Abstract
This chapter discusses the advances of the envelope glycoprotein (Env) structure as related to the interactions of conserved Env structures with receptor molecules and antibodies with implications for the design of vaccine immunogens and inhibitors. The human immunodeficiency virus (HIV) Env binds to cell surface–associated receptor (CD4) and coreceptor (CCR5 or CXCR4) by one of its two non-covalently associated subunits, gp120. The induced conformational changes activate the other subunit (gp41), which causes the fusion of the viral with the plasma cell membranes resulting in the delivery of the viral genome into the cell and the initiation of the infection cycle. As the only HIV protein exposed to the environment, the Env is also a major immunogen to which neutralizing antibodies are directed and a target that is relatively easy to access by inhibitors. A fundamental problem in the development of effective vaccines and inhibitors against HIV is the rapid generation of alterations at high levels of expression during long chronic infection and the resulting significant heterogeneity of the Env. The preservation of the Env function as an entry mediator and limitations on size and expression impose restrictions on its variability and lead to the existence of conserved structures.
I. Chapter Overview
The HIV envelope glycoprotein (Env) binds to cell surface–associated receptor (CD4) and coreceptor (CCR5 or CXCR4) by one of its two noncovalently associated subunits, gp120. The induced conformational changes activate the other subunit (gp41), which causes fusion of the viral with the plasma cell membranes resulting in delivery of the viral genome into the cell and initiation of the infection cycle. As the only HIV protein exposed to the environment, the Env is also a major immunogen to which neutralizing antibodies are directed, and a target which is relatively easy to access by inhibitors. A fundamental problem in the development of effective vaccines and inhibitors against HIV is the rapid generation of alterations at high levels of expression during long chronic infection and the resulting significant heterogeneity of the Env. The preservation of the Env function as entry mediator and limitations on size and expression impose restrictions on its variability and lead to existence of conserved structures. In this chapter, we discuss advances in our understanding of the Env structure as related to interactions of conserved Env structures with receptor molecules and antibodies with implications for the design of vaccine immunogens and inhibitors.
II. Introduction
Viral membrane–associated glycoproteins have diverse functions in the life cycle of an enveloped virus (Dimitrov 2004, Smith 2004). They attach virions to cells by binding to host cell receptors, mediate membrane fusion and some of the subsequent steps of virus entry, direct progeny virion morphogenesis during budding, and in some cases have receptor‐destroying enzymatic activity for virion release and prevention of superinfection. HIV is no exception. Its envelope glycoprotein (Env) serves at least two functions that are critical for the HIV replication cycle—binding to a receptor (CD4) and a coreceptor (CCR5 or CXCR4) by using one of its two noncovalently associated subunits, gp120, and fusing the viral with the plasma cell membranes, which is mediated by the other subunit gp41. It is also a major antigen and immunogen to which all known neutralizing antibodies are directed. In this chapter, we focus on advances in our knowledge of the Env structure and function as related to its interaction with CD4, coreceptors, and neutralizing antibodies emphasizing conservation of Env structural elements that could be used in the design of vaccine immunogens and inhibitors. A number of excellent reviews have been published, which can provide more details of various aspects of the Env and serve as a source of additional citations (Broder 1996, Burton 1997, Burton 2005, Dimitrov 1997, Douek 2006, Fox 2006, Freedman 2003, Gallo 2003, Hunter 1990, Liu 2004, Markovic 2004, Mc Cann 2005, Mitchison 2005, Pierson 2003a, Rawat 2003, Ray 2006, Reeves 2002, Root 2004, Sodroski 1999, Wyatt 1998, Zolla‐Pazner 2004).
III. Structure of the Env (gp120–gp41)
Like many other viral envelope glycoproteins the HIV Env consists of two subunits, the surface glycoprotein (SU), which is responsible for binding to receptor molecules, and the transmembrane glycoprotein (TM), which mediates fusion of the viral membrane with the plasma cell membrane. Initially synthesized as a nonfusogenic polyprotein precursor, gp160, the Env is cleaved by host cell proteases (furin) into the SU (gp120) and the TM (gp41) subunits, which remain noncovalently associated. We will refer to this complex as gp120‐gp41 but will also use interchangeably the abbreviation Env to designate a functional fusogenic HIV envelope glycoprotein. Like other viral envelope glycoproteins the Env is oligomeric; the currently accepted view is that it is a trimer of heterodimers consisting of gp120 and gp41. It is heavily glycosylated resulting in a relatively high molecular weight of about 160 kDa for a monomer, about half of its mass is due to carbohydrates.
A. Primary Structure and Sequence Variation
A monomeric Env molecule consists of about 840–860 amino acids depending on the isolate in which about 480 residues belong to gp120. The sequence analysis of gp120 from various isolates suggests the existence of five relatively conserved regions (C1–C5) and five regions (V1–V5) with significantly higher sequence variability—up to 60–80% (Figure 1, Figure 2 ); (Myers 1994, Starcich 1986). Four of these variable regions (V1–V4) have disulfide bridges at the two ends. The TM glycoprotein (gp41) is more conserved than the SU protein (gp120) as is commonly the case with other viral envelope glycoproteins likely related to its major role in fusion of the viral with the cell membranes. It includes a fusion domain (FD), also known as fusion peptide, which consists of a hydrophobic stretch of about 20 amino acid residues at the N‐terminus, two heptad repeats HR1 and HR2, transmembrane domain (TM), three stretches of residues between these four major regions, and a cytoplasmic tail. The FD, the heptad repeats, and the TM are highly conserved. The total number of potential glycosylation sites, most of which are functional, varies for gp120 but is close to 20 and 4 for gp41. The extent of conservation of each of these sites is also variable. The gp41 glycosylation sites are more conserved than those on gp120. The primary structural features of the Env with approximate amino acid numbering are summarized in Fig. 1A.
Figure 1.
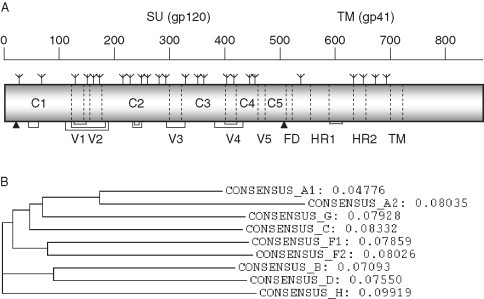
Primary structure of HIV‐1 Env glycoprotein and sequence variations in different regions of the Env lead to several HIV‐1 subtypes. (A) A schematic diagram representing different regions of HIV‐1 Env glycoprotein. Approximate locations of the cleavage sites (arrowheads), glycosylation sites (branched symbols), constant (C1–C5) and variable (V1–V5) regions, fusion domain (FD), heptad repeats (HR1 and HR2), and transmembrane domain (TM) are shown along with the numbering scheme of amino acids. The cross‐linking disulfide bonds connecting various segments are indicated as brackets. (B) The phylogenetic tree constructed by using consensus sequences of HIV‐1 M group subtypes A1, A2, B, C, D, F1, F2, G, and H is shown along with evolutionary distances with the maximum value of 0.1.
Figure 2.
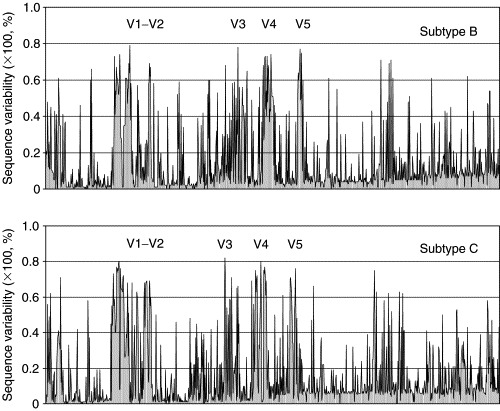
Sequence variability at each amino acid position of the Env of prominent HIV‐1 subtypes B and C. The x‐axes indicate the positions of amino acids as well as allowed gaps from multiple sequence alignments while the y‐axes denote the value of sequence variation at each position. The variable loops apparently have larger sequence variations comparing to other portions of the Env (see the text).
Phylogenetic analysis of envelope sequences revealed the existence of clusters that are approximately equidistant from one another. These were named clades or subtypes. Initially six clades, [A–F], with the prototypic “North‐American/European” strains relabeled subtype B, were found (Myers et al., 1992). Five of these six Env‐based subtypes/clades [A, B, C, D, and F, subtype E″ is now designated as a circulating recombinant form (CRF01_AE)] were also identified from the gag gene (Louwagie et al., 1993). Based on phylogenetic comparisons of partial sequences subtypes G to J were added (Janssens 1994, Leitner 1995). These subtypes together were designated as a group called M which stands for “main,” distinguishing from the groups O (outlier) (Gurtler et al., 1994) and N (non‐M/non‐O) (Simon et al., 1998). Figure 1B shows the phylogenetic relationships among the HIV‐1 M group members. The tree was constructed by using M group consensus sequences which were downloaded from the HIV Sequence Database, August 2004 (http://www.hiv.lanl.gov). To demonstrate the sequence variations of HIV‐1 Env, samples of 100 Env sequences from subtype B and C were obtained from the HIV Sequence Database, aligned, and the amino acid variability at each position was calculated (Korber et al., 1994) (Fig. 2). Note that although the level of variation is very high in the variable regions (up to 60–80%), other regions of the Env are relatively conserved in some cases containing invariant residues. It is tempting to speculate that those regions with close to 100% conservation have important functions and if targeted by antibodies or small molecule drugs may not mutate without significant loss of fitness of the virus.
B. Secondary Structure Elements
The Env sequence was used for prediction of its secondary structure by computer modeling. Perhaps the most popular model was developed by Gallaher 1989, Gallaher 1995 before any Env three‐dimensional (3D) structures were available. The model predicted predominantly helical structures for gp120 but later the crystal structure analysis of the gp120 revealed mostly β‐sheet structures. However, the model correctly predicted essential features of gp41, specifically the two heptad repeats for gp41 that form helical structures. The gp41 model is useful because of lack of available 3D structure of the native gp41. In addition to the prediction of the localization of the heptad repeats, it is also useful for other applications including localization of the antibody epitopes.
C. Tertiary (3D) Structures of gp120 at Atomic Resolution
The determination of the crystal structure of a deglycosylated gp120 core from IIIB complexed with a two‐domain fragment from CD4 and the Fab 17b (Figure 3, Figure 4, Figure 5 ) at a resolution of 2.5 Å in 1998 by Kwong et al. (1998) was a major breakthrough which is still a paradigm for research on the Env structure and function. Later the resolution was improved to 2.2 Å, and the structure of the gp120 core from another (primary) isolate, YU2, was solved (Kwong et al., 2000). The 3D structure of gp120 with any of the variable regions (V1–V5) was not available until recently when the crystal structure of the JR‐FL gp120 core with the V3 was determined in complex with CD4 and the broadly neutralizing antibody Fab X5 at 3.5‐Å resolution (Fig. 5B) (Huang et al., 2005b). The fully glycosylated unliganded gp120 core structure from an SIV isolate was also recently solved at 4 Å despite resolution‐limiting problems (Figure 3, Figure 4). The structural details derived from these four published crystal structures have provided a wealth of information on the interactions with receptors and antibodies as described in more detail below.
Figure 3.
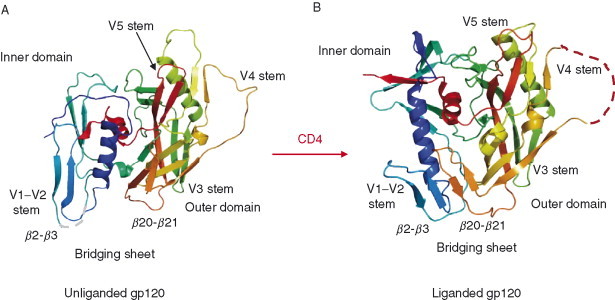
Crystal structures of gp120 core in the unliganded and liganded states. (A) Ribbon diagram of the unliganded SIV gp120 core is shown as in the same orientation of the liganded HIV gp120 structure. The color codes are in rainbow representation from colors blue to red for the N‐ to C‐terminus. The positions of variable loops and bridging sheets are labeled. (B) Ribbon diagram depicting the 3D‐structure of HIV gp120 core complexed with the first two domains (D1, D2) of CD4 receptor and the Fab fragment of human monoclonal neutralizing antibody 17b (CD4 and 17b are not shown here). The outer domains (in green and yellow) of liganded and unliganded gp120 are relatively conserved while a dramatic change in the inner domain (blue and cyan) occurs. The bridging sheet that connects inner and outer domains is not formed in the unliganded gp120.
Figure 4.
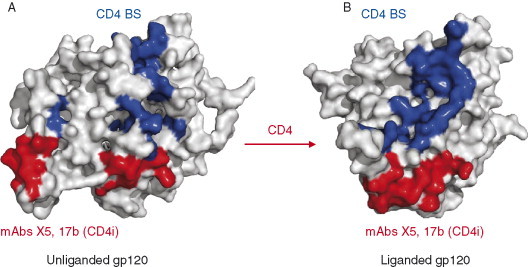
Molecular surface diagrams of unliganded (A) and liganded (B) gp120 cores are rendered as viewed from the perspective of CD4 receptor binding. The residues in direct contact with CD4 are in blue; residues contacting the CD4i antibodies, namely, 17b and X5 are in red. The contact residues were selected by limiting interatomic distance of 3.8 Å between gp120 core to the CD4 and CD4i antibodies.
Figure 5.
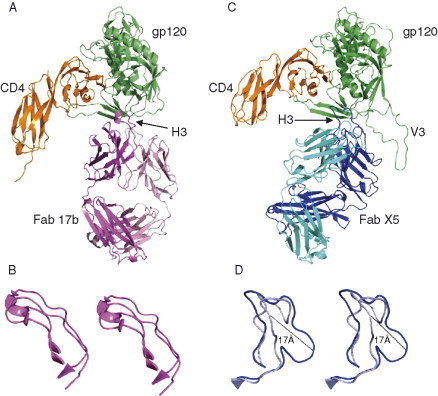
Structures of HIV‐1 gp120 complexes with CD4 receptor and CD4i antibodies, 17b and X5. (A) HIV‐1 gp120 core (green) is bound to the CD4 (orange) and Fab 17b antibody (magenta for heavy and pink for light chains). (B) CDR H3 conformations of antibodies in the free and bound forms are given in stereoviews as crystal structures of 17b and X5 antibodies were available in isolation (PDB codes: 1RZ8 and 1RHH, respectively). (C) HIV‐1 gp120 core with an intact V3 (green) is bound to the CD4 (orange) and Fab X5 antibody (blue for heavy and cyan for light chains). CDR H3 loops are labeled and indicated by arrows. The CDR H3 conformations of 17b antibody (C) are similar in free and bound forms. Notably, the H3 of X5 (D) undergoes a large conformational change with the maximum displacement up to 17 Å (blue in bound form and light blue in free form).
The gp120 complexed with CD4 and antibody has a unique fold comprising two domains, inner and outer as designated with respect to the locations of the N‐ and C‐termini which are bridged by a four‐stranded antiparallel sheet (Fig. 3B). The inner domain contains two helices and a small five‐stranded β‐sandwich. The outer domain consists of a six‐stranded mixed‐directional β‐sheet which clamps a helix, α2, and a seven‐stranded antiparallel β‐barrel. The location of the V1–V2 stem is near to the inner domain. The V4 and V5 appear to be stemming out from different regions of the outer domain surface. The recently solved structure of gp120 with the V3 suggests a structured V3, which protrudes 3 nm from the core toward the target membrane (Fig. 5B) (Huang et al., 2005b). The CD4‐bound gp120 core structure for three different isolates, IIIB, YU2, and JR‐FL, complexed with two different antibodies, 17b and X5, is essentially the same suggesting not only lack of conformational changes induced by antibodies but also that the core structure is preserved for these three isolates. In addition, since the seven disulfide bridges in the core are conserved and buried, one can expect that the major features of the gp120 core as the existence of inner and outer domains joined by a bridging sheet as well as various structural elements including the predominantly β‐type of structural elements would be preserved in all HIV isolates. The sequences comprising the inner domain are relatively more conserved than those for the outer domain. The topological structure of gp120 was found compatible with results from biochemical studies. However, the unique two‐domain arrangement linked by a bridging sheet that allows large receptor‐induced conformational change has not been anticipated.
The unliganded gp120 (free gp120) has structural arrangements that are remarkably different from those of its CD4‐bound form (Fig. 3). The CD4 binding induces large structural changes in the inner domain. Although the overall inner domain structure in the unliganded gp120 is different from that in the CD4‐bound gp120 structure, the elements of the secondary gp120 structures are preserved but significantly shuffled and reorganized. Indeed in contrast to the liganded state, the inner domain in the unliganded state is not a single domain but a mixer of distinct substructures—an α‐helix, a β‐ribbon from one half of the bridging sheet with the V1–V2 stem, and a three‐stranded β‐sheet with two consecutive strands (Fig. 3A). There are four conserved disulfide bonds in the inner domain that could interlock the structural elements and allow for a large motion with respect to each other. In contrast to the inner domain, the outer domain structure does not change significantly after binding of CD4 except for some local variations as shown for segments colored with green and yellow (Fig. 3). A prominent feature of the unliganded structure is that the bridging sheet is absent and each of its two β‐ribbons is displaced up to 20–25 Å. There are two major differences between the unliganded and liganded gp120 structures in relation to CD4 binding. First, the dislocation of the CD4‐binding loop with a conserved GGDPE sequence motif, which contacts the complementarity determining region (CDR)2‐like loop of CD4. Second, the reorientation of the β20‐β21 loop that forms the β‐ribbons of the bridging sheet. In addition, both the receptor and coreceptor binding sites are not formed in the unliganded conformation (Fig. 4).
D. 3D Structures of gp41 Fragments
The 3D structure of gp41 in its native state complexed with gp120 is currently unknown. However, several structures of fragments from gp41 have been solved which likely correspond to a postreceptor‐binding state. The crystal structures of self‐assembled HIV‐1 (Chan 1997, Weissenhorn 1997) and SIV (Malashkevich et al., 1998) heptad repeats revealed a six‐helix coiled‐coil bundle (Fig. 6 ). This coiled‐coil structural feature was previously noted in the hemagglutinin membrane spanning subunit (HA2) (Bullough 1994, Carr 1993) and in the TM subunit of Moloney murine leukemia virus (Mo‐MLV) (Fass et al., 1996). The heptad repeats HR1 and HR2 are about 40–60 amino acid residues long each with 4–3 hydrophobic repeat sequence and are located between the fusion and the transmembrane domains (Fig. 6A). Complexation of peptides based on these heptad repeats leads to the formation of a thermodynamically stable core of gp41. The gp41 core, the N36–C34 complex, is a six‐stranded helical bundle structure consisting of an internal trimeric coiled coil of three N36 helices running parallel to each other, and of external shell of three C34 helices running antiparallel to the N36 helices in a left‐handed manner around the central coiled‐coil trimer (Fig. 6B). The overall size of the complex in a rectangular shape is about 35 Å in width and 55 Å in height. The 46‐residue fragment which connects N36 with C34 is thought to be highly flexible.
Figure 6.
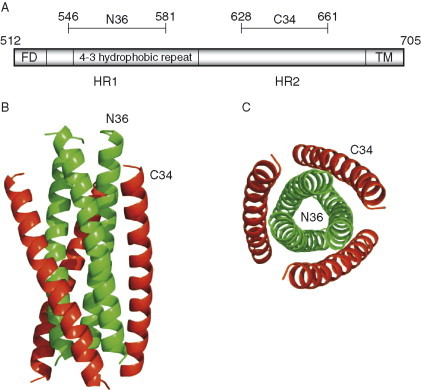
Crystal structure trimeric gp41 fragment. (A) A schematic view of gp41 Env showing the locations of functional regions corresponding to the N36 and C34 peptide fragments. (B) The peptides N36–C34 complex forms a stable α‐helical domain of six‐helix bundle structure. The N36 (green) and C34 (red) helices point to each other in the opposite directions; N36 forms the inner core of the trimeric structure while C34 warps the core. (C) The bottom view of the trimer clearly depicts the arrangement of N36–C34 complex.
The conserved patterns of the amino acid residues in the heptad regions are correlated with the structural and functional properties of the α‐helical core structure of gp41. Most of the N‐peptide amino acid residues make protein–protein interactions in the internal trimer and form grooves on the surface, which interact with the C‐peptide. Thus, N‐peptide residues involved in the interactions are highly conserved among HIV‐1, HIV‐2, and SIV. Similarly, C‐peptide residues interacting with N‐peptide helices are conserved for a broad range of isolates. A key structural feature on the surface of the N36 trimer is a deep and large cavity which is made up of Leu568, Val570, Trp571, Gly572, and Leu576 resulting in a hydrophobic pocket. This pocket accommodates three protruding hydrophobic residues, Ile635, Trp631, and Trp628, from the C34 helix. All N36 residues forming the cavity are identical between HIV‐1 and SIV strains.
The gp41 structure has provided useful information about the membrane fusion mechanism as well as the possibility for its inhibition. Mutations of residues responsible for the gp41 core stabilization affect HIV infectivity and membrane fusion. The positions of some key mutations map to the interaction site between the N36 and C34 helices.
