Highlights
-
•
The SARS-CoV genome encodes eight accessory proteins; none is essential for RNA replication.
-
•
Several appear to be involved in virus–host interactions and to influence the pathogenicity of the virus.
-
•
Some of these accessory proteins modulate the interferon pathway and the production of pro-inflammatory cytokines.
-
•
Five of the accessory proteins are incorporated into virions as minor structural proteins.
-
•
Other coronaviruses including MERS-CoV also encode accessory proteins.
Keywords: SARS-CoV, Other coronaviruses, Accessory proteins, Structure and function
Abstract
The huge RNA genome of SARS coronavirus comprises a number of open reading frames that code for a total of eight accessory proteins. Although none of these are essential for virus replication, some appear to have a role in virus pathogenesis. Notably, some SARS-CoV accessory proteins have been shown to modulate the interferon signaling pathways and the production of pro-inflammatory cytokines. The structural information on these proteins is also limited, with only two (p7a and p9b) having their structures determined by X-ray crystallography. This review makes an attempt to summarize the published knowledge on SARS-CoV accessory proteins, with an emphasis on their involvement in virus–host interaction. The accessory proteins of other coronaviruses are also briefly discussed. This paper forms part of a series of invited articles in Antiviral Research on “From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses” (see Introduction by Hilgenfeld and Peiris (2013)).
1. Introduction
The severe acute respiratory syndrome coronavirus (SARS-CoV) was identified as the etiological agent of severe acute respiratory syndrome during the 2002–2003 outbreak (Peiris et al., 2003). SARS-CoV belongs to the genus Betacoronavirus of the family Coronaviridae (King et al., 2011). Coronaviruses are enveloped viruses with positive-sense, non-segmented, single-stranded RNA genomes, characterized by club-like projections on the virus particles (Masters, 2006). The first two thirds of the coronavirus genome encode the replicase genes, which are translated into two large polyproteins, pp1a and pp1ab, that are processed into 15 or 16 non-structural proteins (nsp) via proteolytic cleavage (Thiel et al., 2003). The remaining one third of the genome contains ORFs for the structural proteins, namely the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins.
In addition to these genomic elements shared by other coronaviruses, the SARS-CoV genome also contains eight ORFs coding for accessory proteins, namely ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b (Fig. 1 ). These proteins are specific for SARS-CoV and do not show significant homology to accessory proteins of other coronaviruses, with the exception of the SARS-like coronavirus SL-CoV-WIV1 that was recently discovered in bats (Ge et al., 2013) and has the same set of accessory proteins as SARS-CoV. The potential connection between these accessory proteins and the high virulence of SARS-CoV has led to detailed structural and functional studies.
Fig. 1.

Schematic diagram showing the genome organization of the severe acute respiratory syndrome coronavirus (SARS-CoV). The eight accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b) are shown as colored boxes. The 5′ leader sequence (black box), open reading frames (ORFs 1a, 1b) encoding components of the replication/transcription complex, the structural genes spike (S), membrane (M), envelope (E), and nucleo-capsid (N) are also indicated (not drawn to scale).
Coronavirus accessory proteins have been generally considered to be dispensable for viral replication in vitro (de Haan et al., 2002, Haijema et al., 2004, Yount et al., 2005, Ontiveros et al., 2001, Shen et al., 2003, Hodgson et al., 2006, Casais et al., 2005). However, several accessory proteins have been shown to exhibit functions in virus-host interactions during coronavirus infection in vivo. For example, deletion of ORF2a (NS2), HE, ORF4, and ORF5a in the mouse hepatitis virus (MHV) led to a significant attenuation of the virus in its natural host (de Haan et al., 2002). On the other hand, continuous passage of infectious bronchitis virus (IBV) in cell culture results in a mutation in the 3b gene coding for a C-terminally truncated 3b protein. Interestingly, compared with wild-type virus, the mutant virus has a growth advantage and increases virulence in the chicken embryo (Shen et al., 2003). Similarly, recent studies have suggested that SARS-CoV accessory proteins may confer biological advantages to the virus in the natural host, and contribute to the pathogenesis of SARS (Narayanan et al., 2008). Extensive functional studies in cell culture and animal models have shown that SARS-CoV accessory proteins are involved in a wide variety of cellular processes, such as cell proliferation, programmed cell death, activation of stress response pathways and cytokine production, to name just a few. Notably, some coronaviral accessory proteins have also been shown to modulate the interferon signaling, which is of paramount importance for host antiviral immunity (Frieman et al., 2007, Dedeurwaerder et al., 2014, Zhang et al., 2013, Cruz et al., 2013).
As part of a series of invited articles in Antiviral Research on “From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses” (Hilgenfeld and Peiris, 2013), this review summerizes our current knowledge on SARS-CoV accessory proteins, including an update on structural and functional studies, with a particular emphasis on the involvement of accessory proteins in virus-host interactions. In addition, we will briefly summarize the current knowledge on accessory proteins of other coronaviruses.
2. Accessory proteins of the SARS-CoV
2.1. Orf3a
Proteins 3a and 3b (p3a and p3b, previously also known as X1 and X2), comprise 274 and 154 amino-acid residues, respectively (Marra et al., 2003, Rota et al., 2003). They are encoded by ORF3a (commonly known as ORF3 or U274) and ORF3b and both make up the second largest sub-genomic RNA in the SARS-CoV genome (Tan et al., 2006). So far, the structures of the p3a and p3b are still not known (Bartlam et al., 2005) (see Table 1 ).
Table 1.
SARS-CoV ORF3a.
| SARS-CoV p3a |
|---|
|
Domains and/or regions with characterized functions
|
Protein 3a was detected in vivo or in vitro when anti-p3a antibodies were used in SARS-infected specimens or patients (Yu et al., 2004). Hence, the protein can be used as a diagnostic marker for patients infected with SARS-CoV. In vivo, the presence of p3a was found in autopsy sections from lung and intestinal tissues of SARS patients (Law et al., 2005). Uniquely, frameshift mutations do not necessarily cause protein 3a to be non-functional (Tan et al., 2005) and even the virus can deliberately make use of frameshift mutations to code for 3a variants by exploiting hepta- and octa-uridine sites (Wang et al., 2006).
It has been shown that SARS-CoV protein 3a is a structural component by using electron microscopy (Ito et al., 2005). Shen et al. supported this finding by demonstrating that the protein can be incorporated into virus-like particles (VLPs) using recombinant baculoviruses expressing E, M, and p3a (Shen et al., 2005). However, for the formation of these VLPs, p3a of SARS-CoV is dispensible (Yount et al., 2005). Being efficiently expressed on the cell surface, the protein was easily detected in a majority of SARS patients due to the triggering of a humoral and cellular immune response in these patients (Lu et al., 2009). In light of immunity, p3a can cause the activation of NF-κB and JNK leading to the upregulation of RANTES and IL-8 in A549 and HEK293T cells (Kanzawa et al., 2006). It has also been demonstrated to up-regulate fibrinogen mRNA and protein levels in A549 cells (Tan et al., 2005).
Protein 3a has been shown to be present in both the plasma membrane as well as intracellularly (Ito et al., 2005, Oostra et al., 2006, Tan et al., 2004, Yuan et al., 2005). It can also be detected by Western blot in SARS-CoV particles purified by sucrose gradient centrifugation. Yu et al. used confocal microscopy and fractionation to show that p3a is distributed in a “punctate pattern” in the cytoplasm while the majority of its expression is concentrated at the Golgi apparatus (Yu et al., 2004). On top of all these findings, Tan et al. have shown that this protein can also be transported to the cell surface and enter the cell through endocytosis (Tan et al., 2004). Additionally, through immunoprecipitation, this protein was shown to interact with SARS-CoV M, S, E, and 7a proteins (Tan et al., 2004, von Brunn et al., 2007).
Being the largest accessory protein (Narayanan et al., 2008), p3a has an extracellular N-terminus and an intracellular C-terminus (Fig. 2 ) Tan et al., 2004, Lu et al., 2006. Another feature of the protein is the presence of a cysteine-rich domain (aa 81–160) Lu et al., 2006.
Fig. 2.

Schematic diagram showing motifs and domains within SARS-CoV accessory proteins p3a and p6. TM, transmembrane domain.
The N-terminal region consists of three transmembrane domains (TMDs) (Tan et al., 2004) (Fig. 3 ). Recently, through computational modeling, Kruger and Fischer proposed a theoretical model of the protein being an ion channel through the use of its transmembrane domains (Kruger and Fischer, 2009). Chien et al. expressed the full-length p3a and reconstituted it into lipid bilayers to characterize the ion-channel activity and suggested that either TMD 2 or 3 lines the pore of the channel (Chien et al., 2013). The central region comprising residues 125–200 of p3a has been shown to be bound to the 5’ UTR of the SARS-CoV genome (Sharma et al., 2007), characterized by Sharma et al. by using yeast-three-hybrid screen, electrophoretic mobility shift assay, and ultraviolet crosslinking (Sharma et al., 2007). Intriguingly, just the N-terminal portion of p3a is able to evoke a strong humoral immune response (Zhong et al., 2006).
