Abstract
Noroviruses are human enteric caliciviruses for which no cell culture is available. Consequently, the mechanisms and factors involved in their replication have been difficult to study. In an attempt to analyze the cis- and trans-acting factors that could have a role in NV replication, the 3′-untranslated region of the genome was studied. Use of Zuker’s mfold-2 software predicted that NV 3′UTR contains a stem-loop structure of 47 nts. Proteins from HeLa cell extracts, such as La and PTB, form stable complexes with this region. The addition of a poly(A) tail (24 nts) to the 3′UTR permits the specific binding of the poly(A) binding protein (PABP) present in HeLa cell extracts, as well as the recombinant PABP. Since La, PTB, and PABP are important trans-acting factors required for viral translation and replication, these RNA–protein interactions may play a role in NV replication or translation.
Keywords: Norwalk virus, Replication, La protein, PTB protein, PAB protein, 3′UTR, RNA–protein interactions
Norwalk virus (NV) is the prototype strain of human caliciviruses (HuCV), a group of agents causing highly transmissible viral gastroenteritis, which spreads rapidly through families, institutions, and communities (reviewed in [1], [2], [3]). Noroviruses are difficult to study because they cannot be cultivated in cell culture; however, molecular techniques have been useful to examine the genome organization and function of the viral proteins. NV has a single stranded positive-polarity RNA genome that contains three open reading frames (ORF), a short 3′ untranslated region (UTR) of 66 nts, and a poly(A) tail [4]. The presence of a 5′UTR has not been established. Sequence analysis of ORF 1 shows the characteristic motifs of the 2C helicase, 3C cystein protease, and 3D RNA polymerase present in the picornavirus genome [4], [5]. ORFs 2 and 3 encode the viral capsid proteins [5], [6].
The mechanisms and factors involved in NV translation and RNA replication remain unknown; however, as a positive-stranded RNA virus, the genomic RNA has to be translated into the viral non-structural proteins and function as a template for negative-strand RNA synthesis. The viral enzyme required for RNA synthesis has to be the viral RNA-dependent RNA polymerase (RdRp) 3Dpol that initiates RNA synthesis at the 3′ end of the genomic and replicative intermediate RNAs. Since viral RdRps are commonly primer-dependent enzymes, preinitiation replication complexes require the presence of structured sequences as cis-acting elements to recruit several other trans-acting factors such as viral and cellular proteins [7], [8], [9], [10].
NV and poliovirus (PV) share a high level of homology between their non-structural proteins, and their polyadenylated tails; thus, it is possible that both RNA viruses use similar mechanisms to replicate their genomes. The poly(A) tail increases infectivity [11] and has been considered an important cis-acting element required for negative-strand RNA synthesis of PV [12]. This element is located at the 3′ end of the genomic RNA, where the initiation of the RNA synthesis takes place and has a critical role in organizing proteins around the start site of the RNA synthesis [12]. This sequence is a specific target for the poly(A) binding protein (PABP), a regulatory factor involved in the control of cellular mRNA stability and translation (reviewed in [13]). The PABP bound to PV poly(A) tail interacts directly with the 5′ end via a protein–protein bridge, promoting the circularization of the genomic RNA, which provides several advantages for viral replication [12].
Besides the cis-acting elements, viral RNA replication also depends on some cellular proteins that form part of the ribonucleoprotein (RNP) complexes formed in the 3′ ends. It has been reported that La antigen, a nuclear protein involved in RNA polymerase III transcription termination [14], binds to the 3′ end of several viral RNAs [15], [16], [17], [18]. The polypyrimidine tract-binding protein (PTB), a 57/60 kDa protein that plays a role in RNA splicing [19], binds to the 3′ end of several viral RNAs [20], [21]. Structural and functional similarities in the replication process among polyadenylated RNA viruses suggest that NV may share some of the elements and follow related strategies to replicate its genome. In order to characterize some elements that could have a role in NV RNA synthesis, the interactions between the 3′UTR, the poly(A) tail, and cellular proteins were analyzed.
