Abstract
Since 2010, outbreaks of variant porcine epidemic diarrhea virus (PEDV) have swept across the world causing substantial economic losses. The development of new, more effective vaccines has been hampered by difficulties in isolating strains and viral genome manipulation. In the present study, we successfully isolated a highly pathogenic field strain AH2012/12, from a pig farm reporting severe diarrhea in China. Phylogenetic analysis revealed that the new isolate belongs to group G2, which represents epidemic and pandemic field strains. Furthermore, we constructed an infectious cDNA clone of the newly isolated strain, rAH2012/12, and the rescued virus displayed phenotypic properties identical to the parental virus in vitro. In vivo experiments demonstrated that the rescued virus displayed similar pathogenicity to the parental isolate, causing high mortality rates in suckling pigs. This study provided a strong basis for the development of live attenuated vaccines and for research into the pathogenic mechanisms of this virus.
Keywords: PEDV, Isolation, Reverse genetics, Highly pathogenic
Highlights
-
•
We successfully isolated one epidemic PEDV strain AH2012/12 with high virulent in newborn pigs.
-
•
We firstly generated the infectious cDNA clone of the virulent PEDV strain AH2012/12 in China.
-
•
The rescued virus has similar biological characteristics with the parent virus in vitro and vivo.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is the causative agent of porcine epidemic diarrhea (PED), a highly contagious disease of pigs characterized by acute watery diarrhea and vomiting. In 2010, an outbreak of PEDV rapidly spread in China and caused substantial economic losses in the swine industry (Sun et al., 2012). Subsequently, the etiologic agent was identified as variant PEDV (Pan et al., 2012). The characteristics of the infection and its epidemiology were quite dramatic with the rates of morbidity and fatality approaching 100% in 1-week old piglets, despite the use of commercial, inactivated vaccines (Pan et al., 2012). In May 2013, PED outbreaks suddenly emerged in the United States and rapidly spread nationwide, as well as to Canada and Mexico, causing high mortality rates in newborn piglets and significant financial concerns (Mole, 2013, Stevenson et al., 2013, Vlasova et al., 2014).
PEDV belongs to the Alphacoronavirus genus within the Coronavirinae subfamily of the Coronaviridae family (Marthaler et al., 2013). It possesses a 28-kb, single-stranded, positive-sense RNA genome, which is predicted to encode at least seven open reading frames (ORFs) in the following order: ORF1a, ORF1b, spike (S), ORF3, envelope (E), membrane (M), and nucleocapsid (N). The S glycoprotein of PEDV plays an important role in the induction of neutralizing antibodies, specific receptor binding, and cell membrane fusion (Bosch et al., 2003, Cruz et al., 2008). According to the phylogenetic analyses of the spike gene, the PEDV strains were divided into two distinct clusters, designated genogroup 1 (G1; classical) and genogroup 2 (G2; field epidemic or pandemic) (Chen et al., 2013, Lee et al., 2010, Li et al., 2013a). Concurrently, several US variant PEDV strains, characterized by insertions and deletions (INDELs) in the spike (S) gene and designated as S-INDEL PEDV, were found to be circulating in US swine farms (Vlasova et al., 2014, Wang et al., 2014; Jarvis et al., 2016). Therefore, the PEDV S glycoprotein is known to be an appropriate viral gene for determining the genetic relatedness among PEDV isolates and for developing diagnostic assays and effective vaccines (Lee et al., 2010, Lee and Lee, 2014, Oh et al., 2014).
The isolation of PEDV in cell culture is the first step toward the development of an attenuated vaccine, as well as a being critical in establishing in vitro virological and immunological assays. However, the cell culture isolation of PEDV has proven to be difficult and even when isolated virus is obtained it may be unable to maintain infectivity upon further passage in cell culture (Chen et al., 2014). Moreover, the necessity of trypsin increases the uncertainty and difficulty of virus isolation. Recently, several PEDV strains have been isolated and successfully grown in cell culture in the US and Korea (Chen et al., 2014, Lee et al., 2015, Oka et al., 2014). However, there are a few reports on the successful propagation of the original highly virulent strains that caused serious outbreaks of PEDV in China (Pan et al., 2012). In this study, we attempted to isolate and propagate PEDV from various PEDV-positive samples in Vero cells, and one highly virulent Chinese strain AH2012/12 was successfully isolated and serially propagated in cell culture for over 50 passages.
Reverse-genetic technology is a powerful tool for researching the molecular biology and pathogenicity of viruses. To date, there have been three reports about the rescue of PEDV using different technological platforms. Li and colleagues recently described the reverse genetics of PEDV based on targeted RNA recombination technology (Li et al., 2013b), and Juggragarn and colleagues constructed a full-length cDNA clone of PEDV by assembling contiguous cDNA fragments in an artificial bacterial chromosome (Jengarn et al., 2015). However, these two rescued PEDV strains belonged to subgroup G1, which represents the classical PEDV strains (Lee, 2015). A more recent study reported the generation of a virulent North American PEDV strain, PC22A, by in vitro transcription (Beall et al., 2016). However, there is still no report on the reverse genetics of a Chinese strain of PEDV. In this study, we generated an infectious cDNA clone of the newly-isolated Chinese virulent PEDV strain, AH2012/12. In vivo experiments in pigs confirmed that the rescued virus displayed similar pathogenicity to the parental isolate. This technical advance will enable the development of effective live attenuated vaccines of this virus and research into its pathogenic mechanisms.
2. Materials and methods
2.1. Cells, clinical samples and virus isolation
The Vero-81 (ATCC No. CCL-81) cell line was used for PEDV isolation experiments. Vero cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with antibiotics (100 units/mL of penicillin, 100 mg/mL of streptomycin, and 0.25 mg/mL of fungizone) (Life Technologies), and 5% heat-inactivated fetal bovine serum (FBS; Life Technologies).
Isolation of PEDV was attempted on Vero cells as described previously with some modifications (Chen et al., 2014, Lee et al., 2015, Pan et al., 2012, Zhang et al., 2015). A small intestine tissue sample was obtained from a piglet that displayed typical PED clinical signs and symptoms, including acute watery diarrhea and vomiting, in Anhui, China, in 2012. The piglet was diagnosed as PEDV-positive using reverse transcription (RT)-polymerase chain reaction (PCR). The intestinal tissue was homogenized and centrifuged. The supernatant was filtered through a 0.22-μm-pore-size syringe filter (Millipore, Billerica, MA, USA) and used to inoculate Vero cells with trypsin in a contrition of 15 μg/mL. After 2 h, the cells were maintained in DMEM containing 10 μg/mL trypsin at 37 °C in a 5% CO2 atmosphere until a cytopathic effect became visible. The infected cells were lysed using a freeze-thaw method, and centrifuged at 2000×g for 10 min. The supernatant was stored at −70 °C until use.
2.2. Electron microscopy (EM) for the detection of PEDV particles
PEDV-infected Vero cell culture media were clarified by centrifugation at 2000×g for 30 min, followed by filtration of the supernatants through 0.22-μm filters. These samples were centrifuged at 106,750×g for 2 h at 4 °C using an ultracentrifuge (Beckman Coulter, Miami, FL, USA). The virus pellets were resuspended and negatively stained with 2% ammonium molybdate, and examined with an electron microscope (Hitachi H7500, Tokyo, Japan).
