Highlights
-
•
Three new rhabdoviruses were detected in bats, China.
-
•
JHBV and BXBV found in Rhinolophus bats could be new members of the genus Vesiculovirus.
-
•
Four strains of YJBV detected in Hipposideros larvatus formed a new lineage.
Keywords: Bat, Rhabdovirus, Vesiculovirus, Genetic diversity
Abstract
The Rhabdoviridae is among the most diverse families of RNA viruses and currently classified into 18 genera with some rhabdoviruses lethal to humans and other animals. Herein, we describe genetic characterization of three novel rhabdoviruses from bats in China. Of these, two viruses (Jinghong bat virus and Benxi bat virus) found in Rhinolophus bats showed a phylogenetic relationship with vesiculoviruses, and sequence analyses indicate that they represent two new species within the genus Vesiculovirus. The remaining Yangjiang bat virus found in Hipposideros larvatus bats were only distantly related to currently known rhabdoviruses.
The family Rhabdoviridae within the order Mononegavirales is a large group of bullet- or rod-shaped viruses with great genetic diversity and complexity (Walker et al., 2015). The virions contain a single or segmented molecule of linear, negative sense single-stranded RNA of size 11–15 kb (Dietzgen et al., 2012, Lyles et al., 2013). Rhabdoviruses have been found replicating in plants, invertebrates and vertebrates, and many members are significant medical, veterinary, or agricultural pathogens, such as lyssaviruses, vesiculoviruses and ephemeroviruses (Lyles et al., 2013). The genus Lyssavirus is represented by the well-known rabies virus (RABV), one of the most wide spread and lethal infectious agents worldwide (Giesen et al., 2015), and also includes other members causing rabies-related human diseases with mortalities approaching 100%, such as Australian bat lyssavirus (ABLV) (Francis et al., 2014), European bat lyssavirus 2 (EBLV-2) (Nathwani et al., 2003) and Irkut virus (IRKV) (Leonova et al., 2009). The genus Vesiculovirus includes Vesicular stomatitis Indiana virus (VSIV), the most intensively studied prototype species for nonsegmented negative-stranded RNA viruses, which infects cattle, horses and pigs causing fever, vesicles in the mucosa of the oral cavity and in the skin of the coronary band and teat, and acute febrile disease in humans (Lyles et al., 2013). Bovine ephemeral fever virus (BEFV) of the genus Ephemerovirus infects cattle and water buffaloes and gives rise to an acute febrile illness, bovine ephemeral fever, which causes severe economic losses by malaise and decreased milk production in cattle (Lyles et al., 2013, Walker and Klement, 2015).
As the second diverse order of mammals (after rodents), bats are important natural reservoirs of viruses of which more than 200 have been discovered, including several agents that are highly pathogenic for humans such as Hendra and Nipah viruses (Halpin et al., 2000, Yob et al., 2001), SARS-related coronavirus (Li et al., 2005), and Ebola virus (Johnson et al., 2010). Bats also harbor and transmit diverse rhabdoviruses such as lyssaviruses, ledanteviruses and vesiculoviruses. Rabies virus of the genus Lyssavirus was the first lethal pathogen recognized to be harbored and spread by bats. Currently, 14 approved species are included within the genus, of which 12 originate from bats, exclusively or partially. Ledanteviruses hold strong ecological associations with bats, with viruses assigned to six of the 14 species discovered in bats, such as Mount Elgon bat virus (MEBV) and Oita virus (OITAV) (Iwasaki et al., 2004, Metselaar et al., 1969). Le Dantec virus (LDV), which is assigned to the type species (Le Dantec ledantevirus) of the genus Ledantevirus, was isolated from the serum of a Senegalese girl with acute febrile illness and signs of hepatosplenomegaly (Cropp et al., 1985), and its RNA was detected in a serotine bat, Eptesicus isabellinus (Vazquez-Moron et al., 2008). Within the genus Vesiculovirus, only one bat-associated member has been reported: American bat vesiculovirus (ABVV), detected in big brown bats (Eptesicus fuscus) in the USA and forming a distinct lineage of vesiculovirus (Ng et al., 2013).
