Abstract
Ribosomal frameshifting is a mechanism of gene expression used by several RNA viruses to express replicase enzymes. This article focuses on frameshifting in two human pathogens, the retrovirus human immunodeficiency virus type 1 (HIV-1) and the coronavirus responsible for severe acute respiratory syndrome (SARS). The nature of the frameshift signals of HIV-1 and the SARS–CoV will be described and the impact of this knowledge on models of frameshifting will be considered. The role of frameshifting in the replication cycle of the two pathogens and potential antiviral therapies targeting frameshifting will also be discussed.
Keywords: Ribosomal frameshifting, HIV-1, Retrovirus, SARS–CoV, Coronavirus, RNA pseudoknot, Translation
1. Introduction
The translation of most eukaryotic mRNAs is initiated in a 5′-cap-dependent mechanism and in principle, this restricts protein synthesis to the first coding sequence on the mRNA. However, many RNA viruses have polycistronic genomes and thus, display a variety of strategies to allow downstream open reading frames (ORFs) to be accessed. Some viruses produce subgenomic-length RNAs in which the relevant downstream ORF is effectively moved to the 5′-end of the RNA from where it can be efficiently translated. Where the replication cycle involves a nuclear step, RNAs can be spliced by the cellular machinery and many cytoplasmically replicating viruses have evolved mechanisms to produce subgenomic mRNAs during transcription. In other viruses, the 5′-end problem is obviated simply by encoding all of the required information in a single ORF and subsequent processing of the encoded polyprotein proteolytically. Viruses have also evolved a number of unconventional translation strategies to express distal ORFs. These include leaky scanning, where the AUG of the 5′-most ORF is inefficiently recognised and ribosomes scan on to initiate at a downstream ORF; ribosomal re-initiation, where a post-termination complex remains associated with the mRNA and re-initiates translation at a downstream ORF and translational fusion, where two (or more) ORFs separated by a stop codon or in an overlapping configuration are translated as a single protein following termination codon suppression or programmed ribosomal frameshifting, respectively (reviewed in Gale et al., 2000, Pe’ery and Mathews, 2000).
Ribosomal frameshifting, the focus of this review, is a process where specific signals in the mRNA instruct the ribosome to change reading frame from the 0 to the −1 frame (movement 5′-wards) at a certain efficiency and to continue translation in the new frame. Frameshift signals are thus found within overlapping coding sequences. Several viruses employ frameshifting during replication, including retroviruses (excepting spumaviruses, gamma- and epsilonretroviruses), several eukaryotic positive-strand RNA viruses, double-stranded RNA viruses of yeast, some plant RNA viruses and certain bacteriophage (see Chandler and Fayet, 1993, Brierley, 1995, Dinman, 1995, Futterer and Hohn, 1996, Farabaugh, 1996, Farabaugh, 2000, Brierley and Pennell, 2001, Atkins et al., 2001, for reviews). In most of the systems studied to date, frameshifting is involved in the expression of replicases. In retroviruses, it allows the synthesis of the Gag-Pol and Gag-Pro-Pol polyproteins from which reverse transcriptase is derived and for most other viruses, frameshifting is required for expression of RNA-dependent RNA polymerases. Eukaryotic ribosomal frameshift signals consist of two essential mRNA elements: a “slippery” sequence, where the ribosome changes reading frame and a stimulatory RNA secondary structure, often an RNA pseudoknot, located a few nucleotides downstream (Jacks et al., 1988a; Brierley et al., 1989, ten Dam et al., 1990). A spacer region between the slippery sequence and the stimulatory RNA is also present and a precise length of this spacer must be maintained for maximal frameshifting efficiency (Brierley et al., 1989, Brierley et al., 1992, Kollmus et al., 1994). The slippery sequence is a heptanucleotide stretch that contains two homopolymeric triplets and conforms in the vast majority of cases to the motif XXXYYYZ (where X can be any nucleotide, Y is A or U and Z is not G). In eukaryotes, frameshifting is thought to occur by “tandem-slippage” of two ribosome-bound tRNAs, presumably peptidyl and aminoacyl tRNAs, which slip from the 0 (X_XXY_YYZ) to the −1 phase (XXX_YYY) (Jacks et al., 1988a). The homopolymeric nature of the sequence seems to be required to allow the tRNAs to remain base-paired to the mRNA in at least two out of three anticodon positions following the slip. Recent work has revealed that the slippery sequence actually forms part of a slightly larger motif, as primary sequences immediately adjacent to the heptanucleotide stretch can influence the efficiency of frameshifting through various mechanisms (Bertrand et al., 2002, Bekaert and Rousset, 2005). In isolation, however, slippery sequences engender very low levels of frameshifting and a stimulatory RNA structure is needed to amplify the signal. There is considerable diversity in the structure of these RNAs and the precise mechanism by which they act is not resolved (reviewed in Giedroc et al., 2000, Brierley and Pennell, 2001). In this article, frameshifting in the retrovirus human immunodeficiency virus type 1 (HIV-1) and the coronavirus responsible for severe acute respiratory syndrome (SARS) will be reviewed. The nature of the frameshift signals of HIV-1 and SARS–CoV will be discussed and the impact of this knowledge on models of frameshifting will be considered. The role of frameshifting in the replication cycle of the two pathogens and potential antiviral therapies targeting frameshifting will also be debated.
2. Frameshifting in HIV-1
HIV-1 is a lentivirus of the Retroviridae and the etiologic agent of acquired immune deficiency syndrome. A description of HIV replication and pathogenesis is far beyond the scope of this review; suffice to say that HIV-1 is responsible for great mortality and morbidity worldwide, has been the subject of intensive study, yet remains a huge global problem. There is an ongoing search for novel targets for antiviral intervention and a detailed molecular understanding of virus gene expression and replication, including frameshifting, may be beneficial in unearthing candidate targets. Although expression of the HIV-1 Gag-Pol polyprotein by ribosomal frameshifting was shown experimentally almost 20 years ago (Jacks et al., 1988b), the structure of the signal and its role in virus replication have remained topics of debate. Without question, the site of frameshifting is the U_UUU_UUA stretch located within the gag/pol overlap some 200 nucleotides upstream of the gag termination codon. Protein sequencing has confirmed frameshifting at this site, although tandem-slippage of both peptidyl-tRNAPhe and aminoacyl-tRNALeu accounts for only about 70% of the frameshift product, with the remaining 30% being derived from single slippage of peptidyl-tRNAPhe on the U-rich stretch (Jacks et al., 1988b, Yelverton et al., 1994). Mutational analysis has established that this region is essential both for frameshifting and virus replication (Biswas et al., 2004). Indeed, in a sequence comparison of one thousand HIV-1 isolates, the U_UUU_UUA sequence is entirely conserved (Biswas et al., 2004). In an early report, it was suggested that the U-rich stretch alone was sufficient for frameshifting in heterologous systems (Wilson et al., 1988) but it is now clear that a stimulatory RNA is necessary, although the precise structure of this element remains somewhat controversial (Fig. 1 ). An examination of the sequences downstream of the HIV-1 slippery sequence and equivalent regions of the genomes of HIV-2 and simian immunodeficiency virus (SIV) suggested the involvement of a stem-loop structure (Jacks et al., 1988b). Subsequently, mutational analysis, frameshift assays in transfected mammalian tissue culture cells and virus-infectivity assays have confirmed that the “original” stem-loop proposed by Jacks et al. is an essential component of the signal (Fig. 1A; Parkin et al., 1992, Kollmus et al., 1994, Stahl et al., 1995, Hill et al., 2002) and that the extent of frameshifting is related to the predicted stability of this stem (Bidou et al., 1997, Hill et al., 2002). A number of more complex models have been proposed, however and these are shown in Fig. 1. Three of the models retain the original stem-loop as a key element, but propose additional interactions. In the model of Le et al. (1991), the stem-loop is elaborated into an H-type pseudoknot by the interaction of three loop nucleotides with a region downstream to give a short stem 2 (Fig. 1B). In a further convolution, Dinman et al. (2002) proposed an intramolecular triplex structure, where in addition to the pseudoknot, four adjacent and consecutive base triples form between bases in stem 1 and loop 2 (Fig. 1D). The model of Dulude et al. (2002) also retained the original stem-loop but suggested an extension at the bottom of the stem by pairing of spacer nucleotides with a region downstream. The resulting two-stem helix is separated by a purine-rich bulge (Fig. 1E). Base-pairing of the spacer region was also proposed in the “short” pseudoknot model of Du et al. (1996) ( Fig. 1C). The structure, although resembling certain other viral frameshift pseudoknots (Brierley and Pennell, 2001) does not retain the original stem-loop element of Jacks and co-workers.
