Abstract
RNA viruses have rapidly evolving genomes which often allow cross-species transmission and frequently generate new virus variants with altered pathogenic properties. Therefore infections by RNA viruses are a major threat to human health. The infected host cell detects trace amounts of viral RNA and the last years have revealed common principles in the biochemical mechanisms leading to signal amplification that is required for mounting of a powerful antiviral response. Components of the RNA sensing and signaling machinery such as RIG-I-like proteins, MAVS and the inflammasome inducibly form large oligomers or even fibers that exhibit hallmarks of prions. Following a nucleation event triggered by detection of viral RNA, these energetically favorable and irreversible polymerization events trigger signaling cascades leading to the induction of antiviral and inflammatory responses, mediated by interferon and NF-κB pathways. Viruses have evolved sophisticated strategies to manipulate these host cell signaling pathways in order to ensure their replication. We will discuss at the examples of influenza and HTLV-1 viruses how a fascinating diversity of biochemical mechanisms is employed by viral proteins to control the NF-κB pathway at all levels.
Abbreviations: ss, single-stranded; ds, double-stranded; HTLV, human T cell leukemia virus; SARS, severe acute respiratory syndrome; IAVs, influenza A viruses; IFN, interferon; IRF3, interferon regulatory factor 3; NF-κB, nuclear factor κB; TLRs, toll-like receptors; PRR, pattern recognition receptor; MyD88, myeloid differentiation primary response gene 88; RIG-I, retinoic acid-inducible gene I; RLRs, RIG-I-like receptors; MDA5, melanoma differentiation factor 5; LGP2, laboratory of genetics and physiology 2; CARD, caspase activation and recruitment domain; ppp, triphosphate; MAVS, mitochondrial antiviral signaling protein; DDX3, DEAD (Asp-Glu-Ala-Asp) box helicase 3; DHX9, DEAH (Asp-Glu-Ala-His) box helicase 9; TRAF2, TNF receptor-associated factor 2; IKK, IκB kinase; NEMO, NF-κB essential modulator; IκB, inhibitor of NF-κB; IKKε/TBK1, IκB kinase ε/TANK-binding kinase; pro-IL-1β, pro-interleukin-1β; ASC, apoptosis-associated speck-like protein containing a carboxy-terminal CARD; NLRP3, NLR (Nod-like receptor) family, pyrin domain containing 3; AIM2, absent in melanoma 2; TAB2, TGF-β activated kinase binding protein 2; TAK1, TGF-β activated kinase 1; TNF, tumor necrosis factor; NIK, NF-κB-inducing kinase; PARP-1, poly(ADP-ribose)-polymerase-1; PIASy, protein inhibitor of activated STAT y; ATM, ataxia-telangiectasia mutated; SUMO, small ubiquitin-related modifier; vRNA, virion RNA; cRNA, complementary RNA; NS1, nonstructural protein 1; PB1-F2, polymerase basic protein 1-frame 2; RIPK2, receptor-interacting protein kinase 2; TRAIL, TNF-related apoptosis inducing ligand; PI3K, phosphoinositide-3-kinase; ATL, adult T-cell leukemia/lymphoma; LTR, long terminal repeat; HBZ, basic leucine zipper factor; TGFβ, transforming growth factor β; Ubc13, ubiquitin-conjugating 13; USP20, ubiquitin-specific peptidase; STAMBPL1, STAM-binding protein-like 1; OPTN, Optineurin; TAX1BP1, Tax-1 binding protein 1; SC35, serine/arginine-rich splicing factor 2; Brd4, bromodomain-containing 4; WWOX, WW domain-containing oxidoreductase; Hsp90, heat shock protein 90 kDa; ChIP-Seq, chromatin immunoprecipitation coupled to deep sequencing
Keywords: NF-κB, RNA virus, Signal transduction, Influenza virus, Interferon signaling
Highlights
-
•
RNA viruses have rapidly evolving genomes.
-
•
Detection of viral RNA and subsequent signal amplification triggers the antiviral response.
-
•
Components of the RNA sensing and signaling machinery inducibly form large oligomers or fibers.
-
•
Virus-induced NF-κB activity has anti- and pro-viral functions.
1. Genome variability of RNA viruses
According to the Baltimore classification, viruses can be grouped depending on their type of genome (DNA, RNA, single-stranded (ss), double-stranded (ds)) and their replication mechanism [1]. RNA viruses are characterized by their pronounced genetic variability which is attributable to various reasons: (i) In contrast to DNA, the spontaneous deamination of cytosine into uracil is not corrected by proofreading enzymes. (ii) The viral RNA polymerases lack proofreading activity, resulting in limited replication fidelity. This leads to a high frequency of nucleotide misincorporation, the rapid accumulation of mutations and the generation of viral quasispecies. (iii) In addition, RNA recombination may occur in cells co-infected by related viruses. Recombination takes place during RNA synthesis and involves a template-switch of the viral polymerase, thus resulting in further changes of the viral genome. (iv) Some RNA viruses such as influenza viruses have segmented genomes that may undergo reassortment upon infection with closely related viruses. These mechanisms lead to a fast evolution of RNA viruses and allow the acquisition of novel pathogenic and biological properties [2], [3]. Thus, many RNA viruses including influenza, human T cell leukemia virus (HTLV) and severe acute respiratory syndrome (SARS) corona viruses are associated with severe or even fatal diseases. The high genetic variability of RNA viruses also allows transmission between different animal species and man. The reassortment between animal and human viruses was mainly responsible for the generation of novel influenza A viruses (IAVs) such as the infamous H1N1 strain that caused the 1918 flu pandemic responsible for the death of millions of infected people [4].
The infection with RNA viruses leads to the induction of signaling cascades in the infected host cell. The RNA viruses are recognized by specialized host cell proteins, thus triggering the activation of kinases and transcription factors which in turn mount an antiviral response as exemplified by the type I interferon (IFN) system [5], [6]. Viruses have developed multitudinous different strategies to disable the production and activities of IFNs. In addition, viruses have learned to usurp host cell signaling pathways to promote their own replication [7].
Recent progress in the identification of proteins recognizing viral RNA and the host cell machinery employed by the virus has broadened our understanding of the molecular mechanisms occurring during virus infection. In addition, host cell factors are ideal targets for therapeutic intervention, as the genetic flexibility of viral target structures frequently allows the generation of escape mutants. We will discuss recent progress at the example of two well-studied viruses with a broad biomedical relevance: IAV as an example of a fast replicating lytic virus that kills the infected cell early after infection and HTLV-1 as a paradigm for a slow growing virus that persists for many years in CD4+ T lymphocytes and may lead to their oncogenic transformation.
2. Detection of RNA viruses by host cell proteins
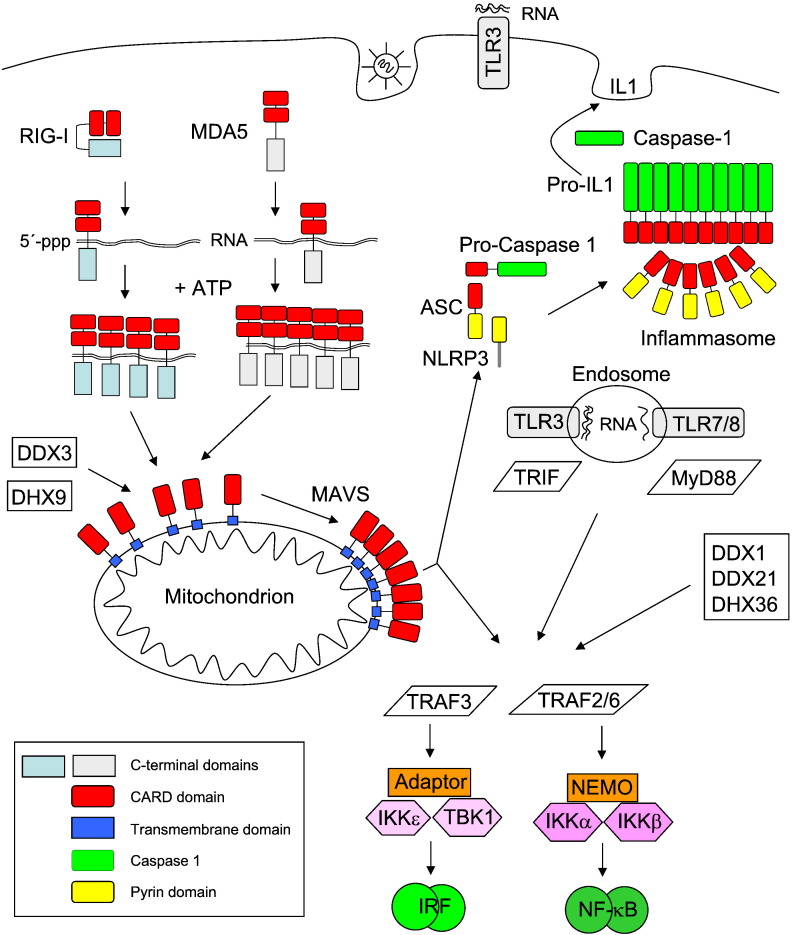
The RNA sensing machinery has a very difficult task, as it must be robust enough to ignore background signals that could lead to receptor misfiring and induction of the innate immune response, which could potentially lead to autoimmune diseases. On the other hand, virus invasion must be recognized at the very early stages in order to ensure an efficient mounting of the IFN response to protect the cells before the viral infection has overwhelmed the cells. Thus, only 20 molecules of viral RNA in a cell are sufficient to trigger the detectable activation of the transcription factor IRF3 (interferon regulatory factor 3) and the induction of the IFN response [8]. This extraordinary sensitivity is due to a cascade of signal multipliers that use a common principle: the generation of homopolymers by prion-like polymerization [8], [9], [10], [11], [12], [13]. This architectural feature allows for ultrasensitive responses by a self-perpetuating mechanism of signal amplification. As schematically displayed in Fig. 1 , this principle is used at several levels of the signaling cascade from sensing of RNAs to the activation of transcription factors such as NF-κB (nuclear factor κB). The activated transcription factors then cooperate to trigger the expression of IFN genes. These cytokines then not only elicit the induced expression of IFN-stimulated genes in the infected cell, but also acts in a paracrine fashion at the surrounding cells. This causes an antiviral state that serves to restrict viral spreading [6], [14].
