Abstract
Many (+)-strand RNA viruses use subgenomic (SG) RNAs as messengers for protein expression, or to regulate their viral life cycle. Three different mechanisms have been described for the synthesis of SG RNAs. The first mechanism involves internal initiation on a (−)-strand RNA template and requires an internal SGP promoter. The second mechanism makes a prematurely terminated (−)-strand RNA which is used as template to make the SG RNA. The third mechanism uses discontinuous RNA synthesis while making the (−)-strand RNA templates. Most SG RNAs are translated into structural proteins or proteins related to pathogenesis: however other SG RNAs regulate the transition between translation and replication, function as riboregulators of replication or translation, or support RNA–RNA recombination. In this review we discuss these functions of SG RNAs and how they influence viral replication, translation and recombination.
Keywords: Subgenomic RNA (SG RNA), Internal initiation, Premature termination, Discontinuous transcription, Riboregulators, Copy-choice RNA recombination
Introduction: definition and occurrence of SG RNAs
Synthesis of subgenomic (SG) messenger RNAs (mRNAs) by (+)-strand RNA viruses allows the differential expression of specific viral genes, both quantitatively and temporally. SG RNAs as considered in this review, have the following properties: (i) they are made in infected cells but do not interfere with the normal course of viral replication; (ii) the SG RNA sequences are shorter than their cognate genomic RNAs; (iii) their sequences are usually co-terminal with the 3′ genomic sequence but sometimes are co-terminal with the 5′ sequences. Yet other viruses make SG RNAs which contain a 5′ co-terminal leader joined to a 3′ co-terminal sequence; (iv) typically, whether a messenger SG RNA contains only one ORF, or multiple ORFs, with some rare exceptions (Dorokhov et al., 2006), only the 5′ ORF is translated (Pasternak et al., 2006, Yang et al., 2009). Although most SG RNAs function as messengers and are translated, other SG RNAs, generally those with 5′ co-terminal sequences, have other functions.
The production of SG RNAs was initially reported in studies of Brome mosaic virus (BMV) and was followed by the discovery of SG RNA in Tobacco mosaic virus (TMV)-infected leaves (Miller and Koev, 2000) and later in Alpha-, Carmo- and Sobemo- families (Strauss and Strauss, 1994, Rico and Hernandez, 2009, McGavin and MacFarlane, 2009). Indeed, among (+) strand RNA viruses of plants, only viruses of the Potyviridae and Comoviridae as well as the Sequiviruses of the Sequiviridae family do not use this strategy (Zaccomer et al., 1995).
Our goal in this review is to describe the major mechanisms by which SG RNAs are generated and to discuss their roles in the life cycle of (+) strand RNA viruses. Since the last reviews discussing that subject (Koev and Miller, 2000, White, 2002, Miller and White, 2006), these RNAs have been shown to function not only as mRNAs, but also as riboregulators of replication and transcription; as well, SG RNAs may participate in genome rearrangements that help maintain genomic integrity and possibly the acquisition of non-self sequences.
Although replicon RNAs made in the laboratory to study the replication of Hepatitis C virus (HCV) and other Flaviviruses have also been referred to as subgenomic RNAs, since they are not seen in a normal infection, they will not be considered in this review.
Mechanisms of SG RNA synthesis and regulation
Internal initiation on the (–)-strand RNA template
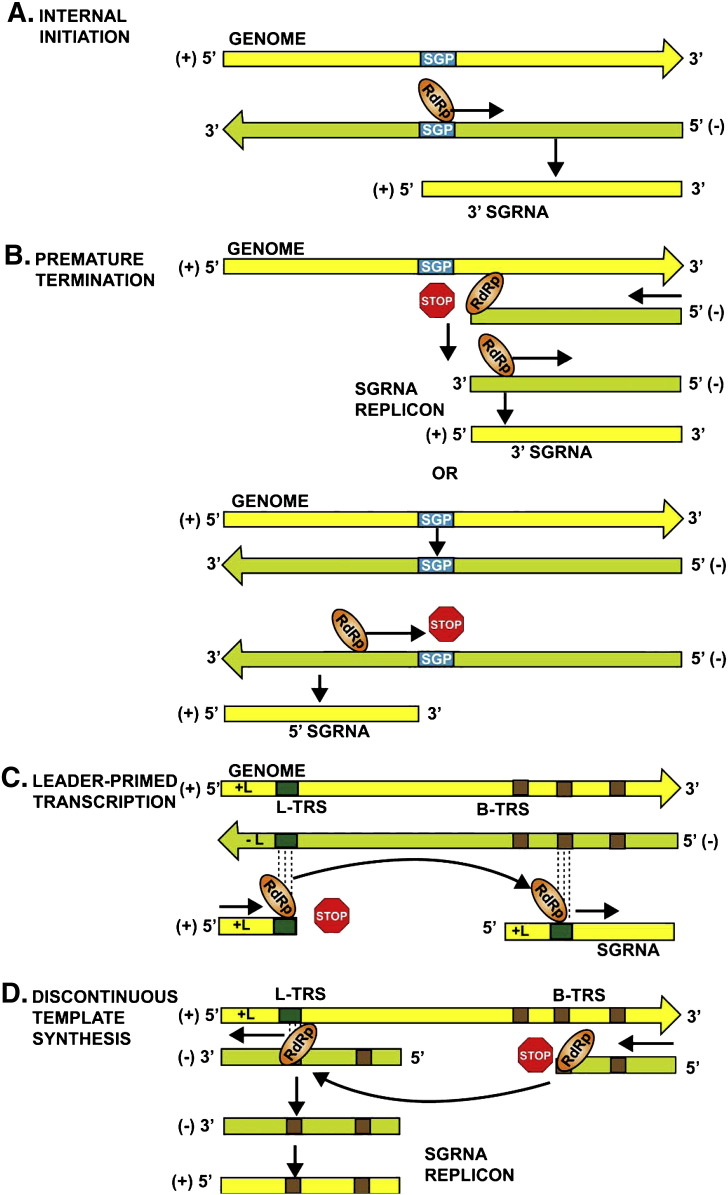
Following the synthesis of the viral RNA-dependent RNA polymerase (RDRP), the (+) strand RNA is copied into a genome-length (−) strand which then serves as a template for the genomic (G) and the SG (+) strand RNAs (Fig. 1A). Thus, the (−) strand RNA contains at least two different promoters, one for the synthesis of G RNA at or near the 3′ end, and one or more internal or subgenomic promoters (SGPs). To synthesize a SG RNA, the viral RDRP recognizes and binds to the SGP and initiates transcription (Koev and Miller, 2000). This mechanism was first found with BMV, a tripartite plant virus. The (−) strand of dicistronic BMV RNA 3 serves as template for transcription of the 3′ co-terminal SG RNA4 due to de novo internal initiation from the SGP. The BMV SGP has been mapped to the sequence from nt − 95 to + 16 (Fig. 2A). Its modular composition includes an AU-rich enhancing region, a poly (U) tract, a core region, the + 1 C transcription initiation site, and a downstream sequence (Wierzchoslawski et al., 2004). The enhancing region (nts − 95 to − 20) greatly increases the amount of SG RNA 4. The core region (nts − 19 to − 1) contains a stem-loop (SL) structure which is responsible for RDRP binding. Adkins and Kao (1998) have suggested an induced fit mechanism whereby the RDRP recognizes key nucleotides, after which nucleotide base-pairing occurs and the SL structure forms. Another model proposes that the SL structure binds to the RDRP complex, after which the key nucleotides stabilize the hairpin (Haasnoot et al., 2000, Haasnoot et al., 2002). Regardless of the mechanism, internal initiation appears to be a multi-step process, which involves both host and viral proteins (Hertz and Huang, 1995a, Hertz and Huang, 1995b, Adkins and Kao, 1998, Haasnoot et al., 2002, Sivakumaran et al., 2004). Other (+)-strand RNA viruses use SGPs with a similar organization (Miller and Koev, 2000, Haasnoot et al., 2000, Morales et al., 2004, Olsthoorn et al., 2004).
Fig. 1.
Schematic representation of different mechanisms for subgenomic RNA synthesis. (A) Internal initiation model (Miller and Koev, 2000), (B) Premature termination model showing termination either during (−) and (+)-strand synthesis (White, 2002), (C) Leader-primed transcription model, (D) Discontinuous template synthesis (Pasternak et al., 2006). Genomic and SG RNA (+)-strands are depicted as horizontal yellow boxes, the (−)-strands are depicted as green boxes, the ovals represent RdRp enzymes capable of starting/stopping at the internal initiation SGP promoters that are depicted as blue SGP boxes. The leader TRS (L-TRS) and the body TRS (B-TRS) are represented by dark green and brown boxes, respectively. Refer to the text for details.
