Abstract
RNA-binding proteins (RNA-BPs) play critical roles in development and disease to regulate gene expression. However, genome-wide identification of their targets in primary human cells has been challenging. Here, we applied a modified CLIP-seq strategy to identify genome-wide targets of the FMRP translational regulator 1 (FMR1), a brain-enriched RNA-BP, whose deficiency leads to Fragile X Syndrome (FXS), the most prevalent inherited intellectual disability. We identified FMR1 targets in human dorsal and ventral forebrain neural progenitors and excitatory and inhibitory neurons differentiated from human pluripotent stem cells. In parallel, we measured the transcriptomes of the same four cell types upon FMR1 gene deletion. We discovered that FMR1 preferentially binds long transcripts in human neural cells. FMR1 targets include genes unique to human neural cells and associated with clinical phenotypes of FXS and autism. Integrative network analysis using graph diffusion and multitask clustering of FMR1 CLIP-seq and transcriptional targets reveals critical pathways regulated by FMR1 in human neural development. Our results demonstrate that FMR1 regulates a common set of targets among different neural cell types but also operates in a cell type–specific manner targeting distinct sets of genes in human excitatory and inhibitory neural progenitors and neurons. By defining molecular subnetworks and validating specific high-priority genes, we identify novel components of the FMR1 regulation program. Our results provide new insights into gene regulation by a critical neuronal RNA-BP in human neurodevelopment.
Human neuronal development, function, and dysfunction rely heavily on translational control of essential genes by RNA-binding proteins (RNA-BPs). Key to understanding the mechanisms and impact of RNA-BPs is to identify their genome-wide targets in cells of the nervous system. High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-seq or CLIP-seq) can isolate RNA-BP targets but requires large numbers of cells and high-quality antibodies (Wheeler et al. 2018). Methods with increased efficiency and specificity have been developed, including irCLIP (Zarnegar et al. 2016) and eCLIP (Van Nostrand et al. 2016), but the difficulties of isolating large numbers of human neurons has still limited our ability to identify genome-wide targets of RNA-BPs. Thus, new strategies are needed to address the function of RNA-BPs in human brain.
One critical RNA-binding protein that regulates the expression of critical genes in neural development, neuronal function, and synaptic plasticity is FMRP translational regulator 1 (FMR1) (Pfeiffer and Huber 2009; Darnell and Klann 2013). Loss of FMR1 results in Fragile X Syndrome (FXS), the most common inherited genetic cause of intellectual disability and the leading genetic contributor to autism (Pieretti et al. 1991; Verkerk et al. 1991; Kaufmann et al. 2017). Studying FMR1 in human neurodevelopment may serve as a gateway for understanding autism, but the identification of RNA targets of FMR1 in humans is largely unexplored, thus limiting our understanding of FMR1 function. To date, most research on FMR1 function and consequences of FMR1 loss has relied on animal models, particularly mouse models. However, recent clinical trials developed based on evidence from animal models failed to correct disease-related phenotypes in FXS patients (Bailey et al. 2016; Berry-Kravis et al. 2016; Zhao and Bhattacharyya 2018). Discrepant impacts of FMR1 deficiency on mouse versus human brains (Kwan et al. 2012) and mouse versus human embryonic stem cells (Doers et al. 2014; Telias et al. 2015; Khalfallah et al. 2017) suggest that interspecies differences in brain development and FMR1 function are significant. Thus, discordance between rodent models and human studies warrants identification of FMR1 targets in human neurons.
Genome-wide binding studies show that FMR1 binds hundreds of mRNAs in the mouse brain (Brown et al. 2001; Darnell et al. 2011; Tabet et al. 2016; Maurin et al. 2018; Sawicka et al. 2019), but only a handful of these targets have been validated in humans. In vitro binding kinetic assays estimate that FMR1 interacts with ∼4% of mRNAs expressed in human fetal brain tissue (Ashley et al. 1993), and a few reports identifying human FMR1 targets have emerged (Ascano et al. 2012; Van Nostrand et al. 2016, 2017; Tran et al. 2019). CLIP-seq using the HEK293 cell line overexpressing tagged FMR1 identified over 6000 RNAs as direct FMR1 targets (Ascano et al. 2012). However, it is unclear how these findings in immortalized non-neural cell lines inform FMR1 functions in the brain. Recent work addressed this challenge by identifying FMR1 targets in post-mortem adult human frontal cortex (Tran et al. 2019) with an emphasis on FMR1's involvement in RNA editing in autism. This study used adult brain tissues containing mixed cell types, making it difficult to attribute the binding of FMR1 to specific cell types that are relevant to neurodevelopment. These genome-wide studies provide a framework for unveiling mechanisms of FMR1 function, but it remains unknown whether the identified RNA targets are regulated by FMR1 in human neuronal cell types and the extent to which FMR1 targets differ across related cell types. Further, FMR1's role is critical in neurodevelopment, and the reports thus far do not interrogate developing neural cells.
Human pluripotent stem cells (hPSCs) provide a paradigm to study aspects of human neurodevelopment because of their ability to generate specific neural cell types in vitro over a period of time corresponding to in vivo human development, thus recapitulating many of the developmental steps relevant and unique to humans (Anderson and Vanderhaeghen 2014; Kim et al. 2014; Zhao and Bhattacharyya 2018). Cortical neurons differentiated from human PSCs resemble fetal neurons (Handel et al. 2016), and so neural progenitor cells (NPCs) and neurons differentiated from human PSCs provide a good model of early cortical development. While a limited number of studies using FXS human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) suggest that FMR1 functions during neural progenitor differentiation and neuronal maturation in human cells (Sheridan et al. 2011; Telias et al. 2013; Doers et al. 2014), the precise role of FMR1 and its mechanisms of function in the human brain are largely unknown, hindering our understanding of how loss of FMR1 causes intellectual disability in FXS patients and reducing our ability to find relevant therapeutic targets.
In this study, we identify FMR1 targets in human neural cells through seamless CRISPR-Cas9 genome editing to tag FMR1, modified CLIP-seq, and transcriptomic analysis of FMR1-deficient cells. We integrate direct targets of FMR1 with differentially expressed genes using graph-based diffusion and multitask graph clustering to define shared and cell type–specific RNA targets. The multilevel data sets we generated serve as important resources for FXS, autism, and neurodevelopmental research communities.
Results
Generation of FMR1-FLAG hPSCs and neural differentiation
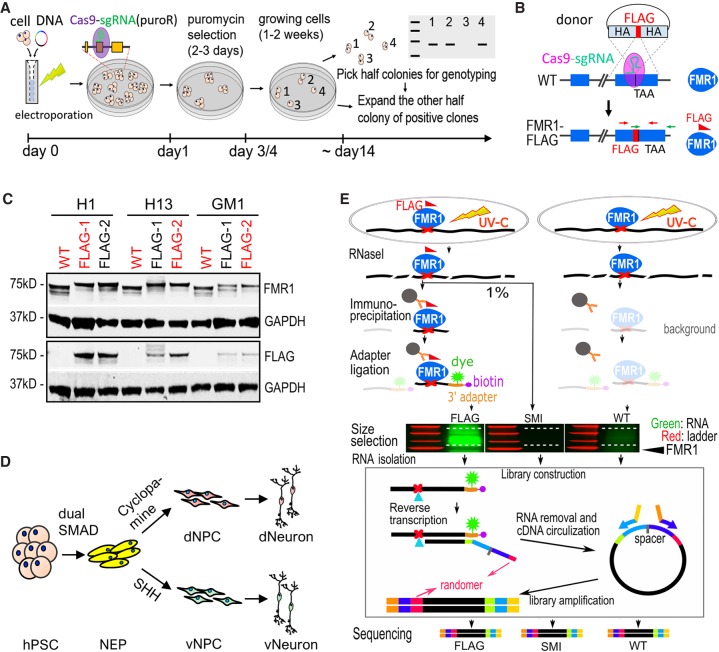
To facilitate isolation of FMR1 for identification of direct RNA targets of FMR1 by CLIP, we tagged FMR1 in hPSCs. We generated FMR1-FLAG hPSCs with endogenous FMR1 tagged by a 3xFLAG tag using a modified CRISPR-Cas9 genome targeting strategy which, through homology-directed recombination with a donor plasmid followed by transient puromycin selection, allowed for fast and seamless genome editing (Fig. 1A,B). In the FMR1-FLAG hPSCs, FMR1 was expressed as a fusion protein with a C-terminal 3xFLAG tag (Fig. 1B). Since FMR1 is an X-linked gene, we generated FMR1-FLAG hPSCs from three male hPSC lines as biological replicates: two hESC lines (H1 and H13) and one hiPSC line (GM1) (Fig. 1C). From each of the three parental hPSC lines (referred to as wild type or WT), at least two isogenic colonies with successful fusion of FLAG tag to FMR1 were confirmed by PCR (Supplemental Fig. S1A), Sanger sequencing (Supplemental Fig. S1B), and immunoblot (Fig. 1C). One FMR1-FLAG line from each parental line was used for subsequent experiments (red in Fig. 1C and Supplemental Fig. S1A). Integrity of the FMR1-FLAG hPSC lines was confirmed by karyotyping and immunofluorescence for stem cell markers, POU5F1 (also known as OCT4), SOX2, and PODXL, as well as absence of the neuroepithelial marker PAX6 (Supplemental Fig. S1C,D). Off-target CRISPR mutations at the top five predicted sites in the FMR1-FLAG hPSC lines were examined by Sanger sequencing, and no off-target mutations were detected (Supplemental Fig. S1E).
Figure 1.
