Highlights
-
•
Review of current knowledge of cis-acting RNA elements essential to coronavirus replication.
-
•
Identification of RNA structural elements in alphacoronavirus terminal genome regions.
-
•
Discussion of intra- and intergeneric conservation of genomic cis-acting RNA elements in alpha- and betacoronaviruses.
Keywords: RNA virus, Coronavirus, Replication, cis-Acting element, RNA structure
Abstract
Coronavirus genome replication is mediated by a multi-subunit protein complex that is comprised of more than a dozen virally encoded and several cellular proteins. Interactions of the viral replicase complex with cis-acting RNA elements located in the 5′ and 3′-terminal genome regions ensure the specific replication of viral RNA. Over the past years, boundaries and structures of cis-acting RNA elements required for coronavirus genome replication have been extensively characterized in betacoronaviruses and, to a lesser extent, other coronavirus genera. Here, we review our current understanding of coronavirus cis-acting elements located in the terminal genome regions and use a combination of bioinformatic and RNA structure probing studies to identify and characterize putative cis-acting RNA elements in alphacoronaviruses. The study suggests significant RNA structure conservation among members of the genus Alphacoronavirus but also across genus boundaries. Overall, the conservation pattern identified for 5′ and 3′-terminal RNA structural elements in the genomes of alpha- and betacoronaviruses is in agreement with the widely used replicase polyprotein-based classification of the Coronavirinae, suggesting co-evolution of the coronavirus replication machinery with cognate cis-acting RNA elements.
1. Introduction
Coronaviruses are enveloped, positive-strand RNA viruses with exceptionally large genomes of approximately 30 kb. They have been assigned to the subfamily Coronavirinae within the family Coronaviridae (de Groot et al., 2012a, Masters and Perlman, 2013). Together with the families Arteriviridae, Roniviridae, and Mesoniviridae, the Coronaviridae form the order Nidovirales (de Groot et al., 2012b). The family Coronaviridae is currently comprised of four genera called Alpha-, Beta-, Gamma- and Deltacoronavirus. Closely related virus species in these genera are grouped together in specific lineages. Coronaviruses infect mammals and birds and include pathogens of major medical, veterinary and economic interest (de Groot et al., 2012a), with severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) and Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) providing two prominent examples of newly emerging, highly pathogenic coronaviruses in humans (Drosten et al., 2003, Ksiazek et al., 2003, Zaki et al., 2012).
Besides their large genome sizes, coronaviruses and related nidoviruses stick out from other plus-strand RNA viruses by the large number of virally encoded nonstructural proteins that are either involved in viral RNA synthesis or interact with host cell functions (reviewed in Masters and Perlman, 2013, Ziebuhr, 2008). Most of the nonstructural proteins (nsp) are expressed as subdomains of two large replicase gene-encoded polyproteins called pp1a (∼450 kDa) and pp1ab (∼750 kDa). Co- and posttranslational cleavage of pp1a/1ab by two types of virus-encoded proteases gives rise to a total of 15–16 mature proteins that (together with the nucleocapsid protein and cellular proteins) form the viral replication-transcription complex (RTC) (Almazán et al., 2004, Schelle et al., 2005, Ziebuhr, 2008, Ziebuhr et al., 2000). This multi-protein complex replicates the viral genome and produces an extensive set of 3′-coterminal subgenomic messenger RNAs (sg mRNAs), the latter representing a hallmark of corona- and other nidoviruses (Pasternak et al., 2006, Sawicki et al., 2007, Ziebuhr and Snijder, 2007). The sg mRNAs are used to express open reading frames located in the 3′-proximal third of the genome. They essentially encode the viral structural proteins, such as the nucleocapsid (N), membrane (M), spike (S) and envelope (E) proteins, and a varying number of accessory proteins, the latter often involved in functions that counteract antiviral host responses (Masters and Perlman, 2013, Narayanan et al., 2008b). Coronavirus sg mRNAs contain a common 5′ leader sequence (approximately 60–95 nt) that is identical to the 5′ end of the genome. The complement of this sequence is attached to the 3′ end of nascent negative strands in a complex process called “discontinuous extension of negative strands” (Sawicki and Sawicki, 1995, Sawicki and Sawicki, 1998, Sawicki et al., 2007, Sethna et al., 1991). This process involves attenuation of negative-strand RNA synthesis at transcription-regulating sequences (TRS) located upstream of the individual ORFs in the 3′-proximal genome region (“body TRSs”, TRS-B). Guided by basepairing interactions between the negative-strand complement of a TRS-B and the TRS located downstream of the 5′ leader on the viral genome (“leader TRS”, TRS-L), the nascent minus strand may be transferred from its downstream position on the template (at the TRS-B) to the TRS-L, where negative-strand RNA synthesis is then resumed and completed by copying the 5′ leader sequence. The set of 3′ antileader-containing sg minus-strand RNAs is subsequently used as templates for the production of the characteristic nested set of 5′ capped, 5′ leader-containing and 3′-polyadenylated sg mRNAs in coronavirus-infected cells (Lai et al., 1983, Sawicki et al., 2001, Sawicki and Sawicki, 1995, Sethna et al., 1989, Spaan et al., 1983). Sg minus-strand RNAs contain a U-stretch at their 5′ end, thus providing a template for 3′ polyadenylation of sg mRNAs (Hofmann and Brian, 1991, Wu et al., 2013).
Similar to other RNA viruses, coronavirus genomes contain important RNA signals in their 5′ and 3′-terminal genome regions, mainly (but not exclusively) in the untranslated regions (UTR). These signals are required for viral RNA synthesis (replication and/or transcription) and are collectively referred to as cis-acting RNA elements (Chang et al., 1994, Dalton et al., 2001, Izeta et al., 1999, Kim et al., 1993, Liao and Lai, 1994, Lin et al., 1994, Lin et al., 1996, Zhang et al., 1994). As indicated above, coronaviruses also contain cis-acting elements at internal positions in the genome, the best documented ones being the leader and body TRSs which play a vital role in the transfer of the nascent minus-strand RNA to an upstream position on the template (see above). Other internal cis-acting elements include specific RNA signals required for genome packing, which have been characterized in a small number of coronaviruses (Chen et al., 2007, Escors et al., 2003, Makino et al., 1990, Morales et al., 2013, Penzes et al., 1994), and a complex RNA pseudoknot structure located in the ORF1a-ORF1b overlap region that mediates a (−1) ribosomal frameshift event and thus controls the expression of the second large ORF on the coronavirus genome RNA (ORF1b) (Brierley et al., 1987, Brierley et al., 1989, de Haan et al., 2002, Namy et al., 2006).
Both viral and cellular proteins, including the viral N protein, heterogeneous ribonucleoprotein (hnRNP) family members, polypyrimidine tract-binding protein, and poly(A)-binding protein (PABP), have been shown to bind to specific coronavirus cis-acting elements and there is evidence to support the biological significance of some of these protein-RNA interactions (reviewed in Shi and Lai, 2005, Sola et al., 2011b).
The coronavirus RTC is a multi-subunit assembly comprised of more than a dozen viral and an unknown number of cellular proteins. The complex is anchored through transmembrane domains present in nsps 3, 4 and 6 to intracellular membranous structures that provide a specialized membrane-shielded compartment in (or at) which viral RNA synthesis takes place (den Boon and Ahlquist, 2010, Gosert et al., 2002, Kanjanahaluethai et al., 2007, Knoops et al., 2008, Oostra et al., 2008, Oostra et al., 2007, Snijder et al., 2006, van Hemert et al., 2008). Over the past years, viral components of the coronavirus RTC have been characterized in considerable detail, providing a wealth of functional and structural information (reviewed in Imbert et al., 2010, Masters, 2006, Ulferts et al., 2010, Ulferts and Ziebuhr, 2011, Ziebuhr, 2008). A large number of virally encoded enzymes, including protease, ADP-ribose-1″-phosphatase, NTPase, 5′-to-3′ helicase, RNA 5′-triphosphatase, RNA polymerase, guanosine-N7 and ribose 2′-O methyltransferases, 3′-to-5′ exoribonuclease and uridylate-specific endoribonuclease, have been identified (Baker et al., 1989, Bhardwaj et al., 2004, Chen et al., 2009, Chen et al., 2011, Decroly et al., 2008, Decroly et al., 2011, Eckerle et al., 2010, Ivanov et al., 2004, Kanjanahaluethai and Baker, 2000, Minskaia et al., 2006, Putics et al., 2005, Saikatendu et al., 2005, Seybert et al., 2000, te Velthuis et al., 2010, Ziebuhr et al., 1995). In some cases, these activities could be linked to specific steps of viral RNA synthesis and/or RNA processing or were shown to interfere with cellular functions (reviewed in Masters and Perlman, 2013, Ziebuhr, 2008). There are multiple interactions between the individual replicase gene-encoded nsps and the structural basis and functional implications of these interactions have been studied in a few cases. For example, it has been shown that the exoribonuclease and ribose 2′ O-methyltransferase activities associated with nsp14 and nsp16, respectively, are each stimulated by specific interactions with nsp10 (Bouvet et al., 2014, Decroly et al., 2011). Also, there is evidence that a heteromultimeric complex formed by nsp7 and nsp8 interacts with (and serves as a processivity factor for) the RNA-dependent RNA polymerase (RdRp, nsp12) (Zhai et al., 2005). Additional interactions between individual subunits of the RTC have been suggested on the basis of two-hybrid screening data (Pan et al., 2008, von Brunn et al., 2007) and there is evidence that a substantial number of coronavirus nsps form homo- and/or heterooligomeric complexes (Anand et al., 2002, Anand et al., 2003, Bouvet et al., 2014, Chen et al., 2011, Ricagno et al., 2006, Su et al., 2006, Xiao et al., 2012, Zhai et al., 2005).
