Highlights
-
•
Deiminated proteins are identified for the first time in a camelid species.
-
•
Extracellular vesicles (EVs) were characterised in llama serum using NTA, protein-specific EV markers and TEM.
-
•
Key immune, metabolic and nuclear proteins are deiminated in llama serum and EVs.
-
•
Comparative studies on deimination may inform inflammatory and metabolic diseases.
Keywords: Peptidylarginine deiminases (PADs), Protein deimination, Llama (Lama glama), extracellular vesicles (EVs), Innate immunity, Adaptive immunity, Metabolism, Complement, Nanobodies, Immunoglobulin, Adiponectin, Histone
Abstract
Peptidylarginine deiminases (PADs) are phylogenetically conserved calcium-dependent enzymes which post-translationally convert arginine into citrulline in target proteins in an irreversible manner, causing functional and structural changes in target proteins. Protein deimination causes generation of neo-epitopes, affects gene regulation and also allows for protein moonlighting. Furthermore, PADs have been found to be a phylogenetically conserved regulator for extracellular vesicle (EVs) release. EVs are found in most body fluids and participate in cellular communication via transfer of cargo proteins and genetic material. In this study, post-translationally deiminated proteins in serum and serum-EVs are described for the first time in camelids, using the llama (Lama glama L. 1758) as a model animal. We report a poly-dispersed population of llama serum EVs, positive for phylogenetically conserved EV-specific markers and characterised by TEM. In serum, 103 deiminated proteins were overall identified, including key immune and metabolic mediators including complement components, immunoglobulin-based nanobodies, adiponectin and heat shock proteins. In serum, 60 deiminated proteins were identified that were not in EVs, and 25 deiminated proteins were found to be unique to EVs, with 43 shared deiminated protein hits between both serum and EVs. Deiminated histone H3, a marker of neutrophil extracellular trap formation, was also detected in llama serum. PAD homologues were identified in llama serum by Western blotting, via cross reaction with human PAD antibodies, and detected at an expected 70 kDa size. This is the first report of deiminated proteins in serum and EVs of a camelid species, highlighting a hitherto unrecognized post-translational modification in key immune and metabolic proteins in camelids, which may be translatable to and inform a range of human metabolic and inflammatory pathologies.
1. Introduction
Lamoids, or llamas, belong to a family of camelids which are economically important livestock and have developed complex features and immunological traits related to their habitat (Wu et al., 2014; Saadeldin et al., 2018a). The llama (Lama glama), Bactrian camel (Camelus bactrianus), dromedary (Camelus dromedarius) and alpaca (Vicugna pacos) differ in their habitat. The Bactrian camel and dromedary are adapted to arid-desert-adapted environments, alpacas to plateaus, and the llama to higher altitudes. Llamas are historically found in the Andean highlands, specifically the Altiplano of southeast Perú and western Bolivia, as well as in Chile and Argentina, which has the third highest population of llamas (Wu et al., 2014). The domesticated llama is closely related to two extant wild South American camelids, the vicuña (Vicugna vicugna) and guanaco (Lama guanicoe). Previous genomic studies have revealed a range of specific adaptions in camelids relating to fat and water metabolism, osmoregulation, blood glucose level regulation, stress responses to heat, aridity, as well as to intense ultraviolet radiation and dust (Wu et al., 2014). Furthermore, a particularly important feature in camelid immunity is the production of small, homodimeric heavy chain-only, antibodies (HCAbs) which are of great value for the biomedical industry (Henry et al., 2019).
Peptidylarginine deiminases (PADs) are phylogenetically conserved calcium-dependent enzymes which post-translationally convert arginine into citrulline in target proteins in an irreversible manner. This can cause functional and structural changes in target proteins (Vossenaar, 2003; György et al., 2006; Wang and Wang, 2013; Bicker and Thompson, 2013). Structures most prone to deimination are beta-sheets and intrinsically disordered proteins, while the position of the arginine is also important; arginines sitting next to aspartic acid residues are most prone to citrullination, arginines next to glutamic acid residues are rarely citrullinated and those flanked by proline are poorly citrullinated (Nomura, 1992; Tarcsa et al., 1996; György et al., 2006). Protein deimination can affect gene regulation, cause generation of neoepitopes (Witalison et al., 2015; Lange et al., 2017) and may also allow for protein moonlighting, an evolutionary acquired phenomenon facilitating proteins to exhibit several physiologically relevant functions within one polypeptide chain (Henderson and Martin, 2014; Jeffrey, 2018; Magnadóttir et al., 2018a). PADs have been identified throughout phylogeny from bacteria to mammals, with 5 tissue specific PAD isozymes in mammals, 3 in chicken, 1 in bony fish and arginine deiminase homologues in bacteria (Vossenaar et al., 2003; Rebl et al., 2010; Magnadóttir et al., 2018a, a; Kosgodage et al., 2019). While studies on PADs in relation to human pathophysiology, including cancer, autoimmune and neurodegenerative diseases (Wang and Wang, 2013; Witalison et al., 2015; Lange et al., 2017; Kosgodage et al., 2017 & 2018) and CNS regeneration (Lange et al., 2011 and 2014) exist, relatively little phylogenetic research has been carried out on PADs in relation to normal physiology and evolutionary acquired adaptions of the immune system. Recent comparative studies focussing on roles for PADs in teleost fish have identified post-translational deimination in key proteins of innate, adaptive and mucosal immunity (Magnadóttir et al., 2018a, b; Magnadottir et al., 2019a; Magnadóttir et al., 2019b). A recent study in shark also revealed novel insights into this post-translational modification in relation to key immune factors, including shark immunoglobulins (Criscitiello et al., 2019). As the camelid family has developed unusual small immunoglobulins similar as to shark through convergent evolution, indicating common factors between shark and camelid immunity, we felt that an investigation of post-translationally deiminated proteins in camelids was warranted.
As PADs have been identified to be a key regulator of extracellular (EV)-release, a mechanism that has been found to be phylogenetically conserved from bacteria to mammals (Kholia et al., 2015; Kosgodage et al., 2017, 2018; Gavinho et al., 2019; Kosgodage et al., 2019), the characterisation of EVs in camelids is of further interest. Extracellular vesicles (EVs) are found in most body fluids and participate in cellular communication via transfer of cargo proteins and genetic material (Inal et al., 2013; Colombo et al., 2014; Lange et al., 2017; Turchinovich et al., 2019; Vagner et al., 2019). EVs in body fluids, including serum, can also be useful biomarkers to reflect health status (Hessvik and Llorente, 2018; Ramirez et al., 2018). Previous work on EVs has hitherto mainly been in the context of human pathologies, while recently comparative studies are growing (Iliev et al., 2018; Yang et al., 2015; Magnadóttir et al., 2019b; Criscitiello et al., 2019; Gavinho et al., 2019; Kosgodage et al., 2019). Few studies have been performed on EVs in camelids but therapeutic effects of EVs isolated from camel milk have been identified in halting cancer progression (Badawy et al., 2018). A recent study in shark identified for the first time deiminated small immunoglobulin proteins as part of EV cargo (Criscitiello et al., 2019). Due to the link between camelid and shark immunity through convergent evolution including the unusual immunoglobulin structure of small heavy chain-only Ig’s, a comparative study on EVs and deiminated EV cargo in a camelid species may provide further insights into shared immunological traits. Camelids have an unusual Ig repertoire and a large diversity of functional nanobodies has been identified in the llama (Harmsen and De Haard, 2007; Deschaght et al., 2017). This has made camelids an important source for small immunoglobulins that can be used for immunotherapy purposes, including for tumour targeting (van Lith et al., 2016), as well as for assessing cancer metastasis (Ramos-Gomes et al., 2018). As these nanobodies can also penetrate the blood-brain barrier (Širochmanová et al., 2018) they are of great value for a range of therapeutic treatment applications, including for brain cancers (Iqbal et al., 2010). Furthermore, as the camelid family has acquired unique metabolic features, they are also of interest as a model species for informing metabolic diseases.
In the current study we assessed post-translationally deiminated proteins in llama serum and serum-derived EVs, and report for the first time EV-mediated export of deiminated key immune, metabolic and nuclear proteins in serum of a camelid species.
2. Materials and methods
2.1. Animals and sampling
Llama (Lama glama L. 1758) serum was shared from excess blood collected in routine health checks of a resident male llama at the Texas A&M Winnie Carter Wildlife Center. Blood collected from the jugular vein of this 21 year old llama was allowed to clot at room temperature for 2 h before serum was collected by centrifuging at 300 g for 10 min. Serum was aliquoted and immediately frozen at −80 °C until further use.
2.2. Extracellular vesicle (EV) isolation and nanoparticle tracking analysis (NTA)
EVs were isolated by step-wise centrifugation according to established protocols using ultracentrifugation and the recommendations of MISEV2018 (the minimal information for studies of extracellular vesicles 2018; Théry et al., 2018). Llama serum was diluted 1:5 in ultrafiltered (using a 0.22 μm filter) Dulbecco’s PBS (DPBS, 100 μl serum added to 400 μl DPBS) and then centrifuged at 4000 g for 30 min at 4 °C for removal of cells and cell debris. The supernatant was collected and centrifuged at 100,000 g for 1 h at 4 °C. The pellet was then resuspended in DPBS and washed again at 100,000 g for 1 h at 4 °C. The resulting EV-enriched pellet was resuspended in 100 μl DPBS, diluted 1/100 in DPBS and analysed by NTA, based on Brownian motion of particles in suspension, using the NanoSight NS300 system (Malvern, UK). The NanoSight was used in conjunction with a syringe pump to ensure continuous flow of the sample, with approximately 40–60 particles per frame and videos were recorded for 5 x 90 s. The replicate histograms generated from the recordings were averaged.
2.3. Transmission electron microscopy (TEM)
EVs were isolated from serum as described above, the EV pellets were fixed with 2.5 % glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.0) for 1 h at 4 °C, resuspended in 100 mM sodium cacodylate buffer (pH 7.0), placed on to a grid with a glow discharged carbon support film, stained with 2 % aqueous uranyl acetate (Sigma-Aldrich) and thereafter viewed in TEM. Imaging was performed using a JEOL JEM 1400 transmission electron microscope (JEOL, Japan) operated at 80 kV at a magnification of 80,000–100,000. Digital images were recorded using an AMT XR60 CCD camera (Deben, UK).
2.4. Western blotting
Llama serum and EV isolates (an EV pellet derived from 100 μl serum, reconstituted in 100 μl DPBS after isolation and purification) were diluted 1:1 in 2x Laemmli sample buffer, boiled for 5 min at 100 °C and separated by SDS-PAGE on 4–20 % gradient TGX gels (BioRad UK). Approximately 5 μg protein was loaded per lane and transferred to nitrocellulose membranes using semi-dry Western blotting. Blocking of membranes was performed in 5 % BSA in TBS-T for 1 h at room temperature (RT) and incubation with primary antibodies, diluted in TBS-T, was carried out at 4 °C overnight (F95 MABN328, Merck, 1/1000; PAD2 ab50257, Abcam, 1/1000; PAD3 ab50246, 1/1000; PAD4 ab50247, 1/1000; citH3 ab5103, 1/1000; CD63 ab216130, 1/1000; Flot-1 ab41927, 1/2000). The membranes were washed in TBS-T for 3 × 10 min at RT and thereafter incubated in the corresponding secondary antibody (anti-rabbit IgG BioRad or anti-mouse IgM BioRad, diluted 1/4000 in TBS-T) for 1 h at RT. Membranes were washed for 6 × 10 min in TBS-T and visualisation performed using electrochemiluminescence (ECL) and the UVP BioDoc-ITTM System (Thermo Fisher Scientific, UK).
2.5. Immunoprecipitation and identification of deiminated proteins in llama serum and EVs
For isolation of total deiminated proteins from llama serum and serum derived EVs, the Catch and Release immunoprecipitation kit (Merck, UK) was used together with the F95 pan-deimination antibody (MABN328, Merck), which has been developed against a deca-citrullinated peptide and specifically detects proteins modified by citrullination (Nicholas and Whitaker, 2002). For F95 enrichment, 50 μl serum was used according to the manufacturer’s instructions (Merck). For EVs, total protein was first extracted from EV-enriched pellets derived from 100 μl serum, using 100 μl radioimmunoprecipitation assay (RIPA) buffer, containing protease inhibitor cocktail (P8340, Sigma, UK), and shaken gently on ice for 2 h. Thereafter proteins were isolated from the EVs by centrifugation at 16,000 g for 30 min, collecting the supernatant containing the proteins. Immunoprecipitation was carried out according to the manufacturer’s instructions (Merck), using a rotating platform overnight at 4 °C. The F95 bound proteins were eluted using denaturing elution buffer, according to the manufacturer’s instructions (Merck). The F95 enriched eluates were then either analysed by Western blotting or by LC—MS/MS (Cambridge Proteomics, Cambridge, UK). For LC—MS/MS, the F95-enriched eluates were run 1 cm into a SDS-PAGE gel and the whole F95-enriched eluate was cut out as one band, whereafter it was processed for proteomic analysis (carried out by Cambridge Proteomics). Peak files were submitted to in-house Mascot (Matrix Science; Cambridge Proteomics). Databases used for protein identification (in house, Cambridge Proteomics) were as follows: Camelidae_family_20190613 (21429 sequences; 9086806 residues) and also specifically for llama: Lama_glama_20190613 (234 sequences; 52757 residues).
3. Results
3.1. EV analysis in llama serum
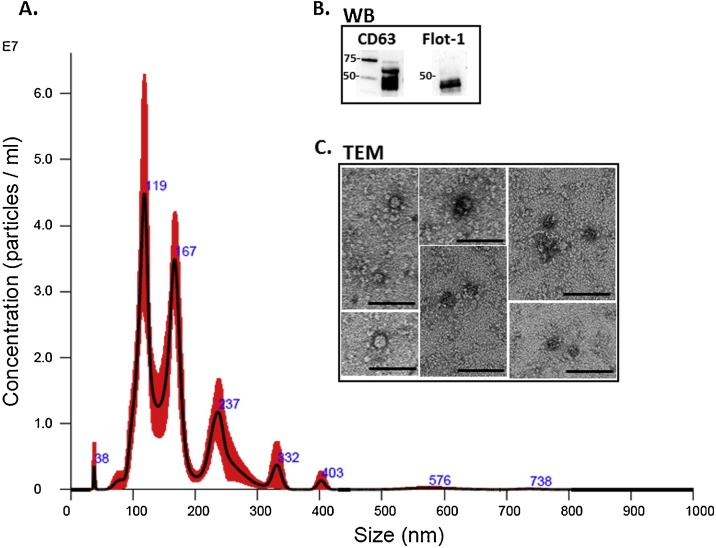
EVs from llama serum were characterised, following step-wise ultracentrifugation, by size exclusion using NTA, by morphological analysis using TEM and by Western blotting using EV-specific markers (Fig. 1 ). A poly-dispersed population of EVs in the size range of 30–576 nm, with main peaks at 38, 119, 167, 237, 323 and 403 nm was identified by NTA analysis (Fig. 1A). Western blotting confirmed that the llama serum EVs were positive for the EV-specific markers CD63 and Flotillin-1 (Fig. 1B). TEM analysis confirmed a poly-dispersed EVs population (Fig. 1C).
Fig. 1.
Extracellular vesicles (EVs) isolated from llama serum. A. Nanoparticle tracking analysis shows a poly-dispersed population of EVs in the size range of 30–576 nm, with main peaks at 38, 119, 167, 237, 323 and 403 nm. B. Llama serum EVs are positive for the EV-specific markers CD63 and Flotillin-1 (Flot-1). C. Transmission electron microscopy (TEM) imaging of EVs isolated from llama serum shows a polydispersed population; scale bar represents 100 nm.
3.2. PAD and deiminated proteins in llama serum
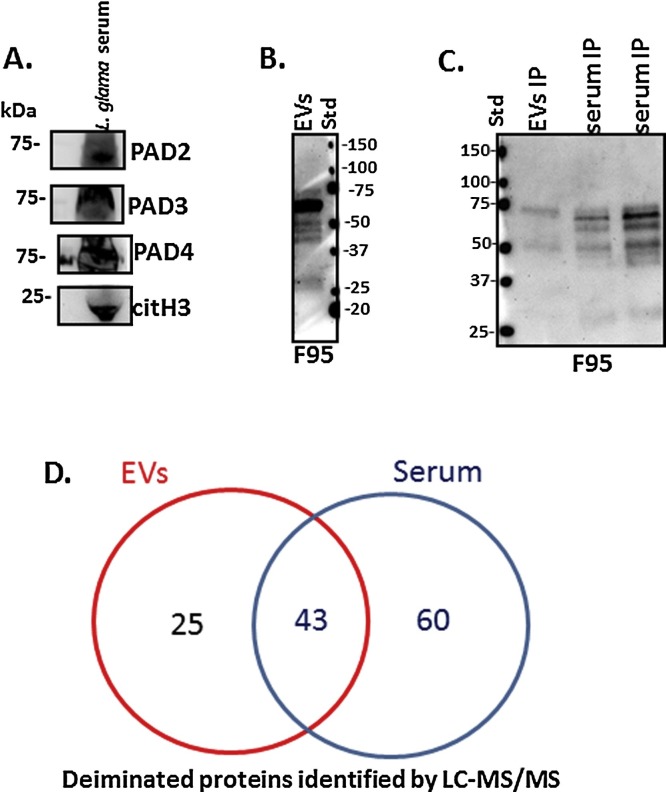
A cross-reaction with human PAD2, 3 and 4 isozyme-specific antibodies was observed in llama serum by Western blotting, at an approximate 70–75 kDa size range as expected for mammalian PADs (Fig. 2 A). Deiminated histone H3 was also detected in llama serum by Western blotting at the expected approximate 20 kDa size (Fig. 2A). Total deiminated proteins in llama serum-EVs were detected by Western blotting using the F95 pan-deimination antibody, revealing a range of proteins between 25–100 kDa in size (Fig. 2B). Deiminated proteins were also assessed by Western blotting after F95 enrichment, using immunoprecipitation (IP) from llama serum and serum-derived EVs (Fig. 2C). Deiminated proteins from the F95 enriched eluates were further identified by LC–MS/MS analysis. In llama serum, 103 hits for camelid proteins were identified, as listed in Table 1 and Supplementary Table 1. Overall, 43 of these deiminated protein hits were common to whole serum and serum-derived EVs, while 60 hits were specific for whole serum (Fig. 2D).
