SEC can compartmentalize P-TEFb via LLPS from the inactive soluble HEXIM1 complex to regulate rapid transcriptional events.
Abstract
Release of paused RNA polymerase II (Pol II) requires incorporation of the positive transcription elongation factor b (P-TEFb) into the super elongation complex (SEC), thus resulting in rapid yet synchronous transcriptional activation. However, the mechanism underlying dynamic transition of P-TEFb from inactive to active state remains unclear. Here, we found that the SEC components are able to compartmentalize and concentrate P-TEFb via liquid-liquid phase separation from the soluble inactive HEXIM1 containing the P-TEFb complex. Specifically, ENL or its intrinsically disordered region is sufficient to initiate the liquid droplet formation of SEC. AFF4 functions together with ENL in fluidizing SEC droplets. SEC droplets are fast and dynamically formed upon serum exposure and required for rapid transcriptional induction. We also found that the fusion of ENL with MLL can boost SEC phase separation. In summary, our results suggest a critical role of multivalent phase separation of SEC in controlling transcriptional pause release.
INTRODUCTION
A general feature of metazoan transcription is that RNA polymerase II (Pol II) is paused 30 to 40 nucleotides downstream from the transcription start site of the immediate early genes (IEGs) and many of the developmentally controlled genes (1, 2). Promoter-proximal pausing of Pol II is currently believed to be the prerequisite for rapid yet synchronous transcriptional induction (3, 4). Acute stress or developmental stimuli can act as a trigger signal for paused Pol II to escape into productive elongation stage (2, 5). The positive transcription elongation factor b (P-TEFb), a heterodimer of the kinase CDK9 and CCNT1, is required for releasing Pol II from promoter-proximal pausing into productive elongation stage (5, 6). The molecular bases and regulatory mechanisms of P-TEFb in stimulating pause release have been extensively explored over the past decades.
In cells, both active and inactive forms of P-TEFb coexist and dynamically cycle between each other reacting to cellular needs (6, 7). Most of the P-TEFb is sequestered in the catalytically inactive HEXIM1/2-containing complex, together with a 7SK small nuclear ribonucleoprotein particle (snRNP) (8). It has long been thought that, upon growth signal stimulation or stress, P-TEFb is freed from the inactive complex and recruited to the chromatin by the active complexes to stimulate transcription (5, 6, 9). However, recent genome-wide studies revealed a broad colocalization of HEXIM1 and CDK9 at gene promoters (10). The mechanism behind the inactive-to-active transition of P-TEFb remains an open question.
The super elongation complex (SEC) is one of the most active P-TEFb complexes required for rapid transcriptional induction of the IEGs and many of the developmentally control genes in the presence, or even absence in certain cases, of paused Pol II (4, 11). SEC was originally identified from in-depth biochemical purifications of some of the most frequently occurring chimera proteins encoded by mixed lineage leukemia (MLL) fusion oncogenes (12). SEC is a large protein complex about 1.5 mDa in size. In addition to P-TEFb, SEC also contains the RNA Pol II elongation factors ELL1-3 and MLL translocation partners including AF4/FMR2 family member 1/4 (AFF1/4), eleven nineteen leukemia (ENL), and ALL1-fused gene from chromosome 9 (AF9) (12). The transcription-activating function of SEC is vital for stress response and normal development (4, 13). Misactivation of SEC by stabilizing SEC components, via either MLL fusion or gain-of-function point mutations, plays a major role in leukemogenesis and the congenital CHOPS syndrome (cognitive impairment and coarse facies, heart defects, obesity, pulmonary involvement, and short stature and skeletal dysplasia) (12, 14). The critical roles of SEC in development and disease pathogenesis exerted from its transcription regulatory activity have been well accepted (5, 15). However, the mechanistic and biophysical bases that determine the assembly of a functional SEC for rapid transcriptional induction still remain unknown.
Heterogeneous mixtures can be partitioned into different immiscible phases via liquid-liquid phase separation (LLPS) (16–18). Increasing evidence showed that formation of membraneless biomolecular condensates through intracellular LLPS provides an efficient way for compartmentalizing bioreactions (19–21). This leads to the proposal that dynamic and multivalent interactions among molecules promoted by LLPS within a biocondensate permit rapid assembly and action of a functional complex, such as SEC, in response to cellular needs. It has been reported recently that a histidine-rich domain in the P-TEFb subunit, CCNT1, promotes LLPS of CCNT1 in vitro (22). Here, we find that LLPS underlies the transition of P-TEFb from the HEXIM1-containing complex to SEC. The SEC components, ENL and AFF4, are able to compartmentalize and concentrate P-TEFb into the LLPS-formed droplets, while P-TEFb together with its inactive partner HEXIM1 is soluble. In addition, we show in real time that SEC fast assembles into phase-separated droplets upon serum exposure, which is required for rapid transcriptional induction of IEGs by SEC. In summary, our results offer a new insight on the mechanism underlying the trigger of rapid gene activation program by SEC and that the functional state of a stable protein complex is related to its physicochemical status on whether the protein complex is multivalent phase separated or not.
RESULTS
The SEC components compartmentalize CCNT1 from HEXIM1 to form a liquid-like droplet
We first asked whether P-TEFb undergoes phase transition when it switches between the inactive and active states through interacting with different factors. Consistent with the previous data, ectopically expressed CCNT1 was displayed as phase-separated droplets (Fig. 1A) (22). However, CCNT1 became ubiquitously diffused once coexpressed with HEXIM1 (Fig. 1A and fig. S1, A and B). Their direct interaction was important for HEXIM1 to dissolve CCNT1 (Fig. 1B). To further explore the effect of HEXIM1 on the cellular pattern of CCNT1, we performed CCNT1 immunofluorescence (IF) staining before and after HEXIM1 knockdown. IF analysis revealed that CCNT1 appeared as large amorphous condensates in control cells, which could reflect the steady-state levels of inactive and active P-TEFb–containing complexes (Fig. 1C) (5). Upon HEXIM1 knockdown, the number of CCNT1 puncta per cell was substantially increased, while the size of individual punctum was reduced (Fig. 1C and fig. S1, C and D).
Fig. 1. The SEC components AFF4 and ENL compartmentalize CCNT1 from HEXIM1 through forming heterotypic phase-separated droplets.
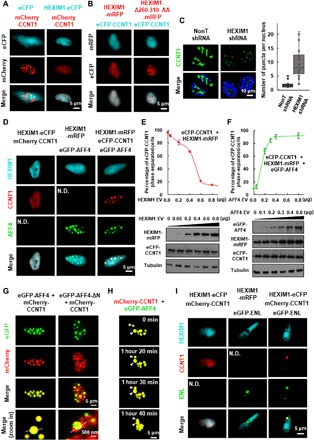
(A) Live cell imaging of HeLa cells expressing mCherry-CCNT1 only or together with HEXIM1-eCFP. (B) Live cell imaging of HeLa cells coexpressing eCFP-CCNT1 together with wild-type HEXIM1 or HEXIM1-Δ260-310 AA-mRFP. (C) IF imaging (left) of CCNT1 in control or HEXIM1 knockdown HCT 116 cells. DNA was counterstained using DAPI (4′,6-diamidino-2-phenylindole). Box plot (right) showing that the number of CCNT1 puncta per nucleus is significantly increased after shRNA-mediated HEXIM1 knockdown. Each n > 20 nuclei; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (D) Live cell imaging of HeLa cells coexpressing HEXIM1 and CCNT1 (left), HEXIM1 and AFF4 (middle), or the three proteins together (right) with different fluorescence tags. N.D., not detected. (E) Percentage of cells with eCFP-CCNT1 phase-separated droplets (upper) and Western analyses (lower) are shown after cotransfecting HeLa cells with eCFP-CCNT1 and increasing amount of the HEXIM1-mRFP construct. Error bars represent SDs. Results are representative of three biological replicates. (F) Percentage of cells with eCFP-CCNT1 phase-separated droplets (upper) and Western analyses (lower) are shown after cotransfecting HeLa cells with eCFP-CCNT1, HEXIM1-mRFP, and increasing amount of the eGFP-AFF4 construct. Error bars represent SDs. Results are representative of three biological replicates. (G) Live cell imaging of HeLa cells coexpressing eGFP-AFF4 and mCherry-CCNT1 (left), and eGFP-AFF4-ΔN and mCherry-CCNT1 (right). (H) Time-lapse fluorescence images of the nucleus of a HeLa cell expressing mCherry-CCNT1 and eGFP-AFF4 subjected to illuminate every 10 min for the times indicated. The two CCNT1 and AFF4 heterotypic droplets underwent spontaneous fusion as indicated by arrows. (I) Live cell imaging of HeLa cells coexpressing HEXIM1 and CCNT1 (left), HEXIM1 and ENL (middle), or the three proteins together (right) with different fluorescence tags.
