models to examine whether relationship status moderates the association between GPS and alcohol misuse. We then tested for sex differences in these interactions. All analyses adjusted for age, sex, educational attainment and current student status. Linear mixed models were adjusted for clustering at the family level. We checked for the robustness of our results by assessing separate models that included interactions between GPS, relationship status and each covariate [41].
Sample
Families in FinnTwin12 were identified from Finland’s Population Registry, permitting comprehensive nationwide ascertainment for twins born from 1983 to 1987. Baseline collection occurred when twins were aged approximately 11–12 years, with a sample of approximately 5600 twins (87% participation) and their families [36]. Follow-up surveys occurred at ages 14, 17.5, and during young adulthood (age range = 20–26). Twin zygosity was determined using items developed for twin children [42]. Confirmation by multiple genetic markers revealed that 97% of same-sex pairs retained the original questionnaire-based zygosity classification [43]. The Helsinki University Central Hospital District’s Ethical Committee and Indiana University’s Institutional Review Board approved the FinnTwin12 study. Of those in the larger sample, a subset of intensively studied individuals also received in-depth clinical interviews (n = 1347) and participated in DNA collection as young adults. In the present study, we limited our analyses to those who had complete information on all relevant study variables and who had initiated alcohol use (n = 1201). The analytical subset did not differ significantly from the full sample in terms of demographic characteristics or alcohol misuse (see Supporting information, Table S1 for more detail).
Genotyping and quality control
Genotyping was conducted using the Human670-QuadCustom Illumina BeadChip at the Wellcome Trust Sanger Institute [36]. Quality control steps included removing SNPs with minor allele frequency (MAF) < 1%, genotyping success rate < 95%, or Hardy–Weinberg equilibrium P < 1 × 10−6, and removing individuals with genotyping success rate < 95%, a mismatch between phenotypical and genotypical gender, excess relatedness (outside known families) and heterozygosity outliers. Genotypes were imputed to the 1000 Genomes Phase 3 reference panel [44] using ShapeIT [45] for phasing and IMPUTE2 [46] for imputation, resulting in 13 688 418 autosomal SNPs for analyses. Prior analyses indicated a single dimension of ancestry in the sample [47]. Although a single dimension of ancestry does not preclude variation along this dimension, we note that fine-scale population substructure is less of an issue for common variants (versus rare variants), especially in the present sample, given the relatively longer LD blocks that make the Finnish population more homogeneous than other populations of mixed European ancestry.
Measures
Alcohol-related behaviors were assessed across increasing levels of severity. Drinking frequency was measured by asking: ‘How often do you use alcohol?’. Responses included ‘never’ (0), ‘once a year’ (1), ‘two to four times a year’ (2), ‘every other month’ (3), ‘once a month’ (4), ‘more than once a month’ (5), ‘once a week’ (6), ‘more than once a week’ (7) and ‘daily’ (8). Intoxication frequency was assessed by asking ‘How often do you use alcohol in such a way that you get really drunk?’. Responses were the same for drinking frequency. We transformed these ordinal measures into pseudo-continuous measures of the frequency of these behaviors in a typical 30-day period [9,48]. Finally, we included a count of life-time DSM-IValcohol dependence (AD) symptoms, assessed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA), a reliable and valid clinical instrument [49]. Each alcohol measure was log-transformed (left anchored at 1) to adjust for positive skew. Relationship status was measured by asking: ‘How long (in years) have you been together with your present partner?’. Respondents that indicated they were not in a relationship were coded as 0. Those who indicated they were in a romantic relationship for any length were coded as 1. We ran sensitivity analyses with a stricter definition of relationship status (those in a relationship ≥ 2 years). Our results did not fundamentally differ from the more inclusive definition and we retained the original measurement of relationship status. Finally, we included age, sex, educational attainment (based on the Finnish education system: basic education; vocational training; secondary education; tertiary education) and whether or not respondents were still in school [50] as covariates.
Genome-wide polygenic scores
We created polygenic scores derived from a large-scale GWAS of number of alcoholic drinks per week in approximately 1 million individuals [15]. As FinnTwin12 was included in the original discovery GWAS, we obtained summary statistics with all Finnish participants, including FinnTwin12 and 23andMe (which are not publicly available) cohorts removed (available n = 534 683). There were 3 707 235 autosomal SNPs in common after quality control. We used the well-established process of clumping and thresholding [51]. SNPs from the discovery GWAS were clumped based on linkage disequilibrium (LD) using the clump procedure in PLINK [52], based on an R2 = 0.25, with a 500 kb window, resulting in 407 604 independent SNPs for creating scores. We then created scores based on differing thresholds of GWAS P-values (P < 0.0001, P < 0.001, P < 0.01, P < 0.05, P < 0.10, P < 0.20, P < 0.30, P < 0.40, P < 0.50). We converted GPS to Z-scores for interpretation.