Fig. 1.
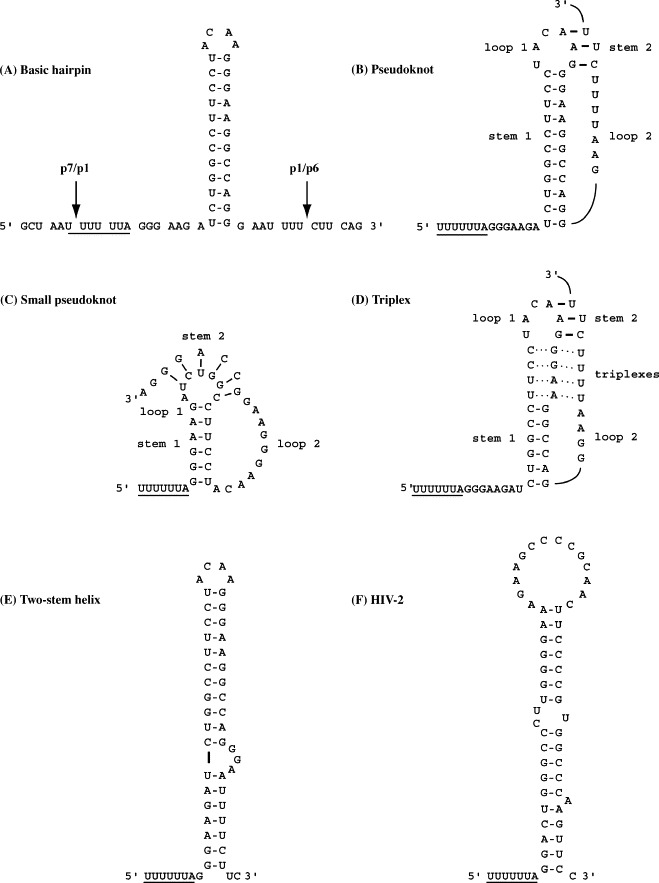
Proposed stimulatory RNAs at the frameshift sites of HIV-1 and HIV-2. The HIV-1 signal is shown in panels A–E and that of HIV-2 in panel F. (A) Basic hairpin. The original stem-loop proposed by Jacks et al. (1988b) is shown with the slippery sequence UUUUUUA upstream (underlined). The gag reading frame is indicated as triplets. Sites of cleavage by the viral protease within the encoded polypeptides are indicated by arrows. (B) The pseudoknot model of Le et al. (1991). In this model, the two pseudoknot stems are connected by single-stranded loops of three (loop 1) and eight nucleotides (loop 2), respectively. (C) The small pseudoknot model of Du et al. (1996). Here, base-pairing of the spacer region immediately downstream of the slippery sequence allows an alternative pseudoknot to be drawn that does not include the original stem-loop of Jacks et al. (1988b). (D) The triplex structure of Dinman et al. (2002). This model is based on the prediction of Le et al. (1991) (panel B) but includes the formation of four triplexes between the 3′-end of loop 2 and the top of stem 1 (dotted lines). (E) The two-stem helix model of Dulude et al. (2002) is the most favoured. Here, a short extension to the bottom of the original stem-loop is proposed, with an unpaired stretch (GGA) in the 3′-arm of the stem delineating the two-stem regions. (F) The HIV-2 (Rod) gag/pro frameshift signal can be folded to resemble the HIV-1 two-stem helix (panel E; see text).
Due to the related complementary pairing schemes of the various models, it has proven difficult to discriminate between them by standard structure probing and structure–function assays (Kang, 1998, Dinman et al., 2002, Dulude et al., 2002) although on balance, the two-stem helix model proposed by the Brakier-Gingras Laboratory is most consistent with these data (Dulude et al., 2002, Baril et al., 2003a; Fig. 1E). Further, the structure appears to be conserved in all subtypes of HIV group M, which constitutes >99% of viral isolates responsible for the worldwide pandemic (Baril et al., 2003a). Recent NMR analysis of the HIV-1 frameshift signal also lends strong support to this model (Staple and Butcher, 2003, Staple and Butcher, 2005, Gaudin et al., 2005).
The NMR data indicate that the HIV-1 stimulatory RNA possesses an apical stem in a continuous helix capped by a non-canonical U-G base-pair and an ACAA tetraloop. There is no evidence to support the formation of a pseudoknot or triplex. A less stable lower stem is present, separated from the upper stem by an asymmetric internal loop (GGA) which introduces a bend between the helices. Interestingly, the internal loop bases show continuous base-stacking, reminiscent of the internal loop of the HIV-1 SL1 stem-loop involved in genome packaging and reverse transcription (Greatorex et al., 2002, Lawrence et al., 2003). SL1 is likely to be involved in binding of the viral Gag polyprotein during packaging; however, there is no evidence to suggest that the HIV frameshift stem-loop binds to Gag. It has been shown that both the lower stem and the internal loop of the HIV-1 frameshift region contribute to frameshift efficiency (Dulude et al., 2002, Baril et al., 2003a), but the mechanism by which they exert their effects is not known. Indeed, this impinges on one of the central questions in the ribosomal frameshifting field and is considered in Section 4. Analysis by mfold indicates that in most HIV-2 strains, the spacer can also be paired to give a similar two-stem structure containing a bulge (an example is shown in Fig. 1F), although the specific asymmetric internal loop of the HIV-1 signal is not apparent in the HIV-2 examples studied (ROD, BEN, D194, MCR35, 96FR12034). Remarkably, the stimulatory RNA of an HIV-1 group O (outlier) strain, MVP5180, is not a stem-loop but a pseudoknot (Baril et al., 2003b). Group O strains are thought to have originated from different monkey to human transmission events, which could explain the dissimilar frameshift signal, although it has been noted that another O strain, ANT70, cannot form the same pseudoknot as MVP5180, as sequence differences are present that would prevent the formation of stem 2 (Baril et al., 2003b).