E. Quaternary (Oligomeric) Structure
The oligomeric 3D structure of the Env is critical for our understanding of the mechanisms of entry and neutralization. The structure remains unknown but there are hopes for progress in the near future. Very recently, cryoelectron microscopy (CEM) provided a glimpse of how an oligomeric Env may look like although not at the atomic level of detail. Two different studies depicted somewhat different trimeric Envs and analyzed their distribution on the virion surface (Fig. 7 ) (Zanetti 2006, Zhu 2006, Zhu 2006 described the structural details of an SIV virion at about 3‐nm resolution in which an individual Env has three monomers of gp120‐gp41 in a tripodlike structure. The overall structure of the Env has two components: “head” and “stalk.” The head is mainly composed of gp120 which is supported by the stalk in the form of three separate gp41 legs. The dimension of the trimeric Env derived from this study, 10.5 nm thickness of the head and 1.9 nm vertical length of the legs are comparable to those derived in an earlier study (Zhu et al., 2003). The open tripodlike leg arrangement is also seen in the Env of Mo‐MLV (Forster et al., 2005). The legs are considerably separated and potentially accessible by antibodies. By using gp120 core structures from the liganded and unliganded states, Zhu et al. performed docking on the tomograms such that the gp120 appears on the top with sugar‐coated facing up and the variable loops along the side of the spike masking critical CD4‐binding site (CD4bs). On the transmembrane glycoprotein side, a lower density was observed between the legs of the stem region, where the highly conserved membrane proximal external region (MPER) is located, causing a gap in the surface‐rendered model which suggests possible interactions for this region with the plasma membrane. The recent CEM study by Zanetti et al. (2006) also focused on the tomographic Env structure of SIV. This study also reveals an Env organization with a three‐lobed membrane‐distal gp120 trimer and tightly interacting monomers in the gp41 trimer leading to a mushroom‐shaped structure with a single stalk. The latter arrangement of the gp41 Env as a single leg contradicts the tripod legs seen by Zhu et al. (2006). Possible reasons for the discrepancy in these models could be due to different data collection and image analysis strategies employed (Subramaniam, 2006). It appears that the CEM imaging is still in a developmental stage, and further refinement of methodologies is needed before the results of this promising technology could be accepted with confidence. However, both models provided new levels of structural knowledge to our understanding of the native trimeric Env conformation. Further advancements in CEM imaging or X‐ray crystallography at higher resolution and analyzing Env complexes with different monoclonal antibodies (mAbs) recognizing various segments of Env could provide more accurate and complete information.
Figure 7.
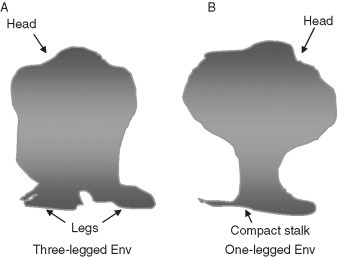
Diagrams illustrate 3D structures of Env spikes as revealed from cryoelectron microscopy. (A) The model obtained at ∼3.2‐nm resolution by Zhu et al. has a head structure comprising trimeric gp120 in three lobes, which is supported by three separate legs in a tripodlike arrangement. The model fitting based on the available gp120 crystal structures suggests carbohydrates on the top; CD4 on the periphery appears closer to the variable loops which may shield the conserved regions of gp120 and gp41. (B) The Env spike model at 2.8‐nm resolution as presented by Zanetti et al. is similar in having a three‐lobed head supported by stalk as seen by Zhu et al. but with a subtly different compact stalk with no obvious separation as three legs at the gp41 stem. Model fitting using the gp120 core structures indicates the exposed receptor binding sites, which are protected by the sugars and variable loops. The bridging sheet is either hidden at the trimer‐g41 interface or protected by the V3 loop.
IV. Env Interactions with CD4 and Coreceptor (CCR5 or CXCR4) Leading to Membrane Fusion
To enter cells, HIV interacts with receptor molecules. Although formally it has not been demonstrated that CD4 and coreceptor are sufficient to mediate membrane fusion after binding to the Env for example, by incorporating them in bilayer membranes and show fusion, it appears that they are the major determinants of the efficiency and kinetics of plasma cell membrane fusion with HIV (Dalgleish 1984, Feng 1996, Klatzmann 1984). Alternative receptors, the most notable being galactosyl ceramide, could mediate fusion of CD4− cells but at very low efficiency, and its biological relevance is not clear (Alfsen 2002, Harouse 1991, Kensinger 2004). Similarly, CCR5 and CXCR4 are the major biologically important coreceptors, although other chemokine receptors can also serve as coreceptors (Coughlan 2000, Puffer 2000, Sharron 2000). A number of other molecules have been found that could enhance the fusion process mostly by enhancing binding but they are not directly involved in the entry process (Broder 1996, Pleskoff 1998). Thus, here we will review advances in our understanding of the Env interactions with CD4 and coreceptor (CCR5 or CXCR4) that are critical for the HIV entry into cells. We will focus mostly on the structural basis of those interactions.
A. CD4 Structure and Biological Function
Human CD4 is a 55–60 kDa type I membrane glycoprotein which consists of 433 amino acids as derived by its cDNA sequence (Littman 1988, Maddon 1985). It contains a 372‐residue extracellular portion linked by a hydrophobic transmembrane domain to a 41‐residue cytoplasmic tail. The extracellular portion can be divided into four immunoglobulin (Ig)‐like domains, designated D1, D2, D3, and D4. Every domain, except D3, contains one disulfide bridge. D1 and D2 are not glycosylated, but D3 and D4 have two N‐linked glycosylation sites. The overall shape of the CD4 extracellular portion is rodlike with a length of about 12.5 nm (Kwong et al., 1990). The transmembrane portion is rich in hydrophobic amino acid residues and forms a helical structure. The short cytoplasmic tail of CD4 associates with p56lck—a tyrosine kinase from the src family. It contains two cysteins, which are essential for the interaction with lck.
The crystal structure of the first two CD4 domains (D1D2) was first solved for human CD4 (Ryu 1990, Wang 1990), and the structure of the membrane proximal domains (D3D4) was later solved for rat CD4 (Lange et al., 1994). Finally, the crystal structure of the whole extracellular portion of CD4 (four‐domain CD4, also known as soluble CD4, sCD4) was solved in 1997 (Wu et al., 1997). Both fragments (D1D2 and D3D4) form rigid, rodlike similar to each other structures. The area buried between the domains allows for a limited flexibility. The first domain, which contains the high‐affinity binding site for gp120, is composed of nine β‐strands following the Ig fold that resemble in many aspects the structure of the variable (V) domains of an Ig. By analogy with the antibody V domains the nine strands are termed A, B, C, C′, C″, D, E, F, G; four of them (ABDE) form an antiparallel β‐sheet, which is packed against another antiparallel β‐sheet formed by CC′C″FG. Also by analogy with the hypervariable CDRs of Ig V domains, the loop between the strands B and C is termed CDR1, that between C′ and C″ termed CDR2, and that between F and G termed CDR3. However, there are two important differences between D1 of CD4 and an Ig V domain: (1) missing the features of an Ig domain, which are involved in the dimerization with another V domain, and (2) the C′/C″ loop (CDR2) protrudes away from the body of the domain; particularly the hydrophobic side chain of F43 is completely exposed to water. That exposure of F43 plays an important role in the interaction with gp120. Domains 1 and 2 have significant overlap, which stabilizes the conformation of the fragment and makes any significant motion at the joint region unlikely. The structure of the fragment from the third and fourth domain of rat CD4 resembles that of the human D1D2 fragment.
The crystal structure of the four‐domain sCD4 molecule suggested that the hinge region between the second and the third domain produces variability in structures suggesting flexibility. It was also found that sCD4 forms dimers and that the dimerization occurs by interactions between the D4 domains. In solution, dimerization occurs at relatively high CD4 concentrations (in the millimolar range), which indicates relatively weak interactions and explains why CD4 dimerization has not been observed in gels. However, at the membrane surface, due to the 2D limitation of CD4 motion and restrictions related to the domain structure of the membrane, the CD4 local concentration could be relatively high leading to formation of dimers. A simple estimation shows that for a typical lymphocyte with a radius of several micrometers, membrane thickness 50 nm and about 104 surface‐associated CD4 molecules, the equivalent bulk CD4 concentration should be in the millimolar range. Earlier observation based on lateral mobility measurements demonstrated that a large portion of membrane‐associated CD4 is dimerized or forms higher order complexes (Pal et al., 1991).
The biological function of CD4 was first studied in rat lymphocytes where it was identified in 1977 by using an mAb—W3/25 (Williams et al., 1977). Its human homologue was identified in human T cells by using the mAb T4 (Reinherz et al., 1979). CD4 is expressed on about 60% of peripheral blood T lymphocytes (Reinherz et al., 1979) and in the cells of the monocyte‐macrophage lineage including microglial cells and dendritic cells, which are antigen‐presenting cells and include Langerhan's cells of the skin and mucous membranes. CD4 plays a central role in the initiation of T cells responses as a coreceptor of the antigen‐dependent and class II major histocompatibility complex (MHC)‐dependent interactions that initiate T‐cell activation through the T‐cell receptor (TCR) (Reinherz and Schlossmann, 1980). According to the coreceptor model both CD4 and TCR bind to the same class II molecule, they physically associate on the cell surface on antigen stimulation, the CD4–TCR complex generates a much stronger signal than TCR alone, and the CD4 molecule can transduce a signal. In addition to its central role in activation of T helper cells, CD4 may have other physiological functions. For example, its interaction with IL‐16 leads to an increase in intracytoplasmic calcium and inositol trisphosphate, and migratory responses.
B. CD4 Binding to gp120
CD4 binds to gp120 with relatively high (nM) affinity, which is highly variable with the isolate tested and does not significantly depend on the temperature suggesting that the binding is entropy determined. The kinetic constant of sCD4 binding to gp120‐gp41 expressing cells depends on temperature suggesting the existence of an energy barrier. The association rate constant at 37 °C was determined to be 1.5 × 105 M−1 s−1 and the respective dissociation rate constant—3.3 × 10−4 s−1 (Dimitrov et al., 1992). The association rate constant decreases with temperature following double hyperbolic dependence with a break at 18 °C. At 4 °C the association constant value reaches 1.1 × 104, which is a 14‐fold decrease in comparison to the value at 37 °C. The equilibrium dissociation constant and the rate constants vary for the different experimental systems used to measure them—binding of sCD4 to gp120‐gp41 expressing cells or to virions, or binding of gp120 to CD4 expressing cells or to sCD4 in solution, thus reflecting changes in the structure of the Env for different virus isolates and the effect of the oligomeric structure. The essential features of the CD4‐gp120 binding process remain consistent to that of binding of two large molecules having binding site areas much smaller than the overall surface area of the molecules—similar to the binding of antibodies to large antigens.
The binding site for gp120 on CD4 was dissected by using mAbs specific for different epitopes of CD4 and by site‐directed mutagenesis of CD4. It was localized on the first domain—amino acids 39–52. The X‐ray crystallography data showed that the binding epitope is a ridgelike structure formed by the C′ and C′′ strands and the loop which connects them, corresponding to the CDR2 of an Ig V domain. At the top of the C′ is a hydrophobic amino acid, F43, which is completely exposed to the water environment and is critical for binding. The exposure of F43 on CD4 suggested that gp120 contains a hydrophobic cleft able to accommodate the protruding F43. The X‐ray crystal structure at 2.5‐Å resolution of an HIV‐1 gp120 core, complexed with a two‐domain fragment of human CD4 and an antigen‐binding fragment of an antibody that blocks chemokine‐receptor binding, revealed a cavity‐laden CD4–gp120 interface, a conserved binding site for the chemokine receptor, evidence for a conformational change on CD4 binding, the nature of a CD4‐induced (CD4i) antibody epitope, and specific mechanisms for immune evasion (Kwong et al., 1998). A more accurate modeling of less‐well‐ordered regions provided conclusive identification of the density in the central cavity at the crux of the gp120–CD4 interaction. The structure of a gp120 core from the primary clinical HIV‐1 isolate, YU2, compared to that of HXBc2 showed that while CD4 binding is rigid, portions of the gp120 core are conformationally flexible; overall differences are minor, with sequence changes concentrated on a surface expected to be exposed on the envelope oligomer (Kwong et al., 2000). Ongoing crystallographic studies of gp120 are revealing how conserved regions involved in CD4 binding, which are the targets of broadly neutralizing antibodies, are concealed from immune recognition (Kwong, 2006).
Binding of CD4 to gp120‐gp41 induces rearrangements in the gp120–gp41 complex resulting in two types of structural changes: (1) dissociation of the CD4–gp120 complex from gp41 (gp120 shedding) and (2) exposure of epitopes on gp120 and gp41 as measured by an increased antibody binding and enhanced cleavage by proteases. While the lack of correlation between sCD4‐induced shedding and membrane fusion argues against gp120 shedding as a fusion intermediate, the possibility remains that shedding represents either an abortive pathway of fusion or a final product of the CD4–gp120–gp41 interaction. Despite the lack of knowledge how shedding is involved in fusion, it is clear that it contributes to the irreversible inactivation of HIV‐1 by sCD4 as well as by neutralizing antibodies. The results of a recent study indicate that the interactions of membrane‐associated oligomeric Env with clusters of membrane‐associated CD4 induce conformational changes that after interactions with coreceptors result in the exposure of helical gp41 structure reactive with antibodies, for example, NC‐1 (Dimitrov et al., 2005). In a parallel reaction, Env‐target complexes dissociate to expose triggered gp120–gp41 on the surface, which further can dissociate to monomers and be inactivated.
C. Interactions of gp120 with Alternative Receptors
Many CD4− cells from neural, epithelial, cervical, and fibroblast origin are infectable by HIV including primary virus isolates. While in some cases the infection still can be mediated by low but undetectable amounts of CD4, in many systems, anti‐CD4 mAbs, for example, Leu3A and OKT4A as well as sCD4 cannot inhibit the infection even at high concentration, clearly demonstrating that the infection is mediated by molecules other than CD4. One of the molecules, which have been implicated in mediating the CD4‐independent infections, particularly in neural, colon epithelial, and possibly sperm cells is the galactosyl ceramide and its derivatives or structural homologues. These molecules are monohexoside glycolipids inserted in the cellular plasma membranes by two aliphatic chains of their ceramide moieties. They contain one galactose residue in β‐glycosidic linkage, which protrudes outside the membrane and is the apparent binding site of gp120 and antibodies. These glycolipids were proposed as alternative HIV receptors based on inhibition of HIV infections by antibodies and binding of gp120 to these galactosyl ceramides as well as the association of greater infectivity with higher expression of those molecules.
Galactosyl ceramides were not detected on lymphoid cells, but are expressed on monocyte‐derived macrophages (MDM). Antibodies to them reduce virion binding, but do not inhibit infection in macrophages. Unlike infection of CD4+ cells, infection of CD4− cells is usually of lower efficiency possibly due to inefficiency of the alternative receptor and the small number of cells expressing it. On the background of this inefficient virus spread, detection of inhibition is difficult. It was demonstrated that the inhibition of HIV‐1 infection of neural cell lines by anti‐galactosyl ceramide antibodies is significant but not complete. However, infection of a colon epithelial cell line (HT29) with such antibodies almost completely prevented infection in contrast to the anti‐CD4 antibody Leu3A, which had no effect. Most of the evidence for the proposed role of galactosyl ceramide as an alternative receptor comes from studies of gp120 (gp160) binding to cells expressing galactosyl ceramide or its derivatives. The binding is specific with relatively high affinity—the equilibrium dissociation constant is in the nanomolar range. While the galactose residue in β‐glycosidic linkage is the likely site of gp120 binding on the glycolipid, the binding of the receptor to gp120 has not been accurately determined, but may require intact 3D structure because gp120 denaturation prevents binding to galactosyl ceramide. A 193‐amino acid fragment from gp120 containing the V3, V4, and V5 regions is probably involved in binding to galactosyl ceramide as shown by generation of infectious chimeric viruses containing that fragment from HIV‐1LAI, which infects galactosyl ceramide expressing cells, in contrast to HIV‐189.6, which does not. The involvement of V3 loop was also shown by anti‐V3 loop antibodies, which blocked the binding of galactosyl ceramides to gp120. Interestingly, the preincubation of gp120 with sCD4 caused an increased binding of gp120 to galactosyl ceramide consistent with the model that CD4 induces conformational changes leading to an increased exposure of epitopes including V3 loop. Whether binding to galactosyl ceramide induces conformational changes in gp120–gp41 needs to be clarified. It has been already shown that galactosyl ceramide mediated entry does not require coreceptor, at least not those that help CD4. Other alternative CD4‐independent infection pathways include Fc‐receptor‐ and CR‐2‐receptor‐mediated virus uptake. Those pathways are not efficient and the receptor nature of the participating molecules is not characterized as extensively as for galactosyl ceramide.
While HIV‐1 infection is generally not so efficient in CD4− cells, some strains of HIV‐2 have the ability to induce rapidly spreading infection and syncytia formation of CD4− cell lines. The highly cytopathic nature of these infections has suggested that these strains are able to utilize an alternative receptor with high efficiency, unlike the case of HIV‐1 infecting galactosyl ceramide expressing cells. It was demonstrated that the receptor for an HIV‐2 strain, termed HIV‐2/vcp, is CXCR4, the coreceptor for the T‐cell line tropic HIV‐1 isolates (Endres et al., 1996). The HIV‐2/vcp strain was derived from the HIV‐2/NIH‐z isolate and was shown to infect a number of CD4− lymphoid cell lines of T‐cell (BC7, HSB, CEMss4‐) and B‐cell (Daudi, Nalm6) origin, as well as the nonlymphoid rhabdomyosarcoma line RD, which cells are not infectable by HIV‐1. The infection with HIV‐2/vcp is rapid with extensive cytophatic and formation of syncytial, which cannot be inhibited by anti‐CD4 antibodies. In this infection, CXCR4 serves as an alternate receptor, which was supported by three lines of evidence: (1) infection of CD4− cells can be inhibited by 12G5, an anti‐CXCR4 specific mAb, (2) cells expressing CXCR4 are able to fuse with HIV‐2/vcp‐infected cells and support viral infection, and (3) CXCR4 was downregulated by the HIV‐2/vcp infection possibly due to direct interaction between the Env and CXCR4 and the Env or other indirect effects. The interaction of the HIV‐2/vcp gp120 with CXCR5 involves residues from the CXCR4 N‐terminus and the second and third extracellular loops (Lin et al., 2003).
The use of an HIV‐1 coreceptor as a primary receptor by isolates of HIV‐2 indicates that whether a molecule will serve as a receptor or coreceptor depends on the virus structure. It is another demonstration of the ability of HIV for rapid accommodation to changing environments. It has been hypothesized that CXCR4 and other chemokine receptors could have been initially used as primary receptors for primate lentiviruses and the adaptation of HIV‐1 to CD4 is a later event (Dimitrov 1997, Dimitrov 1997).
D. Structure and Biological Function of the Chemokine Receptors CXCR4 and CCR5
Available evidence suggests that biologically important coreceptors for HIV are the chemokine receptors CXCR4 and CCR5 (Berger et al., 1999). They consists of an extracellular N‐terminus, an intracellular C‐terminus, seven α‐helical transmembrane domains with several conserved Pro residues, and three intracellular and three extracellular loops composed of hydrophilic amino acids (Dimitrov 1997, Dimitrov 1998). Highly conserved cystein residues form disulfide bonds between the first and the second extracellular loops, and between the N‐terminus and the third extracellular loop. Both CXCR4 and CCR5 are 352‐amino acids long proteins and possess highly acidic N‐termini. CXCR4 contains two potential N‐linked glycosylation sites—one in the N‐terminus, where most G‐protein–coupled receptors also contain such sequence motifs and one in the second extracellular loop. CCR5 possesses only one N‐linked glycosylation site in the third extracellular loop. The C‐termini of both molecules are rich in conserved Ser and Thr residues and represent potential phosphorylation sites by the family of G‐protein–coupled receptor kinases following ligand binding. The highly conserved cysteine residues that are believed to form disulfide bonds may confer a unique barrel shape by bringing the extracellular domains into closer proximity.
E. Env Interactions with CXCR4 and CCR5
CXCR4 can be coimmunoprecipitated with CD4 in the presence of gp120 (Lapham et al., 1996). It can interact with CD4 also in the absence of gp120 (Basmaciogullari 2006, Lapham 1999, Sloane 2005). Gp120 can also interact with CXCR4 in the absence of CD4 but with relatively low affinity—for example, an affinity constant of 86 nM was measured for the interaction between gp120 and CXCR4 expressed on the surface of CD4− neuronal cells (Hesselgesser et al., 1997). Thus, the high‐affinity nanomolar CD4–gp120 interaction significantly increases the affinity of CXCR4 to both gp120 and CD4 on complexation. Similar findings were reported for the binding of gp120 to CCR5‐expressing cells in the presence of competing radiolabeled chemokines—MIP‐1β, MIP‐1α, and RANTES (Trkola 1996a, Wu 1996). It was shown that gp120 binding to CCR5 was 100‐ to 1000‐fold enhanced by soluble or cell surface–associated CD4 measured by inhibition of the chemokine binding to CCR5. Antibodies against CD4i epitopes, V3 and V2 loop epitopes, and a C3‐V4 epitope on gp120, as well as antibodies to the gp120 binding site on CD4 and to lesser extent on the CDR3‐like region of CD4 D1 prevented the enhancement effect. In the absence of CD4 a relatively low‐affinity interaction between gp120 and CCR5 can occur. In the absence of gp120 CCR5 similarly to CXCR4 associates with CD4 (Lapham 1999, Staudinger 2003, Xiao 1999). In some cell lines, association of CD4 with CCR5 was not observed (Basmaciogullari et al., 2006).
