Abstract
Lactococcus lactis has previously been proposed as a vaccine platform for the safe delivery of heterologous antigens. Here we utilized L. lactis as a live vector for expression of listeriolysin O (LLO), a major Listeria monocytogenes antigen and virulence factor. A variety of plasmid constructs were designed to permit either constitutive or nisin-inducible expression of secreted or non-secreted LLO in L. lactis. Recombinant strains were subsequently tested in a murine model for vaccination efficacy against L. monocytogenes infection. CD8+ T lymphocytes specific for the LLO91–99 epitope were detected when strains were administered via the intraperitoneal (IP) but not the oral route. Challenge with live L. monocytogenes revealed different levels of protection among the three vaccine strains tested with the nisin-inducible LLO-secreting L. lactis strain providing the greatest protection against secondary infection. This work highlights the usefulness of the GRAS (Generally Regarded As Safe) organism L. lactis as the basis of a live vaccine vector against L. monocytogenes. The work suggests that LLO-expressing L. lactis strains may also have the potential to act as a platform for directing other co-expressed antigens towards the cytosolic MHC class I pathway for enhanced stimulation of the CD8+ T-cell response.
Keywords: Listeria monocytogenes, Lactococcus lactis, Listeriolysin O, Vaccine, CD8
1. Introduction
Listeria monocytogenes, the causative agent of listeriosis, is a food-borne pathogen which can infect immunocompromised individuals causing meningitis and septicemia [1]. Pregnant women are particularly susceptible, developing flu-like symptoms followed by chorioamnionitis and miscarriage [1]. Although L. monocytogenes infections are generally sporadic in the community, several large-scale common-source outbreaks have occurred worldwide [2]. Once listeriosis is established in an infected host the mortality rate is high (20–30%), and in terms of human mortality L. monocytogenes is considered to be among the most significant bacterial causes of food-borne diseases [3]. Moreover, L. monocytogenes also causes infections of cattle and sheep and can result in appreciable losses through foetal infection and spontaneous abortions in these animals. Infection of farm animals is also considered to pose a zoonotic threat to humans who may be exposed to L. monocytogenes through contaminated milk or meat products [4].
L. monocytogenes is an intracellular pathogen which has a unique mechanism of cellular infection. Upon phagocytosis, the pathogen secretes a hemolysin, listeriolysin O (LLO), that forms pores in the phagosomal membrane enabling the bacterium to access the host cell cytoplasm [3]. Through the expression of other specific virulence factors (including ActA, Hpt, and PlcB), L. monocytogenes replicates and moves within the cytosol and can spread from cell to cell without exposure to the extracellular milieu [5]. Investigation of the immune response against L. monocytogenes showed that in addition to its role in intracellular pathogenesis, LLO is a major immunodominant listerial antigen. Indeed protective immunity against L. monocytogenes is dependent on cytotoxic CD8+ cell-mediated immunity against epitopes of the two major virulence factors LLO and P60 [6]. The central role of LLO in the listerial infection cycle, coupled to the significant antigenicity of this protein indicates that LLO may have potential as the basis of a vaccine against L. monocytogenes. Live bacterial vectors such as attenuated Bacillus anthracis and aroA − Salmonella typhimurium strains have previously been engineered to express LLO for vaccination against L. monocytogenes [7], [8], [9]. In addition, several attenuated L. monocytogenes mutants have been investigated as possible vaccines against listeriosis or as heterologous antigen delivery systems for vaccination against cancer and other pathogens [10], [11], [12], [13]. However, although the efficacy of these approaches was clearly demonstrated, all the abovementioned vectors are based upon live attenuated pathogens which may have the potential for reversion to a virulent state.
Lactococcus lactis is a GRAS (Generally Regarded As Safe) microorganism that is widely used in the food industry. The development of numerous inducible and constitutive expression systems for L. lactis has enhanced the use of this organism as a cellular factory for expression of various heterologous proteins of biotechnological interest [14]. This work has extended into the use of L. lactis as a live vaccine vector for delivery of heterologous antigens. Several antigens such as tetanus toxin fragment C (TTFC) [15], Helicobacter pylori Cag12 antigen [16], Giardia lamblia cyst wall protein 2 [17], and SARS-coronavirus nucleocapsid protein [18] have been successfully expressed in L. lactis with promising immunological outcomes following administration in mice. Moreover, L. lactis has also been used as a vehicle for delivery of therapeutic bioactive substances (including interleukin-10 (IL-10) and trefoil factors) to reduce inflammation and enhance recovery in murine models of colitis [19], [20]. Indeed, an L. lactis strain expressing human IL-10 and utilizing a novel biological containment strategy has recently been the subject of clinical trials in patients with Crohn's disease [19]. Since L. lactis is a non-pathogenic, non-invasive and non-commensal food-grade bacterium, it is particularly attractive as a basis for safe delivery of antigens or bioactive molecules. Furthermore, the Gram-positive bacterium L. lactis has a relatively small genome size with few exoproteins and unlike proposed Gram-negative hosts, does not produce endotoxin [21], [22], [23].
In the present work, we successfully expressed L. monocytogenes LLO in L. lactis to act as a potential live vaccine vector. The P44 constitutive promoter and the PnisA nisin-inducible promoter were employed to express LLO in different compartments (intracellular and secreted). LLO was expressed in an active form in all cases except the constitutive intracellular form. Investigation of the immune response upon vaccination of BALB/c mice via the intraperitoneal (IP) and oral routes revealed different levels of LLO-specific CD8+ T cells, IgG antibodies and protection against challenge with L. monocytogenes. The current work demonstrates the application of L. lactis in the development of potential vaccine platforms against L. monocytogenes.
2. Materials and methods
2.1. Bacterial strains, plasmids, and culture conditions
A summary of bacterial strains and plasmids used in this study is shown in Table 1 . Luria–Bertani (LB) broth was used for Escherichia coli cultures while M17 broth (Oxoid) supplemented with 0.5% glucose (i.e. GM17) was used for Lactococcus. For L. monocytogenes, brain heart infusion (BHI) broth (Oxoid) was used. Technical agar (Merck) was added (1.5%, w/v) when solid media were required. Incubation temperatures were 30 °C for L. lactis and 37 °C for L. monocytogenes and E. coli. When required, ampicillin (Amp) was used at a concentration of 100 μg/ml for E. coli while chloramphenicol (Cm) was used at 10 μg/ml for E. coli, and L. lactis and at 7.5 μg/ml for luminescent L. monocytogenes. All cell culture media and reagents were obtained from Gibco unless otherwise stated.
Table 1.