We note that alcohol consumption and problematic use, though highly correlated, have distinct genetic influences [53]. We ran a series of sensitivity analyses to determine if recent GWAS focused on alcohol problems or dependence [16,17] provided better assessments of genetic liability for alcohol misuse (see Supporting information, Fig. S2). However, in each case, the original scores were the most predictive.
Analytical strategy
First, we estimated the effect of GPS across each P-value threshold to determine the most predictive score (based on model R2) for each alcohol phenotype. We then tested whether relationship status moderated the association of the genome-wide polygenic scores. In the instances where we found evidence for a significant interaction, we fitted a more robust model for evaluating G × E [41], which includes all G × covariate and E × covariate interaction terms. Finally, we tested for sexspecific G × E by including a three-way interaction term. We determined whether estimates were significant using an α of P < 0.05 (two-sided test). Because the FinnTwin12 data is a family-based data set, we evaluated all hypotheses using a linear mixed model with random intercepts for each family in the lme4 [54] package in in R version 3.5.1 [55]. We estimated effect size (ΔR2) using a method designed for mixed effects models [56] with the MuMIn package [57].
RESULTS
Males exhibited higher mean levels of each alcohol measure (Table 1). The alcohol phenotypes were also modestly correlated(rdrinking × intox =0.64,rdrinking × ADsx =0.37, rintox × ADsx = 0.43), with stronger correlations between the consumption items than with the measure of AD symptoms [53].
Table 1.
Descriptive statistics for Finnish Twin Study (FinnTwin12).
| Males (n = 551) |
Females (n = 650) |
Full Sample (n = 1201) |
|||||
|---|---|---|---|---|---|---|---|
| Mean/n | SD/% | Mean/n | SD/% | Mean/n | SD/% | χ2/t-test | |
| Drinking frequency | 5.10 | 4.54 | 3.43 | 3.24 | 4.20 | 3.98 | * |
| Intoxication frequency | 2.02 | 1.97 | 1.12 | 1.44 | 1.54 | 1.76 | * |
| DSM-IV alcohol dependence symptoms | 1.54 | 1.35 | 1.29 | 1.37 | 1.40 | 1.37 | * |
| GPSa | −0.03 | 1.02 | 0.03 | 0.98 | 0.00 | 1.00 | |
| Age | 21.94 | 0.77 | 21.95 | 0.76 | 21.94 | 0.77 | |
| Educational attainment | * | ||||||
| Basic education | 38 | 6.9% | 30 | 4.6% | 68 | 5.7% | |
| Vocational training | 207 | 37.6% | 157 | 24.2% | 364 | 30.3% | |
| Secondary education | 299 | 54.3% | 424 | 65.2% | 723 | 60.2% | |
| Tertiary education | 7 | 1.3% | 39 | 6.0% | 46 | 3.8% | |
| Enrolled in school | 285 | 51.7% | 401 | 61.7% | 686 | 57.1% | * |
| In relationship | 269 | 48.8% | 416 | 64.0% | 685 | 57.0% | * |
P < 0.05 for χ2/t-test difference between males and females.
Standardized (Z-scores) genome-wide polygenic scores (GPS) including single nucleotide polymorphisms (SNPs) with P < 0.50. SD = standard deviation.
Polygenic score performance
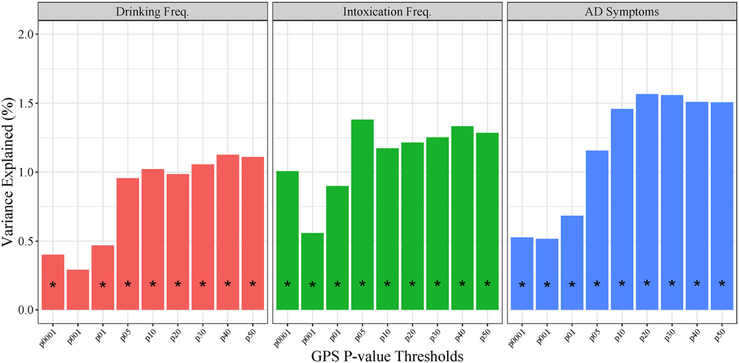
Figure 1 provides the incremental R2 for polygenic scores at different P-value inclusion thresholds. The variance explained at each P-value threshold in GPS represents the change in R2 from the baseline model (age and sex as covariates) after including the GPS at that P-value threshold. GPS were significantly associated with each alcohol-related behavior across almost all of the P-value thresholds, with the exception of the most restrictive scores in relation to drinking frequency. GPS explained more variance as P-value thresholds became more inclusive, peaking and leveling off at thresholds between P < 0.20 and P < 0.50. We decided to use the most liberal threshold (P < 0.50) for all models going forward.