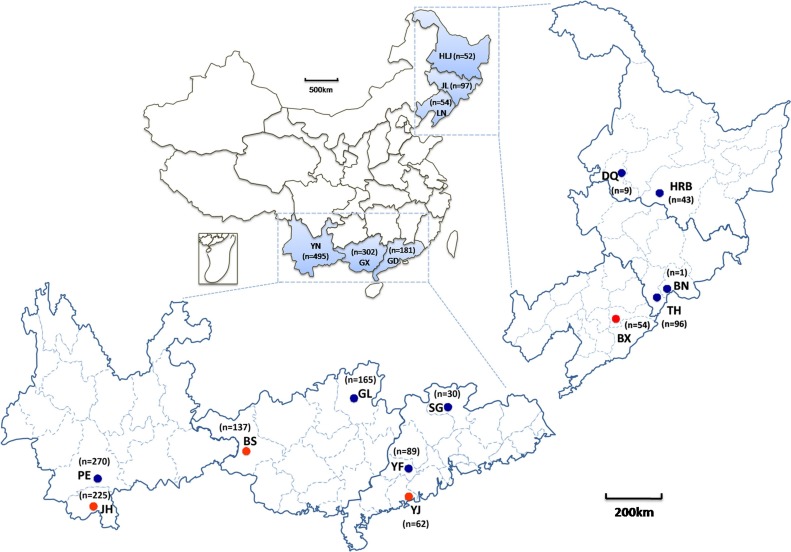
To investigate bat virus ecology in China, 1181 bats were collected at 13 locations in the south (Yunnan Province, Guangdong Province and Guangxi Zhuang Autonomous Region) and northeast (Liaoning Province, Jilin Province and Heilongjiang Province) between 2005 and 2014 (Fig. 1 and Table S1). The collection was comprised of 25 species within 12 genera of 5 families: Rhinolophidae (n = 302), Hipposideridae (n = 483), Vespertilionidae (n = 233), Emballonuridae (n = 63), and Pteropodidae (n = 100). Intestines (with contents) and lungs of bats from each location were pooled and subjected to viral metagenomic analysis as per our published method (He et al., 2013). The procedures for sampling of bats in this study were reviewed and approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary, Academy of Military Medical Sciences, China (Laboratory Animal Care and Use Committee Authorization, permit number: JSY-DW-2015-01). All live bats were maintained and handled according to the Principles and Guidelines for Laboratory Animal Medicine (2006), Ministry of Science and Technology, China.
Fig. 1.
Details of bat collection in Yunnan Province, Guangdong Province, Liaoning Province, Jilin Province, Heilongjiang Province and the Guangxi Zhuang Autonomous Region. Circles: bat-sampling locations. Samples positive for rhabdoviruses are identified in red, with negatives in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A total of 210 reads with lengths of ∼140 nucleotides (nt) were annotated to rhabdovirus, corresponding to the RNA-dependent RNA polymerase domain of the L gene. These reads could be classified into two groups, showing identities of 84% (nt) and 91% (amino acid, aa) with vesiculoviruses and 68% (nt) and 52% (aa) with lyssaviruses. The reads and their phylogenetic neighbors from GenBank were used as a template to design degenerate nested or semi-nested PCR primers targeting the conserved L gene (252–715 nt, shown in Table S2). Viral RNA of each sample was extracted and submitted to RT-PCR screening. Results showed that eight bats were positive for rhabdoviruses: an intermediate horseshoe bat (Rhinolophus affinis) (4.2%, 1/24) in Jinghong City, Yunnan; three greater horseshoe bats (Rhinolophus ferrumequinum) (18.8%, 3/16) in Benxi City, Liaoning; two intermediate roundleaf bats (Hipposideros larvatus) in Baise City, Guangxi (4.1%, 2/49) and two in Yangjiang City, Guangdong (16.7%, 2/12) (Table 1 ).