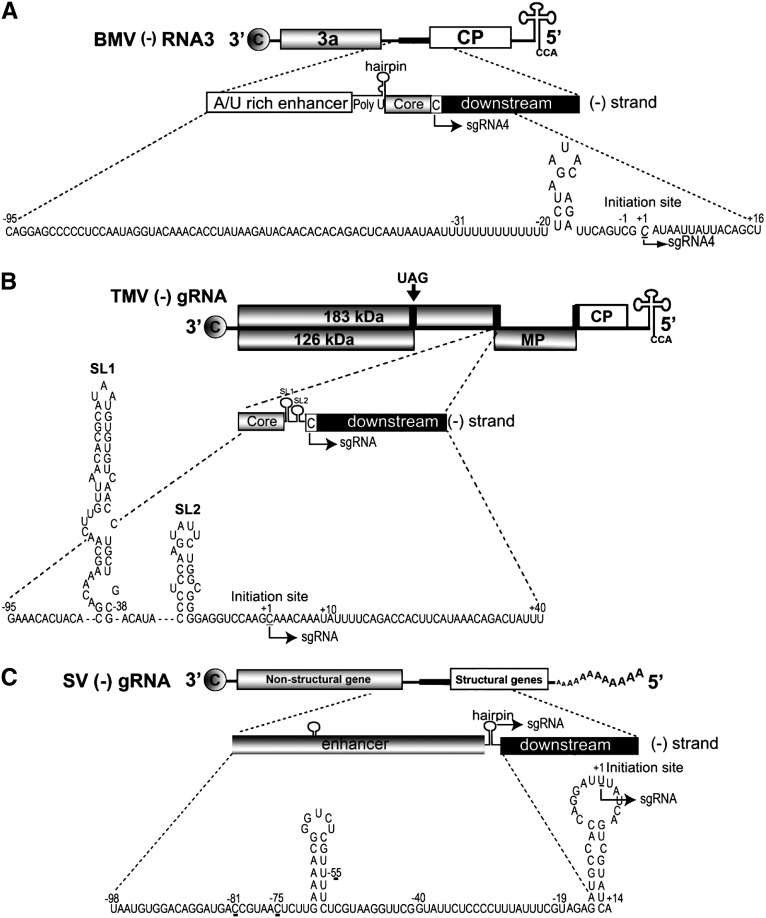
Fig. 2.
Schematic organization of subgenomic (SGP) promoters in RNA viruses. (A) The SGP domain is shown as a solid, black box located between the 3a and CP ORFs on BMV (−) RNA3. The arrow indicates the initiation site and the direction of subgenomic RNA4 synthesis. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 95 to + 16). The SGP subdomains are indicated as follows: the enhancer including the poly U tract (nt − 95 to − 20), the core region (nt − 19 to − 1) including the hairpin, the initiation + 1 cytidilate, and the downstream portion. (B) TMV SGP promoters are indicated by the black rectangles: the first promoter is located between 183-kDa RDRP protein ORF and MP ORFs, and the second one between MP and CP ORFs. The arrow above the G RNA indicates the position of the amber read-through codon, the putative initiation site for I1 SG RNA expressing 54-kDa protein of unknown function. The arrow below (−) G RNA indicates the initiation site for one of the SG RNAs. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 95 to + 40). The promoter core (− 35 to + 10),the two hairpin structures, SL1 and SL2, are indicated,as well as the initiation + 1 cytidilate, and the downstream portion (Grdzelishvili et al., 2000). (C) The SGP promoter of SV (−) strand RNA is shown in the top line as a solid, black box. The arrow on (−) G RNA diagram indicates the initiation site and the direction of SG RNA synthesis. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 98 to + 14). The sequence between − 19 to + 5 represents the minimal sequence with promoter activity. The sequence from − 40 to + 14 was shown to enhance the promoter activity. Mutations at the marked nucleotide positions: − 81, − 75, − 55, down regulate the SGP promoter activity.
TMV has a single-stranded non-segmented RNA genome and represents another example of a virus that synthesizes SG RNAs via internal initiation. Three of these SG RNAs are co-terminal with the 3′ terminus of the genomic RNA; the smallest SG RNA has only one ORF, and encodes CP. The middle-sized SG RNA has two ORFs (for MP and CP), but only the 5′ ORF, that for MP, is expressed. The third and largest SG RNA, referred as the I1, has been detected in TMV-infected tobacco tissue. It contains in addition to ORFs for MP and CP, a 5′ ORF for a 54-kDa protein with a sequence identical to that of the read-through region of the 183-kDa replicase. Interestingly, RNA structures that are able to regulate the activity of TMV promoters can be far removed from the initiation site. Culver et al. (1993) found that the location of the TMV SGP in relation to the 3′ UTR is a crucial factor in enhancing expression. Later, Szecsi et al., 1999, Shivprasad et al., 1999 associated the positive effect of the 3′ UTR with the three pseudoknots contained therein; these were found to redistribute RDRP activity localizing most of it to the nearest SGP promoter.
Both RNA sequence and secondary structures are generally critical for the activity of an SGP (Koev et al., 1999, Haasnoot et al., 2000). In the case of the TMV promoter for the middle-sized SG RNA (Fig. 2B), computer analysis predicted two stem-loop structures (SL1 and SL2) upstream of the transcription start site. The SL1 secondary structure, rather than its sequence, was critical for promoter activity. On the other hand, removal of most of the SL2 region increased accumulation of the SG RNA four-fold (Grdzelishvili et al., 2000). Also, the three SGP promoters of Barley yellow dwarf virus (BYDV) fold into different size stem-loops downstream of their respective initiation sites (Koev and Miller, 2000). In the case of promoters of Barley stripe mosaic virus (BSMV) SG RNAs β1, β2, and γ, the conservation of both the sequences and the secondary structures appear to be important for their activity (Johnson et al., 2003). Here the substantial differences between SG RNAs β1, β2, and γ promoter sequences were postulated to explain competition for the viral RDRP, coordination of the temporal expression and abundance of the proteins, and constitutive expression of the γ b protein.
Viruses producing multiple SG RNAs often contain homologous sequences within their SGP promoters (Koev and Miller, 2000). Also, certain elements in genomic promoters for (−) strand synthesis can share similarities with elements in the internal promoters; see for example, a stem-loop C (SLC) in BMV (Haasnoot et al., 2002) and the triloop hairpin (hpE) in Alfalfa mosaic virus (AMV) (Olsthoorn et al., 2004). Initiation from the 3′ end of the (−) sense genomic BMV RNA obeys different rules than does the initiation of SG RNA (Stawicki and Kao, 1999), likely due to additional factors (Diez et al., 2000) that can adjust the properties of the RDRP (Adkins and Kao, 1998, Ranjith-Kumar et al., 2003, Haasnoot et al., 2002, Sivakumaran et al., 2004).
In the case of Sindbis virus (SV) (the prototype of the genus Alphavirus, family Togaviridae), the minimal sequence of the (−) strand RNA needed for the SGP activity extends from nt − 19 to nt + 5 but a sequence extending from − 98 to + 14 increases promoter activity at least 6 folds. Most of this increase could be accounted for by strongly conserved sequences extending from − 40 to − 20, and from + 6 to + 14 (Wielgosz et al., 2001) (Fig. 2C).
Three classes of mutations down regulate the synthesis of the SV SG RNA: (i) promoter mutations, (ii) nsp2 protein mutations, and (iii) a single mutation in nsP3. SV makes four nonstructural proteins; nsP1 is responsible for capping and methylation of the G and SG RNAs; nsP2 has both a protease activity and an RNA helicase activity; the function of nsP3 is not known, and nsP4 is the viral RDRP. Promoter mutations which affect the synthesis of SV SG RNA have been described by Hertz and Huang, 1995a, Hertz and Huang, 1995b, Lin et al., 2002. These effects are often cell-dependent, suggesting a role for cellular factors in the synthesis of SG RNA.
Mutations in nsP2 of SV and SFV, which mapped to its protease domain, decreased protease activity and thus slowed cleavage of the nonstructural polyproteins, a step necessary for the efficient synthesis of SG RNA. Also, a single insertional mutation in nsP3 lowered the level of the SV SG RNA synthesis, but the mechanism responsible is not known (LaStarza et al., 1994).
By incubating a labeled SGP sequence with an SV RNA transcriptase/replicase complex, distinct protein sites on nsP4 were found to be responsible for the recognition of the SGP and G promoters (Li and Stollar, 2004, Li and Stollar, 2007). It was shown that Arg to Ala changes at positions 331 or 332 knocked out the in vitro synthesis of SG but not G RNA (Li and Stollar, 2007). Conversely, by changing the Arg residue at 545, 546, or 547 to Ala, the synthesis of G RNA but not SG RNA was knocked out (Li et al., 2010).
Viruses in the family Caliciviridae also make SG RNAs by internal initiation on the (–)-strand (Morales et al., 2004). In at least one case, these SG RNAs are packaged into viral particles (Neill, 2002).
Premature termination
In a second mechanism for making a SG mRNA, the RDRP complex, instead of copying the (+) strand RNA genome into a full length (−) strand RNA, terminates “prematurely” at a specific STOP signal, and synthesizes a shortened (−) strand of SG RNA, which then serves as a template for the synthesis of a SG (+) strand RNA (Fig. 1B). These SG (+) strand RNAs, like those made by internal initiation, have sequences which are co-terminal with the 3′ sequence of the viral genome. Premature termination (PT) can, however, also occur during synthesis of (+) strand RNA (White, 2002), thus generating 5′ co-terminal SG RNAs (Fig. 1B) (Wierzchoslawski et al., 2006). Premature termination (PT) supporting formation of SG RNAs is seen with various RNA viruses including Toroviruses (van Vliet et al., 2002), Roniviruses (Cowley et al., 2002), Betanodaviruses (Iwamoto et al., 2005), Dianthoviruses (Guenther et al., 2004), Closteroviruses (Gowda et al., 2001), and Nodaviruses (Lindenbach et al., 2002).
Flock house virus (FHV), the prototype virus of Nodaviridae, uses PT to make a SG RNA. FHV has a bipartite (+) strand RNA genome but both RNAsare packaged into a single viral particle. RNA 1 encodes a replicase protein A, while RNA 2 encodes protein α, the precursor of the capsid proteins. FHV infection also gives rise to a 387 nt SG RNA (RNA 3), the sequence of which is 3′ co-terminal with that of RNA 1. RNA 3 has two overlapping ORFs, B1 and B2. The B1 ORF encodes the C-terminal 102 amino acids of the A protein. No function has been associated with this polypeptide. The B2 protein is a suppressor of RNA interference, both in plants and cultured insect cells. RNAs 1, 2, and 3all have a 5′ cap, but none are polyadenylated.
