Abstract
The highly pathogenic Middle East respiratory syndrome (MERS)-related coronavirus (CoV) is transmitted from dromedary camels, the natural reservoir, to humans. For at present unclear reasons, MERS cases have so far only been observed in the Arabian Peninsula, although MERS-CoV also circulates in African dromedary camels. A recent study showed that MERS-CoV found in North/West- (Morocco) and West-African (Burkina Faso and Nigeria) dromedary camels are genetically distinct from Arabian viruses and have reduced replicative capacity in human cells, potentially due to amino acid changes in one or more viral proteins. Here, we show that the spike (S) proteins of the prototypic Arabian MERS-CoV strain, human betacoronavirus 2c EMC/2012, and the above stated African MERS-CoV variants do not appreciably differ in expression, DPP4 binding and ability to drive entry into target cells. Thus, virus-host-interactions at the entry stage may not limit spread of North- and West-African MERS-CoV in human cells.
Keywords: MERS-coronavirus, Spike, Entry, Dromedary camel, Zoonosis
1. Introduction
The Middle East respiratory syndrome-related coronavirus (MERS-CoV) causes the severe lung disease MERS (Zaki et al., 2012), which takes a fatal course in roughly ~35% of infected patients (WHO, 2019). MERS-CoV is endemic in the Middle East, where the virus is transmitted from dromedary camels, the natural reservoir, to humans (Perera et al., 2013; Reusken et al., 2013). Human-to-human transmission is inefficient but resulted in several hospital outbreaks of MERS (Assiri et al., 2013; Harriman et al., 2013; Memish et al., 2013), and there is concern that the virus may adapt to humans and cause a pandemic.
Infection of dromedary camels with MERS-CoV is not limited to the Middle East. African camels are frequently infected with MERS-CoV (Ali et al., 2017a, 2017b; Chu et al., 2014, 2015, 2018; Corman et al., 2014; Deem et al., 2015; Kiambi et al., 2018; Miguel et al., 2017; Ommeh et al., 2018; Perera et al., 2013; Reusken et al., 2013, 2014; van Doremalen et al., 2017) and the responsible viruses are genetically distinct from those circulating in the Middle East (Chu et al., 2018; Kiambi et al., 2018; Ommeh et al., 2018). Moreover, viruses isolated from animals in Morocco, Nigeria and Burkina Faso form a distinct phylogenetic subclade, C1, and exhibit reduced ability to replicate in human respiratory cells (Chu et al., 2018). In addition, MERS-CoV transmission from camels to humans has not been observed in North- and West-Africa (Munyua et al., 2017; So et al., 2018), although two livestock handlers in Kenya were shown to harbor antibodies against MERS-CoV (Liljander et al., 2016), Moreover, no MERS cases were documented in Africa. At present, the barrier(s) impeding efficient spread of African MERS-CoV in human cells and camel-human transmission of these viruses remain to be identified.
The MERS-CoV spike protein (S) is incorporated into the viral envelope and facilitates viral entry into target cells (Li, 2016). For this, the S protein binds to the cellular receptor dipeptidyl peptidase 4 (DPP4, CD26) (Raj et al., 2013) via its surface unit, S1, and fuses the viral membrane with a target cell membrane via its transmembrane unit, S2 (Li, 2016). Binding of MERS-S to DPP4 is essential for MERS-CoV infection of cells and DPP4 expression and the S protein/DPP4 interface are major determinants of MERS-CoV cell and species tropism (Raj et al., 2013; van Doremalen et al., 2014). The S proteins of North- and West-African MERS-CoV of the C1 clade harbor 6–9 amino acid substitutions relative to MERS-CoV (Fig. 1 A, Table 1 ) and these substitutions might reduce S protein-driven entry into target cells. However, this possibility has not been examined so far.