Fig. 2.
Western blotting of deiminated proteins and PAD in llama serum. A. Llama PAD homologues were identified at the expected size of approximately 70–75 kDa using the anti-human PAD2, PAD3 and PAD4 isozyme specific antibodies respectively. Deiminated histone H3 (citH3), representative of neutrophil extracellular traps (NETs), was verified in llama serum. B. Total deiminated proteins were assessed by Western blotting in llama serum EVs, using the F95 pan-deimination specific antibody. C. Immunoprecipitated (IP) deiminated proteins after F95-enrichment were assessed both in serum-EVs and whole serum of llama by Western blotting. D. The Venn diagram represents deiminated proteins identified in total serum and serum-derived EVs by F95 enrichment and LC–MS/MS analysis. Overall, 43 proteins were identified in common with both samples, while 60 proteins were found deiminated in serum only and 25 proteins were identified as deiminated in EVs only.
Table 1.
Deiminated proteins identified by F95 enrichment and LC–MS/MS in total serum of llama (Lama glama). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody. The F95 enriched eluate was analysed by LC–MS/MS and peak list files were submitted to in-house Mascot (Cambridge Proteomics). Peptide sequence hits scoring with Lama glama (LAMGL) are included as well as hits with other camelids (CAMFR=Camelus ferus; CAMDR=Camelus dromedaries; LAMGU=Lama guanicoe; VICPA=Vicugna pacos (Alpaca)). Hits with uncharacterized proteins are omitted in the list. For a full list of peptide sequences and m/z values see Supplementary Table 1. An asterix (*) indicates that the protein hit is specific to whole serum only.
| Protein name (*unique for serum) | Number of peptide sequences identified | Total score (p<0.05)ⱡ |
|---|---|---|
| *O97643_LAMGL | 6 | 282 |
| Fibrinogen A-alpha chain | ||
| *P01973|HBA_LAMGL | 5 | 189 |
| Hemoglobin subunit alpha | ||
| P68226|HBB_LAMGL | 4 | 166 |
| Hemoglobin subunit beta | ||
| Q865W8_LAMGL | 4 | 109 |
| Beta actin | ||
| A0A1W5VKM5_LAMGL | 2 | 117 |
| Anti-RON nanobody | ||
| *A0A1W5VKM7_LAMGL | 2 | 111 |
| Anti-RON nanobody | ||
| *A0A1W5VKR8_LAMGL | 2 | 96 |
| Anti-RON nanobody | ||
| *A0A1W5VKQ9_LAMGL | 1 | 66 |
| Anti-RON nanobody | ||
| S9XDK9_CAMFR | 34 | 1575 |
| Complement C3-like protein | ||
| S9WI87_CAMFR | 20 | 972 |
| Serum albumin | ||
| S9XAP9_CAMFR | 9 | 522 |
| Keratin, type I cytoskeletal 14-like protein | ||
| S9Y6J1_CAMFR | 11 | 502 |
| Keratin, type II cytoskeletal 5 isoform 13-like protein | ||
| *S9 × 688_CAMFR | 10 | 462 |
| Keratin 6A-like protein | ||
| *S9YD43_CAMFR | 9 | 443 |
| Complement component 4A-like protein | ||
| *S9Y253_CAMFR | 8 | 380 |
| Kininogen-2 isoform I | ||
| *T0MII3_CAMFR | 7 | 371 |
| Alpha-2-macroglobulin-like protein | ||
| S9XI90_CAMFR | 8 | 357 |
| Keratin, type II cytoskeletal 75-like isoform | ||
| S9 × 494_CAMFR | 6 | 285 |
| Keratin, type I cytoskeletal 42 | ||
| S9XBS9_CAMFR | 6 | 263 |
| Ig gamma-3 chain C region | ||
| *S9XYY2_CAMFR | 5 | 246 |
| Hemopexin | ||
| A0A075T9L1_CAMDR | 4 | 174 |
| Dipeptidylpeptidase 4 | ||
| S9XXW2_CAMFR | 3 | 172 |
| Fibrinogen beta chain | ||
| *S9WDV3_CAMFR | 3 | 169 |
| Fibrinogen gamma chain isoform gamma-B | ||
| *A0A1K0GY87_VICPA | 4 | 166 |
| Globin A1 | ||
| S9WB99_CAMFR | 3 | 146 |
| Histone H2B | ||
| S9XNF8_CAMFR | 3 | 142 |
| Xaa-Pro dipeptidase | ||
| *S9YS49_CAMFR | 3 | 132 |
| Putative E3 ubiquitin-protein ligase Roquin | ||
| *S9YGW7_CAMFR | 3 | 123 |
| Heparin cofactor 2 | ||
| S9WPM4_CAMFR | 3 | 115 |
| Adiponectin | ||
| A2V743_CAMDR | 4 | 109 |
| Beta actin | ||
| *S9XP08_CAMFR | 1 | 109 |
| Inter-alpha-trypsin inhibitor heavy chain H1 | 2 | |
| S9XM15_CAMFR | 2 | |
| Ferritin | ||
| T0NNK2_CAMFR | 1 | 93 |
| L-lactate dehydrogenase | ||
| S9 × 3E8_CAMFR | 1 | 90 |
| Ig kappa chain V-II region RPMI 6410-like protein | ||
| A0A0A0PAR2_CAMDR | 2 | 88 |
| Heat shock protein 90 | ||
| *S9XYF2_CAMFR | 2 | 87 |
| Heat shock cognate protein HSP 90-beta-like isoform 3 | ||
| S9XHZ4_CAMFR | 1 | 83 |
| Phosphotriesterase-related protein | ||
| S9XM68_CAMFR | 2 | 73 |
| Xaa-Pro dipeptidase isoform 3 | ||
| S9WT57_CAMFR | 2 | 73 |
| Tubulin beta chain | ||
| *S9YV02_CAMFR | 1 | 69 |
| Non-specific protein-tyrosine kinase | ||
| *S9XC57_CAMFR | 1 | 63 |
| Plasminogen | ||
| *S9YL21_CAMFR | 2 | 62 |
| Apolipoprotein A-I | ||
| *S9WKZ8_CAMFR | 1 | 62 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | ||
| S9WIA5_CAMFR | 1 | 61 |
| Glutathione synthetase | ||
| T0MHN9_CAMFR | 1 | 60 |
| Pyruvate kinase | ||
| *S9Y4U4_CAMFR | 1 | 58 |
| Complement C1q subcomponent subunit C isoform 2 | ||
| S9WAX5_CAMFR | 2 | 57 |
| Unconventional myosin-Va isoform 2 | ||
| S9WVY1_CAMFR | 1 | 56 |
| Actinin, alpha 1 isoform 6-like protein | ||
| S9XA40_CAMFR | 1 | 55 |
| Heat shock cognate protein | ||
| *S9YFM0_CAMFR | 1 | 52 |
| Keratin, type II cytoskeletal 71 | ||
| A0A0U2KTX5_CAMDR | 1 | 52 |
| VHH5 (Fragment) | ||
| S9XR87_CAMFR | 1 | 50 |
| Ig lambda chain C regions isoform 19-like protein | ||
| S9WGH8_CAMFR | 1 | 49 |
| Lysozyme | ||
| S9WMX2_CAMFR | 2 | 49 |
| Dystonin | ||
| *S9 × 8K9_CAMFR | 1 | 46 |
| Transaldolase | ||
| *T0NM23_CAMFR | 2 | 45 |
| Rootletin | ||
| S9W6I0_CAMFR | 1 | 45 |
| Ferritin | ||
| S9WF34_CAMFR | 1 | 43 |
| Tubulin alpha chain | ||
| *S9YSI7_CAMFR | 1 | 43 |
| Triosephosphate isomerase | ||
| *S9XSQ6_CAMFR | 1 | 40 |
| Vitamin d-binding protein-like protein | ||
| *S9YMC0_CAMFR | 2 | 40 |
| Transcription factor 20 isoform 1 | ||
| *S9Y636_CAMFR | 1 | 39 |
| Receptor-type tyrosine-protein phosphatase-like N | ||
| *S9 × 6M4_CAMFR | 1 | 37 |
| Dyslexia-associated protein | ||
| T0MH94_CAMFR | 1 | 36 |
| Rabenosyn-5-like protein | ||
| S9WJW3_CAMFR | 1 | 35 |
| N6-adenosine-methyltransferase subunit | ||
| *A8IY99_LAMGU | 1 | 35 |
| Gamma-fibrinogen | ||
| S9WRI7_CAMFR | 1 | 35 |
| Nuclear receptor coactivator 5 isoform 3-like protein | ||
| S9W421_CAMFR | 1 | 35 |
| Hemoglobin, epsilon 1 | ||
| *S9W711_CAMFR | 1 | 34 |
| Charged multivesicular body protein 4c | ||
| *S9 × 089_CAMFR | 1 | 34 |
| Ig lambda chain V-III region LOI-like protein | ||
| S9WUC8_CAMFR | 1 | 32 |
| Ig kappa chain V-II region RPMI 6410-like protein | ||
| *S9WVI6_CAMFR | 1 | 32 |
| Complement C1q subcomponent subunit A | ||
| *S9Y5S1_CAMFR | 1 | 32 |
| Transcriptional repressor NF-X1 | ||
| *S9YC53_CAMFR | 1 | 32 |
| Alpha-1-antitrypsin-like protein | ||
| *S9Y3F6_CAMFR | 1 | 31 |
| Dual specificity testis-specific protein kinase 1 | ||
| S9WKI8_CAMFR | 1 | 31 |
| HEAT repeat-containing protein 7B1 | ||
| *S9WVS9_CAMFR | 1 | 29 |
| Peroxisome proliferator-activated receptor gamma coactivator-related protein 1 | ||
| *S9XVK5_CAMFR | 1 | 29 |
| Transthyretin | ||
| *S9Y967_CAMFR | 1 | 28 |
| General transcription factor II, i isoform 4 isoform 1-like protein | ||
| *T0MC04_CAMFR | 1 | 28 |
| Spermatogenesis-associated protein 2-like protein | ||
| *S9YSZ6_CAMFR | 1 | 28 |
| Centromere protein J |
Ions score is -10*Log(P), where P is the probability that the observed match is a random event. Individual ions scores > 20 indicated identity or extensive homology (p < 0.05). Protein scores were derived from ions scores as a non-probabilistic basis for ranking protein hits. Cut-off was set at Ions score 20.
3.3. Identification of deiminated proteins in EVs from llama serum
Llama serum-derived EVs showed positive for deiminated proteins by Western blotting, using the pan-deimination F95 antibody (Fig. 2B). Deiminated proteins were further identified by F95 enrichment and LC–MS/MS analysis, revealing 68 deiminated protein hits in total for EVs, with 25 hits unique to EVs (not identified from serum). Peptide sequences of hits with camelid proteins and m/z values are listed in Table 2 and Supplementary Table 2. Overlap with deiminated proteins identified in whole llama serum and EVs is represented in the Venn diagram in Fig. 2D.
Table 2.
Deiminated proteins identified by F95 enrichment and LC–MS/MS in EVs isolated from serum of llama (Lama glama). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody, the F95 enriched eluate was analysed by LC–MS/MS and peak list files were submitted to Mascot. Peptide sequence hits scoring with L. Lama glama (LAMGL) are presented as well as hits with other camelids (CAMFR=Camelus ferus; CAMDR=Camelus dromedaries; LAMGU=Lama guanicoe). Hits with uncharacterised proteins are not listed. For a full list of peptide sequences and m/z values see Supplementary Table 2. An asterix (*) indicates that the protein hit is unique for EVs only.
| Protein name (*unique for EVs) | Number of peptide sequences identified | Total score (p<0.05)ⱡ |
|---|---|---|
| A0A1W5VKM5_LAMGL | 2 | 164 |
| Anti-RON nanobody | ||
| Q865W8_LAMGL | 2 | 85 |
| Beta actin | ||
| *S9XAP9_CAMFR | 9 | 554 |
| Keratin, type I cytoskeletal 14-like protein | ||
| *S9 × 688_CAMFR | 10 | 496 |
| Keratin 6A-like protein | ||
| S9Y6J1_CAMFR | 10 | 438 |
| Keratin, type II cytoskeletal 5 isoform 13-like protein | ||
| S9WI87_CAMFR | 9 | 430 |
| Serum albumin | ||
| *S9YN99_CAMFR | 6 | 417 |
| Keratin, type I cytoskeletal 17-like isoform | ||
| *S9XI90_CAMFR | 6 | 318 |
| Keratin, type II cytoskeletal 75-like isoform | ||
| S9 × 494_CAMFR | 5 | 269 |
| Keratin, type I cytoskeletal 42 | ||
| S9XBS9_CAMFR | 4 | 162 |
| Ig gamma-3 chain C region | ||
| A0A075T9L1_CAMDR | 3 | 153 |
| Dipeptidylpeptidase 4 | ||
| S9 × 684_CAMFR | 2 | 136 |
| Keratin, type II cytoskeletal 8 | ||
| S9WB99_CAMFR | 3 | 133 |
| Histone H2B | ||
| S9YQ51_CAMFR | 3 | 114 |
| Tubulin beta chain | ||
| *S9WX81_CAMFR | 3 | 89 |
| Histone 1, H2ai isoform 3-like protein | ||
| *S9 × 8G9_CAMFR | 2 | 88 |
| Desmoplakin | ||
| A2V743_CAMDR | 2 | 85 |
| Beta actin | ||
| A0A0A0PAR2_CAMDR | 2 | 85 |
| Heat shock protein 90 | ||
| S9XA40_CAMFR | 1 | 85 |
| Heat shock cognate protein | ||
| T0NNK2_CAMFR | 1 | 81 |
| L-lactate dehydrogenase | ||
| S9XNF8_CAMFR | 2 | 74 |
| Xaa-Pro dipeptidase | ||
| T0MHN9_CAMFR | 1 | 71 |
| Pyruvate kinase | ||
| S9WVY1_CAMFR | 1 | 60 |
| Actinin, alpha 1 isoform 6-like protein | ||
| *A0A0E3Z5I3_CAMDR | 1 | 59 |
| Superoxide dismutase | ||
| S9XHZ4_CAMFR | 1 | 57 |
| Phosphotriesterase-related protein | ||
| S9W9Y4_CAMFR | 1 | 57 |
| Ferritin | ||
| S9XR87_CAMFR | 1 | 56 |
| Ig lambda chain C regions isoform 19-like protein | ||
| S9 × 3E8_CAMFR | 1 | 50 |
| Ig kappa chain V-II region RPMI 6410-like protein | ||
| S9WAX5_CAMFR | 2 | 49 |
| Unconventional myosin-Va isoform 2 | ||
| S9WF34_CAMFR | 1 | 44 |
| Tubulin alpha chain | ||
| *S9W806_CAMFR | 2 | 43 |
| Filamin-A isoform 1 | ||
| *S9 × 6X3_CAMFR | 2 | 42 |
| Scaffold attachment factor B-like protein | ||
| T0MH94_CAMFR | 1 | 40 |
| Rabenosyn-5-like protein | ||
| *S9Y0S0_CAMFR | 1 | 39 |
| DNA-directed RNA polymerase subunit beta | ||
| S9WJW3_CAMFR | 1 | 38 |
| N6-adenosine-methyltransferase subunit | ||
| S9XM68_CAMFR | 1 | 38 |
| Xaa-Pro dipeptidase isoform 3 | ||
| *S9YV02_CAMFR | 1 | 38 |
| Non-specific protein-tyrosine kinase | ||
| S9WGH8_CAMFR | 1 | 32 |
| Lysozyme | ||
| S9WRI7_CAMFR | 1 | 32 |
| Nuclear receptor coactivator 5 isoform 3-like protein | ||
| S9W421_CAMFR | 1 | 32 |
| Hemoglobin, epsilon 1 | ||
| *S9WI71_CAMFR | 1 | 32 |
| Metabotropic glutamate receptor 3 | ||
| *S9WB50_CAMFR | 1 | 31 |
| TSC22 domain family protein 3-like protein | ||
| S9WMX2_CAMFR | 1 | 31 |
| Dystonin | ||
| S9WKI8_CAMFR | 1 | 31 |
| HEAT repeat-containing protein 7B1 | ||
| S9WIA5_CAMFR | 1 | 31 |
| Glutathione synthetase | ||
| *S9XC05_CAMFR | 1 | 31 |
| Telomere-associated protein RIF1 isoform 1 | ||
| A0A0U2KTX5_CAMDR | 1 | 30 |
| VHH5 | ||
| *T0MGG7_CAMFR | 1 | 30 |
| Nucleoredoxin | ||
| *S9XET3_CAMFR | 1 | 30 |
| Rac GTPase-activating protein 1 | ||
| S9XDK9_CAMFR | 1 | 30 |
| Complement C3-like protein | ||
| *S9XMI2_CAMFR | 1 | 29 |
| Pseudopodium-enriched atypical kinase 1 | ||
| *S9Y3S9_CAMFR | 1 | 29 |
| Core histone macro-H2A.1 isoform 2 | ||
| S9XXW2_CAMFR | 1 | 29 |
| Fibrinogen beta chain | ||
| *S9YGX6_CAMFR | 1 | 29 |
| PAS domain-containing serine/threonine-protein kinase | ||
| *T0NMU1_CAMFR | 1 | 28 |
| SH2 domain-containing protein 7 | ||
| *T0MIT6_CAMFR | 1 | 28 |
| Serine-tRNA ligase, mitochondrial | ||
| *S9W449_CAMFR | 1 | 28 |
| Fc receptor-like protein 5 |
Ions score is -10*Log(P), where P is the probability that the observed match is a random event. Individual ions scores > 22 indicated identity or extensive homology (p < 0.05). Protein scores were derived from ions scores as a non-probabilistic basis for ranking protein hits. Cut-off was set at Ions score 20.
