Abstract
The Nipah virus fusion (F) protein is proteolytically processed to F1 + F2 subunits. We demonstrate here that cathepsin L is involved in this important maturation event. Cathepsin inhibitors ablated cleavage of Nipah F. Proteolytic processing of Nipah F and fusion activity was dramatically reduced in cathepsin L shRNA-expressing Vero cells. Additionally, Nipah virus F-mediated fusion was inhibited in cathepsin L-deficient cells, but coexpression of cathepsin L restored fusion activity. Both purified cathepsin L and B could cleave immunopurified Nipah F protein, but only cathepsin L produced products of the correct size. Our results suggest that endosomal cathepsins can cleave Nipah F, but that cathepsin L specifically converts Nipah F to a mature and fusogenic form.
Keywords: Nipah, Fusion protein, Cathepsin L, Proteolytic processing
Introduction
Nipah virus was identified as the causative agent of the 1999 Malaysia and Singapore outbreak of febrile viral encephalitis and severe respiratory illness in humans and swine, respectively (Chua et al., 2000). The outbreak was contained with the slaughter of more than one million pigs following the realization that the virus was spread from pigs to humans. Since the initial identification, additional Nipah virus outbreaks in Bangladesh (2001–2004) have been described, with case mortality rates approaching 70% and suspected human-to-human transmission (Hsu et al., 2004). Nipah virus is an enveloped negative-strand RNA virus which together with Hendra virus is classified as a Henipavirus within the Paramyxoviridae family (Harcourt et al., 2000). Although fruit bats are thought to be the natural reservoir for Henipaviruses (Chua et al., 2002), both Nipah and Hendra viruses exhibit broad host tropism (Bossart et al., 2002). It is this broad host range, zoonotic transmission, and high pathogenicity that has led to the classification of these viruses as biosafety level 4 pathogens.
Successful replication and systemic transmission of Nipah virus require viral entry via fusion of the viral envelope with a cellular membrane. This prerequisite first step is facilitated by two viral glycoproteins, namely the attachment (G) protein, which binds the cellular ligand Ephrin-B2 (Bonaparte et al., 2005, Negrete et al., 2005), and the fusion (F) protein. The F protein is synthesized as a precursor protein (F0) that requires posttranslational proteolytic processing to form a disulfide-linked F1 + F2 heterodimer. This essential cleavage event exposes the fusion peptide, thus producing a mature F protein capable of mediating virus–cell and cell–cell membrane fusion. Typically, the paramyxovirus F proteins are processed by either an extracellular, tissue-specific trypsin-like protease after a single basic residue or by furin, a ubiquitous secretory pathway protease, following a multibasic cleavage motif (Garten et al., 1994). Proteolytic processing of Henipavirus F proteins, however, deviates from these established cleavage mechanisms. N-terminal sequencing of the F1 subunits predicted that cleavage occurred after a single basic residue (arginine109 or lysine109 in Nipah and Hendra virus F proteins, respectively) (Michalski et al., 2000, Moll et al., 2004). However, neither these basic residues nor the amino acids immediately upstream of the processing site dictated cleavage specificity (Craft and Dutch, 2005, Moll et al., 2004). Furthermore, activation of the Henipavirus F proteins is furin-independent, requires a low intracellular pH (Diederich et al., 2005, Moll et al., 2004, Pager et al., 2004), and occurs during endocytosis and recycling of the F protein (Diederich et al., 2005, Meulendyke et al., 2005). Recently, we demonstrated that the endosomal protease cathepsin L was involved in the proteolytic processing of the Hendra virus F protein (Pager and Dutch, 2005). However, the protease involved in activation of Nipah F remains to be described.
In this study, we used cysteine protease inhibitors and cell lines deficient in cathepsin L to examine the role of cathepsin L in the proteolytic processing and fusion activity of the Nipah F protein. Nonspecific and specific cathepsin protease inhibitors effectively blocked cleavage of Nipah F. Knockdown of cathepsin L activity via small hairpin RNA (shRNA) reduced processing and cell–cell fusion promoted by Nipah F. Additionally, membrane fusion activity was absent in cathepsin L-deficient mouse embryonic fibroblasts (MEFs) but could be restored upon coexpression of cathepsin L. Finally, purified cathepsin L and B efficiently digested uncleaved Nipah F protein into two fragments, but only the products produced by cathepsin L were the correct size for F1 and F2 subunits. Our data show a role for cathepsin L in maturation and activation of the Nipah virus F protein. Thus, primary proteolytic processing of Henipavirus F proteins occurs via an acid-dependent endosomal protease mechanism.
