Abstract
Porcine epidemic diarrhea virus (PEDV) strains can be divided into non-S-INDEL and S-INDEL strains. PEDV pathogenesis is strain-specific, and studies in neonatal pigs have demonstrated that the PEDV non-S-INDEL strains are more pathogenic than the PEDV S-INDEL strains. RNA viruses, including PEDV, can interact with a large number of pattern recognition receptors (PRRs) in the intestinal mucosa, including toll-like receptors (TLRs) and RIG-I-like receptors (RLRs). We investigated the differential gene modulation of TLRs, RIG-I, and downstream mediators on the intestinal mucosa of neonatal pigs infected with PEDV S-INDEL and non-S-INDEL strains. Ten five-day-old piglets were inoculated orally with 10 ml of 104 TCDI50/ml of either PEDV non-S-INDEL or S-INDEL strains. PEDV S-INDEL infection induced pro-inflammatory cytokines through the non-canonical NF-κB signaling pathway by activating RIG-I. In contrast, PEDV non-S-INDEL infection suppressed the induction of pro-inflammatory cytokines and type 1 interferon production by down-regulation of TLRs and downstream signaling molecules.
Keywords: Porcine epidemic diarrhea virus, Toll-like receptor, RIG-I-like receptors, MyD88, TRAF6, IRF7, NF-κB, Pro-inflamatory cytokines, Type 1 interferon, Canonical pathway
Highlights
-
•
Differential gene modulation of TLR and RIG-I-like receptors and downstream mediators.
-
•
PEDV S-INDEL induces pro-inflammatory cytokines through non-canonical NF-κB signaling pathway.
-
•
PEDV S-INDEL pro-inflammatory cytokines activation is RIG-I dependent.
-
•
PEDV non-S-INDEL suppresses the induction of pro-inflammatory cytokines and type 1 interferon.
-
•
PEDV non-S-INDEL effect is mediated by down-regulation of TLRs and its downstream-signaling molecules.
-
•
PEDV S-INDEL and PEDV non-S-INDEL cause differential modulation on innate immune response pathways.
-
•
Differential modulation could be translated into differences in pathogenesis and clinical outcomes.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) belongs to the order Nidovirales, family Coronaviridae, and genus Alphacoronavirus, and is an enveloped virus with a single-stranded positive RNA genome. It contains four structural proteins-spike (S), membrane (M), nucleocapsid (N), and envelop (E)-responsible for viral infectivity and the induction of immune response (Duarte and Laude, 1994, Duarte et al., 1994, Kocherhans et al., 2001). PEDV causes enteric disease, resulting in significant morbidity and mortality in neonatal pigs, and has been reported as a major source of substantial economic losses in most swine producer countries (Chen et al., 2010, Chen et al., 2013, Chen et al., 2012, Cima, 2014, Song and Park, 2012, Takahashi et al., 1983). In 2014, a less pathogenic PEDV strain was reported in the United States and several other swine producer countries (Chen et al., 2016, Vlasova et al., 2014, Yamamoto et al., 2015). This new strain presented insertions and deletions on the amino terminal region of the S protein. On the basis of differences in the S gene and virulence, emerging PEDV strains can be divided into non-S-INDEL (S gene insertions and deletions) and S-INDEL strains (Vlasova et al., 2014). PEDV pathogenesis is strain-specific, and pathogenesis studies in neonatal pigs have demonstrated that the PEDV non-S-INDEL strain is more pathogenic than the PEDV S-INDEL strain (Chen et al., 2016, Wang et al., 2016, Yamamoto et al., 2015). PEDV pathogenesis is also inversely correlated with the age of the animals. In adult pigs, PEDV infection is self-resolving regardless of previous PEDV immune status. Moreover, PEDV S-INDEL was shown to be clinically relevant in neonates, but clinical disease could not be reproduced in pigs older than three weeks (Annamalai et al., 2015, Chen et al., 2016).
Host pattern recognition receptors (PRRs), such as toll-like receptors (TLRs) and the cytosolic retinoic acid-inducible gene-1 (RIG-1)-like receptors, recognize pathogen-associated molecular patterns (PAMPs) during viral infection (Akira et al., 2006, Alexopoulou et al., 2001b, Takeuchi and Akira, 2010, Uematsu and Akira, 2008). This interaction triggers the interferon regulatory transcription factor (IRF3/7) and activates NF-κB, which modulates the expression of several pro-inflammatory cytokines and chemokines. Type 1 interferon alpha/beta (IFNα/IFNβ) are the two essential cytokines that can control viral infections (Kawai et al., 2005, Seth, 2005, Xu et al., 2005). Within nidoviruses, acute respiratory syndrome coronavirus (SARS-CoV) and Middle-East respiratory syndrome coronavirus (MERS-CoV) can evade the host immune system by interfering with the NF-κB signaling pathway (DeDiego et al., 2014, Matthews et al., 2014), and the infectious bronchitis virus (IBV) inhibits the phosphorylation of kinases that are necessary to activate downstream signaling cascades (Chen et al., 2013, Devaraj et al., 2007, Kint et al., 2015a, Sun et al., 2012, Zhou, 2007).
Two signaling pathways, known as the classical (canonical) pathway and the alternative (non-canonical) pathway (Kawai and Akira, 2010, Loo and Gale, 2011), lead to the activation of NF-κB. The canonical pathway includes the recruitment of the myeloid differentiation primary response gene 88 (MyD88), containing the toll/interleukin-1 receptor (TIR) domain for eventual activation of NF-κB and induction of type I interferons (Kawai and Akira, 2007, Kawai and Akira, 2010, Thompson and Locarnini, 2007). In addition, endosomal receptor TLR3 contains exclusively TIR-domain-containing adapter-inducing interferon-β (TRIF) adapter proteins that interact with TRAF6, which induces IRF3/7 similar to the MyD88 pathway (Yamamoto et al., 2003, Zhengfan, 2004;). Finally, both the MyD88 and TRIF pathways activate NF-κB and induce expression of the antiviral type I interferons (Thompson and Locarnini, 2007). The non-canonical pathway is TLR-independent, and NF-κB can be modulated by RIG-I-TRAF3 mediated through IRF3 (Devaraj et al., 2007).
Several in vitro studies have tried to elucidate the role of PEDV in innate immune response at the cellular level. It has been demonstrated that the nucleocapsid (N) protein of PEDV, during infection of HEK-293T cells, inhibited IFN-β production by annexing the vital interaction between IRF3 and TBK1 (Ding et al., 2014b). Other in vitro studies in porcine intestinal epithelial cells (IECs) determined that PEDV infection impeded the production of IFN-β by inhibiting the RIG-I pathway and hampering the activation of IRF3 (Cao et al., 2015a). Studies in Vero cells showed that PEDV infection degraded STAT-1 and disrupted the IFN response (Li et al., 2016). Hence, it has been shown that PEDV can regulate different immunological pathways in vitro; the main regulatory effect of PEDV on mucosal innate immunity and its strain-dependence on viral pathogenicity is still unknown.
The aim of this study was to investigate the differential gene modulation of pattern recognition TLR and RIG-I-like receptors and downstream mediators on the intestinal mucosa of neonatal pigs infected with PEDV non-S-INDEL and PEDV S-INDEL strains.
2. Materials and methods
2.1. Animal study
Thirty 5-days-old conventional piglets were selected for the molecular evaluation of the gene modulation of pattern recognition TLR and RIG-I-like receptors and downstream mediators on intestinal mucosa. The experimental design as well as information of the virus strains used in this study, kinetic of virus shedding, virus distribution in tissues, and pathogenicity was previously described in a PEDV clinical pathogenesis study (Chen et al., 2016). In brief, pigs were injected intramuscularly with a dose of Excede (Zoetis, Kalamazoo, MI) at time of delivery to Iowa State University, Laboratory Animal Resources facilities (Ames, IA). All pigs were confirmed negative for PEDV, PDCoV, TGEV, and porcine rotaviruses (groups A, B, and C) by virus-specific PCRs on rectal swabs, and seronegatives by PEDV indirect immunofluorescent assay. After one day of acclimation, six-day-old pigs were inoculated orogastrically with 10 ml of 104 TCID50/ ml of PEDV non-S-INDEL (USA/IN19338/2013) or PEDV S-INDEL (USA/IL20697/2014), or 10 ml of virus-negative culture medium. Five pigs from each group were euthanized at three and seven days post-inoculation (dpi), respectively. Sections of distal small intestine of approximately 0.5 cm in length were snap frozen and saved at −80 °C. Frozen intestine tissue samples were aliquoted, placed in an RNAlater™ Stabilization Solution (Life Technologies, Carlsbad, CA), and kept at −80 °C until further use.
