Abstract
The need for new antimicrobial agents is becoming one of the most urgent requirements in modern medicine. The venoms of many different species are rich sources of biologically active components and various therapeutic agents have been characterized including antimicrobial peptides (AMPs). Due to their potent activity, low resistance rates and unique mode of action, AMPs have recently received much attention. This review focuses on AMPs from the venoms of scorpions and examines all classes of AMPs found to date. It gives details of their biological activities with reference to peptide structure. The review examines the mechanism of action of AMPs and with this information, suggests possible mechanisms of action of less well characterised peptides. Finally, the review examines current and future trends of scorpion AMP research, by discussing recent successes obtained through proteomic and transcriptomic approaches.
Keywords: Pore forming peptides, Infection, Scorpion venom, Venomics, Therapeutics
Graphical abstract
Highlights
-
•
In-depth analysis of AMPs from scorpion venom.
-
•
Focus on biological activity and structure – function relationships.
-
•
Discussion of possible mechanisms of action.
-
•
Future strategies for further mining of bioactive compounds from venoms.
1. Introduction
Over the last few decades an increasing number of pathogenic microorganisms have developed resistance to conventional antibiotics. This poses problems in the clinical management of infections, especially in immuno-compromised individuals but also increasingly, at the community level. During the same period, more worryingly, the development of new antibiotics has decreased. Selective pressures caused by the overuse of conventional antibiotics in the healthcare setting, as well as unprecedented increases both in global transport systems and migration has led to a situation of previously isolated forms of resistance being widespread. The ability of mobile genetic elements of bacterial plasmids to spread through a population is increasing the rate of resistance (Kumarasamy et al., 2010). While the media portrayal of an imminent crisis of untreatable infections is misguided, the position in reality is rather more complex. Although treatment options for some pathogens have undoubtedly decreased, for others the position has actually reversed (Livermore et al., 2009). A major problem in modern antibiotic drug development is designing agents that are not easily susceptible to resistance and therefore a new approach to drug development is required (Jenssen et al., 2006).
The need for new antimicrobials agents has been highlighted by the release of a consensus statement following discussion of leading academic, clinical and industrial experts (Chopra et al., 2006) who discussed the growing problem of resistance within Gram negative bacteria and the lack of new compounds readily available to treat such pathogens. The report highlighted that no novel classes of antimicrobial agents has been developed since the 1980's and with no compounds in clinical trials, the threat of untreatable infection in the near future is feasible.
Antimicrobial peptides (AMPs) represent an ancient defence mechanism that transverses the evolutionary spectrum and remains an effective strategy against invading pathogens. Because of their selectivity for prokaryotic membranes and their membrane-disruptive mechanisms for which microbes have little natural resistance, the spotlight in recent years has turned towards the development of novel antibiotics from these peptides (Zasloff, 2002). Previous attempts to develop AMPs into the clinical setting have proven frustrating, with a number of peptides being rejected during the latter stages of clinical trials (Gordon et al., 2005). However a recent commentary (Fox, 2013) examined these failures and provided optimism for future AMP development; in this light, a number of these previous attempts are discussed below. For example, pexiganan (Lamb and Wiseman, 1998), a 22 residue peptide developed for the treatment of foot ulcers was stopped because of manufacturing difficulties and changes in the clinical trial design, leading to a non-approval letter been issued (1999) due to deficiencies with US Chemistry Manufacturing & Controls. The United States Federal Drug Administration (FDA) also indicated that pexiganan was no more effective in treating foot ulcers than conventional antibiotics. Similarly plectasin (NZ2114) (Mygind et al., 2005) developed by Novozymes for the treatment of Gram positive infections and licenced (2008) to Sanofi-Aventis was abandoned because of commercial rather than scientific reasons. In another example, omiganan (Sader et al., 2004), which had good efficacy in inhibiting catheter associated infections, was rejected on the grounds of cost when it was demonstrated to show no clinical statistical difference when compared to the widely used povidone iodine.
Stringent regulations placed on new antimicrobials was also highlighted by Chopra et al., 2006 who argued that, in contrast, other drugs, for example those used to treat cancer, are approved with a high degree of adverse side effects and that this has contributed to the lack of development of new antimicrobials. However recent acceptance of the lipopeptide, Daptomycin (2003) and the vancomycin derived Telavancin (2007), which have relatively low therapeutic indices compared with conventional β-lactams, would suggest a possible change in regulation. In this changing climate, pexiganan is re-entering clinical trials, as Locilex (Fox, 2013).
Scorpion venom has proved a rich source of bioactive molecules, especially ion channels blockers; in recent years it has been increasingly recognized that scorpion venoms also have an abundant supply of AMPs and it has been suggested (see inter alia, Hernández-Aponte et al., 2011) that the presence of AMPs might protect the venom gland from infection and facilitate the action of other neurotoxins. Scorpion venom AMPs are positively charged amphipathic peptides and can be conveniently divided into three structural categories: (1) cysteine containing peptides with disulfide bridges; (2) peptides with an amphipathic α-helix but lacking cysteine residues and (3) peptides rich in certain amino acids such as proline and glycine. This review aims to provide an examination of AMPs from scorpion venom (Table 1 ; Fig. 1 ), with discussions on structure, biological activity and proposed mechanisms of action, together with a final discussion of recent and future strategies for mining new bioactive molecules.
Table 1.
List of antimicrobial peptides (AMPs) derived from the venom of various scorpions.
| Name (UniProtKB) | Sequence | Sequence similarities | Length (A)/molecular weight (Da) | Activity preference | Scorpion species (Family) | Reference |
|---|---|---|---|---|---|---|
| A. Cysteine containing AMPs | ||||||
| Scorpine or Panscorpine (KBX3_PANIM) |
GWINEEKIQKKIDERMGNTVLGGMAKAIV HKMAKNEFQCMANMDMLGNCEKHCQT SGEKGYCHGTKCKCGTPLSY |
Class 3 subfamily | 75/8350; 3DSBs | Against bacteria (B. subtilis& K. pneumoniae) & parasite P. berghei. | P. imperator (Scorpionidae) | Conde et al., 2000 |
| Opiscorpine-1 (KBX31_OPICA) |
KWFNEKSIQNKIDEKIGKNFLGGMAKAV VHKLAKNEFMCVANVDMTKSCDTHCQ KASGEKGYCHGTKCKCGVPLSY |
Class 3 subfamily | 76/8428; 3DSBs | Against the yeasts F. culmorum, F. oxysporum & bacteria P. aeruginosa, E. coli. |
O. carinatus (Scorpionidae) |
Zhu and Tytgat., 2004 |
| Opiscorpine-2 (KBX32_OPICA) |
KWLNEKSIQNKIDEKIGKNFLGGMAKAV VHKLAKNEFMCMANMDPTGSCETHCQK ASGEKGYCHGTKCKCGVPLSY |
Class 3 subfamily | 76/8367; 3DSBs |
Against yeasts & bacteria. |
O. carinatus (Scorpionidae) |
Zhu and Tytgat., 2004 |
| Opiscorpine-3 (KBX33_OPICA) |
KWLNEKSIQNKIDEKIGKNFLGGMAKAV VHKLAKNEFMCVANVDMTKSCDTHCQK ASGEKGYCHGTKCKCGVPLSY |
Class 3 subfamily | 76/8394; 3DSBs |
Against yeasts & bacteria. |
O. carinatus (Scorpionidae) |
Zhu and Tytgat., 2004 |
| Opiscorpine-4 (KBX34_OPICA) |
KWLNEKSIQNKIDEKIGKNFLGGMAKAV VHKLAKNEFMCVANIDMTKSCDTHCQK ASGEKGYCHGTKCKCGVPLSY |
Class 3 subfamily | 76/8408; 3DSBs |
Against yeasts & bacteria. |
O. carinatus (Scorpionidae) |
Zhu and Tytgat., 2004 |
| Heteroscorpine (KBX3_HETLA) | GWINEEKIQKKIDEKIGNNILGGMAKAV VHKLAKGEFQCVANIDTMGNCETHCQK TSGEKGFCHGTKCKCGKPLSY |
Class 3 subfamily | 76/8293; 3DSBs |
Against bacteria (B. subtilis, K. pneumoniae & P. aeruginosa). |
H. laoticus (Scorpionidae) |
Uawonggul et al., 2007 |
| HgeScplp1 (KBX3_HADGE) |
GWMSEKKVQGILDKKLPEGIIRNAAKAIV HKMAKNQFGCFANVDVKGDCKRHCKA EDKEGICHGTKCKCGVPISYL |
Class 3 subfamily | 76/8370; 3DSBs |
Against B. subtilis |
H. gertschi (Caraboctonidae) |
Diego-Garcia et al., 2007 |
| HgeβKTx (KBX2_HADGE) |
KSTVGQKLKKKLNQAVDKVKEVLNKSEVMCPVVSSFCKQHCARLGKSGQCDLLECIC | Class 2 subfamily | 58/6427; 3DSBs |
Against B. subtilis |
H. gertschi (Caraboctonidae) |
Diego-Garcia et al., 2007 |
| HgeScplp2 (KBX32_HADGE) |
GILREKYAHKAIDVLTPMIGVPVVSKIVNNAAKQLVHKIAKNQQLCMFNKDVAGWCEKSCQQSAHQKGYCHGTKCKCGIPLNYK | Class 3 subfamily | 84/9326; 3DSBs |
Against yeasts & bacteria |
H. gertschi (Caraboctonidae) |
Schwartz et al., 2007 |
| Bactridin-1 (AMP1_TITDI) |
KDGYIIEHRGCKYSCFFGTNSWCNTECT LKKGSSGYCAWPACWCYGLPDNVKIFD SNNLKC |
NaCh-inhibitor family (β-subfamily) | 61/6928; 4DSBs |
Against B. subtilis, M. luteus, E. faecalis, P. aeruginosa, Y. enterocolitica, A. calcoaceticus |
T. discrepans (Buthidae) |
Diaz et al., 2009 |
| Bactridine-2 (AMP2_TITDI) |
KDGYLVGNDGCKYSCFTRPGTYCANECSRVKGKDGYCYAWMACYCYSMPNWVKTWNRATNRCGR | NaCh-inhibitor family (β-subfamily) | 64/7374; 4DSBs |
Against B. subtilis, M.luteus, E. faecalis, P. aeruginosa, Y. enterocolitica, A. calcoaceticus |
T. discrepans (Buthidae) |
Diaz et al., 2009 |
| B. Long chain non–cysteine containing AMPs | ||||||
| Opistoporin-1 (OPO1_OPICA) |
GKVWDWIK STAKKLWNSE PVKELKNTA LNAAKNLVAEK IGATPS |
NDBP-3.5 | 44/4836 | Broad Spectrum, against Gram-positive & Gram-negative bacteria; against fungi. |
O. carinatus (Scorpionidae) |
Moerman et al., 2002 |
| Opistoporin-2 (OPO2_OPICA) |
GKVWDWIKSTAKKLWNSEPVKELKNTA LNAAKNFVAEKIGATPS |
NDBP-3.6 | 44/4870 | Broad Spectrum, against Gram-positive & Gram-negative bacteria & fungi. |
O. carinatus (Scorpionidae) |
Moerman et al., 2002 |
| Hadrurin (HADR_HADAZ) |
GILDTIKSIASKVWNSKTVQDLKRKGINWVANKLGVSPQAA | NDBP-3.1 | 41/4436 | Against S. typhimurium, K. pneumoniae, P. aeruginosa, E. coli & S. marcescens |
H. aztecus (Iuroidea) |
Torres-Larios et al., 2000 |
| Pandinin-1 (PAN1_PANIM) |
GKVWDWIKSAAKKIWSSEPVSQLKGQV LNAAKNYVAEKIGATPT |
NDBP-3.4 | 44/4800 | Against Gram-positive bacteria B. subtilis, S. epidermidis, E. faecalis and S.aureus. | P. imperator (Scorpionidae) | Corzo et al., 2001 |
| Parabutoporin (PBPO_PARSC) |
FKLGSFLKKAWKSKLAKKLRAKGKEML KDYAKGLLEGGSE EVPGQ |
NDBP-3.2 | 45/5030.3 | Against Gram-negative bacteria & Fungi |
P. schlechteri (Buthidae) |
Moerman et al., 2002 |
| Vejovine (F1AWB0_9SCOR) | GIWSSIKN LASKAWNSDI GQSLRNKAAG AINKFVADKIGVTPSQAASM TLDEIVDA MYYD |
NDBP-3.4 | 47/4873 | Against P. aeruginosa, K. pneumoniae, E. coli, E. cloacae and A. baumanii |
V. mexicanus (Vaejovidae) |
Hernandez-Aponte et al., 2011 |
| BmKbpp (BPK3_MESMA) |
FRFGSFLKKVWKSKLAKKLRSKGKQLLKDYANKVLNGPEEEAAAPAE | NDBP-3.3 | 47/5321 | Against Gram-negative bacteria |
M. martensii (Buthidae) |
Zeng et al., 2000 |
| Im-1 (BPP_ISOMC) |
FSFKRLKGFAKKLWNSKLARKIRTKGLKYVKNFAKDMLSEGEEAPPAAEPPVEAPQ | NDBP-3.3 | 56/6344.5 | Against E. coli, S. aureus & B. subtilis |
I. maculatus (Buthidae) |
Miyashita et al., 2010 |
| Heterin-1 | GVWDWLKKTAKNVWNSDIVKQLKGKAINAAKNYVAEKIGATPS-NH2 | NDBP-3.14 | 43/4742.54 | Against Gram-positive & Gram-negative bacteria |
H. spinifer (Scorpionidae) |
Wu et al., In press |
| C. Intermediate chain non–cysteine containing AMPs | ||||||
| Meucin-24 (KBX2_MESEU) |
GRGREFMSNLKEKLSGVKEKMKNS | Class 2 subfamily | 24/2753.95 | Against P. berghei ookinetes |
M. eupeus (Buthidae) |
Gao et al., 2010 |
| Meucin-25 (M25_MESEU) |
VKLIQIRIWIQYVTVLQMFSMKTKQ | No similarity | 25/3095.56 | Against P. berghei ookinetes |
M. eupeus (Buthidae) |
Gao et al., 2010 |
| HsAp (JX311701) |
SGTSEKERESGRLLGVVKRLIVCFR SPFP-NH2 |
No similarity | 29/3246 | Against Gram-positive & Gram-negative bacteria; against fungi. |
H. spinifer (Scorpionidae) |
Nie et al., 2012 |
| Pandinin-2 (PAN2_PANIM) |