2.3. Genome sequencing
Total RNAs were extracted from the tissue homogenate or virus cultures using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and the SuperScript III First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA, USA) was used for the RT procedure. The S1 nucleotide sequence of the virus was compared with that of other PEDV isolates available in GenBank database (NCBI) to identify similarities, upon which primers were designed. The whole genome was divided into 14 overlapping fragments (Table S1) that were amplified by PCR using the Phanta Super Fidelity DNA polymerase (Vazyme, China), and the 5′ and 3′ termini of the genomic sequence were synthesized using rapid amplification of the cDNA ends (RACE). The PCR products were purified from the agarose gel using the AxyPrep DNA Gel Extraction Kit (Axygen, Hangzhou, China), and cloned into the pEASY-Blunt Zero vector (Trans, Beijing, China). Three clones were sequenced by a commercial service provider (Invitrogen, Shanghai, China).
2.4. Sequence analysis
The overlapping sequences of the PCR products were combined to obtain the full-length genomic sequence of the AH2012/12 strain. The sequence alignments were generated using the Clustal W program. Phylogenetic trees of the full-length genome and S nucleotide sequences were generated using the distance-based neighbor-joining method in the MEGA, version 5.05, software. The bootstrap values were calculated based on 1000 replicates, and the evolutionary distances were computed using the Jukes-Cantor method.
2.5. Design of the rAH2012/12 clone
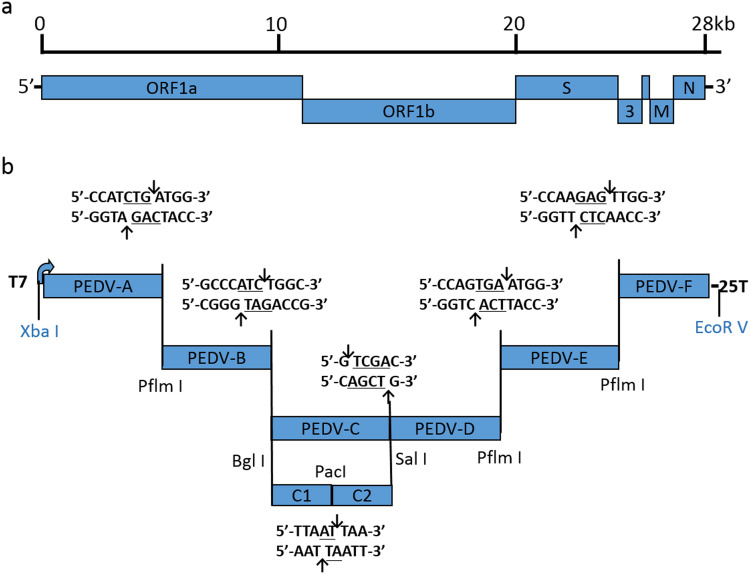
The whole genome of AH2012/12 was separated into six fragments (A, B, C, D, E, and F), and the primer sets used to amplify each fragment are shown in Table 1. The PEDV A fragment contained a T7 start site, whereas the F subclone terminated in 24 T residues, allowing for in vitro transcription of capped, polyadenylated transcripts. The enzyme sites at the two ends of each fragment are shown in Fig. 1. The cDNA obtained from passage 10 of clone AH2012/12 was used as the template. All fragments were amplified from the template using highly-fidelity enzyme (Phanta Max, Vazyme, China), and the PCR products were electrophoresed, isolated from the gel, and then cloned into the low copy cloning vector pSMART (Lucigen, Middleton, USA). The C fragment was unstable in the bacteria and subsequently subdivided into two subclones, C1 and C2, and these two fragments were ligated using the unique enzyme site PacI (Fig. 1). In fragments B, D, and F, three naturally occurring PflmI sites were removed by introducing silent mutations (A6602G, A18052G, and A26077G) to prevent interference with the assembly of the full-length infectious clone (Table 1).
Table 1.
Primer sequences for construction of the subgenomic replicon of PEDV strain AH2012/12, and site-directed mutagenesis.
| Primers | Sequence (5′−3′) | Purpose |
|---|---|---|
| Primers for the construction of infectious, full-length cDNA clones of AH2012/12 | ||
| A-T7-1-F | TAATACGACTCACTATAGGGAGAACTTAAAAAGA | Fragment A |
| TTTTCTATCTACGGA | ||
| A-5060-R | TACCTTGACAACGGCGCGTGCATAA | |
| B-4957-F | TTACTTTTGAGTGTGCAGATATGAT | Fragment B |
| B-9248-R | GTATAACGTGTCGTTGTGCTTAGAA | |
| C-9184-F | TTGTTTTGCCCATCTGGCCAAG | Fragment C |
| C-15144-R | AGCAATACAACTGCATCAGTTTTAA | |
| D-15068-F | TACCCTGATCCTTCAAGAATCCTCT | Fragment D |
| D-19784-R | GTTGTATCTTGTAAATAGAAGGCAT | |
| E-19687-F | GTGGTGTAAGGATCATAAACTCCAG | Fragment E |
| E-24928-R | CCAGTCACATTTGAAGCTTGTCTAA | |
| F-24832-F | TCAAAGATGTCTCTAAGTCTGCCAA | Fragment F |
| F-28032-24T-R | TTTTTTTTTTTTTTTTTTTTTTTTTGTGTATCCATATCAACAC | |
| Primers for the transcription of AH2012/12 N protein | ||
| N-SP6-F | GCCTATTTAGGTGACACTATAGATGGCTTCTGTCAGTTTTC | Generate the AH2012/12 N transcripts |
| N-(21 T)-R | TTTTTTTTTTTTTTTTTTTTTTTAATTTCCAGTATCGAAG | |
| Primers for the construction of site-directed mutations | ||
| 6602-A/G-F | CACCTTAGAGACCCGTTAATTGGTAATG | Site-directed mutagenesis |
| 6602-A/G-R | CATTACCAATTAACGGGTCTCTAAGGTG | |
| 18052-A/G-F | GTCCATCACATACCCGTTTATTGGTAAT | |
| 18052-A/G-R | ATTACCAATAAACGGGTATGTGATGGAC | |
| 26077-A/G-F | CTGCATTCCGGTGCTTGGAGCAC CAACTGGTGTAAC | |
| 26077-A/G-R | TCCAAGCACCGGAATGCAGACCTGTCGGCCCATCAC | |
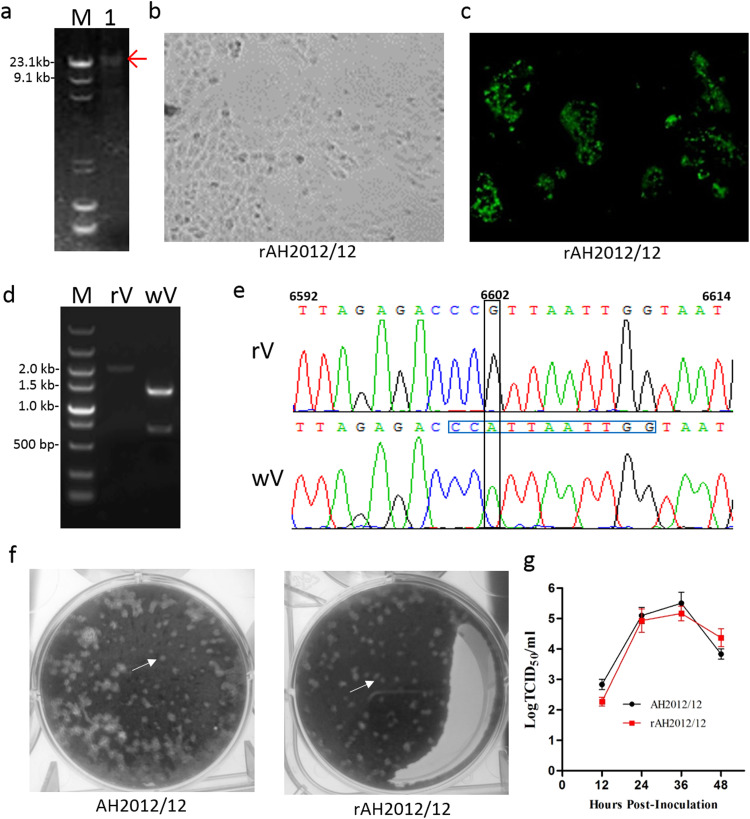
Fig. 1.