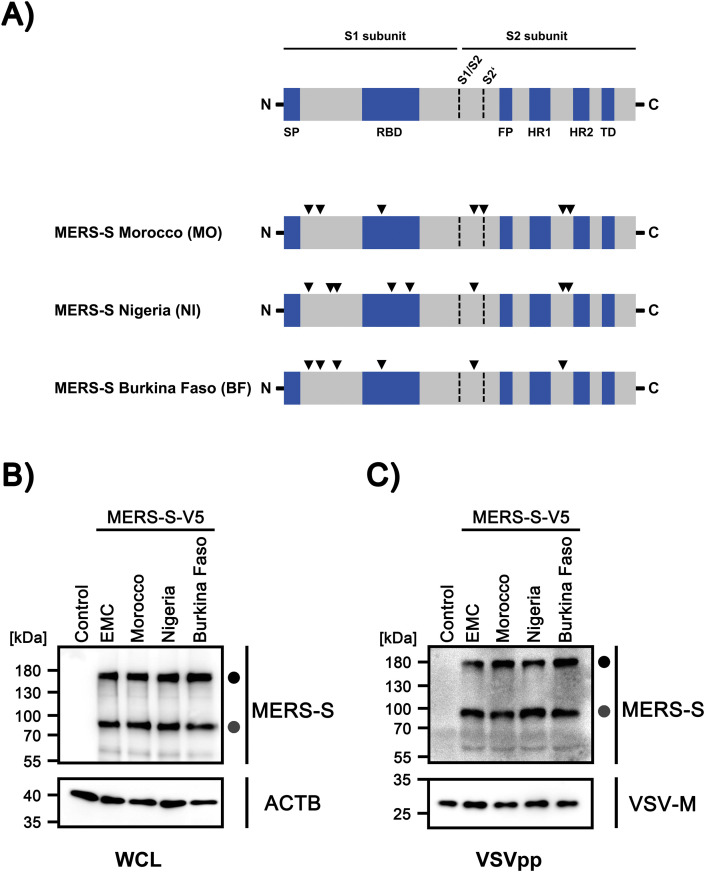
Fig. 1.
S proteins of North/West- and West-African MERS-CoV isolates from dromedary camels are robustly expressed in human cells and efficiently incorporated into viral particles.
(A) Domain organization of the MERS-S proteins studied here. Black arrowheads indicate amino acid variations found in S proteins of African viruses as compared to the S protein of the prototypic Arabian MERS-CoV strain, human betacoronavirus 2c EMC/2012 (GenBank: JX869059.2). Abbreviations: SP = signal peptide, RBD = receptor binding domain, FP = fusion peptide, HR1/HR2 = heptad repeat 1/2, TD = transmembrane domain. (B) The indicated S proteins were transiently expressed in 293T cells, whole cell lysates (WCL) were prepared at 48 h posttransfection and S protein expression was analyzed via Western blot, using an antibody targeting the C-terminal V5-tag. Cells expressing no S protein were used as negative control and detection of β-actin (ACTB) served as loading control. Similar results were obtained in two separate experiments. (C) Rhabdoviral transduction vectors (VSVpp) harboring the indicated S proteins were concentrated by centrifugation and, following lysis, analyzed by Western blot for S protein incorporation, using an antibody targeting the C-terminal V5-tag. Transduction vectors harboring no S protein were used as negative controls and detection of vesicular stomatitis virus matrix protein (VSV-M) served as loading control. Similar results were obtained in a separate experiment. Numbers on the left side of each blot indicate the molecular weight in kilodalton (kDa). Further, bands representing the precursor S protein (S0, black circle) and the S2 subunit of proteolytically processed S protein (grey circle) are indicated.
Table 1.
Amino acid variations between MERS-S EMC and the S proteins of MERS-CoV of North/West- and West-African dromedary camels.
| S protein | Variationa | Localizationb |
|---|---|---|
| MERS-S MO camel/Morocco/CIRAD-HKU213/2015 GenBank: MG923469.1 | V26A | S1 / n/a |
| A89S | S1 / n/a | |
| T424I | S1 / RBD | |
| S856Y | S2 / n/a | |
| R884L | S2 / PS(S2’) | |
| A1158S | S2 / n/a | |
| V1209L | S2 / n/a | |
| MERS-S NI camel/Nigeria/NV1657/2016 GenBank: MG923475.1 | V26A | S1 / n/a |
| H167Y | S1 / n/a | |
| H194Y | S1 / n/a | |
| L495F | S1 / RBD | |
| L588F | S1 / RBD | |
| S856Y | S2 / n/a | |
| A1158L | S2 / n/a | |
| L1200F | S2 / n/a | |
| MERS-S BF camel/Burkina Faso/CIRAD-HKU785/2015 GenBank: MG923471.1 | V26A | S1 / n/a |
| A89S | S1 / n/a | |
| H194Y | S1 / n/a | |
| T424I | S1 / RBD | |
| S856Y | S2 / n/a | |
| A1158S | S2 / n/a |
Amino acid position (numbering according to MERS-S EMC).
