The ability of pathogens such as Staphylococcus aureus to evolve resistance to antibiotics used in the treatment of infections has been an important concern in the last decades. Resistant acquisition usually translates into treatment failure and puts patients at risk of unfavorable outcomes. Furthermore, the laboratory testing of antibiotic resistance does not account for the different environment the bacteria experiences within the human body, leading to results that do not translate into the clinic. In this study, we forced methicillin-resistant S. aureus to develop nafcillin resistance in two different environments, a laboratory environment and a physiologically more relevant environment. This allowed us to identify genetic changes that led to nafcillin resistance under both conditions. We concluded that not only does the environment dictate the evolutionary strategy of S. aureus to nafcillin but also that the evolutionary strategy is specific to that given environment.
KEYWORDS: Staphylococcus aureus, antibiotic resistance, nafcillin, USA300, adaptive laboratory evolution, drug resistance mechanisms
ABSTRACT
Antimicrobial susceptibility testing standards driving clinical decision-making have centered around the use of cation-adjusted Mueller-Hinton broth (CA-MHB) as the medium with the notion of supporting bacterial growth, without consideration of recapitulating the in vivo environment. However, it is increasingly recognized that various medium conditions have tremendous influence on antimicrobial activity, which in turn may have major implications on the ability of in vitro susceptibility assays to predict antibiotic activity in vivo. To elucidate differential growth optimization and antibiotic resistance mechanisms, adaptive laboratory evolution was performed in the presence or absence of the antibiotic nafcillin with methicillin-resistant Staphylococcus aureus (MRSA) TCH1516 in either (i) CA-MHB, a traditional bacteriological nutritionally rich medium, or (ii) Roswell Park Memorial Institute (RPMI), a medium more reflective of the in vivo host environment. Medium adaptation analysis showed an increase in growth rate in RPMI, but not CA-MHB, with mutations in apt, adenine phosphoribosyltransferase, and the manganese transporter subunit, mntA, occurring reproducibly in parallel replicate evolutions. The medium-adapted strains showed no virulence attenuation. Continuous exposure of medium-adapted strains to increasing concentrations of nafcillin led to medium-specific evolutionary strategies. Key reproducibly occurring mutations were specific for nafcillin adaptation in each medium type and did not confer resistance in the other medium environment. Only the vraRST operon, a regulator of membrane- and cell wall-related genes, showed mutations in both CA-MHB- and RPMI-evolved strains. Collectively, these results demonstrate the medium-specific genetic adaptive responses of MRSA and establish adaptive laboratory evolution as a platform to study clinically relevant resistance mechanisms.
IMPORTANCE The ability of pathogens such as Staphylococcus aureus to evolve resistance to antibiotics used in the treatment of infections has been an important concern in the last decades. Resistant acquisition usually translates into treatment failure and puts patients at risk of unfavorable outcomes. Furthermore, the laboratory testing of antibiotic resistance does not account for the different environment the bacteria experiences within the human body, leading to results that do not translate into the clinic. In this study, we forced methicillin-resistant S. aureus to develop nafcillin resistance in two different environments, a laboratory environment and a physiologically more relevant environment. This allowed us to identify genetic changes that led to nafcillin resistance under both conditions. We concluded that not only does the environment dictate the evolutionary strategy of S. aureus to nafcillin but also that the evolutionary strategy is specific to that given environment.
INTRODUCTION
Staphylococcus aureus is a commensal Gram-positive bacteria that colonizes human skin, as well as nasal and respiratory tracts. Upon breaching skin or mucosal barriers, S. aureus can cause infections of skin, blood, and tissues (1). Although historically associated with hospital and health care infections, community-acquired methicillin-resistant S. aureus (CA-MRSA) infections are now widespread globally (2), of which USA300 is the most common clonal lineage in North America (3). MRSA TCH1516 is a well-studied representative USA300 strain isolated from an adolescent at the Texas Children’s Hospital in Houston with severe sepsis (4).
In vitro methods for evaluating antibiotic activity against bacterial pathogens were developed and standardized in 1961 as a “one size fits all” screen (5). This method has been paramount in antibiotic research, but translation to in vivo efficacy has been increasingly questioned (6, 7). Determination of the MIC for potential drugs has also varied considerably between “standard” testing media from different manufacturers (8) and with additionally supplemented cations (9). Differential susceptibility is even more pronounced between traditional testing media and more physiologically relevant medium conditions (i.e., taking factors such as supplemented cations, interaction with host factors, and nutrient availability into consideration) (10–12). In this study, differential antibiotic response was examined between standard bacteriological testing medium cation-adjusted Mueller-Hinton broth (CA-MHB) and Roswell Park Memorial Institute (RPMI) medium, a medium used in cell and tissue culture for mammalian cells, supplemented with 10% Luria-Delbruck (LB) (RPMI + 10%LB).
Despite the implications of medium-specific susceptibility of important pathogens, little work has been done to understand any medium-specific differential genetic response to tolerance under an antibiotic stress. Adaptive laboratory evolution (ALE) is an appropriate tool that can be utilized to meet this challenge and study the adaptive capabilities of microorganisms in vitro, as mutants that have differential resistance properties can be identified in a straightforward manner. ALE has been applied to study the adaptive response to a number of external stressors such as temperature (13, 14) or antibiotics (15–17). Specifically relevant to S. aureus, previous studies have utilized ALE to study the adaptive capabilities to various antibiotics (18–22). These studies have enabled an assessment of current and potential treatment strategies via identification of mutational targets and associated phenotypic changes that confer resistance to antibiotics of interests. Antistaphylococcal beta-lactams (e.g., nafcillin, oxacillin. flucloxacillin, cloxacillin) are the treatment of choice against serious methicillin-susceptible S. aureus (MSSA) infections (23, 24). A representative of this class, nafcillin, has been identified as one of the antibiotics with medium-dependent efficacies (11), making it an ideal candidate for this study. The Clinical and Laboratory Standards Institute (CLSI) breakpoint for nafcillin in CA-MHB is greater than or equal to 4 μg/ml, and susceptible is less than or equal to 2 μg/ml.
In this work, ALE was applied to uncover medium-specific mechanisms of resistance to nafcillin in a controlled setting. First, ALE was implemented to adapt S. aureus TCH1516 to both medium conditions (CA-MHB and RPMI + 10%LB), in order to optimize cellular performance and establish a suitable baseline with which to compare any further evolutionary work. Second, ALE was harnessed to study nafcillin resistance of such medium-adapted strains in order to gain insights into the genetic basis for adaptation in differing medium conditions. Finally, resistant strains were assessed for growth rate, effective nafcillin resistance, and virulence capabilities, so phenotypic trade-offs could be identified.
RESULTS
Laboratory evolution for adaptation to medium environments.
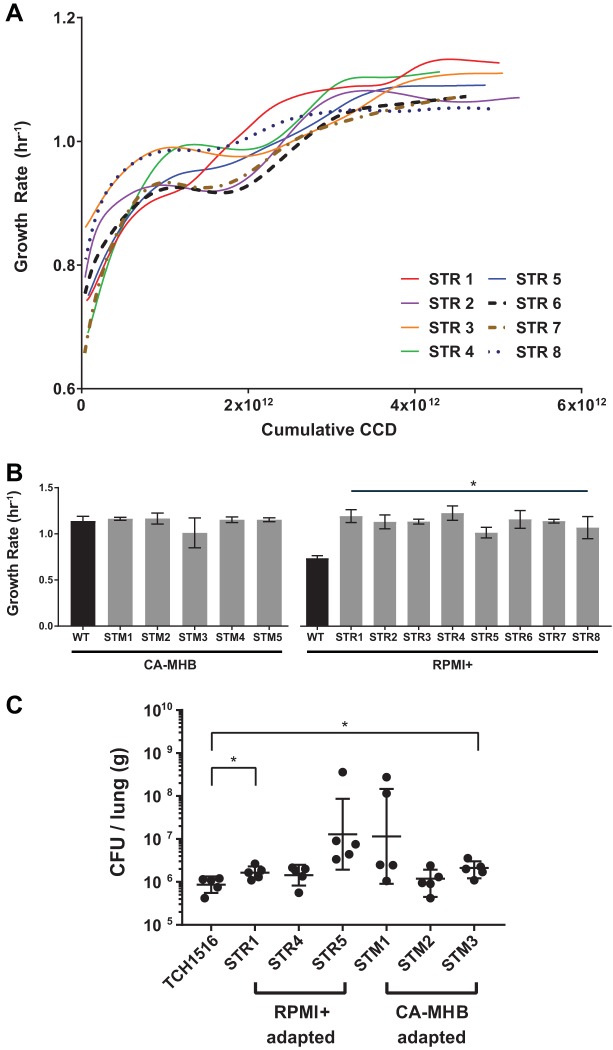
S. aureus TCH1516 was forced to evolve under two medium conditions to understand how it adapts under growth rate selection to different nutritional environments. The two chosen medium types were CA-MHB and RPMI + 10%LB (referred to as RPMI+), since differential susceptibility to nafcillin was observed across both conditions (see Table S1 in the supplemental material) (11). Five independent populations of S. aureus TCH1516 were forced to evolve on CA-MHB, while eight independent populations were forced to evolve on RPMI+ for an average of 108 and 100 batch flask transfers, respectively (Table S2). Flask transfers were performed when an optical density at 600 nm (OD600) of 0.3 ± 0.02 or 0.434 g (dry weight [DW])/liter was achieved to prevent the cells from entering stationary phase, thus selecting for advantages in growth rate. Although no growth rate improvements were observed for evolutions performed in CA-MHB, population growth rates for S. aureus on RPMI+ increased from a starting wild-type growth rate of 0.75 ± 0.1 h−1 to 1.1 ± 0.1 h−1, an ∼1.5-fold increase, during a range of 4.52 × 1012 to 5.26 × 1012 cumulative cell cycle divisions (CCD) (Fig. 1A and B). CCD has previously been shown to effectively represent the time scale for ALEs in contrast to elapsed time or generations (25). It should be noted that the overall growth rate of the population at the end of the evolution on RPMI+ (1.06 ± 0.10 h−1) was similar to that of the starting growth rate on CA-MHB (1.12 ± 0.083 h h−1) (Table S2). Clonal isolates were selected from each of the final flasks of the independently evolved populations of the medium adaptation ALEs (i.e., endpoint clones) to RPMI+ (eight clones) and CA-MHB (five clones) in order to explore the phenotypes from the isolated evolved genotypes. Growth rates were measured for each of the endpoint clones, and there was concordance between the values observed for the populations at the end of the evolutions. The increase in growth rate of S. aureus TCH1516 through adaptation to RPMI+, but not to CA-MHB, was confirmed on the clonal level. Similar work has been performed forcing S. aureus to evolve in various medium conditions, although growth rates were not reported (26, 27). The identical growth rate between the two conditions evaluated here indicates an apparent maximum achievable growth rate for strain TCH1516 in a batch growth rich medium environment, given the stated evolution times. Following medium adaptation, the medium-adapted strains were evaluated for their virulence capabilities and sequenced to explore the genetic mechanisms behind observed fitness improvements.