Organization of the PEDV strain AH2012/12 molecular clone. (a) The organization of the AH2012/12 genome. (b) The full-length AH2012/12 genome was divided into seven contiguous cDNAs designated PEDV A–F and flanked by enzyme sites that allowed for directed assembly of a full-length cDNA. The restriction enzyme site Xba I on the 5′-end of fragment A and the enzyme site EcoR V on the 3′-end of fragment F were existed in the enzyme sites area of the cloning vector pSMART.
2.6. Assembly of full-length recombinant PEDV
The rAH2012/12 clone was designed using six separate fragments flanked with two class II restriction sites PflmI and BglI that leave nonpalindromic overhangs, and two conventional restriction sites PacI and SalI. The purified recombinant plasmids were then digested as shown in Fig. 1. After digestion, fragments were electrophoresed on 0.8% (wt/vol) agarose gel, and appropriate bands were excised and gel purified using a Qiaex II gel extraction kit (Qiagen). Purified fragments rAH2012/12-A through to rAH2012/12-F were mixed and ligated using T4 DNA ligase (Fermentas, Vilnius, Lithuania) overnight at 4 °C, followed by phenol/chloroform extraction and precipitation with isopropyl alcohol. The obtained full-length cDNAs were transcribed using a T7 transcription kit (mMessage mMachine; Ambion, Austin, TX, USA). For rAH2012/12 N transcripts, 1 μg of linearized plasmid DNA encoding the N gene (amplified using the primers in Table 1) was transcribed by SP6 RNA polymerase with a 4:1 ratio of cap analog to GTP (Ambion).
2.7. In vitro transfection
The transfection of full-length transcripts was performed as previously described with the following modifications (Beall et al., 2016, Scobey et al., 2013, Yount et al., 2003). Briefly, about 20 μg genome-length and 5 μg N RNA transcripts were mixed with 500 μL of Vero cells (1×107 cells/mL) in opt-MEM (Gibco, Gaithersburg, MD, USA) and then added to an electroporation cuvette. Three pulses of 250 V at 50 μF were used to transfect the cells with ECM630 (BXT, Holliston, USA). The cells were allowed to recover for 10 min at room temperature and were then transferred to a 12-well flask in growth medium at 37 °C for 2 h, after which time the cells were washed and incubated in cell culture medium. Trypsin was added to the culture at 7 μg/mL 24 h post-electroporation to assist in virus recovery and spread. After four days, the virus was harvested from the flask. To obtain high titers, the progeny were passaged another two times. Then the virus was plaque purified and propagated in Vero cells.
2.8. Detection of marker mutations in the infectious clone rAH2012/12
Intracellular RNA was isolated from either wild-type (WT) or the plaque purified rescued virus-infected cells 24 h after infection. After RT-PCR, a 2048-nt amplicon (nucleotide positions 5922–7969) was obtained spanning the PflmI site at position 6602 that had been ablated in the rescued component clones, but not in the WT virus. The fragment amplified from the WT virus could be digested into two fragments (684 and 1363 bp) by PflmI, whereas the fragment amplified from the rescued virus could not be digested by PflmI. And the amplicons were used to sequence for genotype verification.
2.9. Viral plaque assay
Vero cells in 12-well plates were inoculated with 500 μL of 10-fold serially-diluted parent (passage 5) or rescued (passage 3) PEDV viruses. After 1 h adsorption at 37 °C, cell monolayers were washed with phosphate-buffered saline (PBS) and overlaid with 1% low melting agarose in DMEM (Invitrogen) containing 7 μg/mL trypsin. After the gel overlay solidified, the plates were inverted (top side down) and placed into an incubator at 37 °C with 5% CO2. At 3 days post-infection (dpi), plaques were picked for cell infecting or visualized by crystal violet staining.
2.10. Growth curves of viruses
For the growth curve analysis, Vero cells were inoculated with PEDV parent and rescued strains at a multiplicity of infection (MOI) of 1 in 24-well plates for 1 h, followed by three washes with PBS. Then, the supernatants and the infected cells were collected at 12, 24, 36, and 48 h post-infection, and stored at −70 °C for virus titration. The virus titers for each time point were determined in Vero cells by TCID50.
2.11. Immunofluorescence microscopy
Vero cells were grown to 70–80% confluence on 24-well plate and inoculated with parent or recombinant virus at a MOI of 0.1, the control was inoculated with medium alone. At 36 h post-infection, the cells were fixed with 4% formaldehyde and blocked with PBS containing 5% bovine serum albumin. After being blocked, the cells were incubated with a PEDV-specific monoclonal antibody (produced in our laboratory, and the immune-purified prokaryotic expression protein was the N protein of PEDV JS2008 strain, GenBank: KC109141), diluted 1:1000, for 1 h. The cells were then washed and incubated with secondary antibody (goat anti-mouse FITC, diluted 1:500) for 45 min. Finally, the cells were washed and then visualized by fluorescent microscopy.