The 3D structures of gp120 complexes with CXCR4 or CCR5 are currently unknown and therefore the exact localization of the interaction sites is not known. However, a number of studies provided data that allow to approximately localize the binding sites on gp120 and on CXCR4 and CCR5. After the identification of CXCR4 as the long‐sought fusion cofactor by E. Berger and associates (Feng et al., 1996), it has been hypothesized that CXCR4 forms a trimolecular complex with CD4 and gp120, and was speculated that the second extracellular loop of CXCR4 is likely to make a contact with gp120 because it is the longest one, and that V3 is likely to be involved in binding to coreceptors because it is a major determinant of the HIV‐1 tropism (Dimitrov, 1996). This model proposed a decade ago continues to be essentially correct but much more information has been accumulated that has provided important clues how gp120 interacts with coreceptors and how these interactions could be inhibited. A first indication that the coreceptor N‐terminus is important for the interaction with gp120 was obtained in the same study that first reported the discovery of an HIV‐1 fusion cofactor—a polyclonal rabbit antiserum to the CXCR4 N‐terminus inhibited HIV‐1 Env‐mediated fusion and virus infection (Feng et al., 1996). Subsequent studies confirmed and extended this initial observation to CCR5 and also discovered the critical role of the coreceptor second extracellular loop in the interaction with gp120. By using chimeras between CCR5 and CCR2b, it was shown that the first 20 amino acids at the N‐terminus of CCR5 were critical for coreceptor activity and that the N‐terminal domain of CCR5 could confer coreceptor function when placed into the CCR2b background (Rucker et al., 1996). A parallel study obtained similar results utilizing the N‐terminus of human CCR5 and the murine CCR5 background (Atchison et al., 1996). Viruses that use only CCR5 as a coreceptor also interact with the extracellular loops and could tolerate substitution of the N‐terminal domain with the corresponding N‐terminal domain from divergent chemokine receptors including CCR2b, CCR1, CXCR2, and CXCR4 (Doranz 1997, Rucker 1996). Recently, mAb directed to the second extracellular loop of CCR5 were detected in long‐term nonprogressing HIV‐1 positive individuals (Pastori et al., 2006). The loss of antibodies in these cases correlated with progression of the disease, which is an indication that the second extracellular loop of CCR5 is a possible target for inhibitors with an in vivo efficacy. Changes in individual residues of CCR5 resulted in different effects on Env‐mediated fusion by an R5‐tropic versus dual‐tropic Env, which indicates that HIV‐1 isolates differ in the way they interact with their coreceptors—CCR5 restricted viruses can interact with two binding sites on CCR5, one in the N‐terminal domain and one in the second extracellular loop, while a dual‐tropic Env exhibited a reduced ability to utilize the second extracellular loop and are more sensitive to mutations in the N‐terminal domain (Doranz 1997, Rucker 1996). Similarly to CCR5 chimeras, chimeras based on CXCR4 and CXCR2 were examined for their ability to support Env‐mediated cell fusion. CXCR4 and CXCR2 share ∼35% amino acid identity. In contrast to the observations with CCR5, the N‐terminal domain of CXCR4 did not confer coreceptor function to CXCR2 or CCR5 (Lu 1997, Picard 1997). The CXCR4 N‐terminus could be substituted by the corresponding region from CXCR2 and still retains the coreceptor function for four of the five examined Env proteins, albeit with lower efficiency than the wild‐type CXCR4. Because of this lower efficiency, it was proposed that the N‐terminus may be contributing directly to the binding or indirectly by promoting conformation that favors interactions with particular Envs. It was also found that the role of the N‐terminus depends on the virus isolate, but does not clearly correlate with the virus tropism. As noted above using an HIV‐2 Env as a tool to identify residues of CXCR4 involved in binding to gp120 suggested that both the second and the third extracellular loops of CXCR4 in addition to its N‐terminus contribute to the gp120 binding (Lin et al., 2003).
Studies with CCR5 show that 10 variants out of 16 natural CCR5 mutations, described in various human populations, responding to chemokines, are able to act as coreceptors, are efficiently expressed at the cell surface, and bind [(125)I]‐MIP‐1beta with affinities similar to wtCCR5 (Blanpain et al., 2000). In addition to Delta32 mutations, only C101X is totally unable to mediate entry of HIV‐1. The fact that nonfunctional CCR5 alleles are relatively frequent in various human populations reinforces the hypothesis of a selective pressure favoring these alleles (Blanpain et al., 2000). Polymorphisms of the chemokine receptor CCR5 genes have been implicated in HIV disease progression, resistance, or nonprogressive infection. There are two distinct forms of the CCR5 protein, 62 and 42 kDa, that are present in human lymphocytic cells and monkey peripheral blood mononuclear cells. The ratio of these two forms of CCR5 changes with cell growth. Localization studies indicate that the 62‐kDa CCR5 resides mainly on the cell membrane and the 42‐kDa CCR5 is present solely in the cytoplasm of the cells and therefore cannot function as HIV coreceptor (Suzuki et al., 2002).
The HIV‐1 Env and SDF‐1α share functional sites on the extracellular domains of CXCR4. Recent data, however, show that there are also four mutations of the second extracellular loop, D182A, D187A, F189A, and P191A, that can reduce HIV‐1 entry without impairing either ligand binding or signaling (Tian et al., 2005). Another study shows that CXCR4 can differ both structurally and functionally between cells, with HIV‐1 infection and chemotaxis apparently mediated by different isoforms (Sloane et al., 2005). A comparison of wild‐type (wt) and dual N‐linked glycosylation site, N11A/N176A, mutant CXCR4 expressed in 3T3 and HEK‐293 cells demonstrated variability in glycosylation and oligomerization in almost half of the isoforms. Immunoprecipitation of CXCR4 revealed monomer and dimer nonglycosylated forms of 34 and 68 kDa from the N11A/N176A mutant, compared with glycosylated 40 and 47 kDa and 73 and 80 kDa forms from wt. The functional specificity of these isoforms was also demonstrated by the fact that of the 11 different isoforms only an 83 kDa form was found to bind gp120 from HIV‐1 IIIB.
F. HIV Entry into Cells Mediated by the Env Interactions with CD4 and Coreceptor
The Env binding to CD4 induces major conformational changes that lead to reorganization of the structural elements comprising the coreceptor binding site (Fig. 4) and enhanced binding to coreceptor (CCR5 or CXCR4) by gp120. The coreceptor binding induces additional conformational changes in gp120 that are transmitted to gp41, which undergoes major conformational changes required for fusion of the viral with the cell membrane. Currently, there are no 3D structures available of the complex of gp120 with coreceptors and the nature of the conformational changes induced by coreceptors in gp120 remains largely unknown. However, several 3D structures of complexes of gp41 fragments are available that are thought to play a major role in the gp41 conformational changes that cause the merging of the viral with the plasma cell membrane. The most prominent of these structures is the so‐called six‐helix bundle which is thought to be a postfusion structure, a result of conformational changes of a pre‐hairpin intermediate (Fig. 6) (Chan 1998, Lu 1995, Weissenhorn 1997). It has been suggested that the formation of this six‐helix coiled‐coil drives the membrane fusion (Markosyan 2003, Melikyan 2000), although there are indications that six‐helix bundles could form prior to fusion (Golding et al., 2002). A parallel pathway is possible that involves the generation of gp41 monomers coexisting with trimers during the fusion process (Dimitrov et al., 2005). The structural basis of the HIV entry mechanism is an active area of research and new exciting developments are expected in the near future.
V. Env Interactions with Antibodies
Infection with HIV or immunization with Env‐based immunogens elicits antibodies which can be divided in six major classes in dependence on the location and properties of their epitopes (Choudhry et al., 2006a): (1) antibodies that bind to the region containing the CD4bs on gp120, (2) antibodies binding better to gp120 complexed with CD4 than to gp120 alone (CD4i antibodies), (3) carbohydrate‐binding antibodies, (4) gp120 V2‐ or V3‐binding antibodies, (5) gp41 antibodies targeting the MPER, and (6) antibodies binding to other epitopes on gp41. Most of these antibodies are isolate specific. HIV uses various strategies to escape immune responses, including rapid generation of mutants that outpaces the development of neutralizing antibodies (Garber 2004, Richman 2003, Wei 2003) and hiding conserved structures of its envelope glycoprotein (Env) that are important for replication (Burton 2002, Johnson 2002, Poignard 2001, Wei 2003). These conserved structures are hidden by variable loops, extensive glycosylation, transient exposure, occlusion within the oligomer, and conformational masking; thus elicitation of broadly cross‐reactive neutralizing antibodies (bcnAbs) in vivo is rare and usually occurs after relatively long periods of maturation (Burton 1997, Zolla‐Pazner 2004). Only several Env‐specific human monoclonal antibodies (hmAbs) have been found (Zolla‐Pazner, 2004) to exhibit neutralizing activity to primary isolates from different clades, including the anti‐gp120 antibodies b12 (Burton 1994, Roben 1994), 2G12 (Sanders 2002, Scanlan 2002, Trkola 1996b), m14 (Zhang et al., 2004b), m18 (Bouma et al., 2003), F105 (Cavacini et al., 1998), 447‐52D (Gorny et al., 1992) and Fab X5 (Moulard et al., 2002), and the anti‐gp41 antibodies 2F5 (Muster et al., 1993), 4E10 (Stiegler 2001, Zwick 2001) and Fab Z13 (Zwick et al., 2001). Recently, several novel gp41‐specific hmAbs were identified that exhibit broad neutralizing activity and bind to conformational epitopes that are distinct from those of 2F5 and 4E10 (Zhang 2007, Zhang 2006). These rare cross‐reactive antibodies are of particular importance because their epitopes can be used as templates for design of vaccine immunogens and as target for inhibitors. The antibodies themselves have potential as therapeutics. Here we will focus on the latest advances in our understanding of such antibodies targeting gp120 or gp41 mostly from a structural point of view.
A. Antibody Interactions with gp120
The epitopes of many anti‐gp120 antibodies have been characterized in the past mostly by site‐directed mutagenesis and competitive binding. Here we will focus on two major classes of gp120‐specific antibodies that recognize receptor binding sites: CD4bs antibodies which compete with CD4 and so‐called CD4i (induced) antibodies that compete with coreceptor for binding to gp120. The binding of the CD4i antibodies to gp120 is typically enhanced to various degrees by complexation of gp120 with CD4.
Perhaps the best‐characterized anti‐HIV antibody is b12, which binds to gp120s of many (but not all) primary isolates and competes with CD4. Therefore, the b12 epitope significantly overlaps the CD4bs. The structure of IgG1 b12 was determined (Saphire et al., 2001) and biochemical studies were carried out to explore the fine mapping of the interaction of many mAbs including b12 with the CD4bs of gp120 (Pantophlet et al., 2003). Further mutagenesis experiments of b12 and the analysis of its structure identified several residues from the heavy chain CDR3 (H3) and CDR2 (H2) that play a role in the binding to gp120 (Zwick et al., 2003). The unique binding ability of b12 to the gp120 core in a partially stabilized CD4‐bound conformation has been recently confirmed by the crystal structure of gp120 core in complex with b12 (Kwong 2006, Zhou 2007). In addition to the b12 structure, the crystal structures of three other CD4bs antibodies in isolation, m18 (Prabakaran et al., 2006b), F105 (Wilkinson et al., 2005) and m14 (Dimitrov and Ji, 2006), have been recently determined. The major structural feature of these antibodies is the existence of long protruding H3s with hydrophobic residues at the tips. The structures are similar at the bases but vary along the torso and the tip regions related to their differences in specificities and neutralizing activities (Fig. 8 ). It was thought that the long protruding H3s of the CD4bs antibodies are required to reach cavities on CD4bs on gp120. However, the recently determined structure of a stabilized (in CD4‐bound state) gp120 core in complex with Fab b12 suggests that actually the b12 H3 does not contact a cavity (Kwong, 2006), and indeed may not contribute significantly to the contact area directly on the CD4bs on gp120 and to the energy of interactions. It remains to be seen whether this is also true for the other CD4bs antibodies or b12 is unique also in this aspect of its interaction with gp120. The epitopes of these antibodies are likely to share some of the gp120 structures because they overlap with the CD4bs. However, their exact localization is currently unknown except for the b12 epitope that was recently determined by solving the crystal structure of its complex with gp120 stabilized in a conformation corresponding to the CD4‐bound gp120 conformation.
Figure 8.
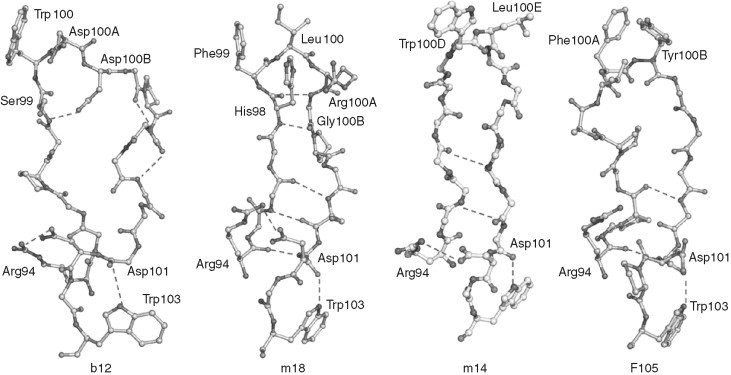
Conformations of CDR H3s from b12, m18, m14, and F105 antibodies. Residues Arg94 and Trp103 from the framework regions play critical role in maintaining the H3 conformations by involving specific salt bridges at the bases. The differences in H3s are markedly noticed along the torso and tip regions.
The coreceptor binding site is highly conserved and a target for broadly neutralizing antibodies. The exact localization of the coreceptor binding site on gp120 is not known because of lack of crystal structure of the complex of gp120 with a coreceptor but extensive mutagenesis studies allowed its location around the bridging sheet (Fig. 4). Prior to CD4 binding the elements contributing to the binding site are dispersed over gp120 surface (Fig. 4A) and masked by the V1–V2 variable loops, therefore, are not easily accessible by neutralizing antibodies. The CD4i conformational changes in gp120 lead to the formation of the coreceptor binding site and to enhanced binding of CD4i antibodies which typically compete with the coreceptor for binding to gp120. A number of CD4i antibodies including 17b, X5, 48d, 47e, E51, and 412d recognize highly conserved CD4i epitopes which overlap to various extents with the coreceptor binding site. The epitopes of 17b and X5 are now known after the determination of the gp120 structure complexed with Fab 17b or Fab X5 (Fig. 5).
The epitope of 17b overlaps significantly with the coreceptor binding site. The long H3 dominates the 17b binding to gp120; H2 and residues from the light chain also contribute (Fig. 5A). The antibody–antigen interface for the gp120–17b interactions buries only 455 Å2 on gp120 and 445 Å2 on 17b. The epitope spans across the four‐stranded bridging sheet (Fig. 5A) and has hydrophobic core flanked by basic residues. Although the 17b paratope is highly acidic, it does not make significant salt bridges with the basic residues of gp120. In the 17b complex structure, a large gap is seen between the V3 base and tips of the light chain. The H3 of 17b appears to be rigid as can be seen only the minor changes between the free (Huang et al., 2004) and bound (Kwong et al., 1998) H3 structures of 17b (Fig. 5B). Importantly, the 17b epitope is well conserved among several HIV‐1 isolates. Of the 18 gp120 contact residues, 12 residues are conserved among all HIV‐1 isolates (Kwong et al., 1998).
The potent broadly neutralizing CD4i Fab X5 was selected from an immunge phage display antibody library and binds with high‐affinity gp120s and gp140s from primary isolates from different clades even in the absence of CD4; however, its binding is significantly (10‐ to 100‐fold) increased in the presence of CD4 (Moulard et al., 2002). Similar to 17b X5 contacts several residues from the bridging sheet but also residues from other regions, which are highly conserved (Fig. 5C, Table I ). Notably, the highly conserved Ile423 residue from β20, which was previously identified as a hotspot (Darbha et al., 2004), shows a loss of 110 Å2 in solvent‐accessible area on contact with X5. In contrast to 17b, the H3 of X5 undergoes large conformational change on binding to gp120 with the maximum of 17 Å displacement for Cα position at Gly100H (Fig. 5D). This is one of the largest induced fits ever observed for an antibody utilizing the flexibility of its H3 loop. The H3 buries 440 Å2 of solvent‐accessible area when X5 binds to gp120; the corresponding loss for the 17b H3 is only 270 Å2. The long highly flexible H3 of X5 may tolerate less‐conserved contact residues, for example, Lys432, but at the same time make a tight binding with functional hotspot residues, for example Ile423 as facilitated by the induced fit. This perhaps might contribute to the broad and potent neutralizing ability of the X5. Table I shows a list of 15 different isolates from three major clades (A–C) that were potently neutralized by X5 antibody along with aligned gp120 epitope residues. The gp120 residues that bind to X5 are highly conserved and exposed as marked by asterisks and buried surface areas at the bottom of each epitope residue in Table I. The very long H3 (22 residues) contains four glycines, several charged (mainly acidic from 6 Asp residues) and hydrophobic residues that could reach the parts of CD4i epitopes which are hidden or sterically restricted to other CD4i nonneutralizing antibodies. The acidic surface of the H3 of X5 may mimic the acidic N‐terminal portion of CCR5 that is necessary for the gp120 binding. 17b exhibits similar acidic properties due to three Asp and three Glu residues. The gp120 X5 epitope residues at positions Arg327, Lys421, and Lys432 are basic, which are not only compensated by the acidic surface of X5 but also form strong salt bridges. Arg327 and Lys421 are conserved and make direct salt bridges with Asp100G and Asp100D residues of H3, respectively, in the donor–acceptor distances range between 2.6 and 2.9 Å. The less‐conserved Lys432 side chain contacts the carbonyl group of the bulky Trp100 which is the perfect candidate for making polar, charged, or stacking interactions with the Lys/Gln/Arg residues at position 432. Though 17b is also acidic no salt bridges are made between gp120 residues and 17b. In addition, the significant role of glycine residues in the H3 of X5 was explored by molecular dynamic simulations. The glycine residues were found to contribute to the H3's flexibility. Taken together, the H3 of X5 appears to be the unique in the mechanism and level of binding activity among known CD4i antibodies. Figure 9A clearly shows how the long H3 of X5 can reach its epitope. An alternative antibody binding mechanism to an exposed receptor binding site of the SARS coronavirus was recently demonstrated (Prabakaran et al., 2006a). The antigen combining site of the anti‐SARS Fab m396 forms a canyon to interact with the exposed parts of the receptor binding site (Fig. 9B). It appears that b12 binds to its binding site on gp120 by a mechanism similar to that of Fab m396 and not of Fab X5.
Table I.
Comparison of gp120 Epitope Residues from 15 Different Isolates for which scFv m9 Derived from the Fab X5 Antibody Exhibits Potent Neutralization
| 15 isolates | X5 contacting gp120 residues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119 | 120 | 122 | 319 | 322 | 323 | 327 | 421 | 422 | 423 | 432 | 434 | 436 | 437 | |
| 2B4C(gp120 in X5 complex) | C | V | L | T | E | I | R | K | Q | I | K | M | A | P |
| QH0692.42 (B) | C | V | L | A | D | I | R | K | Q | I | K | M | A | P |
| SF162.LS(B) | C | V | L | A | D | I | R | K | Q | I | K | M | A | P |
| SC422661.8(B) | C | V | L | – | E | I | R | K | Q | I | K | M | A | P |
| AC10.0.29(B) | C | V | L | T | D | I | R | K | Q | F | K | M | A | P |
| PVO.4(B) | C | V | L | A | D | I | R | K | Q | I | K | M | A | P |
| Q168.a2(A) | C | V | L | A | – | I | R | K | Q | I | Q | I | A | P |
| Q461.e2(A) | C | V | L | A | D | I | R | K | Q | I | Q | M | A | P |
| Q769.d22(A) | C | V | L | A | D | I | R | K | Q | I | Q | I | A | P |
| Q259.d2.17(A) | C | V | L | A | D | I | R | K | Q | I | Q | I | A | P |
| Q23.17(A) | C | V | L | A | D | I | R | K | Q | I | Q | M | A | P |
| Du151.2(C) | C | V | L | A | E | I | R | K | Q | I | R | M | A | P |
| Du422.1(C) | C | V | L | A | E | I | R | K | Q | I | R | M | A | P |
| Du123.6(C) | C | V | L | A | D | I | R | K | Q | I | R | M | A | P |
| Du156.12(C) | C | V | L | A | D | I | R | K | Q | I | R | M | A | P |
| Du172.17(C) | C | V | L | A | D | I | R | K | Q | I | Q | M | A | P |
| * | * | * | : | : | * | * | * | * | : | : | : | * | * | |
| Buried surface area (Å) | 34.8 | 33 | 40 | 37 | 48 | 72 | 46.6 | 29.6 | 38.3 | 110 | 53.8 | 83.9 | 10 | 64 |
Residues forming the epitope are highly conserved as shown by asterisks. The mutation sites with similar amino acids are shown by colons.
Figure 9.
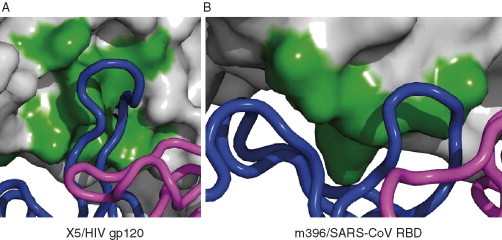
Two different antigen‐binding sites and binding modes CDRs. (A) In gp120–Fab X5 antibody interaction, the long CDR H3 protrudes into the CD4i binding site. (B) Conversely, in the SARS Env–Fab m396 antibody interaction, the antibody CDRs form like a canyon around the protruding binding site.
The neutralizing activity of CD4i antibodies could be significantly reduced because of the steric restriction of access to their epitopes. The conserved discontinuous segments of gp120 overlapping with the coreceptor‐binding site are recognized by CD4i mAbs, which efficiently bind to gp120 on CD4 binding. But, once the CD4 docks on to the receptor site, the space needed for the antibody binding to its epitope is significantly reduced. It was found that the size restriction effect leads to an inverse correlation between the antibody neutralizing activity and its size (Labrijn et al., 2003). As shown in Fig. 10 , the available space between the CD4i epitope and the target cell membrane after CD4 attachment is estimated to be about 85 Å in the highest dimension (Labrijn et al., 2003). While comparing the dimensions of different formats of CD4i antibodies, IgG, Fab, and scFv, as shown in Fig. 10 the antibody fragments in either Fab or scFv are more effective than the whole IgG antibody molecule for getting into the restricted binding site needed for neutralization. However, it should be noted that other factors including avidity effects due to bivalency could contribute to binding. For example, in some cases IgG1 X5 is more potent neutralizer of some isolates than scFv X5 (Labrijn et al., 2003) and in vivo could have much greater neutralizing activity due to the effector functions of its Fc.