The present results indicate that NV 3′UTR with a 24 poly(A) tail (3′UTR(A)) is able to form a stable stem-loop structure of 44 nts as predicted by the mfold-2 software [22]. Using a UV-induced crosslinking assay, 10 proteins from HeLa cells were found to bind to the polyadenylated NV 3′UTR. La from HeLa cells and a recombinant PTB specifically interact with the NV 3′UTR (3′UTR) while the PABP exclusively binds to the poly(A) tail.
Materials and methods
Cells. HeLa cells were grown in Dulbecco’s minimal essential medium supplemented with 10% newborn calf serum, 5000 U penicillin, and 5 μg streptomycin, in a 5% CO2 incubator at 37 °C. The culture medium was changed every other day until the cells reached confluence.
Computer prediction of the secondary structure of NV 3′UTR. Computer analysis of the 3′UTR and 3′UTR(A) of NV was performed using the mfold-2 software [22] in http://www.rpi.edu/~zukerm/.
In vitro transcription of 3′ NV genomic RNAs. Two RNA molecules were synthesized, both containing nt 7588–7654 of the NV 3′UTR genome, (3′UTR), and only one of them having a poly(A) tail of approximately 24 nts (3′UTR(A)). The RNAs were produced by in vitro transcription using T7 RNA polymerase from the two PCR-amplified cDNAs containing the respective regions. The PCR was performed using the complete NV cDNA as template. The sense primer used in the PCR contained the bacteriophage T7 promoter sequence, while the antisense poly(A) primer was used for the addition of the 24 A to the 3′UTR PCR product. The PCR was performed for 35 cycles of 94 °C for 1 min, 42 °C for 1 min, and 75 °C for 30 s, using a Perkin–Elmer Cetus DNA thermocycler. The resulting PCR products were purified by a QIAquiq gel extraction G-50 kit (Qiagen) before they were used as templates for RNA synthesis. After transcription reaction, the DNA template was removed by treating the samples with DNase RQ1 (Promega). Unincorporated nucleotides in the reaction mixture were removed by gel filtration. For synthesis of radiolabeled RNA transcripts, [α-32P]UTP or [α-32P]ATP (Dupont) was included in the transcription reaction.
Preparation of HeLa cell extracts. HeLa cells were prepared using a method previously described [24].
Mobility shift electrophoresis assay. Mobility shift electrophoresis assay was performed using a method previously described [23]. The amount of S10 extract from HeLa cells varied from 5 to 20 μg, while 500 nM of the recombinant PABP (rPABP) was used.
Mobility supershift electrophoresis assay. As much as 1.5 μl of polyclonal anti-PABP and anti-GADPH antibodies was incubated separately with 10 μg of S10 extracts. The antigen-antibody reaction was allowed to proceed for 30 min on ice before addition of the labeled RNA. The RNA–protein supercomplexes were analyzed in a 6% native gel as described before.
UV cross-linking of RNA–protein complex. UV-induced cross-linking assay of RNA-protein complexes was performed using a method previously described [24] in the presence of 40 or 60 μg of S10 extract from HeLa cells and 100 or 500 ng of the recombinant PTB protein.
Immunoprecipitation of UV-induced cross-linked La–protein complexes. Immunoprecipitation of the cross-linked La–protein complex was performed using a method previously described [23]. Five hundred nanograms of monoclonal anti-La or anti-actin antibodies was used.
Results
HeLa cell proteins interact with the 3′UTR of NV
The 66 nt long 3′UTR from NV was able to form a secondary structure as predicted by the mfold-2 program (Fig. 1A ) [22]. The stem-loop structure formed with the last 47 nts has a ΔG=−9.6, suggesting its stability and was not altered in the presence of the 24 nts long poly(A) tail (Fig. 1B). In order to determine if the 3′UTR was able to interact with cellular proteins present in HeLa cell extract, mobility shift assays were performed using [α-32P]UTP labeled RNA representing the complete 3′UTR as a probe. Under this condition, a major RNA–protein complex was observed as a detectable smear (Fig. 2A ). It was also observed that the amount of free RNA decreased when the concentrations of S10 extract increased (Fig. 2A, lanes 2–5). Furthermore, when the RNA–protein complex was treated with RNase before electrophoresis through the native gel, two main complexes were observed (Fig. 2B, lanes 2–5). The major complex, with the fastest migration, was called complex I.
Fig. 1.
