Abstract
Talin and vinculin, both actin-cytoskeleton-related proteins, have been documented to participate in establishing bacterial infections, respectively, as the adapter protein to mediate cytoskeleton-driven dynamics of the plasma membrane. However, little is known regarding the potential role of the talin–vinculin complex during spotted fever group rickettsial and Ebola virus infections, two dreadful infectious diseases in humans. Many functional properties of proteins are determined by their participation in protein–protein complexes, in a temporal and/or spatial manner. To resolve the limitation of application in using mouse primary antibodies on archival, multiple formalin-fixed mouse tissue samples, which were collected from experiments requiring high biocontainment, we developed a practical strategic proximity ligation assay (PLA) capable of employing one primary antibody raised in mouse to probe talin–vinculin spatial proximal complex in mouse tissue. We observed an increase of talin–vinculin spatial proximities in the livers of spotted fever Rickettsia australis or Ebola virus-infected mice when compared with mock mice. Furthermore, using EPAC1-knockout mice, we found that deletion of EPAC1 could suppress the formation of spatial proximal complex of talin–vinculin in rickettsial infections. In addition, we observed increased colocalization between spatial proximity of talin–vinculin and filamentous actin-specific phalloidin staining in single survival mouse from an ordinarily lethal dose of rickettsial or Ebola virus infection. These findings may help to delineate a fresh insight into the mechanisms underlying liver specific pathogenesis during infection with spotted fever rickettsia or Ebola virus in the mouse model.
Subject terms: Infection, Infectious diseases
The authors observed an increase of talin-vinculin spatial proximities in the livers of spotted fever Rickettsia australis or Ebola virus-infected mice when compared to mock mice. Furthermore, using EPAC1 knockout mice, they found that deletion of EPAC1 suppresses the formation of spatial proximal complex of talin-vinculin in rickettsial infections.
Introduction
Dynamic plasma membrane remodeling is a vital part of various cell biological processes and is achieved by the interplay between the plasma membrane and the underlying actomyosin network mediated by protein adapters [1]. Talin and vinculin, both as adapter proteins for the dynamic interaction between the transmembrane receptor integrin and actin cytoskeleton, play important roles, in particular in transmitting mechanosignals to regulate cell structure and function [2–5]. Talin is a ubiquitous cytosolic protein, predominantly detected at regions of cell–substratum or cell–cell adhesions [2–4]. Talin functions as an adapter for integrin in forming mechanosensitive intercellular or cell-extracellular matrix (ECM) tensions, generating cell–cell junctions or focal adhesions between cell and ECM, respectively [6–8]. Via the dynamic correlation with its binding partner, vinculin, talin has been documented to participate in establishing bacterial infections via its role as the adapter to mediate the cytoskeleton-driven dynamics of the plasma membrane [5, 9–12]. Bacteria have been shown to directly manipulate host subcellular actin-mediated pulling force to promote intercellular spread [5, 13, 14], including the spotted fever group (SFG) of Rickettsia (R.) spp [12]. To promote cell–cell spread, R. parkeri secrets surface cell antigen 4 to interact with vinculin and block the association between vinculin and another binding partner, α-catenin, reducing vinculin-dependent mechanotransduction at cell–cell junctions [5]. Therefore, SFGRs have evolved a strategy to target vinculin-based host cytoskeletal force generation to enable spread. However, little is known regarding the potential role of the complex of talin–vinculin during SFG rickettsioses (SFGR) infections.
Many functional properties of a protein are determined by participation in protein–protein complexes, in a temporal and/or spatial manner [15–17]. Protein–protein interactions (PPIs) include highly specific physical contacts/linkages between two or more protein molecules. In a specific biomolecular context, physical contacts are established spatially between special chains of protein molecules [18–20]. Technologically, the association between proteins can be clarified by immunoprecipitation, which is performed coupling with western blot (WB) to test target proteins, requiring homogenization of sample and resulting in losing spatial information about the targets, both at intra- and intercellular levels [21]. Colocalization of different proteins with fluorescence microscopy is frequently used to assess their potential association in situ. However, colocalization under epifluorescent/confocal fluorescent microscope does not necessarily mean a physical interaction [7]. A practical step toward unraveling the complex molecular relationship, temporal and spatial, of protein-to-protein physical interaction is essential for accomplishing a comprehensive knowledge of the functional outcomes of a PPI [8]. The concept of spatial proximity between two protein targets is merging to be a practical approach for endogenous quantitative detection of spatial proximal complex of proteins at the single molecule level in fixed cells and tissue samples. Currently, in situ application of proximity ligation assay (PLA) is used to visualize the spatial proximal signals of two protein targets in tissue samples [22, 23], directly examining proximal interaction between the proteins, in responses to different stimuli [24]. However, the commercially available PLA assay (Sigma Aldrich) restricted the use of primary antibody raised in mouse on murine samples due to the antibody incompatibility between the kit and the sample. Furthermore, traditional histological fixatives (e.g., formaldehyde or glutaraldehyde) cause strong chemical crosslinking of proteins, which impede detection of many sensitive antigens [25–27]. Our formalin-fixed paraffin-embedded (FFPE) tissue samples were collected during experiments requiring high biocontainment, for which multiple 10% formalin fixation steps are mandatory, leading to heavy antigen masking. The technical guideline for antigen retrieval for PLA assay, as provided in the manufacturer’s instructions or publications [22–24], resulted in an ineffective antigen retrieval on our archival FFPE tissue samples collected from the experiments requiring high biocontainment.
In the present study, based on the methodological mechanism of the PLA, we developed a special antigen retrieval protocol and a panel of quality controls by manipulating primary antibodies, normal mouse IgG, and oligonucleotide probes to establish the capacity of employing primary antibodies raised in mouse to probe protein–protein spatial proximity in mouse tissue during PLA. Taking advantage of established mouse models of SFGR infections [28–30], we examined the spatial proximal complex of talin–vinculin in mouse liver tissues. During lethal SFGR infections, significant bacterial loads are detected in liver tissues coupled with various pathological manifestations making it ideal to study this interaction [31, 32]. We observed, compared with mock infection, increased talin–vinculin spatial proximities (TVSPs) in liver in response to lethal R. australis infection. Interestingly, retrospective studies showed that similar increased TVSPs in liver in response to lethal Ebola virus (EBOV) infection in mouse. Given that the exchange proteins directly activated by cAMP 1 (EPAC1) plays critical role during SFGR infections [30], using EPAC1-knockout (KO) mice, we further found that deletion of EPAC1 could suppress TVSPs in mice infected with 2 LD50 of R. australis. These findings may help to delineate a fresh insight into the mechanisms underlying the pathogenesis targeting the liver during SFGR and EBOV infections.
Materials and methods
Mice
Archival FFPE blocks of liver tissues of ordinarily lethal dose of R. australis-infected mice and EBOV-infected mice were from previous studies, respectively [29, 33]. The EPAC1-KO mice were generated as previously described [34]. All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch.
