Abstract
Homeobox D 10 (HOXD10) is important in cell differentiation and morphogenesis and serves as a tumor suppressor gene (TSG) in a number of malignancies. The present study investigated its promoter methylation status and association with the clinicopathological features of endometrial cancer (EC), and measured HOXD10 protein expression levels. EC samples (n=62), including 50 endometroid adenocarcinoma (EA) and 12 mucinous endometrial carcinoma samples (EC) and 70 non-cancerous samples were collected. All samples were evaluated for the methylation status of several TSGs, including HOXD10, using methylation-specific PCR. HOXD10 expression level was evaluated using immunohistochemistry. 5-Aza-2-deoxycytidine treatment was performed in the EC cell line Ishikawa to observe the change in HOXD10 expression levels. HOXD10 promoter methylation was more frequent in cancer samples (P<0.001). Downregulation of HOXD10 in EC samples was confirmed at the protein level using immunohistochemistry (P<0.001) and immunohistochemical staining was negatively associated with methylation status (P<0.05). Less HOXD10 protein was expressed in MEC compared with EA samples (P<0.001). The HOXD10 promoter was hypermethylated in both EA and MEC, causing decreased HOXD10 protein expression levels in EC cells. HOXD10 expression levels were partially reversed by 5-Aza-2-deoxycytidine treatment. The results of the present study demonstrated that epigenetic silencing of HOXD10 putatively contributed to the tumorigenesis of EA. Although there was no significant difference in HOXD10 methylation between EA and MEC, HOXD10 protein expression levels differed between these two diseases, indicating that it may be a useful protein biomarker for distinguishing between these two lesions.
Keywords: endometroid carcinoma, mucinous endometrial carcinoma, DNA methylation, HOXD10
Introduction
Endometrial carcinoma (EC) is one of the most common gynecological malignancies in developed countries, accounting for 20–30% of female malignancies, with a prevalence that is increasing annually (1,2). Early diagnosis and treatment has allowed a possible 5-year survival rate of 90% (3,4). Mucinous endometrial carcinoma (MEC) is an independent and infrequent pathological pattern of EC and accounts for 1–9% of all uterine carcinomas (5). Previous studies describing its pathomorphism and diagnostic criteria have suggested that MEC and endometroid adenocarcinoma (EA) differ with respect to patient age and lymphatic metastasis (6–9). MEC is more frequent in older women, and is more likely to involve lymphatic metastasis (6–9). Compared with EA, MEC samples exhibit decreased paired box 2 expression levels, low TP53 mutation rates and increased CD10, estrogen receptor (ER), progesterone receptor (PR), K-ras, p16, c-MET, epidermal growth factor receptor (EGFR), PTEN and PD-L1 expression levels and sporadic promoter hypermethylation of the mutL homolog 1 gene (10–15).
Like many malignancies, endometrial cancer is a complex disease driven by genetic, epigenetic and environmental factors (16–18). DNA methylation at the 5-C of a cytosine ring of a CpG island is one of the most important epigenetic alterations in cancer initiation (19). Studies have demonstrated that aberrant DNA methylation is associated with malignant formation (20–23). Tumor suppressor genes (TSG) may be hypermethylated, which leads to decreased expression levels and alteration of cell growth, differentiation, proliferation and apoptosis, resulting in the development of tumors (20–23). Therefore, assaying DNA methylation status may be a method for early diagnosis (24,25).
Hypermethylation of promoters of a number of TSGs, such as EGF-containing fibulin extracellular matrix protein 1, glutathione S-transferase P1, suppressor of cytokine signaling 3, 3OST2, basic helix-loop-helix family member E22/cysteine deoxygenase 1/CUGBP Elva-like family member 4, SHP1 and transmembrane protein with EGF-like and two follistatin-like domains 2, have been studied in certain EC tissues and cell lines (such as Ishikawa and KLE) and the methylation frequency is high (26–31). The present study investigated five TSGs (HOXD10, SHH, ZNF545, PCDH17 and MEIS1) whose promoters are reported to be hypermethylated in other malignancies but have not been evaluated for methylation in EC (32–40). For example, HOXD10 is hypermethylated and lowly expressed in colon adenocarcinoma cells, which downregulates the RHOC/AKT/MAPK pathway to enhance apoptosis and restrict the proliferation, migration and invasion of colon adenocarcinoma (32,33). Methylation of the Shh promoter and reduced expression of SHH is frequent in basal cell carcinoma (34,35). ZNF545 functions as a tumor suppressor in colorectal cancer and is frequently inactivated by promoter methylation (36,37). Hypermethylation of PCDH17 is correlated to poor prognosis in acute lymphoblastic leukemia (38,39). MEIS1 genes are frequently hypermethylated in different types of leukemia (40). The present study measured the methylation statuses of these genes, and then selected the most hypermethylated genes and measured protein expression levels in tumor and control samples. The present study aimed to analyze the methylation status of the TSG in EC and to elucidate the association between methylation, expression levels and clinicopathological characteristics. In order to further validate the association between the methylation status of a gene and its expression level, the present study detected the expression level of HOXD10 before and after DNA methylation transferase inhibitor treatment.