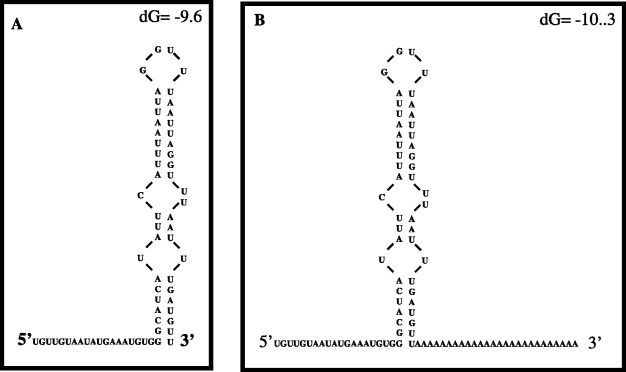
Schematic representation of the NV 3′UTR secondary structure. (A) Predicted secondary structure of the 66 nt long 3′UTR, which forms the complete 3′UTR of NV genome. (B) Predicted secondary structure of the same region plus a 24 nt long poly(A) tail. Both predictions were performed using the mfold-2 software (http://mfold2.wustle.edu).
Fig. 2.
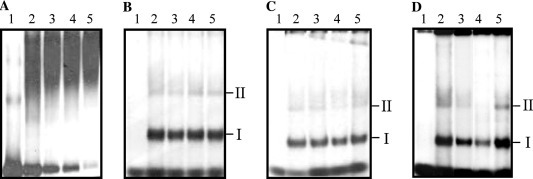
Mobility-shift analysis of the complete 3′UTR of NV and S10 extract from HeLa cell. (A) [α-32P]UTP labeled 3′UTR incubated without (lane 1) or with 5, 10, 15, and 20 μg of S10 extract from HeLa cells (lanes 2–5, respectively). (B) [α-32P]UTP labeled 3′ UTR was incubated without (lane 1) or with 5, 10, 15, and 20 μg of HeLa S10 extract (lanes 2–5, respectively) followed by RNase treatment. (C) [α-32P]UTP labeled 3′UTR RNA was incubated without (lane 1) or with 20 μg S10 extract from HeLa cells (lanes 2–5), in the absence (lanes 2) or, the presence of 0.6, 0.9, and 1.2 M KCl (lanes 3–5, respectively). (D) [α-32P]UTP labeled 3′UTR RNA was incubated without (lane 1) or with 20 μg of S10 extract from HeLa cells (2–5) in the absence (lane 2) or presence of 10- and 20-fold molar excess of homologous (lanes 3 and 4, respectively) or 20-fold molar excess of non-related heterologous competitor (lane 5). Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Mobility of complexes I and II is indicated.
To determine the stability of both RNA–protein complexes, complex formation was performed in the presence of increasing KCl concentrations. Both complexes maintained their migration up to 1.2 M of KCl (Fig. 2C, lanes 2–5), which represents more that 10 times the salt concentration under physiological conditions (approximately 120 nM NaCl). These results suggest that both complexes, I and II, are stable.
The specificity of the RNA–protein binding was further demonstrated in competition experiments (Fig. 2D), using 10- and 20-fold molar excess of unlabeled homologous or a non-related heterologous (referred to as nts 111–191 from NV that do not interact with cellular proteins as has been described before [23]) RNAs as competitors. A 10-fold molar excess of the unlabeled homologous 3′UTR strongly reduced complexes I and II formation, while a 20-fold molar excess of the same competitor completely inhibited complex II formation. However, a 20-fold molar excess of unlabeled non-related heterologous RNA transcript was an inefficient competitor (Fig. 2D, lane 4). These results strongly suggest that the proteins present in HeLa cell S10 extract bound specifically to the 3′UTR RNA.