2.2. RNA extraction from porcine intestine tissue
RNA was extracted from 5 mg of porcine intestinal tissue using the Ambion® MagMAX™ total RNA isolation kit (Life Technologies) and a Kingfisher® 96 magnetic particle processor (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's instructions. RNA was eluted into 70 µL of elution buffer and stored at −80 °C.
2.3. Expression of mRNA for TLRs, inflammatory signaling pathways, and cytokines on porcine intestinal mucosa
The relative quantification of gene expression of toll-like receptors TLR2, TLR3, TLR4, TLR7, TLR8, and TLR9 was evaluated on total RNA extracted from porcine intestinal tissues. Gene expression of inflammatory signaling pathway molecules, including RIG-1, TRIF, MyD88A, MyD88B, IRF7, TRAF6, NF-κB1 (p105) NF-κB1 (p50), and RelA (p65), was also conducted by SYBR-green RT- PCR on the RNA extracted from intestinal pig mucosa. Gene expression of cytokines IFN-α, IL-6, IL-12, and TNF-α in intestinal mucosa was also investigated following the same protocol described herein for the rest of the genes. All reactions were performed in triplicate and the relative gene expression of each target gene was evaluated in reference to the expression of housekeeping genes GAPDH and beta-actin. All primers were custom-synthesized (Integrated DNA Technologies, Inc., Coralville, IA) to target amplicons, with sizes ranging between 95 and 120 nt base pairs according to the cDNA sequence of each target gene, collected from the National Center for Biotechnology Information (NCBI) database ( Table 1).
Table 1.
Primers used for real-time PCR analysis of genes expression of pattern-recognition receptor TLR and RIG-I-like and downstream mediators on pig intestinal mucosa.
| Gene Name | Primer Name | Primer Sequence (5’−3”) | Amplicon Length | Accession number |
|---|---|---|---|---|
| GAPDH | Por-GAPDH-40F | GGAAAGGCCATCACCATCTT | 85 bp | XM_021091114.1 |
| Por-GAPDH−125R | CATGGTCGTGAAGACACCAG | |||
| Β-Actin | Por-βActin−598 F | CCCAGCACCATGAAGATCAA | 88 bp | XM_005670976.2 |
| Por-βActin−686 R | GATCCACATCTGCTGGAAGG | |||
| TLR2 | Por-TLR2-1601F | GAGTCTGCCACAACTCAAAGA | 68 bp | XM_005653576.3 |
| Por-TLR2–1669R | CAGAACTGACAACATGGGTAGAA | |||
| TLR3 | Por-TLR3-1658F | GCGGTCCTGTTCAGTTTCT | 72 bp | KT735340.1 |
| Por-TLR3–1730R | AAGGCATCTGCTGGGATTT | |||
| TLR4 | Por-TLR4-263F | AACTGCAGGTGCTGGATTTAT | 74 bp | AB078418.1 |
| Por-TLR4–337R | CCGTCAGTATCAAGGTGGAAAG | |||
| TLR7 | Por-TLR7-1376F | CCCAGGTCCTCGAATCATTAC | 77 bp | DQ647699.2 |
| Por-TLR7–1453R | CATTAAGAGGCAAGGAGGAAGA | |||
| TLR8 | Por-TLR8-1980F | CTTTGATGATGACGCTGCTTTC | 77 bp | KF019635.1 |
| Por-TLR8–2057R | GGTGTGTCACTCCTGCTATTC | |||
| TLR9 | Por-TLR9-597F | CCTCACACATCTCTCACTCAAG | 82 bp | KC860785.1 |
| Por-TLR9–679R | GGTGACAATGTGGTTGTAGGA | |||
| RIG-I | Por_RIG-I_1922F | GAGCCCTTGTGGATGCTTTA | 91 bp | KC011279.1 |
| Por_ RIG-I_2013R | GGGTCATCCCTATGTTCTGATTC | |||
| TRIF | Por_TRIF_1027F | CTCCGGTGCAGTCAAACA | 91 bp | KC969185.1 |
| Por_TRIF_1118R | GGTAGTGTGTGCTGGTTTCT | |||
| MyD88A | Por_MyD88_1852F | GGCAGCTGGAACAGACCAA | 41 bp | EU056736.1 |
| Por_ MyD88_1893R | GGCAGGACATCTCGGTCAGA | |||
| MyD88B | Por_MyD88_2501F | TGCAGGTGCCCATCAGAAG | 64 bp | EU056737.1 |
| Por_ MyD88_2565R | TGATGAACCGCAGGATGCT | |||
| NF-ƙB1 (p105) | Por_ NF-ƙB1_1387F | GAGGTGCATCTGACGTATTC | 118 bp | NM_001048232.1 |
| Por_ NF-ƙB1_1505R | CACATCTCCTGTCACTGCAT | |||
| NF-ƙB1 (p50) | Por_ NF-ƙB1_1249F | AAGCACGGAACTGTAGACAC | 107 bp | KC316024.1 |
| Por_NF-ƙB1_1355R | TCTGTGGTTTCTGTGACTTTCC | |||
| RELA (p65) | Por_RELA_1616F | ACATGGACTTCTCAGCCCTTCTGA | 285 bp | CN155798.1 |
| Por_RELA_1331R | CCGAAGACATCACCCAAAGATGCT | |||
| IRF7 | Por_IRF−7_1193F | CACTACACAGAGAAGCTGCT | 91 bp | HQ026022.1 |
| Por_IRF−7_1284R | ACCTCCCAGTAGACTTTGC | |||
| TRAF6 | Por_TRAF−6_1338F | GGGAACGATACGCCTTACAA | 156 bp | NM_001105286.1 |
| Por_TRAF−6_1494R | CTCTGTCTTAGGGCGTCC | |||
| IL−6 | Por_IL−6_260F | CAAGGAGGTACTGGCAGAAA | 164 bp | JQ839263.1 |
| Por_IL−6_424F | CAGCCTCGACATTTCCCTTAT | |||
| IL12-p35 | Por_IL12-p35_200F | CAGGCCCAGGAATGTTCAAA | 166 bp | NM_213993.1 |
| Por_IL12-p35_366R | CGTGGCTAGTTCAAGTGGTAAG | |||
| TNF-α | Por_TNFα_157F | CCTACTGCACTTCGAGGTTATC | 158 bp | JF831365.1 |
| Por_TNFα_315R | GCATACCCACTCTGCCATT | |||
| IFN-α | Por_IFNα_372F | TTCTGCACTGGACTGGATC | 103 bp | KF414740.1 |
| Por_IFNα_475R | TCTGTGGAAGTATTTCCTCACAG |
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH); Β-Actin; Toll-like receptor (TLR); retinoic acid-inducible gene-1 (RIG-1)-like receptor; TIR-domain-containing adapter-inducing interferon-β (TRIF); myeloid differentiation primary response gene 88 (MyD88); nuclear factor (NF)-κB; nuclear factor NF-kappa-B p65 subunit (RELA); interferon regulatory transcription factor (IRF); TNF receptor associated factor (TRAF); interleukin (IL); tumor necrosis factor (TNF); Interferons (IFN).