Fig. 3.
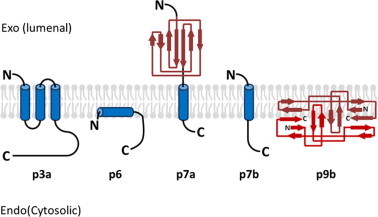
Schematic diagram showing the known topological and structural features of SARS-CoV accessory proteins. N-terminus (N) and C-terminus (C) are indicated. α-Helices are represented by blue columns and β-strands are represented by red arrows. The topological or structural features of p3b, p8a, and p8b are not well understood. Ribbon diagrams for p7a and p9b are adapted from Nelson et al. (2005) and Meier et al., 2006, respectively.
The cysteine-rich domain of p3a serves a multitude of functions and some have suggested its role in the p3a ion channel. The p3a is able to interact with the SARS-CoV S protein by formation of disulfide linkages through its cysteine-rich regions, mainly the CWLCWKC region (aa 127–133) (Zeng et al., 2004). Another important feature is the oligomerization of p3a as it is able to form homodimers and homotetramers (Lu et al., 2006). Through inducing point mutations, Lu et al. showed that Cys133 is important for oligomerization of the protein and confirmed the formation of dimers and tetramers through FRET (Lu et al., 2006). Utilizing Xenopus oocytes, their results were suggestive of the tetramers being able to form a potassium-permeable ion channel and modulating release of SARS-CoV particles (Lu et al., 2006, Shi et al., 2006). Chan et al. further validated that blocking these ion channels suppressed SARS-CoV-mediated apoptosis (Chan et al., 2009).
The C-terminal domain is hydrophilic in nature and contains both the YxxΦ (where x represents any aa and Φ is an amino acid with a hydrophobic, bulky side-chain) and the diacidic motifs (ExD, Asp-x-Glu, where x represents any aa). This domain can play a role in G1 cell-cycle arrest by depletion of cyclin D3 (Yuan et al., 2007). Interestingly, p3a seems to share a similar topology with the M protein (Marra et al., 2003). The YxxΦ motif plays a role in internalization of proteins into various intracellular components, including proteins from the plasma membrane into the endosomes (Bonifacino and Traub, 2003, Trowbridge et al., 1993). The diacidic motif is required for ER export (Nishimura and Balch, 1997). Tan et al. demonstrated that deletion of the C-terminal domain would prevent the protein from being expressed on the cell surface (Tan et al., 2004). Furthermore, they also hypothesized that the YxxΦ motif presented in p3a would allow the internalization of the S protein and prevent its presentation on the cell surface (Tan, 2005). This could be involved in evading host immunity systems or even facilitate the assembly of SARS-CoV.
p3a can be detected in the supernatant in two different forms from infected Vero E6 cells or Caco2 cells (Lu et al., 2006, Huang et al., 2006). Huang et al. demonstrated by sucrose-gradient centrifugation and densitometric scans the presence of a 37-kDa form (protein 3a-1) and a 31-kDa form (protein 3a-2). The 37-kDa form was detected in fractions with similar densities (1.18–1.20 g/ml) to the SARS-CoV particles, as opposed to fractions containing the 31-kDa form (1.13–1.15 g/ml) Tan et al., 2005, Yuan et al., 2007, suggesting that the 31-kDa form may be present as extracellular membrane structures while the 37-kDa form may be assembled into the mature virons. The presence of these two forms is due to the protein 3a being able to be modified post-translationally. This protein can be O-glycosylated as demonstrated by Oostra et al. through electrophoretic mobility-shift assays, transforming the unmodified 31-kDa form into a 33-kDa form (Oostra et al., 2006). p3a resembles the M protein of other coronaviruses, such as MHV, in terms of (1) the subcellular localization pattern of M protein (Tan et al., 2004, Klumperman et al., 1994, Yuan et al., 2005), (2) having an extracellular N-terminus and a C-terminal endodomain, (3) spanning the membrane three times (Rottier et al., 1984, Rottier et al., 1986, Braakman and Van Anken, 2000), and (4) being considered a structural protein (Ito et al., 2005, Shen et al., 2005). Despite both M and 3a proteins being glycosylated, p3a in Ost-7 cells is neuraminidase-sensitive but not O-glycosidase sensitive, in contrast to the M protein where it is sensitive to both (Oostra et al., 2006).
Recombinant viruses with ORF3a deleted (i.e., SARS-CoV Δ3a) showed significant reduction in virus titres after infecting Vero, MA104, and Caco2 cells (Yount et al., 2005). Interestingly, Yount et al. demonstrated that deletion of ORF3 and ORF6 decreased the overall virus titres as compared to just deleting ORF 3a alone, suggesting that ORF3a is non-essential (Yount et al., 2005). In contrast, there is conflicting evidence that ORF3a may be essential, because of the reduction of viral titres in SARS-CoV-infected cells transfected with a siRNA targeting p3a (Åkerström et al., 2007). We propose that this may be due to the subsequent knockdown of p3b as well, resulting in the lower virus production.
Using transmission electron microscopy (TEM), Freundt et al. observed in Vero cells that only in the presence of p3a, intracellular vesicles will be formed, as observed in the pathology in SARS patients (Freundt et al., 2010). The presence of Lamp-1 through immunofluorescence can also be detected in these vesicles (Freundt et al., 2010), indicating the formation of late endosomes. Expression of p3a is also required for the Golgi fragmentation for its acquisition of a viral envelope. Through yeast-2-hybrid and FRET assays, Padhan et al. have demonstrated caveolin-1 binding to p3a, which may play a role in virus uptake and release (Padhan et al., 2007).p3a has also been associated with ER stress through the activation of the PERK pathway but not the IRE-1 and ATF6 pathway (Minakshi et al., 2009). Minakshi et al. analyzed this by transfecting Huh7 cells with p3a and luciferase-tagged ER stress-related molecules, showing that it can enhance protein folding, but not activate Endoplasmic Reticulum Associated Degradation (ERAD) Minakshi et al., 2009. The long-term triggering of the PERK pathway can also induce virus-related apoptosis through the expression of its downstream mediators of ATF4 and CHOP, similar to IBV infection (Liao et al., 2013). Padhan et al. have shown that protein 3a can lead to increased activation of the p38 MAP kinase pathway and induce the mitochondria to leak cytochrome c to induce apoptosis (Padhan et al., 2008).
2.2. Orf3b
Protein 3b (p3b) is translated from ORF3 using an IRES (Rota et al., 2003), overlapping the SARS-CoV E gene as well as the ORF3a. Similarly to the SARS-CoV p3a, p3b does not share any homology with other known proteins (Marra et al., 2003). Moreover, p3b is not expressed in SARS-CoV isolates from bats due to the presence of a stop codon (Ren et al., 2006). Anti-p3b antibodies can also be detected in the sera of infected SARS patients (Guo et al., 2004). Yuan et al. have shown using immunofluorescence that the SARS-CoV p3b is localized in the nucleolus of infected Vero E6 cells (Yuan et al., 2005) as well as in the mitochondria (Yuan et al., 2006, Freundt et al., 2009). The shuttling behavior may be due to the presence of a nuclear export sequence and determined by a leptomycin B-sensitive mechanism. Furthermore, involvement of p3b has also been demonstrated in the induction of necrosis and apoptosis independent of its subcellular localization (Khan et al., 2006, Yuan et al., 2005). p3b is also able to inhibit the antiviral response by down-regulating type-I interferon (IFN-β) as well as the mitochondrial antiviral response (Freundt et al., 2009, Spiegel et al., 2005). Using flow cytometry, Yuan et al., showed that COS-7 cells transfected with p3b are arrested at the G0/G1 phase (Yuan et al., 2005). The protein may play a role in immunomodulation through binding to RUNX1b, as seen from yeast-two-hybrid and co-immunoprecipitation by Varshney et al. Varshney et al. (2012). Additionally, p3b can act as an interferon antagonist through inhibition of IRF3 (Kopecky-Bromberg et al., 2007) and activation of AP-1 through upregulating the expression levels of JNK and ERK (Varshney and Lal, 2011) (see Table 2 ).
Table 2.
SARS-CoV ORF3b.
| SARS-CoV p3b |
|---|
|
Domains and/or regions with characterized functions
|
2.3. Orf6
Protein 6 (p6) is detected in lung and ileum tissues of patients and in SARS-CoV-infected Vero E6 cells (Geng et al., 2005). The protein is found to associate with cellular membranes and is mainly localized in the ER and Golgi apparatus, either when expressed via a recombinant murine coronavirus or when it is transfected as an EGFP-fusion protein (Geng et al., 2005, Pewe et al., 2005). By sucrose gradient centrifugation and a virus-capture assay, Huang et al. have demonstrated that p6 is incorporated into mature virions (Huang et al., 2007). Moreover, when co-expressed with SARS-CoV S, M, and E proteins, p6 is incorporated into VLPs. However, p6 is not detected in the virions when expressed via a recombinant murine coronavirus (MHV) (Pewe et al., 2005). These results indicate that specific physical interactions between p6 and other SARS-CoV structural proteins may be required for recruiting p6 into the virus particles (see Table 3 ).
