Abstract
The multi-domain non-structural protein 3 (Nsp3) is the largest protein encoded by the coronavirus (CoV) genome, with an average molecular mass of about 200 kD. Nsp3 is an essential component of the replication/transcription complex. It comprises various domains, the organization of which differs between CoV genera, due to duplication or absence of some domains. However, eight domains of Nsp3 exist in all known CoVs: the ubiquitin-like domain 1 (Ubl1), the Glu-rich acidic domain (also called “hypervariable region”), a macrodomain (also named “X domain”), the ubiquitin-like domain 2 (Ubl2), the papain-like protease 2 (PL2pro), the Nsp3 ectodomain (3Ecto, also called “zinc-finger domain”), as well as the domains Y1 and CoV-Y of unknown functions. In addition, the two transmembrane regions, TM1 and TM2, exist in all CoVs. The three-dimensional structures of domains in the N-terminal two thirds of Nsp3 have been investigated by X-ray crystallography and/or nuclear magnetic resonance (NMR) spectroscopy since the outbreaks of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) in 2003 as well as Middle-East Respiratory Syndrome coronavirus (MERS-CoV) in 2012. In this review, the structures and functions of these domains of Nsp3 are discussed in depth.
Keywords: Ubiquitin-like domain, Papain-like protease, Macrodomain, Nucleic-acid binding domain, Innate immunity, Structural biology
Abbreviations: 3CLpro, 3C-like protease; 3Ecto, Nsp3 ectodomain; ADPr, ADP-ribose; ADRP, ADP-ribose-1″-phosphate phosphatase; ARTD, ADP-ribosyltransferases diphtheria toxin-like; βSM, betacoronavirus-specific marker; CHIKV, Chikungunya virus; CM, convoluted membrane; CoV, coronavirus; DMV, double-membrane vesicle; DPUP, Domain Preceding Ubl2 and PL2pro; DUB, deubiquitinating; ER, Endoplasmic Reticulum; GST, glutathione S-transferase; hISG15, human interferon-stimulated gene 15; HCoV, human coronavirus; HEV, hepatitis E virus; HKU, Hong Kong University; HVR, hypervariable region; IBV, infectious bronchitis virus; IFN, interferon; IL-6, Interleukin 6; IRF, interferon regulatory factor; mISG15, mouse interferon-stimulated gene 15; Mpro, main protease; Mac, macrodomain; MARylation, mono-ADP-ribosylation; MDM2, mouse double minute 2 homolog; MERS, Middle-East respiratory syndrome; MHV, mouse hepatitis virus; MKRN, makorin ring finger protein; N, nucleocapsid; NAB, nucleic-acid binding domain; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; Nsp, non-structural protein; NTD, N-terminal domain; ORF, open reading frame; PABP, poly(A)-binding protein; PARP, poly(ADP-ribose) polymerase; PARylation, poly-ADP-ribosylation; RCHY1, RING finger and CHY zinc-finger domain-containing protein 1; RID, Ras-interacting domain; PLpro, papain-like protease; R.M.S.D., root-mean-square deviation; RTC, replication/transcription complex; SARS, severe acute respiratory syndrome; SUD, SARS-unique domain; TGEV, transmissible gastroenteritis virus; TM, transmembrane; TNF, tumor necrosis factor; TRS, transcriptional regulatory sequence; Ubl, ubiquitin-like; Ub, ubiquitin; USP, ubiquitin-specific protease; UTR, untranslated region; VEEV, Venezuelan equine encephalitis virus
Highlights
-
•
Nonstructural protein 3 (∼200 kD) is a multifunctional protein comprising up to 16 different domains and regions.
-
•
Nsp3 binds to viral RNA, nucleocapsid protein, as well as other viral proteins, and participates in polyprotein processing.
-
•
The papain-like protease of Nsp3 is an established target for new antivirals.
-
•
Through its de-ADP-ribosylating, de-ubiquitinating, and de-ISGylating activities, Nsp3 counteracts host innate immunity.
-
•
Structural data are available for the N-terminal two thirds of Nsp3, but domains in the remainder are poorly characterized.
1. Introduction
This review of published research on the coronavirus non-structural protein 3 (Nsp3) forms part of a series in Antiviral Research on “From SARS to MERS: research on highly pathogenic human coronaviruses” (Hilgenfeld and Peiris, 2013). Two excellent earlier papers dealt with aspects of Nsp3. The first described the state of knowledge of the papain-like protease (PLpro) (Báez-Santos et al., 2015), while the second adopted a bioinformatics viewpoint when describing Nsp3 and other non-structural proteins involved in anchoring the coronavirus replication/transcription complex (RTC) to modified membranous structures originating from the endoplasmic reticulum (ER) (Neuman, 2016). We build on these fine reviews, focusing on recent results and discussing the structures and functions of the individual Nsp3 domains in sequential order.
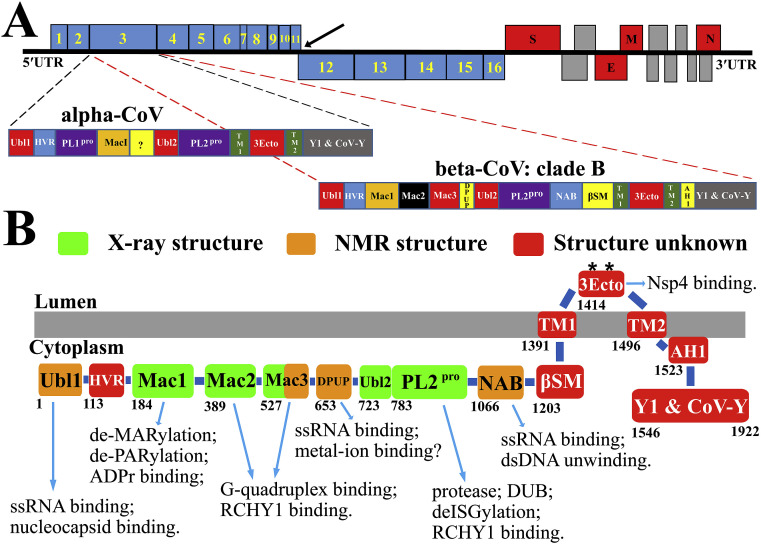
Coronavirus (CoV) is a member of the subfamily Coronavirinae within the family Coronaviridae of the order Nidovirales. It is the enveloped positive-sense single-stranded RNA (+ssRNA) virus with the largest genome of all known RNA viruses thus far (Brian and Baric, 2005, Gorbalenya et al., 2006). The genomes of different CoVs comprise between 26 and 32 kilobases; however, the overall organization of the genomes is similar. The 5′-terminal two thirds of the genome include two open reading frames (ORFs), 1a and 1b, that together encode all non-structural proteins for the formation of the RTC, whereas the 3′-proximal third encodes the structural and accessory proteins (Fig. 1 A; Brian and Baric, 2005). ORF1a encodes polyprotein (pp) 1a containing Nsp1-11, while ORF1a and ORF1b together produce pp1ab containing Nsp1-16 through a (-1) ribosomal frameshift overreading the stop codon of ORF1a (Fig. 1A; Brierley et al., 1989). Coronaviruses are divided into four genara: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Adams and Carstens, 2012). CoVs can infect many species (Fehr and Perlman, 2015); currently, the coronaviruses infecting humans are all from the genera alpha-CoV or beta-CoV. HCoV 229E and HCoV NL63 belong to the former (Tyrrell and Bynoe, 1965, van der Hoek et al., 2004), whereas HCoV OC43, HKU1, SARS-CoV, and MERS-CoV belong to the latter genus (Hamre and Procknow, 1966, Woo et al., 2005, Drosten et al., 2003, Ksiazek et al., 2003, Kuiken et al., 2003, Peiris et al., 2003, Zaki et al., 2012). Furthermore, HCoV OC43 and HKU1 belong to clade A of beta-CoV, while the two highly pathogenic human CoVs, SARS-CoV and MERS-CoV, are from clades B and C, respectively.
Fig. 1.
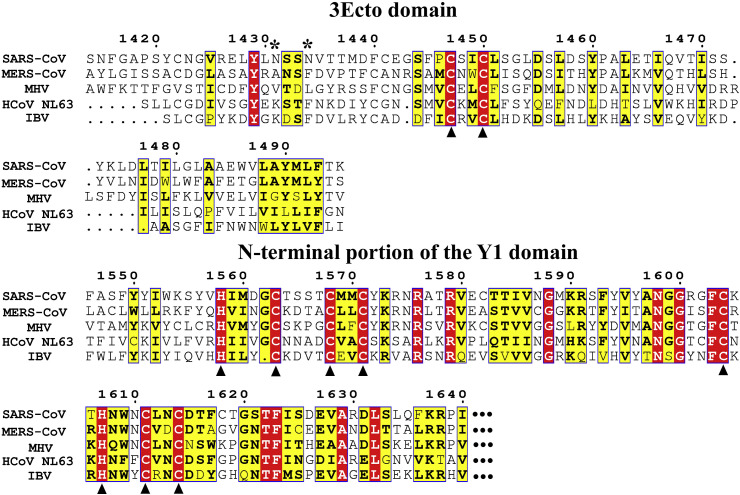
Genome organization of coronaviruses; Nsp3 domains and their functions. (A) The 5′-terminal two thirds of the CoV genome comprise ORF1a and ORF1b. ORF1a encodes the polyprotein 1a (Nsp1-11) while ORF1a plus ORF1b produce the polyprotein 1ab (Nsp1-16) through a ribosomal frameshift overreading the stop codon of ORF1a (indicated by a black arrow). The 3′-proximal third encodes the structural proteins S, E, M, and N as well as accessory proteins. The polyproteins pp1a and pp1ab are processed by the viral proteases PL1pro, PL2pro (both domains of Nsp3), and Mpro (3CLpro, Nsp5). The domain organization of Nsp3 is different in different CoV genera. The Nsp3 of HCoV NL63 as a representative of alpha-CoVs, and of SARS-CoV in clade B of the genus beta-CoV, are zoomed out. The question mark within HCoV-NL63 Nsp3 indicates a region of unknown function and structure. (B) Summary of the functions and domain organization of SARS-CoV Nsp3. Nsp3 is bound to double-membrane vesicles recruited from the endoplasmic reticulum (ER) membrane. The protein passes through this membrane twice, via the two transmembrane regions TM1 and TM2. AH1 is possibly an amphipathic helix attached to the ER membrane, next to TM2. Except for the 3Ecto domain, all other Nsp3 domains are located in the cytosol. All domains with known three-dimensional structures are indicated in light green (X-ray structures) or orange (NMR structures), whereas parts with unknown structure are in red. The best characterized functions of each domain of Nsp3 are shown.*: glycosylation sites in the 3Ecto domain (Asn1431 and Asn1434; Harcourt et al., 2004).
Nsp3 is the largest multi-domain protein produced by coronaviruses (Fig. 1A). It features a somewhat different domain organization in different CoV genera. The individual coronaviruses can possess 10 to 16 domains of which eight domains and two transmembrane regions are conserved, according to a recent bioinformatic analysis (Neuman, 2016). The domain organization of Nsp3 from HCoV NL63 as a representative of alpha-CoVs, and from SARS-CoV in clade B of the genus beta-CoV are displayed in Fig. 1A. Nsp3 is released from pp1a/1ab by the papain-like protease domain(s), which is (are) part of Nsp3 itself (Fig. 1A; Ziebuhr et al., 2000). Nsp3 plays many roles in the viral life cycle (Fig. 1B). It can act as a scaffold protein to interact with itself and to bind other viral Nsps or host proteins (von Brunn et al., 2007, Pan et al., 2008, Imbert et al., 2008, Pfefferle et al., 2011, Ma-Lauer et al., 2016). In particular, Nsp3 is essential for RTC formation (van Hemert et al., 2008, Angelini et al., 2013). The RTC is associated with modified host ER membranes that produce convoluted membranes (CMs) and double-membrane vesicles (DMVs) in SARS-CoV-, MHV (mouse hepatitis virus)- as well as MERS-CoV-infected cells (Snijder et al., 2006, Knoops et al., 2008, Hagemeijer et al., 2011, de Wilde et al., 2013). Nsp3 and Nsp5 were detected on the CMs in SARS-CoV-infected cells by immunogold electron microscopy (Knoops et al., 2008). Co-expression of Nsp3, Nsp4, and Nsp6 can induce DMV formation in SARS-CoV-infected cells but the same result was not observed when Nsp3 lacking its C-terminal third (residues 1319–1922) was co-expressed with Nsp4 and Nsp6 (Angelini et al., 2013). Correspondingly, co-expression of only the C-terminal third of Nsp3 (residues 1256–1922) and Nsp4 induces the occurrence of the zippered ER and membrane curvature in SARS-CoV- or MHV-infected cells, which is likely to enhance DMV formation (Hagemeijer et al., 2014). Above all, Nsp3 is a key component for coronavirus replication; however, many functions of Nsp3 remain to be investigated. In this review, the current knowledge on the structures and functions of the individual Nsp3 domains is summarized and discussed.
2. Ubiquitin-like domain 1 and the Glu-rich acidic region
The ubiquitin-like domain 1 (Ubl1) and the Glu-rich acidic region are located at the N-terminus of Nsp3. These two regions together are also named “Nsp3a” (Neuman et al., 2008). Nsp3a exists in all CoVs in spite of no more than 15% amino-acid sequence identity between the domains in CoVs from different genera.