Bacterial strains and plasmid vectors
| Strain or plasmid name | Description | Reference or source |
|---|---|---|
| Escherichia coli Top10 | Chemically competent intermediate host, plasmid free | Invitrogen |
| E. coli M15 (pREP4) | Expression host for pQE30-cloned genes, containing the repressor plasmid pREP4 (Kanamycin resistant; KmR) for suppression of basal protein expression under uninduced conditions | Qiagen |
| E. coli BL21 | Chemically competent E. coli, used in this study as an intermediate host for pQE30, plasmid free | Novagen |
| Listeria monocytogenes EGDe serovar 1/2a | Wild type L. monocytogenes | [65] |
| Luminescent L. monocytogenes EGDe | Constitutively luciferase-expressing L. monocytogenes EGDe (carrying pPL2luxPhelp on its chromosome through single cross-over integration), CmR | [42] |
| Lactococcus lactis subspecies cremoris MG1363 | Plasmid free Lactococcus strain | [66] |
| L. lactis NZ9700 | Nisin producer strain | [29] |
| L. lactis NZ9000 | L. lactis subsp. cremoris MG1363 carrying nisRK on the chromosome | [29] |
| L. lactis NZ9000 ΔhtrA | L. lactis NZ9000 with a chromosomal deletion in the htrA promoter and the 5′-end region of htrA | [32] |
| pQE30 | Expression vector using phage T5 promoter and adding an N-terminal six-His tag to the expressed protein, AmpR | Qiagen |
| pNZ8048 | E. coli–L. lactis shuttle vector containing PnisA promoter and start codon in NcoI site, CmR | [29] |
| pNZ44 | pNZ8048 derivative containing P44 promoter instead of PnisA promoter, CmR | [31] |
| pNZP44:CYTO-LLO | Modified pNZ8048 containing P44 promoter with downstream His-tagged hly, CmR | This study |
| pNZP44:SEC-LLO | Modified pNZ8048 containing P44 promoter with downstream secretion signal of Usp45 protein and His-tagged hly, CmR | This study |
| pNZPnisA:CYTO-LLO | Modified pNZ8048 containing PnisA promoter (NcoI site eliminated) with downstream His-tagged hly, CmR | This study |
| pNZPnisA:SEC-LLO | Modified pNZ8048 containing PnisA promoter (NcoI site eliminated) with downstream secretion signal of Usp45 protein and His-tagged hly, CmR | This study |
2.2. Cloning of LLO in modified pNZ8048 plasmid vectors
Four plasmids were constructed for the expression of LLO in L. lactis: the constitutive pNZP44:SEC-LLO and pNZP44:CYTO-LLO plasmids, and the nisin-inducible pNZPnisA:SEC-LLO and pNZPnisA:CYTO-LLO plasmids (Fig. 1 ). Plasmids pNZP44:SEC-LLO and pNZPnisA:SEC-LLO were designed to secrete LLO using the N-terminal secretion signal of the Usp45 protein which is a secreted lactococcal protein [24]. The use of the Usp45 secretion signal has previously been described to direct the secretion of heterologous proteins in L. lactis [25], [26]. Plasmids pNZP44:CYTO-LLO and pNZPnisA:CYTO-LLO lack a secretion signal and were thus designed to produce LLO intracellularly. For constitutive antigen expression, the lactococcal P44 promoter was used [27] while for inducible expression PnisA was used [28].
Fig. 1.
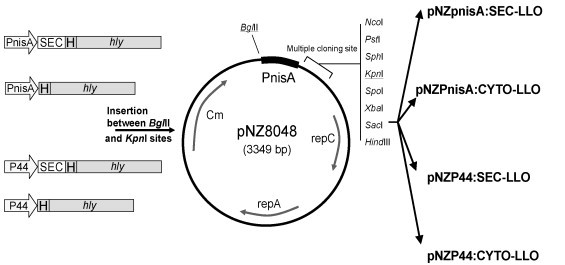
Diagrammatic representation of the four created constructs. PnisA and P44 are nisin-inducible and constitutive promoters respectively. SEC: the secretion signal of Usp45 protein; H: six-His tag; hly: gene of LLO. The four constructs to the left were made by fusing the promoters and His-tagged hly precisely (with or without SEC) using the splicing by overlap extension (SOE) technique. These constructs were cloned between BglII and KpnI sites in pNZ8048 resulting in the corresponding plasmid vectors to the right. Details are described in Section 2.
Plasmid pNZ8048 [29] was used as a platform to create the four plasmids. pNZ8048 contains the PnisA promoter with a downstream NcoI restriction site (CCATGG) containing the first start codon (ATG). However, since the hly gene (encoding LLO) contains an internal NcoI restriction site, we modified pNZ8048 to eliminate this NcoI site and added a new start codon in frame with the hly gene. To achieve this we eliminated the PnisA promoter (along with the NcoI site) from pNZ8048 by enzymatic digestion (BglII and KpnI) and replaced it with our promoter (PnisA or P44)/insert constructs without the need for NcoI digestion (Fig. 1). The promoter/insert constructs were created using the splicing by overlap extension (SOE) technique [30]. The primers used in the PCR reactions are shown in Table 2 . All PCR reactions were performed using the high fidelity KOD hot start DNA polymerase (Novagen) following manufacturer's instructions.
Table 2.
Oligonucleotide primers
| Primer code | Primer sequence (5′–3′)a |
|---|---|
| pQE30 forward primer | GAAGGATCCGATGCATCTGCATTCAATAAAG (BamhI) |
| pQE30 reverse primer | ACGCCTGCAGTTCGATTGGATTATCTACTTTATTA (PstI) |
| 1 | CCAAGATCTAGTCTTATAACTATACTG (BglII) |
| 2 | TCTTTTTTTTCATTTTGAGTGCCTCCTTATAATTTATTTTG |
| 3 | GGAGGCACTCAAAATGAAAAAAAAGATTATCTC |
| 4 | TGATGTGTATCAGCGTAAACACCTGACAACG |
| 5 | TGTTTACGCTGATACACATCACCATCACCATCACGGA |
| 6 | AGTCGGTACCTTATTCGATTGGATTATCTAC (KpnI) |
| 7 | GGTGATGTCCCATTTTGAGTGCCTCCTTATAATTTATTTTG |
| 8 | AGGCACTCAAAATGGGACATCACCATCACCATCACGGA |
| 9 | CCAAGATCTAACAATTGTAACCCATACAG (BglII) |
| 10 | TCTTTTTTTTCATAAAAGCGACTCCTTTCCCTCACACATCA |
| 11 | AGGAGTCGCTTTTATGAAAAAAAAGATTATCTC |
| 12 | TGATGTCCCATAAAAGCGACTCCTTTCCCTCACACAT |
| 13 | AGGAGTCGCTTTTATGGGACATCACCATCACCATCACGGA |
When applicable, recognition sites of restriction enzymes are underlined and enzyme name is mentioned between parentheses.
The pQE30 plasmid was used as an intermediate cloning vector to create a six-Histidine-tagged (i.e. His-tagged) antigen for subsequent cloning into modified pNZ8048. The hly gene of L. monocytogenes EGDe (accession number AL591824) minus the secretion signal was PCR amplified from chromosomal DNA using pQE30 forward and reverse primers (Table 2). This hly gene was digested by BamHI and PstI and ligated using T4 DNA ligase (Roche) to a similarly digested pQE30 plasmid (Qiagen) which introduced an N-terminal six-Histidine tag to hly. The ligation reaction mixture was transformed to chemically competent E. coli BL21 (Novagen) by heat shock following manufacturer's instructions and plated onto LB agar containing ampicillin. Colony PCR was used to identify positive colonies and plasmid (pQE30/hly) was extracted from E. coli BL21 using Qiagen Miniprep Kit (Qiagen). The integrity of the DNA sequence was confirmed by sequencing (Lark Technologies Inc., UK).
For the nisin-inducible secreted LLO construct, primers 1 and 2 were used to amplify promoter PnisA using pNZ8048 as a template while primers 3 and 4 were used to amplify the Usp45 secretion signal (hereafter designated SEC) from L. lactis MG1363 chromosomal DNA preparation. The resulting two PCR products were spliced together by the SOE technique using primers 1 and 4 and a 1:1 molar ratio of the two PCR products. This SOE product (PnisA + SEC) was further spliced to the His-tagged hly gene (amplified from pQE30/hly using primers 5 and 6) using primers 1 and 6.
Similarly, for the nisin-inducible non-secreted LLO construct, primers 1 and 7 were used to amplify PnisA using pNZ8048 as a template. Subsequently, this resulting PnisA was SOE-spliced to the His-tagged hly (amplified from pQE30/hly using primers 6 and 8) using primers 1 and 6.
For the constitutive secreted LLO construct the P44 promoter was amplified from plasmid pNZ44 [31] using primers 9 and 10, SEC signal was amplified from the L. lactis MG1363 chromosome with primers 11 and 4. These two PCR products were first spliced by SOE technique using primers 9 and 4 and the resulting PCR product was further spliced to the His-tagged hly (previously amplified from pQE30/hly template using primers 5 and 6) using primers 9 and 6.
