Abstract
RNA–RNA recombination salvages viral RNAs and contributes to their genomic variability. A recombinationally-active subgenomic promoter (sgp) has been mapped in Brome mosaic bromovirus (BMV) RNA3 (Wierzchoslawski et al., 2004. J. Virol.78, 8552–8864) and mRNA-like 5′ sgRNA3a was characterized (Wierzchoslawski et al., 2006. J. Virol. 80, 12357–12366). In this paper we describe sgRNA3a-mediated recombination in both in vitro and in vivo experiments. BMV replicase-directed co-copying of (−) RNA3 with wt sgRNA3a generated RNA3 recombinants in vitro, but it failed to when 3′-truncated sgRNA3a was substituted, demonstrating a role for the 3′ polyA tail. Barley protoplast co-transfections revealed that (i) wt sgRNA3a recombines at the 3′ and the internal sites; (ii) 3′-truncated sgRNA3as recombine more upstream; and (iii) 5′-truncated sgRNA3 recombine at a low rate. In planta co-inoculations confirmed the RNA3–sgRNA3a crossovers. In summary, the non-replicating sgRNA3a recombines with replicating RNA3, most likely via primer extension and/or internal template switching.
Keywords: Subgenomic RNA, RNA replication, RNA recombination, Primer extension, Strand switching, (+) RNA viruses
Introduction
Genetic RNA recombination contributes significantly to the high genetic variability of RNA viruses, and RNA–RNA crossovers have been observed both in natural RNA virus sequences (Allison et al., 1989, Becher et al., 2001, Bujarski and Figlerowicz, 1998, Molenkamp et al., 2000), and by using experimental systems (Bujarski and Kaesberg, 1986, Figlerowicz et al., 1998, Lai, 1992, Adair and Kearney, 2000, Bruyere et al., 2000, Hu et al., 2003, Mikkelsen et al., 2000). While some viruses can recombine by re-joining of genomic RNA fragments (Gmyl et al., 2003), most of the RNA virus systems are thought to recombine via replicase switching mechanisms (reviewed in Agol, 2006, Alejska et al., 2001, Hu et al., 2003). Specifically, primer extension have been demonstrated during RNA replication for flaviviruses (Ranjith-Kumar et al., 2002a), tombusviruses (Nagy et al., 1999b, Shapka and Nagy, 2004), Poliovirus (Kirkegaard and Baltimore, 1986, Guillot et al., 2000), nidoviruses (Lai et al., 1985), and for retroviruses (Hu et al., 2003, Mikkelsen et al., 2000, Zhang and Ma, 2001).
Brome mosaic virus (BMV) is a tripartite, (+) stranded representative of the Bromoviridae family that has been used as a model RNA virus (Noueiry and Ahlquist, 2003). The multiparticism of the BMV RNA genome evolved to favor recombination (Bujarski and Kaesberg, 1986), which salvages damaged/mutated viral RNAs and/or secures genome variability. We have observed both inter- and intra-segmental crossovers among BMV RNAs, and we have suggested mechanisms where both RNA sequences and replicase proteins direct the distribution of crossover sites (Nagy et al., 1994, Nagy et al., 1999a, Nagy and Bujarski, 1994, Nagy and Bujarski, 1996, Nagy and Bujarski, 1997, Nagy and Bujarski, 1998).
To further study the molecular mechanism of homologous recombination, we have focused on the multifunctional intergenic region of BMV RNA3. This region includes the subgenomic promoter (sgp), which was shown previously to participate in de novo initiation of sgRNA4 and was mapped as an efficient recombination hot spot (Bruyere et al., 2000, Wierzchoslawski et al., 2003, Wierzchoslawski et al., 2004, Wierzchoslawski and Bujarski, 2006). Similarly, recombinationally-active sgps have been predicted in other RNA viruses (Koev et al., 1999, Miller and Koev, 2000, Rohayem et al., 2005). Yet an additional function of the intergenic region in BMV RNA3 is in the formation of 5′ sgRNA3a by premature termination (Wierzchoslawski et al., 2006). SgRNA3a covers the 5′ part of RNA3 including the 3a ORF and a short 3′ oligo(A) tail, so to a certain extent it resembles a cellular mRNA. Indeed, it accumulates in polysomes and it actively synthesizes BMV 3a protein (Wierzchoslawski et al., 2006).
Because sgRNA3a was shown to occur only as (+) strands (Wierzchoslawski et al., 2006), another potential function of sgRNA3a could be in priming homologous recombination on (−) RNA3 strands. In this work, we show that sgRNA3a recombines with RNA3 both in vitro and in vivo, either in protoplasts or in whole plants. The 3′ polyA tail makes possible the incorporation of sgRNA3a, probably via primer extension, whereas more upstream sequences can support recombination via strand switching, possibly at the packaging signal. Overall, our data indicate that sgRNA3a can mediate homologous BMV RNA–RNA crossovers through various mechanisms, and that similar means might operate during recombination in other RNA viruses.
Results
Incorporation of sgRNA3a in vitro: primer extension on (−) strands
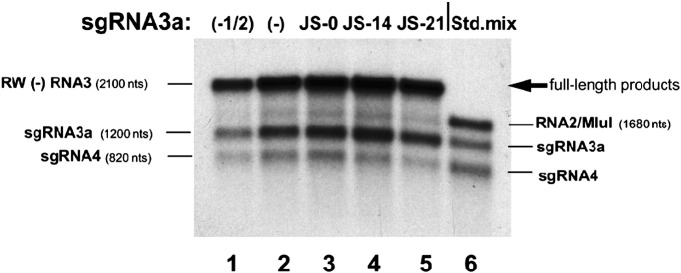
To test if sgRNA3a recombines with RNA3, RW(−) RNA3 and a three-fold excess of sgRNA3a were copied in vitro by BMV replicase (Dzianott et al., 2001). SgRNA3a variants JS-14 or JS-21 (Fig. 1 ) carried the sequence corresponding to BamHI and PstI marker sites at flanking position 121 and 1190 nts, and either 14 or 21 3′ A residues, respectively. In addition, because JS-14 and JS-21 RNAs were transcribed from BglII-digested plasmid, both carried four extra 3′ bases (GAUC). JS-0 did not carry the 3′ polyA tail along with twelve upstream 3′ nts. A band corresponding to the full-length radioactive product was observed, along with those for both subgenomic RNAs: sgRNA3a and sgRNA4 (Fig. 2 ). The reaction profiles of RW(−) RNA3 alone (lanes 1 and 2) were similar to those for the mixtures of RW(−) RNA3 with either JS-0, JS-14 or JS-21 RNAs (lanes 3 to 5). Thus, the excess of sgRNA3a did not prevent copying of either RW(−) RNA3 or both sgRNAs. The full-length RNA3 products with JS-21 (lane 5) were cut out from the gel, purified, subjected to RT-PCR amplification, and the cDNA material was cloned (see Materials and methods). Based on a restriction enzyme analysis of sixty clones, five clones (8%) acquired marker restriction sites from JS-21 sgRNA3a (Table 2). All five clones carried both restriction markers, indicating that the entire JS-21 sequence recombined. In contrast, no recombinant clones were identified for JS-14 RNA, indicating that the shorter polyA tail did not support recombination. Also, JS-0 sgRNA3a did not generate recombinants (Table 2), indicating that the polyA tail alone and/or the 12 upstream bases were important. Finally, RT-PCR of the full-length RNA copying products (cut from the gel, lanes 1 or 2, Fig. 2) did not generate marker-bearing cDNA recombinants, reflecting the lack of contaminating sgRNA3a molecules. Overall, these results demonstrate the incorporation of (+) sgRNA3a during RNA3 copying.
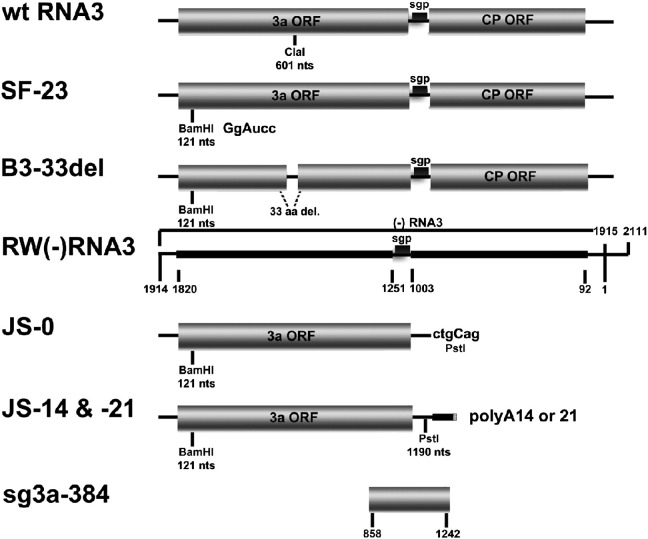
Fig. 1.
BMV RNA3 variants used in the in vitro and whole-plant experiments. Wt RNA3 is shown on the top with the ClaI restriction site indicated. An intermediate SF-23 construct carries a sequence corresponding to the BamHI marker site, whereas B3-33del carries a 33-aa deletion in the 3a ORF. JS-14 and JS-21 are sgRNA3a constructs carrying 3′ polyA tails of 14 and 21 A residues, respectively, plus RNA sequences corresponding to BamHI and PstI marker sites, whereas JS-0 does not carry a 3′ polyA tail. sg3a-384 is a (+) strand RNA primer corresponding to a middle region of the RNA3 sequence. Thin lines mark non-coding regions, whereas shaded rectangles represent ORFs for the 3a movement and coat (CP) proteins. The positions of marker sites are indicated by small vertical bars. To construct RW (−) RNA3, a portion of (−) RNA3 between nts 1 and 1914 (the nucleotide positions correspond to those in wt genomic (+) RNA3) was fused immediately downstream of the (+) RNA3 promoter (nts 1915–2111). The sequences complementary to the 3a and CP ORFs are shown as thick lines with the corresponding nucleotide positions below (for more details, see Wierzchoslawski and Bujarski, 2006).
Fig. 2.
Characterization of BMV RdRp in vitro copying products of mixtures of RW(−) RNA3 and sgRNA3a templates. Template RNA mixtures were copied as described in Materials and methods, and the radioactive RdRp products were separated in a 1% denaturing agarose gel. The sgRNA3a variants are specified above (lanes 3 to 5). Lanes 1 and 2 contain copying products of RW(−) RNA3 alone [symbolized by (−)] with half the amount loaded in lane 1. Lane 6 contains a mixture of three radioactive in vitro transcripts as migration standards (the RNA2 marker was synthesized from plasmid pB2TP5 after linearization with MluI).
Table 2.