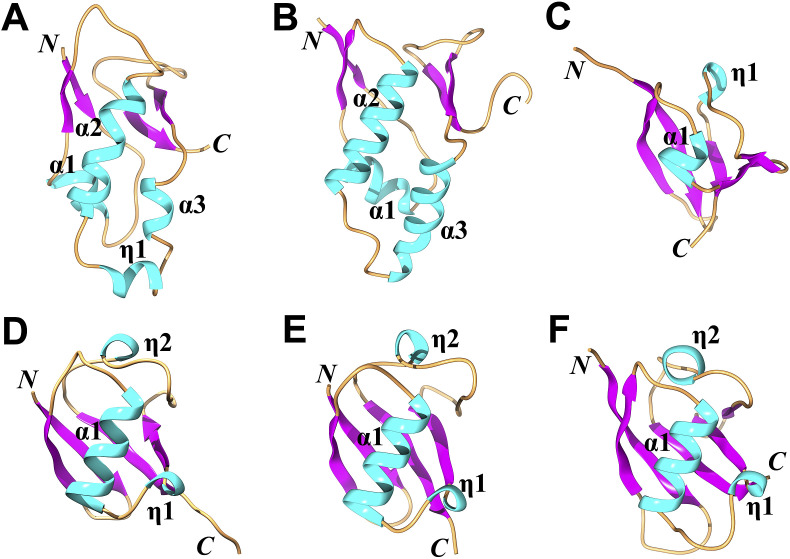
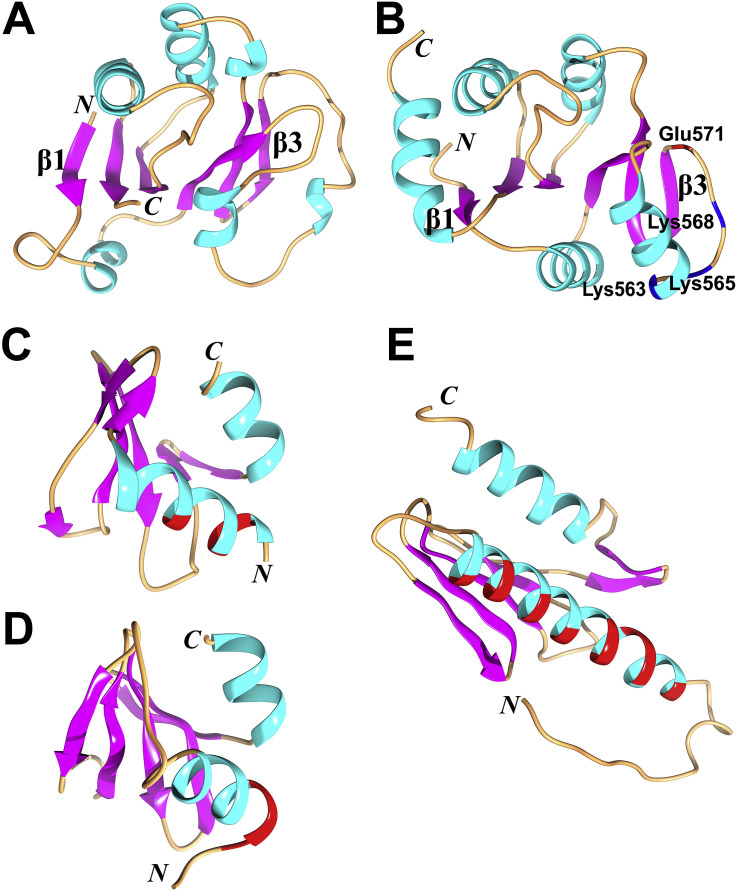
Two Ubl1 structures from betacoronaviruses of different clades have been determined by NMR spectroscopy so far (Table 1 ); one is from SARS-CoV in clade B (Serrano et al., 2007) and the other from MHV in clade A (Keane and Giedroc, 2013). In SARS-CoV, the Ubl1 comprises residues 1–112; the core residues 20–108 form a typical ubiquitin-like fold with secondary-structure elements in the following order: β1−α1−β2−α2−η1−α3−β3−β4 (η: 310 helix; Fig. 2 A; Serrano et al., 2007); residues outside this core are flexible. The well-defined structure of MHV Ubl1 (residues 19–114) with the secondary-structure elements β1−α1−β2−α2−α3−β3−β4 is similar to that of SARS-CoV Ubl1 (Fig. 2B), with a root-mean-square deviation (R.M.S.D.) of 2.8 Å (for 85 out of 95 Cα atoms; Z-score: 7.4) according to the Dali server (Holm and Rosenström, 2010). A structural difference between the two Ubl1 domains is that the two disjoined helices η1-α3 in SARS-CoV Ubl1 are replaced by one long continuous helix (α3) in MHV Ubl1 (Fig. 2A and B).
Table 1.
Structural information on CoV Nsp3 domains and regions.
| Domain/region | Res. no.#/MW% | Method | Coronavirus | Reference |
|---|---|---|---|---|
| Ubl1 | 1-112/12.6 | NMR | SARS-CoV | Serrano et al. (2007) |
| NMR | MHV | Keane and Giedroc (2013) | ||
| Acidic domain (HVR) | 113-183/8.3 | n. d. | SARS-CoV | Serrano et al. (2007) |
| PL1pro *1 | n. a./23.6 | X-ray | TGEV | Wojdyla et al. (2010) |
| Mac1 (X domain) | 184-365/19.5 | X-ray | SARS-CoV | Saikatendu et al. (2005) |
| X-ray | SARS-CoV | Egloff et al. (2006) | ||
| X-ray | HCoV-229E, IBV | Xu et al. (2009) | ||
| X-ray | HCoV-229E, IBV | Piotrowski et al. (2009) | ||
| X-ray | FCoV | Wojdyla et al. (2009) | ||
| X-ray | MERS-CoV | Cho et al. (2016) | ||
| Mac2 (SUD-N) | 389-524/15.2 | X-ray | SARS-CoV*2 | Tan et al. (2009) |
| Mac3 (SUD-M) | 525-652/14.0 | NMR | SARS-CoV | Chatterjee et al. (2009) |
| X-ray | SARS-CoV*2 | Tan et al. (2009) | ||
| NMR | SARS-CoV*3 | Johnson et al. (2010) | ||
| DPUP (SUD-C) | 653-720/7.8 | NMR | SARS-CoV | Johnson et al. (2010) |
| X-ray | MHV*4 | Chen et al. (2015) | ||
| NMR | HKU9 | Hammond et al. (2017) | ||
| Ubl2−PL2pro | 723-1036/35.2 | X-ray | SARS-CoV | Ratia et al. (2006) |
| X-ray | SARS-CoV + human Ub | Chou et al. (2014) | ||
| X-ray | SARS-CoV + human Ub | Ratia et al. (2014) | ||
| X-ray | SARS-CoV + diUb | Békés et al. (2016) | ||
| X-ray | SARS-CoV + hISG15*5 | Daczkowski et al. (2017a) | ||
| X-ray | SARS-CoV + mISG15*6 | Daczkowski et al. (2017a) | ||
| X-ray | MERS-CoV | Lei et al. (2014) | ||
| X-ray | MERS-CoV | Lee et al. (2015) | ||
| X-ray | MERS-CoV + human Ub | Bailey-Elkin et al. (2014) | ||
| X-ray | MERS-CoV + human Ub | Lei and Hilgenfeld (2016) | ||
| X-ray | MERS-CoV + hISG15*5 | Daczkowski et al. (2017b) | ||
| PL2pro | n. a./28.6 | X-ray | MERS-CoV | Clasman et al. (2017) |
| Ubl2−PL2pro | X-ray | IBV | Kong et al. (2015) | |
| Ubl2−PL2pro | X-ray | MHV*4 | Chen et al. (2015) | |
| NAB | 1066-1180/13.0 | NMR | SARS-CoV | Serrano et al. (2009) |
| βSM (G2M) | 1203-1318/12.5 | n. d. | ||
| TM1 | 1391-1413†/2.4 | n. d. | ||
| 3Ecto | 1414-1495/9.0 | n. d. | ||
| TM2 | 1496-1518†/2.7 | n. d. | ||
| AH1 | 1523-1545†/2.7 | n. d. | ||
| Y1 + CoV-Y | 1546-1922/41.9 | n. d. |
#: Nsp3 of the SARS-CoV strain TOR2 (Genbank: AY274119.3); %: molecular mass (kD); n. d.: structure is not determined; *1: absent in SARS-CoV; n. a.: does not apply (residue numbers are only given for SARS-CoV); *2: Mac2−Mac3 structure; *3: Mac3−DPUP structure; *4: DPUP−Ubl2−PL2pro structure; *5: Ubl2−PL2pro−C-terminal Ubl domain of human ISG15 structure; *6: Ubl2−PL2pro−C-terminal Ubl domain of mouse ISG15 structure; †: regions predicted by TMHMM server v. 2.0 (Krogh et al., 2001). TM1 and TM2 are transmembrane regions while AH1 is not (Oostra et al., 2008).
Fig. 2.
Structures (in cartoon view) of the ubiquitin-like domain 1 (Ubl1) and Ubl2 in SARS-CoV, Ubl1 in MHV, as well as their structural homologues. (A) Ubl1 (residues 20–108) of SARS-CoV (PDB entry: 2IDY; Serrano et al., 2007). (B) Ubl1 (19–114) of MHV (PDB entry: 2M0A; Keane and Giedroc, 2013). (C) Ubl2 (residues 1–60) of SARS-CoV (PDB entry: 2FE8; Ratia et al., 2006). (D) human ubiquitin (PDB entry: 1UBQ; Vijay-Kumar et al., 1987). (E) human interferon-stimulated gene 15 (hISG15; PDB entry: 1Z2M; Narasimhan et al., 2005). hISG15 contains two linked ubiquitin-like domains; here, the N-terminal Ubl domain is shown. (F) the Ras-interacting domain of RalGDS (PDB entry: 1LFD; Huang et al., 1998). The N and C termini of all structures are marked. All α and 310 (η) helices are labeled and shown in cyan. β strands are in purple and loops are in brown. This figure and Fig. 3, Fig. 5, as well as 8 were generated by using Chimera (Pettersen et al., 2004).
The known functional roles of Ubl1 in CoVs are related to single-stranded (ssRNA) binding and interacting with the nucleocapsid (N) protein (Fig. 1B; Serrano et al., 2007, Hurst et al., 2010, Hurst et al., 2013). The Ubl1 of SARS-CoV binds ssRNA containing AUA patterns. Surprisingly, many negatively charged regions (such as the 310 helix, η1) show obvious conformational changes in the NMR spectra when RNA is added to the protein solution (Serrano et al., 2007), indicating that RNA binding has long-range effects on the protein conformation. In view of the presence of several AUA repeats in the 5′-untranslated region (UTR) of the SARS-CoV genome, the Ubl1 likely binds to this region.
In MHV, the Ubl1 domain efficiently binds the cognate nucleocapsid (N) protein; thus it seems to be important for virus replication as well as initiation of viral infection. There is a critical relationship between Nsp3 interaction with the N protein and infectivity, as this interaction serves to tether the viral genome to the newly translated RTC at an early stage of coronavirus infection (Hurst et al., 2010, Hurst et al., 2013). Deletion of the Ubl1 core (residues 19–111) of MHV abrogates viral replication (Hurst et al., 2013). The major interface regions of the complex Ubl1−N involve acidic residues of Ubl1 helix α2 and the serine- and arginine-rich region (SR-rich region) of the N protein, as shown by NMR titration experiments (Keane and Giedroc, 2013). However, the acidic residues in helix α2 are not absolutely conserved among different CoVs, implying that the details of the interactions between Ubl1 and N protein will not be the same. In addition, the binding affinity between the bovine coronavirus (BCoV) N (residues 57–216) and MHV Ubl1 is about 260-fold lower compared to MHV N (residues 60–219) and its cognate Ubl1 (Keane and Giedroc, 2013). A structure of the Ubl1−N complex would help understand why non-cognate Ubl1 and N protein bind weakly to each other. Thus far, only a computer docking model of the MHV Ubl1−N complex was reported (Tatar and Tok, 2016). This model proposes that residues of β1, α1, the loop between β1 and α1, β3, and β4 of MHV Ubl1 interact with the N-terminal domain (NTD) as well as the SR-rich region of the N protein. Differently from what was suggested above, most acidic residues of Ubl1 helix α2 do not interact with the SR-rich region of N in the docking model (Tatar and Tok, 2016).
The interaction between the N protein and nucleic acid is essential for CoV genome transcription (Chang et al., 2014). The NTD plus the SR-rich region (residues 60–219) of MHV N play an important role in interacting with transcriptional regulatory sequence (TRS) RNA (Grossoehme et al., 2009). The N−TRS RNA complex prevents the formation of the Ubl1−N complex (Keane and Giedroc, 2013). The competition between N protein binding to either the TRS or the Ubl1 might be connected to the switch between viral transcription and replication. It has been shown that the SR region of N protein can be phosphorylated (Peng et al., 2008). Each of two phosphomimetic substitutions of serine residues predicted to be phosphorylated (S207D and S218D) in the SR region of MHV N decreases the binding affinity to Ubl1 by about 3-fold, compared to wild-type N (Keane and Giedroc, 2013).
The overall structure of the SARS-CoV Ubl1 domain is similar to human ubiquitin (Ub) and that of each of the two ubiquitin-like domains of human or mouse interferon-stimulated gene 15 (hISG15 or mISG15) (Fig. 2D and E; Vijay-Kumar et al., 1987, Narasimhan et al., 2005, Daczkowski et al., 2017a). In human Ub as well as in the ISG15s, only a short 310 helix is found at the position of η1-α3 or α3 in Ubl1 of SARS-CoV or MHV (Fig. 2D and E). Ub and ISG15 are important for innate antiviral immunity (Heaton et al., 2016, Morales and Lenschow, 2013); therefore, viruses tend to not only inhibit the conjugation of Ub or ISG15 to targets but also remove Ub or ISG15 from ubiquitinated or ISGylated proteins, respectively (Yuan and Krug, 2001, Bakshi et al., 2013, Yang et al., 2014). Thus, in CoVs, one or two papain-like protease (PLpro) domain(s) within Nsp3 possess deubiquitinating (DUB) and deISGylating activities (see below; for a recent review on the role of viral proteases in counteracting the host-cell's innate immune system, see Lei and Hilgenfeld (2017)). Interestingly, two ubiquitin-like domains (Ubl1 and Ubl2) exist in all CoVs (see below; Neuman, 2016). Considering that ubiquitin-like modules are often involved in protein−protein interactions to regulate various biological processes (Hochstrasser, 2009), such as the MHV Ubl1−N interaction mentioned above, a novel possible function of Ub-like domains in CoVs might be the interaction with target proteins of Ub (or ISG15) by mimicking the shape of these two molecules. The purpose of such mimicry could be to somehow interfere with pathways involving ubiquitinated or ISGylated host targets, thereby leading to disruption of host anti-viral signal transduction or protein degradation.
The Ubl1 of SARS-CoV is also similar to the Ras-interacting domain (RID) of RalGDS (Ral guanine nucleotide dissociation stimulator; Fig. 2F; Serrano et al., 2007). Ras regulates cell-cycle progression via binding to the RID of Ras-interacting proteins (Hofer et al., 1994, Huang et al., 1998, Coleman et al., 2004). By mimicking the RID, the Ubl1 might interrupt the interactions between Ras and its effectors, thus affecting the cell cycle to support virus replication. In agreement with this, it is known that both MHV and SARS-CoV induce cell-cycle arrest in the G0/G1 phase (Chen and Makino, 2004, Yuan et al., 2005).
Following the Ubl1, the second subdomain of Nsp3a in CoVs is the Glu-rich acidic region. It comprises residues 113–183 of SARS-CoV Nsp3, with more than 35% Glu and 10% Asp (Serrano et al., 2007). Because of the non-conserved amino-acid sequence, this region is also designated as “hypervariable region (HVR)” (Neuman, 2016). The HVR region is intrinsically disordered in SARS-CoV and in MHV (Serrano et al., 2007, Keane and Giedroc, 2013) and does not affect the conformation of the globular Ubl1 domain in SARS-CoV (Serrano et al., 2007). Currently, the function of HVR in CoVs is unknown. Glu/Asp-rich proteins are often involved in many biological roles, such as DNA/RNA mimicry, metal-ion binding, and protein−protein interactions (Chou and Wang, 2015). The Ubl1+HVR region has been demonstrated via a yeast-two-hybrid (Y2H) assay to interact with SARS-CoV Nsp6, whereas a GST pull-down study identified Nsp8, Nsp9, and NAB–βSM–TM1 of Nsp3 (NAB: nucleic-acid binding domain; βSM: betacoronavirus-specific marker; TM1: transmembrane region 1; see below) as binding partners (Imbert et al., 2008). Does the HVR play any role in these protein−protein interactions? This question is yet to be answered. Furthermore, the acidic region is dispensable for MHV replication (Hurst et al., 2013). On the other hand, this region does exist in all CoVs. It is conceivable that it may have regulatory rather than essential roles in the coronavirus replication process. However, the exact role(s) of the acidic region in CoVs should be further investigated.
