Abstract
The porcine reproductive and respiratory syndrome virus (PRRSV) glycoprotein 4 (GP4) resembles a typical type I membrane protein in its structure but lacks a hydrophilic tail at the C-terminus, suggesting that GP4 may be a lipid-anchored membrane protein. Using the human decay-accelerating factor (DAF; CD55), a known glycosyl-phosphatidylinositol (GPI) lipid-anchored protein, chimeric constructs were made to substitute the GPI-anchor domain of DAF with the putative lipid-anchor domain of GP4, and their membrane association and lipase cleavage were determined in cells. The DAF-GP4 fusion protein was transported to the plasma membrane and was cleaved by phosphatidylinositol-specific phospholipase C (PI-PLC), indicating that the C-terminal domain of GP4 functions as a GPI anchor. Mutational studies for residues adjacent to the GPI modification site and characterization of respective mutant viruses generated from infectious cDNA clones show that the ability of GP4 for membrane association corresponded to virus viability and growth characteristics. The residues T158 (ω − 2, where ω is the GPI moiety at E160), P159 (ω − 1), and M162 (ω + 2) of GP4 were determined to be important for virus replication, with M162 being of particular importance for virus infectivity. The complete removal of the peptide–anchor domain in GP4 resulted in a complete loss of virus infectivity. The depletion of cholesterol from the plasma membrane of cells reduced the virus production, suggesting a role of lipid rafts in PRRSV infection. Remarkably, GP4 was found to co-localize with CD163 in the lipid rafts on the plasma membrane. Since CD163 has been reported as a cellular receptor for PRRSV and GP4 has been shown to interact with this receptor, our data implicates an important role of lipid rafts during entry of the virus.
Keywords: PRRS, GP4, Infectious clone, GPI, Lipid rafts, Membrane association, CD163
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a recently emerged viral disease and causes significant economic losses to the swine industry today. The etiological agent is the PRRS virus (PRRSV), which is a member of the family Arteriviridae (Meulenberg et al., 1993, Meulenberg et al., 1994, Wensvoort et al., 1991, Nelson et al., 1993) that includes other viruses such as equine arteritis virus (EAV), lactate dehydrogenase-elevating virus of mice (LDV), and simian hemorrhagic fever virus (SHFV). The family Arteriviridae together with the families Coronaviridae and Roniviridae form the order Nidovirales (Cavanagh, 1997, Cowley et al., 2000, González et al., 2003, Smits et al., 2003). PRRSV contains a single-stranded positive-sense RNA genome of approximately 15 kb that encodes two large non-structural polyproteins (PP1a and PP1a/1b) in the 5′-terminal 12 kb region and 8 structural proteins in the 3′-terminal 3 kb region: GP2 (glycoprotein 2), E (small envelope), GP3, GP4, GP5, ORF5a, M (membrane), and N (nucleocapsid) proteins in order (Firth et al., 2011, Johnson et al., 2011, Meulenberg et al., 1993, Nelsen et al., 1999, Snijder and Meulenberg, 1998, Snijder et al., 1999, Wootton et al., 2000). While N protein associates with the genomic RNA and makes up the viral capsid, the other proteins are membrane-associated. Of these, GP5 and M form a disulfide-linked heterodimer (Mardassi et al., 1996) that is essential for virus infectivity (Faaberg et al., 1995, Snijder et al., 2003). The E protein is a myristoylated protein (Du et al., 2010) and likely functions as an ion-channel protein embedded in the viral envelope facilitating the uncoating of virus and release of the genome into the cytoplasm (Lee and Yoo, 2006). GP2, GP3, and GP4 are minor glycoproteins and form a disulfide-linked heterotrimer essential for viral infectivity (Wieringa et al., 2003a, Wieringa et al., 2003b). Co-expression of E, GP2, GP3, and GP4 results in the transport of these proteins from the endoplasmic reticulum (ER) through the Golgi, suggesting an important role of the hetero-multimerization for virus assembly and maturation (Wissink et al., 2005). ORF5a is a newly identified membrane protein encoded in the internal ORF within ORF5 with an unknown function (Firth et al., 2011, Johnson et al., 2011).
GP4 protein is of 178 and 183 amino acids for the North American (Type II) and European (Type I) PRRSV, respectively (Meulenberg et al., 1995, Murtaugh et al., 1995). Amino acid sequence analysis of GP4 reveals two distinct hydrophobic domains, one at the extreme N-terminus at positions 1–17 and the other at the C-terminus at 165–183, which likely functions as the signal peptide and a membrane anchor, respectively (Meulenberg et al., 1995). GP4 however has a unique structural feature, not commonly seen in the class I-type integral membrane protein since GP4 does not contain a cytoplasmic tail which normally protrudes into the lumen once it associates with the membrane. The reason(s) for the lack of the cytoplasmic tail in GP4 is unknown.
Glycosyl-phosphatidylinositol (GPI) anchor is a C-terminal post-translational modification found in some eukaryotic proteins residing in the outer leaflet of the cell membrane. Genes encoding GPI-anchored proteins specify two signal sequences in the primary translation product: an N-terminal signal sequence for ER targeting and a C-terminal hydrophobic sequence that directs its association to the membrane via the lipid (Orlean and Menon, 2007). The process for GPI biosynthesis takes place in the ER (Takeda and Kinoshita, 1995), and the proteins subjected to GPI-modification enter the ER lumen via the N-terminal ER targeting signal which is cleaved off by a signal peptidase in the lumen (Gerber et al., 1992). In addition to the N-terminal signal sequence, GPI-anchored proteins contain a C-terminal hydrophobic sequence that directs the cleavage of the signal sequence from the protein and the replacement with a preformed GPI-anchor by action of the transamidase complex (Ikezawa, 2002). The GPI-linked proteins are then targeted to the membranes. The structural feature of PRRSV GP4 resembles that of a GPI-anchored protein.
Hundreds of functionally and structurally diverse proteins have been identified as GPI-anchored proteins including the NS1 protein of Dengue virus (Jacobs et al., 2000), prion proteins for transmissible spongiform encephalopathies (Taylor and Hooper, 2006), adhesion molecules (Dustin et al., 1987), decay accelerating factor (DAF; Nickells et al., 1994), just to name a few (see a review; Ikezawa, 2002). GPI modification serves a variety of functions including directing proteins to the cell surface (Lisanti et al., 1988), association with lipid rafts (Taylor and Hooper, 2006), lymphocytes activation (Robinson et al., 1989), and signal transduction (Cary and Cooper, 2000, Jacobs et al., 2000). The GPI anchor in particular has the propensity to target proteins to lipid rafts (Brown and London, 1998, Metzner et al., 2008). Lipid rafts are dynamic assemblies of the lipid-ordered phase of microdomains that are highly enriched with cholesterol and sphingolipids in the exoplasmic leaflet of the plasma membrane (Ikonen, 2001, Simons and Toomre, 2000). Lipid rafts compartmentalize cellular processes by serving as organizing centers for assembly of signaling molecules, influencing membrane fluidity and protein trafficking, endocytosis, transcytosis as well as for host–pathogen interactions (Ikonen, 2001, Pike, 2009, Simons and Toomre, 2000, van der Goot and Harder, 2001). For viruses, lipid rafts play an important role in viral entry (Ewers and Helenius, 2011); Norkin, 1999, Parton and Lindsay, 1999, Vieira et al., 2010, assembly (Manié et al., 2000, Ono and Freed, 2001) and budding (Chazal and Gerlier, 2003, Scheiffele et al., 1999). Some viral membrane-associated and also envelope proteins such as the Gag protein of human immunodeficiency virus type 1 (HIV-1) (Ono and Freed, 2001), the hemagglutinin of influenza virus (Takeda et al., 2003), the tegument protein of herpes simplex virus (Lee et al., 2003), the NS1 protein of dengue virus (Noisakran et al., 2008), and the spike protein of severe acute respiratory syndrome coronavirus (SARS-CoV) (Lu et al., 2008) associate with the lipid rafts. The involvement of lipid rafts in PRRSV infection has not been examined. In the present study, we provide evidence that the viral GP4 protein is a GPI-anchored protein, which co-localizes with the PRRSV receptor, CD163 in the lipid rafts and may be involved in the viral entry process.
Results
GPI-anchored membrane association of PRRSV GP4
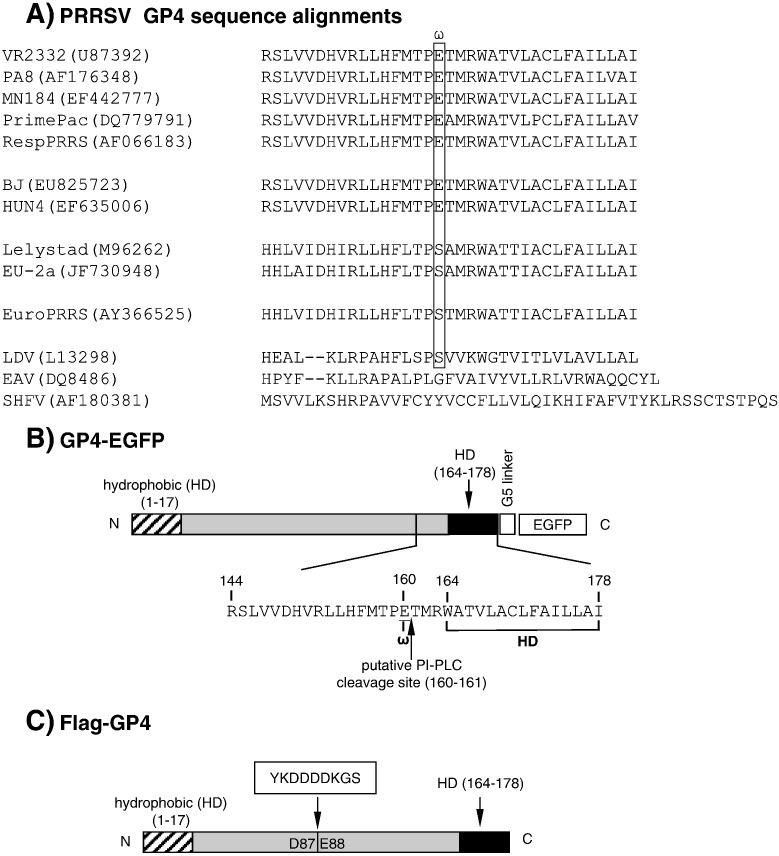
The topology of GP4 predicts two hydrophobic domains on the protein; one at residues 1–17 and the other at 164–178 (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred; Claros and von Heijne, 1994). The N-terminal hydrophobic domain likely functions as the signal peptide to direct the protein to ER membrane and the C-terminal hydrophobic domain to anchor the protein to membrane, but unlike the common structure seen in type I membrane glycoproteins, GP4 does not possess a hydrophilic cytoplasmic tail following the hydrophobic anchor domain at the C-terminus (Fig. 1 ). The cytoplasmic tail generally protrudes into the lumen when the protein is associated with the ER membrane, and the unusual feature of GP4 resembles the topology of GPI-anchored protein, for example, DAF which is a well-characterized GPI-anchored protein (Nickells et al., 1994). When GP4 sequences were analyzed using the GPI prediction program (http://gpi.unibe.ch; Fankhauser and Mäser, 2005), a GPI anchor signal was readily detectable. When a large number of GP4 sequences were examined including the European and North American genotypes, the European-like PRRSV circulating in the USA, and the highly virulent PRRSV emerged in China (Fig. 1A), the GPI anchor signal became more prominent and appeared to be highly conserved across the genotypes including LDV and EAV. Thus, we hypothesized that the PRRSV GP4 protein might be modified by GPI attachment and anchored with the membrane through the GPI.
