Abstract
Background
Borna disease virus (BDV) replicates in the nucleus and establishes persistent infections in mammalian hosts. A human BDV strain was used to address the first time, how BDV infection impacts the proteome and histone lysine acetylation (Kac) of human oligodendroglial (OL) cells, thus allowing a better understanding of infection-driven pathophysiology in vitro.
Methods
Proteome and histone lysine acetylation were profiled through stable isotope labeling for cell culture (SILAC)-based quantitative proteomics. The quantifiable proteome was annotated using bioinformatics. Histone acetylation changes were validated by biochemistry assays.
Results
Post BDV infection, 4383 quantifiable differential proteins were identified and functionally annotated to metabolism pathways, immune response, DNA replication, DNA repair, and transcriptional regulation. Sixteen of the thirty identified Kac sites in core histones presented altered acetylation levels post infection.
Conclusions
BDV infection using a human strain impacted the whole proteome and histone lysine acetylation in OL cells.
Keywords: Borna disease virus, Human BDV, Oligodendroglia cells, Proteomic, Histone, Lysine acetylation, Bioinformatics-assisted analysis
Highlights
-
•
A human strain of BDV (BDV Hu-H1) was used to infect human oligodendroglial cells (OL cells).
-
•
This study is the first to reveal the host proteomic and histone Kac profiles in BDV-infected OL cells.
-
•
BDV infection affected the expression of many transcription factors and several HATs and HDACs.
Introduction
Borna disease virus (BDV), a member of the family Bornaviridae in the order Mononegavirales, is a neurotropic, enveloped virus with a non-segmented, negative-strand (NNS) ribonucleic acid (RNA) genome (de la Torre, 1994, Schneemann et al., 1995). BDV replicates in the cell nucleus and persistently infects a wide variety of mammal species including humans (Bode and Ludwig, 2003, de La Torre et al., 1996a, Iwata et al., 1998, Kinnunen et al., 2013, Ludwig et al., 1988). One of the first human BDV strains, Hu-H1, was isolated from a bipolar patient׳s white blood cells and differs genetically and biologically from the laboratory reference BDV strains V and C6BV (Bode et al., 1996, de la Torre et al., 1996b).
Oligodendroglia (OL) cells, a cell line derived from human fetal oligodendrocytes, are a major component of the brain white matter that play a pivotal role in maintaining neurological function. OL cells support both natural and experimental infection with several neurotropic NNS RNA viruses including BDV (Ibrahim et al., 2002, Koster-Patzlaff et al., 2007), canine distemper virus (Muller et al., 1995), and measles virus (Baczko et al., 1988). Laboratory-adapted and wild-type (e.g. human) BDV strains have been shown to differentially affect various host cell types (Li et al., 2013, Poenisch et al., 2009, Williams et al., 2008, Wu et al., 2013). Using a natural human BDV strain appeared particularly advantageous to provide better insight into the pathological consequences of human OL cell infection than laboratory strains from animal origin. Moreover, using a virus which had been isolated during an acute depressive episode of a bipolar patient should eventually support our understanding of BDV׳s impact on neuropathogenesis.
BDV affects the expression of several host mRNA transcripts (Carbone et al., 2001), but the mechanisms remain unclear. The recent discovery of histone lysine acetylation (Kac) as a modulator of gene expression in response to virus infection has brought fresh insight into viral epigenetic regulation (Ferrari et al., 2012). BDV infection has been shown to affect site-specific histone acetylation in cortical neurons in vitro (Suberbielle et al., 2008); however, the histone Kac profile of BDV-infected OL cells remains unknown.
In this study, we hypothesized that BDV infection epigenetically impacts the OL cell proteome through histone Kac. Therefore, using an integrated quantitative proteomic approach assisted by bioinformatic analysis, we comprehensively investigated the proteome profile and constructed a histone Kac atlas of BDV-infected OL cells.
Results
BDV infection of OL/BDV cells
To confirm successful BDV infection of OL/BDV cells, RT-PCR, Western blotting, and immunofluorescence assays were performed to examine the major markers of successful BDV infection – p24 and p40 RNA and protein levels (de la Torre, 1994). Rapid spread of the virus infection was observed in tissue culture; 100% of the cells were infected 14 days post-infection, while 100% of control cells remained non-infected (Supplemental Fig. 1).
Proteomic profiling
We applied SILAC labeling-based proteomics to comparatively quantify the host proteome of OL/BDV cells and control cells. A scheme of the experimental workflow is shown in Fig. 1. In total, 4436 non-redundant proteins were identified, of which 4383 proteins displayed quantifiable differential expression levels in response to BDV infection (Supplemental Table 1). Among these, 1572 proteins displayed a greater than or equal to 1.5-fold increased expression and 165 proteins displayed a lesser than or equal to 1.5-fold decreased expression in response to BDV infection. The proteomic dataset was divided into four quantiles (Q1-Q4) based on the cumulative distribution of SILAC L/H ratios: Q1, less than 15%; Q2, 15–50%; Q3, 50–85%; and Q4, greater than 85%. Enrichment analyses were separately performed in each quantile, and the overrepresented annotations were clustered through one-way hierarchical clustering for comparative analysis.
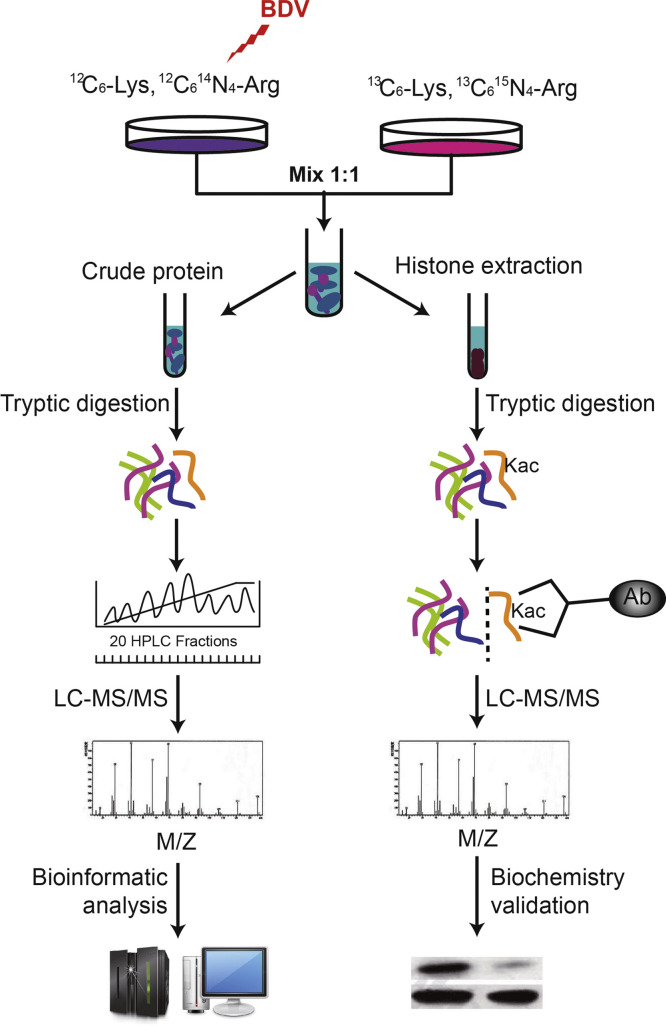
Fig. 1.