RNA3–sgRNA3a recombination from in vitro reactions and from C. quinoa inoculations: summary of restriction analysis of the RT-PCR products.
| Experiment |
|||||
|---|---|---|---|---|---|
| In vitro (RdRp reaction)a | Total number of cDNA clones analyzed | Number of recombination | Recombinants frequency (%) | ||
| 1. RW(−) RNA3 + JS-21 sgRNA3a | 60 | 5 | 8 | ||
| 2. RW(−) RNA3 + JS-14 sgRNA3a | 60 | 0 | 0 | ||
| 3. RW(−) RNA3 + JS-0 sgRNA3a | 60 | 0 | 0 | ||
| In whole plants (C. quinoa)b | Total number of cDNA clones analyzed | Distribution of restriction sitesc | Recombination frequency (%) | ||
| 1. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a (0.2×) | No infection | No infection | No infection | ||
| 2. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a (0.5×) | No infection | No infection | No infection | ||
| BamHI/PstI | PstI | BamHI | |||
| 3. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a (1.5×) | 22 | 20 | 0 | 1 | 100 |
| 4. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a (3.0×) | 21 | 21 | 0 | 0 | 100 |
| 5. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a (5.0×) | 35 | 32 | 1 | 2 | 100 |
| 6. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-0 sgRNA3a (1.5×) | No infection | No infection | No infection | ||
| 7. Wt RNAs 1 and 2 + B3-33del RNA3 + JS-0 sgRNA3a (5.0×) | No infection | No infection | No infection | ||
| 8. Wt RNAs 1 and 2 + B3-33del RNA3 | No infection | No infection | No infection | ||
See Materials and methods for the in vitro recombination assays.
The co-inoculation with wt RNAs 1 and 2 + B3-33del RNA3 + JS-21 sgRNA3a generated, on average, from one to three local lesions per each inoculated leaf (three leaves of two C. quinoa plants) from two independent experiments. The lesion tissue was cut out, combined, and the virus extracted, followed by cDNA cloning of viral RNA, and analysis of cDNA clones by sequencing (see also Materials and methods and Fig. 8). The numbers in parentheses: 0.2×, 0.5×, 1.5×, 3.0× and 5.0× indicate the molar ratio of sgRNA3a to RNA3 in the used RNA inoculation mixtures.
The numbers show the amount of cDNA clones carrying the restriction markers as listed.
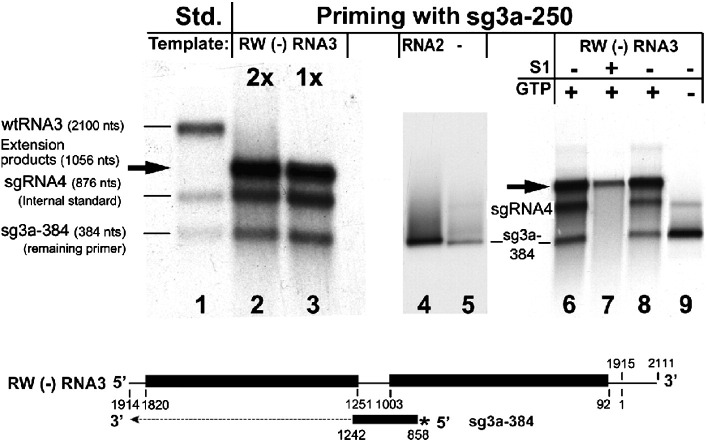
To confirm that BMV RdRp supports primer extension, a radioactive 384-nt sgRNA3a fragment (named sg3a-384, see Fig. 1 and Table 1 ) was co-copied in vitro with RW(−) RNA3. This generated extension products of the expected size, and the more sg3a-384 that was input the more efficient was the synthesis (Fig. 3 , lanes 2 and 3). A distinct extension product was observed neither with the noncomplementary BMV RNA2 template (lane 4) nor without template (lane 5). To test the double-stranded (ds) character of the reaction products, half of the incubation mixture was treated with S1 nuclease (see Materials and methods). The full-length extension products remained undigested, whereas bands corresponding to both the internal ss RNA4 standard and the unincorporated ss sg3a-384 primer disappeared (compare lanes 6 and 7). Our data not only demonstrate that BMV RdRp generated the dsRNA extension products but also suggest that the radioactive products did not emerge via terminal nucleotidyl transferase (TNT) activity. To further analyze TNT activity, the extension reactions were carried out with and without GTP. The latter did not generate the extension products (compare lanes 8 and 9). Altogether, these experiments show that BMV RdRp extended the sg3a-384 primer on the (−) RNA3 template.
Table 1.
List of primers, plasmids and constructs used in this work.
| Constructa | Primer # | nt positionb |
Deoxyoligonucletide sequence (5′–3′)c | Purpose | |
|---|---|---|---|---|---|
| Start | Finish | ||||
| pJS-21 | 1 | 1 | 38 | TGTAATACGACTCACTATAGGTAAAATACCAACTAATTCTCGTTCGATTCCGGCGAAC | To create sgRNA3a (JS-21) that carries 21 A residues at the 3′ end and an additional (to BamHI at 120 nts) marker restriction site PstI at position 1190 nts (underlined). T7 promoter is in bold. |
| 2 | 1232 | 1166 | CGCTGAATTAGGACATAGATCTTTTTTTTTTTTTTTTTTTTTAATAATAACTGCAGACACACAACATAGAATATCC | ||
| pJS-14 | 1 | See above | See above | To create sgRNA3a (JS-14) that carries 14 A residues at the 3′ end and an additional (to BamHI at 120 nts) marker site PstI at position 1190 nts (underlined). | |
| 3 | 1232 | 1166 | CGCTGAATTAGGACATAGATCTTTTTTTTTTTTTTAATAATAACTGCAGACACACAACATAGAATATCC | ||
| pJS-22 | 1 | See above | See above | To create sgRNA3a (sg-21) that carries 21 A residues at the 3′ end and an additional (to BamHI at 120 nts and PstI at 1190 nts) marker restriction site: HindIII at position 780 nts (underlined). | |
| 4 | 824 | 767 | GATCCACAGACTGGTTAGAAATACCTCTAATATAATTTTTTAAGCTTTTCTTATCGAG | ||
| sgRNA3a-5′ | 5 | 32 | 73 | GGCGAACATTCTATTTTACCAACATCGGTTTTTTCAGTAGTG | To clone the full-length segment of sgRNA3a carrying restriction marker sites as a result of RNA3–sgRNA3a recombination. |
| sgRNA3a-3′ | 6 | 1279 | 1238 | GTCATCTTACCAGTTCCTGAAGTCGACATTATTAATACGCTG | |
| pB3-33del | 7 | 70 | 100 | GTGATACTGTTTTGGATCCCGATGTCTAAC | 5′ primer, BamHI site (underlined). |
| 8 | 530 | 377 | GTAAAGCCGACAACTGAATTGTGGCCTCCTG (500)—(400)G AGAAAACAAACGATGCGTGGAACG | Reverse primer with flanking sequences securing the 33-aa deletion within the 3a ORF | |
| 9 | 872 | 843 | CTTCCTGGGCAACCTGATCAACAGATTGTAACGGTC | 3′ primer, BclI site (underlined) | |
| 10 | 890 | 863 | CAACTAACAAATCTTCCTGGGCAACCTG | 3′ primer, for down- stream to PflMI site. |
|
| sg3a-384 | 2 | See above | See above | To amplify the RNA3 cDNA from positions 858 to 1242 (T7 promoter is in bold) and to use it as RNA transcription template. | |
| 11 | 1242 | 858 | TGTAATACGACTCACTATAGGTTGATCAGGTTGCCCAGGAAG | ||
| 5′-580 | 1 | See above | See above | To create 3′-nested deletion fragment 580 bp carrying an additional (to BamHI at 120 nts) AvaI marker sites at position 560 nts (underlined). | |
| 12 | 602 | 545 | ACATCGATTCCTACCGCTATCACCGCCGATGACTTCCATCGGACAATCATAGCTCGGGGTCAA | ||
| 5′-380 | 1 | See above | See above | To create 3′-nested deletion fragment 380 bp carrying an additional (to BamHI at 120 nts) SmaI marker site at position 360 nts (underlined). | |
| 13 | 365 | 333 | GAATCCCCTCCCGGGTAACTCTCCTTTATCATACTTGG | ||
| 5′-150 | 1 | See above | See above | To create 3′-nested deletion fragment 150 bp carrying BamHI restriction site at 120 nts. | |
| 14 | 142 | 123 | GCCAACGTCAGACGTAGTTCGTGAGG | ||
| 3′-860 | 15 | 1258 | 1221 | CCTGAAGTCGACATTATTAATACGCTGAATTAGGACATAGATC | To create 5′-nested deletion fragment 860 bp, carrying an additional (to PstI at 1190 nts) SmaI marker site at position 360 nts (underlined). T7 promoter is in bold. |
| 16 | 352 | 373 | TGTTAATACGACTCACTATAGGAGTTACCCGGGAGGGGATTCATG | ||
| 3′-680 | 15 | See above | See above | To create 5′-nested deletion fragment 680 bp, carrying an additional (to PstI at 1190 nts) AvaI marker site (underlined). T7 promoter is in bold. | |
| 17 | 542 | 564 | TGTTAATACGACTCACTATAGGAGCTTTGACCCCGAGCTATGATTG | ||
| 3′-440 | 15 | See above | See above | To create 5′-nested deletion fragment 440 bp, carrying an additional (to PstI at 1190 nts) HindIII marker site at position 780 nts (underlined). T7 promoter is bold. | |
| 18 | 757 | 798 | TGTTAATACGACTCACTATAGGA CTGAGACAACTCGATAAGAAAAGCTTAAAAAATTATATTAG | ||
| Neg-RNA3 | 19 | 1 | 35 | GTAAAATACCAACTAATTCTCGTTCGATTCCGGCG | To create template for in vitro transcription of (−)-strand RNA3 construct. Two rounds of PCR included (i) amplification of RNA3 fragment without T7 promoter with primers 19 and 20 followed by (ii) creation of (−)-strand RNA3 bearing T7 promoter (bold) with primers 19 and 21. |
| 20 | 2111 | 2066 | TGGTCTCTTTTAGAGATTTACAGTGTTTTTCAACACTGTACGGTACC | ||
| 21 | 1 | 35 | TGTAATACGACTCACTATAGGGTGGTCTCTTTTAGAGATTTACAGTGTTTTTCAACACTGTACG | ||
| mut-RNA3 | 22 | 1 | 51 | TGTTAATACGACTCACTATAGGAAAAATACCAACTAATTCTCGTTCGATTCCGGCGAACATTCTATTTTACC | To generate mut-RNA3 template carrying the 5′ A → U substitution (underlined) at position 2 (Hema and Kao, 2004). |
| 23 | 2111 | 2066 | GGCCTTAAGTGGTCTCTTTTAGAGATTTACAGTGTTTTTCAACACTGTACGGTACC | ||
See Materials and methods for detailed description of each construct.