Table 3.
SARS-CoV ORF6.
| SARS-CoV p6 |
|---|
|
Domains and/or regions with characterized functions
|
Initial reverse genetic studies by Yount et al. have shown that recombinant SARS-CoV with ORF6 deleted (rSARS-CoV-Δ6) replicates similarly to wild-type virus in cell cultures as well as in infected BALB/c mice (Yount et al., 2005). However, a later study by Zhao et al. has demonstrated that, at a low MOI (0.01), rSARS-CoV-Δ6 replicates slower and to lower titres in cell cultures (Zhao et al., 2009). Compared with wild-type virus, lower viral titres as well as lower levels of viral RNA and protein were observed for rSARS-CoV-Δ6 at the early stage of infection, but the differences gradually diminished as the infection progressed. Importantly, when transgenic mice expressing the SARS-CoV receptor hACE2 were used, rSARS-CoV-Δ6-infected mice demonstrated a lower morbidity and mortality, with slightly lower virus titres in lung and brain tissue compared with wild-type virus (Zhao et al., 2009). Moreover, when ORF6 was introduced into an otherwise sub-lethal strain of MHV, the recombinant virus displayed enhanced growth in cell cultures and resulted in lethal encephalitis in infected mice (Pewe et al., 2005, Tangudu et al., 2007). Taken together, these results indicate that SARS-CoV p6 enhances viral replication in vitro and in vivo, and it may serve an important role in the pathogenesis during SARS-CoV infection.
SARS-CoV protein 6 is a 63-aa polypeptide with an amphipathic N-terminal portion (aa 1–40) and a highly polar C-terminal portion. An initial study using GFP-fusion protein and digitonin permeabilization supported an N-endo, C-endo membrane topology (Netland et al., 2007) (Fig. 3). Later, it was confirmed that residues 2–37 in the N-terminal region form an α-helix and are embedded in the cellular membrane (Zhao et al., 2009). On the other hand, the C-terminal part contains two signal sequences. The sequence YSEL (aa 49–52) is known to target proteins for internalization into endosomes, while the acidic tail signals ER export (Fig. 2) (Netland et al., 2007).
Utilizing the recombinant MHV expressing SARS-CoV p6 as mentioned above, Netland et al. demonstrated that the enhancement effect of p6 is dependent on the N-terminal amphipathic region but not the C-terminal polar region (Netland et al., 2007). Using transmission electron microscopy, Zhou et al. showed that full-length p6 or the N-terminal domain modified the cellular membrane, producing perinuclear vesicles that resemble the double-membrane vesicles (DMVs) known to be associated with coronavirus replication (Zhou et al., 2010). The ability to modify intracellular membranes may also explain the observation that SARS-CoV p6 induces ER stress in transfected cells (Ye et al., 2008). The p6-induced perinuclear vesicles, similar to the virus-induced DMVs, may derive and reorganize the membrane from the ER, thus leading to perturbation of ER function and induction of ER stress (Ye et al., 2008, Knoops et al., 2008). Notably, p6 is found to partially co-localize with nonstructural protein 3 (nsp3), which contains the papain-like viral protease and is a marker of the replication complex, as well as nonstructural protein 8 (nsp8), the primer-independent, non-canonical RNA polymerase (Tangudu et al., 2007, Zhou et al., 2010, Kumar et al., 2007). Moreover, physical interaction between p6 and nsp8 in SARS-CoV-infected Vero E6 cells has been confirmed by Kumar et al. using yeast two-hybrid and co-immunoprecipitation (Kumar et al., 2007). These results indicate that the N-terminal domain of p6 may play a role in the formation of the replication complex in SARS-CoV-infected cells and serve auxiliary or modulatory functions during SARS-CoV genome replication.
Protein 6 was identified as a β-interferon antagonist by Kopecky-Bromberg et al. with the intriguing finding that overexpression of p6 inhibits nuclear import of STAT1 in cells treated with IFN-β (Kopecky-Bromberg et al., 2007). Further studies by the same group revealed that p6 can physically interact with karyopherin alpha 2 (KPNA2) and tether it to the rough ER (Frieman et al., 2007). The immobilized KPNA2 interacts with karyopherin beta 1 (KPNB1) and prevents it from forming a complex with STAT1 to facilitate nuclear import (Zhao et al., 2009). The KPNA2-interacting region in p6 is mapped to 10 aa in the C-terminal region (Frieman et al., 2007). Later, Hussain et al. confirmed that p6 but not its C-terminally truncated mutant (p6ΔC) impeded the nuclear import of proteins harboring a classical nuclear localization signal (NLS) (Hussain et al., 2008). Further studies also demonstrated that the C-terminal tail is necessary but not sufficient to impede nuclear import, and a lipophilic N-terminus context is required (Hussain and Gallagher, 2010). Taken together, these results point to a role of the C-terminal domain of p6 in modulating host protein nuclear transport and type-I interferon signaling, which may be important for immune evasion during SARS-CoV infection.
One recent study has demonstrated a physical interaction and co-localization between SARS-CoV p6 and another accessory protein, 9b (Calvo et al., 2012). Other studies have also shown that when overexpressed, SARS-CoV p6 can stimulate DNA synthesis in cell culture and suppress the expression of co-transfected constructs (Geng et al., 2005, Hussain et al., 2008, Gunalan et al., 2011). The physiological significances of these findings in SARS-CoV replication and pathogenesis are currently unknown.
2.4. Orf7a
SARS-CoV accessory protein 7a (p7a, also known as ORF8, U122, or X4) is detected in SARS-CoV-infected Vero E6 cells and in lung specimens from SARS patients (Chen et al., 2005, Fielding et al., 2004, Nelson et al., 2005). The protein is localized in the ER and ER-Golgi intermediate compartment (ERGIC) in SARS-CoV-infected cells and in transfected cells (Fielding et al., 2004, Nelson et al., 2005). Using sucrose gradient centrifugation and a virus capture assay, Huang et al. have shown that SARS-CoV p7a is a structural protein incorporated into mature virions (Huang et al., 2006). Although p7a physically interacts with the SARS-CoV S protein and accessory protein 3a, neither S nor p3a is required for recruiting p7a into the virus particle; since p7a-containing VLPs can be produced in cells expressing only SARS-CoV M, E, and 7a (Tan et al., 2004, Huang et al., 2006) (see Table 4 ).
Table 4.
SARS-CoV ORF7a.
| SARS-CoV p7a |
|---|
|
Domains and/or regions with characterized functions
|
An early study based on reverse genetics has shown that deletion of ORF7a from the SARS-CoV genome does not significantly affect viral RNA synthesis and replication efficiency in cell culture (Yount et al., 2005). Schaecher et al. have also found that deletion of both ORF7a and 7b does not significantly affect SARS-CoV replication in transformed cells and in infected golden Syrian hamsters (Schaecher et al., 2007, Pekosz et al., 2006). Later, Dediego et al. confirmed that recombinant SARS-CoV with ORF6, 7a, 7b, 8a, 8b, and 9b deleted produced viral particles with similar morphology as wild-type SARS-CoV and replicated similarly in transgenic mice expressing the SARS-CoV receptor (DeDiego et al., 2008). These results indicate that p7a is not essential for SARS-CoV replication in vitro and in vivo.
SARS-CoV p7a is a type-I transmembrane protein with 122 amino-acid residues in length (Fig.3). The 15 N-terminal residues constitute a signal peptide, which is cleaved by signal peptidase in both SARS-CoV-infected cells and in transfected cells (Fielding et al., 2004, Nelson et al., 2005). The aa residues 16–96 comprise the luminal domain, which folds into a compact seven-stranded β sandwich that resembles members of the Ig superfamily (Nelson et al., 2005). The sequence and structural homology between SARS-CoV p7a and ICAM-1 suggested that p7a may interact with the lymphocyte function-associated antigen 1 (LFA-1), which was later confirmed by direct in vitro binding experiments in Jurkat cells (Hänel et al., 2006, Hänel and Willbold, 2007). These data suggest LFA-1 to be an attachment factor or even receptor for SARS-CoV on human leukocytes, although a majority of p7a is found to remain intracellular in SARS-CoV-infected cells (Nelson et al., 2005, Hänel and Willbold, 2007). Residues 97–117 of SARS-CoV p7a are highly hydrophobic and transverse the cellular membrane. The last 5 residues of the C-terminal tail (KRKTE) form a typical ER retention motif (KKXX or KXKXX, where X is any amino acid). When this motif is mutated from KRKTE to ERETE, p7a fails to be retro-transported to the ER and accumulates in the Golgi, where it undergoes rapid proteolytic processing (Fielding et al., 2004). Moreover, when the transmembrane domain and the short C-terminal tail of SARS-CoV p7a are fused with the cell-surface protein CD4, the chimera protein (CD4/orf7a-TM-tail) is found to be partially retained in the Golgi (Nelson et al., 2005). Fielding et al. have demonstrated physical interaction between SARS-CoV p7a and the host protein small glutamine-rich tetratricopeptide repeat-containing protein (SGT) (Fielding et al., 2006). Previous studies have shown that SGT interacts with the HIV-1 accessory protein vpu, which is essential in modulating viral particle release (Bour and Strebel, 2003). These results, together with the fact that SARS-CoV p7a is localized in the ER and ERGIC where coronaviruses generally assemble, and that p7a physically interacts with SARS-CoV M and E proteins, indicate that p7a may serve a function in the assembly stage during SARS-CoV replication (Fielding et al., 2006).