The mRNA expression levels were quantified according to the ΔΔCt method (Livak and Schmittgen, 2001). Briefly, the difference in cycle times, ΔCt, was determined as the difference between the tested gene and the reference housekeeping genes. The ΔΔCt was obtained by finding the difference between groups. The fold change was calculated as 2-∆∆Ct. Real-time RT-PCR was performed with eluted RNA and primers, mixed with the commercial Power SYBR Green RNA-to-CT™ 1-Step Kit (Applied Biosystems, Foster City, CA), following the manufacturer's recommendations. The reverse transcription reaction was conducted at standard mode for 30 min at 48 °C, enzyme activation was conducted at 95 °C for 10 min using an Applied Biosystems™ 7500 real-time PCR instrument. The strands were denatured at 95 °C for 15 s, then annealed and extended at 60 °C for 1 min (40 cycles). A dissociation curve was obtained for each quantitative PCR run to assess its target specificity. The real-time RT-PCR was analyzed with a threshold fixed at a 0.1 setting. Cycle threshold (CT) values ≤ 35 were considered positive for the housekeeping and TLR gene expressions. All samples were tested in triplicate and the results were expressed as fold changes relative to the control animals.
2.4. Statistical analysis
Data were analyzed for normality by Kolmogorov-Smirnov test. The statistical significance between the two treatment groups was determined by non-parametric statistical analysis using the Mann-Whitney test. Significance was assessed at p < 0.05. Data analysis was performed using GraphPad Prism® (GraphPad Software Inc., La Jolla, CA).
3. Results
3.1. PEDV non S-INDEL and S-INDEL infection showed differential modulation on PRRs genes in intestinal mucosa
RNA viruses, including PEDV, can interact with a large number of pattern recognition receptors (PRRs) in the intestinal mucosa, such us toll-like receptors (TLRs) and RIG-I-like receptors (RLRs). This interaction plays a critical role in the activation of the innate immune response. TLRs are normally classified based on their anatomical location in membranes or cytoplasmic receptors. Numerous TLRs are expressed in porcine enterocytes with different functions in molecule recognition. In addition, other intracytoplasmic PRRs, such as retinoic acid-inducible gene (RIG)-I-like receptors (RLRs) can sense and recognize double-stranded or single-stranded RNA from a variety of pathogens. In this study, we evaluated gene modulatory effects on a group of PRRs, including TLRs and RLRs, in the intestinal mucosa during early infection by PEDV non-S-INDEL and S-INDEL strains.
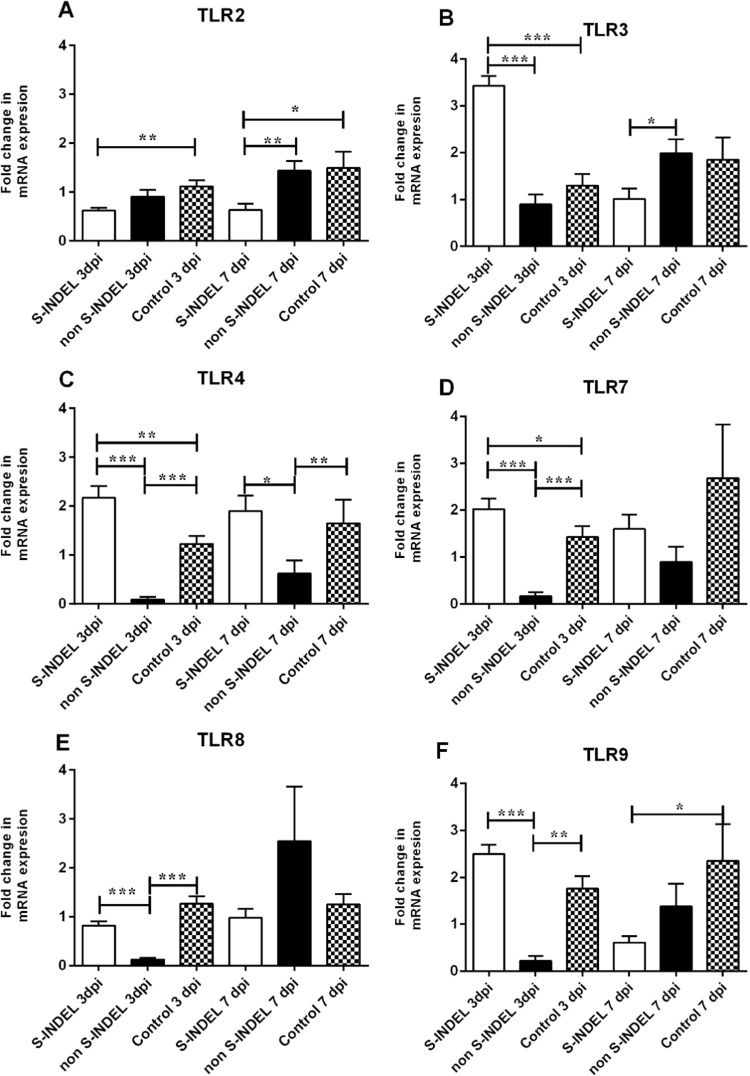
Pigs infected with PEDV S-INDEL showed a significant up-regulatory effect (p < 0.05) on TLR3, TLR4, and TLR7 gene expression by 3 dpi compared to non-infected control pigs and pigs infected with the PEDV non-S-INDEL strain ( Fig. 1B–D). The regulatory effect observed in the PEDV S-INDEL-infected group was transient, returning to basal control levels by 7 dpi. PEDV S-INDEL infection had a significant down-regulatory effect (p < 0.05) on TLR2 gene expression at 3 and 7 dpi (Fig. 1A). However, PEDV S-INDEL infection did not affect gene modulation of TLR8 and TLR9 compared to the control group (Fig. 1E, F). In contrast, infection with PEDV non-S-INDEL was shown to have a down-regulatory effect on the expression of TLR4, TLR7, TLR8, and TLR9 (p < 0.05) compared to the non-infected control and PEDV S-INDEL-infected groups consistently at 3dpi (Fig. 1C–F). However, no statistically significant differences in TLR2 and TLR3 gene modulation levels (p > 0.05) were observed between the PEDV non-S-INDEL and control groups at either 3 or 7 dpi (Fig. 1A, B).
Fig. 1.
Changes in Toll Like receptor (TLRs) mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa (A-F). Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of TLR2 (A), TLR 3 (B), TLR4 (C), TLR7 (D), TLR8 (E), and TLR9 (F) at the intestinal mucosa was determined individually in each animal by SYBR- green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
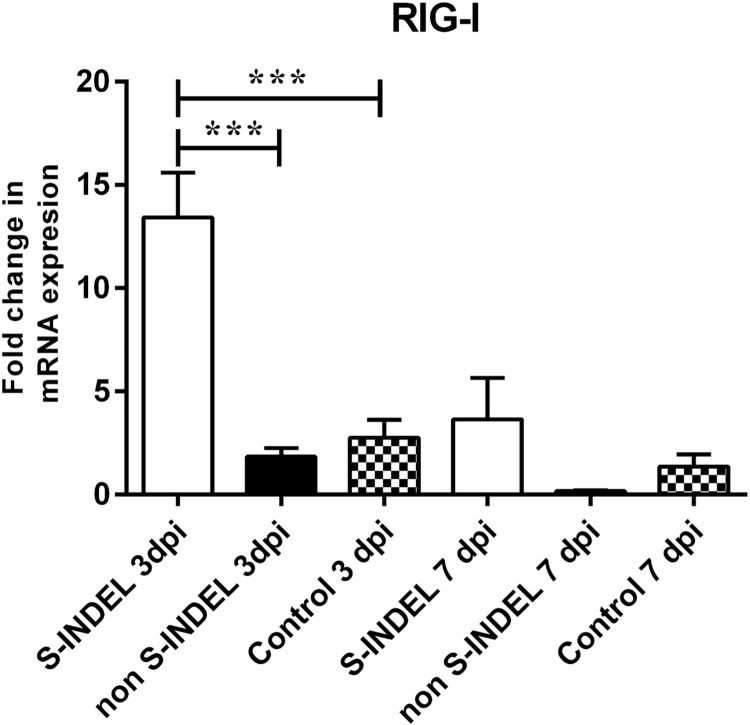
The modulation of RIG-I was only affected in response to PEDV S-INDEL infection at 3 dpi, with a significant increment in gene expression compared to the control and non-S-INDEL groups (p < 0.05). However, this modulatory effect was transient, with expression levels returning to the same levels observed for the control and PEDV non-S-INDEL groups by 7 dpi ( Fig. 2). Fig. 7 summarize the differential modulatory effect on PRRs genes induced by PEDV S-INDEL vs non S-INDEL at dpi 3.