FWGALAKGALKLIPSLFSSFSKKD | NDBP-4.1 | 24/2612 | Against Gram-positive bacteria B. subtilis, S. epidermidis, E. faecalis and S. aureus | P. imperator (Scorpionidae) | Corzo et al., 2001 |
| Heterin-2 | FWGALAKGALKLIPSLVSSFTKKD-NH2 | NDBP4.2 | 24/2576.47 | Against Gram-positive bacteria |
H. spinifer (Scorpionidae) |
Wu et al., In press |
| D. Short chain non–cysteine containing AMPs | ||||||
| IsCT (NDB52_OPIMA) |
ILGKIWEGIKSLF-NH2 | NDBP-5.2 | 13/1502 | Against Gram-positive & Gram-negative bacteria. |
O. madagascariensis (Hemiscorpiidae) |
Dai et al., 2001 |
| IsCT2 (NDB53_OPIMA) |
IFGAIWNGIKSLF-NH2 | NDBP-5.3 | 13/1463.92 | Against Gram-positive & Gram-negative bacteria. |
O. madagascariensis (Hemiscorpiidae) |
Dai et al., 2002 |
| BmKb1 (KB1_MESMA) |
FLFSLIPSAISGLISAFK-NH2 | NDBP-4.2 | 18/1910 | Against Gram-positive bacteria S. aureus, M. luteus, B. subtilis & Gram-negative E. coli, and P. aeruginosa. |
M. martensii (Buthidae) |
Zeng et al., 2004 |
| Meucin-13 (NDB5Y_MESEU) |
IFGAIAGLLKNIF-NH2 | NDBP-5 | 13/1375.82 | Potent against Gram-positive bacteria. |
M. eupeus (Buthidae) |
Gao et al., 2009 |
| VmCT1 (NDB5D_VAEMS) |
FLGALWNVAKSVF-NH2 | NDBP-5.13 | 13/1450.8 | Against Gram-positive & negative bacteria |
V. mexicanus (Vaejovidae) |
Ramirez-Carreto et al., 2012 |
| VmCT2 (NDB5E_VAEMS) |
FLSTLWNAAKSIF-NH2 | NDBP-5.14 | 13/1496.8 | Against Gram-positive & negative bacteria |
V. mexicanus (Vaejovidae) |
Ramirez-Carreto et al., 2012 |
| Mucroporin (MUCR_LYCMC) |
LFGLIPSLIGGLVSAFK-NH2 | Short cationic antimicrobial peptide family | 17/1731 | Potent against Gram-positive bacteria. |
L. mucronatus (Buthidae) |
Dai et al., 2008 |
| AamAP1 (VAP1_ANDAM) |
FLFSLIPHAIGGLISAFK-NH2 | Short cationic antimicrobial peptide family | 18/1931.94 | Against S. aureus, E. coli & C. albicans |
A. amoreuxi (Buthidae) |
Almaaytah et al., 2012 |
| AamAP2 (VAP2_ANDAM) |
FPFSLIPHAIGGLISAIK-NH2 | Short cationic antimicrobial peptide family | 18/1880.93 | Against S. aureus, E. coli & C. albicans |
A. amoreuxi (Buthidae) |
Almaaytah et al., 2012 |
| Imcroporin (IMCR_ISOMC) |
FFSLLPSLIGGLVSAIK-NH2 | Short cationic antimicrobial peptide family | 17/1761 | Against Gram-positive bacteria M. luteus, B. thuringiensis, S. aureus and B. subtilis. |
I. maculatus (Buthidae) |
Zhao et al., 2009 |
| StCT1 (NDB5X_SCOTI) |
GFWGSLWEGVKSVV-NH2 | NDBP-5 | 14/1549.79 | Against Gram-positive bacteria S. aureus, M. luteus. |
S. tibetanus (Euscorpiidae) |
Yuan et al., 2010 |
| StCT2 (NDB5Y_SCOTI) |
GFWGKLWEGVKSAI-NH2 | NDBP-5 | 14/1576.84 | Against Gram-positive bacteria. |
S. tibetanus (Euscorpiidae) |
Cao et al., 2012 |
| Meucin-18 (NDB59_MESEU) |
FFGHLFKLATKIIPSLFQ | NDBP-5 | 18/2106 | Against Gram-positive bacteria, lethal to the fungus Beauveria sp. |
M. eupeus (Buthidae) |
Gao et al., 2009 |
| Ctriporin (VAP_CHATC) |
FLWGLIPGAISAVTSLIKK-NH2 | Short cationic antimicrobial peptide family | 19/2015.2 | Against Gram-positive & fungi pathogens: S. aureus M. luteus, B. thuringiensis B. subtilis, C. albicans. |
C. tricostatus (Chaeriloidea) |
Fan et al., 2011 |
| Hp1090 (NDB59_HETPE) |
IFKAIWSGIKSLF-NH2 | NDBP-5.9 | 13/1508.88 | Against Gram-positive bacteria, against HCV. |
H. petersii (Scorpionidae) |
Yan et al., 2011 |
| Pantinin-1 (KC538864) |
GILGKLWEGFKSIV-NH2 | NDBP-5 | 13/1545.90 | Against Gram-positive bacteria & fungi. | P. imperator (Scorpionidae) | Zeng et al., 2013 |
| Pantinin-2 (KC538865) |
IFGAIWKGISSLL-NH2 | NDBP-5 | 14/1403.71 | Against Gram-positive bacteria & fungi. | P. imperator (Scorpionidae) | Zeng et al., 2013 |
| Pantinin-3 (KC538866) |
FLSTW NGIKSLL-NH2 | NDBP-5 | 14/1490.80 | Against Gram-positive bacteria & fungi. | P. imperator (Scorpionidae) | Zeng et al., 2013 |
| TsAP-1 (HF677516) |
FLSLIPSLVGGSISAFK-NH2 | Short cationic antimicrobial peptide family | 17/1733.32 | Against Gram-positive S. aureus and the yeast, C. albicans. |
T. serrulatus (Buthidae) |
Guo et al., in press |
| TsAP-2 (HF677517) |
FLGMIPGLIGGLISAFK-NH2 | Short cationic antimicrobial peptide family | 17/1735.20 | Against Gram-positive S. aureus and the yeast, C. albicans |
T. serrulatus (Buthidae) |
Guo et al., in press |
Fig. 1.
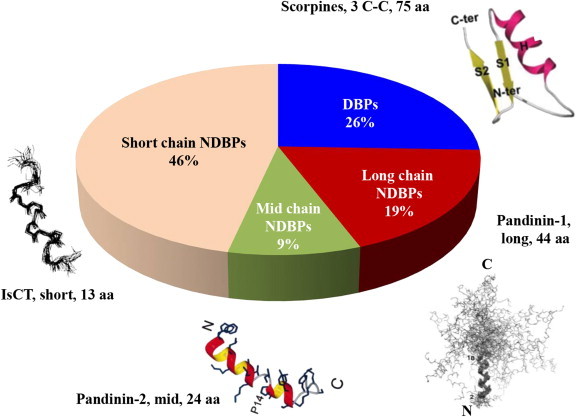
Different classes of antimicrobial peptides (∼45) derived from scorpion venoms and relative proportion of each category.
2. Cysteine containing antimicrobial peptides
Cysteine containing peptides are ubiquitous in scorpion venoms and usually contain 3 or 4 disulphide bridges. These peptides have been characterised as interacting with ion channels, namely Na+, K+, Ca2+ and Cl− channels, and make up the largest family of peptides in the venom with non-disulphide bridged peptides being a smaller family. Whilst AMPs from scorpions have increasingly been found to belong to the non-disulphide bridged family, disulphide bridged peptides having interesting biologically activities that more than warrants mention.
The noted similarity between insect defensins and scorpion toxins (Bontems et al., 1991) led to the isolation of the first scorpion defensin from the haemolymph of the North African scorpion Leiurus quinquestriatus. This 4.3 KDa peptide contained 38 residues, with the characteristic 6 cysteines of many ion channel scorpion toxins and showed a high degree of homology to insect defensins within the order Odonata. This peptide was active against Gram-positive M. luteus but inactive against Gram-negative E. coli (Cociancich et al., 1993) (Details of MIC values for all bacteria and fungi tested, against all peptides mentioned in this review are provided in Table 2 ). The first cysteine-constrained AMP from scorpion venom (scorpine) was isolated by Possani and colleagues (Conde et al., 2000) from the venom of the African scorpion Pandinus imperator. Scorpine (8.3 KDa, 75 residues, 3 disulphide bridges) had a unique structure, with N-terminal similarity to some insect cecropins and C-terminal similarity to some scorpion defensins.
Table 2.
MIC values of scorpion venom antimicrobial peptides against Grame-positive and Grame-negative bacteria.

Scorpine was active (MIC 1–10 μM) against both Gram-positive (B. subtilis) and Gram-negative (K. pneumonia) bacteria in agar diffusion assays. The anti-malarial properties of scorpine were also investigated against the causative parasite P. berghei (ED50 0.7 μM and 10 μM against ookinete and gamete stages, respectively). A second putative peptide (BmTXKS2) was identified during this same period, as a full length cDNA clone isolated from the venom gland of the Chinese scorpion, Buthus martensii Karsch (Zhu et al., 2000). BmTXKS2 was predicted to have 39 residues including 6 cysteines and have similarity to haemolymph defensins. However there have been no subsequent reports of the translated peptide (either native or recombinant) being isolated and no biological data is available. In subsequent years, two more members of the scorpine family members have been identified – opiscorpine from Opistophtalmus carinatus (Zhu and Tytgat, 2004) and heteroscorpine-1 from Heterometrus laoticus (Uawonggul et al., 2007).
Six peptides have been isolated from the venom of Tityus discrepans termed the bactridines (Bacts) (Diaz et al., 2009), which unlike the scorpine family contain 4 disulphide bridges. Bacts 1 and 2 have been characterised more extensively with Edman degradation and in silico analysis revealing charged peptides containing 61 and 64 residues respectively.
Bacts 1 showed an MIC range of 22–77 μM whilst Bacts 2 had an MIC range of 27–65 μM (Appendix Table 1). In hemolytic assays, Bacts 1 showed only 0.2% at 90 μM and 1.1% at 180 μM with Bacts 2 showing 7.6 and 21% at the same concentrations, respectively. Whilst these peptides do not have potent antimicrobial activity, their mechanism of action is of most interest. Using the pathogen Y. enterocolitica loaded with 1 μM of the Na+ fluorescent indicator CoroNa™ red the efflux of sodium ions was observed upon bacts 1 and 2 interactions. Furthermore this could be blocked in the presence of the sodium channel blockers amiloride (10 μM) and mibefradil (25 μM) although no inhibition was observed in the presence of 30 μM tetrodotoxin; furthermore these interactions were shown to be Na+ Channel specific with no interactions observed with K+ or Ca2+ channels.
Bacts 1 showed no toxicity towards mice (0.4 μM/g) however was toxic to cockroaches (2.8 μM) induced sialorrhoea in crabs at 0.1 μM/g and death in 3 out of 7 crabs at 0.2 μM/g. Bacts 2 however was toxic to mice at the same concentration suggesting specificity towards different classes of sodium channels. Peigneur et al. (2012) showed no modulation of the mammalian sodium channels Nav1.2-Nav1.8 with bacts 1, however no modulation of the insect DmNav1 or the bacterial NaChBac was seen either suggesting further research needs to be carried out. Bacts 2 showed modulation of Nav1.2, Nav1.4 and Nav1.6. Bacts 1 shares 78% homology to ardiscretin (D'Suze et al., 2004) which is an insect sodium channel blocker from the same venom whilst Bacts 2 shares 98% homology to the first 60 residues of Tz1 and Td4 from Tityus zulianus and Tityus discrepans respectively (Borges et al., 2004). Structural modelling of the two peptides suggested a classical α-β-motif in which the peptides have an alpha helical region and conserved cysteine constrained beta sheet structure.
The lesser characterised Bacts peptides (3–6) have molecular masses of 6916, 7362, 7226, and 7101 respectively and their antimicrobial activities are shown in Supplementary Table 1. In hemolytic assays at 90 μM Bacts 3 showed only 0.1% hemolysis whilst bacts 5 showed 0.9%; Bacts 4 and 6 showed 4.3 and 3.1%. Sodium efflux studies were positive and again inhibited with amiloride whilst tetrodotoxin had no effect, interestingly bacts 3 and 6 showed only partial inhibition with mibefradil and no inhibition observed with bacts 5 (Appendix Table 2). Toxicity testing revealed no effect on crabs with bacts3, 4 and 5 and transient effects with 6 whilst toxicity was observed with 3, 4 and 6 with mice and transient toxicity observed with 6 (Appendix Table 3). Bacts 1 represents a novel basis for an AMP although further work is required before a sodium channel specific mechanism of action can be truly revealed however it would be of interest to the field of venom antimicrobials if these questions were answered and more bacteridine like AMPs were isolated (Kuhn-Nentwig, 2003, Zeng et al., 2005).
3. Antimicrobial peptides without cysteine residues
Whereas the original antimicrobial peptides isolated from scorpion venoms contained cysteine residues, various non-disulphide bridged peptides have subsequently been isolated (e.g. Kuhn-Nentwig, 2003, Zeng et al., 2005, Zeng et al., 2013, Gao et al., 2010) and these are now in the majority. In this review, peptides have been divided into three groups – long-chain peptides (>35 amino acids), intermediate-chain peptides (20–35 amino acids) and short chain peptides (<20 amino acids).
3.1. Long chain peptides
Parabutoporin, isolated from the venom of the South African scorpion Parabuidethus schlechteri is a highly basic peptide (45 amino acids, overall charge of +7). A structurally similar, basic molecule (charge +4), existing as two isoforms, opistoporin 1 and 2 was isolated from another South African scorpion Opistophtalmus carinatus (Moerman et al., 2002). These two isoforms (44 amino acids) differ at position 34 where a phenylalanine replaces a leucine. Parabutoporin was predicted to have an α-helical structure between amino acids 3–35 in comparison to opistoporin 1 and 2 with two α-helical regions between residues 3–14 and 20–39 which are connected by a short random coil WNSEP (Moerman et al., 2002). Circular dichroism (CD) spectra of parabutoporin in 40% 2,2,2-trifluoroethanol (TFE), dimyristoylglycerophosphocholine (Myr2Gro-pCho) showed the peptide adopts an α-helical structure in membrane mimicking conditions. Under the same solvent conditions, opistoporin re-organises from an unordered state into a continuous α-helical structure (Moerman et al., 2002). The amphipathic natures of both parabutoporin and opistoporin 1 were also predicted with α-helical wheel projections, which clearly show distinct hydrophobic and hydrophilic regions within each peptide, although comparison of these projections clearly shows that parabutoporin has a larger polar surface.