Generation of FMR1-FLAG hPSCs and neural cells for CLIP. (A) Schematic diagram of one-step seamless genome editing using CRISPR-Cas9. A Cas9-sgRNA plasmid also confers puromycin resistance (puroR) which allows for a temporary selection to obtain seamless genome editing in ∼2 wk. (B) Diagram of generation of FMR1-FLAG hPSCs using CRISPR-Cas9 and a donor plasmid for FLAG knock-in. Exons of FMR1 are shown as blue boxes. TAA indicates the position of a stop codon. HA represents homology arm, and crossed dash lines depict homology-directed recombination. (C) Western blot detecting FMR1 or FLAG in FMR1-FLAG and WT hPSCs, GAPDH as a loading control. hPSC lines in red were used for experiments. (D) Diagram of neural differentiation of hPSCs. Neuroepithelial cells (NEP) were differentiated from hPSCs by dual SMAD inhibition followed by patterning to forebrain dorsal NPCs (dNPC) and ventral MGE-like NPCs (vNPC) by modulating the SHH pathway. Patterned NPCs were further differentiated to neurons. (E) Flow chart of CLIP-seq. Both FMR1-FLAG and control (WT) cells were subjected to UV crosslinking. RNAs were partially digested by RNase I and FMR1-bound RNAs were immunoprecipitated using a FLAG antibody. Immunoprecipitated RNAs (and background levels of RNAs) were ligated to a fluorescent 3′ adapter for visualization (green) and reverse transcription. Randomers were incorporated in the RT primer to allow for removal of duplicated Illumina reads from individual cDNAs. A size-matched input (SMI) control, RNAs from FMR1-FLAG cells that were not immunoprecipitated but size-selected, was also used as a control.
We differentiated the three parental hPSCs (parental) and isogenic FMR1-FLAG hPSCs (FLAG) into dorsal forebrain and ventral forebrain neural progenitor cells and then to dorsal and ventral forebrain neurons, hereafter referred to as dNPC, vNPC, dNeuron, and vNeuron, respectively. Differentiation was carried out using established methods of dual SMAD inhibition followed by patterning with a sonic hedgehog (SHH) inhibitor (cyclopamine) for dNPC and SHH for vNPC (Fig. 1D; Chambers et al. 2009; Li et al. 2009; Maroof et al. 2013). Successful patterning of the two lineages was confirmed by expression of PAX6 in dNPCs and NKX2-1 in vNPCs (Supplemental Fig. S2A–C). The dNPCs and vNPCs were then differentiated to highly enriched populations of dNeurons and vNeurons (Supplemental Fig. S2D). dNeurons and vNeurons expressed SLC17A7 (also known as VGLUT1) and GABA, respectively (Supplemental Fig. S2E,F). The FLAG tag was detected by immunofluorescence in NPCs and neurons differentiated from FMR1-FLAG hPSCs but not parental cells (Supplemental Fig. S2G,H). Therefore, our FMR1-FLAG hPSCs express endogenously FLAG-tagged FMR1 and can be patterned into highly enriched forebrain NPCs and neurons, enabling isolation of FMR1 through the FLAG tag.
Identification of FMR1 binding targets in human neural cells using an incorporated CLIP strategy
Identification of FMR1 targets by CLIP required large quantities (>108) of cells in both mouse brain (Darnell et al. 2011; Maurin et al. 2018) and a human transformed nonneural cell line (Ascano et al. 2012). However, the limited number of neurons differentiated from human hPSCs in culture prohibited us from using similar protocols. We adapted two recently developed CLIP methods, irCLIP and eCLIP, that demonstrated robust CLIP from relatively small numbers of HeLa or HEK293 cells (<106) (Van Nostrand et al. 2016; Zarnegar et al. 2016), with additional modifications to improve efficiency (2.6-fold 16 h+ vs. 1.5 h−) (Supplemental Fig. S2I). As in irCLIP, a near-infrared dye-labeled 3′ adapter was ligated to RNAs isolated by CLIP to facilitate visualization of RNAs on gels for size selection (Fig. 1E). Pilot CLIP experiments with FMR1-FLAG NPCs demonstrated better isolation of RNAs by a FLAG antibody than several FMR1 antibodies and successful enrichment of RNAs in <106 NPCs (Supplemental Fig. S2J,K). Moreover, RNAs were successfully isolated from the FMR1-FLAG neurons (Supplemental Fig. S2L).
To identify FMR1-bound RNAs, CLIP using a FLAG antibody was performed on NPCs and neurons (Fig. 1D) differentiated from the three FLAG hPSCs (red in Fig. 1C; FLAG in Fig. 1E) with their parental hPSCs serving as negative controls (red in Fig. 1C; WT in Fig. 1E). To increase the rigor of target selection, two negative controls were included for each FMR1-FLAG-CLIP: (1) CLIP on WT NPCs or neurons (WT in Fig. 1E); and (2) size-matched input (SMI as described in eCLIP) of the FMR1-FLAG NPCs or neurons (SMI in Fig. 1E). RNAs isolated from CLIP and SMI were converted to stranded cDNA libraries for Illumina sequencing (Fig. 1E).
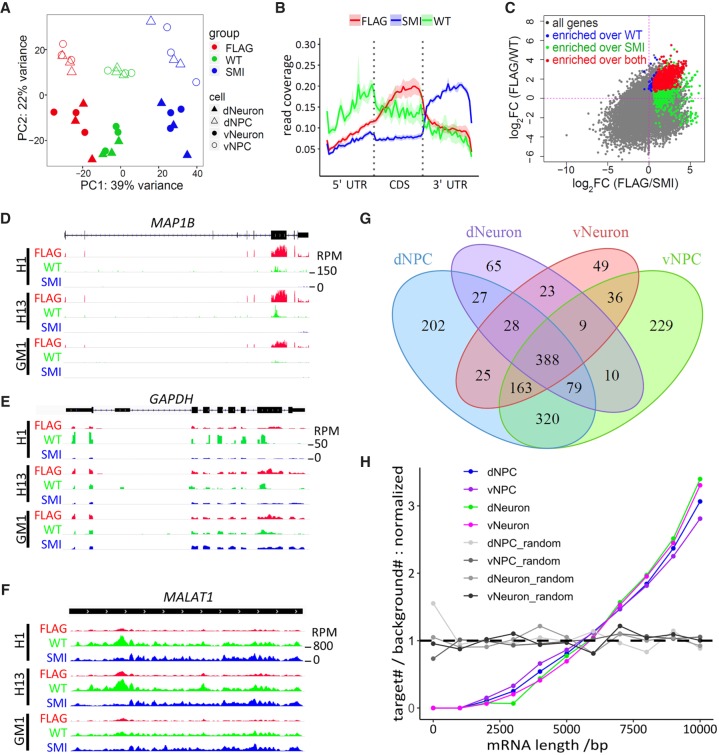
Principal component analysis of the sequencing data shows that samples were clustered by experimental groups (CLIP, WT, and SMI) in both NPCs and neurons (Fig. 2A) and that there was a high correlation between replicates of CLIP samples but not between differential experimental groups (Supplemental Fig. S3A–C), demonstrating the robustness of our neural differentiation and modified CLIP method for isolation of FMR1 targets. Analysis of read distribution across gene subregions revealed enriched binding of FMR1 in coding sequences (CDSs) of genes compared to untranslated regions (5′ UTR and 3′ UTR), whereas a similar pattern was not observed in either the SMI control or the WT control samples (Fig. 2B). These results are consistent with previous characterization of FMR1 targets in mice (Darnell et al. 2011), supporting successful isolation of FMR1-bound RNAs from human neural cells.
Figure 2.
Identification of FMR1 targets by CLIP in human neural cells. (A) PCA plot of CLIP-seq data. Color and shapes represent experimental conditions and cell types. (B) Line plots show relative distribution of reads over gene elements. (5′ UTR / 3′ UTR) 5′ and 3′ untranslated region, (CDS) coding sequence. Reads mapped to protein-coding genes of all samples were used for the analysis. Lines and shades represent mean ± SE. (C) Representative scatter plot of log2(fold change) (cell: dNPC) shows that FMR1 targets (red) were defined as significantly enriched in the FLAG group over both WT control (blue) and SMI control (green). Gray genes were not significantly enriched in the FLAG group over either control. (D–F) Visualization of reads by Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al. 2013) on representative FMR1 target, MAP1B (D), and nontargets, GAPDH (E) and MALAT1 (F). Tracks of dNPC are shown. Scale of height (RPM) is the same for all tracks in the same panel. (G) Venn diagram showing overlaps of FMR1 targets identified in four cell types analyzed. (H) Analysis of length distribution of FMR1 targets in various cell types. Line plots show normalized ratio of number of targets to number of background genes at 1000-bp windows of mRNA lengths. The random sets were the same number of genes randomly picked from the background genes (protein-coding genes with a RPKM > 0.1). P < 2.2 × 10−16 for each of the four cell types, two-sample Kolmogorov–Smirnov test comparing targets to background genes.
FMR1 targets were defined as significantly enriched (P < 0.05 and fold change>1.3) (Supplemental Table S1) in FLAG samples over both WT and SMI control samples (Fig. 2C). Published CLIP experiments often lack the inclusion of CLIP of cells that do not have the antigen recognized by the antibody used for CLIP. We found that our WT control (CLIP of parental lines without FLAG tag) served as an important control for reducing false positives (Fig. 2C), demonstrating the importance of including these rigorous controls for CLIP-seq (Van Nostrand et al. 2017). The use of both WT and SMI controls allowed us to stringently select FMR1 targets, such as MAP1B (Fig. 2D), and reduce false positives, such as the highly expressed coding gene GAPDH (Fig. 2E) and noncoding gene MALAT1 (Fig. 2F). RNAs of 1653 genes were identified as FMR1 targets with 1232 genes from dNPC, 1234 from vNPC, 629 from dNeuron, and 721 from vNeuron groups (Fig. 2G; Supplemental Table S1). Of the 1653 targets, 1650 were protein-coding genes, consistent with the function of FMR1 as a translation regulator. FMR1 targets were validated by RNA immunoprecipitation-qPCR (Supplemental Fig. 3D,E), and dysregulation of some FMR1 target protein levels was also observed in neurons (Supplemental Fig. S3F–I).