Subunit / Functional domain (if applicable); Abbreviations: S1 = S1 subunit; S2 = S2 subunit; RBD = receptor binding domain, PS(S2’) = priming site at the S2’ position (884-RSAR-887), n/a = not applicable.
2. Results
We employed a previously described vesicular stomatitis virus (VSV)-based pseudotyping system to study MERS-S-driven host cell entry (Kleine-Weber et al., 2018, 2019). Pseudotyping systems are known to adequately model key aspects of the coronavirus entry process. In order to study host cell entry driven by S proteins from the C1 subclade, we employed PCR-based mutagenesis to generate expression constructs for the S proteins of MERS-CoV from Morocco (camel/Morocco/CIRAD-HKU213/2015, MO), Nigeria (camel/Nigeria/NV1657/2016, NI) and Burkina Faso (camel/Burkina Faso/CIRAD-HKU785/2015, BF), using a published expression construct for MERS-CoV EMC S protein as template (Kleine-Weber et al., 2018, 2019). Moreover, expression constructs for all S proteins were generated that encoded a C-terminal V5 antigenic tag. Western blot analysis of cells transfected to express the S proteins under study revealed that MERS-S EMC, MO, NI and BF were expressed and proteolytically processed to comparable levels (Fig. 1B). Moreover, these S proteins were incorporated into VSV particles with similar efficiency (Fig. 1C). These results suggest that mutations present in North- and West-African MERS-S of the C1 subclade do not reduce S protein expression and proteolytic processing in human cells.
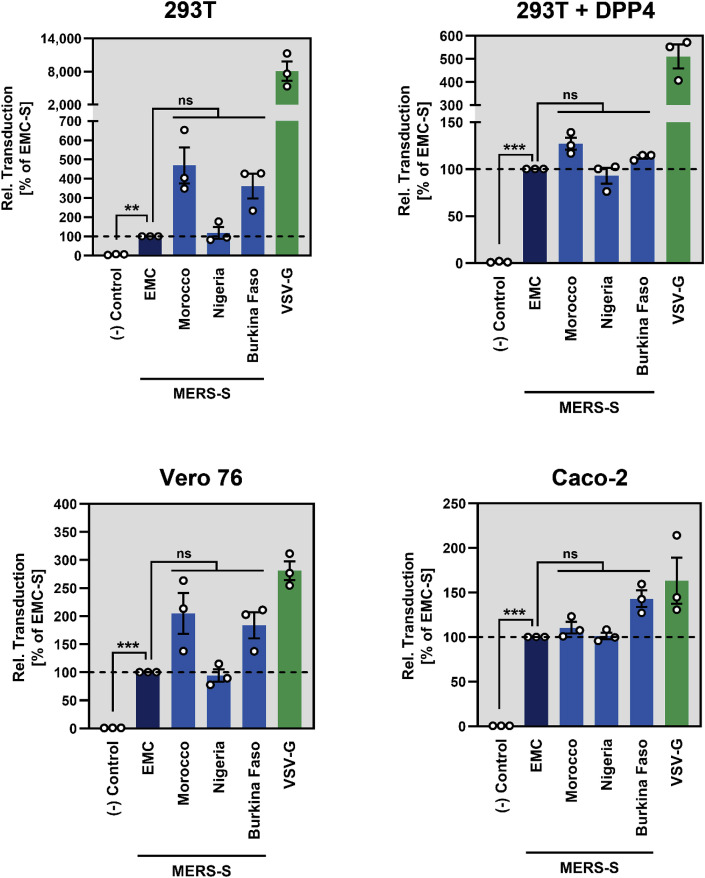
We next asked whether DPP4 binding of North- and West-African MERS-S was altered. For this, 293T cells transfected to express the S proteins under study were incubated with soluble DPP4 fused to the Fc portion of human immunoglobulin and binding was quantified by flow cytometry, as described previously (Kleine-Weber et al., 2019). The results showed that MERS-S EMC, MO, NI, and BF bound to DPP4 robustly and with comparable efficiency while DPP4 binding to cells expressing no S protein was within the background range (Fig. 2 ). Finally, we tested whether the robust binding to DPP4 translated into efficient S protein-driven entry. For this, cell lines were selected that were shown to express low levels (293T), intermediate levels (Vero 76) or high levels of DPP4 (Caco-2, 293T + DPP4) (Kleine-Weber et al., 2019). MERS-S MO, NI and BF mediated entry into all cell lines with at least the same efficiency as MERS-S EMC (Fig. 3 ). Moreover, under conditions of low or medium DPP4 expression, entry mediated by MERS-S MO and BF was even more efficient than entry mediated by MERS-S EMC (Fig. 3), although these differences were not statistically significant.