In studies of FHV RNA replication in yeast (Lindenbach et al., 2002), two cis-acting elements were found in RNA 1 (Fig. 3C): (i) a proximal subgenomic control element (PSCE) extends from nt 2282 to nt 2777 (the start site for the synthesis of SG RNA 3 is at nt 2721), and (ii) a distal subgenomic control element (DSCE) from nt1229 to nt 1239. Base pairing between the nt sequence 1229 to 1239 in the DSCE and both a 6 nt sequence and a 4 nt sequence (almost 200 nt apart) in the PSCE is required for the synthesis of RNA 3. For the most efficient synthesis of RNA 3, the PSCE sequence between nt 2302 and 2777 was needed; however some synthesis was seen with the 5′ limit of the PSCE at nt 2518. Thus, the region from nt 2302 to 2518 serves as an enhancer. Disruption of PSCE/DSCE base pairing increased the amount of RNA 1 made, but then neither (+) nor (−) strand RNA 3 was made. Apparently, the long distance interaction between DSCE and PSCE gives rise to a secondary or tertiary structure that results in premature termination when (−) strand RNA 1 is made. In addition, work by Eckerle et al. (2003), strongly suggests that the (+) and (−) strands of RNA 3 participate in a full-fledged RNA 3 replication even in the absence of RNA 1. Furthermore, as with the synthesis of RNA 1 and RNA 2, the synthesis of (+) strand FHV RNA 3 great exceeds that of (−) strand RNA. Finally, in this system, RNA 3 is required for the synthesis of RNA 2, and once RNA 2 is made, it suppresses the synthesis of RNA 3.
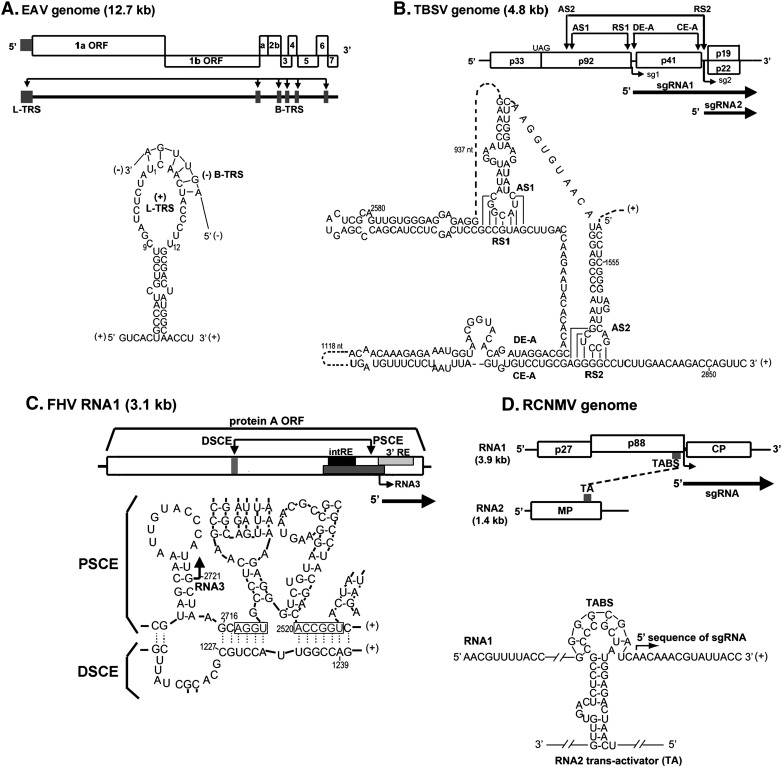
Fig. 3.
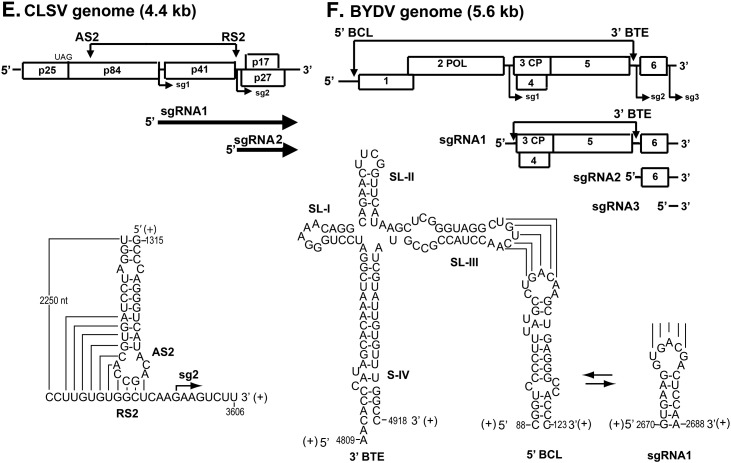
Long distance RNA–RNA interactions that regulate SG RNA synthesis in RNA viruses. (A) Schematic overview of the genome organization and expression of EAV. The gray boxes in the genomic RNA indicate the positions of the leader TRS and (major) body TRSs. Below is shown the proposed base pairing interaction between the sense leader TRS (L-TRS) and the antisense body TRS (B-TRS) in the 3′ end of the nascent minus strand (van Marle et al., 1999); only the loop and the top of the stem of the predicted hairpin structure are presented (B) Linear representation of the TBSV RNA genome showing its coding organization. The relative positions of interacting RNA elements involved in SG RNA transcription are shown above the genome and are indicated by arrows. Initiation sites for SG RNA transcription are labeled sg1 and sg2, and corresponding structures of the two SG RNAs are represented by bold arrows below the genome. Below, the long-distance RNA-RNA interactions that regulate SG RNA transcription in TBSV are shown in detail. Relevant sequences of the TBSV genome are presented with corresponding genomic coordinates. The AS1/RS1 base-pairing interaction is essential for the efficient transcription of SG RNA1 (Choi and White, 2002), while the AS2/RS2 and DE-A/CE-A base-pairing interactions promote SG RNA2 transcription (Choi et al., 2001, Lin and White, 2004); (C) Representation of genomic RNA1 of Flock house virus (FHV). Interaction between the distal subgenomic control element (DSCE) and the proximal subgenomic control element (PSCE) is required for RNA3 synthesis. The internal replication element (intRE) and 3′ replication element (3′ RE) are required for RNA1 replication. Below, potential base pairing of the DSCE to regions proximal to the subgenomic region start site. Gray boxes mark PSCE residues that potentially base pair to the DSCE. Putative helices 1 and 2 are bracketed. Arrow, RNA3 start site at nt 2721 (Lindenbach et al., 2002); (D) Schematic of the RCNMV genome showing the relative positions of the in trans interacting RNA elements: the loop portion of a stem-loop in RNA2 (termed trans-activator or TA) and a complementary sequence in RNA1 (termed TA binding site or TABS) located just upstream from the initiation site for SG mRNA transcription (Guenther et al., 2004); (E) Representation of CLSV genome and the proposed AS2 and RS2 interaction during regulation of SG RNA2 synthesis (Xu and White, 2008); (F) Overview of the BYDV genome and the in cisinteraction between genomic 5′-UTR stem-loop (BCL) and the genomic 3′-BTE, and the in trans interaction between the genomic 3′ BTE and the 5′ UTR of SG RNA1 (Rakotondrafara et al., 2006). Numbers depict the nucleotide positions in the viral genomic RNA. The nucleotides participating in the long-range base-pairing are joined by lines.
Tomato bushy stunt virus (TBSV) uses a similar strategy to make SG RNAs (Fig. 3B) (White and Nagy, 2004, Zhang et al., 2004). While making (−) strand RNA, the RDRP complex terminates at the promoter regions for (+) sense SG RNAs, generating the subviral-length (−) sense 3′ SG RNAs (White, 2002), which then act as templates to amplify the (+) sense 3′ SG RNA. Substitution of the transcription-initiating nucleotides for SG RNA 1 or 2 inhibits the accumulation of these RNAs, but not of the corresponding (−) sense SG RNA templates (White, 2002). Additionally, TBSV RDRP can be considered as the essential catalytic complex regulating viral replication and SG RNA transcription. Wu and White (2007) have shown that both processes can be effectively uncoupled in vivo by deletion of up to five C-terminal RDRP residues. Subsequently, the replicase C-terminus was proposed to function at an early step of the PT transcriptional pathway mediating (i) efficient production of minus-strand templates for SG RNA production; (ii) accurate termination of minus strands, and (iii) efficient utilization of the SGP promoter (Wu and White, 2007). The RDRP termination depends on a multicomponent RNA attenuation signal that includes: (i) long distance RNA–RNA interactions (Fig. 3C), (ii) the spacer segments between the 5′ receptor sequences (RS) and transcriptional initiation sites and (iii) the corresponding downstream sequences predicted to contain transcriptional promoter elements (Lin and White, 2004, Lin et al., 2007). The long-distance base pairings between 5′ receptor sequences (RS1 and RS2) in SGP promoters of RNA1 and 2, and the activator sequences (AS1 and AS2) in the coding region of p92, as well as between the 5′ distal element (DE) of SGP promoter of RNA2 and the complementary downstream core element A (CE), form a physical barrier during (−) strand synthesis (Fig. 3B) (Wang et al., 2008). The first two of these interactions span over 1 and 2 kb, respectively, and the distance between these various structures affects the termination efficiency (Lin et al., 2007).