We then compared the target genes identified in human NPCs and neurons with published CLIP data sets from human HEK293 cells (referred to hereafter as Ascano) (Ascano et al. 2012), human post-mortem brain (referred to hereafter as Tran) (Tran et al. 2019), and mouse brains (referred to hereafter as Darnell and Maurin) (Darnell et al. 2011; Maurin et al. 2018). Because NPCs are proliferative, we compared the targets identified in dNPC and vNPC with Ascano data on proliferating HEK293 cells. Although there is significant overlap with the Ascano data (Supplemental Fig. S4A,B), most FMR1 targets in HEK293 cells were not identified as FMR1 targets in the NPCs, and the FMR1 targets identified in the NPCs but not HEK293 cells were enriched in genes associated with neural differentiation (Supplemental Fig. S4C). Hence, FMR1 targets identified in human NPCs provide a more relevant reference for studying FMR1 functions during neurodevelopment, and so we excluded the Ascano data in subsequent analyses.
To examine the patterns of overlap of FMR1 targets identified in our study and published studies on brain tissues, we applied Gaussian mixture model clustering on the union of hits identified in our human neural cells and published mouse (Darnell et al. 2011; Maurin et al. 2018) and human (Tran et al. 2019) brain tissues, resulting in a total of 2620 genes (Supplemental Fig. S4D). Clusters 1–8 were largely composed of the CLIP targets identified in our study and were enriched for both known functions of FMR1, such as neuronal development, dendritic transport, and synaptic processes, and more general processes of histone modification and cytoskeleton organization. Cluster 9 and 10 specifically comprised previously identified FMR1 targets in mouse and human brains (Darnell et al. 2011; Maurin et al. 2018; Tran et al. 2019) and were enriched primarily for neuronal synapse processes (Supplemental Fig. S4E). Hence, we successfully identified FMR1-bound RNAs from human NPCs and neurons using FMR1-FLAG hPSCs and CLIP.
Motif analysis of high-confidence FMR1-binding sites in our identified targets revealed a top motif, UGGA, which is consistent with previously identified FMR1-binding consensus sequences (Supplemental Fig. S4F; Ascano et al. 2012; Maurin et al. 2018). We also observed higher FMR1 binding of mRNAs containing G-quadruplex in three of the four cell types (Supplemental Fig. S4G), consistent with reports that FMR1 binds G-quadruplex in RNAs (Guo and Bartel 2016). Length distribution analysis showed that FMR1 targets were significantly longer compared to all expressed protein-coding genes (P < 2.2 × 10−16, Kolmogorov–Smirnov test) (Fig. 2H), corroborating a recent ribosome footprinting study showing that FMR1 preferentially regulates the translation of large proteins (Greenblatt and Spradling 2018). Taken together, the results support the idea that FMR1 preferentially binds long mRNAs to regulate their translation.
FMR1-KO NPCs exhibit delayed neural differentiation
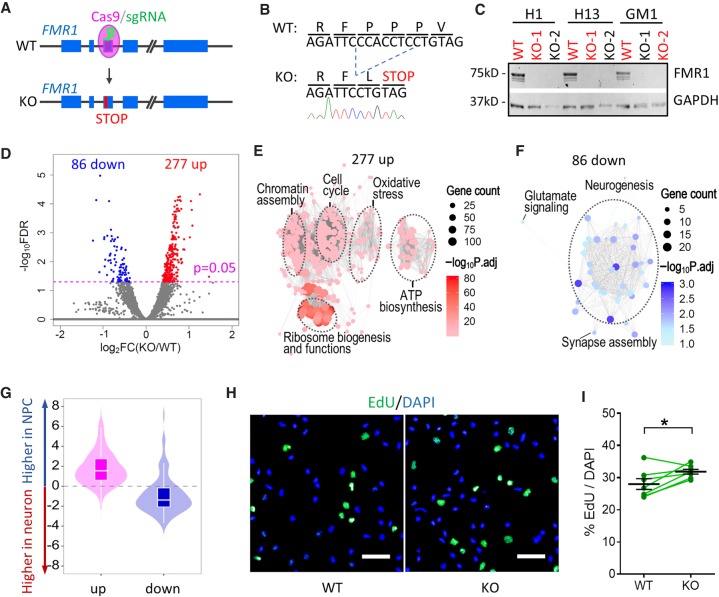
To integrate identification of FMR1 targets in human NPCs and neurons with the impact of FMR1 on downstream gene expression, we analyzed the transcriptome of cells lacking FMR1. We generated isogenic FMR1 knockout (FMR1 KO) hPSC lines from the same WT hPSC lines used to create FMR1-FLAG lines (H1, H13, and GM1). FMR1 KO lines were generated using CRISPR-Cas9 with a sgRNA targeting the third exon of the FMR1 gene to induce frameshift mutations (Fig. 3A). We selected clones with the same 7-bp deletion at the sgRNA targeting site in all three hPSC lines resulting in premature stop codons (Fig. 3B) for subsequent experiments. KO hPSCs had normal morphology, normal karyotypes, expressed stem cell markers, and had no detectable off-target mutations (Supplemental Fig. S5A–C). FMR1 levels were undetectable in all three KO hPSC lines (Fig. 3C). Following differentiation as described above (Fig. 1), highly enriched forebrain NPCs and neurons were obtained from the three pairs of isogenic KO and parental (WT) hPSCs, and the absence of FMR1 was confirmed by immunofluorescence in the KO hPSC differentiated cells (Supplemental Fig. S5D–H).
Figure 3.
Transcriptomic analysis of NPCs and neurons derived from WT and FMR1-KO hPSCs. (A) Schematic diagram of generation of isogenic FMR1 KO hPSCs using a seamless CRISPR-Cas9 strategy. (B) Sanger sequencing shows a 7-bp deletion in the FMR1 gene and a premature stop codon of FMR1 KO cells. (C) Western blot confirming knockout of FMR1 in FMR1 KO hPSCs, GAPDH as a loading control. hPSC lines in red were used for experiments. (D) Volcano plot of gene expression in WT and FMR1 KO dNPCs. (E) GO enrichment of the 277 genes up-regulated in FMR1 KO dNPCs. (F) GO enrichment of the 86 genes down-regulated in FMR1 KO dNPCs. (G) Violin and box plots of up-regulated (up) and down-regulated (down) genes in KO dNPCs with respect to their expression levels in dNPCs and dNeurons (Box plot: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range). (H) Representative images of EdU incorporation assay of dNPCs derived from H1 WT and KO hPSCs (scale bar, 50 µm). (I) Quantitative analysis of the percentage of EdU+ dNPCs (mean ± SE; dots and lines show batches of differentiation; P = 0.02, paired t-test, two tails).
Transcriptomic analysis of KO NPCs and neurons showed clear clustering of the samples by cell types by principal component analysis (PCA) (Supplemental Fig. S6A). The distinct gene expression patterns in the four cell types confirmed successful dorsal/ventral patterning and neuronal differentiation. We identified 363 differentially expressed genes (DEGs) in KO dNPCs compared to isogenic parental dNPCs (Fig. 3D) and 287 DEGs in vNPCs (Supplemental Fig. S6B,C; Supplemental Table S2). The 277 up-regulated genes in KO dNPCs showed enrichment in pathways related to cell cycle regulation, chromatin assembly, oxidative stress, ATP production, and ribosomal functions (Fig. 3E), while the 86 down-regulated genes were enriched for neuronal differentiation and synaptic functions (Fig. 3F), as assessed by Gene Ontology (GO). The results suggest that the KO dNPCs are more proliferative and less differentiated compared to controls. Indeed, most of the up-regulated genes were expressed higher in dNPCs than in dNeurons (dNPC enriched genes), while most of the down-regulated genes were dNeuron-enriched genes (Fig. 3G). We then assessed proliferation of KO dNPCs using pulse labeling by a thymidine analog EdU (Fig. 3H) and found that, indeed, KO dNPCs incorporated more EdU compared to isogenic WT control dNPCs, confirming the increased proliferation and DNA replication in KO NPCs (Fig. 3I). Compared to NPCs, relatively few genes were differentially expressed in KO neurons; 6 DEGs were identified in dNeuron and 55 in vNeuron (Supplemental Fig. S6D,E; Supplemental Table S2). Although the number of DEGs in KO neurons is small, their changes can still provide insight into the effect of FMR1 KO on the transcriptome. Therefore, we applied a GMM-based clustering approach to the 30,644 measured genes in our RNA-seq data (Methods). The DEG genes are contained in clusters (Supplemental Fig. S6F) exhibiting significant up-regulation (Cluster 1) and high (Cluster 2) and moderate (Cluster 3, 5) down-regulation in NPCs compared to neurons (two-sided t-test, P value < 0.05) (Supplemental Methods). Up-regulated clusters are enriched in pathways associated with transcription, DNA damage and replication, and cell cycle, while the down-regulated clusters were enriched for functions specific to the nervous system (e.g., nervous system development, synaptic transmission, neurotransmitter processes). Taken together and consistent with previous findings that FXS NPCs display a more immature phenotype (Doers et al. 2014; Motanis and Buonomano 2015; Telias et al. 2015), our results suggest a delayed differentiation from NPCs to neurons in the absence of FMR1.