Reagents and antibodies
Information for special reagents: mouse anti-talin antibody, clone TA205 (Millipore Sigma, 05-385), rabbit anti-vinculin antibody (42H89L44, Invitrogen, 700062), rabbit anti-rickettsia antibody (provided by Dr David Walker), rabbit anti-Ebola virus antibody (provided by Dr Thomas Ksiazek), mouse anti-Rab5 (BD transduction, 610724) and rabbit anti-von Willebrand factor (vWF, Thermo Fisher, PA5-16634), Alexa Fluor 488-conjugated goat anti-mouse IgG (Thermo Fisher, A-11017), Alexa Fluor 594-conjugated goat anti-rabbit IgG (Thermo Fisher, A-11012), Duolink® In Situ PLA® Probe Anti-Rabbit PLUS, Affinity purified Donkey anti-Rabbit IgG(H+L) (Millipore Sigma, DUO92002-30RXN), Duolink® In Situ PLA® Probe Anti-Mouse MINUS, Affinity purified Donkey anti-Mouse IgG(H+L) (Millipore Sigma, DUO92004-30RXN), Duolink® In Situ Detection Reagents Red (Millipore Sigma, DUO92008-30RXN), Duolink® In Situ Mounting Medium with DAPI (Millipore Sigma, DUO82040-5ML), Duolink® Blocking Solution (Millipore Sigma, DUO82007-4ML), Duolink® Antibody Diluent (Millipore Sigma, DUO82008-2.5ML), Duolink® In Situ Washing Buffer A (Millipore Sigma, DUO82046-1EA), Duolink® In Situ Washing Buffer B (Millipore Sigma, DUO82048-1EA), TritonX-100 (Fisher BioReagents, BP151-100), Phalloidin iFluor 488 (Abcam, ab176753).
Cell culture
To establish signal positive controls for the PLA, human umbilical vein endothelial cells (HUVECs, Cell Applications, Atlanta, GA) were cultured in Endothelial Cell Growth Medium (Cell Applications, Atlanta, GA) with humidity in 5% CO2 at 37 °C. The media were changed every 48 h. Cells were allowed to grow until 90% confluency was achieved. Cell passages 2–5 were used for all experiments, and cells were fixed by paraformaldehyde solution 4% in phosphate-buffered saline (PBS) for 30 min at room temperature (RT) before PLA.
Immunofluorescence (IF)
After antigen retrieval and blocking, the sample was incubated with mouse anti-talin antibody (1:500) and vinculin recombinant rabbit monoclonal antibody (1:500) for 2 h before incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:2000) and Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:2000) for 1 h. The rickettsia infected sample was incubated with rickettsia rabbit antibody (1:2000) for 2 h before incubation with Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:2000) for 1 h. The Ebola infected sample was incubated with Ebola rabbit antibody (1:500) for 2 h before incubation with Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:2000) for 1 h. A mouse monoclonal IgG (Thermo Fisher) or Rabbit Polyclonal IgG (Thermo Fisher) served as a negative control [29]. Fluorescent images were taken and analyzed using an Olympus BX51-microscope or using a Nikon A1R MP ECLIPSE Ti Confocal, with NIS-Elements imaging software version 4.50.00 (Nikon, Tokyo, Japan).
Proximity ligation assay
During preliminary test on tissue sections from archival, multiple FFPE tissue samples, different published or commercially available methods were tested for antigen retrievers (i.e., heat-mediated epitope retrieval in pH 6.0/10 citrate buffer or pH 8.0 EDTA buffer, and enzymatic antigen retrieval using proteinase K) with minimal success obtaining promising signals. Tissue sections from BSL3/4 samples were lysed on slides using proteinase K (Thermo Fisher) digestion during incubation at 98 °C in citrate buffer (pH 6, Thermo Fisher, 005000). Optimized antigen retrieval for the archival, multiple FFPE tissue samples were achieved by incubating tissue sections in pH 6.0 citrate buffer and heated in a steamer (Black and Decker, New Britain, Connecticut) for 10 min. After washing in PBS, samples were incubated with proteinase K for 5 min at RT. For permeabilization (only for HUVECs), the samples were treated with 0.25% TritonX-100 (diluted in PBS) for 10 min at RT, and then blocked with Duolink® blocking solution in shaker incubator at 37 °C for 1 h. Then incubated with anti-talin (human) mouse monoclonal antibody (1:500 diluted with Duolink® antibody diluent) and vinculin recombinant rabbit monoclonal antibody (1:500 diluted with Duolink® antibody diluent) for 2 h, rinsed twice with 200 µl RT-equilibrated washing buffer A, 5 min each after removing the antibody. Samples were then incubated with MINUS probe and PLUS probe at 37 °C for 1 h and rinsed twice using RT-equilibrated wash buffer A. 1 × ligase (1 unit) added in 1 × ligation (39 units) was added to the sample immediately and incubated in a shaker incubator at 37 °C for 45 min, before Duolink amplification (1 unit 1 × polymerase and 79 units 1 × Duolink amplification) for 2 h at 37 °C. The slides were mounted with a coverslip with a minimal volume of Duolink® In Situ Medium with DAPI. All slides were analyzed with Olympus BX51 fluorescence microscope or using Nikon A1R MP ECLIPSE Ti Confocal, with NIS-Elements imaging software version 4.50.00 (Nikon, Tokyo, Japan).
Phalloidin staining in tissue after PLA
To couple PLA and phalloidin staining assay for FFPE tissue samples, before mounting the Duolink® In Situ Medium with DAPI as described above, the slides were analyzed with a fluorescence microscope to record the PLA (red) and background green auto fluorescence. Then, PLA-stained samples were rinsed thrice with RT-equilibrated washing buffer before adding phalloidin for 30 min at RT in a dark chamber on the shaker. After the slides were rinsed three times, a minimal volume of Duolink® In Situ Medium with DAPI was added onto the slides covered by a coverslip. All slides were analyzed with an Olympus BX51 fluorescence microscope. PBS was used as negative reagent control.
Quantitative analysis of spatial proximal signals
PLA-stained liver tissue samples from mice were examined and fluorescent images were captured with an Olympus BX51 image system using a final 40× optical zoom. The number of dot signals of the TVSPs (Figs. 1 and 2c) in each microscopic field were manually enumerated [35, 36]. Twenty microscopic fields were examined for each case. The results were expressed as total dot signals enumerated in each field. Data are representative of at least three experiments. P values were determined using a standard Student’s t test or two-way ANOVA.
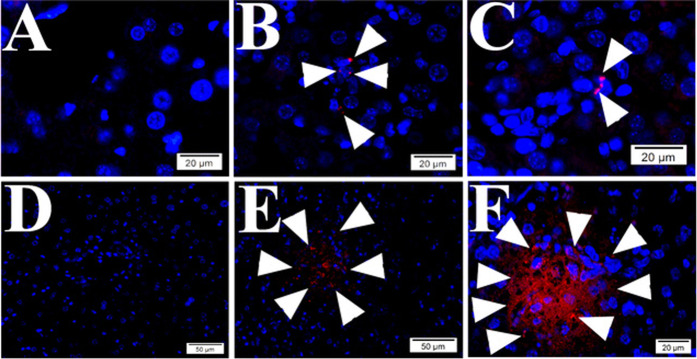
Fig. 1. Positive and negative controls for the PLAs.
Signal positive (a–c) and signal negative (d) controls of the proximity ligation assays (PLA) were established using HUVECs. Mouse anti-talin antibodies were titrated at 1:50 (a), 1:500 (b), and 1:2000 (c) during PLA assay. There is no positive signal visualized during PLA using mouse anti-Rab5 and rabbit anti-vWF in HUVECs (d) and mouse liver tissue (h). Normal mouse and rabbit IgGs were used as reagent negative controls during PLA assay in HUVECs (e). IF to von Willebrand factor (arrowheads in f) and Rab5 (arrowheads in g) were processed in HUVECs. Nuclei were counter-stained with DAPI (blue). Scale bars indicate 20 µm.
