Abstract
Avian infectious bronchitis virus (IBV) infection is one of the major viral respiratory diseases of chickens. Better understanding of the molecular basis of viral pathogenesis should contribute significantly towards the development of improved prophylactic, therapeutic and diagnostic reagents to control infections. In the present investigation, transcriptional profiles were analyzed by using RNA recovered from the lung tissue of IBV infected 18-day-old chicken embryos at 6, 24, 48 and 72 h post IBV infection. This microarray analysis was completed using avian cDNA arrays comprised of fragments of 1191 unique chicken and turkey gene transcripts. These arrays were generated from normalized cDNA subtraction libraries that were derived from avian pneumovirus (APV) infected chicken embryo fibroblast (CEF) cultures and tissues obtained from APV infected turkeys subtracted with their respective uninfected cultures and tissues. Of the 1191 unique genes represented on the array, the expression of a total of 327 genes (27% of total) were altered by two-fold or more from 6 through 72 h post-infection. A comparative analysis of IBV regulated genes with genes previously reported to change in expression following infection with other avian respiratory viruses revealed both conserved and unique changes. Real-time qRT-PCR was used to confirm the regulated expression of genes related to several functional classes including kinases, interferon induced genes, chemokines and adhesion molecules, vesicular trafficking and fusion protein genes, extracellular matrix protein genes, cell cycle, metabolism, cell physiology and development, translation, RNA binding, lysosomal, protein degradation and ubiquitination related genes. Microarray analysis served as an efficient tool in facilitating a comparative analysis of avian respiratory viral infections and provided insight into host transcriptional changes that were conserved as well as those which were unique to individual pathogens.
Keywords: Infectious bronchitis virus, Avian pneumovirus, Newcastle disease virus, Host genes expression analysis
1. Introduction
Avian viral respiratory infections caused by avian influenza virus, avian infectious bronchitis virus (IBV), avian pneumovirus (APV) and Newcastle disease virus (NDV) result in considerable economic losses to the poultry industry worldwide (Cavanagh, 2003). IBV is one of the major respiratory viruses of chickens that is endemic in all chicken producing countries (Cavanagh, 2003), causing an acute and highly contagious upper respiratory tract infection. The economic impact of the disease is primarily due to poor weight gain, reduced feed efficiency, a drop in egg production and quality and mortality (Kapczynski et al., 2002). Clinically the infection is characterized by coughing, rales, sneezing, nasal discharge and mortality (up to 10%) in young birds (<4 weeks). The virus has been recovered from trachea, bronchus, lungs, esophagus, proventriculus, duodenum, jejunum, caecal tonsils, kidney and cloaca of experimentally infected birds with the most persistent infection in kidney and caecal tonsils. The endemic nature of infection, lack of effective vaccines due to the absence of inter-strain cross protection and circulation of various IBV strains and genotypes in poultry flocks makes the control of IBV infection a challenging task (Farsang et al., 2002). It is well recognized that characterization of in vivo survival mechanisms of the invading organism and identification of the components of host response leading to the elimination of the invading pathogen may provide the basis of pathogenesis and immunity and may lead to improvements in control strategies (Qureshi et al., 1999). The recent introduction of microarray technology has facilitated the large scale analysis of the molecular events occurring during avian respiratory viral infections (Munir and Kapur, 2003, Munir et al., 2005). In order to understand the host cellular determinants involved in the pathogenesis and immunity to IBV, we initiated experiments involving the study of post-IBV infection gene transcriptional responses. Microarray analyses were performed with RNA isolated from lung tissue at various times following IBV infection of 18-day-old chick embryos. We selected 18-day-old embryos for this study because they proved an excellent model to investigate the molecular elements governing host responses to a single pathogen while excluding the potential for co-infection by other pathogens. Furthermore, chicken embryos are immunologically competent at this age and support IBV replication with pathological lesions in epithelial cells of various tissues including lungs, trachea, kidney and bursa of fabricius following IBV inoculation (Kapczynski et al., 2002, Lee et al., 2002). Recently, even younger chicken embryos (8 and 14 days old) have successfully been used as models to study the tissue tropism and kinetics of other avian viral infections (Qureshi et al., 1999, Worthington et al., 2003). Thus, chicken embryos provide a physiologically relevant model system for studying host responses to virus infection. In the current study, we identified several unique genes and pathways that may be involved in IBV infection. Further characterization of the role of these host factors in IBV pathogenesis will help reveal the commonalities and unique differences in the host response to avian viral pathogens and will offer better strategies for disease control.
2. Materials and methods
2.1. Virus and host systems
The clinical IBV isolate (B8358) used in these studies was kindly provided by Dr. Keith West of the Prairie Diagnostic Services (PDS), University of Saskatchewan, Canada. The IBV isolate B8358 was isolated from 28-week-old layers during an IBV infection outbreak at a local poultry farm. The clinical signs associated with virus infection include subdued activity, hoarse sounding noises and squeak and mouth breathing of birds in barn. Sick birds submitted for examination and diagnosis 24 h after the appearance of clinical signs, showed mild catarrhal exudates and mildly hyperemic areas in trachea. Histopathological examination of tissues showed loss of cilia, sloughing of epithelial cells, mucosal congestion, edema with mild hemorrhage and infiltration of heterophils and lymphocytes and severe cuboidal metaplasia in trachea. No significant changes were seen in lung, heart, spleen, and kidney tissues. The virus was isolated from tracheal tissues, identified by electron microscopy and propagated in embryonated chicken eggs at the virology laboratory of PDS. The phylogenetic analysis of the S1 gene of IBV B8358 showed 80.3% sequence homology to the IBV serotype California (CA/633/85) (PDS unpublished data). Further, intra-tracheal inoculation of IBV (B8358) in 4 weeks old layers at VIDO showed similar disease symptoms and pathology as described above. Two hundred microliters of choroallontoic fluid containing IBV was inoculated into 18-day-old chick embryos via the chorioallontoic route [175 embryo lethal dose 50 (ELD50)]. This dose was selected as it was known to be sufficient to produce moderate clinical symptoms in 4-week-old chicks. IBV genes were not present on the array but to verify the presence of productive infection of embryonic lung tissue we quantified IBV N gene transcripts in RNA used for microarray analysis and gene expression. The virus present in IBV infected lung tissue collected at 6 h PI was not detectable by real-time PCR. However, 4.6 × 105 and 5.3 × 106-fold increase in N gene transcript was detected in RNA samples derived from IBV infected embryonic lung tissue collected at 24 and 72 h PI, respectively. Moreover, pathological lesions including curling of toes, and congestion of embryos and embryonic lung tissue was observed at 24 h PI. Furthermore, at 72 h PI there was substantial difference in growth and development of virus infected and non-infected embryos. These data confirm a progressive increase in IBV viral gene expression within infected embryonic lung tissue.
In each set of experiment (n = 2), the lung tissues from virus infected (n = 3) and sham inoculated (n = 3) (sham inoculum was 200 μl of sterile chorioallontoic fluid) embryos were pooled and collected in ice cold TRIZOL (Invitrogen, Life technologies, Carlsbad, CA) at 6, 24, 48 and 72 h post infection (PI). Within 30 min of collection, the tissues were homogenized (Polytron, PT MR 3100, Kinematica, AG, Switzerland) and stored at −80 °C until used.
3. cDNA array
The chicken-turkey array used for hybridization in these studies was an extended version of the avian array used by Munir and Kapur (2003). In brief, the array was generated from normalized cDNA subtraction libraries derived from avian pneumovirus (APV, sub type C) infected chicken embryo fibroblast (CEF) cultures and bursa and thymus tissues obtained from APV infected turkeys subtracted with uninfected cultures and tissues, respectively. The chicken-turkey cDNA array is composed of 1247 chicken and turkey genes (with 3741 spots on array, i.e. each transcript was spotted in triplicate). After subtracting the redundancy of cDNAs derived from 56 genes, these arrays represent 1191 unique chicken and turkey genes. As per a recent public database blast search of the 1191 unique expressed sequence tags (ESTs) spotted onto the array there are 753 genes with no known orthologs in public DNA databases (Munir, unpublished data).
4. cDNA synthesis, probe labeling, and array hybridization
Total cellular RNA was extracted from tissue homogenized in TRIZOL according to the manufacturer's instruction. The RNA was quantified and checked for quality, using an Agilent RNA 6000 nano kit 5065-4476 (Agilent Technologies, 7637 Waldbronn, Germany). Five to ten micrograms of good quality total RNA (with 28s/18s ratio > 1.7) was used to synthesize cDNA probes for hybridization onto a chicken array. The cDNA was prepared by using oligo dT primers and superscript II (Invitrogen, Life technologies, Carlsbad, CA) reverse transcriptase. The cDNA from infected and control samples were labeled with the monofunctional dyes Cy3 and Cy5 (Amersham, Piscataway, NJ). Probes were hybridized onto arrays at 65 °C for 8 h. The buffers and the protocols for hybridization and the post-hybridization processing were reported previously (http://www.agac.umn.edu/microarray/protocols/protocols.htm, Munir and Kapur, 2003). All hybridizations were performed under optimized and uniform conditions. The complete experiment including embryo infection and RNA isolation was performed twice. Probe synthesis and microarray hybridization was repeated at least three to four times per time point within each replicate experiment.
5. Data analysis
The spot intensities of the hybridized images were obtained by using an Affymetrix scanner (428 Array scanner) and the Jaguar software version 2.0. Before importing the gene identifiers and the gene description, the scanned data files were analyzed for their quality by observing individual image spot quality and by subtracting the local background fluorescence from fluorescent intensity of each spot for both the Cy3 and Cy5 channels. Spots with background intensity higher than either dye were flagged as absent and discarded. The replicate spots that had infected/control intensity ratios of 2 or more standard deviations higher than the mean intensity ratio were also discarded. For quantitative analysis and further normalization, the data sets were analyzed by GeneSpring (Silicon Genetics, Redwood city, CA) software version 6.1. The entire data set was normalized by intensity dependent (Lowess) normalization.
6. Data validation
Genes with known critical functions in host pathogen interaction and genes previously characterized in similar studies (Munir and Kapur, 2003) were selected for further validation. Differential expression of selected genes was validated for all four time points PI by real-time quantitative RT-PCR (qRT-PCR) using the SYBR Green based detection system. The specificity of the amplified products was determined by visualizing on 2% agarose gel.