When transfected in mammalian cells, SARS-CoV p7a has been shown to induce caspase-dependent apoptosis as evidenced by the cleavage of poly(ADP-ribose) polymerase (PARP), a well-characterized apoptotic marker (Tan et al., 2004). This is consolidated by the finding that recombinant SARS-CoV with 7a and 7b deleted is not as efficient as wild-type virus in inducing DNA fragmentation, although the induction of early apoptotic markers such as annexin-V binding and caspase-3 activation are similar (Schaecher et al., 2007). These results indicate that SARS-CoV-induced apoptosis is only partially contributed by p7a, and other viral factors may be involved in the process. Later, Tan et al. demonstrated that the pro-apoptotic property of SARS-CoV p7a depends on its transmembrane domain, which is also required for its interaction with Bcl-XL, an anti-apoptotic protein belonging to the Bcl-2 family (Tan et al., 2007). Interestingly, SARS-CoV p7a interacts strongly with the pro-survival members of the Bcl-2 family (such as Bcl-XL, Bcl-w, Mcl-1, and A1), but not the pro-apoptotic Bcl-2 family proteins (such as BAD, BID, BAX and BAK) (Tan et al., 2007). The selective interactions may hinder the pro-survival functions of these proteins and result in apoptosis induction in SARS-CoV-infected cells.
When overexpressed, protein 7a can inhibit the growth of Balb/c 3T3 cells in a dose-dependent and time-dependent manner (Chen et al., 2005). A later study by Yuan et al. confirmed that overexpression of 7a led to cell cycle arrest at the G0/G1 phase, possibly by inhibiting the phosphorylation of retinoblastoma (Rb) protein (Yuan et al., 2006). Deletion-mutant experiments narrowed down the domain spanning aa 44–82 to be essential for cytoplasmic localization and cell-cycle arrest (Yuan et al., 2006). Overexpression of 7a has also been shown to activate nuclear factor kappa B (NF-κB) and c-Jun N-terminal kinase (JNK), which in turn induce the production of pro-inflammatory cytokines such as interleukin 8 (IL-8) and RANTES (Kanzawa et al., 2006). Moreover, using EGFP fusion protein, Kopecky-Bromberg et al. showed that overexpression of 7a activates the p38 mitogen-activated protein kinase (MAPK) and inhibits cellular protein synthesis at the translation level (Kopecky-Bromberg et al., 2006). However, p7a-induced apoptosis is not inhibited in the presence of a p38 inhibitor, indicating that p7a induces apoptosis via mechanisms independent of the MAPK pathways (Kopecky-Bromberg et al., 2006).
Taken together, although SARS-CoV p7a is not essential for viral replication, it is a minor structural protein and may serve certain functions during viral assembly. Notably, p7a is actively involved in virus–host interaction. By inhibiting cellular translation, activating stress-induced MAP kinases, suppressing cell-cycle progression and inducing caspase-dependent apoptosis, p7a may be an important player in SARS-CoV pathogenesis.
2.5. Orf7b
Accessory protein 7b was predicted to be translated from a second ORF of SARS-CoV sgRNA7 (Snijder et al., 2003). The expression of 7b was later confirmed in Vero cells infected with SARS-CoV (Schaecher et al., 2007). No experiments have been performed to detect the expression of 7b in tissue samples from SARS-CoV patients, although the presence of anti-p7b antibodies in SARS-convalescent patient sera indicates that the 7b protein is likely expressed in vivo (Guo et al., 2004). Using sucrose gradient fractionation and immune-gold labeling, Schaecher et al. demonstrated that p7b is associated with intracellular viruses and incorporated into purified virions, although more stringent virus-capture assays have not been performed (Schaecher et al., 2007) (see Table 5 ).
Table 5.
SARS-CoV ORF7b.
| SARS-CoV p7b |
|---|
|
Domains and/or regions with characterized functions
|
SARS-CoV 7b is proposed to be translated via a ribosome leaky scanning mechanism, because the expression of 7b is significantly reduced when the upstream 7a start codon is mutated to a strong Kozak sequence or when an additional AUG is introduced upstream of the 7b start codon (Schaecher et al., 2007). Protein 7b is a 44-aa, highly hydrophobic polypeptide (Schaecher et al., 2007). It is an integral transmembrane protein with a luminal N-terminus and a cytoplasmic C-terminus (Fig. 3). Similar to p7a, the 7b protein is localized throughout the Golgi compartment in both SARS-CoV-infected cells and in cells transfected with 7b cDNA (Schaecher et al., 2007). The Golgi-restricted localization of p7b was attributed to the transmembrane domain, specifically aa 21–23 and 27–30 of p7b (Schaecher et al., 2008). Notably, when the Golgi-targeting sequence of SARS-CoV p7b is used to replace the transmembrane domain of the cell-surface protein CD4, the chimeric protein is retained in the Golgi complex (Schaecher et al., 2008). Thus, the transmembrane domain of p7b is both necessary and sufficient for its Golgi localization.
Various reverse genetic experiments have shown that the SARS-CoV ORF7b is not essential for viral replication in vitro and in vivo (Yount et al., 2005, Schaecher et al., 2007, DeDiego et al., 2008). Interestingly, a prototype virus (strain Frankfurt-I) isolated during the 2003 SARS outbreak has a 45-nt deletion in the transmembrane domain of ORF7b upon propagation in cell culture for 3 passages (Thiel et al., 2003). Pfefferle et al. have performed extensive investigations on the consequence of this deletion using reverse genetics (Pfefferle et al., 2009). It was found that compared with the parental virus, the recombinant SARS-CoV with ORF7b deleted had a replicative advantage in Caco-2 and Huh7 cells, but not in Vero cells (Pfefferle et al., 2009). Moreover, the virus titre and viral RNA levels were significantly higher in Syrian Golden Hamsters infected with the 7b-deleted virus, indicating that SARS-CoV p7b may be an attenuating factor in vivo (Pfefferle et al., 2009). In a different approach, Åkerström et al. showed that siRNA specific for SARS-CoV sgRNA7 can inhibit sgRNA7 and sgRNA8 (thus silencing the expression of 7a, 7b, 8a, 8b) without affecting the full-length genomic mRNA and other sgRNAs (Åkerström et al., 2007). In Vero E6 cells expressing this siRNA, the production of progeny viruses was significantly reduced, indicating that p7a/p7b (and p8a/p8b) may play certain roles during the replication cycle of SARS-CoV. The discrepancies regarding the functions of p7b during SARS-CoV replication may be due to the different systems and animal models used. Further studies are required to validate these results and characterize the detailed mechanisms.
2.6. ORF8a and ORF8b
In SARS-CoV isolated from animals and early human isolates, sgRNA8 encodes a single protein 8ab. Interestingly, in SARS-CoV isolated from humans during the peak of the epidemic, there is a 29-nt deletion in the middle of ORF8, resulting in the splitting of ORF8 into two smaller ORFs, namely ORF8a and ORF8b (He et al., 2004, Guan et al., 2003). During the late stage of the SARS epidemic, larger deletions (82-nt and 415-nt deletion) were also identified in clusters of viruses from human isolates (He et al., 2004, Chiu et al., 2005). Initially, this dramatic genomic change in ORF8 has been proposed to contribute to the zoonotic transition of SARS-CoV from palm civets to humans and thus has attracted particular attention. However, it has been shown that palm civets are equally susceptible to human SARS-CoV isolates with or without the 29-nt deletion (Wu et al., 2005). Moreover, further studies using reverse genetics showed that the ORF8 of SARS-CoV is not essential for virus replication in vitro and in vivo (Yount et al., 2005, DeDiego et al., 2008). Thus, whether the mutations in ORF8 are due to genomic instability or adaptive evolution remains to be investigated (see Table 6 ).
Table 6.