Results
Cleavage of Nipah F is abrogated in the presence of nonspecific and specific cathepsin inhibitors
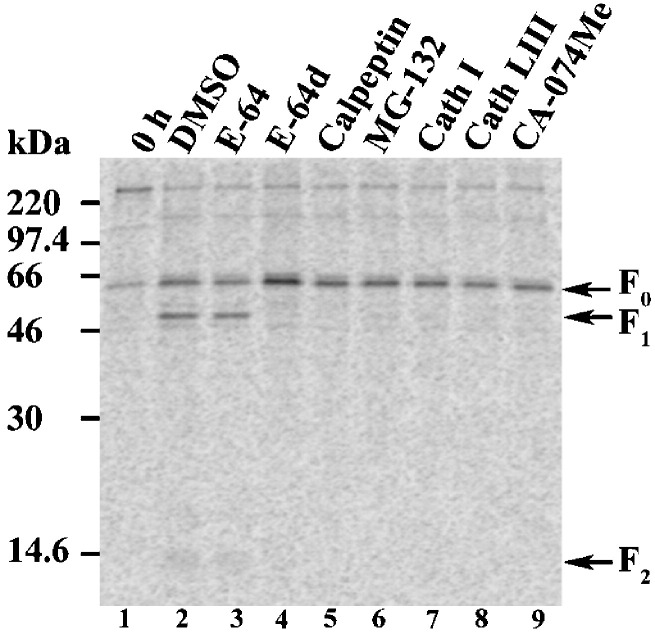
To examine whether an endosomal cysteine cathepsin protease was involved in proteolytic processing of Nipah F, Vero cells transfected with pCAGGS-Nipah F were metabolically labeled and chased in the absence or presence of both nonspecific and specific cathepsin inhibitors. Nipah F was immunoprecipitated with a rabbit polyclonal antibody to inactivated Nipah virus, which efficiently detects the mature Nipah F protein. Immunoprecipitated F proteins were analyzed by 15% SDS-PAGE and visualized using the Typhoon imaging system. Immediately after metabolic labeling, a weak precursor F0 band was seen (Fig. 1 , lane 1), which was subsequently converted during the 2 h chase to the mature F1 + F2 heterodimer (Fig. 1, lane 2). Cleavage was not blocked by the addition of a membrane-impermeable cysteine protease inhibitor, E-64 (Fig. 1, lane 3). However, inhibition of intracellular calpain and cathepsins B, H, and L with E-64d and calpeptin; proteosome and cathepsin L with MG-132; or endosomal cathepsins with general (cathepsin inhibitor I [Cath I]) or cathepsin B/L inhibitors (cathepsin L inhibitor III [Cath LIII] and CA-074Me), effectively abolished proteolytic processing of Nipah F (Fig. 1, lanes 4–9). Correct intracellular trafficking of Nipah F in the absence and presence of select cysteine protease inhibitors was confirmed by endoglycosidase H digestion and biotinylation of cell surface Nipah F (data not shown). These data indicate that inhibition of cathepsin proteases specifically blocked Nipah F processing. Cathepsin L was recently shown to be involved in cleavage of the Hendra virus F protein (Pager and Dutch, 2005). The sensitivity of Nipah F processing to changes in intracellular pH (Diederich et al., 2005) and the similar profile of inhibition with both the nonspecific and specific cathepsin inhibitors strongly suggest that cathepsin L is involved in activation of the Nipah F protein.
Fig. 1.
The addition of nonspecific and specific cysteine cathepsin inhibitors ablate proteolytic processing of Nipah F. Vero cells expressing pCAGGS-Nipah F were starved, labeled with Tran35S label, and chased for 2 h in the absence (DMSO) or presence of 10 μM E-64, E-64d, calpeptin, cathepsin L inhibitor III (Cath LIII), and CA-074Me, 1 μM MG-132, and 50 μM cathepsin inhibitor I (Cath I). Samples were analyzed by immunoprecipitation, 15% SDS-PAGE, and the Typhoon imaging system. Positions of F0, F1, and F2 are indicated.