Finally, the constitutive non-secreted LLO construct was created by a SOE reaction between P44 promoter (amplified from pNZ44 template with primers 9 and 12) and the His-tagged hly (amplified from pQE30/hly as a template with primers 13 and 6) by the use of primers 9 and 6.
Each construct was digested sequentially with BglII then KpnI enzymes. Similarly, plasmid pNZ8048 was digested by BglII and KpnI and gel extracted by the Qiagen gel-extraction kit (Qiagen) to remove the intervening fragment which contained PnisA along with the undesired NcoI site (Fig. 1). Ligation was performed using T4 DNA ligase (Roche) and the ligation reaction was transformed to a commercial chemically competent E. coli Top10 (Invitrogen) following the manufacturer's instructions and plated onto LB agar containing chloramphenicol. After incubation at 37 °C for 24–48 h, positive colonies were detected by colony PCR. Plasmids were extracted from E. coli Top10 using the Qiagen Miniprep Kit (Qiagen) and DNA sequence was confirmed (Lark Technologies Inc., UK).
2.3. Transformation of Lactococcus strains with hly-containing vectors
L. lactis MG1363 and L. lactis NZ9000 (a derivative of MG1363) were used as host strains for pNZP44:CYTO-LLO and pNZPnisA:CYTO-LLO, respectively. For the LLO secretory plasmids, pNZPnisA:SEC-LLO and pNZP44:SEC-LLO, L. lactis NZ9000 ΔhtrA (kindly provided by Prof. Airi Palva, University of Helsinki) was used as a host [32]. HtrA is an extracellular housekeeping protease [33] and HtrA-deficient Lactococcus mutants have been reported to be more efficient in expression of secreted protein [25], [32]. Electro-competent Lactococcus strains, prepared as previously described [34], were transformed with the corresponding hly-containing vectors using Gene Pulser (Biorad) and plated onto GM17 agar containing chloramphenicol and incubated at 30 °C for 24 h. Colonies were checked by colony PCR and positive clones were preserved in glycerol stocks at −80 °C.
2.4. Investigation of LLO production by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Western blot
Overnight cultures of the inducible Lactococcus strains (i.e. containing pNZPnisA:SEC-LLO or pNZPnisA:CYTO-LLO) were sub-cultured (5%, v/v) in fresh GM17 broth and grown statically at 30 °C to an optical density at 600 nm (OD600) of 0.5. At this point nisin, used as filter-sterilized culture supernatant of the nisin-secreting strain Lactococcus NZ9700 [22], was added at a concentration of 0.2% (v/v). These strains were allowed to grow for a further 3 h and then cells were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS) and then pellets were frozen at −20 °C until analysis. For the constitutive strains (containing pNZP44:SEC-LLO or pNZP44:CYTO-LLO), overnight cultures were inoculated into fresh GM17 broth and grown to an OD600 of about 0.8 at which point cells were similarly harvested, washed and preserved at −20 °C. For SDS–PAGE analysis, cells were thawed on ice, resuspended in cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) containing 30 mg/ml lysozyme (Sigma) and kept on ice for 30 min. This was followed by sonication and centrifugation at 10,000 × g for 30 min at 4 °C. Aliquots of the bacterial lysate (supernatant) were mixed with SDS–PAGE 2× sample buffer and kept at −20 °C. The remaining lysate was used for purification of the His-tagged LLO using Ni-NTA affinity chromatography.
Culture supernatants of the LLO-secreting strains, L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO and pNZP44:SEC-LLO) were also collected using cold trichloroacetic acid (TCA) precipitation (15%, w/v final concentration) as previously described [35].
For Western blot analysis, gels were blotted against a nitrocellulose membrane (Hybond-ECL™, Amersham Biosciences UK Ltd.) and the membrane was then blocked overnight at 4 °C in 5% skimmed milk in TBS buffer (0.8% sodium chloride, 20 mM Tris–HCl, pH 7.6). Primary rabbit anti-LLO antibody (Diatheva, Italy) and secondary anti-rabbit antibody (ECL western blotting system) were used at 1/1000 and 1/1500 dilutions in 5% and 10% skimmed milk, respectively. Western blot detection was performed using the Amersham ECL western blotting system (Amersham Biosciences UK Ltd.) according to the protocol recommended by the manufacturer.
Since animal vaccination with the nisin-induced vaccine strains requires continued in vivo expression of LLO after the in vitro induction period, we examined the kinetics of LLO production following removal of the nisin inducer. The two nisin-inducible strains were induced with nisin for 1 h, and then washed three times with PBS and resuspended in fresh GM17 broth. LLO production was analyzed over time by Western blot.
2.5. Purification of LLO and assessment of LLO hemolytic activity
His-tagged LLO was purified from the cell lysate (pNZP44:CYTO-LLO and pNZPnisA:CYTO-LLO) or acellular supernatant (pNZPnisA:SEC-LLO and pNZP44:SEC-LLO) by means of Ni-NTA gel affinity chromatography. Cell lysate was passed through Ni-NTA gel (Qiagen) packed in chromatography columns (Biorad), and LLO was purified using the procedure described by Qiagen (QIAexpressionist™). A similar procedure was used for the secreted LLO (pNZPnisA:SEC-LLO and pNZP44:SEC-LLO) but the supernatant pH was first adjusted to pH 8 using NaOH before passing through the Ni-NTA gel.
To assess if LLO is produced in active form, the hemolytic activity of preparations was checked using the method described by Kohda et al. [36] with some modifications. Briefly, aliquots of 100 μl of 0.5% (v/v) sheep red blood cells (RBCs) suspended in PBS (pH 5.5) were distributed in 1.5 ml Eppendorf tubes. Twofold serially diluted (in PBS pH 5.5) purified LLO (for non-secreted LLO) or dialyzed culture supernatant (for secreted LLO) was added to each tube to a final volume of 1 ml. A positive control (distilled water) and a negative control (PBS, pH 5.5) were also included. Tubes were incubated statically at 37 °C for 45 min after which they were centrifuged and supernatants collected. Absorbance was measured at 415 nm and hemolytic units were calculated. For purified LLO from NZ9000 (pNZPnisA:CYTO-LLO), one hemolytic unit (HU) was defined as the amount of LLO required to cause 50% hemoglobin release from sheep RBCs as compared to the 100% hemoglobin release of the positive control (distilled water) [36]. For secreted LLO, hemolytic activity was expressed in terms of complete hemolytic units (CHU) defined as the reciprocal of the highest dilution of supernatant showing complete hemolysis [37].
It is noteworthy that we failed to detect any LLO production in the Lactococcus MG1363 (pNZP44:CYTO-LLO) strain by any of the previous detection techniques. Consequently, all subsequent experiments involved only the three remaining LLO-producing strains.
2.6. Intracellular growth of the LLO-producing strains
Intracellular growth of the constructed L. lactis strains was investigated in J774 macrophage-like cells as described previously [38] with some modifications. Briefly, J774 cells were normally grown in DMEM glutamax® medium (Gibco) containing 10% foetal calf serum FCS at 37 °C and 5% CO2. Before the test, J774 cells were grown for 24 h in 96-well tissue culture plates (Sarstedt). Lactococcus strains were washed twice with PBS after 1 h-nisin induction from OD600 of 0.5 (for the inducible strains) or at OD600 of 0.8 for the control L. lactis NZ9000 ΔhtrA strain and the constitutive NZ9000 ΔhtrA (pNZP44:SEC-LLO) strain. L. monocytogenes EGDe was also included as a positive control. After washing, strains suspended in culture medium (i.e. DMEM glutamax® plus 10% FCS) were added to wells containing the J774 cells at a multiplicity of infection (MOI) of about 2–5 bacterial cells per each J774 cell. Plates were incubated for 1 h before removal of growth medium followed by a single wash with fresh warm medium and addition of fresh medium containing 20 μg/ml gentamicin to kill extracellular bacteria [39]. Plates were then incubated for a further 1 h. J774 cells were lysed (to release intracellular bacteria) at this timepoint (2 h sample, T2) using 200 μl cold sterile distilled water followed by dilution and plating onto appropriate agar media. Gentamicin-containing medium was removed from remaining wells, fresh medium was added and plates incubated for another 6 h following which time the bacterial count was similarly determined (T8 sample).