Fig. 1.
Sequence alignments of the C-terminal region of GP4 protein from various types of PRRSV and other arteriviruses (A). VR2332, PA8, MN184, Prime Pac, and RespPRRS represent North American PRRSV genotype II. BJ and HUN4 represent the highly virulent PRRSV recently emerged in China. Lelystad and EU-2a represent European PRRSV genotype I. EuroPRRS represents the prototype European PRRSV identified in the USA. LDV, lactate dehydrogenase-elevating virus of mice; EAV, equine arteritis virus; SHFV, simian hemorrhagic fever virus. GenBank accession numbers are indicated in parenthesis. Putative GPI anchor attachment (omega ω) sites are indicated in box. (B) Structural presentations of the GP4 protein of PRRSV PA8. GP4 contains two hydrophobic domains (HD), one at the N-terminus and the other at the C-terminus. GP4 was cloned to express as a fusion protein with EGFP using a linker of five glycine (5G) residues. ‘ω’ at position 160 (E160) indicates the putative GPI modification site, and the arrow between positions 160 and 161 indicates the potential cleavage site specific for PI-PLC lipase. Numbers indicate amino acid positions. (C) The Flag-tag sequence of YKDDDDKGS was inserted between positions D87 and E88 (nucleotide sequence positions 261 and 262) of GP4 gene cloned in pXJ41, and pXJ41-Flag-GP4 was constructed.
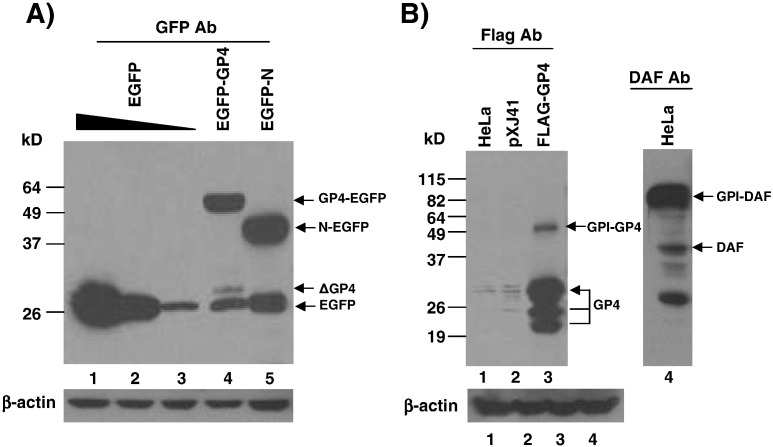
Since GP4 is a minor protein and thus its expression level is low, and because a suitable antibody for GP4 is not available, a GP4–EGFP fusion construct was made using a linker of five glycine residues inserted between the EGFP and GP4 sequences to facilitate the detection of GP4 in cells (Fig. 1B; Tsuneki et al., 2007). When HeLa cells were transfected with the pEGFP-GP4 plasmid, the GP4–EGFP fusion protein of about 56 kDa was expressed (Fig. 2A, lane 4). In addition, a minor band of 30 kDa which was slightly larger than EGFP alone was identified (Fig. 2A, lane 4, ΔGP4), suggesting that the C-terminal portion of GP4 might have been cleaved and the cleaved portion which was fused with EGFP was detectable by EGFP antibody. Such band was absent in cells transfected with pEGFP-N which is a fusion construct of the PRRSV N protein with EGFP (Fig. 2A, lane 5). This result suggests that GP4 is possibly a GPI-anchored protein and not all but some of the GP4 molecules are modified by GPI attachment.
Fig. 2.
Expression and detection of GP4 in HeLa cells. (A) Cells were transfected with pEGFP-GP4 for 24 h (lane 4) and the total cell lysates were subjected to western-blot using anti-GFP rabbit antibody. For GFP expression, 2.0, 1.0, 0.5 μg of pEGFP-N1 (Clontech) was individually transfected for differential expression (lanes 1, 2, and 3). pcDNA3.0 (Invitrogen) which contains the identical promoter to that of pEGFP-N1 (Clontech) was used to make up the total DNA of 2.0 μg. The PRRSV N gene fused with EGFP (pEGFP-N; Rowland et al., 2003) served as an additional control (lane 5) and β-actin served as the loading control. (B) Western blot of the total lysates of HeLa cells transfected with pXJ41-Flag-GP4 (lane 3). HeLa cells were mock-transfected (lane 1) or transfected with the empty vector pXJ41 (lane 2) as controls. HeLa cells constitutively express human DAF readily detectable by anti-DAF MAb (Sigma-Aldrich), and this protein served as a positive control of GPI-anchored DAF (lane 4).
A GPI-anchored protein can be distinguished from its un-modified form by its mobility (Ikezawa, 2002), and thus the migration pattern of GP4 was examined by SDS-PAGE and western blot. HeLa cells naturally express human DAF as a 41 kDa protein before GPI-modification (Karnauchow et al., 1996) and after glycosylation and GPI-modification, it becomes approximately 82–88 kDa (Nickells et al., 1994), which can readily be detectable by DAF MAb from cell lysates (Fig. 2B, lane 4). To facilitate the detection of GP4 while maintaining its structural integrity as much as possible, the Flag sequence tag of ‘YKDDDDK’ was inserted between positions D87 and E88 of GP4, and pXJ41-FLAG-GP4 was constructed (Fig. 1C). HeLa cells transfected with pXJ41-FLAG-GP4 produced the GP4 protein of approximately 29 kDa (Fig. 2B, lane 3, lower arrow). This form of GP4 was predominant among two other forms of GP4 produced in these cells. GP4 is an integral membrane protein with multiple glycans added onto it (Meulenberg et al., 1995, Wissink et al., 2005), and the 29 kDa protein and two additional bands of smaller sizes likely represent the fully mature and the partially glycosylated forms of GP4 as previously observed (Das et al., 2010). A 50 kDa band was identified (Fig. 2B, lane 3, upper arrow), and this is likely the fully modified GPI-anchored form of GP4.
To determine the GPI modification of GP4, phosphatidylinositol-specific phospholipase C (PI-PLC) digestion was conducted. PI-PLC is a lipase known to cleave and release a GPI-linked protein from the cellular membrane, and therefore the release of a soluble protein by PI-PLC digestion is considered standard biochemical evidence for GPI-anchored proteins (Ikezawa, 2002). For this experiment, HEK-293 cells were transfected with pXJ41-FLAG-GP4 and at 24 hpt, trypsinized and digested with PI-PLC in suspension. The supernatants were then analyzed by western blot using anti-FLAG Ab. In parallel, PI-PLC-digested cells were subjected to FACS analysis after staining with anti-FLAG Ab. Unexpectedly, no protein band equivalent to digested GP4 was observed from the supernatants, and the intensity of fluorescence was not diminished in PI-PLC-digested cells as compared to the untreated cells (data not shown). Total lysates of PI-PLC-digested cells and untreated cells were also examined by western blot, but no difference was found in their migration patterns. The 50 kDa band did not change its migration after PI-PLC digestion (data not shown), and we concluded that the GPI modification of GP4 was resistant to PI-PLC digestion.
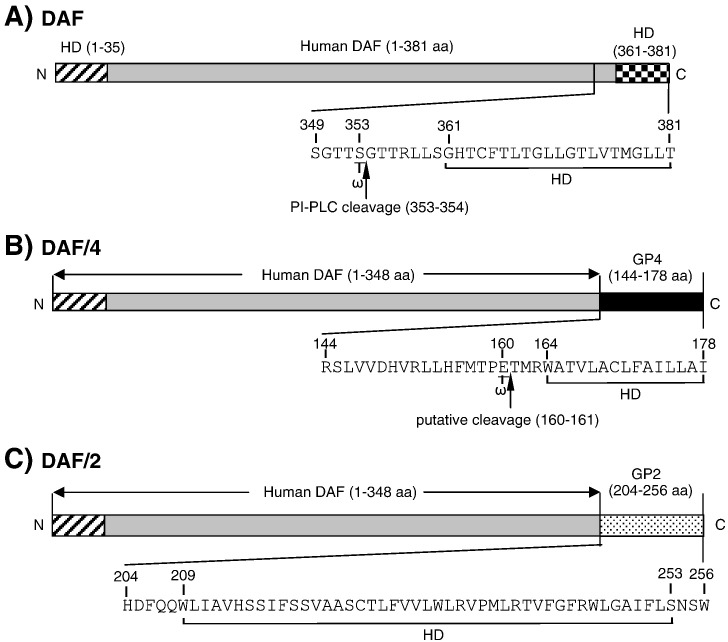
When intact cells are treated with PI-PLC, partial or total resistance to digestion may occur due to the inaccessibility of an enzyme to the cleavage site, expression of the protein on the cell surface in both GPI-modified and GPI-unmodified hydrophobic peptide-anchored forms, or tight association of the GPI-modified protein with a PLC-non-susceptible protein on the cell surface (Rosenberry, 1991). The PRRSV GP4 protein may exist as the GPI-modified form and the GPI-unmodified hydrophobic peptide-anchored form, which may have caused the inability of PI-PLC digestion. Thus to further determine if GP4 was a GPI-modified protein, two chimeric constructs were made such that the known GPI-anchor domain of human DAF was substituted with the putative GPI-modification domain of GP4 protein (Figs. 3A, B) and the C-terminal hydrophobic region of GP2 protein of PRRSV (Fig. 3C). The residue cross-linked to the lipid moiety is termed ω site, and upstream residues are designated ω-minus while downstream residues are designated ω-plus with respect to their positions from the ω site (Ikezawa, 2002). The ω site of human DAF is serine at position 353 (S353) (Nickells et al., 1994; Fig. 3A), which is identical to the computer prediction (GPI modification prediction; http://mendel.imp.ac.at/gpi/gpi_server.html). By using the same program, the potential ω site for GP4 was predicted to be at E160, and based on this prediction the region of 144–178 of GP4 was chosen to replace with the 349–381 fragment of human DAF to construct pXJ41-DAF/4 (Fig. 3B). The PRRSV GP2 protein was chosen to serve as a GPI-negative control and the 204–256 fragment of GP2 was used to replace the 349–381 region of DAF. This construct was designated pXJ41-DAF/2 (Fig. 3C).
Fig. 3.
Construction of human DAF and PRRSV GP4 chimeric proteins. (A) Human DAF served a positive control as a GPI-anchored protein. The C-terminal amino acid sequence is presented with indications of the GPI modification site (ω) at S353 and the lipase cleavage site between S353 and G354 (arrow). HD denotes hydrophobic domain. (B) The C-terminal 33 amino acids containing the GPI domain of human DAF were replaced with the putative GPI domain representing the 35 amino acids at positions 144–178 of GP4, and DAF/4 was generated. Numbers for 144–178 indicate amino acid positions with respect to GP4. For GP4, the ω site is predicted at E160, and the putative lipase cleavage site is predicted between E160 and T161 (arrow). (C) GP2 is type I integral membrane protein, and the C-terminal 33 amino acids of human DAF were replaced with the C-terminal 53 amino acids representing positions 204 and 256 of PRRSV GP2 to generate DAF/2. This region of GP2 contains a hydrophobic trans-membrane sequence plus a short hydrophilic cytoplasmic tail. Numbers for 204–256 indicate amino acid positions with respect to GP2.
