Abstract
Coronavirus spike proteins mediate host-cell-attachment and virus entry. Virus replication takes place within the host cell cytosol, whereas assembly and budding occur at the endoplasmic reticulum-Golgi intermediate compartment. In this study we demonstrated that the last 39 amino acid stretches of Alphacoronavirus spike cytoplasmic domains of the human coronavirus 229E, NL63, and the porcine transmissible gastroenteritis virus TGEV interact with tubulin alpha and beta chains. In addition, a partial co-localization of TGEV spike proteins with authentic host cell β-tubulin was observed. Furthermore, drug-induced microtubule depolymerization led to changes in spike protein distribution, a reduction in the release of infectious virus particles and less amount of spike protein incorporated into virions. These data demonstrate that interaction of Alphacoronavirus spike proteins with tubulin supports S protein transport and incorporation into virus particles.
Keywords: Coronavirus, Spike protein, Microtubule, Assembly, Viral infectivity, Intracellular transport, Interaction, Incorporation, TGEV
Highlights
-
•
The cytoplasmic domain of coronavirus S proteins interacts with tubulin.
-
•
Microtubule depolymerization influences S protein distribution.
-
•
Viral titers are reduced after microtubule depolymerization.
-
•
S protein incorporation into virus particles depends on intact microtubule.
1. Introduction
Coronaviruses (CoVs) are positive single-stranded RNA viruses. They infect birds and mammals, especially their respiratory and gastrointestinal systems. Due to high mutation and recombination rates in coronaviruses frequent host-shifting events from animal-to-animal and animal-to-human have occurred (Chinese, 2004, Guan et al., 2003, Lau et al., 2005, Woo et al., 2009). Bats were identified as a natural reservoir during the severe acute respiratory syndrome (SARS) outbreak in China in the year 2002–2003 (Ge et al., 2013, Li et al., 2005). Similarly, a strain closely related to human CoV 229E was found in bats in Ghana (Corman et al., 2015, Pfefferle et al., 2009). Thus, it is speculated that several human and animal CoVs originated from bats (Liu et al., 2015, Shi and Hu, 2008, Woo et al., 2009).
The CoV spike (S) glycoprotein, a key determinant for host range, is necessary for receptor binding and membrane fusion (Graham and Baric, 2010). The S protein contains a large ectodomain, a transmembrane domain, and a C-terminal cytoplasmic tail. The cytoplasmic domain consists of a cysteine-rich and a charge-rich region and mediates S incorporation into virions resulting in infectious virus particles (Bosch et al., 2005, Godeke et al., 2000). Some CoVs like the transmissible gastroenteritis virus (TGEV), as well as the HCoVs NL63 and 229E contain a tyrosine-based sorting signal within their charge-rich region which – in the case of the TGEV S protein – was shown to be important for intracellular retention (Schwegmann-Wessels et al., 2004).
Free movement of virus particles through the host cell cytoplasm is restricted. The cytosol is highly viscous and contains structural barriers like organelles as well as cytoskeletal elements (Leopold and Pfister, 2006, Luby-Phelps, 2000, Verkman, 2002). Therefore, diffusion of virus-sized particles is very unlikely and the arrival at specific cellular regions or compartments is nearly impossible (Sodeik, 2000). Consequently, many viruses use the cytoskeleton network of the host cell as a transport system to reach the compartment where replication processes take place or to find their way out of the cell (Leopold and Pfister, 2006, Ploubidou and Way, 2001, Radtke et al., 2006). Furthermore, it is known that the cytoskeleton plays a crucial role during virus attachment, internalization, endocytosis, transcription, replication, assembly, exocytosis as well as cell-to-cell spread. For this purpose viruses rearrange cellular filaments and use them as tracks (Radtke et al., 2006). Regarding TGEV, the actin-binding protein filamin A is a putative interaction partner of the TGEV S protein (Trincone and Schwegmann-Wessels, 2015) and in the case of TGEV-infected IPEC-J2 cells actin filaments were shown to be important for viral replication and release (Zhao et al., 2014). Additionally, an interaction of TGEV nucleocapsid protein with the type 3 intermediate filament vimentin was shown to be required for viral replication as well (Zhang et al., 2015). Another major component of the dynamic cytoskeletal matrix is represented by microtubules. Those polarized structures are built of α/β-tubulin heterodimers and are important for cell shape, transport, motility, and cell division (Heald and Nogales, 2002, Nogales, 2000). Several viruses are known to interact with tubulin or their molecular motors like kinesin or dynein (Biswas and Das Sarma, 2014, Han et al., 2012, Hara et al., 2009, Henry Sum, 2015, Hsieh et al., 2010, Hyde et al., 2012). CoVs like a demyelinating strain of mouse hepatitis virus (MHV) use microtubules for neuronal spread and the feline infectious peritonitis virus (FIPV) is transported via microtubules towards the microtubule organizing center (Biswas and Das Sarma, 2014, Dewerchin et al., 2014). For TGEV an up-regulation of microtubule-associated α- and β-tubulin was detected in swine testis (ST) cells after infection (Zhang et al., 2013). In the study presented here, we analyzed the interaction of tubulin with the last 39 amino acid stretches of the S protein cytoplasmic tail of Alphacoronaviruses like TGEV, HCoV NL63, and HCoV 229E. Our results show that tubulins interact with the cytoplasmic domain of α-CoVs spike proteins. Reduced release of infectious virus particles as well as differentially distributed S proteins was observed after drug-induced tubulin depolymerization. Therefore, tubulin may help the S protein to be properly transported, localized, and assembled into virions.
2. Material and methods
2.1. Cell lines and virus strains
Human embryonic kidney cells (HEK-293) were used for co-immunoprecipitation via GFP Trap® pull down assay and ST cells were used for immunofluorescence analysis and plaque assay. Both cell lines were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. Hypsignathus monstrosus kidney cells (HypNi/1.1) (Kuhl et al., 2011) and Pipistrellus pipistrellus kidney cells (PipNi/1) (Muller et al., 2012) (provided by Marcel Müller) were used for immunofluorescence analysis and were grown in DMEM supplemented with 5% fetal calf serum.
The Purdue strain of TGEV (PUR46-MAD, provided by L. Enjuanes) was propagated in ST cells. After 24 h of incubation at 37 °C the supernatants were harvested, centrifuged and after addition of 1% fetal calf serum stored at −80 °C.
2.2. Plasmids
Fusion proteins of the last 39 amino acid (aa) stretches of TGEV-S, SARS-CoV-S, HCoV-NL63-S, and HCoV-229E-S cytoplasmic domains with GFP were constructed (S-39aa-GFP-CT and S-39aa-GFP-NT) by using Invitrogen/Life Technologies Gateway cloning ( Table 1).
Table 1.
Last 39 amino acid stretches of coronavirus cytoplasmic domains linked to GFP.
| TGEV | CCCSTGCCGCIGCLGSCCHISCSRRQFENYEPIEKVHVH |
| HCoV NL63 | CLSTGCCGCCNCLTSSMRGCCDCGSTKLPYYEFEKVHVQ |
| HCoV 229E | LCCCSTGCCGFFSCFASSIRGCCESTKLPYYDVEKIHIQ |
| SARS-CoV | CCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT |
Transmissible gastroenteritis virus (TGEV), human coronavirus (HCoV), severe acute respiratory syndrome coronavirus (SARS-CoV).
Full length TGEV S wildtype was fused to GFP named TGEV Swt-GFP. The full length mutant of TGEV S where the tyrosine at position 1440 is exchanged by an alanine fused to GFP is named TGEV S Y/A-GFP. Tubulins C-terminally tagged with an HA (YPYDVPDYA) peptide (TUBB2-HA, TUBB4A-HA, TUBB6-HA, TUBA4A-HA) were used for co-immunoprecipitation. As compartment markers a GFP-tagged ERGIC-53 and galactosyltransferase were used (Winter et al., 2008). Full length TGEV M cDNA was transfected for co-localization studies. Full length TGEV nucleocapsid (N) cDNA and empty GFP plasmid served as negative control.
2.3. GFP Trap® pull down assay and SDS-PAGE
To determine interaction partners of the S protein cytoplasmic domain, HEK-293 cells were seeded on 10 cm dishes and transfected with empty GFP vector or with S-39aa-GFP-CT/NT fusion protein one day later by using Lipofectamine® 2000 (Life Technologies) following the manufacturer's instructions.