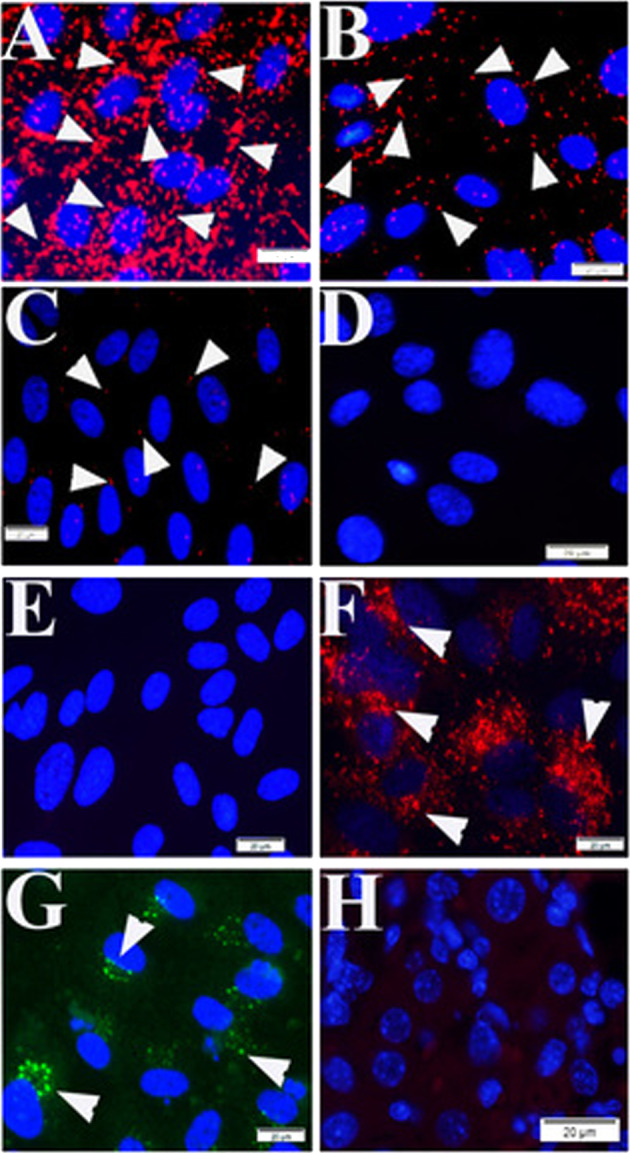
Fig. 2. Quality controls for PLA in mouse tissue using primary antibodies raised in mouse.
During PLA assay on mouse liver tissue samples from SFGR infection mouse on day 19, there is no specific signals if no primary antibody employed (only normal mouse and rabbit IgGs) (a), using rabbit anti-vinculin antibodies and normal mouse IgG employed (b), using rabbit anti-vinculin antibodies and mouse anti-talin, but no oligonucleotide probes (e). There is specific signal detected when rabbit anti-vinculin and mouse anti-talin antibodies were employed during PLA on mouse liver tissue (c, d). Nuclei were counter-stained with DAPI (blue). Scale bars indicate 20 µm.
Statistical analysis
Values are reported as mean ± SEM. The statistical significance was determined using Student’s t test or two-way ANOVA analysis (Sigmaplot, Sigma Stat, Jandel Scientific Software, San Rafael, CA). P values are as follows: **P < 0.01 and *P < 0.05. Statistical significance was considered as P < 0.05.
Results
TVSP in HUVECs was established as the positive control for the PLAs in the present study
The first critical step in developing the PLA was to establish signal positive and negative controls. The TVSPs in cells are well documented [7]. Therefore, we developed the PLA result of TVSPs in HUVECs as a signal positive control. For reagent negative control, mouse normal IgG and rabbit normal IgG were used as the primary antibodies. In Fig. 1, red dot signals from the TVSPs were visualized under fluorescent microscope only in signal positive controls (Fig. 1a–c), but not in the reagent negative controls (Fig. 1e). Xie and Perrino [37] reported improper antibody dilution and insufficient washing between steps during staining can lead to false positives. In addition to routine standard rinse procedures between incubations (see “Materials and methods”), we performed antibody titration assay for optimal working concentrations (Fig. 1). Based upon titration of the primary antibodies (Fig. 1a–c), we used 1:500 as working dilutions for the mouse anti-talin and rabbit anti-vinculin antibodies in PLAs (Fig. 1b). vWF, recognized as an EC marker, is synthesized and stored in a storage granule called as Weibel–Palade body (WPB) in the endothelial cytosol [38, 39]. Our IF staining visualized that vWF was present in rod-shaped cytoplasmic WPBs (Fig. 1f). Rab5 is a cytosolic protein and localizes to early endosomes, and the complex of host Rab5 and vinculin is required for bacterial entry [9]. Our IF staining visualized that Rab5 was also present in the cytosol (Fig. 1g). As expected from a signal negative control, we did not detect potential PLA signals in HUVECs (Fig. 1d) and tissue (Fig. 1h) when using mouse anti-Rab5 and rabbit anti-vWF during the PLA although they both located in the cytosol.
IF assay of talin and vinculin in mouse tissues using mouse anti-talin antibody
To visualize talin and vinculin antigens in mouse tissues with IF assay, we used a published protocol to block endogenous IgG with casein for 15 min before incubation with primary antibodies composed of mouse anti-talin and rabbit anti-vinculin antibodies [30]. Under IF microscope, compared with the rabbit IgG group, specific signals from vinculins were detected (Supplementary Fig. 1A, B). However, we were unable to detect promising signals of talin, possibly due to nonspecific background or too weak signals after dilution of the secondary tracer. These nonspecific signals are from the FITC-conjugated goat anti-mouse secondary antibodies in mouse tissue after incubation with normal mouse IgG (Supplementary Fig. 1C, D). We attributed these to the technical issues resulting from “mouse on mouse” during immunodetection [40].
Rationales for designing the panel of quality controls for PLA on mouse tissue sample using one primary antibody raised in mouse
In the present study, we developed a panel of quality controls by manipulating primary antibodies, normal mouse IgG, and oligonucleotide probes to establish the capacity of employing primary antibodies raised in mouse to probe protein–protein spatial proximity in mouse tissue during PLA.
Based on the mechanism of PLA and the above results, there was a possibility of proximity formation between talin and vinculin in mouse tissue. Meanwhile, there was risk of nonspecific signals formed by reaction between oligonucleotide-traced rabbit antibodies-linked vinculin and oligonucleotide-traced nonspecific endogenous mouse IgGs in mouse tissue when using mouse anti-talin antibody. In order to exclude such nonspecific signals during PLA in mouse tissue, we set up three different negative quality control groups. The first group was composed of both rabbit and mouse normal IgGs (Fig. 2a); the second group was composed of rabbit anti-vinculin antibody and mouse normal IgGs (Fig. 2b); the third group was similar as the first group, but no oligonucleotide probes (Fig. 2e) (Table 1). There was no signal detected during PLA assays using these three groups of negative quality controls (Fig. 2a, b, e), supporting positive PLA signals we detected when using mouse anti-talin and rabbit anti-vinculin antibodies in liver tissues from a mouse infected with lethal dose of SFGRs (Fig. 2c, d) (Supplementary Fig. 2).
Table 1.
