Abstract
Objective:
To characterize the ecological effects of biologic therapies on the gut bacterial and fungal microbiome of psoriatic arthritis (PsA)/spondyloarthritis (SpA) patients.
Methods:
Fecal samples from PsA/SpA patients pre- and post-treatment with tumor necrosis factor inhibitors (TNFi; n=15) or an anti-interleukin (IL)-17A monoclonal antibody inhibitor (IL-17i; n=14) underwent sequencing (16S, ITS and shotgun metagenomics) and computational microbiome analysis. Fecal levels of fatty acid metabolites and cytokines/proteins implicated in PsA/SpA pathogenesis or intestinal inflammation were correlated with sequence data. Additionally, ileal biopsies obtained from SpA patients who developed clinically overt Crohn’s disease (CD) after treatment with IL-17i (n=5) were analyzed for expression of IL-23/Th-17 related cytokines, IL-25/IL-17E-producing cells and type-2 innate lymphoid cells (ILC2s).
Results:
There were significant shifts in abundance of specific taxa after treatment with IL-17i compared to TNFi, particularly Clostridiales (p=0.016) and Candida albicans (p=0.041). These subclinical alterations correlated with changes in bacterial community co-occurrence, metabolic pathways, IL-23/Th17-related cytokines and various fatty acids. Ileal biopsies showed that clinically overt CD was associated with expansion of IL-25/IL-17E-producing tuft cells and ILC2s (p<0.05) compared to pre-IL-17i treatment levels.
Conclusion:
In a subgroup of SpA patients, the initiation of IL-17A blockade correlated with features of subclinical gut inflammation and intestinal dysbiosis of certain bacterial and fungal taxa, most notably C. albicans. Further, IL-17i-related CD was associated with overexpression of IL-25/IL-17E-producing tuft cells and ILC2s. These results may help to explain the potential link between inhibition of a specific IL-17 pathway and the (sub)clinical gut inflammation observed in SpA.
Keywords: Microbiome, Spondyloarthritis, Psoriatic arthritis, IL-17 inhibitor, Candida
INTRODUCTION
The microbiome, the collection of microorganisms that co-inhabit human surfaces, serves many important functions, ranging from carbohydrate metabolism and synthesis of essential vitamins to immune system modulation and protection from pathogens(1). Intestinal dysbiosis (i.e. the imbalance in the composition of gut microbiota), has been implicated in the development of autoimmune disorders and chronic inflammatory arthritis, including axial and peripheral spondyloarthritis (SpA)(2), ankylosing spondylitis (AS)(3), psoriatic arthritis (PsA)(4), reactive arthritis(5) and inflammatory bowel disease (IBD)(6).
Notably, the microbiome can affect the metabolism, bioavailability and efficacy of drugs(7). Recent examples include novel checkpoint inhibitors(8, 9), whose anti-cancer efficacy is also microbiome-dependent. Conversely, several drugs, including those that lack known antibiotic properties, can alter the gut microbial composition(10). Of direct relevance to autoimmune and chronic inflammatory arthritis, interleukin (IL)-17A blockade has been associated with candidiasis(11) and exacerbation of Crohn’s disease (CD)(12), a condition linked to higher frequencies of Candida albicans colonization(13). This is not unexpected since IL-17A is known to provide protection against extracellular pathogens and maintain intestinal epithelial health(14).
In this study, we characterized the ecological effects of the most commonly prescribed biologic therapies in PsA and SpA on the gut bacterial and fungal microbiome. We hypothesized that biologic therapy, particularly IL-17A inhibition, would perturb the gut bacterial and fungal communities, altering their interrelationships and causing downstream changes in gut metabolite and local cytokine production. We further examined biopsies of patients who developed gut inflammation post-treatment with IL-17i and characterized the changes in adhesive/invasive bacterial scores and local immune responses.
MATERIALS AND METHODS
Subject recruitment:
Adult PsA/SpA subjects were recruited from the NYU clinics prior to initiating biologic therapy. Fifteen subjects analyzed in the TNFi cohort were either naïve to prior biologic treatment (93.3%) or with brief exposure more than one year prior to enrollment (6.7%). They were compared to fifteen age, gender, and ethnicity-matched controls to determine differences between healthy and PsA/SpA subjects. A different set of fourteen subjects in the IL-17i cohort were also compared with the TNFi cohort, of which 9 (64.3%) were inadequate responders to TNFi, warranting the switch to IL-17i therapy (secukinumab). This allowed us to observe the natural history of microbiome fluctuation in a typical clinical progression of biologic therapy for SpA and psoriatic disease. Ileal biopsies were analyzed from an additional cohort (University of Palermo) of five HLA-B27+ SpA subjects (all diagnosed with AS) who developed CD after treatment with IL-17i (secukinumab) compared to five who did not.
Subject evaluation and sample collection:
For the TNFi and IL-17i NYU cohorts, demographic and clinical data were recorded. Fecal samples were collected for microbiome, metagenomic, metabolomic and cytokine analyses. Blood was collected for basic laboratory testing and human leukocyte antigen (HLA)-typing. In the TNFi cohort, samples were collected at baseline (pre) and 6 months after starting therapy (maintenance; mean 27±10 weeks). In the IL-17i cohort, samples were collected at baseline (pre), 5 weeks after starting therapy (loading; mean 7±2 weeks) and 3 months after starting therapy (maintenance; mean 17±5 weeks). All 14 subjects provided samples at the pre and loading visits and 9 provided samples at the maintenance visit. For the SpA (AS) cohorts, ileal biopsies were collected before (baseline) and after initiation of IL-17i therapy (post IL-17i), and used for histologic, immunohistochemical and mRNA quantification analyses.
DNA extraction, 16S rRNA and internal transcribed spacer (ITS) gene sequencing, and data analysis:
Gut bacterial and fungal DNA was extracted and sequenced as previously described(15). Bacterial microbiota composition was determined by sequencing the V4 hypervariable region of bacterial 16S rRNA (Illumina MiSeq platform)(5). Fungal microbiota composition was determined by sequencing the fungal ITS1 region using published primers(16). Obtained sequences were demultiplexed and clustered into operational taxonomic units (OTUs) with closed-reference OTU picking using Quantitative Insights into Microbial Ecology (QIIME) v1.9.1(17). QIIME(17) and R v3.4.3(18) were used to calculate alpha and beta diversity, taxa summary and the change in taxa relative abundance in the NYU cohorts.
Linear discriminant analysis (LDA) effect size (LEfSe):
LEfSe v1.0.7(19) was used to identify differentially abundant bacterial taxa between groups.
Co-occurrence networks:
Taxa were summarized at the genus level and pruned to ≥0.1% relative abundance in at least two samples. Co-occurrence of taxa was calculated using SparCC(20), and results were validated with 10 rarefactions of the input table. Significantly co-occurring taxa (p<0.05) were kept for further analysis. Cytoscape v3.2.1(21) was used to visualize the co-occurrence networks.
Shotgun sequencing and metagenomic analysis:
DNA libraries were generated following the Nextera XT Illumina protocol and sequenced on the Illumina NextSeq Sequencer platform. Raw sequencing reads were processed using Humann2(22). The Kruskal-Wallis test was used to identify differentially abundant enzymatic pathways in the TNFi and IL-17i cohorts.
Measurement of fecal cytokines/proteins and fatty acid metabolites:
Fecal protein was extracted(4) and used to measure PsA/SpA-related cytokines and inflammatory proteins with multiplex and ELISA assays. Short, medium and long-chain fatty acid (FA) levels were measured using gas chromatography mass spectrometry (GC-MS) and liquid chromatography mass spectrometry (LC-MS) methods.
Metadata correlation analysis:
Spearman correlations were performed on pooled pre- and post-treatment samples to identify bacterial and fungal taxa that significantly correlate with cytokines/proteins and FAs. Using QIIME(17), OTUs were grouped at the genus level and tables pruned to contain only OTUs with >0.5% relative abundance in at least 2 samples, followed by Spearman correlations with FDR correction. Associations with genera that achieved FDR <0.1 and FDR <0.2 were plotted as correlograms based on previously published thresholds(4).
Biopsy specimen collection, processing and analysis:
Ileal mucosal biopsy specimens were collected from ten additional SpA (AS) patients before and after IL-17i therapy—5 who developed CD and 5 controls who did not. Presence of adherent and invasive bacteria was assessed with Warthin starry staining and antibodies directed against bacterial lipopolysaccharide (LPS), as previously described(23). mRNA levels of tight junction proteins Occludin and Claudin 1 were assessed with RT-PCR. In the five subjects who developed CD, biopsies were histologically evaluated for inflammation using hematoxylin & eosin (H&E) and immunoperoxidase staining, as previously published (24). Expression of IL-25/IL-17E, IL-25R/IL-17RB, IL-17A, IL-23p19 and IL-13 was assessed with RT-PCR and immunohistochemical staining. Type-2 innate lymphoid cells (ILC2s) were identified and quantified by techniques based on confocal microscopy and flow cytometry of lamina propria mononuclear cells (LPMCs).
Detailed methods for analyses, sequencing, cytokine/protein measurement, GC-MS, LC-MS and biopsy specimen collection/processing, as well as links to publicly available data can be found in the supplementary methods section.