Refers to the exact nt positions on the RNA3 template.
All restriction marker sites introduced by primers are underlined, T7 promoter sequence is in bold.
Fig. 3.
Analysis of the products of in vitro RdRp extension reactions of radioactive sg3a-384 RNA on the RW (−) RNA3 template. The products were separated in denaturing 1.2% agarose gels. Lane 1, mixture of in vitro-transcribed radioactive wt BMV RNA3, sgRNA4, and sg3a-384 RNA as size standards; lanes 2 and 3, primer extension reaction with a single (lane 3) or double (lane 2) amount of the radioactive sg3a-384 RNA. In addition, lanes 2 and 3 contain 32P-labeled sgRNA4 as an internal loading standard that was added after the reaction. The reaction in lane 2 contains a double amount of the sg3a-384 primer. Lanes 4 and 5, control extension reactions of sg3a-384 primer on the BMV RNA2 template (lane 4) or without an RNA2 template (lane 5). Lanes 6 and 7, digestion of sg3a-384 extension products without (lane 6) or with (lane 7) S1 nuclease. Both, sgRNA4 (added post-reaction) and ss sg3a-384 RNA disappeared after S1 nuclease treatment (lane 7). Lanes 8 and 9, analysis of TNT activity of BMV RdRp, with the extension reactions completed with (lane 8) or without (lane 9) GTP. Both the RW(−) RNA3 template and the corresponding sg3a-384 RNA primer (* symbolizes the radioactivity) are shown schematically below (see Fig. 1 for more details).
To further examine if a polyA tail could initiate RNA copying, JS-21 RNA was subjected to the in vitro RdRp reaction as the only template. Full-sized radioactive products (Fig. 4 ) demonstrated that BMV RdRp initiated at the 3′ tail (lanes 2 to 4). The reaction was much more efficient when using an equimolar amount of JS-21 as the template (lane 4), whereas JS-14 was less efficient (lane 3) and JS-0 was copied only marginally (lane 2); shorter sgRNA3a fragments 5′-580 and 5′-380 were not copied (lanes 5 and 6). Therefore, RdRp is able to initiate RNA synthesis preferentially at the polyA tail; however its presence is not indispensable. The ds nature of the RNA products was further confirmed by their resistance to S1 nuclease treatment (lanes 7 through 10).
Fig. 4.
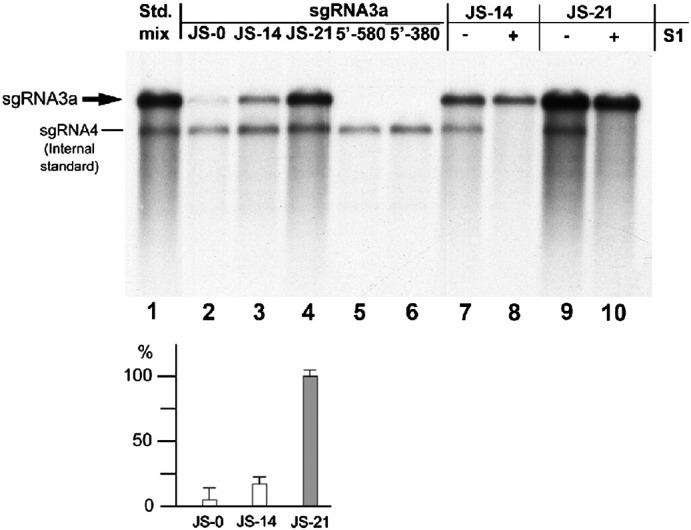
In vitro copying of sgRNA3a templates with BMV RdRp. Equimolar amounts of sgRNA3a constructs JS-0 (lane 2), JS-14 (lane 3), JS-21 (lane 4), 5′-580 (lane 5) and 5′-380 (lane 6) were copied in standard BMV replicase reactions as the only templates. The radioactive products were separated in a 1.0% formamide/formaldehyde agarose gel and blotted to nylon membrane followed by autoradiography. Bands representing the copying products are marked by thick arrows. Lane 1, 32P-labeled mixture of JS-21 and sgRNA4 transcripts as size standards. In addition, lanes 2 to 10 contain the sgRNA4 loading marker, added post-copying reaction. The densitometric quantification results are shown below. The open bars represent the relative density of bands corresponding to the in vitro copying products of JS-0 to JS-21 (which was taken as 100%) after normalizing to the internal sgRNA4 standard. Lanes 7 to 10, analysis of the ds nature of the copying products of JS-14 and JS-21 RNAs by S1 nuclease digestion (lanes 8 and 10, respectively) versus undigested products (lanes 7 and 9).
Recombination of BMV RNA3 with sgRNA3a in barley protoplasts
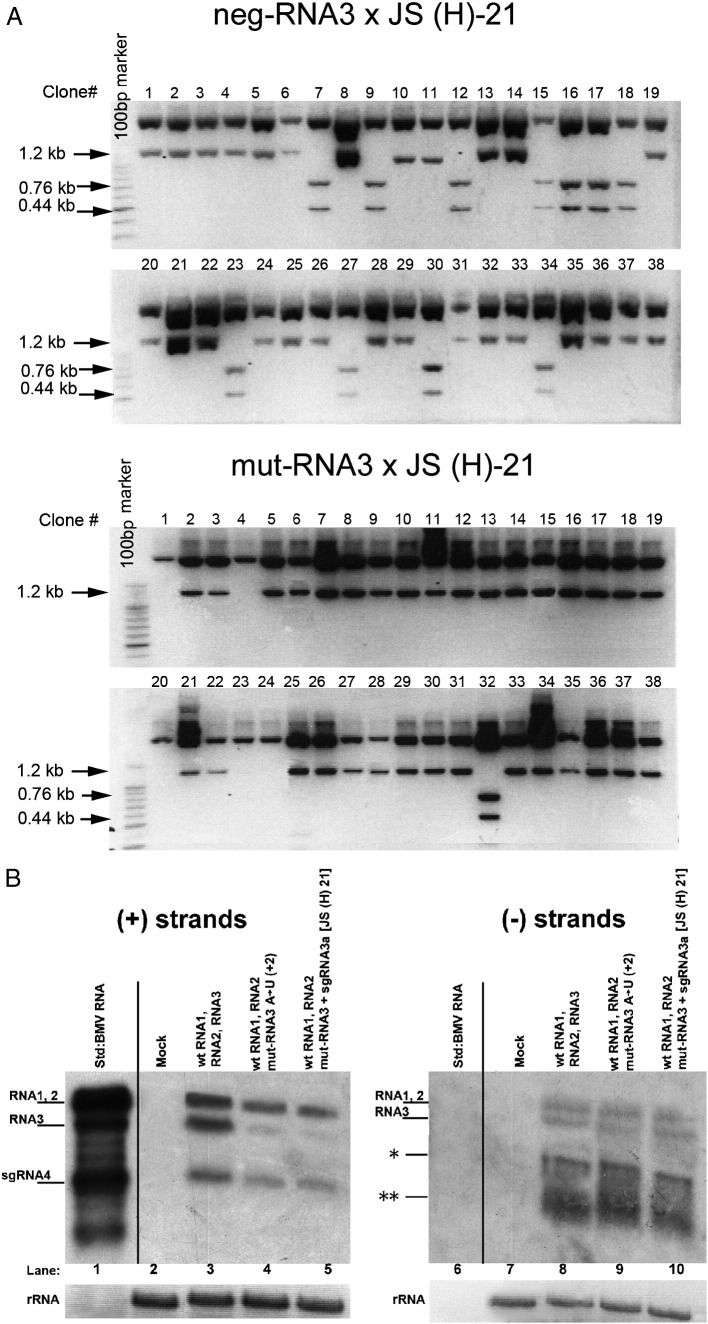
To test if sgRNA3a would recombine with RNA3 in host cells under reduced selection pressure, barley protoplasts were co-transfected with a mixture of transcribed BMV RNA1, 2, 3 and the full-length sgRNA3a variant JS(H)-21. As compared to JS-21, the JS(H)-21 RNA carried an additional HindIII marker site at nt position 780 (see Materials and methods). All the designed marker restriction sites were silent and stable during infection, and they did not create a codon usage discrepancy (not shown). After incubating for 48 h, total RNA was extracted, the sgRNA3a region was amplified by RT-PCR, followed by cloning and restriction digestion at designed marker positions. Among one-hundred-and-two clones there were fourteen clones that carried at least one sgRNA3a marker, setting the recombination frequency at 14% (Fig. 5 ). Most of the recombinant RNA3 variants carried a nucleotide sequence corresponding to the BamHI marker, suggesting that efficient crossovers occurred at upstream positions.
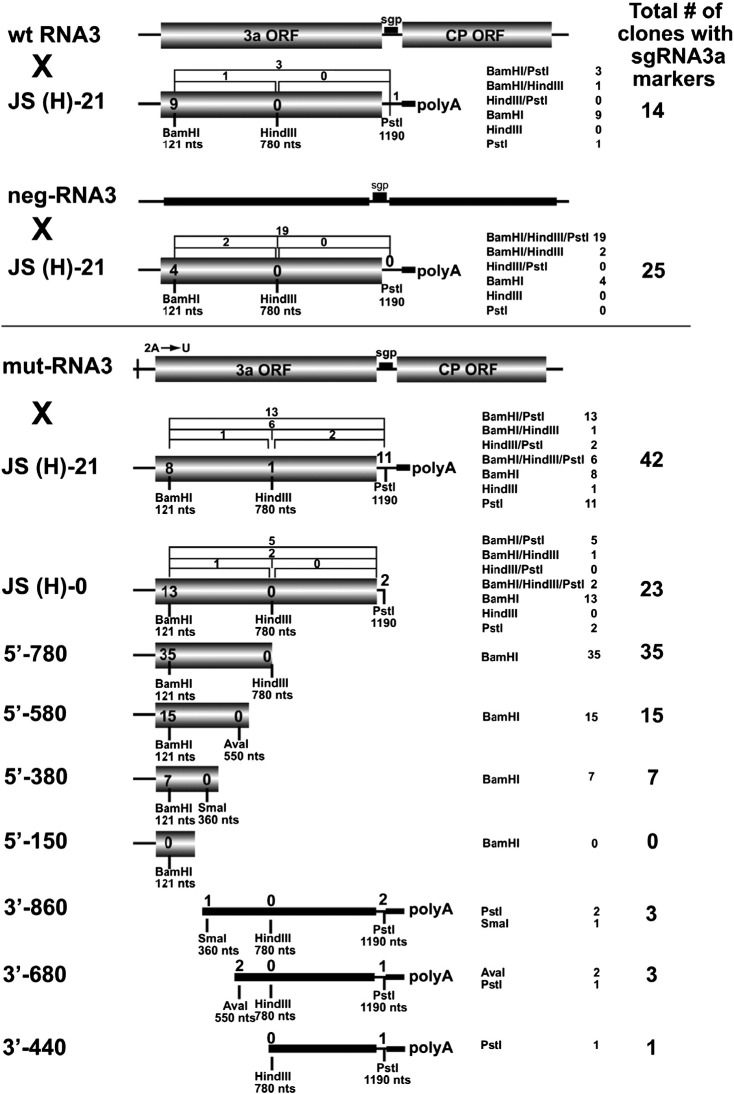
Fig. 5.