Nipah F cleavage and fusion activity is suppressed in cathepsin L-deficient cells
Previously, we described the design and utilization of two cathepsin L-specific shRNA oligonucleotides (Pager and Dutch, 2005). In Cath L98-expressing Vero cells, the cathepsin L protein level was undetectable, and only minimal enzyme activity levels remained. In contrast, a small amount of cathepsin L protein and moderately reduced enzyme activity was retained following suppression with the Cath L662 shRNA oligonucleotide. Furthermore, both Cath L98- and Cath L662-expressing Vero cells demonstrated similar cathepsin B enzyme activities as seen in Scramble-expressing and nontransfected Vero cells (Pager and Dutch, 2005). To determine if maturation of Nipah F was effected in Vero cells expressing cathepsin L shRNA oligonucleotides, Nipah F was expressed in both untransfected and shRNA-transfected Vero cells, and F proteolysis assessed by metabolic labeling and immunoprecipitation. Similar to untransfected Vero cells, Nipah F was efficiently processed in cells expressing the Scramble shRNA oligonucleotide (Fig. 2A, lanes 1 and 3). In contrast, Nipah F cleavage was undetectable in Cath L98-expressing Vero cells (Fig. 2A, lane 4). However, compared to untransfected or Scramble shRNA-expressing cells, Nipah F processing was reduced in Cath L662-expressing cells (Fig. 2A, lane 5). These results clearly support a role for cathepsin L in proteolytic processing of the Nipah F protein.
Fig. 2.
Proteolytic processing and membrane fusion activity of Nipah F in cathepsin L shRNA-expressing Vero cells. (A) Expression of Nipah F in shRNA-expressing Vero cells. Vero cells expressing either no shRNA or Scramble-, Cath L98-, or Cath L662-shRNA were transfected with Nipah F, and expression and proteolytic processing analyzed by metabolic labeling and immunoprecipitation as previously described. (B) Luciferase reporter gene assay. Nipah F, Nipah G, and a T7 luciferase reporter gene plasmid were transfected into shRNA-expressing Vero cells. BSR cells, which stably express T7 polymerase, were overlaid onto the Nipah F and G-transfected Vero cells for 3 h at 37 °C. Cells were lysed and luciferase activity was measured. The results presented are representative of three separate experiments, with Nipah G alone set as the background value. Cell–cell fusion values from Cath L98- and Cath L662-expressing Vero cells were normalized to fusion values within Scramble shRNA-expressing cells.
The decreased cathepsin L levels in shRNA knockdown cells impeded proteolytic processing of Nipah F. Therefore, using a quantitative luciferase reporter gene assay, we examined the ability of Nipah F to promote cell–cell fusion in cathepsin L shRNA-expressing Vero cells. ShRNA-expressing Vero cells were transfected with pCAGGS-Nipah F and G and a luciferase reporter gene under the control of a T7 promoter, and fusion quantified (via luciferase production) following overlay of these cells with T7 polymerase-expressing BSR cells (Buchholz et al., 1999). Nipah F and G efficiently promoted cell–cell fusion in Vero cells expressing the Scramble shRNA oligonucleotide (Fig. 2B). In contrast, membrane fusion was significantly reduced in Cath L98 shRNA-expressing Vero cells, in agreement with undetectable levels of Nipah F processing (Fig. 2A, lane 4) and a small amount of cathepsin L enzyme activity remaining (Pager and Dutch, 2005). Consistent with the intermediate level of proteolytic processing of Nipah F in Cath L662-expressing Vero cells (Fig. 2A, lane 5), cell–cell fusion was increased compared to that in Cath L98-expressing cells, but not to the same level as in Scramble-expressing cells (Fig. 2B). Therefore, inhibition of Nipah F proteolytic processing and fusion activity correlated with suppression of cathepsin L expression.