Fig. 2.
Changes in cytosolic retinoic acid-inducible gene-1 (RIG-I) gene mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa. Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of RIG-I at the intestinal mucosa was determined individually in each animal by SYBR-green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
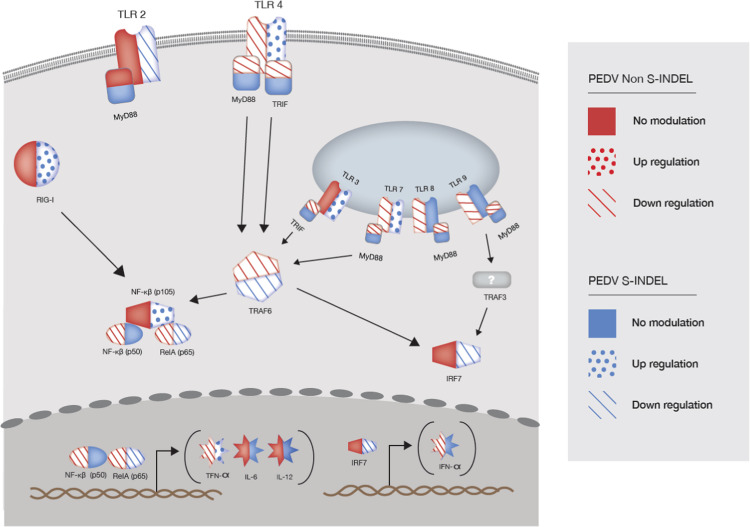
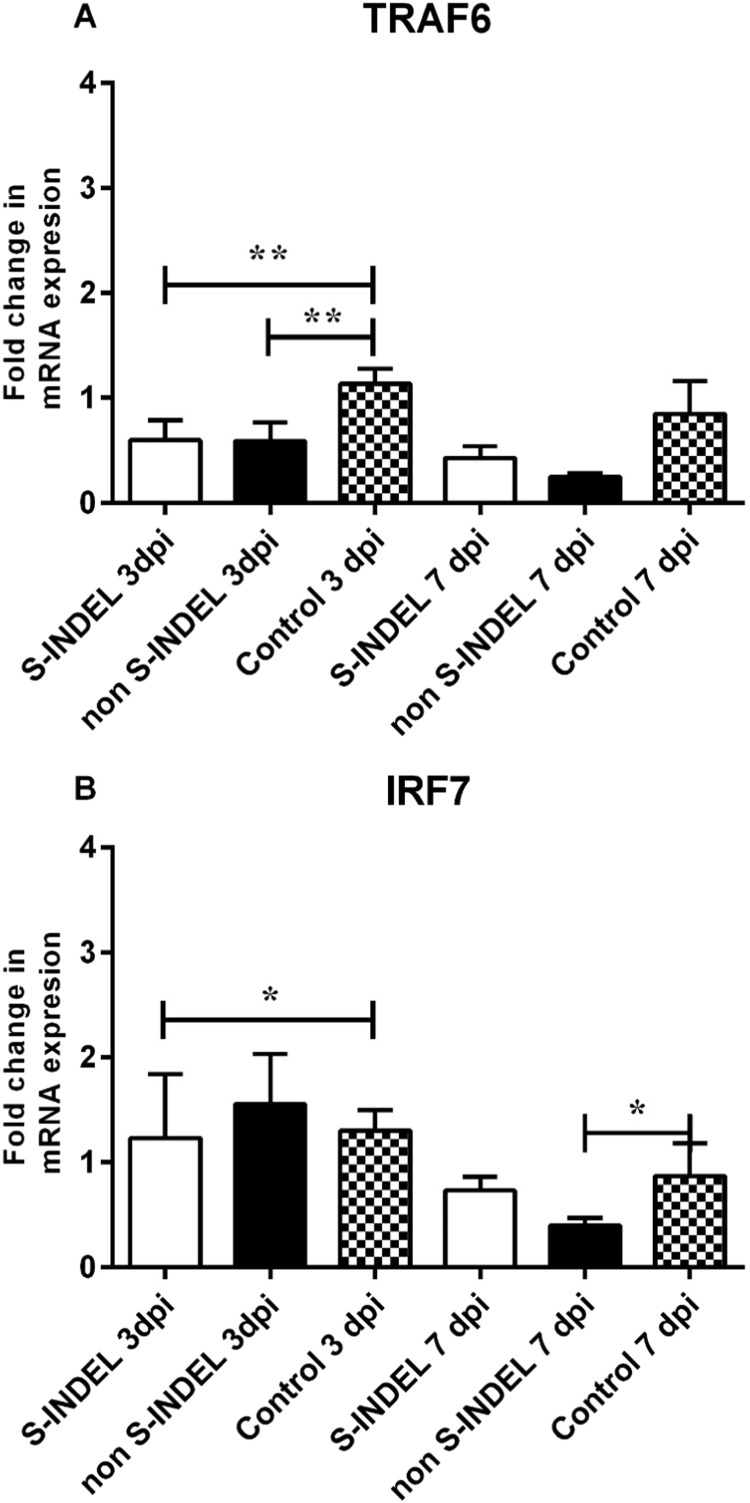
Fig. 7.
Differential gene modulation of pattern-recognition receptor TLR and RIG-I-like, and downstream mediators on intestinal mucosa of pigs infected with PEDV non S-INDEL and PEDV S-INDEL strains. This figure present differential gene modulation at day post-infection (dpi) 3. PEDV non-S-INDEL infection suppressed the induction of the pro-inflammatory cytokine tumor necrosis factor alpha (TNF-α), and type 1 interferon production (IFN- α) through the down regulation of the cytoplasmic membrane and endosomal TLRs (TLR4, TLR7/8, TLR9), and TLR-downstream signaling molecules (MyD88/TRIF and TRAF6). Although the expression levels of both p50 and p65 were down-regulated after infection with the PEDV non-S-INDEL strain, the down-regulatory effect on MyD88 and TRIF gene pathways did not negatively affect the expression of NF-κB ( ). Contrary, PEDV S-INDEL infection induced a positive modulatory effect on TLR3, TLR4, and TLR7 gene expression. However, no significant modulatory effect was observed in the levels of TRIF, and MyD88, genes. PEDV S-INDEL infection induced the pro-inflammatory cytokines TNF-α, and interleukin (IL)−12 through the non-canonical NF-κB signaling pathway by the activation of the intracytoplasmic RIG-I receptor (
). Contrary, PEDV S-INDEL infection induced a positive modulatory effect on TLR3, TLR4, and TLR7 gene expression. However, no significant modulatory effect was observed in the levels of TRIF, and MyD88, genes. PEDV S-INDEL infection induced the pro-inflammatory cytokines TNF-α, and interleukin (IL)−12 through the non-canonical NF-κB signaling pathway by the activation of the intracytoplasmic RIG-I receptor ( ).
).