For co-immunoprecipitation, the cells were additionally co-transfected with the tubulin candidates tagged with the HA-peptide by using Polyethylenimine (PEI 1 µg/µl, Polysciences). DNA (24 µg) was mixed with 3 ml Opti-MEM (Life Technologies) and incubated for 5 min. Following this, 20 µl of PEI was added and incubated for 15 min. Then this mixture was added dropwise to the cells and incubated overnight.
HEK-293 cells were lysed in NP-40 lysis buffer in the presence of complete™ (protease inhibitor cocktail, Roche Diagnostics, Mannheim) for 30 min on ice followed by centrifugation for 10 min at 20,000g and 4 °C. ChromoTek GFP-Trap® which consists of agarose beads coated with antibodies derived from alpaca against GFP was used for both, the general screening method as well as specific interaction studies between tubulins and S proteins. The purification was done as described in the manufacturer's protocol. For the interaction experiments, the NaCl concentration of the washing buffer was increased from 150 mM to 300 mM to avoid unspecific pull down of tubulins. Instead of 100 µl 2×SDS sample buffer 25 µl 5×SDS sample buffer was used. Cell lysates and eluates were subjected to SDS-PAGE. By a semi-dry technique (Kyhse-Andersen, 1984) the proteins in the gel were transferred to nitrocellulose membranes (GE Healthcare) which were subsequently blocked in 5% milk powder in TBS for one hour and then incubated with the first antibody overnight at 4 °C (rat-anti-HA antibody 1:300 provided by E. Kremmer; rat-anti-GFP antibody 1:1000, Chromotek). The next day, the membrane was washed 3 times with TBS-T for 10 min and treated with the secondary antibody (HRP-conjugated donkey-anti-rat antibody 1:10,000, Sigma-Aldrich) for 1–2 h. Afterwards, the membrane was washed 3 times in TBS-T for 10 min and once in TBS for 5 min. The blot was exposed to Millipore Immobilon™ Western Chemiluminescent HRP substrate (Fischer Scientific) and visualized in a GelDoc documentation system. Regarding the general screening for interaction partners of the S protein, the SDS gel was fixed and then stained with Coomassie solution. After destaining of the gel, visible bands were cut out, digested, and analyzed by mass spectrometry.
2.4. In gel digestion and mass spectrometry
The excised gel pieces were equilibrated with 25 mM NH4HCO3 and subsequently incubated for 16 h with trypsin (25 ng/µl) as described previously (Thyrock et al., 2013). The resulting proteolytic peptides were extracted by sequential agitation with 200 µl of 25 mM NH4HCO3, 50% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA), 80% ACN/0.1% TFA, and neat ACN. The supernatants were combined and dried in vacuo. Desalting was performed by use of ZipTip C18-tips as per description (Thyrock et al., 2013).
Analysis of purified peptides was performed by use of Synapt GS2 instrument (Waters, Manchester, UK) equipped with a Z-spray source in the positive ion sensitivity mode. Source parameters were: source temperature: 80 °C, capillary voltage: 0.8 kV, sampling cone voltage: 20 V, and source offset voltage: 50 V. Peptide sequences were deduced from fragment ion spectra derived from nanoESI ion mobility spectrometry (IMS) low energy CID experiments (Thyrock et al., 2013).
2.5. Immunofluorescence
ST cells used for immunofluorescence were transfected with TGEV Swt-GFP, TGEV S Y/A-GFP or with the S-39aa-GFP-CT fusion protein of TGEV, NL63, 229E or SARS-CoV by the help of ICAfectin™ 441 (Eurogentec) following the manufacturer's instructions (with 0.75 µl ICAfectin™ 441 per well) or alternatively using Lipofectamine® 2000 (Life Technologies).
HypNi/1.1 and PipNi/1 cells were seeded on 24-well plates and transfected with TGEV Swt-GFP or S-39aa-GFP-CT of TGEV and SARS-CoV using Lipofectamine® 2000 (Life Technologies) following the manufacturer's instructions. Cells expressing GFP alone or the TGEV nucleocapsid (N) protein served as negative control. Three hours post transfection (hpt) the cells were mock-treated with 0.2% dimethylsulfoxide (DMSO) or with 10 µg/ml Nocodazole (NOC, 5 mg diluted in 1 ml DMSO) for 3 h, following medium change. One hour later, i.e. 7 hpt, the cells were fixed with 3% paraformaldehyde (PFA) in PBS. Cells were permeabilized with 0.2% Triton/PBS for 5 min. To detect the authentic tubulin a monoclonal Cy3 conjugated anti-β-tubulin antibody produced in mice (1:500, Sigma-Aldrich) was used.
ST, HypNi/1.1, and PipNi/1 cells were infected by TGEV (MOI of 1.5) for 1 h at 37 °C. In the case of HypNi/1.1 and PipNi/1 cells, transfection with cDNA encoding the cellular receptor for TGEV, porcine aminopeptidase N (pAPN), was performed one day prior to infection. One hour post infection, the cells were mock-treated with 0.2% DMSO or with 10 µg/ml NOC for 3 h followed by medium change. Seven hours post infection (hpi) the cells were fixed with 3% PFA and permeabilized. TGEV S protein was detected with monoclonal antibody 6 A.C3 (1:200, provided by L. Enjuanes). As secondary antibody Alexa Fluor® 568 or Alexa Fluor®488 anti-mouse (1:1000, Life Technologies) was used. Nuclei were stained with DAPI. ST cells expressing the compartment markers were transfected with the specific cDNAs one day before infection. TGEV M protein encoding cDNA was transfected and detected 22 hpt with monoclonal antibody 9D.B4 (1:200, provided by L. Enjuanes). As secondary antibody Cy3-conjugated anti-mouse (1:1000, Sigma Aldrich) was used. Immunofluorescence analyses were done by confocal microscopy using Leica TCS SP5.
2.6. Plaque assay
ST, HypNi/1.1, and PipNi/1 cells were seeded on 6-well plates and the chiropteran cells were transfected with cDNA encoding for pAPN one day post seeding. Cells were infected by TGEV (MOI 1.5) one day after transfection. After incubation for 1 h at 37 °C, cells were washed tree times with medium and then cultured with DMEM plus 3% fetal calf serum at 37 °C. Supernatants (100 µl) were collected at time point 0 and 24 hpi. Additionally, the cells were either treated with 0.2% DMSO or incubated with 10 µg/ml NOC at different time points (1 h before infection, during infection, directly after infection). Regarding HypNi/1.1 and PipNi/1 cells, NOC treatment was only done after infection. ST cells were seeded on 96-well plates and inoculated with 40 µl of the collected supernatants for 1–2 h at 37 °C. Afterwards, inoculum was discarded and cells were treated with methylcellulose overnight. The next day cells were fixed with 3% PFA and treated with antibodies against viral proteins (same as for IFA). By using the Nikon Eclipse Ti microscope plaque forming units were counted for each time point.
2.7. Virus particle assay
ST cells were seeded on 100 mm cell culture dishes. One day later cells were infected by TGEV (MOI 0.1 for 1 h at 37 °C). After washing, the cells were mock-treated with 0.2% dimethyl sulfoxide (DMSO) or treated with 10 or 50 µg/ml Nocodazole to depolymerize the microtubuli. Cell culture supernatant was ultracentrifuged 24 hpi at 200,000g in a SW 41 rotor (Beckman Coulter) for 1 h at 4 °C. The virus particle pellet was solubilized in 50 µl 2×SDS sample puffer (non-reducing conditions, no heating) and subjected to SDS-PAGE. In parallel, ST cells were lysed in NP-40 lysis puffer in the presence of cOmplete™ (protease inhibitor cocktail, Roche Diagnostics, Mannheim) and subjected to SDS-PAGE as well (non-reducing conditions). Separated proteins were transferred to nitrocellulose membranes (GE Healthcare) which were subsequently blocked with 1% blocking reagent (Roche) in blocking buffer (100 mM maleic acid, 150 mM NaCl, pH 7.5) overnight at 4 °C. After washing the membrane 3 times with phosphate buffered saline +0.1% Tween (PBS+T) and once with PBS the nitrocellulose membrane was treated with the antibody against the TGEV S protein (mAb 6 A.C3, 1:200), the TGEV M protein (mAb 9D.B4, 1:200) or the TGEV N protein (mAb FIPV3-70, 1:1000, Thermo Scientific) for 1 h at 4 °C. After washing, the membranes were treated with anti-mouse peroxidase-conjugated antibody (Dako; 1:1000) for 1 h at 4 °C. Chemiluminescent peroxidase substrate (Thermo Scientific) and the Chemi Doc system (Biorad) were used for chemiluminescence signal detection.