RESULTS
Baseline characteristics.
Characteristics of the TNFi and IL-17i cohorts are shown in Supplementary Table 1. The IL-17i cohort tended to be older (p=ns) and female-predominant (p=ns) compared to the TNFi cohort. Body mass index was comparable in both cohorts (p=ns). Phenotypically, the majority of subjects had PsA, with a small minority diagnosed as axial SpA. All subjects from the TNFi cohort had psoriasis (mostly mild) and either peripheral PsA (53.3%) or a combination of peripheral/axial PsA (46.7%). The average tender and swollen joint counts were 5.5 and 3.6, respectively. In the IL-17i cohort, nine subjects had psoriasis (64.3%) and were diagnosed with either peripheral PsA (35.7%), axial PsA (7.1%), a combination of peripheral/axial PsA (28.6%), or axial SpA (21.4%). One subject had predominantly skin disease. Very few subjects in both cohorts were HLA-B27+ and about a third in each cohort had elevated CRP (28.6% IL-17i vs 33.3% TNFi; p=ns).
PsA/SpA is characterized by intestinal bacterial dysbiosis.
Using 16S rRNA gene sequencing, we compared the intestinal microbiota of biologic-naïve PsA/SpA subjects (TNFi baseline) to age, gender, race and ethnicity-matched healthy controls. We noted an expansion of Clostridiales (order) and Erysipelotrichales (order), as well as a reduction of Bacteroidales (order) relative abundance in PsA/SpA compared to healthy subjects (Supplementary Figure 1A). Furthermore, and as previously shown by our group, several low abundance taxa were overrepresented in healthy controls, the majority of which belong to the Bacteroidales order (Supplementary Figure 1B).
Intestinal bacterial perturbations have distinctive features after TNFi and IL-17i therapies.
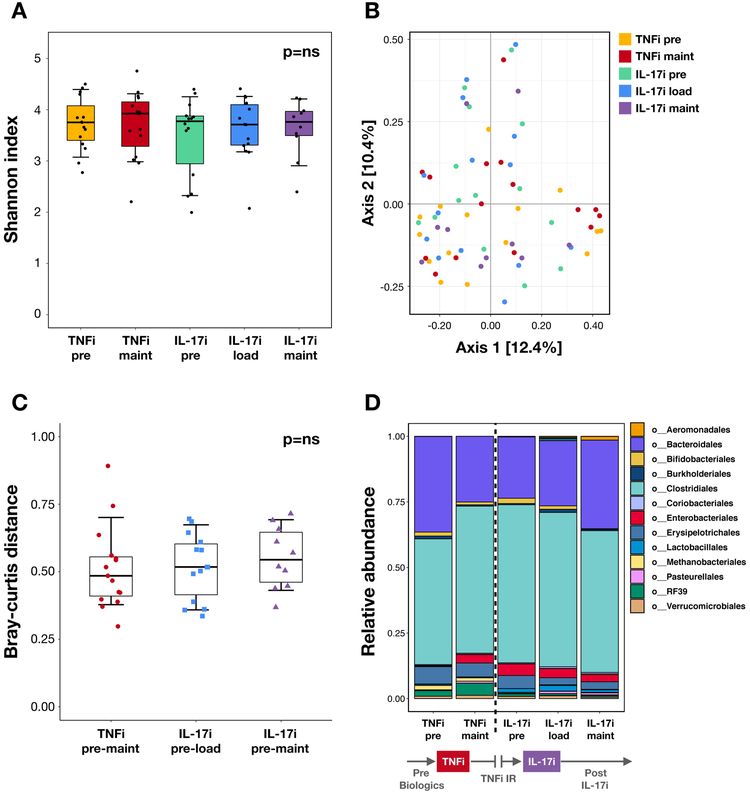
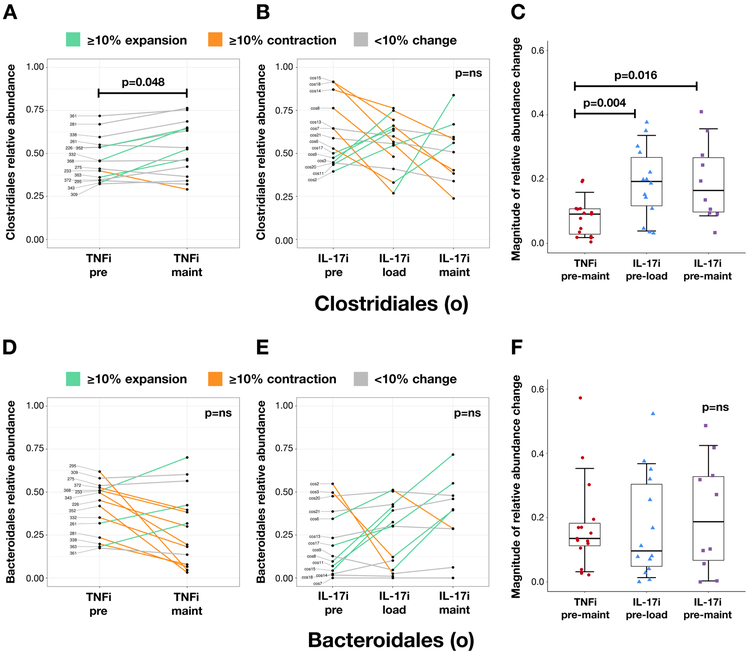
We next analyzed the microbiota of fecal samples from subjects pre- and post-treatment with TNFi and IL-17i. Neither alpha (Figure 1A; Supplementary Figure 2) nor beta (Figure 1B) diversity were significantly different within each cohort. We then analyzed whether there were community dissimilarities over time between the TNFi and IL-17i cohorts. While changes in beta diversity did not show significant differences (Figure 1C), there were noticeable perturbations in specific taxa. In particular, there was an expansion of Clostridiales and a reduction of Bacteroidales at the TNFi maintenance visit compared to baseline, and an opposite reduction of Clostridiales with concomitant expansion of Bacteroidales at the IL-17i maintenance visit (Figure 1D). Furthermore, we observed high variability in therapy-response in specific subjects. There were prominent shifts in Clostridiales relative abundance (both expansion and reduction) with IL-17i therapy, which were not pronounced with TNFi therapy (Figure 2A-C, p<0.005 and p<0.05). We found a similar pattern at higher taxonomic levels, particularly with Firmicutes (phylum; Supplementary Figure 3A-C) and Clostridia (class; Supplementary Figure 3D-F). In contrast, most subjects had a reduction of Bacteroidales relative abundance with TNFi and about a third showed expansion with IL-17i (Figure 2D-F). Additional taxa of interest are summarized in Supplementary Table 2.
Figure 1: Bacterial composition at different stages of TNFi and IL-17i therapy.
(A) Alpha diversity as measured by the Shannon diversity index. There are no significant differences pre-to-post treatment in the TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) cohorts using the Wilcoxon signed-rank test. (B) Principal coordinate analysis plot of beta diversity as measured by the Bray-Curtis distance. No distinct clustering patterns are identified. (C) Community dissimilarity pre-to-post treatment with TNFi (distance measured between pre and maintenance visits) and IL-17i (distance measured between pre and loading visits and pre and maintenance visits) using Bray-Curtis. No significant differences are seen when comparing TNFi to IL-17i using the Mann-Whitney U test. (D) Mean relative abundance of taxa at the order level pre-TNFi to post-IL-17i treatment. Relative abundance is in parts per unit. Legend lists only the top taxa.
Figure 2: Changes in Clostridiales and Bacteroidales relative abundance with TNFi and IL-17i therapy.
Changes in Clostridiales (order) and Bacteroidales (order) relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) therapy. Each line represents an individual subject. Relative abundance is in parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
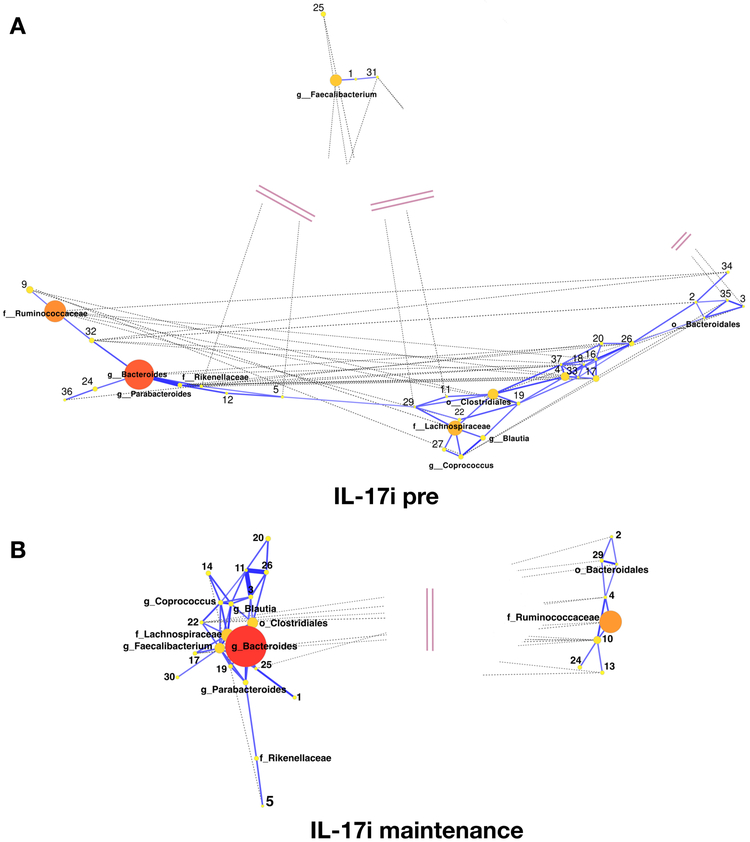
Figure 3: Co-occurrence networks of bacterial taxa pre- and post-treatment with IL-17i.