Fig. 1.
Sensing of viral RNA and induction of the host cell defense. The inducible aggregation of RLRs, MAVS and the inflammasome is displayed. While NF-κB activation proceeds via TRAF2/6, TRAF3 activates the kinases TBK1 and IKKε. These kinases phosphorylate IRF transcription factors, thus allowing for their dimerization and DNA-binding. Important domains in the proteins are shown in colors. The inflammasome can be also activated upon association with the mitochondrial MAVS protein, but this process is not displayed for the sake of clarity.
In a first step, viral RNAs need to be detected by receptors that survey the extracellular, vacuolar, and cytosolic compartments for signs of virus infection. Membrane-bound toll-like receptors (TLRs) comprise the first identified pattern recognition receptor (PRR) family that is specialized in the recognition of RNA [15], [16]. TLR3 and TLR7/8 are inserted in membranes in a way that the RNA-recognition domains face towards the extracellular space or to the endosomal lumen. This orientation allows the sensitive detection of viral RNA at the cell surface or of pathogen-derived RNA in the endosomes. TLR3 recognizes specifically not only viruses with a dsRNA genome or dsRNA intermediates generated during virus replication, but also mRNAs [15], [17], [18]. The stretches of virus-derived dsRNA leading to TLR3 activation are much longer than the short dsRNA structures (such as secondary clover-leaf structure of transfer RNA) that are present in uninfected cells. Therefore, this receptor is a bona fide PRR that can discriminate between viral and host cell RNA [19]. In contrast to TLR3, the TLR7 and TLR8 receptors bind ssRNA that is rich in guanosines/uridines or uridines [20], [21] and are thus unable to distinguish between pathogen and self-nucleic acids. However, as the endosomes of uninfected cells do not contain significant amounts of RNA, the limited specificity of TLR7/8 does not result in receptor misfiring. TLR 1, 2 and 5–9 signal via MyD88 (myeloid differentiation primary response gene 88), an adapter protein that contributes to the protection from primary IAV infection [22].
The detection of viral RNA in the cytosol is mediated by the family of RIG-I (retinoic acid-inducible gene I)-like receptors (RLRs) and also by further cytosolic proteins. As the cytosolic localization of these receptors frequently exposes them to host cell RNA, signaling from these proteins is restricted by two mechanisms: (i) their basal expression is further induced and (ii) they have mechanisms to discriminate between viral and host cell RNA. The mammalian RLR family consists of three central members, namely RIG-I, MDA5 (melanoma differentiation factor 5), and LGP2 (laboratory of genetics and physiology 2) [23]. They belong to the DExD/H-box family of helicases and harbor two helicase domains followed by a C-terminal domain which contributes to ligand specificity. Only RIG-I and MDA5 have two adjacent CARDs (caspase activation and recruitment domains), which allow coupling to downstream signaling adaptors [24]. In uninfected cells, intramolecular interactions between the CARD and the C-terminal domain maintain RIG-I in a closed, inactive conformation. RIG-I binds preferentially to dsRNA with blunt ends and a triphosphate (ppp) moiety at the 5′ end [25], [26]. The requirement for a 5′ppp allows to discriminate between self and non-self, as the phosphates of host cell mRNAs are either masked by a 7-methyl-guanosine cap (mRNA) or removed in the case of tRNA and rRNA. In addition, RIG-I can sense RNAs with complementary 5′ and 3′ ends that hybridize to form a “panhandle” structure as they occur for many viral genomes, including those of IAV [26]. After binding of RIG-I to its cognate RNA, the CARDs are released from their inhibitory association with the helicase domain [27]. The RNA-bound RIG-I monomers then hydrolyze ATP to assemble into a filament that propagates from the dsRNA ends to the interior [12], [13]. RIG-I is further activated upon K63-linked polyubiquitination, where non-covalent binding of three chains of K63-branched ubiquitin to the tandem CARDs allows the formation of RIG-I tetramers which are required for downstream signal activation [28]. Activated and ubiquitinated RIG-I can then bind to its downstream effector MAVS (mitochondrial antiviral signaling protein) to induce activation of IRF and NF-κB transcription factors, ultimately resulting in the expression of IFN encoding genes. The agonistic RNAs leading to MDA5 activation are not as clearly defined as in the case of RIG-I. A determining factor is the length of the activating dsRNA with longer nucleic acids being more efficient in MDA5 activation [29]. Structural studies with dsRNA-bound MDA5 show that the receptor cooperatively assembles in a head-to-tail fashion along the length of dsRNA to form a filament-like structure [30]. These multiple protein–protein interactions are mainly mediated by the tandem CARD. Accordingly, dsRNA serves as a signaling platform allowing the organized assembly of MDA5 filaments to activate the downstream signaling molecule MAVS. Another RNA sensor is LGP2, but the nature of the agonistic RNAs is not well studied and its regulatory role in antiviral immune responses does not provide a coherent picture [31].
Cytosolic RNA sensing also employs DExD/H-box helicases outside the RLR helicase subfamily. This group comprises DDX3 (DEAD (Asp-Glu-Ala-Asp) box helicase 3), DHX9 (DEAH (Asp-Glu-Ala-His) box helicase 9) and the DDX1–DDX21–DHX36 complex [32]. DDX3 binds RNA and regulates replication of human immunodeficiency virus [33], but this protein also plays important functions that are independent from its ability to bind RNA. For example, DDX3 has been identified as a regulatory subunit of casein kinase 1 in the Wnt-β-catenin signaling pathway [34]. DDX3 and DHX9 sense viral RNA and then couple to MAVS in order to induce IFN expression [35], [36]. The DDX1–DDX21–DHX36 complex functions as a RNA sensor and associates with further adaptor proteins to activate the type I IFN response [37].
3. Virus-induced activation of host cell transcription factors and the inflammasome
An important signal integrator for the activated RNA receptors is the mitochondrial adaptor protein MAVS, which relays the signals obtained from the RNA sensors to the host cell signaling cascades. The majority of this protein is found at the outside of mitochondria, while small amounts have also been described in association with peroxisomes [38] and the ER [39]. In vitro experiments showed that RNA-bound RIG-I activates the adaptor protein MAVS by inducing its aggregation by prion-like polymerization in the presence of K63-linked polyubiquitin [10], [28]. Viral infection causes the formation of detergent-resistant and high-molecular-weight MAVS polymers. The polymerization process critically depends on the presence of an N-terminal CARD in MAVS that serves as a prion domain and is thus required for the polymerization process. Polymerized MAVS then triggers activation of various downstream responses such as the inflammasome and the transcription factors IRF and NF-κB.
An initial step in MAVS-mediated NF-κB activation is the recruitment of multiple ubiquitin E3 ligases including TRAF2 (TNF receptor-associated factor 2) and TRAF6 [40]. TRAF6-mediated K63-ubiquitination creates docking sites for proteins with a ubiquitin-binding domain, thus fostering protein/protein interactions and allowing for the induced proximity of signaling proteins mediating NF-κB activation [32]. The central step in the canonical NF-κB pathway is the activation of the IκB kinase (IKK) complex, which is composed of the kinases IKKα and IKKβ and the scaffold protein NEMO (NF-κB essential modulator). The activated IKK complex phosphorylates the inhibitory IκB (inhibitor of NF-κB), leading to its subsequent proteolytic elimination, which then allows the DNA-binding subunits of NF-κB to enter the nucleus and to bind to DNA [41]. MAVS polymerization leads to a TRAF3 dependent activation of the kinases IKKε/TBK1 (IκB kinase ε/TANK-binding kinase), which are activated by transautophosphorylation [42], [43], [44], [45]. The activated IKKε/TBK kinases in turn phosphorylate IRFs, thus causing their dimerization, nuclear translocation and the induction of target gene transcription [46]. The IRF and NF-κB transcription factors bind cooperatively to the IFN-β enhancer and recruit accessory proteins to trigger IFN-β expression [47].
Besides the activation of IRF/NF-κB transcription factors, MAVS can also activate the inflammasome, a multi-protein complex that catalyzes the conversion of pro-caspase-1 to active caspase-1, which then converts pro-interleukin-1β (pro-IL-1β) into the mature cytokine to trigger inflammation [48], [49]. Inflammasomes are typically composed of the adaptor protein ASC (apoptosis-associated speck-like protein containing a carboxy-terminal CARD) which bridges pro-caspase-1 and danger sensors such as NLRP3 (NLR family, pyrin domain containing 3) or AIM2 (absent in melanoma 2). Many viruses including IAV activate the inflammasome typically via NLRP3 [50], [51]. While the ASC adaptor contains a CARD and a pyrin domain, the NLRP3 sensor contains a pyrin domain that allows interaction with ASC. RIG-I can activate the inflammasome not only by a MAVS-independent pathway [52], but also via NLRP3 recruitment of MAVS to mitochondria [53], [54]. The inducible attachment to mitochondria depends on the N-terminal part of NLRP3, reinforcing the notion that homotypic interactions between pyrin domains or CARDs are essential for virus-induced signaling cascades. Two recent studies show that pyrin domain and CARDs of AIM2 and NLRP3 can form filaments which, in turn, assemble the CARD of the ASC adapter. At the end of this cascade the CARD filaments of caspase-1 cluster and lead to cleavage of the pro-caspase [9], [11]. Formal proof for the formation of these ASC polymers in virus-infected cells is still missing. In summary, inducible filament formation occurs at several steps of the RNA virus-induced signaling cascade. A more general question relates to the reversibility of aggregate formation, which may be achieved by proteolysis or autophagy. It also remains unknown whether there is a “molecular memory” for the signaling complexes. Also the exact composition and stoichiometry of the multi-protein aggregates that are inducibly formed during these signaling processes are not well understood. For example it is known that RIG-I binding to MAVS initiates the recruitment of > 30 interacting partners [55]. However, the exact order of binding and the occurrence of distinct interactomes have not been studied.