2.12. Experimental design of infection
Fifteen 3-day-old piglets, randomly selected from two sows that were PEDV RNA and antibody negative, were divided into three groups and housed in separate rooms. Pigs were fed a mixture of liquid milk replacer and yogurt and had free access to water. Groups A and B, which contained five piglets each, were challenged orally with strain AH2012/12 (passage 10) or rAH2012/12 (passage 5) at 1.0×105 TCID50 (2 mL), respectively. The viruses used for inoculation were confirmed negative for transmissible gastrointestinal virus (TGEV), porcine rotavirus and porcine deltacoronavirus (PDCoV) by virus-specific RT-PCRs. The five control piglets (Group C) were inoculated with cell culture media. All animals were monitored daily for clinical signs of disease, including diarrhea and vomiting. Rectal swabs were collected for scoring fecal denseness (scores: 0 normal; 1 pasty stool; 2 semiliquid diarrhea; and 3 liquid diarrhea) and for enumerating fecal viral RNA shedding by RT-quantitative PCR (RT-qPCR). The primers and probes targeting conserved regions of the PEDV nucleocapsid protein gene were as described before (Chen et al., 2014, Kim et al., 2007), with modifications to obtain a closer match to the nucleotide sequence from PEDV AH2012/12 strain (accession no. KU646831; and the sequences of primers and probe were as follows: forward, 5′-CGCAAAGACTGAACCCACTAACTT-3′; reverse, 5′-TTGCCTCTGTTGTTACTCGGGGAT-3′; probe, 5′-TGTTGCCATTGCCACGACT ATAC-3′). At necropsy, intestinal tissues and contents were grossly evaluated. Fresh jejunum and ileum were collected. Additionally, a portion of the jejunum and ileum were fixed in 10% neutral buffered formalin for histopathology and immunohistochemistry examinations. The villous atrophy is a typical histopathological change of the infection of a highly virulent PEDV (Jung et al., 2014, Liu et al., 2015, Madson et al., 2016, Stevenson et al., 2013). And the villous height and crypt depth (VH:CD) ratios would be calculated using a computerized image system with as previous described (Jung et al., 2014). The fixed tissue sections were evaluated for PEDV antigen by immunohistochemistry (IHC) using a PEDV-specific monoclonal antibody (1:200 dilution), produced in our laboratory using the antigen retrieval method described previously (Madson et al., 2014). If the piglets died sooner, necropsy was performed as described above. All of the animal experiments were performed with the approval of the Jiangsu Academy of Agricultural Sciences Experimental Animal Ethics Committee (NKYVET 2015-0126). All efforts were made to minimize animal suffering and to reduce the number of animals used.
2.13. Statistical analysis
All data were analyzed using GraphPad Prism (Version 5.03, San Diego, CA, USA) software. Differences among groups were examined using one-way analysis of variance (ANOVA), followed by Tukey's tests.
3. Results
3.1. Virus isolation and in vitro characterization
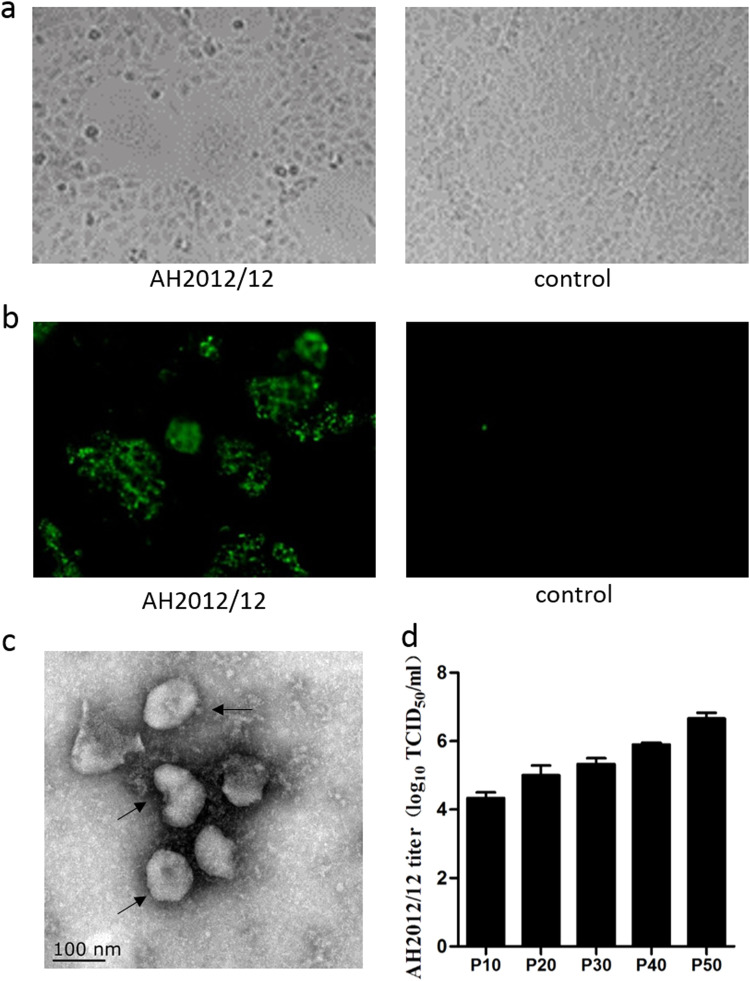
New PEDV isolate AH2012/12 was successfully isolated from the intestine of a naturally-infected piglet from a commercial farm located in Anhui Province, China, in 2012. The typical cytopathic effects (CPEs) of PEDV infection, such as cell fusion, syncytium, and detachment, appeared on the third day after tissue supernatant inoculation in Vero cells ( Fig. 2a). Infected cells were detected by fluorescent antibody staining. As shown in Fig. 2b, AH2012/12-infected cells exhibited fluorescence correlating to the PEDV-N protein as detected by an anti-N monoclonal antibody. Moreover, electron microscopy of a negatively-stained sample revealed typical coronavirus particles, i.e. medium-sized viral particles of approximately 100–120 nm in diameter and characteristic surface projections (Fig. 2c). To investigate whether the isolate could be efficiently cultivated and maintained in cell culture, we serially passaged isolate AH2012/12 for a total of 50 passages in Vero cells. Prominent CPEs were observed within 48 h post-infection in passages three and four. In later passages, prominent CPEs appeared within 24 h post-infection. Meanwhile, we selected five passages to infect Vero cells at the same time. The culture supernatants were collected at 24 or 48 h post-infection at which a 70% CPE is commonly developed, and the virus titers were measured. The results showed that the infectious titer of the isolate ranged from 104.5 to 106.5 TCID50/mL up to passage 50 (Fig. 2d).
Fig. 2.
Virus isolation and identification of PEDV strain AH2012/12. (a) The CPEs of PEDV strain AH2012/12 on Vero cells. (b) Fluorescence microscopy of AH2012/12 on Vero cells. (c) Electron microscopy images of PEDV virions of PEDV strain AH2012/12 infecting Vero cells in cell culture media. Scale bars=50 nm. (d) Growth kinetics of AH2012/12 with different passages in Vero cells.
3.2. Complete genomic characterization of AH2012/12
Sequence analysis revealed that the full-length genomic sequence of AH2012/12 (GenBank accession number: KU646831) was 28,032 nucleotides in length, excluding the polyadenylated sequences, and exhibited the genomic organization typical of all previously sequenced PEDV strains, consisting of a 292-nt 5ʹ UTR, the 20,345-nt ORF1a/1b gene (nt 293 to 12,601 for 1a and nt 12,601 to 20,637 for 1b), the 4158-nt S gene (nt 20,634 to 24,791), the 672-nt ORF3 gene (nt 24,791 to 25,462), the 231-nt E gene (nt 25,443 to 25,673), the 681-nt membrane (M) gene (nt 25,681 to 26,361), the 1326-nt N gene (nt 26,373 to 27,698), and a 334-nt 3ʹ UTR. The entire genome sequence of PEDV determined in this study was further compared with those of three Chinese epidemic strains (AH2012, CH-GDGZ-2012, and PEDV-7C), four vaccine related strains (CV777, DR13, Attenuated DR13, and JS2008) and two American strains (USA-Iowa107-2013 and USA-Minnesota61-2013) available in the GenBank database, and the nucleotide similarities are described in Table 2. Isolate AH2012/12 showed the highest nucleotide identity (99.4%) with the Chinese re-emergent strains CH-GDGZ-2012 and PEDV-7C. The percentage identity between isolate AH2012/12 and isolates AH2012 and USA-Minnesota61-2013 was 98.2% and 98.1%, respectively. AH2012/12 was genetically distinct from the vaccine strains, CV777 and Attenuated DR-13, exhibiting relatively low percentages of nucleotide identity ranging from 96.7% to 96.8%. In addition, we also compared the amino acid identities of the S gene of AH2012/12 with that of other strains (data not shown). The S protein of AH2012/12 was 4158 nucleotides long, encoding a protein of 1385 amino acids. Compared with the PEDV reference strain CV777, it shared low amino acid identity (92.8%), whereas it showed the highest level of identity (99.5%) with the Chinese strain GD-A.