Despite major progress in the characterization of proteins and cis-acting RNA elements involved in coronavirus RNA synthesis, the molecular mechanisms that mediate specific steps of coronavirus RNA replication and transcription are far from being understood. Important information on cis-acting RNA elements has been obtained from studies using defective interfering (DI) RNAs and, more recently, genetically engineered coronavirus mutants (reviewed in Brian and Baric, 2005, Masters, 2007, Sola et al., 2011b).
In this article, we will summarize previous work on (beta)coronavirus genomic cis-acting RNA elements and will then move on to present conclusions from our recent bioinformatic and RNA structure probing studies on cis-acting RNA elements conserved in alphacoronaviruses.
2. Identification and delineation of coronavirus cis-acting elements
Historically, cis-acting RNA elements required for coronavirus RNA synthesis have mainly been studied by using naturally occurring and genetically engineered defective interfering RNAs (DI RNAs) (reviewed in Brian and Baric, 2005, Brian and Spaan, 1997, Masters, 2007, Sola et al., 2011b). DI RNAs are replication-competent genome-derived RNA molecules that contain extensive internal deletions but retained all the cis-acting RNA signals required for replication by functional replicase complexes provided by a helper virus that replicates in the same cell (Levis et al., 1986, Weiss et al., 1983). Essentially, these cis-acting elements comprise the untranslated 5′- and 3′-terminal genome regions but, in some cases, may also extend into coding regions. In some cases, they also contain noncontiguous sequences derived from internal genome regions. Coronavirus DI RNAs were first described for the betacoronaviruses MHV and BCoV (Chang et al., 1994, de Groot et al., 1992, Hofmann et al., 1990, Luytjes et al., 1996, Makino et al., 1985, Makino et al., 1988a, Makino et al., 1988b, Makino et al., 1984). Subsequently, these studies were extended to alpha- and gammacoronaviruses (Izeta et al., 1999, Mendez et al., 1996, Penzes et al., 1994, Penzes et al., 1996). Over the years, studies of defective genomes proved to be very useful for identifying coronavirus RNA elements required for replication (and packaging). However, DI RNAs also have disadvantages, a major one being homologous recombination between the RNA replicon and the helper virus genome. Thus, for example, artificial DI RNAs containing mutant 5′ leader sequences rapidly acquire the leader sequence of the helper virus, a process commonly referred to as “leader switching” (Chang et al., 1994, Chang et al., 1996, Makino and Lai, 1990). The latter occurs very often if poorly replicating mutant DI RNAs are to be characterized which generally require amplification steps by serial passaging to determine their phenotype. With the development of reverse-genetic systems suitable to produce and manipulate full-length coronavirus cDNA copies, an attractive alternative for studying cis-acting RNA elements at the genome level (including long range RNA-RNA interactions) is now available that overcomes some of the limitations of DI RNA-based systems (Almazán et al., 2000, Casais et al., 2001, Scobey et al., 2013, Tekes et al., 2008, Thiel et al., 2001, van den Worm et al., 2012, Yount et al., 2000, Yount et al., 2003).
2.1. Delineation of 5′ cis-acting elements
DI RNAs studies on representative betacoronaviruses revealed that less then 500 nt from the 5′ end of the genome (466 nt in MHV and 498 nt in BCoV) are required for replication (Chang et al., 1994, Kim et al., 1993, Luytjes et al., 1996). In subsequent studies on alpha- and gammacoronaviruses, minimal 5′ cis-acting signals of 649 and 544 nt were determined for TGEV (Escors et al., 2003) and IBV, respectively (Dalton et al., 2001). The 5′-terminal genome regions of 466 nt (MHV) to 649 nt (TGEV) comprise the entire 5′ UTR ranging in size from 210 nt (MHV, BCoV and HCoV-OC43) to 314 nt (TGEV) and thus extend into the nsp1-coding region of ORF 1a (see below). By contrast, the gammacoronavirus IBV features a much larger 5′ UTR (528 nt) (Boursnell et al., 1987) and lacks a counterpart of nsp1 (Ziebuhr et al., 2001). It therefore appears that the gammacoronavirus 5′ UTR (alone) contains all the 5′ RNA signals required for genome replication.
2.2. Functional and structural features of coronavirus 5′ cis-acting elements
Cis-acting RNA structures in the 5′-terminal region of the coronavirus genome have first been studied for BCoV using DI RNA-based systems (Brown et al., 2007, Chang et al., 1994, Chang et al., 1996, Gustin et al., 2009, Raman et al., 2003, Raman and Brian, 2005). In the 5′-terminal 215 nts of the BCoV genome, four stem-loops (designated I [comprised of Ia and Ib], II, III, and IV) were defined. Enzymatic probing and mutational analysis of both naturally occurring and genetically engineered DI RNAs were used to (i) corroborate the predicted RNA secondary structures and (ii) determine their functional significance in DI RNA replication. More recently, two further stem-loops (called SL-V and SL-VI) were identified in the BCoV nsp1-coding region of which SL-VI was confirmed to be essential for DI RNA replication (Brown et al., 2007).
Subsequent studies suggested (a varying degree of) conservation of 5′ cis-acting elements among betacoronaviruses and even the more distantly related alpha- and gammacoronaviruses (Chen and Olsthoorn, 2010, Kang et al., 2006). To facilitate the discussion of data obtained in studies of different viruses by different laboratories, we will use in this article a uniform nomenclature for the main RNA structural elements in the 5′-proximal genome region (SL1, SL2, [SL3 if present], SL4 and SL5). The nomenclature is based on SL designations used by the Leibowitz and Giedroc laboratories and in predictions of genus- and lineage-specific conservation patterns of 5′ cis-acting elements in alpha-, beta- and gammacoronaviruses (Chen and Olsthoorn, 2010, Kang et al., 2006, Liu et al., 2006, Liu et al., 2007). The proposed functional and structural conservation of 5′ cis-acting elements among betacoronaviruses received strong support by reverse-genetic data demonstrating that SARS-CoV SL1, SL2, and SL4 can functionally replace their counterparts in the MHV genome when introduced individually (Kang et al., 2006). By contrast, replacement of the entire MHV 5′ UTR with that of SARS-CoV did not produce a viable MHV mutant, suggesting a requirement for additional stable or transient long-range RNA-RNA interactions of the 5′ UTR with other genome regions. Evidence to support this hypothesis was obtained in subsequent studies. For example, the energetically unstable lower part of MHV SL1 was found to be involved in long-range RNA interactions with the 3′ UTR (Li et al., 2008) (see below). Also, in a study using MHV/BCoV chimera, a region downstream of SL4 was revealed to be engaged in long-range interactions with the nsp1-coding region, thus possibly forming an extensive higher-order RNA structure (Guan et al., 2012). A subsequent BCoV DI RNA mutagenesis study (Su et al., 2014) further suggested that this multipartite RNA structure may involve several stem-loop (sub)structures identified in earlier studies (Gustin et al., 2009, Raman and Brian, 2005) but require refolding of other RNA structures suggested earlier to be essential for DI RNA replication (Brown et al., 2007). The study by Su et al. (2014) also identified an intriguing requirement in cis of an oligopeptide sequence in the N-proximal part of nsp1, suggesting that nsp1 may be an essential cis-acting protein factor in betacoronavirus replication, in addition to its multiple other functions (Brockway and Denison, 2005, Huang et al., 2011a, Huang et al., 2011b, Kamitani et al., 2006, Kamitani et al., 2009, Lei et al., 2013, Lokugamage et al., 2012, Narayanan et al., 2008a, Tanaka et al., 2012, Tohya et al., 2009, Wathelet et al., 2007, Züst et al., 2007). Possible interacting partners for nsp1 remain to be identified.