Figure 1.
Predictive power of genome-wide association studies (GWAS) and Sequencing Consortium of Alcohol and Nicotine Use (GSCAN) polygenic scores. Vertical bars represent change in model R2 from base model (age and sex as covariates) to model including polygenic scores at various P-value inclusion thresholds (determined by P-value from discovery GWAS) for drinking frequency (left), intoxication frequency (center), and alcohol dependence symptoms (right). *Association P< 0.05.
In order to ensure the GPS were predictive of alcohol problems above and beyond levels of consumption, which are genetically correlated but distinct phenotypes [53], we estimated the effect of GPS while accounting for either drinking or intoxication frequency. GPS were significantly related to AD symptoms after statistically controlling for drinking frequency (b = 0.085, P < 0.01) or intoxication frequency (b = 0.075, P < 0.01; see Supporting information, Table S2). Finally, we estimated the polyserial correlation between GPS and relationship status (ρ = 0.005, P > 0.05) to assess the possibility of gene–environment correlation.
Main effects of polygenic score and relationship status
Table 2 provides the estimates for the linear mixed models evaluating the joint effect of GPS and relationship status. In the model for main effects (model 1), those currently in a relationship had lower levels of intoxication frequency, but not drinking frequency or AD symptoms. GPS remained significantly associated with each of these alcohol-related behaviors.
Table 2.
Linear mixed models for alcohol related behaviors (n = 1201).
| Model 1 |
Model 2 |
Model 3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b | 95% CI | b | 95% CI | b | 95% CI | |||||||
| Drinking frequency | ||||||||||||
| Female | −0.456 | −0.572 | −0.340 | *** | −0.458 | −0.574 | −0.342 | *** | −0.437 | −0.606 | −0.268 | *** |
| In relationship | −0.090 | −0.198 | 0.018 | −0.087 | −0.195 | 0.021 | −0.069 | −0.224 | 0.086 | |||
| GPS | 0.109 | 0.050 | 0.168 | *** | 0.169 | 0.085 | 0.253 | *** | 0.193 | 0.085 | 0.301 | ** |
| In relationship × GPS | – | – | – | −0.105 | −0.211 | −0.001 | * | −0.158 | −0.307 | −0.009 | * | |
| Female × GPS | – | – | – | – | – | – | −0.061 | −0.230 | 0.108 | |||
| Female × in relationship | – | – | – | – | – | – | −0.038 | −0.254 | 0.178 | |||
| Female × in | – | – | – | – | – | – | 0.109 | −0.107 | 0.325 | |||
| relationship × GPS | ||||||||||||
| Pseudo-R2 | 0.073 | 0.076 | 0.077 | |||||||||
| Intoxication frequency | ||||||||||||
| Female | −0.543 | −0.657 | −0.429 | *** | −0.544 | −0.658 | −0.430 | *** | −0.535 | −0.700 | −0.370 | *** |
| In relationship | −0.178 | −0.284 | −0.072 | ** | −0.176 | −0.282 | −0.070 | ** | −0.171 | −0.322 | −0.020 | * |
| GPS | 0.111 | 0.054 | 0.168 | *** | 0.179 | 0.097 | 0.261 | *** | 0.239 | 0.133 | 0.345 | *** |
| In relationship × GPS | – | – | – | −0.118 | −0.220 | −0.016 | * | −0.222 | −0.365 | −0.079 | ** | |
| Female × GPS | – | – | – | – | – | – | −0.149 | −0.314 | 0.016 | |||
| Female × in relationship | – | – | – | – | – | – | −0.016 | −0.226 | 0.194 | |||
| Female × in | – | – | – | – | – | – | 0.223 | 0.013 | 0.433 | * | ||
| relationship × GPS | ||||||||||||
| Pseudo-R2 | 0.110 | 0.114 | 0.117 | |||||||||
| AD symptoms | ||||||||||||
| Female | −0.197 | −0.317 | −0.077 | ** | −0.199 | −0.319 | −0.079 | ** | −0.123 | −0.297 | 0.051 | |
| In relationship | −0.097 | −0.209 | 0.015 | −0.095 | −0.207 | 0.017 | −0.028 | −0.189 | 0.133 | |||
| GPS | 0.123 | 0.064 | 0.182 | *** | 0.191 | 0.105 | 0.277 | *** | 0.196 | 0.084 | 0.308 | ** |
| In relationship × GPS | – | – | – | −0.119 | −0.229 | −0.009 | * | −0.154 | −0.309 | 0.001 | ||
| Female × GPS | – | – | – | – | – | – | −0.018 | −0.192 | 0.156 | |||
| Female × in relationship | – | – | – | – | – | – | −0.134 | −0.359 | 0.091 | |||
| Female × in | – | – | – | – | – | – | 0.069 | −0.154 | 0.292 | |||
| relationship × GPS | ||||||||||||
| Pseudo-R2 | 0.029 | 0.032 | 0.034 | |||||||||
Linear mixed models for each of the alcohol phenotypes. Each model includes age, educational attainment, and student status as covariates. Clustering at the family level modeled by including random intercepts. GPS and alcohol phenotypes were standardized for easier interpretation.