Antimicrobial assays for parabutoporin showed growth inhibition of all Gram-negative organisms assayed with MIC's generally varying from 1 to 6 μM. The peptide showed less activity towards Gram-positive organisms with MIC's generally greater than 25 μM. Interestingly, a decrease in growth inhibition was observed in the presence of Mg2+ ions (Moerman et al., 2002). Opistoporin 1 showed less inhibitory activity against Gram-negative organisms (MIC values generally ranging from 6 to 50 μM) than parabutoporin. On the other hand, differences between the effects of opistoporin and parabutoporin on Gram-positive organisms were less marked, although Gram-positive bacteria did appear slightly more sensitive to the former. In antifungal studies, both parabutoporin and opistoporin inhibited growth of a range of fungi (B. cinerea, F. culmorum and S. cerevisiae) at similar concentrations (50% growth inhibition between 0.3 and 3.5 μM). Parabutoporin was more than twice as effective as opistoporin in disrupting eukaryotic cell membranes. Using human erythrocytes, parabutoporin induced 50% haemolysis at 37 μM; in comparison, opistoporin 1 induced only 30% haemolysis at 100 μM.
Hadrurin is the prototype of another cysteine-free AMP family purified from the venom of the Mexican scorpion Hadrurus aztectus. Hadurin (4436 Da, 41 amino acids), carries an overall positive charge at physiological pH and has the same structural profile as opistoporin, namely two α-helical regions connected by an undefined region between amino acid residues 12–16 (Torres-Larios et al., 2000). The plot of an α-helical wheel shows opposite hydrophobic and hydrophilic regions and these two distinct regions allow the peptide to adopt an amphipathic conformation, as the carboxyl-terminal helix can rotate 100° with respect to the amino-terminal helix. In antimicrobial activity studies with Gram-negative organisms, several E. coli strains were very sensitive with MICs less than 10 μM; in comparison, Pseudomona strains required 50 μM for full inhibition (Torres-Larios et al., 2000). Haemolytic activity was observed at 30 μM. Studies with synthetic analogues have demonstrated that the (unnatural) D-isoform of hadrurin has a different activity profile from the native l-isomer, suggesting activity is a consequence of membrane disruption (Torres-Larios et al., 2000).
Studies on two antimicrobial peptides isolated from Pandinus imperator (Corzo et al., 2001) have shed further light on the contribution of the two α-helical regions discussed above, to biological function. Pandinin 1 (Pin1) has two α-helical regions between residues 3–18 and 20–39, separated by a random coil region containing a Pro-19 kink. In contrast, NMR analysis of pandinin 2 (Pin2) in a 60% TFE solvent containing DPC micelles, showed that this second peptide had a singular helical structure between residues 2–18 with no significant kink at the equivalent proline residue (Pro-14) (Nomura et al., 2005). The structural differences found between these two peptides are significant. While the antimicrobial activity of Pin 1and Pin 2 against a range of Gram-positive and Gram-negative organisms was broadly similar, it is the haemolytic studies on eukaryotic membranes that are of most interest. Pin2 lysed 51% of sheep erythrocytes at 22 μM whilst Pin1 only lysed 1.4% at the same concentration (Corzo et al., 2001). The authors suggest that the ability of Pin 2 to lyse both eukaryotic and prokaryotic membranes may be related more to the venom toxicity, in a similar manner to the bee venom peptide melittin, than a specific antimicrobial action. In contrast, the activity of Pin1 is more in keeping with magainin1, the prototype AMP isolated from the skin of the South American toad, Xenopus. A flexible hinge region is also important in successful AMP design. Replacing (structurally rigid) Pro-14 in Pin2 with a (flexible) GVG tripeptide reduces the haemolytic activity of Pin2 without altering its antimicrobial activity (Rodriguez et al., 2011). This substitution creates a more flexible region while retaining the essential amphipathic nature of Pin 2 which is more in tune with the dual helical structure of Corzo, Nakajima and colleagues have subsequently demonstrated that the haemolytic activity of Pin1, Pin2 and other cationic amphipathic peptides (e.g. IsCt1, see Section 3.3) depends on the animal species studied (guinea pig > pig > sheep) and this is turn is related to the phosphatidylcholine:sphingomyelin ratio in the different erythrocyte membranes (Belokoneva et al., 2003).
With such contrasting effects on prokaryotic and eukaryotic membranes, a comparison of the mechanisms of action of these two peptides is of particular interest and it is instructive to consider these mechanisms in greater detail. On examination of the effects of Pin2 on phosphatidylcholine (PC) lipid vesicles using a calcian dye release assay, it was seen that the dose response curve for dye efflux was sigmoidal, suggesting peptide oligomerisation during the time course of the experiment. The effects of Pin2 were seen at very low peptide: phospholipid concentrations (Belokoneva et al., 2004). When phosphatidylethanolamine (PE), known to promote negative membrane curvature (Matsuzaki et al., 1998), was incorporated into the PC vesicles, no change in Pin2 activity was observed, supporting the hypothesis of a barrel-stave mechanism of pore formation (see Fig. 2 for a background explanation to current hypotheses for the mechanism of action of AMPs). Dextran-loaded liposome assays also showed great variability in the size (1.8–5 nm) of the membrane pores generated by Pin2, dependent on peptide: phospholipid ratios (Belokoneva et al., 2004). Based on NMR studies of Pin2, Nomura and colleagues (Nomura et al., 2005) proposed a pore forming mechanism of action, as a consequence of Pin2 inducing clustering of hydrophobic residues which allows further incursion into the membrane hydrophobic core and interaction with the inner membrane. This process can be conveniently visualized in a step-wise fashion as shown in Fig. 3 . Pin2 inserts into the membrane at a 45° angle however is seen to be only at a 25° angle around the Leu 12 residue. This causes a slight kink in the peptide when in the membrane, and is a phenomena seen in other pore forming AMPs, thus thought to be essential for activity. After insertion, oligomerisation occurs between the Pin2 monomers and then pore oligomerisation is seen leading to membrane being trapped between these pores to be ‘pinched off’. Thus Pin2 induces a 2 fold attack on the target with loss of intracellular constituents through the pore lumen and loss of membrane through pore association.
Fig. 2.
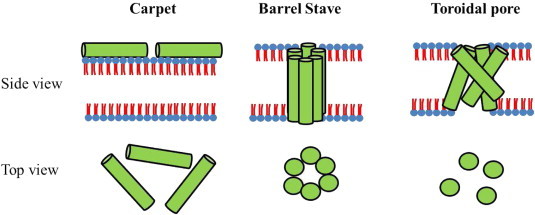
The mechanism of action of AMP's is postulated to occur in a number of stages (Zasloff, 2002): (1) Electrostatic interaction onto the membrane surface; (2) a peptide threshold concentration is reached before membrane disruption can occur after which a number of models have been proposed. In the Carpet model (Pouny and Shai, 1992), peptides remain parallel to the bilayer causing a detergent like effect. In the Barrel stave pore model peptides insert perpendicularly into the bilayer and self-association occurs forming a pore containing peptide–peptide interactions (Baumann and Mueller, 1974). The more recent Toroidal model (Ludtke et al., 1996) indicates a pore forming mechanism in which the pore lumen is lined with both peptides and phospholipid in a less rigid association.
Fig. 3.
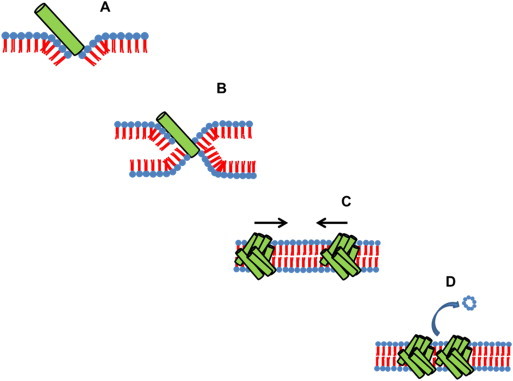
Pandinin 2 interaction causes negative membrane curvature and insertion of the peptide at a 45°angle (A) Electrostatic attraction of the inner leaflet allows negative membrane curvature and formation of a trans membrane helices (B). Oligomerisation of the peptide occurs within the membrane allowing formation of the barrel stave pore. The pores are further attracted to each other (C), causing pinching off of the membrane between pores (D) This mechanism causes a twofold attack; loss of membrane through pore attraction and loss of intracellular contents through the pores.
In contrast, NMR studies with Pin1 (Nomura et al., 2005) showed a radically different mode of action, with a detergent-like effect due to the formation of cubic phase structures. It was suggested (Nomura et al., 2005) that Pin1 sits within the interface of the membrane between the hydrophobic core and the polar phospholipid head groups. This can also be conveniently visualised (Fig. 4 ). The Pin1 N-terminal helices are tilted at a 30° angle with respect to the horizontal bilayer plane and the C-terminal helices. Interactions between tryptophan residues at position 4, 6 and 15 on the tilted helices and the polar head groups cause the membrane disruption and the formation of cubic phase structures. The N-terminal helices rotates around the average helical axis (Fig. 4).
Fig. 4.
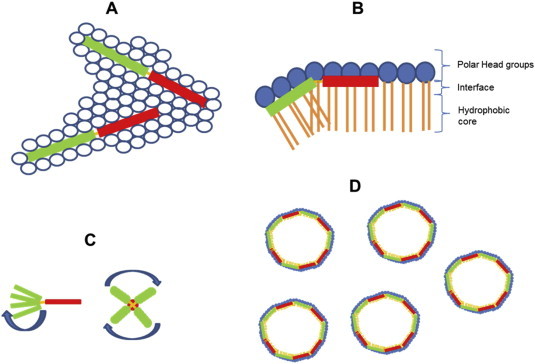
Interaction of Pandinin 1 interaction with phospholipid head groups. Pin1 sits between the head groups (A) within the interface between the head groups and HC core (B). Interactions between pin1 1–18 (green) and the head groups causes negative membrane curvature (A) as pin1 1–18 is at a 30° tilt with respect to pin1 20–44 (red) due to the presence of a proline at position 19. Membrane disruption accrues when pin1 1–18 rotates around the average helical axis, which is parallel to the lipid long axis (C) causing membrane disruption and dispersion of the lipid into the cubic phase (D). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Using primers originally designed to detect a bradykinin-potentiating peptide (K-12), Zeng and colleagues have identified a full length cDNA encoding a putative AMP (BmKbpp) from the venom glands of the Chinese scorpion, Buthus martensii Karsch. BmKbpp (NDBP-3.3 family) is predicted to encode a 47 amino acid mature peptide, the C-terminal region showing 57% homology to peptide K-12 (Zeng et al., 2000). BLAST analysis of BmKbpp revealed high homology with a number of scorpion toxins such as NDB9.6 (98%) from Lychas murconatus, Tx297 (96%) from Buthus occintanus israelis and parabutoporin (68%), suggesting a novel family of AMPs (Zeng et al., 2012).
Secondary structure predictions of BmKbpp show a similar structure as Pin1, with two α-helical regions (between residues 3–35 and 39–47) separated by a random coil region. Also, α-helical wheel projections reveal a highly amphipathic molecule. As with Pin1 low haemolytic activity is seen with only 39.9% lyses of human red blood cells at a concentration of 50 μM. Antimicrobial assays of BmKbpp in liquid cultures showed preferential activity against Gram-negative organisms (MIC's typically in the range 2–5 μM) in comparison to Gram-positive organisms (MIC's typically > 20 μM). Using a bioassay-guided fractionation strategy, Miyashita et al. (2010) isolated Im-1 (6.3 kDa, 56 amino acids) from the venom of Isometrus maculatus. Im-1 belongs to the Bpp family and showed 43% homology toward parabutoporin, especially at the N-terminus. Secondary structure analysis indicated an α-helical structure between residues 3–40 which was confirmed by CD spectra in 50% TFE. Synthetic Im-1 had potent antimicrobial effects in liquid culture assays against both Gram-negative and Gram-positive bacteria (typical MIC values 0.4–0.8 μM and 0.8–1.6 μM, respectively) (Miyashita et al., 2010).
Another long chain peptide, vejovine (4.8 kDa, 47 amino acids), has been isolated from the venom of Vaejovis mexicanus (Hernandez-Aponte et al., 2011), with 52% homology to hadrurin. In antimicrobial assays performed in liquid broth cultures, both native and synthetic vejovine showed preferential activity towards Gram-negative organisms (E. coli MIC values 4.4–20 μM) with no activity towards Gram-positive Staphylococcus aureus. Vejovine gradually breaks down when freshly milked venom is stored, producing a truncated derivative (Vm36), lacking the first eight N-terminal amino acids of vejovine. In comparison to vejovine, Vm36 showed no antimicrobial activity, demonstrating the importance of the N-terminal region of this and presumably other AMPs with similar structure. In CD studies, vejovine adopted an α-helical secondary structure in 60% TFE although it remained unordered in aqueous solution. In contrast, Vm36 peptide adopted an α-helical structure in aqueous solution which may indicate that the increased flexibility of vejovine is a key factor in promoting antimicrobial activity. Vejovine (50 μM) induced 40% haemolysis of human erythrocytes in eukaryotic cytotoxicity studies.
Using a cDNA cloning strategy, three peptides have been identified from the venom gland of Heterometrus spinifer termed Heterin-1, Heterin-2 and spiniferin (Wu et al., in press). Heterin-1 is 43 residues in length and shares 73% homology with the opistoporins and 68% with pandinin 1 with secondary structure prediction indicative of a mono-helical structure between residues 3–38 flanked by two random coil regions, however, unlike these other peptides Heterin-1 has an amidated C-terminal. In antimicrobial assays it had good activity against the Gram positive pathogens of B. megaterium and M. luteus (4.0 μM) whilst its potency against S. aureus and a range of Gram-negative organisms was less so (15.0–42.0 μM) in cytotoxicity assays 54% hemolysis was seen at 10.0 μM.
3.2. Intermediate chain peptides
A completely new class of scorpion AMPs has been reported from the venom gland of the Asian scorpion Heterometrus spinifer (Nie et al., 2012). Clones encoding four highly homologous peptides were characterised and one peptide (HsAp) was synthesized by solid state methods. HsAp (29 amino acids) showed no significant homology to any other class of scorpion AMPs. It has broad spectrum antibacterial activity against both Gram-positive (MICs typically 11–50 μM) and Gram-negative organisms (MICs typically 25–50 μM), without clear target-cell specificity as well as anti-fungal activity (MIC approx. 50 μM). HsAp was postulated to have an α-helical structure (residues 7–24) with coiled N and C terminals. It has classical amphipathic characteristics although its hydrophobic face is disrupted by two hydrophilic residues (Ser and Glu). Nie and colleagues indicated that HsAp is a bacterial infection-responsive peptide because the genomic sequence of its gene is intronless. At present HsAp has limited therapeutic potential because of its potent haemolytic activity against human erythrocytes (95% haemolysis at 3.2 μM).