Recombination of BMV RNA3 templates with sgRNA3a constructs in barley protoplasts. wt RNA3 × JS(H)-21 indicates an experiment with wt genomic RNA3 (wt RNA3) and full-length sgRNA3a (JS(H)-21). neg-RNA3 × JS(H)-21 indicates an experiment with negative-strand RNA3 (neg-RNA3) and full-length sgRNA3a (JS(H)-21). mut-RNA3 × JS(H)-21 indicates an experiment with an RNA3 derivative bearing a point mutation (A → U) at position 2 near the 5′ end. The remaining sgRNA3a-derivatives that were tested for recombination with mut-RNA3 are shown below, including JS(H)-0: an sgRNA3a derivative without a polyA tail; 5′-780, 5′-580, 5′-380, 5′-150: the 5′-nested deletion sgRNA3a fragments; and 3′-860, 3′-680, 3′-440: the 3′-nested deletion sgRNA3a fragments. The positions of marker restriction sites are indicated below each construct. The column on the right shows the general recombination frequency (per-cent) based on an analysis of one-hundred insert-bearing cDNA clones. The frequency of recombinants bearing single restriction markers is shown inside the shaded rectangles, whereas that of those carrying double or triple markers is represented above the constructs as brackets. The 3′nested RNAs do not carry the 3a ORF initiation codon and they are therefore represented by thick black lines. The other elements are as in Fig. 1. The numbers summarize the results from two independent transfection experiments with each sgRNA3a derivative.
The moderate frequency of JS(H)-21-mediated recombination (14%) could be due to limited access to the replicating RNA3 molecules. This was tested by using a negative-strand RNA3 template dubbed neg-RNA3. Here, co-transfection of JS(H)-21 with neg-RNA3 increased the recombination frequency to 25% (Fig. 5, Fig. 6A). Apparently, the use of the negative RNA3 template enhanced the crossovers, probably by increased interaction with the JS(H)-21 primer at the initial stages of infection.
Fig. 6.
Recombination of BMV RNA3 with sgRNA3a in barley protoplasts. A. Restriction enzyme digestion of recombinant RNA3 cDNA clones obtained from barley protoplasts that were transfected with a mixture of wt BMV RNAs 1 and 2 and (top) Neg-RNA3 plus JS(H)-21 sgRNA3a or (bottom) mut-RNA3 and JS(H)-21 sgRNA3a. The RNA3 region representing the sgRNA3a sequence was amplified from total RNA extracts by RT-PCR, and the cDNA products were cloned into the pGEM-T Easy system. The insert sequences were released by HindIII/EcoRI digestion and separated by electrophoresis in a 1.5% agarose gel. An intact 1.2-kb fragment reflected the lack of the HindIII marker site at nt position 780, whereas the double 0.44 kb and 0.76 kb bands indicate the presence of the HindIII marker. B. Northern blot analysis showing the accumulation in protoplasts of either (+) or (−) strands (left and right panels, respectively) of BMV RNAs. Total protoplast RNA was separated in a 1.2% denaturing agarose gel, blotted, and probed with a 3′-specific RNA probe detecting either (+) or (−) strands (see Materials and methods). Lanes 1 and 6, virion BMV RNA used as size standards; lanes 2 and 7, negative controls from mock inoculated protoplasts; lanes 3 and 8, protoplasts transfected with equimolar amounts of BMV RNAs 1 and 2 and wt RNA3; lanes 4 and 9, protoplasts transfected with BMV RNAs 1 and 2 and (A → U) RNA3; lanes 5 and 10, protoplasts transfected with BMV RNAs 1 and 2, (A → U) RNA3, and sgRNA3a. Ribosomal RNA (rRNA) bands (after staining with ethidium bromide) are shown below. The position corresponding to the (−) RNA4 band is marked with a single asterisk on the right panel, whereas the migration position of degradation products (likely because minus strands are not encapsidated and thus less protected) is marked with two asterisks.
The role of negative strands was further supported by using a (+) RNA3 construct (dubbed mut-RNA3) that carried a 5′ A → U substitution at nt position 2. According to Hema and Kao (2004), this mutation decreases the accumulation of (+) RNA3 strands without affecting minus strands (both confirmed by northern blotting, Fig. 6B, left and right panels, respectively). Mut-RNA3 was co-transfected with wt RNAs 1 and 2 and JS(H)-21 RNA, and analysis of progeny cDNA clones revealed a three-fold increase in the recombination frequency to 42% (Figs. 5B and 7 ). This revealed that the lower rate of (+) strand synthesis enhanced the crossovers, likely due to reduced competition with RNA3 replication and/or better access to (−) strands (see Discussion).
Fig. 7.
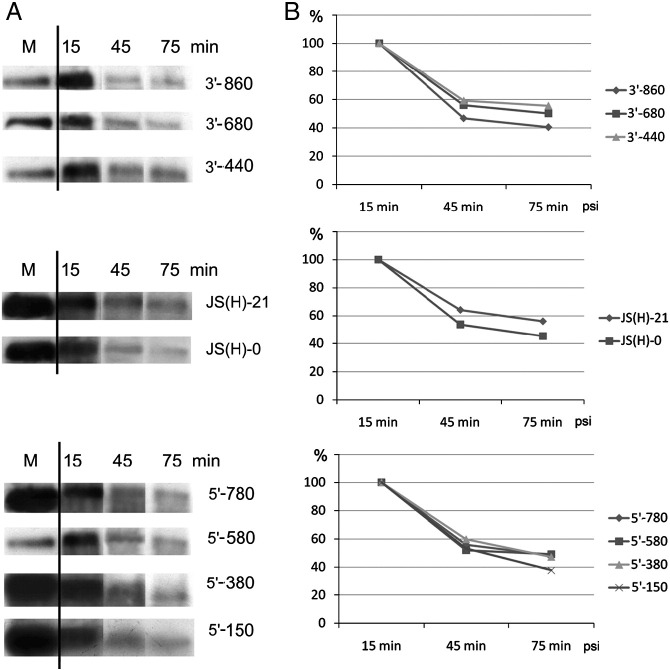
The stability of the sgRNA3a constructs used for transfection experiments in barley protoplasts. A. Autoradiogram representing the intensity of bands corresponding to individual radioactive sgRNA3as that were transfected into protoplasts and extracted after incubation for 15, 45, or 75 min. M stands for the size marker; the names of the constructs are indicated on the right hand side. B. The results of densitometric analysis of the bands shown in A, calculated as the percentage of the initial band intensity.
To test the role of the 3′ polyA tract, the 3′ truncated construct JS(H)-0 was co-transfected with mut-RNA3 (Fig. 5). This decreased the RNA3-sgRNA3a recombination frequency to 23% and the crossovers occurred mainly at internal positions (see below).
The role of 3′ sequences was further examined by co-transfecting protoplasts with a set of 5′ nested sgRNA3a fragments, including 5′-780, 5′-580, 5′-380 and 5′-150 nt transcripts (Fig. 5). Among 100 clones analyzed, 35, 15, 7 and 0 acquired the nucleotide sequence corresponding to the 5′ BamHI marker when using the 5′-780, 5′-580, 5′-380, and 5′-150 sgRNA3a derivatives, respectively. Altogether, our results suggest that in addition to the recombinationally-active 3′ polyA tail, the inner regions of sgRNA3a also support homologous crossovers (see Discussion).
Closer analysis of the restriction markers' distribution revealed that for JS(H)-21 RNA, the downstream PstI site occurred in 46%, the middle HindIII site in 14%, and the upstream BamHI site in 40% of the recombinants (Fig. 5). Thus, nearly half of the crossovers occurred near the JS(H)-21 3′ end, whereas the others took place more upstream. The 3′-side crosses (RNA sequence corresponding to the PstI marker) dropped to 27% for JS(H)-0, demonstrating that the lack of the 3′ polyA tail favored the upstream sites. This upstream preference was even higher for the 5′-780, 5′-580, and 5′-380 RNAs, which generated only BamHI-carrying recombinants. Overall, these data show the existence of at least two recombination regions, one at the 3′ terminus and an additional upstream site possibly between nts 550 and 780. The upstream site can be folded into strong stem–loop structures (data not shown), and indeed Choi and Rao (2003) have shown that hairpins between nts 601 and 817 act as the RNA3 packaging signal dubbed PE. The 5′-380 and 5′-150 RNAs do not fold into comparable structures. We hypothesize that two independent mechanisms operate during RNA3–sgRNA3a recombination: polyA-mediated 3′-terminal primer extension and PE-mediated strand switching (see Discussion).
The 3′-nested fragments, including the 3′-860, 3′-680, and 3′-440 RNAs (Fig. 5), were used to determine the role of 5′ sequences; their respective recombination frequencies were 3%, 3%, and 1%. This reveals the importance of the 5′ region and suggests that the infrequent recombinants arose via double crossovers (only double crosses could rescue the missing 5′ end in the replicable RNA3). The 5′-side crossovers likely contribute to maintaining the essential 5′ cis-acting signals on viral RNAs.
To assess whether the recombination frequencies were due to altered RNA stability, the degradation of sgRNA3a variants was studied in barley protoplasts (Fig. 7). Individual radioactive transcripts were co-transfected into barley protoplasts along with unlabeled RNAs 1 and 2 and mutated RNA3. The protoplasts were thoroughly washed to remove the un-transfected radioactive material, and total RNA was extracted at three time points post-transfection. These experiments revealed that all the transcript variants were stable to a similar degree within the 75 min assay time (Fig. 7). Our data indicate that the observed frequencies are at the level of recombinant formation.
The in planta repair of internally-truncated RNA3
To test recombination with sgRNA3a in whole plants, a 33-codon deletion was engineered within the RNA3 3a ORF (construct B3-33del, Fig. 1). Two leaves of Chenopodium quinoa were co-inoculated with wt BMV RNAs 1 and 2, the B3-33del RNA3 (2.5 μg each RNA per leaf in 50 μl inoculation mixture, i.e. 50 μg each/ml) and five different amounts of JS-21 sgRNA3a (0.2; 0.5; 1.5; 3.0 and 5.0 molar ratio of sgRNA3a to RNA3). This generated, on average (from two independent experiments), one, three and three local lesions per leaf (three leaves of two plants), respectively, for the three highest sgRNA3a concentrations (experiments 1 to 5, Table 2 ). By contrast, in the presence of JS-0 at two different molar ratios (experiments 6 and 7) or in the absence of any sgRNA3a variant (experiment 8), no local lesions emerged. This demonstrates that JS-21 but not JS-0 sgRNA3a restored the function of the RNA3 segment and points to the role of the 3′ polyA tail.