The 5′-proximal SL1 and SL2 are predicted to be conserved across all genera of the Coronavirinae (Chen and Olsthoorn, 2010, Liu et al., 2007). Nuclear magnetic resonance (NMR) spectroscopy studies of MHV and HCoV-OC43 SL1 RNAs supported the predicted stem-loop and revealed 2–3 noncanonical base pairs in the middle of the stem. The fully base-paired SL1 was proposed to exist in equilibrium with higher-energy (partially unfolded) conformers. Characterization of MHV mutants containing specific replacements in SL1 and sequence analysis of second-site revertants supports a “dynamic SL1” model in which SL1 is structurally and functionally bipartite (Li et al., 2008). While the upper part of SL1 is (required to be) stable, the lower part is (required to be) unstable, possibly indicating the requirement for an optimized stability to permit or fine-tune transient long-range (RNA- or protein-mediated) interactions between the 5′ and 3′ UTRs required for sgRNA transcription and genome replication, respectively.
SL2 is the most conserved structure in the coronavirus 5′ UTR (Chen and Olsthoorn, 2010, Liu et al., 2007). It is comprised of a 5-bp stem and a highly conserved loop sequence, 5′-CUUGY-3′, that was shown to adopt a 5′-uYNMG(U)a or uCUYG(U)a-like tetraloop structure (Lee et al., 2011, Liu et al., 2009). Reverse genetics data confirmed that SL2 is required for MHV replication and may have a specific role in sgRNA synthesis. Within certain structural constraints, nucleotide replacements were found to be tolerated or could be rescued by increasing the stem stability, suggesting a limited plasticity of this important cis-acting RNA element (Liu et al., 2009).
The predicted SL3 (named SL-II in BCoV DI RNA studies) appears to be conserved only in a subset of beta- and gammacoronaviruses (Chen and Olsthoorn, 2010). For BCoV and closely related viruses, the TRS-L core sequence (CS) has been proposed to be exposed in the SL3 loop region, a structure similar to the TRS-L hairpin structure reported for the related arterivirus equine arteritis virus (EAV) (Chang et al., 1996, van den Born et al., 2004, van den Born et al., 2005). By contrast, in most other coronaviruses, the TRS-L CS and flanking regions were suggested to be located in nonstructured regions (Chen and Olsthoorn, 2010, Stirrups et al., 2000, Wang and Zhang, 2000).
SL4 is a long hairpin located downstream of the TRS-L CS and suggested to be conserved in all coronavirus genera (Chen and Olsthoorn, 2010, Raman et al., 2003, Raman and Brian, 2005). In the vast majority of coronaviruses, SL4 contains a short ORF comprised of only a few codons. Because of its position in the genome upstream of ORF1a it is generally referred to as the uORF. Recent reverse genetics work in the MHV system (Wu et al., 2014, Yang et al., 2011) showed that disruption of the uORF yields viable mutants that, however, acquire alternate uORFs upon serial passaging in cell culture. In vitro, uORF-disrupted RNAs showed enhanced translation of the downstream ORF. The available data suggest that the uORF represses ORF1a/1b translation and has a beneficial but non-essential role in coronavirus replication in cell culture. SL4 may be further subdivided into SL4a and SL4b. Interestingly, despite its conservation in coronaviruses, SL4 tolerates extensive mutations. Thus, for MHV, it was shown that base pairing in SL4a is not required for replication and separate deletions of SL4a and SL4b are tolerated. By contrast, complete deletion of SL4 and a 3-nt deletion immediately downstream of SL4 abolished or profoundly impaired viral RNA synthesis. The characterization of second-site mutations and a viable MHV mutant in which SL4 was replaced with a shorter sequence-unrelated stem-loop led to a model in which SL4 was proposed to function as a spacer element that controls the orientation of SL1, SL2, and TRS-L and thereby directs subgenomic RNA synthesis (Yang et al., 2011). The SL4 sequence overlaps with the ‘hotspot’ of the 5′-proximal genomic acceptor required for BCoV discontinuous transcription (Wu et al., 2006), thus further supporting a role of the region immediately downstream of TRS-L in subgenomic RNA synthesis.
Chen & Osthoorn (2010) predicted that variations of another RNA structure called SL5 may be conserved in specific coronaviruses genera and/or lineages. In alpha- and betacoronaviruses, SL5 extends into ORF1a. Depending on the lineage studied, conserved loop sequences could be identified in substructural hairpins of SL5. Sequence conservation was found to be more pronounced in alpha- compared to betacoronaviruses. Thus, for example, in alphacoronaviruses, three hairpins, called SL5a, 5b, and 5c, each containing a 5′-UUCCGU-3′ loop sequence, were identified. Similar structures were only partly conserved in betacoronaviruses and significant lineage-specific variations in the substructural hairpins and their loop sequences were identified. A possible SL5 equivalent in gammacoronaviruses was predicted to adopt a rod-like structure that lacks conserved loop sequences (Chen and Olsthoorn, 2010). As outlined above, possible betacoronavirus SL5 substructures located within (or extending into) the nsp1-coding region (termed SLs IV, V, VI, and VII) have been characterized structurally and functionally using BCoV DI RNA and MHV reverse genetics systems (Brown et al., 2007, Guan et al., 2011, Guan et al., 2012, Raman and Brian, 2005).
2.3. Characterization of alphacoronavirus 5′-proximal RNA structures
To a large extent, previous work on coronavirus cis-acting elements has focused on only two species of closely related lineage A betacoronaviruses (represented by BCoV and MHV), while there is limited information on functionally and structurally related elements in other coronaviruses. We therefore decided to embark on a more detailed characterization of putative alphacoronavirus cis-acting elements located in the 5′ and 3′ genome regions. As a starting point, we used 20 coronavirus genomes representing all the currently approved species from the four genera of the Coronavirinae and the newly identified MERS-CoV (de Groot et al., 2013) and calculated alignment-based secondary structures using LocARNA (v.1.7.2) (Will et al., 2012). Although LocARNA considers both sequence and secondary structure to calculate these multiple alignments, we failed to detect RNA secondary structures conserved across all coronavirus genera when using these highly diverse sequences. We therefore resorted to producing separate genus-wide alignment-based secondary structure predictions for coronaviruses. To do this, we used complete 5′ UTR regions and 20 nts from the ORF1a 5′ end to calculate consensus structures with RNAalifold -r -p --color --noLP --MEA (v.2.0.7) (Lorenz et al., 2011) and -C, respectively, if constraints were used. The subgroups (lineages) obtained in these sequence alignments were consistent with the previously recognized subgroups of alpha- and betacoronaviruses (de Groot et al., 2012a, de Groot et al., 2013) (not shown). As shown in Fig. 1 , all the functionally relevant RNA secondary structure elements identified previously in the betacoronavirus 5′ genome regions, SL1, SL2 and SL4, could be identified. In line with previous predictions and despite the pronounced sequence diversity in the 5′-terminal genome regions, these RNA structures are suggested to be conserved among all currently approved betacoronavirus lineages and species. This conclusion is also supported by the large number of covariations, suggesting a strong selection pressure to retain these base-pairing interactions. Fig. 1 also shows the lack of conservation of a stable hairpin structure containing the TRS-L. It should be noted that, for several betacoronaviruses, it is possible to force a stem-loop containing the TRS-L, but this stem-loop is only supported by two conserved base pairs. In line with previous reports, bovine coronavirus (and other viruses belonging to the species Betacoronavirus 1) appear(s) to be an exception in that a more stable SL3 containing the 5′-UCUAAAC-3′ sequence in the loop region can be predicted in this case. Overall, the structure prediction shown in Fig. 1 turned out to be in perfect agreement with previous studies (see chapter 2.2).
Fig. 1.
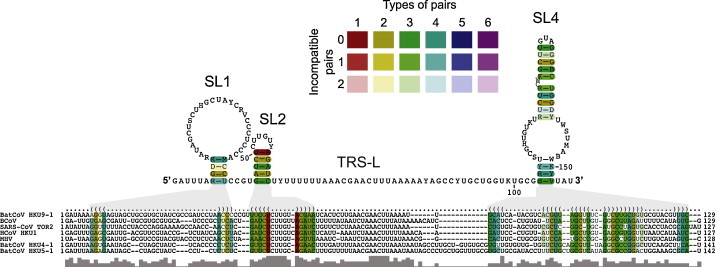
Alignment-based secondary structure prediction of 5′ genome regions of betacoronaviruses. The viruses included in this analysis represent all currently recognized lineages and species in the genus Betacoronavirus. The alignment was generated using LocARNA and the structure was calculated using RNAalifold. The consensus sequence is represented using the IUPAC code: A (adenine), C (cytosine), G (guanine), U (uracil), R (purine [A or G]), Y (pyrimidine [C or U]), M (C or A), K (U or G), W (U or A), S (C or G), B (C, U, or G [not A]), D (A, U, or G [not C]), H (A, U, or C [not G]), V (A, C, or G [not U], N (any base). Colors are used to indicate conserved base pairs: from red (conservation of only one base-pair type) to purple (all six base-pair types are found); from dark (all sequences contain this base pair) to light colors (1 or 2 sequences are unable to form this base pair). The gray bars below the alignment indicate the extent of sequence conservation. Gray shadows are used to link RNA structures with the corresponding dot-bracket notations above the alignment. To refine the alignment, an anchor at the highly conserved apical loop of SL2 was used.