Two antimalarial peptides (meucin-24 and meucin-25) have been identified by screening a venom gland cDNA library from the Asian scorpion Mesobuthus eupeus (Gao et al., 2010). The two peptides have been synthesized by solid state methods. CD spectral analysis of synthetic meucin-24 (2.8 kDa, 24 amino acids) showed a disordered conformation in water; however α-helical formation increased in 50% TFE and NMR spectra revealed an α-helical structure between residues 4–20 and random coil regions at the N- and C- terminals (Gao et al., 2010). Although meucin-24 can be considered amphipathic, the hydrophobic face of the molecule contains only six residues; this reduction in hydrophobicity has been postulated to be responsible for the lack of antibacterial activity (Gao et al., 2010). The charged residues in this amphipathic molecule are unusually, predominantly anionic and therefore not thought to interact with negatively charged membrane phospholipids. The authors provide an excellent structural comparison to the classical AMP, magainin 2, which has a larger hydrophobic face and a predominantly cationic hydrophilic domain (Fig. 5 ).
Fig. 5.
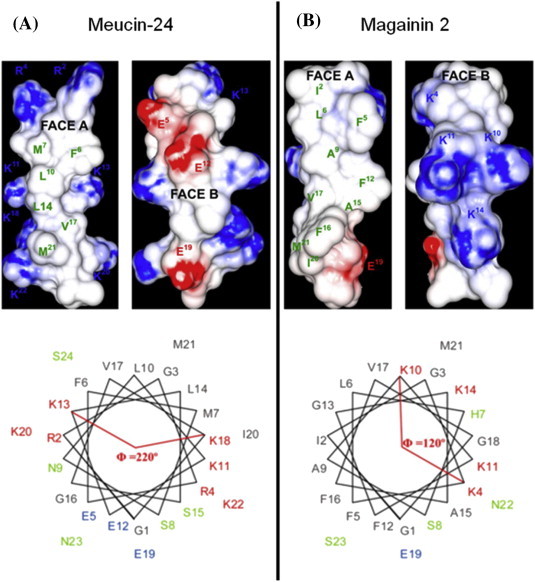
Surface charge distribution (A) and helical wheel projections (B) of the linear antimicrobial peptides meucin-24 and magainin 2 (Gao et al., 2010).
Secondary structure analyses of meucin-25 (3.1 kDa, 25 amino acids) by CD and NMR revealed a radically different structure to that of meucin-24 (Gao et al., 2010). In water, meucin-25 revealed a β-sheet motif with 39% β-sheet and 11% α-helical content. However in 50% TFE, the proportion of α-helical structure increased to 56%; in contrast, the proportion of β-sheet was reduced to 21%. Molecular modelling of meucin-25 was ambiguous, suggesting that meucin-25 may be able to fold in more than one way. Both meucin-24 and meucin-25 (at 100 μM) showed neither haemolytic activity nor antimicrobial activity against either fungi or Gram-positive and Gram-negative bacteria. Only meucin −24 showed any cytotoxicity (17% reduction viability of GC-2 cells at 50 mM). However both peptides (20 μM) produced a 40–50% reduction in the development of malarial parasite Pandinus berghei ookinetes and at 10 μM, completely eradicated Pandinus falciparum ookinetes in 72 h (Gao et al., 2010). The potent effects on intraerythrocytic P. falciparum yet little haemolytic or cytotoxic effects and no effect on bacterial and fungal membranes, is intriguing. One possible explanation that we suggest is an affinity-driven transfer from the erythrocyte membrane to the parasitic membrane.
Heterin-2 from the scorpion Heterometrus spinifer is a 24 residue C-terminally amidated peptide which shares 92% homology to pandinin 2 secondary structure predictions revealed a single helical domain between residues 2–21 flanked by two random coil regions and is amphipathic in nature. Moderate antimicrobial potency was observed against Gram-positive pathogens (5.6–30.0 μM) and it was broadly less potent against Gram-negative organisms (15.8–>45.0 μM), it also exhibited high cytotoxicity with 6.4 μM causing over 90% hemolysis. Interestingly when the C-terminal random coil region is removed (KKD) the therapeutic index is improved with a halving of the haemolytic potential and an overall increase in the MIC against Gram-positive organisms; however a reduction is seen against Gram-negative pathogens (Wu et al., in press).
3.3. Short chain peptides
The first short chain AMPs were isolated from the venom of the African scorpion Opisthacanthus madagascarienis. IsCT and IsCT2 (Dai et al., 2001, Dai et al., 2002) share 78% homology (both approx. 1.5 kDa, 13 amino acids), have the same net charge (+2) and are both amidated at the C-terminus. The precursor of each peptide consists of a signal peptide and a C-terminal pro-sequence which contains the typical C-terminal processing signals Gly-Arg-Arg or Gly-Lys-Arg. Both peptides showed a broad spectrum profile against most Gram-positive and Gram-negative bacteria on solid agar plates (MICs typically 0.6–16 μM) although Gram-negative Pseudomonas strains were unusually resistant (MICs typically > 66 μM). Both peptides were weakly haemolytic (10% lysis of sheep erythrocytes at 100 μM) (Dai et al., 2002) although IsCT (approx. 3 μM) was extremely effective, in comparison to mastoparan, at degranulating rat peritoneal mast cells, as measured by a histamine release assay (Dai et al., 2001). A pore-forming mechanism of action was postulated (Dai et al., 2002) due to the threshold concentration kinetics during antimicrobial assays and calcian release assays with synthetic membranes. The latter revealed a membrane disruptive mechanism for both peptides. At low peptide concentrations a preferential affinity towards phosphatidic acid (PA) compared with phosphatidylcholine (PC) was noticed; however this selectivity was not seen with high concentrations of peptides.
Extensive structure-function analysis has been carried out on IsCT and IsCT2 (Dai et al., 2001, Dai et al., 2002, Lee et al., 2004). CD spectra and helical wheel predictions of both peptides revealed an α-helical organisation in membrane-mimicking environments (40 and 70% TFE) which adopted a classical amphipathic structure. A sequence comparison (Fig. 6 ) between the two peptides shows differences at Phe-2 (Leu), Ala-4 (Lys) and Asn-7 (Gln) (IsCT2 residues in brackets). Helical wheel analysis demonstrates that these differences cause changes within the hydrophilic face of IsCT2, although there is no overall change in charge; this suggests that both overall ionic charge and amphipathic nature, as distinct from specific amino acid residues, are critical for AMP function. Mutation studies on IsCT (Lee et al., 2004) showed the importance of the hydrophobic residue at position 6. Substitution of Trp6 for Ala6 resulted in a remarkable decrease in both antimicrobial and haemolytic activities. Dai and colleagues (Dai et al., 2002) also identified two other peptides in the crude venom, analogous to IsCT and IsCT2 (IsCTf and IsCT2f, respectively). These latter two peptides are thought to be proteolytic products of the parent molecules devoid of the two penultimate amino acids (Leu and Phe-NH2). IsCTf and IsCT2f have no antimicrobial activity and have no α-helical structure as evidenced by CD spectral analysis, implicating these two C-terminal residues as critical for function.
Fig. 6.
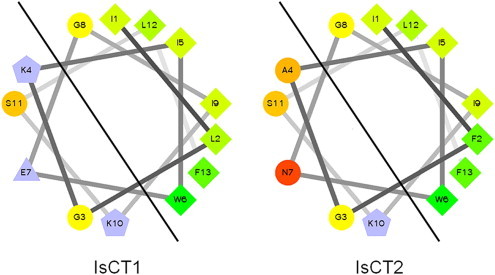
Helical wheel comparison of IsCT and IsCT2, two short cytolytic peptides from the venom of Opisthanthus madagascarienis.
Two AMPs, BmKb1 and BmKn2, have been identified from the venom gland cDNA library of Buthus martensii (Zeng et al., 2004). These peptides are both basic (BmKb1, charge +1; BmKn2, charge +2) and like the IsCT peptides discussed earlier, also have amidated C-terminals. BmKb1 has a 52 amino acid pre-pro sequence containing a 22 amino acid signal sequence, which after post translational modification gives an 18 amino acid mature peptide. In comparison, BmKn2 has a 47 amino acid pre-pro sequence containing a 23 amino acid signal sequence, finally resulting in a 13 amino acid mature peptide. Secondary structure predictions suggest that both BmKb1 and BmKn2 have α-helical regions (amino acids 2–16 and 2–11 respectively) flanked by two random coils; helical wheel projections demonstrate their amphipathic natures. The shorter and more highly charged BmKn2 is considerably more active than BmKb1 (MICs for Gram-positive organisms were typically 0.4–6 μM for BmKn2 as compared with 9–45 μM for BmKb1; MICs for Gram-negative organisms were typically 1–15 μM for BmKn2, as compared with 10–50 μM for BmKb1). The activity of BmKn2 against 18 strains of multidrug resistant N.gonorrhoeae has recently been analysed via an MTT assay with these strains having an MIC50 value of between 6.9 and 27.6 μM (Arpornsuwan et al., In Press). This study also noted the C-terminally amidated residue was critical for function with a dramatic decrease in activity observed upon deletion.
Based on these findings, PCR primer sets were designed to detect BmK1 homologues in other venoms, for example mucroporin from Lychas mucronatus (Dai et al., 2008) and imcroporin from Isometrus maculates (Zhao et al., 2009). Mucroporin shows 51% homology with BmKb1 and consists of a 35-amino acid pre-pro peptide which gives rise to a 17 residue mature peptide amidated at the C-terminal. Mucroporin (charge +1) and a mucroporin-M1 analogue (charge +5) were both active against Gram-positive bacteria (MIC values 3–29 μM), although the difference in charge did not have a consistent effect on bacterial susceptibility. In contrast, both peptides had much weaker activity against Gram-negative bacteria, with MIC values greater 50 μM (Dai et al., 2008).
Because S. aureus was particularly sensitive to mucroporin M1, this peptide was further tested against clinically important isolates of this particular bacteria. The data revealed that mucroporin-M1 can inhibit a range of antibiotic-resistant pathogens of both methicillin- and penicillin- resistant strains (4–10 μM), as well as penicillin- sensitive strains (4–20 μM). Scanning electron microscopy (SEM) studies showed rapid lyses of bacterial cells after mucroporin or mucroporin-M1 treatment, suggesting a membrane disruptive mechanism. Mucroporin M1 has been shown to specifically inhibit RNA viruses, including SARS-corona virus, influenza and measles viruses, presumably by targeting the viral membrane (Li et al., 2011). DNA viruses were not inhibited. The same groups have more recently shown (Zhao et al., 2012) that the peptide inhibits hepatitis B virus replication, both in vitro and in vivo, by activating the mitogen-activated protein kinase (MAPK) pathway, which resulted in the down-regulation of the nuclear hormone transcription factor, HNF4α.
Imcroporin, the second peptide identified as a BmKb1 homologue through a PCR screen, has the same number of amino acids and charge, as mucroporin. Secondary structure prediction and helical wheel analysis suggested that imcroporin has a highly amphipathic nature with a 100% alpha-helical conformation (Zhao et al., 2009). The antimicrobial activity profile of imcroporin was very similar to mucroporin; growth of Gram-negative organisms was weakly inhibited (MICs typically >57 μM) whereas Gram-positive organisms, including methicillin- and penicillin- resistant strains of clinical isolates of S. aureus were more sensitive to the peptide (MICs typically 11–30 μM). For comparison, the MICs of all clinical isolates against vancomycin were 3 μM. Imcroporin was also tested in an in vivo mouse model (Zhao et al., 2009). The peptide (single dose, 60 μg/g, given one hour after intraperitoneal infection with S. aureus) was as effective as vancomycin (60 μg/g) in curing mice; all mice survived, seven days after treatment. However imcroporin also exhibited significant cytolytic properties against mammalian cell lines and red blood cells, as evidenced by MTT assays and haemolysis assays respectively. Imcroporin (57 μM) killed between 25 and 100% of kidney and hepatoma cells and haemolysed 70% of human erythrocytes. Kill kinetics and enzyme activity assays of cell supernatants revealed, as with mucroporin, a rapid membrane disruptive mechanism of action.
The great majority of scorpions are considered harmless and probably as a consequence, the characterisation of peptides found in their venom glands has been very limited. The Asian scorpion, Scorpiops tibetanus is one such example and Cao, Li and colleagues (Yuan et al., 2010) have cloned a new antimicrobial peptide gene (StCT1) from S. tibetanus by screening a venom gland cDNA library with IsCT primers. The precursor of StCT1 is characterized by a signal peptide (24 amino acids) followed by a putative mature peptide of 14 residues and finally, an unusual 37-amino acid acidic propeptide at the C-terminus, suggesting that the mature peptide is C-terminally amidated. A synthetic amidated peptide corresponding to the putative mature peptide of StCT1 showed preferential activity towards S. aureus with weak activity towards Gram-negative organisms, in a similar manner to mucroporin and imcroporin. MIC values against methicillin-resistant S. aureus strains ranged between 85 and 135 μM whilst penicillin-resistant strains were more sensitive (MIC 34 μM). It is interesting to note that although the amidated StC1 peptide has approx 40% homology with IsCT, the latter peptide (in contrast to StC1) has activity against both Gram-positive and Gram-negative organisms.Using the same approach, a StCT2 gene has also been cloned (Cao et al., 2012). A synthetic, amidated 14 amino acid StCT2 peptide, corresponding to the predicted mature peptide, also showed potent activity against S. aureus and methicillin resistant S. aureus (MICs values 3–13 μM). Both StCT1 and StC2 peptides belong to the NDBP -5 family (see Table 1).
Two structurally divergent peptides (meucin-18; meucin-13) have been isolated from a venom gland library of the Asian scorpion Mesobuthus eupeus by a random sequencing strategy (Gao et al., 2009). Based on MIC values, most Gram-positive organisms were 3–4 fold more susceptible to meucin-18 than meucin-13, while Gram-negative organisms were 2–5 fold more susceptible to the longer peptide. Meucin-18 was also 2–14 fold more potent than meucin-13 against a range of fungi and yeast.
Both meucin-18 and meucin-13 were cytotoxic toward rabbit erythrocytes; however meucin-18 (6.25 μM) was twice as haemolytic as meucin-13 at the same concentration (74 vs 38% hemolysis, respectively). Whole cell patch clamp experiments identified a sharp decrease in ion currents followed by a rapid, partial recovery, when rat dorsal root ganglion (DRG) cells were exposed to either peptide. These electrophysiological experiments were supported by morphological studies, when the surface of DRG cells was transformed from a smooth to a rough state after incubation with peptides. Evidence for the peptides causing membrane permeabilization in microbial cells (bacteria, yeast and fungi) was obtained with the fluorescent DNA-binding dye, propidium iodide. Taken together, these studies suggest that meucins cause permanent cell damage.