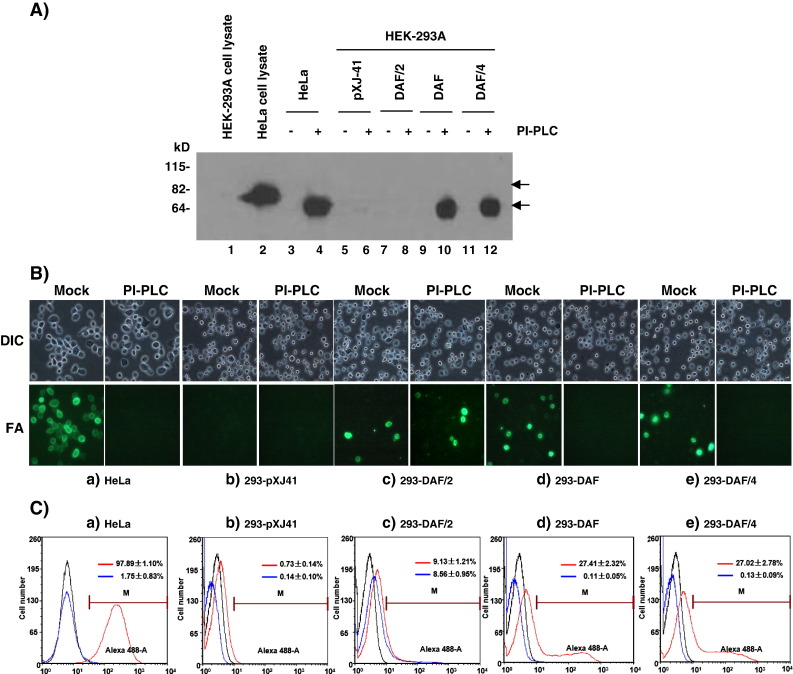
Using the DAF/4 fusion construct, GPI modification of GP4 was re-examined by PI-PLC digestion. The endogenous expression of human DAF was high in HeLa cells whereas negligible and undetectable in HEK-293 cells (data not shown). Thus, HEK-293 cells were used in this study to express DAF/4. DAF of orangutan erythrocytes (EOr) migrates as an 88 kDa protein, and after digestion with PI-PLC, its migration reduces to 67 kDa (Nickells et al., 1994). In our study, the molecular migration of human DAF in HeLa cells decreased from 82–88 kDa to 65–70 kDa when digested with PI-PLC (Fig. 4A, lanes 2, 4), which was consistent with the orangutan report. After PI-PLC digestion, fluorescent cells by DAF staining were also reduced by 96% in HeLa cells (Figs. 4, Ba, Ca), confirming the efficient digestion of DAF by PI-PLC. In contrast, HEK-293 cells transfected with the empty vector pXJ41 showed no fluorescence for DAF (Figs. 4, Bb, Cb), and no cleavage product was released by PI-PLC digestion (Fig. 4A, lane 6). Thus, HEK-293 cells were transfected with pXJ41-DAF, pXJ41-DAF/4, or pXJ41-DAF/2, and analyzed by PI-PLC digestion followed by western blot (Fig. 4A), fluorescence staining (Fig. 4B), and FACS (Fig. 4C) analyses. DAF/2, DAF, and DAF/4 were all expressed on the cell surface and detectable by DAF-specific MAb EVR1 (Figs. 4B, C; panels c, d, e). After digestion with PI-PLC, the fluorescence was lost in DAF and DAF/4 expressing cells, and DAF and DAF/4 proteins cleaved by PI-PLC were released to the supernatants, which were then detectable by western blot (Fig. 4A, lanes 10, 12). In contrast, the intensity of fluorescence and the percentage of fluorescence-positive cells were unaffected for DAF/2 by PI-PLC treatment (Fig. 4B, panel c; Fig. 4C, panel c), and the DAF/2 protein was not detectable in the supernatant (Fig. 4A, lane 8). These results show that the C-terminal region of GP4 was modified for PI-PLC-specific GPI-attachment, whereas GP2 protein did not undergo such a modification, which is consistent with previous reports that GP2 anchors on the cell membrane through the C-terminal hydrophobic transmembrane domain (Meulenberg et al., 1995, Wissink et al., 2005).
Fig. 4.
PI-PLC digestion of DAF/4.HeLa or 293 cells were transfected with the empty vector pXJ41, pXJ41-DAF/2, pEF6/V5-DAF, and orpXJ41-DAF/4. One day post-transfection, cells were collected by trypsinization and washed in PI-PLC buffer. Cells were then divided into two equal fractions, and one fraction was digested with PI-PLC at 1.5 unit/ml (+), while the other fraction was mock-digested (−). Following incubation at 37 °C for 30 min, cells were centrifuged at 2000 rpm for 10 min. Then, supernatants were subjected to western blot analysis (A; lanes 3 through 12), whereas collected cells were subjected to immunofluorescence (B) and FACS analyses (C) using anti-DAF MAb (Sigma-Aldrich)as the primary antibody, followed by staining with Alexa Fluor 488® conjugated goat anti-mouse IgG (H + L) secondary antibody. DIC, differential interference contrast; FA, fluorescence antibody staining; blue plot, PI-PLC treated cells; red plot, untreated cells; black plot, mock-stained cells with DAF-specific MAb EVR1as a negative control. Marker (M) was used to analyze the data created using the software FCS Express Version 3 Research Edition. Gated cells by the marker represent positive cells.
Mutational studies on the GPI-anchor region
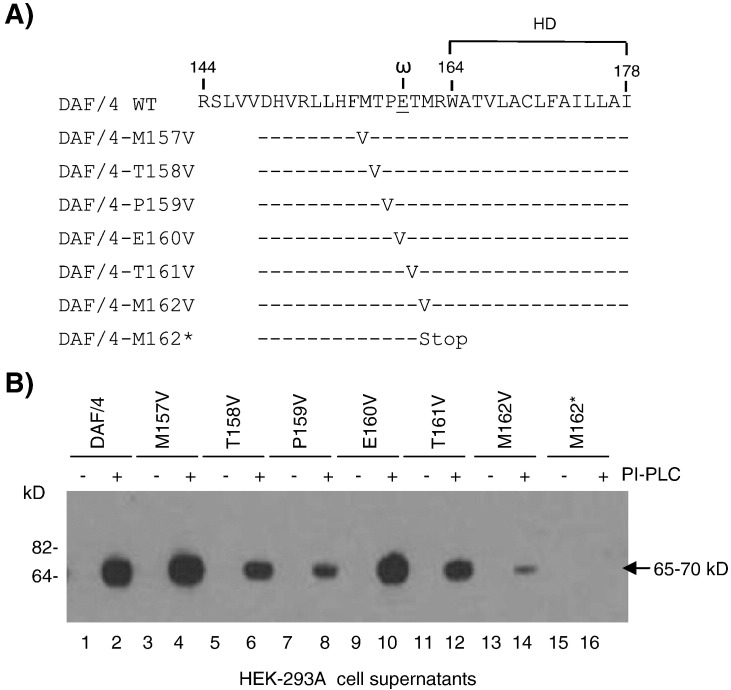
Since our data showed that PRRSV GP4 was a GPI-modified protein, the importance of individual amino acids surrounding the GPI attachment site (ω site) was examined for PI-PLC cleavage. Using pXJ41-DAF/4 as the parental construct, six mutants were constructed such that the residues adjacent to E160 (the ω site) were individually changed to valine (Fig. 5A; Gerber et al., 1992, Furukawa et al., 1997). Following expression in transfected cells and after PI-PLC digestion of wild-type DAF/4, the cleaved product released into the supernatant was identified as a 65–70 kDa band (Fig. 5B, lane 2). Mutants DAF/4-M157V (ω − 3) and DAF/4-E160V (ω) showed similar cleavage properties to those of wild-type DAF/4 as inferred from the intensity of the cleaved product (Fig. 5B, lanes 4, 10) and the percentage of positive cells (Fig. 5C, panels a, d; Fig. 5D, panels a, d).
Fig. 5.
(A) Mutagenesis for individual amino acids adjacent to the ω site (E160) of GP4 to valine (V) using pXJ41-DAF/4 as the parental plasmid. Constructs were designated based on the mutagenized amino acid and its position with respect to the GP4 sequence (A). HEK-293 cells were transfected with pXJ41-DAF/4 wild-type (WT) or individual mutant plasmids and 1 day post-transfection, were collected by trypsinization. Cells were then washed in PI-PLC buffer and divided into two equal fractions. One fraction was digested with PI-PLC at 1.5 unit/ml (+) while the other fraction was mock-digested (−), followed by incubation at 37 °C for 30 min. Cells were centrifuged at 2000 rpm for 10 min, and their supernatants were subjected to western blot (B), whereas pelleted cells were subjected to immunofluorescence using a fluorescent microscope (model AX70, Olympus) (C) and FACS for surface expression of DAF (D) analyses using anti-DAF MAb. DIC, differential interference contrast microscopy.
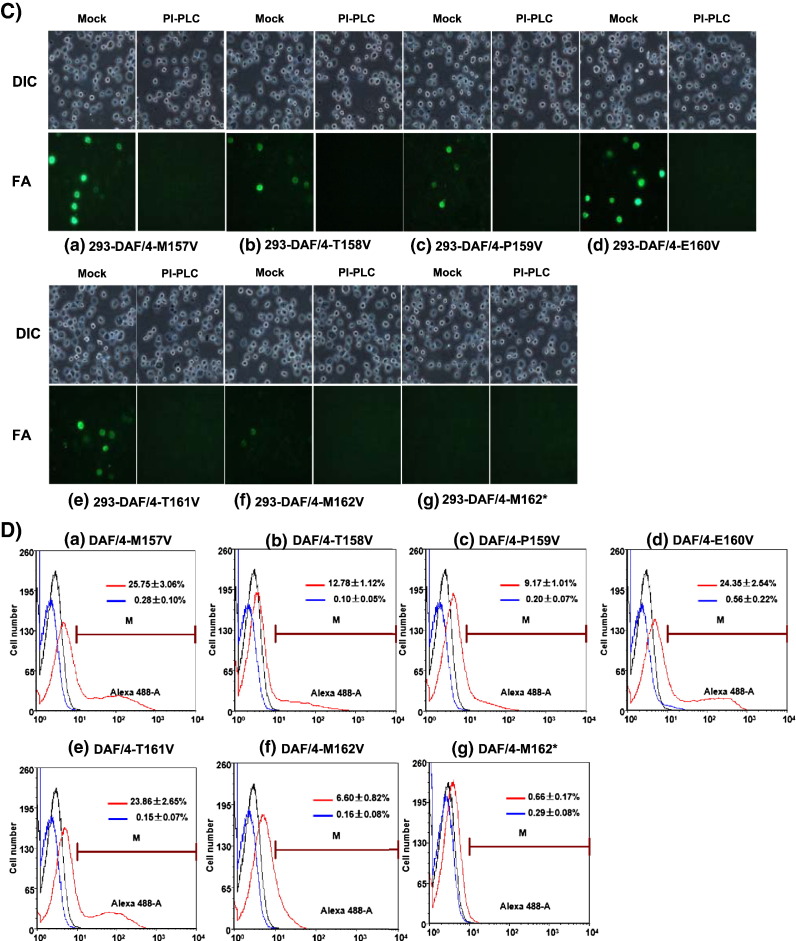
For DAF/4-T161V (ω + 1), the fluorescence intensity was 6 × 102 as compared to 1 × 103 for DAF/4 (Fig. 5C, panel e; Fig. 5D, panel e). The release of DAF in the supernatant reduced slightly (Fig. 5B, lane 12), but the percentage of positive cells for DAF/4-T161V was similar to that of DAF/4 (Fig. 4C, panel e; Fig. 5D, panel e). The percentage of positive cells for DAF/4-T158V (ω − 2) was significantly lower (P < 0.05) than those of DAF/4, followed by DAF/4-P159V (ω − 1), and DAF/4-M162V (ω + 2) (Fig. 5D, panels b, c, f). Their intensity of fluorescence also decreased in order (Fig. 5C, panels b, c, f; Fig. 5D, panels b, c, f), and DAF released to the supernatant decreased accordingly (Fig.5B, lanes 6, 8, 14). DAF/4-M162*, whose the C-terminal hydrophobic domain was truncated by mutating M162 to the TAG stop codon (Fig. 5A), was anticipated not to anchor the GP4 protein on the cell membrane, and as expected no band was released in the supernatant of the digestion reaction (Fig. 5B, lane 16; Fig. 5C, panel g; Fig. 5D, panel g). These results show that the residues surrounding the ω site of GP4 contribute to the ability of GPI to anchor to the membrane. These experiments were repeated three times for confirmation, and the representative results are shown in Figs. 5C and D. In summary, all mutations but M162* retained the ability of GPI-anchorage to various extents.