4. Discussion
For the first time deiminated proteins are described in a camelid, using the llama (Lama glama) as a model species. Post-translational deimination was identified in key immune, nuclear and metabolic proteins. A llama PAD homologue was identified at an expected 70–75 kDa size similar as for human PADs by Western blotting via cross reaction with anti-human PAD2, PAD3 and PAD4 antibodies. PAD2 is known to be the phylogenetically most conserved PAD form (Vossenaar et al., 2003; Magnadóttir et al., 2018a) and has also been seen in shark (Criscitiello et al., 2019). Deiminated histone H3, a marker of neutrophil extracellular trap formation (NETosis), was also detected in llama serum by Western blotting and is described for the first time in a camelid species but was recently described in shark (Criscitiello et al., 2019). NETosis is driven by PADs (Li et al., 2010), is conserved throughout phylogeny and is important in innate immune defences against a range of pathogens including bacteria, viruses and helminths (Brinkmann et al., 2004; Branzk et al., 2014; Schönrich and Raftery, 2016). NETosis has also been associated with clearance of apoptotic cells and during tissue remodelling in teleosts (Magnadóttir et al., 2018a, 2019a). Furthermore, NETosis is linked to a range of autoimmune diseases, due to NET activation via neo-epitopes (O’Neil and Kaplan, 2019), which can also lead to organ damage (Lee et al., 2017). NETosis is also associated with cancer (Gonzalez-Aparicio and Alfaro, 2019) and neurodegenerative diseases (Pietronigro et al., 2017).
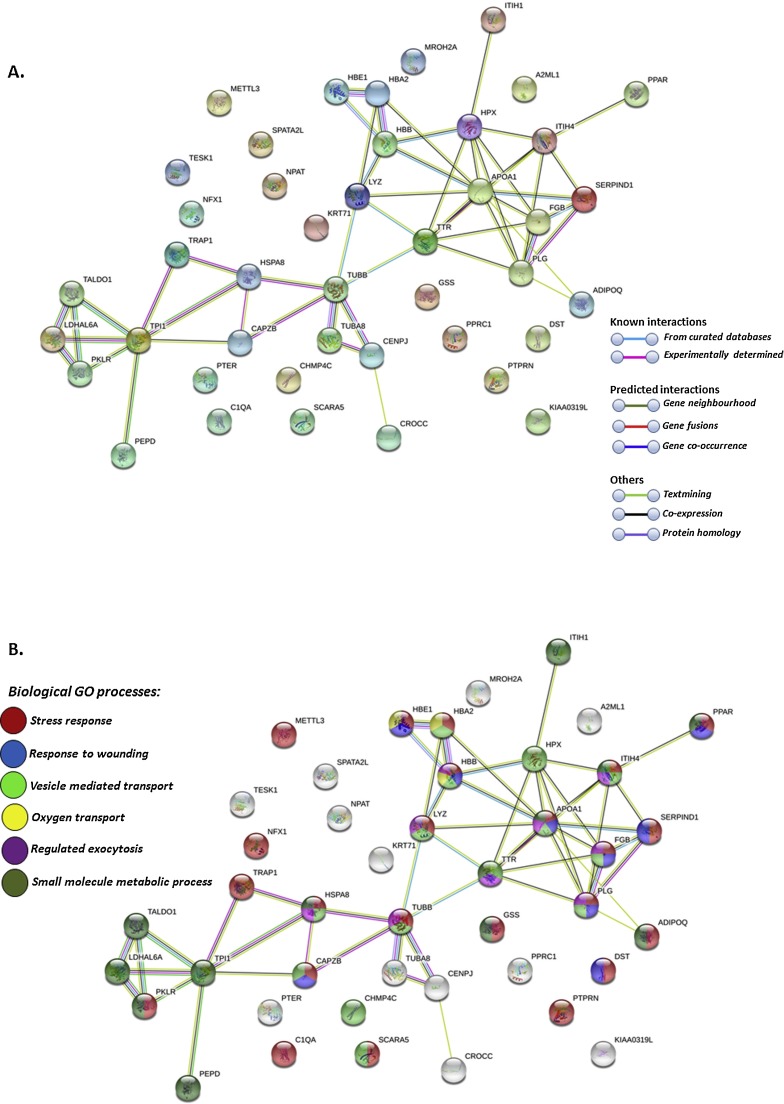
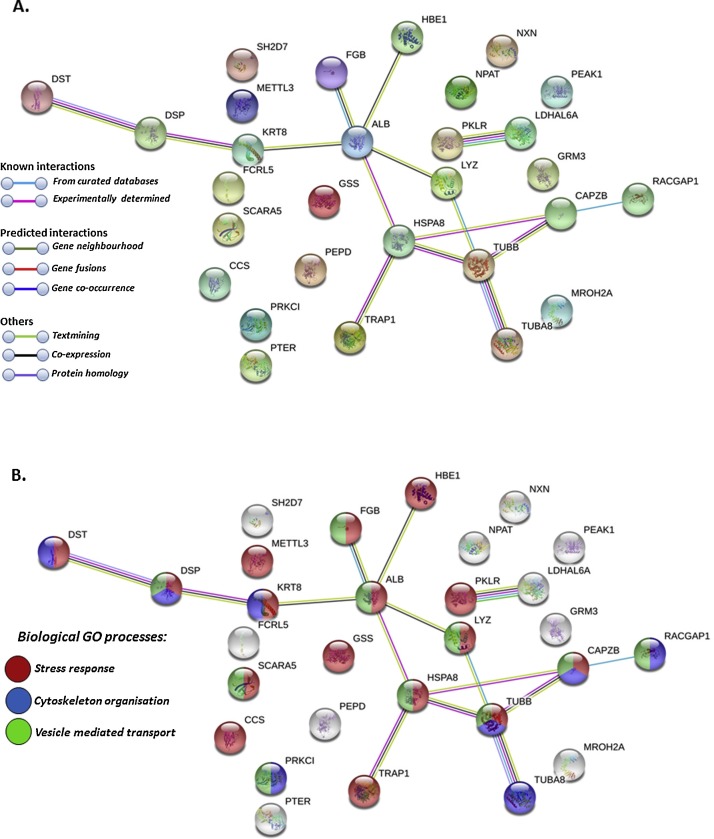
Further deiminated proteins identified in llama serum and serum-derived EVs by F95 enrichment and LC—MS/MS analysis included key proteins of camelid innate and adaptive immunity, nuclear proteins, as well as proteins involved in metabolic function. Using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis (https://string-db.org/) for protein-protein interaction, the PPI enrichment value for deiminated proteins in whole llama serum was found to be p < 1.0e-16 for 43 deiminated proteins out of 103 identified in serum, which indicates that these proteins have more interactions among themselves than what would be expected for a random set of proteins of similar size, drawn from the genome (STRING analysis, see Fig. 3 A). For deiminated proteins in 29 out of the 68 deiminated EV cargo proteins, the PPI enrichment value was found to be p = 0.0193 (STRING analysis, see Fig. 4 A). Such enrichment indicates that the proteins are at least partially biologically connected, as a group. As the camelid proteins are not present in the STRING database, corresponding protein homologues for human were chosen to create the protein-protein interaction networks shown in Fig. 3, Fig. 4. Out of the 103 camelid proteins identified as deiminated in serum, protein IDs for 43 were present for Homo sapiens to perform the assessment of protein-protein interactions and identification of main biological GO pathways (response to stress, response to wounding, oxygen transport, small molecule metabolic process, vesicle mediated transport and regulated exocytosis; Fig. 3B). For llama EVs, out of 68 proteins identified as deiminated, 29 corresponding protein homologues were found in the STRING database for Homo sapiens and were analysed highlighting main biological GO pathways (response to stress, cytoskeleton organisation and vesicle-mediated transport; Fig. 4B).
Fig. 3.
Protein-protein interaction networks of deiminated protein hits identified in whole llama (Lama glama) serum. Reconstruction of protein-protein interactions based on known and predicted interactions using STRING analysis. Due to annotations for camelids not being present in STRING, proteins are based on corresponding human protein identifiers. A. Coloured nodes represent query proteins and first shell of interactors; white nodes are second shell of interactors. B. Biological GO processes are highlighted for the same protein network as follows: red = response to stress; blue = response to wounding; green = vesicle mediated transport; yellow = oxygen transport; purple = regulated exocytosis; dark green = small molecule metabolic process. Coloured lines indicate whether protein interactions are identified via known interactions (curated databases, experimentally determined), predicted interactions (gene neighbourhood, gene fusion, gene co-occurrence) or via text mining, co-expression or protein homology (see colour key for connective lines in A).
Fig. 4.
Protein-protein interaction networks of deiminated protein hits identified in EVs of llama (Lama glama) serum. Reconstruction of protein-protein interactions based on known and predicted interactions using STRING analysis. Due to annotations for camelids not being present in STRING, proteins are based on corresponding human protein identifiers. A. Coloured nodes represent query proteins and first shell of interactors. B. Biological GO processes are highlighted as follows: red = response to stress; blue = cytoskeleton organisation; green = vesicle mediated transport. Coloured lines indicate whether protein interactions are identified via known interactions (curated databases, experimentally determined), predicted interactions (gene neighbourhood, gene fusion, gene co-occurrence) or via text mining, co-expression or protein homology (see colour key for connective lines in A).
Deimination protein candidates identified here in llama serum and EVs, which are involved in immune, nuclear and metabolic functions, are further discussed below, including where appropriate in a comparative context with relevant human diseases. Proteins that have previously been identified as deiminated in other species are listed first.
Nanobodies are based on immunoglobulin single variable domains, derived from the variable domains of heavy chain-only antibodies, which occur naturally in camelids (Deschaght et al., 2017). In the llama the heavy chain-only antibodies are comprised of at least two subclasses (Henry et al., 2019). These types of homodimeric, light chain-less antibodies have evolved through convergent evolution (Brooks et al., 2018), in at least two groups (camelids and cartilaginous fish), and their variable binding domains (VHHS) are of great value for therapeutic and diagnostic applications (Muyldermans, 2013; Criscitiello, 2014; Steeland et al., 2016; Könning et al., 2017; Henry et al., 2019). Nanobodies can act against challenging targets such as small molecules and toxins (Wesolowski et al., 2009; Bever et al., 2016), viruses (Wei et al., 2011; Hassiki et al., 2016; Vanlandschoot et al., 2011; Cohen, 2018), enzymes (Muyldermans, 2013), ion channels (Wei et al., 2011; Danquah et al., 2016) and G protein-coupled receptors GPCRs (Cromie et al., 2015). Llama nanobodies have been shown to tether to early endosomes and to mitochondria (Traub, 2019), be used for diagnostics (Shriver-Lake et al., 2018), be used for design of cancer immunotherapeutics (Hussack et al., 2018; Bannas and Koch-Nolte, 2018; Rossotti et al., 2019) and have been approved for passive immunotherapy (Sheridan, 2019). Our current finding, that llama nanobodies can be post-translationally deiminated may shed some light on their observed structural variation which still remains to be fully explained as sequence alignment does not fully elucidate their diversity (Mitchell and Colwell, 2018). As a structurally analogous immunoglobulin in shark, new antigen receptor (NAR) (Greenberg et al., 1995; Barelle et al., 2009; Flajnik and Dooley, 2009; De Silva et al., 2019) was recently also found to be deiminated (Criscitiello et al., 2019), our current finding may provide novel insights into function of these immune proteins and be useful for refinement in therapeutic nanobody development. Llama single-chain antibodies were here found to be deimination candidates both in llama whole serum and in serum derived EVs, highlighting also their EV-mediated export.
Ig proteins were identified here as being deiminated in llama serum and in serum-derived EVs, scoring with Ig components from other camelids. Immunoglobulins (Ig) are key molecules in adaptive immunity but post-translational deimination of Ig’s has hitherto received little attention. Deimination of the IgG Fc region in patients with bronchiectasis and RA has been identified (Hutchinson et al., 2017). Furthermore, deimination of Ig’s in teleost fish was recently described (Magnadóttir et al., 2019a), as well as in shark (Criscitiello et al., 2019). Given the increased focus on understanding of Ig diversity throughout phylogeny (Smith et al., 2012; Zhang et al., 2013; de los Rios et al., 2015; Zhang et al., 2016, 2017; Stanfield et al., 2018) and the unique features of camelid immunoglobulins (Plasil et al., 2019), our current finding of deimination of llama Ig’s highlights a novel concept that may further understanding of Ig diversity throughout evolution. Of additional interest is the finding that some deiminated Ig proteins were also found to be exported in EVs.
Complement components identified to be deiminated in llama serum included C3, C4 and C1q, while only C3 was identified to be deiminated in serum derived EVs. Complement component C3 plays a central role in all pathways of complement activation and can also be directly activated by self- and non-self surfaces via the alternative pathway without a recognition molecule (Dodds and Law, 1998; Dodds, 2002). In camelids, the alternative and classical pathway haemolytic activity of serum has been assessed in the dromedary camel with respect to age and gender (Olaho-Mukani et al., 1995a; and 1995b). Hitherto little is known about roles for post-translational deimination of complement components throughout phylogeny. In teleost fish, C3 has been identified in serum in deiminated form (Magnadóttir et al., 2019a) and also found to be deiminated in mucosal and serum EVs of teleost fish (Magnadóttir et al., 2019b, 2019c), while post-translationally deiminated C3 was identified in shark total serum but not EVs (Criscitiello et al., 2019). Other complement components, including C4 and C1q, which belong to the classical pathway of complement activation were here identified as deiminated in llama whole serum and some of those have also recently been reported to be deiminated in teleost fish (Magnadóttir et al., 2019a). The C1q subcomponent can bind to the Fc region of immunoglobulins that are bound to antigen and activate the classical part of the complement pathway (Reid et al., 2002; Reid, 2018). Interestingly, an essential role for arginine in C1q has previously been suggested for C1q-IgG interaction (Kojouharova et al., 2004). C1q also serves as a potent pattern recognition molecule which recognises self, non-self and altered self-signals (Nayak et al., 2012; Reid, 2018) and may therefore also bind to deiminated neo-epitopes (Magnadóttir et al., 2019a). The complement system has multifaceted roles. It forms part of the first line of immune defence against invading pathogens, acting in clearance of necrotic or apoptotic cells (Dodds and Law, 1998; Sunyer and Lambris, 1998; Fishelson et al., 2001; Carroll and Sim, 2011). Complement also has roles in regeneration (Del Rio-Tsonis et al., 1998; Haynes et al., 2013) and tissue remodelling (Lange et al., 2004a, 2004b; Lange et al., 2005, 2006, 2019). Furthermore, C1 is also implicated in multiple non-complement functions including binding of apoptotic cells, cleavage of nuclear antigens and cleavage of MHC class I (Lu and Kishore, 2017). Post-translational deimination of complement components may possibly influence their function including cleavage ability, binding, deposition and generation of the convertase.
Apolipoprotein A–I is primarily involved in lipid metabolism where conformational plasticity and flexibility are regarded as key structural features (Arciello et al., 2016). Apo A–I is associated with regulation of mitochondrial function and bioenergetics (White et al., 2017). Furthermore, Apo A–I has been shown to have a regulatory role in the complement system by affecting membrane attach complex (MAC) assembly and thus the final lytic pathway (Hamilton et al., 1993; Jenne et al., 1991; French et al., 1994). Given the diverse roles of Apo A–I, the current finding of deiminated forms in serum may be of quite some relevance and has previously been identified in teleost fish (Magnadóttir et al., 2019a). Apo A–I was here found to be deiminated in whole llama serum only.
Serum albumin is a major acidic plasma protein in vertebrates and serves as a transport molecule for fatty acids, bilirubin, steroids, amino acids and copper, as well as having roles in maintaining the colloid osmotic pressure of blood (Peters, 1996). In camelids, total albumin levels have been assessed, for example in relation to reproductive efficiency (El-Malky et al., 2018) and as biomarkers of oxidative stress (El-Deeb and Buczinski, 2015). While albumin has been identified as a glycoprotein in some species (Metcalf et al., 2007) investigation of post-translational deimination has been limited, but was recently identified in teleost fish (Magnadóttir et al., 2019a). Serum albumin was identified as deiminated in both whole llama serum and EVs.
Hemopexin is a scavenger protein of haemoglobin and a predominant heme binding protein, which contributes to heme homeostasis (Smith and McCulloh, 2015; Immenschuh et al., 2017). Hemopexin also associates with high density lipoproteins (HDL), influencing their inflammatory properties (Mehta and Reddy, 2015). Hemopexin is a plasma glycoprotein that has been previously identified as a deimination candidate in teleost fish (Magnadóttir et al., 2019a) and shark (Criscitiello et al., 2019). Here, hemopexin was found deiminated in whole llama serum only.
Inter-alpha-trypsin inhibitor (heavy chain H1) belongs to the serpin family of proteins, which have protease-inhibitory functions and are involved in diverse physiological and pathological processes including fertilisation, ovulation, coagulation, inflammation, as well as tumorigenesis, metastasis and dementia (Zhuo and Kimata, 2008; Weidle et al., 2018). Inter-alpha-trypsin inhibitor is synthesised in the liver, circulates in the blood and has two chains, a light and heavy chain, whereof the heavy-chain (ITIH) includes a von Willebrand domain and can interact with the extracellular matrix (Bost et al., 1998). ITIH is downregulated in tumours via methylation and ITIH2 is strongly reduced in invasive cancers (Hamm et al., 2008). While ITIH was recently identified as a deimination candidate in teleost fish (Magnadóttir et al., 2019a) further studies on the regulation of ITIH2 via post-translational deimination have not been carried out. ITIH was here identified as deiminated in whole llama serum only.