(A) IL-17i pre (baseline) visit. (B) IL-17i post (maintenance) visit. Nodes represent individual taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
IL-17i disrupts bacterial interactions in the gut.
To further explore perturbations in the bacterial community, we performed a network analysis to identify groups of intestinal bacteria that co-occur with one another before and after biologic therapy with TNFi (Supplementary Figures 4A, 4B, 14; Supplementary Table 3A) or IL-17i (Figure 3, Supplementary Figures 15A, 15B; Supplementary Table 3B). At the pre-TNFi phase, there were two clusters dominated by Bacteroides and Ruminococcaceae (family) that became less prominent (nodes arranged in a linear structure rather than in clusters) with treatment (Supplementary Figures 4A, 4B, 14; Supplementary Table 3A). By contrast, in the pre-IL-17i phase, there was a strong positive correlation between Bacteroides and Ruminococcaceae, with nodes arranged in a linear structure. However, post-IL-17i Bacteroides and Ruminococcaceae separated into two negatively correlated clusters (Figure 3, Supplementary Figures 15A, 15B; Supplementary Table 3B). Of note, the pre-IL-17i network differed markedly from the pre-TNFi network, likely reflecting prior exposure to TNFi in this group (i.e., altered baseline). In addition, the pre-IL-17i network also showed differences with the post-TNFi network, which could be partially due to different exposure durations to biologic agents. Overall, however, these findings suggest that biologic therapy is not only associated with perturbations at the level of the individual taxon but also with changes in the inter-connectivity and co-occurrence among specific bacteria, especially with IL-17i, where positive symbiotic correlations became negative associations after treatment.
Intestinal Candida expansion is predominantly observed with IL-17i treatment.
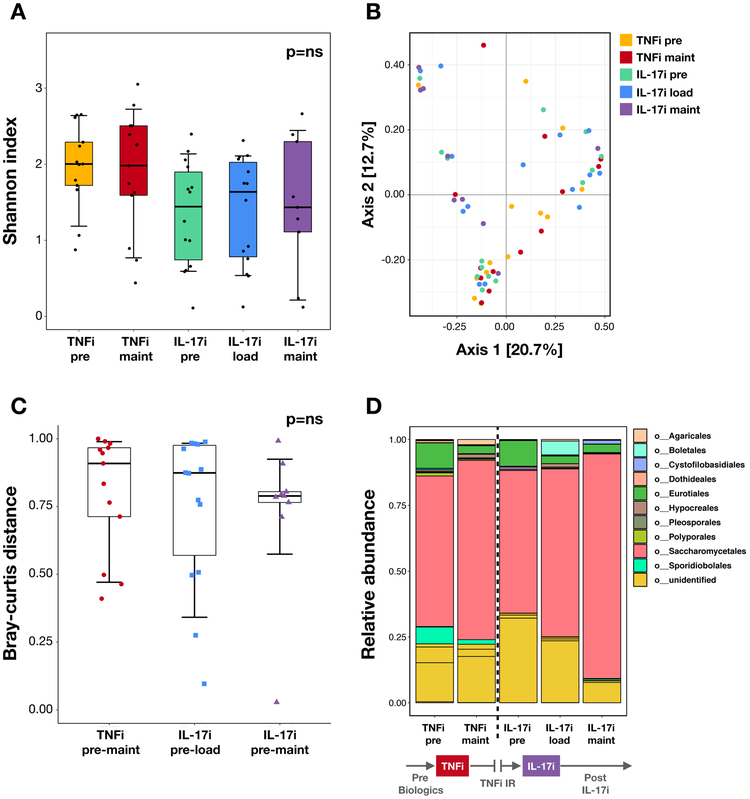
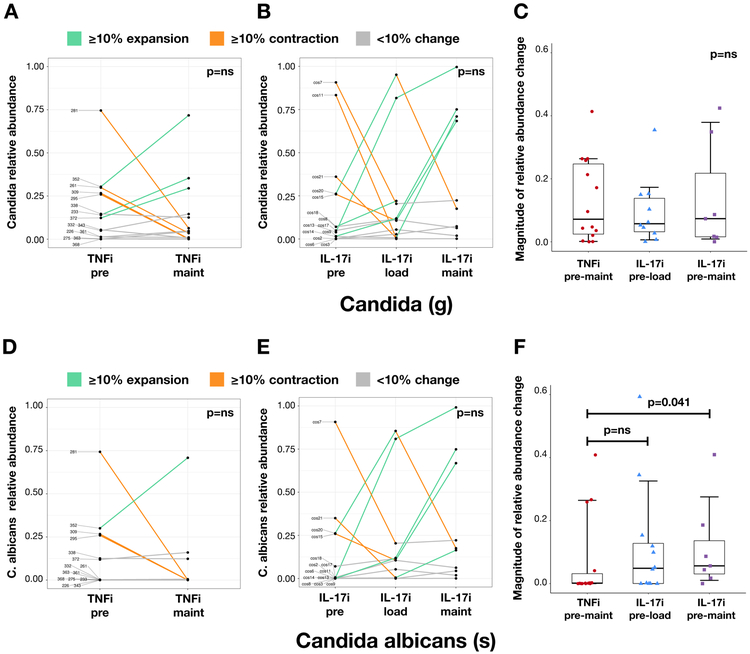
Using ITS sequencing, we subsequently analyzed changes in fungal microbiota following biologic therapy. Akin to results from the bacterial data, alpha and beta diversity were not significantly different in the TNFi- or IL-17i-treated cohorts (alpha: Figure 4A, Supplementary Figure 5; beta: Figure 4B-C). However, there were notable perturbations in specific fungal taxa. These included an expansion of Saccharomycetales (order) at the TNFi and the IL-17i maintenance visits relative to baseline, both on average (Figure 4D) and in the majority of subjects in each cohort (Supplementary Figure 6A-C). Importantly, there were notable IL-17i-treatment specific differences in a subset of patients, including a robust expansion of Candida (29% of cohort; Figure 5B) and C. albicans (21% of cohort; Figure 5E). A smaller subset of subjects demonstrated a similar magnitude in reduction of Candida (14% of cohort; Figure 5B) and C. albicans (7% of cohort; Figure 5E). Moreover, the magnitude of change in C. albicans relative abundance (expansion or reduction) was significantly higher with IL-17i compared to TNFi between baseline and maintenance visits (Figure 5F, p<0.05). In contrast, several subjects showed a robust expansion of Saccharomyces cerevisiae, mostly in the TNFi cohort (40% of cohort; Supplementary Figure 6D). Correspondingly, the magnitude of S. cerevisiae relative abundance change was significantly higher in TNFi compared to IL-17i between baseline and maintenance visits (Supplementary Figure 6F, p<0.05). Additional taxa of interest are summarized in Supplementary Table 4.
Figure 4: Fungal composition at different stages of TNFi and IL-17i therapy.
(A) Alpha diversity as measured by the Shannon diversity index. There are no significant differences pre-to-post treatment in the TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) cohorts using the Wilcoxon signed-rank test. (B) Principal coordinate analysis plot of beta diversity as measured by the Bray-Curtis distance shows no distinct clustering patterns. (C) Community dissimilarity pre-to-post treatment with TNFi (distance measured between pre and maintenance visits) and IL-17i (distance measured between pre and loading visits and pre and maintenance visits) using Bray-Curtis. No significant differences are seen when comparing TNFi to IL-17i using the Mann-Whitney U test. (D) Mean relative abundance of taxa at the order level pre-TNFi to post-IL-17i treatment. Relative abundance is in parts per unit. Legend lists only the top taxa.
Figure 5: Changes in Candida and C. albicans relative abundance with TNFi and IL-17i therapy.
Changes in Candida and C. albicans relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) treatment. Each line represents an individual subject. Relative abundance is in parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
We then performed differential enrichment analysis to identify bacterial taxa that could set apart subjects with high Candida and C. albicans expansion (“expanders”) from all other subjects pre- and post-treatment with IL-17i (Supplementary Figures 7 and 8, respectively; none achieved FDR). Pre-treatment, Candida expanders associated with a higher abundance of taxa from the Clostridiales order, most notably Faecalibacterium (Supplementary Figure 7A). At the maintenance visit, Candida expanders were distinguished by higher abundance of Bacteroidetes and taxa within the Bacteroidetes phylum (i.e. Bacteroides; Supplementary Figure 7B). In the case of C. albicans, the most prominent taxon distinguishing high expanders at the pre-treatment visit was the Veillonellaceae family, also belonging to the Clostridiales order (Supplementary Figure 8A). Post-treatment, expanders were again characterized by higher abundance of Bacteroidaceae and Bacteroides (Supplementary Figure 8B).