Table 1.
Bat rhabdoviruses detected by PCR.
| Sampling location | Hosta | Tissue | Virus | Positive rate (positive No./total No.) |
|---|---|---|---|---|
| Jinghong, Yunnan | Rh. affinis | intestines | JHBV | 4.2% (1/24) |
| Benxi, Liaoning | Rh. ferrumequinum | intestines & lungs | BXBV | 18.8% (3/16) |
| Yangjiang, Guangdong | Hi. larvatus | intestines | YJBV | 16.7% (2/12) |
| Baise, Guangxi | Hi. larvatus | intestines | YJBV | 4.1% (2/49) |
Rh.: Rhinolophus; Hi.: Hipposideros.
We tried several times to isolate the rhabdoviruses by BHK-21 and Vero E6 cells as well as 3-day-old Kunming mice, however, results of RT-PCR detection of the cell culture and mouse internal organs were negative, resulting in a failure to isolate any virus.
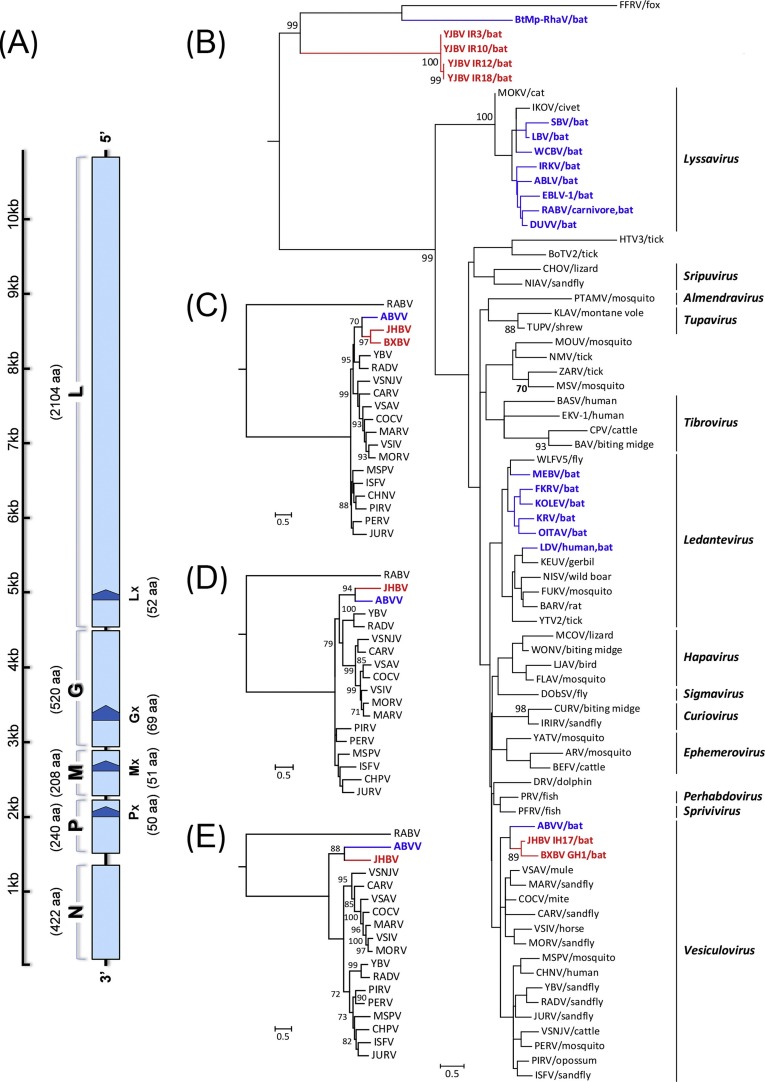
The amplicons obtained (252 bp) were aligned by ClustalW with representatives of all rhabdoviruses except five genera of plant and fish rhabdoviruses (Cytorhabdovirus, Dichorhavirus, Nucleorhabdovirus, Varicosavirus and Novirhabdovirus) as well as some currently unclassified rhabdoviruses, such as Bat rhabdovirus (BtMp-RhaV/SX2013) (Wu et al., 2016), Muir Springs virus (MSV) (Walker et al., 2015) and Huangpi tick virus 3 (HTV3) (Li et al., 2015). Details of reference sequences were shown in Table S3. Identities were calculated using MegAlign (Table S4) and the phylogenetic tree was constructed by MEGA7 using the Maximum Likelihood method with 1000 bootstrap values under the General Time Reversible model (Fig. 2 B) (Kumar et al., 2016). Rhabdoviruses from Jinghong (Jinghong bat virus, JHBV) and Benxi (Benxi bat virus, BXBV) showed 82.7% nt and 96.5% aa identities between each other and clustered together within the genus Vesiculovirus sharing the highest 87.2% aa identity with ABVV; whereas four strains of Yangjiang and Baise formed an individual clade with 95.7% nt and 97.7% aa identity, revealing they are likely to be the same virus and named Yangjiang bat virus (YJBV), they showed only distant phylogenetic relationships with currently known rhabdoviruses, and shared the highest nt identity of 60.4–62.2% with an unclassified fox fecal rhabdovirus (FFRV) (Bodewes et al., 2014), and only 49.5–53.8% (nt) with lyssaviruses.
Fig. 2.