To determine whether the recovery of infection was due to complementation or genetic recombination, the progeny RNA3 sequences from combined local lesion tissue were amplified by RT-PCR and cloned, and the presence of restriction markers was analyzed. The analysis of 21–35 cDNA clones from each infection did reveal that all the sequences carried the JS-21 restriction markers, with over 90% carrying both markers and the rest carrying the upstream (BamHI) marker site. Only one clone carried the downstream (PstI) marker (experiment 5, Table 2). This demonstrates that the repair of the 3a ORF was by RNA recombination rather than due to complementation and suggests 3′ extension as the main mechanism of recombination (see Discussion). A control inoculation with wt BMV RNAs 1, 2 and 3 plus JS-21 sgRNA3a revealed a 20-fold increase in the number of local lesions (data not shown). This further emphasizes the requirement for the functional 3a ORF within the RNA3 molecule. Northern blot analysis (Fig. 8 ) confirmed normal profiles of BMV RNAs in the leaves with local lesions (shown in lanes 3, 4 and 5) but not in the symptomless plants (lanes 1 and 2), demonstrating the regenerated virus in the infected plants.
Fig. 8.
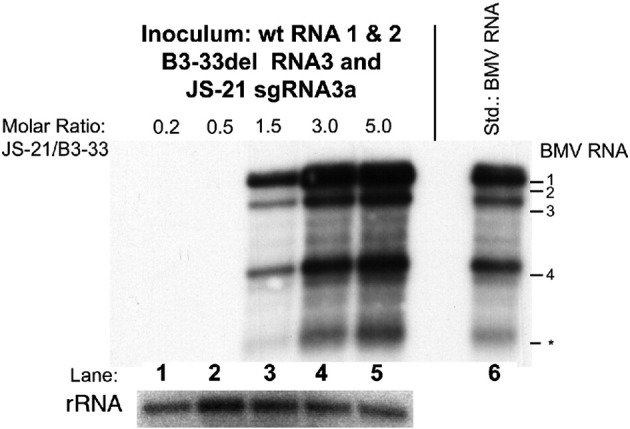
Recombination in planta between 3a-truncated BMV RNA3 and sgRNA3a. C. quinoa seedlings were co-inoculated with wt RNAs 1 and 2, B3-33del RNA3, and various amounts of JS-21 sgRNA3a (the molar ratios of JS-21 to B3-33del are indicated on the top, lanes 1–5). Total RNA was extracted from the inoculated leaves seven days later, separated by electrophoresis in a denaturing agarose gel, blotted onto nylon membrane, and probed with the BMV (+) strand 3′ probe (see Materials and methods). BMV RNAs were detected in plants infected with higher molar ratios of JS-21 to B3-33 del RNAs (lanes 3 to 5). Lane 6 shows the migration of marker BMV RNAs extracted from a viral preparation that was isolated from local lesion tissue after infection with wt transcript BMV RNAs. The lower panel shows the concentration of ribosomal RNA (rRNA) after staining with ethidium bromide. The positions of individual BMV RNAs are indicated on the right. The asterisk marks possible degradation product (in necrotic lesions of C. quinoa tissue).
RT-PCR controls
To verify that the recombinant RNAs emerged during RNA replication rather than during RT-PCR (Cocquet et al., 2006), protoplasts were co-transfected with a mixture of wt BMV RNA3 and JS(H)-21 sgRNA3a, but without wt RNAs 1 and 2. Another control involved RT-PCR amplification of the RNA3 sequences from a mixture of wt BMV RNAs and JS(H)-21 RNA, omitting the protoplast transfection. All the RNA transcripts used in these experiments were pre-treated with an excess of RNase-free DNase (Ambion) in order to remove the plasmid DNA template and to prevent DNA-amplified PCR products. Subsequent analyses of cDNA clones did not detect RNA3 recombinants (not shown). In yet another control, total RNA was extracted from protoplasts that were transfected with wt (virion) BMV RNA. No false positive recombinants were identified (not shown), signifying the lack of cross-contamination in the protoplast assays. For the in vitro assays, an RT-PCR negative control involved cloning of the cDNA products from mixtures of RW(−) RNA3 and either JS-0 or JS-21 RNAs without copying by BMV RdRp. Among the 55 cDNA clones analyzed, none carried the JS-0 or JS-21 markers (not shown). Thus, the RNA templates did not recombine during the RT-PCR reactions.
Discussion
Previous studies have suggested a role for 5′ sgRNA3a in BMV RNA recombination (Wierzchoslawski et al., 2006). This work provides supporting experimental evidence from both in vitro and in vivo systems. The in vitro BMV replicase assays demonstrate recombination between (+) sgRNA3a and (−) RNA3 and its dependence upon a 3′ polyA tract (Table 2, Fig. 3, Fig. 4). These recombinants likely arose via a primer extension mechanism. Subsequent protoplast transfections revealed that (i) sgRNA3a recombines efficiently with RNA3, and a reduction in (+) strands (mut-RNA3 variant) or their lack at the initial stages of infection (neg-RNA3 variant) increase the frequency of crossovers; (ii) 3′-truncated sgRNA3a fragments recombine preferably at more central and upstream positions, whereas (iii) 5′-truncated sgRNA3a fragments recombine at a very low rate (Fig. 5). Moreover, whole-plant co-inoculations led a deletion within the 5′ 3a ORF to be repaired through recombination with sgRNA3a (Table 2). Overall, our results demonstrate viral recombination events between subgenomic and genomic RNAs that likely occur via two different mechanisms: primer extension and strand switching.
In vitro RdRp copying: the primer extension model
The in vitro RdRp assays (Table 2, top part) confirm the previously suggested (Wierzchoslawski et al., 2006) role of the polyA tail in the primer extension mechanism. Because the polyA tail (or the U-track in the (−) strand) is likely unpaired (assuming it is not PABP-bound), it might serve as a preferred re-initiation/recombination site. This is supported not only by our observation of RdRp initiation at the polyA tail (Fig. 4) but also by similar observations made in BMV (Karran and Hudak, 2008, Choi et al., 2004), Bamboo mosaic potexvirus (Cheng et al., 2002, Tsai et al., 1999), picornaviruses (Barton et al., 2001, Herold and Andino, 2001, Svitkin et al., 2008), Sindbis virus (Hill et al., 1997), and Encephalomyocarditis virus (Cui et al., 1993). Some (+) sense RNA viruses use the poly(A) tail for priming of (−) strand RNA synthesis e.g., Coxsackie B virus (Melchers et al., 1997) and Bamboo mosaic potexvirus (Cheng et al., 2002) or to enhance template recruitment (Herold and Andino, 2001, Wang et al., 2000, Leonard et al., 2004). Also, in Cucumber mosaic cucumovirus (CMV), which is closely related to BMV, the RNA3 component lacks the polyA tract and it does not result in formation of homologous recombinants (de Wispelaere et al., 2005). Yet another effect might be due to the close proximity of the sgRNA4 transcription site, which may enhance RdRp recruitment (Sztuba-Solińska and Bujarski, 2008).
Our evidence for BMV RdRp initiating at the polyA tail (Fig. 4) suggests that (−) sgRNA3a strands could accumulate; nevertheless they were not detected in the infected tissue (Wierzchoslawski et al., 2006). The (−) strands may be at undetectable levels or access to the polyA 3′ tail could be blocked by translation (Wierzchoslawski et al., 2006) or by other protein factors (Perales et al., 2003, Welnowska et al., 2009) (e.g., by PABP; Bradrick et al., 2006, Khan et al., 2008, Zhang et al., 2007). Future studies on sgRNA3a will address the link between replication, translation, and recombination.
Primer extension represents a more widely-employed mechanism of RNA virus recombination. It has been demonstrated in vitro for Hepatitis C virus protein NS5B (Ranjith-Kumar et al., 2002a, Ranjith-Kumar et al., 2002b, Ranjith-Kumar et al., 2004, Rodriguez-Wells et al., 2001), Turnip crinkle carmovirus (Nagy et al., 1999b), and Cucumber necrosis tombusvirus (Panaviene et al., 2004).
Recombination in protoplasts: the internal strand transfer model
The co-transfection experiments revealed a moderate frequency (14%) of recombination between wt RNA3 and sgRNA3a (JS(H)-21) in protoplasts (Fig. 5). We hypothesize that this reflects limited access to the replicating (−) strands. Indeed, the use of either mut-RNA3 (Choi et al., 2004 and Fig. 6A) or neg-RNA3 increased the recombination frequency to 42% or 25%, respectively. The subsequent use of an sgRNA3a construct without a polyA tail (JS(H)-0) decreased the frequency to 23% and shifted the crossovers to upstream positions, verifying the role of the polyA tail (Fig. 5). For Infectious Bronchitis coronavirus or Equine rhinovirus, the cross sites clustered at the exposed polyA tails (Jonassen et al., 1998). Suzuki et al., 2003, Galli et al., 2003 have shown that secondary structures play a role in cross-site selection (Artsimovitch and Landick, 2009). Our MFOLD-based modeling (data not shown) revealed exposure of the unpaired 3′ polyA tail within sgRNA3a. Also, for shorter sgRNA3a fragments (Fig. 5), some correlation could be drawn between the AU content at their 3′ ends and their recombination frequency (not shown). Previously, the AU content was proposed to facilitate the detachment of the 3′ end from the donor template (Wierzchoslawski et al., 2006, Nagy and Bujarski, 1996). Here, the highest AU pair content (64%) was calculated for the JS(H)-21 and 5′-780 constructs, and these also yielded the highest proportions of recombinants, 42% and 35%, respectively. The 5′-580 fragment had the lowest number of AU pairs (49%), and the crossover frequency was reduced to 15%.
In addition to the effects on recombination frequency, another feature that impacts our interpretation of the protoplast results is the diverse distribution of marker mutations among the tested sgRNA3a constructs. For instance, the mut-RNA3 × JS(H)-21 experiment generated only six recombinants carrying all three markers; thirteen carrying BamHI–PstI double markers; one and two carrying (respectively) BamHI–HindIII or HindIII–PstI double markers; and eleven, one, and eight carrying (respectively) PstI, HindIII, or BamHI single markers (Fig. 5). The apparent deficiency of the central HindIII marker (also observed for JS(H)-0) suggests additional crosses within this region. Also, a noticeable drop in the recombination activity between the 5′-780 and 5′-580 RNAs (from 35% to 15%) suggests an additional recombination site between nts 550 and 780. This region has been mapped as the position-dependent RNA3 packaging element PE (nts 601–817), holding strong affinity to the multimeric CP complexes (Choi and Rao, 2003). Following the initial binding of CP subunits to the 3′ tRNA-like structure, a cis interaction with PE forms a ribonucleoprotein (RNP) complex consisting of CP and RNA3 that then interacts in trans with sgRNA4 to facilitate the co-packaging of both RNAs into virions (Annamalai and Rao, 2006). Thus, it is possible that CP binding at the PE element might bridge over the recombining RNA substrates and/or might create a roadblock for progressing RdRp, thereby facilitating template switching. Clearly, RNA packaging is not the only function of BMV CP (Yi et al., 2009a, Yi et al., 2009b). CP co-purifies with BMV replicase (Bujarski et al., 1982), and it binds at several sites to BMV RNAs, including the B-box, the PE, and the 3′ tRNA-like structure (Yi et al., 2009a, Yi et al., 2009b, Choi and Rao, 2003).