FMR1 regulates gene networks important for neural cell functions
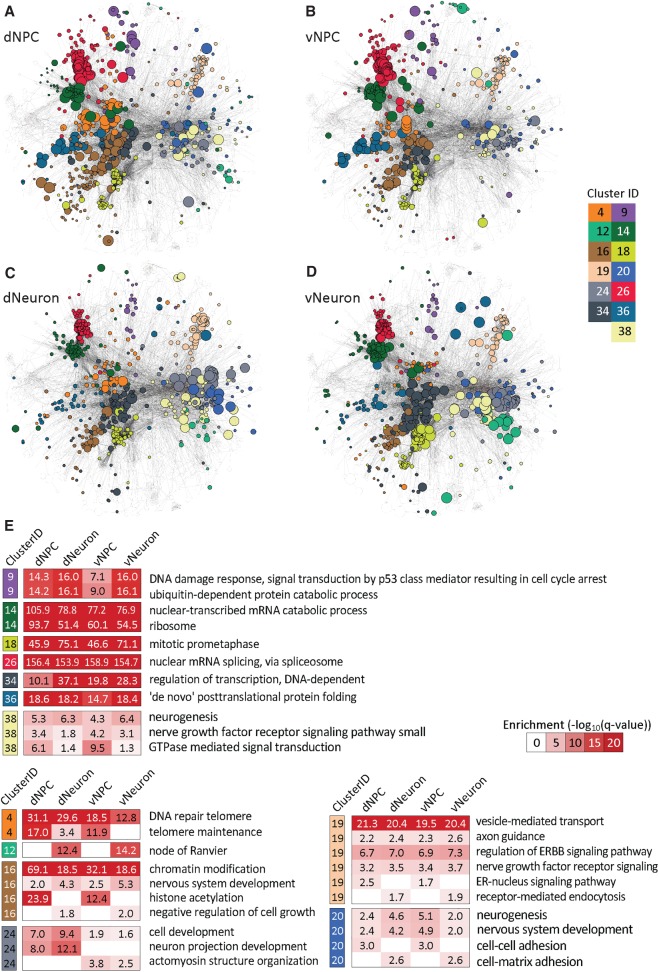
We next combined our FMR1 CLIP-seq and FMR1 KO RNA-seq data to assess the utility of analyzing both the binding and KO data together as opposed to considering each measurement alone for understanding FMR1's overall function in human developing neural cells. A direct comparison of these two sets of NPC data showed limited overlap between CLIP-seq and RNA-seq data (Supplemental Fig. S7), and we hypothesized that this lack of overlap could be because specific FMR1 gene targets are different from differentially expressed genes, but they are functionally related as gene sets. FMR1 could regulate translation of its direct targets, and some of these targets, for example, transcription factors, may affect transcription of other genes. To test this hypothesis and investigate cell type–specific regulation by FMR1, we applied a new network-based analysis framework, comprising graph diffusion (Yeger-Lotem et al. 2009; Huang et al. 2013; Chasman et al. 2016a) and graph clustering to identify subnetworks that could better explain the relationship among the CLIP-seq and RNA-seq data. Within each cell type, we used graph diffusion to obtain the top 5% of genes that were most connected to the CLIP and RNA hits. We created a weighted network that included these top 5% nodes and the CLIP and RNA hits across all cell types spanning a total of 1810 genes. We next applied a multitask graph clustering approach to identify 45 subnetworks in each of the four cell types simultaneously (Fig. 4A–D; Supplemental Fig. S8; Supplemental Table S3). With the exception of dNeuron, which had very few mRNA hits, many of the clusters included both mRNA and CLIP hits (dNPC: 37 clusters, vNPC: 32, vNeuron: 17) (Supplemental Fig. S8; Supplemental Table S3A). Some clusters exhibited cell type–specific patterns (Fig. 4E; Supplemental Fig. S9A,B; Supplemental Table S3A; Supplemental Methods), while others were shared across the neurons (15 clusters), NPCs (nine clusters), or all four cell types (15 clusters). The common clusters were enriched in functions such as neurogenesis, mRNA splicing, DNA damage response, protein modification, and catabolism, which likely reflect common functions of FMR1 across different cell types and developmental stages (Supplemental Table S3B). Cluster #12, enriched in nodes of Ranvier, was uniquely identified in neurons, suggesting distinct functions of FMR1 in neurons compared to NPCs. Likewise, cluster #24 was specific to ventral cells and exhibited enrichment for “actomyosin structure organization,” implicating a role of FMR1 in early development and migration of ventral inhibitory neurons that require actomyosin-dependent cytoskeleton remodeling (Tielens et al. 2016; Silva et al. 2018). The clusters are significantly different from those obtained from randomized inputs and from CLIP- and RNA-seq alone, suggesting that, although there is low overlap between the mRNA and CLIP hits, the clusters reveal related relevant pathways, demonstrating the importance of network-based integration (Supplemental Table S5A).
Figure 4.
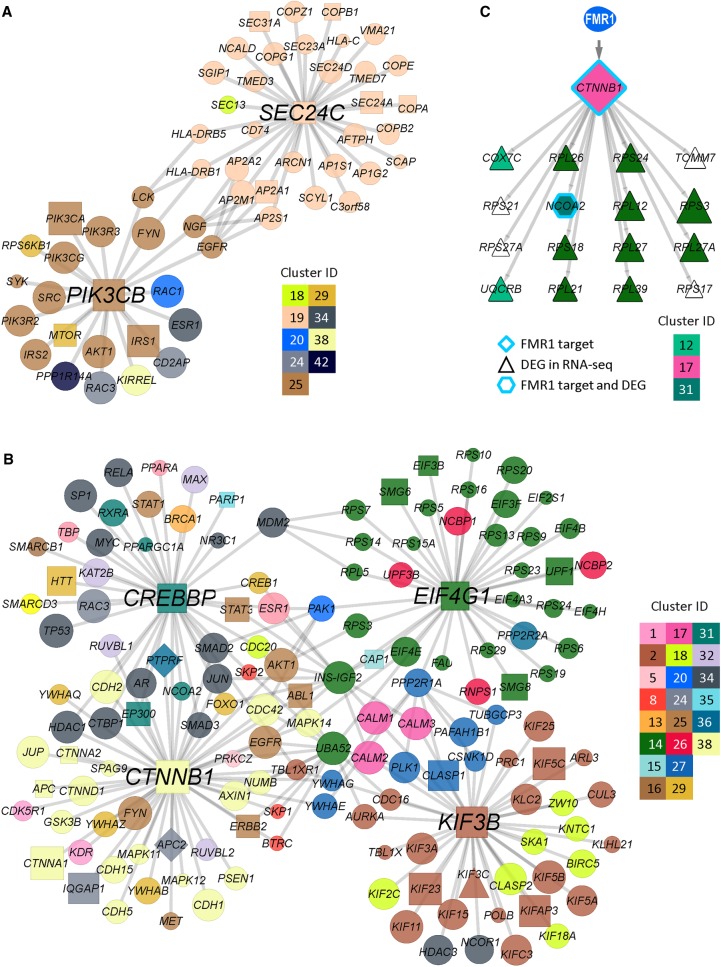
Cell type–specific network clusters of CLIP- and RNA-seq integrative network analysis. (A–D) Cell type–specific network clusters for dNPC (A), vNPC (B), dNeuron (C), and vNeuron (D), identified from our integrative network clustering analysis approach using CLIP and RNA-seq data simultaneously. Node colors correspond to a cluster assignment, and the size of the node is proportional to node diffusion values, which assess the network proximity to the CLIP- and RNA-seq hits. Colors correspond to 13 clusters, seven of which are conserved across all cell types (9, 14, 18, 26, 34, 36, 38) and six clusters that exhibit cell type–specific patterns either among the NPC versus neurons or dorsal versus ventral cell types (4, 12, 16, 19, 20, 24). (E) Enriched Gene Ontology (GO) processes in the 13 clusters depicted in A–D. Cluster IDs and color coding are the same as in A–D. Red-white heat maps show significance of enrichment in a cluster from a particular cell type (−log10(FDR), hypergeometric test).
To prioritize gene hubs in the CLIP-seq and RNA-seq data, we ranked each gene based on their proximity to the CLIP- and RNA-seq hits in each cell type. We used a combination of the graph diffusion score (described above), CLIP-seq signal, and the number of edges associated with a gene. We selected the top 1% of genes (193 genes across all four cell types) as gene hubs for further interpretation and experimental validation (Supplemental Table S4). We identified 17 gene hubs that are shared across all four cell types, 20 genes that are common in NPCs only, and nine genes that are common in neurons only. Gene hubs prioritized using both RNA-seq and CLIP-seq data have significantly lower overlap than when diffusing from random gene sets or from RNA-seq and CLIP-seq hits alone (Supplemental Table S5B). These hubs also more accurately reveal genes associated with neuronal processes (Supplemental Fig. S10) than when using RNA-seq or CLIP-seq alone, further highlighting the importance of considering both types of measurements for understanding FMR1 function. The hubs fell in the common clusters (Cluster #16: RB1, Cluster #19: AP2A1, and Cluster #25: PTPN11, FYN) as well as in cell type–specific clusters (e.g., NPC-specific hubs, Cluster #4: ATM, Cluster #15: HSPA4, and neuron-specific hubs, Cluster #12: ANK1, Cluster #34: AR, Cluster #20: PKN1). Several of these hubs have also been associated with neurodegenerative or cognitive diseases (e.g., ABL1, CREBBP, CTNNB1, MED12, PPP2R1A, SMC1A, PIK3CA) (Supplemental Table S4), providing support for the relevance of identified genes for the human brain (Piñero et al. 2017).