The frameshift process is crucial to HIV replication as it allows expression of the Gag-Pol polyprotein and thus targeting of replicative enzymes to the particle core during assembly. It also sets a precise ratio of Gag:Gag-Pol polyproteins, with some 5–10% of translational events yielding Gag-Pol. Maintenance of this ratio appears to be essential. Expression of HIV Gag alone, although sufficient for assembly and release of virus-like particles, leads to non-infectious virions lacking indispensable viral enzymes (discussed in Cen et al., 2004). Similarly, expression of the Gag-Pol polyprotein alone is detrimental, resulting in intracellular activation of the HIV-1 protease (PR) and inhibition of assembly and budding of virus-like particles (Park and Morrow, 1991, Karacostas et al., 1993, Cherry et al., 1998). Overexpression of Gag-Pol in T lymphocytes does lead to the secretion of virus-like particles, but these are non-infectious, not the least because they are devoid of genomic RNA (Kaye and Lever, 1996). Even subtle modulations of the Gag-Pol ratio can have profound effects on virus-infectivity. Shehu-Xhilaga et al. (2001) co-expressed Gag and Gag-Pol to generate an intracellular gradient of Gag:Gag-Pol ratios from 20:1 to 20:21 and found that genome RNA dimerisation was progressively inhibited as the concentration of Gag-Pol was increased and at approximately equimolar concentrations, HIV-1 infectivity was reduced about 1000-fold. Similarly, plasma virion isolates carrying stem-loops associated with greater than 60% reduction in frameshift efficiency were shown to have profound defects in replication (Telenti et al., 2002). Thus, even subtle modulations of the Gag:Gag-Pol ratio can substantially reduce virus fitness and this has implications for antiviral therapies (see below). More work is needed before a complete molecular explanation of the reduced viral fitness seen in such experiments is available. This is partly due to our incomplete understanding of the function of the proteins encoded in the frameshift region and also that the region is dual coding, which complicates mutational analysis. The organisation of the HIV-1 genome is shown in Fig. 2 , focusing on the proteins encoded in the Gag and Gag-Pol polyproteins in the vicinity of the frameshift signal. Frameshifting in HIV-1, as in most retroviruses, allows expression of the viral PR required for polyprotein cleavage during maturation. In Gag, PR cleavage sites almost precisely flank the frameshift signal (see Fig. 1, Fig. 2) and yield, in addition to N-terminal proteins (MA, CA, p2, NC), the p1 and p6 (p6Gag) proteins. In Gag-Pol, the equivalent overlapping coding sequences are cleaved to produce the transframe octapeptide (TFP) and p6* (p6Pol). All of the proteins encoded by the frameshift region have important roles in the virus life cycle. Changing specific residues within spacer peptide p1 affects infectivity, processing and dimer stability, probably by influencing activity of p15-NC, the nucleocapsid-p1-p6Gag precursor involved in genomic RNA binding (Hill et al., 2002). p6Gag, which includes a late budding domain (PTAP), is essential for virus assembly and release (Gottlinger et al., 1991; reviewed in Demirov and Freed, 2004, Morita and Sundquist, 2004). The transframe region of Gag-Pol is critical for PR regulation. The TFP-p6Pol-PR intermediate has low dimer stability with TFP-p6Pol functioning to inhibit PR until the appropriate point in maturation. Intramolecular cleavage at the p6Pol-PR site frees the N-terminus of PR, a critical step in the formation of a stable tertiary structure of PR and enzymatic activity (Louis et al., 1999a, Louis et al., 1999b). TFP-p6Pol can also inhibit PR directly, allowing further regulation (Paulus et al., 1999).
Fig. 2.
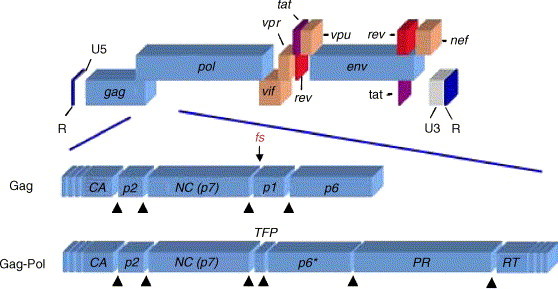
Genomic organisation of HIV-1. Key features of the HIV-1 genome are shown. Non-coding regions include the terminal repeat regions (R) and the 5′- (U5) and 3′- (U3) untranslated regions. The coding regions (not to scale) comprise the gag, pol and env genes common to all retroviruses, plus the HIV-1 accessory genes vif, vpr, tat, rev, vpu and nef. The lower portion focuses on the proteins encoded at and adjacent to the gag/pol overlap. Black triangles indicate cleavage sites recognised by the viral protease (PR), which releases the capsid (CA), p2, nucleocapsid (NC), p1 and p6 (p6Gag) polypeptides from within the portion of the Gag polyprotein shown and the CA, p2, NC, transframe peptide (TFP), p6* (p6Pol), PR and RT (reverse transcriptase) proteins from within the portion of the Gag-Pol polyprotein shown. The site of divergence between the polyproteins encoded by gag and gag-pol is indicated by an arrow (labelled fs [frameshift]).
The region of the genome that includes the frameshift region is likely to be of considerable importance in the development of resistance to antiviral drugs, particularly reverse transcriptase and PR inhibitors. There are several potential mechanisms that can be envisaged, including greater PR and reverse transcriptase production through frameshift regulation, enhanced packaging of viral enzymes via changes in p6Gag and control of activation of the viral PR, via p6Pol (Peters et al., 2001). Regarding frameshifting, a novel Gag-Pol frameshift site has been documented in HIV-1 variants resistant to PR inhibitors (Doyon et al., 1998). In these isolates, the p1/p6Gag cleavage site is changed from Phe-Leu to Phe-Phe, which at the nucleotide sequence level generates an additional homopolymeric and potentially slippery sequence (U_UUU_CUU to U_UUU_UUU) that could act to increase levels of Pol-encoded enzymes to counteract the PR deficit. This change is located within the lower stem of the Dulude structure (see above), yet the C to U transition is not predicted to destabilise the helix greatly (GU replaces GC pair). Despite lacking an obvious downstream stimulatory RNA, the novel sequence is functional in frameshifting (perhaps, because it contains seven consecutive U residues and may be more effective as a slippery sequence in the absence of a stimulatory RNA (Jacks et al., 1988a)) and supports Gag-Pol synthesis and PR activity in HIV molecular clones in which the authentic slippery sequence has been inactivated. The cleavage site mutation also generates a more effective cleavage site for both wild-type and PR-inhibitor-resistant proteases (Doyon et al., 1996). Thus, the reduced activity of drug-resistant proteases may be compensated for both by improved cleavage rates and increased levels of PR. It will be interesting to see whether other drug resistant isolates emerge that show stimulation of frameshifting independent of cleavage site changes, for example, by stabilisation of the stem-loop. There is no evidence that the virus itself can regulate frameshifting during replication however. In HIV-1 reporter constructs, for example, the level of frameshifting is not changed by co-infection with HIV-1 (Cassan et al., 1994, Reil et al., 1994). Similarly, the development of an infectious recombinant gammaretrovirus that expresses Gag-Pol by virtue of the HIV-1 frameshift signal rather than its natural readthrough signal argues against a specific role for HIV-encoded proteins in the frameshift process (Brunelle et al., 2003, Gendron et al., 2005).
3. Frameshifting in the SARS–CoV
The etiological agent of SARS is a novel coronavirus (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Kuiken et al., 2003, Peiris et al., 2003, Poutanen et al., 2003) with the first documented cases being traced to Guangdong Province, China, in November 2002 (Zhong et al., 2003, Xu et al., 2004). In February 2003, the disease was introduced to Hong Kong (Chim et al., 2003, Guan et al., 2004) and subsequently spread across 25 countries. Over 8000 cases and 774 deaths were attributed to this single outbreak (reviewed in Poon et al., 2004). To date, the source of this virus remains unidentified. Epidemiological data suggest that it is zoonotic; some of the earliest community-infected patients had a history of either trading and slaughtering of wild animals or contact with urban rodents (Zhong et al., 2003, Xu et al., 2004, Song et al., 2005).
Furthermore, viruses highly homologous (>99% nucleotide identity) to SARS–CoV have been isolated from Himalayan palm civets (Paguma larvata) and a raccoon dog (Nyctereutes procyonoides) from live animal markets in Guangdong Province (Guan et al., 2003, Song et al., 2005). Characteristically, these isolates are phylogenetically related to those from the earliest independent human cases of SARS (Chinese SARS Molecular Epidemiology Consortium, 2004, Song et al., 2005). Ferrets (Mustela furo), domestic cats (Felis domesticus) and Himalayan palm civets have also been shown to shed virus after experimental infection with SARS–CoV (Martina et al., 2003, Wu et al., 2005). Animal to human transmission of this virus has not yet been proven and the animal reservoir remains to be established.
Like all coronaviruses, the SARS–CoV genome is a single-stranded, non-segmented RNA of positive polarity. On the basis of phylogeny clustering, species of this genus are classified into three groups (1–3). Specifically, the SARS–CoV lineage has been proposed to cluster with group 2 (Eickmann et al., 2003, Gibbs et al., 2004, Snijder et al., 2003). Other human viruses monophyletically related are human coronaviruses OC43 (St-Jean et al., 2004, Vijgen et al., 2005a, Vijgen et al., 2005b) and HKU1 (Woo et al., 2005). The SARS–CoV genome is approximately 29.7 kb in length, with major genes arranged in a characteristic coronavirus pattern: 5′-replicase, spike, envelope, membrane, nucleocapsid-3′, flanked by untranslated regions (UTRs; see Fig. 3 ). Additionally, nine ORFs in the intergenic regions between the spike and nucleocapsid genes are present and predicted to encode nine proteins of unknown function (Marra et al., 2003).