Design strategy of the panel of negative quality controls, compared with the signal positive control group.
| Groups | The 1st negative group | The 2nd negative group | The 3rd negative group | The positive control group |
|---|---|---|---|---|
| Mouse anti-talin | − | − | + | + |
| Rabbit anti-vinculin | − | + | + | + |
| Mouse normal IgG | + | + | − | − |
| Rabbit normal IgG | + | − | − | − |
| MINUS probe | + | + | − | + |
| PLUS probe | + | + | − | + |
| Signal in PLA | − | − | − | + |
TVSP increases in liver during lethal R. australis infection and EBOV infections
Before processing PLA of talin–vinculin on adjacent tissue sections from same tissue blocks, we validated R. australis infection (n = 4) and EBOV infections (n = 3), respectively, using archival liver tissues samples with IF staining (Fig. 3). PLA was applied to liver samples from groups of mock (n = 3) or lethal R. australis infections (n = 4) on day 6–7 post infection (p.i.) at BSL3 level, and mock (n = 3) and lethal EBOV infections (n = 3) on day 7–8 p.i. at BSL4. PLA dot signals in liver tissues (Fig. 4) were enumerated by fluorescent microscopy. Compared with the mock group, respectively, PLA signals of TVSP significantly increased in both SFGR infection and Ebola infection groups (P < 0.01) (Fig. 5). These data suggest increased TVSPs in liver in response to acute intracellular pathogen infections.
Fig. 3. The IF assay of Rickettsia and Ebola virus antigens.
Representative IF staining (red) of rickettsiae (b, c) and Ebola virus (e, f) in livers from archived mouse samples of fatal SFGR and Ebola virus infection. Mock infection mouse samples processed in ABSL3 (a) and ABSL4 (d) were employed as mock controls. Nuclei were counter-stained with DAPI (blue). Scale bars, 20 µm (a–c, f) and 50 µm (d, e).
Fig. 4. PLA signals of TVSPs in the tissue of mouse liver.
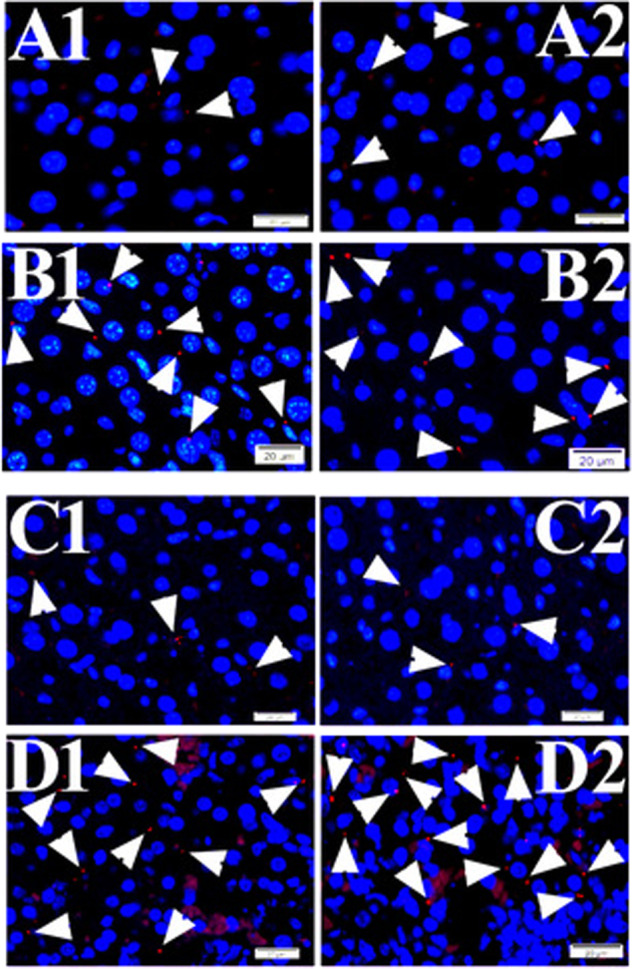
Representative PLA signals (red) of TVSPs (arrowheads) in liver tissues from mock at BSL3 (a1, a2), mock at BSL4 (c1, c2), rickettsial infection (b1, b2), and Ebola infection (d1, d2) groups. Nuclei were counter-stained with DAPI (blue). Scale bars indicate 20 µm.
Fig. 5. Quantitative analysis of TVSPs in the liver samples of mice infected with mock, R. australis (R), or Ebola virus.

The number of dot signals of the TVSPs in liver tissues were enumerated by fluorescent microscopy. Twenty microscopic fields were examined for each case. The results were expressed as total dot signals enumerated in each field. The data presented are representative of at least three individual mice. The error bar is standard error of the mean. **P < 0.01. WT wild-type mice. KO EPAC1-knockout mice.
Deletion of EPAC1 gene suppresses the upregulation of TVSP in liver during rickettsial infections
Given that we have reported that EPAC1 plays a critical role during fatal rickettsioses and inhibition of EPAC1 can protect mice from an ordinary lethal dose of rickettsiae [30], we now propose the question: what effect will inhibition of EPAC1 induce on the TVSP during SFGR infections? PLA was applied to murine liver tissues from groups of WT-mock (n = 3), EPAC1-KO mock (n = 3), lethal WT-R. australis (n = 4), and surviving EPAC1-KO-R. australis (n = 3) mice (Fig. 6). Quantitative analysis found that, compared with the lethal WT-R. australis group, the PLA signals of TVSP were reduced in the livers of EPAC1-KO-R. australis-infected survivals (P < 0.01) (Fig. 5). These data suggest that TVSPs increase in livers in response to SFGR infection, in an EPAC1-dependent manner.
Fig. 6. PLA signals of TVSPs in liver tissues from WT and EPAC1 KO mice.
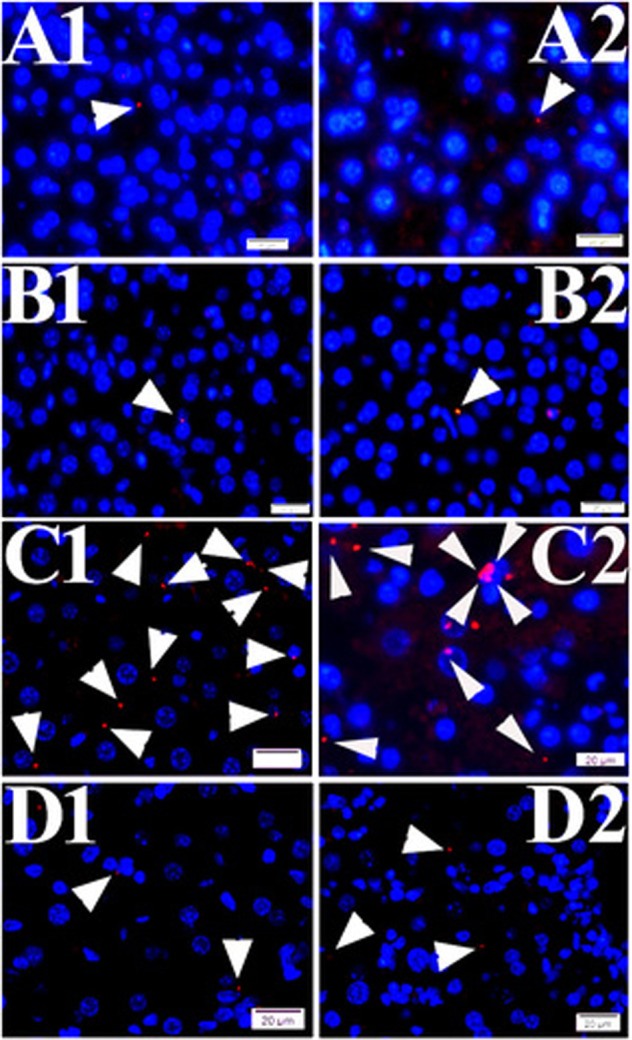
PLA signals (red) of TVSPs (arrowheads) in liver tissues from wild-type (WT) mock (a1, a2), EPAC1-KO mock (b1, b2), WT-R. australis (c1, c2), and EPAC1-KO R. australis-infected mice (d1, d2). Nuclei were counter-stained with DAPI (blue). Scale bars indicate 20 µm.