FIG 1.
Medium adaptation of S. aureus TCH1516. (A) Fitness trajectories depicting growth rate increase throughout the course of the medium adaptation ALE in RPMI+. Strains STR 1, 4, and 5 served as progenitors for the medium-adapted starting points in the tolerance evolution. (B) Clonal growth rates for single clones isolated by streaking endpoint populations. Measurements were determined from biological duplicates and an average of two consecutive flasks. STR strains are S. aureus RPMI+-adapted strains. STM strains are S. aureus CA-MHB-adapted strains. Values that are significantly different (P < 0.0001) from the value for the wild type (WT) by two-way ANOVA are indicated by a bar and asterisk. (C) ALE-derived strains maintain parental lineage virulence in a murine pneumonia model of infection. Values that are significantly different (P < 0.05) by t test with Welch’s correction are indicated by a bar and asterisk.
MIC susceptibility testing of S. aureus TCH1516. Dagger indicates differential susceptibility greater than or equal to 4× across CA-MHB and RPMI + 10%LB. Download Table S1, DOCX file, 0.02 MB (18.3KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth phenotypes for S. aureus TCH1516 populations that evolved on CA-MHB and RPMI+. Download Table S2, DOCX file, 0.02 MB (19.8KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Medium-adapted strain virulence in a murine model of pneumonia.
Continued passaging of pathogenic strains of bacteria in vitro can lead to attenuation, yielding derived laboratory strains that are disparate to those seen within patients (28). As multiple dedicated pathways are essential for virulence within a host, it is often the case that laboratory-evolved strains become nonpathogenic, due to the disruption of those pathways, leading to attenuation, therefore decreasing their clinical relevance. A murine pneumonia infection model was used to examine the virulence of medium-adapted strains in comparison to the pathogenic TCH1516 parental lineage. This model utilizes an intratracheal injection to establish a bacterial pneumonia and has been previously used to assess lung bacterial burdens (29). Surprisingly, despite RPMI+ serving as a better mimic for physiological conditions, strains that were adapted to RPMI+ did not have a virulence advantage within the host compared to those adapted to the standard laboratory growth medium (Fig. 1C). It was determined that medium-adapted strains maintain their pathogenicity and had no gross virulence defects in comparison with the TCH1516 parental lineage, indicating that these strains were not attenuated.
Mutation analysis of whole-genome resequencing for medium adaptation.
Whole-genome sequencing was performed on evolved populations and selected clones from the ALE experiments on the two selected medium types to explore whether mutations could be linked to the observed fitness improvements. Sequences were analyzed to determine mutations from the multiple replicates under each condition (30, 31). For the CA-MHB condition, only endpoint clones were sequenced given the lack of an apparent fitness change during the course of the evolution, and a total of three unique mutations were found across all five replicates (two clones had no mutations detected [Table S3]). For the RPMI+ condition, there were 261 unique mutations across all of the intermediate and endpoint populations and clones selected during the experiment, with the clones having between 4 and 12 mutations each (Table S4). To focus the analysis, mutations were labeled as “key mutations” if a gene or genetic region contained multiple unique mutations across replicates or if an identical mutation appeared across independent ALE replicates. For RPMI+, there were eight genes or genetic regions that met these criteria, with three having greater than two instances. A summary of the RPMI+ medium adaptive mutations is shown in Table 1. For CA-MHB, there was no gene which shared mutations across two of the endpoint clones.
TABLE 1.
Key reproducibly occurring mutations detected in the final populations and clones of S. aureus TCH1516 after adaptive laboratory evolution in RPMI+
| Genea | Specific function | Mutation typeb | Protein and nucleotide changec |
Strain(s)d |
|---|---|---|---|---|
| apt | Adenine phosphoribosyl transferase | SNP | D119E (GAT→GAA) | 8 |
| SNP | H104Y (CAC→TAC) | 1 | ||
| SNP | P76S (CCT→TCT) | 6p | ||
| SNP | G73D (GGC→GAC) | 4 | ||
| SNP | A67V (GCT→GTT) | 2p, 6, 7 | ||
| SNP | A67T (GCT→ACT) | 5 | ||
| SNP | V66L (GTA→CTA) | 1p | ||
| SNP | V41L (GTA→TTA) | 2 | ||
| mntA (znuC_1) | Manganese ABC transporter | SNP | L11I (TTA→ATA) | 4 |
| SNP | L2I (TTA→ATA) | 2p | ||
| SNP | M1M (TTG→ATG)† | 3, 5, 6p, 7 | ||
| mntA, mntR (znuC_1, ideR) | Manganese ABC transporter/Mn-dependent transcriptional regulator MntR | SNP | A→G, intergenic (−1/−121) | 4 |
| SNP | C→T, intergenic (−2/−120) | 2p | ||
| INS | (GTTTAGGCTAACCTAATTAA)1→2, intergenic (−43/−79) | 3, 5, 6p, 7 | ||
| stk1 (prkC) | Serine/threonine-protein kinase | SNP | A124P (GCG→CCG) | 4 |
| SNP | V470D (GTT→GAT) | 1 | ||
| cspA_2 | Cold shock protein CspA | SNP | A60V (GCT→GTT) | 2 |
| DEL | Δ1 bp, coding (34/201 nt) | 4 | ||
| dynA (RS07370) | Bacterial dynamin-like protein | SNP | Q1098E (CAA→GAA) | 2 |
| SNP | S618T (TCT→ACT) | 6 | ||
| recJ | Single-stranded-DNA-specific exonuclease RecJ | SNP | S757S (TCG→TCT) | 3 |
| SNP | A348V (GCA→GTA) | 8 | ||
| lyrA | Lysostaphin resistance protein A | SNP | L48L (CTA→CTT) | 1 |
| DEL | Δ1 bp, coding (1210/1260 nt) | 5 | ||
| SUB | 2 bp→AT, coding (1216 − 1217/1260 nt) | 5 | ||
The gene locus tag corresponds to USA300HOU_RSXXXXX. The gene nomenclature provided by prokka annotation, reflected in the mutation analysis, is shown in the parentheses.
SNP, single nucleotide polymorphism; INS, insertion; DEL, deletion; SUB, substitution.
nt, nucleotide; †, mutation led to formation of a start codon.
p denotes population.
Mutations identified for S. aureus TCH1516 after medium adaptation to CA-MHB. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S3, XLSX file, 0.01 MB (11KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 after medium adaptation to RPMI+. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S4, XLSX file, 0.06 MB (66.2KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In RPMI+ medium conditions, the most prevalent gene that mutated was apt, with all independent replicates containing at least one mutation in this gene, which remained present in the majority of endpoint clones (Table 1). The apt gene encodes an adenine phosphoribosyltransferase which enables nucleotide salvage reactions converting adenine to AMP (32). Mutations in this gene have also been discovered after in vitro passaging of S. aureus after exposure to increasing concentrations of vancomycin (18). Constructed apt deletion mutants experienced significant reduction in extracellular DNA (eDNA) release, a major constituent for biofilm stability and formation, low production of extra polymeric substances (33, 34), as well as increased resistance to Congo red (35).
An additional highly mutated region for growth rate optimization on RMPI+ was the mntA gene and its intergenic region upstream of both mntA and its regulator mntR. The mntA gene encodes a manganese permease subunit of an ATP binding transporter, while mntR encodes a metal-dependent transcriptional regulator (36). An identical mutation was identified in the start codon of mntA across three independent ALEs, modifying the initiation site from a suboptimal form (UUG) to AUG, which is the optimal start codon in prokaryotes (37, 38). Mutations in the intergenic region include two single nucleotide polymorphisms (SNPs) occurring 1 and 2 nucleotides upstream of mntA, likely affecting its promoter. The other intergenic change was an insertion of a 20-nucleotide sequence, 43 bp upstream of mntA. Acquisition of manganese is important for cell survival and replication of pathogens and is crucial for cell detoxification of reactive oxygen species (39). Inactivation of the MntABC transporter complex in another USA300 strain has been shown to attenuate virulence in in vivo mouse models (40). Manganese acquisition appears to be particularly relevant in endovascular infections. Disruption of mntA, mntH, mntR, or both mntA and mntH also significantly reduces intracellular survival in human endothelial cells. Bioavailable Mn is utilized by S. aureus to detoxify reactive oxygen species and protect against neutrophil killing, enhancing the ability to cause endocardial infections (41, 42).