Generation of GPI-anchor mutant viruses
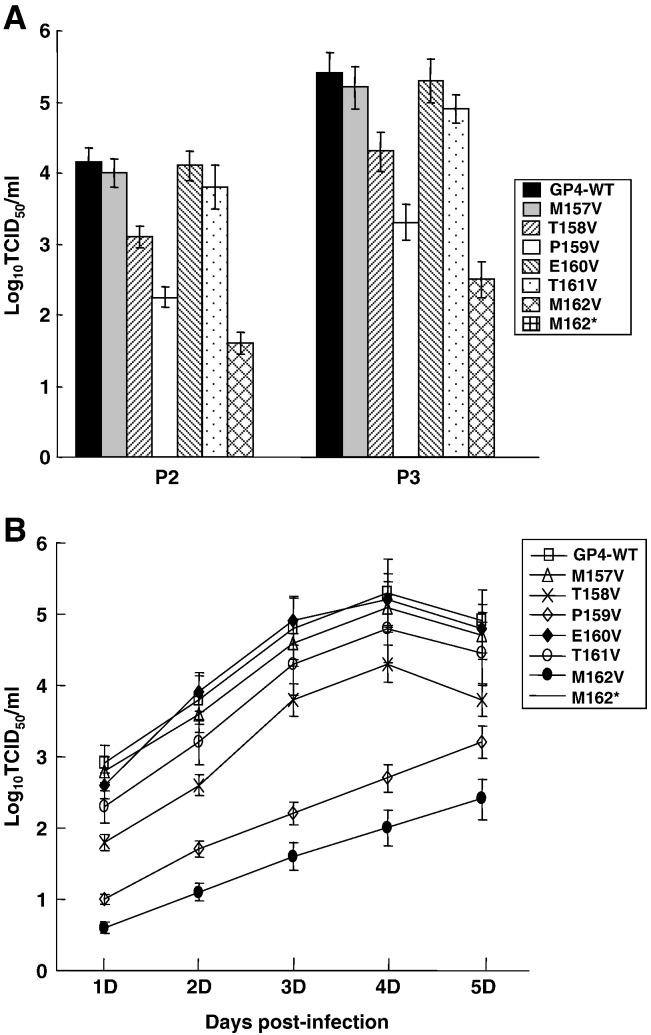
To determine the significance of residues adjacent to the ω site of GP4 for virus infectivity, the respective mutations were introduced into the PRRSV infectious cDNA clone and a total of 7 mutant viral genomic clones were generated. The PRRSV infectious cDNA clone was modified to place the full-length genomic sequence under the eukaryotic promoter and thus DNA transfection of the full-length genomic clones can produce infectious progeny in transfected cells. Thus, the wild type and mutant genomic clones were transfected into MARC-145 cells and the cells were incubated for 5 days. Cytopathic effects (CPE) were evident for PRRSV-GP4-WT and some of the mutants. Culture supernatants were collected and designated ‘passage-1’. For mutants that did not produce visible CPE, extracellular and intracellular RNAs were examined for the presence of viral genome by RT-PCR for N gene at passages 3 and 5. The mutant viruses PRRSV-GP4-M157V (ω − 3), PRRSV-GP4-E160V (ω), and PRRSV-GP4-T161V (ω + 1) grew to titers similar to that of PRRSV-GP4-WT (Fig. 6A), and these were the mutants that did not impair the ability for GPI-anchorage in HEK-293 cells. These mutant viruses grew normally and induced CPE typical for PRRSV. Mutation at ω − 2 (T158V), ω − 1 (P159V), and ω + 2 (M162V) appeared to be important for virus growth as the titers for PRRSV-GP4-T158V, PRRSV-GP4-P159V, and PRRSV-GP4-M162V decreased in ‘passage 2’ and ‘passage 3’ (Fig. 6A). Their growth kinetics was also slower compared to that of PRRSV-GP4-WT (Fig. 6B), and their characteristics were consistent with their fluorescence staining. In particular, PRRSV-GP4-M162V (ω + 2) appeared to affect the virus infectivity most notably. PRRSV-GP4-M162V grew slowly and its titer was also low. The truncation mutant PRRSV-GP4-M162* did not exhibit any sign of infectivity (Figs. 6A, B). Furthermore, no viral RNA was detectable at passages 3 and 5 and thus it was concluded that the lack of GPI-anchor, thus the lack of membrane association, was lethal for PRRSV infectivity.
Fig. 6.
Production of ‘passage-2 (P2)’ and ‘passage-3 (P3)’ mutant viruses in MARC-145 cells (A) and their growth kinetics (B).Fresh MARC-145 cells were infected with ‘passage-2’ virus at a multiplicity of infection (MOI) of 5, and their supernatants were harvested at indicated times. Virus titers were determined by a microtitration infectivity assay and recorded as tissue culture infectious dose (TCID)50/ml. Experiments were conducted in duplicate and repeated three times. The data are shown as mean titers ± standard error.
Inhibition of virus entry by cholesterol depletion in MARC-145 and PAM cells
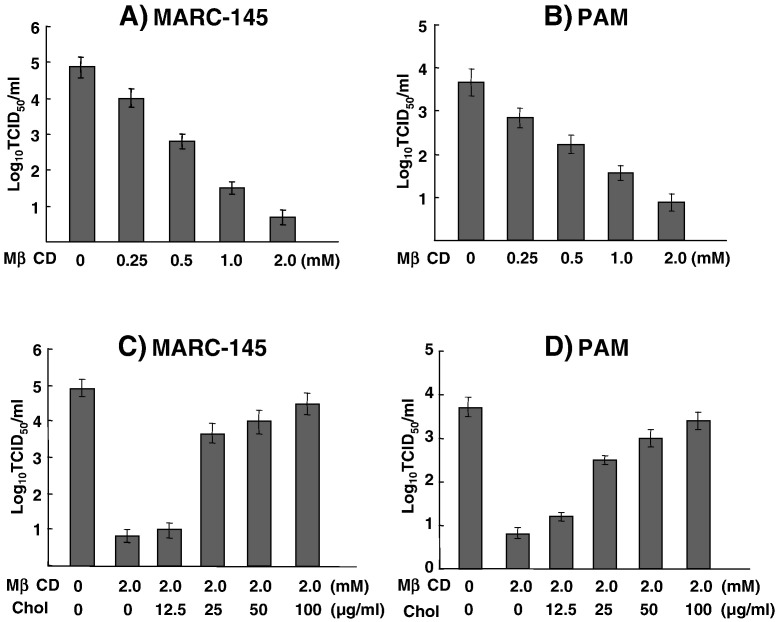
For mouse hepatitis coronavirus, a member virus in the Order Nidovirales, the cholesterol levels on the cell membrane determine the susceptibility of cells to virus infection, and lipid rafts are required for virus entry and membrane fusion (Choi et al., 2005). Since cholesterol is a major component residing in the lipid rafts of the cell membrane, we examined whether lipid rafts were involved in the entry of PRRSV in permissive cells. A cholesterol-depletion experiment was conducted using methyl-β-cyclodextrin (MβCD) in both MARC-145 and PAM cells. MβCD is not incorporated into membranes but extracts cholesterol selectively from membranes by binding it in the central non-polar cavity and thus depleting from the plasma membrane (Ilangumaran and Hoessli, 1998). To avoid the possibility that the newly synthesized cholesterol and/or cholesterol from internal compartments may restore the rafts and affect the entry of virus, incubation of MβCD-treated cells with PRRSV was limited to 1 h at 37 °C. It was apparent that the MβCD treatments of MARC-145 and PAM cells impaired the production of PRRSV in a dose-dependent manner (Figs. 7A, B), suggesting an essential role of cholesterol for PRRSV entry.
Fig. 7.
Reduction of virus production by MβCD in MARC-145 (A) and PAMs (B) cells. Cells were pretreated with 0.25–2.0 mM of MβCD for 1 h at 37 °C followed by infection with PRRSV PA8 for 1 h at a MOI of 5. Supernatants were collected 3 days post-infection from MARC-145 cells or 20 h post-infection from PAMs and subjected to virus titration in MARC-145 cells by microtitration infectivity assays. For cholesterol replenishment assays, MARC-145 (C) or PAM (D) cells were pretreated with 2.0 mM of MβCD for 1 h at 37 °C followed by supplementation with cholesterol at indicated concentrations for 1 h at 37 °C, and then infected with PRRSV for 1 h at a MOI of 5. Culture supernatants were collected 3 days post-infection and titrated as described above. Experiments were conducted in duplicate and repeated three times. The data are shown as mean titers ± standard error.
To confirm the involvement of cholesterol in PRRSV infectivity, a depletion reversion study was conducted. Cells were treated with MβCD for 1 h and then, the depletion was reversed by supplementing with cholesterol at various concentrations, followed by PRRSV infection and examination of virus yield from these cells. After replenishment with cholesterol, the virus titer increased dramatically, and at 100 μg/ml of cholesterol supplementation, the production of virus was almost fully restored in both MARC-145 and PAM cells (Figs. 7C, D).These data indicate that the reduction of virus production by MβCD was due to the depletion of cholesterol from the cells and this effect was reversible, suggesting a role of lipid rafts in PRRSV infection.