Materials and methods
Patients and samples
A total of 132 well-conserved paraffin-embedded tissue blocks were obtained from the Department of Pathology, Shandong University Qilu Hospital (Shandong, China), which were initially taken between 2006 and 2015, including endometrial adenocarcinoma (n=50), mucinous endometrial carcinoma (n=12), simple hyperplasia (n=22) and complex hyperplasia (n=48). Patient data, such as age, tumor differentiation, depth of myometrial invasion and lymph node metastasis, were collected. Case diagnoses were made according to World Health Organization Classification of Tumors of Female Reproductive Organs (4th edition) (41). The present study was approved by the Ethical Research Committee at Shandong University (approval no. mecsdums 2012032).
Cell lines and culture
The human endometrial carcinoma cell line Ishikawa was obtained from the European Collection of Cell Cultures (Sigma-Aldrich; Merck KGaA) and maintained at 37°C in Dulbecco's modified Eagle's medium/Ham's F-12 medium (DMEM/F12; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). Cell lines were treated at 37°C for 72 h with 5-Aza-2′-deoxycytidine (5-Aza-CdR; Sigma-Aldrich; Merck KGaA) at a concentration of 1, 2, 5 µM as a demethylation treatment. Media and 5-Aza-CdR were replaced every 24 h.
Methylation-specific PCR (MSP)
DNA samples were extracted from paraffin-embedded tissue blocks using a genomic DNA purification kit (Qiagen GmbH), and treated to convert unmethylated cytosine to uracil using a cpGenome DNA modification kit (InterGen Co.). Primer sequences, conditions and product length for MSP are presented in Table SI. MSP reactions were performed in a 10 µl system, using 1 µl (20 ng) template DNA for each reaction. H2O was used as a negative control.
Immunohistochemistry
A two-step method was used according to the manufacturer's instructions regarding primary antibody and secondary antibody described below. Sections (5-µm thick) were pre-treated using sodium citrate buffer (pH 6.0; 0.01 mol/l; Beijing Solarbio Science & Technology Co., Ltd.) at 98°C for 5 min to retrieve cell antigens and blocked with goat serum (1:10; cat. no. ZLI-9021; OriGene Technologies, Inc.) at room temperature for 20 min. Sections were incubated overnight at 4°C with primary antibody (anti-HOXD10; 1:150; cat. no. ab172865; Abcam). Colonic carcinoma tissue sections (5-µm thick) were used as the positive control. The primary antibody was replaced with phosphate-buffered saline and was used as a negative control. The sections were incubated with horseradish peroxidase-labeled secondary antibody (1:100; Universal PV9000 kit; OriGene Technologies) for 30 min at 37°C and visualized using DAB (1:20; Beijing Solarbio Science & Technology Co., Ltd.). The sections were stained with hematoxylin for 2 min at room temperature. Optical microscope (magnification, ×100 and ×400; Carl Zeiss AG) was used for visualization.
A semi-quantitative scoring system was used to obtain a staining score for HOXD10 expression levels. A total of five high power fields from each section were randomly selected, and scores were assigned according to intensity and percentage of stained cells. Scores were averaged to yield a final score. Intensity scores were classified according to staining intensity: Negative staining (staining intensity score, 0); positive yellow staining (staining intensity score, 1); positive brown staining (staining intensity score ≤2); and positive dark brown staining (staining intensity score, 3). Percentage score was classified according to percentage of stained cells: No staining (score =0); positive staining 0–10% (score, 1); positive staining 11–50% (score, 2); positive staining 50–80% (score, 3); 80–100% (score, 4). A two-score average was used as the score of the fields. Final scores of 0–3 were considered negative, while scores >4 were considered positive.
Western blot analysis
Ishikawa cells were harvested and subjected to protein extraction with RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.). The concentration of protein was measured using the BCA method (BCA Protein Assay kit; cat. no. PC0020; Beijing Solarbio Science & Technology Co., Ltd.). Equal amounts of protein (50 µg) were separated by 10% SDS-PAGE gel and transferred to PVDF membranes (EMD Millipore). After blocking with 5% skim milk (Beijing Solarbio Science & Technology Co., Ltd.) at room temperature for 1 h, the membranes were incubated overnight at 4°C with primary antibodies as follows: Anti-HOXD10 (1:100; cat. no. ab172865; Abcam) and anti-β-actin (1:1,000; cat. no. 4970T; Cell Signaling Technology, Inc.). The immune complexes were incubated with horseradish peroxidase-conjugated secondary antibody. The blots were developed using chemiluminescence (EMD Millipore) with the Las-4000 Imaging system (Fujifilm).
Statistical analysis
SPSS software (version 22.0; IBM, Corp.) was used for statistical analysis. A Pearson χ2 test was used to compare gene methylation status with clinical data among patients with cancer. P<0.05 was considered to indicate a statistically significant difference.