3. Papain-like protease 1 domain
The papain-like protease domain(s) is/are responsible for releasing Nsp1, Nsp2, and Nsp3 from the N-terminal region of polyproteins 1a/1ab in CoVs (Harcourt et al., 2004, Barretto et al., 2005). The papain-like protease 1 domain (PL1pro) follows the HVR region (see Fig. 1A) in the alpha-CoVs and in clade A of beta-CoVs (Graham and Denison, 2006, Ziebuhr et al., 2001, Chen et al., 2007, Wojdyla et al., 2010, Neuman, 2016). Interestingly, the PL1pro is not complete in the gamma-CoV infectious bronchitis virus (IBV; Ziebuhr et al., 2001) and in Hipposideros pratti bat CoV, a virus relating to clade B of the beta-CoVs (Genbank code NC_025217.1; Neuman, 2016). In these latter viruses, some parts (such as the zinc-finger motif; see below) and the residues of the catalytic triad of the PL1pros are missing. Furthermore, the PL1pro is totally absent in beta-CoV clades B, C, and D as well as in delta-CoVs. Both the two highly human-pathogenic SARS-CoV (Fig. 1A) and MERS-CoV thus do not have a PL1pro domain; they only possess the other papain-like protease, the PL2pro domain that is conserved in all coronaviruses (see below). It is still not clear why certain CoVs encode two PLpros.
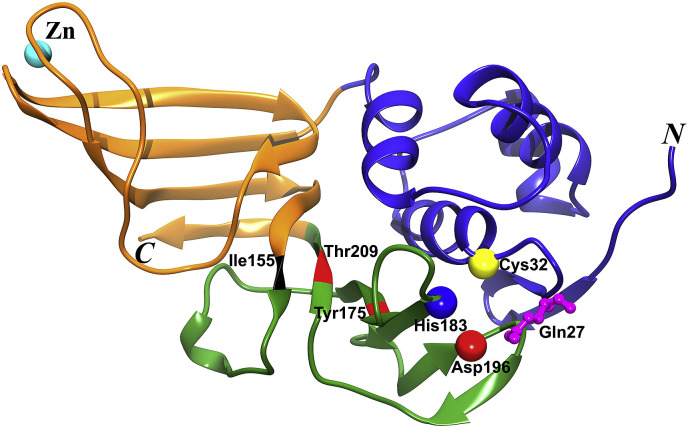
Thus far, only one structure of a PL1pro domain has been determined, that from the alpha-CoV Transmissible Gastroenteritis Virus (TGEV) (Table 1; Wojdyla et al., 2010). The PL1pro resembles an extended right-hand scaffold with thumb, palm, and fingers subdomains (Fig. 3 ). It contains a zinc finger in the fingers subdomain as well as a catalytic triad, Cys32−His183−Asp196. A canonical oxyanion hole as known from papain (Ménard et al., 1991) is present in TGEV PL1pro, with the main-chain amide of the catalytic cysteine residue and the side-chain of a glutamine residue (Gln27) 5 residues N-terminal to the cysteine contributing to the stabilization of the oxyanion transition state of peptide hydrolysis (Fig. 3; Wojdyla et al., 2010). The fold of the PL1pro is similar to that of the PL2pro of SARS-CoV (see below; R.M.S.D. 3.1 Å, for 202 out of 211 Cα atoms; Dali Z-score = 18.4) and MERS-CoV (R.M.S.D. 3.1 Å, for 198 out of 211 Cα atoms; Z-score = 18.7) as well as to several human ubiquitin-specific proteases (USPs, such as USP 2, 7, 14, 21 etc., Z-scores from 11.2 to 12.6) (Ratia et al., 2006, Wojdyla et al., 2010, Lei et al., 2014). The biggest difference between these PLpro domains is found in the zinc-finger regions, which are obviously flexible (Wojdyla et al., 2010, Lei et al., 2014). Furthermore, the electrostatic surface potential of TGEV PL1pro features two negative patches which are absent in SARS-CoV PL2pro (Wojdyla et al., 2010). One patch is located at the opposite side of the active site, between the thumb and palm subdomains, and the other is near the active-site groove and the surrounding region, between the thumb and fingers subdomains (Wojdyla et al., 2010). The latter patch is related to the substrate binding and specificity of TGEV PL1pro (Wojdyla et al., 2010).
Fig. 3.
Crystal structure of the papain-like protease domain 1 (PL1pro) of TGEV. Cartoon view of the overall structure (PDB entry: 3MP2; Wojdyla et al., 2010). The thumb, fingers, and palm subdomains are shown in blue, brown, and green, respectively. The Cα atoms of the catalytic triad residues (Cys32−His183−Asp196) are displayed as yellow, blue, and red spheres. Residue Gln27 contributing to the oxyanion hole is shown in ball & stick style. Ile155, Thr209, and Tyr175 forming the S4 pocket are labeled; Ile155 is in black and the latter two are in red. The N and C termini of the PL1pro are indicated.
The PL1pro of TGEV has been demonstrated to process the cleavage site Nsp2↓3 (↓: cleavage site) and to exhibit DUB activity to remove ubiquitin from Lys48-/Lys63-linked Ub chains in vitro (Putics et al., 2006, Wojdyla et al., 2010). The P4−P1 residues of the cleavage site between Nsp2 and 3 are Lys−Met−Gly−Gly in TGEV (Table 2 ), while the last four residues of ubiquitin are Leu−Arg−Gly−Gly. Therefore, the S4 pocket of TGEV PL1pro should be able to accommodate residues as different as Lys and Leu. In contrast, the P4−P1 residues in the polyprotein substrates of PL2pro are Leu−Xaa−Gly−Gly (Xaa is Asn or Lys) in SARS-CoV (Harcourt et al., 2004, Barretto et al., 2005). P4 is the same residue as in the ubiquitin substrate; thus, the corresponding pocket of SARS-CoV PL2pro is tailor-made for leucine. Residues Ile155, Tyr175, and Thr209 form the S4 subsite in TGEV PL1pro (Fig. 3; Wojdyla et al., 2010), whereas the corresponding residues in SARS-CoV PL2pro are Pro249, Tyr265, and Thr302 (Ratia et al., 2006). When superimposing the structures, Wojdyla et al. (2010) found that the Cα atom of Ile155 of TGEV PL1pro is 3 Å away from the Cα atom of the corresponding Pro249 in SARS-CoV PL2pro, thereby creating a larger S4 pocket in TGEV PL1pro, so that it can bind lysine, in addition to leucine.
Table 2.
Cleavage sites of PL1pro and PL2pro in CoVs and the P5-P2′ residues for each cleavage site.
| Nsp1↓2 | Nsp2↓3 | Nsp3↓4 | Reference | |
|---|---|---|---|---|
| TGEV | RTGRG↓AI | NKMGG↓GD | PKSGS↓GF | Putics et al. (2006) |
| n. d. | PL1pro | n. d. | ||
| HCoV NL63 | GHGAG↓SV | TKLAG↓GK | AKQGA↓GF | Chen et al. (2007) |
| PL1pro | PL2pro | PL2pro | ||
| HCoV 229E | KRGGG↓NV | TKAAG↓GK | AKQGA↓GD | Ziebuhr et al. (2007) |
| PL1pro > PL2pro | PL1pro < PL2pro | n. d. | ||
| MHV | KGYRG↓VK | RFPCA↓GK | SLKGG↓AV | Bonilla et al. (1997) |
| PL1pro | PL1pro | PL2pro | Kanjanahaluethai and Baker (2000) | |
| SARS-CoV#1 | ELNGG↓AV | RLKGG↓AP | SLKGG↓KI | Harcourt et al. (2004) |
| PL2pro | PL2pro | PL2pro | ||
| MERS-CoV#1 | KLIGG↓DV | RLKGG↓AP | KIVGG↓AP | Yang et al. (2014) |
| PL2pro | PL2pro | PL2pro | ||
| IBV#2 | / | VCKAG↓GK | EKKAG↓GI | Lim et al. (2000) |
| / | PL2pro | PL2pro |
↓: cleavage site; n. d., not determined; #1: absence of PL1pro; #2: partial presence of PL1pro; /: absence of the cleavage site.
As mentioned above, for reasons unknown so far, many CoVs contain two PLpros. Both PL1pro and PL2pro are involved in releasing Nsp1, Nsp2, and Nsp3 in these CoVs. However, the two PLpros in different CoVs show varying substrate specificity. The PL1pro of MHV cleaves Nsp1↓2 and Nsp2↓3, while the PL2pro cleaves Nsp3↓4 (Table 2; Bonilla et al., 1997, Kanjanahaluethai and Baker, 2000). Human coronavirus NL63 (HCoV-NL63) PL1pro processes Nsp1↓2 while the PL2pro processes the other two cleavage sites, Nsp2↓3 and Nsp3↓4 (Table 2; Chen et al., 2007). Both PL1pro and PL2pro of HCoV 229E can cleave Nsp1↓2 and 2↓3 (Table 2); however, the PL1pro is more efficient in cleaving Nsp1↓2 while the PL2pro is more efficient with respect to the latter site (Ziebuhr et al., 2007). Some viruses, such as SARS-CoV, MERS-CoV, and IBV, comprise only one functional PL2pro to process all three cleavage sites (Table 2). The residues (P5−P2′) of the three cleavage sites are diversified in MHV, HCoV NL63, and HCoV 229E, although the P1 is conserved as a small residue (Gly or Ala) (Table 2). In contrast, the P1 and P2 residues (Gly−Gly or Ala−Gly) are absolutely identical in all the cleavage sites of SARS-CoV, MERS-CoV, and IBV; furthermore, the P4−P1 residues are − to a certain extent − conserved in each of these three viruses (Table 2). Therefore, the presence of two PLpros with slightly different substrate specificity in some CoVs may be required to cleave native substrates that deviate from the uniform ones processed by SARS-CoV, MERS-CoV, or IBV PLpros. Unfortunately, studies on the details of recognition of different substrates by PL1pro and PL2pro are hampered by the fact that no crystal structures of the two enzymes from the same virus are available.
4. Macrodomains and the "Domain Preceding Ubl2 and PL2pro (DPUP)"
4.1. Macrodomain I (Mac1, X domain)
A conserved macrodomain (also called “X domain”, Nsp3b) follows the HVR or the PL1pro domain in all coronaviruses (Fig. 1A; Gorbalenya et al., 1991, Neuman et al., 2008, Neuman, 2016). Macrodomains widely exist in bacteria, archaea, and eukaryotes (Han et al., 2011). In addition, these conserved domains are also present in several positive-sense ssRNA (+ssRNA) viruses of the families Hepeviridae, Togaviridae, and Coronaviridae, such as hepatitis E virus (HEV), alphavirus, rubella virus, and all coronaviruses (Koonin et al., 1992, Snijder et al., 2003). Our group has shown that the X domain (Mac1) is dispensable for RNA replication in the context of a SARS-CoV replicon (Kusov et al., 2015). Recently, evidence accumulated showing that the X domain plays a role in counteracting the host innate immune response (Eriksson et al., 2008, Kuri et al., 2011, Fehr et al., 2015, Fehr et al., 2016).
The first crystal structure of an Nsp3 domain of any coronavirus was the unliganded X domain of SARS-CoV (Table 1; Saikatendu et al., 2005). A little later, the structure of the SARS-CoV X domain in complex with ADP-ribose (ADPr) was determined (Table 1; Egloff et al., 2006). Subsequently, structures of the unliganded X domain and/or its complex with ADPr from HCoV 229E, IBV, HCoV NL63, Feline CoV (FCoV), and MERS-CoV were reported (Table 1; Piotrowski et al., 2009, Xu et al., 2009, Wojdyla et al., 2009, Cho et al., 2016). All structures show that the X domain adopts a conserved three-layered α/β/α sandwich fold (Fig. 4 ). The domain with this fold is called a macrodomain because of its similarity to the extra domain in the MacroH2A variant of human histone 2A (Pehrson and Fried, 1992, Saikatendu et al., 2005). Typically, the X domain includes a central β sheet with seven β strands in the order β1−β2−β7−β6−β3−β5−β4, with β1 and β4 being antiparallel to the rest (Fig. 4). Only the X domain of IBV is an exception, since it lacks the first strand, β1 (Piotrowski et al., 2009, Xu et al., 2009). Six helices are located on the two sides of this β sheet, with helices α1, α2, and α3 on one side and α4, α5, and α6 on the other (Fig. 4).
Fig. 4.
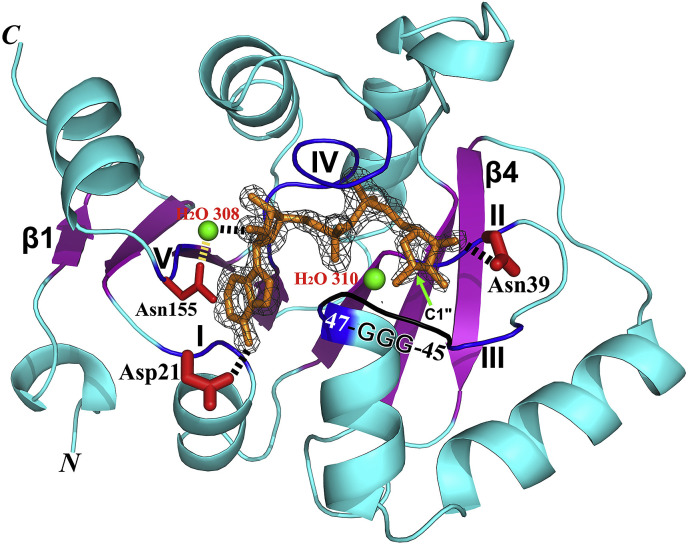
Structure of the MERS-CoV macrodomain I (Mac1, X domain) in complex with ADP-ribose (ADPr) (PDB entry: 5HOL). The protein features an α/β/α sandwich fold. The central β sheet with the strand order β1−β2−β7−β6−β3−β5−β4 is shown in purple, β1 and β4 are labeled. An Fo-Fc omit difference map of ADPr is shown in black (contoured at 4.0 σ). The ADPr itself is displayed as brown sticks. The five regions (blue) relating to ADPr binding are marked by Roman numbers I – V. Fixing the two ends of the ADPr, Asp21 and Asn39 are displayed by thicker red sticks. The O2′ of ADPr forms a hydrogen bond with a water molecule (H2O 308; green sphere) being stabilized by the side-chain of Asn155. The “GGG” triple-glycine motif is displayed in black. H2O 310 (green sphere) corresponds to a water molecule that has been proposed to mediate a nucleophilic attack onto the C1″ atom of the ADPr in the de-MARylation reaction catalyzed by the VEEV X domain (Li et al., 2016a). The N and C termini of the X domain are marked. This figure and Fig. 6 were prepared using Pymol (Schrödinger; http://www.pymol.org/).