Fibrinogen is a glycoprotein, synthesised in liver (Tennent et al., 2007) and forms part of the acute phase response as part of the coagulation cascade (Tiscia and Margaglione, 2018). In camelids, fibrinogen is a biomarker for stress and infection (Greunz et al., 2018; El-Bahr and El-Deeb, 2016; El-Deeb and Buczinski, 2015). Impaired mechanism of fibrinogen formation and fibrin polymerization are implicated with various pathologies including coagulopathies and ischemic stroke (Weisel and Litvinov, 2013), while acquired fibrinogen disorders can be associated with cancer, liver disease or post-translational modifications (Besser and MacDonald, 2016). Fibrinogen is indeed a known deimination candidate and this post-translational modification contributes to its antigenicity in autoimmune diseases (Hida et al., 2004; Muller and Radic, 2015; Blachère et al., 2017). Fibrinogen was here identified as deiminated both in whole llama serum and EVs.
Tubulin beta-chain participates in cytoskeletal rearrangement and its deimination has previously been linked to EV release (Kholia et al., 2015). Deimination of tubulin may therefore be crucial for facilitating EV-mediated processes in homeostasis, immune responses and in relation to pathologies. Here, tubulin was identified as deiminated in both whole llama serum and EVs.
Histone H2B was identified as being deiminated in both llama serum and EVs and in addition, Histone 1 (H2ai isoform 3-like protein) and core histone macro-H2A.1 isoform 2 were identified as deiminated in EVs only. Histones undergo various posttranslational modifications that affect gene regulation and can also act in concert (Latham and Dent, 2007; Bird, 2007). In addition to acetylation, phosphorylation and ubiquitination, histones are known to undergo deimination, including H2B (Sohn et al., 2015) and H2A (Hagiwara et al., 2005), as identified in this study in llama. Other histones that are known to undergo deimination include H3 and H4 (Chen et al., 2014; Kosgodage et al., 2018).
Heat shock protein 90 (Hsp90) was here found to be deiminated both in llama whole serum and EVs. HSP90 has been described in camelid (Saeed et al., 2015). Hsp90 is a phylogenetically highly conserved chaperone protein involved in protein folding, stabilisation of proteins against heat stress, and aids in protein degradation (Buchner, 1999; Picard, 2002). Hsp90 also stabilizes a number of proteins required for tumour growth and is therefore important in anti-cancer drug investigations (Goetz et al., 2003). Hsp90 is responsible for most of the ATPase activity of the proteasome (Imai et al., 2003) and has an ATP binding region, which also is the main binding site of drugs (Chiosis et al., 2006). In camelids, Hsp90 has been related to adaptive tolerance of camel somatic cells to acute and chronic heat shock (up to 20 h at 45 degrees Celsius), which is lethal to many mammalian cells (Saadeldin et al., 2018b). Hsp90 has previously been described to be post-translationally deiminated in rheumatoid arthritis, allowing deimination-induced shifts in protein structure to generate cryptic epitopes capable of bypassing B cell tolerance (Travers et al., 2016). It is of some interest to find that Hsp90 is also found deiminated in llama serum, further highlighting protein moonlighting functions of Hsp90 in physiological and pathophysiological context via post-translational deimination. In addition, finding post-translational deimination of the same protein throughout phylogeny also supports translational value between species to further understanding of this post-translational modification in human pathologies.
The following deimination protein candidates identified in the current study in llama serum and serum-EVs have to our knowledge not been previously reported as deiminated:
Alpha 2-macroglobulin is closely related to other thioester containing proteins, such as complement proteins C3, C4 and C5 (Sottrup-Jensen et al., 1985; Davies and Sim, 1981). Alpha-2-M is phylogenetically conserved from arthropods to mammals and found at high levels in mammalian plasma. Alpha-2-M forms part of the innate immune system and clears active proteases from tissue fluids (Armstrong and Quigley, 1999). Here, Alpha-2-M was found deiminated in whole llama serum only and has not reported as deiminated before, to our knowledge.
Adiponectin is the most abundant secreted adipokine with pleiotrophic roles in physiological and pathophysiological processes (Fiaschi, 2019). It has received considerable interest in the field of metabolic and obesity research (Frankenberg et al., 2017; Spracklen et al., 2019), as well as in diabetes (Yamauchi et al., 2003), due to its key function in regulating glucose (Yamauchi et al., 2002). Adiponectin is furthermore linked to longevity (Chen et al., 2019a, 2019b), regenerative functions (Fiaschi et al., 2014) and has roles in myopathies, such as Duchenne muscular dystrophy and collagen VI-related myopathies (Gamberi et al., 2019). Adiponectin is also implicated in a range of cancers, often in relation to obesity (Parida et al., 2019). Furthermore, adiponectin plays roles in reproduction, embryo pre-implantation and embryonic development (Barbe et al., 2019). Due to the range of functions in relation to key pathophysiologies there is great interest in drug development for modulating adiponectin signalling (Fiaschi, 2019). Given the unique metabolic adaptive features of camelids, the identification of post-translational deimination of this key metabolic protein may be of some interest. Recent studies in rheumatoid arthritis made a correlation between inflammation, autoantibodies and adiponectin levels (Hughes-Austin et al., 2018; Liu et al., 2019), while adiponectin itself has not been previously identified to be deiminated to our knowledge. Post-translational deimination may be a hitherto unrecognized mechanism for adiponectin, also in humans, to adapt moonlighting functions via changes in protein folding and therefore interaction with other proteins. Adiponectin is a small 244 aa protein (NP_001171271.1) in humans and contains 2 unfolded regions and 7 arginine sites, while camel adiponectin has 8 arginine sites. These could be subjected to PAD-mediated deimination and therefore modulate adiponectin folding and function, depending on which arginine is deiminated. Here, deiminated adiponectin was identified in whole llama serum only.
Dystonin is a plakin-family adhesion junction plaque protein and was here identified as deiminated in both llama serum and EVs. Dystonin, along with XVII collagen, form hemidesmosomes and both proteins are autoantigens believed to be responsible for the Type II hypersensitivy pathologic in the pruritic skin disease bullous pemphigoid (Bağcı et al., 2017; Basseri et al., 2018). Dystonin has also been linked to Sjögren’s syndrome and linked to a hypermethylation status (Gonzalez et al., 2011), but post-translational deimination of dystonin has not been previously described to our knowledge, although deimination is associated with a range of autoimmune diseases, including Sjögren’s syndrome (Konsta et al., 2014; Selmi et al., 2015).
Xaa-Pro dipeptidase, also known as prolidase, is an enzyme that in humans is encoded by the PEPD gene. Post-translational modifications of prolidase regulate its enzymatic abilities. Deficiency in prolidase leads to a rare, severe autosomal recessive disorder (prolidase deficiency) that causes many chronic, debilitating health conditions in humans (Viglio et al., 2006). These phenotypic symptoms vary and may include skin ulcerations, mental retardation, splenomegaly, recurrent infections, photosensitivity, hyperkeratosis, and unusual facial appearance. Furthermore, prolidase activity was found to be abnormal compared to healthy levels in various medical conditions including: bipolar disorder, breast cancer, endometrial cancer, keloid scar formation, erectile dysfunction, liver disease, lung cancer, hypertension, melanoma, and chronic pancreatitis (Kitchener and Grunden, 2012). In some cancers with increased levels of prolidase activity, such as melanoma, the differential expression of prolidase and its substrate specificity for dipeptides with proline at the carboxyl end suggests the potential of prolidase in becoming a viable, selective endogenous enzyme target for proline prodrugs (Mittal et al., 2005). Serum prolidase enzyme activity is also currently being explored as a biomarker for diseases including chronic hepatitis B and liver fibrosis (Duygu et al., 2013; Şen et al., 2014; Stanfliet et al., 2015). Phosphorylation of prolidase has been shown to increase its activity while dephosphorylation leads to a decrease in enzyme activity. Post-translational deimination of prolidase has not been described before to our knowledge and may add to understanding of how this enzyme is regulated. Prolidase was here identified as deiminated in both llama serum and EVs.
Dipeptidylpeptidase 4 (DPP4, also known as CD26) was here identified to be deiminated in both llama whole serum and EVs. DPP4 controls glucose homeostasis and has complex roles in inflammation and homeostasis, including in liver cytokine expression, while its activity in plasma has been shown to correlate with body weight and fat mass (Varin et al., 2019). Interestingly, in camel milk, DDP4 inhibitory peptides have been identified and suggested to play roles in the regulation of glycaemia in humans (Nongonierma et al., 2018). Furthermore, roles for DDP4 in cancer have been found to relate to its post-translational processing of chemokines, thereby limiting lymphocyte migration to sites of inflammation and tumours (Barreira da Silva et al., 2015). As the success of antitumour immune responses depends on the infiltration of solid tumours by effector T cells, a process which is guided by chemokines, DPP4 inhibitors have been suggested as a strategy to enhance tumour immunotherapy (Barreira da Silva et al., 2015). Furthermore, serum DDP4 activity levels in primary HIV infection were found to be significantly decreased and to correlate with inflammation and HIV-induced intestinal damage (Ploquin et al., 2018). Middle East respiratory syndrome coronavirus (MERS-CoV) has been found to utilize dipeptidyl peptidase 4 (DPP4) as an entry receptor, via glycosylated sites (Peck et al., 2017). Therefore the identification of DPP4 as a deimination candidate may be of some relevance as such post-translational modification can affect DPP4 structure and function, allowing for moonlighting functions which may vary in pathological compared to pathophysiological milieus.
E3 ubiquitin-protein ligase Roquin, belongs to the Roquins which are a family of highly conserved RNA-binding proteins involved in ubiquination, are crucial for T-cell-dependent B-cell responses (Athanasopoulos et al., 2016) and play important roles in modulating T-cell activity (Akef and Muljo, 2018). Roquins repress constitutive decay elements containing mRNAs and play a critical role in RNA homeostasis and immunological self-tolerance (Zhang et al., 2015; Essig et al., 2018; Mino and Takeuchi, 2018). Roquin plays multifaceted roles both in the generation of a homeostatic immune response, as well as during chronic inflammation and autoimmunity (Schaefer and Klein, 2016; Lee et al., 2019). While roquin causes post-translational ubiquination of target proteins, post-translational deimination of roquin itself may modulate is function and is described here for the first time. Roquin was here identified as deiminated in whole llama serum only.
Serine-tRNA ligase, mitochondrial was here identified as a deimination candidate protein in llama EVs only. It has been linked to HUPRA syndrome which is a rare mitochondrial disease characterized by hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis (Rivera et al., 2013). It has furthermore been linked to progressive spastic paresis (Linnankivi et al., 2016). Post-translational deimination of this mitochondrial protein has not been described before to our knowledge.
Nucleoredoxin (Nrx) was here identified to be deiminated in EVs only. It is an oxidoreductase of the thioredoxin family of proteins with numerous functions in the redox regulation of metabolic pathways, cellular morphology, and signal transduction (Urbainsky et al., 2018). Nrx has been shown to inhibit Wnt-beta-catenin signalling (Funato et al., 2006) and is linked to Ca2+-mediated mitochondrial reactive oxygen species metabolism (Rharass et al., 2014). Nrx has furthermore been identified as an epigenetic cancer marker related to the oxidative status of human blood (Schöttker et al., 2015), but hitherto not been described as deiminated.
Dyslexia-associated protein was here identified to be deiminated in total serum of llama. It is associated with developmental dyslexia (Levecque et al., 2009) and has been shown to have roles in cell-cell interactions and signalling and neuronal migration (Velayos-Baeza et al., 2008), as well as in axon guidance (Poon et al., 2011). Dyslexia-associated protein has previously been found to follow the classical clathrin-mediated endocytosis pathway and its surface expression seems regulated by endocytosis, indicating that the internalization and recycling of the protein may be involved in fine-tuning its role in neuronal migration (Levecque et al., 2009). It has been descried to be highly glycosylated in different mammalian cell lines (Velayos-Baeza et al., 2008), while deimination has not been described before.
Metabotropic glutamate receptor 3 was identified here as deiminated in llama EVs only. It has, like other components of the glutamatergic system, a widespread distribution outside the central nervous system (CNS) and has been linked to regulation of the brain-gut axis (Julio-Pieper et al., 2013). It has also been linked to pain (Acher and Goudet, 2015) and psychotic disorders (Matrisciano et al., 2016). The group III mGlu receptors have been described in human stomach and colon, revealing a huge potential for these receptors in the treatment of peripheral disorders, including gastrointestinal dysfunction (Julio-Pieper et al., 2013). As post-translational deimination has not been described before in this receptor, but may well affect is tertiary structure, our current finding may be of some relevance in relation to strategies for developing antagonistic probes (Wenthur et al., 2019). Such pharmacological tools originally designed for mGlu receptors in the CNS may also be directed towards new disease targets in the periphery. Ulcerative colitis and Crohn’s disease are potential targets, as irritable bowel diseases can be co-morbid with anxiety and depression (Julio-Pieper et al., 2013).
Glutathione synthetase (GSS) was here identified to be deiminated in both llama whole serum and EVs. GSS is the second enzyme in the glutathione (GSH) biosynthesis pathway involved in homeostasis and cellular maintenance and also acts as a potent antioxidant (Njålsson and Norgren, 2005). In camelids, glutathione peroxidase, which also belongs to the GSH biosynthesis pathway and is a potent anti-oxidant, has been identified as a seminal plasma fertility biomarker (Waheed et al., 2015). Furthermore, camel milk has been shown to boost glutathione and total anti-oxidant capacity in sera and exudates in animal studies (Arab et al., 2017). In a diabetic mouse model, the activities of GSH, alongside glucose insulin and ROS levels, were restored after camel whey protein treatment (Sayed et al., 2017). In humans, GSS deficiency is an autosomal recessive disorder with varyingly severe clinical manifestations that include metabolic acidosis, hemolytic anemia, hyperbilirubinemia, neurological disorders and sepsis (Guney Varal et al., 2019). Deimination of GSS identified here has not been assessed before and may possibly affect the GSH biosynthesis pathway and such post-translational regulation remains to be further investigated.
Rootletin, also known as ciliary rootlet coiled-coil protein (CROCC), is a protein that is required for centrosome cohesion and is therefore important in mitosis (Bahe et al., 2005; Graser et al., 2007). It was identified here as deiminated in whole llama serum only. Rootletin has been shown to be phosphorylated and to have the ability to form centriole-associated fibers, suggesting a dynamic model for centrosome cohesion based on entangling filaments (Bahe et al., 2005). Deletion of rootletin in mouse models causes photoreceptor degeneration and impaired mucociliary clearance, supporting its key function in rootlet structures (Yang et al., 2005). Post-translational deimination of rootletin, as identified here, may possibly facilitate its dynamic functions.
Centromere protein J is is a highly conserved and ubiquitiously expressed centrosomal protein, involved in microtubule disassembly and plays a structural role in the maintenance of centrosome integrity, genome stability and normal spindle morphology during cell division (Tang et al., 2009; McIntyre, 2012). It was here identified as deiminated in whole llama serum only. Knockout mouse models targeting the gene encoding this protein have phenotypes of impaired glucose tolerance, hypoalbuminemia and increased micronuclei, indicative of genomic instability (Gerdin, 2010). In humans, it is associated with primary autosomal recessive microcephaly (Gul et al., 2006) and the microcephalic primordial dwarfism disorder Seckel syndrome (Al-Dosari et al., 2010; McIntyre et al., 2012). Deimination has not bee described before in this protein to our knowledge.
Telomere-associated protein RIF1 isoform 1 is involved in the repair of double-strand DNA breaks in response to DNA damage (Silverman et al., 2004; Escribano-Díaz et al., 2013; Drané et al., 2017) and protects teleomeres (Fontana et al., 2018). It was identified here as deiminated in EVs only. RIF1 has been associated with cancer via activation of human telomerase reverse transcriptase expression (Liu et al., 2018). It is also required for immunoglobulin class-switch recombination during the germinal centre reaction for humoral antibody immunity (Di Virgilio et al., 2013). RIF1 is involved in telomere homeostasis and was recently found to be post-translationally S-acylated, identifying a novel posttranslational modification regulating DNA repair (Fontana et al., 2019). Post-translational deimination has hitherto not been described.
Rabenosyn-5-like protein was here identified as a deimination protein candidate in both llama whole serum and EVs. It acts in early endocytic membrane fusion and membrane trafficking of recycling endosomes (Naslavsky et al., 2004; Rai et al., 2017). It plays a role in the lysosomal trafficking of CTSD/cathepsin D from the Golgi to lysosomes and also promotes the recycling of transferrin directly from early endosomes to the plasma membrane (Navaroli et al., 2012). Rabenosyn-5-like protein also binds phospholipid vesicles and plays roles in regulating protein sorting and recycling to the plasma membrane (Nielsen et al., 2000; de Renzis et al., 2002; Naslavsky et al., 2004). Rabenosyn-5 was recently identified as an upregulated urinary biomarker associated with malignant upper gastrointestinal cancer (Husi et al., 2019) and found to be increased in diabetic kidney disease (Dumont et al., 2017). Rabenosyn-5 has been shown to be phosphorylated, regulating its recruitment to membranes (Macé et al., 2005). Post-translational deimination has hitherto not been described..
Spermatogenesis-associated protein 2 (SPATA2) was here identified as deiminated in whole llama serum only. It is expressed in testis and to a lesser extent in spleen, thymus, and prostate (Graziotto et al., 1999). SPATA2 has been identified as a bridging factor that regulates TNF-alphA–Induced necroptosis and is instrumental for TNF-induced cell death (Elliott et al., 2016; Kupka et al., 2016; Schlicher et al., 2016; Wagner et al., 2016; Schlicher et al., 2017). SPATA2 ensures normal secretory function of Sertoli cells (Zhao et al., 2017) and has recently been identified as a novel predictor in ovarian cancer outcome (Wieser et al., 2019). While SPATA2 has been linked to ubiquination (Schlicher et al., 2016), post-translational deimination has not been invesigated.