Results
HOXD10 promoter hypermethylation in EC
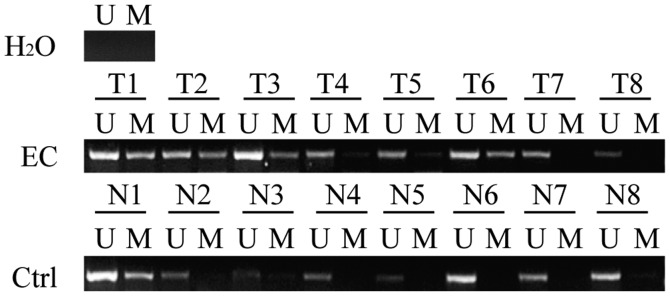
HOXD10 promoter methylation measured by MSP was greater in cancer samples compared with non-cancerous samples (Fig. 1 and Table I). Hypermethylation of SHH, ZNF545, PCDH17 and MEIS1 in EA and MEC occurred, but a similar methylation frequency was observed in non-cancerous lesions (Fig. S1).
Figure 1.
HOXD10 promoter hypermethylation in EC. Methylation statuses of eight out of 62 cancer samples and eight out of 70 non-cancerous samples. U, unmethylated HOXD10; M, methylated HOXD10; EC, endometrial carcinoma; N, non-cancerous tissue; T, tumor tissue. H2O were used as negative control.
Table I.
MSP data for HOXD10 in primary endometrial lesions and clinical pathological correlations.
| HOXD10 | |||
|---|---|---|---|
| Category | Methylated | Unmethylated | P-value |
| Simple hyperplasia | 4 | 18 | 0.4750 |
| Complex and atypical hyperplasia | 7 | 41 | |
| Endometroid adenocarcinoma | 35 | 15 | 0.5380 |
| Mucinous endometrial | 8 | 4 | |
| Total endometrial carcinoma | 43 | 19 | <0.0001 |
| Total control group | 11 | 59 | |
HOX, homeobox.
Decreased expression levels of HOXD10 in EC
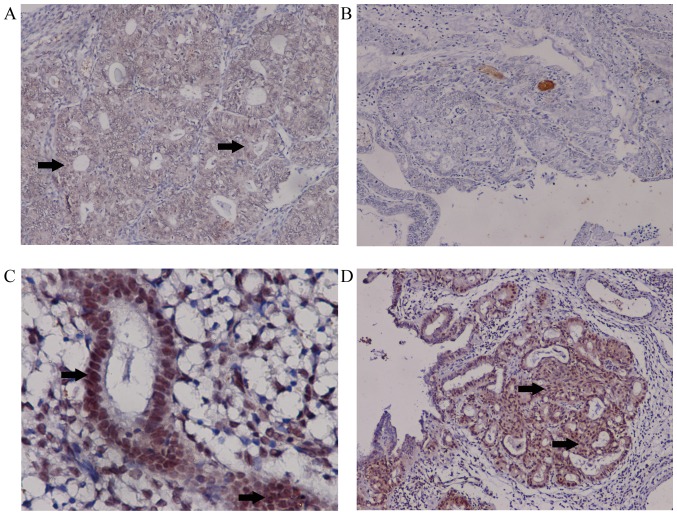
HOXD10 expression levels were measured in cancer and control samples. Downregulation of HOXD10 in cancer samples was confirmed at the protein level using immunohistochemistry. HOXD10 expression levels were decreased in cancer cases (EA and MEC) compared with the controls (37/62 vs. 14/70; P<0.01). Compared with EA, MEC samples did not express HOXD10 (P<0.001; Table II). In EC, positive staining for HOXD10 occurred in the cytoplasm. For simple and complex hyperplasia, positive staining was evident in the nucleus and cytoplasm (Fig. 2). Staining of colonic carcinoma tissues was provided as positive control (Fig. S2).
Table II.
Immunohistochemistry data of HOXD10 in primary endometrial lesions.
| HOXD10 | |||
|---|---|---|---|
| Category | Positive | Negative | P-value |
| EA | 25 | 25 | 0.0010 |
| MEC | 0 | 12 | |
| Cancer samples | 25 | 37 | <0.0001 |
| Control samples | 56 | 14 | |
HOX, homeobox; EA, endometroid adenocarcinoma; MEC, mucinous endometrial carcinoma.
Figure 2.
Decreased expression levels of HOXD10 in EC. (A) Classic HOXD10 staining in EC (×100). (B) No HOXD10 staining was observed in MEC samples (×100). (C) For simple hyperplasia, EC tissue was strongly stained and HOXD10 is positively stained in the nucleus and cytoplasm (×400). (D) In complex hyperplasia, EC tissue was strongly stained, and HOXD10 was positively stained in the nucleus and cytoplasm (×100). HOX, homeobox; EC, endometrial carcinoma; MEC, mucinous endometrial carcinoma. Arrows indicate positively stained cells within the figure.
Association between HOXD10 methylation, gene expression levels and clinicopathological features in EC
The present study compared methylation of the HOXD10 gene and its expression levels in cancer and control samples. Staining intensity of HOXD10 was negatively associated with promoter methylation (P<0.05). The majority of samples with promoter methylation lacked HOXD10 protein expression levels, indicating a significant association between promoter hypermethylation and transcriptional silencing (P<0.05).
There was no significant difference in HOXD10 methylation between EA and MEC samples (P>0.05). No association between HOXD10 methylation and clinical characteristics (patient age, tumor differentiation, tumor size, depth of myometrial invasion and lymph node metastases) was observed in EA or MEC cases (Table III).