Similar cis-acting regulations of the 3′ co-terminal SG RNAs production operate in Cucumber leaf spot virus (CLSV) (Fig. 3E) (Xu and White, 2008, Xu and White, 2009), Pothos latent aureusvirus (PoLV) and Potato virus X (PVX, Potexvirus) (Kim and Hemenway, 1999). The PVX SG RNA production is regulated in cis by conserved octanucleotide sequences located upstream of the two PVX SGP promoters, as well as by complementary elements in the genomic 5′ UTR (Kim and Hemenway, 1999). On the other hand, Red clover necrotic mosaic virus (RCNMV) exploits an in trans bimolecular interaction to stall and dislodge the replicase complex during (–)-strand synthesis (Fig. 3D) (Guenther et al., 2004, Tatsuta et al., 2005). RCNMV possesses a bipartite genome with SG RNA being synthesized from RNA1. The synthesis of the SG RNA requires a trans-activation by an RNA hairpin structure located in RNA2 (see below) (Guenther et al., 2004). Specifically, this activation involves the loop portion of an SL structure in RNA2 (trans-activator or TA) which base-pairs with a complementary sequence in RNA1 (TA binding site or TABS) that is located upstream from the SG RNA transcription initiation site (Fig. 3D) (Guenther et al., 2004). It has been suggested that protein factors may bind to and stabilize the bimolecular RNA–RNA contact. Interestingly, the disruption of the RNA–RNA interaction does not affect replication of RCNMV genomic components (Tatsuta et al., 2005). Thus, intermolecular communication secures the switching between replication and transcription, resolving the problem of their mutual interference.
Also, TCV has recently been found to use a premature termination mechanism for making SG RNA2 (bearing CP ORF) (Wu et al., 2010). Analyses revealed that (−) strand SG RNA2 accumulation can be uncoupled from that of its (+) strand counterpart. An extended SL RNA structure positioned 5′ to the initiation site for SG RNA2 was found to mediate PT mechanism by functioning in the (+) strand of the viral genome. As with other viruses that use a PT mechanism, the high degree of identity between the SGP promoter for (+) strand genome and that for SG RNA2 transcription, support the idea that TCV uses a PT mechanism for SG RNA2 transcription.
As noted above, in addition to making the 3′ co-terminal SG RNA, RNA 4, BMV also makes a 5′ co-terminal SG RNA, RNA 3a. In this case, the RDRP likely pauses during (+) strand synthesis at the internal oligo U tract resulting in formation of the 5′ co-terminal 3′ polyadenylated (+) SG RNA3a (Fig. 1A) (Wierzchoslawski et al., 2006). The binding of another RDRP molecule to the near-by SGP promoter core hairpin which initiates the SG RNA4 transcription might pose an additional obstacle to the progression of the RDRP during (+) strand synthesis (Wierzchoslawski et al., 2006, Sztuba-Solińska and Bujarski, 2008).
Other examples of elements in SGPs that act as road blocks for the progressing RDRP have been reported for Citrus leaf blotch virus (CLBV) (Vives et al., 2002), Grapevine vitivirus A (GAV) (Galiakparov et al., 2003), Citrus tatter leaf virus (CTLV) (Tatineni et al., 2009), and Citrus tristeza virus (CTV) (Che et al., 2001). CTLV contains two overlapping 5′ ORFs, the expression of which requires the production of two 5′ (+) strand co-terminal SG RNAs, which result from premature termination. The 3′-ends of both SG RNAs terminate at two SL structures: SL1 and SL2, with the transcription initiation site for the 3′ SG RNAs located in the loop region of SL2 (Tatineni et al., 2009). Likewise, CTV terminates one of its 5′ SG RNAs (referred as low-molecular-weight tristeza LMT1) at the two SL structures located upstream of the internal initiation site for the 3′ SG RNA (Gowda et al., 2003).
Discontinuous transcription
Discontinuous transcription (DT) as a mechanism for the synthesis of 3′ co-terminal SG RNAs is exemplified by viruses in the families Coronaviridae (prototype virus: Murine hepatitis virus or MHV) and Arteriviridae (prototype virus: Equine arteritis virus or EAV) of the order Nidovirales (Pasternak et al., 2001, Sawicki and Sawicki, 2005). A special feature of these (+)- strand RNA viruses is that they encode a nested set of SG RNAs which vary in size, but all of which have a 3′ sequence co-terminal with the 3′ end of the G RNA. These SG RNAs encode the structural proteins and in the case of the Coronaviruses, also several proteins accessory to the replicase proteins. Except for the smallest SG RNAs, all contain multiple open ORFs, but in each case only the 5′ ORF is translated. Thus, although most of these SG RNAs are structurally polycistronic, functionally they are monocistronic.
Also of note, in contrast to the SG RNAs made by internal initiation or by premature termination (see above), those generated by MHV or EAV contain, in addition to coding sequences derived from the 3′ portion of the G RNA, a short sequence that is identical to a 5′ leader sequence of the G RNA. The explanation for how in a SG RNA a 5′ leader is joined to a 3′ terminal sequence remained controversial for some years. Models were proposed that suggested discontinuous synthesis of RNA transcripts, but this then raised the question as to whether the discontinuous synthesis occurred during (+) or (−) strand syntheses (Pasternak et al., 2006). The first model, known as the leader-primed transcription model (Fig. 1C), proposed that transcription would start by copying the 3′ end of the G RNA (−) strand RNA thereby giving rise to a 5′ leader sequence; it followed that the discontinuous step would occur during the synthesis of (+) strand RNA. However, the finding of multiple species of (−) strand RNAs, the sizes of which corresponded to the sizes of the SG RNAs, provided strong support for the proposal by Sawicki and Sawicki (1995) that discontinuous synthesis of SG RNA occurred at the level of (−) strand RNA synthesis (Fig. 1D). These (−) strand RNAs would then serve as the templates for the SG RNAs. This is now the generally accepted model.
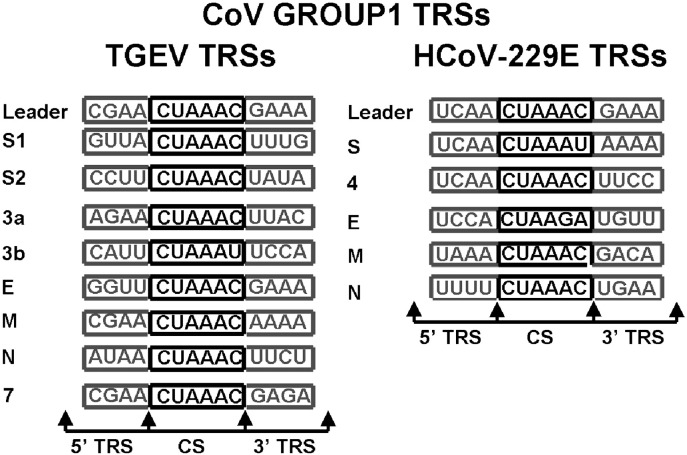
The key to this model is the finding of transcription-regulatory sequences (TRSs) in the viral genome. The TRSs containthree sequence blocks: the core sequence (CS) and the 5′-TRS and 3′-TRS flanking sequences (Fig. 4 ) (Alonso et al., 2002). The CS includes six nucleotides that are highly conserved in each TRS of any given Coronavirus. The most frequently used core sequences of Coronaviruses, i.e. those belonging to Group 1 (hexamer 5′-CUAAAC-3′) and Group 2 (heptamer 5′-UCUAAAC-3′) share homology, while the CS of Coronavirus belonging to Group 3 (e.g. infectious Bronchitis virus coronavirus [IBV]) has the most divergent sequence (5′-CUUAACAA-3′). The 5′ and 3′ flanking regions, which are partially conserved in the different genes of related viruses, influence the activity of the CS (Alonso et al., 2002). The TRS at the 3′ end of the 5′ leader sequence is referred to as the leader TRS (L-TRS), whereas TRSs located at the 5′ end of each gene downstream of the genes coding for the replicase proteins are referred to as body TRSs (B-TRSs). These findings suggested that the TRS elements are involved in discontinuous transcription.
Fig. 4.
Coronavirus transcription-regulating sequences (TRSs). TRSs from two Group1 CoVs: TGEV and HCoV-229E are shown. CSs sequences are represented inside black boxes, 5′-TRS and 3'-TRS flanking sequences are indicated at both 5′ and 3′ ends inside gray boxes (modified from Coronavirus Replication and Reverse Genetics. L. Enjuanes (Ed.). Curr. Top. Microbiol. Immunol. 287. Publisher: Springer, edition 2004).
According to the current model (Fig. 1D), following the synthesis of the MHV replicase proteins, pp1a and pp1ab, synthesis of (−) strand RNA begins. In some cases, e.g. for purposes of replication, the RDRP complex makes a complete (−) strand copy of the genome which serves as template for the synthesis of full-length (+) strand G RNA. In other cases, i.e. for making SG RNAs, (−) strand RNA synthesis begins, but stalls after copying one of the body TRSs in the viral genome. This (−) strand TRS at the 3′ end of the nascent RNA then base pairs with the (+) strand TRS at the 3′ end of the leader sequence, facilitating translocation of the nascent (−) strand RNA to the 5′ end of the genome where the RDRP complex copies the leader sequence to the end of the genome (Sawicki et al., 2001, Pasternak et al., 2001, Zúñiga et al., 2004). The various (−) strand SG RNAs made in this fashion serve as templates for the synthesis of the same-length (+) strand SG mRNAs and/or the production of shorter internally nested SG RNAs (Wu and Brian, 2010). The latter mechanism would likely contribute to the greater abundance of the 3′ co-terminal SG RNAs.