Experimental workflow. OL/BDV cells and control cells were maintained in SILAC medium. An aliquot of crude proteins from the mixed cells was digested in solution by trypsin. Protein identification and quantification were performed using LC-MS/MS and data inquiry. In parallel, histones from the cell mixture were extracted, digested, and affinity enriched followed by LC-MS/MS analysis. Further biochemistry was applied to validate the MS analysis results.
BDV infection affects transcription factors
We found 201 transcription factors in the whole OL cell proteome (Supplemental Table 2), of which 84 transcription factors showed significantly increased expression and 11 showed significantly decreased expression in response to BDV infection. Only 30 of these significantly differentiated transcription factors were mapped onto the KEGG database. The highest-ranking canonical KEGG pathways are listed in Table 1.
Table 1.
Canonical KEGG pathways associated with the differential transcription factors.
| KEGG pathway | Mapping | P-value |
|---|---|---|
| hsa05202 Transcriptional misregulation in cancer | 6 | 1.84E-06 |
| hsa04380 Osteoclast differentiation | 5 | 4.59E-05 |
| hsa04917 Prolactin signaling pathway | 3 | 2.04E-03 |
| hsa04668 TNF signaling pathway | 3 | 8.18E-03 |
| hsa04630 Jak-STAT signaling pathway | 2 | 1.07E-02 |
| hsa04920 Adipocytokine signaling pathway | 2 | 4.05E-02 |
| hsa04062 Chemokine signaling pathway | 3 | 4.14E-02 |
Bioinformatic analysis
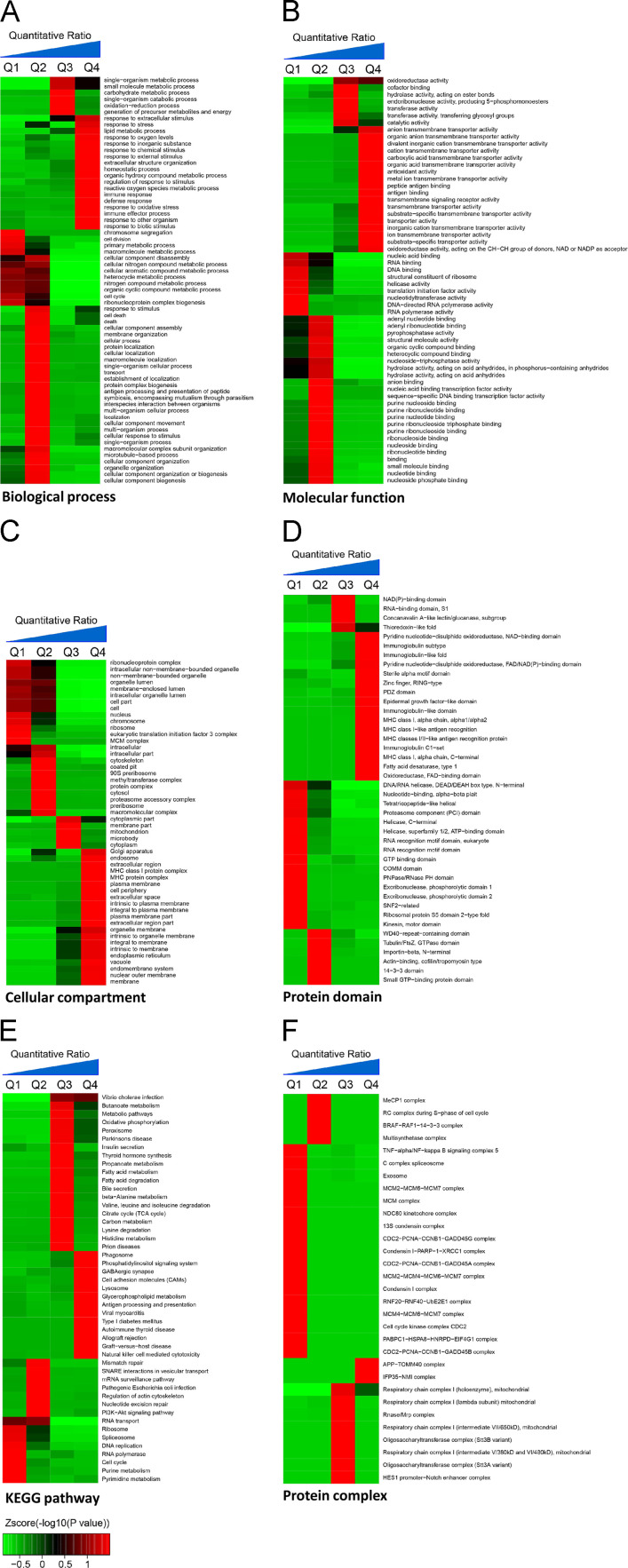
We analyzed the quantifiable proteome data set for three enrichment gene ontology (GO) categories: biological process, molecular function, and cellular compartment ( Fig. 2A–C). Using Interpro domain enrichment analysis, we analyzed the domain features of those enriched proteins dysregulated by BDV infection (Fig. 2D). We performed a pathway clustering analysis through the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Fig. 2E). Using a manually curated CORUM database, we performed enrichment analysis on protein complexes and used k-means clustering to identify the specific subgroups that were most impacted by BDV infection (Fig. 2F). We obtained two protein complexes (the amyloid precursor protein mitochondrial translocase APP-TOMM40 and IFP35-NMI) enriched in Q4 that are primarily related to mitochondrial protein transport and interferon signaling, respectively (Lee et al., 2012, Zhou et al., 2000), and 17 protein complexes enriched in Q1 that are primarily related to DNA replication, DNA repair response, and chromosome shape regulation. The results are summarized in Table 2.
Fig. 2.
Enrichment and clustering analysis of the quantifiable proteomic data set. Quantifiable proteins were classified by gene ontology annotation based on (A) molecular function, (B) cellular compartment, and (C) biological process. In each category, the differential quantifiable proteins were divided into four quantiles based on the cumulative distribution of SILAC L/H ratios: Q1, less than 15%; Q2, 15–50%; Q3, 50–85%; and Q4, greater than 85%. An enrichment analysis was separately performed in each quantile for diverse categories, and the overrepresented annotations were clustered through one-way hierarchical clustering for comparative analysis. Quantifiable proteins were also annotated based on (D) the PFAM domain database, (E) the KEGG pathway database, and (F) the CORUM protein complex database.
Table 2.