Potential association between TVSP and filamentation of actin
Talin and vinculin both play regulatory roles in actin-cytoskeleton dynamics in cells [2–5]. Taking advantage of the filamentous actin-specific fluorescent phalloidin staining provided the opportunity to examine the association between TVSP and the activation of filamentation of actin after SFGR infection. PLA assay coupling fluorescent phalloidin staining found no significant difference in colocalization of TVSP and filamentous actin signals in livers between WT-mock and lethal WT-R. australis mice (Supplementary Fig. 3A1-2, B1-2, C1-2). Interestingly, we observed completely different pattern of colocalization between TVSP and phalloidin signals in liver from one WT survivor on day 19 after SFGR infection (Supplementary Fig. 3A3-4, B3-4, C3-4). We used this case for PLA quality controls.
Of note, we also examined liver tissue of one WT survivor on day 21 after EBOV infection and observed, compared with the WT-mock (Supplementary Fig. 3A5, B5, C5), increased colocalization between TVSP and phalloidin signals (Supplementary Fig. 3A6, B6, C6). This preliminary information encourages us to evaluate and validate the potential association between TVSP and filamentation of actin in future.
Discussion
In this study, we designed and tested a new strategy using primary antibodies raised in mouse to detect TVSP in mouse tissues. A specifically designed panel of quality controls warrants promising readouts of the PLA. Histology-based quantitative data from the PLA indicate increased TVSP complexes in response to both SFGR and EBOV infections. To our knowledge, these are the first reported direct images of signals of TVSP in liver during acute infections. We further observed that TVSP in liver increased after infections with R. australis in an EPAC1-dependent manner, suggesting a potential new molecular basis underpinning this aberration in liver.
Talin, one of the most abundant components in cells, is a ubiquitous cytosolic protein concentrated at regions of cell–substratum contact in cellular environments [2–4]. It is a long rod-like protein, one end of which contains an integrin binding site, while the other end contains an actin-binding domain. At rest, Rap1-GTP-interacting adapter molecule (RIAM) binds to the closed R3 domain of talin and anchors it to the cell membrane. When actin filaments are pulled, the R3 domain will undergo a conformational change, which causes RIAM to dissociate and vinculin to bind [7, 41]. Talin is a mechanosensitive molecule and this mechanosensitivity relates to the interaction of talin with vinculin [42, 43]. Vinculin is a major talin-binding partner that facilitates crosstalk between talin and actin through its talin-binding head domain (V-head) and its actin-binding tail domain (V-tail) [44, 45]. Vinculin is also a central component of mechanosensitive adhesive complexes that forms between cells and the ECM [8]. The force-dependent interaction between talin and vinculin plays a crucial role in the initiation and growth of focal adhesions [6, 7]. Given identification of a talin-binding site localized within the N-terminal vinculin head domain [44, 45] and characterization of the functional role of talin–vinculin interplay to regulate actin-based cell membrane dynamics in response to mechanosignals [7, 41], it is important to delineate the mechanism(s) targeting talin–vinculin interaction, in particular their dynamic spatial proximity, in pathophysiological context. Evidence from developmental studies suggests a high level of spatial proximity between talin and vinculin in response to mechnosignals [46]. Nevertheless, the potential role of the dynamics of TVSP during inflammation and infections remains completely unknown.
Host cytoskeleton plays a key role in many stages of infection and inflammation [47–49]. Due to its role as a filter, the liver is exposed to many systemic infectious pathogens. These pathogens may directly or indirectly affect the liver depending on the characteristics of the pathogens [50]. Liver inflammation due to various etiologies can result in liver fibrosis, leading to cirrhosis and carcinogenesis [51]. Vinculin participates in enhancing bacterial infection and the inflammatory response through underpinning integrin, which binds to ECM proteins, many bacterial pathogens and viruses [9–12]. Besides this, talin and vinculin, two interacting proteins that localize in focal adhesions to mediate integrin linkage to the actin cytoskeleton, function during viral infection [48]. However, whether talin or vinculin are involved in the pathophysiological processes in the liver during systemic infections is not yet clear. EBOV, a member of the family Filoviridae, is a major human pathogen and causes severe EBOV disease in humans and non-human primates with case-fatality rates in humans of up to 90% [52–56]. On July 17, 2019, WHO declared global health emergency as the current Ebola outbreak in the Democratic Republic of the Congo (https://www.bbc.com/news/health-49025298). Rickettsioses represent devastating human infections [57]. Untreated or misdiagnosed SFGR infections are frequently associated with severe morbidity and mortality [58–62]. It has been forecasted that increased ambient temperature under conditions of global climate change is a driver in rickettsial epidemiology, leading to more widespread distribution of rickettsioses [63]. The intracellular pathogen Rickettsia manipulates intercellular tension via a secreted effector protein to promote engulfment of protrusions and cell-to-cell spread [5]. Whether these interacting proteins, talin and vinculin, play a role in the progress of these deadly infectious diseases still needs further exploration. In order to explore the TVSP in liver during these dread diseases, we chose to utilize our archival sample from Ebola and SFGR infected mice. This is the first report of enhanced TVSP in liver during systemic infections with SFGR and EBOV. The PLA fluorescence displayed higher intensity in infected tissues than that in normal tissues, in a manner dependent of EPAC1.
Cyclic adenosine monophosphate (cAMP) is an important molecular switch that translates environmental signals into regulatory effects in cells [64]. As such, a number of microbial pathogens have evolved a set of diverse virulence-enhancing strategies that exploit the cAMP signaling pathways of their hosts [65]. In multicellular eukaryotic organisms, the effects of cAMP are transduced by two ubiquitously expressed intracellular cAMP receptors, the classic cAMP-dependent protein kinase A and the more recently discovered exchange proteins directly activated by cAMP (EPAC) [66]. To date, two isoforms, EPAC1 and EPAC2, have been identified in humans. They are produced by independent genes, and predominantly expressed in different cell types, but both act on the same immediate downstream effectors [67, 68]. EPAC proteins function by responding to increased intracellular cAMP levels and activating the immediate downstream effectors, the Ras superfamily small GTPases Rap1 and Rap2. We have been interested in host cAMP signaling pathways, EPAC1 in particular, for their possible roles in rickettsial adhesion and invasion. We performed in vivo analyses using EPAC1-KO mouse [69] and EPAC-specific inhibitors (ESIs). We found that the deletion of the EPAC1 gene protected mice from an ordinarily lethal dose of rickettsiae (2 × 106 PFU of R. australis) [30]. Most importantly, pharmacological inhibition of EPAC1 by ESI09, an ESI, in mice [70] mimicked the EPAC1-KO phenotype. ESI09 treatment dramatically decreased the morbidity and mortality associated with fatal spotted fever rickettsioses [30]. In addition, in vitro investigations demonstrated that EPAC1 inhibition suppressed rickettsial adherence to and invasion into ECs. However, aside from what we have shown, little is known regarding the precise mechanism underlying the role of EPAC1 in fatal rickettsioses. Observations from the present study provide a fresh insight into the mechanism(s) underlying the pathogenesis of SFGR infections, regarding the potential role of the spatial proximal complex of talin–vinculin. For instance, cytokines were documented to regulate talin-relevant [71] and vinculin-relevant [72] cytoskeletal dynamics in cells. “Cytokine storm” has been well known as central host responses during SFGR [32] or EBOV [54–58] infections. We do not know the increased TVSP induced by infection itself or a form of outcome from host cytokine response, which will be investigated in future studies.