The leader and body TRSs do not show complete sequence conservation. For instance, with Murine hepatitis virus (MHV) DI RNAs, it was found that there were 2–4 copies of the L-TRS with the core sequence 5′ UCUAA, whereas the different B-TRSs had a core sequence centering around 5′ AAUCUAAAC. More important, it has been demonstrated that formation of a duplex between the (+)-strand L-TRS sequence in the genome and the (–)-strand copy of a B-TRS is required for synthesis of the SG RNA having that TRS at its 5′ end (Fig. 1, Fig. 3).
The relative contributions of the L-TRS and the B-TRS to the different 5'-TRS in the SG RNA are variable. In some cases, the entire sequence of the junction TRS in the SG RNA is derived from the B-TRS; in other instances, both the L- and the B-TRS contribute to the junction TRS. In an interesting experiment, it was found that when cells were infected with two different strains of MHV, many of the SG RNAs had a leader sequence of the co-infecting virus, indicating that the B-TRS of one strain can base pair with the L-TRS of the second strain (Pasternak et al., 2006).
How is it determined at which B-TRS the synthesis of the (−) strand RNA stalls, leading to duplex formation with an L-TRS? The level of SG RNA production by both Arteriviruses and Coronaviruses was shown to depend upon the efficiency of interactions between L-TRS and B-TRS (Pasternak et al., 2001, Pasternak et al., 2003, Zúñiga et al., 2004, Sola et al., 2005). Furthermore, it has been proposed that the L–B TRS junctions occur at multiple sites, with a preference for 3′ proximal nucleotides within the B-TRS (van der Most et al., 1994). With Transmissible gastroenteritis coronavirus (TGEV), it has been shown that sequences flanking the core TRS influence transcription of the SG RNAs (Curtis et al., 2004, Sola et al., 2005). Among Nidovirales, the 3′ end of the G RNA is critical for SG RNA transcription. In MHV, the 300 nt 3′ UTR promotes transcription (Lin et al., 1996), but only the 3′ 55 nts are required for (−) strand RNA synthesis. It was also shown that mutations disrupting the U-turn motif of the 5′ UTR stem-loop 2 (SL2) affected SG RNA synthesis, suggesting that SL2 mediates specific interactions with viral and/or cellular proteins involved in the synthesis of SG RNAs (Liu et al., 2007). In general, the 5′ UTRs of Coronavirus RNAs fold into similar secondary structures containing three to four SL structures that include a highly conserved 5 nt hairpin loop SL2 with a U-turn motif; SL1 and SL2 are close to each other (shown in Liu et al., 2007). The polypyrimidine-tract binding (PTB) protein hnRNPI may play a role in the regulation of SG RNA transcription due to its ability to interact with short pyrimidine-rich tracts, e.g. UCU, or CUCU. Since the 5′ UTR SL2 contains UCUAA repeats, PTB binding might assist circularization of the viral genome and aid template switching during discontinuous SG RNA synthesis (Choi et al., 2002, Oberstrass et al., 2005).
Related to the discontinuous mechanism is the question of how the synthesis of SG RNAs is regulated, and how the ratios of the different SG RNAs to each other are maintained. To address this problem, studies were carried out on the nsp1 protein of both EAV and MHV (Tijms et al., 2001, Donaldson et al., 2007). This protein contains a zinc-binding domain that assists in RNA-protein interactions and regulates the replication/transcription balance (Tijms et al., 2001). The EAV nsp1 proteininteracts with p100-binding polypeptide (p100BP) which co-functions with the RNA polymerase II transcription factor c-Myb. It has been speculated that the nsp1-p100BP interaction is important for SG RNA synthesis, either directly or by recruiting another protein to the viral RDRP complex. Alternatively, nsp1 might modulate transcription in the infected cell, explaining its targeting to the nucleus (Tijms and Snijder, 2003). Similarly, a deficiency in nucleocapsid protein (N) of Human coronavirus (HCoV-229E) impaired RNA replication, but not transcription, demonstrating that N protein regulates the equilibrium between these two processes (Schelle et al., 2005).
Also, it has been proposed that a long range interaction regulates TGEV SG RNA transcription (Moreno et al., 2008b). Here, a 9-nucleotide (nt) sequence located 449 nt upstream of the N gene TRS core sequence (CS-N) interacts with a complementary sequence immediately upstream of CS-N. The complementarity between these two 9-nt elements in TGEV was functionally relevant in the transcriptional activation of the N gene. Moreover, a positive correlation between the predicted stability of the base-pairing interaction and the accumulation levels of SG RNA N was observed (Moreno et al., 2008b).
Functions of subgenomic RNAs
Role in translation
Most of the SG RNAs generated by (+)-strand RNA viruses function as mRNAs. This is consistent with their composition, i.e. the presence of a 5′ cap (van Vliet et al., 2002) and a 3′ polyA tail (e.g. Wierzchoslawski et al., 2006). Some SG RNAs lack the 5′ cap but contain an IRES structure. For instance, the uncapped SG RNAs in TMV strain U1 (Grdzelishvili et al., 2000) and in Crucifer-infecting tobamovirus (crTMV) (Dorokhov et al., 2006) harbor an IRES, enabling ribosomes to initiate translation at a distant 5′-site.
With other viruses, e.g. Turnip crinkle virus (TCV), or Barley yellow dwarf virus (BYDV), the 3′ UTR cap-independent translation elements (CITEs) are important for launching translation of SG RNAs. In some cases a CITE acts cooperatively with an IRES. Among eight distinct structural classes of CITEs described to date, all contain an SL structure which base-pairs to a 5′ UTR sequence. The 3′ location of CITEs favors the translation of SG RNAs over that of genomic RNAs (Qu and Morris, 2000, Scheets and Redinbaugh, 2006, Shen et al., 2006).
Although the 5′-UTR of Potato leafroll virus (PLRV) SG RNA lacks a 3′ translational enhancer (Juszczuk et al., 2000), an IRES signal for translation of replication-associated protein 1 (Rap1) has been identified internally within the PLRV RNA genome 1500 nt downstream of its 5′ end (Jaag et al., 2003). The presence of CA-rich motifs, which increase the length but reduce the secondary structure of the SG RNA 5′UTR boost the protein expression by the SG RNAs of TBSV, Carnation mottle virus (CMV), Cardamine chlorotic fleck virus (CCFV) (Skotniki et al., 1993), and TCV (Qu and Morris, 2000).
Role in replication
One of the first demonstrations that an SG RNA could control viral RNA replication was shown with FHV RNA3, a SG RNA derived from RNA1 (see above) (Eckerle and Ball, 2002). RNA1 mutants deficient in the SG RNA3 synthesis failed to make RNA2. However, when RNA3 was supplied in trans, synthesis of RNA 2 resumed (Eckerle and Ball, 2002, Eckerle et al., 2003). Precisely how RNA3 exerts its effect is not understood. However, the RNA3-dependent replication signal has been mapped to the RNA2 3′ end (Albariño et al., 2003).
The influence of an SG RNA on viral replication is also seen with BYDV (Shen and Miller, 2004). BYDV makes three 3' co-terminal SG RNAs by internal initiation. SG RNA3 has no coding capacity, and its function is not known. Translation of BYDV G RNA and SG RNA1 is mediated by BTE: barley yellow dwarf (like) translation elementin the 5′end of the 3′ UTR (Fig. 3F) (Shen et al., 2006, Rakotondrafara et al., 2006). BTEs as cap-independent translation elements can be characterized by two structural features: (i) a conserved 17 nt sequence, that includes a stem loop (SL-I) with a GN RNA loop motif, and (ii) a loop (not in SL-I) that can base pair to a loop in the 5′UTR of the RNA (Kneller et al., 2006). It has been proposed that the premature addition of SG RNA2, which contains the 5′ UTR BTE, regulates BYDV replication by inhibiting translation of the viral polymerase from genomic RNA (G RNA). Thus, the G RNA ceases to function as a messenger and instead functions as a template for replication (Shen and Miller, 2004).
A role in separation of replication from translation has been proposed for the 5′ co-terminal SG RNAs of CTV, BMV and CTLV (Gowda et al., 2003, Wierzchoslawski et al., 2006, Tatineni et al., 2009). The 5′ SG RNA can serve as a template for expression of viral protein, e.g. movement protein in BMV, which sets the genomic RNA free for more efficient replication, recombination and packaging (Gowda et al., 2003). As with BYDV SG RNA 2, the early addition of the 5′ co-terminal SG RNA3a reduced BMV RNA replication in barley protoplasts (J. Sztuba-Solińska and J.J. Bujarski, unpublished). In contrast, the in planta experiments showed a dose–dependent bell-shaped response in which high concentrations of SG RNA3a reduced virus yield. We speculate that the translation of minute amounts of initial SG RNA3a leads to an excess of MP which facilitates the viral spread. At higher doses the SG RNA3a may, however, act as a molecular decoy that sequesters translational factors, reducing translation of replicase proteins (J. Sztuba-Solińska and J.J. Bujarski, unpublished).
Role in recombination
RNA recombination can salvage damaged or mutated viral RNAs and can contribute to genome variability (Cheng and Nagy, 2003, Chetverin et al., 2005). The exchangeable subgenomic components can facilitate the production of rearranged viral RNA genomes. For instance, the transcription mechanism in Nidovirales, involving leader–body TRS duplex formation together with distinct cis-acting signals that affect the nascent strand transfer during SG RNAs production, resembles the copy-choice RNA recombination (Figs. 1C and D) (van Marle et al., 1999, Pasternak et al., 2006). The high recombination frequencies in Coronaviruses and Arteriviruses have been associated with the highly structured 3′ UTRs in the genomes of these viruses (Molenkamp et al., 2000, Pasternak et al., 2000, Pasternak et al., 2001). Here, the base-pairing between the donor and acceptor molecules at consensus TRS motifs plays a role in the production of Nidoviral SG RNAs, suggestive of similarity-assisted recombination (Yuan et al., 2004, Pasternak et al., 2006). Apparently, these viruses frequently use a transcription strategy for recombination.