SARS-CoV ORF8a and ORF8b.
| SARS-CoV p8a, p8b, and p8ab |
|---|
|
Domains and/or regions with characterized functions
|
The undeleted ORF8 (or ORF8ab) encodes a 122-amino-acid protein with an N-terminal hydrophobic signal sequence. ORF8a encodes a 39-amino-acid polypeptide, and residues 1–35 are identical to the N-terminal of 8ab. ORF8b encodes an 84-aa polypeptide and residues 9–84 are identical to the C-terminal of 8ab (Oostra et al., 2007). Whereas Keng et al. have demonstrated by indirect immunofluorescence that 8a and 8b are expressed in SARS-CoV-infected Vero E6 cells, Oostra et al. were unable to detect 8b in SARS-CoV-infected Vero E6 cells (Oostra et al., 2007, Keng et al., 2006). Although there is no data regarding the expression of 8a or 8b in vivo, Chen et al. have identified anti-p8a antibodies in two out of 37 patients with SARS (Chen et al., 2007). Also, different subcellular localizations have been reported for p8a, p8b, and p8ab. Using an EGFP fusion protein, Keng et al. have shown that p8a and p8b are found in cytoplasmic punctuated vesicle-like structures, whereas p8ab localizes diffusely to the cytoplasm (Keng et al., 2006). On the other hand, Law et al. have observed EGFP-fused p8b to localize diffusely in both the cytosol and the nucleus (Law et al., 2006). Finally, using a T7 promoter-driven expression system, Oostra et al. have demonstrated a reticular pattern and ER localization for p8a and p8ab, but a diffuse cytoplasmic localization for p8b (Oostra et al., 2007). These discrepancies should be addressed, possibly by using more specific monoclonal antibodies, or expressing 8a and 8b fusion proteins under a more physiologically relevant background, such as using recombinant coronaviruses.
Oostra et al. have characterized the biochemical properties of p8a, p8b, and p8ab (Oostra et al., 2007). Owing to its N-terminal signal sequence, SARS-CoV p8ab enters the ER, where it is N-glycosylated at asparagine residue 81. The mature p8ab remains associated with the ER and possibly undergoes multimerization, but it is not transported to the Golgi for secretion (Oostra et al., 2007). Inside the ER, the protein up-regulates the synthesis of ER chaperons and activates the ATF6 branch of unfolded-protein response (UPR), but shows no effect on the PERK and IRE1 branches of the UPR (Sung et al., 2009). The 29-nt deletion disrupts the functional expression of ORF8. Although ORF8a contains the signal sequence, the protein is too short to be transported into the ER. For ORF8b, due to the lack of a signal sequence, it cannot enter the ER and thus is not glycosylated at the same asparagine residue as in 8ab (Oostra et al., 2007).
Proteins 8b and 8ab have been shown to undergo ubiquitination when expressed in both in vitro expression systems and in cell culture (Le et al., 2007). Moreover, p8b and p8ab physically interact with monoubiquitin as well as polyubiquitin, suggesting potential involvement of these proteins in the host ubiquitin–proteasome system. Interestingly, the N-linked glycosylation on residue Asn81 in p8ab stabilizes it and protects it from degradation by the proteasome. In sharp contrast, in the absence of the p8a region, p8b, which is not N-glycosylated at the same asparagine residue, undergoes rapid proteasomal degradation (Le et al., 2007).
Chen et al. have shown that SARS-CoV replication is enhanced in cells stably expressing 8a and reduced in cells transfected with siRNA targeting 8a (Chen et al., 2007). However, the results were based on detection of virus RNA and assessment of virus-induced CPE only, and a direct measure of infective virus titre should have been performed. Moreover, overexpression of 8a also induces caspase-dependent apoptosis (Chen et al., 2007). Ectopic expression of 8b reduces the level of co-transfected SARS-CoV E protein, but not that of other structural proteins. Strikingly, in SARS-CoV-infected cells, the expression of 8b and E are found to be mutually exclusive, indicating a functional antagonism between the two proteins (Keng et al., 2006). Similar to SARS-CoV p6, overexpression of 8b has been shown to stimulate DNA synthesis, although co-expression of p6 and 8b does not produce a synergistic effect (Law et al., 2006). The significance of these findings in SARS-CoV pathogenesis remains to be determined.
2.7. Orf9b
Accessory protein 9b is translated from a second ORF of SARS-CoV sgRNA9 via a ribosomal leaky scanning mechanism (Xu et al., 2009). The sgRNA9 also encodes the SARS-CoV N protein, and in fact ORF9b is a complete internal ORF within the N gene in an alternate reading frame (Xu et al., 2009). The 9b protein is detected in SARS-CoV-infected cells and the lung and ileum tissues from SARS patients (Nal et al., 2005). Moreover, anti-p9b antibodies are also detected in the serum of SARS patients (Qiu et al., 2005). These results indicate that SARS-CoV 9b is expressed in vitro and in vivo. Using sucrose gradient fractionation, Xu et al. have shown that p9b is incorporated into mature virions and packaged into VLPs when co-expressed with E and M proteins (Xu et al., 2009). These data indicate that p9b may be a structural component of SARS-CoV virions, but it is also possible that p9b is included during virus assembly due to interaction with E protein, as demonstrated by von Brunn et al. von Brunn et al. (2007). The SARS-CoV 9b protein is also shown to self-interact and interact with nsp5, nsp14, and the accessory protein p6, although the functional significances are not fully understood (von Brunn et al., 2007, Calvo et al., 2012). Several reverse genetic studies have demonstrated that p9b is not essential for SARS-CoV replication in vitro and in vivo (von Brunn et al., 2007, DeDiego et al., 2008) (see Table 7 ).
Table 7.
SARS Co-V ORF9b.
| SARS-CoV p9b |
|---|
|
Domains and/or regions with characterized functions
|
Protein 9b is a 98-amino-acid polypeptide with no sequence homology to any known proteins. Meier et al. have determined the crystal structure of p9b and shown that the protein has a novel fold with seven β-strands (Meier et al., 2006). The β-strands from two molecules form two adjacent twisted β-sheets, resulting in a highly interlocked handshake structure (Meier et al., 2006) (Fig. 3). Interestingly, the structure contains a hydrophobic central cavity, which binds lipid and stabilizes the molecule. Together with its membrane-association nature, this suggested that p9b may have a function during virus assembly (Meier et al., 2006).
Protein 9b contains no known NLS and enters the nucleus by passive diffusion (Sharma et al., 2011). However, amino-acid residues 46 to 54 (LRLGSQLSL) of p9b belong to a nuclear export signal (NES) motif LX1-3LX2-4LXL known to be transported by exportin 1 via an energy-dependent mechanism (Moshynskyy et al., 2007). Indeed, SARS-CoV p9b has been shown to physically interact with exportin 1 and moreover, mutation of the leucine-rich NES or inhibition of exportin 1 by leptomycin B have been shown to inhibit nuclear export of the 9b protein (Sharma et al., 2011, Moshynskyy et al., 2007). The nuclear export of p9b facilitates its degradation in the cytoplasm, whereas accumulation of p9b in the nucleus can induce caspase-dependent apoptosis (Sharma et al., 2011).
3. Accessory proteins of other betacoronaviruses
SARS coronavirus and its likely precursor, bat coronavirus SL-CoV WIV1 (Ge et al., 2013), as well as bat coronavirus HKU3 belong to lineage b of the genus Betacoronavirus ( Lau et al., 2005). Lineage a (see Fig. 4 for a phylogenetic tree) comprises a number of viruses that probably did not originate from bat coronavirus precursors, such as Bovine Coronavirus (BCoV), Murine Coronavirus (Mouse Hepatitis Virus, MHV), Human Coronavirus HKU1, and Human Coronavirus OC43. The genomes of BCoV, MHV, and Human Coronavirus OC43 encode the accessory protein NS2 (See Fig. 5 ), which, at least in case of MHV, has been shown to have phosphodiesterase activity specific for 2′,5′-oligoadenylate (2-5A) (Zhao et al., 2011, Zhao et al., 2012). Synthesis of the latter by the host enzyme oligo-2′,5′-A synthetase (OAS) is induced by double-stranded RNA intermediates occurring during viral replication, and leads to activation of the host’s RNase L, which in turn degrades viral and cellular mRNA (Silverman, 2007). As a result, viral replication and protein synthesis are interrupted. Thus, reduction of 2-5A levels by NS2 constitutes a mechanism of subversion of the interferon-inducible OAS – RNase L pathway. Interestingly, this process is organ- and cell-type specific; NS2 is essential for replication of MHV A59 only in the liver, but not in the brain (Zhao et al., 2013). This seems to depend on the level of the IFN-induced expression of the OAS gene, which differs between cell types. The NS2 protein belongs to the superfamily of 2H phosphodiesterases that is characterized by the presence of a pair of conserved His-X-Thr/Ser motifs (Mazumder et al., 2002).
Fig. 4.