Vitamin D-binding protein (VDBP) belongs to the albumin gene family, together with human serum albumin and alpha-fetoprotein. It is a multifaceted protein mainly produced in the liver, where its regulation is influenced by estrogen, glucocorticoids and inflammatory cytokines (Bikle and Schwartz, 2019). It is secreted into the blood circulation and is able to bind the various forms of vitamin D including ergocalciferol (vitamin D2) and cholecalciferol (vitamin D3), the 25-hydroxylated forms (calcifediol), and the active hormonal product 1,25-dihydroxyvitamin D (calcitriol) (Verboven et al., 2002; Norman, 2008). It transports vitamin D metabolites between skin, liver and kidney, and then on to various target tissues (Norman, 2008). VDBP is a macrophage activating factor (MAF) and has been tested as an anti-cancer agent via activation of macrophages against cancer cells (Yamamoto et al., 2008). Some association has also been made between polymorphisms of VDBP and the risk of coronary artery disease (Tarighi et al., 2017). Post-translational modifications (which still remain to be identified) of VDBP have been associated with multiple sclerosis (MS) (Perga et al., 2015), while protein deimination is well known to be associated with MS (Moscarello et al., 2013). VDBP has previously been identified to be glycosylated (Kilpatrick and Phinney, 2017) but was here identified as a deimination candidate in whole llama serum only. Post-translational deimination may contribute to various functions of VDBP in physiological as well as pathophysiological processes.
Pseudopodium-enriched atypical kinase 1 (PEAK1) was here identified as deiminated in EVs only. It is a tyrosine kinase and scaffold protein that transmits integrin-mediated extracellular matrix (ECM) signals to facilitate cell movement and growth. While aberrant expression of PEAK1 has been linked to cancer progression, its normal physiological role in vertebrate biology is still relatively unknown. Deletion of the PEAK1 gene in zebrafish, mice and human endothelial cells (ECs) induced severe defects in new blood vessel formation due to deficiencies in EC proliferation, survival and migration. PEAK1 seems therefore to play roles in modulation of cell adhesion and growth factor cues from the extracellular environment necessary for new vessel formation during vertebrate development and cancer (Wang et al., 2018). PEAK1 has not been described as deiminated before.
Desmoplakin was here identified to be deiminated in llama EVs only. Desmoplakin is a unique and critical component of desmosomal cell-cell junctions and involved in integrity of the cytoskeletal intermediate filament network (Bendrick et al., 2019). It has been shown to be required for epidermal integrity and morphogenesis in the Xenopus laevis embryo (Bharathan and Dickinson, 2019) and novel roles in coordination of cell migration were recently established (Bendrick et al., 2019). Mutations in desmoplakin have been linked to multiple allergies, severe dermatitis and metabolic wasting (SAM) syndrome (Liang et al., 2019). It is also linked to Carvajal syndrome, involving altered skin and hair abnormalities, and heart diseases (Yermakovich et al., 2018), including cardiomyopathies (Reichl et al., 2018; Chen et al., 2019a, 2019b). Desmosomal proteins have been shown to have both tumour-promoting and tumour-suppressive functions, depending of cancer types and can regulate cell proliferation, differentiation, migration, apoptosis, and impact treatment sensitivity in different types of cancers (Zhou et al., 2017). As the roles of desmosomal proteins in cancer and metastasis are not fully understood, the identification of deiminated desmoplakin in llama EVs here, and not described before, may be of some interest and add to understanding of diverse functional ability via such post-translational modification.
TSC22 domain family protein 3, also called glucocorticoid-induced leucine zipper protein (GILZ), was here identified as a deimination candidate in llama EVs only. It is a glucocorticoid-responsive molecule involved in immune regulation and glucocorticoid actions. Its interactions with signal transduction pathways, many of which are operative in RA and other inflammatory diseases, suggest that it is a key endogenous regulator of the immune response including a key glucocorticoid-induced regulator of inflammation in rheumatoid arthritis (RA) (Beaulieu and Morand, 2011). GILTZ is a small, 135-amino acid protein with anti-inflammatory properties and has been shown to inhibit NF-κB and MAPK pathways (Bereshchenko et al., 2019; Ricci et al., 2019). It has also been shown to be induced in response to hypoxia by a HIF1α-dependent mechanism and in response to cholesterol starvation, leading to downstream shedding of procoagulant EVs in ovarian cancer (Koizume et al., 2019). Post-translational deimination of GILZ has not been described before but may indeed affect its multifaceted functions, including in inflammation and cancer.
In the current study we report deiminated proteins in llama serum and serum-derived EVs. Due to the fact that the llama genome is not yet fully annotated, the hits identified here may underestimate the amount of deiminated proteins present in llama serum and EVs. Therefore a wider protein-hit analysis was carried out including other members of the camelid family, using known sequences from Camelus ferus, Camelus dromedaries, Lama guanicoe and Vicugna pacos. Deimination of key immune factors of innate and adaptive immunity and key metabolic proteins is identified here for the first time in a camelid species, highlighting putative protein moonlighting functions via post-translational deimination. It must be noted that in relation to previously observed increases of deiminated proteins with age (Ding et al., 2017), the llama used in this study would be considered relatively old at 21 years of age, as typical llama lifespans are 15–25 years, with some individuals surviving 30 years or more.
Our findings presented here furthermore complement expanding research in the comparative EV research field. Previous studies on the camel urinary proteome revealed enriched proteins from EVs and relevance to stress and immune responses as well as antimicrobial activities (Alhaider et al., 2012). Research on EVs is a relatively new field in comparative immunology and to our knowledge; this is the first description of EVs in serum of a camelid species. Previous studies on EVs in camelids focussed on EVs in camel milk, which were shown to have anti-cancerous properties (Badawy et al., 2018). As PADs have been identified to play major roles in the regulation of EV release (Kholia et al., 2015; Kosgodage et al., 2017 and 2018), including in host-pathogen interactions (Gavinho et al., 2019), such EV-mediated communication may be of great relevance also for addressing diverse zoonotic diseases identified in camels (El-Alfy et al., 2019; Zhu et al., 2019).
In continuation of the current pilot study, the assessment of changes in deiminated proteins in camelid serum, and their lateral transfer via EVs, will be of great interest to assess animal health in response to infection and environmental stress. Our findings will further current understanding of the roles for EVs, PADs and posttranslational deimination throughout phylogeny and in relation to adaption to a range of, including extreme, environments. Furthermore, novel identification of post-translational deimination in key proteins of metabolism and immunity may reveal hitherto unrecognized moonlighting function of these proteins throughout phylogeny, in relation to physiological and pathological processes, as well as being translational to and informing inflammatory and metabolic diseases. PAD-mediated contribution to protein moonlighting and in EV-mediated communication in response to physiological and pathophysiological changes remains therefore a field of further studies.
5. Conclusion
This is the first report of deiminated proteins in serum and serum-EVs of a camelid species, using the llama as a model animal. Our findings highlight a hitherto unrecognized post-translational modification in key immune and metabolic proteins in camelids, which may be translatable to and inform a range of human metabolic and inflammatory pathologies.
CRediT authorship contribution statement
Michael F. Criscitiello: Resources, Funding acquisition, Validation, Writing - review & editing. Igor Kraev: Formal analysis, Resources, Validation, Visualization. Sigrun Lange: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Yagnesh Umrania and Michael Deery at the Cambridge Centre for Proteomics for the LC–MS/MS analysis, as well as Alice Blue-McLendon of Texas A&M University's Winnie Carter Wildlife Center for sharing llama serum. This study was funded in part by a University of Westminster start-up grant to SL and U.S. National Science Foundation grant IOS-1656870 to MFC. Thanks are due to The Guy Foundation for funding the purchase of equipment utilised in this work.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.molimm.2019.10.017.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Acher F., Goudet C. Therapeutic potential of group III metabotropic glutamate receptor ligands in pain. Curr. Opin. Pharmacol. 2015;20:64–72. doi: 10.1016/j.coph.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Akef A., Muljo S.A. How T cells go rogue in the absence of roquins. Noncoding RNA Investig. 2018;2:20. doi: 10.21037/ncri.2018.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dosari M.S., Shaheen R., Colak D., Alkuraya F.S. Novel CENPJ mutation causes Seckel syndrome. J. Med. Genet. 2010;47(6):411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- Alhaider A.A., Bayoumy N., Argo E., Gader A.G., Stead D.A. Survey of the camel urinary proteome by shotgun proteomics using a multiple database search strategy. Proteomics. 2012;12(22):3403–3406. doi: 10.1002/pmic.201100631. [DOI] [PubMed] [Google Scholar]
- Arab H.H., Salama S.A., Abdelghany T.M., Omar H.A., Arafa E.A., Alrobaian M.M., Maghrabi I.A. Camel milk attenuates rheumatoid arthritis via inhibition of mitogen activated protein kinase pathway. Cell Physiol. Biochem. 2017;43(2):540–552. doi: 10.1159/000480527. [DOI] [PubMed] [Google Scholar]
- Arciello A., Piccoli R., Monti D.M. Apolipoprotein a-I: the dual face of a protein. FEBS Lett. 2016;590(23):4171–4179. doi: 10.1002/1873-3468.12468. 2016. [DOI] [PubMed] [Google Scholar]
- Armstrong P.B., Quigley J.P. Alpha2-macroglobulin: an evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 1999;23:375. doi: 10.1016/s0145-305x(99)00018-x. [DOI] [PubMed] [Google Scholar]
- Athanasopoulos V., Ramiscal R.R., Vinuesa C.G. ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 2016;46(5):1082–1090. doi: 10.1002/eji.201545956. [DOI] [PubMed] [Google Scholar]
- Badawy A.A., El-Magd M.A., AlSadrah S.A. Therapeutic effect of camel milk and its exosomes on MCF7 cells in vitro and in vivo. Integr. Cancer Ther. 2018;17(4):1235–1246. doi: 10.1177/1534735418786000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bağcı I.S., Horváth O.N., Ruzicka T., Sárdy M. Bullous pemphigoid. Autoimmun. Rev. 2017;16(5):445–455. doi: 10.1016/j.autrev.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Bahe S., Stierhof Y.D., Wilkinson C.J., Leiss F., Nigg E.A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171(1):27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannas P., Koch-Nolte F. Perspectives for the development of CD38-specific heavy Chain antibodies as therapeutics for multiple myeloma. Front. Immunol. 2018;9:2559. doi: 10.3389/fimmu.2018.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan N.K., Dickinson A.J.G. Desmoplakin is required for epidermal integrity and morphogenesis in the Xenopus laevis embryo. Dev. Biol. 2019;450(2):115–131. doi: 10.1016/j.ydbio.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe A., Bongrani A., Mellouk N., Estienne A., Kurowska P., Grandhaye J., Elfassy Y., Levy R., Rak A., Froment P. Mechanisms of adiponectin action in fertility: an overview from gametogenesis to gestation in humans and animal models in normal and pathological conditions. Int. J. Mol. Sci. 2019;20:1526. doi: 10.3390/ijms20071526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle C., Gill D.S., Charlton K. Shark novel antigen receptors—the next generation of biologic therapeutics? Adv. Exp. Med. Biol. 2009;655:49–62. doi: 10.1007/978-1-4419-1132-2_6. [DOI] [PubMed] [Google Scholar]
- Barreira da Silva R., Laird M.E., Yatim N., Fiette L., Ingersoll M.A., Albert M.L. Dipeptidylpeptidase 4 inhibition enhances lymphocyte trafficking, improving both naturally occurring tumor immunity and immunotherapy. Nat. Immunol. 2015;16(8):850–858. doi: 10.1038/ni.3201. [DOI] [PubMed] [Google Scholar]
- Basseri S., Ly T.Y., Hull P.R.J. Dyshidrotic bullous pemphigoid: case report and review of literature. Cutan. Med. Surg. 2018;22(6):614–617. doi: 10.1177/1203475418763544. [DOI] [PubMed] [Google Scholar]
- Beaulieu E., Morand E.F. Role of GILZ in immune regulation, glucocorticoid actions and rheumatoid arthritis. Nat. Rev. Rheumatol. 2011;7(6):340–348. doi: 10.1038/nrrheum.2011.59. [DOI] [PubMed] [Google Scholar]
- Bendrick J.L., Eldredge L.A., Williams E.I., Haight N.B., Dubash A.D. Desmoplakin harnesses Rho GTPase and p38 mitogen-activated protein kinase signaling to coordinate cellular migration. J. Invest. Dermatol. 2019;139(6):1227–1236. doi: 10.1016/j.jid.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereshchenko O., Migliorati G., Bruscoli S., Riccardi C. Glucocorticoid-induced leucine zipper: a novel anti-inflammatory molecule. Front. Pharmacol. 2019;10:308. doi: 10.3389/fphar.2019.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser M.W., MacDonald S.G. Acquired hypofibrinogenemia: current perspectives. J. Blood Med. 2016;7:217–225. doi: 10.2147/JBM.S90693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever C.S., Dong J.-X., Vasylieva N., Barnych B., Cui Y., Xu Z.-L. VHH antibodies: emerging reagents for the analysis of environmental chemicals. Anal. Bioanal. Chem. 2016;408:5985–6002. doi: 10.1007/s00216-016-9585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker K.L., Thompson P.R. The protein arginine deiminases: structure, function, inhibition, and disease. Biopolymers. 2013;99(2):155–163. [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bikle D.D., Schwartz J. Vitamin D binding protein, total and Free vitamin D levels in different physiological and pathophysiological conditions. Front. Endocrinol. (Lausanne) 2019;10:317. doi: 10.3389/fendo.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachère N.E., Parveen S., Frank M.O., Dill B.D., Molina H., Orange D.E. High-titer rheumatoid arthritis antibodies preferentially bind fibrinogen citrullinated by peptidylarginine deiminase 4. Arthritis Rheumatol. 2017;69(5):986–995. doi: 10.1002/art.40035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost F., Diarra-Mehrpour M., Martin J.P. Inter-alpha-trypsin inhibitor proteoglycan family – a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 1998;252:339–346. doi: 10.1046/j.1432-1327.1998.2520339.x. [DOI] [PubMed] [Google Scholar]
- Branzk N., Lubojemska A., Hardison S.E., Wang Q., Gutierrez M.G., Brown G.D., Papayannopoulos V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014;15(11):1017–1025. doi: 10.1038/ni.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Rossotti M.A., Henry K.A. Immunological functions and evolutionary emergence of heavy-chain antibodies. Trends Immunol. 2018;39(12):956–960. doi: 10.1016/j.it.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Buchner J. Hsp90 & Co. - a holding for folding. Trends Biochem. Sci. 1999;24(4):136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- Carroll M.V., Sim R.B. Complement in health and disease. Adv. Drug Deliv. Rev. 2011;63(12):965–975. doi: 10.1016/j.addr.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Chen R., Kang R., Fan X.-G., Tang D. Release and activity of histone in diseases. Cell Death Dis. 2014;5:e1370. doi: 10.1038/cddis.2014.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.L., Tao J., Zhao P.J., Tang W., Xu J.P., Zhang K.Q., Zou C.G. Adiponectin receptor PAQR-2 signaling senses low temperature to promote C. elegans longevity by regulating autophagy. Nat. Commun. 2019;10(1):2602. doi: 10.1038/s41467-019-10475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song J., Chen X., Chen K., Ren J., Zhang N., Rao M., Hu Z., Zhang Y., Gu M., Zhao H., Tang H., Yang Z., Hu S. A novel genotype-based clinicopathology classification of arrhythmogenic cardiomyopathy provides novel insights into disease progression. Eur. Heart J. 2019;40(21):1690–1703. doi: 10.1093/eurheartj/ehz172. [DOI] [PubMed] [Google Scholar]
- Chiosis G., Caldas Lopes E., Solit D. Heat shock protein-90 inhibitors: a chronicle from geldanamycin to today’s agents. Curr. Opin. Investig. Drugs. 2006;7(6):534–541. [PubMed] [Google Scholar]
- Cohen J. Llama antibodies inspire gene spray to prevent all flus. Science. 2018;362(6414):511. doi: 10.1126/science.362.6414.511. [DOI] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Criscitiello M.F. What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin. Drug Discov. 2014;9(7):725–739. doi: 10.1517/17460441.2014.920818. [DOI] [PubMed] [Google Scholar]
- Criscitiello M.F., Kraev I., Lange S. Deiminated proteins in extracellular vesicles and plasma of nurse shark (Ginglymostoma cirratum) - novel insights into shark immunity. Fish Shellfish Immunol. 2019;92:249–255. doi: 10.1016/j.fsi.2019.06.012. [DOI] [PubMed] [Google Scholar]
- Cromie K.D., Van Heeke G., Boutton C. Nanobodies and their use in GPCR drug discovery. Curr. Top. Med. Chem. 2015;15:2543–2557. doi: 10.2174/1568026615666150701113549. [DOI] [PubMed] [Google Scholar]