3. Results
Potential interaction partners of the cytoplasmic domain of TGEV-S were identified using a screening method involving a GFP Trap pull down assay followed by SDS-PAGE and Coomassie staining. In addition to the GFP fusion protein three clear bands were visible. These detected bands were identified by mass spectrometry. The first band at around 70 kDa contained fragments that matched to heat shock 70 kDa protein 1A and 8. Fragments of the second band at around 55 kDa referred to the pro-alpha chain of collagen type I, although this size did not fit to the expected one for COL1A1. A band at around 50 kDa matched to the group of tubulin beta chains (TUBB1, TUBB2A, TUBB2B, TUBB3, TUBB4A, TUBB4B, TUBB5, TUBB6 and TUBB8). As representatives of tubulin beta chains the three genes TUBB2A, TUBB4A, and TUBB6 were chosen for further analysis. For comparison one gene encoding a tubulin alpha chain, TUBA4A, was enclosed in the study. The ORF's of the chosen genes were cloned and tagged with HA.
3.1. Co-immunoprecipitation of tubulins with the TGEV S protein cytoplasmic domain
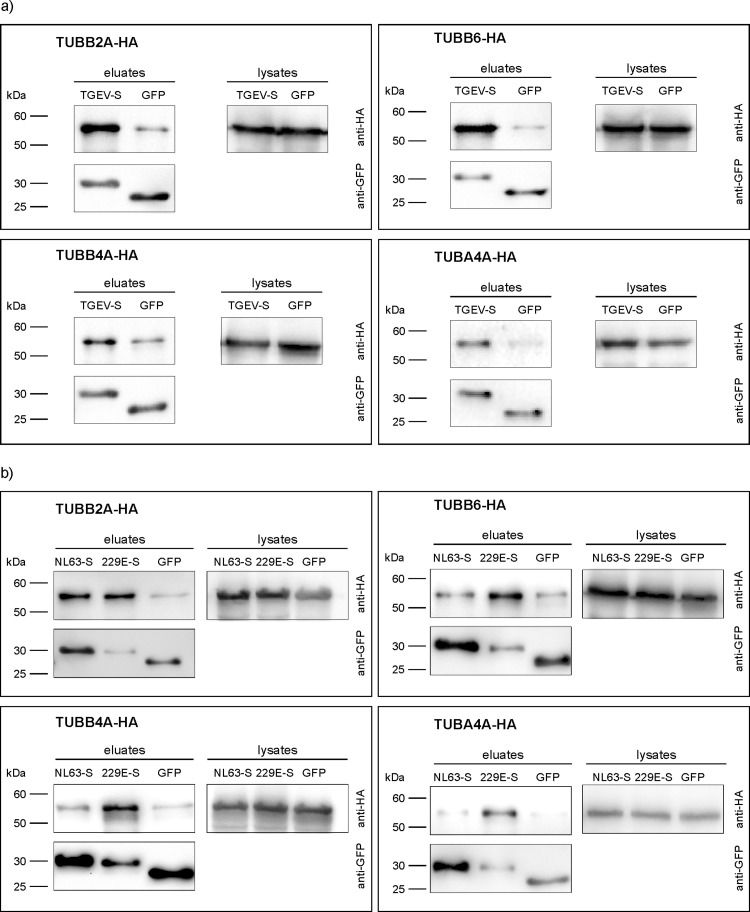
To validate the interaction of the 39 amino acid stretch of the TGEV S protein cytoplasmic domain with the different tubulin chain proteins a co-immunoprecipitation experiment was performed ( Fig. 1a). After co-transfection of HA-tagged tubulins and TGEV-S-39aa-GFP-NT purification was done via GFP Trap® pull down assay. As negative control, cells co-expressing tubulin candidates and the empty GFP vector were used. All four tubulin-HA proteins were detected in the lysates at similar quantity at the expected sizes of 50–55 kDa. Considering the eluates, strong signals for tubulin-HA were detected in cases of the co-expression of TGEV-S-39aa-GFP-NT and tubulins. In the negative control weak bands of unspecific binding were visible. Similar protein quantities of HA-tagged tubulins in the cell lysates as well as similar quantities of GFP in the eluates at 25–30 kDa strengthen the specific interaction between TGEV-S and tubulin.
Fig. 1.
Co-immunoprecipitation of alphacoronavirus S fusion proteins and different tubulins via GFP-Trap®. S constructs were fused to GFP and precipitated by the help of anti-GFP-coated beads and detected by anti-GFP antibodies (lower panels, at around 30 kDa). Tubulins were tagged with HA peptide and detected by anti-HA antibodies in the cell lysates and the precipitated eluates (upper panels, at around 55 kDa). Co-immunoprecipitation with TGEV-S-GFP-NT (TGEV-S) fusion protein (a); co-immunoprecipitation with human coronavirus NL63-S-GFP-NT (NL63-S) or 229E-S-GFP-NT (229E-S) fusion protein (b).
3.2. Co-immunoprecipitation of tubulins with corresponding parts of the human CoV-229E and CoV-NL63 S protein cytoplasmic domains
Similar to TGEV-S-39aa-GFP-NT, the 39aa stretches of HCoV-229E-S and HCoV-NL63-S cytoplasmic tail were fused to GFP. Afterwards, cells were co-transfected with the GFP fusion constructs and the HA-tagged tubulins followed by GFP Trap® pull down assay, SDS-PAGE, and Western blot (Fig. 1b). TUBB2A was purified by these methods and clearly confirmed as interaction partner. TUBB6, TUBB4A, and TUBA4A were co-precipitated in cells co-transfected with 229E S-39aa-GFP-NT but not with NL63 S-39aa-GFP-NT. Here, the band for TUBB6-HA, TUBB4A, and TUBA4A in the NL63 S-39aa-GFP-NT eluate was as weak as for the negative control.
3.3. The TGEV Swt full length protein partly co-localizes with authentic cellular β-tubulins
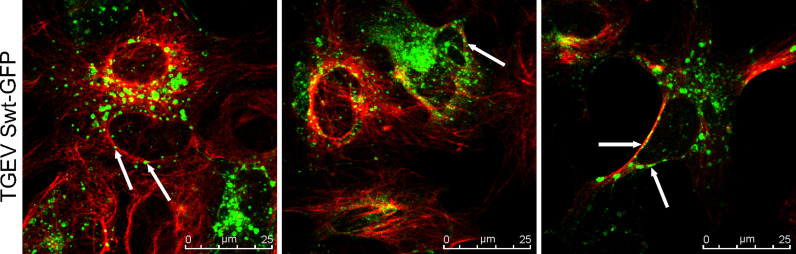
As the interaction of the TGEV S cytoplasmic tail with tubulins was confirmed by co-immunoprecipitation, confocal microscopy was done to localize TGEV Swt full length protein and host cell β-tubulin ( Fig. 2). ST cells expressing the TGEV Swt-GFP were fixed and incubated with an antibody against the authentic β-tubulins. A partial co-localization of the TGEV Swt-GFP protein with β-tubulin was observed (Fig. 2, white arrows).
Fig. 2.
Co-localization study of TGEV Swt proteins and authentic β-tubulin. ST cells transfected with TGEV-Swt full length fused to GFP, white arrows point at TGEV Swt-GFP proteins; Expression of authentic β-tubulin (red), TGEV-Swt (green). The figure shows three representative image sections out of one experiment. Immunofluorescence analysis was done by confocal microscopy using Leica TCS SP5.