The 3′-nested sgRNA3a fragments recombined only marginally (1% to 3%, Fig. 5). The recovery of the replicable full-length RNA3 requires double crosses between RNA3 and sgRNA3a. Whether CP or other factors can mediate BMV RNA recombination at this or other cross sites will be the subject of further studies.
Recombination in whole plants
We have previously demonstrated efficient crossovers between BMV RNA3 variants in doubly-infected local lesions on C. quinoa leaves (Bruyere et al., 2000). This system utilized two co-replicating RNA3 variants that crossed within the restriction marker sites. In our current paper we describe recombination in C. quinoa between a replicating RNA3 construct (PB3-33del) that carries a disabled movement protein ORF and a non-replicable sgRNA3a carrying two marker sites (JS-21). The incorporation of JS-21 sequences into the progeny RNA3 is evident by the two sgRNA3a markers being found in the vast majority of the RNA3 recombinants (Table 2, lower panel). The appearance of sgRNA3a–RNA3 crosses reflects inefficient complementation between sgRNA3a and PB3-33del. Indeed, the number of local lesions was about twenty-fold smaller with mutated RNAs as compared with the same amount of wt BMV RNA transcripts (not shown). Furthermore, all local lesions accumulated the recombinants. This apparent lack of complementation has been observed not only with this and previous BMV four-component systems (this work and Bruyere et al., 2000, respectively), but also after co-inoculation of cowpeas with deletions in both movement and coat protein genes of Cowpea chlorotic mottle bromovirus (Allison et al., 1990). However, a low-level of complementation cannot be excluded, as it might help at the initial stages of local lesion formation. In fact, sgRNA3a was found to support efficient translation of movement protein (Wierzchoslawski et al., 2006). The lack of replication of sgRNA3a likely imposes critical constraints on the four-component system, favoring the accumulation of RNA3 recombinants.
Elucidation of the exact recombination mechanism will require further experiments, and our protoplast data suggest that both 3′ primer extension and internal strand switching occur. The fact that all restriction markers were found in the majority of in planta recombinants emphasizes the 3′ primer extension mechanism. It is possible that the location of the deletion in the PB3-33del input RNA (nts 401–499) affected the folding of the nearby PE element, and consequently disabled the role of CP binding during internal template switching, as postulated above. The lack of recombination activity with JS-0 further points to primer extension via the 3′ polyA tail. The polyA tail may interact more easily with the exposed intercistronic region on the (−) RNA3 template than with other regions along the RNA3 sequence. However, one may speculate that at least some of the observed effects were due to higher stability of the polyA-carrying JS-21 than of JS-0 RNAs. Increased stability has been reported for the polyadenylated RNAs of Tobacco mosaic virus (Gallie and Kobayashi, 1994), potexviruses (Guilford et al., 1991, Tsai et al., 1999), herpesviruses (Conrad et al., 2007), and for eukaryotic mRNAs (Grosset et al., 2000, Anderson, 2005). It has been shown that the polyA tail blocks the RNA/exonuclease complex (Meyer et al., 2004).
In summary, we show that ss non-replicating sgRNA3a can recombine with replicating viral RNA. Our data suggest that this process can occur either by 3′ primer extension or by internal replicase switching. The specific features that define potentially useful RNA substrates might be the presence of a 3′ polyA tail, high AU content of the 3′ ends, and the presence of target secondary structures. The strand switching mechanism might utilize protein binding sites (e.g., CP or RdRp) that may bridge over the recombining RNAs and/or serve as roadblocks during RNA replication. All these features enhance the generation of progeny mosaic RNAs, as previously suggested (Urbanowicz et al., 2005). Crossover events have been mapped at highly structured 5′-sides (Che et al., 2001, Gowda et al., 2003, Gowda et al., 2009) and at oligo A regions (Jonassen et al., 1998). The RNAs of other bromoviruses and togaviruses carry internal polyA tracts (Fujisaki et al., 2003, Iwahashi et al., 2005, Fauquet et al., 2005), and sgRNA3a-like RNAs were observed in Broad bean mottle virus- and Cowpea chlorotic mottle virus-infected hosts (Dzianott and Bujarski, unpublished results). Further experiments are required to unravel more details about the role of subgenomic components in the RNA virus life cycle.
Our data have implications for RNA virus evolution. We show that recombination with non-replicating RNAs can rescue a functional RNA genome when plants are inoculated with defective viruses. For instance, this confirms previous findings that cellular RNAs were incorporated into some viral RNAs (Mayo and Jolly, 1991, Sano et al., 1992). The observed crossovers demonstrate that such genome rescue is potentially significant in natural infections. Because plant RNA viruses can generally replicate in primarily inoculated cells, the recombinational repair of movement protein likely plays an important role in virus survival. Both the ability to compensate for high mutation rates and the possibility of in planta recombination between a not properly adapted virus and a co-infecting RNA may have potential roles during virus adaptation to new hosts, and they may also facilitate the divergence of new viral species. The results described here are likely relevant to other (+) stranded RNA viruses.
Materials and methods
Materials
Plasmids pB1TP3, pB2TP5, and pB3TP7 (Janda et al., 1987) were used as templates to synthesize in vitro the capped full-length transcripts of BMV (Russian strain) RNA1, 2, and 3, respectively, by using the MEGAscript T7 kit (Ambion, Austin, TX). Maloney murine leukemia virus (MMLV) reverse transcriptase, Taq DNA polymerase, restriction enzymes, and dNTPs were from Promega Corp. or from New England Biolabs, Inc.
Generation of RNA3 and sgRNA3a variants
Plasmid pB3TP7 was used as a template to generate mut-RNA3 that carries the 5′ A → U substitution at nt position 2 by using oligomers 22 and 23 (Hema and Kao, 2004); and to generate the neg-RNA3 construct that transcribes the (−) RNA strand by using two consecutive rounds of PCR reactions with primers 19 and 20, followed by amplification with primers 19 and 21 (Table 1).
The generation of the RW (−) RNA3 template was described in Wierzchoslawski and Bujarski (2006). The previously published pB3TP7-derived plasmid SF-23 (Bruyere et al., 2000) was used as a template to design pJS-21 carrying a new PstI restriction site (C insertion at 1190 nts). This was accomplished by PCR amplification of SF-23 with primers 1 and 2 (Table 1). The PCR products were purified with a Qiaquick® PCR Purification Kit (Qiagen), digested with BglII (1221 nts) and PflMI (816 nts) enzymes and re-ligated between BglII and PflMI sites into SF-23.
The resulting pJS-21 plasmid was 3′ linearized with BglII (1221 nts), and the capped RNAs were synthesized in vitro by run-off transcription with the MEGAscript kit (Ambion) followed by phenol/chloroform extraction and ethanol precipitation. Since BglII digestion led to a 5′-protruding overhang, and the transcribed JS-21 RNAs carried four extra bases (GAUC) at the 3′ terminus (underlined), i.e. 5′-AAAAAAAAGAUC-3′. After centrifugation, the RNA pellet was air dried and dissolved in RNase-free water. The unincorporated ribonucleotides were removed on MicroSpin™ G-25 Columns (GE Healthcare) and the integrity of the RNA was determined by electrophoresis in denaturing (formamide–formaldehyde) agarose gels.
Alternatively, the PCR products were used as templates for direct in vitro transcription reactions. For instance, JS-0 sgRNA3a was synthesized by run-off in vitro transcription from PstI-cut (nt position 1187, Table 1) PCR product of JS-21. This removes the polyA tail plus the 12 upstream nucleotides.
To delete a 33-amino acid stretch in the 3a ORF, a 5′ portion was amplified with primers 7 and 8 (Table 1), and the resulting PCR product was used for the second PCR reaction along with either primer 9 (3′R BclI3a) or 10 (3′R PflM13adel). Then, the final PCR products were digested with BamHI and either BclI or PflMI restriction enzymes, followed by cloning into (respectively) BamHI/BclI or BamHI/PflMI-digested plasmid SF-23. Both approaches generated the final plasmid pB3-33del, which was linearized with EcoRI and used to synthesize B3-33del RNA3.
To generate sg3a-384 RNA, primers 2 and 11 (Table 1) were used for PCR amplification from pB3TP7, and the resulting cDNA product was purified and used as a template for in vitro transcription.
Plasmid pJS-21 was used to introduce (via a silent mutation) a HindIII restriction site marker at nt position 780 with primers 1 and 4 (Table 1). The PCR product was purified with a Qiaquick® PCR Purification Kit (Qiagen) and digested with BamHI (121 nts) and PflMI (816 nts) enzymes followed by religation into BamHI/PflMI-cut pJS-21. The resulting pJS-22 was linearized either with BglII to transcribe JS(H)-21 RNA carrying the polyA tail and four extra bases (GAUC); with PstI to transcribe JS(H)-0 RNA missing the polyA tail; or with PflMI to transcribe the 5′-780 bp RNA (see Fig. 5).
Plasmid pJS-22 was used as a template to design three 5′-nested RNAs. The 5′-580 DNA (with BamHI and AvaI markers) was generated with primers 1 and 12; the 5′-380 DNA (with BamHI and SmaI markers) with primers 1 and 13; and the 5′-150 DNA (with BamHI marker) with primers 1 and 14 (Table 1). Three 3′-nested constructs were also obtained from pJS-22: 3′-860 DNA (with SmaI and PstI markers) was made by amplification with primers 15 and 16; 3′-680 DNA (with AvaI and PstI markers) with primers 15 and 17; and 3′-440 (with HindIII and PstI markers) with primers 15 and 18.
All the above PCR-amplified cDNA templates were transcribed in vitro with the MEGAscript T7 System (Ambion), and the resulting capped RNAs were used in subsequent experiments.