4. The NF-κB pathways
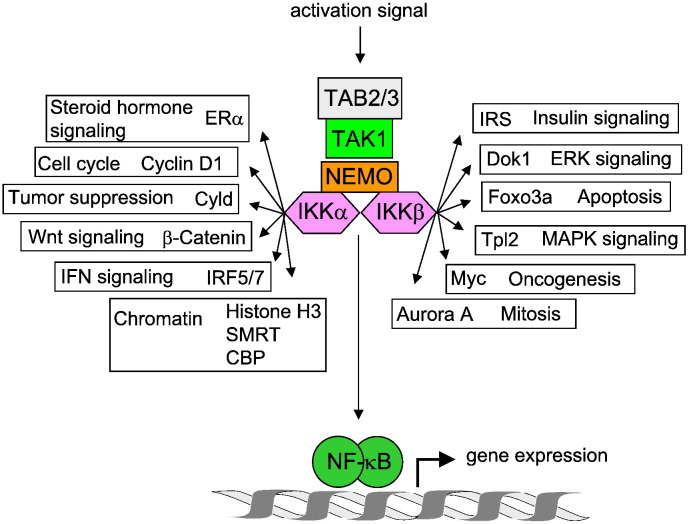
Exposure of cells to perilous agents such as damage-associated and pathogen-associated molecular patterns leads to the induction of signaling cascades which ultimately leads to NF-κB activation [41]. Three major pathways mediating NF-κB activation have been identified: the so-called canonical and noncanonical pathways and the atypical NF-κB activation pathway [41]. All these NF-κB activating pathways have in common that they lead to the generation of DNA-binding dimers, which are retained in the cytosol of unstimulated cells by association with inhibitory IκB proteins. The canonical NF-κB activation pathway is activated by the IKK complex [56]. Upstream signals lead to the attachment of K63-branched polyubiquitin chains to NEMO and further proteins, which enhances association of proteins containing ubiquitin-binding domains such as TAB2 (TGF-β activated kinase binding protein 2). This in turn allows the recruitment of the TAB2 interacting kinase TAK1 (TGF-β activated kinase 1), which activates the IKKs by trans-phosphorylation of serines in the activation loop [57]. The active IKKs phosphorylate IκBα in order to allow its subsequent phosphorylation-dependent ubiquitination and proteolytic degradation, thus releasing the DNA-binding NF-κB dimer from its inhibitor. While the relevance of IKKβ for IκBα phosphorylation has been demonstrated in knockout animals [58], a number of studies have revealed that the canonical IKKs can phosphorylate many more cytoplasmic and nuclear substrate proteins that are distinct from IκB and NF-κB proteins. Thus the IKKs serve to relay and coordinate NF-κB with other signaling pathways such as insulin and Wnt signaling [59], [60]. This intricate crosstalk serves to shape the diverse biological functions of NF-κB into context-specific responses [61]. A representative selection of IKK substrates and the affected signaling pathways is represented in Fig. 2 .
Fig. 2.
Schematic display of IKK substrates. Arrows point to the different IKKα/IKKβ substrates; the biological functions of the substrates are also indicated.
The noncanonical NF-κB pathway is induced by specific members of the TNF (tumor necrosis factor) cytokine family, including the CD40 ligand. The key feature of the noncanonical pathway is the processing of the precursor p100 protein by the ubiquitin/proteasome system [62], [63]. In comparison to the canonical pathway, this pathway is independent of IKKβ or NEMO, whereas the stabilization of NIK (NF-κB-inducing kinase) and IKKα is essential. Phosphorylation of p100 at several serines in the C-terminus allows for subsequent ubiquitination of p100 at K855 and processing of the precursor to the NF-κB subunit p52 [64], [65]. The released p52 subunit is then dimerizing with the RelB subunit to form p52/RelB heterodimers [66]. The transcriptional response by p52/RelB dimers is distinct from that induced by the canonical, IκBα-regulated pathway which typically leads to the generation of p50/p65 dimers.
The atypical NF-κB activation pathway is activated with DNA damage where lesions are sensed by poly(ADP-ribose)-polymerase-1 (PARP-1). This enzyme synthesizes poly(ADP-ribose), thus allowing the dynamic assembly of a protein complex containing NEMO, PIASy (protein inhibitor of activated STAT y), and the DNA damage-responsive kinase ataxia-telangiectasia mutated (ATM) [67]. PIASy then triggers the attachment of SUMO (small ubiquitin-related modifier) to NEMO [68], followed by phosphorylation and consequent ubiquitination [69]. A fraction of activated ATM is also traveling to the cytosol where TRAF6-dependent activation of the TAK1/TAB2 and the IKK complex takes place. This activation depends on monoubiquitination of NEMO [70]. As a result of these processes which are reviewed in more detail elsewhere [71], the activated IKKs lead to the generation of DNA-binding NF-κB dimers which can be further regulated in the DNA damage response by IKKε-mediated phosphorylations [72].
Active NF-κB acts as a transcription factor that induces hundreds of genes as part of an adjustment program serving to cope with the danger and stress signals leading to NF-κB activation. Among the NF-κB target genes are regulators of inflammatory cytokines (e.g. IL-8), cell survival, proliferation and cell surface proteins [73]. Accordingly, NF-κB-induced gene products are important to build a first line of defense against invading pathogens. As all viruses inevitably induce NF-κB activity, they have developed strategies to control NF-κB activity or even to abuse its activity to sustain transcription of viral genes by using NF-κB DNA binding sites in their promoters [74], [75].
5. The diverse functions of NF-κB in IAV infection
IAVs primarily infect not only lung epithelial cells, but also macrophages and recruited leukocytes can be infected [76]. IAVs have a segmented genome consisting of eight negative strand RNAs. The virion RNA (vRNA) is used as a template for transcription to generate viral mRNA and complementary RNA (cRNA), respectively. While the cRNA is used as a template for vRNA synthesis, the mRNA can be translated to generate viral proteins [77], [78]. IAVs encode at least 10 viral proteins including the regulatory nonstructural protein 1 (NS1), while some strains express the additional proteins PB1-F2 (polymerase basic protein 1-frame 2) and PB1 N40. It is now known for almost two decades that IAVs activate NF-κB signaling [79], [80], [81], [82], [83], [84], [85], but the precise role of NF-κB for IAV replication and spreading is still not understood.
5.1. IAV supporting functions of NF-κB
A number of studies suggest that IAV-elicited NF-κB activity helps in virus replication and spreading, based on the finding that NF-κB inhibition also impairs IAV propagation [81], [82], [83], [84], [85]. A seminal paper showed that inhibition of IKK activity by the small molecule inhibitors BAY11-7085 and BAY11-7082 severely impaired IAV infection of human lung carcinoma cell lines [86]. This study also described that an infection with vaccinia virus was not NF-κB-dependent, demonstrating that this transcription factor is specifically required for IAV infection. This basic finding was confirmed by several independent follow-up studies which showed that other IKK inhibitors such as acetyl salicylic acid and SC75741 efficiently block IAV propagation [81], [84]. Along this line, inhibition of NF-κB by expression of a dominant-negative IKKβ mutant or a non-degradable IκBα mutant also resulted in diminished IAV replication in lung A549 cells, further indicating that NF-κB activity promotes efficient IAV production [87]. Since NF-κB is responsible for the majority of IAV-elicited gene expression [88], the beneficial effect of IKK inhibition will be relevant not only for IAV-infected cells but also for neighboring cells which are exposed to exaggerated concentrations of cytokines and chemokines that can cause severe lung damage. The IKK inhibitor BAY11-7082 impairs IAV replication not only in cultured cells but also in mouse models. Intraperitoneal administration of the inhibitor causes impaired cytokine expression and reduced virus titers in the bronchoalveolar lavage of IAV-infected mice [85]. Mice lacking the cytosolic protein kinase RIPK2 (receptor-interacting protein kinase 2) show signs of hyperinflammation along with increased NF-κB activity and expression of cytokines and chemokines. Consistent with the notion of virus-supportive NF-κB signaling these animals are hypersusceptible to IAV infection [89]. The molecular mechanisms employed by NF-κB to support IAV infection are not well understood. One report shows that SC75741 efficiently impairs IAV-induced expression of cytokines and pro-apoptotic factors such as TNF-related apoptosis inducing ligand (TRAIL) or the CD95 ligand. As these ligands trigger the activation of caspases, also the caspase-mediated nuclear export of viral ribonucleoproteins is downregulated [81]. Another study using the IKK inhibitor BAY11-7082 did not observe effects on nucleocytoplasmic trafficking of the viral ribonucleoprotein complex [83]. These authors rather reported that the NF-κB inhibitor pyrrolidinedithiocarbamate impairs vRNA synthesis, but the limited specificity of this compound raises the need for further experiments to substantiate these results. This work also showed that overexpression of p65 activates IAV transcription from the cRNA promoter [83], but the molecular mechanisms employed by p65 are not clear. It would be interesting to know whether p65 has the ability to bind to RNA, as it was previously shown for the NF-κB subunit p50 [90]. Members of the NF-κB activation pathway were also found in RNAi-based genome-wide screens that were conducted to identify host factors involved in virus replication. From the large number of proteins that are involved in NF-κB activation only the kinases IKKα and IKKε were detected in more than one screen [91]. While these papers collectively show a proviral function of NF-κB, another study failed to support this notion. Inhibition of NF-κB in A549 cells upon expression of a non-degradable IκBα mutant reduced IAV-induced expression of proinflammatory cytokines, but did not affect virus replication [92].