Microbial gene pathway perturbations after biologic therapy.
Shotgun sequencing and metagenomic analysis were performed to identify functional changes that occur in response to biologic therapy. We identified several differentially abundant microbial pathways in both cohorts. In the TNFi-treated patients, the majority of pathways that changed with therapy were related to nucleotide metabolism (Supplementary Figure 9; Supplementary Table 5). In contrast, pathways that changed with IL-17i therapy were related to cell wall, vitamin, carbohydrate and amino acid metabolism, particularly tryptophan (Supplementary Figure 10; Supplementary Table 6).
Changes in key bacterial and fungal taxa correlate with IL-23/Th17 cytokines and immunomodulatory metabolites.
We then measured levels of cytokines and proteins that have been implicated in either PsA/SpA pathogenesis and/or intestinal inflammation, including IL-12, IL-23, the IL-17-family, calprotectin, CCL20 and secretory IgA (sIgA). We also measured short, medium and long-chain FAs. We found several correlations between these metadata and the intestinal microbiota at the genus level in TNFi (Supplementary Figures 11A, 12A; Supplementary Tables 7, 8) and IL-17i-treated (Supplementary Figures 11B, 12B; Supplementary Tables 7, 8) patients.
Interestingly, the TNFi cohort had mostly negative correlations with cytokines and other metadata, whereas the IL-17i cohort had mostly positive correlations. In the TNFi cohort, Bacteroides positively correlated with CCL20, sIgA and several cytokines involved in the IL-23/Th17 axis. In contrast, Prevotella and Catenibacterium negatively correlated with the same cytokines and inflammatory proteins. In addition, several taxa from the Clostridiales and Bacteroidales orders negatively correlated with calprotectin (Supplementary Figures 11A, 12A). In the IL-17i cohort, members of the Bacteroidales order had strong positive correlations with IL-25/IL-17E, while members of the Clostridiales order showed weak positive correlations with IL-21 and/or IL-23. Of interest, Faecalibacterium correlated positively with propionic acid and negatively with octanoic acid, while taxa of the Clostridiales order positively correlated with several long-chain fatty acids. Of the fungal taxa, Candida positively correlated with sIgA and negatively correlated with valeric acid and stearic acid (Supplementary Figures 11B, 12B).
IL-17i-induced Crohn’s disease is characterized by decreased bacterial counts and simultaneous expansion of both IL-25/IL-17E-producing tufts cells and ILC2s in the lamina propria.
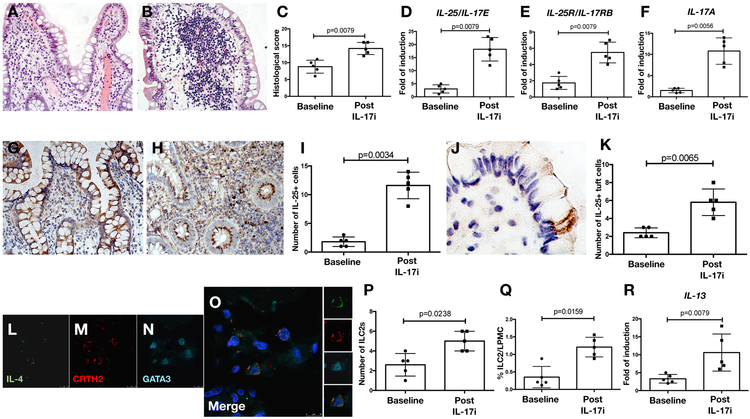
The described observations in gut microbiota dynamics following TNFi and IL-17i were seen in subjects exposed to their respective therapies for relatively short periods of time. Despite correlations with intestinal inflammatory mediators, none of the participants developed clinically evident gut inflammation. Therefore, and to further characterize the effects of IL-17A inhibition on gut immunity and inflammation, we examined ileal biopsies from an additional cohort of SpA (AS) patients who developed clinically overt CD after IL-17i exposure. All five subjects demonstrated reduction in adherent and invasive bacteria in non-lesional biopsies via Warthin starry staining and LPS immunostaining (Supplementary Figure 13A-E), as well as increased expression of two tight junction proteins, Occludin and Claudin 1 (Supplementary Figure 13F-K). Curiously, the same pattern was observed in five control SpA (AS) subjects who were also treated with IL-17i but did not develop CD. In those with colitis, histological evaluation of ileal samples pre- and post-IL7i clearly demonstrated the presence of a Crohn’s-like inflammation after therapy (Figure 6A-C). This inflammation was characterized by increased expression of IL-25/IL-17E, IL-25R/IL-17RB and IL-17A post-IL-17i, as evaluated by RT-PCR (Figure 6D-F) and immunohistochemistry (Figure 6G-K), and increased numbers of IL-25/IL-17E+ tuft cells, which are chemosensory cells lining the intestinal epithelium, known to secrete IL-25/IL-17E in response to parasitic infections(25). There were no changes in IL-23 levels. The higher expression of IL-25/IL-17E was accompanied by expansion of ILC2s (p<0.05), defined as CD3−IL4+CRHT2+GATA3+ cells by confocal microscopy (Figure 6L-P) or lin−(ST2−)CD45+IL-25R+KLRG1+Thy1+ cells by flow cytometry (Figure 6Q), as well as increased production of IL-13 (Figure 6R).
Figure 6: IL-25 driven inflammation characterizes colitis seen in in SpA (AS) subjects after IL-17i therapy.
A-B: Representative hematoxylin and eosin staining images demonstrating histologic changes in the ileum of SpA (AS) subjects at baseline (A) and after the onset of clinically evident Crohn’s disease (CD) (B) during treatment with IL-17i (secukinumab). C: Histological score in SpA (AS) ileal samples at baseline and after the onset of CD. D-F: Relative mRNA levels of IL-25/IL-17E (D), IL-25R/IL-17RB (E) and IL-17A (F) in five SpA (AS) subjects at baseline and after the onset of CD as assessed by RT-PCR. G-H: Representative images showing IL-25 immunostaining in SpA (AS) ileal samples at baseline and after the onset of CD. I: Number of IL-25 positive cells the gut of SpA (AS) subjects. J: Representative image showing the expression of IL-25/IL-17E in the context of specialized epithelial cells that were morphologically identified as tuft cells in the gut of SpA (AS) subjects after the onset of CD during treatment with IL-17i (secukinumab). K: Number of IL-25 positive tuft cells at baseline and after the onset of CD. L-O: Representative confocal microscopy images of IL-4 (L), CRTH2 (M) and GATA-3 (N) in SpA (AS) ileal tissue after the onset of CD during treatment with IL-17i (secukinumab). O: Merge triple staining showing IL-4/CRTH2/GATA3 positive ILC2 co-localization in the gut of AS (SpA) patients. P: Number of type-2 innate lymphoid cells (ILC2s) in the gut of SpA (AS) subjects. Q: Percentage of ILC2s evaluated by flow cytometry in the gut of SpA (AS) subjects at baseline and after the onset of CD during IL-17i (secukinumab) treatment. R: Relative mRNA levels of IL-13 in the ileum of five SpA (AS) subjects at baseline and after the onset of CD as assessed by RT-PCR. Statistical comparisons calculated using the Student’s t-test and the Mann-Whitney U test.
DISCUSSION
We describe, for the first time, the effects of two biologic therapies (i.e. TNFi and IL-17i) on the intestinal microbiome of PsA and SpA patients. Through our longitudinal study approach, we were able to directly discern changes in the microbiome before and after therapy. Consistent with our previous findings, we observed intestinal dysbiosis in PsA/SpA patients prior to the initiation of biologic therapy. Although variations in specific taxa differed from our previous study(4), this was unsurprising given that microbiome sequencing was performed using different platforms. After biologic therapy, patients demonstrated further bacterial and fungal perturbations, which were more prominent with IL-17i and characterized by significant changes in Clostridiales and related taxa. This is of relevance because Clostridia contribute to the maintenance of intestinal homeostasis(26), with many species producing butyrate, a short-chain FA (SCFA) that can induce the differentiation of colonic regulatory T-cells (Treg)(27), which are critical for self-tolerance and prevention of autoimmune disease. Moreover, Clostridia spp. play a physiologic role in gut protection since early inoculation with Clostridium renders mice resistant to the development of colitis(28). In humans, low abundance of Faecalibacterium prausnitzii not only associates with IBD, but also correlates with both ulcerative colitis (UC) disease activity(29) and postoperative recurrence of CD(30).
In our study, IL-17i led to a robust expansion of Candida and C. albicans in a subgroup of patients of varying clinical phenotypes, with a few subjects demonstrating a reduction. We also found that at the pre-treatment stage, these Candida and C. albicans expanders were associated with a higher abundance of taxa from the Clostridiales order. Similarly, Candida expanders were associated with a higher abundance of Bacteroides (and related taxa) post-treatment. A variant of this relationship was seen in a recent study showing that commensal Clostridia and Bacteroidetes were critical for the prevention of C. albicans intestinal colonization in mice(31).