P < 0.05
P < 0.01;
P < 0.001. GPS = genome-wide polygenic scores.
G × E interaction models
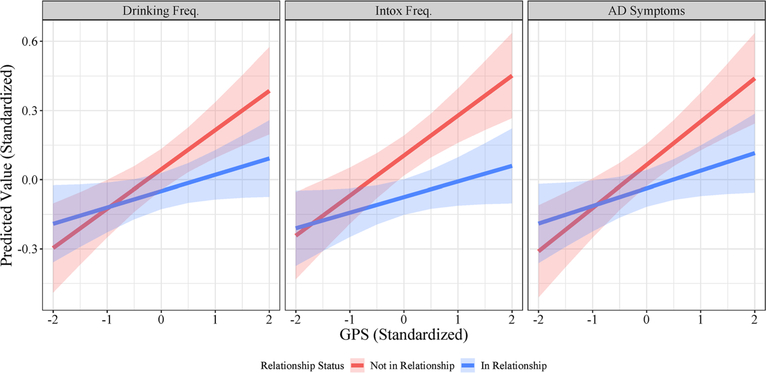
Model 2 (Table 2) presents the estimates for G × E. There was a significant interaction between relationship status and polygenic scores for each alcohol behavior. We refitted each of these models with interactions between relationship status and each covariate and interactions between GPS and each covariate (plotted in Fig. 2; see Supporting information, Tables S3–S5 for full results) to account for possible confounding [41]. P-values were attenuated, especially in the models for drinking frequency and AD symptoms, but the nature of the interactions remained unchanged for the other phenotypes. The shape of the interaction was similar among all phenotypes, but most pronounced for intoxication. In the case of intoxication frequency, there was a stronger association between genetic risk score and intoxication frequency among individuals who are not in romantic relationships, and a relatively weaker association between genetic risk score and intoxication frequency among those who were in romantic relationships.
Figure 2.
Gene–environment interaction across relationship status and polygenic risk. Standardized predicted values of drinking frequency (left), intoxication frequency (center), and alcohol dependence symptoms (right) across the observed range of polygenic scores for those in a relationship (blue) and those not in a relationship (red). Shaded areas represent 95% pointwise confidence intervals of estimates.
Sex differences in G × E
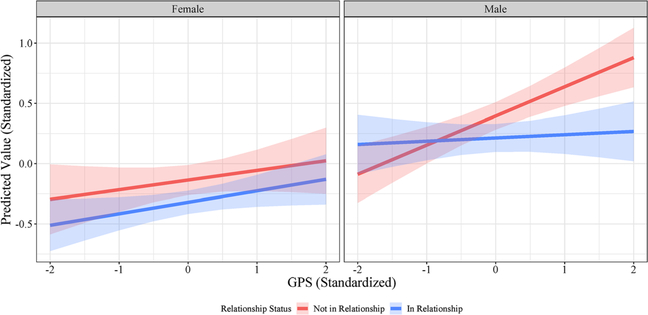
Finally, we tested for sex differences in the interaction between relationship status and GPS. We found no evidence of a significant three-way interaction between sex, relationship status and GPS for either drinking frequency or AD symptoms. However, we did find a significant three-way interaction in the models for intoxication frequency. This interaction remained significant even after adjusting for possible confounding in the G × E interactions. Figure 3 displays the predicted values from this model. For intoxication frequency, the G × E effect appears to be driven by the effect in males.