Fig. 2.
S proteins of North/West- and West-African MERS-CoV isolates from dromedary camels efficiently bind to DPP4. 293T cells expressing the indicated S proteins or no S protein at all (Control) were successively incubated with soluble DPP4 containing a C-terminal Fc tag (sol-DPP4-Fc) and AlexaFluor488-conjugated anti-human antibody, before DPP4 binding to the respective S protein was analyzed by flow cytometry. Presented are the combined data of three independent experiments for which sol-DPP4-Fc binding to MERS-S EMC was set as 100%. Error bars indicate the standard error of the mean (SEM). Statistical significance was tested by one-way analysis of variance with Sidak's posttest (p > 0.05, not significant, ns; p ≤ 0.01, **).
Fig. 3.
Host cell entry driven by the S proteins of North/West- and West-African MERS-CoV isolates from dromedary camels is robust. 293T, 293T transfected to express DPP4, Vero 76 and Caco-2 cells were inoculated with equal volumes of rhabdoviral transduction vectors harboring the indicated S proteins or no S protein (Control). At 18 h posttransduction, the activity of the virus-encoded luciferase, which served as an indicator for transduction efficiency, was measured in cell lysates. Presented are the combined data of three independent experiments for which transduction mediated by MERS-S EMC was set as 100%. Error bars indicate SEM. Statistical significance was tested by one-way analysis of variance (ANOVA) with Sidak's posttest (p > 0.05, ns; p ≤ 0.01, **; p ≤ 0.005, ***).
3. Discussion
Our results show that amino acid substitutions present in North- and West-African MERS-S proteins relative to MERS-S EMC do not compromise S protein expression in human cells, at least when transfected cells are examined. Similarly, proteolytic processing of the S proteins in the constitutive secretory pathway, which is known to be carried out by furin (Gierer et al., 2015; Millet and Whittaker, 2014), was not appreciably altered. Moreover, binding of North- and West-African S proteins to DPP4 was not diminished as compared to MERS-S EMC, despite the presence of at least one substitution in the receptor binding domain (RBD) in each S protein tested. This finding might not be unexpected since the substituted amino acid residues do not make direct contact with residues in DPP4 (Lu et al., 2013). In keeping with these observations, all African S proteins mediated robust viral entry into non-human primate (Vero 76) and human cell lines (293T, Caco-2) expressing different levels of DPP4 (Kleine-Weber et al., 2019). In fact, MERS-S MO- and BF-driven entry into cell lines expressing low or intermediate levels of DPP4 was augmented as compared to MERS-S EMC, in keeping with these S proteins showing slightly enhanced DPP4 binding as compared to MERS-S EMC. Finally, it is noteworthy that MERS-S activation in Caco-2 cells mainly depends on the cellular serine protease TMPRSS2 while activation in 293T and Vero 76 cells is mediated by the cellular cysteine protease cathepsin L (Kleine-Weber et al., 2018, 2019). Thus, North- and West-African MERS-S proteins seem to be able to use both pathways available for S protein activation in human cells.
Confirmation of our findings with authentic viruses is pending and we cannot exclude that, for instance, the S protein modulates recognition of the virus by sensors of the interferon system, which cannot be measured with the assays available to us. Moreover, we note that a recent study examining two MERS-S sequences (C2 subclade) from camels in Ethiopia demonstrated that these sequences, when inserted into MERS-CoV EMC, reduced viral entry and replication and increased sensitivity to antibody-mediated neutralization (Shirato et al., 2019). The reduction in entry was observed for Vero and to a lesser degree for Vero-TMPRSS2 cells and was generally modest. Nevertheless, these results suggest that S proteins from viruses circulating in Ethiopia might harbor mutations that diminish entry into human cells and that are not present in the MERS-S proteins studied here. Amino acid residues I139, L515, E851 and S1302 in the spike protein are unique to Ethiopian MERS-CoV and warrant further analysis.