Figure 10.
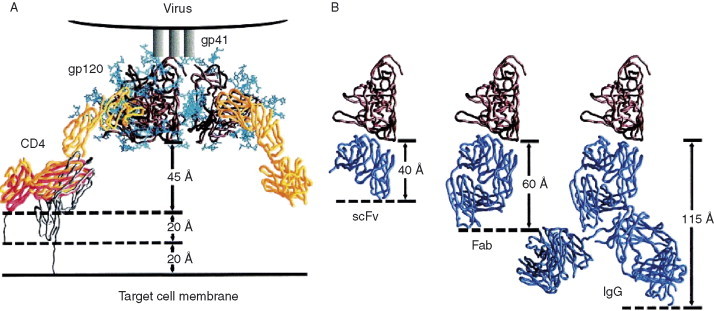
Steric restriction of access to CD4i epitopes on CD4 binding. (A) The sketch with molecules shown describes the attachment of HIV‐1 from viral membrane to the cell surface CD4 receptor. The binding of CD4 induces conformational changes resulting into the exposure of coreceptor binding site, which is sterically restricted for the CD4i antibodies. Taken into considerations of the dimensions derived from structures of gp120, CD4, and possible flexibility of CD4 molecule, a total distance of about 85 Å between the gp120 and target cell membrane is measured. (B) Dimensions of antibodies in different formats, Fv, Fab, and IgG molecules, are also shown. This clearly shows that CD4i antibodies of scFvs and Fabs have better access to the restricted binding site for competing with the coreceptor than IgGs have.
B. Antibody Interactions with gp41
Two most prominent gp41 antibodies are 2F5 and 4E10, which have been isolated almost two decades ago by H. Katinger and his associates by EBV immortalization of B lymphocytes from an HIV‐1‐infected individual. On average 2F5 appears to be more potent than 4E10 but 4E10 exhibits broader neutralizing activity when tested in cell line/pseudovirus assays (Binley et al., 2004). 2F5 and 4E10 recognize almost the same contiguous but adjacent segments ELDKWA and NWF[D/N]IT, respectively, in the Trp‐rich environment of the MPER of gp41 (Fig. 11A ). A 36‐mer gp41 peptide, DP178 (T20) (aa 638–673) contains the ELDKWA region near to its C‐terminal region. This peptide plays an essential role in the fusogenic structure formation and is a potent inhibitor of HIV infection in patients, currently the only entry inhibitor in clinical use. The MPER, which includes the epitopes of 2F5 and 4E10, is highly conserved and mutations of the hydrophobic residues Trp666, Trp670, and Trp672 in this region largely affect the viral entry. However, attempts to use the MPER for elicitation of 2F5‐ or 4E10‐like antibodies have met limited success. To understand better the interactions of these antibodies with their epitopes, which could provide some clues for development of effective vaccine immunogens, the crystal structures of both 2F5 (Ofek et al., 2004) and 4E10 (Cardoso et al., 2005) complexes with peptides from the MPER have been determined. Below these structures are discussed in detail.
Figure 11.
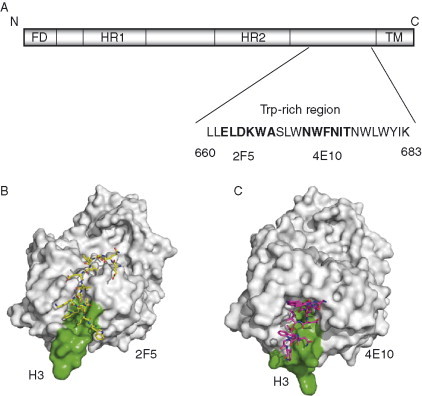
Antibody interactions at the membrane‐proximal region of gp41. (A) Schematic diagram of gp41 shows the different important regions, FD, fusion domain, HR1, HR2‐heptad repeats, and TM, transmembrane domain. The location of membrane‐proximal region containing the core 2F5 and 4E10 epitopes on the Trp‐rich region of gp41 is indicated along with amino acids sequence. Sequence numbering corresponds to HXB2 scheme. Crystal structures of Fab 2F5 (B) and 4E10 (C) in complex with peptides from the MPER. The H3s of the antibodies are shown in green.
The crystal structure of 2F5 in complex with a 17‐mer peptide is shown in Fig. 11B where 2F5 is in surface representation (Ofek et al., 2004). The peptide (residues 654–670) lies at the CDR interface between the heavy and light chains. It is in a relatively extended conformation and spans around 25 Å measured from Glu659 to Trp670 (the leading residues up to Glu659 are disordered in the structure). Two of the three turns, Asp664‐Ala667 and Trp666‐Leu669, in the 2F5‐bound gp41 peptide that belong to type I β‐turn are overlapping. Interestingly, three intrapeptide hydrogen bonds constrain the conformations of only six residues from 664 to 669. The total surfaces of 635 Å2 on 2F5 and 563 Å2 on the gp41 peptide buried in the antibody–gp41 peptide interactions are typical for an antibody–antigen interaction. Most of the residues between Gln657 and Trp670, except Leu660 and Ser668, directly bind to the antibody. Strikingly the contact region is not only restricted to the CDRs of 2F5, but also includes nonpolymorphic region such as the N‐terminus of the light chain. The 2F5 binding site on the peptide is only on one exclusive face which accounts for 41% of the total peptide area available for binding. This indicates that the unbound part of gp41 may interact with other portions of the Env. An analysis of the gp41 peptide surface reveals two major regions: one region which is bound to 2F5 is charged while the other region which is occluded from 2F5 is hydrophobic. The latter property of the surface further suggests for possible protein–protein interactions that occlude from the 2F5 binding. The failures to elicit 2F5‐like antibodies by peptides may be related to the lacking of appropriate occlusion. Another hint for the mechanism of 2F5 binding is inferred form the binding mode of the H3 itself. The length of the 2F5 H3 is 22 amino acids which is the same as the length of the H3 of the CD4i antibody X5. Unlike Fab X5, the 2F5 does not make any contact through the H3 tip but only at the base (Fig. 11B, H3 is shown in green). The H3 tip has several hydrophobic residues that present a protruding flat surface. This surface aligns with the hydrophobic indole side chain from Trp670, the terminal residue of the gp41 peptide. The arrangement involving the 2F5 H3 and the gp41 peptide terminal residue in a hydrophobic plane indicates a possibility that the apex of H3 could interact directly with the viral membrane or to accommodate 2F5 to recognize the epitope closer to membrane proximal region. In agreement with other biochemical and NMR studies, it appears that the 2F5 epitope is relatively flexible, probably assuming different conformations depending on the state of gp41. Interestingly, there is no evidence for any access restriction due to size for 2F5.
The interaction of 4E10 with a 13‐residue peptide containing the sequence NWFDIT is topologically similar to that of 2F5 with its epitope but differs in details (Cardoso et al., 2005 (Fig. 11C). The 4E10‐bound 13‐residue peptide has a helical conformation, in contrast to the 2F5‐bound peptide, and is similar to the 19‐residue peptide structure from the Trp‐rich MPER determined by NMR. The key residues Trp672, Phe673, Ile675, and Thr676 appear on the one side of the helix rendering a hydrophobic surface, which interacts with the 4E10 antibody. The residues Trp672 and Phe673 use their side chains to plunge into a hydrophobic pocket created by the CDRs at the antibody‐combining site of 4E10. The total surfaces of 580 and 529 Å2 are buried on 4E10 and the peptide, respectively, on the binding. The 4E10 H3 does not make any contacts through its tip similarly to 2F5 (Fig. 11B and C). As is in the case of 2F5, this indicates a possibility that the H3 tip contacts the viral membrane or other portions of the ectodomain of the intact virus. In agreement with this possibility is biochemical analysis using Env on proteoliposomes demonstrating enhanced binding of 2F5 and 4E10 in presence of lipid membrane (Ofek et al., 2004). An interesting feature of the 4E10 are the five glycines in the 18‐residue long H3, which could certainly contribute to flexibility that may be required for epitope recognition, particularly, two tryptophan residues at the tip, at positions 100 and 100B, to reach the membrane.
These results suggest that conserved and steric constrains‐free regions are available as potential epitopes on gp41, for example the epitopes of 2F5 and 4E10. The two antibodies 2F5 and 4E10 share some of the structural features and interaction patterns with the core gp41 epitopes, and also specific features related to their distinct epitopes. How useful will be the information for the MPER structures that are part of their epitopes for the design of effective vaccine immunogens remains to be seen.
Recently, six novel gp41‐specific hmAbs were identified that exhibit broad neutralizing activity and bind to conformational epitopes that are distinct from those of 2F5 and 4E10 (Zhang 2007, Zhang 2006). They do not compete significantly with 2F5 and 4E10 indicating that the localization of their epitopes is likely outside the MPER. The conserved structures containing these epitopes are being characterized.
C. Mimicry of Receptors by Miniproteins and Antibodies
The conserved CD4bs on gp120 and structurally contiguous segments including the β‐hairpin rigid motif of CD4 prompted for the rational design of CD4 mimics that could block the HIV entry (Huang 2005a, Martin 2003a, Vita 1999b, Zhang 1999). The CD4–gp120 binding interactions mainly involve contiguous segments rendered by the CD4 residues 31–35, 40–48, and 58–64 in which about 40% contribution is from the CDR2‐like β‐hairpin region containing the Phe43 hotspot. A 31‐amino acid long CD4 mimic specific for gp120 was initially designed by grafting the major contributor of the CD4‐binding component, the CDR2‐like loop of CD4 with a major hotspot Phe43, on a small structural scaffold stabilized by a disulfide bond from scorpion toxin charybdotoxin (Drakopoulou et al., 1998). Later, a mini‐CD4 protein called CD4M9 with 28 amino acids using the scyllatoxin scaffold was designed, and its three‐dimension structure was solved by NMR (Vita et al., 1999b). Based on the structural information derived from the CD4–gp120–17b complex, CD4M9, CD4M32, and CD4M33 miniproteins were designed, and their applications as possible therapeutics were tested by determining several thermodynamic and neutralization parameters. The NMR structure of CD4M9 showed a well‐defined β‐hairpin with a phenylalanine residue at the position 23, which is equivalent to CDR2 region of CD4, appeared to retain some of the conserved gp120–CD4 interactions in the miniprotein–gp120 docked complex. Finally, the crystal structures of CD4M33 and its analogue F23 in complex with gp120 were determined and the extent of molecular mimicry and neutralization breadth were analyzed (Huang et al., 2005a). In spite of the highly flexible envelope, the conformation of gp120 in these mimic complexes are very similar to that induced by CD4 (Fig. 12 ). Interestingly, the β‐hairpin CD4M33 engages in hydrogen bonding to the strand β15 of gp120 in a similar way as CD4 does. This demonstrates the successful attempt of grafting CD4–gp120 binding interface on to a smaller scaffold. Thermodynamic characterization of gp120 binding to these mimics showed that only half of the associated entropic changes occur compared to CD4 binding. Nonetheless, these mimics induce the same conformational change in gp120 as CD4 that are required for enhanced binding of 17b to gp120. The difference between CD4M33 and F23 mimics is only that the phenyl ring in CD4M33 is replaced with a biphenyl side chain of residue 23. This substitution significantly enhances the structural mimicry of CD4 at this specific position (Huang et al., 2005a). The successful structural mimicry by these miniproteins will prompt researchers to further attempt to design native CD4‐like mimics with greater antiviral activity against HIV.
Figure 12.
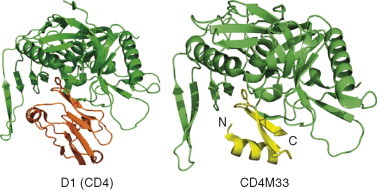
Mimicry of receptor CD4 by miniprotein CD4M33. The binding of gp120 (green) to the CD4 (first domain, D1 is only shown) on left and the miniprotein CD4M33 on right are depicted in ribbon diagrams.
In the giant struggle between the HIV and the immune system, antibodies with unique properties have evolved some of which mimic CD4 and coreceptors but do not induce the same conformational changes as receptors because that could lead to enhancement of infection. In addition, CD4 binds to gp120 through its first domain, which is similar to the V domain of an antibody. For example, the comparison between D1 domain of CD4 and VH domain of b12 is shown in Fig. 13A and B . The molecular views were generated by translating the superposition of the two molecules based on the disulfide bridge locations. Tyr53 residue positioned at H2 of b12 as labeled in Fig. 13B was found critical—a Y53G point mutation greatly diminished the binding of b12 to gp120 (Zwick et al., 2003). Based on the footprint data, two fingers, H3 (Trp100) and H2 (Tyr53), were speculated to occupy the hydrophobic pocket on gp120 surface. Since the H2 of b12 is the equivalent of the C′C′′ or CDR2‐like region of CD4, the b12 could be used as a receptor mimic by further protein engineering of its H2.
Figure 13.
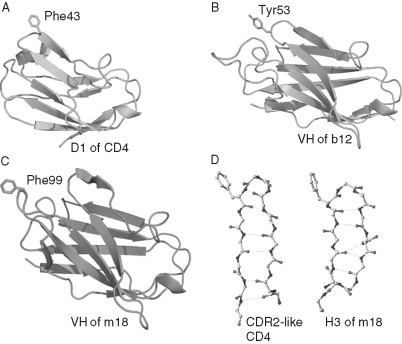
Comparisons of CD4 D1 domain with VH domains of CD4bs antibodies b12 and m18. (A) D1 domain CD4 (green) with Phe43 in sticks. (B) VH domain of b12 antibody (cyan) with Tyr53 at the CDR H2 in sticks. (C) VH domain of m18 antibody (blue) with Phe99 at the CDR H2 in sticks. (D) Backbone skeletal views of the CDR2‐like region of CD4 and the H3 of m18 indicate a common β‐hairpin structure with a phenylalanine residue at the tip.
The Fab m18 is another CD4bs antibody with broad neutralizing activity which was recently identified (Zhang et al., 2003) and its crystal structure solved (Prabakaran et al., 2006b). Its VH domain that is comparable to D1 of CD4 is shown in Fig. 13C. The most remarkable feature of this antibody is the H3 structure which highly resembles the CDR2‐like part of CD4 (Fig. 13D). The m18 H3 adopts not only a β‐hairpin but forms a rigid structure with cross‐linking hydrogen bonds throughout the torso region of H3, and importantly has a Phe residue at the position 99 analogous to the hotspot Phe43 of CDR2‐like loop in CD4. The unexpected structural similarities including hotspot Phe residues and robust β‐sheet features observed (Fig. 13D) suggest for possible protein grafting of H3 to mimic the CDR2‐like C′C′′ of CD4, which might provide a useful strategy for developing antibody‐based CD4 mimetic to inhibit HIV entry.
Not only CD4bs antibodies mimic CD4 but also CD4i antibodies, which mostly bind to the coreceptor binding site, mimic certain features of the HIV coreceptors. Unlike the case of CD4 and its mimics, there is no 3D structure available of a coreceptor. However, the crystal structures of several CD4i Fabs were solved and they revealed mechanisms and atomic‐level details for three interesting features: posttranslational mimicry of coreceptor by tyrosine sulfation of antibody, an alternative molecular mechanism controlling such sulfation, and highly selective V(H)‐gene usage (Choe 2003, Huang 2004). This is another demonstration of the adaptive capabilities of the immune system when confronted by extraordinary viral defenses.
VI. The Env as Vaccine Immunogen and Target for Inhibitors
The development of an effective vaccine against HIV is an international public health priority and the role HIV envelope plays in infection makes it a primary target for such efforts. A multitude of approaches have been attempted that are eloquently summarized in numerous recent reviews (D'Souza 2004, Duerr 2006, Koff 2006, Letvin 2005, McMichael 2006, Singh 2005, Slobod 2005, Spearman 2006, Wang 2006). From this work, it is clearly possible to create vaccines that induce cellular responses that will protect against disease progression by suppressing viral loads once infection occurs. However, none have been able to achieve the penultimate goal of preventing infection entirely. This goal will require an Env‐based vaccine that induces a protective antibody response. Here, we will summarize the prevailing approaches how to use the Env for eliciting broadly neutralizing antibodies and their epitopes as targets for inhibitors with special emphasis on the common challenges.
A. The Relationship Between Viral Neutralization and Protection
A guiding principle of current HIV vaccine efforts is that antibodies that neutralize HIV in vitro can protect animals from HIV infection in vivo. This principle is based on three observations:
-
i
In an expanding number of passive challenge studies, macaques treated with mAbs or pooled, high‐titered antisera that neutralized in vitro were protected from cell‐free SHIV virus challenge (Baba 2000, Emini 1992, Ferrantelli 2003, Ferrantelli 2004, Hofmann‐Lehmann 2001a, Hofmann‐Lehmann 2001b, Hofmann‐Lehmann 2002, Mascola 1999, Mascola 2000, Mascola 2003a, Nishimura 2002, Parren 2001, Putkonen 1991, Ruprecht 2001, Shibata 1999, Van Rompay 1998, Zhang 2004a). The antibodies that have demonstrated the best efficacy are IgG1b12, 2G12, 2F5, and HIVIg. Strong cellular responses do not enhance the protection to SHIV challenge provided by passively transferred antibodies (Mascola et al., 2003b).
-
ii
Second, in natural infection, autologous neutralizing responses exert selective pressure on HIV evolution in vivo (Albert 1990, Arendrup 1992, Bradney 1999, Eichberg 1992, Montefiori 1991, Parren 1999, Reitz 1988, Richman 2003, Watkins 1993, Wei 2003). HIV evades this selective pressure by several mechanisms such as introducing new N‐linked glycosylation residues to present a protective glycan shield (Back 1994, Chackerian 1997, Derdeyn 2004, Kolchinsky 2001, Quinones‐Kochs 2002, Reitter 1998, Wei 2003), conformational or entropic masking of vulnerable epitopes (Kwong et al., 2002), shedding monomeric envelope proteins that enhance the dominance of nonneutralizing epitopes (Burton and Montefiori, 1997), or by simple epitope variation. While it is discouraging that the antibodies cannot control the infection, the observation that HIV has to evade these antibodies suggests that they have an impact.
-
iii
Long‐term nonprogression is associated with the presence of high‐titered broadly neutralizing responses (Cao 1995, Cecilia 1999, Hutto 1996, Kloosterboer 2005, Pilgrim 1997, Scarlatti 1996, Zhang 1997). Although others have postulated that this connection is more tenuous (Montefiori et al., 1996) because a direct link between the levels of neutralizing antibodies and disease progression cannot be established (Cecilia et al., 1999), the recent studies connecting evasion with neutralization suggest that perhaps the antibody responses in these patients may be better able to contain the virus long term.
Based on these observations, it is easy to conclude that vaccines that stimulate such neutralizing responses would be highly desirable. Thus, significant efforts have been directed to developing immunogens that induce antibodies with broadly neutralizing specificities.
B. Nonneutralizing Antibodies and Protection?
Antibodies that score positive in an in vitro neutralization assay can mediate protection in vivo. However, can an antibody that scores negative in such an assay be as effective? The literature suggests that there is a significant subset of these binding but nonneutralizing antibodies that impact HIV disease progression and possibly even transmission. Binding/nonneutralizing antibodies against virus surface glycoproteins have been shown to protect against infection in other virus systems, including Sindbis virus (Stanley et al., 1986), Venezuelan equine encephalomyelitis virus (Mathews and Roehrig, 1982), herpesvirus (Dix et al., 1981), and vesicular stomatitis virus (Lefrancois, 1984). For HIV, the most compelling evidence of nonneutralizing but protective antibodies comes from tests of DNA/MVA vaccines in macaques. Here, vaccines containing pol, gag, and env sequences (encoding the first 270 amino acid residues of the ADA envelope) afforded stronger protection against mucosal SHIV 89.6P challenge than matched vaccines containing only gag and pol (Amara et al., 2002). Since the more effective vaccine containing env did not raise neutralizing antibodies against the challenge virus, the enhanced protection was attributed to high titers of anti‐envelope binding antibodies. A more recent macaque study showed that the viral containment and immune preservation conferred by a DNA/adenovirus vaccine was significantly enhanced by inclusion of chimeric gp140 sequences that were heterologous with respect to the challenge SHIV (Letvin et al., 2004). Although the investigators attributed this protection to cross‐reactive cellular responses raised against conserved HIV envelope sequences, this study did not rule out a role for nonneutralizing antibodies. In sum, these nonneutralizing antibodies appear to protect against disease via mechanisms overlooked by conventional in vitro viral neutralization assay.
The most common mechanism attributed to “nonneutralizing” control is Fc receptor‐mediated or complement‐mediated inhibitory or cytolytic activity. For instance, antibody‐dependent cell‐mediated cytotoxicity (ADCC) (reviewed in Ahmad 1996, Gomez‐Roman 2006) has been associated with improved control of viremia and CD4+ counts in HIV‐infected patients (Ahmad 2001, Forthal 1999, Forthal 2001) and slower disease progression in SIV‐infected macaques (Banks et al., 2002). One recent study observed a correlation between ADCC activity and reduced viral load in rhesus macaques after mucosal challenge with SIV (Gomez‐Roman et al., 2005). Fc‐mediated effector mechanisms have also been attributed to the enhanced neutralizing efficacy of HIV+ or SIV+ serum observed when MDM or immature dendritic cells (iDC) are used as cellular targets instead of activated PBMCs (Holl 2004, Holl 2006a, Holl 2006b). In fact, mAbs that present minimal neutralizing activity in PBMC‐based assays can be highly inhibitory in MDM‐ or iDC‐based assays (Holl et al., 2006a). These mAbs recognize portions of the native envelope spike that are exposed on “dead” spikes that are expressed on native virions but are unable to mediate fusion because they are uncleaved or have lost the gp120 portion (Moore 2006, Zanetti 2006, Zhu 2006). How these nonneutralizing antibodies may impact HIV transmission remains unanswered. Although a clear‐cut protective mechanism for nonneutralizing anti‐HIV antibodies is not established, it is nevertheless prudent to consider Env‐based vaccine candidates that may stimulate such inhibitory activities.