PPIs regulate a variety of cellular functions, which are central to understanding the functional relationships between proteins [71]. PPIs have been studied from different perspectives: biochemistry, quantum chemistry, molecular dynamics, and signal transduction, among others [72, 73], and a multitude of methods have been applied to detect the PPIs [20]. The most conventional methods are yeast two-hybrid screening and affinity purification coupled to mass spectrometry [74]. However, these approaches have a common limitation that all work on cells or tissue homogenates, thus missing critical compartmental information in cell biology or histology. Moreover, these methods have their own disadvantages, respectively, limiting wider applications accurately. The main criticism of the yeast two-hybrid screen of PPIs is the possibility of a high number of false-positive (and false-negative) identifications [75]. This partly clarifies the overlap, often found very minimal, in results when using a (high throughput) two-hybrid screening, especially when using different experimental systems [76–78]. In addition, the colocalization of proteins can be clarified by immunoprecipitation, which is performed with beads to isolate proteins, followed by WB to test target proteins [79]; fluorescence colocalization microscopy is frequently used to assess potential links between distinct molecules; however, this method can lead to striking false-positive results and erroneous conclusions [80, 81]. Furthermore, colocalization does not necessarily mean a physical interaction [80]. A practical step toward unraveling the complex molecular relationship, temporal and spatial, of protein-to-protein physical interaction is essential for accomplishing a comprehensive knowledge of the functional outcomes [82]. Since detection of protein–protein proximity has been developed to probe the proximal PPI in native environment [16, 83, 84], PLA is being more and more widely used in tissues and cells [22, 23]. In a typical PLA experiment, two antibodies are used to recognize two proteins of interest. Each antibody is conjugated to a short sequence of oligonucleotides that are complementary to each other. When these probes are in the proximity of less than 40 nm, the addition of two connector oligonucleotides that hybridize with the probes will lead to the formation of a closed circle template in the presence of a ligase [24, 83]. It can probe spatial proximity of endogenous proteins directly in individual cells and tissues to reveal the cellular and molecular architecture and its responses to perturbations [24].
However, a major factor limiting wide applications of PLA is the high cost for commercial kits. Furthermore, the currently commercially available secondary antibodies conjugated to the PLA probes can only bind to rabbit, mouse, or goat IgG. Hence, whether a PLA experiment can be performed depends on the availability of the primary antibodies that are from two of the above sources. For mouse samples, employing non-mouse original primary antibody was the technical principle from the manufacture of the PLA kit (https://www.sigmaaldrich.com/technical-documents/protocols/biology/duolink-troubleshooting-guide.html), consequently causing additional cost for purchasing other primary antibodies and probe marker kits that were used to conjugate the primary antibodies to the PLA probes. Mouse antibodies are the most widely used primary antibodies and very popular for routine assays (WB, ELISA, IP, IHC, and IF). Combining the results of PLA with one of these routine assays could provide more robust information, especially if identical primary antibodies are employed in these assays, suggesting the importance of developing strategies to perform PLA on mouse samples using primary antibodies raised in mouse. In this study, the infected samples were all of mouse origin. Through setting up a practical and scientific strategy, we explored the feasibility of using primary antibodies from sample species in PLA. Outcomes from this mouse-on-mouse PLA exhibited promising signal intensities and clean backgrounds. This means, the host species of primary antibodies using for PLA were not restricted by the species of the model. In addition, taking advantage of fluorescent-laser capture microdissection, we can extend the applications of PLA in biological/biomolecular science research.
An additional attractive feature of our optimized PLA procedure is the ability to perform this technique on multiple FFPE samples using optimized crucial effective antigen retrieval. Many pathogens that threaten human health, including rickettsia ssp, coronaviruses like SARS and SARS-Cov-2, and filoviruses like Ebola, require high-containment BSL3 and BSL4 laboratories. Many of these pathogens are also select agents, a designation that requires extra steps to ensure proper sample inactivation. Assays like PLA are performed at BSL2, thereby requiring sample inactivation prior to removal from the BSL3 or 4. Inactivation typically requires long periods in and multiple changes of fixative, leading to possible overfixation, which is not ideal for the preservation of features in tissues. Here, we show that PLA is achievable even in these less-than-ideal fixation conditions.
We observed the potential association between TVSP and filamentation of actin in single case of WT survivor on day 19 after SFGR infection and WT survivor on day 21 after EBOV infection, respectively. The observation window was out of our primary scope and we had limited access to more samples at similar time p.i.s. However, this information raised several questions: is there a correlation between TVSP and the activation of actin filamentation? Is TVSP a potential actin-cytoskeleton-mediated healing mechanism or pathogenic as it is found in surviving mice of both infections? Is TVSP possibly protective during hepatic healing from infections? These questions will be delineated in future studies.
Through studying the talin–vinculin spatial interaction in liver of mice infected with SFGR and EBOV, we found that talin–vinculin formed spatial proximity during these infections in a host EPAC1-dependent manner. In the light of these findings, TVSP may serve as a target for deeper insights into the pathological mechanism(s) underlying these important human diseases.
Supplementary information
Acknowledgements
We thank Addie Smith for technical support. This work was supported by NIH grant R01AI121012 (BG) and R21AI137785 (BG); R01-NS-079166, R01-NS-095747, R01-DA-036165, and R01-DA-050530 (SJT). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
YL, JX, BZ, ZS, QC, BJ, XL, YZ, JZ, and MW performed experiments. JB and AD analyzed data. YL, JX, TRS, YJ, SR, SJT, TS, TK, and BK analyzed data and wrote the manuscript. BG designed the study, performed experiments, analyzed data, and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contribute equally: Yakun Liu, Jie Xiao
Supplementary information
The online version of this article (10.1038/s41374-020-0420-9) contains supplementary material, which is available to authorized users.