Table 2.
Nucleotide comparison of the full-length genomes of AH2012/12 isolate to other PEDV strains.
| Strains | Nucleotide identity (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AH2012/12 | AH2012 | CH-GDGZ-2012 | PEDV-7C | CV777 | DR13 | Attenuated DR13 | JS2008 | USA-Iowa107-2013 | USA-Minnesota61-2013 | |
| AH2012/12 | 98.2 | 99.4 | 99.4 | 96.7 | 97.5 | 96.8 | 96.9 | 97.5 | 98.1 | |
| AH2012 | 98.0 | 98.0 | 96.8 | 97.7 | 96.9 | 97.1 | 98.4 | 99.4 | ||
| CH-GDGZ-2012 | 99.2 | 96.5 | 97.3 | 96.6 | 96.7 | 97.3 | 97.9 | |||
| PEDV-7C | 96.5 | 97.3 | 96.6 | 96.8 | 97.3 | 97.8 | ||||
| CV777 | 97.7 | 97.7 | 97.7 | 96.9 | 96.6 | |||||
| DR13 | 97.7 | 97.8 | 97.7 | 97.5 | ||||||
| Attenuated DR13 | 99.6 | 97.1 | 96.7 | |||||||
| JS2008 | 97.2 | 96.9 | ||||||||
| USA-Iowa107-2013 | 98.9 | |||||||||
| USA-Minnesota61–2013 | ||||||||||
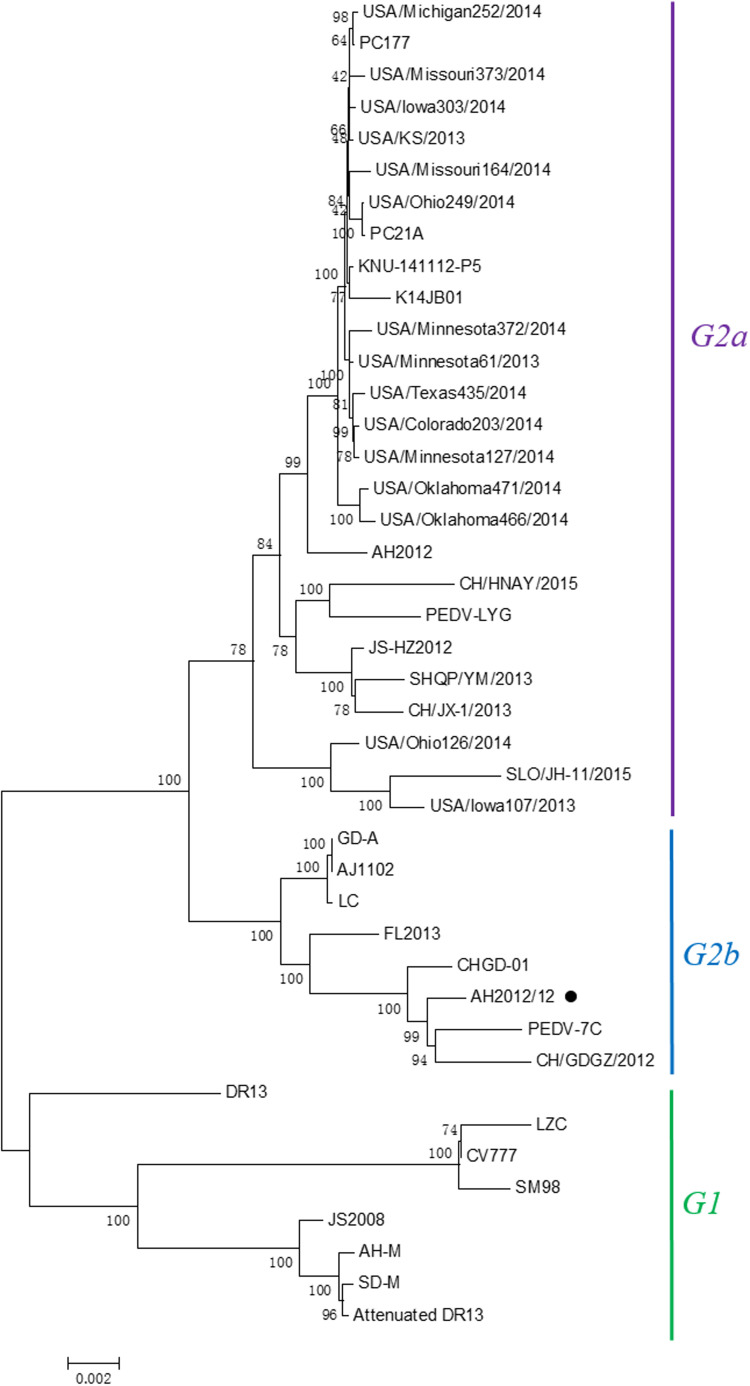
3.3. Phylogenetic analysis of isolate AH2012/12
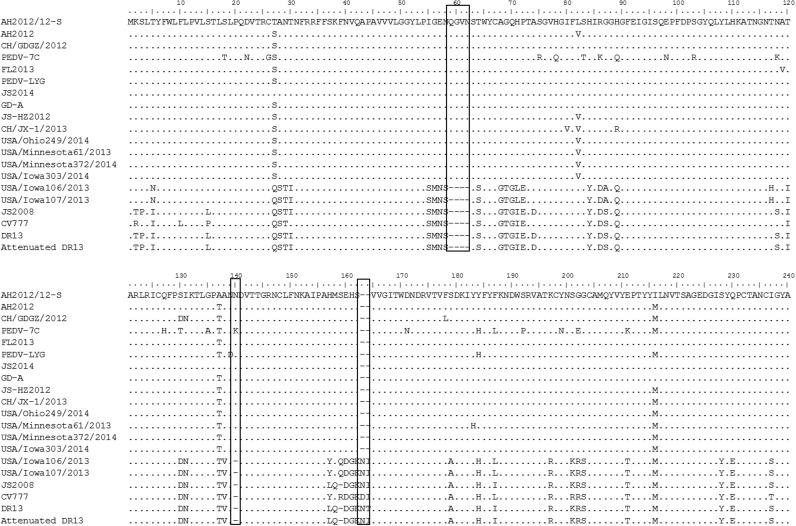
Phylogenetic analysis based upon the complete genome demonstrated that PEDV isolate AH2012/12 clustered with the original China PEDV strains CHGD-01 and CH/GDGZ/2012, belonging to the genogroup G2b ( Fig. 3). The S gene alignment showed that AH2012/12 harbors two significant insertions at amino acids 59–62 (QGVN) and 140 (N), and a deletion of two amino acids (DI) between positions 163 and 164 at the N terminus compared with the prototype CV777 strain ( Fig. 4).