C. Antibodies against CD4i Epitopes: Perceptions, Realities, and Opportunities
Humoral responses against gp120 epitopes exposed during viral entry may provide new opportunity for vaccine development. These responses warrant attention because they recognize some of the most conserved and functionally important regions of the HIV envelope. The question at hand is whether these responses (and the antigens that raise them) are worth pursuing as vaccines. Unfortunately, the view of CD4i epitopes as a vaccine target has been colored by the recent findings discussed below.
1. Potency of CD4i Antibodies and CCR5 Expression
Using computational models based on the crystal structure of CD4‐bound gp120, it has been suggested that CD4i epitopes are actually occluded during entry (Labrijn et al., 2003). Support for this model was generated by showing that IgG1 X5, which recognizes a CD4i epitope, is profoundly less effective than the smaller Fab or scFv fragments of the same antibody at neutralizing a small panel of primary R5 using isolates (Labrijn et al., 2003). However, for some isolated IgG1 X5 is more potent than the smaller fragments likely due to the avidity effect because of its bivalency. In addition, the neutralizing activities of antibodies to CD4i epitopes are dependent on assay conditions. We have found over 100‐fold differences in the levels of CCR5 expression between TZM‐bl cell line commonly used in neutralization assays and PHA‐activated PBMCs (Choudhry 2006b, Platt 1998, Platt 2005 have shown that the entry kinetics of R5 isolates are exquisitely sensitive to the levels of CCR5 expressed on the target cell. In fact, using artificial cell lines as targets, they demonstrated that viruses become increasingly susceptible to entry inhibitors, such as T‐20, as the levels of CCR5 drop below <104 molecules/cell. Binley et al. (2004) further emphasized this point by showing that the neutralizing efficacy of Fab X5 is significantly improved in PBMC‐based neutralization assays as compared to cell line–based assays. While it is not known how much CCR5 is expressed on the cells initially targeted by HIV, numerous studies have determined the expressed levels of CCR5 on various mucosal and lymphoid tissues to be significantly lower than 104 per cell, well in the range that CD4i antibodies may be effective.
2. CD4i Epitopes and HIV/SIV Infection
It is clear that CD4i epitopes are raised during HIV infection, since hmAbs that recognize CD4i epitopes in the coreceptor binding site have been derived from HIV+ persons (Robinson 1992, Xiang 2002a). Last year, it was reported (Decker et al., 2005) that sera from most HIV‐infected persons contain antibodies that were extremely potent and cross‐reactive in the presence of small amounts of sCD4, which presumably stabilizes the exposure of CD4i epitopes. It was further shown that the titers of broadly neutralizing antibodies that are detected in the CD4‐triggered neutralization assay correlated strongly with the abilities of the sera to block the binding of a biotinylated human mAb (19e) to a CD4i epitope on gp120–CD4 complexes in ELISA. These findings have led to the perception that high‐tittered responses to CD4i epitopes are found in all HIV‐infected persons and are therefore meaningless and irrelevant to vaccine design.
Unfortunately, this perception may be a misinterpretation of these results. First, it was demonstrated that increases in neutralizing potency can be observed in HIV+ serum in the presence of sCD4 (Decker et al., 2005). They also showed that HIV+ sera competed with mAb 19e for binding to gp120–CD4 complexes using an assay that tested percentage blocking with a single dilution of serum. Such assays, however, do not demonstrate the presence of a high binding titer of anti‐CD4i antibodies. Second, it was proposed that the responses to the highly conserved domains may constrain the breadth of the viral quasispecies that occur during natural infection and drive the evolution of the virus to protect these epitopes (Decker et al., 2005). This hypothesis is consistent with observations from SIVMneCL8 infection of rhesus macaques where the neutralization sensitive and mildly pathogenic strain becomes resistant and highly pathogenic in part by introducing N‐ and O‐linked glycosyl residues in the V1 region that occludes its coreceptor binding (Chackerian 1997, Kimata 1999a, Kimata 1999b).
This perception also begs the question whether lead vaccine candidates that are intending to target responses to the CD4‐binding domain (CD4BD) should be abandoned given the evidence that responses directed to the CD4BD are highly prevalent in HIV infection and are associated with progression to AIDS (Hioe et al., 2001). CD4BD antibodies have also been shown to inhibit antigen presentation (Hioe 2000, Hioe 2001, Tuen 2005). We would argue that in the absence of well‐designed safety and animal protection studies that would exclude one epitope or another, it is prudent to consider immunogens designed to effect responses toward any of these epitopes. Therefore, it is reasonable to propose and to test whether a preexisting humoral or mucosal response directed to CD4i epitopes could afford protection against primate lentiviral infection. Several indirect observations from the literature suggest that the answer is yes.
-
i
Infection of macaques with macrophage tropic SIV strains, such as SIVmac1A11 (Luciw et al., 1992), SIVmac17E‐Cl, or SIVmac316 (Puffer et al., 2002), leads to a transient or attenuated viremia. These CD4‐independent isolates (Puffer et al., 2002) can generate potent neutralizing responses that may have a role in controlling the observed viremia and in the protection generally observed in subsequent challenge of infected macaques with highly pathogenic SIV strains.
-
ii
SIV strains that are deficient in either variable loops or specific glycosylation sites that occlude the coreceptor interacting domain in the SIV spike protein are CD4 independent, and highly susceptible to neutralization by SIV immune sera and mAbs that recognize these CD4i epitopes (Johnson 2002, Johnson 2003a, Johnson 2003b). Attenuated versions of these SIV isolates induce antibody responses capable of neutralizing the “neutralization resistant” SIVmac239 in vitro (Reitter et al., 1998), and protecting against SIVmac239 challenge (Mori et al., 2001).
-
iii
Protection in cohorts of individuals that were exposed to HIV but remain uninfected has been associated with the presence of mucosal or serum antibodies to HIV that, in some studies, exhibited neutralizing activity. In one cohort, this protection was associated with antibody titer to epitopes expressed on CD4–gp120 complexes but not HIV‐specific T cell responses (Nguyen et al., 2006).
-
iv
While passive protection studies have delineated a clear correlation between the neutralizing efficacy of antibodies in vitro and their ability to passively protect against SHIV challenge in vivo, formulations of polyclonal HIVIg (which contains antibodies to these CD4i epitopes, among others), 2G12, and 2F5 demonstrated protective efficacy from vaginal challenge while similar infusions with mixtures of 2G12, and 2F5 did not (Mascola et al., 2000). This is despite the observations that HIVIg/2G12/2F5 and 2G12/2F5 formulations demonstrated equivalent neutralization titer in vitro.
In addition, it appears that fitness during transmission is enhanced if the virus expresses/exposes the coreceptor binding domain on the viral spike. Taken together, we believe the preponderance of evidence suggests that vaccines that target such CD4i epitopes may have provided some level of protection against transmitted virus and logically coincides with ongoing vaccine development efforts in the field.
D. The Hunt for the Right Immunogen
The daunting part of this challenge is the evasive power provided by the sequence diversity of the Env. It was quickly apparent that standard vaccine approaches using killed virus or soluble monomeric gp120 or gp160 as immunogens generated only “type‐specific” immunity, and neutralized only the source virus of the immunogen or its very close relatives (Burton et al., 2004). Today, the effort is to identify immunogens or immunization strategies that induce antibody responses that exhibited a broader neutralizing and/or protective phenotype. The immunogen approaches can be grouped in two broad overlapping categories based on whether the respective antibodies target epitopes on the virion spike or epitopes (such as CD4i) that appear during entry.
1. Generating Antibodies That Target the Virion Spike Before Binding to Receptors
Five of the most broadly neutralizing antibodies, 2G12, b12, 447–52D, 2F5, and 4E10, recognize conserved epitopes that are expressed on the viral spike before binding to receptors, leading some investigators to suggest that the optimal vaccine candidate should induce antibodies that preferentially bind to the viral spike (Burton 2004, Fouts 1997, Parren 2001). Efforts to reach this goal have focused on the following approaches:
-
i
Adding or removing Asn residues to alter the level of N‐linked glycosylation that shields the CD4bs. By exposing the deep CD4BS pocket, the hope is the resulting immunogen will induce broadly neutralizing antibodies like b12 (Koch et al., 2003).
-
ii
Expressing the outer domain of gp120 (Yang et al., 2004). The outer domain is exposed on the envelope spike and contains binding surfaces for 2G12, IgG1b12, and broadly neutralizing anti‐V3 loop antibodies.
-
iii
Expressing soluble forms of the oligomeric envelope trimer to mimic the spike as it appears on the HIV virion. Typically, investigators have expressed these proteins as fusions between gp120 and the ectodomain of gp41 (reviewed in Cho, 2003). These constructs typically generated preparations consisting of mixtures of monomeric and oligomeric forms. Recent efforts have improved consistency and yields of trimeric forms by introducing disulfide links between the proximal domains of gp120 and gp41 (called SOS or SOSIP envelopes) (Beddows 2005, Binley 2000), using envelope genes derived from HIV strains with highly stable spikes (Lian 2005, Sharma 2006, Srivastava 2002, Zhang 2001), creating HIV‐SIV envelope chimeras (Center et al., 2004), or fusing the envelope ectodomain to non‐HIV sequences that preferentially form trimers (Pancera 2005, Yang 2000, Yang 2002). In an effort to produce soluble spikes with an antigenic profile more consistent with the native virion, investigators have produced the SOS and SOSIP variants in cell lines that overexpress furin, a protease which cleaves gp120–gp41 fusion into its respective domains.
-
iv
Minimizing the conformational or entropic masking of the conserved neutralizing domains by introducing mutations that restrain the movement of gp120 (Kwong 2002, Xiang 2002b). This concept is derived from studies showing that the binding of broadly neutralizing antibodies such as b12 and 2G12 consistently realized minimal entropic change in gp120. This contrasts sharply with other less effective antibodies such as F105 which generate significantly larger entropic changes. Given the range of movement possible between the inner and outer domains of envelope indicated by the crystal structures, it was proposed that the virus may evade neutralization by using an entropic mask or a conformational barrier that antibodies must overcome to actually bind. Introducing mutations that limit this movement, such as replacing the tryptophan with a serine in position 375, and reduce the entropic requirements for antibody binding may improve the chances of inducing the preferred antibody specificities.
-
v
Mimicking the high‐mannose‐type oligosaccharides that are presented on HIV envelope and recognized by 2G12 (reviewed in Wang, 2006). Recently, constructs have been described that mimic the binding site of 2G12 using organic scaffolds decorated with synthetic oligomannose structures. These structures are recognized by 2G12 to varying degrees. The immunogenicity of these constructs has not been described.
-
vi
Mimicking the MPER of the envelope spike recognized by 2F5 and 4E10 (reviewed in Zwick, 2005). It has been recently appreciated that the lipid membrane and hydrophobic context of the epitope is critical for antibody binding (Haynes et al., 2005a). This observation may explain the dearth of success using peptide‐based mimics of the eptiope to induce 2F5‐ and 4E10‐type responses and has rejuvenated efforts to develop new mimics. Several novel constructs have been presented (Brunel 2006, Luo 2006); however, immunogenicity data are limited.
Unfortunately, where they have been evaluated, the immunogen strategies described above typically fail to elicit antibodies capable of neutralizing more than a minor fraction of primary isolates (Beddows 2005, Graham 2002, Selvarajah 2005). To make matters worse, it was recently hypothesized that B cells responding to MPER epitopes are deleted because the lipid portion of their target epitope is considered “self” (Haynes et al., 2005b). This hypothesis would explain why such responses are so infrequently observed. It also suggests that induction of a response directed to the MPER may require immunogens capable of breaking one's natural tolerance to the cell membrane. Whether IgG1b12 and 2G12 recognized similarly tolerized epitopes is unclear. Certainly, designing vaccine immunogens that would target responses against these epitopes represents a daunting immunological, structural, and potentially, regulatory challenge.
2. Generating Antibodies That Target Entry Intermediates
As summarized earlier, the coreceptor‐binding domain is a structure that is highly conserved among HIV, SIV, and HIV‐2. This has prompted investigators to develop immunogens that induce antibodies that target this structure. One such immunogen is the gp120–CD4 complex, which forms when the virus attaches to cell surface receptor CD4. Studies in mice, goats, and more recently rhesus macaques have all shown that broadly neutralizing antibody responses are elicited by immunization with various forms of a gp120–CD4 complex (Bower 2004, Celada 1990, Devico 1996, Fouts 2002, Gershoni 1993, Kang 1994). Three groups of immunogens have been developed that attempt to represent gp120–CD4 complex. The first group consists of complexes between soluble envelope protein subunits (gp120) and soluble human CD4 (Bower 2004, Celada 1990, Devico 1996, Fouts 2000, Fouts 2002, Gershoni 1993, He 2003, Varadarajan 2005). These immunogens are produced by simply mixing the two soluble components together to allow them to bind and form complexes or expressing the gp120 and sCD4 as a genetically tethered chimeric molecule. An example of such a chimera is the full‐length single chain (FLSC) which is genetic fusion between gp120BaL and the D1‐D2 domain of CD4 (Fouts et al., 2000). The second group consists of soluble envelope proteins complexed with human mAb, A32 (Liao et al., 2004). A32 binds to an epitope defined by the C1‐C4 region on HIV envelope subunit, gp120 and, like CD4, is known to induce the expression of the coreceptor binding domain within gp120 (Liao 2004, Wyatt 1995). Again these complexes are produced by admixing the two components to allow them to bind in solution to form complexes (Liao et al., 2004). The third are complexes between gp120 and a CD4 mimic peptide, CD4M9 (Fouts 2002, Varadarajan 2005, Vita 1999a). Unfortunately, the affinity of the CD4M9 for gp120 is insufficient to permit formation of a stable complex in solution from the two components (Vita et al., 1999a). Stable complexes can be expressed, however, as chimeric fusion protein, SCBaL/M9 (Fouts et al., 2000) or gp120‐M9 (Varadarajan et al., 2005), where the gp120BaL or gp120JRFL, respectively, are genetically tethered to CD4M9 by a short amino acid linker. Stable complexes have also been created using CD4M33 (Martin et al., 2003b), a modified CD4M9 that exhibits an affinity for gp120 closer to that of CD4 (Huang et al., 2005a). The presumption is that these complexes all exhibit the antigenic features presented when the HIV envelope spike interacts with cell surface CD4. Thus far, only the gp120–CD4 admixed or cross‐linked complexes have been shown to elicit neutralizing antibody response. The others are still being evaluated. Whether the resulting neutralizing response arises from the exposure of cryptic epitopes either enhancing their own immunogenicity or alter the immunogenicity of other extant epitopes on gp120 (Celada 1990, DeVico 1995) is still unclear. Either instance would enhance the potential for gp120–CD4 complexes to elicit broadly cross‐reactive CD4i antibodies.
Three recent studies have generated rather different results using various forms of gp120–CD4 complex immunogens (He 2003, Liao 2004, Varadarajan 2005). However, these studies suffer from one of either two major flaws. First, they evaluate the immunogenicity of their constructs in animal models (mice or guinea pigs) that are heterologous to the CD4 moiety used in their immunogens (human sCD4). It has been known since 1990, when the first studies of gp120–CD4 complexes appeared, that substantial levels of anti‐CD4 antibodies are elicited when the CD4 used in the complex is from a species (i.e., humans) different from the one (i.e., rodents) that is immunized (Celada et al., 1990). Human CD4 is highly immunogenic in rodents and biases responses to a gp120–CD4 complex away from the conserved gp120 epitopes. This obscures the immunogenic properties of the constrained HIV envelope moiety in favor of anti‐CD4 responses. That said, two independent reports (Bower 2004, Srivastava 2004) including one single‐chain gp120‐CD4 immunogen tested in mice (Bower et al., 2004) show that broadly neutralizing antibody fractions can be isolated from animals immunized with heterologous gp120–CD4 complexes that do not recognize CD4.
Second, immunogens which do not truly mimic structure presented by the gp120–CD4 complex were used. One study (Liao et al., 2004) used gp120 conformationally constrained by the A32 mAb as an immunogen. It has been proposed that A32 binds to gp120 in such a way that it is a “CD4 mimic” as judged by the exposure of CD4i epitopes recognized by the mAbs 17b and 48d (Wyatt et al., 1995). Guinea pigs immunized with covalent conjugates of gp120(BaL) and A32 mounted neutralizing antibody responses that were by and large indistinguishable from those elicited by gp120(BaL) alone. We have found that the gp120 conformational changes induced by A32 and CD4 are distinct as judged by differential reactivity with CD4i antibodies such as 19e that recognize epitopes in the bridging sheet of envelope (Fouts et al., unpublished data). Notably, serum antibody responses to this epitope are thought to constrain viral diversity in vivo (Decker et al., 2005). This observation may explain why the A32–gp120(BaL) complexes elicited a different pattern of reactivity than our gp120–CD4 complexes.
Other approaches are also being utilized to target the CD4i epitopes. The most common is to remove the hypervariable V1, V2, and/or V3 regions of the envelope that are the primary target of the “type‐specific” antibody responses and that shield the conserved neutralizing epitopes (reviewed in Cho, 2003). More recently, investigators have developed constructs derived from the envelope sequences of CD4‐independent isolates that have been adapted to grow on cell lines devoid of CD4 (Hoffman 1999, Kolchinsky 2001) or isolated from a patient with high level of broadly neutralizing antibodies (Quinnan 1999, Vujcic 1995, Zhang 2002). These envelopes contain structural alterations that provide more receptive interactions with the coreceptor such as fewer N‐linked glycosylation or shifting the V1–V2 loops.
As the full spectrum and potential of CD4i epitopes is only recently becoming apparent, it is difficult to argue that CD4i epitopes are poor targets for vaccine development (Labrijn et al., 2003). In addition, it is not known whether mAbs specific for CD4i epitopes are protective in passive transfer studies in rhesus macaques. In this regard, until passive transfer studies are carried out using mixtures of mAbs specific for CD4i epitopes and shown to be negative, it is premature to exclude this strategy based on in vitro neutralization data alone. It should also be recognized that immunization with gp120–CD4 complexes can dramatically change the immunodominance profile of gp120 (Denisova 1996, Fouts 2002, Kang 1994; Fouts et al., unpublished data), and it is possible that this leads to the immunogenicity of previously silent epitopes that elicit protective responses in vivo.
E. The Env as Target for Inhibitors
The entry process for HIV is also a prime target for therapeutics. Early efforts to interfere with entry utilized polyclonal antibody preparations developed from HIV+ patients (HIVIg) or mAbs (2G12, 2F5, 4E10), each of which exhibited exceptional neutralizing capacity in vitro (reviewed in Choudhry et al., 2006a). Unfortunately, these preparations did not provide much in the way therapeutic utility despite their safety. When they did impact viral load, the effect was transient with resistant viruses quickly emerging. A key breakthrough came with the licensure of T‐20, or enfuvirtide. Targeting the HR1 of gp41 (reviewed in Weiss, 2003), this drug was the first in its class to reach the marketplace and is at the forefront of an army of other inhibitors making their way through clinical development. These drugs generally fall within four broad groups and are being developed for both therapeutic and vaginally or rectally applied microbicidal indications.
-
i
Antibodies. Given their safety record, clinical development of antibodies for HIV therapy continues. Promising new candidates target CD4 and CCR5, attempting to minimize the chances of evasion by targeting cellular receptors instead of the envelope (Dimitrov, 2004).
-
ii
Peptides. This group is populated by Fuzeon and a variety of follow‐on candidates, each targeting the helical region of gp41. The main challenge with this group is delivery. Fuzeon requires intramuscular administration twice daily making it a rather unfavorable choice for patients. Newer delivery methods and formulations are being developed that may help solve this problem (Markovic 2006, Pierson 2003a, Pierson 2003b).
-
iii
Lectins. A variety of lectins have been shown to inhibit HIV by binding to the mannose structures that cover the envelope spike. These drugs are currently being developed for vaginal or rectal use as microbicides; however, they have potential therapeutic utility (De 2005, Pierson 2003a). Cyanovirin (CV‐N) is currently the furthest in clinical development.
-
iv
Small compounds. This group is where the bulk of the new inhibitors fall. Thus far, only Maraviroc, a small molecule antagonist that targets CCR5 has reached Phase III. As a group, these drugs are proving to be highly effective at reducing viral load but are falling out of development because of a variety of safety problems (De 2005, Pierson 2003a, Kadow 2006, Markovic 2006).
The drugs and their respective targets are eloquently described in many recent reviews some of which are cited above. Given the wealth of research directed to understanding the nuances of HIV entry, we do not anticipate that this pipeline of drug candidates will dry anytime soon.
VII. Conclusions
HIV has evolved a number of strategies to escape host immune surveillance, prominently by modifications to its Env. Latest advances in our understanding of its structure at atomic level of detail promise to provide us with new tools to design effective vaccines and inhibitors. In spite of the significant progress, the contribution to the development of vaccines and therapeutics of the wealth of information about the Env structure is still relatively small. However, current developments promise to revolutionize the way therapeutics and vaccines will be designed in the future. It remains to be seen whether this promise will materialize.
Acknowledgments
We thank the members of our group Protein Interactions, and Anthony DeVico, Robert Blumenthal, Peter Kwong, Xinhua Ji and Hana Golding for helpful discussions. This project was supported by the NIH Intramural AIDS Targeted Antiviral Program (IATAP), the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the Gates Foundation to DSD, and CRADA between NCI and Profectus BioSciences, Inc.