- Danquah W., Meyer-Schwesinger C., Rissiek B., Pinto C., Serracant-Prat A., Amadi M. Nanobodies that block gating of the P2X7 ion channel ameliorate inflammation. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aaf8463. [DOI] [PubMed] [Google Scholar]
- Davies S.G., Sim R.B. Intramolecular general acid catalysis in the binding reactions of alpha 2-macroglobulin and complement components C3 and C4. Biosci. Rep. 1981;1(6):461–468. doi: 10.1007/BF01121579. [DOI] [PubMed] [Google Scholar]
- de los Rios M., Criscitiello M.F., Smider V.V. Structural and genetic diversity in antibody repertoires from diverse species. Curr. Opin. Struct. Biol. 2015;33:27–41. doi: 10.1016/j.sbi.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis S., Sönnichsen B., Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 2002;4(2):124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- De Silva D.P.N., Tan E., Mizuno N., Hosoya S., Reza M.S., Watabe S., Kinoshita S., Asakawa S. Transcriptomic analysis of immunoglobulin novel antigen receptor (IgNAR) heavy chain constant domains of brownbanded bamboo shark (Chiloscyllium punctatum) Fish Shellfish Immunol. 2019;84:370–376. doi: 10.1016/j.fsi.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Del Rio-Tsonis K., Tsonis P.A., Zarkadis I.K., Tsagas A.G., Lambris J.D. Expression of the third component of complement, C3, in regenerating limb blastema cells of urodeles. J. Immunol. 1998;161(12):6819–6824. [PubMed] [Google Scholar]
- Deschaght P., Vintém A.P., Logghe M., Conde M., Felix D., Mensink R., Gonçalves J., Audiens J., Bruynooghe Y., Figueiredo R., Ramos D., Tanghe R., Teixeira D., Van de Ven L., Stortelers C., Dombrecht B. Large diversity of functional nanobodies from a camelid immune library revealed by an alternative analysis of next-generation sequencing data. Front. Immunol. 2017;8:420. doi: 10.3389/fimmu.2017.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Enriquez-Algeciras M., Bhattacharya S.K., Bonilha V.L. Protein deimination in aging and age-related diseases with ocular manifestations. In: Nicholas A., Bhattacharya S., Thompson P., editors. Protein Deimination in Human Health and Disease. Springer; Cham: 2017. [Google Scholar]
- Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T., Nussenzweig A., Casellas R., Robbiani D.F., Nussenzweig M.C. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339(6120):711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds A.W. Which came first, the lectin/classical pathway or the alternative pathway of complement? Immunobiology. 2002;205(4-5):340–354. doi: 10.1078/0171-2985-00137. [DOI] [PubMed] [Google Scholar]
- Dodds A.W., Law S.K. The phylogeny and evolution of the thioester bond-containing proteins C3, C4 and alpha 2-macroglobulin. Immunol. Rev. 1998;166:15–26. doi: 10.1111/j.1600-065x.1998.tb01249.x. [DOI] [PubMed] [Google Scholar]
- Drané P., Brault M.E., Cui G., Meghani K., Chaubey S., Detappe A., Parnandi N., He Y., Zheng X.F., Botuyan M.V., Kalousi A., Yewdell W.T., Münch C., Harper J.W., Chaudhuri J., Soutoglou E., Mer G., Chowdhury D. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature. 2017;543(7644):211–216. doi: 10.1038/nature21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont V., Tolvanen T.A., Kuusela S., Wang H., Nyman T.A., Lindfors S., Tienari J., Nisen H., Suetsugu S., Plomann M., Kawachi H., Lehtonen S. PACSIN2 accelerates nephrin trafficking and is up-regulated in diabetic kidney disease. FASEB J. 2017;31(9):3978–3990. doi: 10.1096/fj.201601265R. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duygu F., Aksoy N., Cicek A.C., Butun I., Unlu S. Does prolidase indicate worsening of hepatitis B infection? J. Clin. Lab. Anal. 2013;27(5):398–401. doi: 10.1002/jcla.21617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Alfy E.S., Abu-Elwafa S., Abbas I., Al-Araby M., Al-Kappany Y., Umeda K., Nishikawa Y. Molecular screening approach to identify protozoan and trichostrongylid parasites infecting one-humped camels (Camelus dromedarius) Acta Trop. 2019;197 doi: 10.1016/j.actatropica.2019.105060. [DOI] [PubMed] [Google Scholar]
- El-Bahr S.M., El-Deeb W.M. Trypanosoma evansi in naturally infected dromedary camels: lipid profile, oxidative stress parameters, acute phase proteins and proinflammatory cytokines. Parasitology. 2016;143(4):518–522. doi: 10.1017/S0031182016000123. 2016. [DOI] [PubMed] [Google Scholar]
- El-Deeb W.M., Buczinski S. The diagnostic and prognostic importance of oxidative stress biomarkers and acute phase proteins in urinary tract infection (UTI) in camels. PeerJ. 2015;3:e1363. doi: 10.7717/peerj.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Malky O.M., Mostafa T.H., Abd El-Salaam A.M., Ayyat M.S. Effect of reproductive disorders on productivity and reproductive efficiency of dromedary she-camels in relation to cytokine concentration. Trop. Anim. Health Prod. 2018;50(5):1079–1087. doi: 10.1007/s11250-018-1533-7. [DOI] [PubMed] [Google Scholar]
- Elliott P.R., Leske D., Hrdinka M., Bagola K., Fiil B.K., McLaughlin S.H., Wagstaff J., Volkmar N., Christianson J.C., Kessler B.M., Freund S.M., Komander D., Gyrd-Hansen M. SPATA2 Links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol Cell. 2016;63(6):990–1005. doi: 10.1016/j.molcel.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., Xu D., Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Essig K., Kronbeck N., Guimaraes J.C., Lohs C., Schlundt A., Hoffmann A., Behrens G., Brenner S., Kowalska J., Lopez-Rodriguez C., Jemielity J., Holtmann H., Reiche K., Hackermüller J., Sattler M., Zavolan M., Heissmeyer V. Roquin targets mRNAs in a 3’-UTR-specific manner by different modes of regulation. Nat. Commun. 2018;9(1):3810. doi: 10.1038/s41467-018-06184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishelson Z., Attali G., Mevorach D. Complement and apoptosis. Mol. Immunol. 2001;38(2-3):207–219. doi: 10.1016/s0161-5890(01)00055-4. [DOI] [PubMed] [Google Scholar]
- Fiaschi T. Mechanisms of adiponectin action. Int. J. Mol. Sci. 2019;20(12):E2894. doi: 10.3390/ijms20122894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi T., Magherini F., Gamberi T., Modesti P.A., Modesti A. Adiponectin as a tissue regenerating hormone: more than a metabolic function. Cell. Mol. Life Sci. 2014;71:1917–1925. doi: 10.1007/s00018-013-1537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik M.F., Dooley H. The generation and selection of single-domain, v region libraries from nurse sharks. Methods Mol. Biol. 2009;562:71–82. doi: 10.1007/978-1-60327-302-2_6. [DOI] [PubMed] [Google Scholar]
- Fontana G.A., Reinert J.K., Thomä N.H., Rass U. Shepherding DNA ends: Rif1 protects telomeres and chromosome breaks. Microb. Cell. 2018;5(7):327–343. doi: 10.15698/mic2018.07.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana G.A., Hess D., Reinert J.K., Mattarocci S., Falquet B., Klein D., Shore D., Thomä N.H., Rass U. Rif1 S-acylation mediates DNA double-strand break repair at the inner nuclear membrane. Nat. Commun. 2019;10(1):2535. doi: 10.1038/s41467-019-10349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg A.D.V., Reis A.F., Gerchman F. Relationships between adiponectin levels, the metabolic syndrome, and type 2 diabetes: a literature review. Arch. Endocrinol. Metab. 2017;61(6):614–622. doi: 10.1590/2359-3997000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L.E., Wohlwend A., Sappino A.P., Tschopp J., Schifferli J.A. Human clusterin gene expression is confined to surviving cells during in vitro programmed cell death. J. Clin. Invest. 1994;93(2):877–884. doi: 10.1172/JCI117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y., Michiue T., Asashima M., Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat. Cell Biol. 2006;8(5):501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- Gamberi T., Magherini F., Fiaschi T. Adiponectin in myopathies. Int. J. Mol. Sci. 2019;20:1544. doi: 10.3390/ijms20071544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavinho B., Rossi, Evans-Osses I., Lange S., Ramirez M.I. Peptidylarginine deiminase inhibition abolishes the production of large extracellular vesicles from Giardia intestinalis, affecting host-pathogen interactions by hindering adhesion to host cells. bioRxiv. 2019;2019:586438. doi: 10.1101/586438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdin A.K. The Sanger mouse genetics programme: High throughput characterisation of knockout mice. Acta Ophthalmol. 2010;88:925–927. [Google Scholar]
- Goetz M.P., Toft D.O., Ames M.M., Erlichman C. The Hsp90 chaperone complex as a novel target for cancer therapy. Ann. Oncol. 2003;14(8):1169–1176. doi: 10.1093/annonc/mdg316. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aparicio M., Alfaro C. Influence of interleukin-8 and neutrophil extracellular trap (NET) formation in the tumor microenvironment: Is there a pathogenic role? J. Immunol. Res. 2019;2019 doi: 10.1155/2019/6252138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S., Aguilera S., Alliende C., Urzua U., Quest A.F., Herrera L., Molina C., Hermoso M., Ewert P., Brito M., Romo R., Leyton C., Pérez P., González M.J. Alterations in type I hemidesmosome components suggestive of epigenetic control in the salivary glands of patients with Sjögren’s syndrome. Arthritis Rheum. 2011;63:1106–1115. doi: 10.1002/art.30212. [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Nigg E.A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007;120(24):4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- Graziotto R., Foresta C., Scannapieco P., Zeilante P., Russo A., Negro A., Salmaso R., Onisto M. cDNA cloning and characterization of PD1: a novel human testicular protein with different expressions in various testiculopathies. Exp. Cell Res. 1999;248(2):620–626. doi: 10.1006/excr.1999.4449. [DOI] [PubMed] [Google Scholar]
- Greenberg A.S., Avila D., Hughes M., Hughes A., McKinney E.C., Flajnik M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374(6518):168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- Greunz E.M., Krogh A.K.H., Pieters W., Ruiz O.A., Bohner J., Reckendorf A., Monaco D., Bertelsen M.F. The acute-phase and hemostatic response in dromedary camels (Camelus dromedarius) J. Zoo Wildl. Med. 2018;49(2):361–370. doi: 10.1638/2017-0221.1. [DOI] [PubMed] [Google Scholar]
- Gul A., Hassan M.J., Hussain S., Raza S.I., Chishti M.S., Ahmad W. A novel deletion mutation in CENPJ gene in a Pakistani family with autosomal recessive primary microcephaly. J. Hum. Genet. 2006;51(9):760–764. doi: 10.1007/s10038-006-0017-1. [DOI] [PubMed] [Google Scholar]
- Guney Varal I., Dogan P., Gorukmez O., Dorum S., Akdag A. Glutathione synthetase deficiency: a novel mutation with femur agenesis. Fetal Pediatr. Pathol. 2019;14:1–7. doi: 10.1080/15513815.2019.1627627. [DOI] [PubMed] [Google Scholar]
- György B., Toth E., Tarcsa E., Falus A., Buzas E.I. Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006;38:1662–1677. doi: 10.1016/j.biocel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Hagiwara T., Hidaka Y., Yamada M. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry. 2005;44:5827–5834. doi: 10.1021/bi047505c. [DOI] [PubMed] [Google Scholar]
- Hamilton K.K., Zhao J., Sims P.J. Interaction between apolipoproteins A-I and A-II and the membrane attack complex of complement. Affinity of the apoproteins for polymeric C9. J. Biol. Chem. 1993;268(5):3632–3638. [PubMed] [Google Scholar]
- Hamm A., Veeck J., Bektas N., Wild P.J., Hartmann A., Hendrichs U., Kristiansen G., Werbowetski-Ogilvie T., Del Maestro R., Knüchel R., Dahl E. Frequent expression loss of inter-α-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer. 2008;8:25. doi: 10.1186/1471-2407-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen M.M., De Haard H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007;77(1):13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassiki R., Labro A.J., Benlasfar Z., Vincke C., Somia M., El Ayeb M. Dromedary immune response and specific Kv2.1 antibody generation using a specific immunization approach. Int. J. Biol. Macromol. 2016;93:167–171. doi: 10.1016/j.ijbiomac.2016.06.031. [DOI] [PubMed] [Google Scholar]
- Haynes T., Luz-Madrigal A., Reis E.S., Echeverri Ruiz N.P., Grajales-Esquivel E., Tzekou A., Tsonis P.A., Lambris J.D., Del Rio-Tsonis K. Complement anaphylatoxin C3a is a potent inducer of embryonic chick retina regeneration. Nat. Commun. 2013;4:2312. doi: 10.1038/ncomms3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B., Martin A.C. Protein moonlighting: a new factor in biology and medicine. Biochem. Soc. Trans. 2014;42(6):1671–1678. doi: 10.1042/BST20140273. [DOI] [PubMed] [Google Scholar]
- Henry K.A., van Faassen H., Harcus D., Marcil A., Hill J.J., Muyldermans S., MacKenzie C.R. Llama peripheral B-cell populations producing conventional and heavy chain-only IgG subtypes are phenotypically indistinguishable but immunogenetically distinct. Immunogenetics. 2019;71(4):307–320. doi: 10.1007/s00251-018-01102-9. [DOI] [PubMed] [Google Scholar]
- Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida S., Miura N.N., Adachi Y., Ohno N. Influence of arginine deimination on antigenicity of fibrinogen. J. Autoimmun. 2004;23(2):141–150. doi: 10.1016/j.jaut.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Hughes-Austin J.M., Deane K.D., Giles J.T., Derber L.A., Zerbe G.O., Dabelea D.M., Sokolove J., Robinson W.H., Holers V.M., Norris J.M. Plasma adiponectin levels are associated with circulating inflammatory cytokines in autoantibody positive first-degree relatives of rheumatoid arthritis patients. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0199578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H., Fernandes M., Skipworth R.J., Miller J., Cronshaw A.D., Fearon K.C.H., Ross J.A. Identification of diagnostic upper gastrointestinal cancer tissue type-specific urinary biomarkers. Biomed. Rep. 2019;10(3):165–174. doi: 10.3892/br.2019.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussack G., Raphael S., Lowden M.J., Henry K.A. Isolation and characterization of camelid single-domain antibodies against HER2. BMC Res. Notes. 2018;11(1):866. doi: 10.1186/s13104-018-3955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson D., Clarke A., Heesom K., Murphy D., Eggleton P. Carbamylation/citrullination of IgG Fc in bronchiectasis, established RA with bronchiectasis and RA smokers: a potential risk factor for disease. ERJ. Open Res. 2017;3(3) doi: 10.1183/23120541.00018-2017. pii: 00018-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D., Strandskog G., Nepal A., Aspar A., Olsen R., Jørgensen J., Wolfson D., Ahluwalia B.S., Handzhiyski J., Mironova R. Stimulation of exosome release by extracellular DNA is conserved across multiple cell types. FEBS J. 2018;285(16):3114–3133. doi: 10.1111/febs.14601. [DOI] [PubMed] [Google Scholar]
- Immenschuh S., Vijayan V., Janciauskiene S., Gueler F. Heme as a target for therapeutic interventions. Front. Pharmacol. 2017;8(2017):146. doi: 10.3389/fphar.2017.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22(14):3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inal J.M., Ansa-Addo E.A., Lange S. Interplay of host-pathogen microvesicles and their role in infectious disease. Biochem Soc Trans. 2013;41(1):258–262. doi: 10.1042/BST20120257. 1. [DOI] [PubMed] [Google Scholar]
- Iqbal U., Trojahn U., Albaghdadi H., Zhang J., O’Connor-McCourt M., Stanimirovic D., Tomanek B., Sutherland G., Abulrob A. Kinetic analysis of novel mono- and multivalent VHH-fragments and their application for molecular imaging of brain tumours. Br. J. Pharmacol. 2010;160(4):1016–1028. doi: 10.1111/j.1476-5381.2010.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey C.J. Protein moonlighting: what is it, and why is it important? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2018;373(1738):20160523. doi: 10.1098/rstb.2016.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne D.E., Lowin B., Peitsch M.C., Böttcher A., Schmitz G., Tschopp J. Clusterin (complement lysis inhibitor) forms a high density lipoprotein complex with apolipoprotein A-I in human plasma. J. Biol. Chem. 1991;266(17):11030–11036. [PubMed] [Google Scholar]
- Julio-Pieper M., O’Connor R.M., Dinan T.G., Cryan J.F. Regulation of the brain-gut axis by group III metabotropic glutamate receptors. Eur. J. Pharmacol. 2013;698(1-3):19–30. doi: 10.1016/j.ejphar.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Kholia S., Jorfi S., Thompson P.R., Causey C.P., Nicholas A.P., Inal J., Lange S. A novel role for peptidylarginine deiminases (PADs) in microvesicle release: a therapeutic potential for PAD inhibitors to sensitize prostate cancer cells to chemotherapy. J. Extracell Vesicles. 2015;4:26192. doi: 10.3402/jev.v4.26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick L.E., Phinney K.W. Quantification of total vitamin-D-binding protein and the glycosylated isoforms by liquid chromatography-isotope dilution mass spectrometry. J. Proteome Res. 2017;16(11):4185–4195. doi: 10.1021/acs.jproteome.7b00560. [DOI] [PubMed] [Google Scholar]
- Kitchener R.L., Grunden A.M. Prolidase function in proline metabolism and its medical and biotechnological applications. J. Appl. Microbiol. 2012;113(2):233–247. doi: 10.1111/j.1365-2672.2012.05310.x. [DOI] [PubMed] [Google Scholar]
- Koizume S., Takahashi T., Yoshihara M., Nakamura Y., Ruf W., Takenaka K., Miyagi E., Miyagi Y. Cholesterol starvation and hypoxia activate the FVII Gene via the SREBP1-GILZ pathway in ovarian cancer cells to produce procoagulant microvesicles. Thromb. Haemost. 2019;119(7):1058–1071. doi: 10.1055/s-0039-1687876. [DOI] [PubMed] [Google Scholar]