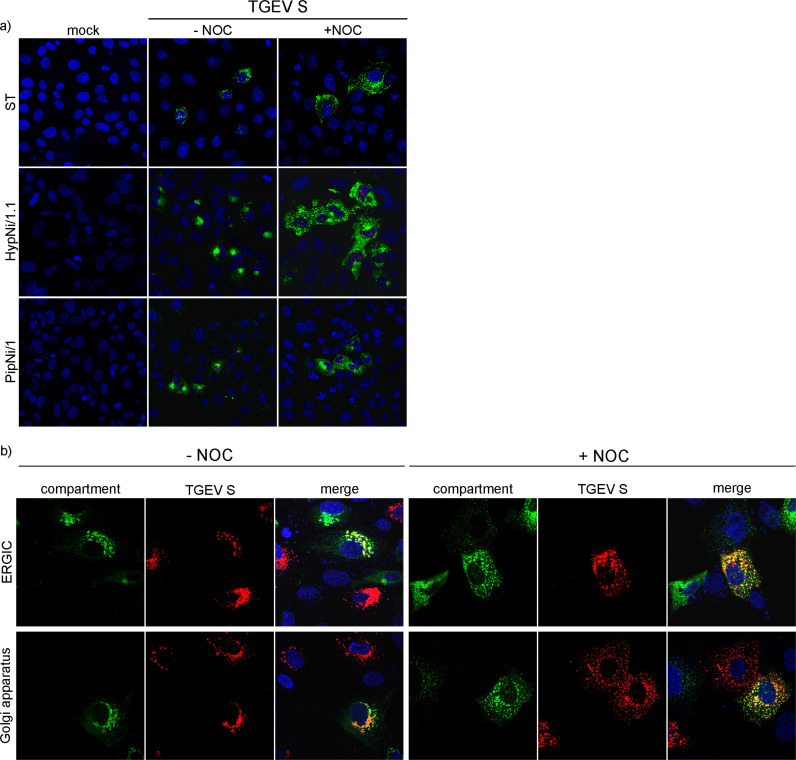
3.4. Spike proteins are differentially distributed after treatment with Nocodazole
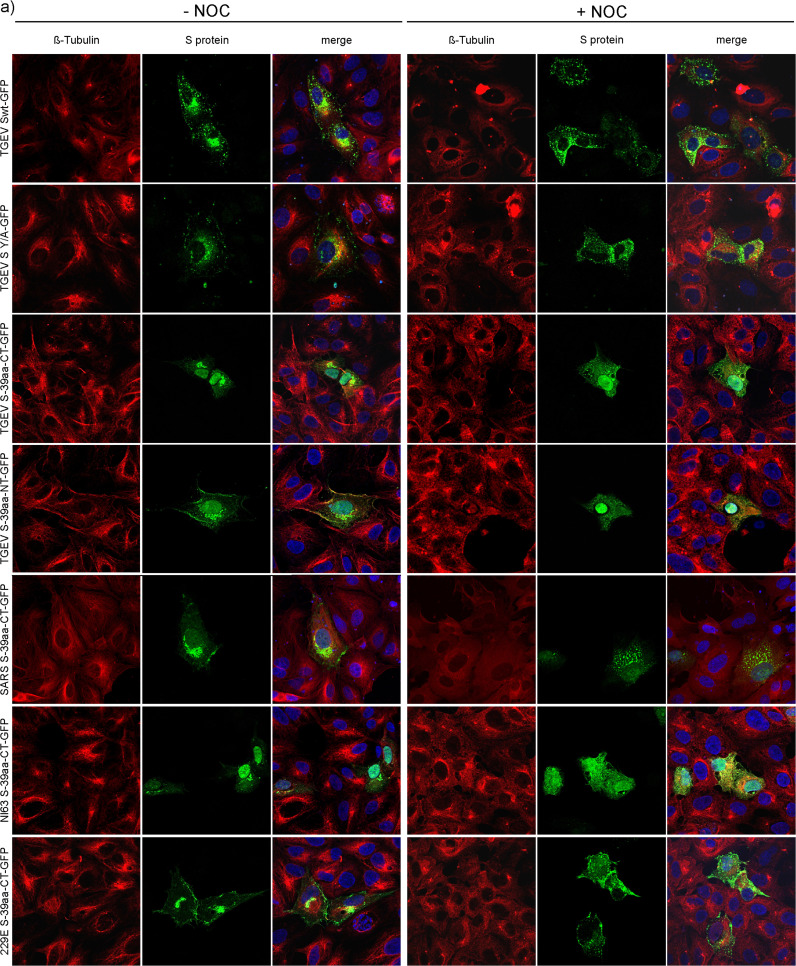
As a next step, a filament depolymerizing drug named Nocodazole (NOC) was used for functional analysis of the S-tubulin-interaction. ST cells were transfected either with full length TGEV Swt or TGEV S Y/A mutant both fused to GFP or with the last 39aa stretches of the S cytoplasmic domain of TGEV, SARS-CoV, 229E or NL63 fused to GFP as well. As negative control cells were transfected with the full length TGEV N or the empty GFP plasmid. The TGEV S Y/A mutant abolishes the retention signal due to mutation of tyrosine into an alanine. This leads to surface expression in single transfected cells while TGEV Swt protein is intracellularly retained (Schwegmann-Wessels et al., 2004). By using the mutant S Y/A protein the importance of the tyrosine-based motif within the cytoplasmic domain of the Alphacoronavirus S proteins of interest was examined. Additionally, a representative of the genus Betacoronavirus (SARS-CoV S), which contains no tyrosine motif was included. Cells were mock-treated or treated with NOC at 3 hpt. To analyze CoV S protein localization during early assembly events, cells were fixed 7 hpt (Risco et al., 1998, Vogels et al., 2011). Next, authentic β-tubulins were stained by using specific antibodies ( Fig. 3a). Cells treated with DMSO showed typically long and filamentous tubulin structures. In NOC treated cells no characteristic filamentous structures of β-tubulin were detected. Here, β-tubulins looked like patches or a camouflage net but not like filaments anymore. Differences in the S protein expression pattern could be observed as well. In DMSO treated cells the S protein accumulated near the nucleus and was less distributed in the cytosol. In contrast, the S protein expression in NOC treated cells was scattered throughout the cytoplasm and looked like vesicles. This phenomenon of differentially distributed S proteins due to NOC treatment was observed for all tested S constructs as well as for the different NOC concentrations of 10 µg/ml and 50 µg/ml. Although, the S cytoplasmic tail fusion protein of TGEV, SARS-CoV and NL63 showed additional GFP expression within the nucleus, the accumulation of S itself was clearly visible as well as the altered localization of the proteins after NOC treatment. Cells expressing GFP alone or the TGEV N protein showed no differences in their expression pattern in DMSO and NOC treated cells (Fig. 3d). Thus, irrespective of the presence or absence of a hydrophobic membrane anchor sequence or of a tyrosine-based signal the proteins associated with β-tubulins. The charge-rich region present in all tested coronavirus S proteins may be important for this interaction. The equal distribution of single GFP as well as TGEV N protein used as control proteins demonstrates that proteins missing a charge-rich region do not interact with tubulin.
Fig. 3.
S protein expression in untreated and NOC treated cells 7 hpt. ST cells transfected with different S constructs fused to GFP (a); S-expressing HypNi/1.1 cells (b); S-expressing PipNi/1 cells (c); GFP- and TGEV N-expressing ST cells as negative control (d); Expression of authentic β-tubulin (red), S proteins or single GFP and TGEV N protein (green), nuclei stained with DAPI (blue). Cells treated with DMSO (−NOC) or treated with NOC (+NOC). Immunofluorescence analysis was done by confocal microscopy using Leica TCS SP5.
The chiropteran cells HypNi/1.1 and PipNi/1 were also transfected with full length TGEV Swt or with the 39 amino acid stretches of TGEV and SARS-CoV S cytoplasmic tail fused to GFP and treated with DMSO or NOC (Fig. 3b, c). Similar results as for the ST cells were observed. Without NOC, S protein was more accumulated near the nucleus whereas in drug-treated cells the S was more dispersed within the cytoplasm. In the chiropteran cell lines a NOC concentration of 50 µg/ml resulted in a cytotoxic effect. A NOC concentration of 10 µg/ml was chosen to compare the effect of tubulin depolymerization on chiropteran and ST cells, respectively.