(A) Schematic genome organization of JHBV with five canonical ORFs (3′-N-P-M-G-l-5′, light blue) and four overlapping accessory ORFs (≥150 nt, Px, Mx, Gx and Lx, dark blue) being predicted. Phylogenetic tree of rhabdoviruses was generated based on 252-nt sequences of the L gene (B); and trees of vesiculoviruses were generated based on 1591-nt sequences of L gene (C), complete sequences of N (D) and G (E) gene, with the RABV as outgroup. The evolutionary history was inferred in MEGA7 by using the Maximum Likelihood method based on the General Time Reversible model. Bootstrap support values of 1000 replicates (≥70%) are shown and the scale bars indicate nucleotide substitutions per site. The sequences obtained in the present study are identified in red, and other bat-associated rhabdoviruses in blue. Definitions of virus abbreviations and their GenBank accession numbers are listed in Table S3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To determine the complete genomes of these viruses, high-throughput sequencing, PCR, genome walking (Genome walking kit, TaKaRa) and Rapid Amplification of cDNA Ends (RACE, Invitrogen) were employed (primer information as shown in Table S2). Results showed that only the complete genome of JHBV (10,866 nt) and partial sequence of the L gene of BXBV (1591 nt) were successfully obtained. The accession numbers of sequences obtained were MF279192-MF279196 and KX343071. The genome of JHBV possesses the typical genomic organization of vesiculovirus comprised of five classical genes and 3′ leader and 5′ trailer sequences (62 nt and 60 nt respectively) (Fig. 2A and Table S5). Matrix (M) gene of JHBV (627 nt, 208 aa) is the shortest in length compared to other vesiculoviruses (633–762 nt, 210–253 aa) and an accessory Mx gene of 156 nt (51 aa) was predicted. Its phosphoprotein (P) gene (723 nt, 240 aa) is the smallest within the genus apart from ABVV (699 nt, 232 aa), and an accessory Px gene (153 nt, 50 aa) was observed. The nucleoprotein (N), glycoprotein (G) and L genes of vesiculoviruses are usually used for genotyping, and those of JHBV show normal sizes of 1269 nt (422 aa), 1563 nt (520 aa) and 6315 nt (2104 aa) respectively, with alternative nested ORFs Gx (210 nt, 69 aa) and Lx (159 nt, 52 aa) observed in the G and L genes. Intergenic sequences of JHBV contained two conserved motifs: the transcription termination/polyadenylation (TTP), 3′-CUUUUUUU-5′, for the upstream gene, and transcription initiation (TI), 3′-UUGU-5′, for the downstream gene.