We then asked whether the SEC components could function as scaffolds to concentrate P-TEFb from the inactive HEXIM1-containing complex via phase separation. AFF4 is the central component of SEC (12). We transfected enhanced green fluorescent protein (eGFP)–AFF4 into the enhanced cyan fluorescent protein (eCFP)–CCNT1 and HEXIM1–monomeric red fluorescent protein (mRFP) coexpressing cells and observed that CCNT1 was incorporated into the AFF4-containing spherical and micrometer-sized condensates, whereas HEXIM1 still remained diffuse (Fig. 1D). Furthermore, HEXIM1 was able to dissolve CCNT1 in a dosage-dependent manner, without affecting CCNT1 expression (Fig. 1E). In contrast, the increase in AFF4 expression led more soluble CCNT1 to be extracted to liquid droplets but had no effects on the expression levels of both CCNT1 and HEXIM1 (Fig. 1F).
The direct interaction between CCNT1 and AFF4 was required for formation of the heterotypic phase-separated droplets, which can fuse and exhibit typical liquid-like property of viscous relaxation to a larger spherical shape to minimize surface areas (Fig. 1, G and H, and fig. S1, E to I). Similarly, ENL, one of the core components of SEC, can also concentrate CCNT1 from HEXIM1 through phase separation (Fig. 1I). In summary, our data demonstrated that CCNT1 could phase separate in the presence of the SEC components but not HEXIM1.
Differential capabilities of the SEC components to phase separate
We then further examined the phase separation kinetics of the SEC components both in vivo and in vitro. Coimmunostaining of AFF4 with ENL, CDK9, or ELL2 revealed nuclear puncta for these tested colocalized SEC components (Fig. 2A). Live cell images also showed that cells with eGFP-AFF4 or eGFP-ENL contained micrometer-sized, spherically shaped, dynamic fluorescent condensates, which exhibited liquid-like fusion behavior (Fig. 2, B and C). In contrast, cells transfected with CDK9-mRFP or ELL2-mRFP mostly displayed a dispersed fluorescent pattern (Fig. 2B). These results suggested the differential capabilities of the SEC components to phase separate in vivo. ENL and AFF4 are capable of functioning as scaffolds that drive the formation of the liquid-like SEC droplets.
Fig. 2. The SEC components exhibit differential phase separation capabilities.
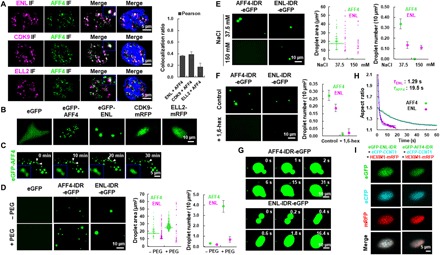
(A) Confocal images (left) showing colocalization of AFF4 with ENL, ELL2, and CDK9 in nuclear puncta in HeLa cells. DNA was counterstained using DAPI. Histogram (right) showing the Pearson correlation coefficient of colocalization ratio. Each n > 20; error bars represent SDs. Results are representative of three biological replicates. (B) Live cell imaging of HeLa cells expressing eGFP-AFF4, eGFP-ENL, CDK9-mRFP, or ELL2-mRFP only. eGFP was used as a negative control. (C) Time-lapse fluorescence images of the nucleus of a HeLa cell expressing eGFP-AFF4 subjected to illuminate every 15 s for the times indicated. The two AFF4 droplets underwent spontaneous fusion as indicated by arrows. (D) Fluorescence microscopy images (left) showing phase-separated droplets formed in 37.5 mM NaCl containing buffer with 20 μM AFF4-IDR-eGFP or ENL-IDR-eGFP in the absence or presence of PEG-8000. Purified eGFP was used as a negative control. Droplet area (middle) and number (right) are also shown. Error bars represent SDs. Results are representative of three biological replicates. (E) Fluorescence microscopy images (left) showing the AFF4-IDR-eGFP or ENL-IDR-eGFP droplets in buffers containing 20 μM purified proteins and different concentrations of NaCl. Droplet area (middle) and number (right) are also shown. Error bars represent SDs. Results are representative of three biological replicates. (F) Fluorescence microscopy images (left) showing that the AFF4-IDR-eGFP or ENL-IDR-eGFP droplets in buffer containing 20 μM purified proteins and 37.5 mM NaCl are sensitive to 3% 1,6-hexanediol. Droplet number (right) is also shown. Error bars represent SDs. Results are representative of three biological replicates. (G) Time-lapse fluorescence images showing that the homotypic AFF4-IDR-eGFP (upper) or ENL-IDR-eGFP (lower) droplets rapidly fuse upon contact into one spherical droplet. The AFF4-IDR-eGFP (upper) and ENL-IDR-eGFP (lower) were subjected to illuminate every second or 200 ms, respectively. The droplet formation buffer contains 10% PEG-8000 and 37.5 mM NaCl. (H) Aspect ratio versus time for droplet fusion of AFF4-IDR-eGFP (green) and ENL-IDR-eGFP (purple). T designates relaxation time of the fusion events, and the blue line indicates nonlinear fitting curve. (I) Live cell imaging of HeLa cells coexpressing eCFP-CCNT1, HEXIM1-mRFP with either eGFP-ENL-IDR (left) or eGFP-AFF4-IDR (right).
Unstructured intrinsically disordered region (IDR) within a protein can promote the formation of phase-separated droplets (23, 24). AFF4 belongs to the AF4/FMR2 family, members of which function as an intrinsically disordered scaffold in recruiting cofactors through their dispersed hydrophobic sites (5). The 760 to 901–amino acid IDR of AFF4 is serine enriched, containing a transcriptional activation domain (Fig. 2A). The YAF9, ENL, AF9, TAF14, SAS5 (YEATS) family member ENL also bears IDRs (fig. S2B). We, therefore, examined whether these IDRs in AFF4 and ENL could phase separate in vitro (fig. S2C). Fluorescence microscopy revealed that both AFF4-IDR-eGFP and ENL-IDR-eGFP fusion proteins were able to form spherical droplets in the absence of the crowding agent polyethylene glycol 8000 (PEG-8000) and that addition of PEG-8000 led to increased droplet numbers and sizes (Fig. 2D and fig. S2D). Both AFF4 and ENL IDR droplets can be observed even when 200 nM purified proteins were added in the reaction system (fig. S2E). Increasing NaCl concentrations in the droplet formation buffers affected the efficiency in forming the AFF4-IDR and ENL-IDR droplets in the presence and absence of PEG-8000 (Fig. 2E and fig. S3, A to C). Furthermore, 1,6-hexanediol, a compound interfering with weak hydrophobic interactions that are essential for assembling these liquid-like condensates, completely inhibited the droplet formation (Fig. 2F and fig. S3D). These droplets were also disrupted when the temperature was increased to 65°C (fig. S3E). Together, our results suggested that both hydrophobic and electrostatic interactions might be important for phase separation of the IDRs of AFF4 and ENL.