GP4 localizes to lipid rafts
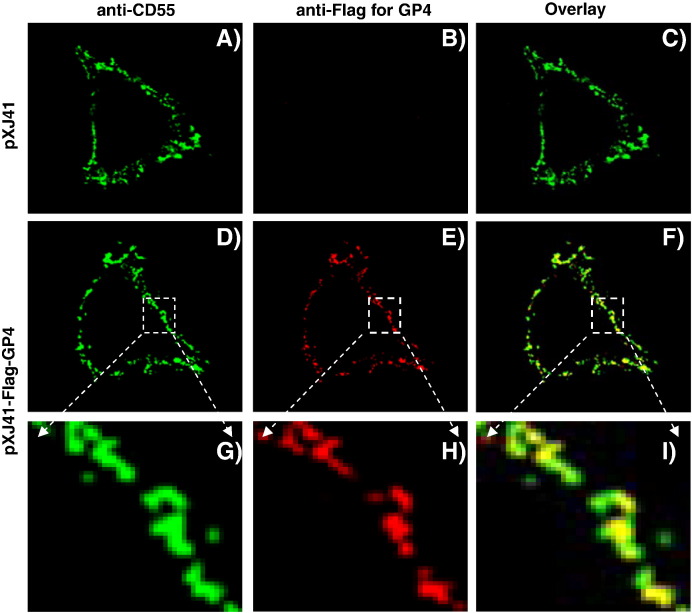
The importance of cholesterol for PRRSV infection suggests that viral proteins may interact with cellular proteins in the lipid rafts during entry. GPI modification has the propensity to target the GPI-proteins to lipid rafts (Brown and London, 1998) and since PRRSV GP4 is a GPI-anchored protein, its localization in the lipid rafts was first examined by dual staining immunofluorescence. DAF (a synonym of CD55) is a resident protein in the lipid rafts and thus is frequently used as a marker for lipid rafts (Stuart et al., 2002). Since HeLa cells constitutively express DAF which is readily detectable by DAF-specific MAb EVR1 (Fig. 2B, lane 4; Fig. 8A), HeLa cells were used to examine co-localization of GP4 and DAF proteins. The GP4 protein fused with a Flag-tag was detectable on the cell membrane (Figs. 8E, H) and was co-localized with CD55 (Figs. 8F, I), demonstrating that the PRRSV GP4 protein is a membrane protein localized in the lipid rafts in HeLa cells.
Fig. 8.
Co-localization of PRRSV GP4 and the DAF (CD55) protein as a lipid raft marker on the plasma membrane of HeLa cell. Cells were transfected with pXJ41-Flag-GP4 (D through F) and at 24 h post-transfection, washed with ice-cold PBS. Cells were then co-stained with DAF-specific MAb EVR1 (anti-CD55; A and D) and rabbit anti-Flag antibody (B and E), followed by staining with Alexa Fluor 488® conjugated goat anti-mouse IgG (H + L) and Alexa Fluor 594® conjugated goat anti-rabbit IgG (H + L) secondary antibodies, respectively. Images were visualized using a laser-scanning confocal fluorescence microscope (model BX50, Olympus). Panels G through I represent the enlargement of the indicated areas in panels D through F, respectively.
Co-localization of GP4 with CD163 in the lipid rafts
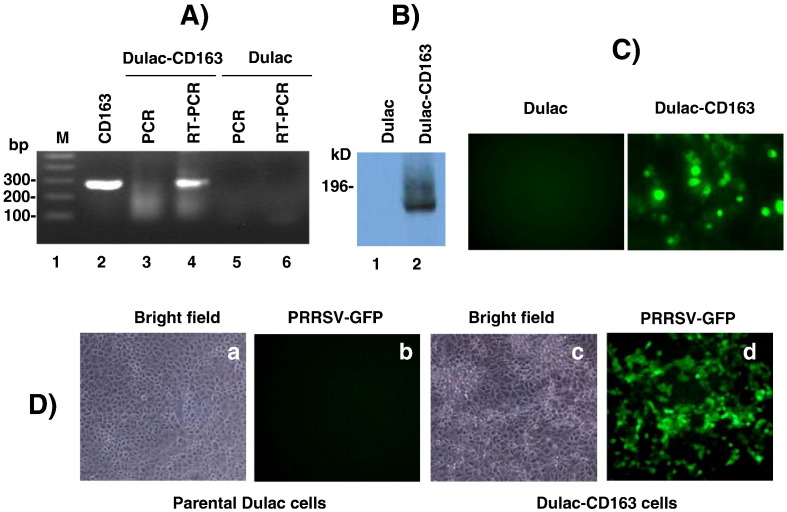
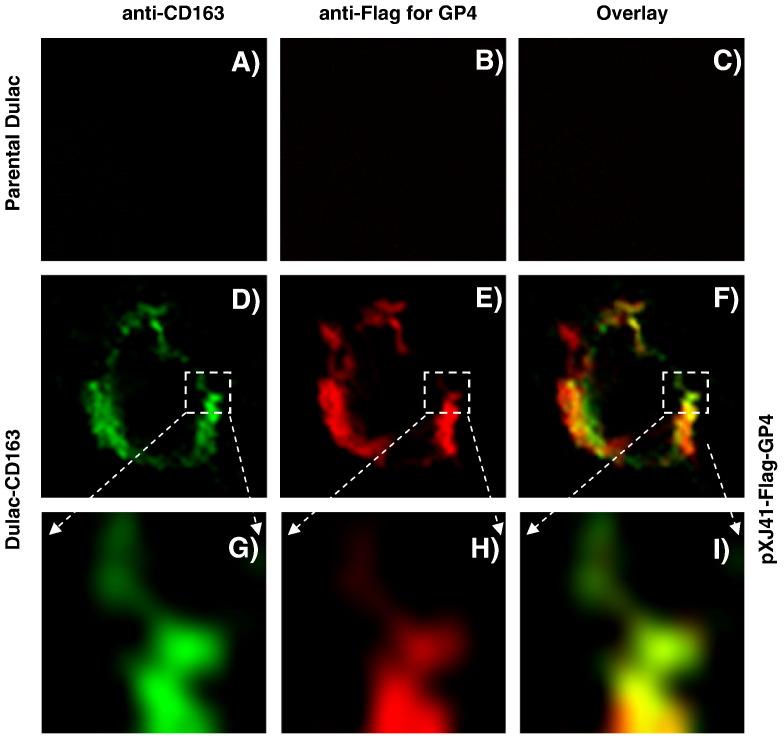
CD163 has been studied as a cellular receptor for PRRSV, and CD163 renders PRRSV non-permissive cells permissive for infection (Calvert et al., 2007, Lee et al., 2010, Patton et al., 2009). CD163 has been shown to localize and form high partition in the lipid rafts (Wolf et al., 2007). Since GP4 co-localizes with DAF (Fig. 8), co-localization of GP4 with CD163 was examined. For this study, cells of the porcine origin were used to co-express both GP4 and CD163. Dulac cells are porcine kidney cells but non-permissive for PRRSV infection, and thus using these cells, a cell line was generated to stably express porcine CD163 which was then designated Dulac-CD163. Dulac-CD163 cells were RT-PCR positive for CD163 transcription (Fig. 9A, lane 4) and the protein was also expressed (Figs. 9B, C). Dulac-CD163 cells became permissive for PRRSV infection and the GFP expression was evident when infected with PRRSV-P129-GFP virus (Fig. 9D), confirming the expression of porcine CD163 and infection of PRRSV in these cells. When Dulac-CD163 cells were transfected with pXJ41-Flag-GP4, CD163 and GP4 were found to be co-localized on the plasma membrane (Figs. 10 , F, I). These results demonstrate the interaction of GP4 with CD163 in the lipid rafts and suggest that this interaction may participate in the initial stage of virus entry into cells.
Fig. 9.
Establishment of Dulac-CD163 porcine kidney cells as a cell line stably expressing porcine CD163. (A) RT-PCR for porcine CD163 mRNA in Dulac-CD163. Total cellular RNA was extracted and treated with DNase I (1 unit/μg of RNA) and PCR was performed after RT (lanes 4 and 6) or without RT (lanes 3 and 5) reactions. Dulac, parental cells; Dulac-CD163, porcine CD163 gene transformed cells. Lane 2, CD163 positive control. (B) Western blot for porcine CD163. Cell lysates were prepared from Dulac-CD163 and resolved by 7.5% SDS-PAGE, followed by blotting to the PVDF membrane and probing with porcine CD163-specific MAb. (C) Immunofluorescence. (D) Permissiveness of Dulac-CD163 cells for PRRSV. Expression of porcine CD163 rendered non-permissive Dulac cells permissive for PRRSV infection. Cells were infected with PRRSV-GFP and the GFP expression was observed by fluorescent microscopy (panel d). PRRSV-specific cytopathic effect was also evident in Dulac-CD163 cells (panel c).
Fig. 10.
Co-localization of PRRSV GP4 with porcine CD163 in Dulac-CD163 cell. Parental Dulac (panels A through C) or Dulac-CD163 (panels D through F) cells were transfected with pXJ41-Flag-GP4 and at 24 hpt, washed with ice-cold PBS. Cells were then incubated with CD163 MAb (A and D) and rabbit anti-Flag antibody (B and E), followed by staining with Alexa Fluor 488® conjugated goat anti-mouse IgG (H + L) and Alexa Fluor 594® conjugated goat anti-rabbit IgG (H + L) secondary antibodies, respectively. Staining images were observed under the laser-scanning confocal fluorescence microscope (model BX50, Olympus). Panels G through I represent the enlarged images of the selected areas in panels D through F, respectively.
Discussion
The signal for GPI modification of a protein consists of three parts: a stretch of three amino acids including the residue where GPI attaches (the ω site), a hydrophobic segment of 12–20 amino acids, and a hydrophilic spacer segment of usually less than 10 amino acids between them (Furukawa et al., 1997). For modification, the hydrophobic segment is first cleaved and the lipid is then attached (Gerber et al., 1992). The GP4 protein satisfies the above conditions with a hydrophobic segment of 15 residues at the C-terminus and a hydrophilic space of 3 amino acids between the hydrophobic segment and the ω site (Fig. 1A). In the present study, we provide evidence that the PRRSV GP4 protein can undergo post-translational modification for GPI attachment and anchors to the membrane via the lipid. The GP4–EGFP fusion protein was cleaved at the ω site and the 30 kDa peptide was likely the cleavage product representing the C-terminal portion of GP4 fused with EGFP. GP4 existed in two forms to anchor to the membrane, and this observation was consistent with other lipid-anchored proteins such as LFA-3 (lymphocyte function-associated antigen 3) and NCAM-1 (neural cell adhesion molecule 1) as they also exist in two forms, a hydrophobic peptide-anchored form and a lipid-anchored form (Arai et al., 2004, Cross, 1990, Dustin et al., 1987). These alternative forms behave differently. The routes (Rothberg et al., 1990) and rates (Keller et al., 1992) for endocytosis of the GPI-anchored and hydrophobic peptide-anchored proteins differ. In addition to the transport differences, the lipid-anchored and peptide-anchored forms of a same protein can also have different biological properties. The lymphocyte surface molecules Ly-6A/E and Qa-2 only activate T cells when present as a GPI-anchored form (Robinson et al., 1989, Su et al., 1991).
The GP4 protein expressed as the Flag-GP4 fusion protein appeared to be resistant to PI-PLC digestion, and this was likely due to the co-existence of lipid-anchored and peptide-anchored forms of GP4. In contrast, the DAF/4 construct became sensitive to PI-PLC digestion, and constructs made in the similar way have been shown to be useful for some proteins including CD4 for the study of HIV. HIV efficiently infected human cells expressing the GPI-anchored CD4 receptor (Diamond et al., 1990, Jasin et al., 1991) and the CD4-GPI fusion protein was efficiently cleaved by PI-PLC (Anderson et al., 1996).