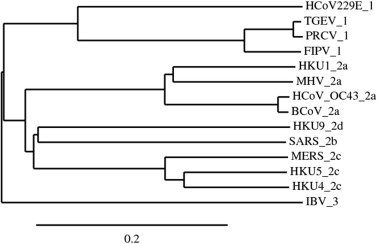
Phylogenetic analysis of coronaviruses based on pp1ab using Kalign multiple sequence alignment and TreeDyn tree viewer (Chevenet et al., 2006, Lassmann and Sonnhammer, 2005). Next to the branch, the virus name is indicated, along with the previously defined viral lineages, i.e. group 1 for Alphacoronavirus members, 2a, 2b, 2c, 2d for Betacoronavirus members, and 3 for the member of the genus Gammacoronavirus. Scale bar indicates nucleotide substitutions per site.
Fig. 5.

Genome organization of members of Betacoronavirus lineages a–d, as well as of the genera Alphacoronavirus and Gammacoronavirus, showing structural and accessory genes downstream of the orf1a/1b gene. The 5′ leader sequence (black box), open reading frames (ORFs 1a, 1b) encoding components of the replication/transcription complex, the genes encoding the structural proteins spike (S), membrane (M), envelope (E), and nucleocapsid (N) are also indicated in gray. Interspersed between (or within) them are the genes coding for putative accessory proteins (not drawn to scale). The following coronavirus genomes are illustrated (GenBank accession numbers in brackets): (1) Bovine coronavirus (NC_003045), (2) Mouse Hepatitis Virus (AC_000192), (3) Human coronavirus HKU1 (NC_006577), (4) Human coronavirus OC43 (NC_005147), (5) Severe Acute Respiratory Syndrome coronavirus (NC_004718), (6) Tylonycteris bat coronavirus HKU4 (NC_009019), (7) Pipistrellus bat coronavirus HKU5 (NC_009020), (8) MERS coronavirus (NC_019843), (9) Rousettus bat coronavirus HKU9 (NC_009021). (10) Feline infectious peritonitis virus (NC_002306), (11) Porcine respiratory coronavirus (DQ811787), (12) Transmissible gastroenteritis virus (DQ811788), (13) Human coronavirus 229E (NC_002645), (14) Infectious Bronchitis Virus (NC_001451).
Betacoronaviruses of lineage a also include a haemagglutinin esterase (HE) gene between orf1ab and the S gene (Fig. 5). The hemagglutinin-esterases (HEs) are a family of viral envelope glycoproteins that mediate reversible attachment to O-acetylated sialic acids by acting both as receptor-binding (“lectins”) and as receptor-destroying enzymes (RDEs) with esterase activity (Woo et al., 2010, Zhang et al., 1992, Vlasak et al., 1988, Schultze et al., 1991). In coronaviruses, this gene is exclusively present in betacoronaviruses of clade a and has probably been acquired after diverging from the ancestors of other betacoronavirus lineages (Zeng et al., 2008), possibly via horizontal gene transfer from Influenza C virus (Langereis et al., 2012). Crystal structures have been determined for the hemagglutinin esterases of BCoV and MHV (Vlasak et al., 1988, Schultze et al., 1991). The E domain (with esterase activity) is found to be highly conserved but the RBD (receptor binding domain) is found to be flexible (Vlasak et al., 1988). HCoV OC43 and BCoV use 9-O-acetylated sialic acids as receptor to initiate infection, and correspondingly possess sialate-9-O-acetylesterases as RDE. Most betacoronaviruses of lineage a attach to 9-O-acetylated sialic acid, but one group of murine coronaviruses has switched to using 4-O-acetylated sialic acid (Schultze et al., 1991). The available crystal structures reveal the adaptations of the RBD to differences in receptor usage (Vlasak et al., 1988, Schultze et al., 1991).
Betacoronaviruses of lineage a also encode an internal ORF within the nucleocapsid gene (Fig. 5). This ORF shares with SARS-CoV ORF9b the property of being completely overlapping with the nucleocapsid gene, but the encoded protein appears to have little in common with protein 9b of the SARS virus. A study performed on the internal ORF within the nucleocapsid gene of MHV suggests that this gene encodes a structural protein that is, however, found to be non-essential for virion formation and viral replication (Fischer et al., 1997).
In the genomes of betacoronaviruses of lineage c, for example in the bat viruses (BtCoV) HKU4, HKU5, and the recently characterized Middle-East Respiratory Syndrome (MERS) coronavirus, there are four ORFs between the S and E genes, which encode proteins p3 (also known as p3a), p4a (also known as p3b), p4b (also known as p3c), and p5 (also known as p3d) (Fig. 5). p3 and p4b have been predicted to each contain one transmembrane helix, whereas p5 has been proposed to comprise three such helices (Siu et al., 2014). p3 and p5 have been found in the ERGIC, whereas p4a and p4b display a diffuse cytoplasmic distribution and are also partly localized to the nucleus (Siu et al., 2014), although in another study, p4b was found to localize exclusively to the nucleus (Matthews et al., 2014). Both the p4a and the p4b protein of MERS coronavirus have been shown to be a type-I interferon antagonists (Siu et al., 2014, Matthews et al., 2014, Niemeyer et al., 2013). The p4a is particularly interesting in this context, as it is a double-stranded (ds) RNA-binding protein that prevents dsRNA (which occurs as an intermediate of viral RNA replication) from binding to the cellular dsRNA-binding protein PACT, which, as a consequence, cannot activate the cellular dsRNA sensors RIG-I and MDA5 (Siu et al., 2014, Niemeyer et al., 2013). In another study, it has been shown that p4a, p4b and p5 are all potent interferon antagonists with p4a being the most powerful in counteracting the antiviral effects of IFN via the inhibition of both interferon production and ISRE (Interferon-Stimulated Response Element) promoter element signaling pathways (Yang et al., 2013). MERS coronavirus also contains an internal ORF within the nucleocapsid gene, which has not yet been characterized (van Boheemen et al., 2012). This ORF was not previously described for BtCoV-HKU4 and BtCoV-HKU5 but is conserved in the genome sequences of both viruses.
In betacoronavirus lineage d, for example in bat coronavirus HKU9, two ORFs, both of which are connected with a transcription regulatory sequence (TRS), are found downstream of the N gene (Fig. 5). The TRSs are located at the 5′ end of coronavirus genes and include a highly conserved AU-rich core sequence that is essential for mediating a 100- to 1000-fold increase in mRNA synthesis when it is located in the appropriate context (Sawicki et al., 2007). Thus, the identification of TRSs before a predicted ORF helps identify new genes. The two accessory ORFs in bat-coronavirus HKU9 encode two putative non-structural proteins, NS7a and NS7b (Woo et al., 2007, Lau et al., 2010). This is in fact the first time that ORFs downstream of the N gene have been observed in a betacoronavirus.
4. Accessory proteins in Alphacoronavirus
In the genome of Feline Infectious Peritonitis Virus (FIPV), there are five ORFs coding for five accessory proteins (Fig. 5). These are the ORF3 proteins (comprising p3a, p3b, and p3c) and the ORF7 proteins, interestingly encoded by two ORFs (7a and 7b) downstream of the N gene. It has been demonstrated that the ORF7 proteins are essential for efficient replication in vitro and for virulence in vivo ( Haijema et al., 2004). The ORF3 proteins were found to have only supportive roles during infection of the target cell by FIPV (Dedeurwaerder et al., 2013). It has also been shown that the FIPV p7a protein is a type-I IFN antagonist but needs the presence of ORF3 proteins to fully exert its antagonistic function (Dedeurwaerder et al., 2014).
The ORF3 and ORF7 proteins of Porcine Respiratory coronavirus (PRCoV) and Transmissible Gastroenteritis Virus (TGEV) are known to play potential roles in determining virulence (Paul et al., 1997, McGoldrick et al., 1999, Tung et al., 1992).
Another notable accessory protein of an alphacoronavirus is the protein encoded by ORF4 in Human Coronavirus 229E (HCoV 229E; Fig. 5). It has been previously shown that the HCoV-229E genome codes for two accessory proteins, p4a and p4b. However, recent sequencing of clinical isolates revealed the presence of a full-length p4, whereas laboratory strains show the presence of a truncated p4 (Farsani et al., 2012). It has been suggested that extensive culturing of HCoV 229E might have resulted in this truncation (Dijkman et al., 2006). The high degree of conservation of the amino-acid sequence of p4 among clinical isolates suggests that the protein plays an important role in vivo.