We therefore used this approach in subsequent studies of conserved RNA structure elements located in the 5′ genome regions of alphacoronaviruses. Predictions were verified and refined by RNA structure probing analyses (Ehresmann et al., 1987, Qu et al., 1983) using in vitro-transcribed RNAs with sequences corresponding to the 5′-terminal genome regions of HCoV-229E and HCoV-NL63, respectively (to be published elsewhere). For the calculation of secondary structures of single sequences, we used RNAfold --noLP (v.2.0.7) (Lorenz et al., 2011) and -C, respectively, if constraints were used.
Using multiple alignments calculated with LocARNA, we were able to identify RNA secondary structures conserved among (all) alpha- and betacoronaviruses, thus confirming and extending earlier studies (Fig. 1, Fig. 2 ) (Chen and Olsthoorn, 2010, Raman et al., 2003). These include (i) SL1, (ii) SL2 (with its short stem and highly conserved loop region [5′-UUUGU-3′ in alphacoronaviruses]), (iii) a poorly structured region containing the TRS-L CS and some flanking sequence, (iv) SL4 (containing the uORF) and (v) SL5, the latter being conserved very well among all alphacoronavirus species (Fig. 2) and significantly more diverse in betacoronaviruses (Chen and Olsthoorn, 2010). The large number of covariant base pairs in the alphacoronavirus SL5 (Fig. 2) suggest significant constraints and a major functional role for this structure in alphacoronavirus (and, possibly, betacoronavirus) replication and there is indeed some experimental evidence to support this hypothesis (Brown et al., 2007, Su et al., 2014). Using RNA structure probing information obtained for the 5′-terminal 600 nts of HCoV-229E and HCoV-NL63, we confirmed and refined our RNA secondary structure predictions for two B-lineage alphacoronaviruses (Fig. 3, Fig. 4 ). The data obtained in these studies (details to be published elsewhere) support a model in which the ∼310-nt 5′ genome regions consistently fold into 4 major RNA structures called SL1, SL2, SL4, and SL5. The latter contains 3 hairpin substructures, SL5a, 5b, and 5c, featuring highly conserved 5(6)-nt loop sequences.
Fig. 2.
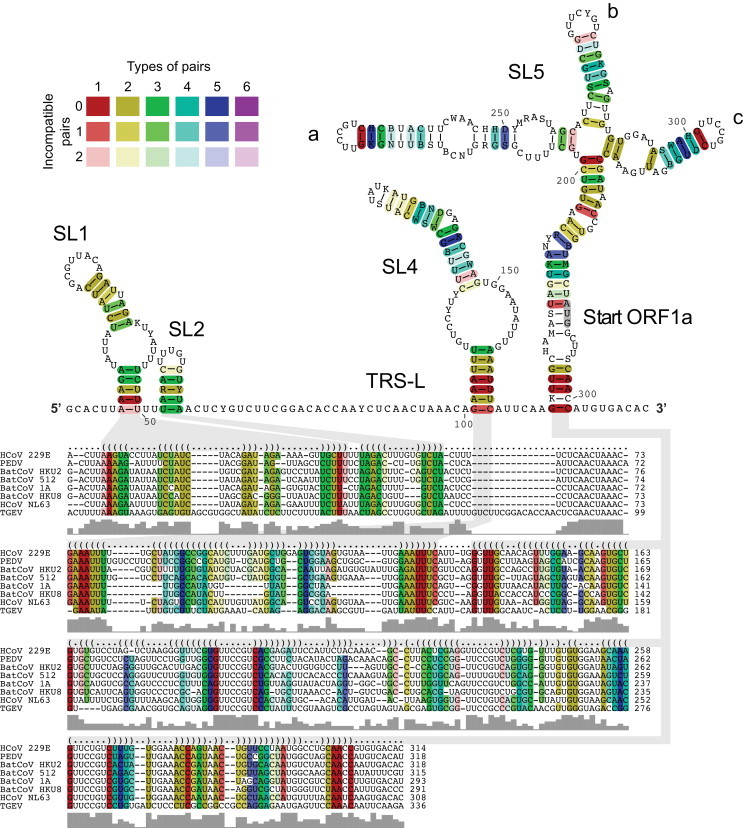
Alignment-based secondary structure prediction of 5′ genome regions of alphacoronaviruses. The viruses included in this analysis represent all currently recognized species in the genus Alphacoronavirus. The alignment was calculated by LocARNA, the structure by RNAalifold. The consensus sequence is represented using the IUPAC code. Colors are used to indicate conserved base pairs: from red (conservation of only one base-pair type) to purple (all six base-pair types are found); from dark (all sequences contain this base pair) to light colors (1 or 2 sequences are unable to form this base pair). The gray bars below the alignment indicate the extent of sequence conservation at a given position. Gray shadows are used to link RNA structures with the corresponding dot-bracket notations above the alignment. To refine the alignment, an anchor at the highly conserved core TRS-L was used.
Fig. 3.
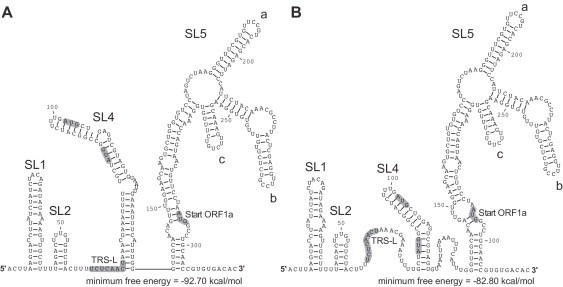
RNA secondary structure of the HCoV-229E 5′ UTR. (A) The RNA secondary structure of the 5′ UTR + 20 nts was predicted using RNAfold --noLP. (B) RNA secondary structure of the 5′ UTR + 20 nts was predicted using RNAfold --noLP -C. Structure probing data were used as constraints. The TRS-L core sequence and translational start codons are indicated.
Fig. 4.
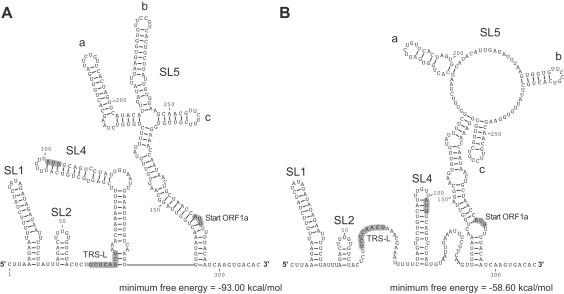
RNA secondary structure of the HCoV-NL63 5′ UTR. (A) The RNA secondary structure of the 5′ UTR + 20 nts was predicted using RNAfold --noLP. (B) RNA secondary structure of the 5′ UTR + 20 nts was predicted using RNAfold --noLP -C. Structure probing data were used as constraints. The TRS-L core sequence and translational start codons are indicated.
The consensus secondary structure predicted for alphacoronaviruses (Fig. 2) was found to fit very well the individual structure predictions for HCoV-229E and HCoV-NL63 (Fig. 3, Fig. 4) and the inclusion of structure probing information as additional constraints required only very few minor adjustments in our structure predictions (Fig. 3, Fig. 4). Most importantly, the basal part of the predicted SL4 was now predicted to be unpaired, thereby extending the single-stranded region downstream of the TRS-L and also affecting the spacing between SL4 and SL5. The (predicted) basal stem of SL4 contains the most conserved sequence within the alphacoronavirus 5′-terminal RNA structural elements (Fig. 2, see the red base pairs). It is therefore reasonable to think that this structurally flexible region is involved in long-range RNA-RNA interactions. In line with this idea, a previous TGEV reverse genetic study showed that mutants permitting additional base-pairing interactions of the copy TRS-B upstream of a reporter sgRNA with the 5′-GAAA-3′ sequence immediately downstream of the TGEV TRS-L CS (5′-ACUAAAC-3′) (see also Fig. 2) enhance the production of this specific reporter sgRNA (Zúñiga et al., 2004). These functional data and our structural analyses of alphacoronavirus 5′-terminal genome regions lead us to suggest that the basal part of SL4 exists in a flexible state, thereby possibly facilitating strand transfer during sg minus-strand RNA synthesis. Both this RNA structural flexibility and the role of proteins that bind in this region and thereby likely affect the SL4 structure remain to be further investigated. Of note, hnRNP family members along with the viral N protein have been shown to bind in this region and the N protein has been suggested to have chaperone and TRS-L/TRS-B unwinding activities (Galán et al., 2009, Grossoehme et al., 2009, Huang and Lai, 1999, Keane et al., 2012, Li et al., 1997, Li et al., 1999, Shi and Lai, 2005, Sola et al., 2011a, Sola et al., 2011b, Zúñiga et al., 2007, Zúñiga et al., 2010). It is therefore tempting to speculate that cellular and/or viral proteins bind and unwind the energetically labile SL4 substructure to facilitate the strand transfer during sg minus-strand RNA synthesis.