Replacement of the GPI modification signal by a hydrophobic peptide segment leads to expression of the protein as a peptide-anchored membrane protein (Takeda and Kinoshita, 1995), and this was consistent with our finding that DAF/2, in which the GPI modification signal in DAF was substituted with the hydrophobic segment of GP2, resulted in peptide-anchored membrane association and thus non-digestible by PI-PLC. This finding confirms that unlike GP4, GP2 is a type I integral membrane protein (Meulenberg et al., 1995, Wissink et al., 2005). In contrast to DAF/2, DAF/4 was successfully digested by PI-PLC, demonstrating that the C-terminal region of GP4 indeed possessed the GPI-modification signal. M157V (ω − 3), E160V (ω), and T161V (ω + 1) mutations of GP4 did not affect the GPI-modification or the growth of mutant viruses, but T158V (ω − 2), P159V (ω − 1), and M162V (ω + 2) were important for both GPI modification and the virus infectivity. The residues at positions ω − 2, ω − 1, and ω + 2 were highly conserved among different PRRSV isolates of the North American and European genotype, and among those three mutations, M162V (ω + 2) contributed the most to the GPI modification and virus infectivity. The M162* mutation, the C-terminal 16 amino acids truncation mutant, was lethal for infectivity, indicating the essential requirement of this hydrophobic segment for PRRSV infection. This is the first report that the GPI-anchor of a viral membrane protein contributes to viral infectivity and growth rates.
For viruses to infect target cells, they first bind to a specific receptor on the cell surface. For PRRSV, CD163 is the cellular receptor (Calvert et al., 2007) and has been recently shown to interact with GP2 and GP4 proteins (Das et al., 2010). CD163 is a macrophage-specific protein in the cysteine-rich scavenger receptor superfamily comprising a large number of cell surface and soluble glycoproteins involved in the recognition of various ligands of proteins, polyribonucleotides, polysaccharides, and lipids (Sarrias et al., 2004). The localization of a receptor in the lipid rafts is crucial for entry of some viruses (Ewers and Helenius, 2011) including SV40 and murine leukemia virus (Lu and Silver, 2000, Pelkmans et al., 2001). For nidoviruses, CD13 is the cellular receptor for human coronavirus 229E and is localized in the lipid rafts (Nomura et al., 2004), and the importance of lipid rafts for virus replication has also been documented (Lorizate and Kräusslich, 2011, Lu et al., 2008, Thorp and Gallagher, 2004). We show in the present study that GP4 localizes to the lipid rafts of the plasma membrane where it associates with CD163, the cellular receptor for PRRSV. The lipid-modification of GP4 contributes to PRRSV infectivity, suggesting an important role of GPI for PRRSV entry and infection. The DAF/4-M162* mutant was lethal for infectivity, which is probably due to the inability of GP4 to anchor to the membrane, and thus unable to associate with the lipid rafts, supporting the notion that the lipid rafts play an important role for PRRSV infection.
Co-localization of GP4 and CD163 in the lipid rafts may mediate the entry of PRRSV. Compared to peptide-anchored GP4, lipid-anchored GP4 is likely to have the priority to locate in lipid rafts where it binds to CD163 to promote the entry of PRRSV. This may explain why certain mutations affecting the ability of lipid-anchor formation of GP4 on the cell membrane affected the titers and growth of PRRSV as seen in other studies (Metzner et al., 2008). Taken together, we have shown here that the PRRSV GP4 protein is a GPI-modified membrane-associated protein. Co-localization of GP4 with CD163 in the lipid rafts on the cell membrane implicates an important role of the complex for PRRSV entry and infection. These findings deepen our understanding on GP4 and establish the cholesterol and lipid rafts as potential targets in the development of control measures against PRRSV infection.
Materials and methods
Cells and viruses
MARC-145 cells (a subline of MA-104 African green monkey kidney cells; Kim et al., 1993) were maintained as described elsewhere (Lee et al., 2005). Porcine alveolar macrophages (PAMs) were kindly provided by Dr. F. Zuckermann (University of Illinois at Urbana-Champaign, Urbana, IL) and grown in RPMI-1640 (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT). HeLa and HEK-293 cells (NIH AIDS Research and Reference Reagent Program, Germantown, MD) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. Dulac porcine kidney cells (obtained from Dr. L. Babiuk, Vaccines and Infectious Disease Organization, Saskatoon, Canada) were free of porcine circovirus type 1 and grown in modified Eagle's medium (MEM) supplemented with 10% FBS. Stocks of PRRSV (North American genotype strain PA8; Wootton et al., 2000) were prepared in MARC-145 cells. PRRSV P129-GFP expressing GFP is described elsewhere (Pei et al., 2009).
Virus titration and one-step growth kinetics
Culture supernatants harvested at 5 days post-transfection with PRRSV infectious clones were designated ‘passage-1’. The ‘passage-1’ virus was used to inoculate fresh MARC-145 cells, and the 5-day harvest was designated ‘passage-2’. The ‘passage-3’ virus was prepared in the same way as for ‘passage-2’. Each passage virus was aliquoted and stored at − 80 °C until use. Virus titers of ‘passage-2’ and ‘passage-3’ were determined in MARC-145 cells by a microtitration infectivity assay and recorded as 50% tissue culture infectious dose per ml (TCID50/ml). To determine the growth kinetics of the mutant viruses, MARC-145 cells were infected in duplicate with ‘passage-2’ virus at a multiplication of infection (MOI) of 5 and incubated for indicated times. Culture supernatants were collected and titrated by a microtitration infectivity assay and recorded as TCID50/ml.
Antibodies, enzymes, and chemicals
Monoclonal antibody (MAb) against human DAF (clone 1G3 anti-DAF [aa:35–135]), anti-FLAG MAb (clone M2), and anti-FLAG rabbit antibody were from Sigma-Aldrich (St. Louis, MO); anti-GFP rabbit antibody from Cell Signaling Technology (Danvers, MA); β-actin MAb (AC-15) from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-porcine CD163 MAb (clone 2A100/11) from AbD Serotec (Raleigh, NC). Tissue culture supernatant containing MAb EVR1 raised against human DAF was kindly provided by K. Dimmock (University of Ottawa, Ottawa, Canada) and described elsewhere (Karnauchow et al., 1996). Alexa Fluor 488®-conjugated goat anti-mouse IgG (H + L), Alexa Fluor 594®-conjugated goat anti-rabbit IgG (H + L), and G418 (neomycin sulfate analog) were purchased from Invitrogen (Carlsbad, CA). Horseradish peroxidase (HRP) conjugated-goat anti-mouse IgG and -goat anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Phosphatidylinositol-specific phospholipase C (PI-PLC) of Bacillus cereus, methyl-β-cyclodextrin (MβCD), and water soluble cholesterol were purchased from Sigma-Aldrich.
Genes and plasmids
The eukaryotic expression vector pXJ41 is a derivative of pXJ40 in which the polylinker region was modified (Xiao et al., 1991). The plasmid pEGFP-N contains the PRRSV N gene fused to EGFP (Rowland and Yoo, 2003). The PRRSV infectious cDNA clone pCMV-S-P129 is described elsewhere (Lee et al., 2005, Lee and Yoo, 2006). To construct pEGFP-GP4, PRRSV GP4 gene was PCR-amplified from pCMV-S-P129 using the GP4-GFP-Fwd and GP4-GFP-Rev primer pair (Table 1 ) and inserted into EcoRI and BamHI sites in pEGFP-N1 (Clontech; Mountain View, CA). To generate pXJ41-FLAG-GP4, the FLAG-tag sequence (YKDDDDKGS) was inserted between nucleotide positions 261 and 262 (corresponding amino acid positions D87 and E88, respectively) of GP4 by three-point ligations of PCR products generated using the primer pairs, FLAG-GP4-A and -B, and FLAG-GP4-C and -D (Table 1) at EcoRI, BamHI, and HindIII sites (Fig. 1F). The human DAF gene was amplified from plasmid pEF6/V5-DAF (provided by K. Dimmock, University of Ottawa, Ottawa, Canada) using DAF-Fwd and DAF-Rev primers (Table 1). The PCR product was digested with EcoRI and BamHI followed by cloning into pXJ41 to generate pXJ41-DAF. pXJ41-DAF/4 was created by overlapping extension PCR (Horton et al., 1989). Briefly, the region representing amino acid positions 1–348 of human DAF was amplified from pEF6/V5-DAF using DAF-Fwd and DAF/4-B primers. The C-terminal region of GP4 (GP4-C, positions 144–179; Fig. 1) was amplified from plasmid pCMV-S-P129 using primers DAF/4-C and DAF/4-D (Table 1). Then, overlapping extension PCR was conducted using the PCR products of DAF (1–348 aa) and GP4-C (144–179 aa) as templates and primers DAF-Fwd and DAF/4-D (Table 1). The DAF/4 fusion gene (1–348 aa fragment of DAF and GP4-C fragment) was inserted at EcoRI and BamHI sites in pXJ41 to generate pXJ41-DAF/4 (Fig. 3). Plasmid pXJ41-DAF/2 was constructed in a similar way using GP2-C representing 204–256 aa of GP2 (Fig. 3). The primer sequences are listed in Table 1.
Table 1.
Primers used in construction and mutagenesis studies.
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| GP4-GFP-Fwd | GAGGAATTCCACCATGGCTGCGTCCCTTC | GP4 amplification |
| GP4-GFP-Rev | TATGGATCCCGACCGCCACCGCCACCAATTGCCAGTAGGA | GP4 amplification |
| Flag-GP4-A | GAGGAATTCCACCATGGCTGCGTCCCTTC | Flag-GP4 first segment amplification |
| Flag-GP4-B | GCGGGATCCCTTATCGTCGTCATCCTTGTAATCTGTCACATTGGC | Flag-GP4 first segment amplification |
| Flag-GP4-C | GAGGGATCCGAGAATTACTTACA | Flag-GP4 second segment amplification |
| Flag-GP4-D | GAGAAGCTTATCAAATTGCCAGTAGGA | Flag-GP4 second segment amplification |
| DAF-Fwd | GTCGAATTCCACCATGGCAACCGTCGCGCGGCC | DAF, DAF/4 and DAF/2 amplification |
| DAF-Rev | GAGGGATCCTTACTAAGTCAGCAAGCCC | DAF amplification |
| DAF/4-B | CGACCACCAAGGAGCGTCCTTTATTTGGGGTTGTTT | DAF/4 first segment amplification |
| DAF/4-C | AAACAACCCCAAATAAAGGACGCTCCTTGGTGGTCG | DAF/4 second segment amplification |
| DAF/4-D | GAGGGATCCTTATCAAATTGCCACCAGAATGG | DAF/4 second segment amplification |
| DAF/2-B | AGCCATTGCTGAAAATCATGTCCTTTATTTGGGGTTGTTT | DAF/2 first segment amplification |
| DAF/2-C | AAACAACCCCAAATAAAGGACATGATTTTCAGCAATGGCT | DAF/2 second segment amplification |
| DAF/2-D | GAGGGATCCTTATCACCATGAGTTCGAAA | DAF/2 second segment amplification |
| DAF/4-157-F | GCGGCTGCTTCATTTCGTCACACCTGAGACCATGA | DAF/4-M157Vmutation |
| DAF/4-157-R | TCATGGTCTCAGGTGTGACGAAATGAAGCAGCCGC | DAF/4-M157V mutation |
| DAF/4-162-F | CATGACACCTGAGACCGTCAGGTGGGCAACCGTT | DAF/4-M162V mutation |
| DAF/4-162-R | AACGGTTGCCCACCTGACGGTCTCAGGTGTCATG | DAF/4-M162V mutation |
| DAF/4-162S-F | CATGACACCTGAGACCTAGAGGTGGGCAACCGTT | DAF/4-M162S mutation |
| DAF/4-162S-R | AACGGTTGCCCACCTCTAGGTCTCAGGTGTCATG | DAF/4-M162Smutation |
Restriction enzyme recognition sites are indicated in boldface italics. Mutagenized codons are underlined in boldface.