Fig. 3.
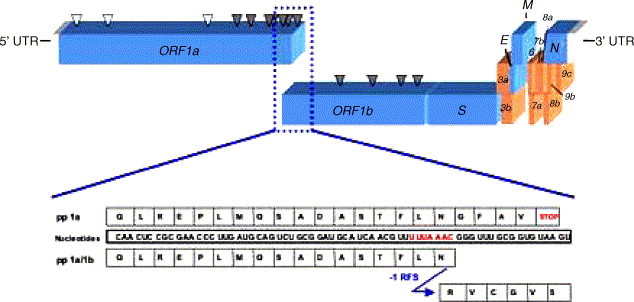
Genomic organisation of the SARS–CoV and the mechanism of translation of open reading frame (ORF) 1b. Key features of the SARS–CoV genome are shown. Untranslated regions (UTRs) flank the genome at both ends. Major ORFs are present in the following order: 5′-replicase (ORF1a, ORF1b), spike (S), envelope (E), membrane (M) and nucleocapsid (N)-3′ (depicted in blue). Other predicted ORFs (in brown) encode proteins of unknown function. ORFs 1a and 1b are translated from the genomic RNA into polyproteins (pps) 1a and 1a/1b. The synthesis of the 1b portion of 1a/1b involves programmed −1 ribosomal frameshifting (−1 RFS). Arrows represent the corresponding polyprotein sites cleaved by the papain-like proteinase 2 (white) and the 3C-like proteinase (grey). The expanded view shows the nucleotide sequence of the overlapping region between ORFs 1a/1b (slippery sequence in red) and the corresponding amino acids of pps 1a and 1a/1b. Tandem-slippage of the peptidyl-tRNAs (codon UUA) and aminoacyl-tRNAs (codon AAC) on the slippery sequence switches the ribosome into the −1 reading frame (frame of ORF 1b) and generates the amino acid sequences depicted.
The SARS–CoV replicase gene is organised into two partially overlapping ORFs (1a and 1b), which encode the polyprotein 1a (nt 265–13,413 isolate Tor2) and the fused polyprotein 1a/1b (nt 265–13,398 and 13,398–21,485, isolate Tor2), synthesised by programmed −1 ribosomal frameshifting (Thiel et al., 2003). The site of frameshifting is a U_UUA_AAC stretch located 12 bases upstream of the 1a stop codon. The overlap region of the SARS–CoV 1a/1b ORF is thus much shorter than that of HIV-1. Another difference is the magnitude of the frameshift. Reporter plasmids encoding the SARS–CoV signal have indicated that the signal is highly efficient, measurements ranging between 14 and 27% in cell-free extracts and mammalian cells (cf. 5–10% for HIV-1) (Thiel et al., 2003, Dos Ramos et al., 2004, Baranov et al., 2005, Plant et al., 2005). This proportion is consistent with the 15–40% described for other coronaviruses in the different phylogenetic groups (Brierley et al., 1987, Bredenbeek et al., 1990, Herold et al., 1993, Eleouet et al., 1995) and in related vertebrate and invertebrate nidoviruses (Snijder et al., 1990, den Boon et al., 1991, Cowley et al., 2000). This suggests that the stoichiometry of the ORF 1a- and 1b-encoded proteins has been evolutionarily conserved and might be critical for viral replication. Remarkably, ORF 1a does not encode major structural protein and thus programmed −1 ribosomal frameshifting in the SARS–CoV (and other nidoviruses) has not evolved to regulate the ratio of structural to replication proteins as occurs with retroviral gag/pol.
Although high resolution structures of coronavirus 1a/1b ribosomal frameshifting signals are not available, extensive mutational analysis, RNA structure probing and preliminary NMR studies indicate that they are composed of a U_UUA_AAC slippery sequence, followed by a single-stranded spacer region and an RNA pseudoknot (reviewed in Brierley, 1995, Brierley and Pennell, 2001; see also Baranov et al., 2005, Plant et al., 2005). In the signal of SARS–CoV, as in other coronaviruses, mutations in the codons corresponding to the ribosomal P-sites (UUA) and A-sites (AAC) of the slippery sequence greatly decrease −1 ribosomal frameshifting (Thiel et al., 2003, Baranov et al., 2005) and changing the identity of bases immediately flanking the U_UUA_AAC stretch has little or no effect (Baranov et al., 2005). Mass spectroscopic analysis of a tagged frameshift product has indicated that the SARS–CoV frameshift occurs solely by tandem-slippage of the peptidyl-tRNALeu and aminoacyl-tRNAAsn to the −1 reading frame codons (Baranov et al., 2005). Deletion analysis of the frameshift site indicates that the signal does not extend beyond 87 nucleotides downstream of the slippery sequence (Dos Ramos et al., unpublished work). Recent work has confirmed that this region contains an RNA pseudoknot structure with some unexpected features (Dos Ramos et al., 2004, Baranov et al., 2005, Plant et al., 2005; see Fig. 4A). The pseudoknot conforms to the H-type organisation in the possession of two base-paired stems (S1 and S2) and two single-stranded loops (L1 and L2), but extensive base-pairing seems to be present in loop 2 (which has alternatively been named SL1 or S3). Like all frameshift-promoting pseudoknots, disruption of base-pairing in either S1 or S2 reduces the efficiency of −1 ribosomal frameshifting substantially (Baranov et al., 2005, Plant et al., 2005). However, although most of loop 2 (SL1, S3) can be deleted without noticeable effect, maintenance of an appropriate conformation of the wild-type loop 2 seems to be required for optimal frameshift efficiency. Certain changes that affect the primary sequence (including bulge A residues) and the potential for the formation of SL1 have measurable effects on the frameshift process, probably by affecting global folding of the pseudoknot (Baranov et al., 2005, Plant et al., 2005). Pseudoknots containing an SL1 motif can be predicted for all group 2 coronaviruses (Plant et al., 2005). In group 3 coronaviruses (e.g. infectious bronchitis virus), however, the region equivalent to SL1 is probably a single-stranded loop (a standard loop 2) (Brierley et al., 1989, Brierley et al., 1991, Plant et al., 2005). The pseudoknots present at the 1a/1b overlap of group 1 coronaviruses (e.g. human coronavirus 229E) appear to form a more “elaborated” pseudoknot that can be viewed as “kissing” stem-loops separated by a very long (∼150 nucleotides) loop 2 (Herold and Siddell, 1993, Eleouet et al., 1995, Baranov et al., 2005). An alternative fold for pseudoknots of this group, with a much shorter loop 2 yet containing an SL1 region close to the junction with stem 2 can be drawn (Plant et al., 2005), but with a relatively unstable stem 2, this conformation seems unlikely and is not supported by earlier mutational analysis (Herold and Siddell, 1993).
Fig. 4.