Fig. 3.
Phylogenetic analysis based on the nucleotide sequence of the full-length genome of AH2012/12. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches. The evolutionary distances were computed using the Jukes-Cantor method and were presented as the number of base substitutions per site. Evolutionary analyses were conducted using MEGA5 software.
Fig. 4.
Alignment of the partial S protein amino acid sequence of AH2012/12 with several representative PEDV strains. The deletions and insertions are shown by grey bars.
3.4. Recovery of recombinant viruses
To build a full-length PEDV cDNA, each plasmid fragment was digested with restriction enzymes, purified, and ligated to create a full-length PEDV cDNA genome ( Fig. 5a). The full-length transcripts were synthesized in vitro using T7 RNA polymerase according to the manual (Beall et al., 2016). As previous swine and human coronavirus infectious clones displayed improved recovery rates and replication efficiencies in the presence of supplemented N gene transcripts (Beall et al., 2016), capped PEDV-N gene transcripts were mixed with full-length genomic transcripts prior to their electroporation into Vero cells. After 4 days, the culture media were collected and passaged another two times. CPEs became evident (Fig. 5b), and the features of the CPEs caused by rAH2012/12 were vacuolation and the formation of syncytia, which was similar to those observed for the parent AH2012/12 (Fig. 2a). In addition, cells tested positive by IFA (Fig. 5c).
Fig. 5.
The recovery and growth characteristics of the recombinant PEDV rAH2012/12. (a) The full-length PEDV cDNA genome was generated by ligation of the digested fragments. The label “M” was the DNA maker λDNA/Hind III, and the label “1” was the ligation product of digested fragments. (b) The CPEs of the rescue strain rAH2012/12 on Vero cells. (c) Fluorescence microscopy of rAH2012/12 on Vero cells. (d) Verification of the marker mutation in the rescued virus from rAH2012/12. One silent mutation A6602G was introduced to remove the naturally occurring PflmI site. Two 2048-nt amplicons (nucleotide positions 5922–7969) were obtained from the rescued (panel rV) and parental virus (panel wV) strains, respectively. (e) The sequence chromatograms of partial fragments (nucleotide positions 6591–6614) of rV and wV. The difference between the two chromatograms was indicated by the black box. The PflmI recognition site is CCANNNNNTGG with blue box. (f) Representative plaques (arrows) of Vero cells infected with the parental AH2012/12 strain and rescued rAH2012/12 strain, from left to right, at 2 dpi. (g) The growth curves of the parental AH2012/12 strain and the rescued rAH2012/12 strain.
3.5. rAH2012/12 marker mutations
In the genome of rescued virus, three silent mutants A6602G, A18052G, and A26077G were introduced to remove the natural PflmI sites. We selected the 6602 nucleotide mutant to identify the rescued virus. After RT-PCR, we obtained two 2048-nt amplicons (nucleotide positions 5922–7969) from the rescued and parental virus strains, respectively. The fragment amplified from the WT virus was digested by PflmI into two fragments (684 and 1363 bp), whereas the fragment amplified from the rescued virus could not be digested (Fig. 5d). Further, this region was sequenced to verify that the marker mutation was present in the rescued virus (Fig. 5e).
3.6. Phenotype of the rescued rAH2012/12 isolate
We also compared the growth phenotypes between rAH2012/12 and AH2012/12. As shown in Fig. 5f, the plaque morphologies of rAH2012/12 and AH2012/12 were similar. Analysis of the viral replication kinetics also showed that the rescued and parental viruses had similar growth curves, reaching a peak titer of 105 TCID50/mL at 36 h post-infection (Fig. 5g).
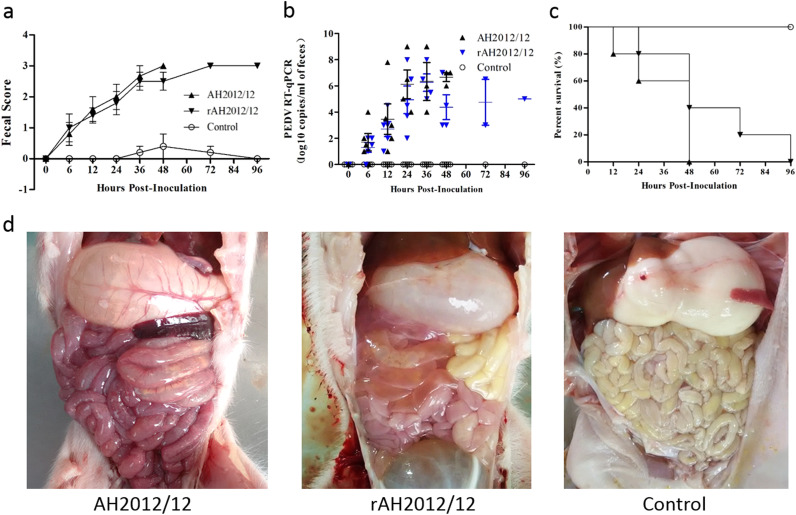
3.7. Rescued rAH2012/12 replication and pathogenesis in newborn piglets
To determine the pathogenicity of AH2012/12 in newborn piglets, and to determine whether the pathology of rAH2012/12 replicated its parental strain AH2012/12 in vivo, suckling piglets were orally inoculated with rAH2012/12 (passage 5) or AH2012/12 (passage 10). While control animals were inoculated with cell culture media.
During the acclimation period, the clinical signs were monitored and piglets in the AH2012/12 and rAH2012/12 challenge groups exhibited lethargy and diarrheic feces by 12 h post-inoculation (hpi), and severe watery diarrhea with vomiting thereafter ( Fig. 6a). Furthermore, RT-PCR of fecal samples revealed that all piglets in the parent and rescued virus inoculated groups were positive for PEDV (3–9 log10 genomic equivalents/mL at 12–48 hpi) (Fig. 6b). The fragments of S1 and a 2048-nt amplicon (nucleotide positions 5922–7969) spanning the PflmI site at position 6602 were amplified and sequenced from the shed PEDV in feces from the parent and rescued virus inoculated groups. These two fragments shared the same sequences as the inoculating viruses, respectively (data not shown). Negative control pigs remained active with normal feces and fecal shedding of PEDV remained undetected throughout the study period. The first piglets died at 12 hpi, and these were from the AH2012/12 challenge group (Fig. 6c). The first piglet from the rAH2012/12 group died at 24 hpi. All piglets in the AH2012/12 and rAH2012/12 challenge groups had died by 3 dpi and 4 dpi, respectively. These results revealed that PEDV strain AH2012/12 could cause significant mortality in newborn piglets (Fig. 6c).
Fig. 6.