Further aspect ratio analysis demonstrated that the newly fused AFF4-IDR and ENL-IDR droplets tended to adopt spherical shapes (Fig. 2, G and H). We also plotted the aspect ratio versus droplet fusion time to determine the relaxation time. Compared with the AFF4-IDR, ENL-IDR droplets had a faster coalescence rate, suggesting that the SEC subunits AFF4 and ENL exhibit different liquid-like viscous relaxation behaviors (Fig. 2H). Furthermore, ENL-IDR only, but not AFF4-IDR, was able to compartmentalize CCNT1 into phase droplets (Fig. 2I). Deletion of IDR from ENL abolished the capability of ENL in forming droplets (fig. S3F). In summary, our results indicated that AFF4-IDR and ENL-IDR are able to phase separate into liquid-like droplets with different kinetics in vitro.
ENL promotes the multivalent phase separation of SEC
We noticed that CDK9, the kinase module of P-TEFb, when overexpressed alone, was not able to form condensates in cells (Fig. 2B). We next examined whether CDK9 can be incorporated into the CCNT1 droplets. Unexpectedly, in addition to HEXIM1, CDK9 is also able to dissolve CCNT1, as shown by live image analyses (Figs. 1 and 3A). This result further suggests that the HEXIM1-containing P-TEFb complex is soluble in cells and that a dynamic phase transition of P-TEFb might occur during its release from the HEXIM1-containing complex into SEC.
Fig. 3. ENL promotes the multivalent phase separation of SEC.
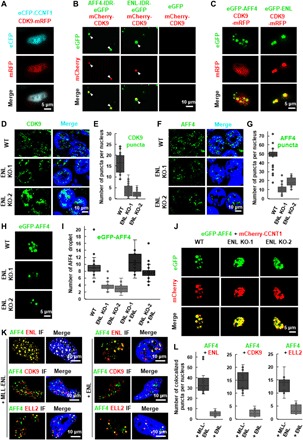
(A) Live cell imaging of HeLa cells coexpressing eCFP-CCNT1 and CDK9-mRFP. (B) Fluorescence microscopy images showing that the purified mCherry-CDK9 proteins can form heterotypic droplets together with AFF4-IDR-eGFP or ENL-IDR-eGFP. The purified eGFP protein was used as a negative control. Purified proteins (10 μM) were used, and the droplet formation buffer contains 10% PEG-8000 and 50 mM NaCl. (C) Live cell imaging of HeLa cells expressing CDK9-mRFP together with eGFP-AFF4 or eGFP-ENL. (D) IF imaging of CDK9 in wild-type and ENL knockout HCT 116 cells. DNA was counterstained using DAPI. (E) Box plot showing that the number of CDK9 puncta per nucleus is significantly decreased after ENL knockout. Each n > 30 nuclei; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (F) IF imaging of AFF4 in wild-type and ENL knockout HCT 116 cells. DNA was counterstained using DAPI. (G) Box plot showing that the number of AFF4 puncta per nucleus is significantly decreased after ENL knockout. Each n > 30 nuclei; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (H) Live cell imaging of HCT 116 wild-type and ENL knockout cells expressing eGFP-AFF4. (I) Box plot showing that the number of eGFP-AFF4 droplets per nucleus is significantly decreased after ENL knockout, which can be rescued by overexpression of mCherry-ENL. Each n > 20; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (J) Live cell imaging of HCT 116 wild-type and ENL knockout cells coexpressing eGFP-AFF4 and mCherry-CCNT1. (K) IF imaging showing costaining of AFF4 with ENL, CDK9, or ELL2 in HeLa cells transfected with MLL-ENL (left) or ENL (right). DNA was counterstained using DAPI. (L) Box plot showing that the number of AFF4/ENL (left), AFF4/CDK9 (middle), and AFF4/ELL2 (right) colocalized puncta per nucleus is significantly increased after transfection with MLL-ENL. Each n > 20; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates.
We further examined whether ENL or AFF4 could incorporate CDK9 into droplets in vitro. Albeit not able to form droplets alone, purified CDK9 can be absorbed by either AFF4-IDR or ENL-IDR and form heterotypic droplets in vitro (Fig. 3B). Notably, the ENL-IDR and CDK9 heterotypic droplets had a recently described core/shell structure (25, 26), with a heterotypic core of CDK9 and ENL-IDR surrounded by a shell of ENL-IDR (Fig. 3B) (27). Live image analyses revealed that only ENL, but not AFF4, was capable of compartmentalizing and concentrating CDK9 in live cells (Fig. 3C). Furthermore, ENL can absorb ELL2 into the phase-separated droplets in live cells, although ELL2 per se, similar to CDK9, lacked the ability to phase separate (fig. S4). Together, these results demonstrated that ENL is critical for compartmentalizing and concentrating P-TEFb and ELL2 via phase separation.
To further evaluate the impact of ENL on SEC phase separation, we generated ENL knockout cell lines (fig. S5, A and B). IF analysis revealed that the deletion of ENL substantially reduced the number of the nuclear CDK9 puncta, without affecting the protein levels of CDK9 and CCNT1 (Fig. 3, D and E, and fig. S5B). Despite the fact that AFF4 can phase separate by itself, we found that the number of nuclear AFF4 puncta was also remarkably decreased in ENL knockout cells (Fig. 3, F and G). Consistently, the formation of eGFP-AFF4 droplets in live cells was less efficient after ENL knockout, which can be rescued by the overexpression of ENL (Fig. 3, H and I, and fig. S5C). Furthermore, we observed obvious disruption of the heterotypic eGFP-AFF4/mCherry-CCNT1 droplets in ENL knockout cells (Fig. 3J). Therefore, ENL could be critical for the formation of SEC droplets.
ENL is frequently translocated to MLL in infant leukemia (28). We costained the tested SEC components in cells overexpressing either wild-type ENL or MLL-ENL fusion protein. Consistent with previous studies showing that the SEC components also exist outside SEC (5), our data demonstrated that the tested SEC components did not fully overlap (Fig. 3K). We also found that the translocation of ENL to MLL led to an increase in the number of the nuclear puncta containing the tested SEC components (Fig. 3, K and L). This observation may provide a biophysical mechanism for MLL-ENL fusion in leukemogenesis.
AFF4 and ENL function together in promoting the fluidity of the SEC puncta
To further shed light on the biophysical properties of the heterotypic droplets formed by P-TEFb and ENL, we performed fluorescence recovery after photobleaching (FRAP) analysis to determine the dynamics within the droplets (29, 30). The eGFP-ENL and CDK9-mRFP heterotypic droplet had a slow FRAP recovery rate, indicating the droplet in a static solid-like state (Fig. 4, A and B). Reacting to environmental stimuli or stress, SEC rapidly activates the transcription of IEGs within minutes (4, 12). It is likely that the SEC droplets will require faster internal reorganization kinetics in vivo. We then asked whether AFF4 could confer a dynamic fluid-like state on SEC. Once AFF4 was overexpressed, eGFP-ENL with CDK9-mRFP or ELL2-mRFP in the heterotypic droplet exhibited faster, liquid-like recovery kinetics (Fig. 4, A and B, and fig. S6, A to C). In addition, cotransfection of AFF4, but not CCNT1, led to a notable increase in the number of eGFP-ENL, and CDK9-mRFP heterotypic droplets formed but reduced in droplet size (Fig. 4, C and D). A possible explanation is that AFF4 might also prevent excessive coalescence of liquid droplet. The eGFP-ENL and CDK9-mRFP droplets exhibited defective liquid-like behaviors, such as irregular shape and incomplete coalescence, after knockdown of AFF4 by short hairpin RNA (shRNA)–mediated RNA interference (RNAi) or inhibition of AFF4 by the small molecule KL-1 (Fig. 4, E and F, and fig. S6, D and E). In summary, our results suggested that AFF4 could function in maintaining proper behaviors of the ENL and P-TEFb heterotypic droplets.