Collectively, our results suggest that amino acid substitutions present in the S proteins of North- and West-African MERS-CoV do not compromise the ability of these viruses to enter human cells. Thus, future efforts to understand why North- and West-African MERS-CoV isolates show reduced replicative potential in human cells should be focused on other aspects of the MERS-CoV lifecycle than S protein-mediated host cell entry.
4. Materials and methods
4.1. Plasmids
Expression plasmids, based on the vector pCAGGS, for VSV-G and MERS-S EMC were previously described (Kleine-Weber et al., 2018, 2019). The MERS-S EMC plasmid was used as template for PCR-based mutagenesis to introduce the mutations found in MERS-S MO (Morocco, camel/Morocco/CIRAD-HKU213/2015, GenBank: MG923469.1), NI (Nigeria, camel/Nigeria/NV1657/2016, GenBank: MG923475.1) and BF (Burkina Faso, camel/Burkina Faso/CIRAD-HKU785/2015, GenBank: MG923471.1) (Table 1). In addition, PCR-based mutagenesis was used to equip the constructs with a C-terminal V5 antigenic tag. The integrity of all sequences was verified using automated sequence analysis.
4.2. Cell culture
293T (human embryonal kidney) and Vero 76 (African green monkey kidney) cells were cultivated in Dulbecco's modified Eagle's medium (DMEM; PAN Biotech). The human colorectal adenocarcinoma cell line Caco-2 was grown in Minimum Essential Media (MEM, Life Technologies). All media were supplemented with 10% fetal bovine serum (FBS, PAN Biotech) and 1x penicillin and streptomycin from a 100x stock solution (Pan Biotech). The cells were incubated under humid conditions at 37°C and 5% CO2. For transfection of 293T cells the calcium-phosphate precipitation method was used.
4.3. Antibodies and DPP4-Fc fusion protein
For Western blot analysis, anti-V5 (mouse, 1:2,500; ThermoFisher Scientific), anti-β-actin (mouse, 1:2,500; Sigma-Aldrich), anti-VSV-M (mouse, 1:2,500; Kerafast) were used as primary antibodies and anti-mouse HRP (horse radish peroxidase) conjugated antibody (goat, 1:2,500; Dianova) was used as secondary antibody. Antibodies were diluted in phosphate buffered saline [PBS] containing 0.5% Tween 20 [PBS-T] supplemented with 5% skim milk powder. For flow cytometry, a recombinant fusion protein of the ectodomain of DPP4 fused to the Fc fragment of human immunoglobulin (sol-DPP4-Fc, 1:200, ACROBiosystems) and an AlexaFlour488-conjugated anti-human antibody (goat, 1:500; ThermoFisher Scientific) were used (ligand and antibody were diluted in PBS containing 1% bovine serum albumin).
4.4. Immunoblot analysis of MERS-S expression and particle incorporation
For analysis of S protein expression, 293T cells were transfected with expression plasmid for MERS-S proteins harboring a C-terminal V5 tag, as described (Kleine-Weber et al., 2018, 2019). To investigate MERS-S incorporation into VSVpp, equal volumes of supernatants containing VSVpp bearing S proteins with V5 tag were centrifuged through a 20% sucrose cushion at 25.000 g for 120 min. Subsequently, cells and VSVpp pellets were lysed and analyzed by immunoblot, following an established protocol (Kleine-Weber et al., 2018, 2019).
4.5. Analysis of DPP4 binding efficiency
DPP4 binding was analyzed as described (Kleine-Weber et al., 2019). In brief, 293T cells were transfected with expression plasmids for MERS-S proteins and empty plasmid as negative control. At 48 h posttransfection, the cells were washed with PBS, pelleted and resuspended in PBS containing 1% BSA and soluble human DPP4-Fc fusion protein at a final dilution of 1:200. After incubation for 1 h at 4 °C, the cells were washed and incubated with AlexaFluor488-conjugated anti-mouse antibody at a dilution of 1:500. Finally, the cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry using an LSR II flow cytometer and the FACS Diva software (both BD Biosciences).