References
- 1.Lamason RL, Welch MD. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol. 2017;35:48–57. doi: 10.1016/j.mib.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burridge K, Connell L. A new protein of adhesion plaques and ruffling membranes. J Cell Biol. 1983;97:359–67. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiler M, Moser M, Mann M. Copy number analysis of the murine platelet proteome spanning the complete abundance range. Mol Cell Proteom. 2014;13:3435–45. doi: 10.1074/mcp.M114.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L, Yang J, Bromberger T, Holly A, Lu F, Liu H, et al. Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation. Nat Commun. 2017;8:1744. doi: 10.1038/s41467-017-01822-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamason RL, Bastounis E, Kafai NM, Serrano R, Del Álamo JC, Theriot JA, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167:670–83.e10. doi: 10.1016/j.cell.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxter NJ, Zacharchenko T, Barsukov IL, Williamson MP. Pressure-dependent chemical shifts in the R3 domain of talin show that it is thermodynamically poised for binding to either vinculin or RIAM. Structure. 2017;25:1856–66.e2. doi: 10.1016/j.str.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Whitewood AJ, Singh AK, Brown DG, Goult BT. Chlamydial virulence factor TarP mimics talin to disrupt the talin-vinculin complex. FEBS Lett. 2018;592:1751–60. doi: 10.1002/1873-3468.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagiwara M, Kokubu E, Sugiura S, Komatsu T, Tada H, Isoda R, et al. Vinculin and Rab5 complex is required [correction of requited] for uptake of Staphylococcus aureus and interleukin-6 expression. PLoS ONE. 2014;9:e87373. doi: 10.1371/journal.pone.0087373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuy AG, Caron E. Integrin-dependent phagocytosis: spreading from microadhesion to new concepts. J Cell Sci. 2008;121:1773–83. doi: 10.1242/jcs.018036. [DOI] [PubMed] [Google Scholar]
- 11.Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr Opin Virol. 2012;2:28–36. doi: 10.1016/j.coviro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Hauck CR, Borisova M, Muenzner P. Exploitation of integrin function by pathogenic microbes. Curr Opin Cell Biol. 2012;24:637–44. doi: 10.1016/j.ceb.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Rajabian T, Gavicherla B, Heisig M, Müller-Altrock S, Goebel W, Gray-Owen SD, et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–8. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumatsu M, Ogawa M, Arakawa S, Suzuki M, Nakayama K, Shimizu S, et al. Shigella targets epithelial tricellular junctions and uses a noncanonical clathrin-dependent endocytic pathway to spread between cells. Cell Host Microbe. 2012;11:325–36. doi: 10.1016/j.chom.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Agapakis CM, Boyle PM, Silver PA. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol. 2012;8:527–35. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Eckert MA, Chang JW, Montgomery JE, Chryplewicz A, Lengyel E, et al. Ultrasensitive, multiplexed chemoproteomic profiling with soluble activity-dependent proximity ligation. Proc Natl Acad Sci USA. 2019;116:21493–500. doi: 10.1073/pnas.1912934116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagci H, Sriskandarajah N, Robert A, Boulais J, Elkholi IE, Tran V, et al. Mapping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mechanisms. Nat Cell Biol. 2020;22:120–34.. doi: 10.1038/s41556-019-0438-7. [DOI] [PubMed] [Google Scholar]
- 18.Ofran Y, Rost B. Analysing six types of protein-protein interfaces. J Mol Biol. 2003;325:377–87.. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
- 19.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–92. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein-protein interaction detection: methods and analysis. Int J Proteom. 2014;2014:147648. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, Montgomery JE, Eckert MA, Chang JW, Tienda SM, Lengyel E, et al. An activity-dependent proximity ligation platform for spatially resolved quantification of active enzymes in single cells. Nat Commun. 2017;8:1775. doi: 10.1038/s41467-017-01854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendez R, Banerjee S. Proximal ligation assay (PLA) on lung tissue and cultured macrophages to demonstrate protein-protein interaction. Bio Protoc. 2017;7:1–16. doi: 10.21769/BioProtoc.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard EL, Loomis KH, Bhosle SM, Vanover D, Baumhof P, Pitard B, et al. Proximity ligation assays for in situ detection of innate immune activation: focus on in vitro-transcribed mRNA. Mol Ther Nucleic Acids. 2019;14:52–66. doi: 10.1016/j.omtn.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 25.Eberhart A, Kimura H, Leonhardt H, Joffe B, Solovei I. Reliable detection of epigenetic histone marks and nuclear proteins in tissue cryosections. Chromosome Res. 2012;20:849–58. doi: 10.1007/s10577-012-9318-8. [DOI] [PubMed] [Google Scholar]
- 26.Strenkert D, Schmollinger S, Schroda M. Protocol: methodology for chromatin immunoprecipitation (ChIP) in Chlamydomonas reinhardtii. Plant Methods. 2011;7:35. doi: 10.1186/1746-4811-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintel BK, Thomas A, Poer DeRaad DE, Slifka MK, Amanna IJ. Advanced oxidation technology for the development of a next-generation inactivated West Nile virus vaccine. Vaccine. 2019;37:4214–21. doi: 10.1016/j.vaccine.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Zhang W, Chang Q, Su Z, Gong D, Zhou Y, et al. A new role for host annexin A2 in establishing bacterial adhesion to vascular endothelial cells: lines of evidence from atomic force microscopy and an in vivo study. Lab Investig. 2019;99:1650–60. doi: 10.1038/s41374-019-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong B, Lee YS, Lee I, Shelite TR, Kunkeaw N, Xu G, et al. Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect Dis. 2013;13:285. doi: 10.1186/1471-2334-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong B, Shelite T, Mei FC, Ha T, Hu Y, Xu G, et al. Exchange protein directly activated by cAMP plays a critical role in bacterial invasion during fatal rickettsioses. Proc Natl Acad Sci USA. 2013;110:19615–20. doi: 10.1073/pnas.1314400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Londoño AF, Mendell NL, Walker DH, Bouyer DH. A biosafety level-2 dose-dependent lethal mouse model of spotted fever rickettsiosis: Rickettsia parkeri Atlantic rainforest strain. PLoS Negl Trop Dis. 2019;13:e0007054. doi: 10.1371/journal.pntd.0007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng HM, Wen J, Walker DH. Rickettsia australis infection: a murine model of a highly invasive vasculopathic rickettsiosis. Am J Pathol. 1993;142:1471–82. [PMC free article] [PubMed] [Google Scholar]
- 33.Drelich A, Judy B, He X, Chang Q, Yu S, Li X, et al. Exchange protein directly activated by cAMP modulates Ebola virus uptake into vascular endothelial cells. Viruses. 2018;10:1–15. doi: 10.3390/v10100563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Drelich A, Yu S, Chang Q, Gong D, Zhou Y, et al. Exchange protein directly activated by cAMP plays a critical role in regulation of vascular fibrinolysis. Life Sci. 2019;221:1–12. doi: 10.1016/j.lfs.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin-nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012;14:1485–95. doi: 10.1111/j.1462-5822.2012.01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YG, Martinez JJ. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect Immun. 2010;78:1895–904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Perrino BA. Quantitative in situ proximity ligation assays examining protein interactions and phosphorylation during smooth muscle contractions. Anal Biochem. 2019;577:1–13. doi: 10.1016/j.ab.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawecki C, Lenting PJ, Denis CV. von Willebrand factor and inflammation. J Thromb Haemost. 2017;15:1285–94. doi: 10.1111/jth.13696. [DOI] [PubMed] [Google Scholar]
- 39.Brandherm I, Disse J, Zeuschner D, Gerke V. cAMP-induced secretion of endothelial von Willebrand factor is regulated by a phosphorylation/dephosphorylation switch in annexin A2. Blood. 2013;122:1042–51. doi: 10.1182/blood-2012-12-475251. [DOI] [PubMed] [Google Scholar]