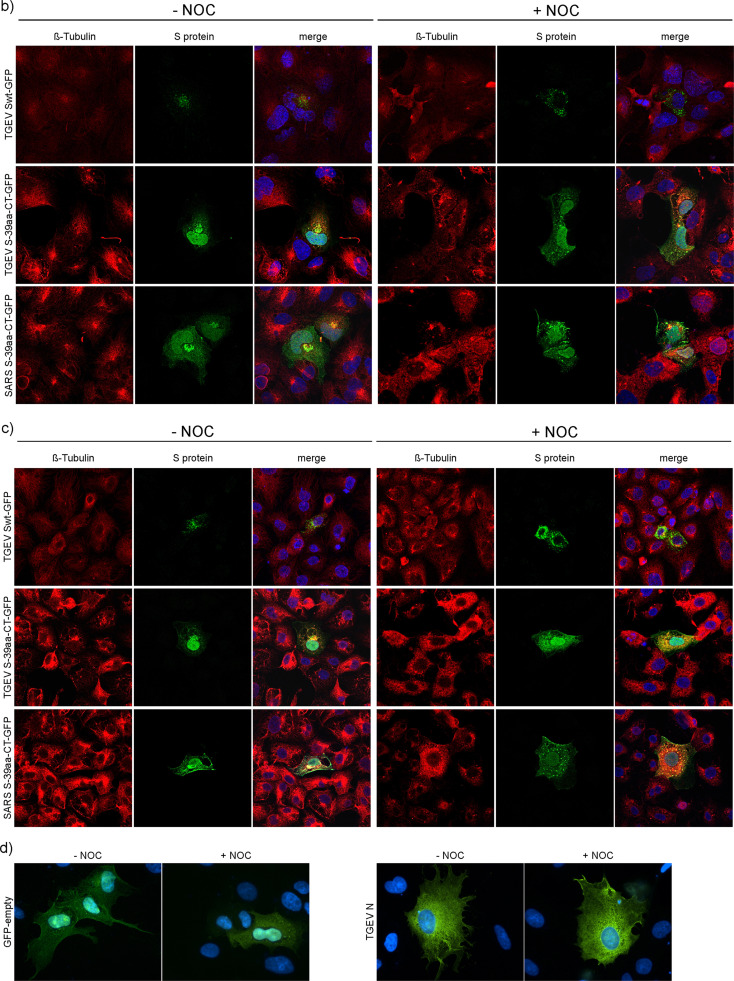
3.5. Differential distribution of TGEV proteins, ERGIC and Golgi compartments after NOC treatment in cultured cells
ST, HypNi/1.1, and PipNi/1 cells were infected with TGEV ( Fig. 4a). After fixation, cells were treated with antibodies against the viral protein S. By immunofluorescence analysis 7 hpi S expression patterns could be observed similar to the S-transfected cells. In infected, non-treated cells, S accumulated stronger near the nucleus as compared to S-transfected cells. The S protein was detected in vesicle-like structures which are distributed all over the cytosol when incubated with NOC.
Fig. 4.
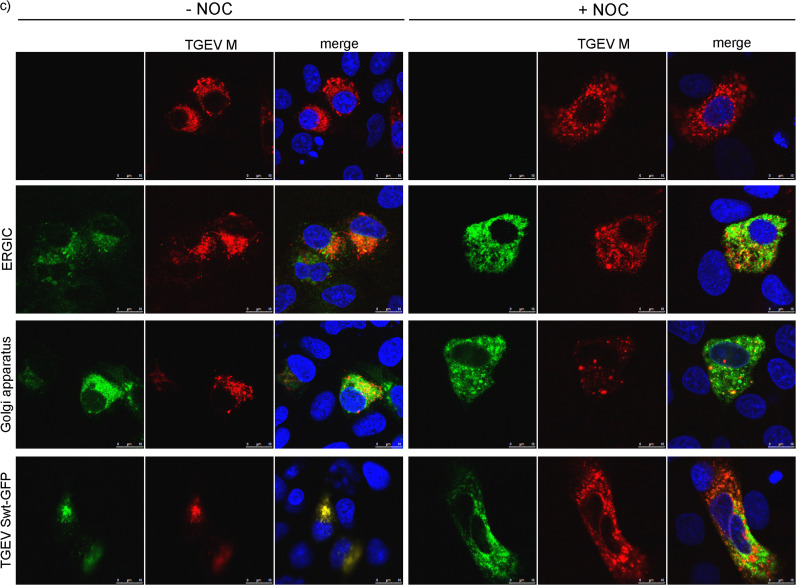
TGEV expression in untreated and NOC treated cells. ST, HypNi/1.1, and PipNi/1 cells were mock-infected or infected by TGEV; S protein was stained in green, 7 hpi (a). ST cells transfected with compartment markers for ERGIC or Golgi compartment (green) were infected by TGEV; TGEV S protein stained in red, 7 hpi (b). ST cells co-transfected with compartment markers for ERGIC or Golgi (green) and with cDNA encoding for the TGEV M protein (red) or with TGEV Swt-GFP (green) and with cDNA encoding for the TGEV M protein (red), 22 hpt (c). Cells treated with DMSO (−NOC), cells treated with NOC (+NOC), nuclei stained with DAPI (blue). Immunofluorescence analysis was performed by confocal microscopy using Leica TCS SP5.
ST cells, first transfected with markers for the ERGIC or Golgi compartment, were infected by TGEV and treated with DMSO (mock) or NOC (Fig. 4b). In mock-treated cells S accumulated near the ERGIC and Golgi compartment 7 hpi. Assembly of CoVs occurs at the ERGIC at around 7 hpi. A partial co-localization of S protein with the compartment markers could be observed. In drug-treated cells the S proteins as well as the ERGIC and the Golgi compartment were scattered throughout the cytoplasm. They were still expressed close to each other and partially co-localized. In addition to the effect on the distribution of the TGEV S protein, NOC had an effect on the cellular organelles themselves.
The effect of NOC treatment on TGEV M protein distribution was analyzed in combination with ERGIC, Golgi, and TGEV Swt-GFP localization 22 h after transfection of the cDNAs. As for the TGEV S protein the M protein was scattered throughout the cytoplasm when the cells were incubated with NOC (Fig. 4c). TGEV M showed a partial co-localization with ERGIC and Golgi compartment and a complete co-localization when co-transfected with TGEV Swt-GFP. Under NOC treatment the part of co-localized TGEV M protein decreased in all co-expression studies (ERGIC, Golgi, TGEV S protein). The reduced co-localization of TGEV S and M protein after depolymerization of tubulins was most prominent and indicated that the mechanism of direct or indirect dependence on tubulin differs between the TGEV S and M protein.
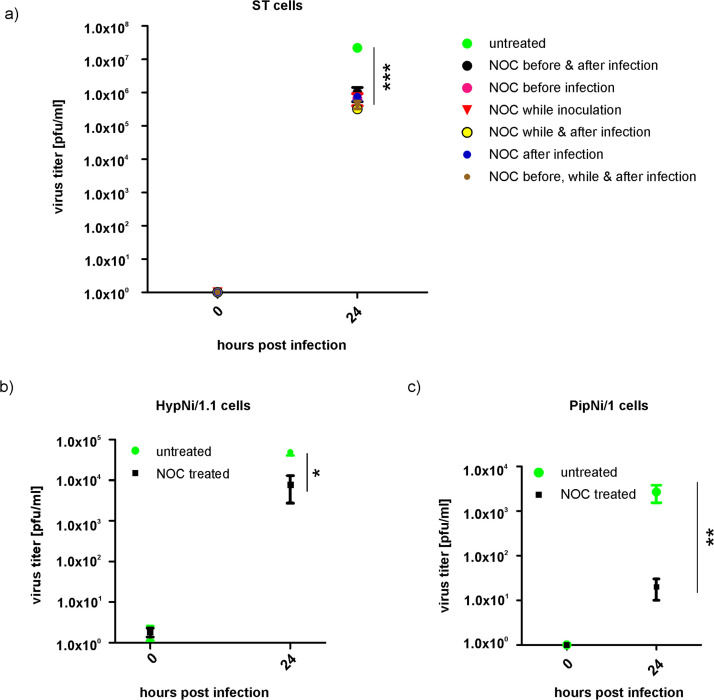
3.6. Release of infectious virus particles is reduced in NOC treated ST cells
ST cells were infected by TGEV and treated with NOC or DMSO at different time points (1 h before infection, during infection or directly after infection). Supernatants were collected (0, 24 hpi) and used for virus quantification on ST cells via plaque assay ( Fig. 5a). Infected ST cells which were not treated with the drug showed a viral titer of about 2×107 pfu/ml 24 hpi. In contrast, for cells treated with NOC at various time points a titer of about 3×105 to 9×105 pfu/ml 24 hpi was measured. A highly significant difference (P≤0.001) in the virus titer of treated versus untreated cells was calculated.
Fig. 5.