Figure 3.
Sex differences in G × E for intoxication frequency. Predicted values of intoxication frequency (standardized) for females (left) and males (right) across the observed range of polygenic scores and sex for those in a relationship (blue) and those not in a relationship (red). Shaded areas represent 95% pointwise confidence intervals of estimates.
We ran a series of supplementary analyses stratified by sex (see Supporting information, Table S6), to further examine whether different G × E patterns emerged across sex. Only the interaction between GPS and relationship status in the model for intoxication frequency in males remained significant after correcting the multiple tests using a 5% false discovery rate [58]. Overall these sexstratified models mirrored the results from the three-way interactions.
DISCUSSION
We tested whether polygenic risk scores derived from a meta-analysis of alcohol consumption were associated with alcohol outcomes in an independent, population-based young adult sample, whether romantic relationship status moderated the association of genetic predispositions with alcohol outcomes and whether observed effects varied between females and males.
Polygenic scores derived from variants associated with consumption are predictive of use, misuse and problems among young adults. As hypothesized, being in a romantic relationship moderated the association between GPS and each alcohol phenotype (drinking frequency, intoxication frequency and AD symptoms). Similar to previous twin research [34,35], among individuals with elevated genetic predisposition levels of misuse were lower in those in a romantic partnership. We posit that the constraints and responsibilities placed on individuals within romantic partnerships limits their ability to express underlying predispositions towards alcohol misuse, fitting with the social control model of gene–environment interaction[5,23]. Additional inspection (Supporting information, Fig. S1) revealed that these interactions did not appear to be driven by outliers at either end of the distribution.
Finally, we examined whether there were sex differences in these G × E effects. We found no evidence of sex differences in the G × E effect for drinking frequency or AD symptoms. However, the G × E effect for intoxication frequency was driven primarily by the effect in males. Simulations revealed modest power (~60%) to detect this three-way interaction. Previous work in social epidemiology has documented how males tend to ‘over-benefit’ from relationships in terms of health [32]. This may reflect the tendency for women in relationships to be the emotional and social support providers, of which men are the receivers [59]. In the current study, we see that this effect may be due in part to limiting genetic liability among a riskier drinking group. This difference does not appear in AD symptoms, which may be due to the fact that these symptoms capture aspects of both consumption and problems. Relationship status may only limit genetic liability in regards to heavy consumption. Additionally, our AD measure was a life-time measure. It is possible that current levels of misuse may differ from life-time symptomology.
Our findings have important practical implications for researchers and clinicians interested in those at greater risk for alcohol misuse. First, the signal for genetic associations may be drastically reduced in young adults in a committed relationship. Future research on gene identification efforts may benefit from the inclusion of important environmental information in order to increase power to detect genetic variants associated with various forms of alcohol misuse. Considering that G × E in the discovery GWAS may be of even more importance with regard to alcohol use phenotypes, as there is consistent evidence of G × E from twin studies [9,34,60,61]. For clinicians, these analyses point to committed relationships as a malleable environmental condition that may help reduce individuals’ level of misuse, in part, by limiting realization of genetic predisposition. Gene–environment correlation (rGE, or when exposure to an environment is influenced by one’s genotype) is always an important consideration, as the presence of rGE can give rise to spurious evidence of G × E [41]. We note that our GPS was uncorrelated with relationship status, increasing the likelihood that the evidence for G × E in the current sample is not due to rGE.
This research has several limitations. First, although the polygenic scores explained more variance in these outcomes than previous iterations using smaller discovery GWAS, the variance explained by the largest meta-analysis of alcohol consumption to date, compiling data from ~1 million individuals, continued to be small (R2 ~ 0.015 in the current study, R2 ~ 0.025 in the original GWAS). Even larger discovery samples with better phenotyping will be necessary to create scores that explain the total SNP-based heritability. Secondly, although we found evidence of G × E, it does not rule out other confounding factors. Larger twin samples with genotypical data that allow for within-family designs will help to further account for possible environmental confounders shared across families (e.g. neighborhood factors, religiosity, socio-economic status; see Supporting information, Fig. S4 for sensitivity analyses). Longitudinal designs will allow us to understand more clearly the direction of effect and rule out explanations other than G × E. For example, those with high genetic propensity for alcohol misuse who experience low alcohol misuse may be more likely to enter a romantic relationship than those with higher levels of misuse. Thirdly, our measure of romantic partnerships did not include relationship characteristics. We examined romantic partnerships status as the moderator of polygenic scores, given the well-established link between relationship status and health in the literature [23]. However, research also suggests that the association between romantic relationship and alcohol use is complex and depends, in part, on relationship characteristics (e.g. relationship quality, partner’s drinking, emotional support) [62]. Identifying data sources that contain phenotypical information on both the respondent and their partner will be important for future research understanding G × E mechanisms. Finally, our measure of AD symptoms was a life-time measure. Supplemental analyses revealed similar patterns between life-time and past 12-month symptoms (see Supporting information, Fig. S3 and Table S7).