GPI anchor prediction and site-directed mutagenesis
The topology of GP4 (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred; Claros and von Heijne, 1994) and the GPI anchor attachment signal (http://gpi.unibe.ch; Fankhauser and Mäser, 2005, Eisenhaber et al., 1998) were predicted using on-line programs available on the Bioinformatics Resource Portal (http://expasy.org/tool). Specific mutations were introduced to plasmids pXJ41-DAF/4 and pCMV-S-P129 using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) with modifications as described elsewhere (Du et al., 2010).
Transfection
Plasmid DNA transfection was carried out using Lipofectamine™ 2000 according to the manufacturer's instruction (Invitrogen, Carlsbad, CA). HeLa cells were seeded in 35-mm diameter dishes and grown to 70% confluency and transfected with 2 μg of DNA diluted in Opti-MEM (Invitrogen, Carlsbad, CA). After 4 h incubation, transfection medium was replaced with DMEM supplemented with 10% FBS. At 24 h post-transfection (hpt), cells were washed with PBS and lysed in the buffer (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% TritonX-100, 1% NP-40 and 1 mM PMSF) for western blot analysis. For transfection of pXJ41-DAF, pXJ41-DAF/2, pXJ41-DAF/4, and their mutant derivatives, HEK-293 cells grown in 60-mm diameter dishes were transfected using 10 μl Lipofectamine 2000 and 4 μg individual plasmid in 2 ml Opti-MEM. After 6 h incubation, the transfection mix was replaced with DMEM supplemented with 10% FBS. At 24 hpt, cells were washed once with PBS and trypsinized for PI-PLC digestion and FACS analysis. For transfection of PRRSV infectious clone pCMV-S-P129 and its derivatives, MARC-145 cells grown in 35-mm diameter dishes were used. MARC-145 cells were transfected with 2 μg of an infectious clone using Lipofectamine 2000 as described above. Transfected cells were incubated at 37 °C for 5 days in DMEM supplemented with 5% FBS prior to collection of supernatant for virus recovery.
PI-PLC digestion
HeLa cells or HEK-293 cells were transfected with pXJ41-DAF, pXJ41-DAF/2, pXJ41-DAF/4 or their derivatives and incubated for 1 day. Cells were washed with PBS, trypsinized, and collected by centrifugation at 2000 rpm for 10 min (Eppendorf 5415R). Then, the cells were washed twice using PI-PLC buffer (10 mM Tris–HCl [pH 7.4], 144 mM NaCl, 0.05% bovine serum albumin [BSA]) and resuspended in PI-PLC buffer. Cells were divided into two equal fractions: one for PI-PLC treatment (1.5 U/ml; Sigma-Aldrich) and another as a control without treatment. Following incubation at 37 °C for 30 min with PI-PLC, both fractions of cells were centrifuged at 2000 rpm for 10 min (Eppendorf 5415R). The supernatants were subjected to western blot analysis whereas the cell pellets were washed twice in ice-cold FACS buffer (1.0% BSA, 0.01% sodium azide in PBS) followed by fluorescein-activated cell sorter (FACS) analysis.
FACS
PI-PLC-treated or untreated cells were stained with DAF-specific MAb EVR1 at 4 °C for 30 min. Cells were washed twice in ice-cold FACS buffer and fixed with 4% paraformaldehyde in PBS (pH 7.4) for 1 h at 4 °C. After wash with FACS buffer, cells were incubated with Alexa Fluor 488®-conjugated goat anti-mouse IgG (H + L) secondary antibody for 1 h, washed twice with FACS buffer, and resuspended in 1% paraformaldehyde in PBS (pH 7.4) for flow cytometry (BD Biosciences LSR II Analyzer). The data were analyzed using the FACS Express software (Ver. 3 Research Edition, De Novo Software; Los Angeles, CA).
Western blot analysis
The supernatants obtained from PI-PLC digested cells were centrifuged to remove cell debris at 13,000 rpm for 20 min in a microcentrifuge (Eppendorf 5415R). The supernatants were collected and boiled in SDS-PAGE loading buffer (60 mM Tris–HCl [PH 6.8], 2% SDS, 0.1% bromophenol blue, 25% glycerol, 5% β-mercapto ethanol) for 5 min, followed by 10% SDS-PAGE and transfer to polyvinylidenedifluoride (PVDF) membranes (Millipore). The membranes were blocked with 5% non-fat dry milk in TBST (10 mM Tris–HCl [PH8.0], 150 mM NaCl, 0.1% Tween 20) for 4 h at room temperature (RT) and incubated with rabbit anti-GFP antibody, anti-FLAG MAb, or anti-DAF MAb overnight at 4 °C. After 5 washes for 10 min each with TBST, the membranes were incubated with HRP-conjugated goat anti-rabbit or goat anti-mouse IgG for 1 h, washed five times for 10 min each with TBST, and developed using Supersignal® West Pico Chemiluminescent Substrate according to the manufacturer's instruction (Pierce, Rockford, IL).
Fluorescent microscopy
Cells resuspended in 1% paraformaldehyde in PBS for FACS analysis were spotted on microscope slides and visualized using a fluorescent microscope (Olympus, model AX70). Dulac-CD163 cells infected with PRRSV P129-GFP were also examined directly by fluorescent microscopy.
Cholesterol depletion, replenishment, and PRRSV infectivity assays
Cell cytotoxicity of MβCD was determined for both MARC-145 cells and PAMs in 96-well plates at concentrations of 0, 0.25,0.5, 1, 2, 5, 10, and 20 mM. Ten millimolar or higher concentration of MβCD was found to be toxic for MARC-145 cells and PAMs, and thus 0.25–2 mM concentrations were chosen for subsequent studies. MARC-145 cells and PAMs were treated with variable concentrations of MβCD for 1 h at 37 °C and infected with PRRSV PA8 at a MOI of 5. After 1 h incubation, cells were washed twice and cultivated in DMEM or RPMI-1640 supplemented with 2% FBS. For cholesterol replenishment, cells were pretreated with 2 mM MβCD for 1 h at 37 °C and then supplemented with variable concentrations of water soluble cholesterol for 1 h at 37 °C (Popik et al., 2002). After two washes with DMEM for MARC-145 cells or RPMI-1640 for PAMs, the cells were infected with PRRSV as above. MARC-145 cells and PAMs were incubated for 3 days and 20 h, respectively, and the culture supernatants were collected and titrated in MARC-145 cells as described above.
Establishment of a stable cell line expressing CD163
Dulac cells were transfected with pcDNA-CD163 which contained the porcine CD163 gene, and selected for neomycin resistance using 1 mg/ml of G418 in the culture medium. G418-resistant cell colonies were picked using cloning cylinders and amplified for screening as described elsewhere (Lee et al., 2004). The gene expression was confirmed by PCR, RT-PCR, and protein assays by FA and western blot, and the cells expressing porcine CD163 were designated Dulac-CD163.
Immunofluorescence and confocal microscopy
HeLa, Dulac, and Dulac-CD163 cells were plated on microscope cover slips and transfected with pXJ41-FLAG-GP4. At 24 hpt, cells were washed once with ice-cold PBS and incubated with respective primary antibodies at 4 °C for 30 min as follows: DAF-specific MAb EVR1 and anti-FLAG rabbit antibody for HeLa cells transfected with pXJ41-FLAG-GP4; CD163-specific MAb and anti-FLAG rabbit antibody for Dulac-CD163 cells transfected with pXJ41-Flag-GP4. Cells were washed twice in ice-cold PBS and fixed with 4% paraformaldehyde in PBS at 4 °C for 1 h. Cells were then washed again three times with ice-cold PBS and incubated with Alexa Fluor 488®-conjugated goat anti-mouse IgG (H + L) or Alexa Fluor 594®-conjugated goat anti-rabbit IgG (H + L) antibodies. The coverslips were washed five times in PBS, mounted on microscope slides in mounting buffer (60% glycerol and 0.1% sodium azide in PBS), and visualized under a laser-scanning confocal fluorescence microscope (model BX50, Olympus).
Acknowledgments
The authors thank Aimee Bachand for construction of DAF/4 and DAF/2 and Federico Zuckermann for providing Z-mac cells. This study was supported by the National Research Initiatives of the US Department of Agriculture Cooperative State Research Education and Extension Service, grant number 2008-35204-04634 awarded to DY.