To further analyze the role of cathepsin L in Nipah F-mediated fusion, MEFs generated from either cathepsin L+/+ or cathepsin L−/− mice (Nakagawa et al., 1998) were transfected with pCAGGS-Nipah F and G and the T7 luciferase vector, as well as with an empty vector or a cathepsin L-expressing plasmid (Chapman et al., 1997). To ensure sufficient cathepsin L expression levels, pSG5-cathepsin L was transfected at twice the concentration of Nipah F. Transfected MEFs were overlaid with BSR cells and fusion activity quantified as before. As with cell–cell fusion assays performed in Vero cells, fusion in cathepsin L+/+ MEFs required the presence of both Nipah F and G proteins (Fig. 3 ). In contrast, expression of both Nipah F and G did not lead to fusion in MEFs lacking cathepsin L, yet cell–cell fusion was partially restored upon coexpression of cathepsin L with Nipah F and G (Fig. 3). We additionally verified that both cathepsin L+/+ and cathepsin L−/− MEFs exhibited normal cathepsin B activity (data not shown). These results therefore confirm that cathepsin L is required to facilitate efficient Nipah F-mediated membrane fusion.
Fig. 3.
Nipah F-mediated membrane fusion in cathepsin L+/+ and cathepsin L−/− MEFs. pCAGGS-Nipah F and Nipah G and a T7-luciferase reporter gene plasmid with or without pSG5-cathepsin L vector were transfected into MEFs. MEFs were overlaid with T7 polymerase-expressing BSR cells and the mixed cell populations incubated for 3 h at 37 °C. Cells were lysed and luciferase activity measured on a luminometer. Background values were ascribed to cells expressing only the attachment protein Nipah G and then subtracted from the values for Nipah F and F + G ± cathepsin L. The results presented are representative of six separate experiments.
Endosomal cathepsin proteases can cleave the Nipah virus F protein
Cathepsin protease inhibitors and studies in cathepsin-deficient cells strongly argue that cathepsin L is involved in posttranslational activation of Nipah F. These studies, however, do not directly show that Nipah F is a substrate for cathepsin L. We therefore examined the ability of purified cathepsin L to digest immunopurified uncleaved Nipah F0 protein to F1 + F2 subunits. We previously showed that uncleaved Hendra F was generated by incubating transfected cells at 20 °C. This temperature shift prevents budding of secretory vesicles from the trans-Golgi network and therefore inhibits cleavage of the F protein (Pager et al., 2004). However, this processing block is overcome by returning cells to 37 °C. Similarly, we found that Tran35S-labeled Nipah F was not cleaved when cells were incubated at 20 °C compared to incubation at 37 °C (Fig. 4A, lanes 3 and 2). However, upon returning cells to 37 °C, Nipah F processing was restored (Fig. 4A, lane 4). Therefore, uncleaved Nipah F, produced after metabolic labeling at 20 °C, was immunoprecipitated and used as a substrate for in vitro cleavage with purified cathepsin L. Nipah F0 was not cleaved to F1 and F2 in the absence of cathepsin L (Fig. 4A, lane 5). However, in the presence of cathepsin L, Nipah F0 was efficiently digested to two products consistent in size to the F1 + F2 heterodimer by 4 h, with the amount of processing increasing with time (Fig. 4A, lanes 6–9). Even in the presence of cathepsin L for 8 h, a fraction of immunopurified uncleaved Nipah F remained undigested, suggesting the presence of an uncleavable form of F0 possibly due to aggregation of unfolded protein during sample preparation. Therefore, the Nipah F protein is a physiologically relevant substrate for cathepsin L digestion.
Fig. 4.
Cathepsins L and B can cleave the Nipah F protein. Uncleaved Nipah F was generated by metabolic labeling of Nipah F at 20 °C for 2 h, followed by immunoprecipitation and incubation with 10 nM cathepsin L (A) and 1 nM cathepsin B (B) at 37 °C for 2–8 h. As a control, an uncleaved Nipah F sample was incubated in buffer without enzyme for 8 h at 37 °C. Samples were analyzed by SDS-PAGE and the Typhoon imaging system. Positions of F0, F1, and F2 are indicated.