One function of the conserved macrodomain is the binding of ADP-ribose or poly(ADP-ribose) (Han et al., 2011). The binding characteristics are the same in most X domains of coronaviruses (Egloff et al., 2006, Xu et al., 2009, Wojdyla et al., 2009, Cho et al., 2016). Like Cho et al. (2016), we have determined the crystal structure of the MERS-CoV X domain in complex with ADP-ribose (ADPr) (PDB entry: 5HOL; Fig. 4). Our structure and the ADPr-binding pattern are almost identical to the structure (PDB entry: 5DUS) described by Cho et al. (2016) and the structure of the SARS-CoV X domain in complex with ADPr (PDB entry: 2FAV; Egloff et al., 2006). The R.M.S.D. are 0.4 Å (for 165 out of 165 Cα atoms; Z-score: 34.2) and 1.2 Å (for 163 out of 171 Cα atoms; Z-score: 28.3), respectively, according to the Dali server (Holm and Rosenström, 2010). Here, we describe the structure of the MERS-CoV X domain in complex with ADPr from our own laboratory as an example (Fig. 4). The ADPr is located in a cleft at the top of the central β sheet (β7−β6−β3−β5). Five stretches of amino-acid residues are mainly involved in the binding of ADPr: I, Gly20−Ala22; II, Ala37−Asn39; III, Lys43−Ala49 (including a “45-GGG-47” triple-glycine motif); IV, Pro124−Phe131; V, Val153−Asn155 (Fig. 4). The adenine base is in contact with regions I and V. In particular, the side-chain of Asp21 accepts a hydrogen bond from the exocyclic NH2 group in position 6 of the adenine, thereby fixing the orientation of the base. This Asp residue is conserved in macrodomains from bacteria, archea, and eukaryotes (Saikatendu et al., 2005, Egloff et al., 2006). When the corresponding Asp20 in the macrodomain protein AF1521 of Archeoglobus fulgidus was replaced by alanine, the ADP-ribose binding affinity was reduced almost 90-fold (Karras et al., 2005). The central ribose moiety is located between regions IV and V. The O2′ of ADPr forms a hydrogen bond with a water molecule (H2O 308) that is stabilized by the side-chain of Asn155 (region V). The two phosphate groups accept a total of four hydrogen bonds from Ile48 (region III) and Gly129, Ile130 as well as Phe131 (region IV). The distal ribose is in contact with regions II and III; The O1″ and O2″ of this ribose form hydrogen bonds with the amides of Gly47 and Gly45 (region III), respectively. The O3″ forms a hydrogen bond with the side-chain amide of Asn39 (region II). Thus, Asp21 and Asn39 appear to fix the two ends of the ADP-ribose, thereby stabilizing its binding to the cleft (Fig. 4). Surprisingly, the orientation of the corresponding Asp in the HCoV-229E X domain is different; this Asp does not directly bind ADP-ribose but is in contact with its neighboring residue Thr-22, and not with the N6 atom of adenine (Piotrowski et al., 2009, Xu et al., 2009). This difference could explain why the binding affinity between the X domain of HCoV 229E and ADPr is about 10-fold lower than that of the MERS-CoV homologue (Piotrowski et al., 2009, Cho et al., 2016). Interestingly, the X domain from IBV strain M41 but not of IBV strain Beaudette can bind ADPr (Xu et al., 2009, Piotrowski et al., 2009). The important “Gly−Gly−Gly” motif of the M41 X domain, involved in binding the distal ribose, is mutated to “Gly−Ser−Gly” in the Beaudette virus, thus preventing ADPr interaction with the X domain (Piotrowski et al., 2009). The virulence of IBV strain Beaudette is attenuated compared to that of IBV strain M41 (Geilhausen et al., 1973). It is an interesting hypothesis that the loss of the ability to bind ADPr may be one of the reasons for the lower pathogenicity of the former IBV.
Macrodomains of some CoVs have been shown to exhibit a weak ADP-ribose-1″-phosphate phosphatase (ADRP) activity in vitro (kcat ≈ 5 - 20 min− 1; Saikatendu et al., 2005, Egloff et al., 2006, Putics et al., 2006). The residue Asn41 of SARS-CoV (corresponding to the Asn39 in MERS-CoV mentioned above) is essential for ADRP activity (Egloff et al., 2006). However, the ADRP activity is dispensable for HCoV-229E replication in cell culture (Putics et al., 2005). On the other hand, when the ADRP activity of the HCoV-229E or that of the SARS-CoV X domain is inactivated through replacement of the Asn mentioned above by Ala, mutant viruses exhibit increased interferon α (IFN-α) sensitivity (Kuri et al., 2011). Interestingly, the corresponding mutants in MHV (strains A59 and JHM) and a mouse-adapted SARS-CoV do not show an increased IFN-β sensitivity (Eriksson et al., 2008, Fehr et al., 2015, Fehr et al., 2016).
Fehr et al. (2016) confirmed that the wild-type X domain of SARS-CoV inhibits the expression of innate-immunity genes (such as IFN-β, interleukin 6 (IL-6)) in vitro and thereby blocks the host immune response. At variance with this, Eriksson et al. (2008) and Fehr et al. (2015) reported that the Asn-to-Ala mutation in the MHV (strains A59 and JHM, resp.) X domain reduces the production of inflammatory cytokines (e.g., IL-6) in vitro and in vivo. Eriksson et al. (2008) hypothesized that the X domain aggravates MHV-induced severe liver pathology, likely by inducing the expression of inflammatory cytokines. These results suggest that the main function of the X domain may differ in different CoVs. On the other hand, the expression level of type-I IFN (α or β) is increased in cells infected with SARS-CoV or MHV carrying the Asn-to-Ala mutation in the X domain (Eriksson et al., 2008, Kuri et al., 2011, Fehr et al., 2016). This indicates that suppression of innate immunity by the X domain may be a feature conserved across the coronaviruses.
Recently, it was demonstrated that macrodomains from several +ssRNA viruses (such as HEV, SARS-CoV, HCoV 229E, Venezuelan equine encephalitis virus (VEEV), and Chikungunya virus (CHIKV)) act as hydrolases removing mono- and/or poly(ADP-ribose) from mono- or poly(ADP-ribosyl)ated proteins, activities designated as de-mono-ADP-ribosylation (de-MARylation) and de-poly-ADP-ribosylation (de-PARylation), respectively (Li et al., 2016a, Fehr et al., 2016, Eckei et al., 2017, McPherson et al., 2017). The weak ADRP activity described for the X domain in the literature is most probably just a non-physiological side reaction of de-MARylation and/or de-PARylation.
The ADP-ribosylation (MARylation or PARylation) of proteins is a reversible posttranslational modification involved in various cellular processes (Aravind et al., 2015, Liu and Yu, 2015). Poly(ADP-ribose) polymerases (PARPs, also named ARTDs, ADP-ribosyltransferases diphtheria toxin-like) are responsible for transferring mono- or poly(ADP-ribose) to target proteins (Liu and Yu, 2015). For example, PARP7 (ARTD14), PARP10 (ARTD10), PARP12 (ARTD12), and PARP14 (ARTD8) add mono-ADPr to other proteins and themselves (Bütepage et al., 2015), while PARP1 (ARTD1) and PARP2 (ARTD2) add poly-(ADPr)s (Gibson and Kraus, 2012). Various amino-acid residues have been identified as acceptor sites for ADP-ribosylation; this still seems to be a matter of some debate. Arg and Ser have certainly been shown to accept ADPr(s) (Laing et al., 2011, Leidecker et al., 2016), but the acidic residues are also thought to be important sites of ADP-ribosylation (Feijs et al., 2013). PARP7, 10, and 12 can act as type-I IFN-stimulated genes (ISGs) and inhibit VEEV replication (Atasheva et al., 2014). Also, Verheugd et al. (2013) reported that PARP10 can block the NF-κB pathway via MARylation of NEMO (“NF-κB essential modulator”). Moreover, the mRNA and protein synthesis of PARP14 (ARTD8) and PARP10 are stimulated by IFN-α in vivo (Eckei et al., 2017). Therefore, some PARPs play a role in the host immune defense. Recently, it has been demonstrated that the X domains of SARS-CoV and HCoV 229E possess the ability to de-MARylate the ADP-ribosylated PARP10 catalytic domain in vitro (Fehr et al., 2016, Li et al., 2016a). However, the relationship between the de-MARylation function of viral macrodomains and their anti-innate immunity activity is still unclear. The de-MARylation activity is a common feature of the X domain (i.e., the first of the macrodomains if there is more than one) of all investigated macrodomain-encoding viruses (Li et al., 2016a, Eckei et al., 2017). Interestingly, the macrodomains of VEEV and SARS-CoV can also remove the entire PAR chain from PARylated PARP5a (ARTD5), PARP1, and PARP3 (ARTD3), without releasing free monomeric ADPr (Li et al., 2016a). Therefore, the macrodomains of these two viruses hydrolyze the amino acid−ADPr ester bond but not ribose−ribosyl glycosidic bonds in PAR chains. A similar observation was also made for the macrodomain of CHIKV, although the de-PARylation of PARylated PARP1 was weak (Eckei et al., 2017). Currently it is unknown whether the de-PARylation activity of macrodomains plays any role in the coronavirus life cycle.
The conserved Asn42 residue, the triple-glycine 48-GGG-50 motif, and Gly123 of the HEV macrodomain (corresponding to Asn39, 45-GGG-47, and Gly129 of the MERS-CoV X domain mentioned above) are essential for the de-MARylation activity (Li et al., 2016a). This is not surprising because they are involved in binding ADP-ribose. A putative mechanism for the de-MARylation activity of the VEEV macrodomain has been proposed (Li et al., 2016a). It is assumed that a water molecule performs a nucleophilic attack onto the C1″ atom of the mono(ADP-ribose). An equivalent water molecule (H2O 310) also exists in our structure of the MERS-CoV X domain−ADPr complex (Fig. 4).
Interestingly, the neighboring helicase domain of HEV can increase the de-PARylation activity of the macrodomain by about 11-fold but not the de-MARylation activity, perhaps because the helicase can support binding of the PAR chain (Li et al., 2016a). This observation raises the question whether a similar phenomenon exists in CoVs? Should the neighboring domains indeed have an influence on the de-MARylation/de-PARylation activities of the CoV X domain, this effect should differ between the various viruses, as there is little conservation of the neighboring regions. In addition, other CoV Nsps have ben demonstrated to interact with the X domain. Using a GST pull-down assay, the X domain of SARS-CoV has been shown to bind the RNA-dependent RNA polymerase, Nsp12 (Imbert et al., 2008). If this interaction does exist in the virus life cycle, is it possible that the two proteins affect the enzymatic activity of each other? Although many three-dimensional structures of CoV macrodomains have been determined, more efforts should be made to study the biological functions of this domain.
4.2. Macrodomains II and III, and the DPUP (SUD-N, SUD-M, SUD-C)
Within Nsp3, a non-conserved region follows the X domain (or Mac 1). When the first SARS-CoV genome sequences were analyzed, this region was recognized as a unique domain only existing in SARS-CoV and therefore called “SARS-unique domain” (SUD) (Snijder et al., 2003). An alternative name is “Nsp3c” (Neuman et al., 2008). The three-dimensional structure of this region has been determined by X-ray crystallography and NMR spectroscopy (Table 1; Tan et al., 2009, Chatterjee et al., 2009, Johnson et al., 2010). This region includes three distinct subdomains: two macrodomains and one frataxin-like fold (Fig. 5 A–C). The three subdomains were named SUD-N, SUD-M, and SUD-C, indicating the N-terminal, the middle, and the C-terminal region of SUD, respectively. A region corresponding to parts of SUD was found to exist in other coronaviruses, mostly of clades B, C, and D of the genus Betacoronavirus (Neuman, 2016). For example, domains similar to SUD-M and SUD-C (but not SUD-N) are also encoded by the MERS-CoV genome (Kusov et al., 2015, Ma-Lauer et al., 2016). Thus, it is no longer appropriate to call this domain “SARS-unique”. Recently, the Nsp3 of MHV was shown by X-ray crystallography to contain a SUD-C-like fold (Chen et al., 2015). These authors renamed this region into “Domain Preceding Ubl2 and PL2pro” (DPUP). In this review, we follow the nomenclature proposed by Chen et al. (2015) and Neuman (2016), and use the designations macrodomain II (Mac2), macrodomain III (Mac3), and Domain Preceding Ubl2 and PL2pro (DPUP) for SUD-N, SUD-M, and SUD-C, respectively.
Fig. 5.
Structures (in cartoon style) of the macrodomains II (Mac2) and III (Mac3), of the Domain Preceding Ubl2 and PL2pro (DPUP) of SARS-CoV and MHV, as well as of the frataxin-like fold protein Yfh1. (A) and (B) Mac2 and Mac3 (PDB entry: 2W2G; Tan et al., 2009). Both domains possess the α/β/α sandwich fold. The central six β strands in the order β1−β6−β5−β2−β4−β3 are displayed in purple. A predominantly positively charged surface patch (Lys563+Lys565+Lys568+Glu571; Nsp3 numbering) of Mac3 being involved in binding oligo(G) (Kusov et al., 2015) is labeled. (C) The SARS-CoV DPUP NMR structure (PDB entry: 2KQW; Johnson et al., 2010). (D) The MHV DPUP X-ray crystal structure (PDB entry: 4YPT; Chen et al., 2015). (E) Structure of the yeast frataxin-like protein Yfh1, as determined by NMR spectroscopy (PDB entry: 2GA5; He et al., 2004). All structures shown in (C), (D), and (E) display the typical frataxin-like fold. Two α helices located at the N- and C- terminal of each structure form one plane and the β sheet forms the other plane. The negatively charged residues (Asp or Glu) in the first α helix (α1) are shown in red (in (C), (D), and (E)); they are possibly involved in binding metal ions. The N and C termini of all structures are marked.
Mac2 (SUD-N) has been shown to be dispensable for the SARS-CoV replication/transcription complex within the context of a SARS-CoV replicon, but surprisingly, Mac3 (SUD-M) is essential, even though it is not conserved throughout the coronaviruses (Kusov et al., 2015). Mac2 and Mac3 each display a typical α/β/α macrodomain fold (Fig. 5A and B). The central β sheet with six β strands in the order β1−β6−β5−β2−β4−β3 is flanked by two (or three) helices on either side. Only the last strand, β3, is antiparallel to the other strands. Interestingly, Mac2 and Mac3 have the same number of β strands in the central β sheet as the X domain of IBV (see above for X domain of IBV). The R.M.S.D. values are 2.5 Å - 2.6 Å (for 119/171 Cα atoms) between Mac2, Mac3, and the X domain of SARS-CoV, according to the Dali server (Holm and Rosenström, 2010). The corresponding values are 2.6 Å - 2.7 Å (for 120/165 Cα atoms) when comparing SARS-CoV Mac2 and Mac3 with the X domain of IBV. Although the X-domain and Mac2/3 share the same fold, the sequence identity among them is only about 11% (Tan et al., 2009). All the residues important for binding ADP-ribose and for de-MARylation/de-PARylation activity (such as the Asn residue and the “GGG” triple-glycine motif interacting with the distal ribose, as mentioned above) are not conserved in Mac2/3; therefore Mac2/3 cannot bind ADP-ribose (Tan et al., 2009, Chatterjee et al., 2009).