Additional key mutations were identified in the RPMI+ growth rate adaptation: mutations in two genes encoding regulatory proteins, cspA and stk1, and in the dynA, recJ, and lyrA genes, encoding a GTPase, an exonuclease, and a protease, respectively (Table 1; see also Text S1 in the supplemental material).
RPMI+ medium adaptation additional mutations Download Text S1, DOCX file, 0.02 MB (21.3KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The medium-adapted strains were subsequently used to understand S. aureus’ tolerization to nafcillin, with the goal of identifying the genetic basis of this process in the different medium environments.
Laboratory evolution for adaptation to nafcillin tolerance.
A tolerance adaptive laboratory evolution (TALE) experiment was implemented to force medium-adapted strains of S. aureus TCH1516 to develop resistance to the β-lactam antibiotic nafcillin and identify mutations enabling an elevated growth rate under increasing antibiotic stress concentrations in both CA-MHB and RPMI+ medium environments. The S. aureus strains selected as starting strains of the TALE experiments consisted of the respective medium-adapted strains, denoted STM (CA-MHB) and STR (RPMI+). The starting strains for the TALE experiments were medium-adapted strains with distinct genotypes (Table 2).
TABLE 2.
Tolerance phenotypes for S. aureus USA300_TCH1516 and medium-adapted evolved populations on CA-MHB and RPMI+a
| Ancestor strain and strain |
ALE no. | Initial growth rate (h−1) |
Starting nafcillin concn (μg/ml) |
Final growth rate (h−1) |
Final nafcillin concn (μg/ml) |
No. of flasks | CCD × 1012 |
|---|---|---|---|---|---|---|---|
| RPMI+ TALE (SNFR) |
|||||||
| STR 1 | 7 | 1.17 ± 0.02 | 0.013 | 0.83 ± 0.12 | 65.52 | 184 | 15.2 |
| 9 | 1.20 ± 0.03 | 0.013 | 0.83 ± 0.08 | 50.4 | 174 | 13.4 | |
| STR 4 | 13 | 1.23 ± 0.08 | 0.013 | 0.85 ± 0.09 | 83.16 | 191 | 14.5 |
| 15 | 1.17 ± 0.08 | 0.013 | 0.83 ± 0.06 | 57.96 | 177 | 14.2 | |
| 17 | 1.04 ± 0.07 | 0.013 | 0.74 ± 0.11 | 65.52 | 172 | 13.6 | |
| STR 5 | 19 | 1.11 ± 0.1 | 0.013 | 0.87 ± 0.14 | 52.92 | 166 | 12.8 |
| 21* | 1.19 ± 0.04 | 0.013 | 0.99 ± 0.08 | 4.32 | 117 | 8.5 | |
| 23 | 1.22 ± 0.08 | 0.013 | 0.89 ± 0.14 | 57.96 | 171 | 13.6 | |
| CA-MHB TALE (SNFM) |
|||||||
| STM 1 | 7 | 0.79 ± 0.07 | 0.5 | 0.77 ± 0.17 | 61.2 | 72 | 3.94 |
| 11 | 0.90 ± 0.12 | 0.5 | 0.87 ± 0.07 | 80.33 | 75 | 4.08 | |
| STM 2 | 13 | 0.94 ± 0.11 | 0.5 | 0.70 ± 0.03 | 61.2 | 68 | 3.75 |
| 15 | 0.87 ± 0.14 | 0.5 | 0.76 ± 0.13 | 61.2 | 70 | 3.81 | |
| STM 3 | 19 | 0.97 ± 0.16 | 0.5 | 0.88 ± 0.08 | 61.2 | 74 | 3.95 |
| 23 | 0.93 ± 0.11 | 0.5 | 0.93 ± 0.06 | 61.2 | 74 | 4.16 | |
Population growth rates for independent replicates were calculated by averaging the initial and final three flasks of the medium adaptation ALEs. An asterisk indicates premature end to experiment due to technical errors.
TALE proved to be effective in developing strains with increased resistance to nafcillin in both RPMI+ and CA-MHB. Three medium-adapted starting strains per medium type (STM 1, 2, and 3 and STR 1, 4, and 5) were forced to evolve in duplicate or triplicate to generate a total of 14 independent evolutions (Table 2). Figure 2A details a typical TALE trajectory of the growth rate and the continuously increasing concentration of nafcillin in RPMI+ (Fig. S1 shows a CA-MHB nafcillin typical TALE trajectory). Over the course of evolution, S. aureus populations underwent an average of 3.93 × 1012 CCD and 72 flasks for CA-MHB and 13.81 × 1012 CCs and 175 flasks for RPMI+. Evolutions on RPMI+ were noticeably longer (Table 2) due to the differential susceptibility of S. aureus TCH1516 to nafcillin in the two medium conditions (11) (Table S1). The MIC on RPMI+ was ∼100-fold less compared to the MIC on CA-MHB for the respective starting strains (Table S5). The initial starting concentrations of nafcillin for the TALEs were therefore adjusted to ensure cell viability. Concentrations of nafcillin reached as high as 600× MIC90 on RPMI+ and 8× MIC90 on CA-MHB (Table 2) compared to the wild type on their respective medium.
FIG 2.
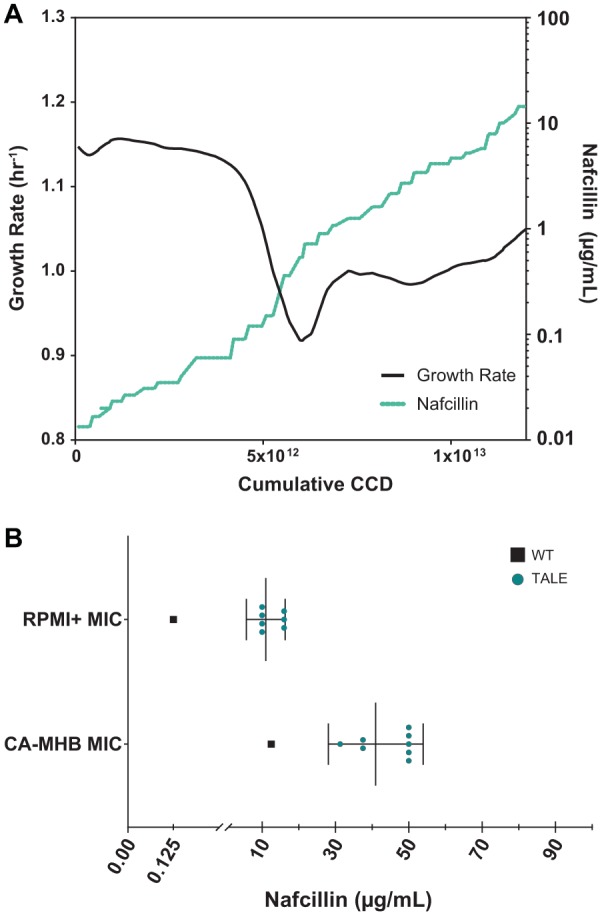
Nafcillin adaptation of medium-adapted strains derived from S. aureus TCH1516. (A) Fitness trajectory for a typical TALE experiment, showing population growth rate and continuously increasing antibiotic concentration. The selected trajectory depicts SNFR9 exposed to nafcillin in RPMI+. (B) A plot of the MICs for selected clones from endpoint populations after nafcillin tolerization. The MICs for the wild-type TCH1516 (black squares) and TALE strains (green circles) on the respective medium are shown.
Fitness trajectory for a typical TALE experiment, showing population growth rate and continuously increasing antibiotic concentration. The selected trajectory depicts SNFM15 exposed to nafcillin in CA-MHB. Download FIG S1, EPS file, 0.1 MB (126KB, eps) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nafcillin sensitivity of medium-adapted strains Download Table S5, DOCX file, 0.02 MB (18.6KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Endpoint clonal isolates from each of the independent TALE replicates were selected to assess and confirm the increased nafcillin resistance phenotype. As expected, nafcillin resistance for the evolved clones was increased. However, the increase in tolerance observed for isolated clones did not quantitatively match the values tolerated by the TALE populations from which they were isolated (Fig. 2B). In RPMI+, an MIC90 ranging from 10 to 20 μg/ml was achieved for isolated clones compared to a range of 45 to 83 μg/ml observed in population endpoints. The same phenomenon was observed in a smaller degree for isolated clones from the TALE in CA-MHB. The MIC90 of nafcillin for CA-MHB TALE isolates ranged between 31.3 and 50 μg/ml compared to 61 to 87 μg/ml measured for TALE final evolved populations (Fig. 2B and Table 2). This can likely be attributed to population dynamics, kin selection (43), “bacterial cheating,” where overproduction of degradative enzymes can inactivate antibiotic molecules (44), or simply due to a difference in culturing methods under which the clonal MICs were determined compared to the culturing conditions during the TALE experiment (see Materials and Methods).