Table III.
Promoter methylation status of HOXD10 in primary endometrial lesions and associations with clinical features.
| HODX10 | |||
|---|---|---|---|
| Clinical feature | Methylated | Unmethylated | P-value |
| Age, years | 50.56±16.17 | 53.32±16.63 | 0.700 |
| Differentiation, n | |||
| Well | 22 | 13 | |
| Moderate | 12 | 3 | 0.630 |
| Poor | 9 | 3 | |
| Depth of myometrial invasion, n | |||
| <1/2 | 38 | 16 | 0.652 |
| ≥1/2 | 5 | 3 | |
| Lymph node metastasis, n | |||
| Yes | 38 | 19 | 0.149 |
| No | 5 | 0 | |
5-Aza-CdR treatment reverses the expression levels of HOXD10 in Ishikawa cells
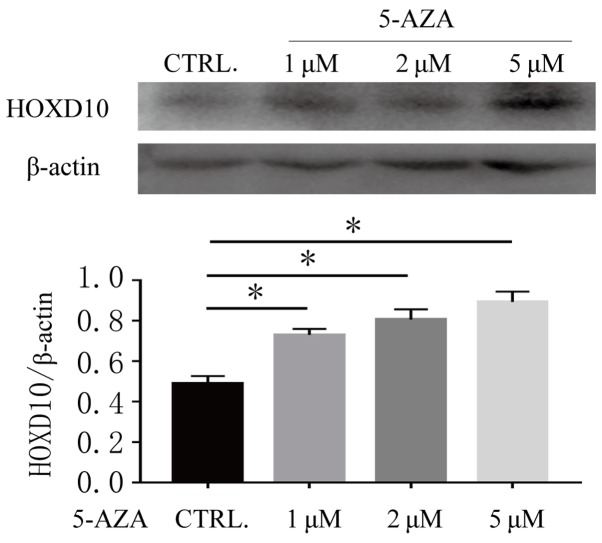
In order to further validate the association between the methylation status of the HOXD10 gene and its expression level, the present study detected the expression level of HOXD10 in the Ishikawa cell line both before and after DNA methylation transferase inhibitor treatment. As presented in Fig. 3, HOXD10 was weakly expressed in Ishikawa cell lines and the expression level of HOXD10 was increased after 72 h treatment with 5-Aza-CdR (P<0.01). These results suggested that DNA methylation may be associated with the promoter methylation status of HOXD10.
Figure 3.
Effects of 5-Aza-CdR on regulation of expression levels of HOXD10 in Ishikawa cell line. β-actin was used as endogenous control. The ratio of HOXD10 to β-actin is shown on the y-axis. *P<0.01. 5-aza-CdR, 5-aza-2′-deoxycytidine; HOX, homeobox.
Discussion
DNA promoter methylation is considered to be a promising diagnostic biomarker for cancer. For certain malignancies, aberrant methylation occurs at the promoter of TSG, which plays a crucial role in the regulation of tumor growth and cell differentiation, proliferation and apoptosis. This causes low gene expression levels, leading to alterations in tumor growth and cell differentiation, proliferation and apoptosis, finally resulting in malignancies that may occur prior to tumor formation. Therefore, DNA methylation may be used as a marker in early cancer screening (24,42–44). Hypermethylation of specific TSG, as well as low mRNA and protein expression levels, has been reported in a number of human malignancies, including EC (28,45,46).
HOXD10 is one of the ANTP homeobox genes located on chromosome 2 (2p31). This gene cluster consists of 39 genes and multiple transcripts and can be divided into four groups (HOX A, B, C and D) (47). HOX regulates stem cell differentiation and embryonic development (48). Abnormal HOX gene expression levels are responsible for certain malignancies, such as breast, thyroid and ovarian cancer (49). The potential use of homeobox genes as diagnostic and prognostic biomarkers has been described in the literature (50,51).
HOXD10 acts as a TSG in a number of malignancies, such as breast cancer (52), gastric carcinogenesis (53) and cholangiocellular carcinoma (54). It inhibits tumorigenesis (55) and angiogenesis and inhibits vascular EGFR, matrix metalloproteinase 14 and uPAR (56). In certain tumor cell lines, upregulation of HOXD10 decreases invasiveness, migration and survival of cancer cells (40,34,44). For example, HOXD10 acts as a tumor-suppressive factor that suppresses proliferation and invasion, promotes G1/S progression arrest and apoptosis via inhibition of the RHOC/AKT/MAPK pathway in human cholangiocellular carcinoma (40). Low or absent HOXD10 expression levels promoted invasion and migration in colorectal cancer (54,57) and inhibited benign transformation of breast tumors (58).
The present study demonstrated that hypermethylation of the HOXD10 promoter occurred more frequently in EC and thus may be a diagnostic tool for EC compared with non-cancerous tissue. In EA and MEC samples, HOXD10 expression levels were negatively associated with methylation status. Aberrant methylation of the HOXD10 promoter may decrease HOXD10 protein expression levels.