References
- Ahmad A., Menezes J. Antibody‐dependent cellular cytotoxicity in HIV infections. FASEB J. 1996;10:258–266. doi: 10.1096/fasebj.10.2.8641559. [DOI] [PubMed] [Google Scholar]
- Ahmad R., Sindhu S.T., Toma E., Morisset R., Vincelette J., Menezes J., Ahmad A. Evidence for a correlation between antibody‐dependent cellular cytotoxicity‐mediating anti‐HIV‐1 antibodies and prognostic predictors of HIV infection. J. Clin. Immunol. 2001;21:227–233. doi: 10.1023/a:1011087132180. [DOI] [PubMed] [Google Scholar]
- Albert J., Abrahamsson B., Nagy K., Aurelius E., Gaines H., Nystrom G., Fenyo E.M. Rapid development of isolate‐specific neutralizing antibodies after primary HIV‐1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- Alfsen A., Bomsel M. HIV‐1 gp41 envelope residues 650–685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J. Biol. Chem. 2002;277:25649–25659. doi: 10.1074/jbc.M200554200. [DOI] [PubMed] [Google Scholar]
- Amara R.R., Smith J.M., Staprans S.I., Montefiori D.C., Villinger F., Altman J.D., O'Neil S.P., Kozyr N.L., Xu Y., Wyatt L.S., Earl P.L., Herndon J.G. Critical role for Env as well as Gag‐Pol in control of a simian‐human immunodeficiency virus 89. 6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 2002;76:6138–6146. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M., Nielsen C., Hansen J.E., Pedersen C., Mathiesen L., Nielsen J.O. Autologous HIV‐1 neutralizing antibodies: Emergence of neutralization‐resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 1992;5:303–307. [PubMed] [Google Scholar]
- Atchison R.E., Gosling J., Monteclaro F.S., Franci C., Digilio L., Charo I.F., Goldsmith M.A. Multiple extracellular elements of CCR5 and HIV‐1 entry: Dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- Baba T.W., Liska V., Hofmann‐Lehmann R., Vlasak J., Xu W., Ayehunie S., Cavacini L.A., Posner M.R., Katinger H., Stiegler G., Bernacky B.J., Rizvi T.A. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian‐human immunodeficiency virus infection. Nat. Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- Back N.K., Smit L., De Jong J.J., Keulen W., Schutten M., Goudsmit J., Tersmette M. An N‐glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- Banks N.D., Kinsey N., Clements J., Hildreth J.E. Sustained antibody‐dependent cell‐mediated cytotoxicity (ADCC) in SIV‐infected macaques correlates with delayed progression to AIDS. AIDS Res. Hum. Retroviruses. 2002;18:1197–1205. doi: 10.1089/08892220260387940. [DOI] [PubMed] [Google Scholar]
- Basmaciogullari S., Pacheco B., Bour S., Sodroski J. Specific interaction of CXCR4 with CD4 and CD8alpha: Functional analysis of the CD4/CXCR4 interaction in the context of HIV‐1 envelope glycoprotein‐mediated membrane fusion. Virology. 2006;353:52–67. doi: 10.1016/j.virol.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Beddows S., Schulke N., Kirschner M., Barnes K., Franti M., Michael E., Ketas T., Sanders R.W., Maddon P.J., Olson W.C., Moore J.P. Evaluating the immunogenicity of a disulfide‐stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2005;79:8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E.A., Murphy P.M., Farber J.M. Chemokine receptors as HIV‐1 coreceptors: Roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Binley J.M., Sanders R.W., Clas B., Schuelke N., Master A., Guo Y., Kajumo F., Anselma D.J., Maddon P.J., Olson W.C., Moore J.P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion‐associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley J.M., Wrin T., Korber B., Zwick M.B., Wang M., Chappey C., Stiegler G., Kunert R., Zolla‐Pazner S., Katinger H., Petropoulos C.J., Burton D.R. Comprehensive cross‐clade neutralization analysis of a panel of anti‐human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lee B., Tackoen M., Puffer B., Boom A., Libert F., Sharron M., Wittamer V., Vassart G., Doms R.W., Parmentier M. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood. 2000;96:1638–1645. [PubMed] [Google Scholar]
- Bouma P., Leavitt M., Zhang P.F., Sidorov I.A., Dimitrov D.S., Quinnan G.V., Jr. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 2003;77:8061–8071. doi: 10.1128/JVI.77.14.8061-8071.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower J.F., Green T.D., Ross T.M. DNA vaccines expressing soluble CD4‐envelope proteins fused to C3d elicit cross‐reactive neutralizing antibodies to HIV‐1. Virology. 2004;328:292–300. doi: 10.1016/j.virol.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Bradney A.P., Scheer S., Crawford J.M., Buchbinder S.P., Montefiori D.C. Neutralization escape in human immunodeficiency virus type 1‐infected long‐term nonprogressors. J. Infect. Dis. 1999;179:1264–1267. doi: 10.1086/314711. [DOI] [PubMed] [Google Scholar]
- Broder C.C., Dimitrov D.S. HIV and the 7‐transmbembrane domain receptors. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- Brunel F.M., Zwick M.B., Cardoso R.M., Nelson J.D., Wilson I.A., Burton D.R., Dawson P.E. Structure‐function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J. Virol. 2006;80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Burton D.R. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- Burton D.R., Montefiori D.C. The antibody response in HIV‐1 infection. AIDS. 1997;11(Suppl. A):S87–S98. [PubMed] [Google Scholar]
- Burton D.R., Pyati J., Koduri R., Sharp S.J., Thornton G.B., Parren P.W., Sawyer L.S., Hendry R.M., Dunlop N., Nara P.L., Lamacchia M., Garraty E. Efficient neutralization of primary isolates of HIV‐1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Burton D.R., Desrosiers R.C., Doms R.W., Koff W.C., Kwong P.D., Moore J.P., Nabel G.J., Sodroski J., Wilson I.A., Wyatt R.T. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton D.R., Stanfield R.L., Wilson I.A. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Qin L., Zhang L., Safrit J., Ho D.D. Virologic and immunologic characterization of long‐term survivors of human immunodeficiency virus type 1 infection [see comments] N. Engl. J. Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- Cardoso R.M., Zwick M.B., Stanfield R.L., Kunert R., Binley J.M., Katinger H., Burton D.R., Wilson I.A. Broadly neutralizing anti‐HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion‐associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Carr C.M., Kim P.S. A spring‐loaded mechanism for the conformational changes of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Cavacini L.A., Samore M.H., Gambertoglio J., Jackson B., Duval M., Wisnewski A., Hammer S., Koziel C., Trapnell C., Posner M.R. Phase I study of a human monoclonal antibody directed against the CD4‐binding site of HIV type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses. 1998;14:545–550. doi: 10.1089/aid.1998.14.545. [DOI] [PubMed] [Google Scholar]
- Cecilia D., Kleeberger C., Munoz A., Giorgi J.V., Zolla‐Pazner S. A longitudinal study of neutralizing antibodies and disease progression in HIV‐1‐infected subjects. J. Infect. Dis. 1999;179:1365–1374. doi: 10.1086/314773. [DOI] [PubMed] [Google Scholar]
- Celada F., Cambiaggi C., Maccari J., Burastero S., Gregory T., Patzer E., Porter J., McDanal C., Matthews T. Antibody raised against soluble CD4‐rgp120 complex recognizes the CD4 moiety and blocks membrane fusion without inhibiting CD4‐gp120 binding. J. Exp. Med. 1990;172:1143–1150. doi: 10.1084/jem.172.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center R.J., Lebowitz J., Leapman R.D., Moss B. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV‐1) Env through the use of HIV‐1/simian immunodeficiency virus chimeras. J. Virol. 2004;78:2265–2276. doi: 10.1128/JVI.78.5.2265-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackerian B., Rudensey L.M., Overbaugh J. Specific N‐linked and O‐linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Cho M.W. Subunit protein vaccines: Theoretical and practical considerations for HIV‐1. Curr. Mol. Med. 2003;3:243–263. doi: 10.2174/1566524033479861. [DOI] [PubMed] [Google Scholar]
- Choe H., Li W., Wright P.L., Vasilieva N., Venturi M., Huang C.C., Grundner C., Dorfman T., Zwick M.B., Wang L., Rosenberg E.S., Kwong P.D. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV‐1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- Choudhry V., Zhang M.Y., Dimitrova D., Prabakaran P., Dimitrov A.S., Fouts T.R., Dimitrov D.S. Antibody‐based inhibitors of HIV infection. Expert. Opin. Biol. Ther. 2006;6:523–531. doi: 10.1517/14712598.6.5.523. [DOI] [PubMed] [Google Scholar]
- Choudhry V., Zhang M.Y., Harris I., Sidorov I.A., Vu B., Dimitrov A.S., Fouts T., Dimitrov D.S. Increased efficacy of HIV‐1 neutralization by antibodies at low CCR5 surface concentration. Biochem. Biophys. Res. Commun. 2006;348:1107–1115. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan C.M., McManus C.M., Sharron M., Gao Z., Murphy D., Jaffer S., Choe W., Chen W., Hesselgesser J., Gaylord H., Kalyuzhny A., Lee V.M. Expression of multiple functional chemokine receptors and monocyte chemoattractant protein‐1 in human neurons. Neuroscience. 2000;97:591–600. doi: 10.1016/s0306-4522(00)00024-5. [DOI] [PubMed] [Google Scholar]
- D'Souza M.P., Allen M., Sheets R., Johnston M.I. Current advances in HIV vaccines. Curr. HIV/AIDS Rep. 2004;1:18–24. doi: 10.1007/s11904-004-0003-1. [DOI] [PubMed] [Google Scholar]
- Dalgleish A.G., Beverley P.C.L., Clapham P.R., Crawford D.H., Greaves M.F., Weiss R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Darbha R., Phogat S., Labrijn A.F., Shu Y., Gu Y., Andrykovitch M., Zhang M.Y., Pantophlet R., Martin L., Vita C., Burton D.R., Dimitrov D.S. Crystal structure of the broadly cross‐reactive HIV‐1‐neutralizing Fab X5 and fine mapping of its epitope. Biochemistry. 2004;43:1410–1417. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- De C.E. Emerging anti‐HIV drugs. Expert. Opin. Emerg. Drugs. 2005;10:241–273. doi: 10.1517/14728214.10.2.241. [DOI] [PubMed] [Google Scholar]
- Decker J.M., Bibollet‐Ruche F., Wei X., Wang S., Levy D.N., Wang W., Delaporte E., Peeters M., Derdeyn C.A., Allen S., Hunter E., Saag M.S. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisova G., Stern B., Raviv D., Zwickel J., Smorodinsky N.I., Gershoni J.M. Humoral immune response to immunocomplexed HIV envelope glycoprotein 120. AIDS Res. Hum. Retroviruses. 1996;12:901–909. doi: 10.1089/aid.1996.12.901. [DOI] [PubMed] [Google Scholar]
- Derdeyn C.A., Decker J.M., Bibollet‐Ruche F., Mokili J.L., Muldoon M., Denham S.A., Heil M.L., Kasolo F., Musonda R., Hahn B.H., Shaw G.M., Korber B.T. Envelope‐constrained neutralization‐sensitive HIV‐1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Devico A., Silver A., Thronton A.M., Sarngadharan M.G., Pal R. Covalently crosslinked complexes of human immunodeficiency virus type 1 (HIV‐1) gp120 and CD4 receptor elicit a neutralizing immune response that includes antibodies selective for primary virus isolates. Virology. 1996;218:258–263. doi: 10.1006/viro.1996.0188. [DOI] [PubMed] [Google Scholar]
- DeVico A.L., Rahman R., Welch J., Crowley R., Lusso P., Sarngadharan M.G., Pal R. Monoclonal antibodies raised against covalently crosslinked complexes of human immunodeficiency virus type 1 gp120 and CD4 receptor identify a novel complex‐dependent epitope on gp 120. Virology. 1995;211:583–588. doi: 10.1006/viro.1995.1441. [DOI] [PubMed] [Google Scholar]
- Dimitrov A.S., Louis J.M., Bewley C.A., Clore G.M., Blumenthal R. Conformational changes in HIV‐1 gp41 in the course of HIV‐1 envelope glycoprotein‐mediated fusion and inactivation. Biochemistry. 2005;44:12471–12479. doi: 10.1021/bi051092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. Fusin—a place for HIV‐1 and T4 cells to meet. Identifying the coreceptor mediating HIV‐1 entry raises new hopes in the treatment of AIDS. Nat. Med. 1996;2:640–641. doi: 10.1038/nm0696-640. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. How do viruses enter cells? The HIV‐1 coreceptors teach us a lesson of complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S., Broder C.C. “HIV and Membrane Receptors.”. Landes Biosciences; Austin, TX: 1997. [Google Scholar]
- Dimitrov D.S., Ji X. Crystal structure of a cross‐reactive HIV‐1 neutralizing CD4 binding site antibody Fab m14. Personal communication. 2006 [Google Scholar]
- Dimitrov D.S., Hillman K., Manischewitz J., Blumenthal R., Golding H. Kinetics of soluble CD4 binding to cells expressing human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 1992;66:132–138. doi: 10.1128/jvi.66.1.132-138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S., Xiao X., Chabot D.J., Broder C.C. HIV coreceptors. J. Membr. Biol. 1998;166:75–90. doi: 10.1007/s002329900450. [DOI] [PubMed] [Google Scholar]
- Dix R.D., Pereira L., Baringer J.R. Use of monoclonal antibody directed against herpes simplex virus glycoproteins to protect mice against acute virus‐induced neurological disease. Infect. Immun. 1981;34:192–199. doi: 10.1128/iai.34.1.192-199.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doranz B.J., Lu Z.H., Rucker J., Zhang T.Y., Sharron M., Cen Y.H., Wang Z.X., Guo H.H., Du J.G., Accavitti M.A., Doms R.W., Peiper S.C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek D.C., Kwong P.D., Nabel G.J. The rational design of an AIDS vaccine. Cell. 2006;124:677–681. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Drakopoulou E., Vizzavona J., Vita C. Engineering a CD4 mimetic inhibiting the binding of the human immunodeficiency virus‐1 (HIV‐1) envelope glycoprotein gp120 to human lymphocyte CD4 by the transfer of a CD4 functional site to a small natural scaffold. Lett. Pept. Sci. 1998;5:241–245. [Google Scholar]
- Duerr A., Wasserheit J.N., Corey L. HIV vaccines: New frontiers in vaccine development. Clin. Infect. Dis. 2006;43:500–511. doi: 10.1086/505979. [DOI] [PubMed] [Google Scholar]
- Eichberg J.W., Murthy K.K., Ward R.H., Prince A.M. Prevention of HIV infection by passive immunization with HIVIg or CD4‐IgG. AIDS Res. Hum. Retroviruses. 1992;8:1515–1519. doi: 10.1089/aid.1992.8.1515. [DOI] [PubMed] [Google Scholar]
- Emini E.A., Schleif W.A., Nunberg J.H., Conley A.J., Eda Y., Tokiyoshi S., Putney S.D., Matsushita S., Cobb K.E., Jett C.M. Prevention of HIV‐1 infection in chimpanzees by gp120 V3 domain‐specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- Endres M.J., Clapham P.R., Marsh M., Ahuja M., Turner J.D., McKnight A., Thomas J.F., Stoebenau‐Haggarty B., Choe S., Vance P.J., Wells T.N., Power C.A. CD4‐independent infection by HIV‐2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- Fass D., Harrison S.C., Kim P.S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV‐1 entry cofactor: Functional cDNA cloning of a seven‐transmembrane, G protein‐coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Ferrantelli F., Hofmann‐Lehmann R., Rasmussen R.A., Wang T., Xu W., Li P.L., Montefiori D.C., Cavacini L.A., Katinger H., Stiegler G., Anderson D.C., McClure H.M. Post‐exposure prophylaxis with human monoclonal antibodies prevented SHIV89. 6P infection or disease in neonatal macaques. AIDS. 2003;17:301–309. doi: 10.1097/00002030-200302140-00003. [DOI] [PubMed] [Google Scholar]
- Ferrantelli F., Rasmussen R.A., Buckley K.A., Li P.L., Wang T., Montefiori D.C., Katinger H., Stiegler G., Anderson D.C., McClure H.M., Ruprecht R.M. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian‐human immunodeficiency virus by human anti‐HIV monoclonal antibodies. J. Infect. Dis. 2004;189:2167–2173. doi: 10.1086/420833. [DOI] [PubMed] [Google Scholar]
- Forster F., Medalia O., Zauberman N., Baumeister W., Fass D. Retrovirus envelope protein complex structure in situ studied by cryo‐electron tomography. Proc. Natl. Acad. Sci. USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthal D.N., Landucci G., Haubrich R., Keenan B., Kuppermann B.D., Tilles J.G., Kaplan J. Antibody‐dependent cellular cytotoxicity independently predicts survival in severely immunocompromised human immunodeficiency virus‐infected patients. J. Infect. Dis. 1999;180:1338–1341. doi: 10.1086/314988. [DOI] [PubMed] [Google Scholar]
- Forthal D.N., Landucci G., Keenan B. Relationship between antibody‐dependent cellular cytotoxicity, plasma HIV type 1 RNA, and CD4+ lymphocyte count. AIDS Res. Hum. Retroviruses. 2001;17:553–561. doi: 10.1089/08892220151126661. [DOI] [PubMed] [Google Scholar]
- Fouts T., Godfrey K., Bobb K., Montefiori D., Hanson C.V., Kalyanaraman V.S., Devico A., Pal R. Crosslinked HIV‐1 envelope‐CD4 receptor complexes elicit broadly cross‐reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2002;99:11842–11847. doi: 10.1073/pnas.182412199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts T.R., Binley J.M., Trkola A., Robinson J.E., Moore J.P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR‐FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts T.R., Tuskan R., Godfrey K., Reitz M., Hone D., Lewis G.K., DeVico A.L. Expression and characterization of a single‐chain polypeptide analogue of the human immunodeficiency virus type 1 gp120‐CD4 receptor complex. J. Virol. 2000;74:11427–11436. doi: 10.1128/jvi.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T.E., Finnegan C.M., Blumenthal R., Kester M. The clinical potential of sphingolipid‐based therapeutics. Cell. Mol. Life Sci. 2006;63:1017–1023. doi: 10.1007/s00018-005-5543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman B.D., Liu Q.H., Del Corno M., Collman R.G. HIV‐1 gp120 chemokine receptor‐mediated signaling in human macrophages. Immunol. Res. 2003;27:261–276. doi: 10.1385/IR:27:2-3:261. [DOI] [PubMed] [Google Scholar]
- Gallaher W.R., Ball J.M., Garry R.F., Griffin M.C., Montelaro R.C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- Gallaher W.R., Ball J.M., Garry R.F., Martin‐Amedee A.M., Montelaro R.C. A general model for the surface glycoproteins of HIV and other retroviruses. AIDS Res. Hum. Retroviruses. 1995;11:191–202. doi: 10.1089/aid.1995.11.191. [DOI] [PubMed] [Google Scholar]
- Gallo S.A., Finnegan C.M., Viard M., Raviv Y., Dimitrov A., Rawat S.S., Puri A., Durell S., Blumenthal R. The HIV Env‐mediated fusion reaction. Biochim. Biophys. Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- Garber D.A., Silvestri G., Feinberg M.B. Prospects for an AIDS vaccine: Three big questions, no easy answers. Lancet Infect. Dis. 2004;4:397–413. doi: 10.1016/S1473-3099(04)01056-4. [DOI] [PubMed] [Google Scholar]
- Gershoni J.M., Denisova G., Raviv D., Smorodinsky N.I., Buyaner D. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J. 1993;7:1185–1187. doi: 10.1096/fasebj.7.12.7690724. [DOI] [PubMed] [Google Scholar]
- Golding H., Zaitseva M., de Rosny E., King L.R., Manischewitz J., Sidorov I., Gorny M.K., Zolla‐Pazner S., Dimitrov D.S., Weiss C.D. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 2002;76:6780–6790. doi: 10.1128/JVI.76.13.6780-6790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Roman V.R., Patterson L.J., Venzon D., Liewehr D., Aldrich K., Florese R., Robert‐Guroff M. Vaccine‐elicited antibodies mediate antibody‐dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J. Immunol. 2005;174:2185–2189. doi: 10.4049/jimmunol.174.4.2185. [DOI] [PubMed] [Google Scholar]
- Gomez‐Roman V.R., Florese R.H., Patterson L.J., Peng B., Venzon D., Aldrich K., Robert‐Guroff M. A simplified method for the rapid fluorometric assessment of antibody‐dependent cell‐mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Gorny M.K., Conley A.J., Karwowska S., Buchbinder A., Xu J.Y., Emini E.A., Koenig S., Zolla‐Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti‐V3 human monoclonal antibody. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Clinical trials of HIV vaccines. Annu. Rev. Med. 2002;53:207–221. doi: 10.1146/annurev.med.53.082901.104035. [DOI] [PubMed] [Google Scholar]
- Gurtler L.G., Hauser P.H., Eberle J., von Brunn A., Knapp S., Zekeng L., Tsague J.M., Kaptue L. A new subtype of human immunodeficiency virus type I (MVP‐5180) from Cameroon. J. Virol. 1994;68:1581–1585. doi: 10.1128/jvi.68.3.1581-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harouse J.M., Bhat S., Spitalnik S.L., Laughlin M., Stefano K., Silberberg D.H., Gonzalez‐Scarano F. Inhibition of entry of HIV‐1 in neural cell lines by antibodies against calactosyl ceramide. Science. 1991;253:320–323. doi: 10.1126/science.1857969. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., StClair W.E., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F., Ortel T.L., Liao H.X. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV‐1 antibodies. Science. 2005;308:1096–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Moody M.A., Verkoczy L., Kelsoe G., Alam S.M. Antibody polyspecificity and neutralization of HIV‐1: A hypothesis. Hum. Antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- He Y., D'Agostino P., Pinter A. Analysis of the immunogenic properties of a single‐chain polypeptide analogue of the HIV‐1 gp120‐CD4 complex in transgenic mice that produce human immunoglobulins. Vaccine. 2003;21:4421–4429. doi: 10.1016/s0264-410x(03)00451-1. [DOI] [PubMed] [Google Scholar]
- Hesselgesser J., Halks‐Miller M., DelVecchio V., Peiper S.C., Hoxie J., Kolson D.L., Taub D., Horuk R. CD4‐independent association between HIV‐1 gp120 and CXCR4: Functional chemokine receptors are expressed in human neurons. Curr. Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- Hioe C.E., Jones G.J., Rees A.D., Ratto‐Kim S., Birx D., Munz C., Gorny M.K., Tuen M., Zolla‐Pazner S. Anti‐CD4‐binding domain antibodies complexed with HIV type 1 glycoprotein 120 inhibit CD4+ T cell‐proliferative responses to glycoprotein 120. AIDS Res. Hum. Retroviruses. 2000;16:893–905. doi: 10.1089/08892220050042837. [DOI] [PubMed] [Google Scholar]