4.6. Production of VSV pseudoparticles (VSVpp) and transduction of target cells
Transduction vectors based on a replication-deficient VSV (Berger Rentsch and Zimmer, 2011) and pseudotyped with the indicated viral glycoproteins (VSVpp) were generated according to a published protocol (Kleine-Weber et al., 2018, 2019). Target cells were transduced with equal volumes of supernatants containing VSVpp and transduction efficiency was quantified at 16 h posttransduction by measuring the activity of virus-encoded firefly luciferase in cell lysates as previously described (Kleine-Weber et al., 2018, 2019).
Acknowledgements
The authors thank Gert Zimmer and Andrea Maisner for providing the replication-deficient VSV vector for pseudotyping and the Vero 76 cell line, respectively. This work was supported, including the efforts of Stefan Pöhlmann, by the Bundesministerium für Bildung und Forschung within the network project RAPID (Risikobewertung bei präpandemischen respiratorischen Infektionserkrankungen; 01KI1723D). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Ali M., El-Shesheny R., Kandeil A., Shehata M., Elsokary B., Gomaa M., Hassan N., El Sayed A., El-Taweel A., Sobhy H., Fasina F.O., Dauphin G., El Masry I., Wolde A.W., Daszak P., Miller M., VonDobschuetz S., Morzaria S., Lubroth J., Makonnen Y.J. Cross-sectional surveillance of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels and other mammals in Egypt, August 2015 to January 2016. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.11.30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.A., Shehata M.M., Gomaa M.R., Kandeil A., El-Shesheny R., Kayed A.S., El-Taweel A.N., Atea M., Hassan N., Bagato O., Moatasim Y., Mahmoud S.H., Kutkat O., Maatouq A.M., Osman A., McKenzie P.P., Webby R.J., Kayali G. Systematic, active surveillance for Middle East respiratory syndrome coronavirus in camels in Egypt. Emerg. Microb. Infect. 2017;6:e1. doi: 10.1038/emi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A., Team K.M.-C.I. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Oladipo J.O., Perera R.A., Kuranga S.A., Chan S.M., Poon L.L., Peiris M. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Euro Surveill. 2015;20 doi: 10.2807/1560-7917.ES.2015.20.49.30086. [DOI] [PubMed] [Google Scholar]
- Chu D.K., Poon L.L., Gomaa M.M., Shehata M.M., Perera R.A., Abu Zeid D., El Rifay A.S., Siu L.Y., Guan Y., Webby R.J., Ali M.A., Peiris M., Kayali G. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Hui K.P.Y., Perera R., Miguel E., Niemeyer D., Zhao J., Channappanavar R., Dudas G., Oladipo J.O., Traore A., Fassi-Fihri O., Ali A., Demissie G.F., Muth D., Chan M.C.W., Nicholls J.M., Meyerholz D.K., Kuranga S.A., Mamo G., Zhou Z., So R.T.Y., Hemida M.G., Webby R.J., Roger F., Rambaut A., Poon L.L.M., Perlman S., Drosten C., Chevalier V., Peiris M. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Said M.Y., Gluecks I., Lattwein E., Bosch B.J., Drexler J.F., Bornstein S., Drosten C., Muller M.A. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg. Infect. Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem S.L., Fevre E.M., Kinnaird M., Browne A.S., Muloi D., Godeke G.J., Koopmans M., Reusken C.B. Serological evidence of MERS-cov antibodies in dromedary camels (Camelus dromedaries) in Laikipia County, Kenya. PLoS One. 2015;10:e0140125. doi: 10.1371/journal.pone.0140125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Muller M.A., Heurich A., Ritz D., Springstein B.L., Karsten C.B., Schendzielorz A., Gnirss K., Drosten C., Pöhlmann S. Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 2015;211:889–897. doi: 10.1093/infdis/jiu407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriman K., Brosseau L., Trivedi K. Hospital-associated Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013;369:1761. doi: 10.1056/NEJMc1311004. [DOI] [PubMed] [Google Scholar]