In conclusion, polygenic scores from a large-scale GWAS of drinks per week predicted levels of alcohol use and misuse among a sample of young adults. However, this association between genetic risk and problematic patterns of use changed as a function of the environment. Individuals at greater genetic risk who were in romantic relationships were less likely to misuse alcohol. For drinking to intoxication, this interaction appears to occur primarily among males. This finding is consistent with previous research findings on social determinants of health that men tend to over-benefit from romantic partnerships [32]. This research underscores the importance of considering the interplay between genes and environment when considering etiology and intervention for problematic alcohol use.
Supplementary Material
Table S1 Descriptive Statistics for Full and Analytic Samples in FinnTwin12.
Table S2 Linear Mixed Models for AD Symptoms (N = 1201).
Table S3 Linear Mixed Models for Drinking Frequency (N = 1201).
Table S4 Linear Mixed Models for Intoxication Frequency (N = 1201).
Table S5 Linear Mixed Models for AD symptoms (N = 1201).
Table S6 Sex-Stratified Linear Mixed Models.
Table S7 Correlations Among Alcohol Phenotypes in FinTwin12.
Figure S1 Levels of Alcohol Misuse Across GPS Quintiles.
Figure S2 Predictive Power of Other Alcohol Related GPS.
Figure S3 Comparisons of Last 12 Month vs. Earlier AD Symptoms.
Figure S4 Between vs. Within Family Effects.
Acknowledgements
Research reported in this publication was supported by the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under award numbers R01AA015416, K02AA018755, K01AA024152 and F32AA022269; the Academy of Finland (grants 100499, 205585, 118555, 141054, 265240, 308248 and 312073); and the Scientific and Technological Research Council of Turkey (TÜBİTAK) under award number 114C117. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Academy of Finland, or the Scientific and Technological Research Council of Turkey. The authors have no conflict of interests to report.
Footnotes
Declaration of interests
None.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Gakidou E, Afshin A, Abajobir AA, Abate KH, Abbafati C, Abbas KM et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet 2017; 390: 1345–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The US Burden of Disease Collaborators The state of us health,1990–2016: burden of diseases, injuries, and risk factors among US states. JAMA 2018; 319: 1444–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO). Global status report on alcohol and health—2018. Geneva, Switzerland; WHO; 2018. [Google Scholar]
- 4.Verhulst B, Neale MC, Kendler KS The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 2015; 45: 1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanahan MJ, Hofer SM Social context in gene– environment interactions: retrospect and prospect. J Gerontol Series B 2005; 60: 65–76. [DOI] [PubMed] [Google Scholar]
- 6.Young-Wolff KC, Enoch M-A, Prescott CA The influence of gene–environment interactions on alcohol consumption and alcohol use disorders: a comprehensive review. Clin Psychol Rev 2011; 31: 800–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick DM, Pagan JL, Viken R, Purcell S, Kaprio J,Pulkkinen L et al. Changing environmental influences on substance use across development. Twin Res Hum Genet 2007; 10: 315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harden KP, Hill JE, Turkheimer E, Emery RE Gene-environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behav Genet 2008; 38: 339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooke ME, Meyers JL, Latvala A, Korhonen T, Rose RJ, Kaprio J et al. Gene–environment interaction effects of peer deviance, parental knowledge and stressful life events on adolescent alcohol use. Twin Res Hum Genet 2015; 18: 507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olfson E, Edenberg HJ, Nurnberger J, Agrawal A, Bucholz KK, Almasy LA et al. An ADH1B variant and peer drinking in progression to adolescent drinking milestones: evidence of a gene-by-environment interaction. Alcohol Clin Exp Res 2014; 38: 2541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virtanen S, Kaprio J, Viken R, Rose RJ, Latvala A Birth cohort effects on the quantity and heritability of alcohol consumption in adulthood: a Finnish longitudinal twin study. Addiction 2019; 114: 836–46. [DOI] [PubMed] [Google Scholar]
- 12.Miles DR, Silberg JL, Pickens RW, Eaves LJ Familial influences on alcohol use in adolescent female twins: testing for genetic and environmental interactions. J Stud Alcohol 2005; 66: 445–51. [DOI] [PubMed] [Google Scholar]