Numerous cathepsin proteases are found within the endosomal/lysosomal pathway. These may be tissue-specific or ubiquitously expressed (Turk and Guncar, 2003). Since both of the ubiquitously expressed cathepsins L and B have been shown to be involved in proteolytic processing of the reovirus outer capsid proteins (Ebert et al., 2002) and the Ebola virus glycoprotein (GP) (Chandran et al., 2005), we examined the ability of purified cathepsin B to digest the Nipah F protein. Cathepsin B cleaved Nipah F0 into two fragments within 2 h. However, the smaller fragment migrated more slowly than Nipah F cleaved endogenously within the cell (Fig. 4B, lanes 2 and 4 versus lanes 6–9), suggesting that cathepsin B does not cleave Nipah F0 correctly. By 8 h, however, this smaller fragment was somewhat reduced in size, possibly due to the carboxypeptidase activity also present in cathepsin B (Turk and Guncar, 2003). This result contrasts with cathepsin B digestion of Hendra F0, where nonspecific products were generated over time (Pager and Dutch, 2005). Although the Nipah F protein may be digested in vitro by cathepsin B, cleavage and fusion ability of Nipah F in cathepsin L-deficient cells and in vitro digestion of Nipah F by cathespin L suggest that cathepsin L correctly cleaves the Nipah F protein into a form that is fusogenically active.
Discussion
Paramyxovirus F proteins may be cleaved by either the secretory protease furin or extracellular trypsin-like enzymes. Furthermore, the ubiquitously expressed endosomal protease cathepsin L was recently demonstrated to be involved in processing the Hendra virus F protein (Pager and Dutch, 2005). Here we show that cathepsin L is also required for proteolytic processing and activation of the Nipah virus F protein. The addition of nonspecific and specific cathepsin protease inhibitors abrogated cleavage of Nipah F (Fig. 1). Expression of Nipah F in cathepsin L shRNA-expressing Vero cells resulted in significant decreases in Nipah F-mediated membrane fusion (Fig. 2B), which corresponded to undetectable to minimal proteolytic processing of Nipah F in these cells (Fig. 2A). Additionally, efficient membrane fusion was promoted by Nipah F and G in wild type MEFs, yet membrane fusion was not observed in cathepsin L−/− MEFs unless cathepsin L was coexpressed with Nipah F and G proteins (Fig. 3). Finally, both purified cathepsin L and B were able to digest immunopurified uncleaved Nipah F protein; however, cathepsin B appeared to cleave Nipah F0 incorrectly (Fig. 4). Identification of cathepsin L as the intracellular protease involved in processing Nipah F correlates with Nipah F cleavage requiring an acid pH within the endosome (Diederich et al., 2005).
Cathepsin L has historically been thought of as a degradative enzyme (Turk and Guncar, 2003). However, recent reports describe cathepsin L as a proteolytic processing enzyme involved in protein maturation. For example, cathepsin L proteolytically cleaves the CDP/CUX transcription factor (Goulet et al., 2004), enkephalin prohormone (Yasothornsrikul et al., 2003), and a number of viral proteins (Chandran et al., 2005, Ebert et al., 2002, Pager and Dutch, 2005, Simmons et al., 2005). Although cathepsin L does not appear to have an identifiable substrate recognition site, the protease is thought to prefer hydrophobic residues at P2 and P3 substrate sites (Turk and Guncar, 2003). Yet, despite this lack of specificity, cathepsin L has been shown to specifically cleave proenkephalin at dibasic and monobasic cleavage sites (Yasothornsrikul et al., 2003). Cleavage of both Nipah F and Hendra F proteins is predicted to occur after a single basic residue. However, site-directed mutagenesis of individual amino acids immediately upstream of the cleavage site within Nipah F and Hendra F proteins did not ablate Henipavirus F processing, demonstrating that no single residue is required for efficient cleavage (Craft and Dutch, 2005, Moll et al., 2004). Indeed, cleavage was only abolished following deletion of six amino acids immediately upstream of the Nipah F cleavage site (Moll et al., 2004). The ability of cathepsin L to cleave Nipah F correlates with a nebulous cleavage motif within Nipah F and proteolytic processing at arginine109.