DNA methylation transferase inhibitor 5-Aza-CdR treatment partly reverted the expression level of HOXD10 in the Ishikawa cell line. These findings suggest that aberrant DNA methylation may be one of the mechanisms underlying HOXD10 downregulation in EC.
In pathological and clinical diagnostic practice, MEC is typically mucinous (5), but better biomarkers to clearly identify MEC are lacking. MEC often co-occurs with EA and lacks clear boundaries, which complicates diagnosis (7). Previous studies have suggested that in MEC, immunohistochemistry has demonstrated diffuse positivity for ER/PR and vimentin, high p16 and low Ki-67 labeling index, has proven useful for the differential diagnosis of EA (12,51). In the present study, immunohistochemistry data revealed a lack of HOXD10 expression levels in MEC, so this may be a reliable diagnostic biomarker.
In the present study, MSP data indicated no differences in methylation of the HOXD10 promoter between EA and MEC (EA samples vs. MEC; 70 vs. 66.6%; P>0.05). Dysregulation of HOXD10 may have mechanisms other than promoter methylation. The precise mechanism of dysregulation of HOXD10 in cancer is not clear. In hepatocellular carcinoma, and colon, gastric and papillary thyroid cancer, it has been demonstrated that promoter methylation of HOXD10 can lead to loss of HOXD10 expression levels (32,33,53,59). MicroRNA (miR) can affect HOXD10 expression levels at the post-transcriptional level in breast, colorectal, ovarian, gastric and non-small cell lung cancer, as well as hepatocellular carcinoma, glioma and hemangioma. miR-10b, miR-224, miR-23a, miR-501, miR-92b-3p and miR-376b reportedly accelerate cancer progression by directly targeting HOXD10 within the 3′UTR (60–65). Other suggestions of mechanisms of HOXD10 dysregulation include the possible role of long non-coding RNAs (lncRNA), such as HOX transcript antisense RNA (HOTAIR). lncRNA HOTAIR expression is dysregulated in breast cancer and can inhibit HOXD10 expression levels (66). No evidence of HOXD10 mutations has been identified in cancer. Further studies are required to understand the pathology underlying MEC.
In summary, as decreased protein expression levels caused by hypermethylation of TSG may result in tumor progression, the present study demonstrated that promoter hypermethylation-mediated silencing of HOXD10 is a frequent event in EC and thus this may be used to diagnose cancers and guide epigenetic treatment for EA and MEC.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- EA
endometroid adenocarcinoma
- EC
endometrial carcinoma
- MEC
mucinous endometrial carcinoma
- ER
estrogen receptor
- PR
progesterone receptor
- PCDHs
protocadherins
- ZNF
zinc finger protein
- TSG
tumor suppressor gene
- MSP
methylation-specific PCR
- 5-aza-CdR
5-aza-2′-deoxycytidine
Funding
The present study was funded by the National Natural Science Foundation of China (grant nos. 81272277 and 81372810).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TZ and CZ designed and supervised experiments. SM, FY and YD wrote the manuscript. SG, JW and HL collected research data. FY and DL performed the MSP and immunohistochemistry. YD and SM conducted the cell line experiments. DL performed clinical evaluations and statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval was obtained from the Committee of Ethical Research at Shandong University (approval no. mecsdums2012032). Written informed consent was obtained from participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Eiriksson L, Cuartero J, Steed H, Pearcey R, Capstick V, Schepansky A, Faught W, Dundas G. Assessment of outcomes in surgically staged I/II endometrial adenocarcinoma patients treated with postoperative vaginal vault radiotherapy only. Int J Gynecol Cancer. 2010;20:1356–1362. doi: 10.1111/IGC.0b013e3181f21c4d. [DOI] [PubMed] [Google Scholar]
- 5.Gungorduk K, Ozdemir A, Ertas IE, Selcuk I, Solmaz U, Ozgu E, Mat E, Gokcu M, Karadeniz T, Akbay S, et al. Is mucinous adenocarcinoma of the endometrium a risk factor for lymph node involvement? A multicenter case-control study. Int J Clin Oncol. 2015;20:782–789. doi: 10.1007/s10147-014-0767-2. [DOI] [PubMed] [Google Scholar]
- 6.Jalloul RJ, Elshaikh MA, Ali-Fehmi R, Haley MM, Yoon J, Mahan M, Munkarah AR. Mucinous adenocarcinoma of the endometrium: Case series and review of the literature. Int J Gynecol Cancer. 2012;22:812–818. doi: 10.1097/IGC.0b013e31824aa877. [DOI] [PubMed] [Google Scholar]
- 7.Rauh-Hain JA, Vargas RJ, Clemmer J, Clark RM, Bradford LS, Growdon WB, Goodman A, Boruta DM, II, Schorge JO, del Carmen MG. Mucinous adenocarcinoma of the endometrium compared with endometrioid endometrial cancer: A SEER analysis. Am J Clin Oncol. 2016;39:43–48. doi: 10.1097/COC.0000000000000015. [DOI] [PubMed] [Google Scholar]