Other groups have demonstrated microbial changes after biologic therapies. For example, CD patients had a significant decrease in Escherichia coli after adalimumab (a TNFi), which was shown to shift their microbiome closer to the composition of healthy individuals(32). Furthermore, CD patients who responded to ustekinumab, an IL-12/23 inhibitor, had higher bacterial diversity pre-treatment while several taxa (e.g. Faecalibacterium) distinguished responders from non-responders(33). In axial SpA, patients who improved with TNFi had a stable microbiome, which was less prone to perturbations with treatment. Additionally, patients who responded to TNFi tended to have higher taxa diversity prior to treatment(34).
Our study showed that bacterial and fungal dysbiosis do not happen in isolation. As indicated by our network analysis, treatment with IL-17i turned positive mutualistic associations between taxa into negative ones, particularly Bacteroides and Ruminococcaceae, a member of the Clostridiales order. In turn, changes in the relative abundance of key taxa correlated with shifts in intestinal levels of IL-23/Th17-related cytokines and beneficial SCFAs such as propionic acid, linked to Treg expansion and abrogation of gut inflammation(27, 35). In the fungal community, changes in Candida correlated positively with sIgA and negatively with valeric acid, which has been associated with IL-10 production and reduction of IL-17A(36). Moreover, metagenomic analysis demonstrated that treatment with IL-17i was associated with upregulation of L-tryptophan biosynthesis. This is of relevance since in vitro binding of a tryptophan-derived photoproduct enhances Th17 cell development and IL-17A expression(37). The reasons for our findings are likely multifactorial but it is possible that, in response to IL-17 blockers, microbial-derived tryptophan is overproduced as a positive forward loop in order to maintain Th17 cell homeostasis in the lamina propria.
Some of the unfavorable associations with Candida are not surprising as prior work suggests that this taxon may be linked to gut inflammation and CD(38). In fact, C. albicans was shown to be an immunogen for anti-Saccharomyces cerevisiae antibodies (ASCA), a sensitive serological marker for IBD(39). Moreover, families with CD (both affected and unaffected relatives) are more frequently colonized by C. albicans(13) and the abundance of a different Candida species, C. tropicalis, was significantly higher in CD compared to non-diseased first-degree relatives, positively correlating with ASCA levels(40). The role of fungal flora in the development of IBD is further supported by the fact that mice lacking dectin-1 have increased susceptibility to dextran sulfate sodium (DSS)-colitis(41). Dectin-1, a receptor for beta-glucan found in fungal cell walls, recognizes fungal organisms and induces a strong Th17 response upon binding(42). In the SKG mouse model, systemic exposure to curdlan (1,3 beta-glucan aggregates) results in the development of a SpA phenotype and CD-like colitis, while inhibition of dectin-1 can prevent the onset of arthritis(43). Because IL-17 confers protection from extracellular pathogens, IL-17 inhibition is likely to allow fungal outgrowth, as in observed cases of candidiasis during IL-17i clinical trials(44, 45). IL-17i has also been shown to exacerbate CD in some patients (12), which has led to the hypothesis that IL-17 inhibition could prompt intestinal C. albicans expansion and downstream intestinal inflammation(42).
In our study, IL-17i resulted in CD-like inflammation in a small cohort of SpA (AS) patients and a reduction in adhesive/invasive bacterial scores in patients who developed CD (non-lesional ileum) as well as those who did not (healthy ileum). This may be related to the normalization of tight junction expression after treatment, which was seen independent of clinically overt CD. Remarkably, ileal biopsies from patients who developed colitis demonstrated significantly higher levels of IL-25/IL-17E-producing tuft cells and concomitant expansion of IL-13-producing ILC2s compared to pre-treatment levels. These data differ from findings in naturally occurring CD, where gut mucosal levels of IL-25/IL-17E are not only reduced, but inversely correlate with endoscopic disease severity(46). Furthermore, a number of studies have demonstrated significant alterations in specific subsets of ILC1 and ILC3 rather than ILC2 cells in naturally occurring CD(47). These differences may be due to the underlying diagnosis of AS in the cohort, which is known to be associated with clinical and subclinical gut inflammation. Therefore, it is possible that patients had a priori subclinical gut inflammation and were predisposed to CD. Alternatively, ILC2 expansion may actually correlate with treatment efficacy as a recent study showed that higher circulating ILC2 counts are associated with PsA remission(48). Taken together, our results suggest that IL-17i-induced CD is driven by divergent pathways compared to typical, ‘idiopathic’ CD. It is, however, essential to note that while IBD exacerbation was frequently observed following IL-17i in CD studies (up to 10%)(12), exacerbations (or de novo) IBD occurred in <1% of SpA patients enrolled in randomized clinical trials(11, 49). Nevertheless, several confirmed cases in routine clinical practice have since been reported(50-52).
Perturbations seen with IL-17i may be specifically related to the inhibition of IL-17A, one of several IL-17 isoforms. This was elegantly demonstrated in IL-17F−/− mice which, unlike IL-17A−/− animals, were resistant to DSS colitis compared to wild-type controls(53). Importantly, the attenuation of colitis in IL-17F−/− mice was linked to over-expansion of Treg in the lamina propria and a higher abundance of commensal Clostridia(53). Pre-treatment with a monoclonal antibody against IL-17F also attenuated colitis(53). Thus, unlike IL-17A inhibition, which may promote deleterious effects on the intestine, blockade of IL-17F or other isoforms (e.g. IL-17E) may have a rather positive outcome and abrogate inflammation in rodents. Whether this is true in human disease will require demonstration in clinical trials.
Limitations of our study include small cohort sizes, subject recruitment from geographically different regions and dietary practices, a phenotypically heterogeneous population, and different drugs used in the TNFi cohort. Although specific microbiota perturbations were pronounced in both bacterial and fungal communities, these results will require validation in larger prospective cohorts, especially since the TNFi cohort was naïve to biologic agents but the majority of subjects in the IL-17i cohort were previously treated with a TNFi. Extending our results would also help us understand whether baseline gut microbiota can be used as predictors of biologic therapy response, as in the case of cancer immunotherapies. In addition, current databases for taxa identification are significantly more comprehensive for bacteria relative to fungi. However, we utilized the same publicly available databases cited by a number of other publications, allowing for data comparison across studies. Improvement in fungal genome sequencing and expansion of taxonomic tools in the near future should enhance our ability to classify these communities in further detail. Given the correlative nature of microbiome studies, it is possible that shifts in Candida (and other bacterial taxa) rather follow changes in immune pressure after local inflammation. Although further work is needed to elucidate directionality, recent human and animal studies imply that candidiasis is likely involved in the pathogenesis of IBD. Finally, the present study is not mechanistic in nature and cannot address how microbial fluctuations correlate with disease outcomes. Future research is therefore needed.
In summary, our findings demonstrate that biologic therapies in PsA and SpA not only modulate immune cell response but are also associated with perturbations in specific bacterial and fungal taxa. IL-17i leads to significant changes in Clostridiales and notable expansion of Candida and C. albicans in over a fourth of patients, which are accompanied by changes in metabolic pathways, levels of PsA/SpA-related cytokines, pro-inflammatory molecules and fatty acid production. Furthermore, IL-17i-induced CD in human SpA differs from ‘idiopathic’ CD and appears to be driven by diverging IL-17 family pathways (i.e. IL25/IL-17E). Moving forward, these IL-17i-induced microbial and immune perturbations should be explored, perhaps through a machine learning model, and incorporated into the clinic by predicting which individuals are susceptible to adverse outcomes from IL-17A and related therapies such as candidiasis and (sub)clinical gut inflammation. Ultimately, understanding the downstream effects of these perturbations could allow for the development of precision medicine approaches in PsA, SpA and related rheumatic diseases.
Supplementary Material
Supplementary Figure 1: PsA/SpA subjects demonstrate intestinal bacterial dysbiosis prior to initiation of biologic therapy. (A) Mean relative abundance of taxa at the order level demonstrates expansion of Clostridiales (order) and Erysipelotrichales (order), and reduction of Bacteroidales (order) in baseline PsA/SpA subjects (prior to TNFi therapy) compared to healthy controls. Relative abundance is in parts per unit. Legend lists only the top taxa. (B) LEfSe analysis identifies bacterial taxa that differentiate healthy and PsA/SpA subjects prior to treatment with TNFi. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. *Indicates taxa that achieve FDR <0.2. **indicates taxa that achieve FDR <0.1. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 2: No change in bacterial alpha diversity with TNFi and IL-17i therapy. Alpha diversity as measured by the (A) number of observed OTUs and (B) the Simpson diversity index. There are no significant differences in diversity pre-to-post treatment with TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) using the Wilcoxon signed-rank test.
Supplementary Figure 3: Changes in Firmicutes and Clostridia relative abundance with TNFi and IL-17i treatment. Changes in Firmicutes (phylum) and Clostridia (order) relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) treatment. Each line represents an individual subject. Relative abundance is represented as parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
Supplementary Figure 4A: Co-occurrence network of bacterial taxa prior to treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 4B: Co-occurrence network of bacterial taxa after treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 5: No changes in fungal alpha diversity with TNFi and IL-17i therapy. Alpha diversity as measured by the (A) number of observed OTUs and (B) the Simpson diversity index. There are no significant differences in diversity pre-to-post treatment with TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) using the Wilcoxon signed-rank test.