Quantification of released infectious virus particles in mock-treated and NOC treated cells via plaque assay. TGEV infected ST cells, NOC treatment at different time points before, while or after infection (a). TGEV infected HypNi/1.1 cells, NOC treatment after infection (b). TGEV infected PipNi/1 cells, NOC treatment after infection (c). (n=3); *p≤0.05; **p≤0.01; ***p≤0.001.
Regarding infected HypNi/1.1 cells a six-fold reduced amount of released infectious virus particles was measured after NOC treatment (~8×104 pfu/ml) compared to untreated (~5×105 pfu/ml) cells (Fig. 5b). PipNi/1 cells infected by TGEV showed a viral titer of about 3×103 pfu/ml, while for infected and NOC treated cells a titer of about 2×101 pfu/ml was measured (Fig. 5c). The amount of bat cells expressing pAPN-GFP at their cell surface was measured by flow cytometry using a monoclonal pAPN antibody (provided by H. Laude). The percentage of pAPN-positive cells for HypNi/1.1 was about 17.1% and for PipNi/1 about 5.6% (mean of 5 experiments).
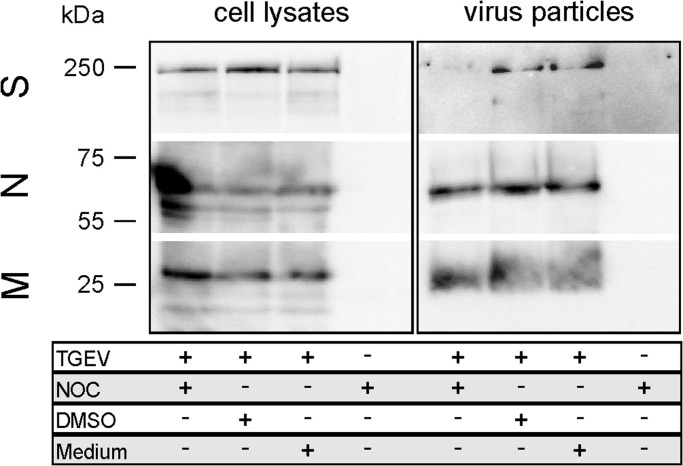
3.7. Less S protein is incorporated into virions after NOC treatment of infected ST cells
ST cells infected with TGEV were treated with DMSO or NOC. After cell lysis as well as virus particle concentration by ultracentrifugation the incorporation of S into virions was examined by SDS-PAGE followed by Western blot ( Fig. 6). Mock-infected cells served as negative control. In infected ST cells, not treated with NOC, S protein was detected in virus particles after ultracentrifugation. In cells treated with NOC (10 as well as 50 µg/ml) a very weak or nearly no signal was observed for the S protein incorporated into viral particles. The signal for TGEV S, M and N proteins were similar in the corresponding cell lysates and comparable amounts of M and N protein were detectable in concentrated virions irrespective of a Nocodazole treatment (10 as well as 50 µg/ml). This indicates a reduced S incorporation into virus particles due to depolymerization of tubulins by NOC treatment of infected ST cells.
Fig. 6.
TGEV particle assay 18 hpi. Detection of viral spike (S), nucleocapsid (N), and membrane (M) proteins in cell lysates and pelleted supernatants of TGEV- or mock-infected ST cells. Cells were either treated with NOC (50 µg/ml), DMSO or medium. Virus particles were concentrated by ultracentrifugation; n=3.
4. Discussion
Viruses rely on the host cell machinery for successful replication (Belov et al., 2007, Beske et al., 2007, Choe et al., 2005, Konan et al., 2003, Moffat et al., 2007, Oostra et al., 2007, Sakaguchi et al., 1996). Nevertheless, more detailed knowledge on virus-host interaction would lead to therapeutic tools for infection control. CoVs use the host secretory pathway during their replication cycle. The vesicular transport on secretory pathways is mostly mediated by microtubules and the corresponding motor proteins (Fokin et al., 2014). In case of MHV it was shown that the early secretory pathway is important during the formation of the replication complex (Knoops et al., 2010, Oostra et al., 2007, Verheije et al., 2008). For the release of infectious progeny the incorporation of CoV S proteins into virus particles during assembly is required. Information about cellular factors interacting with coronaviral S proteins during transport and assembly would be important to understand the virus infection process.
In this study we searched for interaction partners of the last 39aa stretches of TGEV, HCoV NL63 and 229E S cytoplasmic domains (all containing a tyrosine-based motif). Four different tubulin alpha and beta chains (TUBB2A, TUBB4A, TUBB6 and TUBA4A) were detected to interact with TGEV-S and human CoV-229E-S. Regarding HCoV NL63, only an interaction for TUBB2A was noticed. Either NL63-S-39aa-GFP-NT does not bind certain tubulins or just with low affinity. A destroyed or impaired binding of NL63-S to some tubulins due to the GFP-tag could be possible as well. Nevertheless, the negative results for NL63-S demonstrate that the S tail of the coronavirus S proteins is not interacting with tubulins due to a sticky unstructured peptide conformation but due to a specific binding to tubulins.
For many viruses a close association with cytoskeletal elements was shown (Luftig, 1982). Adenovirus type 2 and 5 particles as well as reovirus particles interact with microtubules (Babiss et al., 1979, Luftig and Weihing, 1975, Miles et al., 1980). Poliovirus and newly synthesized viral RNA of Simian virus 40 are associated with actin filaments and intermediate filaments (Ben-Ze’ev et al., 1981, Lenk and Penman, 1979). An interaction is likely necessary for initial infection, transport of viral components as well as for the assembly of new viral particles (Hsieh et al., 2010). Moreover, tubulin was already found packaged into virions of Epstein-Barr virus, human cytomegalovirus, and murine leukemia virus (Johannsen et al., 2004, Varnum et al., 2004, Wang et al., 2003). For several viruses like adenovirus, African swine fever virus, canine parvovirus, herpes simplex virus as well as lyssavirus and rabies virus an interaction either of the whole virus capsid or of single viral proteins with cytoplasmic dynein was examined (Alonso et al., 2001, Douglas et al., 2004, Jacob et al., 2000, Kelkar et al., 2004, Raux et al., 2000, Suikkanen et al., 2003, Ye et al., 2000). Viruses may have evolved microtubule-binding motifs or similar amino acid sequences complementary to motifs in dynein for successful interaction (Leopold and Pfister, 2006, Pasick et al., 1994). An indirect interaction of S proteins with tubulin via motor proteins like dynein or kinesin may be a possible scenario for coronaviruses as well.
In transfected as well as in infected cells coronavirus S protein was scattered throughout the cell after NOC treatment. The same was shown for expression of TGEV M protein. The difference in localization of S and M protein after NOC treatment indicate a difference in interaction with tubulin. It is possible that one or both viral proteins interact in an indirect way with tubulin via other cellular proteins. The redistribution of the compartment markers for ERGIC and Golgi shows a dependence of these two marker proteins on tubulin as well in a direct or indirect manner. This phenomenon demonstrates that it is not possible to exclude that the viral S and/or the M protein interact with cellular proteins that themselves are dependent on tubulin. Nevertheless, our experiments visualize that S and M use different modes of interaction and that this leads to a regular incorporation of M protein into viral particles even under NOC treatment but to a strong reduction of S protein incorporation into viral particles. A redistribution in NOC treated cells was already shown for different viral proteins, like Crimean-Congo hemorrhagic fever virus nucleocapsid protein (Simon et al., 2009) or herpes simplex virus 1 capsids (Sodeik et al., 1997). In the two tested chiropteran cell lines (HypNi/1.1 and PipNi/1) similar results as for S-transfected or TGEV-infected ST cells were obtained. Also here, a redistribution of the S proteins only in NOC treated cells as well as less released infectious virus particles compared to DMSO treated cells were noticed, suggesting a conserved viral strategy to use the host for its own advantage during replication. Due to the assumption that many human and animal CoVs originated from bats and that most eukaryotic cells contain microtubules, a conserved microtubule-dependent CoV replication strategy is likely.