Currently, most known functions of Mac2/3 are connected with RNA binding. Mac2-3 (SUD-NM) preferentially binds oligo(G), which can form G-quadruplexes; as expected for these structural modules, the binding affinity is enhanced by potassium ions (Tan et al., 2007, Tan et al., 2009). According to a mutational study, two positively charged lysine-patches of Mac2 are involved in oligo(G) binding, i.e. Lys476+Lys477 (in the loop between α3 and β5; residue numbering starts at N-terminus of Nsp3) and Lys505+Lys506 (at the end of α4), while the residues Lys563+Lys565+Lys568 (+Glu571) of Mac3 (located between α2 and β3) are absolutely essential for binding (Fig. 5B; Tan et al., 2009). Moreover, working with the SARS-CoV replicon, our laboratory has shown that mutation of the same lysine patch of Mac3 in the context of the replicon completely abolished SARS-CoV replication, indicating that binding of G-quadruplex RNA could be an essential element of RTC activity (Kusov et al., 2015). Also, Mac3 can bind (GGGA)2 and (GGGA)5 as well as (GGGA)2GG (Johnson et al., 2010). In contrast, Mac3−DPUP (SUD-MC; DPUP: SUD-C, see below) only binds (GGGA)2GG but not (GGGA)2 or (GGGA)5. A 3′-terminal G nucleotide is apparently important for binding to Mac3−DPUP (Johnson et al., 2010). These data indicate that the DPUP subdomain may fine-tune the specificity of RNA binding by Mac3 (Johnson et al., 2010).
The SARS-CoV genome contains three G6-stretches and two G5-stretches (Tan et al., 2009, Johnson et al., 2010), but none of them is conserved in all SARS-CoV strains. However, two GGGAGGGUAGG nucleotide segments, located in the Nsp2 and Nsp12 coding sequences, are highly conserved in various SARS-CoV strains (Johnson et al., 2010). These two nucleotide segments differ by only one base from the sequence favored by Mac3−DPUP, (GGGA)2GG. Johnson et al. (2010) therefore proposed that these two sequences could be potential physiological substrates of Mac3−DPUP. Besides specific elements in the genome of SARS-CoV, Mac2-3 might bind G-rich stretches in host mRNAs. In fact, Mac2-3 prefers to bind longer G-stretches, such as (G)10 to (G)14 (Tan et al., 2007). Such long G-stretches exist in several 3′ non-translated regions of host mRNAs, such as the NF-κB signaling pathway-related protein TAB3 mRNA and apoptotic signaling pathway protein Bbc3 mRNA (Tan et al., 2007, Tan et al., 2009). Mac2-3 may regulate the expression of these genes by binding to the poly(G) stretches in the corresponding mRNAs, thereby leading to disruption of the host antiviral response as well as of apoptotic signals.
Mac3 has also been reported to bind oligo(A) (Chatterjee et al., 2009, Johnson et al., 2010). This observation (which is not in agreement with the results reported by Tan et al., 2007, Tan et al., 2009) might suggest that Mac3 binds the poly(A) tail of the viral genome, or of subgenomic mRNAs, or of host mRNAs. Poly(A)-binding protein (PABP) binds the genomic poly(A) tails of BCoV (bovine coronavirus), MHV, and TGEV, thereby enhancing the replication of these viruses (Spagnolo and Hogue, 2000, Galán et al., 2009). Is it possible that Mac3 binding to oligo(A) competes with the binding between PABP and the poly(A) tail? The question is yet to be answered.
Besides binding to nucleic acids, Mac2-3 of SARS-CoV has been shown to interact directly with host proteins, e.g. the E3 ubiquitin ligase RCHY1 (Ma-Lauer et al., 2016). RCHY1 and several other host proteins, Paip1, MKRN2, and MKRN3 etc. were reported to interact with Nsp3 (Pfefferle et al., 2011). However, the detailed binding region(s) on Nsp3 have not been identified. Ma-Lauer et al. (2016) demonstrated that Mac2-3 and the PL2pro of Nsp3 bind RCHY1, thus resulting in down-regulation of the antiviral protein p53 (see below). It is an interesting hypothesis that such interactions, which are absent from other CoVs because they lack Mac2-3, might account for a unique pathogenicity-related pathway utilized by SARS-CoV.
The DPUP (SUD-C) follows the Mac3 domain in SARS-CoV (Fig. 1A). Deletion of the domain within the context of a SARS-CoV replicon leads to a large reduction of RNA synthesis, but some basal RTC activity remains, indicating that the DPUP is not absolutely essential for replication (Kusov et al., 2015). Currently, three DPUP structures are available, one each from SARS-CoV and MHV (Table 1; Fig. 5C and D; Johnson et al., 2010, Chen et al., 2015), and the third one from bat coronavirus HKU9 (Table 1; Hammond et al., 2017). All DPUPs adopt a similar topology and overall structure. The R.M.S.D values between SARS-CoV DPUP and that of MHV or HKU9 are 2.1 Å (for 62 out of 74 Cα atoms; Z-score: 7.1) or 2.0 Å (for 62 out of 77 Cα atoms; Z-score: 7.0), respectively, according to the Dali server (Holm and Rosenström, 2010). The DPUP consists of an anti-parallel β sheet with two α helices located N- and C- terminal to this β sheet (Johnson et al., 2010, Chen et al., 2015). The two α helices form one plane while the β sheet forms the other; this resembles a typical frataxin-like fold (Bencze et al., 2006). Proteins featuring the frataxin-like fold are commonly involved in controlling cellular oxidative stress by binding iron to maintain the iron homeostasis (Bencze et al., 2006). In case of the yeast frataxin homologue Yfh1, cells lacking this gene were demonstrated to be highly sensitive to H2O2 and elevated metal ion levels (such as iron and copper) (Foury and Cazzalini, 1997). Several Glu and Asp residues in the N-terminal α helix of Yfh1 are possibly involved in binding metal ions (Fig. 5E; He et al., 2004, Bencze et al., 2006). Interestingly, “EEXXXE” and “DDD” motifs exist in the first helix of the SARS-CoV and MHV DPUP, respectively, even though the sequence identity of DPUP is only 13% between these two viruses. Neuman et al. (2008) found that SARS-CoV Mac2−Mac3−DPUP can bind cobalt ions, while Mac3 alone and Mac2*−Mac3 (2*: C-terminal half of Mac2) cannot. According to these observations, it is conceivable that the DPUP region binds metal ions. Furthermore, infection with SARS-CoV can induce transcription of oxygen stress-related genes of the host (Hu et al., 2012). Any involvement of DPUP in this biological process is speculative at this time.
The Mac2-3−DPUP oligodomain (SUD) has been shown to interact with Nsp9, Nsp12, and NAB−βSM−TM1 (see below) of Nsp3 by using a GST pull-down assay (Imbert et al., 2008). Using Y2H and co-immunoprecipitation (CoIP) assays, the oligoprotein Ubl1−HVR−Mac1-2-3* (3*, N-terminal third of Mac3) of SARS-CoV Nsp3 has been found to bind Nsp2, ORF3a, and ORF9b (von Brunn et al., 2007); However, with the slightly larger region Ubl1−HVR−Mac1-2-3−DPUP, these interactions were not confirmed in an Y2H assay (Pan et al., 2008). It seems that DPUP might modulate the various binding processes. Furthermore, the DPUP subdomain could also regulate the sequence specificity of RNA binding by Mac3 as mentioned above (Johnson et al., 2010).
The relative orientation of SARS-CoV Mac2 and Mac3 is fixed by an artificial disulfide bond and dimer formation in the crystal (Tan et al., 2009). The NMR structure shows that Mac2 and Mac3 as well as Mac3 and DPUP have no preferred relative orientations to one another (Johnson et al., 2010). However, Mac2, Mac3, and DPUP are surrounded by other domains within Nsp3; it is unclear whether these other domains affect the relative orientation among the three. More multi-domain structures will be needed to answer this question and to elucidate the structural basis of mutual influences of these modules onto each other (see, e.g., above for the influence of the HEV helicase on the macrodomain of this virus).
5. Ubiquitin-like domain 2 and papain-like protease 2
Besides the Mac1 (X domain), the largest number of crystal structures for any Nsp3 domain have been determined for the ubiquitin-like domain 2 (Ubl2) plus the papain-like protease 2 (PL2pro). So far, structures of this region are available for SARS-CoV, MERS-CoV, IBV, and MHV (Table 1; Ratia et al., 2006, Lei et al., 2014, Kong et al., 2015, Chen et al., 2015). Ubl2 and PL2pro are conserved in all CoVs (Neuman et al., 2008, Neuman, 2016). The exact functional role of the Ubl2 domain is not clear so far, while the PL2pro was reported to possess proteolytic, deubiquitinating, and deISGylating activities (Barretto et al., 2005, Lindner et al., 2005, Yang et al., 2014, Mielech et al., 2014).
5.1. Ubiqutin-like domain 2 (Ubl2)
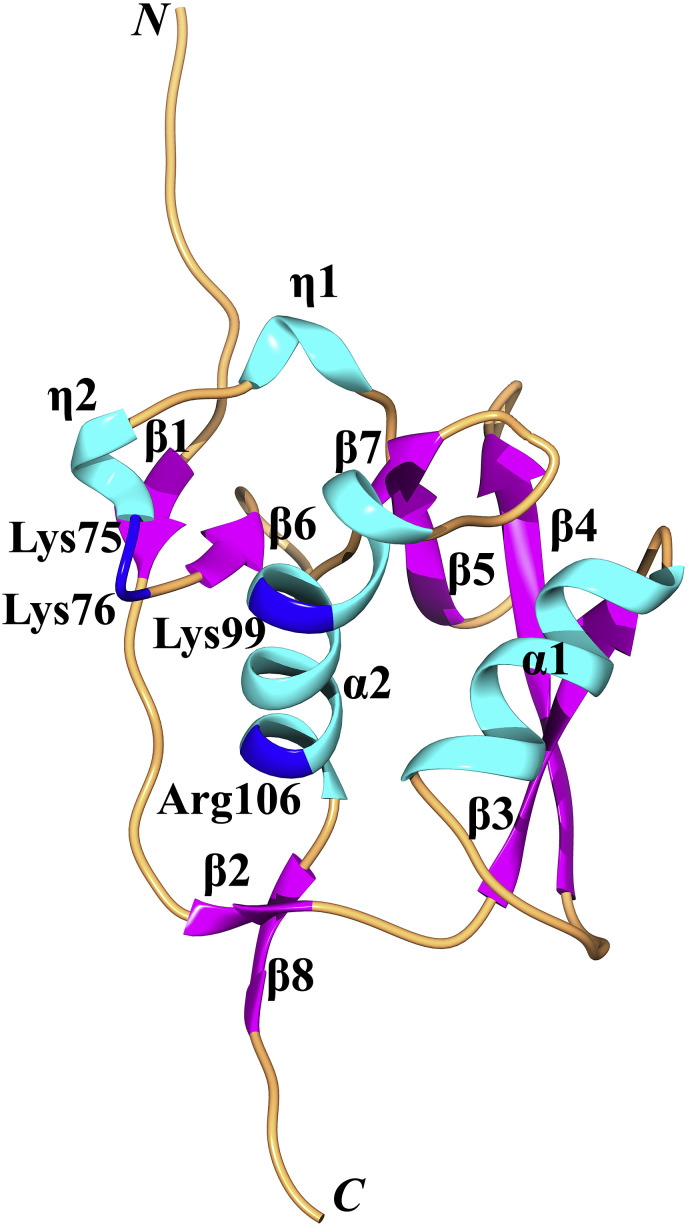
The Ubl2 is the second ubiquitin-like subdomain located within Nsp3 (Fig. 2, Fig. 6 ). The structures of Ubl2 in different CoVs are more conserved compared to the Ubl1. For example, the R.M.S.D. between the Ubl2s of SARS-CoV and MHV is 1.2 Å (for 58 out of 68 Cα atoms; Z-score: 11.1) according to the Dali server (Holm and Rosenström, 2010), whereas the corresponding value for the Ubl1s of the two viruses is 2.8 Å (for 85 out of 93 Cα atoms; Z-score: 7.5).
Fig. 6.
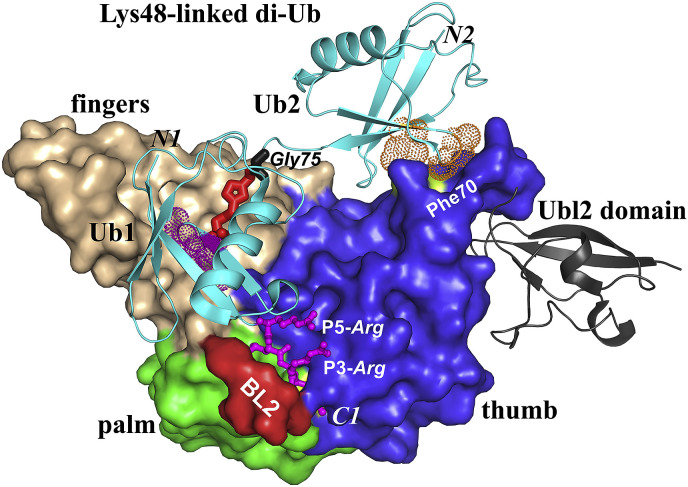
Structure of the SARS-CoV papain-like protease 2 (PL2pro) in complex with Lys48-linked diubiquitin (PDB entry: 5E6J; Békés et al., 2016). The Ubl2 is shown as a grey cartoon. The catalytic domain (PL2pro) is displayed in surface view. The thumb, fingers, and palm subdomains are shown in blue, light brown, and green, respectively. The blocking loop 2 (BL2) is depicted in red. The Lys48-linked diubiquitin is displayed as a light-blue cartoon. Lys48 of Ub1 is linked to the C-terminal Gly75 of Ub2 (black sticks) via a triazole (red sticks). The N and C termini of Ub1 (N1, C1) as well as the N terminus of Ub2 (N2) are marked. The conserved hydrophobic patches (Ile44, Ala46, Gly47) of Ub1 and Ub2 are indicated by purple and orange dots, respectively. The residue Phe70 (yellow) interacting with the hydrophobic patch of Ub2 is labeled. The C-terminal Arg−Leu−Arg−Gly−Gly residues (RLRGG) of Ub1 are shown in ball & stick style (purple). P3-Arg and P5-Arg are marked.