5. Accessory proteins in Gammacoronavirus
The most studied gammacoronavirus to date is IBV. The genomic organization of a classic gammacoronavirus is 5′-UTR-Pol-S-3a-3b-E-M-5a-5b-N-UTR-3′ (UTR = untranslated region). Hence, it contains four accessory genes. The IBV sub-genomic mRNA 3 (sgRNA3) and sgRNA5 are known to be polycistronic (Liu et al., 1991, Liu and Inglis, 1992a, Liu and Inglis, 1992b). The sgRNA3 has been shown to be tricistronic and encodes 3a, 3b and E protein (previously known as 3c protein) (Liu et al., 1991, Liu and Inglis, 1991, Liu and Inglis, 1992a, Liu and Inglis, 1992b), whereas sgRNA5 is bi-cistronic and encodes 5a and 5b (Liu and Inglis, 1992a, Liu and Inglis, 1992b). Serial passages of IBV in Vero cells led to the emergence of a mutant IBV with a truncated 3b gene (Shen et al., 2003). The mutant virus is more virulent in chicken embryos, suggesting that the 3b protein may play a certain function in viral pathogenesis (Shen et al., 2003). Reverse genetics studies have shown that proteins 3a, 3b. 5a, and 5b are dispensable for viral replication (Hodgson et al., 2006, Casais et al., 2005). In non-classical Australian strains of IBV, accessory genes 3a and 3b were found to be replaced by the unrelated X1 gene, and 5a was lacking (Mardani et al., 2008). More recently, it has been demonstrated that a potential ORF situated between the M gene and gene 5a/b of IBV is transcribed through template-switching involving a non-canonical TRS, resulting in a 11-kD-protein of unknown function (Bentley et al., 2013).
6. Conclusions
SARS-CoV encodes eight accessory proteins, namely p3a, p3b, p6, p7a, p7b, p8a, p8b, and p9b. Reverse genetic studies have demonstrated that, whereas none of these proteins is essential for SARS-CoV replication, some of them, in particular p6 and p7b, may contribute to the virulence of SARS-CoV (Pewe et al., 2005, Pfefferle et al., 2009). Five of the SARS-CoV accessory proteins (p3a, p6, p7a, p7b, and p9b) have been shown to be incorporated into mature virions and are thus minor structural proteins. Most SARS-CoV accessory proteins are involved in more than one cellular process, such as interfering with the MAP-kinase pathway, DNA synthesis, induction of pro-inflammatory cytokines, or induction of caspase-dependent apoptosis, to name just a few. The crystal structures of SARS-CoV accessory proteins 7a and 9b have been determined, providing a structural basis for their biological functions (Nelson et al., 2005, Meier et al., 2006). However, the pathophysiological significances of these accessory proteins in the context of SARS-CoV infection are not completely understood, and further structural and functional studies are required.
Knowledge on the accessory proteins of other coronaviruses is even more limited than for SARS-CoV. The best characterized member of betacoronaviruses of lineage a, murine coronavirus (MHV), as well as some related viruses encode a hemagglutinin esterase and a 2’,5’-oligoadenylate phosphodiesterase, the latter of which is involved in reducing the activities of RNase L. Lineage-a betacoronaviruses also encode an “internal” protein through a gene completely overlapping with the nucleocapsid gene. Betacoronaviruses of lineage c, the most prominent example for which is the newly emerging Middle East Respiratory Syndrome coronavirus (MERS-CoV), feature a similar gene “within” the nucleocapsid gene. The bat coronavirus HKU9, a member of betacoronavirus lineage d, and some alphacoronaviruses such as Feline Infectious Peritonitis Virus (FIPV) contain open reading frames downstream of the nucleocapsid gene. Proteins 7a and 7b of FIPV are involved in counteracting the antiviral response exerted by type-I interferons, with support from proteins 3a and 3b. None of the accessory proteins of other alphacoronaviruses or of the gammacoronavirus Infectious Bronchitis Virus appear to have important in vivo functions, but again, further characterization is required to shed more light on their functions.
Acknowledgements
This work was partially supported by a Competitive Research Programme (CRP) grant (R-154-000-529-281), the National Research Foundation, Singapore, and an Academic Research Fund (AcRF) Tier 1 grant (RGT17/13), Nanyang Technological University and Ministry of Education, Singapore.
References
- Åkerström S., Mirazimi A., Tan Y.-J. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antiviral Res. 2007;73:219–227. doi: 10.1016/j.antiviral.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 2005;15:664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K., Keep S.M., Armesto M., Britton P. Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J. Virol. 2013;87:2128–2136. doi: 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes∗. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Bour S., Strebel K. The HIV-1 Vpu protein: a multifunctional enhancer of viral particle release. Microbes Infect. 2003;5:1029–1039. doi: 10.1016/s1286-4579(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Braakman I., Van Anken E. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic. 2000;1:533–539. doi: 10.1034/j.1600-0854.2000.010702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E., DeDiego M.L., Garcia P., Lopez J.A., Perez-Brena P. Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo. Virus Res. 2012;169:282–288. doi: 10.1016/j.virusres.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.-M., Tsoi H., Chan W.-M., Zhai S., Wong C.-O. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Y., Shuang B., Tan Y.X., Meng M.J., Han P. The protein X4 of severe acute respiratory syndrome-associated coronavirus is expressed on both virus-infected cells and lung tissue of severe acute respiratory syndrome patients and inhibits growth of Balb/c 3T3 cell line. Chin. Med. J. 2005;118:267–274. [PubMed] [Google Scholar]
- Chen C.-Y., Ping Y.-H., Lee H.-C., Chen K.-H., Lee Y.-M. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A.-L., Jacq B., Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien T.H., Chiang Y.L., Chen C.P., Henklein P., Hänel K. Assembling an ion channel: ORF 3a from SARS-CoV. Biopolymers. 2013:628–635. doi: 10.1002/bip.22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R.W., Chim S.S., Tong Y.-k., Fung K.S., Chan P.K. Tracing SARS-coronavirus variant with large genomic deletion. Emerg. Infect. Dis. 2005;11:168. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J.L., Becares M., Sola I., Oliveros J.C., Enjuanes L. Alphacoronavirus protein 7 modulates host innate immune response. J. Virol. 2013;87:9754–9767. doi: 10.1128/JVI.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder A., Desmarets L.M., Olyslaegers D.A., Vermeulen B.L., Dewerchin H.L. The role of accessory proteins in the replication of feline infectious peritonitis virus in peripheral blood monocytes. Vet. Microbiol. 2013;162:447–455. doi: 10.1016/j.vetmic.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder A., Olyslaegers D.A., Desmarets L.M., Roukaerts I.D., Theuns S. ORF7-encoded accessory protein 7a of feline infectious peritonitis virus as a counteragent against IFN-α-induced antiviral response. J. Gen. Virol. 2014;95:393–402. doi: 10.1099/vir.0.058743-0. [DOI] [PubMed] [Google Scholar]
- DeDiego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Wilbrink B., Pyrc K., Zaaijer H.L. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol. J. 2006;3:422X–423X. doi: 10.1186/1743-422X-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsani S.M.J., Dijkman R., Jebbink M.F., Goossens H., Ieven M. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes. 2012;45:433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Gunalan V., Tan T.H., Chou C.F., Shen S. Severe acute respiratory syndrome coronavirus protein 7a interacts with hSGT. Biochem. Biophys. Res. Commun. 2006;343:1201–1208. doi: 10.1016/j.bbrc.2006.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Tan Y.-J., Shuo S., Tan T.H., Ooi E.-E. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S.T., Weiss S.R., Masters P.S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H., Liu Y.M., Chan W.S., Lo A.W., Au D.M. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 2005;579:6763–6768. doi: 10.1016/j.febslet.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., Liu X., Zhuang Z. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Gunalan V., Mirazimi A., Tan Y.J. A putative diacidic motif in the SARS-CoV ORF6 protein influences its subcellular localization and suppression of expression of co-transfected expression constructs. BMC Res. Notes. 2011;4:446. doi: 10.1186/1756-0500-4-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.-P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänel K., Stangler T., Stoldt M., Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J. Biomed. Sci. 2006;13:281–293. doi: 10.1007/s11373-005-9043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänel K., Willbold D. SARS-CoV accessory protein 7a directly interacts with human LFA-1. Biol. Chem. 2007;388:1325–1332. doi: 10.1515/BC.2007.157. [DOI] [PubMed] [Google Scholar]
- He J.-F., Peng G.-W., Min J., Yu D.-W., Liang W.-J. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Narayanan K., Ito N., Peters C., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 2006;80:210–217. doi: 10.1128/JVI.80.1.210-217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 2007;81:5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Gallagher T. SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus. Virus Res. 2010;153:299–304. doi: 10.1016/j.virusres.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Perlman S., Gallagher T.M. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J. Virol. 2008;82:7212–7222. doi: 10.1128/JVI.02406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-κB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.-T., Choi Y.-W., Welkers M.R., Chan D.Z., Shen S. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Fielding B.C., Tan T.H., Chou C.-F., Shen S. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, A.M., Adams, M.J., Lefkowitz, E.J., Carstens, E.B., 2011. Virus Taxonomy: IXth Report of the International Committee on Taxonomy of Viruses, Access Online via Elsevier.