Gao et al. (2009) have then gone on to try and rationalize their biological data in terms of structural differences between meucin-13 and meucin-18. Both peptides belong to group 5 (Table 1). Meucin-13 has an amidated C-terminal in contrast to meucin-18 which does not. Extensive structural studies have been carried out on both peptides using CD spectroscopy and NMR spectrometry. The CD spectrum of meucin-13 in 50% TFE characterized the peptide as being primarily α-helical. This was confirmed by NMR, which identified an α-helical structure between residues 5–13 with an unordered N-terminus. Meucin-18 showed similar results to meucin-13, although NMR revealed a more ordered N-terminal structure (Fig. 7 ). This is thought to be due to the presence of alanine at position 6 (meucin-13) instead of phenylalanine (meucin-18). A structurally ordered N-terminus is postulated to be important for biological activity. Gao and colleagues have also suggested that differences in biological potency might be related to differences in the hydrophilic/hydrophobic balance between the two meucins due to the more balanced amphipathicity of meucin-18. The C-terminal extension of meucin-18 with a free (charged) carboxy terminus means that overall, meucin-13 is more hydrophobic. This relative increase in hydrophobicity may reduce the biological activity of the latter due to its increased self-association in aqueous solution (see Jiang et al., 2008 for a useful summary of this proposal). Gao and colleagues also noted that meucin-18 has a positively-charged region (adjacent Lys-6 and Lys-11 residues) that could prove crucial for increased interaction with negatively-charged prokaryotic membranes.
Fig. 7.
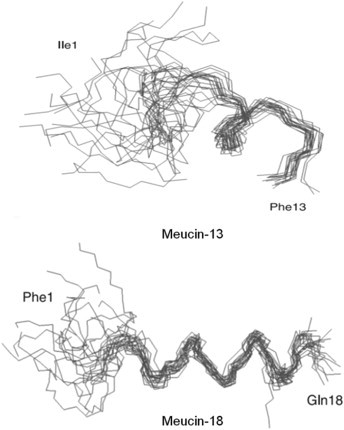
Comparison between NMR structures of meucin-13 and meucin-18 (Gao et al., 2009).
The pattern of α-helical peptides from scorpion venoms showing preferential activity towards Gram-positive organisms has continued with the cloning of Ctriporin by the random screening of a large number of clones from the venom gland cDNA library of Chaerilus tricostatus (Fan et al., 2011). Ctriporin is a 19 amino acid peptide (net charge: +2) with, like many other previously described peptides, an amidated C-terminus that is produced by the post translational cleavage of a 34-residue acidic propeptide. Secondary structure analysis and CD spectra of the peptide in 30 and 70% TFE revealed an amphipathic α-helical structure. However, the ambiguity still remains as to whether the peptide is completely α-helical or contains a random coil region, due to the presence of Gly-4, Gly-7 and Pro-8, residues known to disrupt α-helical structures. MIC values (Gram-positive organisms) ranged from 3 to 5 μM whereas Gram-negative organisms had MICs >49 μM). All clinical isolates of MRSA strains gave MIC values of 5 μM (c.f. vancomycin 3–6 μM). Similarly the penicillin-resistant strain of S. epidermidis had an MIC of 5 μM, analogous to vancomycin. Ctriporin also had antifungal activity against Candida (MIC 10 μM) and in vivo studies with mice demonstrated that topical applications of the peptide were more effective than currently established topical treatments, pexiganan and omiganan pentahydrochloride in treating fungal and S. aureus infections. Time kill kinetics and scanning electron microscope studies suggested that ctriporin caused a rapid disruption of cell membranes, analogous to other α-helical peptides.
Two further members of the NDBP-5 class of short antimicrobial peptide (AamAP1 and AamAP2) have been isolated from the venom of the North African scorpion Androctonus amoreuxi (Almaaytah et al., 2012) and the precursor-encoding cDNAs have been cloned, using a shot-gun approach. They both have 18 amino acids and are amidated at their C-termini. AamAP1 and 2 differ by only 2 residues with leucine being replaced by proline at position 2 and phenylalanine being replaced by isoleucine at position 17. Secondary structure prediction suggested that AamAP2 has a far greater random coil region than AamAP1. However these structural differences did not manifest themselves in changes in biological activity. Although neither peptide was particularly active (MICs 20–150 μM) AamAP1 was slightly more active against Gram-negative organisms while the reverse was true for Gram-positive organisms. Both were equally effective against yeast and caused a significant haemolysis of red blood cells in the same concentration range.
Using a size-selective screening strategy, three new antimicrobial peptides (Pantinin-1, -2 and -3) were characterized from a venom gland cDNA library of the African scorpion Pandinus imperator (Zeng et al., 2013). All three were predicted to be mature peptides of 13–14 amino acid residues with precursor peptides indicating the now-characteristic C-terminal post-translational processing, resulting in C-terminal amidation. Peptides were cationic and secondary structure prediction suggested that pantinins are α-helical, amphipathic structures. Amino acid sequence homology revealed that these peptides belong to the ever-growing 5th family of NDBPs. Pantinins are relatively potent against Gram-positive bacteria (MICs 4–48 μM), including vancomycin- and methicillin-resistant bacteria but are much weaker against Gram-negative bacteria (MICs 36– >87 μM). In addition to their antimicrobial activities, the three peptides showed haemolytic activity on human RBCs in a dose-dependent manner.
Two C-terminal amidated peptides (TsAP-1 and -2) have been isolated from the venom of Tityus serrulatus by shot gun cloning and LC-MS identification (Guo et al., in press). Each peptide contained 17 residues with a net charge of +2. TsAP-1 had very weak antimicrobial activity against yeast and both Gram-positive and Gram-negative organisms (MICs typically 120–160 μM) and showed very little haemolytic activity. TsAP-2 had no effect against Gram-negative organisms at the highest concentrations tested but was surprisingly effective in inhibiting the growth of both yeast and Gram-positive organisms (MICs 5–10 μM). TsAP-2 had weak haemolytic activity (18%) at four times the MIC for Gram-positive bacteria. The α-helical content (76% TsAP-2 vs. 59% TsAP-1) and hydrophobicity (0.51 hydrophobic moment TsAP-2 vs. 0.43 TsAP-1) has been suggested for the divergence in biological activities of these two peptides.
4. Speculations on the mechanism of action of scorpion AMPs
An N-terminal, di-helical region of the scorpine family of cysteine-constrained peptides has been postulated to be responsible for their antimicrobial activity (Zhu and Tytgat, 2004). The first 35 residues of opistoscorpine 1 were noted to have similarity to the cecropins, a family of linear AMP's from the insect Hyalophora cecropia, which have antimicrobial activity and remarkably low haemolytic activity (Holak et al., 1988, Hu et al., 2013). This idea (Zhu and Tytgat, 2004) is supported by the alignment (Fig. 8 ) of cecropin A with the N-terminal regions of 3 other scorpine family peptides (heteroscorpine, panscorpine and opistoscorpine-1). These regions also align well with the two helices of cecropin A determined by NMR (Holak et al., 1988). Mechanistic studies of cysteine constrained peptides such as the scorpines are limited, compared with that of their non-cysteine containing counterparts. To date, no comparable haemolytic or cytolytic studies of the N-terminal regions of scorpines have been published. Because of their noted similarity and low haemolytic activity this should be an attractive and profitable area to revisit.
Fig. 8.

Cecropin A from Hyalophora cecropia, residues highlighted in yellow (A) show the helical regions deduced by NMR. (B) Alignment of the N-terminal regions of 4 scorpine like peptides with Cecropin A. High conservation is seen within areas of the scorpine peptides supporting suggestions made by Zhu and Tytgat, (2004) that the N-terminal region is responsible for the antimicrobial properties of these peptides. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Because mechanistic studies have only been carried out on the long-chain peptide Pin1, it is logical to compare the structures of other long chain peptides to this. Alignment of the sequences of the six biologically active, long-chain AMPs (opistoporins, hadrurin, BmKbpp and vejovine) with Pin1 (Fig. 9 ) reveal a remarkably conserved region within the N-terminal helices, including the Trp corresponding to position 15 in Pin 1.
Fig. 9.
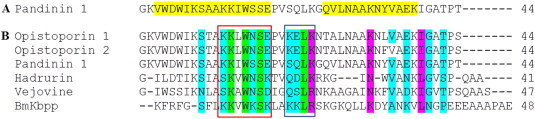
Pandinin 1 from Pandinus imperator-residues highlighted in yellow (A) show the helical regions deduced by NMR. (B) Alignment of the di-helical long chain peptides with Pandinin 1. High conservation is seen within areas of the N-terminal helices (red box) and the hinge region grey box) shown to be critical for function in Pandinin 1 suggesting a shared mechanism of action in this class of peptides. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When compared with the helical regions of Pin1 identified from NMR studies (Nomura et al., 2005) (Fig. 9A, highlighted in yellow), this conservation becomes apparent. There is also high conservation within the hinge region (Fig. 9B). Solid state NMR data of Pin 1 interaction with phospholipid bilayers suggested that the two alpha-helical regions of the peptide cause lipid bilayers to adopt a cubic phase (Nomura et al., 2005). Since the long chain peptides identified in Fig. 9 also have two distinct alpha helices, then it is a likely proposition that these other peptides also promote a cubic phase in lipid bilayers. Taken together, these alignments may provide us with an insight sight into a general mechanism of action of this class of AMP; however studies comparable to that of Pin 1 clearly need to be undertaken before any firm conclusions can be made. Although these long peptides undoubtedly cause membrane disruption, it is very unlikely that they do so by forming pores, simply on the basis of their size. We are therefore left at present with the carpet like model to describe their mechanism of action.
A structural review of diverse antimicrobial peptides highlighted that most of these molecules exhibit imperfect amphipathicity (Wimley, 2010). They are characterized by the presence of polar as well as non-polar faces with the latter containing a “polar pocket”. This has led to the proposal of a new interfacial model for AMP action (Wimley, 2010, Marks et al., 2011), which is a hybrid between the classical carpet model and the more recent toroidal model. The interfacial model consists of three main processes that take place consecutively: (i) an initial electrostatic interaction with the negatively charged bacterial membrane driven by the polar face; (ii) an interaction with the membrane hydrophobic core driven by the peptide hydrophobic face, causing the peptide to insert into the membrane, and (iii) the polar pocket interacts with the phospholipid head groups on the inner membrane leaflet causing negative curvature of this inner membrane (Marks et al., 2011, Bobone et al., 2012).
Analysis of scorpion short chain antimicrobial peptides by helical wheel projection reveals the amphipathic nature of these peptides with no clear “polar pocket” within the structures. However it is well documented that C-terminal amidation increases the biological activity of these short chain cationic peptides and it has been proposed that this is a consequence of an increase in net positive charge (Strandberg et al., 2007). If the helical wheel structures of these peptides are examined (Fig. 10 ), it can be clearly seen that amidation of the negatively charged C-terminal carboxy group lies within the hydrophobic section of the peptide, thus changing the charge distribution of the peptide. However, the slightly longer peptides (e.g imcroporin, mucroporin, ctriporin and BmKb1) have C-terminal amidated residues that lie within the hydrophilic face of the peptide thus changes in charge distribution are less pronounced (Fig. 11 ). This 'rebalancing' of charge between the amphipathic facets of the peptide clearly has implications for biological activity and it poses an interesting question as to what the precise role of this amidation site has on scorpion AMP mechanism of action.
Fig. 10.
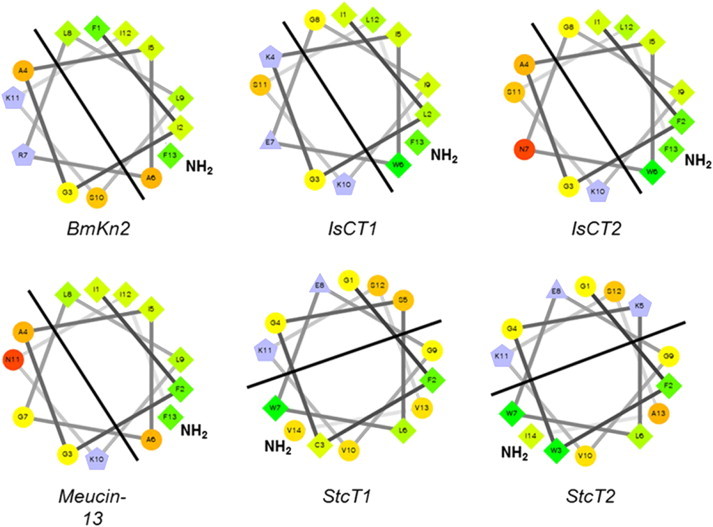
Helical wheel projections of six short chain cytolytic peptides; each peptide exhibits clear amphipathicity with the amidated C-terminal residue laying within the hydrophobic region creating a ‘polar pocket’ that is essential for the interfacial model. Thus this key amidation site is key for the cytolytic mechanism of short chain peptides from scorpions.
Fig. 11.
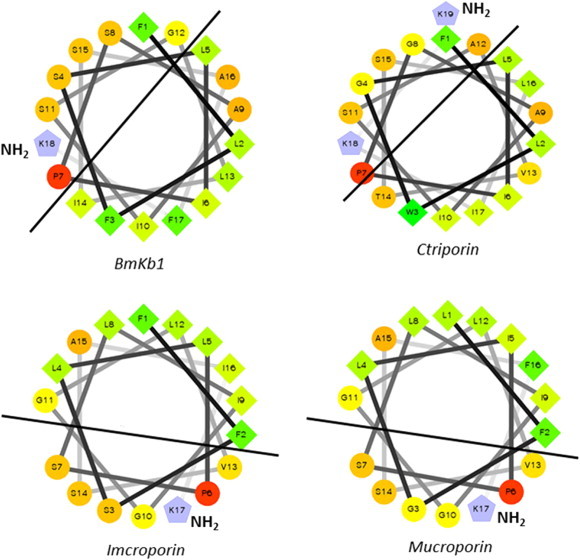
Helical wheel projections of 4 longer chain short cytolytic peptides; each peptide exhibits clear amphipathicity with the amidated C-terminal residue lying within the hydrophilic region. Thus no polar pocket is present which indicates a mechanism of action different from the interfacial model.
Whilst extensive mechanistic studies have yet to be carried out on these peptides with amidation sites within the hydrophilic face, it is instructive to compare them to Pin2 a mid-chain AMP which contains 24 residues (Fig. 12 A). Sequence alignments of Pin2 with the four short chain peptides revealed similar features when a central GALK motif was removed from the middle of the Pin2 sequence (Gly-8 to Lys-11, Fig. 12B). A LIPS motif found within Pin 2 (Leu-12 to Ser-15), thought important for biological activity, is also found in all four peptides with the exception of a Ser→Ala replacement in Ctriporin (Fig. 12C). Finally, another interesting separate alignment of Pin 2 and meucin-18 revealed the same motif as part of a conserved K(L/I)IPSLF sequence (Fig. 12 D). At present there is little biophysical experimental data to verify whether these identified sequence alignments have any structural importance (and therefore biological significance). Further examination at a biophysical level is clearly important if scorpion AMPs are to provide viable templates for therapeutically useful molecules.