- 13.Duncan LE,Keller MC A critical reviewof thefirst 10years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry 2011; 168: 1041–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dick DM, Agrawal A, Keller MC, Adkins A, Aliev F,Monroe S et al. Candidate gene–environment interaction research: reflections and recommendations. Perspect Psychol Sci 2015; 10: 37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019; 51: 237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL,Adams MJ, Howard DM et al. Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry 2018; 10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters RK, Polimanti R, Johnson EC, McClintick JN,Adams MJ, Adkins AE et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 2018; 21: 1656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plomin R, Haworth CMA, Davis OSP Common disorders are quantitative traits. Nat Rev Genet 2009; 10: 872–8. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan PF, Daly MJ, O’Donovan M Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogdan R, Baranger DAA, Agrawal A Polygenic risk scores in clinical psychology: bridging genomic risk to individual differences. Annu Rev Clin Psychol 2018; 14: 119–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudbridge F Polygenic epidemiology. Genet Epidemiol 2016; 40: 268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick DM,Barr PB, Cho SB, Cooke ME, Kuo SI-C, Lewis TJ et al. Post-GWAS in psychiatric genetics: a developmental perspective on the ‘other’ next steps. Genes Brain Behav 2018; 17: e12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umberson D, Crosnoe R, Reczek C Social relationships and health behavior across life course. Annu Rev Sociol 2010; 36: 139–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinescu D, Turkheimer E, Beam CR, Horn EE, Duncan G, Emery RE Is marriage a buzzkill? A twin study of marital status and alcohol consumption. J Fam Psychol 2016; 30: 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Power C, Rodgers B, Hope S Heavy alcohol consumption and marital status: disentangling the relationship in a national study of young adults. Addiction 1999; 94: 1477–87. [DOI] [PubMed] [Google Scholar]
- 26.Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J,Zhang H et al. Epidemiology of DSM-5 alcohol use disorder: results from the National Epidemiologic Survey on alcohol and related conditions III. Epidemiology of DSM-5 alcohol use. Disorder epidemiology of DSM-5 alcohol use disorder. JAMA Psychiatry 2015; 72: 757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staff J, Schulenberg JE, Maslowsky J, Bachman JG,O’Malley PM, Maggs JL et al. Substance use changes and social role transitions: proximal developmental effects on ongoing trajectories from late adolescence through early adulthood. Dev Psychopathol 2010; 22: 917–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn EE, Xu YF, Beam CR, Turkheimer E, Emery RE Accounting for the physical and mental health benefits of entry into marriage: a genetically informed study of selection and causation. J Fam Psychol 2013; 27: 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi K, Kandel DB On the resolution of role incompatibility—a life event history analysis of family roles and marijuana use. Am J Sociol 1985; 90: 1284–325. [Google Scholar]
- 30.Kaplan RM, Kronick RG Marital status and longevity in the United States population. J Epidemiol Community Health 2006; 60: 760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monin JK, Clark MS Why do men benefit more from marriage than do women? Thinking more broadly about interpersonal processes that occur within and outside of marriage. Sex Roles 2011; 65: 320–6. [Google Scholar]
- 32.Kiecolt-Glaser JK, Newton TL Marriage and health: his and hers. Psychol Bull 2001; 127: 472–503. [DOI] [PubMed] [Google Scholar]
- 33.Umberson D Gender, marital status and the social control of health behavior. Soc Sci Med 1992; 34: 907–17. [DOI] [PubMed] [Google Scholar]
- 34.Barr PB, Salvatore JE, Maes HH, Korhonen T, Latvala A,Aliev F et al. Social relationships moderate genetic influences on heavy drinking in young adulthood. J Stud Alcohol Drugs 2017; 78: 817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath AC, Jardine R, Martin NG Interactive effects of genotype and social environment on alcohol consumption in female twins. J Stud Alcohol 1989; 50: 38–48. [DOI] [PubMed] [Google Scholar]
- 36.Kaprio J The Finnish twin cohort study: an update. Twin Res Hum Genet 2013; 16: 157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen P, Jacobson KC Developmental trajectories of substance use from early adolescence to young adulthood: gender and racial/ethnic differences. J Adolesc Health 2012; 50: 154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the national comorbidity survey replication. Arch Gen Psychiatry 2005; 62: 593–602. [DOI] [PubMed] [Google Scholar]