The ubiquitously expressed cathepsin L and B have been described to proteolytically process viral proteins, thus promoting entry of both nonenveloped and enveloped viruses. Cathepsin L and B were first described to facilitate entry of nonenveloped reoviruses after receptor-mediated endocytosis by degrading the outer capsid proteins to form infectious subvirion particles capable of transversing the endosomal membrane (Ebert et al., 2002). During exocytic transport, the Ebola virus GP is proteolytically cleaved by furin to GP1 and GP2 (Volchkov et al., 1998). However, entry of enveloped Ebola virus requires that GP1 is further processed by cathepsin B and/or L (Chandran et al., 2005). Finally, a role for both cathepsin L and extracellular proteases generated by lung inflammatory cells in maturation of the SARS coronavirus spike (S) protein was recently described (Matsuyama et al., 2005, Simmons et al., 2005). An endocytosis motif present within the cytoplasmic tail of Henipavirus F proteins promotes proteolytic activation within the acid environment of the endosome (Diederich et al., 2005, Meulendyke et al., 2005, Vogt et al., 2005), where cathepsin L was recently shown to be involved in proteolytic processing of the Hendra virus F protein (Pager and Dutch, 2005). Our results demonstrate that Nipah F proteolytic maturation requires cathepsin L digestion. Despite endogenous cathepsin B enzyme activity in cathepsin L shRNA-expressing Vero cells and cathepsin L−/− MEFs, proteolytic processing and fusion activity of Nipah F were severely debilitated in these cells, strongly indicating that cathepsin L (and not cathepsin B) is required for maturation and fusion activity of the Nipah virus F protein. Purified cathepsin B can cleave immunopurified Nipah F precursor protein into two fragments, but the smaller digested product migrated slower than an endogenously produced F2 fragment. The cellular mechanism of how Nipah F is localized to endosomal compartments containing cathepsin L and/or B is unknown and it is also unclear how incorrect processing is prevented in these compartments. These questions warrant further investigation.
Until recently, entry mechanisms described for enveloped viruses required receptor interactions at the plasma membrane, the low pH within the endosome, or a combination of receptor binding at neutral pH and low pH exposure to trigger conformational changes within viral fusion proteins to drive viral membrane fusion (Earp et al., 2004). Viral entry of Ebola virus and SARS coronavirus following endocytic uptake and digestion of fusion proteins by endosomal cathepsin proteases may be an additional entry strategy used by enveloped viruses (Chandran et al., 2005, Simmons et al., 2005). Paramyxovirus-mediated fusion is thought to exclusively occur at the plasma membrane (Earp et al., 2004). Nipah F mediates membrane fusion at neutral pH (Moll et al., 2004), yet Nipah virions may be taken up into cells by endocytosis (Vogt et al., 2005), and proteolytic activation of the F protein by cathepsin L occurs within an acidic endosomal compartment (Diederich et al., 2005, Meulendyke et al., 2005, Pager and Dutch, 2005). This dichotomy raises the possibility that Henipaviruses may utilize both the pH-dependent and pH-independent mechanisms of entry, although the exact entry mechanism remains to be dissected.
Nipah virus is a highly pathogenic virus with the dangerous ability to spread from human-to-human (Hsu et al., 2004). However, no approved antiviral therapy currently exists. The antiviral drug ribavirin has been used to treat Nipah virus encephalitis (Chong et al., 2001), and modified peptides directed to the C-terminal heptad repeat have been shown to potently inhibit Henipavirus fusion and infection (Bossart et al., 2005). Since efficient Henipavirus infection and spread requires cathepsin L activation of the F protein, inhibition of cathepsin L activity may prove to be an effective antiviral strategy.
Materials and methods
Cell lines and protease inhibitors
Vero and BSR cells (provided by Karl–Klaus Conzelman, Pettenkofer Institut) and MEFs derived from cathepsin L+/+ and cathepsin L−/− mice (provided by Terence Dermody, Vanderbilt University) were maintained in Dulbecco's modified Eagle's media (DMEM; Gibco Invitrogen) containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. The protease inhibitors E-64 and E-64d were purchased from Sigma, and CA-074Me from Peptides International. Calpeptin, MG-132, cathepsin I, and cathepsin LIII inhibitors were purchased from Calbiochem.
Plasmids
Plasmids containing the Nipah F and G genes were provided by Lin-fa Wang (Australian Animal Health Laboratory). The F gene was excised from pTOPO by digestion with SalI and ligated into XhoI-digested pCAGGS. The G gene was released as a PmeI fragment and ligated into the SmaI site of pCAGGS. Both Nipah F and G were cloned in the correct orientation and gene sequence integrity confirmed by sequencing following subcloning. The pSG5-cathepsin L plasmid was kindly provided by Terence Dermody (Vanderbilt University).