Fig. 4. Fluidity of the ENL heterotypic droplets depends on AFF4.
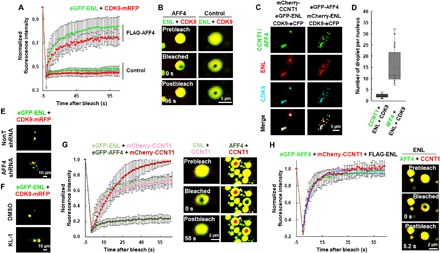
(A) Normalized FRAP recovery curves for eGFP-ENL (green) and CDK9-mRFP (red) in the heterotypic droplet in the presence or absence of FLAG-AFF4. The bleaching events occurred at 0 s. Results shown are from six biological replicates. (B) Live cell confocal images showing FRAP of the heterotypic eGFP-ENL and CDK9-mRFP droplet in the presence or absence of FLAG-AFF4. (C) Live cell imaging of HeLa cells coexpressing ENL and CDK9 together with CCNT1 (left) or AFF4 (right) with different fluorescence tags. (D) Box plot showing the number of the indicated heterotypic droplets per nucleus. Each n > 20; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (E) Live cell imaging of control and AFF4 knockdown HCT 116 cells coexpressing eGFP-ENL and CDK9-mRFP. (F) Live cell imaging of HCT 116 cells coexpressing eGFP-ENL and CDK9-mRFP in the presence or absence of KL-1. (G) Normalized FRAP recovery curves (left) for mCherry-CCNT1 (pink or red), eGFP-ENL (light green), and eGFP-AFF4 (green) in the CCNT1/ENL and CCNT1/AFF4 heterotypic droplets, respectively. The bleaching events occurred at 0 s. Live cell confocal images (right) showing FRAP of the CCNT1/ENL and CCNT1/AFF4 heterotypic droplets. Results shown are from six biological replicates. (H) Normalized FRAP recovery curves (left) for eGFP-AFF4 (green) and mCherry-CCNT1 (red) in the heterotypic droplet in the presence of FLAG-ENL. The bleaching events occurred at 0 s. Live cell confocal images (right) showing FRAP of the heterotypic AFF4/CCNT1 droplet in the presence of FLAG-ENL. Results shown are from six biological replicates.
We also determined the liquid-like condensate features of the AFF4 droplets by FRAP assay. Unexpectedly, eGFP-AFF4 formed droplets with a slow recovery rate after photobleaching, similar to eGFP-ENL (fig. S6, F and G). In contrast, the ENL and AFF4 heterotypic droplet exhibited a much faster recovery rate, even after multiple rounds of photobleaching, with apparent diffusion coefficients of ~0.18 ± 0.018 and 0.075 ± 0.007 μm2/s, respectively (fig. S6, F to H). In addition, when bleaching the entire droplets, we found that the recovery rate of the ENL and AFF4 heterotypic droplet remained unchanged, indicating that the exchange of molecules within the droplets was as fast as that happened between the droplets and the surrounding solution (fig. S7, A to C). These results suggested interdependence between ENL and AFF4 in promoting their own fluidity. Note that AFF4 also promotes the fluidity of CCNT1, although AFF4 per se remained rigid in the droplet (Fig. 4G and fig. S7D). However, when ENL was coexpressed, fluidity of both eGFP-AFF4 and mCherry-CCNT1 in the heterotypic droplet was significantly enhanced, with Dapp of ~0.18 and 0.16 μm2/s, respectively (Fig. 4H and fig. S7E). Likely, as a large disordered scaffold protein, although AFF4 can promote the FRAP recovery rate of CCNT1, the full behavior of AFF4 per se requires other SEC components, such as ENL. Together, our results suggested that AFF4 promotes fluidity of the SEC component droplets and that the dynamics of AFF4 requires ENL. Therefore, AFF4 and ENL are both required for nucleating and fluidizing SEC phase droplets.
Phase separation of SEC is required for its function in rapid gene induction
We further asked if the ability of SEC to phase separate is critical for its function in rapid gene induction. Live imaging analyses indicated that the number of the AFF4 and ENL heterotypic droplets was substantially reduced after serum starvation, while new AFF4-ENL and AFF4-CCNT1 heterotypic droplets formed within 30 min of exposure to serum (Fig. 5A and fig. S8A). Consistently, the increase in the number of endogenous AFF4 puncta after serum treatment was also observed, suggesting a dynamic formation of SEC droplets in vivo (Fig. 5, B and C).
Fig. 5. SEC puncta localize to immediate-response genes in vivo upon serum treatment.
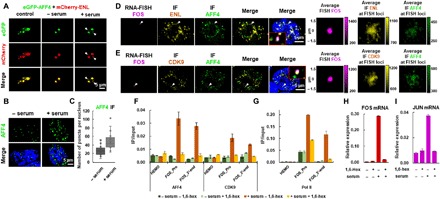
(A) Live cell imaging of HCT 116 cells coexpressing eGFP-AFF4 and mCherry-ENL in control, serum-starved, and serum-treated conditions. The newly formed eGFP-AFF4 and mCherry-ENL heterotypic droplets after serum treatment are indicated by arrows. (B) IF imaging of AFF4 in serum-starved or serum-treated HCT 116 cells. DNA was counterstained using DAPI. (C) Box plot showing that the number of AFF4 puncta per nucleus is significantly increased after serum treatment. Each n > 20 nuclei; error bars represent the distribution between the 90th and 10th percentiles. Results are representative of three biological replicates. (D) Confocal imaging of FOS RNA FISH with concurrent ENL and AFF4 IF showing that ENL and AFF4 co-occupy the FOS loci after serum treatment. DNA was counterstained using DAPI. Zoomed-in views of the white arrow–indicated regions are shown. The three columns on the right show average FOS FISH signal and average ENL (or AFF4) IF signal centered on the FISH foci (see Materials and Methods). Data shown were analyzed from at least four biological replicates. (E) Confocal imaging of FOS RNA FISH with concurrent CDK9 and AFF4 IF showing that CDK9 and AFF4 co-occupy the FOS loci after serum treatment. DNA was counterstained using DAPI. Zoomed-in views of the white arrow–indicated regions are shown. The three columns on the right show the average FOS FISH signal and average CDK9 (or AFF4) IF signal centered on the FISH foci (see Materials and Methods). Data shown were analyzed from at least four biological replicates. (F and G) ChIP-qPCR showing the occupancies of AFF4, CDK9 (F), and Pol II (G) at the promoter and 3′-end of FOS under the indicated conditions. The HEMO gene serves as a negative control for ChIP-qPCR. (H and I) RT-qPCR showing the RNA levels of FOS (H) and JUN (I) under indicated conditions. (F to I) Error bars represent SDs. Results are representative of three biological replicates.
We then examined whether phase separation is required for the function of SEC. We first performed IF with concurrent nascent RNA fluorescent in situ hybridization (FISH) for FOS proto-oncogene (FOS) to determine whether SEC exists in puncta in the vicinity of this gene. The results revealed that the tested SEC components co-occurred in the same puncta at the FOS gene, but not the XIST gene (Fig. 5, D and E, and fig. S8B). We also noticed a rare phenomenon that the FOS and JUN loci could spatially overlap with the same AFF4 puncta (fig. S8, C and D). Further average image analysis revealed that these SEC components’ fluorescence intensities (ENL/AFF4 or CDK9/AFF4) are most enriched at the center of the FISH foci on average (Fig. 5, D and E). The randomly selected nuclear positions were also calculated as a control, and no similar trend was observed (fig. S8E).