Main results of bioinformatic analysis.
| Q4 (the quantile with high L/H ratio) | Q1(the quantile with low L/H ratio) | |
|---|---|---|
| Biological process | Multiple stimuli (e.g., extracellular, stress, oxygen levels, inorganic substances), lipid metabolism, extracellular structure organization, reactive oxygen species and organic hydroxy species metabolic responses, immune and defense responses | Chromosomal segregation and cell divison, primary and macromolecular metabolic processes |
| GO-molecular functions | Various transmembrane transporter activities | Nucleic acid processing |
| GO-cellular compartment | Membranes (e.g., Golgi apparatus, endosomes, MHC class molecules, organelle membranes, plasma membrane, nuclear outer membrane) | Nucleus, chromosomes, ribosomes, and minichromosome maintenance (MCM) complex |
| Protein domain | Immune competence (e.g., MHC class molecules and immunoglobulins) | Nucleic acid processing (e.g., helicase, nucleotide-binding, RNA recognition, exoribonuclease, and transcription regulation [SNF2-related and COMM domains]) |
| KEGG pathway | Phospatidylinositol signaling, GABAergic synapse, cell adhesion molecules (CAMs), glycerophospholipid metabolism, and immune response pathways (e.g., antigen processing and presentation, autoimmune thyroid disease, allograft rejection, graft-versus-host disease) | Nucleic acid, cell cycle, ribosomal processes, purine and pyrimidine metabolism |
| Protein complex | Two protein complexes related to mitochondrial protein transport and interferon signaling | 17 protein complexes related to DNA replication, DNA repair response, chromosome shape regulation |
BDV infection affects histone Kac
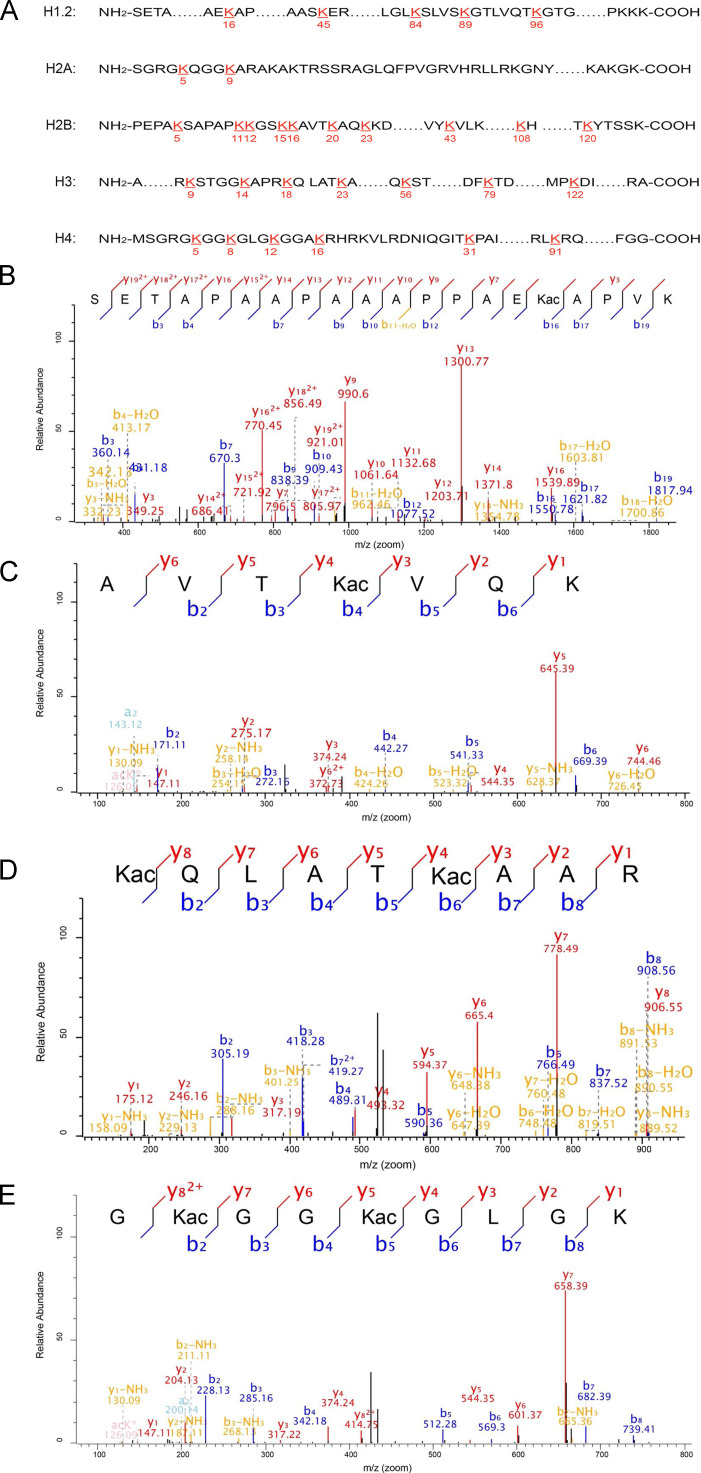
As BDV infection impacts transcription factor expression and histone Kac regulates gene transcription, we investigated the affect of BDV infection on site-specific histone Kac. We applied SILAC-based quantitative proteomics to comparatively profile histone Kac in OL/BDV cells and control cells. A total of 30 Kac sites in core histones were identified ( Fig. 3A), which covers almost all reported Kac sites in mammalian core histones. The representative spectra of well-known histone Kac peptides, including the peptide spectra of H1.2K16ac, H2BK20ac, H3K18ac, and H4K5ac, are provided (Fig. 3B–E). Furthermore, the sequences of identified Kac peptides in core histones and all corresponding quantitative Kac profiles in response to BDV infection are summarized in Table 3. As indicated, the Kac levels in 15 of 30 sites were significantly decreased (e.g., H2AK5ac, H2BK5ac, H3K14ac, and H4K5ac), whereas the Kac level in one site, H2BK15, was significantly increased in response to BDV infection. The Kac levels in the other 14 sites showed no significant changes.
Fig. 3.
Analysis of histone lysine acetylation (Kac). (A) The illustration of identified histone Kac sites in OL cells in response to BDV infection. The identified sites in core histones were numbered and highlighted. (B) MS/MS spectra of a tryptic peptide ion histone H1.2K16 acetylated peptide _ (ac)SETAPAAPAAAPPAEK(ac)APVK. (C) MS/MS spectra of a tryptic peptide ion histone H2BK20 acetylated peptide_AVTK(ac)VQK. (D) MS/MS spectra of a tryptic peptide ion histone H3K18 acetylated peptide_K(ac)QLATK(ac)AAR. (E) MS/MS spectra of a tryptic peptide ion histone H4K5 acetylated peptide _GK(ac)GGK(ac)GLGK.
Table 3.
Summary of quantifiable changes in histone lysine acetylation (Kac) sites.