To determine whether the poly(A) tail was also involved in the complex formation, an [α-32P]UTP labeled 3′UTR(A) was incubated with S10 extract from HeLa cells (Fig. 3A ). Two main complexes, with similar migration patterns to the ones observed with the 3′UTR, were detected (Fig. 3A, lanes 3 and 2, respectively). The complexes formed with both RNAs could be different since the poly(A) tail is a target for the binding of cellular proteins; however, RNP complexes were treated with RNAses, thus neither the poly(A) tail nor the complex formed with it, which were not labeled with [α-32P]UTP, can be detected by autoradiography. To visualize the complexes formed with the poly(A) tail, [α-32P]ATP was used to label both 3′RNAs. Under these conditions, we could detect differences in the complex formation with both RNAs. The 3′UTR and the 3′UTR(A) were able to form five complexes (Fig. 3B, lanes 3 and 2, respectively); complexes I, III, and V formed with the 3′UTR(A) had a similar migration pattern to complexes II, III, and V formed with the 3′UTR, respectively (Fig. 3B, lanes 2 and 3, respectively). However, complex II formed with the 3′UTR(A), which together with complex V are the most prominent complexes formed with this RNA, did not correspond to any complex formed with the 3′UTR. Complexes III and IV were not consistently discernible in subsequent experiments. Complexes formed with the 3′UTR were overexposed with respect to those formed with the 3′UTR(A). These results suggest that the poly(A) tail may influence the formation of the RNA–protein complexes either because it changes the size and conformation of the RNA and/or because it interacts with some other proteins present in the S10 HeLa cell extract.
Fig. 3.
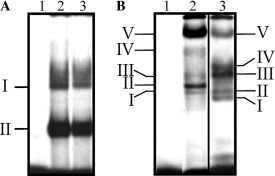
Mobility-shift analysis of the complete 3′UTR and the 3′UTR (A) of NV and S10 extract from HeLa cells. (A) [α-32P]UTP labeled 3′UTR (lanes 1 and 2) or 3′UTR(A) RNAs were incubated in the absence (lane 1) or presence of 20 μg of S10 extract from HeLa cells (lane 2 and 3) followed by RNAse treatment. (B) [α-32P]ATP labeled 3′UTR (lane 3) or 3′UTR(A) RNAs (lanes 1 and 2) were incubated in the absence (lane 1) or presence of 10 μg of S10 extract from HeLa cells (lanes 2 and 3) followed by RNAse treatment. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Mobility of complexes is indicated on both sides of the figure.
To determine if the poly(A) tail was able to interact with some HeLa cell proteins, competition experiments using both unlabeled RNAs and a non-related heterologous RNA were performed. Unlabeled competitor RNAs were preincubated with cytoplasmic extract before the addition of [α-32P]ATP labeled 3′UTR(A). A 25-fold molar excess of unlabeled homologous RNA resulted in an efficient competition of all complexes (Fig. 4A , lane 3), while the addition of 25-fold molar excess of the unlabeled 3′UTR (heterologous RNA) was able to compete efficiently with all complexes, except complex II, which did not compete at all (Fig. 4A, lane 4). Moreover, the amount of radioactivity present in complex II was higher than the amount present in complex II formed without competitor RNAs (Fig. 4A, lanes 4 and 1, respectively). These results suggest that complex II contains proteins, which bind specifically to the poly(A) tail. The addition of a 25-fold molar excess of the unlabeled non-related heterologous RNA was able to compete with complex V, suggesting that this complex is not specific (Fig. 4A, lane 5).
Fig. 4.
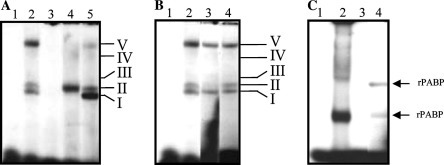
Specificity of RNA–protein complexes formed with the complete 3′UTR and the 3′UTR(A) of NV with S10 extract from HeLa cells. (A) [α-32P]ATP labeled 3′UTR(A) RNA was incubated with 10 μg of S10 extract from HeLa cells in the absence (lanes 1 and 2) or presence of 25-fold molar excess of unlabeled homologous (lane 3), heterologous (lane 4), and non-related heterologous competitors (lane 5). (B) Ten micrograms of S10 extract from HeLa cells was incubated in the absence (lanes 1 and 2) or in the presence of 1.5 μl of polyclonal antibodies to human PAB protein (lane 3) or an anti-GADPH antibody (lane 4) before the addition of [α-32P]ATP labeled 3′UTR(A) RNA. The antibody–RNA–protein supercomplex was further processed under the same conditions described for the RNA–protein complex. (C) [α-32P]ATP labeled 3′UTR RNA (lane 2) or 3′UTR(A) (lanes 1 and 3) was incubated in the absence (lane 1) or presence of 100 ng of recombinant PAB protein (lanes 2 and 3) followed by RNase treatment. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Free RNA was loaded on lane 1. Mobility of complexes formed with S10 HeLa cell extract and with the rPABP is indicated on the right-hand side of the figure.