- 8.Musa F, Huang M, Adams B, Pirog E, Holcomb K. Mucinous histology is a risk factor for nodal metastases in endometrial cancer. Gynecol Oncol. 2012;125:541–545. doi: 10.1016/j.ygyno.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Galic V, Schiavone MB, Herzog TJ, Holcomb K, Lewin SN, Lu YS, Neugut AI, Hershman DL, Wright JD. Prognostic significance of mucinous differentiation of endometrioid adenocarcinoma of the endometrium. Cancer Invest. 2013;31:500–504. doi: 10.3109/07357907.2013.820321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson CL, Hang S, Hansen K, He M, Sung CJ, Quddus MR, Xiong M, Wang Y, Patel NR, Lawrence WD, Xiong J. Endometrial adenocarcinomas with significant mucinous differentiation: A characterization of intratumoral heterogeneity of KRAS mutations in mucinous and endometrioid histologic components. Int J Gynecol Cancer. 2018;28:241–247. doi: 10.1097/IGC.0000000000001168. [DOI] [PubMed] [Google Scholar]
- 11.Sloan EA, Moskaluk CA, Mills AM. Mucinous differentiation with tumor infiltrating lymphocytes is a feature of sporadically methylated endometrial carcinomas. Int J Gynecol Pathol. 2017;36:205–216. doi: 10.1097/PGP.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 12.Lax SF. Pathology of endometrial carcinoma. Adv Exp Med Biol. 2017;943:75–96. doi: 10.1007/978-3-319-43139-0_3. [DOI] [PubMed] [Google Scholar]
- 13.Fadare O, Roma AA, Mhawech-Fauceglia P, Parkash V, Rabban JT. The diagnosis of mucinous lesions in endometrial samplings by gynaecological pathologists: An analysis of diagnostic reproducibility. Pathology. 2018;50:276–285. doi: 10.1016/j.pathol.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Jones NL, Xiu J, Chatterjee-Paer S, Buckley de Meritens A, Burke WM, Tergas AI, Wright JD, Hou JY. Distinct molecular landscapes between endometrioid and nonendometrioid uterine carcinomas. Int J Cancer. 2017;140:1396–1404. doi: 10.1002/ijc.30537. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Sahasrabuddhe VV, Mendoza-Cervantes D, Zhao R, Duggan MA. Tissue-based immunohistochemical biomarker expression in malignant glandular lesions of the uterine cervix: A systematic review. Int J Gynecol Pathol. 2018;37:128–140. doi: 10.1097/PGP.0000000000000406. [DOI] [PubMed] [Google Scholar]
- 16.Stampoliou A, Arapantoni-Dadioti P, Pavlakis K. Epigenetic mechanisms in endometrial cancer. J BUON. 2016;21:301–306. [PubMed] [Google Scholar]
- 17.Zhou JY, Zhang L, Wei LH, Wang JL. Endometrial carcinoma-related genetic factors: Application to research and clinical practice in China. BJOG. 2016;123(Suppl 3):S90–S96. doi: 10.1111/1471-0528.14007. [DOI] [PubMed] [Google Scholar]
- 18.Banno K, Yanokura M, Iida M, Masuda K, Aoki D. Carcinogenic mechanisms of endometrial cancer: Involvement of genetics and epigenetics. J Obstet Gynaecol Res. 2014;40:1957–1967. doi: 10.1111/jog.12442. [DOI] [PubMed] [Google Scholar]
- 19.Men C, Chai H, Song X, Li Y, Du H, Ren Q. Identification of DNA methylation associated gene signatures in endometrial cancer via integrated analysis of DNA methylation and gene expression systematically. J Gynecol Oncol. 2017;28:e83. doi: 10.3802/jgo.2017.28.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caplakova V, Babusikova E, Blahovcova E, Balharek T, Zelieskova M, Hatok J. DNA methylation machinery in the endometrium and endometrial cancer. Anticancer Res. 2016;36:4407–4420. doi: 10.21873/anticanres.10984. [DOI] [PubMed] [Google Scholar]
- 21.Romanek-Piva K, Gałczyński K, Adamiak-Godlewska A, Rechberger T, Postawski K. DNA methylation and DNA methyltransferase (DNMT1) activity pattern in endometrial carcinoma. Ginekol Pol. 2016;87:6–10. doi: 10.17772/gp/60556. [DOI] [PubMed] [Google Scholar]
- 22.Fialkova V, Vidomanova E, Balharek T, Marcinek J, Kudela E, Hanysova S, Visnovsky J, Dobrota D, Hatok J. DNA methylation as mechanism of apoptotic resistance development in endometrial cancer patients. Gen Physiol Biophys. 2017;36:521–529. doi: 10.4149/gpb_2017032. [DOI] [PubMed] [Google Scholar]
- 23.Bartosch C, Lopes JM, Jeronimo C. Epigenetics in endometrial carcinogenesis-part 1: DNA methylation. Epigenomics. 2017;9:737–755. doi: 10.2217/epi-2016-0166. [DOI] [PubMed] [Google Scholar]