2.7. Animals and immunization protocols
Female BALB/c mice, 6–8 weeks in age, were used in all animal experiments. All animal procedures were reviewed and approved by the ethical assessment committee of University College Cork (UCC). Animals were divided into groups of five mice each. Groups were given eight doses of the three LLO-producing strains either by oral gavage or intraperitoneal injection on days 1, 2, 7, 14, 21, 28, 35, and 36. L. lactis NZ9000-treated groups (oral and IP) and non-treated groups were included as negative controls. Groups treated with sub-lethal IP L. monocytogenes EGDe (2 × 103 CFU/mouse for a total of six doses on a weekly basis) were also included as a positive control.
Inoculation doses were 5 × 109 CFU/mouse for the oral experiments while for the IP route, doses of 2 × 104 and 2 × 105 CFU/mouse were given on days 1 and 2, respectively followed by booster doses of 107 CFU/mouse. This gradual increase of the IP inoculation doses was intended to avoid any possible adverse effects associated with parenteral administration of high doses of bacteria to naïve mice. The two inducible strains were grown to an OD600 of 0.5 in GM17 broth (10 μg/ml Cm), induced with nisin 0.2% (v/v) for 1 h, washed twice with PBS and resuspended in PBS prior to inoculation. For the constitutive LLO-secreting strain, L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO), overnight cultures were inoculated into fresh GM17 broth containing 10 μg/ml Cm and grown to an OD600 of 0.8 at which point cells were harvested, washed twice with PBS and resuspended again in PBS for inoculation. Final dose volume per mouse was 200 μl for both oral and IP inoculations.
2.8. Detection of LLO-specific CD8+ T cells by the enzyme-linked immunospot (ELISPOT) test
The ELISPOT test was used as described previously [40] to detect cytotoxic CD8+ cells specific to the H2-Kd-restricted LLO epitope, LLO91–99, GYKDGNEYI (Peptide Protein Research, UK). Mouse mastocytoma line P815-1-1 cells (The European Collection of Cell Culture; ECACC) were used as antigen presenting cells (APCs). P815-1-1 cells pulsed with 10−6 M of LLO91–99 peptide or non-pulsed were used to stimulate splenocytes isolated from mice after the vaccination regimen. Numbers of LLO91–99-specific IFN-γ-secreting cells were counted using a stereomicroscope.
2.9. Detection of LLO-specific IgG1 and IgG2a in sera of immunized mice by the enzyme-linked immunosorbent assay (ELISA)
Titres of LLO-specific IgG1 and IgG2a antibodies were determined in sera of treated mice and control groups using ELISA as previously described [18]. Briefly, 96-well ELISA plates (Costar®, Corning Incorporated) were coated overnight at 4 °C with 100 μl per well of 100 μg/ml LLO [purified by Ni-NTA affinity chromatography from cell lysate of E. coli M15 (pREP4) harboring pQE30/hly after induction with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h at room temperature with shaking]. Plates were washed with PBS containing 0.05% Tween 20 (Sigma) (PBST) and blocked for 3 h with 10% skimmed milk, then washed again. Dilutions of serum samples were added for 1 h at room temperature followed by washing with PBST and addition of horseradish peroxidase (HRP)-conjugated rat anti-mouse IgG1 or IgG2a (BD Biosciences) for 1 h at room temperature. Detection was performed using the HRP substrate, 3,3′,5,5′-tetramethylbenzidine (TMB) and the reaction was stopped by addition of 1 M sulfuric acid. Absorbance was measured at 450 nm and titres were defined as the serum dilution reciprocal, as calculated from the regression equation of the linear part of the curve obtained by plotting serum dilutions against absorbance, that has absorbance equal to 2S.D. (double standard deviation) above the highest absorbance value observed with the negative control groups.
2.10. Challenge test using luminescent L. monocytogenes
Listerial challenge was performed as previously described [41]. Briefly, mice were challenged after a specified period (3 days or 6 weeks) from the last vaccination booster with intraperitoneal injection of 200 μl of 2 × 106 CFU/ml, i.e. 4 × 105 CFU/mouse, luminescent L. monocytogenes EGDe (Table 1). This luminescent strain has been recently characterized and proved to have identical growth characteristics to the wild type [42]. Mice were euthanized 3 days post-challenge and the listerial burden was determined in spleens and livers by measuring the whole-organ luminescence in Xenogen IVIS100 machine (Xenogen, Alameda, CA). This was followed by organ homogenization, serial dilutions and plating on BHI agar plates containing 7.5 μg/ml Cm. Plates were incubated at 37 °C for 2 days and Listeria counts were calculated per organ. The limit of detection (LOD) of Listeria was 50 CFU/organ.
2.11. Interpretation of data and statistical analysis
The Student's t-test was used for statistical analysis of data and results with P values of less than 0.05 were considered statistically significant.
3. Results
3.1. Production of hemolytically active LLO by engineered lactococcal vectors
Ni-NTA gel purification of the His-tagged LLO and SDS–PAGE followed by Western blotting using polyclonal rabbit anti-LLO revealed efficient LLO production in three strains: L. lactis NZ9000 (pNZPnisA:CYTO-LLO), L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO), and L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO) (Fig. 2 ). His-tagged LLO was found in the cell lysate fraction (C) but not in the culture supernatant (secreted fraction; S) (Fig. 2B) of induced NZ9000 (pNZPnisA:CYTO-LLO). For the LLO-secreting strains (NZ9000 ΔhtrA harboring pNZP44:SEC-LLO or pNZPnisA:SEC-LLO), Western blot with anti-LLO antibodies showed positive bands in both the C and S fractions (Fig. 2B). His-tagged LLO could be isolated by Ni-NTA chromatography from the C fraction of NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) indicating its presence along with the precursor SEC-LLO (data not shown). On the contrary, no His-tagged LLO could be isolated from the C fraction of NZ9000 ΔhtrA (pNZP44:SEC-LLO) suggesting the presence of only the precursor intracellularly (data not shown).
Fig. 2.
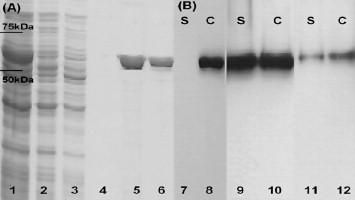
SDS–PAGE and Western blots that confirm LLO production by three of the constructed strains. (A) Lanes 1–6 represent SDS–PAGE of the Ni-NTA purification procedure of the His-tagged LLO produced by Lactococcus lactis NZ9000 (pNZPnisA:CYTO-LLO), where lane 1 is the cell lysate, lane 2 is the flow-through of the Ni-NTA column, lanes 3 and 4 are two washings of the column, and lanes 5 and 6 show the eluted LLO. (B) Lanes 7–12 show a Western blot of LLO using polyclonal rabbit anti-LLO antibodies. C: cell lysate; S: culture supernatant fraction (precipitated by TCA treatment). Results of LLO production assessment are shown for L. lactis NZ9000 (pNZPnisA:CYTO-LLO) (lanes 7 and 8), L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) (lanes 9 and 10) and L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO) (lanes 11 and 12).