We experimentally validated several of the prioritized hubs. Among the top hubs in dNeuron were PIK3CB and SEC24C (Fig. 5A). We validated the regulation of PIK3CB and SEC24C by FMR1 by RIP-qPCR and immunoblot (Supplemental Fig. S3). PIK3CB is novel to our study and was identified as an FMR1 target in both dNPCs and dNeurons. Indeed, up-regulation of PIK3CB activities contributes to both neuronal and behavioral deficits of FMR1-deficient mice, which can be rescued by PI3K inhibitors (Gross et al. 2010, 2015). SEC24C is a component of the coat protein complex II that plays a role in intracellular transport from ER and has been shown to be essential for neuronal homeostasis in both mouse brain and hiPSC-derived neurons (Wang et al. 2018). Sec24c has been identified as a FMR1 target in CLIP from juvenile mouse brains (Maurin et al. 2018) but not adult human brains (Tran et al. 2019). The validated top hub in vNeuron, KIF3B (Fig. 5B), was also identified in a CLIP data set of human cortex (Tran et al. 2019) and plays an important role in axon establishment through polarized trafficking of critical components (Ichinose et al. 2019). Other top validated hubs in vNeuron included CTNNB1, EIF4G1, and CREBBP. CTNNB1 is down-regulated in vNeuron (Supplemental Fig. S3I) while up-regulated in dNPCs (Supplemental Fig. S11B), with no change in other cells, suggesting cell type–specific regulation by FMR1. EIF4G1 is a key component in protein translational initiation in many cell types, including neurons, and its dysregulation is implicated in neurodegenerative Parkinson's disease (Taymans et al. 2015). Although direct association between EIF4G1 and FMR1 has not been shown previously, translational dysregulation is a clear consequence of FMR1 deficiency both in animal models and in humans (Richter et al. 2015). CREBBP is a transcription factor whose expression is critical in neurodevelopment, and its intrinsic histone acetyltransferase activity may contribute to altered histone acetylation seen in FMR1-deficient mouse brains and mouse adult NPCs (Korb et al. 2017; Li et al. 2018).
Figure 5.
Hub genes and subnetworks in neural cells. (A,B) Hubs in dNeuron (A) and vNeuron (B) and the subnetworks consisting of the first neighbor genes of hubs. Node shapes discriminate between hits and nonhits and different types of hits. CLIP-seq target (square), RNA-seq DEG (triangle), both (diamond), and network prioritized genes (circle). Node sizes correspond to diffusion score, and node colors correspond to cluster assignments. (C) A proposed model of the relationship between FMR1 CLIP-seq targets and DEGs. Here, CTNNB1, which has been identified as an FMR1 direct target, encodes a predicted transcription factor regulating 16 DEGs. Node sizes and colors correspond to diffusion score and cluster assignment, respectively.
To investigate the mechanism that FMR1 affects transcriptional changes by regulating protein levels of targets such as transcription factors and signaling proteins, we used an Integer Linear Programming (ILP)-based approach (Chasman et al. 2016b) to identify regulatory networks connecting FMR1 to such transcriptional regulators mediating large-scale transcriptomic changes (Supplemental Table S5C; Supplemental Fig. S11D,E). One such network focused on CTNNB1 (encoding catenin beta 1), which was among the top ranked FMR1 targets that regulate DEGs in dNPC (Fig. 5C). While the RNA level of CTNNB1 did not show a significant change in FMR1 KO dNPC, its protein level was elevated in KO dNPC (Supplemental Fig. S11). Our analysis indicated that catenin beta 1 regulates genes related to ribosome biogenesis (Fig. 5C). The ribosomal genes regulated by catenin beta 1 were elevated in FMR1 KO dNPC (Supplemental Fig. S11C), consistent with a previous report that catenin beta 1 promotes ribosome biogenesis (Davuluri et al. 2019). Taken together, our results show that studying FMR1 at multiple regulatory levels through our network-based approach can identify components of important cellular functions regulated by FMR1 in human neural cells. These network clusters and gene hubs validate some of the animal studies and point to novel functional targets for further mechanistic investigation of FXS.
Human FMR1 targets are associated with FXS and autism phenotypes
FXS is characterized by cognitive disability, anxiety, attention deficit, language delay, and physical features such as large ears and long faces (Kaufmann et al. 2017). To examine how gene regulation by FMR1 may contribute to characteristics of FXS and autism, we compared FMR1 targets, the prioritized gene hubs and the subnetworks comprising FMR1 targets to genes associated with autism risk genes (SFARI) (Abrahams et al. 2013) and a published list of IQ genes (Savage et al. 2018). We discovered significant overlap of our data with both data sets (Fig. 6A; Supplemental Table S6A). For example, high-confidence syndromic autism risk genes, ADNP, ARID1B, KMT2A, and TRIP12, were FMR1 targets in both NPCs and neurons (full list in Supplemental Table S6A). The significant association with IQ genes is consistent with lower IQ scores in the majority of FXS patients. We next performed gene-disease association analysis on the targets, the prioritized gene hubs, and the subnetworks comprising FMR1 targets using a large database of gene-human disease association, DisGeNET (Fig. 6B; Piñero et al. 2017). Intellectual disability is among the top enriched disease features by the FMR1 targets, prioritized gene hubs, and subnetworks and is increased when considering the prioritized hubs in both NPCs and neurons (Fig. 6B; Supplemental Table S6B–F). Other brain function-related FXS features were also significantly enriched, including hyperactive behavior, autistic disorder, ADHD, and aggressive behavior (Fig. 6B). Besides the FXS-related characteristics, FMR1 target genes were also significantly associated with other FXS-related non-neural characteristics, such as muscle hypotonia, implying common molecular regulation by FMR1 in other cell types. Moreover, several of the clusters of FMR1 targets were also enriched for features of FXS diseases in all cell types (Cluster #16 and Cluster #25) (Supplemental Fig. S12) as well as in a cell type–specific manner (Cluster# 4 and #45 in dNPCs, Cluster #34 in neurons). Taken together, analyzing the data at the entire target set level, additional prioritized nodes, and the individual subnetworks, shows that FMR1 targets in human NPCs and neurons are significantly associated with prominent symptoms of FXS patients as well features broadly associated with a number of neurological disorders.
Figure 6.
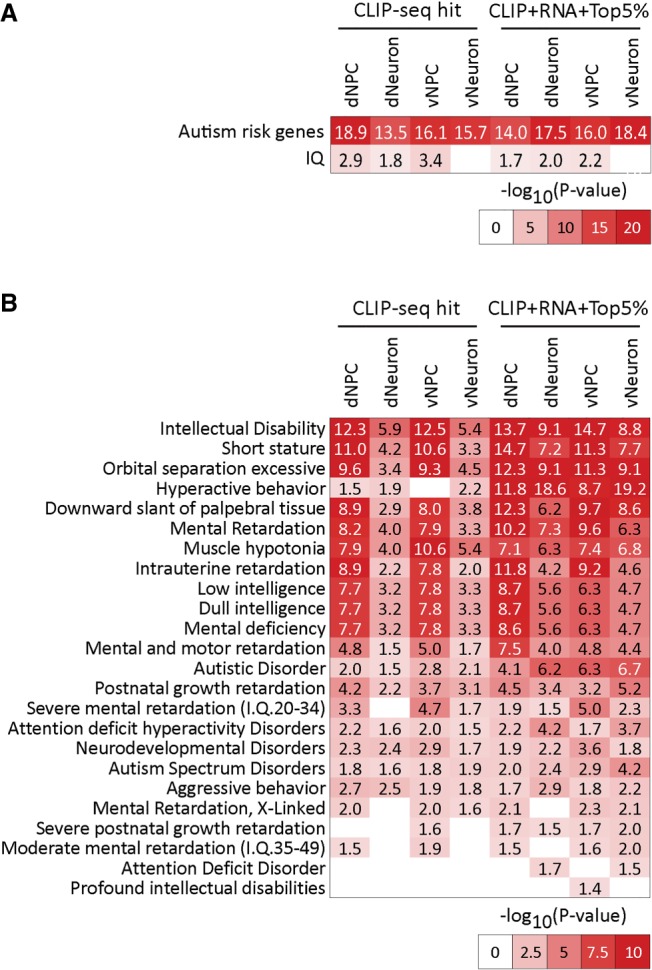
Disease and phenotype enrichment of FMR1 targets and prioritized genes in human neural cells. (A) Heat map of enrichment of autism risk genes and IQ-related in our CLIP-seq target set (left) and network diffusion-based augmented gene sets (right). (B) Heat map of enrichment of genes associated with neuronal diseases and symptoms from DisGeNet, in our CLIP-seq (left) and network diffusion augmented gene sets (right) in each of the four cell types. Enrichment was tested using FDR-corrected hypergeometric test. Numbers indicate −log10(FDR). Network diffusion-based gene sets include genes that were in the top 5% after diffusion.
Discussion
In this study, we addressed the difficulty of identifying genome-wide FMR1 targets in human neural cells and discovered that human FMR1 targets include cell type–specific as well as shared gene sets across dorsal and ventral forebrain NPCs and neurons. Target genes are enriched in pathways critical for neurodevelopment and are associated with clinical phenotypes of FXS and autism. Integrative network analyses of CLIP data together with transcriptomic data identify molecular subnetworks and gene hubs, several of which have an important role in human neurodevelopment.
Several factors have been hypothesized to mediate target selection by FMR1, including binding to consensus sequence motifs, RNA secondary structure (RG4), and subregions of the gene body. While motif analysis shows that FMR1 preferentially binds certain motifs (Ascano et al. 2012; Maurin et al. 2018; and this study), the motifs are more degenerate than for many other RNA-binding proteins and are thus poor predictors of FMR1 targets. Binding of FMR1 to RG4 has also been reported in mouse brain (Maurin et al. 2018), and our analysis also indicates preferential binding of RG4-containing RNAs by FMR1. However, we did not observe a consistent preference of binding to RG4-containing RNAs; binding preference was not observed in dNeuron and was only mildly indicated in the other three cell types. FMR1 binding has also been shown to be preferential to the coding region (CDS) of mRNAs versus to 5′ and 3′ UTRs in mouse (Darnell et al. 2011), and we confirmed this in our study. Binding to CDSs is considered to be related to the translational regulation of FMR1 targets. In addition, our analysis indicates that FMR1 preferentially binds longer RNA targets. Ribosome profiling in Drosophila showed that genes dysregulated at translation are longer (Greenblatt and Spradling 2018), suggesting that dysregulated genes were directly bound by FMR1. Thus, our results corroborate different aspects of FMR1 binding that have been reported and extend the finding by showing for the first time that FMR1 binds long RNAs in human cells.