- 39.Fleming CB, White HR, Catalano RF Romantic relationships and substance use in early adulthood: an examination of the influences of relationship type, partner substance use, and relationship quality. J Health Soc Behav 2010; 51: 153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salvatore JE, Cho SB, Dick DM Genes, environments, and sex differences in alcohol research. J Stud Alcohol Drugs 2017; 78: 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller MC Gene x environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry 2014; 75: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill Goldsmith H A zygosity questionnaire for young twins: are search note. Behav Genet 1991; 21: 257–69. [DOI] [PubMed] [Google Scholar]
- 43.Knaapila A, Silventoinen K, Broms U, Rose RJ, Perola M,Kaprio J et al. Food neophobia in young adults: genetic architecture and relation to personality, pleasantness and use frequency of foods, and body mass index—a twin study. Behav Genet 2011; 41: 512–21. [DOI] [PubMed] [Google Scholar]
- 44.The 1000Genomes Project Consortium A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaneau O, Zagury J-F, Marchini J Improved whole chromosome phasing for disease and population genetic studies. Nat Methods 2012; 10: 5. [DOI] [PubMed] [Google Scholar]
- 46.Howie BN, Donnelly P, Marchini J A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLOS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyers JL. Elucidating Genetic and Environmental Influences on Alcohol-Related Phenotypes ProQuest Dissertations & Theses Global. Richmond, VA: Virginia Commonwealth University; 2012. [Google Scholar]
- 48.Barr PB, Salvatore JE, Maes HH, Aliev F, Latvala A,Viken RJ et al. Education and alcohol use: a study of gene–environment interaction in young adulthood. Soc Sci Med 2016; 162: 158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH,Hesselbrock VM, Nurnberger JI et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 1994; 55: 149–58. [DOI] [PubMed] [Google Scholar]
- 50.Timberlake DS, Hopfer CJ, Rhee SH, Friedman NP,Haberstick BC, Lessem JM et al. College attendance and its effect on drinking behaviors in a longitudinal study of adolescents. Alcohol Clin Exp Res 2007; 31: 1020–30. [DOI] [PubMed] [Google Scholar]
- 51.International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS Measures of current alcohol consumption and problems: two independent twin studies suggest a complex genetic architecture. Alcohol Clin Exp Res 2011; 35: 2152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bates D, Maechler M, Bolker B, Walker S Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67: 1–48. [Google Scholar]
- 55.R Core Development Team A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 56.Nakagawa S, Schielzeth H, O’Hara RB A general and simple method for obtaining R-squared from generalized linear mixed-effects models. Methods Ecol Evol 2013; 4: 133–42. [Google Scholar]
- 57.Bartoń K MuMIn: Multi-Model Inference. R package version 1.43.6; 2019. (Accessed April 9, 2019).
- 58.Benjamini Y, Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 1995; 57: 289–300. [Google Scholar]
- 59.Kawachi I, Berkman LF Social ties and mental health.J Urban Health 2001; 78: 458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dick DM, Bernard M, Aliev F, Viken R, Pulkkinen L,Kaprio J et al. The role of socioregional factors in moderating genetic influences on early adolescent behavior problems and alcohol use. Alcohol Clin Exp Res 2009; 33: 1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. J Abnorm Psychol 2007; 116: 213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhule-Louie DM, McMahon RJ Problem behavior and romantic relationships: assortative mating, behavior contagion, and desistance. Clin Child Fam Psychol Rev 2007; 10: 53–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Descriptive Statistics for Full and Analytic Samples in FinnTwin12.
Table S2 Linear Mixed Models for AD Symptoms (N = 1201).
Table S3 Linear Mixed Models for Drinking Frequency (N = 1201).
Table S4 Linear Mixed Models for Intoxication Frequency (N = 1201).
Table S5 Linear Mixed Models for AD symptoms (N = 1201).
Table S6 Sex-Stratified Linear Mixed Models.
Table S7 Correlations Among Alcohol Phenotypes in FinTwin12.
Figure S1 Levels of Alcohol Misuse Across GPS Quintiles.
Figure S2 Predictive Power of Other Alcohol Related GPS.
Figure S3 Comparisons of Last 12 Month vs. Earlier AD Symptoms.
Figure S4 Between vs. Within Family Effects.