5.2. IAV inhibiting functions of NF-κB
Several reports note an antiviral function of NF-κB. Myeloid cells lacking the gene encoding the NF-κB inhibitory A20 protein show exaggerated NF-κB activation after IAV infection. As expected, these cells produce increased levels of proinflammatory cytokines and type I IFN. However, knockout of A20 in myeloid cells leads to protection of mice against lethal IAV infection, thus pointing to an antiviral role of increased NF-κB activity [93]. The antiviral function of NF-κB most probably relies on its ability to induce the expression of inflammatory mediators that help to clear the infection. This notion is supported by experiments where the administration of 5′ppp RNA to cells or mice causes the RIG-I-mediated induction of inflammatory and IFN-stimulated genes. This induction of innate immunity protected the cells and animals from a subsequent infection with IAVs [94], showing the antiviral effects of inflammatory mediators. These results also imply that an imbalance between the harmful and beneficial effects of inflammatory mediators contributes to the pathogenesis of influenza. How can these various results be reconciled and explained? It is important to note that most NF-κB signaling proteins relay to further information processing pathways. As schematically depicted in Fig. 2, IKKα and IKKβ phosphorylate many other substrate proteins beyond IκBα [59], [60]. These substrate proteins allow a close coordination of NF-κB signaling with other pathways and this crosstalk serves to shape the diverse biological functions of NF-κB [61]. As IKK inhibition will also necessarily impinge on further pathways, the antiviral function of IKK inhibitors may not exclusively be attributable to NF-κB inhibition. In the future it will be important to investigate the contribution of NF-κB-dependent gene expression on IAV infection by targeting downstream effectors such as the DNA-binding subunits. Another important aspect relates to the infection models. In the primary phase of influenza virus infection a limited amount of IAVs infects lung cells including alveolar macrophages that are among the first cells that encounter infectious IAV particles [95]. It is conceivable that NF-κB is antiviral during this initial phase upon generation of inflammatory mediators attracting innate immune cells (e.g. neutrophils and monocytes) and by triggering expression of cytokines and antiviral IFN. It was shown that cytokines and soluble mediators from H5N1-infected human macrophages can activate the expression of RNA-sensing proteins such as RIG-I, MDA5, and TLR3 [96]. This paracrine effect allows uninfected cells to detect IAV infection with higher sensitivity and to produce increased amounts of antiviral mediators. Once the infection is established, the massively released virus progeny will cause an exacerbated NF-κB activation that supports IAV replication by ill-defined mechanisms and causes an excessive proinflammatory response in the lung which contributes to IAV-induced mortality.
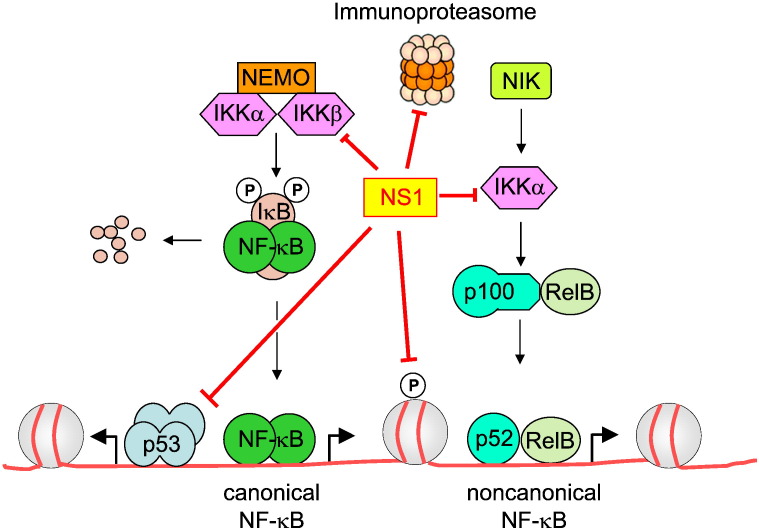
The antiviral function of NF-κB in the early phase of IAV infection also explains why IAVs employ several mechanisms to dampen or inhibit NF-κB activity. Deletion of the NS1 gene in IAV results in enhanced IFN production in infected host cells, pointing to an IFN inhibiting function of the NS1 protein [80]. Accordingly, expression of the NS1 protein antagonizes IAV-induced NF-κB activation and subsequent IFN synthesis. Several studies show that the IFN antagonizing function of the NS1 protein depends on the IAV genotype [97], [98]. For example, a virus containing the 1918 pandemic NS1 gene was more efficient at blocking the IFN response than its parental IAV [97]. The IFN antagonizing function of NS1 is mediated by several mechanisms, which are summarized in Fig. 3 . Co-immunoprecipitation experiments showed the constitutive association of NS1 with the kinase domains of IKKα and IKKβ [99]. This binding interferes with their function, as NS1 expression prohibited their downstream effects. These include IKK-induced IκBα phosphorylation and IKKα-mediated phosphorylation of histone H3 S10 [99]. Besides its antagonizing activity on the classical NF-κB pathway, the NS1 protein also impairs NIK-induced processing of p100 to the DNA-binding p52 protein [99]. The relative contribution of the alternative NF-κB signaling pathway for IAV replication is not clear, as infection with IAV strains expressing the NS1 protein only modestly activates the noncanonical NF-κB pathway [99], [100]. The NS1 protein was also reported to impair the transcriptional activity of other transcription factors such as p53 [101] and immune-proteasome pathways [98]. It is conceivable that NS1 will affect more cellular signaling steps, as interactome screens have shown numerous cellular binding partners for this viral protein including members of the PI3K (phosphoinositide-3-kinase) and AKT signaling pathways [102], [103]. Some IAV strains additionally express the accessory PB1-F2 protein which stems from an alternative reading frame. One report showed that PB1-F2 binds to IKKβ and impairs DNA-binding of NF-κB [104]. In contrast, another study reports on an NF-κB intensifying activity of PB1-F2 [105], so that more work is required to clarify its role on the NF-κB system.
Fig. 3.
Cellular targets inhibited by the IAV protein NS1. The red lines point to the cellular targets that show reduced activity in the presence of NS1.
NF-κB activity will directly influence the IAV-infected host cell, but its ability to trigger expression of chemokines and cytokines will also regulate non-infected neighboring cells in a paracrine manner. This complexity raises the need to study its relevance in suitable animal systems, preferably in animals such as mice that can be genetically manipulated. This will allow one to reveal the role of NF-κB in the broad spectrum of IAV infection that ranges from asymptomatic or subclinical infection to a severe viral pneumonia [106].
6. The role of NF-κB in HTLV-1 infection
HTLV-1 was the first discovered human retrovirus [107] and is the etiological agent for inflammatory diseases including the neurodegenerative disorder tropical spastic paraparesis/HTLV-1-associated myelopathy and adult T-cell leukemia/lymphoma (ATL) [108], [109]. Between 10 and 20 million people worldwide are infected with this virus and less than 5% of the virus carriers develop ATL over decades [110]. At present there is no treatment to cure this aggressive and lethal malignancy of CD4+ T lymphocytes. HTLV-1 has a diploid genome, comprised of two identical strands of positive sense RNA. Similar to other retroviruses the genome contains two long terminal repeat (LTR) sequences that drive expression of the viral structural proteins. The viruses also express further regulatory proteins including Tax-1, Rex, p30, and HTLV-1 basic leucine zipper factor (HBZ) [111]. The most intensively studied protein is Tax-1, as it is required for the transforming ability of HTLV-1 [112]. Further evidence for the importance of Tax-1 came from the analysis of transgenic Tax-1-expressing mice which develop tumors closely resembling the phenotype of HTLV-1-induced ATL [113], [114]. The Tax-1 protein impinges on a multitude of signaling pathways including NF-κB as discussed in more detail below. The other HTLV-1-encoded proteins are less intensively studied, but it is known that Rex, p30 and HBZ negatively regulate the activity or expression of Tax-1. The Tax-1 protein reportedly interacts with more than 100 host cell proteins including transcription factors, transcriptional regulators, chromatin modifiers, protein kinases and proteins involved in cytoskeleton structure and dynamics [115]. In addition, numerous intracellular localizations for Tax-1 were reported including the nucleus [116], microtubule organization center [117], at the centrosome during mitosis [118], [119] and in the cytoplasm. It was also suggested that Tax-1 enters the secretory pathway in a leaderless manner and can migrate from the endoplasmic reticulum to the Golgi complex [120], thus even exerting extracellular functions. The large number of interaction partners and intracellular localizations may help to explain the impact of Tax-1 on almost all cellular signaling pathways including p53 activation and signaling via mitogen-activated protein kinases, G-proteins and TGFβ (transforming growth factor β) signaling [115].
From all these signaling pathways, NF-κB seems to be of special relevance for the process of Tax-1-induced ATL, as suggested by several findings: (i) Tax-1 mutants that are deficient in the activation of NF-κB fail to trigger Tax-1-induced cell proliferation and HTLV-1-induced immortalization of T-cells [121], [122]. (ii) Inhibition of NF-κB by various approaches such as IKK inhibition and overexpression of dominant-negative forms of IκBα or p100 protects cells from Tax-1-mediated transformation in cell culture and animal experiments [123], [124], [125], [126]. (iii) Inhibition of noncanonical NF-κB activation by knockout of the gene coding for the p100/p52 proteins largely prevents tumorigenesis in Tax-1 transgenic mice [127], thus clearly demonstrating the contribution of NF-κB for Tax-1-mediated oncogenesis. While these results collectively show the relevance of the Tax-1–NF-κB axis for the induction of tumorigenesis, it is interesting to note that expression of Tax-1 is lost in the majority of all ATLs during the late stages of leukemogenesis [128], [129], [130]. And also ATLs lacking Tax-1 expression show strong activation of canonical and noncanonical NF-κB pathways, implying the relevance of Tax-1-independent mechanisms mediating chronic NF-κB activity [131], [132]. Proof-of-concept for the idea that ongoing constitutive IKK activity per se is sufficient to induce tumor formation was revealed in a mouse model where constitutively active IKKβ was expressed in intestinal epithelial cells. Expression of the active IKKβ induces spontaneous tumors in aged mice [133], thus revealing that IKK-induced tumors can be generated in the absence of additional pro-tumorigenic events, at least in intestinal epithelial cells. As active IKKs do not only feed in the NF-κB activation pathway (see Fig. 2), a potential contribution of other IKK-regulated pathways for tumor generation remains to be revealed in future studies.