We also challenged the cells with 1,6-hexanediol. The results indicated that treatment with 1,6-hexanediol disrupted the AFF4/ENL and AFF4/CCNT1 heterotypic droplets and caused notable reductions in the occupancies of AFF4, CDK9, and Pol II at the FOS gene, which was accompanied by impaired transcription induction of the IEGs including FOS (Fig. 5, F to I, and fig. S9, A to E). In line with the essential role of ENL in SEC liquid droplet formation, we found that the AFF4 and CDK9 puncta were detached from the FOS FISH foci in ENL knockout cells (Fig. 6, A and B). Single-molecule expression analysis by FISH further revealed that the ENL knockout substantially slowed down the transcriptional induction rate of the FOS gene upon serum treatment, as revealed by the statistical analyses of the averaged FISH intensities over the serum treatment time periods (Fig. 6, C to E, and fig. S9, F and G). Furthermore, only wild-type ENL, but not the ENL IDR deletion mutant, can rescue the defect in FOS transcriptional induction caused by the ENL knockout (Fig. 6, C to E, and fig. S9, F and G). In summary, our data indicated that the capability of ENL to phase separate is key to the function of SEC in rapid transcriptional activation and that SEC could exert its rapid gene induction function in the form of phase-separated droplets.
Fig. 6. Rapid transcriptional induction by SEC depends on its ability to phase separate.
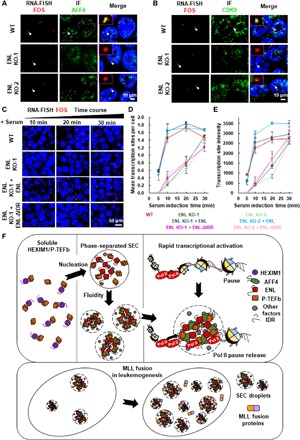
(A and B) Confocal imaging of FOS FISH with concurrent AFF4 (A) or CDK9 (B) IF in wild-type and ENL knockout HCT 116 cells after serum treatment. (C) Confocal imaging of FOS FISH in wild-type and ENL knockout HCT 116 cells after serum stimulation for different time periods. Only wild-type ENL, but not the ENL IDR deletion mutant, can rescue FOS transcriptional induction defect caused by ENL knockout. DNA was counterstained using DAPI. (D) Mean number of locus transcribing FOS per cell after serum stimulation for different time periods in wild-type and ENL knockout HCT 116 cells. Total n > 100 cells. Results are representative of three biological replicates. (E) Median fluorescence intensities of FOS transcribing loci after serum stimulation for different time periods in wild-type and ENL knockout HCT 116 cells. Total n > 100 cells. Results are representative of three biological replicates. (F) Cartoon model showing that the SEC components compartmentalize P-TEFb from HEXIM1 and that the SEC complex form phase-separated droplets at its target gene to promote RNA pol II pause release. The fusion of the SEC subunits, such as ENL, with MLL leads to increased phase separation of SEC.
DISCUSSION
SEC is essential for release of paused RNA Pol II into productive transcription elongation in both developmental and disease contexts (6, 31, 32). Yet, the nature of SEC that drives its rapid yet synchronous response to cellular needs remained unknown. In this study, we found that the transition of P-TEFb from the inactive into the active form involves phase separation. Our data demonstrated that HEXIM1/P-TEFb is widely distributed and soluble in the nucleus, while the multisubunit complex SEC exists and functions in the form of phase-separated droplets. The kinase module P-TEFb and the Pol II processivity stimulator ELL2 are unable to phase separate on their own but can be compartmentalized and fluidized by the intrinsically disordered SEC subunits ENL and AFF4. Reacting to extracellular stimuli, the LLPS-formed SEC droplets are promptly assembled in close proximity to the IEGs, leading to rapid transcriptional induction (Fig. 6F). SEC subunit mutations, such as fusion with MLL, might affect the phase separation of SEC, contributing to disease pathogenesis (Fig. 6F).
Weak and dynamic multivalent protein-protein interactions are believed to be the main driving force in forming the phase-separated biocondensates (33–35). One of the intriguing observations in our study is that the stable protein complex SEC undergoes phase separation within cells. Deletion of direct protein-protein interaction domains in SEC components does not affect phase separation of individual factor but impaired the formation of heterotypic droplets. Thus, phase separation might be dispensable for the SEC assembly, but required for its full function. On the basis of our observations, we propose that phase separation occurs broadly, not limited to factors that weakly and fuzzily interact, within cells to grant phase components high dynamics or establish a functional environment. In the case of SEC, phase separation could provide a physicochemical basis for it to coalesce with its substrate, the phosphorylated Pol II C-terminal repeat domain, which has also been found to phase separate in vivo, to induce rapid yet synchronous transcriptional response (36–39). The transcriptional induction rate of IEGs after serum exposure was greatly slowed down once phase separation of SEC is disrupted by ENL knockout. In addition, we observed the incorporation of two IEGs into the same phase-separated SEC droplet by combined single-molecule RNA FISH and IF analyses (fig. S8E), possibly providing a partial explanation for simultaneous activation of multiple IEGs within minutes of serum exposure.
As a critical part of immediate accommodation to acute stress, rapid transcriptional activation allows immediate, selective, and reversible change of cellular transcriptional capacity for maximizing cell viability and fitness (40). SEC uses multivalent phase separation to ensure its fast response to acute stress at the transcriptional level. ENL, the driver of the phase separation process of SEC, exists in multiple protein complexes, such as SEC-like 2 and 3 (SEC-L2 and SEC-L3), and the DOT1L-complex (DotCom) (11, 41). Furthermore, AFF4 belongs to the AFF family, which also contains AFF2 and AFF3 as the central factors of SEC-L2 and SEC-L3, respectively (11). Thus, phase separation might be a theme common to these transcription- or epigenetics-related protein complexes. It has been demonstrated that these protein complexes have their specific targets and play unique functions in cells (37). Future phase separation exploration on these closely related, but functionally distinct, protein complexes could facilitate to reveal the general mechanisms underlying the specific partition of protein-protein interactions by specific types of phase-separated biocondensates in vivo.
MATERIALS AND METHODS
Cell culture
HeLa, HCT 116, human embyronic kidney (HEK) 293T, and HEK293 Flp-In TRex cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum (Ex-Cell Bio) and 1% penicillin and streptomycin (HyClone), at 37°C with 5% CO2 in a humidified incubator. HEK293T cells were used for the lentiviral packaging used in shRNA-mediated knockdown and single-guide RNA (sgRNA)–mediated knockout experiments. For confocal imaging and FRAP experiment, HeLa and HEK293 Flp-In Trex (FLAG-ENL) cells were grown on glass-bottom cell culture dishes (NEST).
Cell treatments
Transfection
Cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instruction with following modifications. One million cells in 1 ml or 500 μl of growth media were plated in 1 well of a 6- or 12-well plate, Lipofectamine-DNA mix was immediately added on top of the cells, and media containing transfection mix were replaced with fresh growth media 12 hours after transfection. Cells were imaged 24 to 36 hours after transfection.
HCT 116 cell serum stimulation
For serum stimulation, cells were first starved by washing three times with 1×phosphate-buffered saline (PBS) and culturing for 40 hours in serum-free media, and then either left untreated or treated with serum for indicated time before harvesting.
1,6-Hexanediol (Sigma) treatment for live imaging
HCT 116 cells were grown on a glass-bottom culture plate with 1 ml of media, and cells were imaged every 30 s. After the second acquisition, 1 ml of 6% 1,6-hexanediol was added to the plate.
KL-1 treatment for live imaging
After transfection for 24 hours, HCT 116 cells were treated with 20 μM SEC inhibitor KL-1 for 6 hours (42).
Lentivirus-mediated RNAi
Human AFF4 (Sigma, clone ID: TRCN0000015823) and HEXIM1 (Sigma, clone ID: TRCN0000245064) shRNA oligos were cloned into the pLKO.1 vector. Nontargeting shRNA construct (SHC002) was purchased from Sigma. Lentiviral particle preparation and infection were performed as described previously (12). Briefly, around 60% confluent HEK293T cells in 15-cm tissue culture plate were cotransfected with 8 μg of the shRNA construct or nontargeting control shRNA, 6 μg of psPAX2 packaging plasmids, and 2 μg of pMD2.G envelope plasmids using Lipofectamine 2000. The media were replaced with fresh culture media after 16 hours of transfection. The lentiviral supernatants were collected 48 and 72 hours after transfection, filtered through the 0.45-μm filters. Cells were infected with the filtered lentiviral supernatants containing polybrene (8 μg/ml; Sigma). Twenty-four hours after infection, cells were subjected to selection with puromycin (2 μg/ml) for an additional 48 hours.