Furthermore, we demonstrated that NOC leads to a reduced amount of released infectious virus particles. Less virus yield after cytoskeleton disruption was already shown for moloney murine leukemia virus and vaccinia virus (Payne and Kristensson, 1982, Satake and Luftig, 1982). To find out which step during the TGEV replication cycle is mostly influenced by the interaction of S with tubulin, cells were drug-treated at different time points (before, while, or after infection). A significantly lower virus titer in the cell culture supernatant of NOC treated cells compared to untreated cells was detected, whereas no differences in viral titers concerning the various time points could be observed. CoVs uses the host secretory pathway to be transported from ER-to-Golgi apparatus as well as during virus assembly (Knoops et al., 2010, Oostra et al., 2007, Verheije et al., 2008, Vogels et al., 2011). An impaired transport of viral components and an affected assembly of S protein into newly generated virus particles may explain the decrease in virus yield. For MHV, microtubules are important for neuronal transport, replication and viral spread (Biswas and Das Sarma, 2014). Also here, a reduced efficiency of viral infection was observed by using microtubule disrupting drugs (Biswas and Das Sarma, 2014). TGEV was able to enter the cells in the absence or presence of NOC, it was able to replicate and to egress but the infection was less efficient regarding the amount of newly formed infectious virions. Due to the fact that depolymerized microtubules did not inhibit TGEV infection completely, virus components have to use other pathways for trafficking beside tubulin filaments to finish their replication cycle. An actin-dependent strategy for virus movement could be possible (Cudmore et al., 1997, Lanier and Volkman, 1998, Sanlioglu et al., 2000). However, lower virus titers may correlate with the differentially distributed S protein in NOC treated cells, too. Actually, S accumulates near the ERGIC where it interacts with the M protein to be incorporated into newly assembled virions (Nguyen and Hogue, 1997, Opstelten et al., 1995). When microtubules are depolymerized S proteins as well as the ERGIC and Golgi compartment are scattered throughout the cell. Due to the diffuse distribution of S and the two compartments which are important for successful TGEV assembly less interaction of viral proteins during assembly may be possible. An impaired transport of virus particles from the Golgi compartment to the plasma membrane due to NOC treatment may also be a reason for lower viral titers. Concerning this matter, a less organized cytoplasm with dispersed Golgi stacks in NOC treated ST cells was already shown by Risco et al. 1998. TGEV virions were seen in these disrupted Golgi stacks whereas only a few were detected on extracellular surfaces (Risco et al., 1998). A reason may be a microtubule-dependent transport of vesicles between the Golgi compartment and the plasma membrane (Risco et al., 1998). To find out if S incorporation into virions is influenced by NOC treatment, virus particles were concentrated out of the supernatant of NOC-treated infected cells and compared to mock-treated cells. In this case, the amount of S protein was similar in DMSO and NOC treated cell lysates, whereas S protein quantity was strongly reduced in pelleted virus particles when infected cells were treated with NOC. This fact fits to the calculated smaller amount of released infectious virus particles measured in drug treated cells. The minimal requirements for assembly of virus like particles is the expression of TGEV M and E structural proteins (Vennema et al., 1996). The amount of M and N protein in released virus particles presented in Fig. 6 revealed nearly no variation after NOC treatment. This demonstrates that coronavirus assembly in general is not disturbed by depolymerization of microtubules. Thus, the reduction in virus titer is mainly caused by the fact that less S protein is present at the assembly site (due to impaired S protein transport) and/or that the incorporation process of S itself into virions is tubulin-dependent.
Our results show that Alpha- and Betacoronavirus S proteins interact with the charge-rich region of their cytoplasmic domains with tubulin beta chains. An interaction with microtubules facilitates TGEV replication and infection efficiency but the depolymerization of microtubules did not inhibit it completely. Thus, an interaction of S with tubulin supports an efficient generation of infectious virus progeny.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
Financial support was provided by a Grant to CSW (SCHW 1408/1.1) from the German Research Foundation (DFG) and by the “Bundesministerium fuer Bildung und Forschung” of the German Government (Zoonosis Network, Consortium on ecology and pathogenesis of SARS, project code 01KI1005A,F to AvB).
CSW is funded by the Emmy Noether Programme from the DFG. ATR is a recipient of a Georg Christoph Lichtenberg Ph.D. fellowship from the Ministry for Science and Culture of Lower Saxony.
This work was performed by ATR in partial fulfillment of the requirements for a Dr. rer. nat. degree from the University of Veterinary Medicine Hannover.
We are grateful to L. Enjuanes, H.-P. Hauri, E. Kremmer, H. Laude, and E. Snijder for providing antibodies, plasmids, and virus. We thank M. Müller and C. Drosten for the chiropteran cell lines (HypNi/1.1, PipNi/1). Thanks to S. Bauer for technical assistance.
References
- Alonso C., Miskin J., Hernaez B., Fernandez-Zapatero P., Soto L., Canto C., Rodriguez-Crespo I., Dixon L., Escribano J.M. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001;75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiss L.E., Luftig R.B., Weatherbee J.A., Weihing R.R., Ray U.R., Fields B.N. Reovirus serotypes 1 and 3 differ in their in vitro association with microtubules. J. Virol. 1979;30:863–874. doi: 10.1128/jvi.30.3.863-874.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Altan-Bonnet N., Kovtunovych G., Jackson C.L., Lippincott-Schwartz J., Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ze’ev A., Horowitz M., Skolnik H., Abulafia R., Laub O., Aloni Y. The metabolism of SV40 RNA is associated with the cytoskeletal framework. Virology. 1981;111:475–487. doi: 10.1016/0042-6822(81)90350-0. [DOI] [PubMed] [Google Scholar]
- Beske O., Reichelt M., Taylor M.P., Kirkegaard K., Andino R. Poliovirus infection blocks ERGIC-to-Golgi trafficking and induces microtubule-dependent disruption of the Golgi complex. J. Cell Sci. 2007;120:3207–3218. doi: 10.1242/jcs.03483. [DOI] [PubMed] [Google Scholar]
- Biswas K., Das Sarma J. Effect of microtubule disruption on neuronal spread and replication of demyelinating and nondemyelinating strains of mouse hepatitis virus in vitro. J. Virol. 2014;88:3043–3047. doi: 10.1128/JVI.02545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., de Haan C.A., Smits S.L., Rottier P.J. Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements. Virology. 2005;334:306–318. doi: 10.1016/j.virol.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Choe S.S., Dodd D.A., Kirkegaard K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology. 2005;337:18–29. doi: 10.1016/j.virol.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., Vallo P., da Silva Filho L.V., Leroy E.M., Thiel V., van der Hoek L., Poon L.L., Tschapka M., Drosten C., Drexler J.F. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S., Reckmann I., Way M. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 1997;5:142–148. doi: 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- Dewerchin H.L., Desmarets L.M., Noppe Y., Nauwynck H.J. Myosins 1 and 6, myosin light chain kinase, actin and microtubules cooperate during antibody-mediated internalisation and trafficking of membrane-expressed viral antigens in feline infectious peritonitis virus infected monocytes. Vet. Res. 2014;45:17. doi: 10.1186/1297-9716-45-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M.W., Diefenbach R.J., Homa F.L., Miranda-Saksena M., Rixon F.J., Vittone V., Byth K., Cunningham A.L. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 2004;279:28522–28530. doi: 10.1074/jbc.M311671200. [DOI] [PubMed] [Google Scholar]
- Fokin A.I., Brodsky I.B., Burakov A.V., Nadezhdina E.S. Interaction of early secretory pathway and Golgi membranes with microtubules and microtubule motors. Biochemistry. 2014;79:879–893. doi: 10.1134/S0006297914090053. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke G.J., de Haan C.A., Rossen J.W., Vennema H., Rottier P.J. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74:1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Han X., Li Z., Chen H., Wang H., Mei L., Wu S., Zhang T., Liu B., Lin X. Influenza virus A/Beijing/501/2009(H1N1) NS1 interacts with beta-tubulin and induces disruption of the microtubule network and apoptosis on A549 cells. PLoS One. 2012;7:e48340. doi: 10.1371/journal.pone.0048340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y., Hasebe R., Sunden Y., Ochiai K., Honda E., Sakoda Y., Umemura T. Propagation of swine hemagglutinating encephalomyelitis virus and pseudorabies virus in dorsal root ganglia cells. J. Vet. Med. Sci. 2009;71:595–601. doi: 10.1292/jvms.71.595. [DOI] [PubMed] [Google Scholar]