The 1591-nt sequences of the L gene of JHBV and BXBV were used to further determine the genetic relationships (Table S4). Similar to the above result obtained by 252-nt sequences, the 1591-nt (530-aa) sequences of JHBV and BXBV shared the highest identity with each other (nt 68.6% and aa 75.6%) and clustered together in the phylogenetic tree (Fig. 2C). When to other strains, both JHBV and BXBV showed the highest aa identity of 66.6% and 69.8% with ABVV respectively; and at the nucleotide level, JHBV held the highest identity of 64.5% with ABVV, while BXBV shared the closest relationship of 64.9% with Chandipura virus (CHNV) (Gurav et al., 2010). Complete sequences of the whole genome and genes of JHBV was used to analyze the homology with other animal-associated rhabdoviruses, showing its nearest relationship with ABVV either (Table S6); and the N and G gene were also used to determine its phylogenetic relationship with other known vesiculoviruses, results showed its individual clade position and distinct clustering with ABVV (Fig. 2D and E).
Approved members of the Rhabdoviridae have expanded dramatically following recently released virus taxonomy reports of the International Committee on the Taxonomy of Viruses (ICTV), which show that rhabdoviruses are far more diverse than previously thought. In 2011, the ICTV listed 54 species, increasing to 75 in the 2015 report. By 2016, five new genera Almendravirus, Curiovirus, Hapavirus, Ledantevirus and Sripuvirus with 69 novel species had been approved, bringing a total of 131 species into 18 genera with additional numerous unclassified viruses (Adams et al., 2017, Blasdell et al., 2015, Contreras et al., 2017, Li et al., 2015, Walker et al., 2015). According to the species demarcation criteria of Vesiculovirus proposed by ICTV, JHBV, identified here, meets the requirements for a new vesiculovirus species. JHBV possesses high amino acid sequence divergence from other vesiculoviruses in its N, L and G proteins, with minima of 42.5%, 40.9% and 51.8% (to ABVV) respectively (Table S6). This significantly exceeds the proposed minimum divergence of 5.0%, 15.0% and 10.0%; additionally, its accessory ORFs differ from all known species in terms of lengths and locations (Fig. 2A and Table S5). For the same reasons, BXBV might also be considered a new vesiculovirus species even though only a partial L gene (1591 nt) was obtained, since this showed a minimum amino acid sequence divergence of 23.7% to others, including JHBV, far surpassing the proposed minimum divergence of 10.0%. Of note is that we have identified a new clade of rhabdoviruses with distant genetic relationship to other known members. We made every effort, including high-throughput sequencing, to determine their full genomes, but due to lack of reference sequences and insufficient samples, we failed to do so. Although their partial L genes cannot present solid evidence for precise assessment of their phylogenetic status, the limited data obtained here suggest that Chinese bats harbor a lineage of rhabdoviruses that could be prototypes of a new genus.
Among the 25 bat species collected in six provinces with different geographic distributions and climate, only three species were positive for rhabdoviruses. Previous research on bat-borne lyssaviruses screened five bat species (261 bats) in Jilin Province, with only one Murina leucogaster being found positive (Liu et al., 2013). Another study on the bat virome of 40 bat species (4440 individuals) in 29 provinces in China also showed that a single rhabdovirus (BtMp-RhaV/SX2013) was identified in only one species (Myotis pequinius) (Wu et al., 2016). These results indicate that the distribution of rhabdoviruses in Chinese bats probably has a host preference and a geographical constraint. The sampling sites of YJBV, Baise City and Yangjiang City, were in neighboring provinces, Guangxi and Guangdong although 600 km apart, and the close relationship among the strains suggests that a hitherto unknown lineage of rhabdoviruses is circulating in bats in the Guangxi-Guangdong region.