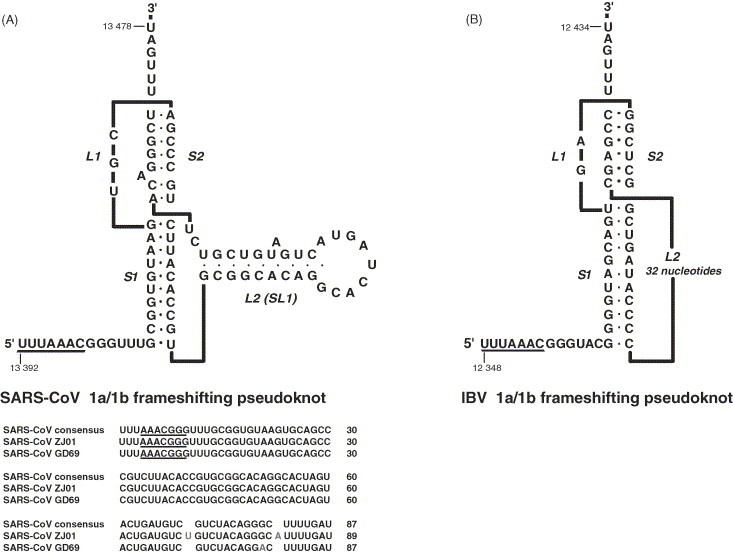
Predicted RNA secondary structures present at the 1a/1b ribosomal frameshifting signals of the SARS Co-V and IBV. (A) SARS coronavirus. Top: predicted secondary structure. The pseudoknot is composed of two double-stranded stems (S1 and S2) connected by a single-stranded loop (L1) and a second loop (L2) which itself folds into a stem-loop of approximately 28 nucleotides (SL1). Numbers correspond to the nucleotide positions in isolate Tor2. Bottom: primary sequence comparisons. The SARS–CoV consensus shows the sequence of 125 isolates (identical in this region). Mutations have been seen in only two isolates (ZJ01 and GD69) and are of 1 or 2 nucleotides (depicted in red) within SL1 or S2. (B) Infectious bronchitis virus. The pseudoknot is composed of two double-stranded stems (S1 and S2) connected by two loops (L1 and L2). Nucleotides of L2 do not obviously base-pair to form a stable stem and were omitted from the graphic. Numbers correspond to the nucleotide position in the Beaudette strain. The “slippery” sequences are underlined.
The investigation of the SARS–CoV frameshift signal carried out in the laboratories of Atkins and Dinman (Baranov et al., 2005, Plant et al., 2005) has led to the discovery of a novel frameshift determinant within or adjacent to stem 2, although the exact motif remains to be defined. As drawn in Fig. 4, stem 2 contains an unpaired adenosine residue whose presence and identity has been proposed to be crucial to frameshifting; replacement by cytosine or deletion of this base reduces the frameshift efficiency to less than 1% (Plant et al., 2005). Similarly, the work of Baranov et al. (2005) has highlighted this region as important, but in their model, stem 2 is set at five base-pairs in length, with the “bulge” A and adjacent CA residues displaced into loop 2. This leaves two unpaired residues (GU) between stems 1 and 2, which these authors suggest are the key element. Unfortunately, the multiple base-pairing possibilities present in this region preclude definitive conclusions with the current data, but both groups have clearly identified an unexpected feature.
The function(s) of many coronavirus 1a/1b-encoded proteins are not known and this has hindered our understanding of the role of frameshifting in this genus. Unlike retroviral Gag and Gag-Pol, the SARS–CoV 1a and 1a/1b polyproteins are cleaved intracellularly and the virus is not thought to package any 1a or 1b components into virions. 1a is processed into 11 products (non-structural proteins nsp1–11), mediated by the cysteine proteinases nsp3 (papain-like proteinase 2) and nsp5 (3C-like proteinase), which display substrate specificity for three (N-terminal) and seven (C-terminal) conserved sites of the polyprotein precursor, respectively (Fig. 3) (Snijder et al., 2003, Thiel et al., 2003, Gao et al., 2003, Fan et al., 2004, Harcourt et al., 2004, Lin et al., 2004, Prentice et al., 2004). Hitherto, the function of the other nsps remains undefined and only nsp9 has been partially characterised as a dimeric protein that binds both single-stranded RNA and single-stranded DNA (Egloff et al., 2004, Sutton et al., 2004). The 1b polyprotein, expressed as a fusion with 1a is trans-cleaved by the 3C-like proteinase at four conserved sites (Fig. 3). This generates five nsps that include an RNA-dependent RNA polymerase (nsp12, which starts with the nine most N-terminal residues of nsp11) (Cheng et al., 2005); a superfamily 1 helicase with associated dNTPase, NTPase and RNA 5′-triphosphatase activities (nsp13) (Thiel et al., 2003, Tanner et al., 2003, Ivanov et al., 2004a); a putative 3′–5′ exonuclease (nsp14) (Snijder et al., 2003); an uridylate-specific endoribonuclease (nsp15) (Bhardwaj et al., 2004, Ivanov et al., 2004b); and a putative S-adenosylmethionine-dependent 2′-O-ribose cap-1 methyltransferase (nsp16) (Snijder et al., 2003; von Grotthuss et al., 2003). At present, we can only speculate that the role of frameshifting is to set the relative levels of 1a- and 1b-encoded proteins. Indeed, for positive-stranded RNA viruses, the exact role of frameshifting is unknown. Presumably, the frameshift allows the required ratio of viral proteins to be produced, but it may also serve to downregulate levels of viral replicases, which may be toxic in high amounts.
4. Frameshift mechanisms
Since −1 frameshifting was first described (Jacks and Varmus, 1985), three inter-related models have circulated in the literature, the factor-binding, pausing and unwinding models. In the first model, the stimulatory RNA acts as a binding site for a protein(s) responsible for promoting or regulating the frameshift process (Jacks et al., 1988a, Brierley et al., 1989). However, despite fairly extensive study, no such proteins have been unearthed, although it cannot be ruled out that integral ribosomal component(s) may interact directly and specifically with the stimulatory RNAs. The second model proposes that ribosomal pausing occurs upon encounter of the stimulatory RNA and is a key element of the frameshift mechanism. In its simplest form, pausing increases the time at which ribosomes are held over the slippery sequence, giving increased opportunity for the tRNAs to realign in the −1 frame (Jacks et al., 1988a). There is good evidence that pausing occurs at pseudoknot-dependent frameshift signals (although specific frameshift-inducing stem-loops have not been tested) and the site of pausing is consistent with placement of the ribosomal P- and A-sites over the slippery sequence (Tu et al., 1992, Somogyi et al., 1993, Lopinski et al., 2000, Kontos et al., 2001). One of the great virtues of the pausing model is its ability to accommodate the variety of stimulatory RNAs that are present at −1 frameshifting signals, including stem-loops. Regardless of the range of secondary and tertiary features presented to the ribosome, as long as pausing occurs, frameshifting results. The idea that pausing alone is sufficient to induce frameshifting is questionable, however. Simple provision of a roadblock to ribosomes in the form of stable RNA hairpins (Brierley et al., 1991, Somogyi et al., 1993), a tRNA (Chen et al., 1995) or even different kinds of RNA pseudoknot (Napthine et al., 1999, Liphardt et al., 1999) is insufficient to bring about frameshifting and in addition, non-frameshifting pseudoknots and stem-loops exist that can still pause ribosomes (Tu et al., 1992, Somogyi et al., 1993, Lopinski et al., 2000, Kontos et al., 2001). Nevertheless, the pausing assays that are currently employed may be insufficiently sensitive to discriminate between a “kinetic pause” of perceivably short duration, yet essential for frameshifting and a “non-productive” pause resulting from a delay in unwinding the stimulatory RNA, but irrelevant to the process itself (Bidou et al., 1997, Kontos et al., 2001). In the unwinding model, it is speculated that the stimulatory RNAs are particularly resistant to the action of a ribosome-associated RNA helicase responsible for unwinding mRNA structures ahead of the decoding centre (Yusupova et al., 2001, Takyar et al., 2005). RNA pseudoknots have been shown to possess unusual structural features which may be refractory to standard helix unwinding, for example triple helical regions formed between pseudoknot stem 1 and loop 2 (Le et al., 1998, Su et al., 1999, Michiels et al., 2001). Modelling studies predict that such triplexes are likely to be one of the first features of the pseudoknot to be encountered by the ribosome (Giedroc et al., 2000), where it could function to stabilise stem 1 and increase the time taken to unwind the structure. As noted several years ago (Draper, 1990) and re-iterated subsequently (Michiels et al., 2001), another potential barrier to unwinding is the unusual topology of the pseudoknot at the beginning of stem 1, where in addition to the two base-paired strands, loop 2 adds a third strand in close association (Fig. 4). Perhaps, the ribosome does not deal effectively with this kind of topological arrangement. Individual features of pseudoknots may also be important. The pseudoknot of the mouse mammary tumour virus (MMTV) gag/pro frameshift signal, for example, possesses a pronounced kink between stems 1 and 2, which may be important for frameshifting (Shen and Tinoco, 1995, Chen et al., 1995, Chen et al., 1996, Kang et al., 1996, Kang and Tinoco, 1997). Similarly, the SARS–CoV pseudoknot seems to possess a frameshift determinant within or adjacent to stem 2 that plays an important role.