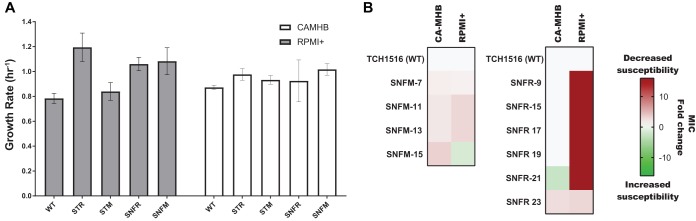
To assess phenotypic trade-offs in the evolved strains, endpoint clonal growth rates were measured in their evolutionary medium as well as the alternate medium type utilized in this study (i.e., a medium swap) under no nafcillin stress. Characterizations were performed with both medium- and nafcillin-adapted clones. As shown above (Fig. 1B), RPMI+ medium-adapted strains (STR) saw a 52% increase in growth rate compared to the wild-type S. aureus TCH1516 (two-way analysis of variance [ANOVA], P < 0.0001) (Table S6). Medium adaptation to CA-MHB (STM) did not confer a fitness advantage in RPMI+ (two-way ANOVA, P = 0.8745). Strains with a higher resistance to nafcillin in RPMI+ (SNFR) resulted in a fitness tradeoff compared to medium-adapted strains in the same medium (STR) with an overall 11% decrease in growth rate (two-way ANOVA, P = 0.0054) (Fig. 3A and Table S6). For the medium swap conditions, there was an unexpected growth rate increase of 29% for strains evolved for resistance to nafcillin in CA-MHB (SNFM) when grown in RPMI+ compared to the progenitor strains that were adapted to CA-MHB (STM) (two-way ANOVA, P < 0.0001) (Fig. 3A and Table S6). There were no significant changes in growth rates observed across any of the strains analyzed in CA-MHB medium (Table S6).
FIG 3.
Phenotypic characterization of TALE strains. (A) Growth rates of wild-type, medium-adapted, and nafcillin-adapted strains. The graph shows the measured growth rates of several selected endpoint clones for strains derived from either RPMI+ (STR and SNFR) or CA-MHB (STM and SNFM) evolutionary conditions. White bars represent clonal growth rates in CA-MHB, and gray bars represent the growth rate for the same clones in RPMI+. The graph shows the growth rates of three tolerization endpoint clones from both medium conditions along a lineage. Data presented are averages from triplicates. A comprehensive ANOVA statistical analysis is provided in Table S5 in the supplemental material. (B) Heat map of the nafcillin MIC fold change of TALE strains compared to the wild-type MIC in both medium types. STM, S. aureus CA-MHB-adapted strain; STR, S. aureus RPMI+-adapted strain; SNFM, S. aureus nafcillin-adapted strain in CA-MHB; SNFR, S. aureus nafcillin-adapted strain in RPMI+.
Comparison of growth rates determined for clonal isolates, using two-way ANOVA analysis in Graphpad Prism 7.04. Download Table S6, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutational analysis for tolerance evolutions.
Whole-genome sequencing was performed on evolved populations and selected clones from TALEs on both medium types to determine shared or unique mutational mechanisms of nafcillin resistance phenotypes. Key mutations were again identified in a similar manner to those from the medium adaptation ALEs (i.e., if the gene or genetic region contained multiple unique mutations or the same mutation across independent ALE replicates). On average, there were fewer mutations in response to nafcillin stress on CA-MHB compared to RPMI+, as represented by the key mutations in the endpoint clones and populations (Tables 3 and 4). Endpoint clones and populations from evolution experiments on CA-MHB led to the identification of 13 unique key mutations across 5 genes, while the ones performed on RPMI+ presented 25 unique key mutations across 10 genes (Tables 3 and 4).
TABLE 3.
Key mutations for final endpoint clones of S. aureus TCH1516 after tolerance adaptive laboratory evolution in CA-MHB to nafcillin (SNFM)
| Genea | Specific function | Mutation type |
Protein and nucleotide changeb |
Strain |
|---|---|---|---|---|
| apt | Adenine phosphoribosyltransferase |
SNP | G59D (GGC→GAC) | 11 |
| SNP | I127N (ATT→AAT) | 19 | ||
| SNP | K82E (AAA→GAA) | 13 | ||
| pbuG | Xanthine/guanine permease |
SNP | Q6* (CAG→TAG) | 7 |
| SNP | A84E (GCA→GAA) | 23 | ||
| vraS | Two-component sensor histidine kinase |
SNP | G330D (GGT→GAT) | 11 |
| SNP | T331I (ACA→ATA) | 19 | ||
| vraT (RS10230) | Transporter associated with VraSR |
SNP | T8K (ACG→AAG) | 13 |
| SNP | V199A (GTT→GCT) | 23 | ||
| SNP | P126S (CCA→TCA) | 19 | ||
| sgtB (mgt) | Monofunctional transglycosylase |
DEL | (T)7→6, coding (109/810 nt) | 11 |
| SNP | Q215* (CAA→TAA) | 15 | ||
| SNP | S121* (TCA→TAA) | 13 | ||
The gene nomenclature provided by prokka annotation, reflected in the mutation analysis, is shown in the parentheses.
An asterisk indicates that a mutation led to a stop codon being formed.
TABLE 4.
Key mutations for final endpoint clones of S. aureus TCH1516 after tolerance adaptive laboratory evolution in RPMI+ to nafcillin (SNFR)
| Genea | Specific function | Mutation | Protein and nucleotide changeb |
Strain(s) |
|---|---|---|---|---|
| mecA | Beta-lactam-inducible penicillin-binding protein |
SNP | D586Y (GAT→TAT) | 7, 9, 13, 15, 17, 23 |
| SNP | V488F (GTT→TTT) | 19 | ||
| rpoD (sigA) | DNA-directed RNA polymerase sigma subunit |
SNP | A187T (GCA→ACA) | 13 |
| SNP | A194V (GCA→GTA) | 19 | ||
| gdpP | Cyclic di-AMP phosphodiesterase |
SNP | N182K (AAC→AAG) | 9 |
| SNP | S222F (TCC→TTC) | 23 | ||
| ywtF | Putative transcriptional regulator |
DEL | Coding (235/1218 nt) | 7 |
| SNP | Y121* (TAC→TAG) | 15 | ||
| SNP | D214N (GAC→AAC) | 19 | ||
| codY | CodY family transcriptional regulator |
SNP | S204L (TCA→TTA) | 9 |
| SNP | K205N (AAA→AAT) | 15 | ||
| cdaA | Cyclic di-AMP synthase | SNP | W76C (TGG→TGC) | 7 |
| SNP | Q55H (CAG→CAT) | 13 | ||
| SNP | A80S (GCT→TCT) | 17 | ||
| ssaA2_4 | Staphylococcal secretory antigen |
SNP | W70* (TGG→TAG) | 9 |
| SNP | C45Y (TGT→TAT) | 15 | ||
| SNP | G65V (GGC→GTC) | 17 | ||
| oatA (oatA_2) | O-Acetyltransferase | SNP | G451S (GGT→AGT) | 7 |
| DEL | Coding (1239-1250/1812 nt) | 9 | ||
| DEL | Coding (29/1812 nt) | 13 | ||
| DEL | Coding (1327/1812 nt) | 15 | ||
| SNP | E341* (GAA→TAA) | 19 | ||
| vraS | Sensor histidine kinase | SNP | G92V (GGC→GTC) | 7 |
| SNP | V66L (GTA→CTA) | 17, 19, 23 | ||
| RS08710 | Heme uptake protein MmpL11 | DEL | Coding (2065-2067/2280 nt) | 7, 19 |
The gene nomenclature provided by prokka annotation, reflected in the mutation analysis, is shown in the parentheses. The gene locus tag corresponds to USA300HOU_RSXXXXX.
An asterisk indicates that a mutation led to a stop codon being formed.
In CA-MHB, the majority of key mutations had been previously identified as being related to a resistance phenotype. One of the most frequently mutated gene sets were those that encoded the regulatory system VraSRT. In fact, vraT is a negative regulator of the vraSR operon which controls transcription of a number of genetic determinants involved in cell wall synthesis and cell division (45). Five of the 13 total key mutations under this condition were SNPs in the genes of this system. This regulatory system has also been shown to be mutated under vancomycin selection pressure in a different USA300 S. aureus strain, which also decreased daptomycin susceptibility (46). Another mutated gene was apt, which also occurred in RPMI+ medium adaptation ALE. This is interesting, as this might be the reason why SNFM clones presented an improved growth rate in RPMI+ conditions (Fig. 3A). As discussed earlier, apt enables nucleotide salvage reactions, a much more energetically favorable pathway than de novo nucleotide synthesis (47), and it has been implicated in the stringent response of bacteria to stressful conditions (32, 48). Mutations resulting in an amino acid substitution and a premature stop codon were discovered in pbuG, which encodes a guanine/xanthine permease. A Bacillus subtilis mutant with defects in pbuG displayed impaired uptake rates of nucleoside sugars guanine and hypoxanthine as well as resistance to toxic purine analog compounds (49). There has also been evidence to suggest a role between purine biosynthesis and increased resistance to vancomycin and daptomycin, two other membrane- and cell wall-targeting antibiotics (50, 51). The last key mutated gene that saw multiple mutations across TALE replicates in CA-MHB was mgt, or sgtB referred to elsewhere, whose gene product is a monofunctional glycosyltransferase responsible for elongation of the glycan strands using lipid-linked disaccharide-pentapeptide as the substrate (52). Each of the mutations in sgtB seems to lead to open reading frame disruption (Table 3), possibly abolishing its transcription. This glycosyltransferase is nonessential in S. aureus (53–56), but it seems to be upregulated upon treatment with cell wall-targeting antibiotics, including oxacillin (57). Furthermore, inactivation of sgtB in the USA300 S. aureus LAC strain has demonstrated increased resistance to several cell wall antibiotics (58).