3.2. Differential modulation of NF-κB through TLR downstream adapters is dependent on PEDV strain
PRRs of the innate immune system initiate signal transduction cascades in response to ligation by microbial-associated molecular patterns (MAMPs), which leads to the transcriptional modulation of downstream signaling molecules. With the exception of TLR3, the myeloid differentiation primary response gene 88 (MyD88) product is the most generally utilized TLR adapter, either by direct interaction (TLR5, 7–9) or via an intermediary interaction (TLR4) with the TIR domain-containing adapter protein (TIRAP)/MAL. Engagement of MyD88 leads to recruitment and assembly of the cytoplasmic IL-1 receptor-associated kinases (IRAK) and the TNF receptor-associated factor (TRAF) 6 to form the IRAK–TRAF6 complex, which leads to the activation of the nuclear factor (NF)-κB pathway. The NF-κB transcriptional factor family is composed of NF-κB1 (p105), NF-κB1 (p50) and RelA (p65), subunits that translocate into nuclei and regulate NF-κB. TRAM and TRIF mediate a signal transduction cascade downstream of both TLR3 and TLR4 (Kawai and Akira, 2010, Thompson and Locarnini, 2007). The MyD88- and TRIF-dependent pathways lead to the activation of interferon regulatory factors (IRFs) and the secretion of type-I interferon (IFN) and pro-inflammatory cytokines by activated NF-κB signaling.
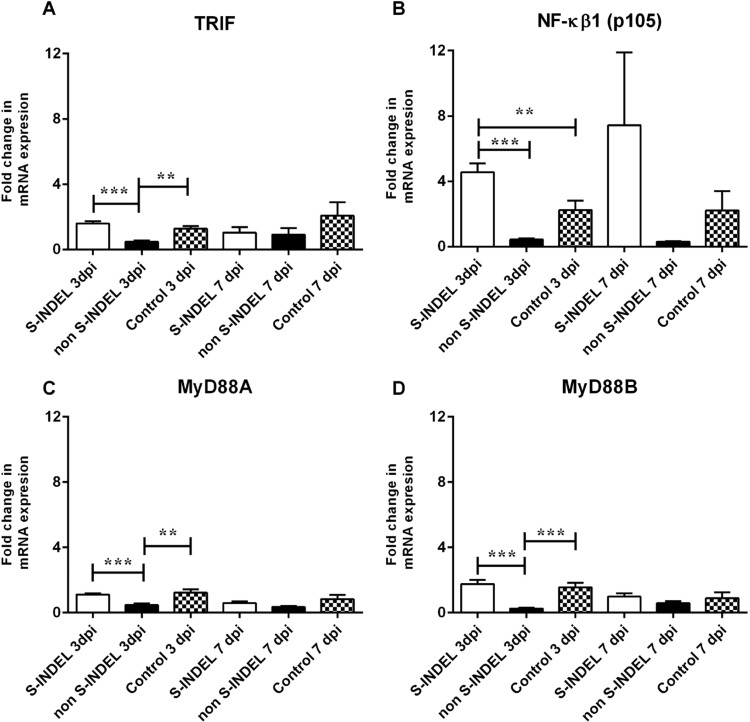
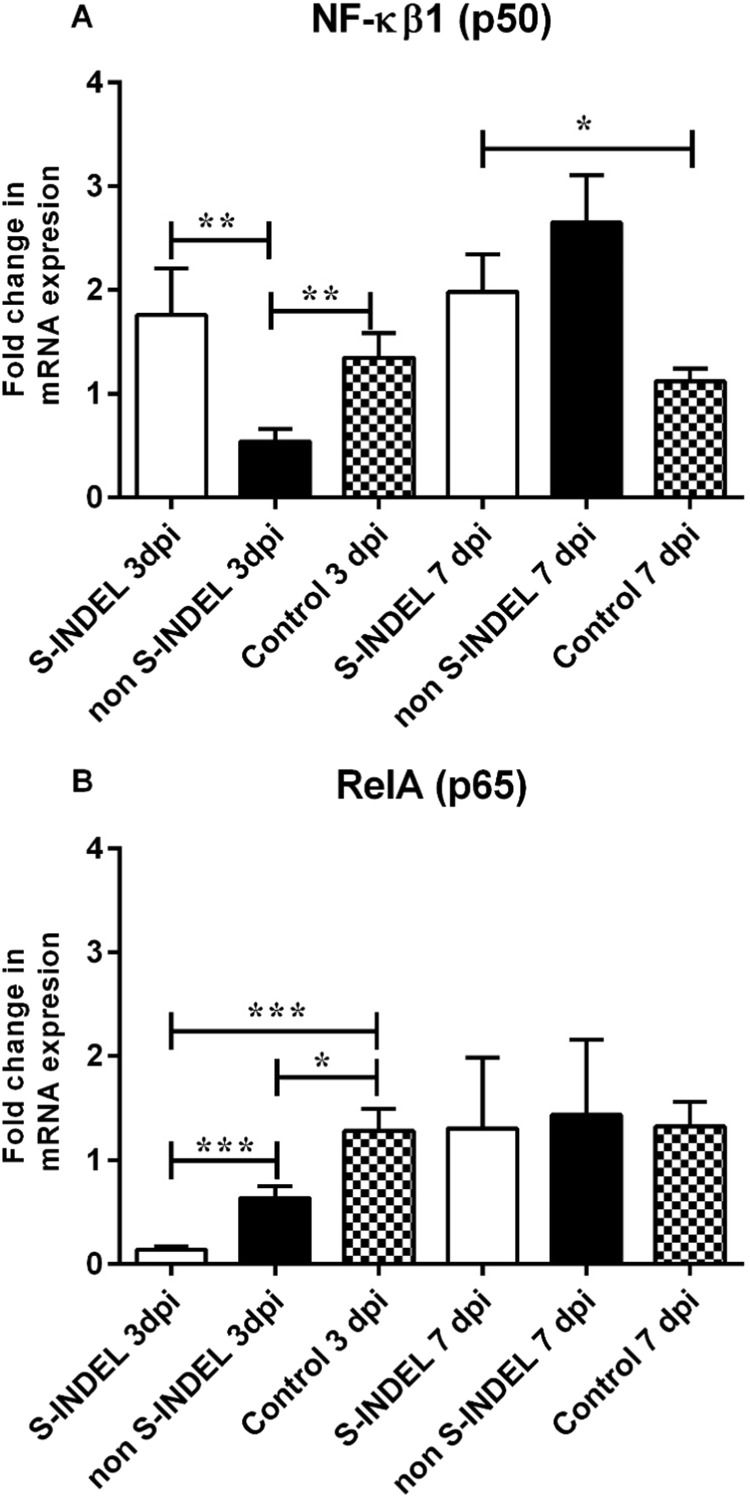
Despite the PEDV S-INDEL strain inducing a positive modulatory effect on TLR3, TLR4, and TLR7 gene expression, no significant differences (p > 0.05) were observed in gene expression levels of TRIF, MyD88 (subunits A and B) (Fig. 3A, C, D). Moreover, the PEDV S-INDEL strain appears to have a down-regulatory effect on TRAF6 and IRF7 genes at 3 dpi (p < 0.05) (Fig. 4A, B). PEDV S-INDEL-infected animals showed a differential gene modulation characterized by a significant up-regulatory effect in expression levels of NF-κB1 and (p50) genes (p < 0.05) at 7 dpi, and a down-modulatory effect on the expression of the RelA/p65 gene compared to both the control and PEDV non-S-INDEL-infected group at 3 and 7 dpi (Fig. 5A, B).
Fig. 3.
Changes in Toll/Interleukin-1 receptor (TIR), myeloid differentiation primary response gene 88 (MyD88), and Nuclear factor (NF)-κB genes mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa (A-D). Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of TRIF (A), NF-κB (B), MyD88A (C), and MyD88B (D) at the intestinal mucosa was determined individually in each animal by SYBR- green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
Fig. 4.
Changes in TNF receptor associated factor (TRAF) 6 and interferon regulatory factor 7 (IRF7) genes mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa (A-B). Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of TRAF 6 (A), and IRF7 (B) at the intestinal mucosa was determined individually in each animal by SYBR-green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
Fig. 5.