The rescued PEDV strain rAH2012/12 mimics wild-type PEDV AH2012/12 infection in newborn piglets. (a) Mean fecal scores after viral inoculation. (b) Mean RT-qPCR titers of the fecal samples. (c) The survival curves of the AH2012/12 and rAH2012/12 infected groups. (d) Macroscopic examinations of the intestine of piglets inoculated with AH2012/12, rAH2012/12, and control medium.
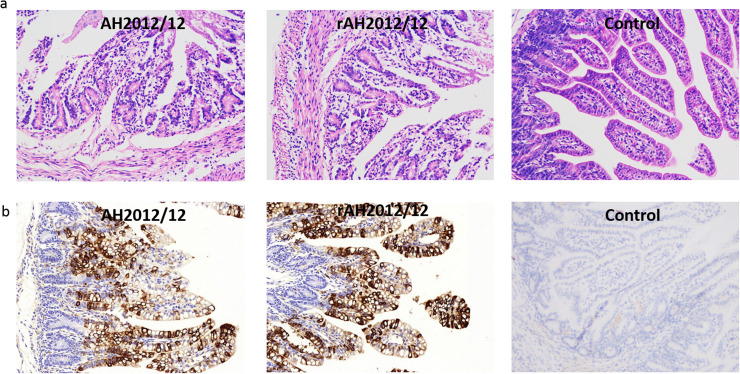
Necropsy examinations were performed when the infected piglets died. All AH2012/12 and rAH2012/12 inoculated piglets displayed typical PED-like lesions. The small intestine was thin-walled and contained soft to watery contents. The stomach was distended and filled with curdled and undigested milk (Fig. 6d). By contrast, the other intestinal organs appeared grossly normal. Histopathological examination showed severe necrosis, vacuolation and villous atrophy of the small intestinal enterocytes in PEDV AH2012/12 and rAH2012/12 inoculated pigs ( Fig. 7a). The mean villous height/crypt depth (VH/CD) ratio of the jejunums of mock-inoculated piglets (5.8±1.0) was higher than those of AH2012/12 (1.5±0.5) and rAH2012/12 (2.1±0.8) inoculated piglets. Immunohistochemistry for PEDV-N confirmed the presence of parent and recombinant virus in the cytoplasm of epithelial cells on atrophied villi in all segments of the small intestines (Fig. 7b).
Fig. 7.
Histology and IHC staining of AH2012/12 and rAH2012/12-infected pig intestines. (a) HE-stained small intestines of piglets inoculated with different PEDV strains. Cell fusion and vacuolation were noted at the villus tips (arrows). (b) For IHC assays, the intestinal tissue sections were stained with a PEDV N monoclonal antibody (1:200 dilution). Positive cells presented in all segments of the small intestines.
4. Discussion
PEDV has now emerged or re-emerged as one of the most devastating viral diseases of swine in the world, leading to significant financial concerns in the global swine industry. Several PEDV vaccines have been developed such as the CV777-attenuated or -inactivated vaccines, live virus vaccine 83P-5, SM98-1 (93 passages), and DR-13 (100 passages), which have been employed in different countries (Kweon et al., 1999, Sato et al., 2011, Song et al., 2007). However, none of these vaccines have significantly reduced the morbidity rate caused by diarrhea, or virus shedding in feces (Lee, 2015). The low to moderate effectiveness of current PEDV vaccines appears to be due to antigenic, genetic, and phylogenetic differences between vaccine and field epidemic strains (Kim et al., 2015, Lee et al., 2010, Lee et al., 2015, Lee and Lee, 2014). Therefore, epidemic PEDV or related strains prevalent in the field should be used for the development of next generation vaccines to control PED. PEDV field isolates need to be isolated and their molecular epidemiology investigated to better control and prevent future PEDV outbreaks. In China, there have only been a few reports of the successful isolation of epidemic PEDV strains (Chen et al., 2015, Pan et al., 2012, Zhang et al., 2015). In our laboratory, we reported and sequenced the Chinese strain AH2012 (KC210145), which is most closely related to emergent strains in the USA. However, we did not successfully culture this strain in cells. In this study, we isolated an epidemic PEDV field strain AH2012/12 from a pig farm reporting severe diarrhea in Anhui province. Phylogenetic analysis revealed that the new isolate belonged to group G2b. We generated an infectious cDNA clone of the virulent Chinese PEDV strain and in vivo experiments in piglets revealed that the rescued virus showed similar pathogenicity to the parental isolate. Our findings will be important in the development of effective live attenuated vaccines and future studies into the pathogenicity of this virus.
The large size of the CoV genome and the instability of some CoV replicase gene sequences during its propagation in bacteria, represent serious obstacles for the development of reverse genetics systems similar to those used for smaller positive sense RNA viruses. However, several alternatives to more conventional plasmid-based approaches have been established in the last thirteen years (Almazan et al., 2014). The same is for PEDV, several reports have described the recovery of infectious particles of PEDV using different technological platforms. Li and colleagues described the reverse genetics of PEDV based on targeted RNA recombination technology that allowed for modification of the 3ʹ-end of the viral genome, which encodes the structural proteins and the ORF3 protein (Li et al., 2013b). The approach based on the use of bacterial artificial chromosomes (BACs) was also overcome the large genome size and toxicity problems. Using this approach, the infectious genome of the TGEV, Middle East respiratory syndrome coronavirus (MERS-CoV) and the severe acute respiratory syndrome coronavirus (SARS-CoV) were generated (Almazan et al., 2006, Almazan et al., 2013, Almazan et al., 2000). Subsequently, Jengarn and colleagues reported that the full-length cDNA clone was constructed by assembling contiguous cDNA fragments encompassing the complete genome of PEDVAVCT12 in a bacterial artificial chromosome (Jengarn et al., 2015). In addition, a research team from the University of North Carolina, Chapel Hill, recently reported a reverse genetics system for PEDV obtained by in vitro transcription of the clone followed by transfection of the full-length RNA (Beall et al., 2016). In fact, this method has been developed to rescue several coronaviruses, including mouse hepatitis virus (Yount et al., 2002), transmissible gastroenteritis virus (Yount et al., 2000), avian infectious bronchitis virus (Casais et al., 2001), SARS-CoV and MERS-CoV (Scobey et al., 2013, Yount et al., 2003). In this study, we also used this method and finally rescued a variant PEDV strain, AH2012/12, isolated from a pig farm reporting severe diarrhea in China.
Reverse genetics technologies have been limited by factors relating to the construction and manipulation of molecular clones and the characterization of recombinant viruses, particularly those with large genomes. As reported in the construction of the other coronavirus infectious clones (St-Jean et al., 2006, Yount et al., 2002), we also encountered many technical difficulties in the construction of the full-length genome of PEDV in this study. Because of the reported phenomenon of bacterial toxicity, we initially used the low-copy number cloning vector pSMART-LC to maintain the six fragments. The results showed that all fragments, with the exception of fragment C, could be accurately maintained in this cloning vector. Fragment C, encompassing nucleotide positions 9,108–15,108, was similar to the previously reported nucleotide sites 12,810–15,657 in PEDV, which were toxic to the bacterial host (Jengarn et al., 2015). Fortunately, when we divided fragment C into two at position 13,014, these two smaller fragments could be maintained in the pSMART-LC vector. This phenomenon revealed that the bacterial toxic domain of PEDV is likely located near to nucleotide position 13,104.