Plant et al. (2003) have recently proposed an elegant model for frameshifting, the 9 Å model, which offers a possible molecular explanation for tRNA movement. Here, ribosomes are paused by their initial failure to unwind the stimulatory RNA whilst in the act of accommodating the A-site tRNA from the A–T state into the A-site proper (A–A state) of the ribosome. During the accommodation process, the anticodon of the A-site tRNA is thought to move about 9 Å (Noller et al., 2002). As the mRNA is essentially held in place by the stimulatory RNA blocking the mRNA channel, the movement of the anticodon, as part of an mRNA codon: anticodon complex, puts strain on the mRNA that may be relieved by −1 slippage of the tRNA. The 9 Å model is consistent with former models in that a failure to unwind the stimulatory RNA is crucial in allowing the A-site anticodon movement to generate tension and pausing of the ribosome would extend the window in which tRNA movement can take place. The resistance to unwinding could also be facilitated by binding of the pseudoknot to proteins in and around the mRNA channel.
As is clear from the above, frameshift models have been tailored mostly towards pseudoknot-containing sites and rely on pseudoknot-specific features. How then do we explain the capacity of stem-loops to induce frameshifting? One possibility is that the stem-loops themselves possess hitherto uncharacterised, novel structural features that can interfere with unwinding or promote the kind of ribosomal pause that leads to frameshifting. The high-resolution structure of the HIV-1 frameshift stem-loop discussed above is an important step forward in this regard. The presence of a kink between the upper and lower base-paired stems of the HIV-1 stimulatory RNA is reminiscent of the inter-stem kink present in the MMTV pseudoknot and may represent an important structural feature. The HIV-1 hairpin is capped by a tetraloop motif which may also be relevant to frameshifting. Of course, it may be that our models of frameshifting are incomplete. Mutational analysis of such sites has already provided hints that the traditional combination of slippery sequence and hairpin may not be the sole defining feature of the signal and other elements may contribute. Kim et al. (2001) measured the frameshift efficiencies evoked in vitro by a series of HIV-1 gag/pol-human T-cell lymphotropic virus type 2 (HTLV-2) gag/pro chimeras. They defined four elements, namely the slippery sequence, spacer, stem-loop and a region upstream of the slippery sequence and combined these in various ways to create a range of hybrid sites. It was found that the regions flanking the slippery sequence and stem-loop could influence frameshifting quite dramatically, possibly by modulating stem-loop unfolding kinetics. Thus, frameshifting at stem-loop structures, like at pseudoknots, is likely to be governed by the rate at which the structure is unfolded. It has recently been discovered that simply annealing an oligonucleotide downstream of a slippery sequence can under certain circumstances promote efficient frameshifting, at least in in vitro translation systems (Howard et al., 2004, Olsthoorn et al., 2004). In this situation, the ribosome encounters a double-stranded region conformationally distinct from a stem-loop, yet similarly, the rate of unwinding of the structure is likely to be the key determinant of frameshifting efficiency.
5. Frameshifting as a target for antiviral intervention
From studies of HIV and other retroviruses, it is clear that modulation of frameshift efficiency can have a dramatic effect on virus viability and the same is likely to be true for positive-strand RNA viruses, since it would alter the levels of non-structural proteins within infected cells. Various antibiotics that target ribosomes have been found to influence frameshifting efficiency at the yeast double-stranded RNA virus (L–A) signal (Dinman and Wickner, 1992) and this has led to the working hypothesis that they could be used as antiviral drugs (Dinman et al., 1997, Dinman et al., 1998, Irvine et al., 1998). However, it remains to be seen whether such compounds (e.g. anisomycin, sparsomycin, cycloheximide) have activity against a broad-spectrum of frameshifting signals. High-throughput screening has also been employed in the search for candidate anti-frameshift drugs active against the HIV-1 stem-loop signal (Hung et al., 1998). One such compound, RG501 (1,4-bis-[N-{3-N,N-dimethylpropyl} amidino] benzene tetrahydrochloride) was found to stimulate frameshifting at the HIV-1 signal about two-fold and inhibited HIV-1 replication in tissue culture. However, this inhibition began at levels of RG501 that did not noticeably affect frameshifting, so the specificity of the effect is questionable and the issue may be complicated by cytotoxicity. The drug was nevertheless clearly able to stimulate frameshifting at the stem-loop-containing signals of HIV-2, SIV and HTLV I gag/pro, but not HTLV-1 pro/pol, which is thought to contain a pseudoknot (ten Dam et al., 1990). It has been speculated (Hung et al., 1998) that RG501 acts by binding to the loop region of hairpins (perhaps by intercalation), stabilising the structure and promoting frameshifting by increasing ribosomal pausing. Targeting of oligonucleotides to frameshift signals may also permit antiviral intervention, for example, through inhibition of frameshifting by interfering with formation of the stimulatory RNA or by stimulation of frameshifting. It is known that the binding of 2′-O-methyl oligoribonucleotides to a region just 3′-wards of the HIV-1 stimulatory RNA leads to an enhancement of frameshift efficiency in vitro, perhaps by (effectively) increasing the length of the stem of the HIV-1 stimulatory RNA (Vickers and Ecker, 1992). Some of the ground rules for oligonucleotide targeting of the HIV-1 frameshift stem-loop have already been worked out (Aupeix et al., 1999, Aupeix-Scheidler et al., 2000, Toulme et al., 2001). So far, however, only one group has demonstrated reduced virus replication upon oligonucleotide-targeting of a frameshift region (Neuman et al., 2005). In this study, a peptide-conjugated phosphorodiamidate morpholino antisense oligomer targeting the pseudoknot stems of the SARS–CoV frameshift signal showed demonstrable antiviral activity. Recent work in our laboratory has also focused on targeting the SARS–CoV frameshifting signal with antisense RNA and chimeric RNA/DNA oligonucleotides (Dos Ramos et al., unpublished work). Specific inhibition of frameshifting independent of mRNA cleavage has been demonstrated in an in vitro translation system using oligonucleotides complementary to different regions of the pseudoknot. Highly active oligonucleotides are currently being tested in vivo. Importantly, the structures of some domains (e.g. S1, SL1) are highly conserved (Fig. 4) raising hopes that such targeting could be broad-spectrum and less susceptible to the generation of escape mutants.
Another issue concerns the occurrence of frameshifting in cellular genes. Recently, a programmed −1 frameshifting signal has been described in the mouse gene Edr (Shigemoto et al., 2001). The Edr signal resembles retroviral examples closely, with a characteristic slippery sequence—spacer-pseudoknot organisation (Manktelow et al., 2005) and the signal is conserved in the human orthologue PEG10 (Lux et al., 2005). Given the possibility that other retrovirus-like motifs have been subsumed into mammalian genes and retained a role for frameshifting, antiviral agents that target this process may have previously unanticipated consequences on cellular metabolism in uninfected cells.