- Kiambi S., Corman V.M., Sitawa R., Githinji J., Ngoci J., Ozomata A.S., Gardner E., von Dobschuetz S., Morzaria S., Kimutai J., Schroeder S., Njagi O., Simpkin P., Rugalema G., Tadesse Z., Lubroth J., Makonnen Y., Drosten C., Muller M.A., Fasina F.O. Detection of distinct MERS-Coronavirus strains in dromedary camels from Kenya, 2017. Emerg. Microb. Infect. 2018;7:195. doi: 10.1038/s41426-018-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., Müller M.A., Drosten C., Pöhlmann S., Hoffmann M. Mutations in the spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 2019;93 doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljander A., Meyer B., Jores J., Muller M.A., Lattwein E., Njeru I., Bett B., Drosten C., Corman V.M. MERS-CoV antibodies in humans, Africa, 2013-2014. Emerg. Infect. Dis. 2016;22:1086–1089. doi: 10.3201/eid2206.160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Al-Tawfiq J.A., Assiri A. Hospital-associated Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013;369:1761–1762. doi: 10.1056/NEJMc1311004. [DOI] [PubMed] [Google Scholar]
- Miguel E., Chevalier V., Ayelet G., Ben Bencheikh M.N., Boussini H., Chu D.K., El Berbri I., Fassi-Fihri O., Faye B., Fekadu G., Grosbois V., Ng B.C., Perera R.A., So T.Y., Traore A., Roger F., Peiris M. Risk factors for MERS coronavirus infection in dromedary camels in Burkina Faso, Ethiopia, and Morocco, 2015. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munyua P., Corman V.M., Bitek A., Osoro E., Meyer B., Muller M.A., Lattwein E., Thumbi S.M., Murithi R., Widdowson M.A., Drosten C., Njenga M.K. No serologic evidence of middle East respiratory syndrome coronavirus infection among camel farmers exposed to highly seropositive camel herds: a household linked study, Kenya, 2013. Am. J. Trop. Med. Hyg. 2017;96:1318–1324. doi: 10.4269/ajtmh.16-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ommeh S., Zhang W., Zohaib A., Chen J., Zhang H., Hu B., Ge X.Y., Yang X.L., Masika M., Obanda V., Luo Y., Li S., Waruhiu C., Li B., Zhu Y., Ouma D., Odendo V., Wang L.F., Anderson D.E., Lichoti J., Mungube E., Gakuya F., Zhou P., Ngeiywa K.J., Yan B., Agwanda B., Shi Z.L. Genetic evidence of Middle East respiratory syndrome coronavirus (MERS-Cov) and widespread seroprevalence among camels in Kenya. Virol. Sin. 2018;33:484–492. doi: 10.1007/s12250-018-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A., Wang P., Gomaa M.R., El-Shesheny R., Kandeil A., Bagato O., Siu L.Y., Shehata M.M., Kayed A.S., Moatasim Y., Li M., Poon L.L., Guan Y., Webby R.J., Ali M.A., Peiris J.S., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.36.20574. pii=20574. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Messadi L., Feyisa A., Ularamu H., Godeke G.J., Danmarwa A., Dawo F., Jemli M., Melaku S., Shamaki D., Woma Y., Wungak Y., Gebremedhin E.Z., Zutt I., Bosch B.J., Haagmans B.L., Koopmans M.P. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg. Infect. Dis. 2014;20:1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Melaku S.K., Kawachi K., Nao N., Iwata-Yoshikawa N., Kawase M., Kamitani W., Matsuyama S., Tessema T.S., Sentsui H. Middle East respiratory syndrome coronavirus in dromedaries in Ethiopia is Antigenically different from the Middle East isolate EMC. Front. Microbiol. 2019;10:1326. doi: 10.3389/fmicb.2019.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So R.T., Perera R.A., Oladipo J.O., Chu D.K., Kuranga S.A., Chan K.H., Lau E.H., Cheng S.M., Poon L.L., Webby R.J., Peiris M. Lack of serological evidence of Middle East respiratory syndrome coronavirus infection in virus exposed camel abattoir workers in Nigeria, 2016. Euro Surveill. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.32.1800175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Hijazeen Z.S., Holloway P., Al Omari B., McDowell C., Adney D., Talafha H.A., Guitian J., Steel J., Amarin N., Tibbo M., Abu-Basha E., Al-Majali A.M., Munster V.J., Richt J.A. High prevalence of Middle East respiratory coronavirus in young dromedary camels in Jordan. Vector Borne Zoonotic Dis. 2017;17:155–159. doi: 10.1089/vbz.2016.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D., Kinne J., McLellan J.S., Zhu J., Munster V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Middle East respiratory syndrome coronavirus (MERS-CoV) 2019. https://www.who.int/emergencies/mers-cov/en/
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]