7. Molecular mechanisms of Tax-1-controlled NF-κB activation
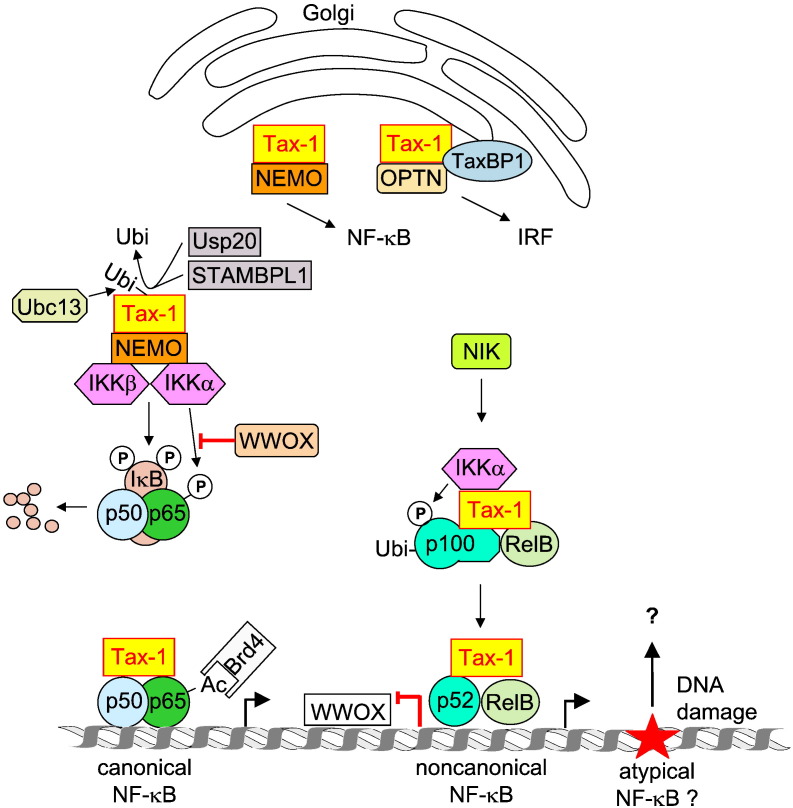
Early and Tax-1-dependent activation of NF-κB occurs by several mechanisms which are schematically summarized in Fig. 4 . The relevance of Tax-1 for the canonical NF-κB activation pathway was seen very early, as NEMO was found to be essential for the NF-κB activating effects of Tax-1 [134]. Further experiments showed that Tax-1 directly binds to the adapter protein NEMO, thus causing the constitutive activation of the IKK complex [135], [136], [137]. A fraction of Tax-1 is found in association with Golgi-associated lipid rafts where this viral protein can recruit the IKK complex to these membrane microdomains [138]. As the activation status of IKKs can be determined with phospho-specific antibodies, it would be interesting to see whether the lipid raft-associated IKKs are active. Tax-1 can be modified by several posttranslational modifications that regulate numerous aspects of its function. Ubiquitination of Tax-1 is important for NF-κB activation, since a Tax-1 mutant unable to be ubiquitinated is impaired in NF-κB activation [139]. Tax-1 polyubiquitin chains are predominantly composed of K63-linked chains, which do not lead to proteasomal degradation but rather allow binding to ubiquitin-binding domains, thus allowing the increase of protein/protein interactions [140]. As NEMO has the ability to bind to K63-linked ubiquitin in its C terminus, this interaction would even strengthen NEMO/Tax-1 interactions [141]. The importance of the NEMO ubiquitin-binding domain was shown for TNF-induced NF-κB activation [142] and it will be interesting to test in the future whether the same holds true for Tax-1-triggered NF-κB activation. The K63-branched ubiquitin is attached to Tax-1 by the E2 ubiquitin-conjugating enzyme Ubc13 (ubiquitin conjugating 13). Deletion of the Ubc13 gene prevents Tax-1/NEMO-binding and Tax-1-mediated NF-κB activation [143], but as Ubc13 also modifies further members of the NF-κB activation pathway such as TRAF6 [144] this defect may not be exclusively attributable to defective Tax-1 ubiquitination. Interestingly, also one of the deubiquitinating enzymes is shared between TRAF6 and Tax-1. The ubiquitin-specific peptidase USP20 (ubiquitin specific peptidase) deubiquitinates Tax-1 and suppresses IL-1β- and Tax-1-induced NF-κB activation [145]. The deubiquitination of Tax-1 can be also mediated by STAM-binding protein-like 1 (STAMBPL1), which was identified in an RNA interference screen [146]. Ubiquitination of Tax-1 also allows binding to the NEMO-related protein Optineurin (OPTN) that functions in various physiological processes including the IFN response. This protein complex also contains the Tax-1 binding protein 1 (TAX1BP1) and the interaction between Tax-1 and OPTN requires the ubiquitin-binding activity of OPTN and the ubiquitination sites of Tax-1 [147]. Immunofluorescence studies show that Tax-1, OPTN and NEMO colocalize in Golgi-associated lipid rafts, but further experiments such as sequential immunoprecipitations must clarify whether the two NEMO family members are contained in the same or in distinct multi-protein complexes. In the nucleus, a fraction of Tax-1 localizes to subnuclear domains which overlap with structures containing the splicing regulator SC35 (serine/arginine-rich splicing factor 2), a protein which is a commonly used marker of spliceosomal speckles [148].
Fig. 4.
Cellular targets regulated by the HTLV-1 Tax-1 protein. The different localizations and interaction partners of Tax-1 are shown; its possible contribution to the induction of the atypical NF-κB pathway is indicated.
The nuclear structures containing Tax-1 also contain the NF-κB subunits p50 and p65 as well as the largest subunit of RNA polymerase II and cyclin-dependent kinase CDK8 [149]. It will be highly interesting to characterize the Tax-1-associated proteins in these nuclear bodies in a more systematic way, although the limited solubility of these chromatin-associated and tightly packed protein complexes is technically challenging. Expression of Tax-1 augments basal p65 acetylation at K310 by an unknown mechanism, which in turn allows binding of Brd4 (bromodomain-containing 4). Knockdown of Brd4 strongly impairs Tax-1-induced NF-κ-driven transcription [150], but it remains to be seen whether Brd4 is a general or stimulus-specific NF-κB coactivator.
Tax-1 also induces the noncanonical NF-κB pathway as revealed by Tax-1-induced IKKα-dependent processing of p100 to the p52 subunit. Mice lacking the p100-encoding gene display a significantly delayed onset of Tax-1-induced tumorigenesis [127], showing the relevance of noncanonical NF-κB signaling in ATL. The binding of Tax-1 to p100 allows the recruitment of IKKα, which in turn leads to phosphorylation-dependent ubiquitination and processing of p100. This Tax-1-dependent process also requires NEMO, as revealed by reconstitution experiments in T cells lacking this adapter protein [151]. The role of NIK was revealed in an animal model using alymphoplasia (aly/aly) mice bearing a NIK mutation [152]. HTLV-1 infection is significantly reduced in aly/aly mice and these animals do not maintain the provirus for 1 year [153], pointing to the importance of functional NIK for HTLV-1 proliferation and provirus maintenance. The protein levels of NIK are strongly restricted by TRAF3-mediated polyubiquitination and proteasomal degradation as well as by miR-31 [154], [155]. Polycomb-mediated silencing of miR-31 in ATL cells leads to the stabilization of NIK, the activation of noncanonical NF-κB activity and apoptosis resistance [155]. These data also show that epigenetic programs impinge on oncogenic signaling, a concept that may contribute to explain why only a fraction of HTLV-1 infected cells develop to ATLs. The p52/RelB dimers suppress expression of the gene encoding WW domain-containing oxidoreductase (WWOX). This tumor suppressor protein specifically inhibits Tax-1-induced activation of the canonical pathway by blocking IKKα recruitment to p65 and subsequent p65 phosphorylation [127], thereby providing a mechanism that links canonical and noncanonical NF-κB pathways in HTLV-1 Tax-1-mediated tumorigenesis.
Our understanding of Tax-1-induced activation of canonical and noncanonical NF-κB signaling has steadily increased over the past years. On the other hand, the mechanisms leading to maintained NF-κB activation at the late stages of HTLV-1 infection after downregulation of Tax-1 expression are still very incomplete. Many NF-κB target genes are themselves strong activators of NF-κB, so that a number of cells that have experienced sustained NF-κB activity maintain this transcription factor active even after disappearance of the initial inducing agent. Such a situation also occurs in rheumatoid arthritis where high levels of cytokines entertain self-stimulatory regulatory circuits [156], [157]. In addition to these mechanisms it can also be assumed that Tax-1-expressing cells undergo epigenetic changes and chromatin remodeling processes resulting in constitutive NF-κB activity. It will be therefore highly interesting to investigate the contribution of these events in the future. Tax-1 also promotes genetic instability by inducing DNA double strand breaks during DNA replication. Tax-1 also inhibits homologous recombination DNA repair and rather favors the error-prone non-homologous end joining pathway [158], [159]. Genomic instability and mutation are enabling characteristics of cancer cells [160], but the ongoing DNA damage might also trigger the atypical NF-κB signaling pathway. It will thus be interesting to study whether characteristic features of the atypical NF-κB pathway such as the occurrence of SUMOylated NEMO in the nucleus are also found in ATLs.
8. Concluding remarks
Virus-encoded regulatory proteins such as Tax-1 and NS1 affect many different host cell signaling pathways. They lack any known enzymatic function, but apparently bind to a large crowd of different cellular proteins. This allows a precise manipulation of protein/protein interactions, which are at the starting point of many signaling cascades. Importantly, the viral proteins also undergo numerous posttranslational modifications which dictate their intracellular localization, function and interaction with further binding partners. Detailed studies on the occurrence and distribution of these modifications will help to explain the characteristic multiplicity of functions displayed by viral proteins. Thus the host cell enzymes controlling the posttranslational modifications of viral proteins are potentially attractive antiviral target structures. The ability of viral proteins to bind to so many interaction partners may also be due to their structural flexibility. A highly dynamic structure and intrinsic disorder were suggested for the Tax-1 protein [115]. Further flexibility can be achieved by its association with chaperones such as Hsp90 (heat shock protein 90) [161] and the prolyl cis–trans-isomerase Pin1 [162], thus increasing the repertoire of binding partners. While the cytosolic events leading to the activation of NF-κB have now reached a level of detailed understanding, the regulation of this transcription factor in the nucleus is still incomplete. It will be highly interesting to study how RNA virus infection controls the (co)recruitment of transcription factors and viral proteins to common or distinct chromatin loci by ChIP-Seq (chromatin immunoprecipitation coupled to deep sequencing) experiments. Also studies on the generation of an epigenetic memory for previous or persistent virus infections will reveal fascinating insights in the future.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft projects SFB 1021 (M.L.S. & M.K.), SCHM1417/9-1 (M.L.S.) and Kr1143/7-1 (M.K.).