In vitro RdRp and TNT assays: analysis of replicase products
BMV replicase enzyme was extracted as described by Dzianott et al. (2001), except that the column buffer additionally contained 0.05% of the nonionic detergent dodecyl-β-d-maltoside (12-M), which stabilizes the enzyme and increases its copying activity (Bujarski et al., 1982). The in vitro RdRp priming reaction included 1 μg of RW (−) RNA3 template, the 32P-labeled sg3a-384 RNA primer, 0.05% of 12-M, and the remaining components, as described in Wierzchoslawski and Bujarski (2006). The radioactive RNA products were separated in 1.0% to 1.2% denaturing agarose gels followed by blotting to a nylon membrane. To detect double-stranded (ds) RNA, the RNA products were treated with S1 nuclease (Promega Corp., cat # M5761) as recommended by the manufacturer, followed by electrophoresis in denaturing agarose gels.
The in vitro RdRp copying reaction included 1 μg of RNA template, 15 μl of BMV replicase, 0.05% of 12-M, 32P-rCTP, and the remaining components as described (Wierzchoslawski and Bujarski, 2006). The control TNT activity assays were performed exactly as for the RdRp activity assays except that a single nucleotide triphosphate (GTP) was omitted in the reaction mixture.
For cDNA cloning of the RdRp products, the full-length RNA3 bands were cut out of the gel and the RNA was eluted followed by ethanol precipitation. The RNA sequences were amplified by RT-PCR with primers 5 and 6 (Table 1) and the PCR products were cloned using the pGEM-T Easy Vector System from Promega, followed by sequencing.
Protoplast assays
Barley mesophyll protoplasts were isolated from five-day-old barley seedlings as described by Rao (2007). On average, one million of protoplast cells were inoculated with 1 μg of each desired combination of the in vitro-capped transcripts by using a PEG-mediated transfection protocol (Rao, 2007). The transfected protoplasts were thoroughly washed three times with mannitol to remove the unincorporated RNA inoculum and incubated for 48 h at 27 °C in the dark with gentle agitation (25 rpm). Protoplasts were then re-suspended in RNA extraction buffer (100 mM glycine, 100 mM NaCl, 10 mM EDTA, and 1% SDS, pH 9.2) (Loesch-Fries and Hall, 1980), followed by phenol/chloroform extraction and RNA precipitation with ethanol in the presence of sodium acetate (Weiland and Dreher, 1989). Total RNA was separated in denaturing 1.2% agarose gels (Sambrook and Russell, 2001). The RNA3-size material was cut out from the gel and purified using spin columns (Ambion, Cat# AM10065) followed by chloroform extraction and ethanol precipitation. The final RNA preparation was subjected to RT-PCR with primers 5 and 6 (Table 1) and the products were purified with the Qiaquick® PCR Purification Kit (Qiagen) and cloned into the pGEM-T Easy Vector System (Promega). The resulting clones were analyzed by restriction digestion. All protoplast experiments were repeated two to three times to validate the accuracy of the results.
Whole-plant inoculations and analysis of progeny RNA
C. quinoa seedlings were inoculated with equal amounts (2.5 μg per leaf, concentration: 50 μg of RNA per ml) of transcribed BMV RNAs 1, 2 and either wt or mutant RNA3, as well as with different amounts of sgRNA3a, depending on the experiment. The inoculated plants were maintained in a greenhouse for ten days, the local lesion tissues were cut off and the virus was extracted from the combined lesion tissue followed by BMV RNA isolation and characterization of RNA3 recombinants (Nagy and Bujarski, 1992, Bruyere et al., 2000, Dzianott et al., 2001). The presence of BMV RNA was detected by northern blotting (Kroner et al., 1989). Blots were hybridized with a probe specific to either the 3′ 200 nts of the positive-strand RNA or to the 3′ 200 nts of the negative-strand RNA in order to visualize the strands for all BMV RNA segments. In contrast, the presence of sgRNA3a (+) strands was detected by northern blotting with a radioactive (32P-labeled) RNA probe complementary to the positive-strand RNA3 sequence between nts 962 and 1111 (described in Wierzchoslawski et al., 2006). Autoradiograms were developed at appropriate exposure times, usually five, two, and two days for the minus-strand, plus-strand, and sgRNA3a probes, respectively. The bands containing the RNA of interest (sgRNA3a or RNA3) were cut out from the ethidium bromide-stained gel, eluted, and amplified by RT-PCR, followed by cloning and sequencing.
Acknowledgments
We thank K. Andrew White for useful comments and suggestions on the manuscript and Barbara Ball for assistance with figures. JJB was supported by grants from the National Science Foundation (MCB-0920617), the National Institutes of Health (G1A62203), and the Plant Molecular Biology Center at Northern Illinois University.
References
- Adair T.L., Kearney C.M. Recombination between a 3-kilobase Tobacco mosaic virus transgene and a homologous viral construct in the restoration of viral and nonviral genes. Arch. Virol. 2000;145:1867–1883. doi: 10.1007/s007050070062. [DOI] [PubMed] [Google Scholar]
- Agol V.I. Molecular mechanisms of Poliovirus variation and evolution. Curr. Top. Microbiol. Immunol. 2006;299:211–259. doi: 10.1007/3-540-26397-7_8. [DOI] [PubMed] [Google Scholar]
- Alejska M., Kurzyńska-Kokorniak A., Broda M., Kierzek R., Figlerowicz M. How RNA viruses exchange their genetic material. Acta Biochim. Pol. 2001;48:391–407. [PubMed] [Google Scholar]
- Allison R., Janda M., Ahlquist P. Sequence of Cowpea chlorotic mottle virus RNAs 2 and 3 and evidence of a recombination event during bromoviral evolution. Virology. 1989;172:321–330. doi: 10.1016/0042-6822(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Allison R., Thompson C., Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl Acad. Sci. USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.T. RNA turnover: unexpected consequences of being tailed. Curr. Biol. 2005;15:R635–R638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Annamalai P., Rao A.L.N. Packaging of Brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 2006;80:10096–10108. doi: 10.1128/JVI.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I., Landick R. Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 2009;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., O'Donnell B.J., Flanegan J.B. 5′ Cloverleaf in Poliovirus RNA is a cis-acting replication element required for negative strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher P., Orlich M., Thiel H.J. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 2001;75:6256–6264. doi: 10.1128/JVI.75.14.6256-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick S.S., Walters R.W., Gromeier M. The Hepatitis C virus 30-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruyere A., Wantroba M., Flasinski S., Dzianott A., Bujarski J.J. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 2000;74:4214–4219. doi: 10.1128/jvi.74.9.4214-4219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J.J., Figlerowicz M. RNA recombination in Brome mosaic virus, a model plus strand RNA virus. Acta Biochim. Pol. 1998;45:847–868. [PubMed] [Google Scholar]
- Bujarski J.J., Kaesberg P. Genetic recombination in a multipartite plant virus. Nature. 1986;321:528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J.J., Hardy S.F., Miller W.A., Hall T.C. Use of dodecyl-β-d-maltoside in the purification and stabilization of RNA polymerase from Brome mosaic virus-infected barley. Virology. 1982;119:465–473. doi: 10.1016/0042-6822(82)90105-2. [DOI] [PubMed] [Google Scholar]
- Che X., Mawassi M., Yang G., Satyanarayana T., Gowda S., Dawson W.O., Bar-Joseph M. 5′-Coterminal subgenomic RNAs in Citrus tristeza virus-infected cells. Virology. 2001;283:374–381. doi: 10.1006/viro.2001.0880. [DOI] [PubMed] [Google Scholar]
- Cheng J.H., Peng C.W., Hsu Y.H., Tsai C.H. The synthesis of minus-strand RNA of Bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. J. Virol. 2002;76:6114–6120. doi: 10.1128/JVI.76.12.6114-6120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.G., Rao A.L.N. Packaging of Brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.K., Hema M., Gopinath K., Santos J., Kao C.C. Replicase-binding sites on plus- and minus-strand Brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 2004;78:13420–13429. doi: 10.1128/JVI.78.24.13420-13429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J., Chong A., Zhang G., Veitia R.A. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Conrad N.K., Shu M.D., Uyhazi K.E., Steitz J.A. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl Acad. Sci. USA. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T., Sankar S., Porter A.G. Binding of Encephalomyocarditis virus RNA polymerase to the 3′-noncoding region of the viral RNA is specific and requires the 3′-poly(A) tail. J. Biol. Chem. 1993;268:26093–26098. [PubMed] [Google Scholar]
- de Wispelaere M., Gaubert S., Trouilloud S., Belin C., Tepfer M. A map of the diversity of RNA3 recombinants appearing in plants infected with Cucumber mosaic virus and Tomato aspermy virus. Virology. 2005;331:117–127. doi: 10.1016/j.virol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Dzianott A., Rauffer-Bruyere N., Bujarski J.J. Studies on functional interaction between Brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology. 2001;289:137–149. doi: 10.1006/viro.2001.1118. [DOI] [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A. Eighth Report of the International Committee on the Taxonomy of Viruses. Elsevier; 2005. Virus Taxonomy. Classification and Nomenclature of Viruses. [Google Scholar]
- Figlerowicz M., Nagy P., Tang N., Kao C.C., Bujarski J.J. Mutations in the N terminus of the Brome mosaic virus polymerase affect genetic RNA–RNA recombination. J. Virol. 1998;72:9980–9989. doi: 10.1128/jvi.72.11.9192-9200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K., Hagihara F., Kaido M., Mise K., Okuno T. Complete nucleotide sequence of Spring beauty latent virus, a bromovirus infectious to Arabidopsis thaliana. Arch. Virol. 2003;148:165–175. doi: 10.1007/s00705-002-0909-0. [DOI] [PubMed] [Google Scholar]
- Galli A., Lai A., Corvasce S., Saladini F., Riva C., Dehò L., Caramma I., Franzetti M., Romano L., Galli M., Zazzi M., Balotta C. Recombination analysis and structure prediction show correlation between breakpoint clusters and RNA hairpins in the pol gene of human immunodeficiency virus type 1 unique recombinant forms. J. Gen. Virol. 2003;89:3119–3125. doi: 10.1099/vir.0.2008/003418-0. [DOI] [PubMed] [Google Scholar]
- Gallie D.R., Kobayashi M. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene. 1994;142:159–165. doi: 10.1016/0378-1119(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Gmyl A.P., Korshenko S.A., Belousov E.V., Khitrina E.V., Agol V.I. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces. RNA. 2003;9:1221–1231. doi: 10.1261/rna.5111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Ayllón M.A., Satyanarayana T., Bar-Joseph M., Dawson W.O. Transcription strategy in a Closterovirus: a novel 5′-proximal controller element of Citrus Tristeza Virus produces 5′- and 3′-terminal subgenomic RNAs and differs from 3′ open reading frame controller elements. J. Virol. 2003;77:340–352. doi: 10.1128/JVI.77.1.340-352.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Tatineni S., Folimonova S.Y., Hilf M.E., Dawson W.O. Accumulation of a 5′ proximal subgenomic RNA of Citrus tristeza virus is correlated with encapsidation by the minor coat protein. Virology. 2009;389:122–131. doi: 10.1016/j.virol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Grosset C., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- Guilford P.J., Beck D.L., Forster R.L. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of white clover mosaic potexvirus. Virology. 1991;182:61–67. doi: 10.1016/0042-6822(91)90648-u. [DOI] [PubMed] [Google Scholar]