Changes in the nuclear translocator NF-κB transcriptional factor family, NF-κB1 (p50) and RelA (p65) genes mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa (A-B). Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of NF-κB1 (p50) (A), and RelA (p65) (B) at the intestinal mucosa was determined individually in each animal by SYBR-green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
Contrary to the PEDV S-INDEL strain, a down-regulatory effect of TLR4, TLR7, TLR8 and TLR9 genes was observed in response to infection with the PEDV non-S-INDEL strain (Fig. 1), which also negatively affected the gene expression of downstream signaling molecules (p < 0.05), including TRIF, MyD88 (subunits A and B), and TRAF6 (Figs. 3A, C, D, 4A). Although the expression levels of both p50 and p65 were down-regulated after infection with the PEDV non-S-INDEL strain (Fig. 5), the down-regulatory effect on MyD88 and TRIF gene pathways did not negatively affect the expression of NF-κB (p105) (Fig. 3B). Fig. 7 summarize the differential modulatory effect of NF-κB through TLR downstream adapters induced by both PEDV strains at dpi 3.
3.3. Mucosal pro-inflammatory cytokine gene regulation is affected by the PEDV S-INDEL but not the PEDV non-S-INDEL strain
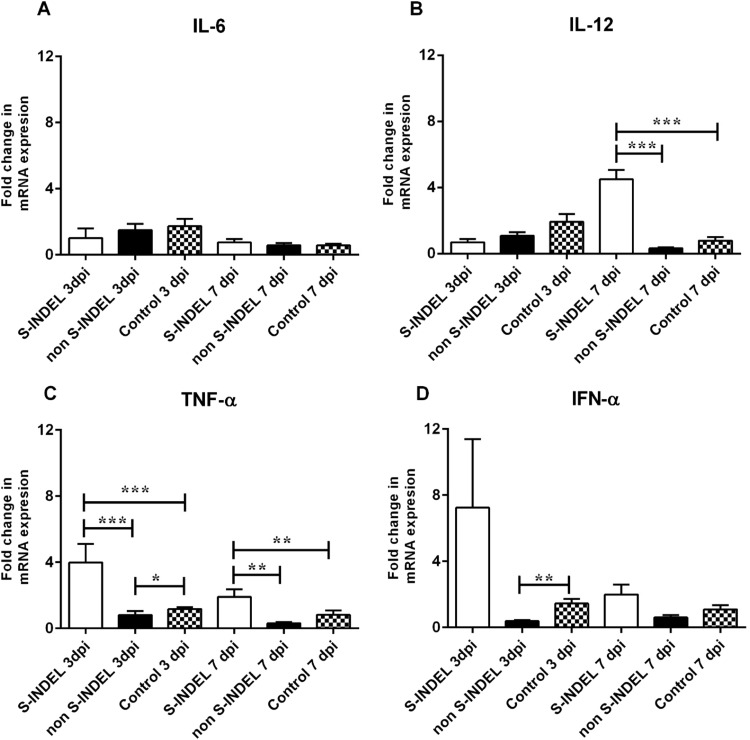
Pro-inflammatory cytokines and type I interferons (IFNs) are produced at the local intestinal mucosal level as part of the innate immune response during the infection process. The modulatory effect of pro-inflammatory interleukins and IFN genes is the result of a long-elaborated pathway that includes the activation of PRRs and downstream mediators, including MyD88 and TRIF, via NF-κB activation. We evaluated whether the differential modulatory effect observed in PRRs and downstream mediators in response to infection with the PEDV S-INDEL versus the non-S-INDEL strain was also translated into a differential modulation in the expression of gene coding for pro-inflammatory cytokines and type I interferons. Neither infection with PEDV S-INDEL nor non-S-INDEL was demonstrated to have a modulatory effect on local production of IL-6 (Fig. 6A). The local expression of IL-12 was not affected at 3 dpi regardless of the PEDV strain. However, a positive modulatory effect on IL-12 gene expression was exerted by PEDV S-INDEL at 7 dpi (Fig. 6B). The PEDV non S-INDEL strain showed a transient negative modulatory effect of the TNF-α gene at 3 dpi, returning to basal control levels by 7 dpi. However, the PEDV S-INDEL strain consistently up-regulated the expression of the TNF-α gene during the study (Fig. 6C). The expression of the IFN-α gene was negatively affected only in response to infection with the PEDV non-S-INDEL strain at 3 dpi (p < 0.05) (Fig. 6D). Fig. 7 summarize the effect of pro-inflammatory cytokine gene regulation by PEDV S-INDEL and PEDV non-S-INDEL strain at dpi 3.
Fig. 6.
Changes in proinflammatory cytokines IL-6, IL-12, and TNF-α, and IFN-α genes mRNA expression induced by porcine epidemic diarrhea virus (PEDV) non S-INDEL and PEDV S-INDEL strains in intestinal mucosa (A-D). Ten pigs in each group were infected with PEDV non S-INDEL, PEDV S-INDEL or media (negative control) and five pigs from each group were necropsied at 3 and 7 days post-infection (dpi). The mRNA levels of IL-6 (A), IL-12 (B), TNF-α (C), and IFN-α (D) at the intestinal mucosa was determined individually in each animal by SYBR-green qRT-PCR. All samples were tested in triplicate and the results are expressed as fold changes relative to the control animals Data are presented as means ± standard errors. Significant difference between PEDV S-INDEL PEDV non S-INDEL and control group are expressed with their p values. *p < 0.05; **p < 0.01, ***p < 0.001.
4. Discussion
Pathogen recognition by TLRs and RLRs activates the innate immune response through signaling pathways, resulting in the production of pro-inflammatory cytokines, type I interferons, and chemokines. Intestinal mucosa are composed of a variety of specialized cells that play specific functions during the disease process (Kawai and Akira, 2006, Kumar et al., 2009). Collectively, intestinal epithelium, dendritic cells, M cells, immune cells in the lamina propria, and peyer patches (e.g., lymphocytes and macrophages) play a role in the intestinal mucosa immune response against pathogens. PRRs are constitutively expressed in all cell populations referred to above and may have differential roles in each cell type and component of the intestinal mucosa. Initial PRR-induced responses are critical in controlling infectious agents, but are also tightly regulated through time-, location-, and cell type-dependent mechanisms. Positive and negative modulation interaction of signaling pathways is the main mechanism for maintaining inmate immune homeostasis. Interferon production, especially type 1 interferon and anti-viral cytokines, is important for host protection against viral invasion. Many viruses can evade the host immune system by regulating signaling pathways, resulting in the blockage of cytokine production (McCartney and Colonna, 2009).
Porcine epidemic diarrhea virus (PEDV) causes enteric diseases, resulting in significant economic losses. PEDV pathogenesis is strain-specific, and pathogenesis studies in neonatal pigs have demonstrated that the PEDV non-S-INDEL strain is more pathogenic than the PEDV S-INDEL strain (Chen et al., 2016, Wang et al., 2016, Yamamoto et al., 2015). PEDV S-INDEL was shown to be clinically relevant in neonates, but clinical disease could not be reproduced in pigs older than three weeks (Annamalai et al., 2015, Chen et al., 2016). The molecular mechanism of innate immune modulation has only been evaluated in vitro, and thus available information is scarce (Cao et al., 2015a, Cao et al., 2015b, Ding et al., 2014a, Gao et al., 2015, Xu et al., 2013). However, it has been demonstrated that in vitro studies are highly dependent on virus strain and cell type, and are not valid for the evaluation of innate immune response against low-virulent strains (Kint et al., 2015b). Therefore, in this study, we evaluated the differential modulation of intestinal mucosa PRR signaling mounted against PEDV non-S-IDEL and S-INDEL in piglets from PEDV-naïve sows. The use of an in vivo model further allowed the observation of the natural TLRs and signaling pathway homeostasis regulated by the intestinal microbiota.