Some host USPs (with a fold similar to the CoV PLpro) also include one or more Ub-like domain(s), which is/are used to regulate the catalytic activity as well as to interact with partners (Komander et al., 2009, Faesen et al., 2012, Pfoh et al., 2015). For example, the N-terminal Ubl domain of USP14 is critical for its recruitment at the proteasome, thereby enhancing its catalytic activity (Hu et al., 2005, Faesen et al., 2012). USP7 (also named “HAUSP”: Herpesvirus-associated USP) includes five Ubl domains (Ubl 1–5), which are located at the C-terminus of the protease domain. Ubl4-5 promote Ub binding and enhance the DUB activity of USP7 by about 100-fold via interacting with the “switching loop” (Trp285-Phe291) in the USP7 catalytic domain (Faesen et al., 2012). Ubl2 of USP7 interacts with the HSV-1 immediate-early protein ICP0 to antagonize the host antiviral response (Pfoh et al., 2015). In contrast to the variable relative orientations of the Ubl domains and the catalytic domain of USP7, the Ubl2 domain is anchored to the CoV PL2pro by two salt-bridges in MERS-CoV and SARS-CoV (Lei et al., 2014), so it is unlikely to regulate the catalytic activity of PL2pro. In agreement with this conclusion, the presence or absence of the Ubl2 of SARS-CoV or MERS-CoV shows almost no effect on the PL2pro activities (Frieman et al., 2009, Clasman et al., 2017).
Currently, several inconsistent roles of Ubl2 are reported. Frieman et al. (2009) demonstrated that the Ubl2 of SARS-CoV is necessary to antagonize the host innate immune response via blocking IRF3 or the NF-κB pathway. In contrast, Clementz et al. (2010) reported that the Ubl2 of SARS-CoV is not necessary for antagonizing IFN production. Also, Mielech et al. (2015) showed that the Val787Ser mutation (Nsp3 numbering) in the MHV Ubl2 reduces the thermal stability of the PL2pro, whereas Clasman et al. (2017) reported that the deletion of Ubl2 in MERS-CoV did not affect PL2pro thermal stability. The former Val residue of MHV is conserved in SARS-CoV and MERS-CoV. It is located in the first strand (β1) and contributes to the hydrophobic core of Ubl2; therefore, the Val-to-Ser change might disrupt the global Ubl2 structure, leading to a decrease in the stability of the PL2pro domain (Mielech et al., 2015).
On the basis of molecular dynamics simulations, the MERS-CoV Ubl2 has recently been proposed to display more molecular flexibility when the PL2pro binds ubiquitin, compared to the situation in the free enzyme. The authors speculate that the difference in flexibility of the Ubl2 might regulate the interaction with downstream targets, thereby modulating the innate immune response (Alfuwaires et al., 2017). Ubiquitination and deubiquitination cannot only regulate the immune response but also the cell-cycle, DNA damage repair, cellular growth etc. (Welchman et al., 2005), and these processes will involve a large number of host proteins. Among these, the coronavirus PL2pro should select its specific targets, such as the host innate-immune system-related proteins TRAF3, STING, TBK1, IRF3 etc. (Chen et al., 2014, Lei and Hilgenfeld, 2017), with the goal of facilitating efficient virus survival. We therefore speculate that the Ubl2 might act as a modulator helping the PL2pro recognize its specific targets during coronavirus infection. However, this idea needs to be verified by future research.
5.2. Papain-like protease 2 (PL2pro)
The PL2pro adopts an extended right-hand fold with thumb, palm, and fingers subdomains, similar to the TGEV PL1pro (Fig. 6; Ratia et al., 2006, Lei et al., 2014, Lee et al., 2015, Kong et al., 2015, Chen et al., 2015, Clasman et al., 2017; see also Table 1) and human USPs (e.g. USP14, USP7; Ratia et al., 2006). A zinc ion is coordinated by four cysteines from two β hairpins in the fingers subdomain and forms a zinc-finger motif. Although the conformations of the zinc finger are variable between different PL2pros (Lei et al., 2014, Lee et al., 2015, Kong et al., 2015, Chen et al., 2015), the motif is essential for structural stability and proteolytic activity (Barretto et al., 2005). The catalytic site of PL2pro comprises the typical Cys–His–Asp triad, just like the PL1pro of TGEV (see above). The catalytic Cys is located in the thumb subdomain (at the N terminus of helix 4 of SARS-CoV and MERS-CoV PL2pro; Ratia et al., 2006, Lei et al., 2014), whereas the His as well as the Asp are located in the palm subdomain. In the free PL2pro, the catalytic triad Cys–His–Asp is pre-formed, different from USP7, where the catalytic residues are only well aligned upon Ub binding to the enzyme (Hu et al., 2002). As we mentioned above, the oxyanion hole of papain-like proteases normally comprises a Gln or Asn side-chain 5 or 6 residues N-terminal to the catalytic Cys. This situation is found in the MHV PL2pro (Chen et al., 2015), but the corresponding residues are Trp, Leu, and Trp in the enzymes of SARS-CoV, MERS-CoV, and IBV, respectively (Ratia et al., 2006, Lei et al., 2014, Kong et al., 2015). Nevertheless, the indole-ring nitrogen of Trp can form a hydrogen bond with the oxyanion intermediate of substrate hydrolysis. The protease activity of the SARS-CoV PL2pro is abolished upon a Trp-to-Ala mutation (Ratia et al., 2006). In contrast, the Leu of MERS-CoV PL2pro totally lacks the ability to contribute to oxyanion stabilization via a hydrogen bond (Lei et al., 2014). The deficient oxyanion hole of MERS-CoV PL2pro causes an about 100-fold lower proteolytic activity compared to that of the SARS-CoV PL2pro when using Arg−Leu−Arg−Gly−Gly-7-amino-4-methylcoumarin (RLRGG-AMC) as a substrate (Báez-Santos et al., 2014). Meanwhile, the corresponding activity of the Leu-to-Trp mutation in MERS-CoV PL2pro is about 50-fold higher than that of the wild-type enzyme, using the same substrate (Lei et al., 2014). As we mentioned before (Lei and Hilgenfeld, 2016), the efficiency of viral proteases does not always have to be optimized during virus evolution. Rather, the creation of temporary intermediates of polyprotein cleavage, in the right temporal order, is necessary for correct virus replication (Kanjanahaluethai and Baker, 2000, Gosert et al., 2002, Harcourt et al., 2004); thus, the proper but not necessarily the highest protease activity is beneficial for virus survival.
In order to investigate the mechanism of the DUB and deISGylating activities of CoV PLpros, the complex of the enzyme with ubiquitin (or ISG15) is important. Until now, structures of SARS-CoV and MERS-CoV PL2pro with mono-Ub as well as of SARS-CoV PL2pro with di-Ub have been obtained (Chou et al., 2014, Ratia et al., 2014, Békés et al., 2016, Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016). Very recently, the structures of both SARS-CoV and MERS-CoV PL2pro in complex with the C-terminal Ubl domain of hISG15 or mISG15 have been reported (Daczkowski et al., 2017a, Daczkowski et al., 2017b). These structures show that the PL2pro of SARS-CoV possesses two ubiquitin-binding sites (named Ub1 and Ub2 sites here; Ratia et al., 2014, Békés et al., 2016). From the prior structure of USP14 in complex with ubiquitin, it is known that two blocking loops (BL1 and BL2) regulate substrate binding (Hu et al., 2005). Different from that, only the BL2 exists in CoV PL2pros and is involved in substrate binding (Fig. 6; Chou et al., 2014, Ratia et al., 2014, Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016), whereas BL1 is absent in CoV PL2pros (Ratia et al., 2006, Lei et al., 2014).
The proximal Ub binding site (Ub1) is, to a certain degree, conserved between the PL2pros of SARS-CoV and MERS-CoV. The region includes the narrow substrate channel between the thumb and the palm subdomains, as well as a hydrophobic patch in the fingers subdomain (Fig. 6). The narrow substrate channel binds the C-terminal RLRGG residues of ubiquitin (Chou et al., 2014, Ratia et al., 2014, Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016; in order to be clear, Ub residues appear in italics here). The C-terminal RLRGG of ubiquitin is similar to the unprimed side of the polyprotein substrates, (R/K)(L/I)XGG in the two viruses. The S1, S2, and S4 sites are well conserved to accommodate the two small glycines (P1, P2) and the hydrophobic P4 residue (Leu or Ile). In contrast, the flexible side-chains in P3 and P5 feature binding patterns that are slightly different between SARS-CoV and MERS-CoV PL2pro. In the SARS-CoV PL2pro(Cys112Ser)−Ub complex, P3-Arg forms a weak salt-bridge with Glu162 (Chou et al., 2014), whereas the corresponding P3-Arg is exposed to solvent in the MERS-CoV complex (Lei and Hilgenfeld, 2016). On the other hand, the P5-Arg is exposed to solvent in the SARS-CoV complex (Chou et al., 2014) but forms a strong salt-bridge with Asp164 in MERS-CoV (Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016). Interestingly, this Asp164 is unique among CoV PL2pros, and the Asp164Ala replacement leads to an about 4.5-fold and 3.5-fold reduction of the proteolytic and DUB activities, respectively (Lei and Hilgenfeld, 2016). As just mentioned, the proteolytic activity of the MERS-CoV PL2pro is not optimized due to the deficient oxyanion hole. On the other hand, the virus requires a strong DUB activity to counteract the host immune response. The suboptimal enzyme activities may be partly compensated by the unique Asp164 (Lei and Hilgenfeld, 2016).
In addition to the binding of the Ub C-terminus to the substrate channel, there is an interaction between a hydrophobic region of the SARS-CoV and MERS-CoV PL2pros in the fingers subdomain and a hydrophobic patch (Ile44, Ala46, Gly47) of Ub (Chou et al., 2014, Ratia et al., 2014, Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016, Békés et al., 2016). This hydrophobic patch of Ub is commonly used to interact with Ub-binding proteins (Dikic et al., 2009). The fingers subdomain residues involved are Tyr208 and Met209 in SARS-CoV, and Tyr209 and Val210 in MERS-CoV (Chou et al., 2014, Ratia et al., 2014, Bailey-Elkin et al., 2014, Lei and Hilgenfeld, 2016, Békés et al., 2016). Moreover, these hydrophobic interactions between the PL2pro and Ub are important for the DUB activity of the enzyme, because disrupting them via a Val210Arg mutation dramatically diminishes the DUB activity in MERS-CoV PL2pro (Bailey-Elkin et al., 2014).
Near the hydrophobic patch of Ub, Arg42 forms a salt-bridge with Glu168 of PL2pro in two structures of the SARS-CoV PL2pro in complex with mono-ubiquitin or Lys48-linked di-Ub (Chou et al., 2014, Ratia et al., 2014, Békés et al., 2016). However, this Glu is replaced by Arg in MERS-CoV PL2pro, resulting in Arg42 instead forming a salt-bridge with Asp165 in the MERS-CoV PL2pro−ubiquitin complex (Lei and Hilgenfeld, 2016). This illustrates that various fine-tuned binding patterns exist between Ub and PL2pros in different CoVs.
Besides the Ub1 binding site, the Ub2 binding site is mapped by the complex of SARS-CoV PL2pro with Lys48-linked di-Ub (Fig. 6; Békés et al., 2016). The Ub2 binding site is located at the first α helix of the thumb subdomain. Phe70 interacts with the common hydrophobic patch (Ile44, Ala46, Gly47) of Ub. Interestingly, MERS-CoV PL2pro seems to lack the corresponding Ub2 binding site. Phe70 of SARS-CoV PL2pro is changed to Lys69 in MERS-CoV (Békés et al., 2016). In addition, Békés et al. (2016) predicted that Trp107 and Ala108 could constitute the Ub1′ binding site in SARS-CoV PL2pro. The Trp107Leu/Ala108Ser double mutation reduces the enzyme's activity towards Lys48-linked tri-Ub-AMC by about 75% (Békés et al., 2016). However, it should be noted that Trp107 contributes to the oxyanion hole of SARS-CoV PL2pro (see above); therefore, the reduced DUB activity upon replacing Trp107 by Leu is perhaps not due to altering the Ub1′ binding site, but rather to destroying the oxyanion hole.
The SARS-CoV PL2pro displays more efficient cleavage activity towards Lys48-linked di-Ub-AMC than Lys63-linked di-Ub-AMC substrates in vitro, demonstrating that the PL2pro preferentially recognizes Lys48-linked polyUb chains (Báez-Santos et al., 2014, Békés et al., 2015, Békés et al., 2016). In contrast, MERS-CoV PL2pro processes Lys48- and Lys63-linked polyUb chains with similar efficiency (Báez-Santos et al., 2014). Lys48-linked Ub chains mainly cause target protein degradation via the 26S proteasome, while Lys63-linked polyUb is mainly related to DNA repair and signal transduction (Ikeda and Dikic, 2008), in particular, in the signal transduction cascades of the host innate immune system (Dikic and Dötsch, 2009). However, the biological significance of the CoV PLpros showing different cleavage activities on Lys48- and Lys63-linked polyUb is still unclear. Furthermore, the SARS-CoV PL2pro cleaves the polyUb chain by removing di-Ubs, not mono-Ub units as in MERS-CoV (Békés et al., 2015). This strongly suggests that MERS-CoV PL2pro possesses the Ub1 and Ub1′ binding sites but not a Ub2 site, consistent with the Phe70 to Lys mutation in MERS-CoV PL2pro as just mentioned.
At the same time, ISG15 utilizes a different Ub2 binding site of SARS-CoV PL2pro, compared to Lys48-linked di-Ub (Békés et al., 2016), but no structure for a full-length ISG15−CoV PL2pro complex is available so far. Daczkowski et al. (2017a) reported that the C-terminal domains of ISG15s (similar to Ub1 mentioned above) from different species have different binding characteristics with SARS-CoV PL2pro according to two structures, the PL2pro in complex with the C-terminal domain of hISG15 and mISG15, respectively. In addition, the structure of mouse USP18 in complex with full-length mISG15 became available this year (Basters et al., 2017). Surprisingly, the N-terminal Ubl domain of mISG15 shows almost no interaction with mUSP18. Does ISG15 behave similarly when binding to the CoV PLpro? How does the N-terminal domain of ISG15 of different species recognize the cognate CoV PLpro? It would be of interest to determine not only the structure of a full-length hISG15−HCoV PLpro complex but also that of mISG15 with MHV PLpro.