CRISPR-Cas9–guided knockout
ENL sgRNA oligos were cloned into the lentiCRISPR v2. Lentiviral particle preparation was performed as described above. HCT 116 cells were infected and selected with puromycin (2 μg/ml) for 48 hours in culture media. The infected cells were maintained in the absence of puromycin until cell clones were ready to be picked. The clones were screened with polymerase chain reaction (PCR) and confirmed by TA cloning plus DNA sequencing and Western blot.
Generation of FLAG-ENL–expressing cell line
pcDNA5/FRT FLAG-ENL expression plasmid was transfected into HEK293 Flp-In TRex cells and selected with hygromycin (100 μg/ml). Expression of FLAG-ENL protein was induced with doxycycline (2 μg/ml) for 48 hours.
Live cell imaging
Cells were grown in a 6- or 12-well plate and imaged using the AxioCam MRm detector on Axio Observer Z1 (Zeiss) in an incubation chamber to maintain culture condition (37°C, 5% CO2). ZEN black edition version 2.3 (Zeiss) was used for acquisition. Images were acquired with the AxioCam MRm Camera with 20× objective. Raw images were processed using ZEN 2.3 (Zeiss). In detail, phase-separated droplets of fluorescence-tagged proteins were imaged and analyzed using the Fiji Is Just ImageJ (FIJI) (43). All images were equally thresholded, and droplet number was identified using the “Analyze Particles” function of FIJI. At least three biological replicates were performed for each experiment.
Recombinant protein purification
pET16b expression vector was first modified to include either eGFP or mCherry following the 10× HIS tag in frame. cDNA encoding AFF4-IDR, ENL-IDR, or full-length CDK9 was inserted to the modified pET16b vectors. All expression constructs were sequenced to ensure the sequence accuracy. The fluorescence-tagged fusion proteins were expressed using the E. coli BL21 expression system. Briefly, recombinant plasmids were transformed into BL21. Mid-log phase of the transformed BL21 cells was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 hours to express the proteins of interest. Cells were harvested by centrifugation and stored at −80°C until needed. To purify the recombinant proteins, cell pellets from 500 ml of culture were resuspended in 40 ml of lysis buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 0.05% Tween 20] in the presence of the protease inhibitor phenylmethanesulfonyl fluoride and homogenized using a high-pressure homogenizer for five cycles at 10,000 mPa. The crude lysate was cleared by centrifugation at 20,000g for 1 hour at 4°C. Ni-nitrilotriacetic acid (NTA) agarose (1 ml; QIAGEN) preequilibrated with lysis buffer was then added to the cleared lysate. After overnight incubation at 4°C, the lysate agarose slurry was washed five times with 10 ml of wash buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 30 mM imidazole, and 0.05% Tween 20]. Protein was eluted with 5 ml of elution buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 250 mM imidazole, and 0.05% Tween 20]. The purified recombinant proteins were analyzed by Coomassie-stained SDS–polyacrylamide gel electrophoresis (PAGE) and dialyzed against droplet formation buffer [50 mM tris-HCl (pH 7.5), 10% glycerol, and 1 mM dithiothreitol].
In vitro droplet assay
Recombinant proteins were concentrated and desalted using Amicon Ultra centrifugal filters (Millipore). Recombinant proteins (10 μM) were then added to droplet formation buffer containing indicated concentration of salt in the presence or absence of the crowding agent PEG-8000. The protein solution was immediately loaded onto a coverslip and imaged with a Zeiss microscope with a 20× objective. For exploring the effects of temperature on in vitro droplet formation, the protein solution containing 10% PEG-8000 was allowed to incubate for 1 hour at the indicated temperature before imaging. Droplets were imaged and analyzed using FIJI. Results shown are representative of two biological replicates.
In vitro droplet quantification
FIJI was used to identify droplets and characterize their number, size, and shape. All images were equally thresholded (>5 pixel minimum droplet size), and droplet area and number were identified using the Analyze Particles function of FIJI. Droplets shape were also identified using the Analyze Particles in the same threshold condition (>20 pixel minimum droplet size); the circularity was analyzed using the function: circularity = 4π (area/perimeter2), and the calculated value 1 being a perfect circle. Hundreds of droplets identified in three independent technical replicates of view were quantified. Results shown are representative of at least two biological replicates.
Droplet aspect ratio analysis
The aspect ratios of AFF4-IDR-eGFP and ENL-IDR-eGFP droplets were determined by ellipse fitting and then calculating the ratio of the long and short axes of the ellipse. For the fusing droplet analysis, the time evolution of the aspect ratio was fit to a function of the form: Ar (t) = 1 + (Ar0 – 1) exp. (−t/τ), in which t is time, τ is the relaxation time, and Ar0 is the initial aspect ratio (44). The nonlinear curve fitting of aspect ratio data was performed using custom-written MATLAB (R2016a) scripts.
Western blot
Cells lysates were resolved in SDS-PAGE gels and transferred to polyvinylidene fluoride membrane. Membrane was then incubated with 1:2000 anti-HEXIM1, 1:5000 anti-AFF4, 1:1000 anti-β-tubulin, 1:5000 anti-FLAG, 1:5000 anti-CCNT1, 1:5000 anti-CDK9, and 1:2000 anti-ENL antibody diluted in TBST (tris-buffered saline with Tween 20) and incubated overnight at 4°C. Horseradish peroxidase–conjugated secondary antibodies (Sigma) were used at a dilution of 1:5000. Enhanced chemiluminescence substrate (Millipore) was applied to the membrane for imaging by autoradiography.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After three washes in PBS for 5 min, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Following rinses with PBS, cells were blocked in blocking buffer (2% bovine serum albumin and 0.3% Triton X-100) for at least 1 hour at room temperature and incubated with diluted primary antibodies (1:800 anti-CCNT1, 1:600 anti-ENL, 1:200 anti-AFF4, 1:600 anti-ELL2, 1:800 anti-CDK9, and 1:800 anti-HEXIM1) in blocking buffer overnight at 4°C. After three washes in PBS, coverslips were incubated at room temperature with secondary antibodies (1:2000 goat anti-rabbit IgG Alexa Fluor 488, 1:2000 goat anti-rabbit IgG Alexa Fluor 647, 1:2000 goat anti-mouse IgG Alexa Fluor 488, and 1:2000 goat anti-mouse IgG Alexa Fluor 555) in the dark for 1 hour, followed by three washes with PBS. Coverslips were mounted on slides using the VECTASHIELD Mounting Medium with DAPI (4′,6-diamidino-2-phenylindole). Three-dimensional images were acquired using a Zeiss LSM 700 confocal microscope with 63×, 1.4 numerical aperture (NA) oil immersion objective lens using Zen Light Edition acquisition software and charge-coupled device (CCD) camera. Images were postprocessed using Zen Light Edition and FIJI. For cell puncta counting, focus calling was performed using the “Object Counter 3D” plugin (see below). For colocalization analysis, the phase-separated puncta were analyzed using “Colocalization_Finder” plugin on FIJI (https://imagej.nih.gov/ij/plugins/colocalization-finder.html). Results shown are representative of at least three biological replicates.