References
- Anderson S.M., Yu G., Giattina M., Miller J.L. Intercellular transfer of a glycosylphosphatidylinositol (GPI)-linked protein: release and uptake of CD4-GPI from recombinant adeno-associated virus-transduced HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5894–5898. doi: 10.1073/pnas.93.12.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M., Itokawa M., Yamada K., Toyota T., Arai M., Haga S., Ujike H., Sora I., Ikeda K., Yoshikawa T. Association of neural cell adhesion molecule 1 gene polymorphisms with bipolar affective disorder in Japanese individuals. Biol. Psychiatry. 2004;55:804–810. doi: 10.1016/j.biopsych.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Brown D.A., London E. Functions of lipid rafts in biologicalmembranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Calvert J.G., Slade D.E., Shields S.L., Jolie R., Mannan R.M., Ankenbauer R.G., Welch S.K. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007;81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary L.A., Cooper J.A. Signal transduction: molecular switches in lipid rafts. Nature. 2000;404:945–947. doi: 10.1038/35010257. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chazal N., Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.S., Aizaki H., Lai M.M. Murine coronavirus requires lipid rafts for virus entry and cell–cell fusion but not for virus release. J. Virol. 2005;79:9862–9871. doi: 10.1128/JVI.79.15.9862-9871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M.G., von Heijne G. TopPred II: an improved software for membrane protein structure predictions. CABIOS. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cross G.A. Glycolipid anchoring of plasma membrane proteins. Annu. Rev. Cell Biol. 1990;6:1–39. doi: 10.1146/annurev.cb.06.110190.000245. [DOI] [PubMed] [Google Scholar]
- Das P.B., Dinh P.X., Ansari I.H., de Lima M., Osorio F.A., Pattnaik A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010;84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond D.C., Finberg R., Chaudhuri S., Sleckman B.P., Burakoff S.J. Human immunodeficiency virus infection is efficiently mediated by a glycolipid-anchored form of CD4. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5001–5005. doi: 10.1073/pnas.87.13.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zuckermann F.A., Yoo D. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth. Virus Res. 2010;147:294–299. doi: 10.1016/j.virusres.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L., Selvaraj P., Mattaliano R.J., Springer T.A. Anchoring mechanisms for LFA-3 cell adhesion glycoproteinat membrane surface. Nature. 1987;329:846–848. doi: 10.1038/329846a0. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Bork P., Eisenhaber F. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 1998;11:1155–1161. doi: 10.1093/protein/11.12.1155. [DOI] [PubMed] [Google Scholar]
- Ewers H., Helenius A. Lipid-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 2011 doi: 10.1101/cshperspect:a004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faaberg K.S., Even C., Palmer G.A., Plagemann P.G. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J. Virol. 1995;69:613–617. doi: 10.1128/jvi.69.1.613-617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser N., Mäser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- Firth A.E., Zevenhoven-Dobbe J.C., Wills N.M., Go Y.Y., Balasuriya U., Atkins J.F., Snijder E.J., Posthuma C.C. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. Gen. Virol. 2011;92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Tsukamoto K., Ikezawa H. Mutational analysis of the C-terminal signal peptide of bovine liver 5′-nucleotidase for GPI anchoring: a study on the significance of the hydrophilic spacer region. Biochim. Biophys. Acta. 1997;1328:185–196. doi: 10.1016/s0005-2736(97)00084-9. [DOI] [PubMed] [Google Scholar]
- Gerber L.D., Kodukula K., Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J. Biol. Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- González J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- Ikonen E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Ilangumaran S., Hoessli D.C. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.G., Robinson P.J., Bletchly C., Mackenzie J.M., Young P.R. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J. 2000;14:1603–1610. doi: 10.1096/fj.14.11.1603. [DOI] [PubMed] [Google Scholar]
- Jasin M., Page K.A., Littman D.R. Glycosylphosphatidylinositol-anchored CD4/Thy-1 chimeric molecules serve as human immunodeficiency virus receptors in human, but not mouse, cells and are modulated by gangliosides. J. Virol. 1991;65:440–444. doi: 10.1128/jvi.65.1.440-444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.R., Griggs T.F., Gnanandarajah J.S., Murtaugh M.P. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded in an alternative open reading frame 5 present in all arteriviruses. J. Gen. Virol. 2011;92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnauchow T.M., Tolson D.L., Harrison B.A., Altman E., Lublin D.M., Dimock K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55) J. Virol. 1996;70:5143–5152. doi: 10.1128/jvi.70.8.5143-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G.A., Siegel M.W., Caras I.W. Endocytosis of glycophospholipid-anchored and transmembrane forms of CD4 by different endocytic pathways. EMBO J. 1992;11:863–874. doi: 10.1002/j.1460-2075.1992.tb05124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Kwang J., Yoon I.J., Joo H.S., Frey M.L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Lee C., Yoo D. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology. 2006;355:30–43. doi: 10.1016/j.virol.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.E., Church G.A., Wilson D.W. A subpopulation of tegument protein vhs localizes to detergent-insoluble lipid rafts in herpes simplex virus-infected cells. J. Virol. 2003;77:2038–2045. doi: 10.1128/JVI.77.3.2038-2045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Bachand A., Murtaugh M.P., Yoo D. Differential host cell gene expression regulated by the porcine reproductive and respiratory syndrome virus GP4 and GP5 glycoproteins. Vet. Immunol. Immunopathol. 2004;102:189–198. doi: 10.1016/j.vetimm.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Calvert J.G., Welch S.W., Yoo D. A DNA-launched reverse genetics system for porcine reproductive and respiratory syndrome virus reveals that homodimerization of the nucleocapsid protein is essential for virus infectivity. Virology. 2005;331:47–62. doi: 10.1016/j.virol.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Park C.K., Nam E., Kim S.H., Lee O.S., Lee du S., Lee C. Generation of a porcine alveolar macrophage cell line for the growth of porcine reproductive and respiratory syndrome virus. J. Virol. Methods. 2010;163:410–415. doi: 10.1016/j.jviromet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Lisanti M.P., Sargiacomo M., Graeve L., Saltiel A.R., Rodriguez-Boulan E. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M., Kräusslich H.G. Role of lipids in virus replication. Cold Spring Harb. Pespect. Biol. 2011 doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Silver J. Ecotropic murine leukemia virus receptor is physically associated with caveolin and membrane rafts. Virology. 2000;276:251–258. doi: 10.1006/viro.2000.0555. [DOI] [PubMed] [Google Scholar]
- Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008;369:344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manié S.N., de Breyne S., Vincent S., Gerlier D. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 2000;74:305–311. doi: 10.1128/jvi.74.1.305-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardassi H., Massie B., Dea S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology. 1996;221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- Metzner C., Salmons B., Günzburg W.H., Dangerfield J.A. Rafts, anchors and viruses-A role for glycosylphosphatidylinositol anchored proteins in the modification of enveloped viruses and viral vectors. Virology. 2008;382:125–131. doi: 10.1016/j.virol.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus belongs to a new virus family, comprising Lactate dehydrogenase-elevating virus, Equine arteritis virus, and Simian hemorrhagic fever virus. Arch. Virol. Suppl. 1994;9:441–448. doi: 10.1007/978-3-7091-9326-6_43. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Petersen-den Besten A., de Kluyver E.P., Moormann R.J., Schaaper W.M., Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh M.P., Elam M.R., Kakach L.T. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 1995;140:1451–1460. doi: 10.1007/BF01322671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen C.J., Murtaugh M.P., Faaberg K.S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E.A., Christopher-Hennings J., Drew T., Wensvoort G., Collins J.E., Benfield D.A. Differentiation of US and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 1993;31:3184–3189. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickells M.W., Alvarez J.I., Lublin D.M., Atkinson J.P. Characterization of DAF-2, a high molecular weight form of decay-accelerating factor (DAF; CD55), as a covalently cross-linked dimer of DAF-1. J. Immunol. 1994;152:676–685. [PubMed] [Google Scholar]
- Noisakran S., Dechtawewat T., Avirutnan P., Kinoshita T., Siripanyaphinyo U., Puttikhunt C., Kasinrerk W., Malasit P., Sittisombut N. Association of dengue virus NS1 protein with lipid rafts. J. Gen. Virol. 2008;89:2492–2500. doi: 10.1099/vir.0.83620-0. [DOI] [PubMed] [Google Scholar]
- Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K., Senda T., Fujimoto T. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 2004;78:8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin L.C. Simian virus 40 infection via MHC class I molecules and caveolae. Immunol. Rev. 1999;168:13–22. doi: 10.1111/j.1600-065x.1999.tb01279.x. [DOI] [PubMed] [Google Scholar]
- Ono A., Freed E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P., Menon A.K. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Parton R.G., Lindsay M. Exploitation of major histocompatibility complex class I molecules and caveolae by simian virus 40. Immunol. Rev. 1999;168:23–31. doi: 10.1111/j.1600-065x.1999.tb01280.x. [DOI] [PubMed] [Google Scholar]
- Patton J.B., Rowland R.R., Yoo D., Chang. K.O. Modulation of CD163 receptor expression and replication of porcine reproductive and respiratory syndrome virus in porcine macrophages. Virus Res. 2009;140:161–171. doi: 10.1016/j.virusres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Pei Y., Hodgins D.C., Wu J., Welch S.K., Calvert J.G., Li G., Du Y., Song C., Yoo D. Porcine reproductive and respiratory syndrome virus as a vector: immunogenicity of green fluorescent protein and porcine circovirus type 2 capsid expressed from dedicated subgenomic RNAs. Virology. 2009;389:91–99. doi: 10.1016/j.virol.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Kartenbeck J., Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Pike L.J. The challenge of lipid rafts. J. Lipid Res. 2009;50:S323–S328. doi: 10.1194/jlr.R800040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popik W., Alce T.M., Au W.C. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J. Virol. 2002;76:4709–4722. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.J., Millrain M., Antoniou J., Simpson E., Mellor A.L. A glycophospholipid anchor is required for Qa-2-mediated T cell activation. Nature. 1989;342:85–87. doi: 10.1038/342085a0. [DOI] [PubMed] [Google Scholar]
- Rosenberry T.L. A chemical modification that makes glycoinositol phospholipids resistant to phospholipase C cleavage: fatty acid acylation of inositol. Cell Biol. Int. Rep. 1991;15:1133–1150. doi: 10.1016/0309-1651(91)90060-v. [DOI] [PubMed] [Google Scholar]
- Rothberg K.G., Ying Y.S., Kolhouse J.F., Kamen B.A., Anderson R.G. The glycophospholipid-linked folate receptor internalizes folate without entering the clathrin-coated pit endocytic pathway. J. Cell Biol. 1990;110:637–649. doi: 10.1083/jcb.110.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95:23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Schneider P., Fang Y., Wootton S., Yoo D., Benfield D.A. Peptide domains involved in the localization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus. Virology. 2003;316:135–145. doi: 10.1016/S0042-6822(03)00482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrias M.R., Gronlund J., Padilla O., Madsen J., Holmskov U., Lozano F. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- Scheiffele P., Rietveld A., Wilk T., Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Pedersen K.W., Raamsman M.J., de Vries A.A. Identification of a novel structural protein of arteriviruses. J. Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Dobbe J.C., Spaan W.J. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 2003;77:97–104. doi: 10.1128/JVI.77.1.97-104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart A.D., Eustace H.E., McKee T.A., Brown T.D. A novel cell entry pathway for a DAF-using human enterovirus is dependent on lipid rafts. J. Virol. 2002;76:9307–9322. doi: 10.1128/JVI.76.18.9307-9322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Waneck G.L., Flavell R.A., Bothwell A.L. The glycosyl phosphatidylinositol anchor is critical for Ly-6A/E-mediated T cell activation. J. Cell Biol. 1991;112:377–384. doi: 10.1083/jcb.112.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda J., Kinoshita T. GPI-anchor biosynthesis. Trends Biochem. Sci. 1995;20:367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]
- Takeda M., Leser G.P., Russell C.J., Lamb R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.R., Hooper N.M. The prion protein and lipid rafts. Mol. Membr. Biol. 2006;23:89–99. doi: 10.1080/09687860500449994. [DOI] [PubMed] [Google Scholar]
- Thorp E.B., Gallagher T.M. Requirements for CEACAMs and cholesterol during murine coronavirus cell entry. J. Virol. 2004;78:2682–2692. doi: 10.1128/JVI.78.6.2682-2692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneki H., Kobayashi S., Takagi K., Kagawa S., Tsunoda M., Murata M., Matsuoka T., Wada T., Kurachi M., Kimura I., Sasaoka T. Novel G423S mutation of human alpha 7 nicotinic receptor promotes agonist-induced desensitization by a protein kinase C-dependent mechanism. Mol. Pharmacol. 2007;71:777–786. doi: 10.1124/mol.106.030866. [DOI] [PubMed] [Google Scholar]
- van der Goot F.G., Harder T. Raft membrane domains: from a liquid-ordered membrane phase to a site of pathogen attack. Semin. Immunol. 2001;13:89–97. doi: 10.1006/smim.2000.0300. [DOI] [PubMed] [Google Scholar]
- Vieira F.S., Corrĕa G., Einicker-Lamas M., Coutinho-Silva R. Host-cell lipid rafts: a safe door for micro-organisms? Biol. Cell. 2010;102:391–407. doi: 10.1042/BC20090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensvoort G., Terpstra C., Pol J.M.A., ter Laak E.A., Bloemraad M., de Kluyver E.P., Kragten C., van Buiten L., den Besten A., Wagenaar F., Broekhuijsen J.M., Moonen P.L.J.M., Zetstra T., de Boer E.A., Tibben H.J., de Jong M.F., van't Veld, P., Groenland G.J.R., van Gennep J.A., Voets M.T., Verheijden J.H.M., Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Wieringa R., De Vries A.A., Post S.M., Rottier P.J. Intra- and intermolecular disulfide bonds of the GP2b glycoprotein of equine arteritis virus: relevance for virus assembly and infectivity. J. Virol. 2003;77:12996–13004. doi: 10.1128/JVI.77.24.12996-13004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa R., de Vries A.A., Rottier P.J. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 2003;77:6216–6226. doi: 10.1128/JVI.77.11.6216-6226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink E.H., Kroese M.V., van Wijk H.A., Rijsewijk F.A.M., Meulenberg J.J., Rottier P.J. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 2005;79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Z., Orsó E., Werner T., Klünemann H.H., Schmitz G. Monocyte cholesterol homeostasis correlates with the presence of detergent resistant membrane microdomains. Cytometry A. 2007;71:486–494. doi: 10.1002/cyto.a.20403. [DOI] [PubMed] [Google Scholar]
- Wootton S.K., Yoo D., Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 2000;145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.H., Davidson I., Matthes H., Garnier J.M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]