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J., Fischer W. Assembly of viral membrane proteins. J. Chem. Theory Comput. 2009;5:2503–2513. doi: 10.1021/ct900185n. [DOI] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T., Druce J. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366:293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., Zeng Q., Heesters B., Huizinga E.G., de Groot R.J. The murine coronavirus hemagglutinin-esterase receptor-binding site: a major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012;8:e1002492. doi: 10.1371/journal.ppat.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E.L. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Poon R.W., Wong B.H., Wang M., Huang Y. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel betacoronavirus subgroup. J. Virol. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.-W. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.Y.P., Liu Y.-M., Geng H., Kwan K.H., Waye M.M.-Y. Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus. FEBS Lett. 2006;580:3643–3648. doi: 10.1016/j.febslet.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T., Wong C.-H., Au T.C., Chuck C.-P., Kong S.-K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Le T.M., Wong H.H., Tay F.P., Fang S., Keng C.T. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007;274:4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Fung T.S., Huang M., Fang S.G., Zhong Y. Up-regulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Cavanagh D., Green P., Inglis S. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Inglis S. Identification of two new polypeptides encoded by mRNA5 of the coronavirus infectious bronchitis virus. Virology. 1992;186:342–347. doi: 10.1016/0042-6822(92)90094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Inglis S. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Tao L., Wang T., Zheng Z., Li B. Humoral and cellular immune responses induced by 3a DNA vaccines against severe acute respiratory syndrome (SARS) or SARS-like coronavirus in mice. Clin. Vaccine Immunol. 2009;16:73–77. doi: 10.1128/CVI.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.-J., Xu K., Schwarz W., Du L. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardani K., Noormohammadi A.H., Hooper P., Ignjatovic J., Browning G.F. Infectious bronchitis viruses with a novel genomic organization. J. Virol. 2008;82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure–function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick A., Lowings J., Paton D. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 1999;144:763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshynskyy I., Viswanathan S., Vasilenko N., Lobanov V., Petric M. Intracellular localization of the SARS coronavirus protein 9b: evidence of active export from the nucleus. Virus Res. 2007;127:116–121. doi: 10.1016/j.virusres.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Ferraro D., Pewe L., Olivares H., Gallagher T. Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J. Virol. 2007;81:11520–11525. doi: 10.1128/JVI.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Balch W.E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- Ontiveros E., Kuo L., Masters P., Perlman S. Analysis of nonessential gene function in recombinant MHV-JHM. Gene 4 knockout recombinant virus. Adv. Exp. Med. Biol. 2001;494:83. doi: 10.1007/978-1-4615-1325-4_13. [DOI] [PubMed] [Google Scholar]
- Oostra M., De Haan C., De Groot R., Rottier P. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80:2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., Rottier P.J. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K., Minakshi R., Towheed M.A.B., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Padhan K., Tanwar C., Hussain A., Hui P.Y., Lee M.Y. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007;88:3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- Paul, P.S., Vaughn, E.M., Halbur, P.G., 1997. Pathogenicity and sequence analysis studies suggest potential role of gene 3 in virulence of swine enteric and respiratory coronaviruses. In: Mechanisms in the Pathogenesis of Enteric Diseases, Springer, pp. 317–321. [DOI] [PubMed]
- Peiris J.S., Yuen K.Y., Osterhaus A.D., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Pekosz A., Schaecher S.R., Diamond M.S., Fremont D.H., Sims A.C. Springer; The Nidoviruses: 2006. Structure, Expression, and Intracellular Localization of the SARS-CoV Accessory Proteins 7a and 7b. (pp. 115–120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangudu C., Olivares H. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 2005;79:11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Krahling V., Ditt V., Grywna K., Muhlberger E. Reverse genetic characterization of the natural genomic deletion in SARS-coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol. J. 2009;6:131. doi: 10.1186/1743-422X-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Li W., Yu M., Hao P., Zhang Y. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Rottier P., Brandenburg D., Armstrong J., Van Der Zeijst B., Warren G. Assembly in vitro of a spanning membrane protein of the endoplasmic reticulum: the E1 glycoprotein of coronavirus mouse hepatitis virus A59. Proc. Natl. Acad. Sci. 1984;81:1421–1425. doi: 10.1073/pnas.81.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J., Welling G.W., Welling-Wester S., Niesters H.G., Lenstra J.A. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Diamond M.S., Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J. Virol. 2008;82:9477–9491. doi: 10.1128/JVI.00784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 2007;81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Wahn K., Klenk H.-D., Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991;180:221–228. doi: 10.1016/0042-6822(91)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Åkerström S., Sharma A.K., Chow V.T., Teow S. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS One. 2011;6:e19436. doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Surjit M., Satija N., Liu B., Chow V.T. The 3a accessory protein of SARS coronavirus specifically interacts with the 5′UTR of its genomic RNA, Using a unique 75 amino acid interaction domain. Biochemistry. 2007;46:6488–6499. doi: 10.1021/bi062057p. [DOI] [PubMed] [Google Scholar]
- Shen S., Lin P.-S., Chao Y.-C., Zhang A., Yang X. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Wen Z., Liu D. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N., Ye S., Alam A., Chen L., Jiang Y. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006;440:570–574. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- Silverman R.H. Viral encounters with 2′, 5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Yeung M.L., Kok K.-H., Yuen K.-S., Kew C. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S.-C., Chao C.-Y., Jeng K.-S., Yang J.-Y., Lai M. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J. The severe acute respiratory syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol. J. 2005;2:5. doi: 10.1186/1743-422X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T., Barkham T., Fielding B.C., Chou C.-F., Shen S. Genetic lesions within the 3a gene of SARS-CoV. Virol. J. 2005;2:51. doi: 10.1186/1743-422X-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Fielding B.C., Goh P.-Y., Shen S., Tan T.H. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Goh P.-Y., Fielding B.C., Shen S., Chou C.-F. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.X., Tan T.H., Lee M.J., Tham P.Y., Gunalan V. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Teng E., Shen S., Tan T.H., Goh P.-Y. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Tham P.-Y., Chan D.Z., Chou C.-F., Shen S. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J. Virol. 2005;79:10083–10087. doi: 10.1128/JVI.79.15.10083-10087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics Á., Hertzig T., Schelle B. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Trowbridge I., Collawn J., Hopkins C. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- Tung F.Y., Abraham S., Sethna M., Hung S.-L., Sethna P. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00412–e00473. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney B., Agnihotram S., Tan Y.-J., Baric R., Lal S.K. SARS coronavirus 3b accessory protein modulates transcriptional activity of RUNX1b. PLoS One. 2012;7:e29542. doi: 10.1371/journal.pone.0029542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney B., Lal S.K. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry. 2011;50:5419–5425. doi: 10.1021/bi200303r. [DOI] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Leider J., Spaan W., Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wong S.-M., Liu D. Identification of hepta-and octo-uridine stretches as sole signals for programmed +1 and −1 ribosomal frameshifting during translation of SARS-CoV ORF 3a variants. Nucleic Acids Res. 2006;34:1250–1260. doi: 10.1093/nar/gkl017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Tu C., Xin C., Xuan H., Meng Q. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 2005;79:2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Zheng B.-J., Zeng R., Lu W., Lin Y.-P. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Wong C.K., Li P., Xie Y. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Biophys. Acta. 2008;1780:1383–1387. doi: 10.1016/j.bbagen.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-J., Chen Y.-C., Hsiao C.-H., Kuo T.-C., Chang S.C. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Li J., Shan Y., Yang Z., Zhao Z. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Yao Z., Li J., Zhao Z. Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol. Cells. 2006;21:186. [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2 doi: 10.1186/1743-422X-2-66. (198-0024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yao Z., Shan Y., Chen B., Yang Z. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 2005;114:70–79. doi: 10.1016/j.virusres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yao Z., Wu J., Zhou Y., Shan Y. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am. J. Respir. Cell Mol. Biol. 2007;37:9–19. doi: 10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R.-F., Shi M.-D., Jiang M.-R., Xie Y.-H. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Jha B.K., Ogden K.M., Dong B., Zhao L. Homologous 2′, 5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Kousoulas K.G., Storz J. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology. 1992;186:318–323. doi: 10.1016/0042-6822(92)90089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Birdwell L.D., Wu A., Elliott R., Rose K.M. Cell-type-specific activation of the oligoadenylate synthetase–RNase L pathway by a murine coronavirus. J. Virol. 2013;87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Falcon A., Zhou H., Netland J., Enjuanes L. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 2009;83:2368–2373. doi: 10.1128/JVI.02371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Rose K.M., Elliott R., Van Rooijen N., Weiss S.R. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J. Virol. 2011;85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Guo Z., Yang H., Peng L., Xie Y. Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients. J. Gen. Virol. 2006;87:369–373. doi: 10.1099/vir.0.81078-0. [DOI] [PubMed] [Google Scholar]
- Zhou H., Ferraro D., Zhao J., Hussain S., Shao J. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J. Virol. 2010;84:3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