6. Conclusions
It is now established that the stimulatory RNAs present at the frameshift signals of HIV-1 and SARS–CoV are examples of stem-loop and pseudoknot stimulators, respectively. However, there remain a number of unanswered questions. Regarding the HIV-1 signal, one of the key uncertainties is whether the stem-loop pauses ribosomes to the same extent as an RNA pseudoknot and at the same place on the mRNA. Pausing assays would confirm that frameshift-stimulating stem-loops can indeed stall ribosomes and may help in judging whether current pausing assays are of much value. Further investigation of the contribution of specific subdomains of the HIV-1 stimulator to the frameshift process is also appropriate, including the role of the GGA bulge (the kink), the base-paired spacer, the potential role of the tetraloop and the occurrence of these motifs in other lentiviruses. The SARS–CoV signal also requires more analysis, especially the role of SL1 and the determinant at the S1–S2 junction. An atomic structure of a pseudoknot of this class would be of immense value in identifying common motifs amongst frameshift-promoting pseudoknots; the preliminary NMR studies of Plant et al. (2005) offer some hope that this can be achieved. The recent development of methods to prepare infectious molecular clones of coronaviruses offers for the first time the opportunity to investigate the role of frameshifting in the replication of the SARS–CoV (reviewed in Baric and Sims, 2005). This will be of broad relevance to frameshifting in positive-strand RNA viruses. The huge public health consequences of the HIV-1 pandemic and concerns over future SARS–CoV outbreaks demands that all avenues be explored in the quest to counteract these agents. In principle, the replication of any virus that uses a frameshift process could be disrupted by modulation of frameshift efficiencies, but a better understanding of the occurrence and the molecular basis of frameshifting will be required before it can be considered as a genuine target.
Acknowledgements
We are grateful to Dr. Louise Pye (nee King) for her contributions to this work and to Dr. Simon Pennell for critical comments on the text.
References
- Atkins J.F., Baranov P.V., Fayet O., Herr A.J., Howard M.T., Ivanov I.P., Matsufuji S., Miller W.A., Moore B., Prere M.F., Wills N.M., Zhou J., Gesteland R.F. Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb. Symp. Q. Biol. 2001;66:217–232. doi: 10.1101/sqb.2001.66.217. [DOI] [PubMed] [Google Scholar]
- Aupeix K., Le Tinevez R., Toulme J.J. Binding of oligopyrimidines to the RNA hairpin responsible for the ribosome Gag-Pol frameshift in HIV-1. FEBS Lett. 1999;449:169–174. doi: 10.1016/s0014-5793(99)00427-5. [DOI] [PubMed] [Google Scholar]
- Aupeix-Scheidler K., Chabas S., Bidou L., Rousset J.P., Leng M., Toulme J.J. Inhibition of in vitro and ex vivo translation by a transplatin-modified oligo (2′-O-methylribonucleotide) directed against the HIV-1 Gag-Pol frameshift signal. Nucleic Acids Res. 2000;28:438–445. doi: 10.1093/nar/28.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS–CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Sims A.C. Development of mouse hepatitis virus and SARS–CoV infectious cDNA constructs. Curr. Top. Microbiol. Immunol. 2005;287:229–252. doi: 10.1007/3-540-26765-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M., Dulude D., Gendron K., Lemay G., Brakier-Gingras L. Efficiency of a programmed −1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA. 2003;9:1246–1253. doi: 10.1261/rna.5113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M., Dulude D., Steinberg S.V., Brakier-Gingras L. The frameshift stimulatory signal of human immunodeficiency virus type 1 group O is a pseudoknot. J. Mol. Biol. 2003;331:571–583. doi: 10.1016/S0022-2836(03)00784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M., Rousset J.P. An extended signal involved in eukaryotic-1 frameshifting operates through modification of the E site tRNA. Mol. Cell. 2005;17:61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Prere M.F., Gesteland R.F., Atkins J.F., Fayet O. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA. 2002;8:16–28. doi: 10.1017/s1355838202012086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Grima B., Liu H., Cassan M., Rousset J.P. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Biswas P., Jiang X., Pacchia A.L., Dougherty J.P., Peltz S.W. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Pachuk C.J., Noten A.F., Charite J., Luytjes W., Weiss S.R., Spaan W.J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59: a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Q. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the ‘slippery-sequence’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle M.N., Brakier-Gingras L., Lemay G. Replacement of murine leukemia virus readthrough mechanism by human immunodeficiency virus frameshift allows synthesis of viral proteins and virus replication. J. Virol. 2003;77:3345–3350. doi: 10.1128/JVI.77.5.3345-3350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M., Delaunay N., Vaquero C., Rousset J.P. Translational frameshifting at the Gag-Pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J. Virol. 1994;68:1501–1508. doi: 10.1128/jvi.68.3.1501-1508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S., Niu M., Saadatmand J., Guo F., Huang Y., Nabel G.J., Kleiman L. Incorporation of pol into human immunodeficiency virus type 1 gag virus-like particles occurs independently of the upstream gag domain in Gag-Pol. J. Virol. 2004;78:1042–1049. doi: 10.1128/JVI.78.2.1042-1049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr., Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Y., Kang H.S., Shen L.X., Chamorro M., Varmus H.E., Tinoco I. A characteristic bent conformation of RNA pseudoknots promotes-1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification and characterisation of the SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry E., Liang C., Rong L., Quan Y., Inouye P., Li X., Morin N., Kotler M., Wainberg M.A. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease–reverse transcriptase fusion proteins. J. Mol. Biol. 1998;284:43–56. doi: 10.1006/jmbi.1998.1968. [DOI] [PubMed] [Google Scholar]
- Chim S.S., Tsui S.K., Chan K.C., Au T.C., Hung E.C., Tong Y.K., Chiu R.W., Ng E.K., Chan P.K., Chu C.M., Sung J.J., Tarn J.S., Fung K.P., Waye M.M., Lee C.Y., Yuen K.Y., Lo Y.M., CUHK Molecular SARS Research Group Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362:1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Demirov D.G., Freed E.O. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Wickner R.B. Ribosomal frameshifting efficiency and gag/Gag-Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Ruiz-Echevarria M.J., Czaplinski K., Peltz S.W. Peptidyl-transferase inhibitors have antiviral properties by altering programmed-1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Ruiz-Echevarria M.J., Peltz S.W. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Richter S., Plant E.P., Taylor R.C., Hammell A.B., Rana T.M. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5331–5336. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Ramos F., Carrasco M., Doyle T., Brierley I. Programmed −1 ribosomal frameshifting in the SARS coronavirus. Biochem. Soc. Trans. 2004;32:1081–1083. doi: 10.1042/BST0321081. [DOI] [PubMed] [Google Scholar]
- Doyon L., Croteau G., Thibeault D., Poulin F., Pilote L., Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon L., Payant C., Brakier-Gingras L., Lamarre D. Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol. 1998;72:6146–6150. doi: 10.1128/jvi.72.7.6146-6150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. Pseudoknots and the control of protein synthesis. Curr. Opin. Cell. Biol. 1990;2:1099–1103. doi: 10.1016/0955-0674(90)90162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Du Z., Giedroc D.P., Hoffman D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: a model for a possible family of structurally related RNA pseudoknots. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- Dulude D., Baril M., Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Eleouet J.F., Rasschaert D., Lambert P., Levy L., Vende P., Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microb. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acids Res. Mol. Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer J., Hohn T. Translation in plants—rules and exceptions. Plant Mol. Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M., Jr., Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS–CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin C., Mazauric M.H., Traikia M., Guittet E., Yoshizawa S., Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Gendron K., Dulude D., Lemay G., Ferbeyre G., Brakier-Gingras L. The virion-associated Gag-Pol is decreased in chimeric Moloney murine leukemia viruses in which the readthrough region is replaced by the frameshift region of the human immunodeficiency virus type 1. Virology. 2005;334:342–352. doi: 10.1016/j.virol.2005.01.044. [DOI] [PubMed] [Google Scholar]