7. Results and discussion
Of the 1191 genes on the array, a total of 327 genes (27%) were altered by two-fold or more at one or more time points between 6 and 72 h PI. Of 200 transcripts that belonged to the category of unknown genes, 177 (88%) and 23 (12%) were up and down-regulated by two-fold or more, respectively. In contrast, of the 127 annotated genes (39% of differentially expressed genes), 111 (87%) and 16 (13%) were two-fold or more up and down-regulation, respectively. The analysis of altered genes revealed expression changes in many vital functional classes including kinase signaling, interferon stimulated genes (ISGs), chemokines and adhesion molecules, vesicular protein trafficking and fusion proteins genes, host genes involved in viral RNA synthesis, extracellular matrix protein genes, and genes belonging to cell cycle, metabolism, physiology and development, RNA binding, transcription and translation regulation, and ubiquitination. A partial list of differentially expressed genes and a graphical representation of kinase gene expression is shown in Table 1 and Fig. 1 , respectively. A comparison of the gene expression profile for IBV infection in ovo (Table 2 ) and APV infection of chicken embryo fibroblast (CEF) cells in vitro (Munir and Kapur, 2003) revealed a list of 14 genes (with >2-fold change in expression) that were common to both viral infections (Table 2). This comparison was completed for genes differentially expressed at 24 and 48 h PI. Gene expression changes at 6 and 72 h PI in IBV infection were not compared with APV since similar time points were not available for APV infection.
Table 1.
Cellular transcripts showing over two fold alterations following IBV infection in ovo
| Accession number | Gene name | Fold change in transcription (standard deviation) PI |
|||
|---|---|---|---|---|---|
| 6 h | 24 h | 48 h | 72 h | ||
| Kinases and phosphatases genes | |||||
| SWP:143Z_HUMAN | kcip-1, factor activating exoenzyme s, fas | 69.76 (41.6) | 26.43 (18.1) | 20.11 (13.7) | 22.02 (11.0) |
| Gb AA23736.1 | Protein kinase [homo sapiens] >gi|7513329|pir | 6.13 (3.05) | 2.53 (0.76) | 1.08 (0.42) | 1.01 (0.34) |
| Ref NP_O67085.1 | Acid phosphatase 1, soluble [rattus norvegicus] | 1.28 (0.38) | 4.58 (0.42) | 1.26 (0.30) | 1.04 (0.17) |
| EMB:CAA83657 | Protein-tyrosine-phosphatase alpha [gallus gallu] | 10.61 (1.62) | 11.76 (0.19) | 4.08 (0.59) | 0.96 (0.18) |
| Inteferon induced genes | |||||
| SWP:INI2_PANTR | Interferon-induced protein 6–16 precursor | 0.85 (0.02) | 3.86 (0.55) | 1.16 (0.01) | 1.12 (0.53) |
| EMB:Q9QXH3 | Alpha-interferon inducible protein (fragment) p27-h | 0.86 (0.02) | 3.22 (1.51) | 1.48 (1.32) | 1.52 (0.66) |
| EMB:AAH02704 | Similar to signal transducer and activator of transcription 1 | 1.77 (1.09) | 1.11 (0.28) | 0.64(0.23) | 1.16 (0.27) |
| WP:STA1_HUMAN | Signal transducer and activator of transcription 1-alpha/beta | 3.03 (0.2) | 1.45 (0.14) | 3.01 (1.07) | 0.96 (0.31) |
| SWP:IFT1_HUMAN | Interferon-induced protein with tetratricopep | 21.94 (7.2) | 35.27 (2.28) | 13.98 (2.18) | 0.99 (0.14) |
| Gb AAH44176.1 | Similar to G protein beta subunit-like [danio rerio] | 17.43 (3.34) | 1.12 (0.59) | 1.07 (0.72) | 1.30 (0.32) |
| Chemoatttractant and adhesion molecules | |||||
| SWP:EMF1_CHICK | Embryo fibroblast protein 1 precursor (emf-1, 9e3, cef4, human il-8 homolog) | 1.21 (0.56) | 4.87 (2.95) | 1.03 (0.20) | 1.19 (0.31) |
| SWP:LSP1_MOUSE | Lymphocyte specific protein-1 | 61.31 (29.35) | 22.84 (13.7) | 4.43 (1.7) | 1.25 (0.64) |
| Signal transduction | |||||
| Gb AAH05446.1 | Adenylyl cyclase-associated cap protein homolog 1 | 1.04 (0.09) | 1.00 (0.05) | 0.46 (0.03) | 0.91 (0.03) |
| Transcription regulation | |||||
| SWP:TYY1_HUMN | Transcriptional repressor protein yy1 | 1.89 (1.2) | 7.21 (1.9) | 0.83 (0.6) | 1.07 (0.3) |
| Gb AAH19668.1 | taf12 rna polymerase ii, tata box binding protein … | 0.78 (0.2) | 7.90 (0.86) | 1.03 (0.06) | 1.32 (0.82) |
| SWP:PTB_PIG | q29099 polypyrimidine tract-binding protein (ptb) (hnrnp1) | 0.82 (0.12) | 8.78 (3.82) | 1.26 (0.28) | 1.03 (0.21) |
| Ref XP_128452.2 | Similar to enhancer factor i chain a-d-rat [m] … | 1.86 (0.75) | 0.63 (0.3) | 3.03 (1.3) | 1.33 (0.74) |
| EMB:CAB62917.1 | E1a binding protein p300 ∼ match: proteins: sw:q09 … | 2.22 (1.3) | 0.51 (0.2) | 1.35 (0.72) | 1.27 (0.21) |
| Gb AAD04629.1 | Pcaf-associated factor 400 [homo sapiens] | 0.43 (0.16) | 0.87 (0.34) | 24.29 (11.2) | 0.93 (0.11) |
| Atigen presentation | |||||
| EMB:O42404 | cd80-like protein precursor. | 0.74 (0.23) | 2.77 (0.40) | 0.96 (0.09) | 0.97 (0.33) |
| Vesicular protein trafficking and ubiqutin proteasome | |||||
| Ref NP_O58040.1| | Clathrin, light polypeptide (lca) [mus musculus… | 3.24 (0.21) | 6.36 (0.44) | 2.92 (1.8) | 1.04 (0.4) |
| SWP:SR54_HUMAN | Signal recognition particle 54 kda protein (s) | 1.32 (0.46) | 6.07 (2.24) | 1.22 (0.65) | 1.01 (0.32) |
| SWP:TERA_HUMAN | p55072 transitional endoplasmic reticulum atpase | 2.20 (0.15) | 1.10 (0.66) | 1.68 (0.86) | 1.08 (0.25) |
| EMB:Q9Y4Z6 | Vacuolar protein sorting. | 1.43 (0.50) | 2.00 (0.56) | 0.47 (0.31) | 1.01 (0.43) |
| Ref NP_003560.1 | Syntaxin 7 [homo sapiens] >gi|2337920|gb|aac518 … | 1.42 (0.08) | 0.46 (0.39) | 0.68 (0.59) | 1.06 (0.19) |
| Gb AAH38447.1 | Similar to coatomer protein complex, subunit alph … | 1.01 (0.08) | 0.93 (0.13) | 1.02 (0.12) | 8.47 (4.23) |
| SWP:RTC0_HUMAN | GTP-binding protein tc10. | 0.16 (0.07) | – | 2.4 (0.4) | 1.00 (0.20) |
| Ref XP_214136.1 | Similar to 26s proteasome non-atpase regulatory … | 2.11 (0.75) | 1.12 (0.78) | 19.87 (2.78) | 0.96 (0.19) |
| SWP:UBPI_HUMAN | Ubiquitin carboxyl terminal hydrolase 18 (Ubp) | 34.74 (10.8) | 21.70 (3.52) | 0.929 (0.14) | 0.890 (0.23) |
| SWP:FUCO_HUMAN | Tissue alpha-1-fucosidase precursor | 1.01 (0.56) | 4.63 (0.77) | 2.46 (0.72) | 0.72 (0.30) |
| Translation | |||||
| SWP:EF1A_CHICK | Elongation factor 1-alpha-1 | 2.24 (0.26) | 1.09 (0.27) | 2.03 (0.76) | 0.94 (0.13) |
| Gb AAO40745.1 | Eukaryotic elongation factor 1-alpha [canis family. | 0.79 (0.31) | 1.53 (0.42) | 4.14 (2.1) | 1.04 (0.01) |
| Gb AAD16874.1 | Peptide elongation factor-1 beta [gallus gallus] | 1.11 (0.58) | 0.93 (0.50) | 0.33 (0.07) | 0.94 (0.18) |
| SWP:IF42_HUMAN | Eukaryotic initiation factor 4a-ii (eif-4a-ii) | 0.83 (0.28) | 7.18 (1.24) | 1.05 (0.30) | 1.13 (0.64) |
| SWP:RL3_BOVIN | 60s ribosomal protein l3. | 3.90 (0.7) | 0.48 (0.65) | 1.09 (0.33) | 1.13 (0.46) |
| Ref XP_110140.2 | Eukaryotic translation initiation factor 3, sub… | 0.34 (0.21) | 0.64 (0.10) | 1.15 (0.17) | 1.26 (0.27) |
| EMB:CAA57493.1 | Ribosomal protein s6 [gallus gallus] >gi|1085253. | 5.04 (1.6) | 0.61 (0.14) | 4.04 (1.6) | 0.75 (0.45) |