2.4. Delineation of 3′ cis-acting elements required for coronavirus replication
Initial information on 3′ cis-acting elements required for RNA replication was (again) derived from betacoronavirus DI RNA studies (Kim et al., 1993, Lin and Lai, 1993, Luytjes et al., 1996). Deletion mutagenesis data suggested that 3′ cis-acting elements encompass the entire 301-nt 3′ UTR plus poly(A) tail, along with a portion of the nucleocapsid (N) protein gene. However, subsequent studies showed that the structural protein genes (including the N protein coding region) tolerate major changes including combinations of single-site mutations and rearrangements of entire genes, suggesting that the 3′-proximal coding regions do not form part of the 3′ cis-acting element (de Haan et al., 2002, Goebel et al., 2004b, Lorenz et al., 2011). Similarly, for members of the species Alphacoronavirus 1 (TGEV, FCoV), it was shown that the N gene was not required for replication (Izeta et al., 1999) and even deletions of the accessory protein genes 7a and 7b were tolerated in FCoV, suggesting that the 3′ cis-acting replication signals do not exceed 283 nts plus poly(A) tail (Haijema et al., 2004). For the gammacoronavirus IBV, a minimal 3′-terminal sequence of 338 nts was reported to be required for DI RNA replication (Dalton et al., 2001), supporting the idea that, across all coronavirus genera, the 3′ UTR contains all the cis-acting elements required for replication. For MHV, it was shown that a significantly smaller fragment of no more than 55 nts suffices for the initiation of negative-strand RNA synthesis (Lin et al., 1994), suggesting differential requirements for plus- versus minus-strand RNA synthesis. Furthermore, using betacoronavirus DI RNA systems, a short poly(A) tract of at least 5–10 nts was found to be required for replication (Spagnolo and Hogue, 2000).
2.5. Structural and functional features of coronavirus 3′ cis-acting elements
Cis-acting RNA elements present in the 3′-terminal genome regions have been studied most extensively in betacoronaviruses. The first essential RNA structural element, called bulged stem-loop (BSL), was discovered in MHV (Hsue and Masters, 1997). It comprises 68 nts immediately downstream of the MHV N gene stop codon and was discovered as a result of attempts to replace the MHV 3′ UTR with that of BCoV (Hsue and Masters, 1997). Despite limited sequence identity in the 3′ UTRs of the two viruses, replacements of the entire 3′ UTR (and specific parts of it) were tolerated, suggesting the presence of conserved structures (rather than sequences) that perform cis-acting functions in betacoronavirus replication (Hsue and Masters, 1997). The conservation of functionally equivalent elements in the 3′ (and 5′) UTR(s) among betacoronavirus genomes was further supported by a study showing that a BCoV-derived reporter DI RNA was efficiently replicated by a range of BCoV- and MHV-related betacoronaviruses (Wu et al., 2003). Interestingly, a possible BSL equivalent was also identified in IBV and other gammacoronaviruses and its functional significance was demonstrated using IBV DI RNA constructs (Dalton et al., 2001). The nearly perfect stem-loop structure in IBV comprises 42 nts and is located at the upstream end of region II, a conserved region in the gammacoronavirus 3′ UTR.
The second essential RNA element within the (betacoronavirus) 3′ UTR is a classical hairpin-type RNA pseudoknot (PK) structure that was first discovered in BCoV and shown to be required for DI RNA replication (Williams et al., 1999). In BCoV, the PK comprises 54 nts and is located immediately downstream of the BSL. Equivalent PK structures were predicted to be conserved in beta- and alphacoronaviruses while gammacoronaviruses were proposed to retain only a few features of this PK or to lack this structure altogether (Williams et al., 1999). In a subsequent study using a reverse genetics approach, the functional significance of the PK in genome replication was demonstrated for MHV (Goebel et al., 2004a). Also the more distantly related betacoronaviruses HCoV-HKU1 (Woo et al., 2005) and SARS-CoV were confirmed to contain this PK structure (Goebel et al., 2004b). The 3′ UTR regions of MHV and SARS-CoV, which only share 38% sequence identity, were shown to be interchangeable, again supporting the conservation of functionally equivalent structures among different betacoronavirus lineages. Apparently, this conservation does not extend to alpha- and gammacoronaviruses because replacements of the MHV 3′ UTR with that of TGEV and IBV, respectively, did not give rise to viable MHV mutants (Goebel et al., 2004b). Together, these data suggest that coronaviruses evolved several genus-specific cis-acting RNA elements. For example, the presence of a BSL followed by a PK structure is limited to betacoronaviruses, while other genera appear to contain only one of these elements, with the PK being conserved in alphacoronaviruses and the BSL in gammacoronaviruses (Dalton et al., 2001, Hsue and Masters, 1997, Williams et al., 1999) (see Section 2.6).
The structures and functionally important substructures of both the BSL and PK have been characterized in significant detail for BCoV and MHV (Goebel et al., 2004a, Hsue et al., 2000, Williams et al., 1999). In the primary structure, the BSL and PK regions overlap by several nucleotides. Formation of the first stem of the PK structure requires base-pairing interactions with the downstream segment F of the BSL, thereby destabilizing the latter structure. In an extensive MHV mutagenesis study, the functional significance of both structures was demonstrated conclusively. Because the two structures cannot exist simultaneously and, yet, each of them is essential for viral replication, it was proposed that the two elements may adopt alternate structures that act as a ‘molecular switch’ controlling the transition between different steps of the viral replication cycle (Goebel et al., 2004a). Initial mechanistic insight into how this ‘molecular switch’ might work was obtained in a subsequent study that provided evidence for a direct interaction between loop 1 of the PK with the extreme 3′ end of the MHV genome (Züst et al., 2008). The characterization of second-site revertants arising from MHV mutants with genetically engineered oligonucleotide insertions in loop 1 revealed distinct replacements at the extreme 3′ end, thereby retaining specific base-pairing interactions with the loop 1 region and thus precluding the formation of stem 1 of the PK. Another set of mutants contained second-site mutations that suggested specific interactions of the PK region with nsp8 and nsp9. Based on this study, a model was proposed in which the formation and disruption of the PK by differential base-pairing interactions with the BSL and 3′-terminal genome sequences, respectively, may lead to alternate structures that govern different steps of the initiation and continuation of negative-strand RNA synthesis (Züst et al., 2008). Further evidence to support this model was obtained in a subsequent MHV reverse genetics study by Liu et al. (Liu et al., 2013). Thermodynamic investigations revealed a limited stability of the PK structure (Stammler et al., 2011), further supporting the structural flexibility of this cis-acting element and, thus, its proposed role as a ‘molecular switch’.
The region downstream of the PK is less well conserved among betacoronaviruses. It is generally referred to as the “hypervariable region (HVR)” and is not identical to the HVR identified at the 5′ end of the 3′ UTR in IBV (Dalton et al., 2001, Williams et al., 1993). The betacoronavirus HVR was predicted to contain a complex RNA structure whose existence and functional significance was supported by enzymatic probing and MHV DI RNA mutagenesis studies (Liu et al., 2001). By contrast, more recent studies showed that large parts or even the entire HVR region can be deleted without causing major defects in MHV replication, arguing against an important role of this genome region in viral replication (Goebel et al., 2007, Züst et al., 2008). Interestingly however, MHV HVR mutants proved to be highly attenuated in vivo, suggesting a possible role in pathogenesis (Goebel et al., 2007).
About 70–80 nts from the 3′ end of the coronavirus genome, there is a conserved octanucleotide sequence, 5′-GGAAGAGC-3′, which was identified in early coronavirus sequence analyses performed in the late 1980s (Boursnell et al., 1985, Lapps et al., 1987, Schreiber et al., 1989) and subsequently found to be universally conserved across all coronavirus genera, with only very few viruses containing single-site replacements in this sequence (Goebel et al., 2007). This strict conservation suggests an important functional role for the octanucleotide sequence. To date, however, the function of the sequence has not been identified. As mentioned above, the entire HVR including the octanucleotide sequence can be deleted from the MHV genome without causing major defects in viral replication in vitro (Goebel et al., 2007). In line with this, replacements of single nucleotides within the octanucleotide motif were tolerated although, in most cases, the octanucleotide mutants exhibited small-plaque phenotypes and/or delayed single-step growth kinetics. In both high- and low-multiplicity-of-infection experiments, octanucleotide and HVR deletion mutants lagged behind the wild-type virus but reached near-wildtype titers at later time points and had no detectable defect in gRNA or sgRNA synthesis (Goebel et al., 2007).