The integrity of non-segmented genomes of Closteroviridae is supported by SG RNA-assisted recombination. In the case of CTV defective interfering (DI) RNAs the junction sites coincide with the SG RNA transcription initiation sites (Yang et al., 1997). This suggested that CTV DI RNAs emerged by recombination of SG RNA with a 5′ region of the G RNA (Bar-Joseph et al., 1997). The proposed model emphasizes the role of intergenic AU-rich sequence located between two SL structures, between ORF10 and ORF11 that might induce the SG RNA11 premature termination followed by template switching (Yang et al., 1997, Bar-Joseph et al., 1997). A similar mechanism might explain the acquisition by viruses in the family Closteroviridae of non-self-sequences, either from co-infecting virus or the host (Cuellar et al., 2008).
It is likely that Norovirus (NoV) recombinant isolates originated from SG RNA-mediated rearrangements (Bull et al., 2005). Here, the high frequency recombination could result from RDRP stalling at the SL structure of the ORF1/ORF2 overlap (Rohayem et al., 2005), forcing the enzyme to hop across to either (–)-strand SG RNA or G RNA species. As a result, the recombinant Norovirus isolates can acquire new ORF2 and ORF3 sequences (Bull et al., 2005).
The multipartite viruses in the family Bromoviridae also utilize SG RNAs for the modular swap of their genomes. The recently described BMV 5′ co-terminal SG RNA3a can prime recombination events on the (−) and (+) RNA3 strands (Wierzchoslawski et al., 2006, Sztuba-Solińska et al., 2011). These and the following studies revealed the presence of several recombination hot-spots within RNA3 including the 5′ UTR, the upstream encapsidation signal (packaging element called PE), the B-box motif and the intergenic polyU track (Sztuba-Solińska et al., 2011; Sztuba-Solińska and Bujarski, unpublished results). It was proposed that the RDRP associated with other protein factors, e.g. coat protein, might bridge over the RNA3 and SG RNA3a molecules in cis and/or in trans at highly structured RNA elements such as B box-like motif, PE element, or 3′ TLS. This can facilitate template switching during (−) and (+) strand synthesis. Apparently, SG RNAs can act as building components that contribute to genomic rearrangements of complete viral genes.
Further perspectives
The activity of transcriptional regulatory elements, their general structure and sequence context, as well as their interactions with protein factors, all affect the production of SG RNA. It is not known, however, how these structures cause stalling/detachment/reattachment of an actively copying RDRP, or how they affect the timing of SG RNA synthesis. Since RDRPs frequently encounter base-paired regions while copying a template (Ng et al., 2008), explaining how RNA structures affect the progress of an RDRP would greatly improve our understanding of regulation of SG RNA synthesis. More work with in vitro systems is needed to shed light on both cis- and trans-acting regulatory signals and their cognate factors that affect the production of SG RNA (Nagy and Pogany, 2000, Li et al., 2005a). One problem is that the replication complexes are membrane bound (Mackenzie, 2005, Denison, 2008). However, by using cell-free extracts from evacuolated plant protoplasts Komoda et al., 2003, Okamoto et al., 2008 have carried out preliminary studies on replication, transcription and translation mechanisms with Tomato mosaic virus (ToMV), BMV, TCV, and RCNMV.
One question concerning SG RNAs that remains to be answered concerns our understanding of how the ratio of genomic to SG RNA synthesis is maintained. Besides the relative strength of the promoters the concentrations of the various NTPs plays a role in determining this ratio (Li et al., 2008, Li et al., 2010). A related question is whether there is competition between the synthesis of G RNA and SG RNA. With respect to viruses with multiple SG RNAs, why are some RNAs made in larger amounts than others? Much also remains to be learned about the particular mechanisms of SG RNAs synthesis and why certain SG RNAs are more robust messengers than other SG RNAs or than the G RNA. A better understanding of SG RNAs and their functions may provide us with new targets for antiviral therapy, specifically concerning important diseases such as SARS, caused by a Coronavirus.
The idea that SG RNAs may be a tempting target for antiviral therapy is supported by the identification of plant-derived compounds (Picard et al., 2005, Li et al., 2007) that inhibit plant and animal viruses, e.g. in Alphaviruses, at nanomolar concentrations. These compounds appear to act by preferential inhibition of synthesis of SG RNAs. Although the exact mechanism is not known, one possibility is that they bind to sites on the RDRP that recognize the SGPs.
Also, the ability to exogenously regulate SG RNAs transcription, e.g., by using a small-molecule ligand and aptamer technology (Wang et al., 2008), offers a promising way to modulate the timing and the levels of SG RNA transcription which, in turn, would provide a means to control viral protein expression. This approach has already found use in regulating the function of the higher-order component of the attenuation signal allowing the control ofTBSV SG RNA transcription (Wang and White, 2007, Wang et al., 2008) and it offers further promising ways for the analysis and modulation of viral processes at either the level of SG RNA transcription or translation.
Since SGPs have been identified as recombination hot spots (Suzuki et al., 2003, Wierzchoslawski et al., 2004, Coyne et al., 2006), the understanding of the mechanism of SG RNA transcription may provide insights into the origin and evolution of viruses. The continual emergence of new recombinant strains, e.g. HCoV-NL63 and HCoVHKU1 isolated in the wake of SARS (van der Hoek et al., 2004), and SARS-like Coronaviruses isolated from animal reservoirs (Li, et al., 2005b), raises the question of the role of SG RNAs in the reshuffling of genome sequences of Corona- and other viruses. Clearly, by incorporating foreign sequences into their genomes, certain viruses acquire new surprising properties (Pasternak et al., 2006, Moreno et al., 2008a). For example, the Closterovirus SG RNA-expressed HSP70h, that facilitates viral assembly and cellular movement, has likely been captured from the host (Prokhnevsky et al., 2005). The Coronavirus SG RNA-expressed hemagglutinin-esterase (HE), acting as a receptor-binding fusion protein, likely originated from Orthomyxoviruses (Zeng et al., 2008).
Viruses that generate SG RNAs are increasingly being used as expression vectors. For example, with Alphaviruses a foreign gene can be placed under the control of the SGP promoter, replacing the genes encoding the structural protein (Rayner et al., 2002, Rausalu et al., 2009). RNAs expressed from this promoter are translated in large amounts, producing proteins which can be used in vaccines, or as therapeutics. Also, by incorporating an RNA sequence encoding an antibody chain that recognizes a ligand on a cell surface, into an SG RNA, one can produce proteins targeted to a specific cell type.
Acknowledgments
This work was supported by grants from National Science Foundation (MCB-0920617) and National Institutes of Health (G1A62203) and by the Plant Molecular Biology Center at Northern Illinois University to JJB, and by grants from the National Institutes of Health (AI49273) and (AI070728) to VS.