| Kac site | Modified sequence | Normalized ratio L/H (↑↓) |
|---|---|---|
| H1.2K16ac | (ac)SETAPAAPAAAPPAEK(ac)APVK | 0.5683↓ |
| H1.2K45ac | ASGPPVSELITK(ac)AVAASK | 0.9335 |
| H1.2K84ac | LGLK(ac)SLVSK | 0.8309 |
| H1.2K89ac | SLVSK(ac)GTLVQTK | 1.2392 |
| H1.2K96ac | GTLVQTK(ac)GTGASGSFK | 0.6828 |
| H2AK5ac | GK(ac)QGGK(ac)AR | 0.4577↓ |
| H2AK9ac | GK(ac)QGGK(ac)AR | 0.4577↓ |
| H2BK5ac | PEPAK(ac)SAPAPK(ac)K(ac)GSK | 0.3748↓ |
| H2BK11ac | PEPAK(ac)SAPAPK(ac)K(ac)GSK | 0.8559 |
| H2BK12ac | K(ac)GSK(ac)K(ac)AVTK | 1.2412 |
| H2BK15ac | GSK(ac)K(ac)AVTK(ac)AQK | 1.5352↑ |
| H2BK16ac | K(ac)AVTK(ac)AQK | 0.8097 |
| H2BK20ac | AVTK(ac)VQK | 0.5944↓ |
| H2BK23ac | KAVTK(ac)AQK(ac)K | 1.1076 |
| H2BK43ac | ESYSVYVYK(ac)VLK | 0.4037↓ |
| H2BK108ac | LLLPGELAK(ac)HAVSEGTK | 0.6612 |
| H2BK120ac | AVTK(ac)YTSSK | 0.5863↓ |
| H3K9ac | K(ac)STGGK(ac)APR | 1.0686 |
| H3K14ac | KSTGGK(ac)APR | 0.5965↓ |
| H3K18ac | K(ac)QLATK(ac)AAR | 0.5660↓ |
| H3K23ac | QLATK(ac)AAR | 0.9600 |
| H3K56ac | YQK(ac)STELLIR | 0.5650↓ |
| H3K79ac | EIAQDFK(ac)TDLR | 0.6652 |
| H3K122ac | VTIMPK(ac)DIQLAR | 0.5615↓ |
| H4K5ac | GK(ac)GGK(ac)GLGK | 0.3468↓ |
| H4K8ac | GGK(ac)GLGK(ac)GGAK | 1.2583 |
| H4K12ac | GLGK(ac)GGAK | 0.9165 |
| H4K16ac | GLGK(ac)GGAK(ac)R | 0.4638↓ |
| H4K31ac | DNIQGITK(ac)PAIR | 0.5159↓ |
| H4K91ac | TVTAMDVVYALK(ac)R | 0.3895↓ |
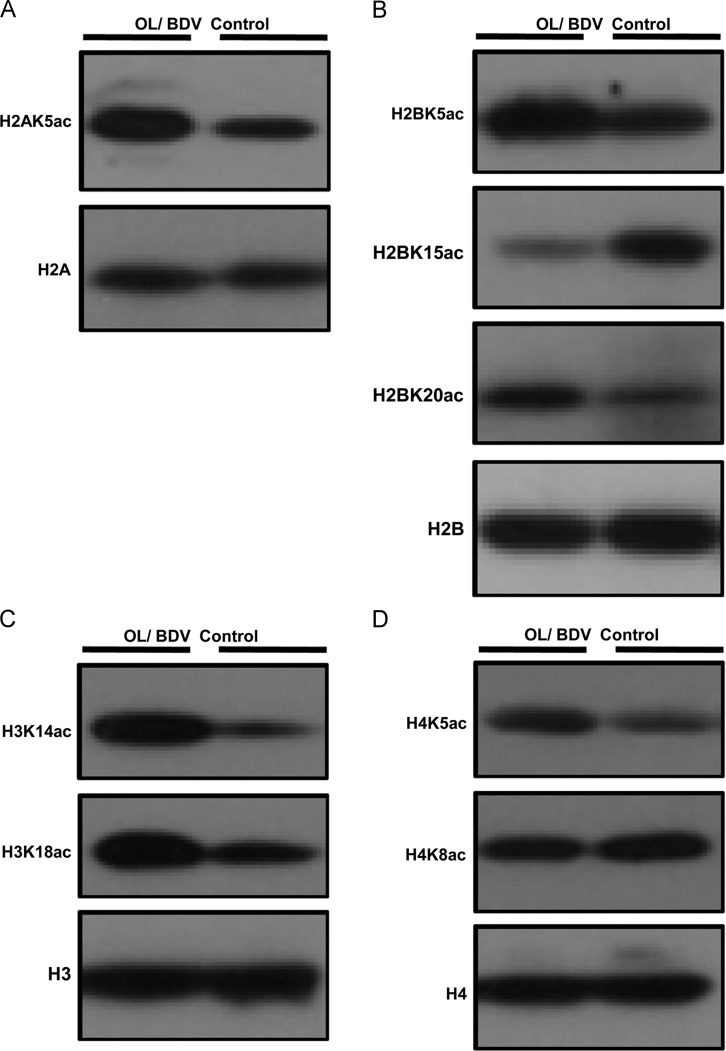
To further validate the different histone acetylation profiles, Western blot analysis was performed with histone Kac sequence-specific antibodies. Consistent with the quantitative results (Table 2), BDV infection significantly decreased the Kac level in H2AK5ac, H2BK5ac, H2BK20ac, H3K14ac, H3K18ac, and H4K5ac ( Fig. 4). Consistent with the MS findings, we observed an increased H2BK15ac level and unchanged H4K8ac level post-BDV infection (Fig. 4).
Fig. 4.
BDV Hu-H1 infection modifies the histone lysine acetylation (Kac) profile. After OL/Hu-H1 cells and control cells were harvested, the core histones from whole cell lysates were extracted. Extracted histones (2 μg) were subject to SDS-PAGE followed by Western blotting analysis to examine histone site-specific Kac in (A) H2A, (B) H2B, (C) H3, and (D) H4 through histone site-specific Kac antibodies.
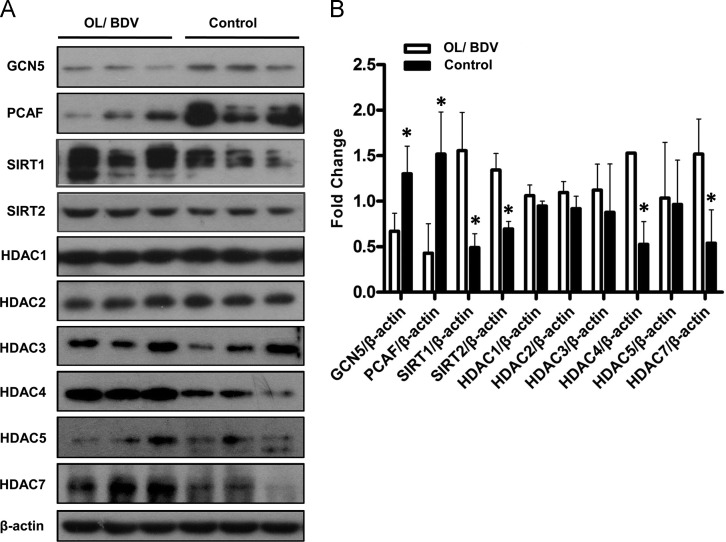
Validation of histone acetyltransferase and deacetylase expression by western blotting
Histone acetylation is dynamic and regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). From the proteomic data, the expression of several HATs (ELP3, BRD4) and HDACs (SIRT1, HDAC7) was significantly altered by BDV infection. To validate these changes, two HATs (GCN5 and PCAF) and eight HDACs (SIRT1, SIRT2, HDACs 1, 2, 3, 4, 5, and 7) were selected for Western blotting ( Fig. 5). Two HATs – GCN5 (p=0.039) and PCAF (p=0.029) – were found to be significantly downregulated, and four HDACs – SIRT1 (p=0.015), SIRT2 (p=0.005), HDAC4 (p=0.002), and HDAC7 (p=0.033) – were found to be significantly upregulated, in OL/BDV cells relative to control cells. There was no significant dysregulation observed in HDACs 1, 2, 3, or 5 (p>0.05).
Fig. 5.
Validation of histone acetyltransferase (HAT) and histone deacetylase (HDAC) expression by Western blotting. (A) Western blots of two HATs (GCN5 and PCAF) and eight HDACs (SIRT1, SIRT2, HDACs 1, 2, 3, 4, 5, and 7) with β-actin used as a control. (B) GCN5 (p=0.039) and PCAF (p=0.029) were found to be significantly downregulated, while SIRT1 (p=0.015), SIRT2 (p=0.005), HDAC4 (p=0.002), and HDAC7 (p =0.033) were found to be significantly upregulated, in OL/Hu-H1 cells relative to control cells. There was no significant dysregulation observed in HDAC1 (p=0.196), HDAC2 (p=0.169), HDAC3 (p=0.520), or HDAC5 (p=0.881).