| EMB:CAA44568.1 | Ribosomal protein homologous to yeast s24 [homo … | 0.33 (0.10) | 3.60 (1.66) | 1.63 (0.2) | 1.15 (0.47) |
| Ref NP_001347.2 | Dead/h (asp-glu-ala-asp/his) box polypeptide 3;(DDX3) … | 0.80 (0.3) | 3.30 (0.88) | 1.00 (0.15) | 0.93 (0.5) |
| Mitochondrial protein | |||||
| Dbj BAC57503 | Cytochrome oxidase subunit ii | 0.64 (0.28) | 2.97 (1.2) | 0.69 (0.40) | 0.96 (0.40) |
| Dbj BAC57502.1 | Cytochrome oxidase subunit I [coturnix chinensis] | 2.07 (0.60) | 1.15 (0.40) | 1.14 (0.1) | 2.75 (0.96) |
| Ref NP_620316.1 | Cytochrome oxidase subunit I | 1.25 (0.10) | 1.89 (1.2) | 0.11 (0.08) | 0.97 (0.27) |
| Cell cycle | |||||
| Ref XP_220119.1 | Similar to G2/mitotic specific cyclin b | 0.84 (0.28) | 1.09 (0.5) | 3.29 (1.02) | 1.06 (0.17) |
| SWP:CGA2_CHICK | Cyclin a2 | 0.84 (0.22) | 0.31 (0.29) | 4.10 (0.95) | 0.96 (0.60) |
| SWP:CCT2_HUMAN | o60583 cyclin t2 | 1.25 (0.94) | 0.99 (0.20) | 0.94 (0.20) | 4.88 (1.02) |
| Cell physiology and development | |||||
| SWP:FKBB_HUMAN | 12.6 kDa fk 506 binding protein | 1.25 (0.57) | 0.91 (0.76) | 0.29 (0.19) | 0.99 (0.13) |
| Gb AAN28379.1 | ABl-interactor 1 [homo sapiens] | 16.61 (7.2) | 0.91 (0.21) | 1.08 (0.11) | 1.42 (0.42) |
| Cytoskeleton and motor proteins | |||||
| SWP:ACT2_HALRO | p27130 actin, muscle 2/4/4a. | 0.12 (0.07) | 1.52 (1.17) | 1.80 (1.7) | 1.19 (0.73) |
| PIR:S06117 | S06117 myosin heavy chain, nonmuscle | 1.23 (0.19) | 3.10 (1.63) | 0.61 (0.22) | 0.79 (0.23) |
| Ref NP_003184.1 | β-Tubulin cofactor e [homo sapiens] >gi|1425.. | 4.81 (1.73) | 1.39 (0.52) | 1.03 (0.19) | 2.16 (1.03) |
| Gb AAK27259.1 | β-Actin [ambystoma mexicanum] | 1.69 (0.58) | 5.71 (2.19) | 0.96 (0.22) | 0.95 (0.06) |
| EMB:Q23882 | Slime mold (d.discoideum) actin mrna itl-1, 3 | 0.81 (0.37) | 0.96 (0.13) | 1.03 (0.15) | 2.22 (0.77) |
| Extracellular matrix proteins | |||||
| SWP:CA11_CHICK | Collagen alpha 1(i) chain precursor. | 0.85 (0.28) | 1.20 (0.33) | 2.17 (0.49) | 0.88 (0.16) |
| SWP:HAS2_CHICK | Hyaluronan synthase 2 | 1.07 (0.12) | 2.59 (0.07) | 0.51 (0.03) | 0.98 (0.18) |
| EMB:O00199 | Integral membrane serine protease seprase | 10.23 (2.6) | 0.83 (0.49) | 0.77 (0.30) | 0.96 (0.13) |
| EMB:Q91002 | Thrombospondin-4 (fragment) | 1.79 (0.47) | 6.88 (3.50) | 1.62 (0.90) | 0.68 (0.06) |
| SWP:PEDF_MOUSE | Pigment epithelium-derived factor precursor | 1.81 (0.52) | 2.57 (0.94) | 2.38 (0.74) | 1.21 (0.27) |
| SWP:ITB1_CHICK | Integrin beta-1 precursor (csat antigen) (jg2) | 0.08 (0.11) | 0.99 (0.02) | 0.15 (0.10) | 5.49 (1.47) |
| SWP:PEDF_BOVIN | Pigment epithelium-derived factor precursor | 0.90 (0.3) | 0.81 (0.22) | 2.56 (1.03) | 1.32 (0.7) |
| Tumor associated proteins | |||||
| EMB:Q9JLL0 | Cysteine-rich repeat-containing protein crim1 | 0.74 | 8.21 (2.30) | 19.33 (9.57) | 1.33 (0.84) |
| Hypothetical proteins | |||||
| EMB:Q9H0P6 | Hypothetical 38.9 kDa protein. | 7.02 (2.53) | 1.51 (0.38) | 1.32 (0.13) | 0.82 (0.26) |
| EMB:Q9Y3V7 | Hypothetical 63.3 kDa protein (fragment) | 1.67 (0.01) | 2.46 (0.94) | 0.925 (0.66) | 0.985 (0.07) |
| EMB:Q9H0I1 | Hypothetical 83.8 kDa protein. | 2.45 (1.12) | 1.21 (0.20) | 1.318 (0.28) | 1.101 (0.23) |
| EMB:CAD38974.1 | Hypothetical protein [homo sapiens] | 0.21 (0.09) | 1.30 (0.04) | 10.977 (4.15) | 0.801 (0.12) |
| Gb AAA27977.1 | Hypothetical protein c50c3.6 [caenorhabditis eleg …] | 1.25 (0.61) | 0.99 (0.96) | 8.675 (2.53) | 1.037 (0.23) |
| Ref XP_043624.2 | Similar to hypothetical protein dkfzp434e1822.1 … | 1.76 (0.26) | 2.11 (1.09) | 1.067 (0.59) | 1.056 (0.42) |
| EMB:CAB43259.1 | Hypothetical protein [homo sapiens] | 4.12 (2.01) | 0.72 (0.36) | 0.956 (0.87) | 0.971 (0.16) |
| Metabolism | |||||
| EMB:AAB34334 | Protein phosphatase 1 gamma 1 (fragment) | 1.56 (0.77) | 0.49 (0.34) | 0.744 | 0.939 (0.56) |
| EMB:AAD02474 | Glyceraldehyde-3-phosphate dehydrogenas | 0.99 (0.51) | 1.85 (0.85) | 4.67 (2.24) | 0.86 (0.60) |
| SWP:SYI_HUMAN | Isoleucyl-trna synthetase, cytoplasmic (ec 6) | 0.94 (0.15) | 0.88 (0.10) | 0.38 (0.14) | 1.69 (0.28) |
| SWP:ATP6_CHICK | Atp synthase a chain (ec 3.6.1.34) (protein 6) | 1.58 (0.38) | 2.29 (0.86) | 0.95 (0.07) | 1.13 (0.21) |
| Gb AAB24816.1 | Farnesyltransferase alpha subunit [human, peptide …] | 2.24 (0.85) | 1.08 (0.92) | 1.01 (0.28) | 0.95 (0.30) |
| SWP:ENOA_CHICK | Alpha enolase (ec 4.2.1.11) (2-phospho-d-glycerate hydro-lyase) | 7.71 (2.44) | 0.68 (0.33) | 1.27 (0.52) | 0.67 (0.17) |
| SWP:PNAD_PIG | Protein n-terminal asparagine amidohydrolase | 3.83 (1.61) | 0.79 (0.3) | 0.92 (0.06) | 1.06 (0.41) |
| SWP:PWP2_HUMAN | Periodic tryptophan protein 2 homolog (pwp2) | 0.39 (0.18) | 1.40 (1.03) | 0.67 (0.33) | 4.46 (1.73) |
| Gb AAC98906.1 | Chaperonin-containing tcp-1 beta subunit homolog … | 0.72 (0.24) | 0.54 (0.33) | 0.73 (0.43) | 0.96 (0.30) |
| Pdb 1AK6 | Destrin, nmr, minimized average structure >gi|2624765 … | 1.20 (0.39) | 0.81 (0.18) | 2.34 (0.50) | 1.07 (0.52) |
| Ref NP_067085.1 | Acid phosphatase 1, soluble [rattus norvegicus] … | 1.28 (0.38) | 4.58 (1.42) | 1.26 (0.30) | 1.04 (0.17) |
| Gb AAA40873.1 | Carboxypeptidase e (ec 3.4.17.10) | 2.59 (0.62) | 1.18 (0.58) | 1.11 (0.53) | 0.93 (0.28) |
| Gb AAA66286.1 | Ornithine decarboxylase | 1.29 (0.57) | 1.50 (0.81) | 10.62 (3.44) | 0.85 (0.46) |
| Nucleic acid binding | |||||
| SWP:N6M1_HUMAN | Putative n6-dna-methyltransferase | 0.57 (0.25) | 0.73 (0.26) | 0.32 | 1.20 (0.34) |
| Unknown function | |||||
| EMB:AAH06414 | Similar to kiaa0952 protein (fragment) | 1.02 (0.03) | 1.21 (0.45) | 9.93 (2.58) | 1.02 (0.47) |
| EMB:Q9D0E4 | 2610021k23rik protein. | 2.59 (0.96) | 1.35 (0.48) | 0.93 (0.58) | 0.92 (0.30) |
| EMB:Q9H7T5 | cdna flj14273 fis, clone place1004913 (fragme | 3.54 (1.03) | 1.49 (0.46) | 1.34 (0.47) | 1.05 (0.45) |
| EMB:Q9NUM1 | cdna flj11277 fis, clone place1009404 | 1.40 (0.59) | 2.58 (0.70) | 1.00 (0.65) | 1.84 (0.86) |
| EMB:Q9ULD7 | kiaa1283 protein (fragment). | 1.24 (0.08) | 2.99 (0.83) | – | 1.03 (0.27) |
| EMB:AAH02380 | Inknown (protein for image:2961244) (fr) | 4.20 (0.56) | 1.01 (0.44) | 2.78 (0.41) | 1.08 (0.46) |
| EMB:Q9H3F0 | mstp034 | 2.21 (1.03) | 1.43 (0.32) | 11.076 (4.56) | 1.11 0.30) |
| EMB:Q9D0V1 | 1110067l12rik protein | 1.10 (0.45) | 2.28 (1.06) | 0.66 (0.37) | 1.16 (0.41) |
| GEN:CAC22485 | Unnamed protein product [homo sapiens] | 5.43 (1.26) | 1.11 (1.04) | 0.37 | 0.96 (0.11) |
| EMB:Q9D9D0 | 1700101g24rik protein | 0.98 (0.36) | 2.36 (1.11) | 0.73 (0.15) | 1.22 (0.23) |
| Dbj BAB30848.1 | Unnamed protein product [mus musculus] | 0.93 (0.43) | 0.83 (0.43) | 4.92 (1.94) | 1.09 (0.15) |