Supplementary Figure 6: Changes in Saccharomycetales and S. cerevisiae relative abundance with TNFi and IL-17i treatment. Changes in Saccharomycetales and S. cerevisiae relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) treatment. Each line represents an individual subject. Relative abundance is in parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
Supplementary Figure 7: LEfSe analysis identifies bacterial taxa that differentiate high Candida expanders pre- and post-IL-17i therapy. (A) Pre-treatment with IL-17i, Candida expanders demonstrate higher abundance of taxa from the Clostridiales order. (B) Post-treatment with IL-17i, Candida expanders are distinguished by higher abundance of the Bacteroidetes phylum. High expansion is defined as the top quartile of positive change in relative abundance from pre to maintenance visits. Taxa that differentiate high Candida expanders are colored in turquoise, and those that differentiate all other subjects are colored in navy. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. None of the identified taxa achieve FDR <0.2. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 8: LEfSe analysis identifies bacterial taxa that differentiate high C. albicans expanders pre- and post-IL-17i therapy. (A) Pre-treatment with IL-17i, C. albicans expanders are distinguished by low-abundance taxa, most notably the Veillonellaceae family. (B) Post-treatment with IL-17i, C. albicans demonstrate higher abundance of the Bacteroidaceae family and Bacteroides. High expansion is defined as the top quartile of positive change in relative abundance from pre to maintenance visits. Taxa that differentiate high C. albicans expanders are colored in turquoise, and those that differentiate all other subjects are colored in navy. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. None of the identified taxa achieve FDR <0.2. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 9: Enzymatic pathways that change with TNFi therapy. Relative abundance of pathways that demonstrate significant changes with TNFi therapy between pre and maintenance visits. Significance determined by the Kruskal-Wallis and Wilcoxon signed-rank tests. Most pathways are related to nucleotide metabolism.
Supplementary Figure 10: Enzymatic pathways that change with IL-17i therapy. Relative abundance of pathways that demonstrate significant changes with IL-17i therapy between pre, loading and maintenance visits. Significance determined by the Kruskal-Wallis and Wilcoxon signed-rank tests. Pathways are related to carbohydrate, amino acid, vitamin and cell wall metabolism.
Supplementary Figure 11: Correlations of gut microbiota with immunologic and biochemical metadata. (A) Correlations from pooled pre- and post-TNFi samples. (B) Correlations from pooled pre- and post-IL-17i samples. Correlograms represent Spearman correlations between 1) bacterial/fungal genera and 2) cytokines/proteins implicated in PsA/SpA pathogenesis and/or intestinal inflammation or fatty acid metabolites. Correlation magnitude is represented by the size and color intensity of each circle. Positive correlations are indicated in blue, while negative correlations are indicated in red. All genera that achieve FDR <0.1 are included. Dotted line visually separates bacterial from fungal taxa.
Supplementary Figure 12: Expanded correlations of gut microbiota with immunologic and biochemical metadata. (A) Correlations from pooled pre- and post-TNFi samples. (B) Correlations from pooled pre- and post-IL-17i samples. Correlograms represent Spearman correlations between 1) bacterial/fungal genera and 2) cytokines/proteins implicated in PsA/SpA pathogenesis and/or intestinal inflammation or fatty acid metabolites. Correlation magnitude is represented by the size and color intensity of each circle. Positive correlations are indicated in blue, while negative correlations are indicated in red. All genera that achieve FDR <0.2 are included. Dotted line visually separates bacterial from fungal taxa.
Supplementary Figure 13:Impact of IL-17i treatment on adherent and invasive intestinal bacteria and tight junction proteins in AS (SpA) patients. A-B: Representative images showing Warthin starry staining in non-inflamed gut mucosa from the same SpA (AS) subject at baseline (A) and after IL-17i treatment (B). C-D: Representative images showing lipopolysaccharide (LPS) immunostaining of gut mucosa from the same SpA (AS) subject at baseline (C) and after IL-17i treatment (D). E: Quantification of adherent and invasive bacteria in ileal biopsies from 10 SpA (AS) subjects at baseline and after IL-17i treatment (secukinumab). Five subjects developed gut inflammation while the other five did not. Bacterial quantification was performed by evaluating ten representative fields. Each subject received a score based on the degree of bacterial infiltration: 0 - no infiltration, 1 - mild focal infiltration, 2 - moderate focal infiltration, or 3 - severe infiltration. F-G: Relative mRNA levels of Occludin (F) and Claudin 1 (G) in ileal biopsies obtained from the same ten SpA (AS) subjects at baseline and after treatment with IL-17i (secukinumab) as assessed by RT-PCR. H-K: Representative images showing Occludin (H-I) and Claudin 1 (J-K) immunostaining in the SpA (AS) ileal biopsies before and after IL-17i (secukinumab) treatment. A-D: Original magnification x400.
Supplementary Figure 14: Expanded co-occurrence network after treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 15A: Expanded co-occurrence network before treatment with IL-17i. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 15B: Expanded co-occurrence network after treatment with IL-17i. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Table 1: Baseline demographic, clinical and laboratory characteristics of the TNFi and IL-17i cohorts. Statistical significance calculated with Fisher’s exact and chi-square tests for dichotomous variables and the independent t-test for continuous variables. ULN signifies upper limit of normal. *Unable to obtain HLA-B27 status for one subject in the IL-17i cohort. #Clinical measures of disease activity prior to the initiation of biologic therapy (TNFi cohort only). P-values <0.05 are shown in bold.
Supplementary Table 2: Bacterial taxa evaluated for changes in relative abundance with TNFi and IL-17i treatment. Taxa chosen based on preliminary data and prior publications. Columns 1 and 2 denote taxa and taxonomic levels, respectively. Columns 3-5 represent p-values when comparing the relative abundance of each taxon pre-to-post treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test. Columns 6-7 represent p-values when comparing the magnitude of change in relative abundance pre-to-post treatment between TNFi and IL-17i cohorts. P-values calculated using the Mann-Whitney U test. P-values <0.05 are shown in bold.
Supplementary Table 3A: Taxa identities of numbered nodes from TNFi co-occurrence networks in Supplementary Figures 4A, 4B and 14.
Supplementary Table 3B: Taxa identities of numbered nodes from IL-17i co-occurrence networks in Figure 3 and Supplementary Figures 15A and 15B.
Supplementary Table 4: Fungal taxa evaluated for changes in relative abundance with TNFi and IL-17i treatment. Taxa chosen based on preliminary data and prior publications. Columns 1 and 2 denote taxa and taxonomic levels, respectively. Columns 3-5 represent p-values when comparing the relative abundance of each taxon pre-to-post treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test. Columns 6-7 represent p-values when comparing the magnitude of change in relative abundance pre-to-post treatment between TNFi and IL-17i cohorts. P-values calculated using the Mann-Whitney U test. P-values <0.05 are shown in bold.
Supplementary Table 5: Enzymatic pathways that are significantly different between TNFi pre and maintenance visits. Column 1 represents pathways. Columns 2-3 represent mean pathway relative abundance values for pre and maintenance visits. Column 4 represents p-values as calculated by the Kruskal-Wallis test. Column 5 represents p-values as calculated by the Wilcoxon signed-rank test. P-values <0.05 are shown in bold.
Supplementary Table 6: Enzymatic pathways that are significantly different between IL-17i pre, loading and maintenance visits. Column 1 represents pathways. Columns 2-4 represent mean pathway relative abundance values for pre, loading and maintenance visits. Column 5 represents p-values as calculated by the Kruskal-Wallis test. Columns 6-7 represent p-values as calculated by the Wilcoxon signed-rank test. P-values <0.05 are shown in bold.
Supplementary Table 7: Changes in levels of fecal cytokines and proteins pre- and post-treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test between TNFi pre and maintenance visits, IL-17i pre and loading visits, and IL-17i pre and maintenance visits. None were statistically significant.
Supplementary Table 8: Changes in levels of fecal fatty acid metabolites pre- and post-treatment with IL-17i. P-values calculated using the Wilcoxon signed-rank test between TNFi pre and maintenance visits, IL-17i pre and loading visits, and IL-17i pre and maintenance visits. P-values <0.05 are shown in bold.
ACKNOWLEDGEMENTS
We would like to acknowledge Michael Colin, Kristen Lee, Gary Zagon and Pamela Rosenthal for recruiting patients to our study. We would also like to thank Rhina Medina and Luz Alvarado for coordinating and collecting patient samples, Yonghua Li for helping set up the 16S rRNA sequencing runs, and Benjamin Wu for his advice with preliminary data analysis. We would like to acknowledge the NYU Langone Genome Technology Center for 16S rRNA sequencing, María Jose Garzón and the Sequencing and Bioinformatics Service of FISABIO for ITS sequencing, as well as the UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences and the NYU Metabolomics Core Resource Laboratory for performing metabolomics. Although not included in this manuscript, we would like to thank Sergei Koralov and Lu Yang for the animal work related to this project.