- Kojouharova M.S., Gadjeva M.G., Tsacheva I.G., Zlatarova A., Roumenina L.T., Tchorbadjieva M.I., Atanasov B.P., Waters P., Urban B.C., Sim R.B., Reid K.B., Kishore U. Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q-IgG interaction. J. Immunol. 2004;172(7):4351–4358. doi: 10.4049/jimmunol.172.7.4351. [DOI] [PubMed] [Google Scholar]
- Könning D., Zielonka S., Grzeschik J., Empting M., Valldorf B., Krah S., Schröter C., Sellmann C., Hock B., Kolmar H. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Konsta O.D., Thabet Y., Le Dantec C., Brooks W.H., Tzioufas A.G., Pers J.O., Renaudineau Y. The contribution of epigenetics in Sjögren’s syndrome. Front Genet. 2014;5:71. doi: 10.3389/fgene.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosgodage U.S., Trindade R.P., Thompson P.T., Inal J.M., Lange S. Chloramidine/Bisindolylmaleimide-I-mediated inhibition of exosome and microvesicle release and enhanced efficacy of cancer chemotherapy. Int. J. Mol. Sci. 2017;18(5):E1007. doi: 10.3390/ijms18051007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosgodage U.S., Onganer P.U., Maclatchy A., Nicholas A.P., Inal J.M., Lange S. Peptidylarginine deiminases Post-translationally deiminate prohibitin and modulate extracellular vesicle release and miRNAs 21 and 126 in glioblastoma multiforme. Int. J. Mol. Sci. 2018;20(1) doi: 10.3390/ijms20010103. p E103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosgodage U.S., Matewele P., Mastroianni G., Kraev I., Brotherton D., Awamaria B., Nicholas A.P., Lange S., Inal J.M. Peptidylarginine deiminase inhibitors reduce bacterial membrane vesicle release and sensitize bacteria to antibiotic treatment. Front. Cell. Infect. Microbiol. 2019;9:227. doi: 10.3389/fcimb.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupka S., De Miguel D., Draber P., Martino L., Surinova S., Rittinger K., Walczak H. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 2016;16(9):2271–2280. doi: 10.1016/j.celrep.2016.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Bambir S., Dodds A.W., Magnadottir B. The ontogeny of complement component C3 in Atlantic cod (Gadus morhua L.)—an immunohistochemical study. Fish Shellfish Immunol. 2004;16:359–367. doi: 10.1016/j.fsi.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Lange S., Bambir S., Dodds A.W., Magnadottir B. An immunohistochemical study on complement component C3 in juvenile Atlantic halibut (Hippoglossus Hippoglossus L.) Dev. Comp. Immunol. 2004;28(6):593–601. doi: 10.1016/j.dci.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Lange S., Dodds A.W., Gudmundsdóttir S., Bambir S.H., Magnadottir B. The ontogenic transcription of complement component C3 and apolipoprotein A-I tRNA in Atlantic cod (Gadus morhua L.)--a role in development and homeostasis? Dev. Comp. Immunol. 2005;29(12):1065–1077. doi: 10.1016/j.dci.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Lange S., Bambir S.H., Dodds A.W., Bowden T., Bricknell I., Espelid S., Magnadottir B. Complement component C3 transcription in Atlantic halibut (Hippoglossus Hippoglossus L.) Larvae. Fish Shellfish Immunol. 2006;20(3):285–294. doi: 10.1016/j.fsi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lange S., Gögel S., Leung K.Y., Vernay B., Nicholas A.P., Causey C.P., Thompson P.R., Greene N.D., Ferretti P. Protein deiminases: new players in the developmentally regulated loss of neural regenerative ability. Dev. Biol. 2011;355(2):205–214. doi: 10.1016/j.ydbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Rocha-Ferreira E., Thei L., Mawjee P., Bennett K., Thompson P.R., Subramanian V., Nicholas A.P., Peebles D., Hristova M., Raivich G. Peptidylarginine deiminases: novel drug targets for prevention of neuronal damage following hypoxic ischemic insult (HI) in neonates. J. Neurochem. 2014;130(4):555–562. doi: 10.1111/jnc.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Gallagher M., Kholia S., Kosgodage U.S., Hristova M., Hardy J., Inal J.M. Peptidylarginine deiminases-roles in cancer and neurodegeneration and possible avenues for therapeutic intervention via modulation of exosome and microvesicle (EMV) release? Int. J. Mol. Sci. 2017;18(6):E1196. doi: 10.3390/ijms18061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Kraev I., Magnadóttir B., Dodds A.W. Complement component C4-like protein in Atlantic cod (Gadus morhua L.) – Detection in ontogeny and identification of post-translational deimination in serum and extracellular vesicles. Dev. Comp. Immunol. 2019;101:103437. doi: 10.1016/j.dci.2019.103437. [DOI] [PubMed] [Google Scholar]
- Latham J.A., Dent S.Y. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Kronbichler A., Park D.D., Park Y., Moon H., Kim H., Choi J.H., Choi Y., Shim S., Lyu I.S. Neutrophil extracellular traps (NETs) in autoimmune diseases: a comprehensive review. Autoimmun. Rev. 2017;16(11):1160–1173. doi: 10.1016/j.autrev.2017.09.012. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Lee S.H., Seo H.B., Ryu J.G., Jung K., Choi J.W., Jhun J., Park J.S., Kwon J.Y., Kwok S.K., Youn J., Park S.H., Cho M.L. Inhibition of IL-17 ameliorates systemic lupus erythematosus in Roquinsan/san mice through regulating the balance of TFH cells, GC B cells. Treg Breg. Sci. Rep. 2019;9(1):5227. doi: 10.1038/s41598-019-41534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levecque C., Velayos-Baeza A., Holloway Z.G., Monaco A.P. The dyslexia-associated protein KIAA0319 interacts with adaptor protein 2 and follows the classical clathrin-mediated endocytosis pathway. Am. J. Physiol. Cell Physiol. 2009;297(1):C160–8. doi: 10.1152/ajpcell.00630.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li M., Lindberg M.R., Kennett M.J., Xiong N., Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010;207(9):1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Li C., Zhang Z., Ni C., Yu H., Li M., Yao Z. Severe dermatitis, multiple allergies and metabolic wasting (SAM) syndrome caused by de novo mutation in the DSP gene misdiagnosed as generalized pustular psoriasis and treatment of acitretin with gabapentin. J. Dermatol. 2019;2019(May 20) doi: 10.1111/1346-8138.14925. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Linnankivi T., Neupane N., Richter U., Isohanni P., Tyynismaa H. Splicing defect in mitochondrial seryl-tRNA synthetase Gene causes progressive spastic paresis instead of HUPRA syndrome. Hum. Mutat. 2016;37(9):884–888. doi: 10.1002/humu.23021. [DOI] [PubMed] [Google Scholar]
- Liu Y.B., Mei Y., Long J., Zhang Y., Hu D.L., Zhou H.H. RIF1 promotes human epithelial ovarian cancer growth and progression via activating human telomerase reverse transcriptase expression. J. Exp. Clin. Cancer Res. 2018;37(1):182. doi: 10.1186/s13046-018-0854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Zhao P., Zhang Q., Che N., Xu L., Qian J., Tan W., Zhang M. Adiponectin promotes fibroblast-like synoviocytes producing IL-6 to enhance T follicular helper cells response in rheumatoid arthritis. Clin. Exp. Rheumatol. 2019;2019(April 11) [Epub ahead of print] [PubMed] [Google Scholar]
- Lu J., Kishore U. C1 complex: an adaptable proteolytic module for complement and Non-complement functions. Front. Immunol. 2017;8:592. doi: 10.3389/fimmu.2017.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macé G., Miaczynska M., Zerial M., Nebreda A.R. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 2005;24(18):3235–3246. doi: 10.1038/sj.emboj.7600799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnadóttir B., Hayes P., Hristova M., Bragason B.Þ., Nicholas A.P., Dodds A.W., Gudmundsdottir S., Lange S. Post-translational protein deimination in cod (Gadus morhua L.) Ontogeny – novel roles in tissue remodelling and mucosal immune defences? Dev. Comp. Immunol. 2018;87:157–170. doi: 10.1016/j.dci.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Magnadóttir B., Hayes P., Gísladóttir B., Bragason B.Þ., Hristova M., Nicholas A.P., Guðmundsdóttir S., Lange S. Pentraxins CRP-I and CRP-II are post-translationally deiminated and differ in tissue specificity in cod (Gadus morhua L.) ontogeny. Dev. Comp. Immunol. 2018;87:1–11. doi: 10.1016/j.dci.2018.05.014. [DOI] [PubMed] [Google Scholar]
- Magnadóttir B., Bragason B.T., Bricknell I.R., Bowden T., Nicholas A.P., Hristova M., Gudmundsdottir S., Dodds A.W., Lange S. Peptidylarginine deiminase and deiminated proteins are detected throughout early halibut ontogeny - complement components C3 and C4 are post-translationally deiminated in halibut (Hippoglossus Hippoglossus L.) Dev. Comp. Immunol. 2019;92:1–19. doi: 10.1016/j.dci.2018.10.016. [DOI] [PubMed] [Google Scholar]
- Magnadóttir B., Kraev I., Guðmundsdóttir S., Dodds A.W., Lange S. Extracellular vesicles from cod (Gadus morhua L.) mucus contain innate immune factors and deiminated protein cargo. Dev. Comp. Immunol. 2019;99 doi: 10.1016/j.dci.2019.103397. [DOI] [PubMed] [Google Scholar]
- Magnadóttir B., Uysal-Onganer P., Kraev I., Dodds A.W., Gudmundsdottir S., Lange S. Extracellular vesicles, deiminated protein cargo and microRNAs are novel serum biomarkers for environmental rearing temperature in Atlantic cod (Gadus morhua L.) Aquacult. Res. 2019 (in press) [Google Scholar]
- McIntyre R.E., Lakshminarasimhan Chavali P., Ismail O., Carragher D.M., Sanchez-Andrade G., Forment J.V. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genetics. 2012;8(11) doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F., Panaccione I., Grayson D.R., Nicoletti F., Guidotti A. Metabotropic glutamate 2/3 receptors and epigenetic modifications in psychotic disorders: a review. Curr. Neuropharmacol. 2016;14(1):41–47. doi: 10.2174/1570159X13666150713174242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N.U., Reddy S.T. Role of hemoglobin/heme scavenger protein hemopexin in atherosclerosis and inflammatory diseases. Curr. Opin. Lipidol. 2015;26(5):384–387. doi: 10.1097/MOL.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf V.J., George P.M., Brennan S.O. Lungfish albumin is more similar to tetrapod than to teleost albumins: purification and characterisation of albumin from the Australian lungfish, Neoceratodus forsteri. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007;147(3):428–437. doi: 10.1016/j.cbpb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Mino T., Takeuchi O. Post-transcriptional regulation of immune responses by RNA binding proteins. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018;94(6):248–258. doi: 10.2183/pjab.94.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L.S., Colwell L.J. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86(7):697–706. doi: 10.1002/prot.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S., Song X., Vig B.S., Landowski C.P., Kim I., Hilfinger J.M., Amidon G.L. Prolidase, a potential enzyme target for melanoma: design of proline-containing dipeptide-like prodrugs. Mol. Pharm. 2005;2(1):37–46. doi: 10.1021/mp049922p. [DOI] [PubMed] [Google Scholar]
- Moscarello M.A., Lei H., Mastronardi F.G., Winer S., Tsui H., Li Z., Ackerley C., Zhang L., Raijmakers R., Wood D.D. Inhibition of peptidyl-arginine deiminases reverses protein-hypercitrullination and disease in mouse models of multiple sclerosis. Dis. Model Mech. 2013;6(2):467–478. doi: 10.1242/dmm.010520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Radic M. Citrullinated autoantigens: from diagnostic markers to pathogenetic mechanisms. Clin. Rev. Allergy Immunol. 2015;49(2):232–239. doi: 10.1007/s12016-014-8459-2. [DOI] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Boehm M., Backlund P.S.Jr., Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol. Biol. Cell. 2004;15(5):2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaroli D.M., Bellvé K.D., Standley C., Lifshitz L.M., Cardia J., Lambright D., Leonard D., Fogarty K.E., Corvera S. Rabenosyn-5 defines the fate of the transferrin receptor following clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(8):E471–80. doi: 10.1073/pnas.1115495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A., Pednekar L., Reid K.B., Kishore U. Complement and non-complement activating functions of C1q: a prototypical innate immune molecule. Innate Immun. 2012;18(2):350–363. doi: 10.1177/1753425910396252. [DOI] [PubMed] [Google Scholar]
- Nicholas A.P., Whitaker J.N. Preparation of a monoclonal antibody to citrullinated epitopes: its characterization and some applications to immunohistochemistry in human brain. Glia. 2002;37(4):328–336. [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 2000;151(3):601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njålsson R., Norgren S. Physiological and pathological aspects of GSH metabolism. Acta Paediatr. 2005;94(2):132–137. doi: 10.1111/j.1651-2227.2005.tb01878.x. [DOI] [PubMed] [Google Scholar]
- Nomura K. Specificity and mode of action of the muscle-type protein-arginine deiminase. Arch. Biochem. Biophys. 1992;293(2):362–369. doi: 10.1016/0003-9861(92)90407-n. [DOI] [PubMed] [Google Scholar]
- Nongonierma A.B., Paolella S., Mudgil P., Maqsood S., FitzGerald R.J. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 2018;244:340–348. doi: 10.1016/j.foodchem.2017.10.033. [DOI] [PubMed] [Google Scholar]
- Norman A.W. A vitamin D nutritional cornucopia: new insights concerning the serum 25-hydroxyvitamin D status of the US population. Am. J. Clin. Nutr. 2008;88(6):1455–1456. doi: 10.3945/ajcn.2008.27049. [DOI] [PubMed] [Google Scholar]
- Olaho-Mukani W., Nyang’ao J.N., Kimani J.K., Omuse J.K. Studies on the haemolytic complement of the dromedary camel (Camelus dromedarius). I. Classical pathway haemolytic activity in serum. Vet. Immunol. Immunopathol. 1995;46(3-4):337–347. doi: 10.1016/0165-2427(94)05367-2. [DOI] [PubMed] [Google Scholar]
- Olaho-Mukani W., Nyang’ao J.N., Kimani J.K., Omuse J.K. Studies on the haemolytic complement of the dromedary camel (Camelus dromedarius). II. Alternate complement pathway haemolytic activity in serum. Vet. Immunol. Immunopathol. 1995;48(1-2):169–176. doi: 10.1016/0165-2427(94)05406-i. [DOI] [PubMed] [Google Scholar]
- O’Neil L.J., Kaplan M.J. Neutrophils in rheumatoid arthritis: breaking immune tolerance and fueling disease. Trends Mol. Med. 2019;25(3):215–227. doi: 10.1016/j.molmed.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Parida S., Siddharth S., Sharma D. Adiponectin, obesity, and cancer: clash of the bigwigs in health and disease. Int. J. Mol. Sci. 2019;20:2519. doi: 10.3390/ijms20102519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Scobey T., Swanstrom J., Jensen K.L., Burch C.L., Baric R.S., Heise M.T. Permissivity of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017;91(19):e00534–17. doi: 10.1128/JVI.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perga S., Giuliano Albo A., Lis K., Minari N., Falvo S., Marnetto F., Caldano M., Reviglione R. Vitamin D binding protein isoforms and apolipoprotein E in cerebrospinal fluid as prognostic biomarkers of multiple sclerosis. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0129291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T., Jr. Academic Press, Inc; San Diego: 1996. All About Albumin. Biochemistry, Genetics, and Medical Applications. [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 2002;59(10):1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietronigro E.C., Della Bianca V., Zenaro E., Constantin G. NETosis in alzheimer’s disease. Front. Immunol. 2017;8:211. doi: 10.3389/fimmu.2017.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasil M., Wijkmark S., Elbers J.P., Oppelt J., Burger P.A., Horin P. The major histocompatibility complex of Old world camelids: class I and class I-related genes. HLA. 2019;93(4):203–215. doi: 10.1111/tan.13510. [DOI] [PubMed] [Google Scholar]
- Ploquin M.J., Casrouge A., Madec Y., Noël N., Jacquelin B., Huot N., Duffy D., Jochems S.P., Micci L. Systemic DPP4 activity is reduced during primary HIV-1 infection and is associated with intestinal RORC+ CD4+ cell levels: a surrogate marker candidate of HIV-induced intestinal damage. J. Int. AIDS Soc. 2018;21(7) doi: 10.1002/jia2.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon M.W., Tsang W.H., Chan S.O., Li H.M., Ng H.K., Waye M.M. Dyslexia-associated kiaa0319-like protein interacts with axon guidance receptor nogo receptor 1. Cell Mol. Neurobiol. 2011;31(1):27–35. doi: 10.1007/s10571-010-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A., Goody R.S., Müller M.P. Multivalency in rab effector interactions. Small GTPases. 2017;27:1–7. doi: 10.1080/21541248.2016.1265700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S.H., Andrews A.M., Paul D., Pachter J.S. Extracellular vesicles: mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS. 2018;15(1):19. doi: 10.1186/s12987-018-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomes F., Bode J., Sukhanova A., Bozrova S.V., Saccomano M., Mitkovski M., Krueger J.E., Wege A.K., Stuehmer W., Samokhvalov P.S., Baty D., Chames P., Nabiev I., Alves F. Single- and two-photon imaging of human micrometastases and disseminated tumour cells with conjugates of nanobodies and quantum dots. Sci. Rep. 2018;8(1):4595. doi: 10.1038/s41598-018-22973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebl A., Köllner B., Anders E., Wimmers K., Goldammer T. Peptidylarginine deiminase gene is differentially expressed in freshwater and brackish water rainbow trout. Mol. Biol. Rep. 2010;37(5):2333–2339. doi: 10.1007/s11033-009-9738-5. [DOI] [PubMed] [Google Scholar]
- Reichl K., Kreykes S.E., Martin C.M., Shenoy C. Desmoplakin variant-associated arrhythmogenic cardiomyopathy presenting as acute myocarditis. Circ. Genom. Precis. Med. 2018;11(12) doi: 10.1161/CIRCGEN.118.002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K.B., Colomb M., Petry F., Loos M. Complement component C1 and the collectins--first-line defense molecules in innate and acquired immunity. Trends Immunol. 2002;23(3):115–117. doi: 10.1016/s1471-4906(01)02164-0. [DOI] [PubMed] [Google Scholar]