Identification of cellular proteins present in the RNA–protein complexes
Since PABP interacts with the poly(A) tail of several mRNAs, the possibility that complex II contained the PABP was analyzed using a supershift assay. Incubation with 1.5 μl of polyclonal anti-PABP antibodies resulted in the inhibition of complex II formation (Fig. 4B, lane 3), while the same amount of anti-GADPH antibodies did not alter any complex formation (Fig. 4B, lane 4). The absence of complex II in the presence of the anti-PABP antibodies demonstrated that PABP was present in complex II.
To corroborate that PABP was able to bind to the 3′UTR(A) tail, a mobility shift assay with rPABP was performed (Fig. 4C). Two complexes with different migrations were observed when rPABP bound to the [α-32P]ATP labeled 3′UTR(A) (Fig. 4C, lane 3), but none of them were formed with [α-32P]ATP labeled 3′UTR (Fig. 4C, lane 2), indicating that the rPABP was able to specifically bind to the poly(A) tail. It has been reported that PABP can bind to the PV 3′UTR with a poly(A) tail containing a minimum of eight nts and that its affinity increases as the poly(A) tail lengthens [12]. The poly(A) tail present in the 3′UTR(A) is 24 nt long, therefore more than one PABP molecule can bind to this sequence. The two complexes observed after the RNAse could contain different defined amounts of rPABP. The expression and purification of the rPABP is described by Herold and Andino [12].
NV 3′UTR interacts with PTB and La proteins
Mobility shift assays demonstrated that both the 3′UTR and 3′UTR(A) RNAs were able to form RNA–protein complexes with some HeLa cell proteins. Next, UV-cross-linking experiments were performed in an attempt to identify some of them. Ten proteins from HeLa cell extract, with apparent molecular masses of 110, 97, 85, 75, 68, 60, 52, 50, 39, and 33 kDa, bound to both RNAs (Fig. 5A , lanes 2 and 3, respectively). All the bands observed in this assay were disrupted after proteinase K treatment (data not shown). It was not possible to detect any differences in the proteins crosslinked to both RNAs; however, a higher amount of radioactivity was observed in the 68 kDa protein crosslinked to the 3′UTR(A) (7%), compared to the same band crosslinked to the 3′UTR RNA. The increased amount of radioactivity in the 68-kDa band, could correspond to the PABP that binds to the poly(A) tail. To examine this possibility, a 50-fold molar excess of the homologous and the heterologous RNAs was used. A strong competition of all the proteins was observed with both RNAs (Fig. 5B, lanes 4 and 3, respectively); however, the 68-kDa protein competed only with the homologous unlabeled RNA (Fig. 5B, lane 4). These results suggest that the presence of the poly(A) tail was responsible for the competition of an extra band of 68-kDa, which co-migrates with the 68 kDa band bound to the [α-32P] ATP labeled 3′UTR(A) RNA. The molecular mass of this protein co-relates with the PABP molecular mass. No competition was observed when the non-related heterologous RNA was used (Fig. 5B, lane 5).
Fig. 5.