References
- 1.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 3.Drake J.W., Holland J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kash J.C., Tumpey T.M., Proll S.C., Carter V., Perwitasari O., Thomas M.J., Basler C.F., Palese P., Taubenberger J.K., Garcia-Sastre A., Swayne D.E., Katze M.G. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Sastre A., Biron C.A. Type 1 interferons and the virus–host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 6.Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Haller O., Kochs G., Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X., Chen J., Xu H., Liu S., Jiang Q.X., Halfmann R., Chen Z.J. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., Schroder G.F., Fitzgerald K.A., Wu H., Egelman E.H. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel J.R., Jain A., Chou Y.Y., Baum A., Ha T., Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peisley A., Wu B., Yao H., Walz T., Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Le B.A., Tough D.F. Type I interferon as a stimulus for cross-priming. Cytokine Growth Factor Rev. 2008;19:33–40. doi: 10.1016/j.cytogfr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 16.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Schulz O., Diebold S.S., Chen M., Naslund T.I., Nolte M.A., Alexopoulou L., Azuma Y.T., Flavell R.A., Liljestrom P., Reis e Sousa Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 18.Kariko K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 19.Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., Davies D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diebold S.S., Massacrier C., Akira S., Paturel C., Morel Y., Reis e Sousa Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 2006;36:3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 21.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 22.Seo S.U., Kwon H.J., Song J.H., Byun Y.H., Seong B.L., Kawai T., Akira S., Kweon M.N. MyD88 signaling is indispensable for primary influenza A virus infection but dispensable for secondary infection. J. Virol. 2010;84:12713–12722. doi: 10.1128/JVI.01675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., Yamaguchi O., Otsu K., Tsujimura T., Koh C.S., Reis e Sousa, Matsuura Y., Fujita T., Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 24.Vitour D., Meurs E.F. Regulation of interferon production by RIG-I and LGP2: a lesson in self-control. Sci. STKE. 2007;2007:e20. doi: 10.1126/stke.3842007pe20. [DOI] [PubMed] [Google Scholar]
- 25.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., Endres S., Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 26.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K.A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 28.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T., Hohng S., Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z., Zhang X., Wang G., Zheng H. The laboratory of genetics and physiology 2: emerging insights into the controversial functions of this RIG-I-like receptor. Biomed. Res. Int. 2014;2014:960190. doi: 10.1155/2014/960190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathinam V.A., Fitzgerald K.A. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garbelli A., Beermann S., Di C.G., Dietrich U., Maga G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS One. 2011;6:e19810. doi: 10.1371/journal.pone.0019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruciat C.M., Dolde C., de Groot R.E., Ohkawara B., Reinhard C., Korswagen H.C., Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 35.Oshiumi H., Sakai K., Matsumoto M., Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur. J. Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Hu M.M., Wang Y.Y., Shu H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z., Kim T., Bao M., Facchinetti V., Jung S.Y., Ghaffari A.A., Qin J., Cheng G., Liu Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., Nibert M.L., Superti-Furga G., Kagan J.C. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zuylen W.J., Doyon P., Clement J.F., Khan K.A., D'Ambrosio L.M., Do F., St-Amant-Verret M., Wissanji T., Emery G., Gingras A.C., Meloche S., Servant M.J. Proteomic profiling of the TRAF3 interactome network reveals a new role for the ER-to-Golgi transport compartments in innate immunity. PLoS Pathog. 2012;8:e1002747. doi: 10.1371/journal.ppat.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y.T., Sun L., Chen Z.J. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayden M.S., Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 43.Larabi A., Devos J.M., Ng S.L., Nanao M.H., Round A., Maniatis T., Panne D. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 2013;3:734–746. doi: 10.1016/j.celrep.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 44.Oganesyan G., Saha S.K., Guo B., He J.Q., Shahangian A., Zarnegar B., Perry A., Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 45.Tu D., Zhu Z., Zhou A.Y., Yun C.H., Lee K.E., Toms A.V., Li Y., Dunn G.P., Chan E., Thai T., Yang S., Ficarro S.B., Marto J.A., Jeon H., Hahn W.C., Barbie D.A., Eck M.J. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura T., Yanai H., Savitsky D., Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 47.Panne D., Maniatis T., Harrison S.C. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 49.Rathinam V.A., Fitzgerald K.A. Inflammasomes and anti-viral immunity. J. Clin. Immunol. 2010;30:632–637. doi: 10.1007/s10875-010-9431-4. [DOI] [PubMed] [Google Scholar]
- 50.Allen I.C., Scull M.A., Moore C.B., Holl E.K., McElvania-TeKippe E., Taxman D.J., Guthrie E.H., Pickles R.J., Ting J.P. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas P.G., Dash P., Aldridge J.R., Jr., Ellebedy A.H., Reynolds C., Funk A.J., Martin W.J., Lamkanfi M., Webby R.J., Boyd K.L., Doherty P.C., Kanneganti T.D. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M., Hannesschlager N., Schlee M., Rothenfusser S., Barchet W., Kato H., Akira S., Inoue S., Endres S., Peschel C., Hartmann G., Hornung V., Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 53.Park S., Juliana C., Hong S., Datta P., Hwang I., Fernandes-Alnemri T., Yu J.W., Alnemri E.S. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J. Immunol. 2013;191:4358–4366. doi: 10.4049/jimmunol.1301170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian N., Natarajan K., Clatworthy M.R., Wang Z., Germain R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belgnaoui S.M., Paz S., Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Hinz M., Scheidereit C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep. 2014;15:46–61. doi: 10.1002/embr.201337983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Deng L., Hong M., Akkaraju G.R., Inoue J., Chen Z.J. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 58.Li Z.W., Chu W., Hu Y., Delhase M., Deerinck T., Ellisman M., Johnson R., Karin M. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J. Exp. Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 61.Oeckinghaus A., Hayden M.S., Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 62.Razani B., Reichardt A.D., Cheng G. Non-canonical NF-kappaB signaling activation and regulation: principles and perspectives. Immunol. Rev. 2011;244:44–54. doi: 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 63.Senftleben U., Cao Y., Xiao G., Greten F.R., Krahn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S.C., Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 64.Amir R.E., Haecker H., Karin M., Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 65.Fong A., Zhang M., Neely J., Sun S.C. S9, a 19 S proteasome subunit interacting with ubiquitinated NF-kappaB2/p100. J. Biol. Chem. 2002;277:40697–40702. doi: 10.1074/jbc.M205330200. [DOI] [PubMed] [Google Scholar]
- 66.Bonizzi G., Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Stilmann M., Hinz M., Arslan S.C., Zimmer A., Schreiber V., Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol. Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 68.Mabb A.M., Wuerzberger-Davis S.M., Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat. Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 69.Huang T.T., Wuerzberger-Davis S.M., Wu Z.H., Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 70.Hinz M., Stilmann M., Arslan S.C., Khanna K.K., Dittmar G., Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol. Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 71.McCool K.W., Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol. Rev. 2012;246:311–326. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Renner F., Moreno R., Schmitz M.L. SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol. Cell. 2010;37:503–515. doi: 10.1016/j.molcel.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 73.Ziesche E., Kettner-Buhrow D., Weber A., Wittwer T., Jurida L., Soelch J., Muller H., Newel D., Kronich P., Schneider H., Dittrich-Breiholz O., Bhaskara S., Hiebert S.W., Hottiger M.O., Li H., Burstein E., Schmitz M.L., Kracht M. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res. 2013;41:90–109. doi: 10.1093/nar/gks916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawakami K., Scheidereit C., Roeder R.G. Identification and purification of a human immunoglobulin-enhancer-binding protein (NF-kappa B) that activates transcription from a human immunodeficiency virus type 1 promoter in vitro. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4700–4704. doi: 10.1073/pnas.85.13.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sambucetti L.C., Cherrington J.M., Wilkinson G.W., Mocarski E.S. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Julkunen I., Melen K., Nyqvist M., Pirhonen J., Sareneva T., Matikainen S. Inflammatory responses in influenza A virus infection. Vaccine. 2000;19(Suppl. 1):S32–S37. doi: 10.1016/s0264-410x(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 77.Barrett T., Wolstenholme A.J., Mahy B.W. Transcription and replication of influenza virus RNA. Virology. 1979;98:211–225. doi: 10.1016/0042-6822(79)90539-7. [DOI] [PubMed] [Google Scholar]