No LLO could be detected in case of L. lactis MG1363 (pNZP44:CYTO-LLO) though the DNA sequence was confirmed to be correct (Lark Technologies Inc., UK). Analysis of both log-phase and stationary-phase cell lysates, cell pellets (for bound LLO) and culture supernatants of L. lactis MG1363 (pNZP44:CYTO-LLO) failed to detect LLO (data not shown). Consequently, all subsequent animal experiments were conducted with only the three strains capable of efficient LLO expression. LLO produced by these three strains showed detectable hemolytic activity which confirmed expression of the active hemolysin in L. lactis. For purified LLO obtained from NZ9000 (pNZPnisA:CYTO-LLO) (Fig. 2A) the specific LLO hemolytic activity ranged from 5 × 105 to 5 × 107 hemolytic units (HU)/mg total protein. For secreted LLO the dialyzed supernatant of L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) demonstrated CHU of 4 while that of L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO) showed a weak hemolytic activity where undiluted dialyzed supernatant caused only 22% hemolysis as compared to the 100% hemolysis of the positive control (distilled water). This latter finding is most likely related to the level of secreted LLO which was much lower in case of L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO) (Fig. 2B). The above values of hemolytic activity are similar to those obtained for LLO expressed in other vectors [37], [43].
The two nisin-inducible strains were examined for continuous LLO production upon the removal of nisin after 1 h induction period. We found that the two strains continued production of LLO in vitro as indicated by the significant accumulation of the protein over a five hour period (data not shown). Similar results were demonstrated by Bermudez-Humaran et al. for nisin-inducible lactococci [44], [45]. Consequently, the inducible strains were treated with nisin for 1 h rather than 3 h in the intracellular growth assay and in animal inoculations.
3.2. Intracellular growth in J774 macrophage-like cells
To assess bacterial growth in J774 cells treated with the bacterial strains, plate counts were performed at T2 and T8 post-treatment. Over the 6-h period between T2 and T8 bacterial counts of wild-type L. monocytogenes increased significantly (P < 0.001) by 100-fold while the counts of the three-LLO producing strains along with the negative control L. lactis NZ9000 ΔhtrA decreased significantly (P < 0.02) by more than twofold (data not shown). These results demonstrate the incapability of Lactococcus strains for long-term survival in macrophage cells.
3.3. Short-term assessment of the immune response 3 days after oral and IP vaccination
3.3.1. CD8+ T lymphocytes specific for the H2-Kd-restricted LLO91–99 epitope are elicited by all strains following IP immunization
The ELISPOT assay was used to analyze the development of LLO-specific CD8+ cells. The mouse mastocytoma cells P815-1-1 were used as antigen presenting cells as they express restricted H2-Kd MHC class I molecules, so any resulting spots would be due to LLO-specific CD8+ cells [40]. The short-term immune response was assessed 3 days after the last booster for both the IP and oral routes. All three strains demonstrated LLO91–99-specific spots following IP inoculations (Fig. 3 ) but no spots were observed following oral inoculations (data not shown). L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) that inducibly secretes LLO showed the greatest propensity to stimulate LLO91–99-specific T cells when compared to the other two vaccine strains (Fig. 3).
Fig. 3.
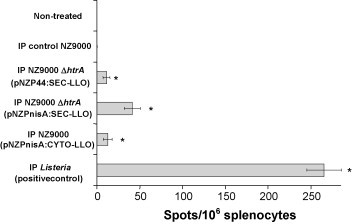
ELISPOT test results 3 days after IP vaccination regimen. Mouse groups (n = 5) were vaccinated by IP injection on days 1, 2, 7, 14, 21, 28, 35, and 36 then examined by ELISPOT on day 39. *P < 0.05 by the Student's t-test as compared to the negative control groups. Error bars represent the mean ± S.E.M. (standard error of the mean).
3.3.2. Serum IgG1 and IgG2a titres in vaccinated mice 3 days after last booster
Serum antibody titres were measured 3 days after the final vaccine booster using LLO-specific ELISA. L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) could elicit LLO-specific serum IgG1 (indicative of a T helper type 2 (Th2) immune response) and IgG2a (indicative of T helper type 1 (Th1) immune response) by both the IP (Fig. 4 ) and oral (data not shown) routes. However, the titres were not statistically significant by the oral route. Although L. lactis NZ9000 ΔhtrA (pNZP44:SEC-LLO) produced detectable IgG2a titres by the IP route and IgG1 titres by both the IP (Fig. 4) and oral (data not shown) routes, only the IgG1 titres by the IP route were statistically significant (Fig. 4). No statistically significant antibody titres were observed with L. lactis NZ9000 (pNZPnisA:CYTO-LLO) by either the oral (data not shown) or IP routes (Fig. 4).
Fig. 4.
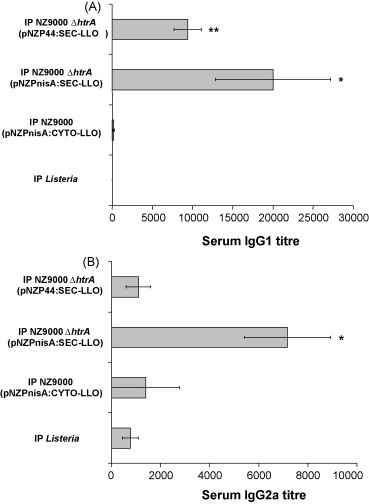
LLO-specific antibody response 3 days (short term) after IP vaccination. Mouse groups (n = 5) were IP vaccinated on days 1, 2, 7, 14, 21, 28, 35, and 36. Murine serum samples were collected on day 39 and analyzed by ELISA for IgG1 (A) and IgG2a (B) titres. *P < 0.05; **P < 0.01 by the Student's t-test as compared to negative control groups. Error bars represent the mean ± S.E.M.
3.3.3. Protection against L. monocytogenes challenge following IP vaccination
It should be noted that the poor oral infectivity of L. monocytogenes in the murine model prevents the use of oral challenge studies [3]. Mice were therefore challenged intraperitoneally with luminescent L. monocytogenes 3 days after the final vaccine booster to mimic the systemic phase of infection. Three days following challenge mice were euthanized and luminescence of individual organs was measured using the Xenogen IVIS100 machine (Xenogen, Alameda, CA) followed by plating and determination of the Listeria counts in livers and spleens. Bacterial count results following IP vaccination revealed significant protection as evidenced by the low listerial count in spleens and livers of mice vaccinated with either of the three LLO-producing strains (Fig. 5 ). On the contrary, strong luminescence and high listerial counts were observed in organs of the non-treated groups or groups treated with the control strain L. lactis NZ9000. However, it was found that mice treated with control strain L. lactis NZ9000 showed significantly lower listerial organ counts (P < 0.05) than the non-treated mice, indicating non-specific immune protection against L. monocytogenes by the L. lactis strain (Fig. 5). No significant difference was observed between the orally vaccinated groups and the negative control groups with any of the three strains (data not shown).
Fig. 5.
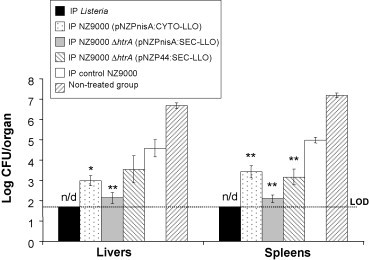
Results of the challenge experiment 3 days after the IP immunization regimen. Mouse groups (n = 5) were IP vaccinated on days 1, 2, 7, 14, 21, 28, 35, and 36 then challenged IP with luminescent Listeria monocytogenes on day 39. Mice were euthanized 3 days later for Listeria count in spleens and livers. *P < 0.02, **P < 0.01 by the Student's t-test as compared to negative control groups. Error bars represent the mean ± S.E.M. LOD: limit of detection of the test. n/d: not detectable.