MHV and BCoV DI RNA studies provided evidence that the 3′ poly(A) tail present at the 3′ end of coronavirus genomes is another essential component of the coronavirus 3′ cis-acting signals, with a minimum of 5 to 10 adenylate residues being required for DI RNA replication (Spagnolo and Hogue, 2000). This requirement corresponds well to the minimal binding site of the poly(A)-binding protein (PABP) on DI RNAs poly(A) sequences (Spagnolo and Hogue, 2000). Recent studies further suggest that, in the course of BCoV infection, 3′ poly(A) tail lengths vary between 30 and 65 nts (Wu et al., 2013). This poly(A) tail length variation was confirmed to occur in beta- and gammacoronavirus infections and in a range of cell types, both in vitro and in vivo (Shien et al., 2014). The biological significance of these observations remains to be determined.
2.6. Identification of 3′ terminal RNA structural elements of HCoV-229E and HCoV-NL63
To identify putative cis-acting elements in the alphacoronavirus 3′ UTR, we used a range of RNA folding algorithms to identify RNA structural elements in the HCoV-229E and HCoV-NL63 3′-terminal genome regions encompassing the last 20 nts from the N gene and the entire 3′-UTR. Because of the length of these sequences (300 nts), many local secondary structures with similar free energies and base pair probabilities were identified and, thus, it proved to be difficult to make reliable predictions on stable RNA secondary structures in this region. As described above for the 5′ UTR, we therefore decided to use a combination of sequence and structural alignments of all currently recognized alphacoronavirus species to identify conserved RNA structures. The predictions were then further refined using structure probing data obtained for HCoV-229E and HCoV-NL63.
The validity of the approach was first tested by analyzing conserved RNA structural elements in the betacoronavirus 3′ UTR for which a large body of information has been obtained in previous structural and functional studies (see above). As shown in Fig. 5 , we were able to detect conserved RNA structural elements in the betacoronavirus 3′ UTR, including the BSL and the two SL structures that form the PK immediately downstream of the BSL. Consistent with previous studies (Goebel et al., 2004a), our predictions suggest that the formation of the PK requires structural rearrangements at the base of the BSL to permit the base-pairing interactions required to form PK stem 1, the latter involving the PK-SL2 loop sequence and the BSL 3′-terminal sequence (Fig. 5A and B). Interestingly, our analyses also revealed another conserved structural element, a short hairpin, immediately upstream of the PK-SL2. Formation of this hairpin would compete with base-pairing interactions required to form the basal part of the BSL and the PK stem 1, respectively (Fig. 5B). Furthermore, the hairpin overlaps partly with the PK loop 1 region that, in a previous study, was suggested to interact with the extreme 3′ end of the genome (Züst et al., 2008). The conservation of both structure and sequence of this hairpin suggest a biological function for this element. In this context, it may be worth mentioning that the hairpin structure is predicted to be disrupted by the 6-nt insertion in loop 1 that was reported previously to cause a poorly replicating and unstable phenotype in MHV (Goebel et al., 2004a). It remains to be seen if the small hairpin represents yet another element in the intricate network of base-pairing interactions between the BSL, the PK, and the 3′ end that together constitute the complex molecular switch proposed by the Masters laboratory (Goebel et al., 2004a). Consistent with previous studies, we identified only one conserved RNA structural element in the HVR downstream of PK-SL2. This stem-loop contained the conserved octanucleotide sequence in a single-stranded region. As pointed out above, the role of this element is currently unclear as both the HVR and octanucleotide sequence proved to dispensable for betacoronavirus (MHV) replication in vitro (Goebel et al., 2007, Liu et al., 2001). Overall, the alignment-based structure prediction algorithms used in our analysis led to conclusions that were consistent with results obtained in previous studies on betacoronavirus 3′ UTRs, suggesting that the approach might also be suitable to make reliable predictions on conserved RNA structural elements in the highly variable alphacoronavirus 3′ UTRs.
Fig. 5.
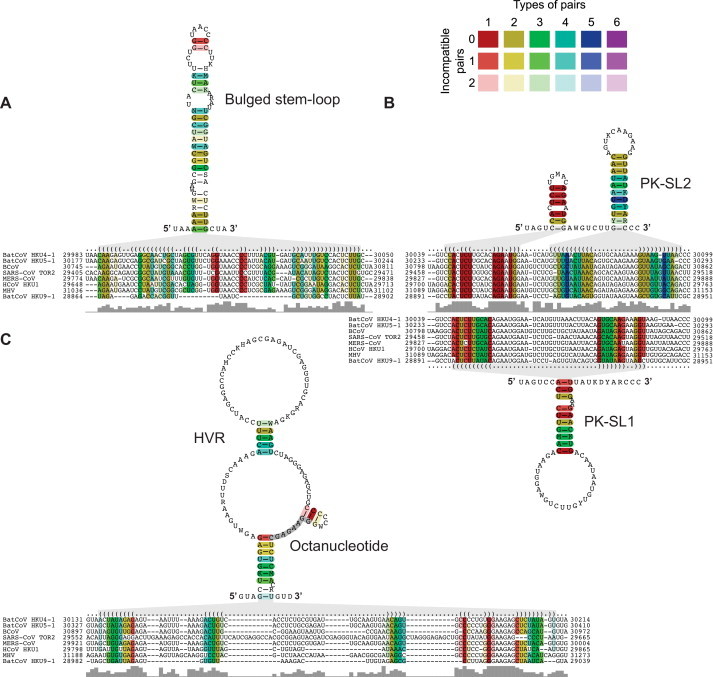
Alignment-based secondary structure prediction of betacoronavirus 3′ genome regions. The viruses included in this analysis represent all currently recognized lineages and species in the genus Betacoronavirus. The alignment was generated using LocARNA and the structure was calculated using RNAalifold. The consensus sequence is represented using the IUPAC code. Colors are used to indicate conserved base pairs: from red (conservation of only one base-pair type) to purple (all six base-pair types are found); from dark (all sequences contain this base pair) to light colors (1 or 2 sequences are unable to form this base pair). Gray bars below the alignment indicate the extent of sequence conservation. Gray shadows are used to link RNA structures with the corresponding dot-bracket notations above the alignment. (A) Alignment-based secondary structure prediction of the bulged stem-loop (BSL) in the 3′ UTR. (B) Alignment-based secondary structure prediction of the pseudoknot (PK) region in the 3′ UTR. Note that PK-SL1 is an alternate structure that requires base-pairing interactions between the loop region of PK-SL2 and the basal part of the BSL shown in (A). Formation of the BSL basal part and PK structure, respectively, are mutually exclusive (see text for details). (C) Alignment-based secondary structure prediction of the hypervariable region (HVR) in the 3′ UTR. To refine the alignment, an anchor at the highly conserved octanucleotide sequence was used.
Our alignment-based secondary structure predictions using representative viruses from all currently recognized alphacoronavirus species revealed the conservation of several RNA structural elements in the alphacoronavirus 3′ UTR. A counterpart of the betacoronavirus BSL structure (Goebel et al., 2004a, Hsue and Masters, 1997) could not be identified in the alphacoronavirus 3′ UTR while structural elements suitable to form a PK structure could be identified in all alphacoronaviruses (Fig. 6A). Interestingly, despite the absence of an upstream BSL in alphacoronaviruses, the formation of this putative PK structure is predicted to require the disruption of a short hairpin immediately upstream of PK-SL2, a scenario that is similar but less complex compared to the situation described above for betacoronaviruses. It remains to be investigated in further studies whether or not alphacoronaviruses employ a molecular switch mechanism similar to what has been described for betacoronaviruses (Goebel et al., 2004a).
Fig. 6.
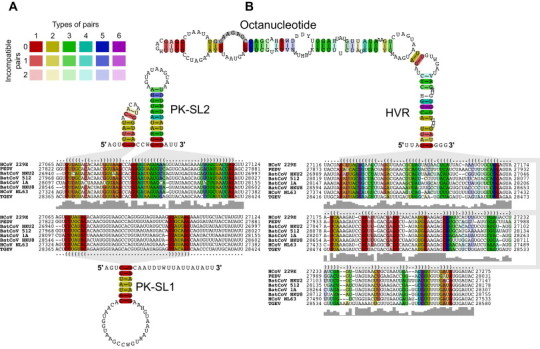
Alignment-based secondary structure prediction of alphacoronavirus 3′-terminal genome regions. The viruses included in this analysis represent all currently recognized species in the genus Alphacoronavirus. The alignment was calculated by LocARNA, the structure by RNAalifold. The consensus sequence is represented using the IUPAC code. Colors are used to indicate conserved base pairs: from red (conservation of only one base-pair type) to purple (all six base-pair types are found); from dark (all sequences contain this base pair) to light colors (1 or 2 sequences are unable to form this base pair). Gray bars below the alignment indicate the extent of sequence conservation at a given position. Gray shadows are used to link RNA structures with the corresponding dot-bracket notations above the alignment. (A) Alignment-based secondary structure prediction of the pseudoknot (PK) region in the 3′ UTR. Formation of the stem of PK-SL1 requires base-pairing interactions with the loop region of SL2. Formation of the PK and the two SL structures shown above the alignment are mutually exclusive (see text for details). (B) Alignment-based secondary structure prediction of the hypervariable region (HVR) in the 3′ UTR. To refine the alignment, an anchor at the highly conserved octanucleotide sequence was used.