We have identified FMR1 targets in four types of human neural cells. Many FMR1 targets (39%–77%) are shared between cell types, suggesting common FMR1 target selection and potentially similar functions. Indeed, GO analysis indicates that FMR1 targets in all four types of neural cells are involved in neurogenesis. Moreover, we also identified FMR1 targets that were specific to certain cell types. For example, there were 137 FMR1 targets identified in neurons that were not identified in NPCs (Fig. 2G), signifying neuron-specific targets. The top enriched GO biological process by the NPC-specific targets included regulation of cell cycle and gene regulation, which may contribute to macrocephaly manifested in some individuals with FXS and autism. FMR1 targets in the ventral (vNPC/vNeuron) but not dorsal (dNPC/dNeuron) neural cells were enriched in cell junctions, ATP binding, and cytoskeleton, suggesting that FMR1 might play a role in the unique tangential migration of the ventrally derived neural cells to the dorsal cortex.
CLIP has been carried out on mouse brains to explore direct targets of FMR1 (Brown et al. 2001; Darnell et al. 2011; Tabet et al. 2016; Maurin et al. 2018). The first attempts at identifying human FMR1 targets using CLIP used human immortalized cell lines derived from peripheral nonneural tissues (Ascano et al. 2012; Van Nostrand et al. 2016, 2017) and, recently, post-mortem adult human frontal cortical tissues (Tran et al. 2019). Although there was overlap between FMR1 targets we identified in the neural cells and those identified in the human post-mortem tissues, the majority of FMR1 targets were not shared between the two types of human samples (Fig. 4). This discrepancy can be attributed to differences in experimental design between studies, different cell types analyzed, and different analysis methods used. It will be useful to use our paradigm to identify FMR1 targets in more mature human neurons generated through long-term culture.
Effects of FMR1 on downstream gene expression have been examined in FXS-iPSC-derived neural cells, in which FMR1 expression is lost or significantly reduced. Microarray and RNA-seq have been performed on cells of different developmental stages during neural differentiation from hPSCs (Bittel et al. 2007; Halevy et al. 2015; Lu et al. 2016; Boland et al. 2017). Although aberrant expression of neural differentiation-related genes was identified in all the studies, inconsistencies were observed between the studies. Halevy et al. (2015) found that genes related to neuronal differentiation were down-regulated in neurons derived from FXS-iPSCs compared to controls and the dysregulation could be attributed to elevated REST expression. Boland et al. (2017) showed that dysregulated genes during neural differentiation were associated with cell signaling and cell migration, also implying defects in neuronal differentiation from FXS-iPSCs. In contrast, Lu et al. (2016) found that genes related to early neuronal differentiation were up-regulated in FXS-iPSC derived neurons. All three studies used controls from different genetic backgrounds that may identify characteristics that are due to inter-individual differences rather than lack of FMR1 specifically. In addition, mosaic FMR1 gene inactivation is frequently found in FXS individuals and cells (Santoro et al. 2012), including the cells used in Boland et al. (2017), making it difficult to define bonafide FMR1 regulation. Using isogenic FMR1-KO neural cells from three hPSC lines, our stringent transcriptomic analysis identified DEGs associated with delayed neural differentiation. A caveat is that FMR1 is lost before neural differentiation in both FXS-iPSCs and our isogenic KO hPSCs, whereas in FXS patients, FMR1 is gradually lost during embryonic brain development (Willemsen et al. 2002). It has been shown that human ESCs derived from FXS embryos recapitulate the gradual loss of FMR1 during neural differentiation and also display deficits in early neural differentiation as manifested by prolonged neuroblast state and delayed neuronal differentiation (Telias et al. 2015). However, limited availability of these FXS hESCs and inability to consistently control methylation prevents extensive characterization and application of these cells (Avitzour et al. 2014).
Network-based integrative approaches are powerful for reconciling complementary gene sets obtained from different omic methods by identifying nodes that are not detected solely by one technology but are important connectors between these gene sets (Chasman et al. 2016a). We provide a novel application of this approach, enabling a more comprehensive view of the FMR1 gene network than that obtained using one omic data type alone. We were able to prioritize the genes based on their connectivity to the genes identified using complementary assays. These genes were more significantly different from random and recovered neuronal genes much better than when using gene sets inferred from each data type alone. In addition, using our innovative approach, we tested the hypothesis that FMR1 may regulate translation of transcriptional regulators which in turn affects transcription of downstream genes observed in RNA-seq data. One example is the FMR1-CTNNB1-ribosome RNA network (Fig. 5C; Supplemental Fig. S11). Dysregulated ribosome biogenesis has recently been implicated in pathogenesis of neurodevelopmental syndromes (Hetman and Slomnicki 2019). Enhanced ribosome biogenesis leads to increased neural progenitor proliferation and macrocephaly (Chau et al. 2018), supporting our observed phenotype in FMR1 KO dNPCs (Fig. 3) and consistent with macrocephaly in FXS. Dysregulated ribosome biogenesis has also been associated with craniofacial dysmorphology (Ross and Zarbalis 2014), a characteristic of FXS. One limitation of our network-based approach is its reliance on existing functional networks, which can be context-unspecific. While our two-step diffusion approach infers a context-specific network, one future direction is to construct a cell type–specific network leveraging public transcriptome data sets as the background networks.
In summary, we have developed several state-of-the-art strategies that can be broadly applied to studies of RNA-BPs. The identification of FMR1 targets specifically in inhibitory neurons is the first of its kind in any species. Our results provide the first genome-wide identification of FMR1 targets in human neurons, which, together with our genetically engineered hPSCs, new CLIP- and RNA-seq data sets, and data analysis methods, serve as much needed resources for FXS, autism, and intellectual disability research communities.
Methods
Culture of human pluripotent stem cells and neural differentiation
Human embryonic cell line H1 (WA01) and H13 (WA13) (WiCell) and GM1 (GM00498-4) iPSC lines were maintained as described (Li et al. 2017). Neural induction was carried out using a dual SMAD method (Chambers et al. 2009) with modifications. Briefly, hPSCs were differentiated to neuroepithelia (NEP) by dual SMAD inhibition for 9 d. NEPs were then patterned to dorsal forebrain or ventral forebrain neural progenitor cells by cyclopamine or SHH, respectively. To obtain neurons, the NPCs were dissociated and plated on Matrigel-coated plates for terminal neuronal differentiation.
Generation of FMR1-FLAG and FMR1-KO hPSC lines by seamless genome editing
FMR1-FLAG and FMR1-KO hPSCs were generated by genome editing as described (Li et al. 2017). In brief, Cas9/sgRNA-expressing plasmids and donor plasmids (plasmid construction described in Supplemental Methods and Supplemental Table S7) were electroporated into hPSCs. Transfected hPSCs were transiently selected with puromycin. In ∼2 wk, individual colonies were screened for desired genome editing and expanded in culture for experiments.
CLIP-seq
CLIP-seq was adapted from published irCLIP and eCLIP methods (Van Nostrand et al. 2016; Zarnegar et al. 2016). Briefly, NPCs and neurons were crosslinked with 254-nm UV-C followed by cell lysis and partial digestion of RNAs. FMR1-bound RNAs were isolated by immunoprecipitation, dephosphorylated, and ligated to a dye-labeled 3′ adapter. Samples were then resolved using SDS-PAGE and transferred to nitrocellulose membranes. FMR1-bound RNAs were visualized and size selected by cutting the membranes. Size-selected RNAs were purified by Proteinase K digestion and ethanol precipitation. The purified RNAs were converted to cDNAs followed by circularization of the cDNAs. cDNA libraries were constructed from the cDNAs by PCR amplification, and libraries were sequenced using Illumina HiSeq 2500.
Analysis of mRNA length of FMR1 targets
Length of genes (sum of exons) was obtained from the human genome sequence and annotation files above. The normalized ratio of targets to background was calculated as number of FMR1 targets in each length window/number of background genes in each length window/(total number of FMR1 targets/total number of background genes). Statistical significance was assessed by a two-sample Kolmogorov–Smirnov test of length distribution of FMR1 targets versus background genes.
Integrative network-based clustering analysis to identify cell type–specific networks from CLIP-seq and RNA-seq data
A novel graph-based clustering algorithm (Shin J, Marx H, Minogue C, et al., in prep.) was used for the integrative analysis of CLIP-seq and RNA-seq data: (1) graph diffusion to define cell type–specific networks; and (2) multitask graph clustering. To define cell type–specific networks, we integrated the FMR1 targets identified in a particular cell type with a two-step graph diffusion approach. This approach requires a skeleton background network and a query set of genes. First, we applied network node diffusion using the FRMP targets identified in a cell type as query nodes. Second, we used the diffused node values to carry out another diffusion, to obtain the diffusion weights on each edge of the graph. The node diffusion process ranks all other genes in the network based on their global connectivity to the input set, thus providing a measure of influence of the input nodes on all other nodes of the network. We use the regularized Laplacian kernel (Smola and Kondor 2003), used previously for network-based ranking of genes (Köhler et al. 2008) and for semisupervised classification tasks (Fouss et al. 2012). After the first node diffusion step, we carried out edge diffusion, enabling us to get cell type–specific weighted graphs where the edge weight between node i and j is proportional to the influence of node i on j. We used the insulated heat diffusion kernel (Vandin et al. 2011; Leiserson et al. 2015) for estimating the effect of one node on its neighbor nodes based on their global connectivity. See Supplemental Methods for details. For both diffusion steps, we used the largest connected component of the STRING network (Szklarczyk et al. 2011) as the skeleton network, which has a total of 8076 genes and 85,721 interactions. At the end of these two diffusion steps, we obtain a cell type–specific weighted network for each cell type where the weights correspond to the similarity values calculated in the last step.