In RPMI+, although there was a higher number of mutations, there was also a higher degree of parallelism, with 36% of key mutations compared to 25.5% in CA-MHB. The most targeted gene for mutation upon exposure to nafcillin in RPMI+ was mecA (Table 4). Mutations in mecA occurred in seven independent lineages, with an SNP at position 586 changing an aspartic acid residue to a tyrosine residue, comprising six of these mutations. Penicillin binding protein 2a (PBP2a) is encoded by mecA and is responsible for catalyzing transpeptidation of peptidoglycan during cell wall synthesis. The binding protein has long been thought to play a vital role in resistance to β-lactamase-resistant semisynthetic β-lactams (nafcillin, oxacillin, methicillin, etc.) due to its lower affinity for these antibiotics (59, 60). Emergence of S. aureus strains containing mecA has been hypothesized to be due in part to horizontal gene transfer from closely related staphylococcal species leading to formation of MRSA precursors (61). It has been discovered that PBP2a is essential for S. aureus survival, although it is able to replace transpeptidation activity by other PBPs, it still requires interaction with the transglycosylase activity of PBP2a (62).
Another highly mutated gene was oatA, which encodes an O-acetyltransferase. Genetic changes included formation of a premature stop codon, in-frame deletion of 12 bp, and two single base pair deletions. OatA encodes the enzyme required for O-acetylation of peptidoglycan by translocation of acetyl groups from a cytoplasmic source across the membrane (61). These results are consistent with previous data showing that exposure of MRSA to methicillin results not only in reduced peptidoglycan cross-linking but also in reduced peptidoglycan O-acetylation (63). O-acetylation is important for resisting autolysis activity from lysozymes (64) and has been shown to increase susceptibility to certain β-lactams (65). Reduction in O-acetylation has great implications for the host-pathogen relationship in S. aureus infections. Strains with mutations in O-acetyltransferase are more effectively killed by macrophages (66). Furthermore, S. aureus oat mutants have been shown to release more interleukin 1β (IL-1β) (66), a critical factor in rapid clearance of S. aureus bacteremia, as shown by the fact that patients with persistent bacteremia on antimicrobial therapy fail to mount a robust IL-1β response (67, 68). In fact, beta-lactam therapy has been shown to elicit a more robust IL-1β response compared to vancomycin therapy in patients with S. aureus bacteremia to potentially explain, at least in part, the more favorable clinical and microbiological data of beta-lactams over vancomycin (69). Coupled with previously cited phenotypic studies, our findings showing oat mutations induced by nafcillin selection pressure in physiological media on MRSA show direct evidence for a specific attenuation of virulence occurring at a genetic locus. These findings lend strong support of the role of nafcillin (and potentially other beta-lactams) as a potentially important adjunct therapy in MRSA bacteremia to enhance bacteremia clearance as previously reported (70).
Similar to what has been observed for the CA-MHB TALEs, the vraSRT system was also mutated, in this case mostly vraS, with a higher preference for amino acid position 66, where a valine was replaced by a leucine. As mentioned earlier, this operon controls transcription of a number of genetic determinants involved in cell wall synthesis and cell division (45).
Two genes involved in regulating levels of cyclic diadenosine monophosphate (c-di-AMP) inside the cell were also mutated: gdpP, encoding a phosphodiesterase, and cdaA (also known as dacA), which encodes an adenylate cyclase. Both DacA and GdpP are involved in nucleotide signaling pathways, while the former produces c-di-AMP, the latter degrades the cyclic dinucleotide molecule (71, 72). Studies suggest that SNP mutations in dacA, distinct from the ones presented here, affect methicillin resistance via nucleotide signaling by reducing c-di-AMP, resulting in faster growing, less resistant, and more virulent strains (73). On the other hand, SNP mutations in gdpP have been observed in S. aureus after repeated exposure to oxacillin concentrations of 200× MIC and insertional mutants revealed increased tolerance to both oxacillin and vancomycin, as well as altered phenotypic signatures (74, 75). Also, clinical isolates from patients with S. aureus lacking mecA determinants were shown to have mutations in gdpP, further implicating the phosphodiesterase in resistance to β-lactams (76). Other mutations included the following: ywtF, encoding a putative transcriptional regulator, belonging to a family of regulators associated with influencing virulence factors, antibiotic resistance, and cell envelope maintenance in various S. aureus species (77–79); codY, encoding a transcriptional regulator that acts as a repressor for more than 100 genes associated with branched-chain amino acid metabolism and virulence production under nutrient limiting conditions (80–82); rpoD, encoding a RNA polymerase subunit; ssaA2_4, encoding a secretory antigen precursor; and RS08710, encoding a heme uptake-related protein.
The overlap between mutations conferring resistance to nafcillin on the genetic level in both medium types was minimal. The key mutation overlap between the two TALE medium conditions was reduced to the vraSTR operon. As previously mentioned, mutations within this operon have previously been shown to increase expression of a cell wall stress stimulon leading to thicker cell wall and envelope (45, 83). From all of the additional key mutations observed in CA-MHB nafcillin tolerized strains, pbuG and sgtB also occurred once in RPMI+ nafcillin tolerized strains, in one replicate each (Table S7). On the other hand, from the other key mutations observed in nafcillin RPMI+ tolerized strains, only the oatA gene was mutated once in one CA-MHB replicate (Table S8). Interestingly, the genetic adaptation observed in the strains tolerized to nafcillin in RPMI+ did not translate to a resistant phenotype in CA-MHB (Fig. 3B), reiterating the medium-specific mechanisms employed toward nafcillin resistance. Nevertheless, the genes that were reproducibly mutated across the independent lineages under the CA-MHB and RPMI+ conditions have, for the most part, been previously identified as being associated with an antibiotic resistance phenotype.
Mutations identified for S. aureus TCH1516 during nafcillin tolerance in RPMI+. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S7, XLSX file, 0.04 MB (36.8KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 during nafcillin tolerance in CA-MHB. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S8, XLSX file, 0.03 MB (33.7KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
For decades, methicillin-resistant S. aureus (MRSA) has been one of the major contributors to community- and hospital-acquired infections with a broad repertoire of infection type, severity, and human hosts (84). In the United States, this common commensal pathogen is responsible for more than 1 million cases of blood infection and close to 200,000 deaths (85). With such alarming figures, it becomes imperative to understand the underlying mechanisms of antibiotic resistance and adaptation to the host environment. Here, we present a method for determining differential mechanisms of resistance on the genetic level under different medium environments utilizing adaptive laboratory evolution, whole-genome sequencing, and phenotypic characterizations of evolved strains. Genotypes of generated strains were characterized to study fundamental underlying differences in how environmental considerations affect susceptibility at a systems level. Insights gained by analyzing repeatedly mutated regions across different medium conditions in tandem with phenotypic assessment can be leveraged to inform more effective treatment strategies and identify novel drug targets. Thus, the major findings from this work include the following: (i) a significant growth rate increase via genetic adaptation to physiological medium (RPMI+) compared to a negligible one observed in CAMHB; (ii) no gross virulence attenuation observed in medium-adapted strains in a pneumonia model of infection; (iii) medium-specific adaptation toward nafcillin tolerance, attributed to parallely mutated genes, mostly related to membrane and cell wall integrity; (iv) key mutated genes previously shown to be associated with clinical resistant strains; (v) mutations in genetic loci under nafcillin selection pressure that could allow for enhanced intracellular survival. These findings support this approach to better understand clinically relevant adaptive strategies of bacteria that may influence not just antibiotic resistance, but also host-pathogen interactions.
Adaptive laboratory evolution was successful in generating medium-adapted strains of S. aureus TCH1516 to a more physiologically relevant medium, RPMI+. Strains adapted to RPMI+ (STR) saw an increase in growth rate, while no such increase was observed in CA-MHB-adapted strains (STM) (Fig. 1B). Mutations identified in RPMI+-adapted strains showed a high degree of evolutionary parallelism with mutations in the apt and mntA genes occurring in almost all of the independent ALE replicates (Table 1). Both gene products have been associated with the SOS stringent response in stressful conditions, while mntA specifically plays a key role in metal acquisition infection when the host limits availability (32, 39, 48, 86). A recent transcriptome analysis has shown that S. aureus TCH1516 is under manganese starvation upon cultivation in RPMI+ (96), strengthening the argument of a transcription and translation optimization of the mnt operon in the RPMI+ medium-adapted strains. Interestingly, mutations in apt, which enables nucleotide salvage reactions, were also identified in tolerance evolution to nafcillin, particularly when evolved on CA-MHB (Table 3). Mutations identified in this phosphoribosyltransferase likely play a crucial role in the improved growth rate in RPMI+ in the presence of no antibiotic for medium- and nafcillin-adapted strains (Fig. 3A).