The DUB and deISGylating activities of CoV PLpros are well established, but the detailed mechanism of the PLpro antagonism of the host innate immune response is still ambiguous (see Lei and Hilgenfeld, 2017, for a recent review). Various cytokines (including interferons (IFNs) and tumor necrosis factors (TNFs)) are produced to inhibit virus replication by two main pathways, the IRF3 pathway and the NF-κB pathway (Seth et al., 2006, Hiscott et al., 2006). For more information on the host innate immune system signaling pathways, the reader should consult other reviews (e.g., Mogensen, 2009, Lei and Hilgenfeld, 2017). Devaraj et al. (2007) found that the SARS-CoV PL2pro can directly bind IRF3 to block its phosphorylation, dimerization, and nuclear translocation, thereby inhibiting IFN-β induction. Furthermore, the PL2pro was found not to block the NF-κB signaling pathway and the protease activity was described as dispensable for antagonizing the IFN response (Devaraj et al., 2007). Clementz et al. (2010) also confirmed that the enzyme activity of HCoV-NL63 PL2pro is not essential for counteracting the antiviral IFN production. In contrast, Frieman et al. (2009) reported that the SARS-CoV PL2pro does not directly bind IRF3 or disrupt its phosphorylation. Instead, the PL2pro was proposed to inhibit the NF-κB signaling pathway by stabilizing its inhibitor, IκBα (Frieman et al., 2009). Furthermore, the protease activity of SARS-CoV PL2pro is important for blocking the TNF-α/NF-κB signaling pathway (Frieman et al., 2009). In addition, the HCoV-NL63 but not the MHV PL2pro has the ability to impede the IRF3 and NF-κB pathways, indicating that the functions of the PL2pro are specific for different CoVs (Frieman et al., 2009). Later, a protein comprising the SARS-CoV PL2pro and the TM (transmembrane region of Nsp3) was demonstrated to inhibit the STING/TBK1/IKKε-mediated signaling pathway (upstream regulators of IRF3; Chen et al., 2014), thereby disrupting IRF3 phosphorylation and dimerization, and blocking the type-I IFN response. SARS-CoV PL2pro plus TM can also physically interact with the STING-TRAF3-TBK1 complex and remove the ubiquitins from ubiquitinated RIG-I, STING, TRAF3, TBK1, as well as IRF3 (Chen et al., 2014). In 2016, it was reported that the SARS-CoV PL2pro can inhibit the Toll-like receptor 7 (TLR7)–mediated type-I IFN response and the NF-κB pathway by removing the Lys63-linked polyUb chain from TRAF3 and TRAF6 (upstream regulators of IRF3 and NF-κB; Li et al., 2016b). Interestingly, the SARS-CoV PL2pro only removes the Lys63-but not the Lys48-linked polyUb chain from TRAF3 and TRAF6 in vivo (Li et al., 2016b). On the other hand, Báez-Santos et al. (2014) and Békés et al., 2015, Békés et al., 2016 have shown that SARS-CoV PL2pro prefers to digest Lys48- over Lys63-linked polyUb chains in vitro (see above). Why does the substrate specificity of PL2pro seem to be different in vivo and in vitro? Does any other factor influence the substrate specificity of PL2pro in vivo when counteracting the cellular innate immune response? These questions are yet to be answered.
In addition, the HCoV-NL63 PL2pro was shown to block the p53-IRF7-IFNβ signaling pathway (Yuan et al., 2015). p53 can induce type-I interferon production via IRF7 (interferon regulatory factor 7; Yuan et al., 2015). Meanwhile, p53 can be degraded via the MDM2- (an E3 ubiquitin ligase) mediated ubiquitin-proteasome system (Haupt et al., 1997). Yuan et al. (2015) found that the HCoV-NL63 PL2pro deubiquitinates and stabilizes MDM2 to augment p53 degradation, thereby antagonizing the host innate immune response. Recently, the PL2pro of SARS-CoV and MERS-CoV as well as the PL1pro/PL2pro of HCoV NL63 were shown to directly interact with the host E3 ubiquitin ligase RCHY1 (also called Pirh2; Ma-Lauer et al., 2016), thereby increasing the stability of the latter. Like MDM2, RCHY1 can induce p53 degradation as well (Leng et al., 2003). Ma-Lauer et al. (2016) found that p53 inhibits the replication of SARS-CoV. Stabilization of RCHY1 by physical interaction with the PL2pro increases the degradation of p53 and supports coronavirus replication (Ma-Lauer et al., 2016). While the HCoV-NL63 PL2pro stabilizes MDM2 by deubiquitinating it (Yuan et al., 2015), the SARS-CoV PL2pro surprisingly does not deubiquitinate RCHY1 (Ma-Lauer et al., 2016). How does the PL2pro stabilize RCHY1? The mechanism has yet to be elucidated.
Besides the functions of PL2pro discussed above, the enzyme was shown to interact with other viral proteins. The region from PL2pro to the C-terminus of Nsp3 in SARS-CoV can interact with the Nsp2, ORF3a, and ORF9b proteins, as identified by Y2H and CoIP assays (von Brunn et al., 2007). Through similar assays, the region PL2pro−NAB−βSM was found to interact with Nsp4 as well as Nsp12 (Pan et al., 2008). The SARS-CoV PL2pro was further shown to bind ORF7a and Nsp6 by using proteomics analysis (Neuman et al., 2008).
Coronavirus PLpro is an important target for developing antiviral drugs. This aspect has been well reviewed by Báez-Santos et al. (2015) within this series; hence, we mention only inhibitors here that have been described since. Two big challenges exist when designing PLpro inhibitors: 1) the S1 and S2 binding sites are tailor-made to accommodate glycine residues and hence they are small; therefore, identifying suitable peptidomimetic chemical structures is difficult; 2) many host USPs feature folds and active sites similar to the PLpros, so specificity of the inhibitors could be an issue. However, there is a good chance that the BL2 loop (mentioned above) of CoV PL2pros could provide sufficient uniqueness to solve the specificity problem. This loop is involved in substrate binding and is different not only between USPs and CoV PLpros but also among different CoVs (Hu et al., 2005, Ratia et al., 2006, Lei et al., 2014, Báez-Santos et al., 2014, Báez-Santos et al., 2015, Lee et al., 2015). For example, this loop comprises 6 amino-acid residues (GNYQCG) in SARS-CoV PL2pro but 7 (GIETAVG) in the enzyme of MERS-CoV, leading to the inability of SARS-CoV PLpro inhibitors to act on MERS-CoV PLpro (Báez-Santos et al., 2014, Hilgenfeld, 2014, Lee et al., 2015). Using a high-throughput assay, the purine derivative 8-(trifluoromethyl)-9H-purin-6-amine (compound 4; Fig. 7 A) was identified as a competitive MERS-CoV PL2pro inhibitor, with an IC50 of about 6 μM in vitro (Lee et al., 2015). Interestingly, this compound is also (moderately) active against SARS-CoV PL2pro (IC50 ≈ 11 μM) but acts as an allosteric inhibitor in this case (Lee et al., 2015). Furthermore, the authors also reported that this inhibitor shows very high selectivity against human ubiquitin C-terminal hydrolase (hUCH-L1; IC50 > 100 μM), which is one of the host proteins most closely related to the CoV PLpro (Lee et al., 2015). In contrast, Clasman et al. (2017) reported that compound 4 features no selective inhibition of CoV PLpros nor host USPs; therefore, this compound could be a pan-assay interference inhibitor (or PAIN). Recently, nine alkylated chalcones (1–9) and four coumarins (10–13), which were isolated from the perennial plant Angelica keiskei, had their inhibitory activities against both the SARS-CoV Mpro (3CLpro, chymotrypsin-like protease) and the PL2pro tested (Park et al., 2016). One of the chalcones, compound 6 (Fig. 7B), exhibited relatively strong inhibition of both the 3CLpro and the PL2pro in vitro, with IC50 values of 11.4 and 1.2 μM respectively (Park et al., 2016). Chalcone 6 uses different inhibition mechanisms for 3CLpro and PL2pro. It is a competitive inhibitor for the former enzyme but a non-competitive one for the latter (Park et al., 2016). Clearly, the large body of structural information available for the CoV PLpros and host DUBs should enable more design of inhibitors specific for the viral enzyme.
Fig. 7.
Recently described inhibitors of the CoV PL2pro. (A) Structural formula of the purine derivative 8-(trifluoromethyl)-9H-purin-6-amine (compound 4). This compound is a competitive MERS-CoV PL2pro inhibitor (Lee et al., 2015). It is also active against SARS-CoV PL2pro but acts as an allosteric inhibitor in this case. (B) A natural-product chalcone, compound 6 from the perennial plant Angelica keiskei, inhibits the SARS-CoV Mpro (3CLpro) and PL2proin vitro (Park et al., 2016).
6. Nucleic acid-binding (NAB) domain and betacoronavirus-specific marker (βSM) domain
The nucleic-acid binding (NAB) and betacoronavirus-specific marker (βSM) domains together are also named “Nsp3e” (Neuman et al., 2008). The latter domain alone was previously called “group 2-specific marker” (G2M) (Neuman et al., 2008). The NAB and βSM domain exist in the genus Betacoronavirus. The corresponding region is absent in alphacoronaviruses and deltacoronaviruses (Neuman, 2016). In gammacoronaviruses, there is a gammacoronavirus-specific marker (γSM) domain at this position (Neuman, 2016).
Structural information on this region is very limited for all coronaviruses. Thus far, only an NMR structure of the NAB domain of SARS-CoV is available (Table 1; Fig. 8 ; Serrano et al., 2009). The structure comprises two antiparallel β sheets (β1+β6; β2+β8) and one parallel β sheet (β3−β4−β5−β7) as well as two α helices and two 310 helices (η1 and η2) in the order β1−β2−β3−α1−β4−β5−η1−η2−β6−β7−α2−β8. Four β strands (β3−β4−β5−β7) and two helices (α1, α2) form a “half-barrel”. The structure of the NAB represents a unique fold (Serrano et al., 2009). The domain has been shown to bind ssRNA as well as to unwind dsDNA (Neuman et al., 2008). When binding to ssRNA, the NAB prefers sequences with repeats of three consecutive Gs (Serrano et al., 2009), such as (GGGA)5 and (GGGA)2. A positively charged surface patch (Lys75, Lys76, Lys99, and Arg106) is involved in RNA binding (Fig. 8). These residues are located in the loop between η2 and β6 as well as in helix α2 (Serrano et al., 2009). The RNA binding behavior of the NAB appears to be similar to that of SARS-CoV Mac3 (SUD-M), which has a specificity for oligo(G) (Tan et al., 2007, Tan et al., 2009), although the latter is also reported to bind oligo(A) (Chatterjee et al., 2009, Johnson et al., 2010, mentioned above). Whether there is a functional relation between Mac3 and NAB, remains to be investigated.
Fig. 8.
NMR structure of the nucleic acid-binding (NAB) domain in SARS-CoV (cartoon style; PDB entry: 2K87; Serrano et al., 2009). The order of secondary-structure elements is β1−β2−β3−α1−β4−β5−η1−η2−β6−β7−α2−β8. The overall structure of NAB represents a unique fold. The residues involved in RNA binding (Lys75, Lys76, Lys99, and Arg106) are displayed in blue. The N and C termini of the NAB domain are labeled.
Currently, no structural information is available concerning the βSM or γSM, and nothing is known about the function of these modules either. A gene encoding the βSM domain of SARS-CoV could not be expressed in E. coli; this module has been predicted to be a nonenzymatic domain (Neuman et al., 2008). In the absence of sequence similarity to any domain of known function, we performed an ab-initio protein structure prediction using the sequence of the SARS-CoV βSM domain and the QUARK online server (Xu and Zhang, 2012). The result indicates that most of this region is intrinsically disordered. This does not preclude that it might adopt a defined structure upon interaction with another Nsp or RNA, or a host protein.
7. Transmembrane regions (TM1 and TM2), Nsp3 ectodomain, Y1 domain, and CoV-Y domain
This part of Nsp3 includes two transmembrane regions as well as three soluble domains, which together constitute about one third of the multidomain protein. The two transmembrane regions are TM1 and TM2, while the three domains are the Nsp3 ectodomain (3Ecto), Y1, and CoV-Y. The sequential order of this part is TM1−3Ecto−TM2−Y1−CoV-Y (Fig. 1A and B). Even though this part exists in all coronaviruses (Neuman et al., 2008, Neuman, 2016), thus far, no three-dimensional structure is available for the entire region nor for a part of it.
Nsp3 of CoVs is thought to pass the ER membrane twice, since there are two predicted transmembrane regions, TM1 and TM2 (Harcourt et al., 2004, Kanjanahaluethai et al., 2007, Oostra et al., 2008). According to the transmembrane region prediction server TMHMM (Krogh et al., 2001), there is a total of three hydrophobic regions in SARS-CoV Nsp3 (Table 1; Fig. 1B). Oostra et al. (2008) proposed that the first two of the three hydrophobic regions span the membrane while the last one (AH1), which has more amphipathic character, does not (Fig. 1B). Thus, the 3Ecto would be the only domain located on the lumenal side of the ER in SARS-CoV Nsp3 (Fig. 1B). The 3Ecto is thought to bind metal ions and has also been designated as a zinc-finger (ZF) domain before (Neuman et al., 2008). Neuman (2016) found that the metal binding Cys-His cluster is not conserved in all CoVs and has renamed this domain into “3Ecto”. In fact, only two cysteine residues are conserved in the CoV 3Ecto domain (Fig. 9 A), hence this domain is unlikely to be a zinc-finger domain. The transmembrane regions plus the 3Ecto are important for the PL2pro to process the Nsp3↓4 cleavage site in SARS-CoV and MHV (Harcourt et al., 2004, Kanjanahaluethai et al., 2007); a possible reason is that the transmembrane part could bring the PL2pro close to the cleavage site between the membrane-associated proteins Nsp3 and Nsp4. Asparagine (N)-linked glycosylation has been found in the 3Ecto domains of SARS-CoV and MHV (Fig. 1, Fig. 9; Harcourt et al., 2004, Kanjanahaluethai et al., 2007). It is unclear if the N-glycan modification affects the 3Ecto conformation or stability. Frequently, N-linked glycans serve as recognition points for partner molecules (Aebi, 2013). It has been shown that interaction of the 3Ecto with the lumenal loop of Nsp4 is essential for the ER rearrangements occurring in cells infected by SARS-CoV or MHV (the 3Ecto is named “lumenal loop of Nsp3” in this paper; Hagemeijer et al., 2014).
Fig. 9.
Multiple sequence alignment of the 3Ecto and the N-terminal portion of the Y1 domain containing two potential zinc fingers. The conserved cysteines in 3Ecto as well as cysteines and histidines in the N-terminal portion of Y1 are marked by triangles. Two glycosylation sites in the 3Ecto domain of SARS-CoV (Asn1431 and Asn1434; Harcourt et al., 2004) are indicated by asterisks. The corresponding sequence accession numbers are: SARS-CoV, Genbank: AY274119.3; MERS-CoV, Genbank: JX869059.2; MHV, Genbank: AY700211.1; HCoV NL63, Genbank: AY567487.2; IBV, Genbank: M95169.1. The figure was generated using the program ESPript (Gouet et al., 1999).
The Y1 and CoV-Y domains are located at the cytosolic side of the ER. The Y1 domain is conserved in all viruses of the order Nidovirales, while CoV-Y is only conserved in all coronaviruses (Neuman, 2016). Since no three-dimensional structure is available for this part, the domain assignment of Y1 and CoV-Y is ambiguous (Neuman, 2016). We found that the sequence identity of Y1+CoV-Y between different CoV genera is above 25% and two Cys-His clusters are present in the N-terminal part of the Y1 domain, possibly binding zinc ions (Fig. 9). However, it is still unclear if the fold and function in this region are conserved. Currently, functional information on this part is limited. It has been shown that the C-terminal third of Nsp3 (βSM (partial) −TM1−3Ecto−TM2−AH1−Y1+CoV-Y) of Nsp3 binds less efficiently to Nsp4 without the Y1 and CoV-Y domains (Hagemeijer et al., 2014), although these two domains are not as important for this process as the 3Ecto.