In summary, the best protection was attained following inoculation with the inducible LLO-secreting strain L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) via the IP route (Fig. 5). These results reflect the previous ELISPOT (Fig. 3) and antibody titre (Fig. 4) results where this particular strain elicited the highest number of LLO91–99-specific spots and LLO-specific serum antibodies respectively. This strain did not elicit statistically significant protection against IP Listeria challenge when given as an oral vaccine, however analysis of the liver counts demonstrated a P value of 0.07 (data not shown) indicating that this approach (oral vaccination) may warrant further study.
3.4. Long-term assessment of the immune response 6 weeks after oral and IP vaccination with L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO)
Since L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) showed the best protection results in the short term by the IP route, this strain was further investigated in longer term vaccination studies (6 weeks following the last booster). Moreover, we assumed that the rapid death of Lactococcus due to gastric acidity (as suggested by our in vitro observations; data not shown) might be a major cause of vaccine failure when given orally. Therefore, we repeated the oral vaccination with the final inoculum resuspended in GM17 broth pH 8.5 (adjusted with 50 mM CO3 2− of sodium bicarbonate) [46]. The L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) strain was given by the oral or IP routes using the previously described vaccination regimens and the ELISPOT test, serum IgG1 and IgG2a titres and challenge test were performed 6 weeks after the last booster. LLO91–99-specific CD8+ lymphocytes were detected following the IP but not the oral (pH 8.5) vaccination regimen (Fig. 6 ). Similarly both statistically significant IgG1 and IgG2a titres were detected following administration of the IP but not the oral vaccine (Fig. 6). Finally, challenge experiments confirmed the protective efficacy of the injected L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) strain (Fig. 7 ). No difference in listerial organ count was observed between the group treated with control L. lactis NZ9000 ΔhtrA and the non-treated group which indicates the lack of non-specific immune stimulation at 6 weeks post-vaccination (Fig. 7). Representative organ luminescence pictures of the challenge experiment are shown in Fig. 7. It is noteworthy that we found the sensitivity of luminescence detection is limited to about 105 CFU/organ below which no luminescence could be measured. This finding confined the use of the luminescence data to qualitative rather than quantitative comparisons in both the short- and long-term challenge experiments. These results confirm that IP inoculation confers longer term protection (6 weeks after last booster) and specific immunity (antibodies and cytotoxic CD8+ cells). Moreover, it is shown that oral inoculation at pH 8.5 did not promote vaccine efficacy and suggests that other gastrointestinal conditions (e.g. the presence of bile) may contribute to the reduced viability of Lactococcus during gastrointestinal transit [47].
Fig. 6.
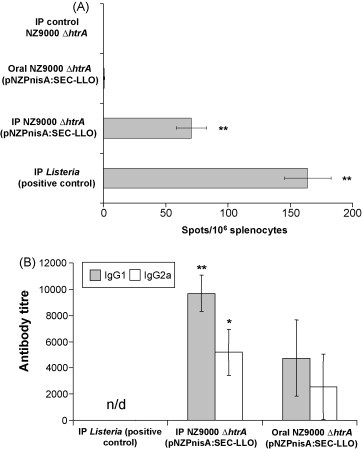
Immune response 6 weeks after immunization with L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO). Mouse groups (n = 5) were IP or orally (pH 8.5) vaccinated on days 1, 2, 7, 14, 21, 28, 35, and 36. (A) LLO91–99-specific CD8+ response examined by the ELISPOT test. (B) Serum antibody titres (IgG1 and IgG2a) examined by the ELISA test. *P < 0.05, **P ≤ 0.02 by the Student's t-test as compared to negative control groups. Error bars represent the mean ± S.E.M. n/d: not detectable.
Fig. 7.
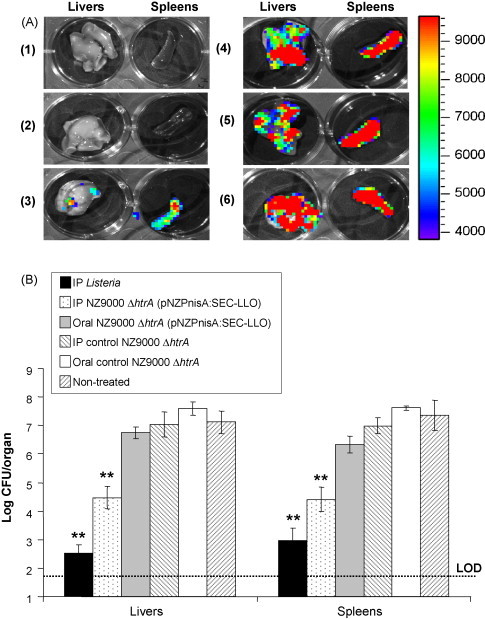
Results of the murine challenge experiment 6 weeks (long term) after the immunization regimen with L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO). Mouse groups (n = 5) were vaccinated on days 1, 2, 7, 14, 21, 28, 35, and 36. Animals were challenged 6 weeks later with luminescent L. monocytogenes via the IP route and mice were euthanized 3 days post-challenge for luminescence detection in the Xenogen IVIS100 machine (A), followed by organ homogenization and plating for Listeria counts in spleens and livers (B). Representative images are shown in (A) showing luminescence detection in isolated livers and spleens of mice vaccinated with (1) sub-lethal IP L. monocytogenes, (2) IP L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO), (3) oral L. lactis NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) (pH 8.5), (4) IP control L. lactis NZ9000 ΔhtrA, and (5) oral control L. lactis NZ9000 ΔhtrA (pH 8.5). (6) represents the non-treated control group. The color bar to the right indicates luminescence signal intensity (in photons s−1 cm−2). **P < 0.01 by the Student's t-test as compared to negative control groups. Error bars represent the mean ± S.E.M. LOD: limit of detection of the test.
4. Discussion
L. lactis has been proposed as a safe and effective vaccine delivery platform for the delivery of heterologous antigens to the immune system [14]. Here we examined a number of approaches to overexpress the major immunodominant antigen, LLO, from L. monocytogenes in L. lactis and analyzed the protective anti-listerial immune response in the murine infection model.
We investigated the efficacy of LLO expression by L. lactis when the gene was placed under the control of constitutive (P44 promoter region) or inducible (PnisA promoter region) promoters. We also examined the effect of protein secretion upon the protective efficacy of the Lactococcus-based vaccine strains. Previous work in our laboratory optimised detection methods and examined expression levels of non-secreted LLO in L. lactis NZ9000 (pNZPnisA:CYTO-LLO) (manuscript submitted). However, intracellular LLO expressed under the relatively weak constitutive P44 promoter [27] (pNZP44:CYTO-LLO) could not be detected in the present study. While LLO is clearly non-toxic in our system and the sequence of this construct was validated (data not shown), the exact reasons for the lack of detectable LLO protein from this construct are unclear. The degradation and toxic effects of intracellularly expressed heterologous proteins in L. lactis have previously been reported [25], [48]. Thus the current cloning and expression work created three potential L. lactis vaccine strains expressing LLO.
We examined the intracellular growth of the three L. lactis vaccine strains in J774 macrophage-like cells by determining bacterial counts at various time intervals. In our model L. lactis was incapable of growth in J774 cells, as evidenced by experimental bacterial counts, irrespective of LLO expression. This contrasted significantly with the L. monocytogenes control which increased in numbers over time. Listeriolysin O is essential for the escape of L. monocytogenes from the macrophage phagosome to the cytoplasmic compartment [3]. It has been shown previously that an engineered LLO-secreting Bacillus subtilis strain can escape from the phagosomal compartment and grow in the J774 cell cytosol [49]. In contrast, Goetz et al. utilized microinjection to introduce a variety of bacteria directly into the cytoplasm of Caco-2 cells and found that bacteria that are not adapted to the cytoplasmic compartment are unable to multiply and grow [50]. Our work supports the latter findings and suggests that L. lactis vaccine vectors engineered to access the cytoplasmic antigen presenting pathway are incapable of further growth in this environment. Unlike L. monocytogenes, Lactococcus is not adapted to multiply and persist inside macrophages and does not possess the molecular factors that enable survival in the host cytosol [51]. It is noteworthy that LLO is rapidly inactivated once in the cytosol of infected cells and does not display toxicity towards the host cytoplasmic membrane [52]. We hypothesized that LLO-expressing L. lactis strains will access the cytoplasmic MHC class I antigen presenting pathway in antigen presenting cells and deliver LLO, an antigen previously determined to provide protective immunity against L. monocytogenes [6].