- Hioe C.E., Tuen M., Chien P.C., Jr., Jones G., Ratto‐Kim S., Norris P.J., Moretto W.J., Nixon D.F., Gorny M.K., Zolla‐Pazner S. Inhibition of human immunodeficiency virus type 1 gp120 presentation to CD4 T cells by antibodies specific for the CD4 binding domain of gp120. J. Virol. 2001;75:10950–10957. doi: 10.1128/JVI.75.22.10950-10957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman T.L., LaBranche C.C., Zhang W., Canziani G., Robinson J., Chaiken I., Hoxie J.A., Doms R.W. Stable exposure of the coreceptor‐binding site in a CD4‐independent HIV‐1 envelope protein. Proc. Natl. Acad. Sci. USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann‐Lehmann R., Rasmussen R.A., Vlasak J., Smith B.A., Baba T.W., Liska V., Montefiori D.C., McClure H.M., Anderson D.C., Bernacky B.J., Rizvi T.A., Schmidt R. Passive immunization against oral AIDS virus transmission: An approach to prevent mother‐to‐infant HIV‐1 transmission? J. Med. Primatol. 2001;30:190–196. doi: 10.1034/j.1600-0684.2001.d01-52.x. [DOI] [PubMed] [Google Scholar]
- Hofmann‐Lehmann R., Vlasak J., Rasmussen R.A., Smith B.A., Baba T.W., Liska V., Ferrantelli F., Montefiori D.C., McClure H.M., Anderson D.C., Bernacky B.J., Rizvi T.A. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian‐human immunodeficiency virus challenge. J. Virol. 2001;75:7470–7480. doi: 10.1128/JVI.75.16.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann‐Lehmann R., Vlasak J., Rasmussen R.A., Jiang S., Li P.L., Baba T.W., Montefiori D.C., Bernacky B.J., Rizvi T.A., Schmidt R., Hill L.R., Keeling M.E. Postnatal pre‐ and postexposure passive immunization strategies: Protection of neonatal macaques against oral simian‐human immunodeficiency virus challenge. J. Med. Primatol. 2002;31:109–119. doi: 10.1034/j.1600-0684.2002.01014.x. [DOI] [PubMed] [Google Scholar]
- Holl V., Hemmerter S., Burrer R., Schmidt S., Bohbot A., Aubertin A.M., Moog C. Involvement of Fc gamma RI (CD64) in the mechanism of HIV‐1 inhibition by polyclonal IgG purified from infected patients in cultured monocyte‐derived macrophages. J. Immunol. 2004;173:6274–6283. doi: 10.4049/jimmunol.173.10.6274. [DOI] [PubMed] [Google Scholar]
- Holl V., Peressin M., Decoville T., Schmidt S., Zolla‐Pazner S., Aubertin A.M., Moog C. Nonneutralizing antibodies are able to inhibit human immunodeficiency virus type 1 replication in macrophages and immature dendritic cells. J. Virol. 2006;80:6177–6181. doi: 10.1128/JVI.02625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holl V., Peressin M., Schmidt S., Decoville T., Zolla‐Pazner S., Aubertin A.M., Moog C. Efficient inhibition of HIV‐1 replication in human immature monocyte‐derived dendritic cells by purified anti‐HIV‐1 IgG without induction of maturation. Blood. 2006;107:4466–4474. doi: 10.1182/blood-2005-08-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.C., Venturi M., Majeed S., Moore M.J., Phogat S., Zhang M.Y., Dimitrov D.S., Hendrickson W.A., Robinson J., Sodroski J., Wyatt R., Choe H. Structural basis of tyrosine sulfation and VH‐gene usage in antibodies that recognize the HIV type 1 coreceptor‐binding site on gp120. Proc. Natl. Acad. Sci. USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.C., Stricher F., Martin L., Decker J.M., Majeed S., Barthe P., Hendrickson W.A., Robinson J., Roumestand C., Sodroski J., Wyatt R., Shaw G.M. Scorpion‐toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure (Camb.) 2005;13:755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Huang C.C., Tang M., Zhang M.Y., Majeed S., Montabana E., Stanfield R.L., Dimitrov D.S., Korber B., Sodroski J., Wilson I.A., Wyatt R., Kwong P.D. Structure of a V3‐containing HIV‐1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E., Swanstrom R. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- Hutto C., Zhou Y., He J., Geffin R., Hill M., Scott W., Wood C. Longitudinal studies of viral sequence, viral phenotype, and immunologic parameters of human immunodeficiency virus type 1 infection in perinatally infected twins with discordant disease courses. J. Virol. 1996;70:3589–3598. doi: 10.1128/jvi.70.6.3589-3598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens W., Heyndrickx L., Fransen K., Motte J., Peeters M., Nkengasong J.N., Ndumbe P.M., Delaporte E., Perret J.L., Atende C. Genetic and phylogenetic analysis of env subtypes G and H in central Africa. AIDS Res. Hum. Retroviruses. 1994;10:877–879. doi: 10.1089/aid.1994.10.877. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Desrosiers R.C. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 2002;53:499–518. doi: 10.1146/annurev.med.53.082901.104053. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Morgan J., Reitter J., Puffer B.A., Czajak S., Doms R.W., Desrosiers R.C. A replication‐competent, neutralization‐sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 2002;76:2075–2086. doi: 10.1128/jvi.76.5.2075-2086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Lifson J.D., Lang S.M., Johnson R.P., Desrosiers R.C. Importance of B‐cell responses for immunological control of variant strains of simian immunodeficiency virus. J. Virol. 2003;77:375–381. doi: 10.1128/JVI.77.1.375-381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Sanford H., Schwall L., Burton D.R., Parren P.W., Robinson J.E., Desrosiers R.C. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 2003;77:9993–10003. doi: 10.1128/JVI.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadow J., Wang H.G., Lin P.F. Small‐molecule HIV‐1 gp120 inhibitors to prevent HIV‐1 entry: An emerging opportunity for drug development. Curr. Opin. Investig. Drugs. 2006;7:721–726. [PubMed] [Google Scholar]
- Kang C.Y., Hariharan K., Nara P.L., Sodroski J., Moore J.P. Immunization with a soluble CD4‐gp120 complex preferentially induces neutralizing anti‐human immunodeficiency virus type 1 antibodies directed to conformation‐dependent epitopes of gp120. J. Virol. 1994;68:5854–5862. doi: 10.1128/jvi.68.9.5854-5862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensinger R.D., Yowler B.C., Benesi A.J., Schengrund C.L. Synthesis of novel, multivalent glycodendrimers as ligands for HIV‐1 gp120. Bioconjug. Chem. 2004;15:349–358. doi: 10.1021/bc034156a. [DOI] [PubMed] [Google Scholar]
- Kimata J.T., Gosink J.J., Kewalramani V.N., Rudensey L.M., Littman D.R., Overbaugh J. Coreceptor specificity of temporal variants of simian immunodeficiency virus Mne. J. Virol. 1999;73:1655–1660. doi: 10.1128/jvi.73.2.1655-1660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata J.T., Kuller L., Anderson D.B., Dailey P., Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- Klatzmann D., Champagne E., Chamaret S., Gruest J., Guetard D., Hercend T., Gluckman J.‐C., Montagnierce L. T‐lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Kloosterboer N., Groeneveld P.H., Jansen C.A., van der Vorst T.J., Koning F., Winkel C.N., Duits A.J., Miedema F., van Baarle D., van Rij R.P., Brinkman K., Schuitemaker H. Natural controlled HIV infection: Preserved HIV‐specific immunity despite undetectable replication competent virus. Virology. 2005;339:70–80. doi: 10.1016/j.virol.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Koch M., Pancera M., Kwong P.D., Kolchinsky P., Grundner C., Wang L., Hendrickson W.A., Sodroski J., Wyatt R. Structure‐based, targeted deglycosylation of HIV‐1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Koff W.C., Johnson P.R., Watkins D.I., Burton D.R., Lifson J.D., Hasenkrug K.J., McDermott A.B., Schultz A., Zamb T.J., Boyle R., Desrosiers R.C. HIV vaccine design: Insights from live attenuated SIV vaccines. Nat. Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P., Kiprilov E., Sodroski J. Increased neutralization sensitivity of CD4‐independent human immunodeficiency virus variants. J. Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B.T., Kunstman K.J., Patterson B.K., Furtado M., McEvilly M.M., Levy R., Wolinsky S.M. Genetic differences between blood‐ and brain‐derived viral sequences from human immunodeficiency virus type 1‐infected patients: Evidence of conserved elements in the V3 region of the envelope protein of brain‐derived sequences. J. Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D. Functional constraints and antibody recognition of the CD4‐binding site on HIV‐1 gp120. Fifth International Conference, AIDS Vaccine 06, 29 August–1 September 2006, Amsterdam, the Netherlands; 2006. [Google Scholar]
- Kwong P.D., Ryu S.‐E., Henrickson W., Axel R., Sweet R., Folena‐Wasserman G., Hensley P. Molecular characteristics of recombinant human CD4 as deduced from polymorphic crystals. Proc. Natl. Acad. Sci. USA. 1990;87:6423–6427. doi: 10.1073/pnas.87.16.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt R., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt R., Majeed S., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structures of HIV‐1 gp120 envelope glycoproteins from laboratory‐adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong P.D., Doyle M.L., Casper D.J., Cicala C., Leavitt S.A., Majeed S., Steenbeke T.D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P.W. HIV‐1 evades antibody‐mediated neutralization through conformational masking of receptor‐binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Labrijn A.F., Poignard P., Raja A., Zwick M.B., Delgado K., Franti M., Binley J., Vivona V., Grundner C., Huang C.C., Venturi M., Petropoulos C.J. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange G., Lewis S.J., Murshudov G.N., Dodson G.G., Moody P.C., Turkenburg J.P., Barclay A.N., Brady R.L. Crystal structure of an extracellular fragment of the rat CD4 receptor containing domains 3 and 4. Structure. 1994;2:469–481. doi: 10.1016/s0969-2126(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Lapham C.K., Ouyang J., Chandrasekhar B., Nguyen N.Y., Dimitrov D.S., Golding H. Evidence for cell‐surface association between fusin and the CD4‐gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- Lapham C.K., Zaitseva M.B., Lee S., Romanstseva T., Golding H. Fusion of monocytes and macrophages with HIV‐1 correlates with biochemical properties of CXCR4 and CCR5. Nat. Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- Lefrancois L. Protection against lethal viral infection by neutralizing and nonneutralizing monoclonal antibodies: Distinct mechanisms of action in vivo. J. Virol. 1984;51:208–214. doi: 10.1128/jvi.51.1.208-214.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T., Alaeus A., Marquina S., Lilja E., Lidman K., Albert J. Yet another subtype of HIV type 1? AIDS Res. Hum. Retroviruses. 1995;11:995–997. doi: 10.1089/aid.1995.11.995. [DOI] [PubMed] [Google Scholar]
- Letvin N.L. Progress toward an HIV vaccine. Annu. Rev. Med. 2005;56:213–223. doi: 10.1146/annurev.med.54.101601.152349. [DOI] [PubMed] [Google Scholar]
- Letvin N.L., Huang Y., Chakrabarti B.K., Xu L., Seaman M.S., Beaudry K., Korioth‐Schmitz B., Yu F., Rohne D., Martin K.L., Miura A., Kong W.P. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J. Virol. 2004;78:7490–7497. doi: 10.1128/JVI.78.14.7490-7497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y., Srivastava I., Gomez‐Roman V.R., Zur Megede J., Sun Y., Kan E., Hilt S., Engelbrecht S., Himathongkham S., Luciw P.A., Otten G., Ulmer J.B. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J. Virol. 2005;79:13338–13349. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Alam S.M., Mascola J.R., Robinson J., Ma B., Montefiori D.C., Rhein M., Sutherland L.L., Scearce R., Haynes B.F. Immunogenicity of constrained monoclonal antibody A32‐human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 2004;78:5270–5278. doi: 10.1128/JVI.78.10.5270-5278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Baribaud F., Romano J., Doms R.W., Hoxie J.A. Identification of gp120 binding sites on CXCR4 by using CD4‐independent human immunodeficiency virus type 2 Env proteins. J. Virol. 2003;77:931–942. doi: 10.1128/JVI.77.2.931-942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman D.R., Maddon P.J., Axel R. Corrected CD4 sequence. Cell. 1988;55:541. doi: 10.1016/0092-8674(88)90211-5. [DOI] [PubMed] [Google Scholar]
- Liu S., Jiang S. High throughput screening and characterization of HIV‐1 entry inhibitors targeting gp41: Theories and techniques. Curr. Pharm. Des. 2004;10:1827–1843. doi: 10.2174/1381612043384466. [DOI] [PubMed] [Google Scholar]
- Louwagie J., McCutchan F.E., Peeters M., Brennan T.P., Sanders‐Buell E., Eddy G.A., Fransen G.G., van der K., Gershy‐Damet G.M., Deleys R. Phylogenetic analysis of gag genes from 70 international HIV‐1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- Lu M., Blacklow S.C., Kim P.S. A trimeric structural domain of the HIV‐1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Lu Z., Berson J.F., Chen Y., Turner J.D., Zhang T., Sharron M., Jenks M.H., Wang Z., Kim J., Rucker J., Hoxie J.A., Peiper S.C. Evolution of HIV‐1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciw P.A., Shaw K.E., Unger R.E., Planelles V., Stout M.W., Lackner J.E., Pratt‐Lowe E., Leung N.J., Banapour B., Marthas M.L. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res. Hum. Retroviruses. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- Luo M., Yuan F., Liu Y., Jiang S., Song X., Jiang P., Yin X., Ding M., Deng H. Induction of neutralizing antibody against human immunodeficiency virus type 1 (HIV‐1) by immunization with gp41 membrane‐proximal external region (MPER) fused with porcine endogenous retrovirus (PERV) p15E fragment. Vaccine. 2006;24:435–442. doi: 10.1016/j.vaccine.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Maddon P., Littman D., Godfrey M., Maddon D., Chess L., Axel R. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: A new member of the immunoglobulin gene family. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- Malashkevich V.N., Chan D.C., Chutkowski C.T., Kim P.S. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: Conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan R.M., Cohen F.S., Melikyan G.B. HIV‐1 envelope proteins complete their folding into six‐helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 2003;14:926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic I. Advances in HIV‐1 entry inhibitors: Strategies to interfere with receptor and coreceptor engagement. Curr. Pharm. Des. 2006;12:1105–1119. doi: 10.2174/138161206776055822. [DOI] [PubMed] [Google Scholar]
- Markovic I., Clouse K.A. Recent advances in understanding the molecular mechanisms of HIV‐1 entry and fusion: Revisiting current targets and considering new options for therapeutic intervention. Curr. HIV Res. 2004;2:223–234. doi: 10.2174/1570162043351327. [DOI] [PubMed] [Google Scholar]
- Martin L., Stricher F., Misse D., Sironi F., Pugniere M., Barthe P., Prado‐Gotor R., Freulon I., Magne X., Roumestand C., Menez A., Lusso P. Rational design of a CD4 mimic that inhibits HIV‐1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Martin L., Stricher F., Misse D., Sironi F., Pugniere M., Barthe P., Prado‐Gotor R., Freulon I., Magne X., Roumestand C., Menez A., Lusso P. Rational design of a CD4 mimic that inhibits HIV‐1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Mascola J.R., Lewis M.G., Stiegler G., Harris D., VanCott T.C., Hayes D., Louder M.K., Brown C.R., Sapan C.V., Frankel S.S., Lu Y., Robb M.L. Protection of macaques against pathogenic simian/human immunodefiency virus 89. 6PD by passive transfer of neutralizing antibodies. J. Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Stiegler G., VanCott T.C., Katinger H., Carpenter C.B., Hanson C.E., Beary H., Hayes D., Frankel S.S., Birx D.L., Lewis M.G. Protection of macaques against vaginal transmission of a pathogenic HIV‐1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- Mascola J.R., Lewis M.G., VanCott T.C., Stiegler G., Katinger H., Seaman M., Beaudry K., Barouch D.H., Korioth‐Schmitz B., Krivulka G., Sambor A., Welcher B. Cellular immunity elicited by human immunodeficiency virus type 1/simian immunodeficiency virus DNA vaccination does not augment the sterile protection afforded by passive infusion of neutralizing antibodies. J. Virol. 2003;77:10348–10356. doi: 10.1128/JVI.77.19.10348-10356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Lewis M.G., VanCott T.C., Stiegler G., Katinger H., Seaman M., Beaudry K., Barouch D.H., Korioth‐Schmitz B., Krivulka G., Sambor A., Welcher B. Cellular immunity elicited by human immunodeficiency virus type 1/simian immunodeficiency virus DNA vaccination does not augment the sterile protection afforded by passive infusion of neutralizing antibodies. J. Virol. 2003;77:10348–10356. doi: 10.1128/JVI.77.19.10348-10356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews J.H., Roehrig J.T. Determination of the protective epitopes on the glycoproteins of Venezuelan equine encephalomyelitis virus by passive transfer of monoclonal antibodies. J. Immunol. 1982;129:2763–2767. [PubMed] [Google Scholar]
- Mc Cann C.M., Song R.J., Ruprecht R.M. Antibodies: Can they protect against HIV infection? Curr. Drug Targets Infect. Disord. 2005;5:95–111. doi: 10.2174/1568005054201580. [DOI] [PubMed] [Google Scholar]
- McMichael A.J. HIV vaccines. Annu. Rev. Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- Melikyan G.B., Markosyan R.M., Hemmati H., Delmedico M.K., Lambert D.M., Cohen F.S. Evidence that the transition of HIV‐1 gp41 into a six‐helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison N.A., Sattentau Q. Fundamental immunology and what it can teach us about HIV vaccine development. Curr. Drug Targets Infect. Disord. 2005;5:87–93. doi: 10.2174/1568005054201562. [DOI] [PubMed] [Google Scholar]
- Montefiori D.C., Zhou I.Y., Barnes B., Lake D., Hersh E.M., Masuho Y., Lefkowitz L.B., Jr. Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology. 1991;182:635–643. doi: 10.1016/0042-6822(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Montefiori D.C., Pantaleo G., Fink L.M., Zhou J.T., Zhou J.Y., Bilska M., Miralles G.D., Fauci A.S. Neutralizing and infection‐enhancing antibody responses to human immunodeficiency virus type 1 in long‐term nonprogressors. J. Infect. Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- Moore P.L., Crooks E.T., Porter L., Zhu P., Cayanan C.S., Grise H., Corcoran P., Zwick M.B., Franti M., Morris L., Roux K.H., Burton D.R. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Yasutomi Y., Ohgimoto S., Nakasone T., Takamura S., Shioda T., Nagai Y. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: Robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild‐type strain. J. Virol. 2001;75:4023–4028. doi: 10.1128/JVI.75.9.4023-4028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M., Phogat S.K., Shu Y., Labrijn A.F., Xiao X.D., Binley J.M., Zhang M.Y., Sidorov I.A., Broder C.C., Robinson J., Parren P.W.H.I., Burton D.R. Broadly cross‐reactive HIV‐1‐neutralizing human monoclonal Fab selected for binding to gp120‐CD4‐CCR5 complexes. Proc. Natl. Acad. Sci. USA. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Ruker F., Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G., MacInnes K., Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res. Hum. Retroviruses. 1992;8:373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- Myers G., Korber B., Wain‐Hobson S., Jeang K.T., Henderson L.E., Pavlakis G.N. “Human Retroviruses and AIDS: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences.”. Los Alamos National Laboratory; Los Alamos, NM: 1994. [Google Scholar]
- Nguyen M., Pean P., Lopalco L., Nouhin J., Phoung V., Ly N., Vermisse P., Henin Y., Barre‐Sinoussi F., Burastero S.E., Reynes J.M., Carcelain G. HIV‐specific antibodies but not T‐cell responses are associated with protection in seronegative partners of HIV‐1‐infected individuals in Cambodia. J. Acquir. Immune Defic. Syndr. 2006;42:412–419. doi: 10.1097/01.qai.0000222289.97825.35. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Igarashi T., Haigwood N., Sadjadpour R., Plishka R.J., Buckler‐White A., Shibata R., Martin M.A. Determination of a statistically valid neutralization titer in plasma that confers protection against simian‐human immunodeficiency virus challenge following passive transfer of high‐titered neutralizing antibodies. J. Virol. 2002;76:2123–2130. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G., Tang M., Sambor A., Katinger H., Mascola J.R., Wyatt R., Kwong P.D. Structure and mechanistic analysis of the anti‐human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R., Nair B.C., Hoke G.M., Sarngadharan M.G., Edidin M. Lateral diffusion of CD4 on the surface of a human neoplastic T‐cell line probed with a fluorescent derivative of the envelope glycoprotein (gp120) of human immunodeficiency virus type 1 (HIV‐1) J. Cell. Physiol. 1991;147:326–332. doi: 10.1002/jcp.1041470219. [DOI] [PubMed] [Google Scholar]