| Dbj BAB28442.1 | Unnamed protein product [mus musculus] >gi|23396 … | 14.17 (6.30) | 6.05 (2.10) | 10.01 (3.63) | 0.88 (0.35) |
| EMB:Q9D8V0 | 1200006o09rik protein | 0.98 (0.54) | 17.24 (5.62) | 0.73 (0.44) | 1.02 (0.44) |
| EMB:Q9H8I2 | CDNA FLJ14005 fis, clone Y79AA1002361 | 1.72 (0.69) | 1.19 (0.78) | – | 0.48 (0.05) |
| Dbj BAC29089.1 | Unnamed protein product [mus musculus] | 6.88 (2.38) | 1.30 (0.90) | 1.08 (0.48) | 0.96 (0.25) |
| Ref XP_032996.1 | Similar to kiaa0819 protein [homo sapiens] | 1.00 (0.75) | 0.47 (0.30) | 1.37 (1.06) | 1.20 (0.48) |
| Dbj BAA07556.1 | kiaa0074∼the ha1438 gene product is related to a … | 16.52 (4.38) | 1.31 (0.77) | 0.70 (0.39) | 1.06 (0.12) |
| Dbj BAA05837.1 | kiaa0045 [homo sapiens] >gi|10863903|ref|np_0042 … | 1.75 (0.38) | 1.66 (0.22) | 4.22 (2.04) | 1.15 (0.26) |
| Ref NP_058040.1 | Clathrin, light polypeptide (lca) [mus musculus …] | 2.64 (0.21) | 1.20 (0.21) | 1.60 (0.23) | 1.04 (0.41) |
| Ref XP_216872.1 | Similar to riken cdna 2410003l22 [mus musculus] … | 36.14 (8.97) | 1.31 (0.77) | 28.83 (6.00) | 1.00 (0.28) |
| Ref XP_214380.1 | Similar to riken cdna 4933405k01 [mus musculus] … | 4.05 (1.31) | 0.92 (0.30) | 1.10 (0.42) | 1.03 (0.30) |
| Dbj BAB28379.1 | Unnamed protein product [mus musculus] | 2.24 (0.33) | 1.13 (0.43) | 0.65 (0.48) | 1.38 (0.58) |
| Ref NP_056371.1 | kiaa0440 protein [homo sapiens] >gi|4151330|gb … | 1.20 (1.03) | 1.67 (0.60) | 10.95 (3.08) | 1.11 (0.40) |
| Dbj BAA91948.1 | Unnamed protein product [homo sapiens] >gi|89228 … | 21.66 (8.86) | 1.08 (0.85) | 1.00 (0.43) | 0.90 (0.20) |
| Dbj BAB14688.1 | Unnamed protein product [homo sapiens] | 2.67 (0.83) | 1.02 (0.59) | 0.93 (0.13) | |
| Dbj BAB26968.1 | Unnamed protein product [mus musculus] >gi|21312 … | 0.70 (0.46) | 0.91 (0.51) | 13.77 (6.58) | 0.900 (0.17) |
| Dbj BAB15047.1 | Unnamed protein product [homo sapiens] | 1.09 (0.43) | 4.48 (2.06) | 0.87 (0.32) | 1.11 (0.21) |
| Ref XP_225174.1 | Similar to riken cdna 4833439l19; expressed seq … | 2.11 (0.26) | 1.16 (0.35) | 0.94 (0.25) | 0.91 (0.39) |
| Dbj BAC11615.1 | Unnamed protein product [homo sapiens] | 24.36 (9.28) | 1.22 (1.27) | 1.40 (0.94) | 0.95 (0.23) |
| Dbj BAC36160.1 | Unnamed protein product [mus musculus] | 1.23 (0.13) | 1.30 (0.29) | 5.48 (2.42) | 1.24 (0.34) |
| Dbj BAC36765.1 | Unnamed protein product [mus musculus] | 1.01 (0.46) | 1.18 (0.22) | 1.10 (0.45) | 5.01 (2.41) |
| Dbj BAC29089.1 | Unnamed protein product [mus musculus] | 6.88 (3.38) | 1.30 (0.90) | 1.08 (0.49) | 0.96 (0.25) |
| Transport protein | |||||
| SWP:NTCH_RAT, P28570 | Sodium-dependent choline transporter (chot1). | 1.75 (0.70) | 2.32 (1.00) | 1.11 (0.82) | 1.06 (0.21) |
| SWP:ABF2_HUMAN | Iron inhibited abc transporter 2; abcf2 | 48.19 (20.75) | 22.79 (8.78) | 15.94 (6.32) | 19.82 (8.47) |
| RNA regulation | |||||
| SWP:RSFR_CHICK | p30374 ribonuclease homolog precursor (rsfr) | 1.54 (0.13) | 5.87 (1.75) | 5.96 (1.66) | 1.93 (1.13) |
Fig. 1.
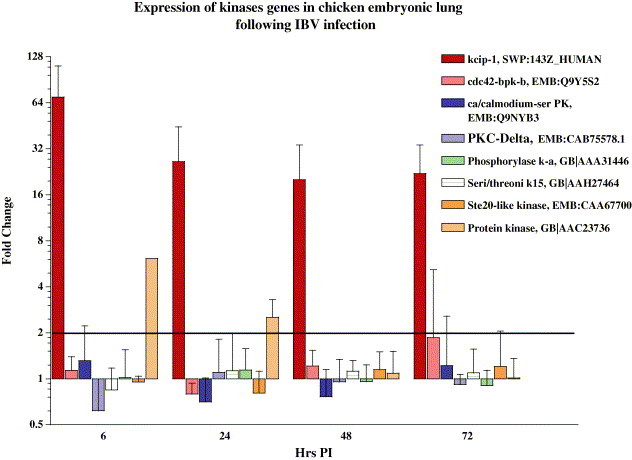
Transcriptional alterations in kinase genes detected by microarray analysis at 6, 24, 48 and 72 h post IBV infection. RNA used in these experiments was derived from lung tissues of IBV infected and sham inoculated 18 days old chick embryo. Procedures for probe synthesis and array hybridization have been described in methods. Fold change in expression of genes represents the mean value derived from two independent experiments with hybridizations performed three to four times with RNA isolated from each experiment.
Table 2.
Genes showing over two fold change in expression and common in IBV and APV infection
| Accession number | Gene name | 24 h PI |
48 h PI |
Functional group | ||
|---|---|---|---|---|---|---|
| IBV (in ovo) | APV (in vitro) | IBV (in ovo) | APV (in vitro) | |||
| SWP:INI2_PANTR | Interferon-induced protein 6–16 precursor | 3.86 | 17.39 | NCb | 13.87 | IFN-induced |
| SWP:IFT1_HUMAN | Interferon-induced protein with tetratricopep (IFI56K) | 35.27 | 32.28 | 13.98 | 19.7 | IFN-induced |
| SWP:STA1_HUMAN | signal transducer and activator of transcription 1-alpha/beta | NC | 16.36 | 3.01 | 33.77 | IFN-induced |
| SWP:EMF1_CHICK | embryo fibroblast protein 1 precursor (emf-1, 9e3 | 4.871 | 7.3 | NC | 39.32 | Chemokine |
| SWP:UBPI_HUMAN | hubp43 ” | 21.7 | 11.94 | NC | 6.23 | Ubiquitin and proteasome |
| EMB:Q9Y4Z6 | Vacuolar protein sorting | 2.00 | NC | 0.47 | 5.18 | Vesicular protein trafficking |
| EMB:O42404 | cd80-like protein precursor | 2.77 | NC | NC | 2.68 | Antigen presentation |
| SWP:PTB_PIGa | Polypyrimidine tract-binding protein (ptb) (hnrnp1) | 8.78 | NC | NC | NC | Trancrip regulation |
| SWP:RSFR_CHICKa | RNase homologue precursor (RSFR) | 5.87 | NC | 5.96 | NC | RNA regulation |
| SWP:EF1A_CHICK | Elongation factor 1-alpha 1 (ef-1-alpha-1) | NC | NC | 2.03 | 0.40 | Translation |
| SWP:CGA2_CHICK | Cyclin a2 (cyclin a) | 0.31 | NC | 4.10 | 2.91 | Cell cycle |
| SWP:FKBB_HUMAN | 12.6 kDa fk506-binding protein (fkbp-12.6) | NC | NC | 0.29 | 0.50 | Cell physiology and deve |
| SWP:ABF2_HUMANa | Iron inhibited abc transporter 2; abcf2 | 30.64 | NC | 15.94 | NC | Transporter protein |
| SWP:FUCO_HUMAN | Tissue alpha-l-fucosidase precursor | 4.63 | NC | 2.46 | 0.40 | Protein degradation |
APV infection in-vitro showed 2.48, 2.54, and 3.29-fold change in hnrnp1, RSFR and abcf2 genes, respectively at 96 h PI.
NC: no change or <2-fold change in the level of expression.
8. Real-time RT-PCR analysis
To confirm results obtained in array experiment by another independent method, 14 genes were selected for real-time RT-PCR assay at all time points PI. The results obtained are shown in Table 3 . The genes selected for real-time PCR belonged to several important functional groups including interferon induced genes, antigen presentation, chemokine and adhesion molecules and transcription factors protein genes. Moreover, the expression of these genes was either up-regulated, down-regulated or unchanged from 6 to 72 h PI. RT-PCR data show 85–90% agreement (except at 72 h PI) with the expression patterns seen with microarray analysis. Thus, confirmation rate is similar to previous reports (Munir and Kapur, 2003). Though the trends of increase and decrease in gene expression at different time points for all genes are consistent for both the assay systems. We noticed a wide gap in fold changes expression of interferon induced protein 6–16 precursor and alpha interferon induciable protein (fragnment) p27-h genes in microarray and RT-PCR assay. We assume that variation in expression levels detected by microarray and RT-PCR may be attributed to differences in the sensitivity and the functional kinetics of both assay systems.
Table 3.