- 40.Yu S, He X, Drelich A, Judy B, Chang Q, Cao S, et al. A practical strategy for immunofluorescent detecting multiple targets in mouse tissues without restrictions on the host specious resources of the primary antibodies. Pathol Res Pract. 2019;215:1049–53.. doi: 10.1016/j.prp.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dedden D, Schumacher S, Kelley CF, Zacharias M, Biertümpfel C, Fässler R, et al. The architecture of Talin1 reveals an autoinhibition mechanism. Cell. 2019;179:120–31. e13. doi: 10.1016/j.cell.2019.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singal SS, Nygard K, Dhruv MR, Biggar K, Shehab MA, Li SS, et al. Co-localization of insulin-like growth factor binding protein-1, casein kinase-2β, and Mechanistic target of rapamycin in human hepatocellular carcinoma cells as demonstrated by dual immunofluorescence and in situ proximity ligation assay. Am J Pathol. 2018;188:111–24.. doi: 10.1016/j.ajpath.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–6. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 44.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–97. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–9. [PubMed] [Google Scholar]
- 46.Huang Y, Day RN, Gunst SJ. Vinculin phosphorylation at Tyr1065 regulates vinculin conformation and tension development in airway smooth muscle tissues. J Biol Chem. 2014;289:3677–88. doi: 10.1074/jbc.M113.508077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell EM, Nunez R, Hope TJ. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:5745–55.. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown C, Morham SG, Walsh D, Naghavi MH. Focal adhesion proteins talin-1 and vinculin negatively affect paxillin phosphorylation and limit retroviral infection. J Mol Biol. 2011;410:761–77. doi: 10.1016/j.jmb.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 49.Rogers KR, Morris CJ, Blake DR. The cytoskeleton and its importance as a mediator of inflammation. Ann Rheum Dis. 1992;51:565–71. doi: 10.1136/ard.51.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talwani R, Gilliam BL, Howell C. Infectious diseases and the liver. Clin Liver Dis. 2011;15:111–30. doi: 10.1016/j.cld.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka M, Miyajima A. Liver regeneration and fibrosis after inflammation. Inflamm Regen. 2016;36:19. doi: 10.1186/s41232-016-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olejnik J, Ryabchikova E, Corley RB, Mühlberger E. Intracellular events and cell fate in filovirus infection. Viruses. 2011;3:1501–31. doi: 10.3390/v3081501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falzarano D, Feldmann H. Vaccines for viral hemorrhagic fevers–progress and shortcomings. Curr Opin Virol. 2013;3:343–51. doi: 10.1016/j.coviro.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–62.. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong G, Qiu X, Olinger GG, Kobinger GP. Post-exposure therapy of filovirus infections. Trends Microbiol. 2014;22:456–63. doi: 10.1016/j.tim.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Hartman AL, Towner JS, Nichol ST. Ebola and marburg hemorrhagic fever. Clin Lab Med. 2010;30:161–77. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Dumler JS, Walker DH. Rocky Mountain spotted fever–changing ecology and persisting virulence. N Engl J Med. 2005;353:551–3. doi: 10.1056/NEJMp058138. [DOI] [PubMed] [Google Scholar]
- 58.Walker DH, Paddock CD, Dumler JS. Emerging and re-emerging tick-transmitted rickettsial and ehrlichial infections. Med Clin North Am. 2008;92:1345–61. doi: 10.1016/j.mcna.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Openshaw JJ, Swerdlow DL, Krebs JW, Holman RC, Mandel E, Harvey A, et al. Rocky mountain spotted fever in the United States, 2000-2007: interpreting contemporary increases in incidence. Am J Trop Med Hyg. 2010;83:174–82. doi: 10.4269/ajtmh.2010.09-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Botelho-Nevers E, Socolovschi C, Raoult D, Parola P. Treatment of Rickettsia spp. infections: a review. Expert Rev Anti Infect Ther. 2012;10:1425–37. doi: 10.1586/eri.12.139. [DOI] [PubMed] [Google Scholar]
- 61.de Sousa R, Nóbrega SD, Bacellar F, Torgal J. Mediterranean spotted fever in Portugal: risk factors for fatal outcome in 105 hospitalized patients. Ann N Y Acad Sci. 2003;990:285–94.. doi: 10.1111/j.1749-6632.2003.tb07378.x. [DOI] [PubMed] [Google Scholar]
- 62.Kim HK, Premaratna R, Missiakas DM, Schneewind O. Rickettsia conorii O antigen is the target of bactericidal Weil-Felix antibodies. Proc Natl Acad Sci USA. 2019. [DOI] [PMC free article] [PubMed]
- 63.Parola P, Socolovschi C, Jeanjean L, Bitam I, Fournier PE, Sotto A, et al. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl Trop Dis. 2008;2:e338. doi: 10.1371/journal.pntd.0000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beavo JA, Brunton LL. Cyclic nucleotide research–still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–8. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 65.McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol. 2012;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng X, Ji Z, Tsalkova T, Mei F. Epac and PKA: a tale of two intracellular cAMP receptors. Acta Biochim Biophys Sin. 2008;40:651–62.. doi: 10.1111/j.1745-7270.2008.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–7. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 68.Kawasaki H, Springett GM, Mochizuki N, Toki S, Nakaya M, Matsuda M, et al. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–9. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 69.Yan J, Mei FC, Cheng H, Lao DH, Hu Y, Wei J, et al. Enhanced leptin sensitivity, reduced adiposity, and improved glucose homeostasis in mice lacking exchange protein directly activated by cyclic AMP isoform 1. Mol Cell Biol. 2013;33:918–26. doi: 10.1128/MCB.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen H, Ding C, Wild C, Liu H, Wang T, White MA, et al. Efficient synthesis of ESI-09, a novel non-cyclic nucleotide EPAC antagonist. Tetrahedron Lett. 2013;54:1546–9. doi: 10.1016/j.tetlet.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Braun P, Gingras AC. History of protein-protein interactions: from egg-white to complex networks. Proteomics. 2012;12:1478–98. doi: 10.1002/pmic.201100563. [DOI] [PubMed] [Google Scholar]
- 72.Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun. 2013;4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cafarelli TM, Desbuleux A, Wang Y, Choi SG, De Ridder D, Vidal M. Mapping, modeling, and characterization of protein-protein interactions on a proteomic scale. Curr Opin Struct Biol. 2017;44:201–10.. doi: 10.1016/j.sbi.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 74.Vidal M, Fields S. The yeast two-hybrid assay: still finding connections after 25 years. Nat Methods. 2014;11:1203–6. doi: 10.1038/nmeth.3182. [DOI] [PubMed] [Google Scholar]
- 75.Serebriiskii I, Estojak J, Berman M, Golemis EA. Approaches to detecting false positives in yeast two-hybrid systems. Biotechniques. 2000;28:328–30. doi: 10.2144/00282rr03. [DOI] [PubMed] [Google Scholar]
- 76.Deane CM, Salwiński Ł, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteom. 2002;1:349–56. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 77.Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7:667–8. doi: 10.1038/nmeth0910-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genom Proteom. 2007;6:302–12. doi: 10.1093/bfgp/elm035. [DOI] [PubMed] [Google Scholar]
- 79.Moresco JJ, Carvalho PC, Yates JR. Identifying components of protein complexes in C. elegans using co-immunoprecipitation and mass spectrometry. J Proteom. 2010;73:2198–204. doi: 10.1016/j.jprot.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lutz MI, Schwaiger C, Hochreiter B, Kovacs GG, Schmid JA. Novel approach for accurate tissue-based protein colocalization and proximity microscopy. Sci Rep. 2017;7:2668. doi: 10.1038/s41598-017-02735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moser B, Hochreiter B, Herbst R, Schmid JA. Fluorescence colocalization microscopy analysis can be improved by combining object-recognition with pixel-intensity-correlation. Biotechnol J. 2017;12:1–8. doi: 10.1002/biot.201600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang C, Freddolino PL, Zhang Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017;45:W291–9. doi: 10.1093/nar/gkx366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weibrecht I, Leuchowius KJ, Clausson CM, Conze T, Jarvius M, Howell WM, et al. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteom. 2010;7:401–9. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 84.Sable R, Jambunathan N, Singh S, Pallerla S, Kousoulas KG, Jois S. Proximity ligation assay to study protein-protein interactions of proteins on two different cells. Biotechniques. 2018;65:149–57.. doi: 10.2144/btn-2018-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.