Antibodies
AFF4, ENL, CDK9, and CCNT1 rabbit polyclonal antibodies were described previously (4, 12, 41). AFF4 mouse monoclonal was purchased from Santa Cruz Biotechnology (sc-390310), β-tubulin from Abcam (ab7291), FLAG from Sigma (F1804), and Pol II from Santa Cruz Biotechnology (N20). HEXIM1 rabbit polyclonal was generated in-house. Human HEXIM1 (1 to 140 amino acids) was expressed as His-tag fusion proteins in PET-16b, purified on NTA agarose according to QIAGEN’s protocol and sent to Wuhan Bioyeargene Biotechnology Co. Ltd. for immunization into rabbits. Human Rpb1 antibody was raised in rabbits against the synthetic peptide ERALRRTLQEDLVKDVLSNGC conjugated via cysteine to KLH.
RNA FISH and data analysis
Stellaris RNA FISH probes targeting FOS, JUN, and XIST were designed and generated by Biosearch Technologies. RNA FISH was performed according to Stellaris’ RNA FISH protocol (Biosearch Technologies). Briefly, cells were grown on 12-mm round coverslips in a 12-well plate, fixed by 3.7% formaldehyde for 10 min, and permeabilized by 70% ethanol for at least 1 hour. Cells were then washed with Wash Buffer A (Biosearch Technologies) and hybridized with 50 μl of hybridization buffer (Biosearch Technologies) containing Stellaris FISH probes for at least 4 hours at 37°C in a humidified chamber. Coverslips were mounted using VECTASHIELD Mounting Medium with DAPI. Images were acquired using a Zeiss LSM 700 confocal microscope with 63×, 1.4 NA oil immersion objective lens using Zen Light Edition acquisition software and CCD camera. Images were postprocessed using Zen Light Edition and FIJI. The distribution fitting of RNA FISH foci intensity data was analyzed using custom-written MATLAB (R2016a) scripts. Results shown are representative of at least three biological replicates.
RNA FISH combined with IF
Immunostaining was performed as described above in ribonuclease (RNase)–free environment. After immunostaining, cells were postfixed with 3.7% formaldehyde in PBS for 10 min at room temperature, and then washed twice with RNase-free PBS. Cells were washed once with wash buffer (20% Stellaris RNA FISH Wash Buffer A, 10% deionized formamide in RNase-free water) for 5 min at room temperature. Cells were hybridized with hybridization buffer (90% Stellaris RNA FISH Hybridization Buffer, 10% deionized formamide) containing Stellaris FISH probes overnight at 37°C. Cells were then washed with wash buffer for 30 min at 37°C. After washing with Stellaris RNA FISH Wash Buffer B at room temperature for 5 min, coverslips were mounted using VECTASHIELD Mounting Medium with DAPI. Images were acquired at Zeiss LSM 700 confocal microscope with 63×, 1.4 NA oil immersion objective lens using Zen Light Edition acquisition software and CCD camera. Images were postprocessed using Zen Light Edition and FIJI. Results shown are representative of at least three biological replicates.
Focus calling (RNA FISH and IF)
The “Object Counter 3D” plugin (https://imagej.nih.gov/ij/plugins/track/objects.html) in FIJI was used for puncta and foci focus calling. The “Threshold” parameter was set such that puncta or foci in close vicinity could be recognized as individual objects. The “Min number of voxels” parameter for RNA FISH foci was set to at least 100. Two adjacent slices were imaged (0.21-μm thickness each) to estimate the number of FOS and JUN RNA FISH foci, or CCNT1, AFF4, ENL, CDK9, and ELL2 IF puncta in fixed cells.
Average image analysis (RNA FISH and IF)
Custom Python scripts were written to analyze 3D images of areas surrounding the local maxima of RNA FISH intensities. RNA FISH foci were manually annotated using the Vaa3D triview mode by traversing Z slices (45). Image blocks were then cropped from the XY plane to get the maximum Z projections of size 101 × 101 pixels. These blocks were aligned by the center, and pixel-wise average was calculated for each IF channel. For negative controls, 200 “pseudo” RNA FISH foci were randomly generated for each imaging experiment. We used Python package matplotlib to generate plots by setting 25 and 99.9% percentiles as minimum and maximum intensities. For RNA FISH combined with IF experiments, at least three independent imaging fields were acquired and performed at least four biological replicates. The exact number of FISH foci analyzed, and the fraction of these foci that overlapped with IF puncta, and the relevant comparative statistics for experiments for which these comparisons were made, are as follows. For Fig. 5D, ENL and AFF4, n = 86 FOS foci; for Fig. 5E, CDK9 and AFF4, n = 69 FOS FISH foci.
FRAP and data analysis
Cells were seeded on glass-bottom cell culture dishes (NEST) and transfected with indicated plasmids. Thirty-six to 48 hours after transfection, fluorescence images were acquired on an inverted Leica TCS SP8 confocal microscope with a 63×, 1.4 NA oil immersion objective lens using LAS V4.4 software and a CCD camera. A 488-nm laser was used for excitation of eGFP-fusion proteins, while a 552-nm laser was used to image the mRFP or mCherry fusion proteins. Imaging conditions were prebleached using 0.5% laser power such that the intensity of the laser did not induce significant bleaching. Bleaching was performed over r (≈1.5 to 2.5 μm; for in-liquid FRAP, r ≈ 1.5 μm; for entire-liquid FRAP, r ≈ 2.5 μm) using 100% laser power, and images were collected every 1.29 s. Fluorescence intensity was measured using LAS V4.4 and was normalized to prebleach intensity.
The nonlinear curve fitting of normalized fluorescence recovery data was performed using custom-written scripts in MATLAB (R2016a). For nonlinear curve fitting and quantitative analysis, data representing the mean fluorescence intensity of the monitored regions were background subtracted and normalized to prebleach intensity. Postbleach FRAP recovery data were averaged over six biological replicates for each experiment.
The FRAP recovery curve was fit to a function of the form: FRAP (t) = a exp (− t/b) + c, where FRAP (t) is the normalized FRAP recovery from the average curve. a, b, and c were inferred through in-built MATLAB functions with 95% confidence intervals. Three independent variables, namely, k*on, koff, and Dapp were extracted from the fitting. The rate of FRAP recovery depends only on the off-rate according to the reaction dominant model (46). So 𝜏off = b, where koff = 1/𝜏off. The fourth parameter, Ceq (or the bound fraction), is derived using the recovered values for k*on and koff, using the relationship Ceq = −a/c = k*on/(k*on + koff). As diffusion is critical for FRAP recovery, an apparent diffusion coefficient was estimated: Dapp = r2bleach/𝜏.
RNA isolation and reverse transcription quantitative PCR
Total RNA was isolated using the E.Z.N.A. Total RNA Kit according to the manufacturer’s protocol, and residual DNA was treated with RNase-free DNase I (New England Biolabs) before reverse transcription quantitative PCR (RT-qPCR). cDNA was generated using RT reagent mix. Real-time qPCR was performed using SYBR Green mix on Bio-Rad CFX96-Real Time System according to the manufacturer’s instructions. Human HEXIM1 RT-qPCR primers were as follows: HEXIM1-F, GCTCGCGTTTCTTTAGCGAG; HEXIM1-R, AAGGGTTAAATCCCCTGCCG, and AFF4, FOS, JUN, JUNB, EGR1, and EGR2 RT-qPCR primers were previously described. The relative expression levels of genes of interest were normalized to the expression of the housekeeping gene GAPDH. Relative fold changes in gene expression were calculated using the ΔΔCT (threshold cycle) method. Results shown are representative of at least three biological replicates.
Chromatin immunoprecipitation
Approximately 1 × 108 HCT 116 cells were cross-linked in PBS containing 1% formaldehyde to the cell culture media at room temperature for 10 min, and cross-linking was quenched by glycine. Fixed chromatin was sonicated into 200– to 800–base pair fragments (Bioruptor, Diagenode) in chromatin immunoprecipitation (ChIP) lysis buffer [10 mM tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, and 0.5% N-lauroylsarcosine] supplemented with protease inhibitor cocktail (Sigma). Chromatin extracts were incubated with a specific antibody and protein A agarose beads at 4°C overnight. Immunoprecipitates were washed with radioimmunoprecipitation assay buffer [50 mM Hepes-KOH (pKa 7.55), 500 mM LiCl, 1 mM EDTA, 1.0% NP-40, and 0.7% Na-deoxycholate] for five times and TE once. After the final wash, DNA was eluted and reverse cross-linked at 65°C. DNA was then purified and used as a template for qPCR. Results shown are representative of at least three biological replicates.