We measured the serum titres of the two IgG isotypes; IgG1 (reflective of the humoral T helper type 2, Th2 CD4+ response) and IgG2a (reflective of the cell-mediated T helper type 1, Th1 CD4+ response) [53], [54]. Although significant antibody titres were elicited by two of the vaccine strains (Fig. 4, Fig. 6), the results of the challenge test suggested that the CD8+ response was more important for protection. Significantly another study found that CD8+ T cells (and not antibodies or CD4+ cells) are the major mediators of immunity against Listeria when LLO was delivered by an attenuated B. anthracis strain [9]. Our data also suggest that future L. lactis vectors expressing LLO combined with other heterologous antigens may be useful in eliciting antibody-mediated as well as cell-mediated immune responses.
Engineered L. lactis strains were capable of eliciting specific CD8+ T cells against the dominant LLO91–99 epitope. While the CD8+ cell responses were significant, vaccine strains induced lower levels of specific CD8+ T cells than the positive control (sub-lethal L. monocytogenes). However, all engineered L. lactis strains provided protective immunity against secondary challenge with L. monocytogenes. It is clear in the literature that robust CD8+ T-cell responses are required for immunity to L. monocytogenes [6]. Here levels of LLO-specific CD8+ cells generated through vaccination with engineered L. lactis were sufficient to elicit enhanced clearance of L. monocytogenes relative to control mice inoculated with control L. lactis.
Overall the greatest response was generated by the LLO-secreting nisin-inducible strain, NZ9000 ΔhtrA (pNZPnisA:SEC-LLO) which also generated the highest LLO expression levels in vitro (Fig. 2). In addition, it is reported in the literature that secreted LLO is more immunogenic than the non-secreted form [7]. We utilized this strain in longer term vaccination studies and analyzed protective immune responses 6 weeks following the final booster inoculation. These studies demonstrated that this vaccine vector was capable of inducing specific protection against subsequent L. monocytogenes infection. The vector was not capable of inducing immunity to the same level as exposure to sub-lethal L. monocytogenes positive control. This is most likely due to the fact that the current L. lactis vectors express only a single L. monocytogenes antigen (i.e. LLO). Previous studies utilizing Salmonella enterica serovar Typhimurium expressing both LLO and P60 listerial antigens showed enhanced protection against listeriosis when compared to vectors expressing single antigens [55].
L. lactis has been repeatedly employed as a mucosal (intranasal or oral) vaccine delivery vehicle [16], [18], [44]. However, the viability of Lactococcus is well-known to be greatly diminished in the gastrointestinal tract [56], thus LLO production and presentation to the immune system is expected to be reduced in this environment. We attempted to maximize the oral dose of Lactococcus, buffer the vaccine inocula to pH 8.5 and increase the number of vaccination doses to compensate for that loss. A similar approach was used by other investigators [54], [57], [58] who used seven to nine oral doses with various vaccination regimens. However, in the present study we did not obtain significant protection against L. monocytogenes following oral vaccination. The reason for this may be the well-reported rapid killing of L. lactis in the gut [56], [59], the vaccination regimen used, the nature of the expressed antigen itself or a combination of all these factors. Further studies and strategies to improve vaccine efficacy via the oral route are ongoing in our laboratory.
A number of host carriers have previously been examined to heterologously express LLO and act as live vaccines against L. monocytogenes. An attenuated B. anthracis strain was used as a live LLO-secreting vaccine vector against Listeria via the subcutaneous route [9]. This could elicit LLO-specific CD8+ responses, anti-LLO antibodies and provided partial protection against L. monocytogenes challenge. Recombinant attenuated aroA− S. typhimurium strains that produced LLO were also investigated as vaccine candidates against listeriosis. The aroA− S. typhimurium strain producing secreted LLO (but not a strain producing intracellular LLO) was found to be protective against Listeria challenge [7]. In another study, S. typhimurium expressed LLO fused to the Yersinia outer protein E (YopE) as a carrier molecule for cytosolic delivery using the S. typhimurium type III secretion system [8]. Oral vaccination with that strain elicited LLO91–99-specific CD8+ T cells and protective immunity upon listerial challenge.
While LLO is a well-described antigen of L. monocytogenes the protein also plays a role in lysis of the phagosomal membrane and can therefore permit entry of other antigens into the cytoplasmic antigen processing pathway [60]. This concept of LLO redirecting accompanying antigens towards a CD8+-mediated immune response is well established in the literature [61], [62]. LLO-secreting Mycobacterium bovis BCG strains were able to increase the protective efficacy of the vaccination against tuberculosis due to better antigen presentation through the cytosolic MHC class I pathway [37]. In addition, E. coli cells expressing both LLO and ovalbumin (OVA) demonstrated enhanced delivery of the OVA Kb-restricted epitope SIINFEKL for MHC class I presentation and could protect 75% of mice upon challenge with an OVA-expressing melanoma cell line [63]. Similarly MHC class I processing of OVA in macrophages could be improved in S. typhimurium dually expressing both LLO and OVA [64].
The use of L. lactis as an LLO-vector has a number of significant advantages over the other proposed platforms described above or attenuated mutants of L. monocytogenes [7], [9], [62]. L. lactis is a food-grade, non-pathogenic bacterium and its safety profile is well-established. Unlike Gram-negative vectors, L. lactis does not produce endotoxins and at the same time it has an innate immunostimulatory effect [54]. As LLO enhances access to the cytoplasmic processing pathway we propose that L. lactis-expressing LLO may also provide a safe platform for the delivery of other heterologous antigens (including viral and tumour antigens) for the generation of specific memory and effector CD8+ cell responses.
In conclusion, we have developed engineered L. lactis strains capable of producing active LLO, an immunodominant protein antigen of L. monocytogenes. One vector strain in particular was capable of secreting prolonged high levels of LLO following induction by nisin and generated protective immunity against L. monocytogenes challenge that was defined by anti-LLO CD8+ T cells. This approach may inform the development of safe anti-listerial vaccines for protection against this zoonotic disease which affects not only humans but also domestic animals where listeriosis results in significant losses [4].
Acknowledgements
Mohammed Bahey-El-Din is funded through a scholarship from the Education and Culture Bureau of the Egyptian Embassy, London. The authors would like to acknowledge the funding received from the Irish Government under the National Development Plan 2000–2006 and the funding of the Alimentary Pharmabiotic Centre by the Science Foundation of Ireland Centres for Science Engineering and Technology (CSET) programme.