The predicted SL2 structure (Fig. 6) could be confirmed by structure probing data obtained for HCoV-229E and HCoV-NL63 (Fig. 7 , detailed structure probing data to be published elsewhere). Furthermore, our structure probing data indicated base-pairing interactions upstream of SL2 in HCoV-NL63, supporting the formation of the predicted small hairpin in this region (Fig. 7A and B). By contrast, no such interactions were seen in HCoV-229E. Also, the structure probing data did not support the formation of a stable PK structure, possibly reflecting a similarly low thermodynamic stability as determined for the equivalent PK in betacoronaviruses (Stammler et al., 2011). Further studies including reverse genetics experiments are required to confirm the existence and biological significance of the predicted alphacoronavirus PK structure.
Fig. 7.
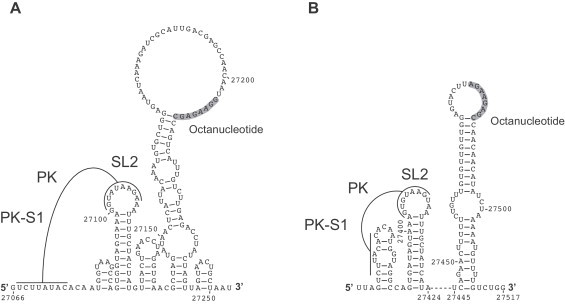
RNA secondary structure predictions of 3′-terminal genome regions of HCoV-229E (A) and HCoV-NL63 (B). Predictions were generated using RNAfold --noLP -C. As constraints, structure probing data were used. Formation of the predicted pseudoknot (PK) requires base-pairing interactions between the loop region of SL2 and an upstream sequence (and, possibly, structural rearrangements), resulting in the formation of PK stem 1 (PK-S1) as indicated. Also shown is the octanucleotide sequence that is conserved across all genera of the Coronavirinae.
With respect to the HVR downstream of PK-SL2, an extensive stem-loop structure was predicted in our analyses of alphacoronavirus 3′ UTRs (Fig. 6B). The structure is supported by a large number of covariant base pairs and contains the conserved octanucleotide sequence in a single-stranded region. The large distal part of the stem-loop structure was further corroborated by structure probing data (Fig. 7, details to be published elsewhere). In both HCoV-229E and HCoV-NL63, the octanucleotide sequence was found to be located in the loop region. Of note, the cell culture-adapted HCoV-NL63 isolate used for our structure probing analysis contained a short deletion apparently acquired upon serial passaging in cell culture, resulting in a significantly smaller loop but retaining the octanucleotide sequence (with one G-to-A replacement) in an identical position in the loop when compared to HCoV-229E (see Fig. 7A and B). This serendipitous deletion shows that the distal part of the extended stem-loop structure is not essential for HCoV-NL63 replication in cell culture. The data also indicate that, despite the deletion, the octanucleotide sequence retains a position in a loop region of the stem-loop structure and tolerates minimal changes, the latter being consistent with MHV reverse genetics data obtained for the HVR/octanucleotide region (Goebel et al., 2007).
3. Conclusions
Based on numerous studies on betacoronaviruses including structural, biochemical, and reverse genetic work (DI RNA and replication-competent virus), a picture of putative cis-acting elements is beginning to emerge (reviewed in Masters, 2007, Sola et al., 2011b). Previous work also identified a growing number of cellular and viral proteins that bind to these structures and likely have important functions at different steps of genomic and/or subgenomic RNA synthesis, genome packaging, genome expression or intracellular targeting of structures engaged in viral RNA synthesis (reviewed in Narayanan and Makino, 2007, Sola et al., 2011b).
Using a combination of bioinformatic and biochemical methods, the present study confirms and extends this previous work. Our study suggests that RNA secondary structure elements may be more conserved than previously thought, both within individual coronavirus genera and across different genera as shown here for the genera Alphacoronavirus and Betacoronavirus. Although significantly more work is needed to further characterize the structures identified in this and previous studies and understand their functional role, the available data suggest a cross-genus conservation of a number of RNA structural elements among alpha- and betacoronaviruses. The conservation pattern is consistent with the replicase gene-based classification of genera within the subfamily Coronavirinae (de Groot et al., 2012a). Conserved elements include stem-loops 1, 2, 4, and, possibly, 5 in the 5′-terminal genome region and a putative PK in the 3′ UTR. The data further suggest that, in both alpha- and betacoronaviruses, the formation of the PK may require structural rearrangements in other regions (upstream of the SL2) and it remains an attractive idea to suggest specific functions for these alternative structures whose mechanistic and functional details remain to be investigated (Goebel et al., 2004a, Züst et al., 2008). Finally, in line with previous observations (Brian and Baric, 2005, Masters, 2007, Sola et al., 2011b), the study confirms a significant degree of variation in the 3′-terminal region of the 3′ UTR, with the conserved octanucleotide sequence being consistently located in a single-stranded region of a stem-loop structure. Interestingly, specific lineages of beta-, gamma- and deltacoronaviruses (as well as other plus-strand RNA viruses) may contain another structural element, called s2m, in the 3′ UTR, thus further adding to the variability of this genome region in coronaviruses (Jonassen et al., 1998, Robertson et al., 2005, Tengs et al., 2013). The conservation pattern of the various lineage-specific structural elements argues against a conserved function in viral RNA synthesis but rather suggests that the 3′ UTR may contain elements that are involved in specific virus-host interactions and/or pathogenesis as has been shown for the MHV HVR (Goebel et al., 2007).
Acknowledgement
The work of R.M. and J.Z. is supported by the Deutsche Forschungsgemeinschaft (SFB 1021, A01).