There is limited information about the role of antiviral innate immunity in the pathogenesis of PEDV infection. However, it is well known that viral infections are usually detected by cell membranes and endosomal-associated TLRs (e.g., TLR4 and TLR3, TLR7/8) and cytosolic RIG-I-like receptors (RLRs), such as RIG-I and MDA5. Activated TLR and RIG-I/MDA-5 signaling pathways initiate effective antiviral innate immune responses, in particular inducing type I INFs (Thompson and Locarnini, 2007). Previous in vitro studies demonstrated that PEDV strain CV777 infection of intestinal epithelial cells (IECs) modulates the NF-κB signaling pathway through up-regulation of TLR2, TLR3, and TLR9, but not RIG-I (Cao et al., 2015a). However, our results showed that in vivo, PEDV S-INDEL infection up-regulated the NF-κB signaling pathway through TLR3, TLR4, TLR7, and RIG-I, resulting in increased expression levels of TNF-α. Although no statistical significance was found, increased levels of IFN-α gene expression were also observed. Interestingly, both PEDV CV777 and PEDV S-INDEL strains belong to genogroup 1, and both are associated with mild clinical disease. Therefore, the differential modulation observed in the NF-κB signaling pathway could be more associated with in vivo-related conditions than genotypic characteristics of the virus strain used in this study. Conversely, PEDV non-S-INDEL down-regulated the NF-κB signaling pathway through a negative modulatory effect of TLR4, TLR7, TLR8, and TLR9, resulting in final attenuation of pro-inflammatory TNF-α and IFN-α gene expression. Moreover, RIG-I gene modulation after PEDV non-S-INDEL remained unaffected. In a previous study, it was demonstrated that PEDV non-S-INDEL had a significantly higher replication rate compared to the less virulent PEDV S-INDEL strain (Chen et al., 2016). Our study demonstrated that the ability of PEDV to induce type I IFNs is strain-specific. Thus, type I IFNs may play an important role in PEDV replication and pathogenesis.
The signaling pathway during ligand binding of single-stranded and double-stranded viral RNA involves the cytoplasmic membrane TLR4 receptor and endosomal TLR7/8 and TLR-9, all of which are used as signaling pathway mediators, (MyD88 and TRIF) for eventual activation of NF-kB (Kawai and Akira, 2007, Kawai and Akira, 2010, Thompson and Locarnini, 2007). However, endosomal receptor TLR3 (Alexopoulou et al., 2001a) contains exclusively TRIF-adapter proteins that interact with TRAF6, which induces phosphorylation of IRF3/7 similar to the MyD88 pathway (Yamamoto et al., 2003, Zhengfan, 2004). Finally, both MyD88 and TRIF pathways activate NF-kB and induce expression of the antiviral type I interferons (Thompson and Locarnini, 2007). In this study, PEDV non-S-INDEL infection had a negative gene modulation on membranes and endosomal TLRs. This negative modulatory effect was translated into a down-regulation of cytoplasmic mediator MyD88 and TRIF genes. In vitro studies demonstrated that silencing TRIF and MyD88 but not RIG-I inhibited PEDV-induced NF-kB activation, suggesting that the TLR signaling pathway is involved in PEDV-induced NF-kB activation (Cao et al., 2015b). In addition, MyD88 is a required component of the innate immune response to mouse-adapted SARS-CoV infection in vivo (Totura et al., 2015). Although PEDV S-INDEL induces NF-kB activation, TLR pathway mediators TRIF and MyD88 were not significantly affected by the positive modulation of the cytoplasmic membrane and endosomal TLR genes in response to infection with PEDV S-INDEL. These contradictory results suggest that TRIF- and MyD88-independent pathways might be involved in NF-κB activation after PEDV S-INDEL infection.
TRAF6 is crucial for both RIG-I- and TLR-mediated antiviral responses. The absence of TRAF6 resulted in enhanced viral replication and a significant reduction in the production of type I IFNs after infection with the RNA virus (Konno et al., 2009). Activation of NF-kB and IRF7, but not IRF3, is normally TRAF6-mediated. In this study, we observed that both PEDV S-INDEL and non-S-INDEL showed a negative modulatory effect on TRAF6, which was translated into a significant down-regulation of the IRF7 gene only in PEDV S-INDEL-infected animals. TRAF6 induced activation of IRF7, while TRAF3 is thought to activate both IRF3 and IRF7 (Konno et al., 2009). However, due to the differential modulation of IRF7 observed in this study, the role of the TRAF6-dependent pathway cannot be fully elucidated, and perhaps IRF3 plays a more important role in type I IFN production in PEDV infection. The role of TRAF3 was not explored in this study but may likely play a role in IRF7 and IFN-α gene modulation. In addition, the down-regulatory effect observed in TRAF6 gene expression for both PEDV strains did not have the same modulatory effect on NF-kB gene expression. This study showed that NF-kB gene expression was PEDV strain-dependent. Although it may need further confirmation, the up-regulation of NF-kB gene expression observed in response to PEDV S-INDEL infection could be linked to the up-regulation of RIG-I gene expression induced after infection with the PEDV S-INDEL strain.
Signaling pathways (canonical and non-canonical) lead to the activation of NF-κB (Kawai and Akira, 2010, Loo and Gale, 2011). Modulation of NF-kB family members (hetero-dimer p50-p65) is affected through th TLRs, which unbound the inhibitory IkB proteins and allowed activation of the NF-kB canonical pathway (Wietek and O’Neill, 2007). We observed that PEDV non-S-INDEL exerted a down-regulatory effect on the p50-p65 hetero-dimer. The result observed during PEDV non-S-INDEL infection is in agreement with a previous in vitro study that demonstrated that PEDV-encoded nucleocapsid (N) protein can impede pro-inflammatory cytokine and type I interferon production by direct interaction with TANK-binding kinase 1 (TBK1), resulting in inhibition of transcription factors, such as IRF3 and IRF7, causing final NF-κB interference (Ding et al., 2014b). Moreover, it has been reported in vitro that PEDV non S-INDEL nsp1 protein induced NF-κB suppression (Zhang et al., 2017). Inhibition of the p50-p65 hetero-dimer blocks activation of the NF-κB canonical pathway and inhibits early TNF-α response (Wietek and O’Neill, 2007). Our results demonstrate that PEDV non-S-INDEL could use this strategy to evade the host immune system in addition to exhibiting an IFN-α down regulatory effect. Although in this study we observed that PEDV S-INDEL down-regulated the p65 gene, the NF-κB pathway was up-regulated. It has been demonstrated that inhibition of the p50-p65 hetero-dimer or the p50 homo-dimer can be compensated for by the p52-RelB hetero-dimer, resulting in the activation of the NF-κB non-canonical pathway (Wietek and O’Neill, 2007). In vitro studies demonstrated that PEDV protein E is capable of inducing p65 in intestinal epithelial cells and the activation of the NF-κB non-canonical pathway (Xu et al., 2013). Moreover, further studies demonstrated that during PEDV CV777 infection, NF-kB p65 was found to be translocated from the cytoplasm to the nucleus, and PEDV-dependent NF-kB activity was associated with viral dose and active replication. In addition, the same study corroborated that upon PEDV N protein overexpression in transfected IECs, p65 was detected in the nucleus (Cao et al., 2015b).
The production of pro-inflammatory cytokines and type I interferon (IFN) at the local intestinal mucosal level is part of the innate immune response during viral infection. Interleukin-6 (IL-6) is associated with improving humoral and mucosal immune response (Meng et al., 2013). In this study, IL-6 mucosal gene expression was not affected by PEDV infection. IL-6 expression levels are not necessarily correlated with the positive gene modulation of other pro-inflammatory cytokines. IL-12 is the major type I cytokine produced by macrophages and dendritic cells. IL-12 is believed to be responsible for enhancing Th1 and S-IgA response at the mucosal level (Boyaka et al., 1999). In this study, we observed that IL-12 was up-regulated at 7 dpi by PEDV S-INDEL. This modulatory effect is in agreement with a previous in vitro study, which also evaluated a low-virulent strain of CV777 (Gao et al., 2015). In this study, we observed a positive TNF-α gene regulatory effect likely associated with the up-regulation of NF-KB in response to PEDV S-INDEL infection. We speculate that this might be the result of the RIG-I non-canonical pathway of NF-κB, perhaps through the mitochondrial antiviral-signaling protein (MAVS) and TRAF3 (not evaluated in this study) (Bowie and Unterholzner, 2008). Other coronaviruses, such as SARS-CoV, also showed a positive modulatory effect of TNF-α through the activation of NF-KB. Conversely, PEDV infection with the high-virulent non-S-INDEL strain induced a down-regulation effect on TNF-α gene expression. This negative modulatory effect on TNF-α could be associated with the severe pathological response characteristic of PEDV non-S-INDEL strains.