Validation of gene regulation by real-time RT-PCR
| Gene name | Fold change in expression at PI time points |
|||||||
|---|---|---|---|---|---|---|---|---|
| 6 h |
24 h |
48 h |
72 h |
|||||
| Array | qRT | Array | qRT | Array | qRT | Array | qRT | |
| Transcriptional repressor protein yy1 | 1.28 | 0.87 | 6.84 | 2.14 | 1.26 | 1.62 | 1.24 | 1.86 |
| Signal transducer and activator of transcription 1-alpha/beta | 3.03 | 2.54 | 1.45 | 0.53 | 2.80 | 2.82 | 0.96 | 0.31 |
| Elongation factor 1-alpha 1 (ef-1-alpha-1) | 3.04 | 1.74 | 1.34 | 1.23 | 3.05 | 2.63 | 0.94 | |
| AAH01874 similar to sequestosome 1 | 1.09 | 2.07 | 1.52 | 2.73 | 1.08 | 1.86 | 0.95 | 0.00 |
| Cd80-like protein precursor | 0.74 | 0.90 | 2.77 | 4.43 | 0.96 | 0.32 | 0.97 | 0.90 |
| Ornithokinin receptor | 0.69 | 0.40 | 1.05 | 0.65 | 0.99 | 0.70 | 1.21 | 1 |
| Interferon-induced protein with tetratricopeptide repeat | 21.9 | 1.8 | 35.2 | 57.6 | 13.9 | 2.5 | 0.99 | 3.2 |
| Embryo fibroblast protein 1 precursor | 2.14 | 3.63 | 1.11 | 1.51 | 1.20 | 2.46 | 0.46 | 0.19 |
| Lumican precursor (lum) (keratan sulfate proteoglycan | 1.10 | 1.10 | 1.14 | 1.31 | 0.85 | 2.29 | 1.11 | 2.14 |
| Complement c3f [fragment] | 0.87 | 2.22 | 1.29 | 1.46 | 1.13 | 2.22 | 0.99 | 3.03 |
| Interferon-induced protein 6–16 precursor | 0.85 | 3.86 | 21.8 | 1.16 | 5.66 | 1.12 | 2.22 | |
| Thrombospondin 2 precursor. | 1.12 | 0.87 | 0.84 | 0.51 | 0.85 | 5.85 | 0.79 | 11.3 |
| Tata element modulatory factor | 0.959 | 0.93 | 0.927 | 1.27 | 0.930 | 8 | 0.963 | 0.34 |
| Alpha-interferon inducible protein (fragment) p27-h | 0.86 | 1.4 | 1.86 | 19.02 | 1.481 | 12.9 | 1.47 | 1.63 |
9. Kinases and phosphatases
The set of kinases and phosphatase genes showing significant and stable changes in expression during IBV infection of embryonic lungs, include 14-3-3 protein zeta/delta, the protein kinase C inhibitor protein-1 (KCIP-1) or factor activating exoenzyme S (Fas), protein kinase (similar to serine/threonine protein kinase (PKR), and protein–tyrosine-phosphatase alpha (Table 1 and Fig. 1). Amongst the eight kinase genes spotted on the array, the gene coding for Fas showed the highest and most stable elevation in expression. The gene was 70-fold up regulated at 6 h PI and gradually declined to 22-fold at 72 h PI. 14-3-3 protein represent a novel class of kinases that recognize phosphoserine motifs on their ligand and play an important role in regulating various cellular processes including signal transduction, cell cycle, apoptosis, stress responses and viral and bacterial pathogenesis. These cellular activities are regulated through an interaction of over100 cellular proteins with Fas protein (van Hemert et al., 2001). Proteins binding to 14-3-3 protein are very diverse and include kinases, phophatases, receptors, structural proteins and transcription factors. In general, most 14-3-3 protein ligands require a phosphorylation step before they can interact with their receptor and thus intracellular kinase/phosphatase signaling networks play a key role in Fas ligand interaction (Fu et al., 2000). The role of 14-3-3 protein in IBV infection is not known. However, recently it has been reported that during hepatitis C virus (HCV) infection, viral C protein directly interacts with 14-3-3 and leads to activation of Raf-1 protein kinase signaling pathway (Aoki et al., 2000). Activation of serine/threonine kinase and phosphatase genes along with the Fas gene in our data set may suggest their important role during IBV infection. The induction of dsRNA dependent PKR as a mediator of antiproliferative and antiviral activity of IFN is well known. However, recently the enhanced expression and increased activity of PKR in certain viral infections has been associated with the activation of specific viral proteins that interact with 14-3-3 protein (Schulze zur Wiesch et al., 2003).
Another kinase related gene that showed 61-fold up-regulation at 6 h and remained up-regulated up to 48 h PI was lymphocyte specific protein 1 (LSP1) gene. Since the regulatory activity of KCIP-1 is mediated through PKC, it is interesting to note that LSP1 acts as a substrate for PKC and MAPKAP kinase 2 (Carballo et al., 1996). LSP1 is an intracellular phosphoprotein that is expressed in B-lineage cells, functional T cells, thymocytes, macrophages and neutrophils derived from fetal liver and bone marrow. LSP1 is an important signaling molecule, regulating the cytoskeletal architecture, cell motility, receptor-induced apoptosis and a negative regulator of neutrophil chemotaxis (Jongstra-Bilen et al., 2000). Alterations in expression of LSP-1, and KCIP-1 genes in our data may suggest their involvement in IBV infection and modulation of immune cell function. However, the precise interaction of these factors with each other or with viral proteins in IBV infection needs further investigation perhaps by array based studies with a larger number of gene sets specifically comprising all or the majority of genes involved in kinase and phosphatase signaling cascades. Further studies are anticipated to provide more information regarding the role of these signaling factors in IBV pathogenesis
10. Viral RNA synthesis genes
RNA synthesis in positive strand RNA viruses is associated with viral replication complexes and is mediated through viral RNA-dependent RNA polymerase (RdRP). RdRP is the key enzyme for coronavirus replication and its sequences is conserved among coronavirus isolates (Lu et al., 2004). However, the formation of the replication complex and recruitment of host cell components to complete viral replication are not fully understood (Brockway et al., 2003). In general, the specificity of RdRP to select viral RNA as a template is dictated by cellular factors including tubulin, actin and heat shock proteins (Oglesbee et al., 1996). The analysis of cellular factors involved in replication of RNA viruses such as Qβ phage, brome mosaic virus (BMV) and tobacco mosaic virus (TMV) has shown that the RdRP holoenzyme can direct the synthesis of positive strand RNA from negative strand RNA. However, the recruitment of RdRP to the transcriptional promoters of positive strand viral RNA to synthesize negative RNA strand requires the interaction of cellular factors like TATA-binding protein (TBP), other general transcription factors and eukaryotic translation initiation factor 3 (eIF3) (Novak and Kirkegaard, 1994). Moreover, the interaction of host elongation factor EF-1 and ribosomal S1 protein with RdRP complex to initiate transcription of polio virus RNA is also well documented (Lai, 1998). Increased gene expression for some of these host factors following IBV infection suggests their involvement in IBV infection. For instance, chicken elongation factor 1 (EF-I) and TBP genes were two- and seven-fold up-regulated, respectively, at different time points following IBV infection (Table 1). In contrast, the eIF3 subunit 6 gene showed marked down regulation (>3-fold) at 6 h PI and then gradual up-regulation (two-fold) at 24 h PI. However, other subunits of eIF3 present on the array (e.g. homolog of mouse eIF3 subunit 7) showed no change of expression during IBV infection. In accordance with findings reported elsewhere (Guo et al., 2000) the down regulation of the eIF3 gene with simultaneous activation of the IFI 56 K gene in our data (Table 1) may possibly be a part of the interferon-induced antiviral pathway. Moreover, four- and five-fold upregulation of the 60s ribosomal protein 13 and ribosomal protein s6, respectively, at 6 h PI in IBV infection may also play a role in viral pathogenesis. There are examples where the antiviral effect of certain plant proteins is mediated through inhibition of ribosomal protein l3 (Rajamohan et al., 2001).
Heterogeneous nuclear ribo-nucleoprotein 1 (hnRNP1) is another cellular protein that is reported to be involved in corona virus replication. Biologically, hnRNP1 is involved in pre-mRNA splicing in the nucleus and translational regulation in the cytoplasm (Shi et al., 2000). The eight-fold up regulation of polypyrimidine tract-binding protein (PTB) or heterogeneous nuclear ribo-nucleoprotein 1 (hnRNP1) gene at 24 h PI suggests a possible role for hnRNP 1 in IBV transcription. Several lines of evidence are consistent with this hypothesis. For instance, the interaction between viral 3′-UTR and cellular hnRNP1 is a necessary requirement for the replication of the viral genome in bovine coronavirus (BCV) and mouse hepatitis virus (MHV) transcription (Lai, 1997, Shi et al., 2000, Spagnolo and Hogue, 2000). Moreover, during MHV infection hnRNPA1 mediates the leader-body fusion event in synthesis of viral sub genomic RNA transcripts (Shen and Masters, 2001). The binding of hnRNP-1 to the internal ribosome entry site on viral RNA of poliovirus, hepatitis A and hepatitis C virus is required for viral mRNA translation (Chung and Kaplan, 1999). However, in a non-segmented negative strand RNA virus infection (e.g. APV) the up-regulation of hnRNP1 gene was observed at later stages (96 h PI) of infection (Munir and Kapur, 2003). We hypothesize that this difference in hnRNP1 gene expression profile for IBV and APV may be due to the different transcriptional strategy (synthesis of nested sets of viral mRNA) of coronavirus.
11. Interferon and interferon induced genes
The induction of chicken interferon (IFN) by different IBV strains greatly varies in different culture environments (Holmes and Darbyshire, 1978). It has been suggested that IBV's ability to induce IFN is linked to the virulence and adaptability of IBV strain for a particular host system (Holmes and Darbyshire, 1978, Otsuki et al., 1987). In the present investigation we were unable to detect the induction of IFN genes due to the absence of IFN coding cDNA on the array. Interferons mediate their effect by binding to cell surface receptors which activate members of the JAK kinase family of proteins. Activated JAK kinases phosphorylate STAT factors. The STAT proteins homo- or heterodimerize and form complexes with other transcription factors to activate transcription of interferon stimulated genes (ISGs) (Haque and Williams, 1998). Amongst the STAT group of ISGs, IBV infection of chicken embryos induced up regulation (two- to three-fold) of STAT 1 and another similar gene in lung tissue at 6 h PI. However, at 48 h PI only the STAT1 gene showed three-fold up-regulation. Similar to many other viral infections the activation of STAT1 seems to be an important component of host innate antiviral activity in coronavirus infection. For instance, STAT-1 null mice show enhanced viral susceptibility, greatly reduced responses to IFN-α and IFN-γ and compromised innate immunity. Moreover, inactivation of the STAT-1 gene following Sendai virus infection (through an interaction to the viral C protein) is believed to be an important viral strategy to evade host innate immune responses (Garcin et al., 2002).
In many viral infections IFI-56K, a member of the interferon induced proteins with tetratricopeptide repeats, is considered to be the most strongly induced gene amongst the known ISGs (Sen, 2001). The marked up regulation of IFI-56K gene, as much as 22-fold in IBV infected chicken embryonic lung (Table 1), is similar to what was observed in many other viral infections including APV infection of CEF cells, HCV and cytomegalovirus infection (Bigger et al., 2001, Munir and Kapur, 2003). The inhibition of protein synthesis by IFI-56K protein is mediated through its interaction with the translation initiation factor eIF3 making it unavailable for protein translation (Guo et al., 2000). Interestingly, the up-regulation of IFI-56K gene and a simultaneous reduction in eIF3 protein gene expression during IBV infection at 6 h PI may implicate strong activation of host innate immune response during the early phase of IBV infection (Table 1). However, subsequent up-regulation of the eIF3 gene with its involvement in viral RNA synthesis, may reveal that effective viral strategies exist for escape from interferon's antiviral effects mediated through activation of IFI-56K gene expression. The overall ISG expression profile in this study and previous studies indicate that stable and marked activation of IFI-56K may be crucial for effective host protection in many viral infections including IBV, APV and HCV (Zhu et al., 2003).
Other ISGs related to IFI-56K, including genes encoding for IFN-induced 58 kDa protein and IFN-induced TPR4 protein, did not show changes in expression following IBV infection. However, these ISGs showed marked up-regulation in other avian respiratory viral infections (Munir and Kapur, 2003). Interferons regulate gene expression in various functional classes including signal transduction proteins, chemokines and adhesion molecules, antigen processing and presentation, signal transduction and protein degradation systems (de Veer et al., 2001).