The tolerance adaptive laboratory evolution (TALE) method proved successful in the generation of S. aureus TCH1516 strains resistant to nafcillin 2.5- to 4-fold higher compared to the wild type in CA-MHB and 80- to 160-fold higher in RPMI+ (Table 2 and Fig. 2B) after continuous exponential growth in the presence of increasing concentrations of nafcillin. The overlap of shared mutations between nafcillin resistance in each medium type point to several previously studied targets for antibiotic resistance (45, 54, 83). Evolution of antibiotic resistance in the tissue culture medium RPMI supplemented with 10% LB appears to enrich for several other mutations, particularly in mecA and other non-mecA genetic determinants (e.g., oatA and vraS). Mutations in mecA were all located in the active site of PBP2a (87), suggesting an alteration in the target for nafcillin, and thus enabling transpeptidase activity to proceed. Mutations affecting synthesis and acquisition of branched-chain amino acids, as well as biosynthesis of peptidoglycan and its precursors potentially suggest a reorganization of metabolic activity more representative of host infection (71, 81, 88). Importantly, mutations in oatA have been previously shown to have significant impact on S. aureus interaction with the host, potentially allowing enhanced intracellular survival to escape from largely extracellularly acting antibiotics like beta-lactams.
In summary, this study describes several mutations involved in adaptation to medium and nafcillin and discusses their possible role in those processes. These hypotheses warrant further investigation into the molecular mechanisms involved in such genetic adaptations, via reintroduction of such mutations into a wild-type strain using targeted genetic engineering approaches (89, 90) or biochemistry elucidation of protein activities and interactions. This study outlines specific mutations that can be tested via these approaches and provides strong contextual evidence of their causality. Furthermore, with a strain-agnostic approach, one could understand if these mutations are strain-specific or general adaptation mechanisms employed by S. aureus (91).
MATERIALS AND METHODS
Adaptive laboratory evolution and tolerance evolution of S. aureus USA300_TCH1516.
The adaptive laboratory evolution (ALE) experiment was begun by streaking the wild-type S. aureus USA300_TCH1516 (taxid 451516) on LB agar plates. Colonies (five for CA-MHB and eight for RPMI+) were then selected and grown overnight at 37°C in the appropriate medium. Each individual flask served as the starting point for independent ALE experiments. An automated liquid handling platform (92) was used to serially propagate the growing cultures and monitor growth rates. Each batch culture was grown in 15 ml of the respective medium at 37°C and well aerated with magnetic stirrers at 1,800 rpm. When the optical density (OD) reached a value of 0.3, 150 μl was inoculated into the next flask, thus maintaining a continuous exponential growth. The automated system measured the OD at 600 nm (OD600) algorithmically on a Tecan Sunrise Absorbance Microplate reader. When the optical density reached a value of 0.3 (Tecan Sunrise plate reader equivalent to an OD600 of 1 on a traditional spectrophotometer with a 1-cm path length), 150 ul was inoculated into the next flask, thus maintaining a continuous exponential growth. The OD600 values were converted to cell dry weight (DW) concentrations using a previously determined OD600-dry cell weight relationship for S. aureus (1.0 OD600 = 0.434 g DW/liter). Last, frozen stocks were taken intermittently throughout the course of the evolution experiments in 50% (vol/vol) glycerol solution and stored at –80°C. Tolerance evolution was performed similarly to medium adaptation as described above with the addition of continuously increasing concentration of nafcillin. The TALE method was adapted from the method in reference 93.
Growth rate calculations were determined and filtered if R2 correlation was less than 0.98. Growth data were then smoothed to minimize noise following methods described in reference 94, by applying a three-median repeat smooth followed by convolution with a symmetrical kernel containing weights (1/4, 1/2, 1/4) and ended with final three-median smooth. Smoothed data were then fit to a piecewise cubic spline. The time scale of cumulative cell cycle divisions (CCD) was computed following methods outlined in reference 25.
MIC determination.
Azithromycin (Fresenius Kabi), ceftazidime (Hospira), clindamycin (Pfizer), colistin (Xellia Pharmaceuticals ApS), daptomycin (Cubist Pharmaceuticals), linezolid (Pfizer), meropenem (Fresenius Kabi), and vancomycin (Mylan International) were purchased from a clinical pharmacy. Ampicillin, ciprofloxacin, gentamicin, and nafcillin were all purchased from Sigma-Aldrich. All drugs were resuspended in 1× Dulbecco’s phosphate-buffered saline (DPBS) (Corning).
The bacterial strains to be used in antibiotic susceptibility testing were first streaked on Luria agar plates from stocks stored at –80°C (in 20% glycerol and 80% Mueller-Hinton broth [MHB]) and grown stationary at 37°C overnight. Isolated colonies were picked from the plate and inoculated into 5 ml of either CA-MHB (MHB [Difco] amended with 20 mg/liter Ca2+ and 10 mg/liter Mg2+) or RPMI+ (phenol-free RPMI 1640 [Gibco] amended with 10% Luria-Delbruck [LB] [Criterion]) medium in a 14-ml Falcon polypropylene round-bottom snap cap tube (catalog no. 352059; Corning) and grown shaking at 100 rpm at 37°C overnight. The following day the overnight cultures were subcultured 1:50 in the desired medium and volume in either the 14-ml snap cap tubes and grown shaking at 100 rpm at 37°C until they reached mid-logarithmic phase (∼OD600 of 0.4). Unless otherwise noted, experiments were conducted in Costar flat-bottom 96 well plates (catalog no. 3370; Corning) with a final volume of 200 μl/well.
For the MIC experiments, the bacteria were cultured in the same medium throughout (CA-MHB or RPMI+) prior to the addition of antibiotics. The mid-logarithmic-phase cultures were diluted to approximately 5 × 105 CFU (∼OD600 of 0.002), and 180 μl was added to each experimental well of the 96-well flat-bottom plate (catalog no. 3370; Costar). Either 20 μl of 1× DPBS or 20 μl of the desired 10× drug stock was added into each culture-containing well. The plates were then incubated shaking at 100 rpm at 37°C overnight. Bacterial growth, as determined by measuring the OD600 of each well, was determined by utilizing an Enspire Alpha multimode plate reader (PerkinElmer). To determine the MIC90, defined as the amount of drug required to inhibit ≥90% of the growth of the untreated controls, the density of each drug-treated well was compared to the density of the untreated control well.
Mouse studies.
All animal experiments were conducted under veterinary supervision and approved by the University of California San Diego (UCSD) IACUC. Bacterial pneumonia was established as previously described (29). In brief, S. aureus strains were grown overnight in cation-adjusted Mueller-Hinton broth (CA-MHB) and then used to inoculate fresh CA-MHB the day of the infection. Cultures were grown to logarithmic phase (∼OD600 of 0.4), washed three times in 1× DPBS (Corning), and resuspended to a concentration of 2.5 × 109 CFU/ml. Juvenile 8-week-old female C57Blk/6J mice were treated with 100 mg of ketamine (Koetis)/kg of body weight and 10 mg of xylazine (VetOne)/kg and then intratracheally infected with 40 μl of the infection culture to give each mouse a 1 × 108 dose using an operating otoscope (Welch Allyn). Mice were allowed to recover on a sloped heating pad and then returned to their home cage. Mice were euthanized 24 h postinfection through CO2 exposure followed by cervical dislocation. All five lobes of the lung were removed, placed into a 2-ml sterile microtube (Sarstedt) with 1 ml of 1× DPBS and 1-mm silica beads (Biospec), and homogenized for three cycles with one cycle consisting of 1 min on a MagNA lyser (Roche) followed by 1 min on ice. Homogenized samples were then serially diluted and spot plated on Luria agar (Criterion) plates, and then grown overnight at 37°C for CFU enumeration.
Whole-genome sequencing and identification of mutations.
A total of 162 samples, including population and clonal samples were submitted for sequencing. Genomic DNA was isolated using Nucleospin Tissue kit (Macherey-Nagel). The resequencing library was constructed from the isolated genomic DNA using Kapa HyperPlus kit (Roche) according to the manufacturer’s instructions. Then, the library was sequenced using a MiSeq reagent kit v3 (Illumina) in 600-cycle paired-end recipe on an MiSeq instrument (Illumina). Resequenced samples were then processed utilizing a modified script of the software breseq v.0.32.1 (30, 31) to map the genomes of the generated strains to the ancestral genome for identification of genetic mutations. All generated strains were mapped to S. aureus USA300_TCH1516 and reannotated using Prokka (95) (NCBI accession number GCA_000017085.1).
Data availability.
Newly determined sequence data were deposited in the NCBI database under accession numbers SRX3480972 to SRX3480983 (STM), SRR10341521 to SRR10341525 (STM), SRX3482887 to SRX3482918 (STR), and SRR8552606 to SRR8552775 (SNFM and SNFR).
ACKNOWLEDGMENT
This research was supported by NIH NIAID grant (U01-AI124316).
We thank Patrick V. Phaneuf, Muyao Wu, Rodolfo A. Salido, Karenina Sanders, Caitriona Brennan, Gregory Humphrey, and Rob Knight for their assistance with whole genome sequencing and analysis.