Fig. 12.

(A) Pandinin 2 from Pandinus imperator-residues highlighted in yellow show the helical regions deduced by NMR. Residues within the red box thought to be critical for function. (B) Alignment of the 4 mid chain helical peptides with Pandinin 2. (C) Alignment of the four peptides with the GALK motif removed from Pandinin 2 showing a high degree of conservation with the LIPS motif conserved throughout the 4 peptides except in ctriporin where LIPG is seen (red box). (D) Alignment of Pandinin 2 with meucin-18, again a high degree of conservation is seen throughout the peptide with a conservative substitution within the LIPS motif to IIPS. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
5. More recent approaches
Recent proteomic studies (Calvete, 2013, Durban et al., 2013, Gibbs et al., 2013) have highlighted the power of collision- and electron transfer-induced dissociation MS/MS techniques to profile the venoms of a number of snakes for the determination of peptide sequences. These techniques have also been extensively used to analyse the venoms of various scorpions (Abdel-Rahman et al., 2014). Recent studies have highlighted the abundance of AMP-like molecules within the transcriptomes of numerous scorpion venoms. 25% of all expressed sequence tags (ESTs) from transcripts of the venom gland of Tityus Stigmurus, were related to AMPs (Almeida et al., 2012). Similar results were obtained from an analysis of the venom gland glands of the Egyptian scorpion Scorpio maurus palmatus (Abdel-Rahman et al., 2013) and the Chinese scorpion, Lychas mucronatus (Ruiming et al., 2010). In this latter study, scorpions were collected from two different and geographically-isolated areas (Yunnan province and Hainan Island) and the data revealed that the relative abundance of ESTs corresponding to AMPs from scorpions collected on Hainan Island (24% total EST population) were double that of scorpions found in Yunnan. This phenomenon has been highlighted previously in mass spectrometry studies where geographical distribution, climate, age and sex have all been integral to the determination of venom components of a number of different scorpion species (e.g. Newton et al., 2007, Abdel-Rahman et al., 2009, Ma et al., 2009).
A combination of transcriptomic and MS/MS approaches have been used to identify a number of AMPs from the venom of Australian scorpion Urodacus yaschenkoi (Luna-Ramírez et al., 2013a, Luna-Ramírez et al., 2013b). Three short chain α-helical peptides (UyCT1, UyCT3 and UyCT5) were characterized, with amidated C-terminii and sharing 43–77% sequence homology with IsCT. Although all three peptides were equally potent against a range of both Gram-positive and Gram-negative bacteria (MICs typically 1–25 μM), they were also cytolytic (50 μM peptides causing > 81% haemolysis of human erythrocytes). Interestingly, when the N-terminal Gly was removed from either UyCT1 or UyCT2, the hemolytic activity dramatically declined to 7%, although the range of MICs increased in concert. Two scorpine-like peptides and another with homology to Pin-1 were also identified, but no antimicrobial assays have been carried out to date.
A more recent study (Luna Ramirez et al., in press) revealed the synergistic effect of these peptides with combinational treatment increasing the therapeutic index. Against A. baumanni UyCT1 has an MIC of 7 μM whilst UyCT5 has an MIC of 14 μM, however combined the MIC decreases to 1 μM for UyCT1 and 5 μM for UyCT5. Interestingly, the haemolytic potency of each peptide is increased when the peptides are administered separately. Structural studies were also carried out on these four peptides (UyCT1, 2, 3 and 5). All four peptides showed unordered structure in aqueous buffer using CD spectra however form helices when in the presence of vesicles mimicking red blood cells (POPC/Cholesterol), Gram-negative E.coli (POPE/POPG) and Gram-positive S. aureus (POPG/TOCL). Helical wheel projections revealed the characteristic amphipathic nature of the peptides. The membrane activity of these peptides was also confirmed by analysing the D-isomeric form which also showed activity.
Dye release assays and isothermal titration calorimetry (ITC) suggests that all the peptides have a greater affinity towards prokaryotic model membrane systems over eukaryotic ones with a K d in the range of 2–30 μM compared with 150–200 μM, however the dye release assays suggest that the peptides lyse neutral membrane systems with greater ease than anionic systems. This apparent contradiction is postulated by the authors to be due to the increased surface interaction of the peptides with ionic lipids thus inhibiting insertion. However there is contraction between the dye release assays and the peptides biological activity which is thought to be due to the relative simplicity of the actual membrane systems.
Finally, several putative AMP-like peptides have been identified on the basis of sequence homologies, although their antimicrobial activities are still to be determined. These include disulphide-bridged, scorpine-like peptides from Opisthacanthus cayaporum (OcyC7; Silva et al., 2009), Tityus costatus Karsch (TcoScp1; Diego-Garcia et al., 2005), Hadrurus gerstchi (Hge Scorpine and Hge Scorpine 2; Diego-Garcia et al., 2008, Schwartz et al., 2008), Heterometrus petersii (HSP017C and HSP039C; Ma et al., 2010), Scorpiops jendeki (SJE005C and SJE056C; Ma et al., 2009), Pandinus cavimanus (Pcav34 and Pcav23; Diego-Garcia et al., 2012) and S. maurus palmatus (Smp76; Abdel-Rahman et al., 2013).
Non-disulphide bridge peptides include peptides from the venoms of Hadrurus gerstchi (Hge027, Hge028, Hge029; Schwartz et al., 2007), Opisthacanthus cayaporum (Ocy1, Ocy2 and Ocy3; Silva et al., 2009) Pandinus cavimanus (Pcav30 and Pcav7; Diego-Garcia et al., 2012), and S. maurus palmatus (Smp13, Smp24 and Smp43; Abdel-Rahman et al., 2013). Hge027, Hge028, Ocy1 and Ocy2 all have homology to IsCT peptides, whereas Hge029 and Smp43 have homology to Pin-1 and Ocy3 as well as Smp24 have homology to pandinin-2. Pcav30 and Pcav7 are similar to Pin-2, IsCT and BmKn2. A large number of other putative NDBPs have been identified in Heterometrus petersii (HSP013C, with homology to opistoporins, HSP028C and HSP049C with homology to Pin-2, as well as six peptides homologous to IsCT (Ma et al., 2010). Two of these peptides, and Hp1239, both 13 residues in length and C-terminally amidated have recently been examined for their anti-viral activity on Herpes simplex virus type 1 (HSV-1). They caused a 60% decrease in plaque forming ability at 10 μM when incubated with African green monkey kidney cells. However cell viability assays on the same kidney cell line revealed around 80% viability at 40 μM with Hp1036, only 30% of the cells were viable at the same concentration with Hp1239. Both peptides had the same hemolytic activity (50%) at 50 μM (Hong et al., 2013).
6. Strategies to develop antimicrobial peptides into the clinical setting
Whilst the field of antimicrobial peptides has developed rapidly over the last two decades, with a plethora of peptides been identified and numerous structural/functional relationship studies undertaken, one fundamental paradigm still remains – how to develop AMPs into the clinical setting?
As discussed at the beginning of this review a number of AMPs have failed to get approval although these have been largely commercial rather than scientific considerations, although as previously noted regulatory bodies may be starting to relax their attitudes towards approval (Fox, 2013). Despite these positive advances in AMP drug development, stability and immunogenicity still remain major problems when considering systemic therapeutic applications (Yount and Yeaman, 2012).
Several groups have examined the protease stability of AMPs. As an example, Carmona et al. (2013) have examined the biological activity of the proteolytic-insensitive D-conformer of the scorpion AMP, Pin 2. A 30–40% reduction in hemolytic activity was observed with the D-conformer, which retained the antimicrobial profile of the native peptide. Crucially, when incubated with either trypsin, elastase, whole human serum or proteases from P.aeruginosa, only the D conformer retained activity, suggesting a feasible strategy for increasing AMP stability. These results are mirrored by previous studies on phyllogenically diverse AMPs including cecropin A (Wade et al., 1990), melittin and paradaxin (Oren et al., 1997), LL-37 (Dean et al., 2011), magainin (Guell et al., 2011) and mastoparan (Jones and Howl, 2012).
Other strategies employed to increase both peptide stability and plasma half life have focussed on the delivery system, for example using liposome encapsulation (Du and Stenzel, 2014) or conjugation to human serum albumin (HAS) as a carrier protein (Xie, 2010). More recent studies have investigated the potential of inorganic nanostructures for AMP delivery (Brandeli, 2012). Interestingly, as well as increasing stability toward proteases, these strategies have also increased the therapeutic index of AMPs in vivo.
The reduction of immunogenicity of a protein/peptide is of great importance not just to AMPs but to protein therapeutics as a whole and consequently a number of strategies has been proposed to achieve this. Although no studies have been directed toward decreasing the immunogenicity of AMPs in particular, various general strategies have been examined and could provide useful areas for further AMP research. For example PEGaylation is an approved non-toxic and non-immunogenic procedure by interfering with antibody and/or HLA epitope binding (Jevsevar et al., 2010). Similarly peptide glycosylation also results in decreased immunogenicity through a similar mechanism (von Delwig et al., 2006). PEGaylation has also been shown to increase the stability of AMPs in the presence of proteases (Falciani et al., 2014) as well as decrease toxicity and increase biocompatibility (Morris et al., 2012). Conjugation of peptides to IgG has also been carried out to successfully evade the adult immune system (Zambidas and Scott 1996) and is a strategy employed within clinical practise for the delivery of anti-inflammatory drugs. Finally modification of a peptide sequence to delete potential T-cell epitopes have also been examined (De Groot 2007). Therefore while a number of different strategies are available to improve stability and reduce immunogenicity, it is important to emphasize that the central goal of any strategy should be to maintain beneficial biological activity.
One approach in the development of AMPs into the clinical setting could be to use them in combinational therapy with conventional antibiotics (Brouwer et al., 2011). Whilst this approach would not rely on the direct killing of bacteria by AMPs they would act as ‘entry peptides’ allowing the conventional antibiotic to gain entry and interact with its intracellular target where resistance has built up at the level of the cell wall. Another attractive strategy would be to use AMP cocktail therapy (Luna-Ramirez et al., 2014). These authors increased the therapeutic index of several UyCT scorpion venom peptides. However in light of the current failure to bring a single AMP to market, this would suggest that any duel AMP cocktail therapies are presently an idealistic goal.
7. Conclusions: gatherers or hunters?- lessons for the 21st century
Over the past two decades, scorpion venoms have proved a rich source of AMPs with a varying range of mechanisms of action. The increased use of proteomic and transcriptomic approaches has already begun to prove fruitful, with a massive acceleration of peptides discovered by these approaches in the last few years. A rhetorical and thought-provoking question was asked during the 1st Oxford world symposium on venoms (September 2012) – what approach is to be taken within future years regarding venom compound research? Are we gatherers or hunters? (Calvete, 2012).
The use of venom transcripts, mass spectrometry and bioinformatics tools leads to the question whether this combined approach is now regarded as paradigm of choice for identifying low abundance, and (perhaps?) more potent, antimicrobial peptides? In terms of both speed and scope of peptide identification through MS/MS techniques and the establishment of dedicated databases of fragmentation patterns, to the prediction of biological activity via bioinformatics, it is certainly an approach to be given serious consideration. However when one considers the many hundreds (over a thousand?) of antimicrobial peptides characterised in the last twenty years, the lack of any really new therapeutic anti-microbial agents is singularly depressing. We must therefore recognize that the approach of the “gatherers”, presently in vogue, has one crucial intellectual limitation- you only get what you are looking for and is in essence a recipe for identifying more of the same. The alternative approach of the “hunter” may be more time consuming but one can argue that an investment in functional assays is the only way to identify truly novel compounds. One particular striking example of this is the identification of the bactridines (Diaz et al., 2009) whose proposed mechanism of action is far removed from membrane active AMPs, indeed would not be considered a conventional target in other streams of antibiotic development. Such serendipitous breakthroughs should be a lesson to us all.
Whether one is a philosophical hunter or gatherer, it cannot be contested that a more detailed biophysical approach to understanding the mechanisms of action of these new peptides is essential if they are to have a positive impact on the clinical management of infection. Techniques such as NMR (Nomura et al., 2004, Nomura et al., 2005), atomic force microscopy (AFM) (García-Sáez et al., 2007, Fernandez et al., 2012, Lam et al., 2012), secondary ionisation mass spectrometry (SIMS) (Rakowska et al., 2013) and X-ray photoemission electron microscopy (Won et al., 2011) can enable us to re-examine and refine our present interpretations of models of peptide interaction with microbial membranes (Marks et al., 2011, Bobone et al., 2012). Such experimental methods, taken together with molecular techniques to determine potential intracellular targets (e.g. Brogden, 2005) and not forgetting that AMPs also have antiviral activities (e.g. Zhao et al., 2012), are crucial if our ultimate goal of using antimicrobial peptides as next generation therapeutics is to be realised.
Ethical statement
All authors declare that there are no ethical issues involved in this work. The requirements approved by Toxicon and the publisher for accepting manuscripts for publication have been followed.
Acknowledgements
This work was supported by an internal grant from the Biomedical Research Centre (BMRC; Sheffield Hallam University, UK) to Patrick L. Harrison, Keith Miller and Peter N. Strong. Mohamed A. Abdel-Rahman acknowledges travel grants from the Arab Fund for Economic and Social Development (AFESD; The State of Kuwait) and the Daniel Turnberg UK/Middle East Travel Fellowship Scheme (The Academy of Medical Sciences, London (AFESD 2-1-72/1884)) enabling him to work at the BMRC (Sheffield Hallam University, UK).
Footnotes
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.toxicon.2014.06.006.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.toxicon.2014.06.006.
Conflict of interest
The authors declare that there are no conflicts of interest.