- 78.McGeoch D., Fellner P., Newton C. Influenza virus genome consists of eight distinct RNA species. Proc. Natl. Acad. Sci. U. S. A. 1976;73:3045–3049. doi: 10.1073/pnas.73.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pahl H.L., Baeuerle P.A. Expression of influenza virus hemagglutinin activates transcription factor NF-kappa B. J. Virol. 1995;69:1480–1484. doi: 10.1128/jvi.69.3.1480-1484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X., Li M., Zheng H., Muster T., Palese P., Beg A.A., Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J. Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ehrhardt C., Ruckle A., Hrincius E.R., Haasbach E., Anhlan D., Ahmann K., Banning C., Reiling S.J., Kuhn J., Strobl S., Vitt D., Leban J., Planz O., Ludwig S. The NF-kappaB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell. Microbiol. 2013;15:1198–1211. doi: 10.1111/cmi.12108. [DOI] [PubMed] [Google Scholar]
- 82.Jin J., Hu H., Li H.S., Yu J., Xiao Y., Brittain G.C., Zou Q., Cheng X., Mallette F.A., Watowich S.S., Sun S.C. Noncanonical NF-kappaB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 2014;40:342–354. doi: 10.1016/j.immuni.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar N., Xin Z.T., Liang Y., Ly H., Liang Y. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 2008;82:9880–9889. doi: 10.1128/JVI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazur I., Wurzer W.J., Ehrhardt C., Pleschka S., Puthavathana P., Silberzahn T., Wolff T., Planz O., Ludwig S. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell. Microbiol. 2007;9:1683–1694. doi: 10.1111/j.1462-5822.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 85.Pinto R., Herold S., Cakarova L., Hoegner K., Lohmeyer J., Planz O., Pleschka S. Inhibition of influenza virus-induced NF-kappaB and Raf/MEK/ERK activation can reduce both virus titers and cytokine expression simultaneously in vitro and in vivo. Antiviral Res. 2011;92:45–56. doi: 10.1016/j.antiviral.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 86.Nimmerjahn F., Dudziak D., Dirmeier U., Hobom G., Riedel A., Schlee M., Staudt L.M., Rosenwald A., Behrends U., Bornkamm G.W., Mautner J. Active NF-kappaB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 2004;85:2347–2356. doi: 10.1099/vir.0.79958-0. [DOI] [PubMed] [Google Scholar]
- 87.Wurzer W.J., Ehrhardt C., Pleschka S., Berberich-Siebelt F., Wolff T., Walczak H., Planz O., Ludwig S. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 2004;279:30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 88.Schmolke M., Viemann D., Roth J., Ludwig S. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 2009;183:5180–5189. doi: 10.4049/jimmunol.0804198. [DOI] [PubMed] [Google Scholar]
- 89.Lupfer C., Thomas P.G., Anand P.K., Vogel P., Milasta S., Martinez J., Huang G., Green M., Kundu M., Chi H., Xavier R.J., Green D.R., Lamkanfi M., Dinarello C.A., Doherty P.C., Kanneganti T.D. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang D.B., Vu D., Cassiday L.A., Zimmerman J.M., Maher L.J., III, Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe T., Watanabe S., Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bernasconi D., Amici C., La F.S., Ianaro A., Santoro M.G. The IkappaB kinase is a key factor in triggering influenza A virus-induced inflammatory cytokine production in airway epithelial cells. J. Biol. Chem. 2005;280:24127–24134. doi: 10.1074/jbc.M413726200. [DOI] [PubMed] [Google Scholar]
- 93.Maelfait J., Roose K., Bogaert P., Sze M., Saelens X., Pasparakis M., Carpentier I., van Loo G., Beyaert R. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 2012;8:e1002570. doi: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goulet M.L., Olagnier D., Xu Z., Paz S., Belgnaoui S.M., Lafferty E.I., Janelle V., Arguello M., Paquet M., Ghneim K., Richards S., Smith A., Wilkinson P., Cameron M., Kalinke U., Qureshi S., Lamarre A., Haddad E.K., Sekaly R.P., Peri S., Balachandran S., Lin R., Hiscott J. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tumpey T.M., Garcia-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Pantin-Jackwood M.J., Schultz-Cherry S., Solorzano A., Van Rooijen N., Katz J.M., Basler C.F. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hui K.P., Lee S.M., Cheung C.Y., Mao H., Lai A.K., Chan R.W., Chan M.C., Tu W., Guan Y., Lau Y.L., Peiris J.S. H5N1 influenza virus-induced mediators upregulate RIG-I in uninfected cells by paracrine effects contributing to amplified cytokine cascades. J. Infect. Dis. 2011;204:1866–1878. doi: 10.1093/infdis/jir665. [DOI] [PubMed] [Google Scholar]
- 97.Geiss G.K., Salvatore M., Tumpey T.M., Carter V.S., Wang X., Basler C.F., Taubenberger J.K., Bumgarner R.E., Palese P., Katze M.G., Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tisoncik J.R., Billharz R., Burmakina S., Belisle S.E., Proll S.C., Korth M.J., Garcia-Sastre A., Katze M.G. The NS1 protein of influenza A virus suppresses interferon-regulated activation of antigen-presentation and immune-proteasome pathways. J. Gen. Virol. 2011;92:2093–2104. doi: 10.1099/vir.0.032060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao S., Song L., Li J., Zhang Z., Peng H., Jiang W., Wang Q., Kang T., Chen S., Huang W. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell. Microbiol. 2012;14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 100.Rückle A., Haasbach E., Julkunen I., Planz O., Ehrhardt C., Ludwig S. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-kappaB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J. Virol. 2012;86:10211–10217. doi: 10.1128/JVI.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li W., Wang G., Zhang H., Xin G., Zhang D., Zeng J., Chen X., Xu Y., Cui Y., Li K. Effects of NS1 variants of H5N1 influenza virus on interferon induction, TNFalpha response and p53 activity. Cell. Mol. Immunol. 2010;7:235–242. doi: 10.1038/cmi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.König R., Stertz S., Zhou Y., Inoue A., Hoffmann H.H., Bhattacharyya S., Alamares J.G., Tscherne D.M., Ortigoza M.B., Liang Y., Gao Q., Andrews S.E., Bandyopadhyay S., De J.P., Tu B.P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., Garcia-Sastre A., Young J.A., Palese P., Shaw M.L., Chanda S.K. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E., Hill D.E., Regev A., Hacohen N. A physical and regulatory map of host–influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reis A.L., McCauley J.W. The influenza virus protein PB1-F2 interacts with IKKbeta and modulates NF-kappaB signalling. PLoS One. 2013;8:e63852. doi: 10.1371/journal.pone.0063852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Le Goffic R., Leymarie O., Chevalier C., Rebours E., Da C.B., Vidic J., Descamps D., Sallenave J.M., Rauch M., Samson M., Delmas B. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog. 2011;7:e1002202. doi: 10.1371/journal.ppat.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thangavel R.R., Bouvier N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods. 2014 doi: 10.1016/j.jim.2014.03.023. (Apr 4, Epub ahead of print) (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poiesz B.J., Ruscetti F.W., Reitz M.S., Kalyanaraman V.S., Gallo R.C. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature. 1981;294:268–271. doi: 10.1038/294268a0. [DOI] [PubMed] [Google Scholar]
- 108.Haynes B.F., Miller S.E., Palker T.J., Moore J.O., Dunn P.H., Bolognesi D.P., Metzgar R.S. Identification of human T cell leukemia virus in a Japanese patient with adult T cell leukemia and cutaneous lymphomatous vasculitis. Proc. Natl. Acad. Sci. U. S. A. 1983;80:2054–2058. doi: 10.1073/pnas.80.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalyanaraman V.S., Sarngadharan M.G., Nakao Y., Ito Y., Aoki T., Gallo R.C. Natural antibodies to the structural core protein (p24) of the human T-cell leukemia (lymphoma) retrovirus found in sera of leukemia patients in Japan. Proc. Natl. Acad. Sci. U. S. A. 1982;79:1653–1657. doi: 10.1073/pnas.79.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verdonck K., Gonzalez E., Van Dooren S., Vandamme A.M., Vanham G., Gotuzzo E. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect. Dis. 2007;7:266–281. doi: 10.1016/S1473-3099(07)70081-6. [DOI] [PubMed] [Google Scholar]
- 111.Romanelli M.G., Diani E., Bergamo E., Casoli C., Ciminale V., Bex F., Bertazzoni U. Highlights on distinctive structural and functional properties of HTLV Tax proteins. Front. Microbiol. 2013;4:271. doi: 10.3389/fmicb.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamaoka S., Tobe T., Hatanaka M. Tax protein of human T-cell leukemia virus type I is required for maintenance of the transformed phenotype. Oncogene. 1992;7:433–437. [PubMed] [Google Scholar]
- 113.Grossman W.J., Kimata J.T., Wong F.H., Zutter M., Ley T.J., Ratner L. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1057–1061. doi: 10.1073/pnas.92.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nerenberg M., Hinrichs S.H., Reynolds R.K., Khoury G., Jay G. The tat gene of human T-lymphotropic virus type 1 induces mesenchymal tumors in transgenic mice. Science. 1987;237:1324–1329. doi: 10.1126/science.2888190. [DOI] [PubMed] [Google Scholar]
- 115.Boxus M., Twizere J.C., Legros S., Dewulf J.F., Kettmann R., Willems L. The HTLV-1 Tax interactome. Retrovirology. 2008;5:76. doi: 10.1186/1742-4690-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith M.R., Greene W.C. Characterization of a novel nuclear localization signal in the HTLV-I tax transactivator protein. Virology. 1992;187:316–320. doi: 10.1016/0042-6822(92)90320-o. [DOI] [PubMed] [Google Scholar]
- 117.Nejmeddine M., Barnard A.L., Tanaka Y., Taylor G.P., Bangham C.R. Human T-lymphotropic virus, type 1, tax protein triggers microtubule reorientation in the virological synapse. J. Biol. Chem. 2005;280:29653–29660. doi: 10.1074/jbc.M502639200. [DOI] [PubMed] [Google Scholar]
- 118.Peloponese J.M., Jr., Haller K., Miyazato A., Jeang K.T. Abnormal centrosome amplification in cells through the targeting of Ran-binding protein-1 by the human T cell leukemia virus type-1 Tax oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18974–18979. doi: 10.1073/pnas.0506659103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ching Y.P., Chan S.F., Jeang K.T., Jin D.Y. The retroviral oncoprotein Tax targets the coiled-coil centrosomal protein TAX1BP2 to induce centrosome overduplication. Nat. Cell Biol. 2006;8:717–724. doi: 10.1038/ncb1432. [DOI] [PubMed] [Google Scholar]
- 120.Alefantis T., Mostoller K., Jain P., Harhaj E., Grant C., Wigdahl B. Secretion of the human T cell leukemia virus type I transactivator protein tax. J. Biol. Chem. 2005;280:17353–17362. doi: 10.1074/jbc.M409851200. [DOI] [PubMed] [Google Scholar]