- Reid K.B.M. Complement component C1q: historical perspective of a functionally versatile, and structurally unusual, serum protein. Front. Immunol. 2018;9:764. doi: 10.3389/fimmu.2018.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rharass T., Lemcke H., Lantow M., Kuznetsov S.A., Weiss D.G., Panáková D. Ca2+-mediated mitochondrial reactive oxygen species metabolism augments Wnt/β-catenin pathway activation to facilitate cell differentiation. J. Biol. Chem. 2014;289(40):27937–27951. doi: 10.1074/jbc.M114.573519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci E., Ronchetti S., Gabrielli E., Pericolini E., Gentili M., Roselletti E., Vecchiarelli A., Riccardi C. GILZ restrains neutrophil activation by inhibiting the MAPK pathway. J. Leukoc. Biol. 2019;105(1):187–194. doi: 10.1002/JLB.3AB0718-255R. [DOI] [PubMed] [Google Scholar]
- Rivera H., Martín-Hernández E., Delmiro A., García-Silva M.T., Quijada-Fraile P., Muley R., Arenas J., Martín M.A., Martínez-Azorín F. A new mutation in the gene encoding mitochondrial seryl-tRNA synthetase as a cause of HUPRA syndrome. BMC Nephrol. 2013;14:195. doi: 10.1186/1471-2369-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossotti M.A., Henry K.A., van Faassen H., Tanha J., Callaghan D., Hussack G., Arbabi-Ghahroudi M., MacKenzie C.R. Camelid single-domain antibodies raised by DNA immunization are potent inhibitors of EGFR signaling. Biochem. J. 2019;476(1):39–50. doi: 10.1042/BCJ20180795. [DOI] [PubMed] [Google Scholar]
- Saeed H., Shalaby M., Embaby A., Ismael M., Pathan A., Ataya F., Alanazi M., Bassiouny K. The Arabian camel camelus dromedarius heat shock protein 90α: cDNA cloning, characterization and expression. Int. J. Biol. Macromol. 2015;81:195–204. doi: 10.1016/j.ijbiomac.2015.07.058. [DOI] [PubMed] [Google Scholar]
- Saadeldin I.M., Abdel-Aziz Swelum A., Alzahrani F.A., Alowaimer A.N. The current perspectives of dromedary camel stem cells research. Int. J. Vet. Sci. Med. 2018;6(Suppl):S27–S30. doi: 10.1016/j.ijvsm.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadeldin I.M., Swelum A.A., Elsafadi M., Mahmood A., Alfayez M., Alowaimer A.N. Differences between the tolerance of camel oocytes and cumulus cells to acute and chronic hyperthermia. J. Therm. Biol. 2018;74:47–54. doi: 10.1016/j.jtherbio.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Sayed L.H., Badr G., Omar H.M., Abd El-Rahim A.M., Mahmoud M.H. Camel whey protein improves oxidative stress and histopathological alterations in lymphoid organs through bcl-XL/Bax expression in a streptozotocin-induced type 1 diabetic mouse model. Biomed. Pharmacother. 2017;88:542–552. doi: 10.1016/j.biopha.2017.01.076. [DOI] [PubMed] [Google Scholar]
- Schaefer J.S., Klein J.R. Roquin--a multifunctional regulator of immune homeostasis. Genes Immun. 2016;17(2):79–84. doi: 10.1038/gene.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicher L., Wissler M., Preiss F., Brauns-Schubert P., Jakob C., Dumit V., Borner C., Dengjel J., Maurer U. SPATA2 promotes CYLD activity and regulates TNF-induced NF-κB signaling and cell death. EMBO Rep. 2016;17(10):1485–1497. doi: 10.15252/embr.201642592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicher L., Brauns-Schubert P., Schubert F., Maurer U. SPATA2: More than a missing link. Cell Death Differ. 2017;24(7):1142–1147. doi: 10.1038/cdd.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrich G., Raftery M.J. Neutrophil extracellular traps go viral. Front. Immunol. 2016;7:366. doi: 10.3389/fimmu.2016.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttker B., Zhang Y., Heiss J.A., Butterbach K., Jansen E.H., Bewerunge-Hudler M., Saum K.U., Holleczek B., Brenner H. Discovery of a novel epigenetic cancer marker related to the oxidative status of human blood. Genes Chromosomes Cancer. 2015;54(9):583–594. doi: 10.1002/gcc.22271. [DOI] [PubMed] [Google Scholar]
- Selmi C., Cantarini L., Kivity S., Dagaan A., Shovman O., Zandman-Goddard G., Perricone C., Amital H., Toubi E., Shoenfeld Y. The 2014 ACR annual meeting: a bird’s eye view of autoimmunity in 2015. Autoimmun Rev. 2015;14(7):622–632. doi: 10.1016/j.autrev.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Şen V., Uluca Ü., Ece A., Kaplan İ., Bozkurt F., Aktar F., Bağlı S., Tekin R. Serum prolidase activity and oxidant-antioxidant status in children with chronic hepatitis B virus infection. Ital. J. Pediatr. 2014;40(1):95. doi: 10.1186/s13052-014-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Llama-inspired antibody fragment approved for rare blood disorder. Nat. Biotechnol. 2019;37(4):33–334. doi: 10.1038/s41587-019-0101-7. [DOI] [PubMed] [Google Scholar]
- Shriver-Lake L.C., Liu J.L., Zabetakis D., Sugiharto V.A., Lee C.R., Defang G.N., Wu S.L., Anderson G.P., Goldman E.R. Selection and characterization of anti-dengue NS1 single domain antibodies. Sci Rep. 2018;8(1):18086. doi: 10.1038/s41598-018-35923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J., Takai H., Buonomo S.B., Eisenhaber F., de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18(17):2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Širochmanová I., Čomor Ľ., Káňová E., Jiménez-Munguía I., Tkáčová Z., Bhide M. Permeability of the blood-brain barrier and transport of nanobdies across the blood-brain barrier. Folia Vet. 2018;62(1):59–66. [Google Scholar]
- Smith L.E., Crouch K., Cao W., Müller M.R., Wu L., Steven J., Lee M., Liang M., Flajnik M.F., Shih H.H., Barelle C.J., Paulsen J., Gill D.S., Dooley H. Characterization of the immunoglobulin repertoire of the spiny dogfish (squalus acanthias) Dev. Comp. Immunol. 2012;36(4):665–679. doi: 10.1016/j.dci.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Smith A., McCulloh R.J. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front. Physiol. 2015;6:187. doi: 10.3389/fphys.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn D.H., Rhodes C., Onuma K., Zhao X., Sharpe O., Gazitt T., Shiao R., Fert-Bober J., Cheng D., Lahey L.J. Local joint inflammation and histone citrullination in a murine model of the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis Rheumatol. 2015;67:2877–2887. doi: 10.1002/art.39283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottrup-Jensen L., Stepanik T.M., Kristensen T., Lønblad P.B., Jones C.M., Wierzbicki D.M., Magnusson S., Domdey H., Wetsel R.A., Lundwall A. Common evolutionary origin of alpha 2-macroglobulin and complement components C3 and C4. Proc. Natl. Acad. Sci. U. S. A. 1985;82(1):9–13. doi: 10.1073/pnas.82.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen C.N., Karaderi T., Yaghootkar H., Schurmann C., Fine R.S. Exome-derived adiponectin-associated variants implicate obesity and lipid biology. Am. J. Hum. Genet. 2019 doi: 10.1016/j.ajhg.2019.05.002. pii: S0002-9297(19)30188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfliet J.C., Locketz M., Berman P., Pillay T.S. Evaluation of the utility of serum prolidase as a marker for liver fibrosis. J. Clin. Lab. Anal. 2015;29(3):208–213. doi: 10.1002/jcla.21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R.L., Haakenson J., Deiss T.C., Criscitiello M.F., Wilson I.A., Smider V.V. The unusual genetics and biochemistry of bovine immunoglobulins. Adv. Immunol. 2018;137:135–164. doi: 10.1016/bs.ai.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeland S., Vandenbroucke R.E., Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov. Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Sunyer J.O., Lambris J.D. Evolution and diversity of the complement system of poikilothermic vertebrates. Immunol. Rev. 1998;166:39–57. doi: 10.1111/j.1600-065x.1998.tb01251.x. [DOI] [PubMed] [Google Scholar]
- Tang C.J., Fu R.H., Wu K.S., Hsu W.B., Tang T.K. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- Tarighi S., Najafi M., Hossein-Nezhad A., Ghaedi H., Meshkani R., Moradi N., Fadaei R., Kazerouni F., Shanaki M. Association between Two Common polymorphisms of vitamin D binding protein and the risk of coronary artery disease: a case-control study. J. Med. Biochem. 2017;36(4):349–357. doi: 10.1515/jomb-2017-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcsa E., Marekov L.N., Mei G., Melino G., Lee S.C., Steinert P.M. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 1996;271(48):30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- Tennent G.A., Brennan S.O., Stangou A.J., O’Grady J., Hawkins P.N., Pepys M.B. Human plasma fibrinogen is synthesized in the liver. Blood. 2007;109:1971–1974. doi: 10.1182/blood-2006-08-040956. [DOI] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscia G.L., Margaglione M. Human fibrinogen: molecular and genetic aspects of congenital disorders. Int. J. Mol. Sci. 2018;19(6):E1597. doi: 10.3390/ijms19061597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M. A nanobody-based molecular toolkit provides new mechanistic insight into clathrin-coat initiation. Elife. 2019;8:e41768. doi: 10.7554/eLife.41768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers T.S., Harlow L., Rosas I.O., Gochuico B.R., Mikuls T.R., Bhattacharya S.K., Camacho C.J., Ascherman D.P. Extensive citrullination promotes immunogenicity of HSP90 through protein unfolding and exposure of cryptic epitopes. J. Immunol. 2016;197(5):1926–1936. doi: 10.4049/jimmunol.1600162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Drapkina O., Tonevitsky A. Transcriptome of extracellular vesicles: State-of-the-art. Front Immunol. 2019;10:202. doi: 10.3389/fimmu.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbainsky C., Nölker R., Imber M., Lübken A., Mostertz J., Hochgräfe F., Godoy J.R., Hanschmann E.M., Lillig C.H. Nucleoredoxin-dependent targets and processes in neuronal cells. Oxid. Med. Cell Longev. 2018;2018 doi: 10.1155/2018/4829872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T., Chin A., Mariscal J., Bannykh S., Engman D.M., Di Vizio D. Protein composition reflects extracellular vesicle heterogeneity. Proteomics. 2019 doi: 10.1002/pmic.201800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandschoot P., Stortelers C., Beirnaert E., Ibañez L.I., Schepens B., Depla E. Nanobodies®: new ammunition to battle viruses. Antiviral Res. 2011;92:389–407. doi: 10.1016/j.antiviral.2011.09.002. [DOI] [PubMed] [Google Scholar]
- van Lith S.A., Roodink I., Verhoeff J.J., Mäkinen P.I., Lappalainen J.P., Ylä-Herttuala S., Raats J., van Wijk E., Roepman R., Letteboer S.J., Verrijp K., Leenders W.P. In vivo phage display screening for tumor vascular targets in glioblastoma identifies a llama nanobody against dynactin-1-p150Glued. Oncotarget. 2016;7(44):71594–71607. doi: 10.18632/oncotarget.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin E.M., Mulvihill E.E., Beaudry J.L., Pujadas G., Fuchs S., Tanti J.F., Fazio S., Kaur K., Cao X., Baggio L.L., Matthews D., Campbell J.E., Drucker D.J. Circulating levels of soluble dipeptidyl peptidase-4 are dissociated from inflammation and induced by enzymatic DPP4 inhibition. Cell Metab. 2019;29(2):320–334. doi: 10.1016/j.cmet.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza A., Toma C., Paracchini S., Monaco A.P. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum. Mol. Genet. 2008;17(6):859–871. doi: 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
- Verboven C., Rabijns A., De Maeyer M., Van Baelen H., Bouillon R., De Ranter C. A structural basis for the unique binding features of the human vitamin D-binding protein. Nat. Struct. Biol. 2002;9(2):131–136. doi: 10.1038/nsb754. [DOI] [PubMed] [Google Scholar]
- Viglio S., Annovazzi L., Conti B., Genta I., Perugini P., Zanone C., Casado B., Cetta G., Iadarola P. The role of emerging techniques in the investigation of prolidase deficiency: from diagnosis to the development of a possible therapeutical approach. J. Chromatogr. B. 2006;832(1):1–8. doi: 10.1016/j.jchromb.2005.12.049. [DOI] [PubMed] [Google Scholar]
- Vossenaar E.R., Zendman A.J., van Venrooij W.J., Pruijn G.J. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25(11):1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- Wagner S.A., Satpathy S., Beli P., Choudhary C. SPATA2 links CYLD to the TNF-α receptor signaling complex and modulates the receptor signaling outcomes. EMBO J. 2016;35(17):1868–1884. doi: 10.15252/embj.201694300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed M.M., Ghoneim I.M., Alhaider A.K. Seminal plasma and serum fertility biomarkers in dromedary camels (camelus dromedarius) Theriogenology. 2015;83(4):650–654. doi: 10.1016/j.theriogenology.2014.10.033. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim. Biophys. Acta. 2013;1829(10):1126–1135. doi: 10.1016/j.bbagrm.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lapek J., Fujimura K., Strnadel J., Liu B., Gonzalez D.J., Zhang W., Watson F., Yu V., Liu C., Melo C.M., Miller Y.I., Elliott K.C., Cheresh D.A., Klemke R.L. Pseudopodium-enriched atypical kinase 1 mediates angiogenesis by modulating GATA2-dependent VEGFR2 transcription. Cell Discov. 2018;4:26. doi: 10.1038/s41421-018-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Meng W., Guo H., Pan W., Liu J., Peng T., Chen L., Chen C.Y. Potent neutralization of influenza a virus by a single-domain antibody blocking M2 ion channel protein. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle U.H., Birzele F., Tiefenthaler G. Potential of protein-based anti-metastatic therapy with serpins and inter α-trypsin inhibitors. Cancer Genomics Proteom. 2018;15(4):225–238. doi: 10.21873/cgp.20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel J.W., Litvinov R.I. Mechanisms of fibrin polymerization and clinical implications. Blood. 2013;121:1712–1719. doi: 10.1182/blood-2012-09-306639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthur C., Daniels J.S., Morrison R., Engers J.L., Niswender C.M., Conn P.J., Lindsley C.W. National Center for Biotechnology Information (US); Bethesda (MD): 2019. Development of a Second Generation mGlu3 NAM Probe. Probe Reports from the NIH Molecular Libraries Program [Internet] pp. 2010–2012. [PubMed] [Google Scholar]
- Wesolowski J., Alzogaray V., Reyelt J., Unger M., Juarez K., Urrutia M. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.R., Datta G., Giordano S. High-density lipoprotein regulation of mitochondrial function. Adv. Exp. Med. Biol. 2017;982:407–429. doi: 10.1007/978-3-319-55330-6_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser V., Tsibulak I., Degasper C., Welponer H., Leitner K., Parson W., Zeimet A.G., Marth C., Fiegl H. Tumor necrosis factor receptor modulator spermatogenesis-associated protein 2 is a novel predictor of outcome in ovarian cancer. Cancer Sci. 2019;110(3):1117–1126. doi: 10.1111/cas.13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witalison E.E., Thompson P.R., Hofseth L.J. Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr. Drug Targets. 2015;16(7):700–710. doi: 10.2174/1389450116666150202160954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Guang X., Al-Fageeh M.B., Cao J., Pan S., Zhou H. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014;5:5188. doi: 10.1038/ncomms6188. [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Suyama H., Yamamoto N. Immunotherapy for prostate cancer with Gc protein-derived macrophage-activating factor, GcMAF. Transl Oncol. 2008;1(2):65–72. doi: 10.1593/tlo.08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- Yang J., Gao J., Adamian M., Wen X.H., Pawlyk B., Zhang L., Sanderson M.J., Zuo J., Makino C.L., Li T. The ciliary rootlet maintains long-term stability of sensory cilia. Mol. Cell. Biol. 2005;25:4129–4137. doi: 10.1128/MCB.25.10.4129-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Martin P., Fogarty B., Brown A., Schurman K., Phipps R., Yin V.P., Lockman P., Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in danio rerio. Pharm. Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermakovich D., Sivitskaya L., Vaikhanskaya T., Danilenko N., Motuk I. Novel desmoplakin mutations in familial carvajal syndrome. Acta Myol. 2018;37(4):263–266. [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Du Pasquier L., Hsu E. Shark IgW C region diversification through RNA processing and isotype switching. J. Immunol. 2013;191(6):3410–3418. doi: 10.4049/jimmunol.1301257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fan L., Hou F., Dong A., Wang Y.X., Tong Y. New insights into the RNA-binding and E3 ubiquitin ligase activities of roquins. Sci. Rep. 2015;5:15660. doi: 10.1038/srep15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Zhang X.J., Song Y.L., Lu X.B., Chen D.D., Xia X.Q., Sunyer J.O., Zhang Y.A. Preferential combination between the light and heavy chain isotypes of fish immunoglobulins. Dev. Comp. Immunol. 2016;61:169–179. doi: 10.1016/j.dci.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang N., Zhang X.J., Chen D.D., Sunyer O.J., Zhang Y.A. Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (oncorhynchus mykiss) Dev. Comp. Immunol. 2017;70:94–105. doi: 10.1016/j.dci.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Xu G., Wang Z., Gao J., Cui S., Liu J. Deletion of Spata2 by CRISPR/Cas9n causes increased inhibin alpha expression and attenuated fertility in male mice. Biol. Reprod. 2017;97(3):497–513. doi: 10.1093/biolre/iox093. [DOI] [PubMed] [Google Scholar]
- Zhou G., Yang L., Gray A., Srivastava A.K., Li C., Zhang G., Cui T. The role of desmosomes in carcinogenesis. Onco Targets Ther. 2017;10:4059–4063. doi: 10.2147/OTT.S136367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L., Kimata K. Structure and function of inter-alpha-trypsin inhibitor heavy chains. Connect. Tissue Res. 2008;49:311–320. doi: 10.1080/03008200802325458. [DOI] [PubMed] [Google Scholar]
- Zhu S., Zimmerman D., Deem S.L. A review of zoonotic pathogens of dromedary camels. Ecohealth. 2019;2019(May 28) doi: 10.1007/s10393-019-01413-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.