According to a Y2H screen, CoIP, as well as GST pull-down assays, different constructs of Nsp3 with different C-terminal regions were identified to interact with various viral non-structural proteins of SARS-CoV (von Brunn et al., 2007, Imbert et al., 2008, Pan et al., 2008). For example, a construct comprising the domains from PL2pro to the end of Nsp3 can bind Nsp2, ORF3a, and ORF9b (see above; von Brunn et al., 2007); the NAB−βSM−TM1 of Nsp3 can interact with Nsp5, Nsp7 − 8, as well as Nsps 12–16, and Y1 plus CoV-Y interacts with Nsp9 and Nsp12 (Imbert et al., 2008); in addition, the NAB−βSM−TM1 of Nsp3 can also interact with other domains within Nsp3, except for Mac1 (X domain) (Imbert et al., 2008); a PL2pro−NAB−βSM−TM1 construct of Nsp3 can bind Nsp4 and Nsp12, while the region from TM1 to the end of Nsp3 only binds Nsp8 (Pan et al., 2008). It has been found that the interaction between the C-terminal region of Nsp3 and Nsp4 is essential for the formation of CMs and DMVs derived from the ER in CoV-infected cells (Angelini et al., 2013, Hagemeijer et al., 2014). The viral RNA and replicase proteins (Nsps) need to be associated with these modified membranes to form the replicative organelles (see Neuman, 2016, for review). In addition, these membranes can protect the viral RNA and Nsps against nucleases and proteases in vitro (van Hemert et al., 2008). Besides the Nsp3−Nsp4 interaction, it is still unclear whether all other interactions really exist or how these interactions affect the viral life cycle. At least, it seems that the membrane-associated region of Nsp3 may regulate the interactions with other viral proteins. It is definitely necessary to put more effort into the structural and functional characterization of this region.
8. Conclusions
Overall, the multi-domain Nsp3 plays various roles in coronavirus infection. It releases Nsp1, Nsp2, and itself from the polyproteins and interacts with other viral Nsps as well as RNA to form the replication/transcription complex. It acts on posttranslational modifications of host proteins to antagonize the host innate immune response (by de-MARylation, de-PARylation (possibly), deubiquitination, or deISGylation). Meanwhile, Nsp3 itself is modified in host cells, namely by N-glycosylation of the 3Ecto domain. Furthermore, Nsp3 can interact with host proteins (such as RCHY1) to support virus survival.
As the largest non-structural protein of CoVs, Nsp3 has also been identified as the major selective target for driving evolution in lineage C betaCoVs on the basis of a high rate of positively selected mutation sites (Forni et al., 2016). Furthermore, the adaptive evolution of Nsp3 of MERS-CoV is still ongoing (Forni et al., 2016). For example, the Arg911Cys mutation (located in the palm subdomain of the PL2pro, corresponding to Arg283 in Lei et al., 2014) of Nsp3 exists in the viral strain KOR/KNIH responsible for the 2015 South Korean outbreak but not in the ancestral strain EMC/2012 (Forni et al., 2016). It is interesting to speculate why coronaviruses keep many essential functions in one protein, while this protein shows high-rate genetic diversity during CoV evolution. In the end, increased research efforts into the structure and function of Nsp3 are needed to achieve a more complete understanding of this protein.
References
- Adams M.J., Carstens E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses. Arch. Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta. 2013;1833:2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Alfuwaires M., Altaher A., Kandeel M. Molecular dynamic studies of interferon and innate immunity resistance in MERS CoV non-structural protein 3. Biol. Pharm. Bull. 2017;40:345–351. doi: 10.1248/bpb.b16-00870. [DOI] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4 doi: 10.1128/mBio.00524-13. pii: e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Zhang D., de Souza R.F., Anand S., Iyer L.M. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 2015;384:3–32. doi: 10.1007/82_2014_414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasheva S., Frolova E.I., Frolov I. Interferon-stimulated poly(ADP-ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 2014;88:2116–2130. doi: 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S., Holzer B., Bridgen A., McMullan G., Quinn D.G., Baron M.D. Dugbe virus ovarian tumour domain interferes with ubiquitin/ISG15-regulated innate immune cell signalling. J. Gen. Virol. 2013;94:298–307. doi: 10.1099/vir.0.048322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basters A., Geurink P.P., Röcker A., Witting K.F., Tadayon R., Hess S., Semrau M.S., Storici P., Ovaa H., Knobeloch K.P., Fritz G. Structural basis of the specificity of USP18 toward ISG15. Nat. Struct. Mol. Biol. 2017;24:270–278. doi: 10.1038/nsmb.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M., Rut W., Kasperkiewicz P., Mulder M.P., Ovaa H., Drag M., Lima C.D., Huang T.T. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem. J. 2015;468:215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M., van der Heden van Noort G.J., Ekkebus R., Ovaa H., Huang T.T., Lima C.D. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol. Cell. 2016;62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencze K.Z., Kondapalli K.C., Cook J.D., McMahon S., Millán-Pacheco C., Pastor N., Stemmler T.L. The structure and function of frataxin. Crit. Rev. Biochem. Mol. Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla P.J., Hughes S.A., Weiss S.R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bütepage M., Eckei L., Verheugd P., Lüscher B. Intracellular mono-ADP-ribosylation in signaling and disease. Cells. 2015;4:569–595. doi: 10.3390/cells4040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein − forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 2009;83:1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 2004;78:5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A.D., Wilkinson K.D., Baker S.C. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Savinov S.N., Mielech A.M., Cao T., Baker S.C., Mesecar A.D. X-ray structural and functional studies of the three tandemly linked domains of non-structural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J. Biol. Chem. 2015;290:25293–25306. doi: 10.1074/jbc.M115.662130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C.C., Lin M.H., Chuang C.Y., Hsu C.H. Macro domain from Middle East respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module: crystal structure and biochemical studies. J. Biol. Chem. 2016;291:4894–4902. doi: 10.1074/jbc.M115.700542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.Y., Lai H.Y., Chen H.Y., Cheng S.C., Cheng K.W., Chou Y.W. Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:572–581. doi: 10.1107/S1399004713031040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.C., Wang A.H. Structural D/E-rich repeats play multiple roles especially in gene regulation through DNA/RNA mimicry. Mol. Biosyst. 2015;11:2144–2151. doi: 10.1039/c5mb00206k. [DOI] [PubMed] [Google Scholar]
- Clasman J.R., Báez-Santos Y.M., Mettelman R.C., O'Brien A., Baker S.C., Mesecar A.D. X-ray structure and enzymatic activity profile of a core papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci. Rep. 2017;7:40292. doi: 10.1038/srep40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M.L., Marshall C.J., Olson M.F. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat. Rev. Mol. Cell Biol. 2004;5:355–366. doi: 10.1038/nrm1365. [DOI] [PubMed] [Google Scholar]
- Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429:1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daczkowski C.M., Goodwin O., Dzimianski J.V., Farhat J.J., Pegan S.D. Structurally guided removal of deISGylase biochemical activity from papain-Like protease originating from the Middle East Respiratory Syndrome Virus. J. Virol. 2017 doi: 10.1128/JVI.01067-17. pii: JVI.01067–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Dötsch V. Ubiquitin linkages make a difference. Nat. Struct. Mol. Biol. 2009;16:1209–1210. doi: 10.1038/nsmb1209-1209. [DOI] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K.J. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eckei L., Krieg S., Bütepage M., Lehmann A., Gross A., Lippok B., Grimm A.R., Kümmerer B.M., Rossetti G., Lüscher B., Verheugd P. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017;7:41746. doi: 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K.K., Cervantes-Barragán L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen A.C., Luna-Vargas M.P., Sixma T.K. The role of UBL domains in ubiquitin-specific proteases. Biochem. Soc. Trans. 2012;40:539–545. doi: 10.1042/BST20120004. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio. 2016;7 doi: 10.1128/mBio.01721-16. pii: e01721–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs K.L., Verheugd P., Lüscher B. Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J. 2013;280:3519–3529. doi: 10.1111/febs.12315. [DOI] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Mozzi A., Pozzoli U., Al-Daghri N., Clerici M., Sironi M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán C., Sola I., Nogales A., Thomas B., Akoulitchev A., Enjuanes L., Almazán F. Host cell proteins interacting with the 3' end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geilhausen H.E., Ligon F.B., Lukert P.D. The pathogenesis of virulent and avirulent avian infectious bronchitis virus. Arch. Gesamte Virusforsch. 1973;40:285–290. doi: 10.1007/BF01242547. [DOI] [PubMed] [Google Scholar]
- Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Graham R.L., Denison M.R. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 2006;80:11610–11620. doi: 10.1128/JVI.01428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS–cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J., de Haan C.A. Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 2011;85:4572–4577. doi: 10.1128/JVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond R.G., Tan X., Johnson M.A. SARS-unique fold in the Rousettus bat coronavirus HKU9. Protein Sci. 2017;26:1726–1737. doi: 10.1002/pro.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Han W., Li X., Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat. Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- He Y., Alam S.L., Proteasa S.V., Zhang Y., Lesuisse E., Dancis A., Stemmler T.L. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton S.M., Borg N.A., Dixit V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016;213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J., Nguyen T.L., Arguello M., Nakhaei P., Paz S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene. 2006;25:6844–6867. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F., Fields S., Schneider C., Martin G.S. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E., Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yen Y.T., Singh S., Kao C.L., Wu-Hsieh B.A. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. 2012;25:277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Hofer F., Martin G.S., Kim S.H. Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol. 1998;5:422–426. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Dikic I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lécine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Chatterjee A., Neuman B.W., Wüthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010;400:724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74:7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Giedroc D.P. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 2013;87:3502–3515. doi: 10.1128/JVI.03112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kong L., Shaw N., Yan L., Lou Z., Rao Z. Structural view and substrate specificity of papain-like protease from avian infectious bronchitis virus. J. Biol. Chem. 2015;290:7160–7168. doi: 10.1074/jbc.M114.628636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stöhr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri T., Eriksson K.K., Putics A., Züst R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V., Ziebuhr J., Weber F. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S., Unger M., Koch-Nolte F., Haag F. ADP-ribosylation of arginine. Amino Acids. 2011;41:257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Lei H., Santarsiero B.D., Gatuz J.L., Cao S., Rice A.J., Patel K., Szypulinski M.Z., Ojeda I., Ghosh A.K., Johnson M.E. Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem. Biol. 2015;10:1456–1465. doi: 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Hilgenfeld R. Structural and mutational analysis of the interaction between the Middle-East respiratory syndrome coronavirus (MERS-CoV) papain-like protease and human ubiquitin. Virol. Sin. 2016;31:288–299. doi: 10.1007/s12250-016-3742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Mesters J.R., Drosten C., Anemüller S., Ma Q., Hilgenfeld R. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antivir. Res. 2014;109:72–82. doi: 10.1016/j.antiviral.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidecker O., Bonfiglio J.J., Colby T., Zhang Q., Atanassov I., Zaja R., Palazzo L., Stockum A., Ahel I., Matic I. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 2016;12:998–1000. doi: 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Li C., Debing Y., Jankevicius G., Neyts J., Ahel I., Coutard B., Canard B. Viral macro domains reverse protein ADP-ribosylation. J. Virol. 2016;90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L., Lin Y.J., Kung S.H., Lin C.W. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17050678. pii: E678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Ng L.F., Liu D.X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Yu X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr. Protein Pept. Sci. 2015;16:491–501. doi: 10.2174/1389203716666150504122435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Müller M.A., Deng W., Lei J., Meyer B., Kusov Y., von Brunn B., Bairad D.R., Hünten S., Drosten C., Hermeking H., Leonhardt H., Mann M., Hilgenfeld R., von Brunn A. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5192–E5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R.L., Abraham R., Sreekumar E., Ong S.E., Cheng S.J., Baxter V.K., Kistemaker H.A., Filippov D.V., Griffin D.E., Leung A.K. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R., Carrière J., Laflamme P., Plouffe C., Khouri H.E., Vernet T., Tessier D.C., Thomas D.Y., Storer A.C. Contribution of the glutamine 19 side chain to transition-state stabilization in the oxyanion hole of papain. Biochemistry. 1991;30:8924–8928. doi: 10.1021/bi00101a002. [DOI] [PubMed] [Google Scholar]
- Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Deng X., Chen Y., Kindler E., Wheeler D.L., Mesecar A.D., Thiel V., Perlman S., Baker S.C. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J. Virol. 2015;89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D.J., Lenschow D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013;425:4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R.I.I.I., Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antivir. Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Schöpf J., Kögl M., Friedel C.C., Müller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Züst R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pöhlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoh R., Lacdao I.K., Georges A.A., Capar A., Zheng H., Frappier L., Saridakis V. Crystal structure of USP7 ubiquitin-like domains with an ICP0 peptide reveals a novel mechanism used by viral and cellular proteins to target USP7. PLoS Pathog. 2015;11:e1004950. doi: 10.1371/journal.ppat.1004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski Y., Hansen G., Boomaars-van der Zanden A.L., Snijder E.J., Gorbalenya A.E., Hilgenfeld R. Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 2009;18:6–16. doi: 10.1002/pro.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1'-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1"-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Chatterjee A., Neuman B.W., Joseph J.S., Buchmeier M.J., Kuhn P., Wüthrich K. Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J. Virol. 2009;83:12998–13008. doi: 10.1128/JVI.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Chen Z.J. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo J.F., Hogue B.G. Host protein interactions with the 3' end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Kusov Y., Mutschall D., Tech S., Nagarajan K., Hilgenfeld R., Schmidt C.L. The "SARS-unique domain" (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 2007;364:877–882. doi: 10.1016/j.bbrc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5:e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar G., Tok T.T. Clarification of interaction mechanism of mouse hepatitis virus (MHV) N and nsp3 protein with homology modeling and protein–protein docking analysis. Curr. Comput. Aided Drug Des. 2016;12:98–106. [PubMed] [Google Scholar]
- Tyrrell D.A., Bynoe M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheugd P., Forst A.H., Milke L., Herzog N., Feijs K.L., Kremmer E., Kleine H., Lüscher B. Regulation of NF-κB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 2013;4:1683. doi: 10.1038/ncomms2672. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., Snijder E.J., Gorbalenya A.E., Coutard B., Piotrowski Y., Hilgenfeld R., Tucker P.A. Structure of the X (ADRP) domain of nsp3 from feline coronavirus. Acta Crystallogr. D. Biol. Crystallogr. 2009;65:1292–1300. doi: 10.1107/S0907444909040074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., van Kasteren P.B., Kikkert M., Snijder E.J., Gorbalenya A.E., Tucker P.A. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J. Virol. 2010;84:10063–10073. doi: 10.1128/JVI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Cong L., Chen C., Wei L., Zhao Q., Xu X., Ma Y., Bartlam M., Rao Z. Crystal structures of two coronavirus ADP-ribose-1"-monophosphatases and their complexes with ADP-ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 2009;83:1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y., Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014;95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- Yuan W., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.X., He F., Zhang L. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Schelle B., Karl N., Minskaia E., Bayer S., Siddell S.G., Gorbalenya A.E., Thiel V. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007;81:3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]