- 121.Akagi T., Ono H., Nyunoya H., Shimotohno K. Characterization of peripheral blood T-lymphocytes transduced with HTLV-I Tax mutants with different trans-activating phenotypes. Oncogene. 1997;14:2071–2078. doi: 10.1038/sj.onc.1201045. [DOI] [PubMed] [Google Scholar]
- 122.Robek M.D., Ratner L. Immortalization of CD4(+) and CD8(+) T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 1999;73:4856–4865. doi: 10.1128/jvi.73.6.4856-4865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kitajima I., Shinohara T., Bilakovics J., Brown D.A., Xu X., Nerenberg M. Ablation of transplanted HTLV-I tax-transformed tumors in mice by antisense inhibition of NF-kappa B. Science. 1993;259:1523. doi: 10.1126/science.8456277. [DOI] [PubMed] [Google Scholar]
- 124.Mori N., Yamada Y., Ikeda S., Yamasaki Y., Tsukasaki K., Tanaka Y., Tomonaga M., Yamamoto N., Fujii M. Bay 11-7082 inhibits transcription factor NF-kappaB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood. 2002;100:1828–1834. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]
- 125.Sanda T., Asamitsu K., Ogura H., Iida S., Utsunomiya A., Ueda R., Okamoto T. Induction of cell death in adult T-cell leukemia cells by a novel IkappaB kinase inhibitor. Leukemia. 2006;20:590–598. doi: 10.1038/sj.leu.2404129. [DOI] [PubMed] [Google Scholar]
- 126.Watanabe M., Ohsugi T., Shoda M., Ishida T., Aizawa S., Maruyama-Nagai M., Utsunomiya A., Koga S., Yamada Y., Kamihira S., Okayama A., Kikuchi H., Uozumi K., Yamaguchi K., Higashihara M., Umezawa K., Watanabe T., Horie R. Dual targeting of transformed and untransformed HTLV-1-infected T cells by DHMEQ, a potent and selective inhibitor of NF-kappaB, as a strategy for chemoprevention and therapy of adult T-cell leukemia. Blood. 2005;106:2462–2471. doi: 10.1182/blood-2004-09-3646. [DOI] [PubMed] [Google Scholar]
- 127.Fu J., Qu Z., Yan P., Ishikawa C., Aqeilan R.I., Rabson A.B., Xiao G. The tumor suppressor gene WWOX links the canonical and noncanonical NF-kappaB pathways in HTLV-I Tax-mediated tumorigenesis. Blood. 2011;117:1652–1661. doi: 10.1182/blood-2010-08-303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koiwa T., Hamano-Usami A., Ishida T., Okayama A., Yamaguchi K., Kamihira S., Watanabe T. 5′-Long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002;76:9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takeda S., Maeda M., Morikawa S., Taniguchi Y., Yasunaga J., Nosaka K., Tanaka Y., Matsuoka M. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer. 2004;109:559–567. doi: 10.1002/ijc.20007. [DOI] [PubMed] [Google Scholar]
- 130.Taniguchi Y., Nosaka K., Yasunaga J., Maeda M., Mueller N., Okayama A., Matsuoka M. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology. 2005;2:64. doi: 10.1186/1742-4690-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hironaka N., Mochida K., Mori N., Maeda M., Yamamoto N., Yamaoka S. Tax-independent constitutive IkappaB kinase activation in adult T-cell leukemia cells. Neoplasia. 2004;6:266–278. doi: 10.1593/neo.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mori N., Fujii M., Ikeda S., Yamada Y., Tomonaga M., Ballard D.W., Yamamoto N. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood. 1999;93:2360–2368. [PubMed] [Google Scholar]
- 133.Vlantis K., Wullaert A., Sasaki Y., Schmidt-Supprian M., Rajewsky K., Roskams T., Pasparakis M. Constitutive IKK2 activation in intestinal epithelial cells induces intestinal tumors in mice. J. Clin. Invest. 2011;121:2781–2793. doi: 10.1172/JCI45349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamaoka S., Courtois G., Bessia C., Whiteside S.T., Weil R., Agou F., Kirk H.E., Kay R.J., Israel A. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 135.Chu Z.L., Shin Y.A., Yang J.M., DiDonato J.A., Ballard D.W. IKKgamma mediates the interaction of cellular IkappaB kinases with the tax transforming protein of human T cell leukemia virus type 1. J. Biol. Chem. 1999;274:15297–15300. doi: 10.1074/jbc.274.22.15297. [DOI] [PubMed] [Google Scholar]
- 136.Harhaj E.W., Sun S.C. IKKgamma serves as a docking subunit of the IkappaB kinase (IKK) and mediates interaction of IKK with the human T-cell leukemia virus Tax protein. J. Biol. Chem. 1999;274:22911–22914. doi: 10.1074/jbc.274.33.22911. [DOI] [PubMed] [Google Scholar]
- 137.Jin D.Y., Giordano V., Kibler K.V., Nakano H., Jeang K.T. Role of adapter function in oncoprotein-mediated activation of NF-kappaB. Human T-cell leukemia virus type I Tax interacts directly with IkappaB kinase gamma. J. Biol. Chem. 1999;274:17402–17405. doi: 10.1074/jbc.274.25.17402. [DOI] [PubMed] [Google Scholar]
- 138.Huang J., Ren T., Guan H., Jiang Y., Cheng H. HTLV-1 Tax is a critical lipid raft modulator that hijacks IkappaB kinases to the microdomains for persistent activation of NF-kappaB. J. Biol. Chem. 2009;284:6208–6217. doi: 10.1074/jbc.M806390200. [DOI] [PubMed] [Google Scholar]
- 139.Lamsoul I., Lodewick J., Lebrun S., Brasseur R., Burny A., Gaynor R.B., Bex F. Exclusive ubiquitination and sumoylation on overlapping lysine residues mediate NF-kappaB activation by the human T-cell leukemia virus tax oncoprotein. Mol. Cell. Biol. 2005;25:10391–10406. doi: 10.1128/MCB.25.23.10391-10406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen Z.J., Sun L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 141.Kensche T., Tokunaga F., Ikeda F., Goto E., Iwai K., Dikic I. Analysis of nuclear factor-kappaB (NF-kappaB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-kappaB. J. Biol. Chem. 2012;287:23626–23634. doi: 10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hadian K., Griesbach R.A., Dornauer S., Wanger T.M., Nagel D., Metlitzky M., Beisker W., Schmidt-Supprian M., Krappmann D. NF-kappaB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-kappaB activation. J. Biol. Chem. 2011;286:26107–26117. doi: 10.1074/jbc.M111.233163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shembade N., Harhaj N.S., Yamamoto M., Akira S., Harhaj E.W. The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation. J. Virol. 2007;81:13735–13742. doi: 10.1128/JVI.01790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 145.Yasunaga J., Lin F.C., Lu X., Jeang K.T. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J. Virol. 2011;85:6212–6219. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lavorgna A., Harhaj E.W. An RNA interference screen identifies the deubiquitinase STAMBPL1 as a critical regulator of human T-cell leukemia virus type 1 tax nuclear export and NF-kappaB activation. J. Virol. 2012;86:3357–3369. doi: 10.1128/JVI.06456-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Journo C., Filipe J., About F., Chevalier S.A., Afonso P.V., Brady J.N., Flynn D., Tangy F., Israel A., Vidalain P.O., Mahieux R., Weil R. NRP/Optineurin cooperates with TAX1BP1 to potentiate the activation of NF-kappaB by human T-lymphotropic virus type 1 tax protein. PLoS Pathog. 2009;5:e1000521. doi: 10.1371/journal.ppat.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Semmes O.J., Jeang K.T. Localization of human T-cell leukemia virus type 1 tax to subnuclear compartments that overlap with interchromatin speckles. J. Virol. 1996;70:6347–6357. doi: 10.1128/jvi.70.9.6347-6357.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bex F., McDowall A., Burny A., Gaynor R. The human T-cell leukemia virus type 1 transactivator protein Tax colocalizes in unique nuclear structures with NF-kappaB proteins. J. Virol. 1997;71:3484–3497. doi: 10.1128/jvi.71.5.3484-3497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu X., Qi J., Bradner J.E., Xiao G., Chen L.F. Bromodomain and extraterminal (BET) protein inhibition suppresses human T cell leukemia virus 1 (HTLV-1) Tax protein-mediated tumorigenesis by inhibiting nuclear factor kappaB (NF-kappaB) signaling. J. Biol. Chem. 2013;288:36094–36105. doi: 10.1074/jbc.M113.485029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiao G., Cvijic M.E., Fong A., Harhaj E.W., Uhlik M.T., Waterfield M., Sun S.C. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shinkura R., Kitada K., Matsuda F., Tashiro K., Ikuta K., Suzuki M., Kogishi K., Serikawa T., Honjo T. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat. Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- 153.Nitta T., Tanaka M., Sun B., Sugihara E., Kimura M., Kamada Y., Takahashi H., Hanai S., Jiang S.W., Fujisawa J., Miwa M. Reduction of human T-cell leukemia virus type-1 infection in mice lacking nuclear factor-kappaB-inducing kinase. Cancer Sci. 2008;99:872–878. doi: 10.1111/j.1349-7006.2008.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liao G., Zhang M., Harhaj E.W., Sun S.C. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 155.Yamagishi M., Nakano K., Miyake A., Yamochi T., Kagami Y., Tsutsumi A., Matsuda Y., Sato-Otsubo A., Muto S., Utsunomiya A., Yamaguchi K., Uchimaru K., Ogawa S., Watanabe T. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21:121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 156.Renner F., Schmitz M.L. Autoregulatory feedback loops terminating the NF-kappaB response. Trends Biochem. Sci. 2009;34:128–135. doi: 10.1016/j.tibs.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 157.Schmitz M.L., Weber A., Roxlau T., Gaestel M., Kracht M. Signal integration, crosstalk mechanisms and networks in the function of inflammatory cytokines. Biochim. Biophys. Acta. 2011;1813:2165–2175. doi: 10.1016/j.bbamcr.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 158.Baydoun H.H., Bai X.T., Shelton S., Nicot C. HTLV-I tax increases genetic instability by inducing DNA double strand breaks during DNA replication and switching repair to NHEJ. PLoS One. 2012;7:e42226. doi: 10.1371/journal.pone.0042226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Majone F., Jeang K.T. Unstabilized DNA breaks in HTLV-1 Tax expressing cells correlate with functional targeting of Ku80, not PKcs, XRCC4, or H2AX. Cell Biosci. 2012;2:15. doi: 10.1186/2045-3701-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 161.Gao L., Harhaj E.W. HSP90 protects the human T-cell leukemia virus type 1 (HTLV-1) tax oncoprotein from proteasomal degradation to support NF-kappaB activation and HTLV-1 replication. J. Virol. 2013;87:13640–13654. doi: 10.1128/JVI.02006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peloponese J.M., Jr., Yasunaga J., Kinjo T., Watashi K., Jeang K.T. Peptidylproline cis–trans-isomerase Pin1 interacts with human T-cell leukemia virus type 1 tax and modulates its activation of NF-kappaB. J. Virol. 2009;83:3238–3248. doi: 10.1128/JVI.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]