REFERENCES
- 1.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jevons MP. 1961. “Celbenin” - resistant Staphylococci. BMJ 1:124–125. doi: 10.1136/bmj.1.5219.124-a. [DOI] [Google Scholar]
- 3.Planet PJ, Diaz L, Kolokotronis S-O, Narechania A, Reyes J, Xing G, Rincon S, Smith H, Panesso D, Ryan C, Smith DP, Guzman M, Zurita J, Sebra R, Deikus G, Nolan RL, Tenover FC, Weinstock GM, Robinson DA, Arias CA. 2015. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis 212:1874–1882. doi: 10.1093/infdis/jiv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez BE, Martinez-Aguilar G, Hulten KG, Hammerman WA, Coss-Bu J, Avalos-Mishaan A, Mason EO Jr, Kaplan SL. 2005. Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics 115:642–648. doi: 10.1542/peds.2004-2300. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. 1961. Standardization of methods for conducting microbic sensitivity tests: second report of the Expert Committee on Antibiotics. World Health Organization Technical Report Series 210. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Hughes D, Andersson DI. 2017. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev 41:374–391. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubicek-Sutherland JZ, Heithoff DM, Ersoy SC, Shimp WR, House JK, Marth JD, Smith JW, Mahan MJ. 2015. Host-dependent induction of transient antibiotic resistance: a prelude to treatment failure. EBioMedicine 2:1169–1178. doi: 10.1016/j.ebiom.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girardello R, Bispo PJM, Yamanaka TM, Gales AC. 2012. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol 50:2414–2418. doi: 10.1128/JCM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry AL, Reller LB, Miller GH, Washington JA, Schoenknect FD, Peterson LR, Hare RS, Knapp C. 1992. Revision of standards for adjusting the cation content of Mueller-Hinton broth for testing susceptibility of Pseudomonas aeruginosa to aminoglycosides. J Clin Microbiol 30:585–589. [1551973] doi: 10.1128/JCM.30.3.585-589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ersoy SC, Heithoff DM, Barnes L V, Tripp GK, House JK, Marth JD, Smith JW, Mahan MJ. 2017. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20:173–181. doi: 10.1016/j.ebiom.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakoulas G, Okumura CY, Thienphrapa W, Olson J, Nonejuie P, Dam Q, Dhand A, Pogliano J, Yeaman MR, Hensler ME, Bayer AS, Nizet V. 2014. Nafcillin enhances innate immune-mediated killing of methicillin-resistant Staphylococcus aureus. J Mol Med (Berl) 92:139–149. doi: 10.1007/s00109-013-1100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H Jr, Corriden R, Rohde M, Hensler ME, Burkart MD, Pogliano J, Sakoulas G, Nizet V. 2015. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deatherage DE, Kepner JL, Bennett AF, Lenski RE, Barrick JE. 2017. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc Natl Acad Sci U S A 114:E1904–E1912. doi: 10.1073/pnas.1616132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, Kishony R. 2011. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahn LJ, Munck C, Ellabaan MMH, Sommer M. 2017. Adaptive laboratory evolution of antibiotic resistance using different selection regimes lead to similar phenotypes and genotypes. Front Microbiol 8:816. doi: 10.3389/fmicb.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R. 2016. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattangady DS, Singh AK, Muthaiyan A, Jayaswal RK, Gustafson JE, Ulanov AV, Li Z, Wilkinson BJ, Pfeltz RF. 2015. Genomic, transcriptomic and metabolomic studies of two well-characterized, laboratory-derived vancomycin-intermediate Staphylococcus aureus strains derived from the same parent strain. Antibiotics (Basel) 4:76–112. doi: 10.3390/antibiotics4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raafat D, Leib N, Wilmes M, François P, Schrenzel J, Sahl H-G. 2017. Development of in vitro resistance to chitosan is related to changes in cell envelope structure of Staphylococcus aureus. Carbohydr Polym 157:146–155. doi: 10.1016/j.carbpol.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 21.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein K, Grønnemose RB, Alm M, Brinch KS, Kolmos HJ, Andersen TE. 2017. Controlled release of plectasin NZ2114 from a hybrid silicone-hydrogel material for inhibition of Staphylococcus aureus biofilm. Antimicrob Agents Chemother 61:e00604-17. doi: 10.1128/AAC.00604-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonow RO, Carabello BA, Chatterjee K, de Leon AC Jr, Faxon DP, Freed MD, Gaasch WH, Lytle BW, Nishimura RA, O’Gara PT, O’Rourke RA, Otto CM, Shah PM, Shanewise JS, Smith SC, Jacobs AK, Adams CD, Anderson JL, Antman EM, Faxon DP, Fuster V, Halperin JL, Hiratzka LF, Hunt SA, Lytle BW, Nishimura R, Page RL, Riegel B. 2006. ACC/AHA 2006 guidelines for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (writing committee to revise the 1998 Guidelines for the Management of Patients With Valvular Heart Disease): developed in collaboration with the Society of Cardiovascular Anesthesiologists: endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. Circulation 114:e84–e231. [DOI] [PubMed] [Google Scholar]
- 24.Bennett JE, Dolin R, Blaser MJ. 2014. Principles and practice of infectious diseases. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 25.Lee D-H, Feist AM, Barrett CL, Palsson BØ. 2011. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS One 6:e26172. doi: 10.1371/journal.pone.0026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez de Evgrafov M, Gumpert H, Munck C, Thomsen TT, Sommer MOA. 2015. Collateral resistance and sensitivity modulate evolution of high-level resistance to drug combination treatment in Staphylococcus aureus. Mol Biol Evol 32:1175–1185. doi: 10.1093/molbev/msv006. [DOI] [PubMed] [Google Scholar]
- 27.Kubicek-Sutherland JZ, Lofton H, Vestergaard M, Hjort K, Ingmer H, Andersson DI. 2017. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J Antimicrob Chemother 72:115–127. doi: 10.1093/jac/dkw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fux CA, Shirtliff M, Stoodley P, Costerton JW. 2005. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol 13:58–63. doi: 10.1016/j.tim.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Dillon N, Holland M, Tsunemoto H, Hancock B, Cornax I, Pogliano J, Sakoulas G, Nizet V. 2019. Surprising synergy of dual translation inhibition vs. Acinetobacter baumannii and other multidrug-resistant bacterial pathogens. EBioMedicine 46:193–201. doi: 10.1016/j.ebiom.2019.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phaneuf PV, Gosting D, Palsson B, Feist A. 2018. ALEdb 1.0: a database of mutations from adaptive laboratory evolution. bioRxiv doi: 10.1101/320747. [DOI] [PMC free article] [PubMed]
- 32.Huang Y-J, Tsai T-Y, Pan T-M. 2007. Physiological response and protein expression under acid stress of Escherichia coli O157:H7 TWC01 isolated from Taiwan. J Agric Food Chem 55:7182–7191. doi: 10.1021/jf071014s. [DOI] [PubMed] [Google Scholar]
- 33.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 35.DeFrancesco AS, Masloboeva N, Syed AK, DeLoughery A, Bradshaw N, Li G-W, Gilmore MS, Walker S, Losick R. 2017. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci U S A 114:E5969–E5978. doi: 10.1073/pnas.1704544114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 37.Hecht A, Glasgow J, Jaschke PR, Bawazer LA, Munson MS, Cochran JR, Endy D, Salit M. 2017. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res 45:3615–3626. doi: 10.1093/nar/gkx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belinky F, Rogozin IB, Koonin EV. 2017. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci Rep 7:12422. doi: 10.1038/s41598-017-12619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diep BA, Phung Q, Date S, Arnott D, Bakalarski C, Xu M, Nakamura G, Swem DL, Alexander MK, Le HN, Mai TT, Tan M-W, Brown EJ, Nishiyama M. 2014. Identifying potential therapeutic targets of methicillin-resistant Staphylococcus aureus through in vivo proteomic analysis. J Infect Dis 209:1533–1541. doi: 10.1093/infdis/jit662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassat JE, Skaar EP. 2012. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin Immunopathol 34:215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juttukonda LJ, Berends ETM, Zackular JP, Moore JL, Stier MT, Zhang Y, Schmitz JE, Beavers WN, Wijers CD, Gilston BA, Kehl-Fie TE, Atkinson J, Washington MK, Peebles RS, Chazin WJ, Torres VJ, Caprioli RM, Skaar EP. 2017. Dietary manganese promotes staphylococcal infection of the heart. Cell Host Microbe 22:531–542.e8. doi: 10.1016/j.chom.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J. 2013. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol 9:683. doi: 10.1038/msb.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. 2013. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 57:83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passalacqua KD, Satola SW, Crispell EK, Read TD. 2012. A mutation in the PP2C phosphatase gene in a Staphylococcus aureus USA300 clinical isolate with reduced susceptibility to vancomycin and daptomycin. Antimicrob Agents Chemother 56:5212–5223. doi: 10.1128/AAC.05770-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moffatt BA, Ashihara H. 2002. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1:e0018. doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson KL, Roux CM, Olson MW, Luong TT, Lee CY, Olson R, Dunman PM. 2010. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol Med Microbiol 60:208–250. doi: 10.1111/j.1574-695X.2010.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxild HH, Nygaard P. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J Bacteriol 169:2977–2983. [3110131] doi: 10.1128/JB.169.7.2977-2983.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongodin E, Finan J, Climo MW, Rosato A, Gill S, Archer GL. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J Bacteriol 185:4638–4643. doi: 10.1128/JB.185.15.4638-4643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaupp R, Lei S, Reed JM, Peisker H, Boyle-Vavra S, Bayer AS, Bischoff M, Herrmann M, Daum RS, Powers R, Somerville GA. 2015. Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. Antimicrob Agents Chemother 59:4226–4238. doi: 10.1128/AAC.00160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang QM, Peery RB, Johnson RB, Alborn WE, Yeh WK, Skatrud PL. 2001. Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J Bacteriol 183:4779–4785. doi: 10.1128/JB.183.16.4779-4785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds PE, Brown DF. 1985. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett 192:28–32. [3850810] doi: 10.1016/0014-5793(85)80036-3. [DOI] [PubMed] [Google Scholar]