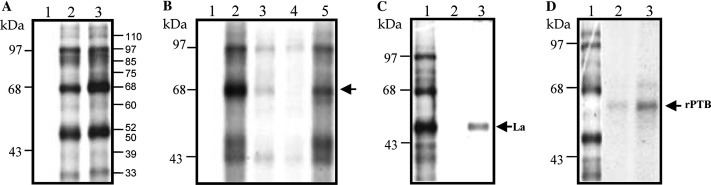
UV-induced cross-linking of the complete 3′UTR and the 3′UTR (A) of NV and S10 extract from HeLa cells. (A) [α-32P]ATP labeled 3′UTR (lanes 1 and 2) and 3′UTR(A) (lane 3) RNAs were UV-crosslinked without (lane 1) or with 60 μg of S10 extract from HeLa cells (lanes 2–3). (B) Forty micrograms of S10 extract from HeLa cells were preincubated 15 min with 25-fold molar excess of heterologous (lane 2), homologous (lane 3), and non-related heterologous (lane 5) RNAs prior to the addition of [α-32P]ATP labeled 3′UTR(A). Free RNA was loaded on lane 1. (C) [α-32P]UTP labeled 3′UTR RNA was cross-linked with 60 μg of S10 extract from HeLa cells (lane 1) and immunoprecipitated with anti-La (lane 3) or anti-actin antibodies (lane 2). (D) [α-32P]UTP labeled 3′UTR RNA was UV-cross-linked with 60 μg of S10 extract from HeLa cells (lane 1) or 100 and 500 ng of recombinant PTB protein (lanes 2 and 3, respectively). After UV-cross-linking, the reaction was followed by RNase treatment. Crosslinked proteins were loaded on an SDS–10% polyacrylamide gel and detected by autoradiography. The migration of the UV-crosslinked 68 kDa, rPTB, and the immunoprecipitated La proteins is indicated by an arrow.
Some of the proteins that crosslinked to both 3′UTR and 3′UTR(A) RNAs have the same molecular weight as a number of factors that interact with the 5′- and 3′UTRs of other RNA viruses. Specifically, the proteins with molecular masses of 60 and 52-kDa co-relate with the cellular proteins PTB and La, respectively.
To investigate if La protein could be one of the proteins that crosslinked to the 3′UTR, an immunoprecipitation after the UV-induced crosslinking assay was performed. A labeled 52 kDa protein, crosslinked to the 3′UTR, was immunoprecipitated by anti-La antibodies (Fig. 5C, lane 3), while anti-actin antibodies were unable to immunoprecipitate any labeled protein (Fig. 5C, lane 2). This fact demonstrated that La was one of the proteins that interacted with the 3′UTR of NV. To analyze the possible interaction of PTB with the NV 3′UTR, labeled 3′UTR was incubated with a recombinant PTB (rPTB) protein (Fig. 5D). rPTB was able to bind to the 3′UTR at 100 and 500 ng (Fig. 5D, lanes 2 and 3, respectively). However, this rPTB showed a migration slightly different from that of the 57/60 kDa protein from HeLa cell extracts crosslinked to the same RNA (Fig. 5D, lanes 1, and 2–3, respectively) as previously described [23]. These results strongly suggest that the 3′UTR contains structural elements that permit the interaction with La and rPTB.
Discussion
Most of the mechanisms and factors involved in NV translation and replication remain unknown; however, as a positive-stranded RNA virus, the viral genome must function as a mRNA for viral protein synthesis and as a template for viral negative-strand RNA synthesis. The initiation of the negative-strand RNA synthesis takes place at the 3′ end of the positive polarity RNA and requires the presence of several elements, such as structured sequences (that often consist of multiple discontinuous sequences of the viral RNA, like the 3′ and 5′UTRs and/or coding regions of the RNA molecule) that serve as cis-acting elements or targets for the interaction of several viral and cellular factors [7], [8], [15], [25], [26]. These RNA–protein complexes have been involved in both viral translation and RNA replication. In the replication process, the presence of cellular proteins could help to recruit and stabilize the RdRp on the initiation sites for viral RNA synthesis [15].
In polyadenylated viruses, such as picornavirus and coronavirus, the poly(A) tail is a specific target sequence for the interaction with the PABP. This poly(A) sequence has been considered one of the major determinants of initiation for negative strand RNA synthesis with a specific role in organizing proteins around the start site and mediating circularization of the RNA molecule [12], [26], [27], [28]. Genome circularization has a critical role in the coordination of translation and RNA synthesis and in the location of the RdRp at the appropriate start site [12].
In this study, evidence that the NV polyadenylated 3′UTR contains the sequences required for the interaction with various cellular proteins is provided.
Using the mfold-2 software, it was possible to predict that the 66 nts present in the complete NV 3′UTR contained a stem-loop structure formed in the last 47 nts. It was demonstrated that in the absence of RNAse, the 3′UTR was able to form a large RNA–protein complex with HeLa cell proteins. When RNP complexes were treated with RNases, two RNA–protein complexes were distinguished. The complete 3′UTR was able to form stable RNA–protein complexes with cellular factors from HeLa S10 extract since they were not disrupted by increasing concentrations of KCl.