Expression of Nipah F proteins, metabolic labeling and immunoprecipitation
The expression and detection of the Nipah F protein were performed as previously described (Pager and Dutch, 2005). In brief, cells were transiently transfected with pCAGGS-Nipah F using the Lipofectamine Plus reagent. Twenty-four-hour posttransfection cells were starved, metabolically labeled with Tran35S (MP Biomedicals), and chased for 2 h with DMEM. The F protein was immunoprecipitated from cell lysates with antibodies to gamma-irradiated Nipah virus (generously provided by Paul Selleck and Chris Morrissy, Australian Animal Health Laboratory), and samples were separated in 15% reducing polyacrylamide gels and visualized using the Typhoon imaging system (Amersham).
Cell fusion assays
A previously described luciferase reporter gene assay was used to measure cell–cell fusion (Pager and Dutch, 2005). Effector cells were transfected with luciferase T7 control DNA and pCAGGS-Nipah F and -Nipah G with or without pSG5-cathepsin L. Nipah F:G and cathepsin L:Nipah F DNA ratios were transfected at 1:1 and 2:1, respectively. At 24 h posttransfection, BSR cells were overlaid onto the effector cells, incubated for 3 h and fusion activity quantified as a measure of luciferase activity. Background values were ascribed to cells expressing only the attachment protein Nipah G and then subtracted from the values for Nipah F + G (± cathepsin L). Luciferase activity of triplicate samples measured in duplicate was reported, and assays were repeated 3–6 times.
In vitro cleavage of immunoprecipitated Nipah F protein
A previously established protocol was used to generate an uncleaved Nipah F product (Pager and Dutch, 2005). Nipah F protein immobilized on protein A-sepharose beads was incubated 0–8 h at 37 °C with 1 nM cathepsin B or 10 nM cathepsin L (Calbiochem) in a sodium acetate-dithiothreitol buffer at pH 6 or pH 5.5, respectively. Digestion reactions were inactivated with an immediate 5 min boil, and samples were resolved by 15% SDS-PAGE and analyzed as described above.
Acknowledgments
We would like to thank Dr. Lin-fa Wang (Australian Animal Health Laboratory) for the Nipah F and G plasmids; Drs. Paul Selleck and Chris Morrissy for the anti-gamma inactivated Nipah virus antiserum (Australian Animal Health Laboratory); Dr. Doug Andres (University of Kentucky) for the pSuper-Scramble plasmid; Karl-Klaus Conzelman (Pettenkofer Institut) for the BSR cells; and Dr. Terence Dermody (Vanderbilt University) for the cathepsin L MEFs and pSG5-cathepsin L vector. We greatly appreciate assistance from Jennifer Strange in the UK Flow Cytometry Core Facility. We are grateful to members of the Dutch lab and Dr. Christopher Broder for critically reviewing the manuscript. C.T.P. and W.W.C. are recipients of an AHA Ohio Valley Affiliate predoctoral fellowship and a Ruth L. Kirshechstein National Research Service Award, respectively. This study was supported by NIAID grant AI063052 to R.E.D.