References
- Almazán F., Galán C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., González J.M., Pénzes Z., Izeta A., Calvo E., Plana-Durán J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Baker S.C., Shieh C.K., Soe L.H., Chang M.F., Vannier D.M., Lai M.M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Binns M.M., Foulds I.J., Brown T.D. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J. Gen. Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lecine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus nsp10: a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Spaan W.J.M. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Denison M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005;340:209–223. doi: 10.1016/j.virol.2005.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.G., Nixon K.S., Senanayake S.D., Brian D.A. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 2007;81:7716–7724. doi: 10.1128/JVI.00549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y., Hofmann M.A., Sethna P.B., Brian D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y., Krishnan R., Brian D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., van den Born E., van den Worm S.H., Pleij C.W., Snijder E.J., Olsthoorn R.C. New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 2007;81:6771–6774. doi: 10.1128/JVI.02231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton K., Casais R., Shaw K., Stirrups K., Evans S., Britton P., Brown T.D., Cavanagh D. cis-Acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 2001;75:125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Family Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus taxonomy. Elsevier; Amsterdam: 2012. pp. 806–828. [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Cowley J.A., Enjuanes L., Faaberg K.S., Perlman S., Rottier P.J.M., Snijder E.J., Ziebuhr J., Gorbalenya A.E. Order Nidovirales. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus taxonomy. Elsevier; Amsterdam: 2012. pp. 785–795. [Google Scholar]
- de Groot R.J., van der Most R.G., Spaan W.J. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J. Virol. 1992;66:5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Volders H., Koetzner C.A., Masters P.S., Rottier P.J. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J.P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 2003;77:7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán C., Sola I., Nogales A., Thomas B., Akoulitchev A., Enjuanes L., Almazán F. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Miller T.B., Bennett C.J., Bernard K.A., Masters P.S. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007;81:1274–1287. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Taylor J., Masters P.S. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 2004;78:7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd, Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B.J., Su Y.P., Wu H.Y., Brian D.A. Genetic evidence of a long-range RNA-RNA interaction between the genomic 5′ untranslated region and the nonstructural protein 1 coding region in murine and bovine coronaviruses. J. Virol. 2012;86:4631–4643. doi: 10.1128/JVI.06265-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B.J., Wu H.Y., Brian D.A. An optimal cis-replication stem-loop IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J. Virol. 2011;85:5593–5605. doi: 10.1128/JVI.00263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.M., Guan B.J., Dziduszko A., Brian D.A. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 2009;83:6087–6097. doi: 10.1128/JVI.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M.A., Brian D.A. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J. Virol. 1991;65:6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M.A., Sethna P.B., Brian D.A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Hartshorne T., Masters P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Lai M.M. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J. Virol. 1999;73:9110–9116. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Ulferts R., Ziebuhr J., Canard B. SARS coronavirus replicative enzymes: structures and mechanisms. In: Lal S.K., editor. Molecular biology of the SARS-Coronavirus. Springer; Berlin/Heidelberg: 2010. pp. 99–114. [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeta A., Smerdou C., Alonso S., Penzes Z., Mendez A., Plana-Durán J., Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen C.M., Jonassen T.O., Grinde B. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Feng M., Schroeder M.E., Giedroc D.P., Leibowitz J.L. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 2006;80:10600–10614. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74:7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Liu P., Leibowitz J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N., Jeong Y.S., Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Patton C.D., Baric R.S., Stohlman S.A. Presence of leader sequences in the mRNA of mouse hepatitis virus. J. Virol. 1983;46:1027–1033. doi: 10.1128/jvi.46.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Li L., Giedroc D.P. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 2011;585:1049–1053. doi: 10.1016/j.febslet.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., Guohui C. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS ONE. 2013;8:e61166. doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B.G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Li H.P., Huang P., Park S., Lai M.M. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.P., Zhang X., Duncan R., Comai L., Lai M.M. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kang H., Liu P., Makkinje N., Williamson S.T., Leibowitz J.L., Giedroc D.P. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 2008;377:790–803. doi: 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.L., Lai M.M. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J. Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J., Lai M.M. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J., Liao C.L., Lai M.M. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J., Zhang X., Wu R.C., Lai M.M. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J. Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Keane S.C., Yang D., Leibowitz J.L., Giedroc D.P. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J. Virol. 2009;83:12084–12093. doi: 10.1128/JVI.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Millership J.J., Kang H., Leibowitz J.L., Giedroc D.P. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Millership J.J., Li L., Giedroc D.P., Leibowitz J.L. A previously unrecognized UNR stem-loop structure in the coronavirus 5′ untranslated region plays a functional role in replication. Adv. Exp. Med. Biol. 2006;581:25–30. doi: 10.1007/978-0-387-33012-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Yang D., Carter K., Masud F., Leibowitz J.L. Functional analysis of the stem loop S3 and S4 structures in the coronavirus 3′UTR. Virology. 2013;443:40–47. doi: 10.1016/j.virol.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Johnson R.F., Leibowitz J.L. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 2001;75:12105–12113. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Gerritsma H., Spaan W.J. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J. Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M.M. Studies of coronavirus DI RNA replication using in vitro constructed DI cDNA clones. Adv. Exp. Med. Biol. 1990;276:341–347. doi: 10.1007/978-1-4684-5823-7_46. [DOI] [PubMed] [Google Scholar]
- Makino S., Shieh C.K., Keck J.G., Lai M.M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C.K., Soe L.H., Baker S.C., Lai M.M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Fujiwara K. Defective interfering particles of mouse hepatitis virus. Virology. 1984;133:9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M.M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Genomic cis-acting elements in coronavirus RNA replication. In: Thiel V., editor. Coronaviruses – molecular and cellular biology. Caister Academic Press; Norfolk, United Kingdom: 2007. pp. 65–80. [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology, 6th ed., Vol. 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. pp. 825–858. [Google Scholar]
- Mendez A., Smerdou C., Izeta A., Gebauer F., Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L., Mateos-Gomez P.A., Capiscol C., del Palacio L., Enjuanes L., Sola I. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5′ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 2013;87:11579–11590. doi: 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. Coronavirus accessory proteins. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2008. pp. 235–244. [Google Scholar]
- Narayanan K., Makino S. Coronavirus genome packaging. In: Thiel V., editor. Coronaviruses – molecular and cellular biology. Caister Academic Press; Norfolk, UK: 2007. pp. 131–142. [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Penzes Z., Tibbles K., Shaw K., Britton P., Brown T.D., Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., Wroe C., Brown T.D., Britton P., Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H.L., Michot B., Bachellerie J.P. Improved methods for structure probing in large RNAs: a rapid ‘heterologous’ sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5′ terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983;11:5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Bouma P., Williams G.D., Brian D.A. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Brian D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79:12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Jr., Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:e5. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D., Wang T., Sawicki S. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 2001;82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.S., Kamahora T., Lai M.M. Sequence analysis of the nucleocapsid protein gene of human coronavirus 229E. Virology. 1989;169:142–151. doi: 10.1016/0042-6822(89)90050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., Grego S., Randell S.H., Baric R.S. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P.B., Hofmann M.A., Brian D.A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P.B., Hung S.L., Brian D.A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Lai M.M. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shien J.H., Su Y.D., Wu H.Y. Regulation of coronaviral poly(A) tail length during infection is not coronavirus species- or host cell-specific. Virus Genes. 2014 doi: 10.1007/s11262-014-1103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Galán C., Mateos-Gómez P.A., Palacio L., Zúñiga S., Cruz J.L., Almazán F., Enjuanes L. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J. Virol. 2011;85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Mateos-Gomez P.A., Almazan F., Zuñiga S., Enjuanes L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011;8:237–248. doi: 10.4161/rna.8.2.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B.A., Siddell S.G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo J.F., Hogue B.G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler S.N., Cao S., Chen S.J., Giedroc D.P. A conserved RNA pseudoknot in a putative molecular switch domain of the 3′-untranslated region of coronaviruses is only marginally stable. RNA. 2011;17:1747–1759. doi: 10.1261/rna.2816711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirrups K., Shaw K., Evans S., Dalton K., Cavanagh D., Britton P. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J. Gen. Virol. 2000;81:791–801. doi: 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.P., Fan Y.H., Brian D.A. Dependence of coronavirus RNA replication on an NH2-terminal partial nonstructural protein 1 in cis. J. Virol. 2014;88:8868–8882. doi: 10.1128/JVI.00738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2008;82:1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengs T., Kristoffersen A.B., Bachvaroff T.R., Jonassen C.M. A mobile genetic element with unknown function found in distantly related viruses. Virol. J. 2013;10:132. doi: 10.1186/1743-422X-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Tohya Y., Narayanan K., Kamitani W., Huang C., Lokugamage K., Makino S. Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 2009;83:5282–5288. doi: 10.1128/JVI.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R., Imbert I., Canard B., Ziebuhr J. Expression and functions of SARS coronavirus replicative proteins. In: Lal S.K., editor. Molecular biology of the SARS-Coronavirus. Springer; Berlin/Heidelberg: 2010. pp. 75–98. [Google Scholar]
- Ulferts R., Ziebuhr J. Nidovirus ribonucleases: Structures and functions in viral replication. RNA Biol. 2011;8:295–304. doi: 10.4161/rna.8.2.15196. [DOI] [PubMed] [Google Scholar]
- van den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Worm S.H., Eriksson K.K., Zevenhoven J.C., Weber F., Zust R., Kuri T., Dijkman R., Chang G., Siddell S.G., Snijder E.J., Thiel V., Davidson A.D. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS ONE. 2012;7:e32857. doi: 10.1371/journal.pone.0032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang X. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 2000;74:10571–10580. doi: 10.1128/jvi.74.22.10571-10580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Levis R., Schlesinger S. Evolution of virus and defective-interfering RNAs in BHK cells persistently infected with Sindbis virus. J. Virol. 1983;48:676–684. doi: 10.1128/jvi.48.3.676-684.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012;18:900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.K., Wang L., Sneed L.W., Collisson E.W. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28:19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Chang R.Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Guan B.J., Su Y.P., Fan Y.H., Brian D.A. Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5′-untranslated-region mutants. J. Virol. 2014;88:846–858. doi: 10.1128/JVI.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Guy J.S., Yoo D., Vlasak R., Urbach E., Brian D.A. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology. 2003;315:174–183. doi: 10.1016/S0042-6822(03)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Ke T.Y., Liao W.Y., Chang N.Y. Regulation of coronaviral poly(A) tail length during infection. PLoS ONE. 2013;8:e70548. doi: 10.1371/journal.pone.0070548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Ozdarendeli A., Brian D.A. Bovine coronavirus 5′-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J. Virol. 2006;80:2183–2193. doi: 10.1128/JVI.80.5.2183-2193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Liu P., Giedroc D.P., Leibowitz J. Mouse hepatitis virus stem-loop 4 functions as a spacer element required to drive subgenomic RNA synthesis. J. Virol. 2011;85:9199–9209. doi: 10.1128/JVI.05092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liao C.L., Lai M.M. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J. Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Coronavirus replicative proteins. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2008. pp. 65–81. [Google Scholar]
- Ziebuhr J., Herold J., Siddell S.G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J. The coronavirus replicase gene: special enzymes for special viruses. In: Thiel V., editor. Coronaviruses – molecular and cellular biology. Caister Academic Press; Norfolk, UK: 2007. pp. 33–63. [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Cruz J.L., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008;82:1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