- Pancera M., Lebowitz J., Schon A., Zhu P., Freire E., Kwong P.D., Roux K.H., Sodroski J., Wyatt R. Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: Characterization and ligand binding analysis. J. Virol. 2005;79:9954–9969. doi: 10.1128/JVI.79.15.9954-9969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R., Ollmann S.E., Poignard P., Parren P.W., Wilson I.A., Burton D.R. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P.W., Burton D.R. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P.W., Moore J.P., Burton D.R., Sattentau Q.J. The neutralizing antibody response to HIV‐1: Viral evasion and escape from humoral immunity. AIDS. 1999;13(suppl. A):S137–S162. [PubMed] [Google Scholar]
- Parren P.W.H.I., Marx P.A., Hessell A.J., Luckay A., Harouse J., Cheng‐Mayer C., Moore J.P., Burton D.R. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C., Weiser B., Barassi C., Uberti‐Foppa C., Ghezzi S., Longhi R., Calori G., Burger H., Kemal K., Poli G., Lazzarin A., Lopalco L. Long‐lasting CCR5 internalization by antibodies in a subset of long‐term nonprogressors: A possible protective effect against disease progression. Blood. 2006;107:4825–4833. doi: 10.1182/blood-2005-06-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard L., Wilkinson D.A., McKnight A., Gray P.W., Hoxie J.A., Clapham P.R., Weiss R.A. Role of the amino‐terminal extracellular domain of CXCR‐4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- Pierson T.C., Doms R.W. HIV‐1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 2003;281:1–27. doi: 10.1007/978-3-642-19012-4_1. [DOI] [PubMed] [Google Scholar]
- Pierson T.C., Doms R.W. HIV‐1 entry inhibitors: New targets, novel therapies. Immunol. Lett. 2003;85:113–118. doi: 10.1016/s0165-2478(02)00235-3. [DOI] [PubMed] [Google Scholar]
- Pilgrim A.K., Pantaleo G., Cohen O.J., Fink L.M., Zhou J.Y., Zhou J.T., Bolognesi D.P., Fauci A.S., Montefiori D.C. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long‐term‐nonprogressive infection. J. Infect. Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt E.J., Durnin J.P., Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 2005;79:4347–4356. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskoff O., Treboute C., Alizon M. The cytomegalovirus‐encoded chemokine receptor US28 can enhance cell‐cell fusion mediated by different viral proteins. J. Virol. 1998;72:6389–6397. doi: 10.1128/jvi.72.8.6389-6397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P., Saphire E.O., Parren P.W., Burton D.R. Gp120: Biologic aspects of structural features. Annu. Rev. Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor‐binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Wu Y.Q., Zhang M.Y., Dimitrov D.S., Ji X. Structural mimicry of CD4 by a cross‐reactive HIV‐1 neutralizing antibody with CDR‐H2 and H3 containing unique motifs. J. Mol. Biol. 2006;357:82–99. doi: 10.1016/j.jmb.2005.12.062. [DOI] [PubMed] [Google Scholar]
- Puffer B.A., Sharron M., Coughlan C.M., Baribaud F., McManus C.M., Lee B., David J., Price K., Horuk R., Tsang M., Doms R.W. Expression and coreceptor function of APJ for primate immunodeficiency viruses. Virology. 2000;276:435–444. doi: 10.1006/viro.2000.0557. [DOI] [PubMed] [Google Scholar]
- Puffer B.A., Pohlmann S., Edinger A.L., Carlin D., Sanchez M.D., Reitter J., Watry D.D., Fox H.S., Desrosiers R.C., Doms R.W. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 2002;76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkonen P., Thorstensson R., Ghavamzadeh L., Albert J., Hild K., Biberfeld G., Norrby E. Prevention of HIV‐2 and SIVsm infection by passive immunization in cynomolgus monkeys. Nature. 1991;352:436–438. doi: 10.1038/352436a0. [DOI] [PubMed] [Google Scholar]
- Quinnan G.V., Zhang P.F., Fu D.W., Dong M., Alter H.J. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retroviruses. 1999;15:561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- Quinones‐Kochs M.I., Buonocore L., Rose J.K. Role of N‐linked glycans in a human immunodeficiency virus envelope glycoprotein: Effects on protein function and the neutralizing antibody response. J. Virol. 2002;76:4199–4211. doi: 10.1128/JVI.76.9.4199-4211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat S.S., Viard M., Gallo S.A., Rein A., Blumenthal R., Puri A. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review) Mol. Membr. Biol. 2003;20:243–254. doi: 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- Ray N., Doms R.W. HIV‐1 coreceptors and their inhibitors. Curr. Top. Microbiol. Immunol. 2006;303:97–120. doi: 10.1007/978-3-540-33397-5_5. [DOI] [PubMed] [Google Scholar]
- Reeves J.D., Doms R.W. Human immunodeficiency virus type 2. J. Gen. Virol. 2002;83:1253–1265. doi: 10.1099/0022-1317-83-6-1253. [DOI] [PubMed] [Google Scholar]
- Reinherz E.L., Schlossmann S.F. The differentiation and function of human T lymphocytes. Cell. 1980;19:821–827. doi: 10.1016/0092-8674(80)90072-0. [DOI] [PubMed] [Google Scholar]
- Reinherz E.L., Kung P.C., Goldstein G., Schlossmann S.F. Separation of functional subsets of human T cells by a monoclonal antibody. Proc. Natl. Acad. Sci. USA. 1979;76:4061–4065. doi: 10.1073/pnas.76.8.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitter J.N., Means R.E., Desrosiers R.C. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- Reitz M.S., Jr., Wilson C., Naugle C., Gallo R.C., Robert‐Guroff M. Generation of a neutralization‐resistant variant of HIV‐1 is due to selection for a point mutation in the envelope gene. Cell. 1988;54:57–63. doi: 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roben P., Moore J.P., Thali M., Sodroski J., Barbas C.F., III, Burton D.R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.E., Yoshiyama H., Holton D., Elliott S., Ho D.D. Distinct antigenic sites on HIV gp120 identified by a panel of human monoclonal antibodies (abstr. Q449) J. Cell. Biochem. 1992;(Suppl. 16E):71. [Google Scholar]
- Root M.J., Steger H.K. HIV‐1 gp41 as a target for viral entry inhibition. Curr. Pharm. Des. 2004;10:1805–1825. doi: 10.2174/1381612043384448. [DOI] [PubMed] [Google Scholar]
- Rucker J., Samson M., Doranz B.J., Libert F., Berson J.F., Yi Y., Smyth R.J., Collman R.G., Broder C.C., Vassart G., Doms R.W., Parmentier M. Regions in beta‐chemokine receptors CCR5 and CCR2b that determine HIV‐1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- Ruprecht R.M., Hofmann‐Lehmann R., Smith‐Franklin B.A., Rasmussen R.A., Liska V., Vlasak J., Xu W., Baba T.W., Chenine A.L., Cavacini L.A., Posner M.R., Katinger H. Protection of neonatal macaques against experimental SHIV infection by human neutralizing monoclonal antibodies. Transfus. Clin. Biol. 2001;8:350–358. doi: 10.1016/s1246-7820(01)00187-2. [DOI] [PubMed] [Google Scholar]
- Ryu S.‐E., Kwong P.D., Truneh A., Porter T.G., Arthos J., Rosenberg M., Dai X., Xuong N., Axel R., Sweet R.W., Hendrickson W.A. Crystal structure of an HIV‐binding recombinant fragment of human CD4. Nature. 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K.O., Kwong P.D., Moore J.P. The mannose‐dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E.O., Parren P.W., Pantophlet R., Zwick M.B., Morris G.M., Rudd P.M., Dwek R.A., Stanfield R.L., Burton D.R., Wilson I.A. Crystal structure of a neutralizing human IGG against HIV‐1: A template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Scanlan C.N., Pantophlet R., Wormald M.R., Ollmann S.E., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., Burton D.R. The broadly neutralizing anti‐human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1‐‐>2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti G., Leitner T., Hodara V., Jansson M., Karlsson A., Wahlberg J., Rossi P., Uhlen M., Fenyo E.M., Albert J. Interplay of HIV‐1 phenotype and neutralizing antibody response in pathogenesis of AIDS. Immunol. Lett. 1996;51:23–28. doi: 10.1016/0165-2478(96)02550-3. [DOI] [PubMed] [Google Scholar]
- Selvarajah S., Puffer B., Pantophlet R., Law M., Doms R.W., Burton D.R. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 2005;79:12148–12163. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.A., Kan E., Sun Y., Lian Y., Cisto J., Frasca V., Hilt S., Stamatatos L., Donnelly J.J., Ulmer J.B., Barnett S.W., Srivastava I.K. Structural characteristics correlate with immune responses induced by HIV envelope glycoprotein vaccines. Virology. 2006;352:131–144. doi: 10.1016/j.virol.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Sharron M., Pohlmann S., Price K., Lolis E., Tsang M., Kirchhoff F., Doms R.W., Lee B. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood. 2000;96:41–49. [PubMed] [Google Scholar]
- Shibata R., Igarashi T., Haigwood N., Buckler‐White A., Ogert R., Ross W., Willey R., Cho M.W., Martin M.A. Neutralizing antibody directed against the HIV‐1 envelope glycoprotein can completely block HIV‐1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- Simon F., Mauclere P., Roques P., Loussert‐Ajaka I., Muller‐Trutwin M.C., Saragosti S., Georges‐Courbot M.C., Barre‐Sinoussi F., Brun‐Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- Singh M., Jeang K.T., Smith S.M. HIV vaccine development. Front. Biosci. 2005;10:2064–2081. doi: 10.2741/1682. [DOI] [PubMed] [Google Scholar]
- Sloane A.J., Raso V., Dimitrov D.S., Xiao X., Deo S., Muljadi N., Restuccia D., Turville S., Kearney C., Broder C.C., Zoellner H., Cunningham A.L. Marked structural and functional heterogeneity in CXCR4: Separation of HIV‐1 and SDF‐1alpha responses. Immunol. Cell Biol. 2005;83:129–143. doi: 10.1111/j.1440-1711.2004.01304.x. [DOI] [PubMed] [Google Scholar]
- Slobod K.S., Bonsignori M., Brown S.A., Zhan X., Stambas J., Hurwitz J.L. HIV vaccines: Brief review and discussion of future directions. Expert Rev. Vaccines. 2005;4:305–313. doi: 10.1586/14760584.4.3.305. [DOI] [PubMed] [Google Scholar]
- Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- Sodroski J.G. HIV‐1 entry inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- Spearman P. Current progress in the development of HIV vaccines. Curr. Pharm. Des. 2006;12:1147–1167. doi: 10.2174/138161206776055859. [DOI] [PubMed] [Google Scholar]
- Srivastava I.K., Stamatatos L., Legg H., Kan E., Fong A., Coates S.R., Leung L., Wininger M., Donnelly J.J., Ulmer J.B., Barnett S.W. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 2002;76:2835–2847. doi: 10.1128/JVI.76.6.2835-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava I.K., Ulmer J.B., Barnett S.W. Neutralizing antibody responses to HIV: Role in protective immunity and challenges for vaccine design. Expert Rev. Vaccines. 2004;3:S33–S52. doi: 10.1586/14760584.3.4.s33. [DOI] [PubMed] [Google Scholar]
- Stanley J., Cooper S.J., Griffin D.E. Monoclonal antibody cure and prophylaxis of lethal Sindbis virus encephalitis in mice. J. Virol. 1986;58:107–115. doi: 10.1128/jvi.58.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcich B.R., Hahn B.H., Shaw G.M., McNeely P.D., Modrow S., Wolf H., Parks E.S., Parks W.P., Josephs S.F., Gallo R.C. Identification and characterization of conserved and variable regions in the envelope gene of HTLV‐III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- Staudinger R., Phogat S.K., Xiao X., Wang X., Dimitrov D.S., Zolla‐Pazner S. Evidence for CD4‐enchanced signaling through the chemokine receptor CCR5. J. Biol. Chem. 2003;278:10389–10392. doi: 10.1074/jbc.M212013200. [DOI] [PubMed] [Google Scholar]
- Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., Katinger H. A potent cross‐clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Subramaniam S. The SIV surface spike imaged by electron tomography: One leg or three? PLoS Pathog. 2006;2(8):e91. doi: 10.1371/journal.ppat.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Miyagi T., Chuang L.F., Yau P.M., Doi R.H., Chuang R.Y. Chemokine receptor CCR5: Polymorphism at protein level. Biochem. Biophys. Res. Commun. 2002;296:477–483. doi: 10.1016/s0006-291x(02)00908-7. [DOI] [PubMed] [Google Scholar]
- Tian S., Choi W.T., Liu D., Pesavento J., Wang Y., An J., Sodroski J.G., Huang Z. Distinct functional sites for human immunodeficiency virus type 1 and stromal cell‐derived factor 1alpha on CXCR4 transmembrane helical domains. J. Virol. 2005;79:12667–12673. doi: 10.1128/JVI.79.20.12667-12673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A., Dragic T., Arthos J., Binley J.M., Olson W.C., Allaway G.P., Cheng‐Mayer C., Robinson J., Maddon P.J., Moore J.P. CD4‐dependent, antibody‐sensitive interactions between HIV‐1 and its co‐receptor CCR‐5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J.P., Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuen M., Visciano M.L., Chien P.C., Jr., Cohen S., Chen P.D., Robinson J., He Y., Pinter A., Gorny M.K., Hioe C.E. Characterization of antibodies that inhibit HIV gp120 antigen processing and presentation. Eur. J. Immunol. 2005;35:2541–2551. doi: 10.1002/eji.200425859. [DOI] [PubMed] [Google Scholar]
- Van Rompay K.K., Berardi C.J., Dillard T., Tarara R.P., Canfield D.R., Valverde C.R., Montefiori D.C., Cole K.S., Montelaro R.C., Miller C.J. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J. Infect. Dis. 1998;177:1247–1259. doi: 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- Varadarajan R., Sharma D., Chakraborty K., Patel M., Citron M., Sinha P., Yadav R., Rashid U., Kennedy S., Eckert D., Geleziunas R., Bramhill D. Characterization of gp120 and its single‐chain derivatives, gp120‐CD4D12 and gp120‐M9: Implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J. Virol. 2005;79:1713–1723. doi: 10.1128/JVI.79.3.1713-1723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita C., Drakopoulou E., Vizzavona J., Rochette S., Martin L., Menez A., Roumestand C., Yang Y.S., Ylisastigui L., Benjouad A., Gluckman J.C. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV‐1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 1999;96:13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita C., Drakopoulou E., Vizzavona J., Rochette S., Martin L., Menez A., Roumestand C., Yang Y.S., Ylisastigui L., Benjouad A., Gluckman J.C. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV‐1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 1999;96:13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic L.K., Quinnan G.V., Jr. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res. Hum. Retroviruses. 1995;11:783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- Wang J., Yan Y., Garrett T.P.J., Liu J., Rodgers D.W., Garlick R.L., Tarr G.E., Husain Y., Reinherz E.L., Harrison S.C. Atomic structure of a fragment of human CD4 containing two immunoglobulin‐like domains. Nature. 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- Wang L.X. Toward oligosaccharide‐ and glycopeptide‐based HIV vaccines. Curr. Opin. Drug Discov. Devel. 2006;9:194–206. [PubMed] [Google Scholar]
- Watkins B.A., Reitz M.S., Jr., Wilson C.A., Aldrich K., Davis A.E., Robert‐Guroff M. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: Evidence for multiple pathways. J. Virol. 1993;67:7493–7500. doi: 10.1128/jvi.67.12.7493-7500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar‐Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., Komarova N.L., Nowak M.A. Antibody neutralization and escape by HIV‐1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Weiss C.D. HIV‐1 gp41: Mediator of fusion and target for inhibition. AIDS Rev. 2003;5:214–221. [PubMed] [Google Scholar]
- Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV‐1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson R.A., Piscitelli C., Teintze M., Cavacini L.A., Posner M.R., Lawrence C.M. Structure of the Fab fragment of F105, a broadly reactive anti‐human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120. J. Virol. 2005;79:13060–13069. doi: 10.1128/JVI.79.20.13060-13069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.F., Galfre G., Milstein C. Analysis of cell surfaces by xenogeneic myeloma‐hybrid antibodies: Differentiation antigens of rat lymphocytes. Cell. 1977;12:663–673. doi: 10.1016/0092-8674(77)90266-5. [DOI] [PubMed] [Google Scholar]
- Wu H., Kwong P.D., Hendrickson W.A. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- Wu L.J., Gerard N.P., Wyatt R., Choe H., Parolin C., Ruffing N., Borsetti A., Cardoso A.A., Desjardin E., Newman W., Gerard C., Sodroski J. CD4‐induced interaction of primary HIV‐1 gp120 glycoproteins with the chemokine receptor CCR‐5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV‐1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Moore J., Accola M., Desjardin E., Robinson J., Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S.H., Doka N., Choudhary R.K., Sodroski J., Robinson J.E. Characterization of CD4‐induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses. 2002;18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- Xiang S.H., Kwong P.D., Gupta R., Rizzuto C.D., Casper D.J., Wyatt R., Wang L., Hendrickson W.A., Doyle M.L., Sodroski J. Mutagenic stabilization and/or disruption of a CD4‐bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Wu L., Stantchev T.S., Feng Y.R., Ugolini S., Chen H., Shen Z., Riley J.L., Broder C.C., Sattentau Q.J., Dimitrov D.S. Constitutive cell surface association between CD4 and CCR5. Proc. Natl. Acad. Sci. USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Farzan M., Wyatt R., Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2000;74:5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Lee J., Mahony E.M., Kwong P.D., Wyatt R., Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Tomov V., Kurteva S., Wang L., Ren X., Gorny M.K., Zolla‐Pazner S., Sodroski J. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 2004;78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs J.A., Grunewald K., Sattentau Q.J., Fuller S.D. Cryo‐Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex in situ. PLoS Pathog. 2006;2(8):e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.W., Chishti Y., Hussey R.E., Reinherz E.L. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 2001;276:39577–39585. doi: 10.1074/jbc.M107147200. [DOI] [PubMed] [Google Scholar]
- Zhang L., Ribeiro R.M., Mascola J.R., Lewis M.G., Stiegler G., Katinger H., Perelson A.S., Davenport M.P. Effects of antibody on viral kinetics in simian/human immunodeficiency virus infection: Implications for vaccination. J. Virol. 2004;78:5520–5522. doi: 10.1128/JVI.78.10.5520-5522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y., Dimitrov D.S. Novel approaches for identification of broadly cross‐reactive HIV‐1 neutralizing human monoclonal antibodies and improvement of their potency. Curr. Pharm. Des. 2007;13:203–212. doi: 10.2174/138161207779313669. [DOI] [PubMed] [Google Scholar]
- Zhang M.Y., Shu Y., Phogat S., Xiao X., Cham F., Bouma P., Choudhary A., Feng Y.R., Sanz I., Rybak S., Broder C.C., Quinnan G.V. Broadly cross‐reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods. 2003;283:17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Zhang M.Y., Xiao X., Sidorov I.A., Choudhry V., Cham F., Zhang P.F., Bouma P., Zwick M., Choudhary A., Montefiori D.C., Broder C.C., Burton D.R. Identification and characterization of a new cross‐reactive human immunodeficiency virus type 1‐neutralizing human monoclonal antibody. J. Virol. 2004;78:9233–9242. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y., Choudhry V., Sidorov I.A., Tenev V., Vu B.K., Choudhary A., Lu H., Stiegler G.M., Katinger H.W.D., Jiang S., Broder C.C., Dimitrov D.S. Selection of a novel gp41‐specific HIV‐1 neutralizing human antibody by competitive antigen panning. J. Immunol. Methods. 2006;317:21–30. doi: 10.1016/j.jim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.F., Bouma P., Park E.J., Margolick J.B., Robinson J.E., Zolla‐Pazner S., Flora M.N., Quinnan G.V., Jr. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4‐independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross‐reactive, primary virus‐neutralizing antibody response. J. Virol. 2002;76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Canziani G., Plugariu C., Wyatt R., Sodroski J., Sweet R., Kwong P., Hendrickson W., Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Fracasso C., Fiore J.R., Bjorndal A., Angarano G., Gringeri A., Fenyo E.M. Augmented serum neutralizing activity against primary human immunodeficiency virus type 1 (HIV‐1) isolates in two groups of HIV‐1‐infected long‐term nonprogressors. J. Infect. Dis. 1997;176:1180–1187. doi: 10.1086/514111. [DOI] [PubMed] [Google Scholar]
- Zhou T., Xu L., Dey B., Hessel A.J., Van Ryk D., Xiang S.H., Yang X., Zhang M.Y., Zwick M.B., Arthos J., Burton D.R., Dimitrov D.S. Structural definition of a conserved neutralization epitope on HIV‐1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Chertova E., Bess J., Jr., Lifson J.D., Arthur L.O., Liu J., Taylor K.A., Roux K.H. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J.D., Grise H., Ofek G.A., Taylor K.A., Roux K.H. Distribution and three‐dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zolla‐Pazner S. Identifying epitopes of HIV‐1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B. The membrane‐proximal external region of HIV‐1 gp41: A vaccine target worth exploring. AIDS. 2005;19:1725–1737. doi: 10.1097/01.aids.0000189850.83322.41. [DOI] [PubMed] [Google Scholar]
- Zwick M.B., Labrijn A.F., Wang M., Spenlehauer C., Saphire E.O., Binley J.M., Moore J.P., Stiegler G., Katinger H., Burton D.R., Parren P.W. Broadly neutralizing antibodies targeted to the membrane‐proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B., Parren P.W., Saphire E.O., Church S., Wang M., Scott J.K., Dawson P.E., Wilson I.A., Burton D.R. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:5863–5876. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