References
- Bonaparte M.I., Dimitrov A.S., Bossart K.N., Crameri G., Mungall B.A., Bishop K.A., Choudhry V., Dimitrov D.S., Wang L.-F., Eaton B.T., Broder C.C. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U.S.A. 2005;102(30):10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart K.N., Wang L.-F., Flora M.N., Chua K.B., Lam S.K., Eaton B.T., Broder C.C. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 2002;76(22):11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart K.N., Mungall B.A., Crameri G., Wang L.-F., Eaton B.T., Broder C.C. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol. J. 2005;2:57. doi: 10.1186/1743-422X-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Finke S., Conzelmann K.-K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.L., Kane S.E., Erickson A.H. Abnormal glycosylation of procathepsin L due to N-terminal point mutations correlates with failure to sort to lysosomes. J. Biol. Chem. 1997;272(13):8808–8816. doi: 10.1074/jbc.272.13.8808. [DOI] [PubMed] [Google Scholar]
- Chong H.-T., Kamarulzaman A., Tan C.-T., Kunjapan S.R., Chew N.-K., Chua K.-B., Lam S.-K. Treatment of acute Nipah encephalitis with ribavirin. Ann. Neurol. 2001;49:810–813. doi: 10.1002/ana.1062. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., Goldsmith C.S., Gubler D.J., Roehrig J.T., Eaton B., Gould A.R., Olson J., Field H., Daniels P., Ling A.E., Peters C.J., Anderson L.J., Mahy B.W. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Koh C.L., Hooi P.S., Wee K.F., Khong J.H., Chua B.H., Chan Y.P., Lim M.E., Lam S.K. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002;4:145–151. doi: 10.1016/s1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- Craft W.W., Jr., Dutch R.E. Sequence motif upstream of the Hendra virus fusion protein cleavage site is not sufficient to promote efficient proteolytic processing. Virology. 2005;341(1):130–140. doi: 10.1016/j.virol.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Diederich S., Moll M., Klenk H.-D., Maisner A. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 2005;280(33):29899–29903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. In: Compans R.W., Cooper M.D., Honjo T., Koprowski H., Melchers F., Oldstone M.B.A., Olsnes S., Potter M., Vogt P.K., Wagner H., editors. vol. 285. Springer-Verlag; New York: 2004. pp. 25–66. (Curr. Top. Microbiol. Immunol.). [Google Scholar]
- Ebert D.H., Deussing J., Peters C., Dermody T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002;277(27):24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., Shaw E., Klenk H.D. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- Goulet B., Baruch A., Moon N.-S., Poirier M., Sansregret L.L., Erickson A., Bogyo M., Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell. 2004;14(2):207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- Harcourt B.H., Tamin A., Ksiazek T.G., Rollin P.E., Anderson L.J., Bellini W.J., Rota P.A. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology. 2000;271:334–349. doi: 10.1006/viro.2000.0340. [DOI] [PubMed] [Google Scholar]
- Hsu V.P., Hossain M.J., Parashar U.D., Ali M.M., Ksiazek T.G., Kuzmin I., Niezgoda M., Rupprecht C., Bresee J., Breiman R.F. Nipah virus encephalitis reemergence, Bangladesh. Emerging Infect. Dis. 2004;10(12):2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U.S.A. 2005;102(35):12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulendyke K.A., Wurth M.A., McCann R.O., Dutch R.E. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005;79(20):12643–12649. doi: 10.1128/JVI.79.20.12643-12649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski W.P., Crameri G., Wang L.-F., Shiell B.J., Eaton B. The cleavage activation and sites of glycosylation in the fusion protein of Hendra virus. Virus Res. 2000;69:83–93. doi: 10.1016/s0168-1702(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Moll M., Diederich S., Klenk H.-D., Czub M., Maisner A. Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. J. Virol. 2004;78(18):9705–9712. doi: 10.1128/JVI.78.18.9705-9712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Roth W., Wong P., Nelson A., Farr A., Deussing J., Villadangos J.A., Ploegh H., Peters C., Rudensky A.Y. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280(5362):450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- Negrete O.A., Levroney E.L., Aguilar H.C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- Pager C.T., Dutch R.E. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005;79(20):12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager C.T., Wurth M.A., Dutch R.E. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 2004;78(17):9154–9163. doi: 10.1128/JVI.78.17.9154-9163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D., Guncar G. Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;59(Pt. 2):203–213. doi: 10.1107/s0907444902021479. [DOI] [PubMed] [Google Scholar]
- Vogt C., Eickmann M., Diederich S., Moll M., Maisner A. Endocytosis of the Nipah virus glycoproteins. J. Virol. 2005;79(6):3865–3872. doi: 10.1128/JVI.79.6.3865-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov V.E., Feldmann H., Volchkova V.A., Klenk H.-D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasothornsrikul S., Greenbaum D., Medzihradszky K.F., Toneff T., Bundey R., Miller R., Schilling B., Petermann I., Dehnert J., Logvinova A., Goldsmith P., Neveu J.M., Lane W.S., Gibson B., Reinheckel T., Peters C., Bogyo M., Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. U.S.A. 2003;100(16):9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]