- 54.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. doi: 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rebets Y, Lupoli T, Qiao Y, Schirner K, Villet R, Hooper D, Kahne D, Walker S. 2014. Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division. ACS Chem Biol 9:459–467. doi: 10.1021/cb4006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. 2011. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol 193:2549–2556. doi: 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 58.Karinou E, Pazos M, Vollmer W, Grundling A. 2018. Inactivation of the monofunctional peptidoglycan glycosyltransferase SgtB allows Staphylococcus aureus to survive in the absence of lipoteichoic acid. bioRxiv doi: 10.1101/311282. [DOI] [PMC free article] [PubMed]
- 59.Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. [6563036] doi: 10.1128/JB.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Utsui Y, Yokota T. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother 28:397–403. [3878127] doi: 10.1128/AAC.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Musser JM, Kapur V. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol 30:2058–2063. [1500513] doi: 10.1128/JCM.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qoronfleh MW, Wilkinson BJ. 1986. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of beta-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob Agents Chemother 29:250–257.[2872855] doi: 10.1128/AAC.29.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pushkaran AC, Nataraj N, Nair N, Götz F, Biswas R, Mohan CG. 2015. Understanding the structure-function relationship of lysozyme resistance in Staphylococcus aureus by peptidoglycan O-acetylation using molecular docking, dynamics, and lysis assay. J Chem Inf Model 55:760–770. doi: 10.1021/ci500734k. [DOI] [PubMed] [Google Scholar]
- 65.Crisóstomo MI, Vollmer W, Kharat AS, Inhülsen S, Gehre F, Buckenmaier S, Tomasz A. 2006. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol Microbiol 61:1497–1509. doi: 10.1111/j.1365-2958.2006.05340.x. [DOI] [PubMed] [Google Scholar]
- 66.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Götz F, Liu GY, Underhill DM. 2010. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome Activation, and IL-1β secretion. Cell Host Microbe 7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rose WE, Eickhoff JC, Shukla SK, Pantrangi M, Rooijakkers S, Cosgrove SE, Nizet V, Sakoulas G. 2012. Elevated serum interleukin-10 at time of hospital admission is predictive of mortality in patients with Staphylococcus aureus bacteremia. J Infect Dis 206:1604–1611. doi: 10.1093/infdis/jis552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose WE, Shukla SK, Berti AD, Hayney MS, Henriquez KM, Ranzoni A, Cooper MA, Proctor RA, Nizet V, Sakoulas G. 2017. Increased endovascular Staphylococcus aureus inoculum is the link between elevated serum interleukin 10 concentrations and mortality in patients with bacteremia. Clin Infect Dis 64:1406–1412. doi: 10.1093/cid/cix157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volk CF, Burgdorf S, Edwardson G, Nizet V, Sakoulas G, Rose WE. 1 August 2019. IL-1β and IL-10 host responses in patients with Staphylococcus aureus bacteremia determined by antimicrobial therapy. Clin Infect Dis doi: 10.1093/cid/ciz686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geriak M, Haddad F, Rizvi K, Rose W, Kullar R, LaPlante K, Yu M, Vasina L, Ouellette K, Zervos M, Nizet V, Sakoulas G. 2019. Clinical data on daptomycin plus ceftaroline versus standard of care monotherapy in the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 63:e02483-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corrigan RM, Bowman L, Willis AR, Kaever V, Gründling A. 2015. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J Biol Chem 290:5826–5839. doi: 10.1074/jbc.M114.598300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dengler V, McCallum N, Kiefer P, Christen P, Patrignani A, Vorholt JA, Berger-Bächi B, Senn MM. 2013. Mutation in the c-di-AMP cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PLoS One 8:e73512. doi: 10.1371/journal.pone.0073512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffiths JM, O’Neill AJ. 2012. Loss of function of the gdpP protein leads to joint β-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 56:579–581. doi: 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung M, Borges V, Gomes JP, de Lencastre H, Tomasz A. 2018. Phenotypic signatures and genetic determinants of oxacillin tolerance in a laboratory mutant of Staphylococcus aureus. PLoS One 13:e0199707. doi: 10.1371/journal.pone.0199707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Argudín MA, Roisin S, Nienhaus L, Dodémont M, de Mendonça R, Nonhoff C, Deplano A, Denis O. 2018. Genetic diversity among Staphylococcus aureus isolates showing oxacillin and/or cefoxitin resistance not linked to the presence of mec genes. Antimicrob Agents Chemother 62:e00091-18. doi: 10.1128/AAC.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hübscher J, Lüthy L, Berger-Bächi B, Stutzmann Meier P. 2008. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan YGY, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr (LCP) family of enzymes release wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siegel SD, Liu J, Ton-That H. 2016. Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol 34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, Brinsmade SR. 2016. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol 101:495–514. doi: 10.1111/mmi.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaiser JC, King AN, Grigg JC, Sheldon JR, Edgell DR, Murphy MEP, Brinsmade SR, Heinrichs DE. 2018. Repression of branched-chain amino acid synthesis in Staphylococcus aureus is mediated by isoleucine via CodY, and by a leucine-rich attenuator peptide. PLoS Genet 14:e1007159. doi: 10.1371/journal.pgen.1007159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 83.McCallum N, Meier PS, Heusser R, Berger-Bächi B. 2011. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother 55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 85.Angus DC, van der Poll T. 2013. Severe sepsis and septic shock. N Engl J Med 369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 86.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. 2013. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lim D, Strynadka N. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol 9:870–876. [DOI] [PubMed] [Google Scholar]
- 88.Sychantha D, Jones CS, Little DJ, Moynihan PJ, Robinson H, Galley NF, Roper DI, Dowson CG, Howell PL, Clarke AJ. 2017. In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA). PLoS Pathog 13:e1006667. doi: 10.1371/journal.ppat.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prax M, Lee CY, Bertram R. 2013. An update on the molecular genetics toolbox for staphylococci. Microbiology 159:421–435. doi: 10.1099/mic.0.061705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Penewit K, Holmes EA, McLean K, Ren M, Waalkes A, Salipante SJ. 2018. Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. mBio 9:e00067-18. doi: 10.1128/mBio.00067-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Machado H, Weng LL, Dillon N, Seif Y, Holland M, Pekar JE, Monk JM, Nizet V, Palsson BO, Feist AM. 2019. Strain-specific metabolic requirements revealed by a defined minimal medium for systems analyses of Staphylococcus aureus. Appl Environ Microbiol 85:e01773-19. doi: 10.1128/AEM.01773-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.LaCroix RA, Palsson BO, Feist AM. 2017. A model for designing adaptive laboratory evolution experiments. Appl Environ Microbiol 83:e03115-16. doi: 10.1128/AEM.03115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mohamed ET, Wang S, Lennen RM, Herrgård MJ, Simmons BA, Singer SW, Feist AM. 2017. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb Cell Fact 16:204. doi: 10.1186/s12934-017-0819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beyer H. 1981. Tukey, John W.: Exploratory Data Analysis. Addison-Wesley Publishing Company Reading, Mass. — Menlo Park, Cal., London, Amsterdam, Don Mills, Ontario, Sydney 1977, XVI, 688 S. Biom J 23:413–414. doi: 10.1002/bimj.4710230408. [DOI] [Google Scholar]
- 95.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 96.Poudel S, Tsunemoto H, Seif Y, Sastry AV, Szubin R, Xu S, Machado H, Olson C, Anand A, Pogliano J, Nizet V, Palsson B. 2020. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators and role in key physiological responses. bioRxiv doi: 10.1101/2020.03.18.997296. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MIC susceptibility testing of S. aureus TCH1516. Dagger indicates differential susceptibility greater than or equal to 4× across CA-MHB and RPMI + 10%LB. Download Table S1, DOCX file, 0.02 MB (18.3KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth phenotypes for S. aureus TCH1516 populations that evolved on CA-MHB and RPMI+. Download Table S2, DOCX file, 0.02 MB (19.8KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 after medium adaptation to CA-MHB. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S3, XLSX file, 0.01 MB (11KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 after medium adaptation to RPMI+. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S4, XLSX file, 0.06 MB (66.2KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RPMI+ medium adaptation additional mutations Download Text S1, DOCX file, 0.02 MB (21.3KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fitness trajectory for a typical TALE experiment, showing population growth rate and continuously increasing antibiotic concentration. The selected trajectory depicts SNFM15 exposed to nafcillin in CA-MHB. Download FIG S1, EPS file, 0.1 MB (126KB, eps) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nafcillin sensitivity of medium-adapted strains Download Table S5, DOCX file, 0.02 MB (18.6KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of growth rates determined for clonal isolates, using two-way ANOVA analysis in Graphpad Prism 7.04. Download Table S6, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 during nafcillin tolerance in RPMI+. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S7, XLSX file, 0.04 MB (36.8KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations identified for S. aureus TCH1516 during nafcillin tolerance in CA-MHB. Nomenclature example A1 F29 I1 R1 = ALE 1 Flask 29 Isolate (I1 = clone, I0 = population) Replicate 1. Download Table S8, XLSX file, 0.03 MB (33.7KB, xlsx) .
Copyright © 2020 Salazar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Newly determined sequence data were deposited in the NCBI database under accession numbers SRX3480972 to SRX3480983 (STM), SRR10341521 to SRR10341525 (STM), SRX3482887 to SRX3482918 (STR), and SRR8552606 to SRR8552775 (SNFM and SNFR).