The G proteins are the class of signaling proteins that mediate many intracellular functions including cytoskeletal remodeling, vesicle transport and growth (Evers et al., 2000, Willard and Crouch, 2000). Activation of G-protein genes and genes related to G-protein signaling in response to IFNs treatment of host cells have been reported in other microarray studies (de Veer et al., 2001). However, the precise mechanism of gene activation is not clear. Moreover, the activation of G proteins in response to the binding of chemokines to transmembrane G-protein receptors and the requirement of JAK/STAT and G-protein interaction for chemotaxis and Ca flux responses in host cells is well characterized (Soriano et al., 2003). In our studies, the up-regulation (17-fold at 6 h PI) of a gene similar to the G protein gene may occur as part of the IFN or chemokine induction pathways mentioned above (Table 1). Further analysis of genes that were altered over two-fold and were common in IBV and other avian viral respiratory infection including APV showed marked differences in the magnitude of expression in the 6–16 precursor protein gene and STAT1 gene (Table 2). Microarray analysis of ISGs in non-infected cells have shown 31- and 21-fold increased expression of MxA protein in response to IFN-β and IFN-α treatment, respectively (Der et al., 1998). Moreover, the Mx protein gene is considered to be a predominant ISG in the majority of negative strand RNA virus infections such as APV, influenza virus, vesicular stomatitis virus and measles virus infection (Atreya and Kulkarni, 1999, Munir and Kapur, 2003, Pavlovic et al., 1990, Schnorr et al., 1993) and a few positive strand RNA viruses such as Toga viruses (Landis et al., 1998). However, there was no change in expression of the Mx protein gene in IBV infected embryos. The absence of change in Mx gene expression is supported by recent findings in other coronavirus studies indicating that the antiviral effect of IFNs in SARS corona virus infection in vitro is not mediated by MxA protein (Spiegel et al., 2004). The induction of STAT-1 and IFI-56K gene expression supports the conclusion that IFN pathway was activated. However, no change in Mx gene expression is intriguing and may possibly be an outcome of an IBV anti-IFN virulence mechanisms.
12. Chemokines and adhesion molecules genes
IBV infection of embryonic chicken lungs resulted in a five-fold increase in 9E3 gene expression at 24 h PI (Table 1). The 9E3 and K60 genes are two of four chicken chemokines known to belong to the CXC family. The 9E3 gene is a chicken homolog of human interleukin 8 (IL8). The role of IL8 gene induction in viral infections is well known. For example, cytomegalovirus (CMV) and Epstein Bar virus (EBV) infections activate the transcription factors NF-kB and AP1 which in turn induce interleukin 8 (IL8) expression and thus enhance the attraction of neutrophils,which serve as the target cell for CMV infection (Murayama et al., 1997, Penfold et al., 1999). In addition to the attraction of heterophils (avian neutrophils), monocytes and lymphocytes, 9E3 is involved in growth regulation, wound healing and angiogenesis (Sick et al., 2000). A five-fold increase in expression of the 9E3 gene in IBV infected embryonic lungs at 24 h PI (Table 1) may be linked to the common patho-physiological and microbiological states observed in IBV infection. For instance, in many respiratory viral infections including IBV, APV and human respiratory syncytial virus, peak accumulation of heterophils in lung lavage has been reported at 24 h PI (Kotani et al., 2000). Similarly, peak IBV replication in the lungs of 18-day-old chicken embryos inoculated with IBV via the chorioallontoic route was observed at 24 h PI (Kapczynski et al., 2002). Elevated 9E3 expression at 24 h post IBV infection in ovo may be correlated to the occurrence of peak IBV titers in lungs at 24 h PI and represent a conserved host response to viral infection. We did not detect significant alterations in other chemokine genes including K60, complement factor C3F and ornithokinin receptor genes which were present on the array. Unaltered expression of these chemokine genes may be an IBV strategy to escape possible adverse effects of chemokines. For example, it is well known for influenza virus infection, that an interaction between regulatory sequences in the viral nucleoprotein gene and host cell factors inhibits neutrophil chemotaxis and activation (Cooper et al., 2001).
Differences in chemokine expression pattern between IBV (Table 1) and APV infection (Munir and Kapur, 2003) may contribute to differences in the patho-physiology of APV (a negative strand RNA virus) and IBV (a positive strand RNA virus) infection. The type of facial edema, infiltration of inflammatory heterophils, exudation and edema of sinuses is much more severe during APV infection of turkeys than during IBV infection. These differences may be the outcome of more extensive chemokine gene upregulation during APV infection. Moreover, unaltered expression of some ISGs, such as RI58, TPR-4, and Mx, and down-regulation of STAT I gene at 24 h post-IBV infection may be attributed to the antagonistic effect of IL-8 on IFN- α induction. Such an antagonistic activity for IL-8 has been reported for a majority of positive stranded RNA viral infections including polio, encephlomyocarditis and some DNA viruses such as herpes simplex virus 1 (HSV-I) infections. Similar inhibitory activity for IL-8 has not been observed in negative strand RNA viruses such as vesicular stomatitis virus and APV (Khabar et al., 1997a, Khabar et al., 1997b, Munir and Kapur, 2003).
Another gene (LSP1) coding for a negative regulator of neutrophil chemotaxis also displayed marked and stable up-regulation during IBV infection. This gene is believed to participate in the inhibition of chemokine and complement gene induction (Jongstra-Bilen et al., 2000). Interestingly, chemokines other than 9E3 and complement factor genes (CF3) did not show any change in expression in our data. Further studies are needed to evaluate the impact of LSP1 on cytokine production and virus replication to determine if LSP1 plays a critical role in IBV pathogenesis
13. Ubiquitin-proteasome and vacuolar trafficking protein genes
In contrast to many well studied enveloped RNA viruses, coronaviruses have a different budding mechanism. Coronaviruses acquire their membrane envelop by budding into the lumen of the Golgi and pre-Golgi compartments with virus release restricted to certain areas of the cell (Tooze et al., 1987). The reason for this intracellular budding and selection of specific sites for budding of the virus is not known (Corse and Machamer, 2003). Viral genomic factors, including Golgi targeting signal sequences on envelop proteins of IBV, involved in the process of virus egress have been well characterized. However, the cellular counterparts interacting in this process are not well known (Corse and Machamer, 2003). In our experiments, alterations in the expression of seven genes with functions in the vesicular proteins (Vps) trafficking including clathrin light chain and transitional endoplasmic reticulum atpase genes, showed greater than two-fold increased expression within 6 h PI. Signal recognition particle 54 kDa protein gene and Vps45 showed six- and two-fold up regulation at 24 PI, respectively, but Vps45 gene showed two-fold down-regulation at 48 h post-IBV infection (Table 1). In addition, the TC10 protein gene and the COP1 gene related to the Vps pathway also showed two- and eight-fold enhanced expression at 48 and 72 h PI, respectively (Table 1). The Vps 45 and signal recognition particle 54 kDa proteins are thought to play important roles in vacuolar protein sorting from golgi to the vacuole (Miller et al., 1995).
The TC 10 protein is a member of the Rho family of GTP binding proteins that is closely related to Cdc42. Recent studies have shown that these proteins function as molecular switches linking cell surface receptors and extracellular signals to the regulation of actin dynamics (Hall, 1998). The interaction between TC10 and coatmer protein COP1 causes massive Golgi membrane actin polymerization and thus play an important role in the control of perinuclear actin dynamics (Kanzaki et al., 2002). The up-regulation of both of these genes (TC10 and COP1) in IBV (Table 1) infection suggests a possible role in the transport of coronavirus proteins through the golgi network (Munir and Kapur, 2003). The involvement of different COP proteins in vacuolar transport of viral proteins of different viruses has been previously reported. For example, upregulation of coatamer complex gene COP9 is reported in APV infection (Munir and Kapur, 2003). The M glycoprotein of coronavirus is one of the viral proteins that interacts with cellular factors and is accumulated in the golgi region and is believed to possess a signal for its retention at this location. The induction of cellular genes including clathrin light polypeptide, COP1 and GTP binding protein TC10 genes in IBV infection may suggest the interaction of these cellular proteins with viral proteins (like the M glycoprotein of the coronavirus) in the process of virus assembly and exocytic pathway (Gottesman and Maurizi, 1992).
The genes related to ubiquitin and the proteasomal degradation pathway that showed over two fold alteration in expression included deubiquitinating enzyme 18 (hubp43) and a gene similar to the 26s proteasome non-ATPase regulatory subunit gene (Table 1). Both genes are over-expressed within 6 h PI. Protein ubiquitinylation is a classical pathway to target proteins for degradation by the 26S proteasome (Khor et al., 2003). The up-regulation of the ubp18 gene and its role in viral replication and evasion of host immunity has also been reported in other viral infections including porcine reproductive and respiratory syndrome (PRRS) virus, CMV and APV. It is believed that the induction of ubiquitin carboxyl-terminal hydrolase 18 (Ubp18) gene in the early phase of infection may be a viral strategy to prevent degradation of its nascent proteins (Munir and Kapur, 2003). However, unchanged expression (during IBV infection) of genes encoding the set of enzymes including E1 (Ub activating), E2 (Ub conjugating) and E3 (Ub substrate ligase), which are found to be regulated in certain other viral infections including APV (Munir and Kapur, 2003), may be consistent with the substrate specificity of these enzymes and may suggest the activity of some other ubiquition pathway during IBV infection. Recently, a number of ubiquitin-like modifier (Ubls) proteins were discovered to have a variety of ubiquitin-like functions, including substrate conjugation (Haas and Siepmann, 1997). In addition, there also exists a class of ubiquitin related proteins in which an ubiquitin domain is built into a larger polypeptide and is not excised or attached to other proteins. These domains may impart properties analogous to those conferred by a transferable Ubl called UbD (Hochstrasser, 2000, Munir and Kapur, 2003). The absence of expression changes in other ubiquitin genes and activation of ubp18 gene and gene similar to 26s proteosome may suggest the involvement of ubiquitin related proteins like Ubl or UbD (that have not yet been identified in avian species) during IBV infection.