Conflict of interest
All authors declare no conflict of interest.
Acknowledgements
This work was supported by General Program of the National Natural Science Foundation of China (31572529), the National Key Basic Research and Development Program of China (2016YFC1200100) and the Science and Technology Basic Work Program from the Ministry of Science and Technology of China (2013FY13600). The funders played no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.virusres.2017.11.028.
Contributor Information
Biao He, Email: heb-001001@163.com.
Changchun Tu, Email: changchun_tu@hotmail.com.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017) Arch. Virol. 2017;162(8):2505–2538. doi: 10.1007/s00705-017-3358-5. [DOI] [PubMed] [Google Scholar]
- Blasdell K.R., Guzman H., Widen S.G., Firth C., Wood T.G., Holmes E.C., Tesh R.B., Vasilakis N., Walker P.J. Ledantevirus: a proposed new genus in the Rhabdoviridae has a strong ecological association with bats. Am. J. Trop. Med. Hyg. 2015;92(2):405–410. doi: 10.4269/ajtmh.14-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Ruiz-Gonzalez A., Schapendonk C.M.E., van den Brand J.M.A., Osterhaus A.D., Smits S.L. Viral metagenomic analysis of feces of wild small carnivores. Virol. J. 2014;11(1):89–101. doi: 10.1186/1743-422X-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M.A., Eastwood G., Guzman H., Popov V., Savit C., Uribe S., Kramer L.D., Wood T.G., Widen S.G., Fish D., Tesh R.B., Vasilakis N., Walker P.J. Almendravirus: a proposed new genus of rhabdoviruses isolated from mosquitoes in tropical regions of the Americas. Am. J. Trop. Med. Hyg. 2017;96(1):100–109. doi: 10.4269/ajtmh.16-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropp C.B., Prange W.C., Monath T.P. LeDantec virus: identification as a rhabdovirus associated with human infection and formation of a new serogroup. J. Gen. Virol. 1985;66(Pt 12):2749–2754. doi: 10.1099/0022-1317-66-12-2749. [DOI] [PubMed] [Google Scholar]
- Dietzgen R.G., Calisher C.H., Kurath G., Kuzman I.V., Rodriguez L.L., Stone D.M., Tesh R.B., Tordo N., Walker P.J., Wetzel T., Whitfield A.E. Rhabdoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. vol. 1. Elsevier; San Diego, CA: 2012. pp. 654–681. (Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses). [Google Scholar]
- Francis J.R., Nourse C., Vaska V.L., Calvert S., Northill J.A., Mccall B., Mattke A.C. Australian Bat Lyssavirus in a child: the first reported case. Pediatrics. 2014;133(4):e1063. doi: 10.1542/peds.2013-1782. [DOI] [PubMed] [Google Scholar]
- Giesen A., Gniel D., Malerczyk C. 30 years of rabies vaccination with Rabipur: a summary of clinical data and global experience. Expert Rev. Vaccines. 2015;14(5):775–783. doi: 10.1586/14760584.2015.1027863. [DOI] [PubMed] [Google Scholar]
- Gurav Y.K., Tandale B.V., Jadi R.S., Gunjikar R.S., Tikute S.S., Jamgaonkar A.V., Khadse R.K., Jalgaonkar S.V., Arankalle V.A., Mishra A.C. Chandipura virus encephalitis outbreak among children in Nagpur division, Maharashtra, 2007. Indian J. Med. Res. 2010;132(4):395–399. [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81(Pt 8):1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- He B., Li Z., Yang F., Zheng J., Feng Y., Guo H., Li Y., Wang Y., Su N., Zhang F., Fan Q., Tu C. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS One. 2013;8(4):e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T., Inoue S., Tanaka K., Sato Y., Morikawa S., Hayasaka D., Moriyama M., Ono T., Kanai S., Yamada A., Kurata T. Characterization of Oita virus 296/1972 of Rhabdoviridae isolated from a horseshoe bat bearing characteristics of both lyssavirus and vesiculovirus. Arch. Virol. 2004;149(6):1139–1154. doi: 10.1007/s00705-003-0271-x. [DOI] [PubMed] [Google Scholar]