- Guillot S., Caro V., Cuervo N., Korotkova E., Combiescu M., Persu A., Aubert-Combiescu A., Delpeyroux F., Crainic R. Natural genetic exchanges between vaccine and wild Poliovirus strains in humans. J. Virol. 2000;74:8434–8443. doi: 10.1128/jvi.74.18.8434-8443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema M., Kao C.C. Template sequence near the initiation nucleotide can modulate Brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 2004;78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Hajjou M., Hu J.Y., Raju R. RNA–RNA recombination in sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 1997;71:2693–2704. doi: 10.1128/jvi.71.4.2693-2704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.S., Rhodes T., Dang Q., Pathak V. Retroviral recombination: review of genetic analyses. Front. Biosci. 2003;8:D143–D155. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- Iwahashi F., Fujisaki K., Kaido M., Okuno T., Mise K. Synthesis of infectious in vitro transcripts from Cassia yellow blotch bromovirus cDNA clones and a reassortment analysis with other bromoviruses in protoplasts. Arch. Virol. 2005;150:1301–1314. doi: 10.1007/s00705-005-0500-6. [DOI] [PubMed] [Google Scholar]
- Janda M., French R., Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned Brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- Jonassen C.M., Jonassen T.Ø., Grinde B. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- Karran R.A., Hudak K.A. Depurination within the intergenic region of Brome mosaic virus RNA3 inhibits viral replication in vitro and in vivo. Nucl. Acids Res. 2008;36:7230–7239. doi: 10.1093/nar/gkn896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Yumak H., Gallie D.R., Goss D.J. Effects of poly(A)-binding protein on the interactions of translation initiation factor eIF4F and eIF4F.4B with internal ribosome entry site (IRES) of Tobacco etch virus RNA. Biochim. Biophys. Acta. 2008;779:622–627. doi: 10.1016/j.bbagrm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in Poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G., Mohan B.R., Miller W.A. Primary and secondary structural elements required for synthesis of Barley yellow dwarf virus subgenomic RNA1. J. Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner P., Richards D., Traynor P., Ahlquist P. Defined mutations in a small region of the Brome mosaic virus 2 gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C.M., Baric R.S., Makino S., Keck J.G., Egbert J., Leibowitz J.L., Stohlman S.A. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 1985;56:449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S., Viel C., Beauchemin C., Daigneault N., Fortin M.G., Laliberte J.F. Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 2004;85:1055–1063. doi: 10.1099/vir.0.19706-0. [DOI] [PubMed] [Google Scholar]
- Loesch-Fries L.S., Hall T.C. Synthesis, accumulation and encapsidation of individual Brome mosaic virus RNA components in barley protoplasts. J. Gen. Virol. 1980;47:323–332. [Google Scholar]
- Mayo M.A., Jolly C.A. The 5′-terminal sequence of potato leafroll virus RNA: evidence of recombination between virus and host RNA. J. Gen. Virol. 1991;72:2591–2595. doi: 10.1099/0022-1317-72-10-2591. [DOI] [PubMed] [Google Scholar]
- Melchers W.J.G., Hoenderop J.G.J., Bruins Slot H.J., Pleij C.W.A., Pilipenko E.V., Agol V.I., Galama J.M.D. Kissing of the two predominant hairpin loops in the Coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative strand RNA synthesis. J. Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Temme C., Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Mikkelsen J.G., Lund A.H., Duch M., Pedersen F.S. Mutations of the kissing–loop dimerization sequence influence the site specificity of Murine leukemia virus recombination in vivo. J. Virol. 2000;74:600–610. doi: 10.1128/jvi.74.2.600-610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Molenkamp R., Greve S., Spaan W.J., Snijder E.J. Efficient homologous RNA recombination and requirement for an open reading frame during replication of Equine arteritis virus defective interfering RNAs. J. Virol. 2000;74:9062–9070. doi: 10.1128/jvi.74.19.9062-9070.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Genetic recombination in Brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 1992;66:6824–6828. doi: 10.1128/jvi.66.11.6824-6828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Efficient system of homologous RNA recombination in Brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 1994;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Homologous RNA recombination in Brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 1996;70:217–225. doi: 10.1128/jvi.70.1.415-426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Engineering of homologous recombination hot-spots with AU-rich sequences in Brome mosaic virus. J. Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Silencing homologous RNA recombination hot spots with GC-rich sequences in Brome mosaic virus. J. Virol. 1998;72:1122–1130. doi: 10.1128/jvi.72.2.1122-1130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Dzianott A., Ahlquist P., Bujarski J.J. Effect of mutations in the helicase-like domain of 1a protein on RNA–RNA recombination in Brome mosaic virus. J. Virol. 1994;69:2547–2556. doi: 10.1128/jvi.69.4.2547-2556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Ogiela C., Bujarski J.J. Mapping sequences active in homologous recombination in Brome mosaic virus: prediction of recombination hot spots. Virology. 1999;254:92–104. doi: 10.1006/viro.1998.9545. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J., Simon A.E. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 1999;18:5653–5665. doi: 10.1093/emboj/18.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A.O., Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- Panaviene Z., Panavas T., Serva S., Nagy P.D. Purification of the Cucumber necrosis virus replicase from yeast cells: role of co-expressed viral RNA in stimulation of replicase activity. J. Virol. 2004;78:8254–8263. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales C., Carrasco L., Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;2:89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Gutshall L., Kim M.J., Sarisky R.T., Kao C.C. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 2002;76:12526–12536. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Gutshall L., Kim M.J., Sarisky R.T., Kao C.C. Mechanism of de novo initiation by the Hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 2002;76:12513–12525. doi: 10.1128/JVI.76.24.12513-12525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Sarisky R.T., Gutshall L., Thompson M., Kao C.C. De novo initiation Pocket mutations have multiple effects on Hepatitis C virus RNA-dependent RNA polymerase activities. J. Virol. 2004;78:12207–12217. doi: 10.1128/JVI.78.22.12207-12217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.L.N. Preparation and inoculation of mesophyll protoplasts from monocotyledonous and dicotyledenous hosts. Curr. Protoc. Microbiol. 2007 doi: 10.1002/9780471729259.mc16d02s4. 16D.2.1–16D.2.8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Wells V., Plotch S.J., DeStefano J.J. Primer-dependent synthesis by Poliovirus RNA-dependent RNA polymerase (3DPol) Nucleic Acids Res. 2001;29:2715–2724. doi: 10.1093/nar/29.13.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohayem J., Münch J., Rethwilm A. Evidence of recombination in the norovirus capsid gene. J. Virol. 2005;79:4977–4990. doi: 10.1128/JVI.79.8.4977-4990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sano Y., van den Vlugt R., de Haan P., Takahashii A., Kawakami M., Goldbach R., Kojima M. On the variability of the 3′ terminal sequence of the Turnip Mosaic virus genome. Arch. Virol. 1992;126:231–238. doi: 10.1007/BF01309697. [DOI] [PubMed] [Google Scholar]
- Shapka N., Nagy P. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J. Virol. 2004;78:2288–2300. doi: 10.1128/JVI.78.5.2288-2300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Hibi T., Masuta Ch. RNA recombination between cucumoviruses: possible role of predicted stem–loop structures and an internal subgenomic promoter-like motif. Virology. 2003;306(1):77–86. doi: 10.1016/s0042-6822(02)00050-8. [DOI] [PubMed] [Google Scholar]
- Svitkin Y., Costa-Mattioli M., Herdy B., Perreault S., Sonenberg N. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP) RNA. 2008;13:2330–2340. doi: 10.1261/rna.606407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztuba-Solińska J., Bujarski J.J. Insights to Bromoviridae single-cell reproduction cycle: lessons from protoplast systems. J. Virol. 2008;82:10330–10340. doi: 10.1128/JVI.00746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Ch.-H., Cheng Ch.-P., Peng Ch.-W., Lin B.-Y., Lin N.-S., Hsu Y.-H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of Bamboo mosaic potexvirus RNA. J. Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz A., Alejska M., Formanowicz P., Blazewicz J., Figlerowicz M., Bujarski J.J. Homologous crossovers among molecules of Brome mosaic bromovirus RNA1 or RNA2 segments in vivo. J. Virol. 2005;79:5732–5742. doi: 10.1128/JVI.79.9.5732-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ullah Z., Grumet R. Interaction between Zucchini yellow mosaic potyvirus RNA-dependent RNA polymerase and host poly-(A) binding protein. Virology. 2000;275:433–443. doi: 10.1006/viro.2000.0509. [DOI] [PubMed] [Google Scholar]
- Weiland J.J., Dreher T.W. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 1989;17:4675–4687. doi: 10.1093/nar/17.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welnowska E., Castello A., Moral P., Carrasco L. Translation of mRNAs from Vesicular stomatitis virus and Vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. J. Mol. Biol. 2009;4:506–521. doi: 10.1016/j.jmb.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Wierzchoslawski R., Bujarski J.J. An efficient in vitro system of homologous recombination in Brome mosaic bromovirus. J. Virol. 2006;80:6182–6187. doi: 10.1128/JVI.02447-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchoslawski R., Dzianott A., Kunimalayan S., Bujarski J.J. A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus. J. Virol. 2003;77:6769–6776. doi: 10.1128/JVI.77.12.6769-6776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchoslawski R., Dzianott A., Bujarski J.J. Dissecting the requirement for subgenomic promoter sequences by RNA recombination of Brome mosaic virus in vivo: evidence for functional separation of transcription and recombination. J. Virol. 2004;78:8552–8864. doi: 10.1128/JVI.78.16.8552-8564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchoslawski R., Urbanowicz A., Dzianott A., Figlerowicz M., Bujarski J.J. Characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of Brome mosaic bromovirus. J. Virol. 2006;80:12357–12366. doi: 10.1128/JVI.01207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Letteney E., Kim Ch.-H., Kao C.C. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009;15:615–626. doi: 10.1261/rna.1375509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Vaughan R.C., Yarbrough I., Dharmaiah S., Kao C.C. RNA binding by the Brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J. Mol. Biol. 2009;391:314–326. doi: 10.1016/j.jmb.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma Y. Evidence for retroviral intramolecular recombinations. J. Virol. 2001;75:6348–6358. doi: 10.1128/JVI.75.14.6348-6358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Morace G., Gauss-Muller V., Kusov Y. Poly(A) binding protein, C-terminally truncated by the Hepatitis A virus proteinase 3C, inhibits viral translation. Nucl. Acids Res. 2007;35:5975–5984. doi: 10.1093/nar/gkm645. [DOI] [PMC free article] [PubMed] [Google Scholar]