Modulation of type I IFN response seems to be a common evasion strategy of viruses in the order Nidovirales (Zhang et al., 2016). SARS-CoV and MERS-CoV, both within the genus Betacoronavirus, do not induce significant IFN response in respiratory cells in vitro. However, transmissible gastroenteritis epidemic virus (TGEV) within the Alphacoronavirus genus induces a high level of IFN-α in newborn pigs. PEDV also exists within the Alphacoronavirus genus; however, we found virus strain-related differences in IFN-α gene modulation. Although no significant IFN-α gene modulation was observed after PEDV S-INDEL infection, there was an increment consistent with previous reports on TGEV. The down-regulation observed in PEDV non-S-INDEL is more consistent with the effect observed with other members of the Betacoronavirus genus. The differential modulatory effect in the TNF gene between PEDV strains observed in this study is coincident with the severity in pathogenicity between PEDV S-INDEL and non-S-INDEL.
In summary, the aim of this study was to investigate the differential gene modulation of pattern recognition TLR and RIG-I-like receptors and downstream mediators on the intestinal mucosa of neonatal pigs infected with PEDV non-S-INDEL and PEDV S-INDEL strains. Our results suggest that PEDV S-INDEL infection induces pro-inflammatory cytokines through the non-canonical NF-κB signaling pathway by activating the RIG-I receptor (Fig. 7). Meanwhile, PEDV non-S-INDEL infection suppresses the induction of pro-inflammatory cytokines and type 1 interferon production by down-regulation of the cytoplasmic membrane and endosomal TLRs, as well as TLR downstream-signaling molecules (canonical NF-κB pathway) (Fig. 7). These data present novel in vivo evidence supporting the notion that the evasion strategy of the PEDV non-S-INDEL strain might provide important insights into the pathogenesis of this strain. Based on our results, both low-virulent PEDV S-INDEL and high-virulent PEDV non-S-INDEL cause differential modulation on innate immune response pathways, which could be translated into differences in pathogenesis and clinical outcomes normally observed after PEDV infection.
Acknowledgements
The authors would like to thank Mary Breuer for preparing Fig. 7, which was modified for use in this manuscript. We sincerely apologize to many scientists whose works were not cited in the reference list because of the space limitation.
Contributor Information
L.G. Gimenez-Lirola, Email: luisggl@iastate.edu.
P.E. Piñeyro, Email: pablop@iastate.edu.
References
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-[kappa]B by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Annamalai T., Saif L.J., Lu Z., Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 2015;168:193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaka P.N., Marinaro M., Jackson R.J., Menon S., Kiyono H., Jirillo E., McGhee J.R. IL-12 Is an effective adjuvant for induction of mucosal immunity. J. Immunol. 1999;162:122–128. [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Herrler G., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D.M., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2013;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Chen X., Yang J., Yu F., Ge J., Lin T., Song T. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) samples from field cases in Fujian, China. Virus Genes. 2012;45:499–507. doi: 10.1007/s11262-012-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima G. PED virus reinfecting U.S. herds:Virus estimated to have killed 7 million-plus pigs. J. Am. Vet. Med. Assoc. 2014;245:166–167. [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Antagonizes Beta Interferon Production by Sequestering the Interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L.R., Jing H.Y., Zeng S.L., Wang D., Liu L.Z., Zhang H., Luo R., Chen H.C., Xiao S.B. Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Antagonizes Beta Interferon Production by Sequestering the Interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J. Gen. Virol. 1994;75(Pt 5):1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- Duarte M., Tobler K., Bridgen A., Rasschaert D., Ackermann M., Laude H. Sequence Analysis of the Porcine Epidemic Diarrhea Virus Genome between the Nucleocapsid and Spike Protein Genes Reveals a Polymorphic ORF. Virology. 1994;198:466–476. doi: 10.1006/viro.1994.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Zhao S., Qin T., Yin Y., Yang Q. Effects of porcine epidemic diarrhea virus on porcine monocyte-derived dendritic cells and intestinal dendritic cells. Vet. Microbiol. 2015;179:131–141. doi: 10.1016/j.vetmic.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. TLR signaling. Semin. Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kint J., Dickhout A., Kutter J., Maier H.J., Britton P., Koumans J., Pijlman G.P., Fros J.J., Wiegertjes G.F., Forlenza M. Infectious Bronchitis Coronavirus Inhibits STAT1 Signaling and Requires Accessory Proteins for Resistance to Type I Interferon Activity. J. Virol. 2015;89:12047–12057. doi: 10.1128/JVI.01057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint J., Fernandez-Gutierrez M., Maier H.J., Britton P., Langereis M.A., Koumans J., Wiegertjes G.F., Forlenza M. Activation of the Chicken Type I Interferon Response by Infectious Bronchitis Coronavirus. J. Virol. 2015;89:1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H., Yamamoto T., Yamazaki K., Gohda J., Akiyama T., Semba K., Goto H., Kato A., Yujiri T., Imai T., Kawaguchi Y., Su B., Takeuchi O., Akira S., Tsunetsugu-Yokota Y., Inoue J.-i. TRAF6 Establishes innate immune responses by activating NF-κB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One. 2009;4:e5674. doi: 10.1371/journal.pone.0005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. Pathogen recognition in the innate immune response. Biochem. J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Li Z., Chen F., Ye S., Guo X., Muhanmmad M.A., Wu M., He Q. Comparative Proteome Analysis of Porcine Jejunum Tissues in Response to a Virulent Strain of Porcine Epidemic Diarrhea Virus and Its Attenuated Strain. Viruses. 2016:8. doi: 10.3390/v8120323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney S.A., Colonna M. Viral sensors: diversity in pathogen recognition. Immunol. Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Ren Y., Suo S., Sun X., Li X., Li P., Yang W., Li G., Li L., Schwegmann-Wessels C., Herrler G., Ren X. Evaluation on the Efficacy and Immunogenicity of Recombinant DNA Plasmids Expressing Spike Genes from Porcine Transmissible Gastroenteritis Virus and Porcine Epidemic Diarrhea Virus. PLos One. 2013;8:e57468. doi: 10.1371/journal.pone.0057468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005 doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xing Y.L., Chen X.J., Zheng Y., Yang Y.D., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z.B. Coronavirus Papain-like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLoS One. 2012:7. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Okada K., Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nihon Juigaku Zasshi. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Locarnini S.A. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio. 2015:6. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S., Akira S. Toll-Like Receptors (TLRs) and their ligands. In: Bauer S., Hartmann G., editors. Toll-Like Receptors (TLRs) and Innate Immunity. Springer Berlin Heidelberg; Berlin, Heidelberg: 2008. pp. 1–20. [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q.H., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct Characteristics and Complex Evolution of PEDV Strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Byrum B., Zhang Y. US variant porcine epidemic diarrhea virus: histological lesions and genetic characterization. Virus Genes. 2016;52:578–581. doi: 10.1007/s11262-016-1334-x. [DOI] [PubMed] [Google Scholar]
- Wietek C., O’Neill L.A.J. Diversity and regulation in the NF-κB system. Trends Biochem. Sci. 2007;32 doi: 10.1016/j.tibs.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z.H., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.-J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Soma J., Nakanishi M., Yamaguchi R., Niinuma S. Isolation and experimental inoculation of an S INDEL strain of porcine epidemic diarrhea virus in Japan. Res. Vet. Sci. 2015;103:103–106. doi: 10.1016/j.rvsc.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ma J., Yoo D. Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology. 2017;510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhengfan, J., 2004. Toll-like Receptor 3-Mediated Activation of NF-B and IRF3 Diverges at Toll-IL-1 Receptor Domain-Containing Adapter Inducing IFN-Beta. [DOI] [PMC free article] [PubMed]
- Zhou H. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded. RNA. 2007 doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]