References
- Adkins S., Kao C.C. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology. 1998;252:1–8. doi: 10.1006/viro.1998.9449. [DOI] [PubMed] [Google Scholar]
- Albariño C.G., Eckerle L.D., Ball L.A. The cis-acting replication signal at the 3′ end of Flock House virus RNA2 is RNA3-dependent. Virology. 2003;311:181–191. doi: 10.1016/s0042-6822(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Alonso S., Izeta A., Sola I., Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteriris virus. J. Virol. 2002;76:1293–1308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph M., Yang G., Gafny R., Mawassi M. Subgenomic RNAs: the possible building blocks for modular recombination of Closteroviridae genomes. Semin. Virol. 1997;8:113–119. [Google Scholar]
- Bull R.A., Hansman G.S., Clancy L.E., Tanaka M.M., Rawlinson W.D., White P.A. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 2005;11:1079–1085. doi: 10.3201/eid1107.041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X., Piestun D., Mawassi M., Yang G., Satyanarayana T., Gowda S., Dawson W.O., Bar-Joseph M. 5′-coterminal subgenomic RNAs in Citrus tristeza virus-infected cells. Virology. 2001;283:374–381. doi: 10.1006/viro.2001.0880. [DOI] [PubMed] [Google Scholar]
- Cheng C.P., Nagy P.D. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J. Virol. 2003;77:12033–12047. doi: 10.1128/JVI.77.22.12033-12047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin A.B., Kopein D.S., Chetverina H.V., Demidenko A.A., Ugarov V.I. Viral RNA-directed RNA polymerases use diverse mechanisms to promote recombination between RNA molecules. J. Biol. Chem. 2005;280:8748–8755. doi: 10.1074/jbc.M412684200. [DOI] [PubMed] [Google Scholar]
- Choi I.R., White K.A. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J. Biol. Chem. 2002;277:3760–3766. doi: 10.1074/jbc.M109067200. [DOI] [PubMed] [Google Scholar]
- Choi I.R., Ostrovsky M., Zhang G., White K.A. Regulatory activity of distal and core RNA elements in tombus virus subgenomic mRNA2 transcription. J. Biol. Chem. 2001;76:41761–41768. doi: 10.1074/jbc.M106727200. [DOI] [PubMed] [Google Scholar]
- Choi K.S., Huang P., Lai M.M. Polypyrimidine-tract binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology. 2002;303:58–68. doi: 10.1006/viro.2002.1675. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeusmonodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- Coyne K.P., Reed F.C., Porter C.J., Dawson S., Gaskell R.M., Radford A.D. Recombination of Feline calicivirus within an endemically infected cat colony. J. Gen. Virol. 2006;87:921–926. doi: 10.1099/vir.0.81537-0. [DOI] [PubMed] [Google Scholar]
- Cuellar W.J., Tairo F., Kreuze J.F., Valkonen J.P.T. Analysis of gene content in sweet potato chlorotic stunt virus RNA1 reveals the presence of the p22 RNA silencing suppressor in only a few isolates: implications for viral evolution and synergism. J. Gen. Virol. 2008;89:573–582. doi: 10.1099/vir.0.83471-0. [DOI] [PubMed] [Google Scholar]
- Culver J.N., Lehto K., Close S.M., Hilf M.E., Dawson W.O. Genomic position affects the expression of tobacco mosaic virus movement and coat protein genes. Proc. Natl Acad. Sci. USA. 1993;90:2055–2059. doi: 10.1073/pnas.90.5.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K.M., Yount B., Sims A.C., Baric R.S. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. J. Virol. 2004;78:6061–6066. doi: 10.1128/JVI.78.11.6061-6066.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R. Seeking membranes: positive-strand RNA virus replication complexes. PLoS Biol. 2008;6(e270):2098–2100. doi: 10.1371/journal.pbio.0060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J., Ishikawa M., Kaido M., Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl Acad. Sci. USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Sims A.C., Graham R.L., Denison M.R., Baric R.S. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 2007;81:6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov Y.L., Ivanov P.A., Komarova T.V., Skulachev M.V., Atabekov J.G. An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. J. Gen. Virol. 2006;87:2693–2697. doi: 10.1099/vir.0.82095-0. [DOI] [PubMed] [Google Scholar]
- Eckerle L.D., Ball L.A. Replication of the RNA segments of a bipartite viral genome is coordinated by a trans activating subgenomic RNA. Virology. 2002;296:165–176. doi: 10.1006/viro.2002.1377. [DOI] [PubMed] [Google Scholar]
- Eckerle L.D., Albariño C.G., Ball L.A. Flock House virus subgenomic RNA3 is replicated and its replication correlates with trans activation of RNA2. Virology. 2003;317:95–108. doi: 10.1016/j.virol.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Galiakparov N., Goszczynski D.E., Che X., Batuman O., Bar-Joseph M., Mawassi M. Two classes of subgenomic RNA of Grapevine virus Aproduced by internal controller elements. Virology. 2003;312:434–448. doi: 10.1016/s0042-6822(03)00239-3. [DOI] [PubMed] [Google Scholar]
- Gowda S., Satyanarayana T., Ayllón M.A., Albiach-Marti M.R., Mawassi M., Rabindran S., Garnsey S.M., Dawson W.O. Characterization of the cis-acting elements controlling subgenomic mRNAs of citrus tristeza virus: production of positive and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology. 2001;286:134–151. doi: 10.1006/viro.2001.0987. [DOI] [PubMed] [Google Scholar]
- Gowda S., Ayllón M.A., Satyanarayana T., Bar-Joseph M., Dawson W.O. Transcription strategy in a closterovirus: a novel 5′-proximal controller element of citrus tristeza virus produces 5′- and 3′- terminal subgenomic RNAs and differs from 3′-open reading frame controller elements. J. Virol. 2003;77:340–352. doi: 10.1128/JVI.77.1.340-352.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grdzelishvili V.Z., Chapman S.N., Dawson W.O., Lewandowski D.J. Mapping of the tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo. Virology. 2000;275:177–192. doi: 10.1006/viro.2000.0511. [DOI] [PubMed] [Google Scholar]
- Guenther R.H., Sit T.L., Gracz H.S., Dolan M.A., Townsend H.L., Liu G., Newman W.H., Agris P.F., Lommel S.A. Structural characterization of an intermolecular RNA–RNA interaction involved in the transcription regulation element of a bipartite plant virus. Nucleic Acids Res. 2004;32:2819–2828. doi: 10.1093/nar/gkh585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot P.C.J., Brederode F.Th., Olsthoorn R.C.L., Bol J.F. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA. 2000;6:708–716. doi: 10.1017/s1355838200992471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot P.C.J., Olsthoorn R.C.L., Bol J.F. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA. 2002;8:110–122. doi: 10.1017/s1355838202012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz J.M., Huang H.V. Evolution of the Sindbis Virus subgenomic mRNA promoter in cultured cells. J. Virol. 1995;69:7768–7774. doi: 10.1128/jvi.69.12.7768-7774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz J.M., Huang H.V. Host-dependent evolution of the Sindbis virus promoter for subgenomic mRNA synthesis. J. Virol. 1995;69:7775–7781. doi: 10.1128/jvi.69.12.7775-7781.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Mise K., Takeda A., Okinaka Y., Mori K., Arimoto M., Okuno T., Nakai T. Characterization of Striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J. Gen. Virol. 2005;86:2807–2816. doi: 10.1099/vir.0.80902-0. [DOI] [PubMed] [Google Scholar]
- Jaag H.M., Kawchuk L., Rohde W., Fischer R., Emans N., Prufer D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl Acad. Sci. USA. 2003;100:8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.A., Bragg J.N., Lawrence D.M., Jackson A.O. Sequence elements controlling expression of Barley stripe mosaic virus subgenomic RNAs in vivo. Virology. 2003;313:66–80. doi: 10.1016/S0042-6822(03)00285-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczuk M., Paczkowska E., Sadowy E., Zagorski W., Hulanicka D.M. Effect of genomic and subgenomic leader sequences of potato leafroll virus on gene expression. FEBS Lett. 2000;484:33–36. doi: 10.1016/s0014-5793(00)02122-0. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Hemenway C.L. Long-distance RNA–RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller E.L.P., Rakotondrafara A.M., Miller W.A. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G., Miller W.A. A positive-strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 2000;74:5988–5996. doi: 10.1128/jvi.74.13.5988-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G., Mohan B.R., Miller W.A. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoda K., Naito S., Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl Acad. Sci. USA. 2003;101:1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaStarza M.W., Grakoui A., Rice C.M. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology. 1994;202:224–232. doi: 10.1006/viro.1994.1338. [DOI] [PubMed] [Google Scholar]
- Li M.L., Stollar V. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc. Natl Acad. Sci. USA. 2004;101:9429–9434. doi: 10.1073/pnas.0400995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Stollar V. Distinct sites on the Sindbis virus RNA dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA. J. Virol. 2007;81:4371–4373. doi: 10.1128/JVI.02672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Lin Y.-H., Stollar V. A cell-free system for the synthesis of Sindbis virus subgenomic RNA: importance of the concentration of the initiating NTP. Virology. 2005;341:24–33. doi: 10.1016/j.virol.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like Coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang L., Li S., Chen X., Shen Y., Zhang Z., He H., Xu W., Shu Y., Liang G., Fang R., Hao X. Seco-pregnane steroids target the subgenomic RNA of alphavirus-like RNA viruses. Proc. Natl Acad. Sci. USA. 2007;104:8083–8088. doi: 10.1073/pnas.0702398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Kwan T.Y., Simmonds H.A., Stollar V. Synthesis of genomic and subgenomic RNA in mosquito cells infected with two Sindbis virus nsP4 mutants: influence of intracellular nucleoside triphosphate concentrations. J. Virol. 2008;82:6880–6888. doi: 10.1128/JVI.00517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Wang H., Stollar V. In vitro synthesis of Sindbis virus genomic and subgenomic RNAs: influence of nsP4 mutations and nucleoside triphosphate concentrations. J. Virol. 2010;84:2732–2739. doi: 10.1128/JVI.01561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.X., White K.A. A complex network of RNA–RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 2004;23:3365–3374. doi: 10.1038/sj.emboj.7600336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J., Zhang X., Wu R.C., Lai M.M.C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J. Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Simmonds H.A., Stollar V. Restriction of a Sindbis virus mutant in BHK cells and relief of the restriction by the addition of adenosine. Virology. 2002;292:78–86. doi: 10.1006/viro.2001.1230. [DOI] [PubMed] [Google Scholar]
- Lin H.X., Xu W., White K.A. A multicomponent RNA-based control system regulates subgenomic mRNA transcription in a tombusvirus. J. Virol. 2007;81:2429–2439. doi: 10.1128/JVI.01969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Sgro J.Y., Ahlquist P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 2002;76:3905–3919. doi: 10.1128/JVI.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Millership J.J., Kang H., Leibowitz J.L., Gierroc D.P. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin W.J., MacFarlane S.A. Rubus chlorotic mottle virus, a new sobemovirus infecting raspberry and bramble. Virus Res. 2009;139:10–13. doi: 10.1016/j.virusres.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Miller W.A., White K.A. Control of plant virus gene expression and replication by long-distance RNA–RNA interactions. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenkamp R., Greve S., Spaan W.J.M., Snijder E.J. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J. Virol. 2000;74:9062–9070. doi: 10.1128/jvi.74.19.9062-9070.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M., Bárcena J., Ramírez M.A., Boga J.A., Parra F., Torres J.M. Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (−)-sense genomic RNA. J. Biol. Chem. 2004;279(17):17013–17018. doi: 10.1074/jbc.M313674200. [DOI] [PubMed] [Google Scholar]