- Heald R., Nogales E. Microtubule dynamics. J. Cell Sci. 2002;115:3–4. doi: 10.1242/jcs.115.1.3. [DOI] [PubMed] [Google Scholar]
- Henry Sum M.S. The involvement of microtubules and actin during the infection of Japanese encephalitis virus in neuroblastoma cell line, IMR32. Biomed. Res. Int. 2015;2015:695283. doi: 10.1155/2015/695283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M.J., White P.J., Pouton C.W. Interaction of viruses with host cell molecular motors. Curr. Opin. Biotechnol. 2010;21:633–639. doi: 10.1016/j.copbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Hyde J.L., Gillespie L.K., Mackenzie J.M. Mouse norovirus 1 utilizes the cytoskeleton network to establish localization of the replication complex proximal to the microtubule organizing center. J. Virol. 2012;86:4110–4122. doi: 10.1128/JVI.05784-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Badrane H., Ceccaldi P.E., Tordo N. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E., Luftig M., Chase M.R., Weicksel S., Cahir-McFarland E., Illanes D., Sarracino D., Kieff E. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar S.A., Pfister K.K., Crystal R.G., Leopold P.L. Cytoplasmic dynein mediates adenovirus binding to microtubules. J. Virol. 2004;78:10122–10132. doi: 10.1128/JVI.78.18.10122-10132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Swett-Tapia C., van den Worm S.H., Te Velthuis A.J., Koster A.J., Mommaas A.M., Snijder E.J., Kikkert M. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J. Virol. 2010;84:833–846. doi: 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konan K.V., Giddings T.H., Jr., Ikeda M., Li K., Lemon S.M., Kirkegaard K. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 2003;77:7843–7855. doi: 10.1128/JVI.77.14.7843-7855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl A., Hoffmann M., Muller M.A., Munster V.J., Gnirss K., Kiene M., Tsegaye T.S., Behrens G., Herrler G., Feldmann H., Drosten C., Pohlmann S. Comparative analysis of Ebola virus glycoprotein interactions with human and bat cells. J. Infect. Dis. 2011;204(Suppl 3):S840–S849. doi: 10.1093/infdis/jir306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Lanier L.M., Volkman L.E. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology. 1998;243:167–177. doi: 10.1006/viro.1998.9065. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk R., Penman S. The cytoskeletal framework and poliovirus metabolism. Cell. 1979;16:289–301. doi: 10.1016/0092-8674(79)90006-0. [DOI] [PubMed] [Google Scholar]
- Leopold P.L., Pfister K.K. Viral strategies for intracellular trafficking: motors and microtubules. Traffic. 2006;7:516–523. doi: 10.1111/j.1600-0854.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Luftig R.B. Does the cytoskeleton play a significant role in animal virus replication? J. Theor. Biol. 1982;99:173–191. doi: 10.1016/0022-5193(82)90397-6. [DOI] [PubMed] [Google Scholar]
- Luftig R.B., Weihing R.R. Adenovirus binds to rat brain microtubules in vitro. J. Virol. 1975;16:696–706. doi: 10.1128/jvi.16.3.696-706.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles B.D., Luftig R.B., Weatherbee J.A., Weihing R.R., Weber J. Quantitation of the interaction between adenovirus types 2 and 5 and microtubules inside infected cells. Virology. 1980;105:265–269. doi: 10.1016/0042-6822(80)90177-4. [DOI] [PubMed] [Google Scholar]
- Moffat K., Knox C., Howell G., Clark S.J., Yang H., Belsham G.J., Ryan M., Wileman T. Inhibition of the secretory pathway by foot-and-mouth disease virus 2 BCE protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J. Virol. 2007;81:1129–1139. doi: 10.1128/JVI.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Raj V.S., Muth D., Meyer B., Kallies S., Smits S.L., Wollny R., Bestebroer T.M., Specht S., Suliman T., Zimmermann K., Binger T., Eckerle I., Tschapka M., Zaki A.M., Osterhaus A.D., Fouchier R.A., Haagmans B.L., Drosten C. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3 doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.P., Hogue B.G. Protein interactions during coronavirus assembly. J. Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Annu. Rev. Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M.J., Wolfs K., Horzinek M.C., Rottier P.J. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J.M., Kalicharran K., Dales S. Distribution and trafficking of JHM coronavirus structural proteins and virions in primary neurons and the OBL-21 neuronal cell line. J. Virol. 1994;68:2915–2928. doi: 10.1128/jvi.68.5.2915-2928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne L.G., Kristensson K. The effect of cytochalasin D and monensin on enveloped vaccinia virus release. Arch. Virol. 1982;74:11–20. doi: 10.1007/BF01320778. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Muller M.A., Annan A., Vallo P., Adu-Sarkodie Y., Kruppa T.F., Drosten C. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A., Way M. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 2001;13:97–105. doi: 10.1016/S0955-0674(00)00180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke K., Dohner K., Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Micro. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- Raux H., Flamand A., Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Muntion M., Enjuanes L., Carrascosa J.L. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J. Virol. 1998;72:4022–4031. doi: 10.1128/jvi.72.5.4022-4031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T., Leser G.P., Lamb R.A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlioglu S., Benson P.K., Yang J., Atkinson E.M., Reynolds T., Engelhardt J.F. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 2000;74:9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake M., Luftig R.B. Microtubule-depolymerizing agents inhibit Moloney murine leukaemia virus production. J. Gen. Virol. 1982;58:339–349. doi: 10.1099/0022-1317-58-2-339. [DOI] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H.Y., Herrler G. A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 2004;279:43661–43666. doi: 10.1074/jbc.M407233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Johansson C., Lundkvist A., Mirazimi A. Microtubule-dependent and microtubule-independent steps in Crimean-Congo hemorrhagic fever virus replication cycle. Virology. 2009;385:313–322. doi: 10.1016/j.virol.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8:465–472. doi: 10.1016/s0966-842x(00)01824-2. [DOI] [PubMed] [Google Scholar]
- Sodeik B., Ebersold M.W., Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suikkanen S., Aaltonen T., Nevalainen M., Valilehto O., Lindholm L., Vuento M., Vihinen-Ranta M. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 2003;77:10270–10279. doi: 10.1128/JVI.77.19.10270-10279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyrock A., Ossendorf E., Stehling M., Kail M., Kurtz T., Pohlentz G., Waschbusch D., Eggert S., Formstecher E., Muthing J., Dreisewerd K., Kins S., Goud B., Barnekow A. A new Mint1 isoform, but not the conventional Mint1, interacts with the small GTPase Rab6. PLoS One. 2013;8:e64149. doi: 10.1371/journal.pone.0064149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincone A., Schwegmann-Wessels C. Looking for a needle in a haystack: cellular proteins that may interact with the tyrosine-based sorting signal of the TGEV S protein. Virus Res. 2015;202:3–11. doi: 10.1016/j.virusres.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum S.M., Streblow D.N., Monroe M.E., Smith P., Auberry K.J., Pasa-Tolic L., Wang D., Camp D.G., 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R.D., Nelson J.A. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Raaben M., Mari M., Te Lintelo E.G., Reggiori F., van Kuppeveld F.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkman A.S. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem. Sci. 2002;27:27–33. doi: 10.1016/s0968-0004(01)02003-5. [DOI] [PubMed] [Google Scholar]
- Vogels M.W., van Balkom B.W., Kaloyanova D.V., Batenburg J.J., Heck A.J., Helms J.B., Rottier P.J., de Haan C.A. Identification of host factors involved in coronavirus replication by quantitative proteomics analysis. Proteomics. 2011;11:64–80. doi: 10.1002/pmic.201000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.Q., Kim W., Gao G., Torrey T.A., Morse H.C., 3rd, De Camilli P., Goff S.P. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 2003;3:4. doi: 10.1186/1475-4924-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C., Schwegmann-Wessels C., Neumann U., Herrler G. The spike protein of infectious bronchitis virus is retained intracellularly by a tyrosine motif. J. Virol. 2008;82:2765–2771. doi: 10.1128/JVI.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Ye G.J., Vaughan K.T., Vallee R.B., Roizman B. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Dong H., Feng L. Identification of the interaction between vimentin and nucleocapsid protein of transmissible gastroenteritis virus. Virus Res. 2015;200:56–63. doi: 10.1016/j.virusres.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H.Y., Chen J.F., Shi D., Lang H.W., Wang Z.T., Feng L. Identification of cellular proteome using two-dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus. Proteome Sci. 2013;11:31. doi: 10.1186/1477-5956-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gao J., Zhu L., Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]