- Johnson N., Vos A., Freuling C., Tordo N., Fooks A.R., Muller T. Human rabies due to lyssavirus infection of bat origin. Vet. Microbiol. 2010;142(3–4):151–159. doi: 10.1016/j.vetmic.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova G.N., Belikov S.I., Kondratov I.G., Krylova N.V., Pavlenko E.V., Romanova E.V., Chentsova I.V., Petukhova S.A. A fatal case of bat lyssavirus infection in Primorye Territory of the Russian Far East. Rabies Bull. Eur. 2009;33(4):5–8. [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li C.X., Shi M., Tian J.H., Lin X.D., Kang Y.J., Chen L.J., Qin X.C., Xu J., Holmes E.C., Zhang Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S., Zhao J., Zhang F., Hu R. Isolation of Irkut virus from a Murina leucogaster bat in China. PLoS Negl. Trop. Dis. 2013;7(3):e2097. doi: 10.1371/journal.pntd.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles D.S., Kuzmin I.V., Rupprecht C.E. Rhabdoviridae. In: David M., Knipe P.M.H., editors. 6th ed. vol. 1. Lippincott, Williams & Wilkins; Philadelphia: 2013. pp. 885–922. (Fields Virology). [Google Scholar]
- Metselaar D., Williams M.C., Simpson D.I., West R., Mutere F.A. Mount elgon bat virus: a hitherto undescribed virus from Rhinolophus hildebrandtii eloquens K. Anderson. Arch. Virol. 1969;26(1):183–193. doi: 10.1007/BF01241186. [DOI] [PubMed] [Google Scholar]
- Nathwani D., Mcintyre P.G., White K., Shearer A.J., Reynolds N., Walker D., Orange G.V., Fooks A.R. Fatal human rabies caused by European bat Lyssavirus type 2a infection in Scotland. Clin. Infect. Dis. 2003;37(4):598. doi: 10.1086/376641. [DOI] [PubMed] [Google Scholar]
- Ng T.F., Driscoll C., Carlos M.P., Prioleau A., Schmieder R., Dwivedi B., Wong J., Cha Y., Head S., Breitbart M., Delwart E. Distinct lineage of vesiculovirus from big brown bats, United States. Emerg. Infect. Dis. 2013;19(12):1978–1980. doi: 10.3201/eid1912.121506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Moron S., Juste J.C., Aznar C., Ruiz-Villamor E., Echevarria J. Asymptomatic rhabdovirus infection in meridional serotine bats (Eptesicus isabellinus) from Spain. Dev. Biol. 2008;131:311–316. [PubMed] [Google Scholar]
- Walker P.J., Klement E. Epidemiology and control of bovine ephemeral fever. Vet. Res. 2015;46:124. doi: 10.1186/s13567-015-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Firth C., Widen S.G., Blasdell K.R., Guzman H., Wood T.G., Paradkar P.N., Holmes E.C., Tesh R.B., Vasilakis N. Evolution of genome size and complexity in the rhabdoviridae. PLoS Pathog. 2015;11(2):e1004664. doi: 10.1371/journal.ppat.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., He G., Zhang J., Yang J., Qian Z., Dong J., Sun L., Zhu Y., Du J., Yang F., Zhang S., Jin Q. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10(3):609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yob J., Field H., Rashd i.A., Morrissy C., van der H.B., Rota P. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001;7(3):439–441. doi: 10.3201/eid0703.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.