Discussion
The goal of this study was to analyze the effects of BDV infection on the proteome and histone Kac profiles of OL cells through a SILAC-based quantitative proteomic approach in order to gain insight into BDV pathogenesis by using a natural human virus strain. To date, several studies have addressed virus-induced changes on the host proteome (e.g., BDV, EBV, HBV, HCV, HIV-1, and SARS-CoV) (Zhou et al., 2011), but minimal data has been published on virus-induced changes in histone Kac. To our knowledge, this is the first SILAC-based quantitative proteomic study to assess the response of human OL cells to human strain BDV Hu-H1 infection and the first comprehensive atlas of host cell histone acetylation change in response to BDV infection. Previous studies have shown that BDV infection impacts gene transcription in OL cells (Carbone et al., 2001, Koster-Patzlaff et al., 2007), while this study found many regulated transcription factors by BDV infection, which linked gene transcription and the differential proteomic expression towards BDV infection. In view that histone acetylation is generally considered a marker involved in activating gene expression, and our data suggested altered histone site-specific acetylation in response to BDV infection, it appears reasonable to consider a link of altered histone acetylation, gene transcription and subsequently proteomics. (Supplemental Fig. 2)
Histone Kac
Previous studies have revealed virus-induced changes in some histone Kac sites. For example, adenovirus small early region 1a (e1a) protein induces H3K18 hypoacetylation by restricting some HATs (Horwitz et al., 2008). With respect to BDV, Suberbielle et al. (2008) revealed histone modifications, including H2BK5 and H2BK20, in BDV laboratory strain He/80 infected-neurons.
Here, 30 histone Kac sites were identified, of which half presented with decreased acetylation and only one site, H2BK15, presented with increased acetylation. Histone modification at the N-terminal plays a central role in chromatin remodeling and transcriptional regulation. Here, most of the quantifiable Kac sites were at N-terminals of core histones, suggesting pronounced epigenetic modulation of BDV-infected OL cells.
Some specific functions of histone site acetylation have already been verified. For example, global hypoacetylation at H3K18 is observed in prostate carcinomas with poor prognosis which suggests that processes resulting in global H3K18 hypoacetylation may be linked to oncogenic transformation (Horwitz et al., 2008). H4K16 acetylation regulates cellular lifespan because the level of H4K16 acetylation increased with age examined in yeast cells (Dang et al., 2009). Deregulated H4K12 acetylation may represent an early biomarker of an impaired genome-environment interaction in the aging mouse brain because aged mice display a specific deregulation of H4K12 acetylation and fail to initiate a hippocampal gene expression program associated with memory consolidation during learning. Moreover, restoration of physiological H4K12 acetylation reinstates the expression of learning-induced genes and leads to the recovery of cognitive abilities (Peleg et al., 2010). Further studies should investigate the biological function of the histone specific-site acetylation altered by BDV infection.
HATs and HDACs
Histone acetylation levels are determined by the combined activities of HATs and HDACs. HATs have been shown to preferentially acetylate specific histones and/or specific lysines (Turner, 2000). HDACs through deacetylation of histones has been associated with decreased transcriptional activation and proteomic expression; for example, non-small cell lung cancer A549 cells treated with suberoylanilide hydroxamic acid (SAHA), a pan HDAC inhibitor, resulted in histone Kac and proteomic changes (Shahbazian and Grunstein, 2007, Wu et al., 2013). Here, the dysregulation of several HATs and HDACs was accompanied by significant proteomic change, suggesting a link between histone Kac and proteomic expression in BDV-infected OL cells (Supplemental Fig. 2).
Two Gcn5 mammalian HAT subclasses have been described: GCN5 and p300/CREB-binding protein-associated factor (PCAF). They are transcriptional coactivators and possess intrinsic histone acetylase activity, providing a direct link between hyperacetylated chromatin and transcriptional activation. GCN5 and PCAF specifically promote acetylation on H3K9, H3K14, H4K8, and H4K16. Deletion of GCN5/PCAF in cells reduces acetylation on H3K9 and correlates with nuclear receptor target gene activation at some level (Jin et al., 2011). The GCN5/PCAF have a preference for H3K14 (Grant et al., 1999), and also acetylate H4K8 and H4K16 to a relatively slight degree (Kuo et al., 1996, Schiltz et al., 1999). By Western blotting, we found that the expression levels of GCN5 and PCAF were downregulated, consistent with the decreased acelytation of H3K14 and H4K16 in response to BDV infection. Interestingly, acetylation of H3K9 and H4K8 was not decreased here, suggesting that other factors possibly impacted acetylation of two sites in BDV-infected OL cells.
HDACs have a highly-conserved domain and are divided into four classes: class I, class II, class III (sirtuins) and class IV (Gregoretti et al., 2004, Yang and Seto, 2007). HDACs 1, 2, and 3 belong to class I and localize to the nucleus. HDACs 4, 5, and 7 belong to class II. Class II HDACs are mainly known for their participation in controlling cell differentiation through regulation of transcription myocyte enhancer factor 2 activity, involved in muscle development and neuronal survival (Lomonte et al., 2004). In addition, HDAC4, HDAC5, and HDAC7 are known to shuttle between the cytoplasm and the nucleus (Kao et al., 2001, McKinsey et al., 2000). In this study, western blotting analysis showed that the expression level of HDACs 4 and 7 were increased in BDV-infected OL cells, while few changes were observed in HDACs 1, 2, 3, and 5. These results suggest that class II HDACs 4 and 7 may pose specific roles in histone lysine acetylation profiles in BDV-infected OL cells. Further studies should primarily investigate whether the shuttle function of HDACs 4 or 7 is related to the nuclear import and export of BDV proteins (Kobayashi et al., 2001). It could be suggested that they both deacetylate non-histone and histone substrates, thereby modulating histone acetylation levels which subsequently impact the proteome profile of the host cell. Sirtuins are structurally and mechanistically different from other HDAC classes; accordingly, their primary substrates do not appear to be the histone proteins (Luo et al., 2001, Vaziri et al., 2001). By Western blotting, SIRT1 and SIRT2 were found with upregulated expression levels in BDV-infected OL cells, suggesting that acetylation profile of non-histone subtrates could be modulated through changing class III HADCs.
Membrane proteins and immune response
BDV enters cells by plasma membrane fusion, replicates and transcribes itself in the nucleus, and then replicates itself through the aforementioned nucleocytoplasmic trafficking of BDV macromolecules (Gonzalez-Dunia et al., 1998, Honda and Tomonaga, 2013). As these processes intimately involve the plasma and nuclear membranes, changes in membrane protein expression should be associated with BDV infection. This study confirms that BDV infection has a profound impact on membrane-associated proteins in Q4, most of which were upregulated in response to BDV infection. Proteins in Q4 were also enriched for immune response; this finding may be related to the aforementioned changes in membrane protein expression considering the pivotal function of membrane proteins in the immune response. As successful viral infection and replication requires evasion from the host immune response, it is reasonable to surmise that BDV would affect host cellular proteins with an antiviral immune response function.