14. Transcription and translation regulation genes
Over two-fold alterations in six genes belonging to the regulation of cellular transcription family were observed. These included transcriptional repressor protein Yin Yang-1 (YY1), TAF12 RNA pol II, human nuclear ribonuclear protein 1 (hnRNP1), similar to enhancer factor I (EF-1), E1a binding protein p300 and PCAF-associated factor 400 (Table 1). Our data indicate a greater than seven-fold up regulation of the YY1 gene at 24 h PI. The YY1 protein is a multifunctional zinc finger protein that regulates the expression of various cellular as well as viral transcription factors including TBP, TFIIB, sequence specific DNA binding transcriptional activator (Sp1), c-myc, ATF/CREB, transcriptional co-regulators such as E1A, CBP, p300, PCAF, histone deacetylase 1 (HDAC1), HDAC2, and HDAC3, c-Fos, p53, α actin, β and γ interferon, P5 of adeno-associated virus, E6 and E7 genes of human papillomavirus, and a number of other viral long terminal repeats including Moloney murine leukemia virus and human immunodeficiency virus type I (Austen et al., 1997, Yao et al., 2001). The role of YY1 protein in inhibition or propagation of virus infection is documented in various virus infections including adeno-associated virus replication where interaction of YY1 protein with E1A at promoter P5 relieves YY1 dependent suppression of the promoter. However, in the absence of E1A, the binding of YY1 to its cis elements represses transcription (Shrivastava and Calame, 1994). The binding of YY1 to the upstream region of the MuLV promoter down-regulates MuLV promoter activity (Flanagan et al., 1992). Moreover, it has been shown that an interaction between c-myc and YY1 protein suppressess the activity of the CXCR4 promoter where, CXCR4 serves as a major co-factor for T-cell tropism of HIV-I (Bleul et al., 1996). However, mutations in the YY1 binding site removes suppression and facilitates HIV propagation (Moriuchi et al., 1999). The role of the YY1 protein in coronavirus infection remains to be investigated. On the basis of alterations in expression of YY1 gene (>7-fold upregulation at 24 h PI) and other transcription factors including p300, PCAF, TBP and hnRNP we hypothesize that YY1 protein interacts with different cellular and viral elements during IBV infection. In addition to the role of hnRNP1 protein discussed earlier the alteration in hnRNP1 gene expression is reported to be involved in cellular immunobiology. It represses the expression of c-fos and zif268 genes and activates transcription of Ha-ras and SP6 kappa promoters that participate in host immune responses (Bemark et al., 1998, Mikheev et al., 2000).
Transcription initiation by RNA polymerase II largely depends on TBP and associated factors (TAFIIs). Proteins like p300 and PCAF are transcriptional co-activators that are said to be involved in various cellular activities including cell growth, cellular transformation and differentiation, transcription initiation and developmental processes (Mitsiou and Stunnenberg, 2003). On the basis of p300 interaction with cellular transcription factor TFIIB and RNA pol II, this protein is thought to play a direct role in transcription initiation thus serving as a molecular bridge between upstream transcriptional activators and basal machinery (Felzien et al., 1999). The CH3 region of p300 has been reported to interact with many viral and cellular transcription factors like adenovirus E1A, SV40 large antigen and transcription co-activator PCAF (Arany et al., 1995, Bhattacharya et al., 1996). The interaction between the CHI region of p300 and or CREB-binding proteins and the carboxy terminal region of STAT2 (an ISG) is supposed to be essential for the antiviral activity of STAT2. Hence, the targeting of p300 by EIA of adenovirus abolishes this antiviral effect of STAT2 and indicates a significant role for this protein in cellular antiviral responses. Moreover, it was recently reported that interaction between P300 and TBP plays an important role in transcription and embryogenesis in yeast (Walker et al., 2004). In some recent studies p300 and PCAF have been found to have intrinsic histone acetyltransferase (HAT) activity and the ability to recruit additional HAT factors (Ji et al., 2003, Schiltz and Nakatani, 2000). HATs have important role in chromatin remodeling and transcription (Baluchamy et al., 2003). The activation of p300, TBP and PCAF genes at 6, 24 and 48 h PI during IBV infection (Table 1) may play such a role in the infection process.
Alteration of host translational machinery to optimize viral protein synthesis is an important viral strategy to establish a productive viral infection. For this purpose multiple biochemical mechanisms operating at the initiation phase of mRNA translation are altered in virus infected cells. For example, eukaryotic initiation factor 4a (eIF4a) is one among the DEAD box family of proteins that has shown marked induction during IBV infection (Table 1). The DEAD box proteins are highly conserved from bacteria to human and are thought to be involved in a wide variety of cellular processes including translation, ribosome biogenesis, RNA splicing, RNA maturation and degradation (Owsianka and Patel, 1999, Wassarman and Steitz, 1991). The eIF4A is an ATP dependent RNA helicase that facilitates binding of the 40S ribosome to mRNA by melting 5′ proximal secondary structure on cellular mRNA (Pause and Sonenberg, 1992). We found seven- and three-fold up regulation of the eIF4a and DDX gene at 24 h PI, respectively during IBV infection. The ATPase and helicase activities of these proteins are essential for replication of some viruses and thus represent attractive targets for intervention (Du et al., 2002). The role of eIF3 down regulation in conjunction to the upregulation of IFI-56K has already been discussed with reference to our data and have also been well studied in other viral infections (Guo et al., 2000).
15. Conclusion
Substantial changes in expression profiles of a number of unique cellular pathways and genes including the activation of 14-3-3 protein kinase mediated signaling pathway, cellular factors like hNRNP1, TBP, eIF3, and PTB that are supposed to interact with RdRP enzyme gene, exclusive and stable alteration of IFI-56 k gene expression in interferon induced genes, enhanced expression of 9E3 and LSP1 genes as chemoattractant regulators, up-regulation of YY1, P300, PCAF associated factor 400, TBP, TAFII genes as transcriptionnal regulators, EF1a, ribosomal proteins l3, ribosomal protein s6, eIF4a and DDX 3 genes in translation regulation suggest the pathobiological significances of these genes and pathways during IBV infection process. However, the biological involvement of these cellular factors during IBV infection has been based on assumptions derived from previously known cellular functions for these genes. Further detailed characterization of these factors, using other molecular approaches will validate their role in IBV infection and will enhance our understanding of respiratory viral pathogenesis and immunity. In addition, a search of regulatory motifs (such as TPR and GTP binding motifs) in unknown but significantly altered genes identified in this study (data not shown here) may lead to the identification of more IFN related genes in the chicken. Moreover, precise determination of the role of viral cis acting elements (like UTR and 3′-poly A tail) and their interaction with cellular factors such as hnRNP1 and kinase C may be helpful in identifying potential targets to interrupt the viral replication cycle.
Acknowledgements
This work was supported by the Functional Pathogenomics of Mucosal Immunity (FPMI) project funded by Genome Prairie, Genome BC and their industry partners, Inimex Pharmaceuticals and Pyxis Genomics Inc. We are thankful to Dr. Andrew Ficzycz for providing information about YY1 protein from his PH.D thesis. We owe thanks to the scientific staff of the FPMI research group, adenovirus research group and avian research group at VIDO for their valuable suggestions and co-operation in designing and conducting the experiments and the data analysis. We are thankful to Stacy Miskolczi and Barry Carrol from the Animal Care facilities at VIDO. Published with permission of the director of VIDO as VIDO journal series number 382.
References
- Aoki H., Hayashi J., Moriyama M., Arakawa Y., Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase Raf-1. J. Virol. 2000;74(4):1736–1741. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Newsome D., Oldread E., Livingston D.M., Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374(6517):81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Atreya P.L., Kulkarni S. Respiratory syncytial virus strain A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology. 1999;261(2):227–241. doi: 10.1006/viro.1999.9835. [DOI] [PubMed] [Google Scholar]
- Austen M., Luscher B., Luscher-Firzlaff J.M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 1997;272(3):1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- Baluchamy S., Rajabi H.N., Thimmapaya R., Navaraj A., Thimmapaya B. Repression of c-Myc and inhibition of G1 exit in cells conditionally overexpressing p300 that is not dependent on its histone acetyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 2003;100(16):9524–9529. doi: 10.1073/pnas.1633700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M., Olsson H., Heinegard D., Leanderson T. Purification and characterization of a protein binding to the SP6 kappa promoter A potential role for CArG-box binding factor-A in kappa transcription. J. Biol. Chem. 1998;273(30):18881–18890. doi: 10.1074/jbc.273.30.18881. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Eckner R., Grossman S., Oldread E., Arany Z., D’Andrea A., Livingston D.M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383(6598):344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- Bigger C.B., Brasky K.M., Lanford R.E. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 2001;75(15):7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul C.C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., Springer T.A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382(6594):829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77(19):10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Colomer D., Vives-Corrons J.L., Blackshear P.J., Gil J. Characterization and purification of a protein kinase C substrate in human B cells Identification as lymphocyte-specific protein 1 (LSP1) J. Immunol. 1996;156(5):1709–1713. [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32(6):567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung R.T., Kaplan L.M. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3’ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 1999;254(2):351–362. doi: 10.1006/bbrc.1998.9949. [DOI] [PubMed] [Google Scholar]