Transparency document
Appendix A. Supplementary data
References
- Abdel-Rahman M.A., Quintero-Hernandez V., Possani L.D. Venom proteomic and venomous glands transcriptomic analysis of the Egyptian scorpion Scorpio maurus palmatus (Arachnida: Scorpionidae) Toxicon. 2013;74:193–207. doi: 10.1016/j.toxicon.2013.08.064. [DOI] [PubMed] [Google Scholar]
- Abdel-Rahman M.A., Harrison P.L., Strong P.N. Snapshots of scorpion venomics. J. Arid Environ. 2014 (in press) [Google Scholar]
- Abdel-Rahman M.A., Omran M., Abdel-Nabi M., Ueda H., McVean A. Intraspecific variation in the Egyptian Scorpion Scorpio maurus palmatus venom collected from different biotopes. Toxicon. 2009;53:349–359. doi: 10.1016/j.toxicon.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Almaaytah A., Zhou M., Wang L., Chen T., Walker B., Shaw C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides. 2012;35:291–299. doi: 10.1016/j.peptides.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Almeida D.D., Scortecci K.C., Kobashi L.S., Agnez-Lima L.F., Medeiros S.R., Silva-Junior A.A. Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genomics. 2012;13:362. doi: 10.1186/1471-2164-13-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann G., Mueller P. A molecular model of membrane excitability. J. Supramol. Struct. 1974;2(5–6):538–557. doi: 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- Belokoneva O.S., Satake H., Maltseva E.L., Palmina N.P., Villegas E., Nakajima T., Corzo G. Pore formation of phospholipid membranes by the action of two hemolytic arachnid peptides of different size. Biochim. Biophys. Acta. 2004;1664:182–188. doi: 10.1016/j.bbamem.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Belokoneva O.S., Villegas E., Corzo G., Dai L., Nakajima T. The hemolytic activity of six arachnid cationic peptides is affected by the phosphatidylcholine-to-sphingomyelin ratio in lipid bilayers. Biochim. Biophys. Acta. 2003;1617:22–30. doi: 10.1016/j.bbamem.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Bobone S., Roversi D., Giordano L., Zotti M., Formaggio F., Toniolo C., Park Y., Stella L. Lipid dependence of antimicrobial peptide activity is an unreliable experimental test for different pore models. Biochemistry. 2012;51(51):10124–10126. doi: 10.1021/bi3015086. [DOI] [PubMed] [Google Scholar]
- Bontems F., Roumestand C., Gilquin B., Ménez A., Toma F. Refined structure of charybdotoxin: common motifs in scorpion toxins and insect defensins. Science. 1991;254:1521–1523. doi: 10.1126/science.1720574. [DOI] [PubMed] [Google Scholar]
- Borges A., Alfonzo M.J., Garcia C.C., Winand N.J., Leipold E., Heinemann S.H. Isolation, molecular cloning and functional characterization of a novel beta-toxin from the Venezuelan scorpion, Tityus zulianus. Toxicon. 2004;43:671–684. doi: 10.1016/j.toxicon.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Brandeli A. Nanostructures as promising tools for delivery of antimicrobial peptides. Mini Rev. Med. Chem. 2012;12(8):731–741. doi: 10.2174/138955712801264774. [DOI] [PubMed] [Google Scholar]
- Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Brouwer C.P., Rahman M., Welling M.M. Discovery and development of a synthetic peptide derived from lactoferrin for clinical use. Peptides. 2011;32(9):1953–1963. doi: 10.1016/j.peptides.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. The 1st Oxford World Symposium on Venoms, Venoms 2012. Oxford University; UK: 2012. (Prote) omics in snake venom research-A mere academic exercise? [Google Scholar]
- Calvete J. Snake venomics: from the inventory of toxins to biology. Toxicon. 2013;75:44–62. doi: 10.1016/j.toxicon.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Cao L., Li Z., Zhang R., Wu Y., Li W., Cao Z. StCT2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides. 2012;36:213–220. doi: 10.1016/j.peptides.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Carmona G., Rodriguez A., Juarez D., Corzo G., Villegas E. Improved protease stability of the antimicrobial peptide Pin2 substituted with D-amino acids. 2013;32(6):456–466. doi: 10.1007/s10930-013-9505-2. [DOI] [PubMed] [Google Scholar]
- Chopra I., Schofield C., Everett M., O'Neill A., Miller K., Wilcox M., Frere J.M., Dawson M., Czaplewski L., Urleb U., Courvalin P. Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect. Dis. 2006;8(2):133–139. doi: 10.1016/S1473-3099(08)70018-5. [DOI] [PubMed] [Google Scholar]
- Cociancich S., Goyffon M., Bontems F., Bulet P., Bouet F., Menez A., Hoffmann J. Purification and characterization of a scorpion defensin, a 4kDa antibacterial peptide presenting structural similarities with insect defensins and scorpion toxins. Biochem Biophys. Res. Commun. 1993;194:17–22. doi: 10.1006/bbrc.1993.1778. [DOI] [PubMed] [Google Scholar]
- Conde R., Zamudio F.Z., Rodriguez M.H., Possani L.D. Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett. 2000;471:165–168. doi: 10.1016/s0014-5793(00)01384-3. [DOI] [PubMed] [Google Scholar]
- Corzo G., Escoubas P., Villegas E., Barnhjam K.J., He W., Norton R.S., Nakajima T. Characterization of unique amphipathic antimicrobial peptides from venom of the scorpion Pandinus imperator. Biochem. J. 2001;359:35–45. doi: 10.1042/0264-6021:3590035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Ma Y., Zhao Z., Zhao R., Wang Q., Wu Y.L., Cao Z.J., Li W.X. Mucroporin, the first cationic host defense peptide from the venom of Lychas mucronatus. Antimicrob. Agents Chemother. 2008;52:3967–3972. doi: 10.1128/AAC.00542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Corzo G., Naoki H., Andriantsiferana M., Nakajima T. Purification, structure-function analysis, and molecular characterization of novel linear peptides from scorpion Opisthacanthus madagascariensis. Biochem Biophys. Res. Commun. 2002;293:1514–1522. doi: 10.1016/S0006-291X(02)00423-0. [DOI] [PubMed] [Google Scholar]
- Dai L., Yasuda A., Naoki H., Corzo G., Andriantsiferana M., Nakajima T. IsCT, a novel cytotoxic linear peptide from scorpion Opisthacanthus madagascariensis. Biochem Biophys. Res. Commun. 2001;286:820–825. doi: 10.1006/bbrc.2001.5472. [DOI] [PubMed] [Google Scholar]
- Dean S.N., Bishop B.M., van Hoek M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Micrcobiol. 2011:111–114. doi: 10.1186/1471-2180-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot A.S., Rivera D.S., McMurry J.A., Buus S., Martin W. Identification of immunogenic HLA-B7 “Achilles' heel” epitopes within highly conserved regions of HIV. Vaccine. 2007;26(24):3059–3071. doi: 10.1016/j.vaccine.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P., D'Suze G., Salazar V., Sevcik C., Shannon J.D., Sherman N.E., Fox J.W. Antibacterial activity of six novel peptides from Tityus discrepans scorpion venom. A fluorescent probe study of microbial membrane Na+ permeability changes. Toxicon. 2009;54:802–817. doi: 10.1016/j.toxicon.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Diego-Garcia E., Abdel-Mottaleb Y., Schwartz E.F., Rodriguez de la Vega R.C., Tytgat J., Possani L.D. 2008. Cytolytic and K+ Channel Blocking Activities of Beta-KTx and Scorpine-like Peptides Purified from Scorpion Venoms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diego-Garcia E., Batista C.V., Garcia-Gomez B.I., Lucas S., Candido D.M., Gomez-Lagunas F., Possani L.D. The Brazilian scorpion Tityus costatus Karsch: genes, peptides and function. Toxicon. 2005;45:273–283. doi: 10.1016/j.toxicon.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Diego-Garcia E., Peigneur S., Clynen E., Marien T., Czech L., Schoofs L., Tytgat J. Molecular diversity of the telson and venom components from Pandinus cavimanus (Scorpionidae Latreille 1802): transcriptome, venomics and function. Proteomics. 2012;12:313–328. doi: 10.1002/pmic.201100409. [DOI] [PubMed] [Google Scholar]
- Diego-Garcia E., Schwartz E.F., D'Suze G., Gonzalez S.A., Batista C.V., Garcia B.I., de la Vega R.C., Possani L.D. Wide phylogenetic distribution of Scorpine and long-chain beta-KTx-like peptides in scorpion venoms: identification of “orphan” components. Peptides. 2007;28:31–37. doi: 10.1016/j.peptides.2006.06.012. [DOI] [PubMed] [Google Scholar]
- D'Suze G., Sevcik C., Corona M., Zamudio F.Z., Batista C.V.F., Coronas F.I., Possani L.D. Ardiscretin a novel arthropod-selective toxin from Tityus discrepans scorpion venom. Toxicon. 2004;43:263–272. doi: 10.1016/j.toxicon.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Du A.W., Stenzel M.H. Drug carriers for the delivery of therapeutic peptides. Biomacromolecules. 2014;15(4):1097–1114. doi: 10.1021/bm500169p. [DOI] [PubMed] [Google Scholar]
- Durban J., Pérez A., Sanz L., Gómez A., Bonilla F., Rodríguez S., Chacón D., Sasa M., Angulo Y., Gutiérrez J.M., Calvete J.J. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciani C., Lozzi L., Scali S., Brunetti J., Bracci L., Pini A. Site-specific pegylation of an antimicrobial peptide increases resistance to Pseudomonas aeruginosa elastase. Amino Acids. 2014;46(5):1403–1407. doi: 10.1007/s00726-014-1686-2. [DOI] [PubMed] [Google Scholar]
- Fan Z., Cao L., He Y., Hu J., Di Z., Wu Y., Li W., Cao Z. Ctriporin, a new anti-methicillin-resistant Staphylococcus aureus peptide from the venom of the scorpion Chaerilus tricostatus. Antimicrob. Agents Chemother. 2011;55(11):5220–5529. doi: 10.1128/AAC.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.L. Antimicrobial peptides stage a comeback. Nat. Biotechnol. 2013;31(5):379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- Fernandez D.I., Le Brun A.P., Whitwell T.C., Sani M.A., James M., Separovic F. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys. Chem. Chem. Phys. 2012;14:15739–15751. doi: 10.1039/c2cp43099a. [DOI] [PubMed] [Google Scholar]
- Gao B., Sherman P., Luo L., Bowie J., Zhu S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009;23:1230–1245. doi: 10.1096/fj.08-122317. [DOI] [PubMed] [Google Scholar]
- Gao B., Xu J., Del Carmen Rodriguez M., Lanz-Mendoza H., Hernandez-Rivas R., Du W., Zhu S. Characterization of two linear cationic antimalarial peptides in the scorpion Mesobuthus eupeus. Biochimie. 2010;92:350–359. doi: 10.1016/j.biochi.2010.01.011. [DOI] [PubMed] [Google Scholar]
- García-Sáez A.J., Chiantia S., Salgado J., Schwille P. Pore formation by a Bax-derived peptide: effect on the line tension of the membrane probed by AFM. Biophys. J. 2007;93:103–112. doi: 10.1529/biophysj.106.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs H.L., Sanz L., Sovic M.G., Calvete J.J. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.) PLoS One. 2013;8:e67220. doi: 10.1371/journal.pone.0067220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon Y.L., Romanowski E.G., McDermott A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005;30(7):505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell I., Cabrefiga J., Badosa E., Ferre R., Talleda M., Bardají E., Planas M., Feliu L., Montesinos E. Improvement of the efficacy of linear undecapeptides against plant-pathogenic bacteria by incorporation of D-amino acids. Appl. Environ. Microbiol. 2011;77(8):2667–2675. doi: 10.1128/AEM.02759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Ma, C., Du, Q., Wie, R., Wang, L., Zhou, M., Chen, T., Shaw, C., Two peptides, TsAP-1 and TsAP-2, from the venom of the Brazilian yellow scorpion, Tityus serrulatus: evaluation of their antimicrobial and anticancer activities. Biochimie. (in press). [DOI] [PubMed]
- Hernandez-Aponte C.A., Silva-Sanchez J., Quintero-Hernandez V., Rodriguez-Romero A., Balderas C., Possani L.D., Gurrola G.B. Vejovine, a new antibiotic from the scorpion venom of Vaejovis mexicanus. Toxicon. 2011;57:84–92. doi: 10.1016/j.toxicon.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Holak T.A., Engström A., Kraulis P.J., Lindeberg G., Bennich H., Jones T.A., Gronenborn A.M., Clore G.M. The solution conformation of the antibacterial peptide cecropin A: a nuclear magnetic resonance and dynamical simulated annealing study. Biochemistry. 1988;27:7620–7629. doi: 10.1021/bi00420a008. [DOI] [PubMed] [Google Scholar]
- Hong W., Li T., Song Y., Zhang R., Zeng Z., Han S., Zhang X., Wu Y., Li W., Cao Z. Inhibitory activity and mechanism of two scorpion venom peptides against herpes simplex virus type 1. Antivir. Res. 2013;102:1–10. doi: 10.1016/j.antiviral.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Wang C., Guo X., Li W., Wang Y., He Q. Broad activity against porcine bacterial pathogens displayed by two insect antimicrobial peptides moricin and cecropin B. Mol. Cells. 2013;35:106–114. doi: 10.1007/s10059-013-2132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen H., Hamill P., Hancock R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevsevar S., Kunstelj M., Porekar V.G. PEGylation of therapeutic proteins. Biotechnol. J. 2010;5(1):113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Vasil A.I., Hale J.D., Hancock R.E., Vasil M.L., Hodges R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers. 2008;90:369–383. doi: 10.1002/bip.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Howl J. Enantiomer-specific bioactivities of peptidomimetic analogues of mastoparan and mitoparan: characterization of inverso mastoparan as a highly efficient cell penetrating peptide. Bioconjug. Chem. 2012;23(1):47–56. doi: 10.1021/bc2002924. [DOI] [PubMed] [Google Scholar]
- Kuhn-Nentwig L. Antimicrobial and cytolytic peptides of venomous arthropods. Cell. Mol. Life Sci. 2003;60:2651–2668. doi: 10.1007/s00018-003-3106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy K.K., Toleman M.A., Walsh T.R., Bagaria J., Butt F. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K.L., Wang H., Siaw T.A., Chapman M.R., Waring A.J., Kindt J.T., Lee K.Y. Mechanism of structural transformations induced by antimicrobial peptides in lipid membranes. Biochim. Biophys. Acta. 2012;18:194–204. doi: 10.1016/j.bbamem.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Lamb H.M., Wiseman L.R. Pexiganan acetate. Drugs. 1998;56(6):1047–1052. doi: 10.2165/00003495-199856060-00011. [DOI] [PubMed] [Google Scholar]
- Lee K., Shin S.Y., Kim K., Lim S.S., Hahm K.S., Kim Y. Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs. Biochem Biophys. Res. Commun. 2004;323:712–719. doi: 10.1016/j.bbrc.2004.08.144. [DOI] [PubMed] [Google Scholar]