Nuclear proteins and DNA replication
As the host cellular chromatin inhibits viral gene expression and replication by suppressing DNA accessibility, viruses that enter and persist in the nucleus (such as BDV) have evolved chromatin-associated mechanisms to efficiently propagate the viral genome (Lieberman, 2006). For example, BDV׳s viral ribonucleoprotein (RNP) directly interacts with the mitotic host chromosome using core histones as a docking platform (Matsumoto et al., 2012). BDV infection was found to differentially impact on nuclear and chromosomal proteins enriched in DNA replication and repair, transcription regulation, and chromosomal shape regulation in Q1, most of which were downregulated in response to BDV infection.
Metabolic pathways
Through KEGG analysis, a wide variety of pathways (e.g., phospatidylinositol signaling, GABAergic synapse, CAMs, glycerophospholipid metabolism, immune response pathways, nucleic acid processes, cell cycle, ribosomal processes, purine metabolism, and pyrimidine metabolism) were identified as the most significantly altered set of host biological pathways. Consistent with the current KEGG findings in phospatidylinositol signaling, glycerophospholipid metabolism, and purine/pyrimidine metabolism, our previous metabonomic profiling study in BDV-infected OL cells revealed significant perturbations in myo-inositol, α-glucose, acetate, pyruvate, and nicotinamide adenine dinucleotide (NAD) (Huang et al., 2012). Thus, these proteomic data and the previous metabonomic data, also based on human BDV Hu-H1 strain, mutually support each other.
Conclusions
Using SILAC-based quantitative proteomics coupled with bioinformatic analysis, this study is the first to reveal the host proteomic and histone Kac profiles in BDV-infected OL cells using a natural human virus strain. BDV infection appears to preferentially dysregulate membrane, nuclear, and chromosomal host protein expression while affecting metabolic pathways, immune response, DNA replication, DNA repair, and transcription regulation. BDV infection was found to affect histone acetylation of specific lysine residues. Moreover, BDV infection affected the expression of many transcription factors, several HATs and HDACs. As histone Kac epigenetically regulates gene transcriptional activation, the differential acetylation of specific lysine residues may have impacted the changes in the host proteome profile.
Funding
This work was supported by the National Basic Research Program of China (973 Program) (grant number 2009CB918300 to P.X.), the National Natural Science Foundation of China (grant number 31300137 to R.H.), and the National Natural Science Foundation of China (grant number 31271362 to Z.C.).
Materials and methods
Virus strain
The human strain BDV Hu-H1 was used throughout the study and kindly provided by Professor Hanns Ludwig (Free University of Berlin, Berlin, Germany). This strain dated back to 1996 and could be recovered from freshly isolated white blood cells of a female bipolar I disorder patient in Germany who was admitted to hospital during a severe depressive episode. The isolation procedure had required laborious co-cultivating of patient׳s PBMCs with human oligodendroglia (OL) cells. After at least 10–12 blind passages which did not exhibit viral growth, infectious human virus had finally been harvested and subsequently passaged to further characterization and storage. The isolation protocol of BDV strain Hu-H1 and two other isolates have been described in full detail in the original publication (Bode et al., 1996). Strain Hu-H1 and other two isolates were originating from PBMC samples which were testing positive for BDV RNA and protein. Further molecular characterization had revealed that corresponding RNAs (of PBMC and isolate) displayed identical nucleotide sequences, proving the authenticity of the human isolates. Moreover, the isolates displayed few but meaningful point mutations which differentiated them from each other and from laboratory strain V and a second lab strain (He/80) (de la Torre et al., 1996b). Further biological characterization had revealed that strain Hu-H1 and other two human strains displayed differing pathogenicity in animal experiments, namely inducing behavioral changes in rabbits but no deadly disease as compared to str. V (Bode et al., 1996).
Own recent experiments of our research group could demonstrate that the above described human strain Hu-H1 inhibited proliferation and supported apoptosis, whereas lab. Strain V did quite the opposite (Li et al., 2013). This remarkable difference was pivotal to again choose strain Hu-H1 for this study, apart from the fact that wild-type viruses are supposed to reflect natural pathogenicity patterns at the cellular level which might be lost in highly adapted laboratory strains.
Cell line and viral infection
Human OL cells (a cell line derived from fetal human oligodendrocytes, 112 passages) and BDV Hu-H1 strain (77 passages in OL cells) (Bode et al., 1996) were used. Like the virus strain, human OL cells were kindly supplied by Professor Hanns Ludwig. Persistently-infected OL cells and non-infected OL cells were cultured in Dulbecco׳s modified Eagle׳s medium (DMEM; Hyclone, Logan, Utah, USA) with 10% fetal bovine serum (FBS, Hyclone), 100 U/ml penicillin, and 100 μg/ml streptomycin (Hyclone) within a humidified incubator (5% CO2, 37 °C) and were passaged when they reached 90% confluence by trypsinization (Hyclone).
Stock viral solution was prepared and titrated as previously described (Huang et al., 2012). Briefly, OL cells washed twice with serum-free DMEM were sub-divided in one 10-cm dish and infected with BDV Hu-H1 at a multiplicity of infection (MOI) of 1.0 (800 μl of virus stock). Cells were then stored in a humidified incubator (5% CO2, 37 °C) for two hours with gentle shaking for 15 min. Excess virus was removed by washing with 5 ml of serum-free DMEM before bathing the cells in 10 ml of culture medium (10% FBS in DMEM). Thereafter, OL cells were passaged 5 to 6 times until all cells were infected with BDV Hu-H1. The now persistently-infected cells (OL/BDV cells) and non-infected OL cells (control cells) were kept under the same conditions for the remainder of the study.
RNA extraction and RT-PCR for BDV detection
Total RNA subjected to reverse transcription polymerase chain reaction (RT-PCR) using avian myeloblastosis virus reverse transcriptase (AMV RT) (Promega, Madison, WI, USA) was extracted from OL/Hu-H1 cells and control cells with Trizol (Trizol LS; Invitrogen, Carlsbad, CA, USA). For BDV phosphoprotein 24 (p24) and nucleoprotein (p40) gene amplification, PCR was performed according to the manufacturer׳s protocols except for the modified annealing temperatures of 50 °C and 61.5 °C (Go Taq® Green Master Mix, Promega, Madison, WI, USA). The PCR products were separated by 2% agarose gel electrophoresis and stained with ethidium bromide. The primer sequences were as follows: p40, OR (5′–GCTGGGTTTCCTTGACACTTG-3′), OF (5′–GATCAAAGCAGGAGCCGAGCAG-3′) (size: 545 bp); and p24, OR (5′–CTCCATCATTGTCTTCATG-3′), OF (5′–TGATCTCAGACCCAGACC-3′) (size: 224 bp).
Imunofluorescence assay for BDV detection
Both OL/BDV cells and control cells were grown on six-well dishes for 30 min at room temperature and fixed with 4% paraformaldehyde followed by permeabilization for 5 min in 0.4% Triton X-100. Thereafter, both lines were rinsed with PBS and blocked with 5% (w/v) skimmed milk solution for one hour at 37 °C. Overnight incubation with anti-recombinant BDV-specific p24 or p40 protein primary monoclonal antibody (both diluted 1:500; GenScript, Piscataway, New Jersey, USA) at 4 °C was followed by one hour of incubation with secondary antibodies at room temperature. After three PBS washes, immunofluorescence was detected by phase-contrast microscopy.