To identify subnetworks in each of the cell types individually, we applied a multitask graph-based clustering algorithm, Arboretum-HiC, that we previously developed to cluster Hi-C matrices (Fotuhi Siahpirani et al. 2016), that finds the network clusters in multiple cell types by simultaneously applying spectral clustering (von Luxburg 2007) to each cell type–specific network while incorporating the relatedness of the cell types. To obtain the relatedness, we used the pairwise Jaccard coefficients between the sets of FMR1 targets from each of four cell types and applied hierarchical clustering to draw a dendrogram. For each cell type, the graph corresponded to a similarity matrix spanning nodes including the original CLIP- and RNA-seq targets and the union of the top 5% of the noninput set nodes from each of the four cell types, resulting in a total of 1810 genes and 17,490 interactions (see Supplemental Methods for details). Once clusters were defined, we annotated them as common or cell type–specific based on the patterns of gene content similarity (Supplemental Methods). We tested these clusters for enrichment of Gene Ontology terms and DisGeNET disease terms. Code is available in Supplemental Code provided with this paper.
Prioritization of gene hubs for each cell type
For each of our four cell types, we ranked all genes using four criteria: (1) global proximity to CLIP-seq hits; (2) global proximity to RNA-seq hits; (3) number of edges which are connected to CLIP-seq hits; and (4) log fold change values of CLIP-seq signal of the genes. Global proximity is measured on the network by node diffusion using the Laplacian kernel (Smola and Kondor 2003) described above. For input genes, we used their diffusion score when they were left out in the leave-one-out step of the λ parameter selection of the Laplacian kernel. To combine the four different values, first we transformed each of the four values into percentile ranks and then took an average of the percentile ranks. We defined hub genes by taking the top 1% from the rank in each cell type, which resulted in 193 genes in total. Code is available in Supplemental Code provided with this paper.
Integer Linear Programming subnetwork searching analysis
To identify plausible network paths connecting FMR1 CLIP-seq targets to the RNA-seq DEGs, first we tested DEGs for enrichment of binding sites of TFs (FDR < 0.05), using transcriptional regulatory networks inferred specifically for neuronal cell types (Chasman et al. 2019; Pearl et al. 2019). Next, we combined the transcriptional networks with the STRING network and identified all possible paths from CLIP targets to the TFs of DEGs allowing for 0, 1, or 2 intermediate nodes. The paths started from a CLIP-seq target that was a TF or a signaling protein (306 genes in dorsal NPC, 278 genes in ventral NPC, 157 genes in dorsal neuron, and 163 in ventral neuron), and the end point was a TF enriched in the DEGs. This resulted in a candidate network comprising 2866 nodes and 910,241 edges. Then, we applied an Integer Linear Programming approach to select the minimal number of intermediate nodes to obtain a list of filtered, directed paths starting from a CLIP target, which is a TF/signaling protein, and ending in a TF enriched in the DEGs for each cell type. We further prioritized the paths for interpretation by scoring them based on their average percentile ranks. From the prioritized list of paths, we took top paths such that the union of the nodes in the selected paths is no more than M (10 or 15), a prespecified threshold (Fig. 5; Supplemental Fig. S11). Code is available in Supplemental Code provided with this paper.
External gene set data sets
Ascano et al. (2012) and Darnell et al. (2011) sets of FMR1 targets were from their published gene lists. The Maurin set of FMR1 targets used in this study was defined as genes with 15 or more reads from the mouse cortex samples, and the list was converted to 1483 human genes (Maurin et al. 2018). The Tran set of FMR1 targets used in this study (Tran et al. 2019) was defined as genes overlapping between the two brain samples. The SFARI human autistic gene set (SFARI-Gene_genes_export18-12-2017.csv) was downloaded from https://gene.sfari.org/. Gene lists of IQ were from Savage et al. (2018).
Data access
The CLIP-seq and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE128860.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Y. Xing and J. Hunt in the Zhao laboratory; K. Knobel at the Waisman IDD Models Core; S. Splinter-BonDurant, D. Pavelec, and M.E. Berres at the UW-Madison Biotechnology Center for technical assistance; A.J. Peterson at the Waisman Center Human Stem Cell Core for generating the H13-KO line; J. Zimbric for early attempts at identification of FMR1 targets; and G. Yeo for advice during optimization of the ireCLIP method. This work was supported by grants from the National Institutes of Health (National Institute of Neurological Disorders and Stroke) (R01MH116582, R01NS105200, R01MH078972, R21NS098767, and R56MH113146 to X.Z., R21HD085288 and R03HD083538 to A.B., R21NS105339 to A.B. and X.Z., U54HD090256 to the Waisman Center, and R01GM117339 to S.R., UW-Madison and Wisconsin Alumni Research Foundation to X.Z. and A.B., UW Vilas Trust (MidCareer Award) to X.Z., Jenni and Kyle Professorship to X.Z., John Merck Fund to X.Z. and A.B., and Brain Research Foundation to X.Z. and S.R.
Author contributions: X.Z., M.L., and A.B. conceived the concept, designed the experiments, interpreted data, and wrote the manuscript. M.L. designed and performed cell culture, gene editing, differentiation, and ireCLIP and seq experiments, and performed some bioinformatics analysis. J.S. and S.R. performed bioinformatics and gene network analysis, interpreted data, created figures, and wrote the manuscript. D.A.C. performed initial gene network analyses. S.L. performed CLIPer analysis. M.P., J.W., and, R.D.R. performed validation assays of gene edited cell lines.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.251405.119.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. 2013. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4: 36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Vanderhaeghen P. 2014. Cortical neurogenesis from pluripotent stem cells: complexity emerging from simplicity. Curr Opin Neurobiol 27: 151–157. 10.1016/j.conb.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT Jr, Wilkinson KD, Reines D, Warren ST. 1993. FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262: 563–566. 10.1126/science.7692601 [DOI] [PubMed] [Google Scholar]
- Avitzour M, Mor-Shaked H, Yanovsky-Dagan S, Aharoni S, Altarescu G, Renbaum P, Eldar-Geva T, Schonberger O, Levy-Lahad E, Epsztejn-Litman S, et al. 2014. FMR1 epigenetic silencing commonly occurs in undifferentiated fragile X-affected embryonic stem cells. Stem Cell Reports 3: 699–706. 10.1016/j.stemcr.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DB Jr, Berry-Kravis E, Wheeler A, Raspa M, Merrien F, Ricart J, Koumaras B, Rosenkranz G, Tomlinson M, von Raison F, et al. 2016. Mavoglurant in adolescents with fragile X syndrome: analysis of Clinical Global Impression-Improvement source data from a double-blind therapeutic study followed by an open-label, long-term extension study. J Neurodev Disord 8: 1 10.1186/s11689-015-9134-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E, Des Portes V, Hagerman R, Jacquemont S, Charles P, Visootsak J, Brinkman M, Rerat K, Koumaras B, Zhu L, et al. 2016. Mavoglurant in fragile X syndrome: results of two randomized, double-blind, placebo-controlled trials. Sci Transl Med 8: 321ra5 10.1126/scitranslmed.aab4109 [DOI] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Butler MG. 2007. Whole genome microarray analysis of gene expression in subjects with fragile X syndrome. Genet Med 9: 464–472. 10.1097/GIM.0b013e3180ca9a9a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Nazor KL, Tran HT, Szucs A, Lynch CL, Paredes R, Tassone F, Sanna PP, Hagerman RJ, Loring JF. 2017. Molecular analyses of neurogenic defects in a human pluripotent stem cell model of fragile X syndrome. Brain 140: 582–598. 10.1093/brain/aww357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487. 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27: 275–280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D, Fotuhi Siahpirani A, Roy S. 2016a. Network-based approaches for analysis of complex biological systems. Curr Opin Biotechnol 39: 157–166. 10.1016/j.copbio.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Chasman D, Walters KB, Lopes TJS, Eisfeld AJ, Kawaoka Y, Roy S. 2016b. Integrating transcriptomic and proteomic data using predictive regulatory network models of host response to pathogens. PLoS Comput Biol 12: e1005013 10.1371/journal.pcbi.1005013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D, Iyer N, Fotuhi Siahpirani A, Estevez Silva M, Lippmann E, McIntosh B, Probasco MD, Jiang P, Stewart R, Thomson JA, et al. 2019. Inferring regulatory programs governing region specificity of neuroepithelial stem cells during early hindbrain and spinal cord development. Cell Syst 9: 167–186.e12. 10.1016/j.cels.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau KF, Shannon ML, Fame RM, Fonseca E, Mullan H, Johnson MB, Sendamarai AK, Springel MW, Laurent B, Lehtinen MK. 2018. Downregulation of ribosome biogenesis during early forebrain development. eLife 7: e36998 10.7554/eLife.36998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. 2013. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 16: 1530–1536. 10.1038/nn.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G, Giusto M, Chandel R, Welch N, Alsabbagh K, Kant S, Kumar A, Kim A, Gangadhariah M, Ghosh PK, et al. 2019. Impaired ribosomal biogenesis by noncanonical degradation of β-catenin during hyperammonemia. Mol Cell Biol 39: e00451-18 10.1128/MCB.00451-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang S-C, Abbeduto L, Bhattacharyya A. 2014. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev 23: 1777–1787. 10.1089/scd.2014.0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotuhi Siahpirani A, Ay F, Roy S. 2016. A multi-task graph-clustering approach for chromosome conformation capture data sets identifies conserved modules of chromosomal interactions. Genome Biol 17: 114 10.1186/s13059-016-0962-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouss F, Francoisse K, Yen L, Pirotte A, Saerens M. 2012. An experimental investigation of kernels on graphs for collaborative recommendation and semisupervised classification. Neural Networks 31: 53–72. 10.1016/j.neunet.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Greenblatt EJ, Spradling AC. 2018. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 361: 709–712. 10.1126/science.aas9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, Ye K, Warren ST, Bassell GJ. 2010. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci 30: 10624–10638. 10.1523/JNEUROSCI.0402-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Raj N, Molinaro G, Allen AG, Whyte AJ, Gibson JR, Huber KM, Gourley SL, Bassell GJ. 2015. Selective role of the catalytic PI3K subunit p110β in impaired higher order cognition in fragile X syndrome. Cell Rep 11: 681–688. 10.1016/j.celrep.2015.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Bartel DP. 2016. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353: aaf5371 10.1126/science.aaf5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy T, Czech C, Benvenisty N. 2015. Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Reports 4: 37–46. 10.1016/j.stemcr.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel AE, Chintawar S, Lalic T, Whiteley E, Vowles J, Giustacchini A, Argoud K, Sopp P, Nakanishi M, Bowden R, et al. 2016. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum Mol Genet 25: 989–1000. 10.1093/hmg/ddv637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, Slomnicki LP. 2019. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J Neurochem 148: 325–347. 10.1111/jnc.14576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-s, Clarke DC, Gosline SJC, Labadorf A, Chouinard CR, Gordon W, Lauffenburger DA, Fraenkel E. 2013. Linking proteomic and transcriptional data through the interactome and epigenome reveals a map of oncogene-induced signaling. PLoS Comput Biol 9: e1002887 10.1371/journal.pcbi.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose S, Ogawa T, Jiang X, Hirokawa N. 2019. The spatiotemporal construction of the axon initial segment via KIF3/KAP3/TRIM46 transport under MARK2 signaling. Cell Rep 28: 2413–2426.e7. 10.1016/j.celrep.2019.07.093 [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Kidd SA, Andrews HF, Budimirovic DB, Esler A, Haas-Givler B, Stackhouse T, Riley C, Peacock G, Sherman SL, et al. 2017. Autism spectrum disorder in fragile X syndrome: cooccurring conditions and current treatment. Pediatrics 139(Supplement 3): S194–S206. 10.1542/peds.2016-1159F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfallah O, Jarjat M, Davidovic L, Nottet N, Cestèle S, Mantegazza M, Bardoni B. 2017. Depletion of the fragile X mental retardation protein in embryonic stem cells alters the kinetics of neurogenesis. Stem Cells 35: 374–385. 10.1002/stem.2505 [DOI] [PubMed] [Google Scholar]
- Kim DS, Ross PJ, Zaslavsky K, Ellis J. 2014. Optimizing neuronal differentiation from induced pluripotent stem cells to model ASD. Front Cell Neurosci 8: 109 10.3389/fncel.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Bauer S, Horn D, Robinson PN. 2008. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 82: 949–958. 10.1016/j.ajhg.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Gresack J, Allis CD, Darnell RB. 2017. Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell 170: 1209–1223.e20. 10.1016/j.cell.2017.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rašin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. 2012. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149: 899–911. 10.1016/j.cell.2012.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. 2015. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47: 106–114. 10.1038/ng.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Zhang X, Johnson MA, Wang ZB, LaVaute T, Zhang SC. 2009. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development 136: 4055–4063. 10.1242/dev.036624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Ananiev GE, Musser MT, Ness KH, Maglaque DL, Saha K, Bhattacharyya A, Zhao X. 2017. Establishment of reporter lines for detecting fragile X mental retardation (FMR1) gene reactivation in human neural cells. Stem Cells 35: 158–169. 10.1002/stem.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stockton ME, Eisinger BE, Zhao Y, Miller JL, Bhuiyan I, Gao Y, Wu Z, Peng J, Zhao X. 2018. Reducing histone acetylation rescues cognitive deficits in a mouse model of Fragile X syndrome. Nat Commun 9: 2494 10.1038/s41467-018-04869-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Chen X, Feng Y, Zeng Q, Jiang C, Zhu X, Fan G, Xue Z. 2016. Integrated transcriptome analysis of human iPS cells derived from a fragile X syndrome patient during neuronal differentiation. Sci China Life Sci 59: 1093–1105. 10.1007/s11427-016-0194-6 [DOI] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. 2013. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 12: 559–572. 10.1016/j.stem.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, Grossi M, Rage F, Bardoni B. 2018. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res 46: 6344–6355. 10.1093/nar/gky267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motanis H, Buonomano D. 2015. Delayed in vitro development of Up states but normal network plasticity in Fragile X circuits. Eur J Neurosci 42: 2312–2321. 10.1111/ejn.13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl JR, Colantuoni C, Bergey DE, Funk CC, Shannon P, Basu B, Casella AM, Oshone RT, Hood L, Price ND, et al. 2019. Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. Cell Syst 8: 122–135.e7. 10.1016/j.cels.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. 2009. The state of synapses in Fragile X syndrome. Neuroscientist 15: 549–567. 10.1177/1073858409333075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. 1991. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66: 817–822. 10.1016/0092-8674(91)90125-I [DOI] [PubMed] [Google Scholar]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. 2017. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 45: D833–D839. 10.1093/nar/gkw943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Bassell GJ, Klann E. 2015. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci 16: 595–605. 10.1038/nrn4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AP, Zarbalis KS. 2014. The emerging roles of ribosome biogenesis in craniofacial development. Front Physiol 5: 26 10.3389/fphys.2014.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7: 219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, Nagel M, Awasthi S, Barr PB, Coleman JRI, et al. 2018. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 50: 912–919. 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Hale CR, Park CY, Fak JJ, Gresack JE, Van Driesche SJ, Kang JJ, Darnell JC, Darnell RB. 2019. FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. eLife 8: e46919 10.7554/eLife.46919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. 2011. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of Fragile X syndrome. PLoS One 6: e26203 10.1371/journal.pone.0026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva CG, Peyre E, Adhikari MH, Tielens S, Tanco S, Van Damme P, Magno L, Krusy N, Agirman G, Magiera MM, et al. 2018. Cell-intrinsic control of interneuron migration drives cortical morphogenesis. Cell 172: 1063–1078.e19. 10.1016/j.cell.2018.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola AJ, Kondor R. 2003. Kernels and regularization on graphs In Learning theory and kernel machines (ed. Schölkopf B, Warmuth MK), Vol. 2777, pp. 144–158. Springer, Berlin/Heidelberg: 10.1007/978-3-540-45167-9_12 [DOI] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet R, Moutin E, Becker JAJ, Heintz D, Fouillen L, Flatter E, Krężel W, Alunni V, Koebel P, Dembélé D, et al. 2016. Fragile X mental retardation protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc Natl Acad Sci 113: E3619–E3628. 10.1073/pnas.1522631113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Nkiliza A, Chartier-Harlin MC. 2015. Deregulation of protein translation control, a potential game-changing hypothesis for Parkinson's disease pathogenesis. Trends Mol Med 21: 466–472. 10.1016/j.molmed.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Telias M, Segal M, Ben-Yosef D. 2013. Neural differentiation of fragile X human embryonic stem cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol 374: 32–45. 10.1016/j.ydbio.2012.11.031 [DOI] [PubMed] [Google Scholar]
- Telias M, Mayshar Y, Amit A, Ben-Yosef D. 2015. Molecular mechanisms regulating impaired neurogenesis of fragile X syndrome human embryonic stem cells. Stem Cells Dev 24: 2353–2365. 10.1089/scd.2015.0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens S, Huysseune S, Godin JD, Chariot A, Malgrange B, Nguyen L. 2016. Elongator controls cortical interneuron migration by regulating actomyosin dynamics. Cell Res 26: 1131–1148. 10.1038/cr.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran SS, Jun HI, Bahn JH, Azghadi A, Ramaswami G, Van Nostrand EL, Nguyen TB, Hsiao YE, Lee C, Pratt GA, et al. 2019. Widespread RNA editing dysregulation in brains from autistic individuals. Nat Neurosci 22: 25–36. 10.1038/s41593-018-0287-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. 2011. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol 18: 507–522. 10.1089/cmb.2010.0265 [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Gelboin-Burkhart C, Wang R, Pratt GA, Blue SM, Yeo GW. 2017. CRISPR/Cas9-mediated integration enables TAG-eCLIP of endogenously tagged RNA binding proteins. Methods 118: 50–59. 10.1016/j.ymeth.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65: 905–914. 10.1016/0092-8674(91)90397-H [DOI] [PubMed] [Google Scholar]
- von Luxburg U. 2007. A tutorial on spectral clustering. arXiv:0711.0189 [cs.DS]. [Google Scholar]
- Wang B, Joo JH, Mount R, Teubner BJW, Krenzer A, Ward AL, Ichhaporia VP, Adams EJ, Khoriaty R, Peters ST, et al. 2018. The COPII cargo adapter SEC24C is essential for neuronal homeostasis. J Clin Invest 128: 3319–3332. 10.1172/JCI98194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Van Nostrand EL, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip Rev RNA 9: e1436 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Bontekoe C, Severijnen L-A, Oostra B. 2002. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet 110: 601–605. 10.1007/s00439-002-0723-5 [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to α-synuclein toxicity. Nat Genet 41: 316–323. 10.1038/ng.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do B T, Chang HY, Khavari PA. 2016. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods 13: 489–492. 10.1038/nmeth.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bhattacharyya A. 2018. Human models are needed for studying human neurodevelopmental disorders. Am J Hum Genet 103: 829–857. 10.1016/j.ajhg.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.