- Moreno P., Ambrós S., Albiach-Marti M.R., Guerri J., Pena L. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008;9:251–268. doi: 10.1111/j.1364-3703.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.L., Zúñiga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Partial purification and characterization of cucumber necrosis virus and tomato bushystunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology. 2000;276:279–288. doi: 10.1006/viro.2000.0577. [DOI] [PubMed] [Google Scholar]
- Neill J.D. The subgenomic RNA of feline calicivirus is packaged into viral particles during infection. Virus Res. 2002;87:89–93. doi: 10.1016/s0168-1702(02)00086-2. [DOI] [PubMed] [Google Scholar]
- Ng K.K., Arnold J.J., Cameron C.E. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., Allain F.H.-T. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Nagano H., Iwakawa H., Mizumoto H., Takeda A., Kaido M., Mise K., Okuno T. Cis-preferential requirement of a − 1 frameshift product p88 for the replication of red clover necrotic mosaic virus RNA1. Virology. 2008;375:205–212. doi: 10.1016/j.virol.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R.C.L., Haasnoot P.C.J., Bol J.F. Similarities and differences between the subgenomic and minus-strand promoters of an RNA plant virus. J. Virol. 2004;78:4048–4053. doi: 10.1128/JVI.78.8.4048-4053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Gultyaev A.P., Spaan W.J.M., Snijder E.J. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 2000;74:11642–11653. doi: 10.1128/jvi.74.24.11642-11653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J.M., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J.M., Snijder E.J. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirussubgenomic mRNA synthesis. J. Virol. 2003;77:1175–1183. doi: 10.1128/JVI.77.2.1175-1183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Picard D., Kao C.C., Hudak K.A. Pokeweed antiviral protein inhibits Brome mosaic virus replication in plant cells. J. Biol. Chem. 2005;280(20):20069–20075. doi: 10.1074/jbc.M413452200. [DOI] [PubMed] [Google Scholar]
- Prokhnevsky A.I., Peremyslov Valera V., Dolja V.V. Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 2005;79:14421–14428. doi: 10.1128/JVI.79.22.14421-14428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Morris T.J. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara A.M., Polacek Ch., Harris E., Miller W.A. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Zhang X., Kao C.C. Enhancer-like activity of a Brome mosaic virus RNA promoter. J. Virol. 2003;77:1830–1839. doi: 10.1128/JVI.77.3.1830-1839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausalu K., Iofik A., Ülper L., Karo-Astover L., Lulla V., Merits A. Properties and use of novel replication-competent vectors based on Semliki Forest virus. Virol. J. 2009;6:33. doi: 10.1186/1743-422X-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner J.O., Dryga S.A., Kamrud K.I. Alphavirus vectors and vaccination. Rev. Med. Virol. 2002;12:279–296. doi: 10.1002/rmv.360. [DOI] [PubMed] [Google Scholar]
- Rico P., Hernandez C. Characterization of the subgenomic RNAs produced by Pelargonium flower break virus: identification of two novel RNAs species. Virus Res. 2009;142:100–107. doi: 10.1016/j.virusres.2009.01.018. [DOI] [PubMed] [Google Scholar]
- Rohayem J., Münch J., Rethwilm A. Evidence of recombination in the Norovirus capsid gene. J. Virol. 2005;79:4977–4990. doi: 10.1128/JVI.79.8.4977-4990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Wang T., Sawicki S.G. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 2001;82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- Scheets K., Redinbaugh M.G. Infectious cDNA transcripts of Maize necrotic streak virus: infectivity and translational characteristics. Virology. 2006;350:171–183. doi: 10.1016/j.virol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Miller W.A. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology. 2004;327:196–205. doi: 10.1016/j.virol.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Shen R., Rakotondrafara A.M., Miller W.A. Trans-regulation of cap-independent translation by a viral subgenomic RNA. J. Virol. 2006;80:10045–10054. doi: 10.1128/JVI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad S., Pogue G.P., Kewandowski D.J., Hidalgo J., Donson J., Grill L.K., Dawson W.O. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology. 1999;255:312–323. doi: 10.1006/viro.1998.9579. [DOI] [PubMed] [Google Scholar]
- Sivakumaran K., Choi S.-K., Hema M., Kao C.C. Requirements for Brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 2004;78:6091–6101. doi: 10.1128/JVI.78.12.6091-6101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotniki M.L., Mackenzie A.M., Torronen M., Gibbs A.J. The genomic sequence of cardaminechlorotic fleck virus. J. Gen. Virol. 1993;74:1933–1937. doi: 10.1099/0022-1317-74-9-1933. [DOI] [PubMed] [Google Scholar]
- Sola I., Moreno J.L., Zuniga S., Alonso S., Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawicki S.S., Kao C.C. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRp) reveal that RdRp can adjust its promoter binding sites. J. Virol. 1999;73:198–204. doi: 10.1128/jvi.73.1.198-204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Hibi T., Masuta C. RNA recombination between cucumoviruses: possible role of predicted stem-loop structures and an internal subgenomic promoter-like motif. Virology. 2003;306:77–86. doi: 10.1016/s0042-6822(02)00050-8. [DOI] [PubMed] [Google Scholar]
- Szecsi J., Ding S.X., Lim C.O., Bendahmane M., Cho M.J., Nelson R.S., Beachy R.N. Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 1999;12:143–152. [Google Scholar]
- Sztuba-Solińska J., Bujarski J.J. Insights into the single-cell reproduction cycle of members of the family Bromoviridae: lessons from the use of protoplast systems. J. Virol. 2008;82(21):10330–10340. doi: 10.1128/JVI.00746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztuba-Solińska J., Dzianott A., Bujarski J.J. Recombination of 5′ subgenomic RNA3a with genomic RNA3 of Brome mosaic bromovirus in vitro and in vivo. Virology. 2011;410:129–141. doi: 10.1016/j.virol.2010.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni S., Afunian M.R., Gowda S., Hilf M.E., Bar-Joseph M., Dawson W.O. Characterization of the 5′- and 3′-terminal subgenomic RNAs produced by a capillovirus: evidence for a CP subgenomic RNA. Virology. 2009;385:521–528. doi: 10.1016/j.virol.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Tatsuta M., Mizumoto H., Kaido M., Mise K., Okuno T. The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 2005;79:978–986. doi: 10.1128/JVI.79.2.978-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., Snijder E.J. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol. 2003;84:2317–2322. doi: 10.1099/vir.0.19297-0. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive stranded RNA virus. Proc. Natl Acad. Sci. USA. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., de Groot R.J., Spaan W.J.M. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl Acad. Sci. USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A.L., Smits S.L., Rottier P.J., de Groot R.J. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives M.C., Galipienso L., Navarro L., Moreno P., Guerri J. Characterization of two kinds of subgenomic RNAs produced by citrus leaf blotch virus. Virology. 2002;295:328–336. doi: 10.1006/viro.2001.1349. [DOI] [PubMed] [Google Scholar]
- Wang S., White K.A. Riboswitching on RNA virus replication. Proc. Natl Acad. Sci. USA. 2007;104:10406–10411. doi: 10.1073/pnas.0704178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Mortazavi L., White K.A. Higher-order RNA structural requirements and small-molecule induction of Tombus virus subgenomic mRNA transcription. J. Virol. 2008;82:3864–3871. doi: 10.1128/JVI.02416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology. 2002;304:147–154. doi: 10.1006/viro.2002.1732. [DOI] [PubMed] [Google Scholar]
- White K.A., Nagy P.D. Advances in the molecular biology of tombusviruses: gene expression, genome replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Wielgosz M.M., Raju R., Huang H.V. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J. Virol. 2001;75:3509–3519. doi: 10.1128/JVI.75.8.3509-3519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchoslawski R., Dzianott A., Bujarski J.J. Dissecting the requirement for subgenomic promoter sequences by RNA recombination of Brome mosaic virus in vivo: evidence for functional separation of transcription and recombination. J. Virol. 2004;78:8552–8564. doi: 10.1128/JVI.78.16.8552-8564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchoslawski R., Urbanowicz A., Dzianott A., Figlerowicz M., Bujarski J.J. Characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of Brome mosaic bromovirus. J. Virol. 2006;80:12357–12366. doi: 10.1128/JVI.01207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-Y., Brian D.A. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl Acad. Sci. USA. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., White K.A. Uncoupling RNA virus replication from transcription via the polymerase: functional and evolutionary insights. EMBO J. 2007;26:5120–5130. doi: 10.1038/sj.emboj.7601931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Oliveri S., Mandic J., White K.A. Evidence for a premature termination mechanism of subgenomic mRNA transcription in a carmovirus. J. Virol. 2010;84:7904–7907. doi: 10.1128/JVI.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., White K.A. Subgenomic mRNA transcription in an aureusvirus: down-regulation of transcription and evolution of regulatory RNA elements. Virology. 2008;371:430–438. doi: 10.1016/j.virol.2007.09.035. [DOI] [PubMed] [Google Scholar]
- Xu W., White K.A. RNA-based regulation of transcription and translation of Aureus virus subgenomic mRNA1. J. Virol. 2009;83:10096–10105. doi: 10.1128/JVI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Mawassi M., Gofman R., Gafny R., Bar-Joseph M. Involvement of a subgenomic mRNA in the generation of a variable population of defective Citrus tristeza virus molecules. J. Virol. 1997;71:9800–9802. doi: 10.1128/jvi.71.12.9800-9802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Hussain S., Wang H., Ke M., Guo D. Translational control of the subgenomic RNAs of severe acute respiratory syndrome coronavirus. Virus Genes. 2009;39:10–18. doi: 10.1007/s11262-009-0357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Murtaugh M.P., Schumann F.A., Mickelson D., Faaberg K.S. Characterization of heteroclite subgenomic RNAs associated with PRRSV infection. Virus Res. 2004;105:75–87. doi: 10.1016/j.virusres.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Zaccomer B., Haenni A.-L., Macaya G. The remarkable variety of plant RNA virus genomes. J. Gen. Virol. 1995;76:231–247. doi: 10.1099/0022-1317-76-2-231. [DOI] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L.W., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl Acad. Sci. USA. 2008;105(26):9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zhang J., Simon A.E. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 2004;78:7619–7633. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]