- 24.Lai HC, Wang YC, Yu MH, Huang RL, Yuan CC, Chen KJ, Wu CC, Chiang KJ, Chao TK. DNA methylation as a biomarker for the detection of hidden carcinoma in endometrial atypical hyperplasia. Gynecol Oncol. 2014;135:552–559. doi: 10.1016/j.ygyno.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang T, Qiu H, Bao W, Li B, Lu C, Du G, Luo X, Wang L, Wan X. Epigenetic inactivation of EFEMP1 is associated with tumor suppressive function in endometrial carcinoma. PLoS One. 2013;8:e67458. doi: 10.1371/journal.pone.0067458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiolka R, Zubor P, Janusicova V, Visnovsky J, Mendelova A, Kajo K, Lasabova Z, Plank L, Danko J. Promoter hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and CDH1 in endometrial cancer. Oncol Rep. 2013;30:2878–2886. doi: 10.3892/or.2013.2752. [DOI] [PubMed] [Google Scholar]
- 28.Sheng Y, Wang H, Liu D, Zhang C, Deng Y, Yang F, Zhang T, Zhang C. Methylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carcinoma. Gynecol Oncol. 2016;140:145–151. doi: 10.1016/j.ygyno.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Chen YC, Tsao CM, Kuo CC, Yu MH, Lin YW, Yang CY, Li HJ, Yan MD, Wang TJ, Chou YC, Su HY. Quantitative DNA methylation analysis of selected genes in endometrial carcinogenesis. Taiwan J Obstet Gynecol. 2015;54:572–579. doi: 10.1016/j.tjog.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Huang RL, Su PH, Liao YP, Wu TI, Hsu YT, Lin WY, Wang HC, Weng YC, Ou YC, Huang TH, Lai HC. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017;23:263–272. doi: 10.1158/1078-0432.CCR-16-0863. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Zhang C, Sheng Y, Yao S, Liu Z, Zhang C, Zhang T. Frequent SOCS3 and 3OST2 promoter methylation and their epigenetic regulation in endometrial carcinoma. Am J Cancer Res. 2015;5:180–190. [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Y, Peng Y, Gao D, Zhang M, Yang W, Linghu E, Herman JG, Fuks F, Dong G, Guo M. Silencing HOXD10 by promoter region hypermethylation activates ERK signaling in hepatocellular carcinoma. Clin Epigenetics. 2017;9:116. doi: 10.1186/s13148-017-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan YH, Wang HY, Lai Y, Zhong W, Liang WL, Yan FD, Yu Z, Chen JK, Lin Y. Epigenetic inactivation of HOXD10 is associated with human colon cancer via inhibiting the RHOC/AKT/MAPK signaling pathway. Cell Commun Signal. 2019;17:9. doi: 10.1186/s12964-018-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang LH, Choi YL, Hua XY, Shin YK, Song YJ, Youn SJ, Yun HY, Park SM, Kim WJ, Kim HJ, et al. Increased expression of sonic hedgehog and altered methylation of its promoter region in gastric cancer and its related lesions. Mod Pathol. 2006;19:675–683. doi: 10.1038/modpathol.3800573. [DOI] [PubMed] [Google Scholar]
- 35.Brinkhuizen T, van den Hurk K, Winnepenninckx VJ, de Hoon JP, van Marion AM, Veeck J, van Engeland M, van Steensel MA. Epigenetic changes in basal cell carcinoma affect SHH and WNT signaling components. PLoS One. 2012;7:e51710. doi: 10.1371/journal.pone.0051710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Wang Y, Fu S, Liu D, Lin S. Methylation-regulated ZNF545 inhibits growth of the p53-mutant KYSE150 cell line by inducing p21 and Bax. Exp Ther Med. 2019;18:1563–1570. doi: 10.3892/etm.2019.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang S, Xiang T, Xiao Q, Li Y, Shao B, Luo T. Zinc-finger protein 545 is inactivated due to promoter methylation and functions as a tumor suppressor through the Wnt/β-catenin, PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer. Int J Oncol. 2017;51:801–811. doi: 10.3892/ijo.2017.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YL, Wang YP, Li HZ, Zhang X. Aberrant promoter methylation of PCDH17 (Protocadherin 17) in serum and its clinical significance in renal cell carcinoma. Med Sci Monit. 2017;23:3318–3323. doi: 10.12659/MSM.902077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uyen TN, Sakashita K, Al-Kzayer LF, Nakazawa Y, Kurata T, Koike K. Aberrant methylation of protocadherin 17 and its prognostic value in pediatric acute lymphoblastic leukemia. Pediatr Blood Cancer. 2017;64 doi: 10.1002/pbc.26259. [DOI] [PubMed] [Google Scholar]
- 40.Musialik E, Bujko M, Kober P, Grygorowicz MA, Libura M, Przestrzelska M, Juszczyński P, Borg K, Florek I, Jakóbczyk M, Siedlecki JA. Promoter DNA methylation and expression levels of HOXA4, HOXA5 and MEIS1 in acute myeloid leukemia. Mol Med Rep. 2015;11:3948–3954. doi: 10.3892/mmr.2015.3196. [DOI] [PubMed] [Google Scholar]
- 41.Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO classification of tumours of female reproductive organs. International Agency for Research on Cancer. 2014;307 [Google Scholar]