The specificity of the complex formation was analyzed in competition experiments performed in the presence of an excess of homologous or heterologous RNAs. A total reduction of complex II and a significant reduction of complex I formed with the 3′UTR and HeLa cell S10 extract were observed when samples were incubated in the presence of homologous, but not heterologous competitor, strongly suggesting that both complexes are specific.
Since the poly(A) tail is an important element for organizing proteins around the start site of negative strand RNA synthesis [12], and it is the binding site for the PABP, we wanted to find out if the presence of the poly(A) tail added to the 3′UTR modified its interaction with HeLa cell proteins. We could not detect any difference in the RNP complexes formed with the [α-32P]UTP 3′UTR or 3′UTR(A), even though the poly(A) tail is a target for the binding of cellular proteins. This finding could be the result of the RNAse treatment, where the poly(A) tail, which was not labeled with the [α-32P]UTP, alone or bound to cell proteins, could be separated from the labeled complexes and consequently was not detected by autoradiography.
To visualize the complexes formed with the poly(A) tail, [α-32P]ATP was used to label both 3′UTR and 3′UTR(A) RNAs. Under this condition, we detected the formation of five complexes with both RNAs, some of them with the same migration patterns, but complex II, formed with the 3′UTR(A), did not correspond to any complex formed with the 3′UTR. Then complex II could be formed by the poly(A) tail bound to proteins present in HeLa cell extract. Since the poly(A) tract is the target site for the PABP interaction, competition experiments of complexes formed with [α-32P]ATP labeled 3′UTR(A) were performed. We could demonstrate that complex II contains proteins that specifically bind to the poly(A) tail, since this complex was not completed with the 3′UTR RNA. Moreover, the presence of the PABP in complex II was confirmed in mobility gel supershift assays using anti-PABP antibodies. In addition, an rPABP was also able to specifically bind to the 3′UTR(A) RNA; but it did not bind to the 3′UTR RNA. The two bands in the mobility shift assay can be explained due to the presence of the 24 nts long tail that could permit the formation of a RNP complex with more than one PABP molecule [12]. Then RNAse treatment could generate two different complexes containing different defined amounts of rPABP.
Using crosslinking assays, 10 proteins from 110 to 33 kDa bound to both 3′UTR and 3′UTR(A) RNAs were detected. However, no difference between both patterns at the usual electrophoretic migration of PABP (68–72 kDa) was found. Even so, the 68 kDa band that crosslinks with the NV3′UTR(A) contains more that one protein, since the competition experiments using an excess of homologous RNA competed completely with this band; while the heterologous RNA, which does not contain the poly(A) tail, only competed partially, suggesting that the 68 kDa band contains at least one protein that specifically binds to the poly(A) tail sequence. Experiments to identify the 97 and the 68-kDa protein (other than the PAPB) are currently being performed.
Besides the finding of PABP bound to the 3′UTR(A), two other cellular proteins, La and rPTB, were identified by immunoprecipitation and crosslinking assays, respectively. This is an interesting finding because both proteins are also associated to some viral functions: La has an essential role in PV translation [29] while PTB is necessary for PV and hepatitis C (HCV) translation [16], [30] and mouse hepatitis virus RNA synthesis [31].
Even though there is no evidence about the mechanisms that operate in NV replication, the work reported here provides new data pointing towards the identification of some of the elements that could have a role in these processes, such as the presence of a stem-loop structure within the polyadenylated 3′UTR of NV genomic RNA, and the identification of some of the cellular proteins that specifically bind to this region, including La, PTB, and PABP. The identification of these cis- and trans-elements, considered molecules that have essential roles in translation and replication of other positive-sense RNA viruses, can help understand the specific mechanisms that take place in NV replication.
Further analyses, including functional studies, are currently being performed to assess the elements and strategies followed by NV to translate and replicate its RNA genome.