Epidemic PEDV infection is characterized by watery diarrhea, dehydration, variable vomiting, high mortality in neonatal piglets, and high morbidity but low mortality in weaned pigs (Liu et al., 2015). Recently, Sunhee Lee and colleagues isolated a highly enteropathogenic Korean strain KNU-141112, which caused fatalities in suckling piglets at one dpi (Lee et al., 2015). A highly pathogenic strain of PEDV, PC22A, isolated from an outbreak in Ohio, USA, in June 2013, caused 100% mortality in neonatal gnotobiotic pigs (Beall et al., 2016, Oka et al., 2014). In this study, phylogenetic analysis of the complete genome revealed that AH2012/12 belongs to genogroup G2b of PEDV along with the highly pathogenic strains KNU-14112 and PC22A (Fig. 3). In vivo experiments also exhibited that isolate AH2012/12 caused 100% mortality in newborn piglets, and the rescued virus showed similar pathogenicity to the parental strain. Although one piglet in the rAH2012/12 challenge group survived for two days longer than the piglets in the parental group, this was likely due to individual differences among the piglets. In addition, the mean PEDV RNA fecal shedding titers in rectal swab samples in this study were determined to be approximately 2 log10 lower than that of highly virulent strain-inoculated piglets in the previous report by using the described real-time RT-PCR method (Chen et al., 2016). The possible reasons causing this different results might be the differences in modified special primers and the time point of sample collection between these two researches. Besides, the mean fecal shedding titers were reduced by the inconsistent disease progressions. Therefore, the individual difference of piglet was also one reason causing the different results. This will be improved in the future study.
The identification of virulence genes is key to understanding the pathogenesis of microorganisms. Due to the limitations of reverse genetics, previous studies have analyzed the virulence genes of PEDV by comparing sequence differences between the parent and attenuated virus, and found that the ORF3 gene might play a role in the pathogenicity of PEDV. Notably, Sato and colleagues found that the S genes of the two attenuated PEDV strains, Korean DR13 (100th) and Japanese 83P-5 (100th), showed remarkable similarity with comparable nucleotide mutations and amino acid substitutions in their parental viruses. This finding revealed that mutations in the S gene of PEDV were associated with the attenuation of virulence in vivo (Sato et al., 2011). Using reverse genetics technology, future studies can investigate the other potential virulence genes of PEDV, as well as the S gene, to further our understanding of PEDV pathogenesis.
In conclusion, an epidemic field strain of PEDV, AH2012/12, was isolated from a pig farm in China reporting severe diarrhea. Animal experiments revealed that this PEDV strain was highly pathogenic to newborn piglets. Furthermore, we performed reverse genetics of AH2012/12, and the rescued virus showed similar biological characteristics to the parental isolate both in vitro and in vivo. This is the first report of the generation of an infectious cDNA clone of a virulent Chinese PEDV strain. These advances will be key to the study of PEDV pathogenesis and, importantly, in the development of live attenuated vaccines.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0500101), the Special Fund for Independent Innovation of Agricultural Science and Technology in Jiangsu province (CX(15)1056), the National Natural Sciences Foundation of China (31472204) and the Jiangsu province Natural Sciences Foundation (BK20131330).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2016.10.011.
Contributor Information
Kongwang He, Email: kwh2003@263.net.
Bin Li, Email: libinana@126.com.
Appendix A. Supplementary material
Supplementary material
.
References
- Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuniga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., DeDiego M.L., Sola I., Zuniga S., Nieto-Torres J.L., Marquez-Jurado S., Andres G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4 doi: 10.1128/mBio.00650-13. (e00650-00613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Sola I., Zuniga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, A., Yount, B., Lin, C.M., Hou, Y., Wang, Q., Saif, L., Baric, R., 2016. Characterization of a Pathogenic Full-Length cDNA Clone and Transmission Model for Porcine Epidemic Diarrhea Virus Strain PC22A. mBio vol. 7. [DOI] [PMC free article] [PubMed]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhu Y., Wu M., Ku X., Ye S., Li Z., Guo X., He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7:5525–5538. doi: 10.3390/v7102891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu X., Shi D., Shi H., Zhang X., Li C., Chi Y., Feng L. Detection and molecular diversity of spike gene of porcine epidemic diarrhea virus in China. Viruses. 2013;5:2601–2613. doi: 10.3390/v5102601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M.C., Lam H.C., Zhang Y., Wang L., Hesse R.A., Hause B.M., Vlasova A., Wang Q., Zhang J., Nelson M.I., Murtaugh M.P., Marthaler D. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev. Vet. Med. 2016;123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lee J.M., Jung J., Kim I.J., Hyun B.H., Kim H.I., Park C.K., Oem J.K., Kim Y.H., Lee M.H., Lee K.K. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch. Virol. 2015;160:1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon C.H., Kwon B.J., Lee J.G., Kwon G.O., Kang Y.B. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine. 1999;17:2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Park C.K., Kim S.H., Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim Y., Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015;208:215–224. doi: 10.1016/j.virusres.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liu H., He K., Guo R., Ni Y., Du L., Wen L., Zhang X., Yu Z., Zhou J., Mao A., Lv L., Hu Y., Yu Y., Zhu H., Wang X. Complete genome sequence of a recombinant porcine epidemic diarrhea virus strain from eastern china. Genome Announc. 2013;1:e0010513. doi: 10.1128/genomeA.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lin C.M., Annamalai T., Gao X., Lu Z., Esseili M.A., Jung K., El-Tholoth M., Saif L.J., Wang Q. Determination of the infectious titer and virulence of an original US porcine epidemic diarrhea virus PC22A strain. Vet. Res. 2015;46:109. doi: 10.1186/s13567-015-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Arruda P.H., Magstadt D.R., Burrough E.R., Hoang H., Sun D., Bower L.P., Bhandari M., Gauger P.C., Stevenson G.W., Wilberts B.L., Wang C., Zhang J., Yoon K.J. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013 infection in 1-day-old cesarean-derived colostrum-deprived piglets. Vet. Pathol. 2016;53:44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- Madson D.M., Magstadt D.R., Arruda P.H., Hoang H., Sun D., Bower L.P., Bhandari M., Burrough E.R., Gauger P.C., Pillatzki A.E., Stevenson G.W., Wilberts B.L., Brodie J., Harmon K.M., Wang C., Main R.G., Zhang J., Yoon K.J. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014;174:60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Otterson T., Goyal S., Rossow K., Collins J. Complete genome sequence of porcine epidemic diarrhea virus strain USA/Colorado/2013 from the United States. Genome Announc. 2013;1 doi: 10.1128/genomeA.00555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole B. Deadly pig virus slips through US borders. Nature. 2013;499:388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- Oh J., Lee K.W., Choi H.W., Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Li W., Zhou Q., Wang D., Bi Y., Chen F., Song Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., Grego S., Randell S.H., Baric R.S. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Desforges M., Almazan F., Jacomy H., Enjuanes L., Talbot P.J. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 2006;80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig.: Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Pan Y., Wang D., Tian X., Song Y., Cao Y. Identification and pathogenicity of a variant porcine epidemic diarrhea virus field strain with reduced virulence. Virol. J. 2015;12:88. doi: 10.1186/s12985-015-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material