- Gibbs A.J., Gibbs M.J., Armstrong J.S. The phylogeny of SARS coronavirus. Arch. Virol. 2004;149:621–624. doi: 10.1007/s00705-003-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc D.P., Theimer C.A., Nixon P.L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger H.G., Dorfman T., Sodroski J.G., Haseltine W.A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greatorex J., Gallego J., Varani G., Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002;322:543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guan Y., Peiris J.S., Zheng B., Poon L.L., Chan K.H., Zeng F.Y., Chan C.W., Chan M.N., Chen J.D., Chow K.Y., Hon C.C., Hui K.H., Li J., Li V.Y., Wang Y., Leung S.W., Yuen K.Y., Leung F.C. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Siddell S.G. An ‘elaborated’ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Raabe T., Schelle-Prinz B., Siddell S.G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M.K., Shehu-Xhilaga M., Crowe S.M., Mak J. Proline residues within spacer peptide p1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 2002;76:11245–11253. doi: 10.1128/JVI.76.22.11245-11253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.T., Gesteland R.F., Atkins J.F. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M., Patel P., Davis S., Green S.R. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine J.H., Horsfield J.A., McKinney C.Z., Tate W.P. A novel strategy to interfere with human immunodeficiency virus type 1 propagation. N. Z. Med. J. 1998;111:222–224. [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H.E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus Gag-Pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Power M.D., Masiarz F.R., Luciw P.A., Barr P.J., Varmus H.E. Characterization of ribosomal frameshifting in HIV-1 Gag-Pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Kang H. Direct structural evidence for formation of a stem-loop structure involved in ribosomal frameshifting in human immunodeficiency virus type 1. Biochim. Biophys. Acta. 1998;1397:73–78. doi: 10.1016/S0167-4781(98)00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., Tinoco I. A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucleic Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., Hines J.V., Tinoco I. Conformation of a non-frame shifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Karacostas V., Wolffe E.J., Nagashima K., Gond M.A., Moss B. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- Kaye J.F., Lever A.M. trans-Acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J. Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Maas S., Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H., Honigman A., Panet A., Hauser H. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tarn J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D.C., Stover C.C., Noznitsky J., Wu Z., Summers M.F. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- Le S.Y., Shapiro B.A., Chen J.H., Nussinov R., Maizel J.V. RNA pseudoknots downstream of the frameshift sites of retroviruses. Genet. Anal. Tech. Appl. 1991;8:191–205. doi: 10.1016/1050-3862(91)90013-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Y., Chen J.H., Pattabiraman N., Maizel J.V. Ion–RNA interactions in the RNA pseudoknot of a ribosomal frameshifting site: molecular modeling studies. J. Biomol. Struct. Dyn. 1998;16:1–11. doi: 10.1080/07391102.1998.10508221. [DOI] [PubMed] [Google Scholar]
- Lin C.W., Tsai C.H., Tsai F.J., Chen P.L., Lai C.C., Wan L., Chiu H.H., Lin K.H. Characterization of trans- and cis-cleavage activity of the SARS coronavirus 3CLpro protease: basis for the in vitro screening of anti-SARS drugs. FEBS Lett. 2004;574:131–137. doi: 10.1016/j.febslet.2004.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J., Napthine S., Kontos H., Brierley I. Evidence for an RNA pseudoknot loop–helix interaction essential for efficient-1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopinski J.D., Dinman J.D., Bruenn J.A. Kinetics of ribosomal pausing during programmed-1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J.M., Clore G.M., Gronenborn A.M. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- Louis J.M., Wondrak E.M., Kimmel A.R., Wingfield P.T., Nashed N.T. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- Lux A., Beil C., Majety M., Barron S., Gallione C.J., Kuhn H.-M., Berg J.N., Kioschis P., Marchuk D.A., Hafner M. Human retroviral gag- and gag-pol-like proteins interact with the transforming growth factor-β receptor activin receptor-like kinase 1. J. Biol. Chem. 2005;280:8482–8493. doi: 10.1074/jbc.M409197200. [DOI] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels P.J., Versleijen A.A., Verlaan P.W., Pleij C.W., Hilbers C.W., Heus H.A. Solution structure of the pseudoknot of SRV−1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sundquist W.I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Napthine S., Liphardt J., Bloys A., Routledge S., Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient-1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Stein D.A., Kroeker A.D., Churchill M.J., Kim A.M., Kuhn P., Dawson P., Moulton H.M., Bestwick R.K., Iversen P.L., Buchmeier M.J. Inhibition, escape and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H.F., Yusupov M.M., Yusupova G.Z., Baucom A., Cate J.H. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R.C., Laurs M., Sohet F., Hilbers C.W., Heus H.A., Pleij C.W. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:1702–1703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Morrow C.D. Overexpression of the Gag-Pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T., Chamorro M., Varmus H.E. Human immunodeficiency virus type 1 Gag-Pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus C., Hellebrand S., Tessmer U., Wolf H., Krausslich H.G., Wagner R. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 1999;274:21539–21543. doi: 10.1074/jbc.274.31.21539. [DOI] [PubMed] [Google Scholar]
- Pe’ery T., Mathews M.B. Viral translational strategies and host defense mechanisms. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2000. pp. 371–424. [Google Scholar]
- Peiris I.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S., Munoz M., Yerly S., Sanchez-Merino V., Lopez-Galindez C., Perrin L., Larder B., Cmarko D., Fakan S., Meylan P., Telenti A. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J. Virol. 2001;75:9644–9653. doi: 10.1128/JVI.75.20.9644-9653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Jacobs K.L., Harger J.W., Meskauskas A., Jacobs J.L., Baxter J.L., Petrov A.N., Dinman J.D. The 9-A solution: how mRNA pseudoknots promote efficient programmed-1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Perez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Guan Y., Nicholls J.M., Yuen K.Y., Peiris J.S. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect. Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J., National Microbiology Laboratory Canada, Canadian Severe Acute Respiratory Syndrome Study Team Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Hoxter M., Moosmayer D., Pauli G., Hauser H. CD4 expressing human 293 cells as a tool for studies in HIV-1 replication: the efficiency of translational frameshifting is not altered by HIV-1 infection. Virology. 1994;205:371–375. doi: 10.1006/viro.1994.1655. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M., Crowe S.M., Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.X., Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Shigemoto K., Brennan J., Walls E., Watson C.J., Stott D., Rigby P.W., Reith A.D. Identification and characterisation of a developmentally regulated mammalian gene that utilises-1 programmed ribosomal frameshifting. Nucleic Acids Res. 2001;29:4079–4088. doi: 10.1093/nar/29.19.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Bredenbeek P.J., Horzinek M.C., Rijnbrand R., Spaan W.J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990;18:4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P., Jenner A.J., Brierley I., Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl G., Bidou L., Rousset J.P., Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Chen L., Egli M., Berger J.M., Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure (Camb.) 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Martinez R., Munoz M., Bleiber G., Greub G., Sanglard D., Peters S. Analysis of natural variants of the human immunodeficiency virus type 1 Gag-Pol frameshift stem-loop structure. J. Virol. 2002;76:7868–7873. doi: 10.1128/JVI.76.15.7868-7873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij C.W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Toulme J.J., Di Primo C., Moreau S. Modulation of RNA function by oligonucleotides recognizing RNA structure. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:1–46. doi: 10.1016/s0079-6603(01)69043-3. [DOI] [PubMed] [Google Scholar]
- Tu C., Tzeng T.H., Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers T.A., Ecker D.J. Enhancement of ribosomal frameshifting by oligonucleotides targeted to the HIV Gag-Pol region. Nucleic Acids Res. 1992;20:3945–3953. doi: 10.1093/nar/20.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Moes E., Li S., Vandamme A.M., Van Ranst M. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology. 2005;337:85–92. doi: 10.1016/j.virol.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S.E., Rathjen P.D., Kingsman S.M., Kingsman A.J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Tu C., Xin C., Xuan H., Meng Q., Liu Y., Yu Y., Guan Y., Jiang Y., Yin X., Crameri G., Wang M., Li C., Liu S., Liao M., Feng L., Xiang H., Sun J., Chen J., Sun Y., Gu S., Liu N., Fu D., Eaton B.T., Wang L.F., Kong X. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 2005;79:2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., He J.F., Evans M.R., Peng G.W., Field H.E., Yu D.W., Lee C.K., Luo H.M., Lin W.S., Lin P., Li L.H., Liang W.J., Lin J.Y., Schnur A. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelverton E., Lindsley D., Yamauchi P., Gallant J.A. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol. Microbiol. 1994;11:303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu L., Li D.X., Yuen K.Y., Peiris J.S.M., Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