- Li Q., Zhao Z., Zhou D., Chen Y., Hong W., Cao L., Yang J., Zhang Y., Shi W., Cao Z., Wu Y., Yan H., Li W. Virucidal activity of a scorpion venom peptide variant mucroporin-M1 against measles, SARS-CoV and influenza H5N1 viruses. Peptides. 2011;32(7):1518–1525. doi: 10.1016/j.peptides.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D.M., Mushtaq S., Warner M. Activity of the anti-MRSA carbapenem razupenem (PTZ601) against Enterobacteriaceae with defined resistance mechanisms. J. Antimicrob. Chemother. 2009;64:330–335. doi: 10.1093/jac/dkp187. [DOI] [PubMed] [Google Scholar]
- Ludtke S.J., He K., Heller W.T., Harroun T.A., Yang L., Huang H.W. Membrane pores induced by magainin. Biochemistry. 1996;35(43):13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- Luna-Ramírez K., Sani M.A., Silva-Sanchez J., Jiménez-Vargas J.M., Reyna-Flores F., Winkel K.D., Wright C.E., Possani L.D., Separovic F. Membrane interactions and biological activity of antimicrobial peptides from Australian scorpion. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbamem.2013.10.022. (in press) [DOI] [PubMed] [Google Scholar]
- Luna-Ramírez K., Quintero-Hernandez V., Vargas-Jaimes L., Batista C.V., Winkel K., Possani L.D. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon. 2013;63:44–54. doi: 10.1016/j.toxicon.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Luna-Ramírez K., Sani M.A., Silva-Sanchez J., Jiménez-Vargas J.M., Reyna-Flores F., Winkel K.D., Wright C.E., Possani L.D., Separovic F. Membrane interactions and biological activity of antimicrobial peptides from Australian scorpion. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbamem.2013.10.022. [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhao R., He Y., Li S., Liu J., Wu Y., Cao Z., Li W. Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: implication for the evolution of the scorpion venom arsenal. BMC Genomics. 2009;10:290. doi: 10.1186/1471-2164-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Zhao R., Zhang W., He Y., Wu Y., Cao Z., Guo L., Li W. Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics. 2010;10:2471–2485. doi: 10.1002/pmic.200900763. [DOI] [PubMed] [Google Scholar]
- Marks J.R., Placone J., Hristova K., Wimley W.C. Spontaneous membrane-translocating peptides by orthogonal high-throughput screening. J. Am. Chem. Soc. 2011;133:8995–9004. doi: 10.1021/ja2017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K., Sugishita K., Ishibe N., Ueha M., Nakata S., Miyajima K., Epand R.M. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- Miyashita M., Sakai A., Matsushita N., Hanai Y., Nakagawa Y., Miyagawa H. A novel amphipathic linear peptide with both insect toxicity and antimicrobial activity from the venom of the scorpion Isometrus maculatus. Biosci. Biotechnol. Biochem. 2010;74:364–369. doi: 10.1271/bbb.90723. [DOI] [PubMed] [Google Scholar]
- Moerman L.F.A., Bosteels S., Noppe W., Willems J., Clynen E., Schoofs L., Thevissen K., Tytgat J., Van Eldere J., van der Walt J., Verdonck F. Antibacterial and antifungal properties of alpha-helical, cationic peptides in the venom of scorpions from southern Africa. Eur. J. Biochem. 2002;269:4799–4810. doi: 10.1046/j.1432-1033.2002.03177.x. [DOI] [PubMed] [Google Scholar]
- Morris C.J., Beck K., Fox M.A., Ulaeto D., Clark G.C., Gumbleton M. Pegylation of antimicrobial peptides maintains the active peptide conformation, model membrane interactions, and antimicrobial activity while improving lung tissue biocompatibility following airway delivery. Antimicrob. Agents Chemother. 2012;56(6):3298–3308. doi: 10.1128/AAC.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mygind P.H., Fischer R.L., Schnorr K.M., Hansen M.T., Sönksen C.P., Ludvigsen S., Raventós D., Buskov S., Christensen B., De Maria L., Taboureau O., Yaver D., Elvig-Jørgensen S.G., Sørensen M.V., Christensen B.E., Kjaerulff S., Frimodt-Moller N., Lehrer R.I., Zasloff M., Kristensen H.H. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437(7061):975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- Newton K.A., Clench M.R., Deshmukh R., Jeyaseelan K., Strong P.N. Mass fingerprinting of toxic fractions from the venom of the Indian red scorpion, Mesobuthus tamulus: biotope-specific variation in the expression of venom peptides. Rapid Commun. Mass Spectrom. 2007;21:3467–3476. doi: 10.1002/rcm.3240. [DOI] [PubMed] [Google Scholar]
- Nie Y., Zeng X.C., Yang Y., Luo F., Luo X., Wu S., Zhang L., Zhou J. A novel class of antimicrobial peptides from the scorpion Heterometrus spinifer. Peptides. 2012;38:389–394. doi: 10.1016/j.peptides.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Nomura K., Corzo G., Nakajima T., Iwashita T. Orientation and pore-forming mechanism of a scorpion pore-forming peptide bound to magnetically oriented lipid bilayers. Biophys. J. 2004;87:2497–2507. doi: 10.1529/biophysj.104.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Ferrat G., Nakajima T., Darbon H., Iwashita T., Corzo G. Induction of morphological changes in model lipid membranes and the mechanism of membrane disruption by a large scorpion-derived pore-forming peptide. Biophys. J. 2005;89:4067–4080. doi: 10.1529/biophysj.105.070292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Z., Hong J., Shai Y. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J. Biol. Chem. 1997;272(23):14643–14649. doi: 10.1074/jbc.272.23.14643. [DOI] [PubMed] [Google Scholar]
- Peigneur S., Sevcik C., Tytgat J., Castillo C., D'Suze G. Subtype specificity interaction of bactridines with mammalian, insect and bacterial sodium channels under voltage clamp conditions. FEBS J. 2012;279:4025–4038. doi: 10.1111/j.1742-4658.2012.08808.x. [DOI] [PubMed] [Google Scholar]
- Pouny Y., Shai Y. Interaction of D-amino acid incorporated analogues of pardaxin with membranes. Biochemistry. 1992;31(39):9482–9490. doi: 10.1021/bi00154a022. [DOI] [PubMed] [Google Scholar]
- Rakowska P.D., Jiang H., Ray S., Pyne A., Lamarre B., Carr M., Judge P.J., Ravi J., Gerling U.I., Koksch B., Martyna G.J., Hoogenboom B.W., Watts A., Crain J., Grovenor C.R., Ryadnov M.G. Nanoscale imaging reveals laterally expanding antimicrobial pores in lipid bilayers. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8918–8923. doi: 10.1073/pnas.1222824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carreto S., Quintero-Hernandez V., Jimenez-Vargas J.M., Corzo G., Possani L.D., Becerril B., Ortiz E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides. 2012;34:290–295. doi: 10.1016/j.peptides.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Villegas E., Satake H., Possani L.D., Corzo G. Amino acid substitutions in an alpha-helical antimicrobial arachnid peptide affect its chemical properties and biological activity towards pathogenic bacteria but improves its therapeutic index. Amino Acids. 2011;40:61–68. doi: 10.1007/s00726-009-0449-y. [DOI] [PubMed] [Google Scholar]
- Ruiming Z., Yibao M., Yawen H., Zhiyong D., Yingliang W., Zhijian C., Wenxin L. Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genom. 2010;11:452. doi: 10.1186/1471-2164-11-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H.S., Fedler K.A., Rennie R.P., Stevens S., Jones R.N. Omiganan pentahydrochloride (MBI 226), a topical 12-amino-acid cationic peptide: spectrum of antimicrobial activity and measurements of bactericidal activity. Antimicrob. Agents Chemother. 2004;48(8):3112–3118. doi: 10.1128/AAC.48.8.3112-3118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E.F., Diego-Garcıa E., Rodrıguez de la Vega R.C., Possani L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones) BMC Genom. 2007;16(8):119. doi: 10.1186/1471-2164-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E.F., Camargos T.S., Zamudio F.Z., Silva L.P., Bloch C., Jr., Caixeta F., Schwartz C.A., Possani L.D. Mass spectrometry analysis, amino acid sequence and biological activity of venom components from the Brazilian scorpion Opisthacanthus cayaporum. Toxicon. 2008;51:1499–1508. doi: 10.1016/j.toxicon.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Silva E.C.N., Camargos T.S., Maranhão A.Q., Silva-Pereira I., Silva L.P., Possani L.D., Schwartz E.F. Cloning and characterization of cDNA sequences encoding for new venom peptides of the Brazilian scorpion Opisthacanthus cayaporum. Toxicon. 2009;54:252–261. doi: 10.1016/j.toxicon.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Strandberg E., Deniz T., Ieronimo M., Kanithasen N., Wadhwani P., Ulrich A. Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl. Chem. 2007;79(4):717–728. [Google Scholar]
- Torres-Larios A., Gurrola G.B., Zamudio F.Z., Possani L.D. Hadrurin, a new antimicrobial peptide from the venom of the scorpion Hadrurus aztecus. Eur. J. Biochem. 2000;267:5023–5031. doi: 10.1046/j.1432-1327.2000.01556.x. [DOI] [PubMed] [Google Scholar]
- Uawonggul N., Thammasirirak S., Chaveerach A., Arkaravichien T., Bunyatratchata W., Ruangjirachuporn W., Jearranaiprepame P., Nakamura T., Matsuda M., Kobayashi M., Hattori S., Daduang S. Purification and characterization of Heteroscorpine-1 (HS-1) toxin from Heterometrus laoticus scorpion venom. Toxicon. 2007;49:19–29. doi: 10.1016/j.toxicon.2006.09.003. [DOI] [PubMed] [Google Scholar]
- von Delwig A., Altmann D.M., Isaacs J.D., Harding C.V., Holmdahl R., McKie N., Robinson J.H. The impact of Glycosylation on HLA-DR1-restricted T Cell Recognition of Type II Collagen in a Mouse Model. 2006;54(2):482–491. doi: 10.1002/art.21565. [DOI] [PubMed] [Google Scholar]
- Wade D., Boman A., Wåhlin B., Drain C.M., Andreu D., Boman H.G., Merrifield R.B. All-D amino acid-containing channel-forming antibiotic peptides. PNAS. 1990;87(12):4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won A., Khan M., Gustin S., Akpawu A., Seebun D., Avis T.J., Leung B.O., Hitchcock A.P., Ianoul A. Investigating the effects of L- to D-amino acid substitution and deamidation on the activity and membrane interactions of antimicrobial peptide anoplin. Biochim. Biophys. Acta. 2011;1808:1592–1600. doi: 10.1016/j.bbamem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Xie D., Yao C., Wang L., Min W., Xu J., Xiao J., Huang M., Chen B., Liu B., Li X., Jiang H. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob. Agents Chemother. 2010;54(1):191–196. doi: 10.1128/AAC.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhao Z., He Y., Wu L., Cai D., Hong W., Wu Y., Cao Z., Zheng C., Li W. A new natural alpha-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides. 2011;32:11–19. doi: 10.1016/j.peptides.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Yount N.Y., Yeaman M.R. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 2012;52:337–360. doi: 10.1146/annurev-pharmtox-010611-134535. [DOI] [PubMed] [Google Scholar]
- Yuan W., Cao L., Ma Y., Mao P., Wang W., Zhao R., Wu Y., Cao Z., Li W. Cloning and functional characterization of a new antimicrobial peptide gene StCT1 from the venom of the scorpion Scorpiops tibetanus. Peptides. 2010;31:22–26. doi: 10.1016/j.peptides.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Zambidis E.T., Scott D.W. Epitope-specific tolerance induction with an engineered immunoglobulin. PNAS. 1996;93(10):5019–5024. doi: 10.1073/pnas.93.10.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zeng X.C., Corzo G., Hahin R. Scorpion venom peptides without disulfide bridges. IUBMB Life. 2005;57:13–21. doi: 10.1080/15216540500058899. [DOI] [PubMed] [Google Scholar]
- Zeng X.C., Li W.X., Peng F., Zhu Z.H. Cloning and characterization of a novel cDNA sequence encoding the precursor of a novel venom peptide (BmKbpp) related to a bradykinin-potentiating peptide from Chinese scorpion Buthus martensii Karsch. IUBMB Life. 2000;49:207–210. doi: 10.1080/713803610. [DOI] [PubMed] [Google Scholar]
- Zeng X.C., Wang S., Nie Y., Zhang L., Luo X. Characterization of BmKbpp, a multifunctional peptide from the Chinese scorpion Mesobuthus martensii Karsch: gaining insight into a new mechanism for the functional diversification of scorpion venom peptides. Peptides. 2012;33:44–51. doi: 10.1016/j.peptides.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Zeng X.C., Wang S.X., Zhu Y., Zhu S.Y., Li W.X. Identification and functional characterization of novel scorpion venom peptides with no disulfide bridge from Buthus martensii Karsch. Peptides. 2004;25:143–150. doi: 10.1016/j.peptides.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Zeng X.C., Zhou L., Shi W., Luo X., Zhang L., Nie Y., Wang J., Wu S., Cao B., Cao H. Three new antimicrobial peptides from the scorpion Pandinus imperator. Peptides. 2013;45:28–34. doi: 10.1016/j.peptides.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Ma Y., Dai C., Zhao R., Li S., Wu Y., Cao Z., Li W. Imcroporin, a new cationic antimicrobial peptide from the venom of the scorpion Isometrus maculates. Antimicrob. Agents Chemother. 2009;53:3472–3477. doi: 10.1128/AAC.01436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Hong W., Zeng Z., Wu Y., Hu K., Tian X., Li W., Cao Z. Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J. Biol. Chem. 2012;287(36):30181–30190. doi: 10.1074/jbc.M112.370312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li W., Jiang D., Zeng X. Evidence for the existence of insect defensin-like peptide in scorpion venom. IUBMB Life. 2000;50:57–61. doi: 10.1080/15216540050176601. [DOI] [PubMed] [Google Scholar]
- Zhu S., Tytgat J. The scorpine family of defensins: gene structure, alternative polyadenylation and fold recognition. Cell. Mol. Life Sci. 2004;61:1751–1763. doi: 10.1007/s00018-004-4149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Borges A., Garcia C.C., Lugo E., Alfonzo M.J., Jowers M.J., Op den Camp H.J.M. Diversity of long-chain toxins in Tityus zulianus and Tityus discrepans venoms (Scorpiones, Buthidae): molecular, immunological, and mass spectral analyses. Comp. Biochem Physiol. 2006;142C:240–252. doi: 10.1016/j.cbpc.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Morgenstern D., Rohde B.H., King G.F., Tal T., Sher D., Zlotkin E. The tale of a resting gland: transcriptome of a replete venom gland from the scorpion Hottentotta judaicus. Toxicon. 2011;57:695–703. doi: 10.1016/j.toxicon.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Valdez-Velázquez L.L., Quintero-Hernández V., Romero-Gutiérrez M.T., Coronas F.I., Possani L.D. Mass fingerprinting of the venom and transcriptome of venom gland of scorpion Centruroides tecomanus. PLoS One. 2013;8:e66486. doi: 10.1371/journal.pone.0066486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D., Yao C., Wang L., Min W., Xu J., Xiao J., Huang M., Chen B., Liu B., Li X., Jiang H. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob. Agents Chemother. 2012;54(1):191–196. doi: 10.1128/AAC.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