Reproducibility and quantification
To ensure data quality and reproducibility, all the live imaging experiments were biologically repeated at least three times, and at least five independent imaging fields were acquired from each biological replicate. For Fig. 1 (E and F), at least 20 independent fields of view from three biological replicates were acquired, quantified, and analyzed. For Figs. 3 (H and I) and 4 (C and D), at least 15 independent fields of view from three biological replicates were acquired, quantified, and analyzed. For all the in vitro droplet experiments, at least 10 independent fields of view from at least two biological replicates were acquired, which totally contained about 50 to 2500 droplets. Quantified imaging areas were approximately 30 μm by 20 μm, and droplet data were displayed as droplet area (μm2) and droplet number (per 10 μm2). For all the RNA FISH and IF experiments, at least nine independent fields of view from at least three biological replicates were acquired and analyzed. For all the RNA FISH combined with IF experiments, at least 12 independent fields of view from four biological replicates were acquired. For Fig. 5D and fig. S8E, 86 FOS FISH foci were counted and analyzed; for Fig. 5E and fig. S8E, 69 FOS FISH foci were counted and analyzed. For all the imaging experiments, data were obtained, analyzed, and averaged from different areas in each biological replicate. Data shown in the figures are representative images of the biological replicates.
Supplementary Material
Acknowledgments
We are grateful to H. Fang for the critical discussion and C. Wang for technical assistance in this study. We also thank A. Shilatifard and K. Liang for the SEC small molecule inhibitors KL-1. Funding: Studies in this manuscript were supported by funds provided by the National Natural Science Foundation of China (31671343 and 3197040262 to C.L.; 31970626 to Z.L.; and 91632201 to W.X.), the National Key R&D Program of China to C.L. (2018YFA0800100), the Thousand Young Talents Plan of China to Z.L. (6231000011), the Natural Science Foundation of Jiangsu Province of China to C. L. (BK20160026) and Z. L. (BK20160666 and BK20170020), and the Fundamental Research Funds for the Central Universities to C.L. (3231008409) and Z.L. (3231008201). Author contributions: C.L. and Z.L. conceived the idea. C.G., Z.C., J.Y., and S.H. performed the experiments. C.G. and P.X. analyzed the imaging data. Z.L., C.L., and W.X. discussed and wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from C.L. (cqlin@seu.edu.cn).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaay4858/DC1
REFERENCES AND NOTES
- 1.Adelman K., Lis J. T., Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M., Paused RNA polymerase II as a developmental checkpoint. Cell 145, 502–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettiger A. N., Levine M., Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C., Garrett A. S., De Kumar B., Smith E. R., Gogol M., Seidel C., Krumlauf R., Shilatifard A., Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 25, 1486–1498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Z., Lin C., Shilatifard A., The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q., Li T., Price D. H., RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterlin B. M., Price D. H., Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Yik J. H. N., Chen R., Nishimura R., Jennings J. L., Link A. J., Zhou Q., Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Brès V., Yoh S. M., Jones K. A., The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20, 334–340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan J. L., Fogley R. D., Flynn R. A., Ablain J., Yang S., Saint-André V., Fan Z. P., Do B. T., Laga A. C., Fujinaga K., Santoriello C., Greer C. B., Kim Y. J., Clohessy J. G., Bothmer A., Pandell N., Avagyan S., Brogie J. E., van Rooijen E., Hagedorn E. J., Shyh-Chang N., White R. M., Price D. H., Pandolfi P. P., Peterlin B. M., Zhou Y., Kim T. H., Asara J. M., Chang H. Y., Young R. A., Zon L. I., Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol. Cell 62, 34–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z., Lin C., Guest E., Garrett A. S., Mohaghegh N., Swanson S., Marshall S., Florens L., Washburn M. P., Shilatifard A., The Super Elongation Complex family of RNA Polymerase II elongation factors: Gene target specificity and transcriptional output. Mol. Cell. Biol. 32, 2608–2617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A., AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith E., Lin C., Shilatifard A., The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25, 661–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izumi K., Nakato R., Zhang Z., Edmondson A. C., Noon S., Dulik M. C., Rajagopalan R., Venditti C. P., Gripp K., Samanich J., Zackai E. H., Deardorff M. A., Clark D., Allen J. L., Dorsett D., Misulovin Z., Komata M., Bando M., Kaur M., Katou Y., Shirahige K., Krantz I. D., Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 47, 338–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohan M., Lin C., Guest E., Shilatifard A., Licensed to elongate: A molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10, 721–728 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Chong P. A., Forman-Kay J. D., Liquid–liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 41, 180–186 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boija A., Klein I. A., Sabari B. R., Dall'Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., Narlikar G. J., Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q.-X., Nixon B. T., Rosen M. K., Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q., Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky V. N., Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wei M.-T., Elbaum-Garfinkle S., Holehouse A. S., Chen C. C.-H., Feric M., Arnold C. B., Priestley R. D., Pappu R. V., Brangwynne C. P., Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., Brangwynne C. P., Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., Parker R., ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeynaems S., Holehouse A. S., Weinhardt V., Kovacs D., Van Lindt J., Larabell C., Van Den Bosch L., Das R., Tompa P. S., Pappu R. V., Gitler A. D., Spontaneous driving forces give rise to protein–RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U.S.A. 116, 7889–7898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura T., Alder H., Gu Y., Prasad R., Canaani O., Kamada N., Gale R. P., Lange B., Crist W. M., Nowell P. C., Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc. Natl. Acad. Sci. U.S.A. 90, 4631–4635 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Francisco J. C., Dai Q., Luo Z., Wang Y., Chong R. H.-H., Tan Y. J., Xie W., Lee G.-H., Lin C., Transcriptional elongation control of hepatitis B virus covalently closed circular DNA transcription by super elongation complex and BRD4. Mol. Cell. Biol. 37, e00040-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C., Garruss A. S., Luo Z., Guo F., Shilatifard A., The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152, 144–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson B. A., Doolittle L. K., Schneider M. W. G., Jensen L. E., Gamarra N., Henry L., Gerlich D. W., Redding S., Rosen M. K., Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harlen K. M., Churchman L. S., The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A. S., Yu T., Marie-Nelly H., McSwiggen D. T., Kokic G., Dailey G. M., Cramer P., Darzacq X., Zweckstetter M., RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Guo Y. E., Manteiga J. C., Henninger J. E., Sabari B. R., Dall'Agnese A., Hannett N. M., Spille J.-H., Afeyan L. K., Zamudio A. V., Shrinivas K., Abraham B. J., Boija A., Decker T.-M., Rimel J. K., Fant C. B., Lee T. I., Cisse I. I., Sharp P. A., Taatjes D. J., Young R. A., Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., McKnight S. L., Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vihervaara A., Duarte F. M., Lis J. T., Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 19, 385–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohan M., Herz H.-M., Takahashi Y.-H., Lin C., Lai K. C., Zhang Y., Washburn M. P., Florens L., Shilatifard A., Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24, 574–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang K., Smith E. R., Aoi Y., Stoltz K. L., Katagi H., Woodfin A. R., Rendleman E. J., Marshall S. A., Murray D. C., Wang L., Ozark P. A., Mishra R. K., Hashizume R., Schiltz G. E., Shilatifard A., Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175, 766–779.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brangwynne C. P., Mitchison T. J., Hyman A. A., Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng H., Bria A., Zhou Z., Iannello G., Long F., Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 9, 193–208 (2014). [DOI] [PubMed] [Google Scholar]
- 46.McNally J. G., Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol. 85, 329–351 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaay4858/DC1