Cell culture and labeling
To profile protein expression and histone Kac in response to BDV infection, OL/BDV cells and control cells were labeled by the SILAC Protein Quantitation Kit (Invitrogen, Carlsbad, CA) according to manufacturer׳s instructions. OL/BDV and control cells were separately grown in DMEM supplemented with 10% FBS and either the light isotopic forms of [U-12C6]-L-lysine and [U-12C6 14N4]-L-arginine or the heavy isotopic forms of [U-13C6]-L-lysine and [U-13C6 15N4]-L-arginine, respectively. After six generations of passaging, the heavy labeling efficiency of [U-13C6]-L-lysine and [U-13C6 15N4]-L-arginine in control cells was evaluated by mass spectrometer analysis to confirm a greater than 99% labeling efficiency. Then, cells were continuously expanded in SILAC medium until reaching the desired population size. Finally, cells in each heavy- or light-labeled pool were harvested separately and combined in equal amounts.
Affinity enrichment of histone lysine acetylated peptides
Prior to affinity enrichment of histone Kac peptides, anti-acetyllysine antibody agarose beads (PTM Biolabs, Chicago, IL) were washed twice with ice-cold PBS. To enrich lysine acetylated peptides, histone trypic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, 0.5% NP-40, pH 8.0) were incubated with prewashed antibody agarose beads at 4 °C overnight with gentle shaking. The beads were washed four times with NETN buffer and twice with ddH2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. The eluted fractions were combined and vacuum-dried and then analyzed by high-performance liquid chromatography with tandem mass spectrometry (HPLC−MS/MS).
LC-ESI-MS/MS analysis
Peptides were dissolved in solvent A (0.1% formic acid in 2% acetonitrile), directly loaded onto a reversed-phase column (360 μm OD×75 μm ID), packed in-house with 3 μm C18 beads (Reprosil-Pur C18-AQ, Dr. Maisch), and eluted with a linear gradient of 5−30% solvent B (0.1% formic acid in 98% acetonitrile) for 35 min at a constant flow rate of 300 nL/min on an EASY-nLC 1000 Ultra performance liquid chromatography (UPLC) system. Eluted peptides were electrosprayed into LTQ-Orbitrap Elite spectrometers (Thermo Scientific) operating in a data-dependent mode that acquired MS/MS spectra for 20 most intense ions. Full MS (m/z range: 350–1300) was acquired using Fourier transform MS (FTMS) in the Orbitrap at 240,000 resolution at 400 m/z, and MS/MS was acquired using collision-induced dissociation (CID) in the LTQ-Orbitrap Elite at 35% normalized collision energy. A lockmass ion from ambient air (m/z 536.165369) was used for internal calibration of all full-scan measurements with an Orbitrap detector. Dynamic exclusion was enabled with a repeat count of 1, repeat duration of 5 s, exclusion duration of 60 s, and exclusion mass width of ±10 ppm relative to the reference mass.
Western blotting
Cell extracts and Western blots were performed as previously described (Liu et al., 2014). Briefly, monolayers of OL/BDV and control cells were lysed in standard lysis buffer and sonicated on ice. Then, 10 μg lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane. The membranes were incubated overnight at 4 °C with primary antibodies (i.e., anti-recombinant BDV p24 antibody, anti-recombinant BDV p40 antibody, both diluted 1:500, GenScript; anti- H2AK5ac, H2BK5ac, H2BK15ac, H2BK20ac, H3K14ac, H3K18ac, H4K5ac and H4K8ac antibodies, all diluted 1:2000, PTM Biolabs, Chicago, IL; anti-GCN5, PCAF, SIRT1, SIRT2, HDACs 1, 2, 3, 4, 5, and 7 antibodies, all diluted 1:1000, CST, Beverly, MA, USA). All membranes were washed and incubated with their respective horseradish peroxidase-coupled immunoglobulin G (IgG). After extensive washing, antibody-detected protein bands were visualized by enhanced chemiluminescence (ECL) and exposed to autoradiography film.
Acknowledgments
We thank Xiaojun Peng (Bioinformatics Center, PTM Biolab Co. Ltd., Hangzhou, Jiangsu, China) for his technical assistance with the bioinformatic analysis and Dr. N.D. Melgiri for editing and proofreading the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.06.040.
Appendix A. Supporting information
Supplemental Figure 1. BDV p24 and p40 Detection. (a) Detection of BDV p24 and p40 RNA by reverse transcription polymerase chain reaction (RT-PCR). Detection of BDV p24 and p40 proteins by (b) Western blotting and (c) immunofluorescence assay. 100% of OL/BDV cells were infected, and 100% of control cells were non-infected.
Supplementary Material
Supplementary Material
Supplemental Figure 2. The link among histone acetylation, gene transcription and proteomics in response to BDV infection.
References
- Baczko K., Liebert U.G., Cattaneo R., Billeter M.A., Roos R.P., ter M.V. Restriction of measles virus gene expression in measles inclusion body encephalitis. J. Infect. Dis. 1988;158(1):144–150. doi: 10.1093/infdis/158.1.144. [DOI] [PubMed] [Google Scholar]
- Bode L., Durrwald R., Rantam F.A., Ferszt R., Ludwig H. First isolates of infectious human borna disease virus from patients with mood disorders. Mol. Psychiatry. 1996;1(3):200–212. [PubMed] [Google Scholar]
- Bode L., Ludwig H. Borna disease virus infection, a human mental-health risk. Clin. Microbiol. Rev. 2003;16(3):534–545. doi: 10.1128/CMR.16.3.534-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone K.M., Rubin S.A., Nishino Y., Pletnikov M.V. Borna disease: virus-induced neurobehavioral disease pathogenesis. Curr. Opin. Microbiol. 2001;4(4):467–475. doi: 10.1016/s1369-5274(00)00237-x. [DOI] [PubMed] [Google Scholar]
- Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., Kaeberlein M., Kennedy B.K., Berger S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JC. Molecular biology of borna disease virus: prototype of a new group of animal viruses. J. Virol. 1994;68(12):7669–7675. doi: 10.1128/jvi.68.12.7669-7675.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Torre JC, Gonzalez-Dunia D., Cubitt B., Mallory M., Mueller-Lantzsch N., Grasser F.A., Hansen L.A., Masliah E. Detection of borna disease virus antigen and RNA in human autopsy brain samples from neuropsychiatric patients. Virology. 1996;223(2):272–282. doi: 10.1006/viro.1996.0479. [DOI] [PubMed] [Google Scholar]
- de la Torre JC, Bode L., Durrwald R., Cubitt B., Ludwig H. Sequence characterization of human borna disease virus. Virus Res. 1996;44(1):33–44. doi: 10.1016/0168-1702(96)01338-x. [DOI] [PubMed] [Google Scholar]
- Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22(7):1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti I.V., Lee Y.M., Goodson H.V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Dunia D., Cubitt B., de la Torre JC. Mechanism of borna disease virus entry into cells. J. Virol. 1998;72(1):783–788. doi: 10.1128/jvi.72.1.783-788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Eberharter A., John S., Cook R.G., Turner B.M., Workman J.L. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999;274(9):5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Honda T., Tomonaga K. Nucleocytoplasmic shuttling of viral proteins in borna disease virus infection. Viruses. 2013;5(8):1978–1990. doi: 10.3390/v5081978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz G.A., Zhang K., McBrian M.A., Grunstein M., Kurdistani S.K., Berk A.J. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321(5892):1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Gao H., Zhang L., Jia J., Liu X., Zheng P., Ma L., Li W., Deng J., Wang X., Yang L., Wang M., Xie P. Borna disease virus infection perturbs energy metabolites and amino acids in cultured human oligodendroglia cells. PLoS One. 2012;7(9):e44665. doi: 10.1371/journal.pone.0044665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.S., Watanabe M., Palacios J.A., Kamitani W., Komoto S., Kobayashi T., Tomonaga K., Ikuta K. Varied persistent life cycles of borna disease virus in a human oligodendroglioma cell line. J. Virol. 2002;76(8):3873–3880. doi: 10.1128/JVI.76.8.3873-3880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Takahashi K., Peng X., Fukuda K., Ohno K., Ogawa T., Gonda K., Mori N., Niwa S., Shigeta S. Detection and sequence analysis of borna disease virus p24 RNA from peripheral blood mononuclear cells of patients with mood disorders or schizophrenia and of blood donors. J. Virol. 1998;72(12):10044–10049. doi: 10.1128/jvi.72.12.10044-10049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.Y., Verdel A., Tsai C.C., Simon C., Juguilon H., Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 2001;276(50):47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- Kinnunen P.M., Palva A., Vaheri A., Vapalahti O. Epidemiology and host spectrum of borna disease virus infections. J. Gen. Virol. 2013;94(Pt 2):247–262. doi: 10.1099/vir.0.046961-0. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Kamitani W., Zhang G., Watanabe M., Tomonaga K., Ikuta K. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J. Virol. 2001;75(7):3404–3412. doi: 10.1128/JVI.75.7.3404-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster-Patzlaff C., Hosseini S.M., Reuss B. Persistent borna disease virus infection changes expression and function of astroglial gap junctions in vivo and in vitro. Brain Res. 2007;1184:316–332. doi: 10.1016/j.brainres.2007.09.062. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Brownell J.E., Sobel R.E., Ranalli T.A., Cook R.G., Edmondson D.G., Roth S.Y., Allis C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383(6597):269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Lee T.S., Goh L., Chong M.S., Chua S.M., Chen G.B., Feng L., Lim W.S., Chan M., Ng T.P., Krishnan K.R. Downregulation of TOMM40 expression in the blood of Alzheimer disease subjects compared with matched controls. J. Psychiatr. Res. 2012;46(6):828–830. doi: 10.1016/j.jpsychires.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Li D., Lei Y., Deng J., Zhou C., Zhang Y., Li W., Huang H., Cheng S., Zhang H., Zhang L., Huang R., Liu X., Ma L., Wang X., Li J., Xie P. Human but not laboratory borna disease virus inhibits proliferation and induces apoptosis in human oligodendrocytes invitro. PLoS One. 2013;8(6):e66623. doi: 10.1371/journal.pone.0066623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P.M. Chromatin regulation of virus infection. Trends Microbiol. 2006;14(3):132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Liu X., Yang Y., Zhao M., Bode L., Zhang L., Pan J., Lv L., Zhan Y., Liu S., Zhang L., Wang X., Huang R., Zhou J., Xie P. Proteomics reveal energy metabolism and mitogen-activated protein kinase signal transduction perturbation in human borna disease virus Hu-H1-infected oligodendroglial cells. Neuroscience. 2014;268:284–296. doi: 10.1016/j.neuroscience.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte P., Thomas J., Texier P., Caron C., Khochbin S., Epstein A.L. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 2004;78(13):6744–6757. doi: 10.1128/JVI.78.13.6744-6757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H., Bode L., Gosztonyi G. Borna disease: a persistent virus infection of the central nervous system. Prog. Med. Virol. 1988;35:107–151. [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Hayashi Y., Omori H., Honda T., Daito T., Horie M., Ikuta K., Fujino K., Nakamura S., Schneider U., Chase G., Yoshimori T., Schwemmle M., Tomonaga K. Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe. 2012;11(5):492–503. doi: 10.1016/j.chom.2012.04.009. [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Olson E.N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. U.S.A. 2000;97(26):14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C.F., Fatzer R.S., Beck K., Vandevelde M., Zurbriggen A. Studies on canine distemper virus persistence in the central nervous system. Acta Neuropathol. 1995;89(5):438–445. doi: 10.1007/BF00307649. [DOI] [PubMed] [Google Scholar]
- Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L., Salinas-Riester G., Dettenhofer M., Kang H., Farinelli L., Chen W., Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Poenisch M., Burger N., Staeheli P., Bauer G., Schneider U. Protein X of borna disease virus inhibits apoptosis and promotes viral persistence in the central nervous systems of newborn-infected rats. J. Virol. 2009;83(9):4297–4307. doi: 10.1128/JVI.02321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen C.A., Vassilev A., Cook R.G., Allis C.D., Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274(3):1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Schneemann A., Schneider P.A., Lamb R.A., Lipkin W.I. The remarkable coding strategy of borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology. 1995;210(1):1–8. doi: 10.1006/viro.1995.1311. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Suberbielle E., Stella A., Pont F., Monnet C., Mouton E., Lamouroux L., Monsarrat B., Gonzalez-Dunia D. Proteomic analysis reveals selective impediment of neuronal remodeling upon borna disease virus infection. J. Virol. 2008;82(24):12265–12279. doi: 10.1128/JVI.01615-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. Histone acetylation and an epigenetic code. Bioessays. 2000;22(9):836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng E.E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Williams B.L., Hornig M., Yaddanapudi K., Lipkin W.I. Hippocampal poly(ADP-Ribose) polymerase 1 and caspase 3 activation in neonatal bornavirus infection. J. Virol. 2008;82(4):1748–1758. doi: 10.1128/JVI.02014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Xu W., Cao L., Li X., He T., Wu Z., Li W. SAHA treatment reveals the link between histone lysine acetylation and proteome in nonsmall cell lung cancer A549 cells. J. Proteome Res. 2013;12(9):4064–4073. doi: 10.1021/pr4004079. [DOI] [PubMed] [Google Scholar]
- Wu Y.J., Schulz H., Lin C.C., Saar K., Patone G., Fischer H., Hubner N., Heimrich B., Schwemmle M. Borna disease virus-induced neuronal degeneration dependent on host genetic background and prevented by soluble factors. Proc. Natl. Acad. Sci. U.S.A. 2013;110(5):1899–1904. doi: 10.1073/pnas.1214939110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26(37):5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- Zhou S., Liu R., Zhao X., Huang C., Wei Y. Viral proteomics: the emerging cutting-edge of virus research. Sci. China Life Sci. 2011;54(6):502–512. doi: 10.1007/s11427-011-4177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao J., Meyerdierks A., Feng L., Naumovski L., Bottger E.C., Omary M.B. Interferon-alpha induces nmi-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J. Biol. Chem. 2000;275(28):21364–21371. doi: 10.1074/jbc.M003177200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. BDV p24 and p40 Detection. (a) Detection of BDV p24 and p40 RNA by reverse transcription polymerase chain reaction (RT-PCR). Detection of BDV p24 and p40 proteins by (b) Western blotting and (c) immunofluorescence assay. 100% of OL/BDV cells were infected, and 100% of control cells were non-infected.
Supplementary Material
Supplementary Material
Supplemental Figure 2. The link among histone acetylation, gene transcription and proteomics in response to BDV infection.