Acknowledgements
We thank Rosa M. del Angel for many helpful suggestions, and Leopoldo Flores for critical comments on the manuscript, Fernando Medina for the cell cultures, and Mónica DeNova for the recombinant PTB protein. We gratefully acknowledge Y. Svitkin and N. Sonenberg (McGill University, Montreal, Quebec, Canada) for the anti-La and anti-PABP antibodies, M. Hernández (CINVESTAV, México) for the anti-actin antibodies, R. Andino (University of California, San Francisco, USA) for the recombinant PABP protein, and X. Jiang (Cincinnati Children’s Medical Hospital Center, Ohio) for the NV cDNA. Melina Várzquez Ochoa and Javier Hernńandez-Acosta had a scholarship from Consejo Nacional de Ciencia y Tecnología. This work was supported by Grant 34983-N from Consejo Nacional de Ciencia y Tecnología.
References
- 1.Fields N.B., Knipe D.M., Howley P.M., editors. Human Caliciviruses. Lippincott, Williams, and Wilkins (Ed.); Philadelphia, USA: 2001. pp. 841–874. (Virology, fourth ed.). [Google Scholar]
- 2.Frankhouser R.L., Noel J.S., Monroe S.S., Ando T., Glass R.I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 3.Subekti D.S., Tjaniadi P., Lsmana M., McArdle J., Iskandriati D., Budiarsa I.N., Walujo P., Suparto H.I., Winoto I., Campbell J.R., Porter K.R., Sajuthi K.D., Ansari A.A., Oyofo B.A. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 2002;66:400–406. doi: 10.1002/jmv.2159. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X., Wang X.M., Wang K., Estes M.K. Sequence and genomic organization of norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 5.Clarke N., Lambden P.R. The molecular biology of caliciviruses. J. Gen. Virol. 1997;78:291–301. doi: 10.1099/0022-1317-78-2-291. [DOI] [PubMed] [Google Scholar]
- 6.Glass P.J., White J.L., Ball J.M., Leparc-Goffart I., Hardy M.E., Estes M.K. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 2000;74:6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diez J.M., Ishikawa, Kaido M., Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA. 2000;97:3918–3931. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamarnik V., Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 9.Hill K.R., Hajjou M., Hu J.Y., Raju R. RNA–RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 1997;71:2693–2704. doi: 10.1128/jvi.71.4.2693-2704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao C., Singh P., Ecket D.J. De Novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 11.Spector D.H., Boltimore D. Requirement of 3′ terminal poly (adenylic acid) for the infectivity of poliovirus RNA. Proc. Natl. Acad. Sci. USA. 1974;71:2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. Biol. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sachs, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 14.Gottlieb E., Stelitz J.A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M.M.C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;213:12–22. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 16.Kurilla M.G., Keene J.D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1983;34:837–845. doi: 10.1016/0092-8674(83)90541-x. [DOI] [PubMed] [Google Scholar]
- 17.Wilusz J., Kurilla M.G., Keene J.D. A host protein (La) binds in a unique species of minus-sense leader RNA during replication of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 1983;80:5827–5831. doi: 10.1073/pnas.80.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurilla M.G., Cebradilla C.D., Holloway B.P., Keene J.D. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J. Virol. 1984;50:773–778. doi: 10.1128/jvi.50.3.773-778.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C.H., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 20.Huang P., Lai M.M. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA confirmation. J. Virol. 1999;71:7567–7578. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T., Lai M.M. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Computer prediction of RNA structure. RNA processing. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez-Escolano L., Uribe-Brito Z., del Angel R.M., Jiang X. Interaction of cellular proteins with the 5′ end of Norwalk virus genomic RNA. J. Virol. 2000;74:8558–8562. doi: 10.1128/jvi.74.18.8558-8562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutiérrez-Escolano L., DeNova-Ocampo M.A., Racaniello V.R., del Angel R.M. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J. Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilipenko E.V., Poperechny K.V., Maslova S.V., Melchers W.J., Slot H.J., Agol V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by ternary (“kissing”) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 26.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spagnolo J., Hogue B. Host protein interactions with the 3′ end of Bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold J., Andino R. Poliovirus requires a precise 5′ end for efficient positive strand RNA synthesis. J. Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowics F., Kenan D.J., Chan E.K.L., Agol V.I., Keene J.D., Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellen U., Witherell G.W., Schmid M., Shin H.S., Pestova T.V., Gil A., Wimmer E. A cytoplasmic 57 kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA. 1993;90:7642–7644. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H.P., Huang P., Park S., Lai M.M.C. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