Supported by:
NIH/NIAMS R03AR072182, The Colton Center for Autoimmunity, Rheumatology Research Foundation, The Riley Family Foundation and The Snyder Family Foundation (Scher); NIH/NIDDK R01 DK114038 (Clemente, Wallach); 2017 Group for Research and Assessment of Psoriasis and Psoriatic Arthritis (GRAPPA) Pilot Research Grant and NIH/NIAMS T32AR069515 (Manasson); NIH/NCI Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center (Heguy); NIH/NIAMS R01 AR073324 (Guma)
Footnotes
Conflicts of interest: UCB, Janssen, Novartis (Scher); AbbVie (Solomon); Amgen, Novartis, Pfizer, UCB, Janssen (Reddy)
REFERENCES
- 1.Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, et al. Brief Report: Dialister as a Microbial Marker of Disease Activity in Spondyloarthritis. Arthritis Rheumatol. 2017;69(1):114–21. [DOI] [PubMed] [Google Scholar]
- 3.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief Report: Intestinal Dysbiosis in Ankylosing Spondylitis. Arthritis Rheumatol. 2015;67(3):686–91. [DOI] [PubMed] [Google Scholar]
- 4.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manasson J, Shen N, Garcia Ferrer HR, Ubeda C, Iraheta I, Heguy A, et al. Gut Microbiota Perturbations in Reactive Arthritis and Postinfectious Spondyloarthritis. Arthritis Rheumatol. 2018;70(2):242–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koppel N, Maini Rekdal V, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356(6344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McInnes IB, Mease PJ, Kirkham B, Kavanaugh A, Ritchlin CT, Rahman P, et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (FUTURE 2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2015;386(9999):1137–46. [DOI] [PubMed] [Google Scholar]
- 12.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61(12):1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Standaert-Vitse A, Sendid B, Joossens M, Francois N, Vandewalle-El Khoury P, Branche J, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol. 2009;104(7):1745–53. [DOI] [PubMed] [Google Scholar]
- 14.Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126(2):177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scher JU, Joshua V, Artacho A, Abdollahi-Roodsaz S, Ockinger J, Kullberg S, et al. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome. 2016;4(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toju H, Tanabe AS, Yamamoto S, Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;7(7):e40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 19.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciccia F, Guggino G, Zeng M, Thomas R, Ranganathan V, Rahman A, et al. Proinflammatory CX3CR1+CD59+Tumor Necrosis Factor-Like Molecule 1A+Interleukin-23+ Monocytes Are Expanded in Patients With Ankylosing Spondylitis and Modulate Innate Lymphoid Cell 3 Immune Functions. Arthritis Rheumatol. 2018;70(12):2003–13. [DOI] [PubMed] [Google Scholar]
- 24.Baert FJ, D’Haens GR, Peeters M, Hiele MI, Schaible TF, Shealy D, et al. Tumor necrosis factor alpha antibody (infliximab) therapy profoundly down-regulates the inflammation in Crohn’s ileocolitis. Gastroenterology. 1999;116(1):22–8. [DOI] [PubMed] [Google Scholar]
- 25.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529(7585):221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–50. [DOI] [PubMed] [Google Scholar]
- 28.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275–83. [DOI] [PubMed] [Google Scholar]
- 30.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21(7):808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busquets D, Mas-de-Xaxars T, Lopez-Siles M, Martinez-Medina M, Bahi A, Sabat M, et al. Anti-tumour Necrosis Factor Treatment with Adalimumab Induces Changes in the Microbiota of Crohn’s Disease. J Crohns Colitis. 2015;9(10):899–906. [DOI] [PubMed] [Google Scholar]
- 33.Doherty MK, Ding T, Koumpouras C, Telesco SE, Monast C, Das A, et al. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. MBio. 2018;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bazin T, Hooks KB, Barnetche T, Truchetet ME, Enaud R, Richez C, et al. Microbiota Composition May Predict Anti-Tnf Alpha Response in Spondyloarthritis Patients: an Exploratory Study. Sci Rep. 2018;8(1):5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. 2019;10(1):760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453(7191):106–9. [DOI] [PubMed] [Google Scholar]
- 38.Gerard R, Sendid B, Colombel JF, Poulain D, Jouault T. An immunological link between Candida albicans colonization and Crohn’s disease. Crit Rev Microbiol. 2015;41(2):135–9. [DOI] [PubMed] [Google Scholar]
- 39.Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130(6):1764–75. [DOI] [PubMed] [Google Scholar]
- 40.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. MBio. 2016;7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombel JF, Sendid B, Jouault T, Poulain D. Secukinumab failure in Crohn’s disease: the yeast connection? Gut. 2013;62(5):800–1. [DOI] [PubMed] [Google Scholar]
- 43.Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, et al. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. 2005;201(6):949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mease PJ, McInnes IB, Kirkham B, Kavanaugh A, Rahman P, van der Heijde D, et al. Secukinumab Inhibition of Interleukin-17A in Patients with Psoriatic Arthritis. N Engl J Med. 2015;373(14):1329–39. [DOI] [PubMed] [Google Scholar]
- 45.van der Heijde D, Gladman DD, Kishimoto M, Okada M, Rathmann SS, Moriarty SR, et al. Efficacy and Safety of Ixekizumab in Patients with Active Psoriatic Arthritis: 52-week Results from a Phase III Study (SPIRIT-P1). J Rheumatol. 2018;45(3):367–77. [DOI] [PubMed] [Google Scholar]
- 46.Su J, Chen T, Ji XY, Liu C, Yadav PK, Wu R, et al. IL-25 downregulates Th1/Th17 immune response in an IL-10-dependent manner in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19(4):720–8. [DOI] [PubMed] [Google Scholar]
- 47.Sonnenberg GF. Regulation of intestinal health and disease by innate lymphoid cells. Int Immunol. 2014;26(9):501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soare A, Weber S, Maul L, Rauber S, Gheorghiu AM, Luber M, et al. Cutting Edge: Homeostasis of Innate Lymphoid Cells Is Imbalanced in Psoriatic Arthritis. J Immunol. 2018;200(4):1249–54. [DOI] [PubMed] [Google Scholar]
- 49.Mease P, Roussou E, Burmester GR, Goupille P, Gottlieb A, Moriarty SR, et al. Safety of Ixekizumab in Patients with Psoriatic Arthritis: Results from a Pooled Analysis of Three Clinical Trials. Arthritis Care Res (Hoboken). 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fobelo Lozano MJ, Serrano Gimenez R, Castro Fernandez M. Emergence of Inflammatory Bowel Disease During Treatment with Secukinumab. J Crohns Colitis. 2018. [DOI] [PubMed] [Google Scholar]
- 51.Philipose J, Ahmed M, Idiculla PS, Mulrooney SM, Gumaste VV. Severe de novo Ulcerative Colitis following Ixekizumab Therapy. Case Rep Gastroenterol. 2018;12(3):617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrlich D, Jamaluddin N, Pisegna J, Padua D. A Challenging Case of Severe Ulcerative Colitis following the Initiation of Secukinumab for Ankylosing Spondylitis. Case Rep Gastrointest Med. 2018;2018:9679287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang C, Kakuta S, Shimizu K, Kadoki M, Kamiya T, Shimazu T, et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat Immunol. 2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: PsA/SpA subjects demonstrate intestinal bacterial dysbiosis prior to initiation of biologic therapy. (A) Mean relative abundance of taxa at the order level demonstrates expansion of Clostridiales (order) and Erysipelotrichales (order), and reduction of Bacteroidales (order) in baseline PsA/SpA subjects (prior to TNFi therapy) compared to healthy controls. Relative abundance is in parts per unit. Legend lists only the top taxa. (B) LEfSe analysis identifies bacterial taxa that differentiate healthy and PsA/SpA subjects prior to treatment with TNFi. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. *Indicates taxa that achieve FDR <0.2. **indicates taxa that achieve FDR <0.1. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 2: No change in bacterial alpha diversity with TNFi and IL-17i therapy. Alpha diversity as measured by the (A) number of observed OTUs and (B) the Simpson diversity index. There are no significant differences in diversity pre-to-post treatment with TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) using the Wilcoxon signed-rank test.
Supplementary Figure 3: Changes in Firmicutes and Clostridia relative abundance with TNFi and IL-17i treatment. Changes in Firmicutes (phylum) and Clostridia (order) relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) treatment. Each line represents an individual subject. Relative abundance is represented as parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
Supplementary Figure 4A: Co-occurrence network of bacterial taxa prior to treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 4B: Co-occurrence network of bacterial taxa after treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 5: No changes in fungal alpha diversity with TNFi and IL-17i therapy. Alpha diversity as measured by the (A) number of observed OTUs and (B) the Simpson diversity index. There are no significant differences in diversity pre-to-post treatment with TNFi (pre vs maintenance visits) and IL-17i (pre vs loading and pre vs maintenance visits) using the Wilcoxon signed-rank test.