- Cooper J.A., Jr., Ridgeway A.L., Pearson J., Culbreth R.R. Attenuation of interleukin 8-induced nasal inflammation by an inhibitor peptide. Am. J. Respir. Crit. Care Med. 2001;163(5):1198–1205. doi: 10.1164/ajrccm.163.5.2008017. [DOI] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer M.J., Holko M., Frevel M., Walker E., Der S., Paranjape J.M., Silverman R.H., Williams B.R. Functional classification of interferon-stimulated genes identified using microarrays. J. Leuk. Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M.X., Johnson R.B., Sun X.L., Staschke K.A., Colacino J., Wang Q.M. Comparative characterization of two DEAD-box RNA helicases in superfamily II: human translation-initiation factor 4A and hepatitis C virus non-structural protein 3 (NS3) helicase. Biochem. J. 2002;363(Pt 1):147–155. doi: 10.1042/0264-6021:3630147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers E.E., Zondag G.C., Malliri A., Price L.S., ten Klooster J.P., van der Kammen R.A., Collard J.G. Rho family proteins in cell adhesion and cell migration. Eur. J. Cancer. 2000;36(10):1269–1274. doi: 10.1016/s0959-8049(00)00091-5. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstrom L.H., Baule C., Soos T., Belak S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31(3):229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felzien L.K., Farrell S., Betts J.C., Mosavin R., Nabel G.J. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol. Cell Biol. 1999;19(6):4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J.R., Becker K.G., Ennist D.L., Gleason S.L., Driggers P.H., Levi B.Z., Appella E., Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell Biol. 1992;12(1):38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Subramanian R.R., Masters S.C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Garcin D., Marq J.B., Strahle L., le Mercier P., Kolakofsky D. All four Sendai Virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology. 2002;295(2):256–265. doi: 10.1006/viro.2001.1342. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Maurizi M.R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 1992;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hui D.J., Merrick W.C., Sen G.C. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19(24):6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A.L., Siepmann T.J. Pathways of ubiquitin conjugation. FASEB J. 1997;11(14):1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Haque S.J., Williams B.R. Signal transduction in the interferon system. Semin. Oncol. 1998;25(1 Suppl. 1):14–22. [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000;2(8):E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Holmes H.C., Darbyshire J.H. Induction of chicken interferon by avian infectious bronchitis virus. Res. Vet. Sci. 1978;25(2):178–181. [PubMed] [Google Scholar]
- Ji A., Dao D., Chen J., MacLellan W.R. EID-2, a novel member of the EID family of p300-binding proteins inhibits transactivation by MyoD. Gene. 2003;318:35–43. doi: 10.1016/j.gene.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Jongstra-Bilen J., Misener V.L., Wang C., Ginzberg H., Auerbach A., Joyner A.L., Downey G.P., Jongstra J. LSP1 modulates leukocyte populations in resting and inflamed peritoneum. Blood. 2000;96(5):1827–1835. [PubMed] [Google Scholar]
- Kanzaki M., Watson R.T., Hou J.C., Stamnes M., Saltiel A.R., Pessin J.E. Small GTP-binding protein TC10 differentially regulates two distinct populations of filamentous actin in 3T3L1 adipocytes. Mol. Biol. Cell. 2002;13(7):2334–2346. doi: 10.1091/mbc.01-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczynski D.R., Sellers H.S., Rowland G.N., Jackwood M.W. Detection of in ovo-inoculated infectious bronchitis virus by immunohistochemistry and in situ hybridization with a riboprobe in epithelial cells of the lung and cloacal bursa. Avian Dis. 2002;46(3):679–685. doi: 10.1637/0005-2086(2002)046[0679:DOIOII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Khabar K.S., Al-Zoghaibi F., Al-Ahdal M.N., Murayama T., Dhalla M., Mukaida N., Taha M., Al-Sedairy S.T., Siddiqui Y., Kessie G., Matsushima K. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 1997;186(7):1077–1085. doi: 10.1084/jem.186.7.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar K.S., Al-Zoghaibi F., Murayama T., Matsushima K., Mukaida N., Siddiqui Y., Dhalla M., Al-Ahdal M.N. Interleukin-8 selectively enhances cytopathic effect (CPE) induced by positive-strand RNA viruses in the human WISH cell line. Biochem. Biophys. Res. Commun. 1997;235(3):774–778. doi: 10.1006/bbrc.1997.6872. [DOI] [PubMed] [Google Scholar]
- Khor R., McElroy L.J., Whittaker G.R. The ubiquitin-vacuolar protein sorting system is selectively required during entry of influenza virus into host cells. Traffic. 2003;4(12):857–868. doi: 10.1046/j.1398-9219.2003.0140.x. [DOI] [PubMed] [Google Scholar]
- Kotani T., Shiraishi Y., Tsukamoto Y., Kuwamura M., Yamate J., Sakuma S., Gohda M. Epithelial cell kinetics in the inflammatory process of chicken trachea infected with infectious bronchitis virus. J. Vet. Med. Sci. 2000;62(2):129–134. doi: 10.1292/jvms.62.129. [DOI] [PubMed] [Google Scholar]
- Lai M.M. RNA-protein interactions in the regulation of coronavirus RNA replication and transcription. Biol. Chem. 1997;378(6):477–481. doi: 10.1515/bchm.1997.378.6.477. [DOI] [PubMed] [Google Scholar]
- Lai M.M. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244(1):1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- Landis H., Simon-Jodicke A., Kloti A., Di Paolo C., Schnorr J.J., Schneider-Schaulies S., Hefti H.P., Pavlovic J. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 1998;72(2):1516–1522. doi: 10.1128/jvi.72.2.1516-1522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Brown C., Jackwood M.W. Tissue distribution of avian infectious bronchitis virus following in ovo inoculation of chicken embryos examined by in situ hybridization with antisense digoxigenin-labeled universal riboprobe. J. Vet. Diagn. Invest. 2002;14(5):377–381. doi: 10.1177/104063870201400503. [DOI] [PubMed] [Google Scholar]
- Lu A., Zhang H., Zhang X., Wang H., Hu Q., Shen L., Schaffhausen B.S., Hou W., Li L. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology. 2004;324(1):84–89. doi: 10.1016/j.virol.2004.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheev A.M., Mikheev S.A., Zhang Y., Aebersold R., Zarbl H. CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res. 2000;28(19):3762–3770. doi: 10.1093/nar/28.19.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Tajima S., Lauffer L., Walter P. The beta subunit of the signal recognition particle receptor is a transmembrane GTPase that anchors the alpha subunit, a peripheral membrane GTPase, to the endoplasmic reticulum membrane. J. Cell Biol. 1995;128(3):273–282. doi: 10.1083/jcb.128.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiou D.J., Stunnenberg H.G. p300 is involved in formation of the TBP-TFIIA-containing basal transcription complex. TAC EMBO J. 2003;22(17):4501–4511. doi: 10.1093/emboj/cdg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriuchi M., Moriuchi H., Margolis D.M., Fauci A.S. USF/c-Myc enhances, while Yin-Yang 1 suppresses, the promoter activity of CXCR4, a coreceptor for HIV-1 entry. J. Immunol. 1999;162(10):5986–5992. [PubMed] [Google Scholar]
- Munir S., Kapur V. Regulation of host cell transcriptional physiology by the avian pneumovirus provides key insights into host-pathogen interactions. J. Virol. 2003;77(8):4899–4910. doi: 10.1128/JVI.77.8.4899-4910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S., Sharma J.M., Kapur V. Transcriptional response of avian cells to infection with Newcastle disease virus. Virus Res. 2005;107(1):103–108. doi: 10.1016/j.virusres.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Murayama T., Ohara Y., Obuchi M., Khabar K.S., Higashi H., Mukaida N., Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF-κB-binding sites of the interleukin-8 gene. J. Virol. 1997;71(7):5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J.E., Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8(14):1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- Oglesbee M.J., Liu Z., Kenney H., Brooks C.L. The highly inducible member of the 70 kDa family of heat shock proteins increases canine distemper virus polymerase activity. J. Gen. Virol. 1996;77(Pt 9):2125–2135. doi: 10.1099/0022-1317-77-9-2125. [DOI] [PubMed] [Google Scholar]
- Otsuki K., Sakagami Y., Tsubokura M. Serological relationship among ten strains of avian infectious bronchitis virus. Acta Virol. 1987;31(2):138–145. [PubMed] [Google Scholar]
- Owsianka A.M., Patel A.H. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. 1999;257(2):330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11(7):2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J., Zurcher T., Haller O., Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 1990;64(7):3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold M.E., Dairaghi D.J., Duke G.M., Saederup N., Mocarski E.S., Kemble G.W., Schall T.J. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. U.S.A. 1999;96(17):9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S.T., Skamene E., Malo D. Comparative genomics and host resistance against infectious diseases. Emerg. Infect. Dis. 1999;5(1):36–47. doi: 10.3201/eid0501.990105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan F., Ozer Z., Mao C., Uckun F.M. Active center cleft residues of pokeweed antiviral protein mediate its high-affinity binding to the ribosomal protein L3. Biochemistry. 2001;40(31):9104–9114. doi: 10.1021/bi002851p. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L., Nakatani Y. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta. 2000;1470(2):M37–M53. doi: 10.1016/s0304-419x(99)00037-2. [DOI] [PubMed] [Google Scholar]
- Schnorr J.J., Schneider-Schaulies S., Simon-Jodicke A., Pavlovic J., Horisberger M.A., ter Meulen V. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J. Virol. 1993;67(8):4760–4768. doi: 10.1128/jvi.67.8.4760-4768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze zur Wiesch J., Schmitz H., Borowski E., Borowski P. The proteins of the Hepatitis C virus: their features and interactions with intracellular protein phosphorylation. Arch. Virol. 2003;148(7):1247–1267. doi: 10.1007/s00705-003-0115-8. [DOI] [PubMed] [Google Scholar]
- Sen G.C. Viruses and interferons. Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Shen X., Masters P.S. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. U.S.A. 2001;98(5):2717–2722. doi: 10.1073/pnas.031424298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Huang P., Li H.P., Lai M.M. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19(17):4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava A., Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22(24):5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sick C., Schneider K., Staeheli P., Weining K.C. Novel chicken CXC and CC chemokines. Cytokine. 2000;12(3):181–186. doi: 10.1006/cyto.1999.0543. [DOI] [PubMed] [Google Scholar]
- Soriano S.F., Serrano A., Hernanz-Falcon P., Martin de Ana A., Monterrubio M., Martinez C., Rodriguez-Frade J.M., Mellado M. Chemokines integrate JAK/STAT and G-protein pathways during chemotaxis and calcium flux responses. Eur. J. Immunol. 2003;33(5):1328–1333. doi: 10.1002/eji.200323897. [DOI] [PubMed] [Google Scholar]
- Spagnolo J.F., Hogue B.G. Host protein interactions with the 3’ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74(11):5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Muhlberger E., Haller O., Weber F. The antiviral effect of interferon-beta against SARS-coronavirus is not mediated by MxA protein. J. Clin. Virol. 2004;30(3):211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S.A., Fuller S.D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J. Cell Biol. 1987;105(3):1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., Steensma H.Y., van Heusden G.P. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23(10):936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- Walker A.K., Shi Y., Blackwell T.K. An extensive requirement for transcription factor IID-specific TAF-1 in Caenorhabditis elegans embryonic transcription. J. Biol. Chem. 2004;279(15):15339–15347. doi: 10.1074/jbc.M310731200. [DOI] [PubMed] [Google Scholar]
- Wassarman D.A., Steitz J.A. RNA splicing Alive with DEAD proteins. Nature. 1991;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Willard F.S., Crouch M.F. Nuclear and cytoskeletal translocation and localization of heterotrimeric G-proteins. Immunol. Cell Biol. 2000;78(4):387–394. doi: 10.1046/j.1440-1711.2000.00927.x. [DOI] [PubMed] [Google Scholar]
- Worthington K.J., Sargent B.A., Davelaar F.G., Jones R.C. Immunity to avian pneumovirus infection in turkeys following in ovo vaccination with an attenuated vaccine. Vaccine. 2003;21(13–14):1355–1362. doi: 10.1016/s0264-410x(02)00689-8. [DOI] [PubMed] [Google Scholar]
- Yao Y.L., Yang W.M., Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell Biol. 2001;21(17):5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhao H., Collins C.D., Eckenrode S.E., Run Q., McIndoe R.A., Crawford J.M., Nelson D.R., She J.X., Liu C. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37(5):1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]