- 42.Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: Molecular mechanism. Cell Biochem Biophys. 2013;67:501–513. doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- 43.Ding WJ, Yang Y, Chen ZX, Wang YY, Dong WL, Cen JN, Qi XF, Jiang F, Chen SN. Methylation level of Rap1GAP and the clinical significance in MDS. Oncol Lett. 2018;16:7287–7294. doi: 10.3892/ol.2018.9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DS, Lee WK, Park JY. Promoter methylation of Wrap53α, an antisense transcript of p53, is associated with the poor prognosis of patients with non-small cell lung cancer. Oncol Lett. 2018;16:5823–5828. doi: 10.3892/ol.2018.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, Mould T, West J, Zikan M, Cibula D, Fiegl H, et al. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 2013;10:e1001551. doi: 10.1371/journal.pmed.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meršaková S, Holubeková V, Grendár M, Višňovský J, Ňachajová M, Kalman M, Kúdela E, Žúbor P, Bielik T, Lasabová Z, Danko J. Methylation of CADM1 and MAL together with HPV status in cytological cervical specimens serves an important role in the progression of cervical intraepithelial neoplasia. Oncol Lett. 2018;16:7166–7174. doi: 10.3892/ol.2018.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbasi AA. Diversification of four human HOX gene clusters by step-wise evolution rather than ancient whole-genome duplications. Dev Genes Evol. 2015;225:353–357. doi: 10.1007/s00427-015-0518-z. [DOI] [PubMed] [Google Scholar]
- 48.Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of Hox genes in stem cell differentiation. World J Stem Cells. 2015;7:583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 50.Javed S, Langley SE. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. 2014;113:535–540. doi: 10.1111/bju.12269. [DOI] [PubMed] [Google Scholar]
- 51.Chekmareva M, Ellenson LH, Pirog EC. Immunohistochemical differences between mucinous and microglandular adenocarcinomas of the endometrium and benign endocervical epithelium. Int J Gynecol Pathol. 2008;27:547–554. doi: 10.1097/PGP.0b013e318177eadc. [DOI] [PubMed] [Google Scholar]
- 52.Vardhini NV, Rao PJ, Murthy PB, Sudhakar G. HOXD10 expression in human breast cancer. Tumour Biol. 2014;35:10855–10860. doi: 10.1007/s13277-014-2324-z. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L, Lam EK, Liu X, Zhang J, Zhou T, et al. Homeobox D10 gene, a candidate tumor suppressor, is downregulated through promoter hypermethylation and associated with gastric carcinogenesis. Mol Med. 2012;18:389–400. doi: 10.2119/molmed.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Zhou J, Mi J, Ma K, Fan Y, Ning J, Wang C, Wei X, Zhao H, Li E. HOXD10 acts as a tumor-suppressive factor via inhibition of the RHOC/AKT/MAPK pathway in human cholangiocellular carcinoma. Oncol Rep. 2015;34:1681–1691. doi: 10.3892/or.2015.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joo MK, Park JJ, Chun HJ. Impact of homeobox genes in gastrointestinal cancer. World J Gastroenterol. 2016;22:8247–8256. doi: 10.3748/wjg.v22.i37.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N. Sustained expression of homeobox d10 inhibits angiogenesis. Am J Pathol. 2002;161:2099–2109. doi: 10.1016/S0002-9440(10)64488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Li Z, Zhao X, Zuo X, Peng Z. miR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol Lett. 2016;12:488–494. doi: 10.3892/ol.2016.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005;65:7177–7185. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
- 59.Cao YM, Gu J, Zhang YS, Wei WJ, Qu N, Wen D, Liao T, Shi RL, Zhang L, Ji QH, et al. Aberrant hypermethylation of the HOXD10 gene in papillary thyroid cancer with BRAFV600E mutation. Oncol Rep. 2018;39:338–348. doi: 10.3892/or.2017.6058. [DOI] [PubMed] [Google Scholar]
- 60.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 61.Li S, Zhang J, Zhao Y, Wang F, Chen Y, Fei X. miR-224 enhances invasion and metastasis by targeting HOXD10 in non-small cell lung cancer cells. Oncol Lett. 2018;15:7069–7075. doi: 10.3892/ol.2018.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yachi K, Tsuda M, Kohsaka S, Wang L, Oda Y, Tanikawa S, Ohba Y, Tanaka S. miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition. Signal Transduct Target Ther. 2018;3:33. doi: 10.1038/s41392-018-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng Z, Liu S, Cai J, Li Z, Wu H, Chen H, Huang Y. miR-501 promotes hemangioma progression by targeting HOXD10. Am J Transl Res. 2019;11:2439–2446. [PMC free article] [PubMed] [Google Scholar]
- 64.Li C, Huo B, Wang Y, Cheng C. Downregulation of microRNA-92b-3p suppresses proliferation, migration, and invasion of gastric cancer SGC-7901 cells by targeting Homeobox D10. J Cell Biochem. 2019;120:17405–17412. doi: 10.1002/jcb.29005. [DOI] [PubMed] [Google Scholar]
- 65.An N, Luo X, Zhang M, Yu R. MicroRNA-376b promotes breast cancer metastasis by targeting Hoxd10 directly. Exp Ther Med. 2017;13:79–84. doi: 10.3892/etm.2016.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.