Supplementary Figure 6: Changes in Saccharomycetales and S. cerevisiae relative abundance with TNFi and IL-17i treatment. Changes in Saccharomycetales and S. cerevisiae relative abundance with TNFi (A and D, respectively) and IL-17i (B and E, respectively) treatment. Each line represents an individual subject. Relative abundance is in parts per unit. Turquoise denotes ≥10% taxa expansion, orange denotes ≥10% taxa contraction, and gray denotes <10% change in either direction. Magnitude of change in relative abundance pre-to-post treatment with TNFi (measured between pre and maintenance visits) and IL-17i (measured between pre and loading visits and pre and maintenance visits) (C and F, respectively). Statistical comparisons within each cohort calculated using the Wilcoxon signed-rank test. Statistical comparisons between TNFi and IL-17i cohorts calculated using the Mann-Whitney U test.
Supplementary Figure 7: LEfSe analysis identifies bacterial taxa that differentiate high Candida expanders pre- and post-IL-17i therapy. (A) Pre-treatment with IL-17i, Candida expanders demonstrate higher abundance of taxa from the Clostridiales order. (B) Post-treatment with IL-17i, Candida expanders are distinguished by higher abundance of the Bacteroidetes phylum. High expansion is defined as the top quartile of positive change in relative abundance from pre to maintenance visits. Taxa that differentiate high Candida expanders are colored in turquoise, and those that differentiate all other subjects are colored in navy. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. None of the identified taxa achieve FDR <0.2. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 8: LEfSe analysis identifies bacterial taxa that differentiate high C. albicans expanders pre- and post-IL-17i therapy. (A) Pre-treatment with IL-17i, C. albicans expanders are distinguished by low-abundance taxa, most notably the Veillonellaceae family. (B) Post-treatment with IL-17i, C. albicans demonstrate higher abundance of the Bacteroidaceae family and Bacteroides. High expansion is defined as the top quartile of positive change in relative abundance from pre to maintenance visits. Taxa that differentiate high C. albicans expanders are colored in turquoise, and those that differentiate all other subjects are colored in navy. Left side of figure shows the linear discriminant analysis (LDA) score on a log10 scale. Log10 LDA is >2 for all taxa. None of the identified taxa achieve FDR <0.2. Right side of figure shows the relative abundance of taxa identified by LEfSe on a square-root scale.
Supplementary Figure 9: Enzymatic pathways that change with TNFi therapy. Relative abundance of pathways that demonstrate significant changes with TNFi therapy between pre and maintenance visits. Significance determined by the Kruskal-Wallis and Wilcoxon signed-rank tests. Most pathways are related to nucleotide metabolism.
Supplementary Figure 10: Enzymatic pathways that change with IL-17i therapy. Relative abundance of pathways that demonstrate significant changes with IL-17i therapy between pre, loading and maintenance visits. Significance determined by the Kruskal-Wallis and Wilcoxon signed-rank tests. Pathways are related to carbohydrate, amino acid, vitamin and cell wall metabolism.
Supplementary Figure 11: Correlations of gut microbiota with immunologic and biochemical metadata. (A) Correlations from pooled pre- and post-TNFi samples. (B) Correlations from pooled pre- and post-IL-17i samples. Correlograms represent Spearman correlations between 1) bacterial/fungal genera and 2) cytokines/proteins implicated in PsA/SpA pathogenesis and/or intestinal inflammation or fatty acid metabolites. Correlation magnitude is represented by the size and color intensity of each circle. Positive correlations are indicated in blue, while negative correlations are indicated in red. All genera that achieve FDR <0.1 are included. Dotted line visually separates bacterial from fungal taxa.
Supplementary Figure 12: Expanded correlations of gut microbiota with immunologic and biochemical metadata. (A) Correlations from pooled pre- and post-TNFi samples. (B) Correlations from pooled pre- and post-IL-17i samples. Correlograms represent Spearman correlations between 1) bacterial/fungal genera and 2) cytokines/proteins implicated in PsA/SpA pathogenesis and/or intestinal inflammation or fatty acid metabolites. Correlation magnitude is represented by the size and color intensity of each circle. Positive correlations are indicated in blue, while negative correlations are indicated in red. All genera that achieve FDR <0.2 are included. Dotted line visually separates bacterial from fungal taxa.
Supplementary Figure 13:Impact of IL-17i treatment on adherent and invasive intestinal bacteria and tight junction proteins in AS (SpA) patients. A-B: Representative images showing Warthin starry staining in non-inflamed gut mucosa from the same SpA (AS) subject at baseline (A) and after IL-17i treatment (B). C-D: Representative images showing lipopolysaccharide (LPS) immunostaining of gut mucosa from the same SpA (AS) subject at baseline (C) and after IL-17i treatment (D). E: Quantification of adherent and invasive bacteria in ileal biopsies from 10 SpA (AS) subjects at baseline and after IL-17i treatment (secukinumab). Five subjects developed gut inflammation while the other five did not. Bacterial quantification was performed by evaluating ten representative fields. Each subject received a score based on the degree of bacterial infiltration: 0 - no infiltration, 1 - mild focal infiltration, 2 - moderate focal infiltration, or 3 - severe infiltration. F-G: Relative mRNA levels of Occludin (F) and Claudin 1 (G) in ileal biopsies obtained from the same ten SpA (AS) subjects at baseline and after treatment with IL-17i (secukinumab) as assessed by RT-PCR. H-K: Representative images showing Occludin (H-I) and Claudin 1 (J-K) immunostaining in the SpA (AS) ileal biopsies before and after IL-17i (secukinumab) treatment. A-D: Original magnification x400.
Supplementary Figure 14: Expanded co-occurrence network after treatment with TNFi. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 15A: Expanded co-occurrence network before treatment with IL-17i. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Figure 15B: Expanded co-occurrence network after treatment with IL-17i. Nodes represent taxa, with the size and color denoting relative abundance (large/red nodes – high abundance; medium/orange nodes – medium abundance; small/yellow nodes – low abundance). Edges represent significant correlations between taxa, with solid blue lines corresponding to positive correlations and dashed gray lines corresponding to negative correlations. Edge lengths are inversely proportional to correlation strength. Pink double lines represent truncated edge distances.
Supplementary Table 1: Baseline demographic, clinical and laboratory characteristics of the TNFi and IL-17i cohorts. Statistical significance calculated with Fisher’s exact and chi-square tests for dichotomous variables and the independent t-test for continuous variables. ULN signifies upper limit of normal. *Unable to obtain HLA-B27 status for one subject in the IL-17i cohort. #Clinical measures of disease activity prior to the initiation of biologic therapy (TNFi cohort only). P-values <0.05 are shown in bold.
Supplementary Table 2: Bacterial taxa evaluated for changes in relative abundance with TNFi and IL-17i treatment. Taxa chosen based on preliminary data and prior publications. Columns 1 and 2 denote taxa and taxonomic levels, respectively. Columns 3-5 represent p-values when comparing the relative abundance of each taxon pre-to-post treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test. Columns 6-7 represent p-values when comparing the magnitude of change in relative abundance pre-to-post treatment between TNFi and IL-17i cohorts. P-values calculated using the Mann-Whitney U test. P-values <0.05 are shown in bold.
Supplementary Table 3A: Taxa identities of numbered nodes from TNFi co-occurrence networks in Supplementary Figures 4A, 4B and 14.
Supplementary Table 3B: Taxa identities of numbered nodes from IL-17i co-occurrence networks in Figure 3 and Supplementary Figures 15A and 15B.
Supplementary Table 4: Fungal taxa evaluated for changes in relative abundance with TNFi and IL-17i treatment. Taxa chosen based on preliminary data and prior publications. Columns 1 and 2 denote taxa and taxonomic levels, respectively. Columns 3-5 represent p-values when comparing the relative abundance of each taxon pre-to-post treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test. Columns 6-7 represent p-values when comparing the magnitude of change in relative abundance pre-to-post treatment between TNFi and IL-17i cohorts. P-values calculated using the Mann-Whitney U test. P-values <0.05 are shown in bold.
Supplementary Table 5: Enzymatic pathways that are significantly different between TNFi pre and maintenance visits. Column 1 represents pathways. Columns 2-3 represent mean pathway relative abundance values for pre and maintenance visits. Column 4 represents p-values as calculated by the Kruskal-Wallis test. Column 5 represents p-values as calculated by the Wilcoxon signed-rank test. P-values <0.05 are shown in bold.
Supplementary Table 6: Enzymatic pathways that are significantly different between IL-17i pre, loading and maintenance visits. Column 1 represents pathways. Columns 2-4 represent mean pathway relative abundance values for pre, loading and maintenance visits. Column 5 represents p-values as calculated by the Kruskal-Wallis test. Columns 6-7 represent p-values as calculated by the Wilcoxon signed-rank test. P-values <0.05 are shown in bold.
Supplementary Table 7: Changes in levels of fecal cytokines and proteins pre- and post-treatment with TNFi and IL-17i. P-values calculated using the Wilcoxon signed-rank test between TNFi pre and maintenance visits, IL-17i pre and loading visits, and IL-17i pre and maintenance visits. None were statistically significant.
Supplementary Table 8: Changes in levels of fecal fatty acid metabolites pre- and post-treatment with IL-17i. P-values calculated using the Wilcoxon signed-rank test between TNFi pre and maintenance visits, IL-17i pre and loading visits, and IL-17i pre and maintenance visits. P-values <0.05 are shown in bold.