References
- 1.Meng J., Doyle M.P. Emerging issues in microbiological food safety. Annu Rev Nutr. 1997;17:255–275. doi: 10.1146/annurev.nutr.17.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy V., Cresence V.M., Rejitha J.S., Lekshmi M.U., Dharsana K.S., Prasad S.P. Listeria—review of epidemiology and pathogenesis. J Microbiol Immunol Infect. 2007;40(1):4–13. [PubMed] [Google Scholar]
- 3.Vazquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14(3):584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czuprynski C.J. Listeria monocytogenes: silage, sandwiches and science. Anim Health Res Rev. 2005;6(2):211–217. doi: 10.1079/ahr2005111. [DOI] [PubMed] [Google Scholar]
- 5.Pizarro-Cerda J., Cossart P. Subversion of cellular functions by Listeria monocytogenes. J Pathol. 2006;208(2):215–223. doi: 10.1002/path.1888. [DOI] [PubMed] [Google Scholar]
- 6.Pamer E.G. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4(10):812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 7.Hess J., Gentschev I., Miko D., Welzel M., Ladel C., Goebel W. Superior efficacy of secreted over somatic antigen display in recombinant Salmonella vaccine induced protection against listeriosis. Proc Natl Acad Sci USA. 1996;93(4):1458–1463. doi: 10.1073/pnas.93.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russmann H., Igwe E.I., Sauer J., Hardt W.D., Bubert A., Geginat G. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing hybrid Yersinia type III proteins. J Immunol. 2001;167(1):357–365. doi: 10.4049/jimmunol.167.1.357. [DOI] [PubMed] [Google Scholar]
- 9.Sirard J.C., Fayolle C., de Chastellier C., Mock M., Leclerc C., Berche P. Intracytoplasmic delivery of listeriolysin O by a vaccinal strain of Bacillus anthracis induces CD8-mediated protection against Listeria monocytogenes. J Immunol. 1997;159(9):4435–4443. [PubMed] [Google Scholar]
- 10.Angelakopoulos H., Loock K., Sisul D.M., Jensen E.R., Miller J.F., Hohmann E.L. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect Immun. 2002;70(7):3592–3601. doi: 10.1128/IAI.70.7.3592-3601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockstedt D.G., Giedlin M.A., Leong M.L., Bahjat K.S., Gao Y., Luckett W. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci USA. 2004;101(38):13832–13837. doi: 10.1073/pnas.0406035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang S., Rasmussen R.A., Nolan K.M., Frankel F.R., Lieberman J., McClure H.M. Live attenuated Listeria monocytogenes expressing HIV Gag: immunogenicity in rhesus monkeys. Vaccine. 2007;25(42):7470–7479. doi: 10.1016/j.vaccine.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson R.J., Bouwer H.G., Portnoy D.A., Frankel F.R. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect Immun. 1998;66(8):3552–3561. doi: 10.1128/iai.66.8.3552-3561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouaille S., Ribeiro L.A., Miyoshi A., Pontes D., Le Loir Y., Oliveira S.C. Heterologous protein production and delivery systems for Lactococcus lactis. Genet Mol Res. 2003;2(1):102–111. [PubMed] [Google Scholar]
- 15.Robinson K., Chamberlain L.M., Schofield K.M., Wells J.M., Le Page R.W. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat Biotechnol. 1997;15(7):653–657. doi: 10.1038/nbt0797-653. [DOI] [PubMed] [Google Scholar]
- 16.Kim S.J., Jun do Y., Yang C.H., Kim Y.H. Expression of Helicobacter pylori cag12 gene in Lactococcus lactis MG1363 and its oral administration to induce systemic anti-Cag12 immune response in mice. Appl Microbiol Biotechnol. 2006;72(3):462–470. doi: 10.1007/s00253-005-0285-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee P., Faubert G.M. Expression of the Giardia lamblia cyst wall protein 2 in Lactococcus lactis. Microbiology. 2006;152(Pt 7):1981–1990. doi: 10.1099/mic.0.28877-0. [DOI] [PubMed] [Google Scholar]
- 18.Pei H., Liu J., Cheng Y., Sun C., Wang C., Lu Y. Expression of SARS-coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Appl Microbiol Biotechnol. 2005;68(2):220–227. doi: 10.1007/s00253-004-1869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braat H., Rottiers P., Hommes D.W., Huyghebaert N., Remaut E., Remon J.P. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease. Clin Gastroenterol Hepatol. 2006;4(6):754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 20.Vandenbroucke K., Hans W., Van Huysse J., Neirynck S., Demetter P., Remaut E. Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology. 2004;127(2):502–513. doi: 10.1053/j.gastro.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Bermudez-Humaran L.G., Langella P., Miyoshi A., Gruss A., Guerra R.T., Montes de Oca-Luna R. Production of human papillomavirus type 16 E7 protein in Lactococcus lactis. Appl Environ Microbiol. 2002;68(2):917–922. doi: 10.1128/AEM.68.2.917-922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunji E.R., Slotboom D.J., Poolman B. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim Biophys Acta. 2003;1610(1):97–108. doi: 10.1016/s0005-2736(02)00712-5. [DOI] [PubMed] [Google Scholar]
- 23.Wegmann U., O’Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007;189(8):3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Asseldonk M., Rutten G., Oteman M., Siezen R.J., de Vos W.M., Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990;95(1):155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 25.Le Loir Y., Azevedo V., Oliveira S.C., Freitas D.A., Miyoshi A., Bermudez-Humaran L.G. Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb Cell Fact. 2005;4(1):2. doi: 10.1186/1475-2859-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steidler L., Neirynck S., Huyghebaert N., Snoeck V., Vermeire A., Goddeeris B. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21(7):785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- 27.van der Vossen J.M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mierau I., Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68(6):705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers O.P., de Ruyter P., Kleerebezem M., de Vos W.M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64(1):15–21. [Google Scholar]
- 30.Horton R.M., Cai Z.L., Ho S.N., Pease L.R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8(5):528–535. [PubMed] [Google Scholar]
- 31.McGrath S., Fitzgerald G.F., van Sinderen D. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol. 2001;67(2):608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindholm A., Smeds A., Palva A. Receptor binding domain of Escherichia coli F18 fimbrial adhesin FedF can be both efficiently secreted and surface displayed in a functional form in Lactococcus lactis. Appl Environ Microbiol. 2004;70(4):2061–2071. doi: 10.1128/AEM.70.4.2061-2071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poquet I., Saint V., Seznec E., Simoes N., Bolotin A., Gruss A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35(5):1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- 34.Holo H., Nes I.F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piard J.C., Hautefort I., Fischetti V.A., Ehrlich S.D., Fons M., Gruss A. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J Bacteriol. 1997;179(9):3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohda C., Kawamura I., Baba H., Nomura T., Ito Y., Kimoto T. Dissociated linkage of cytokine-inducing activity and cytotoxicity to different domains of listeriolysin O from Listeria monocytogenes. Infect Immun. 2002;70(3):1334–1341. doi: 10.1128/IAI.70.3.1334-1341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess J., Grode L., Gentschev I., Fensterle J., Dietrich G., Goebel W. Secretion of different listeriolysin cognates by recombinant attenuated Salmonella typhimurium: superior efficacy of haemolytic over non-haemolytic constructs after oral vaccination. Microbes Infect. 2000;2(15):1799–1806. doi: 10.1016/s1286-4579(00)01333-2. [DOI] [PubMed] [Google Scholar]
- 38.Wisniewski J., Krawczyk-Balska A., Bielecki J. Associated roles of hemolysin and p60 protein for the intracellular growth of Bacillus subtilis. FEMS Immunol Med Microbiol. 2006;46(3):330–339. doi: 10.1111/j.1574-695X.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- 39.Guimaraes V.D., Gabriel J.E., Lefevre F., Cabanes D., Gruss A., Cossart P. Internalin-expressing Lactococcus lactis is able to invade small intestine of guinea pigs and deliver DNA into mammalian epithelial cells. Microbes Infect. 2005;7(5/6):836–844. doi: 10.1016/j.micinf.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Carvalho L.H., Hafalla J.C., Zavala F. ELISPOT assay to measure antigen-specific murine CD8(+) T cell responses. J Immunol Methods. 2001;252(1/2):207–218. doi: 10.1016/s0022-1759(01)00331-3. [DOI] [PubMed] [Google Scholar]
- 41.Stack H.M., Sleator R.D., Bowers M., Hill C., Gahan C.G. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl Environ Microbiol. 2005;71(8):4241–4247. doi: 10.1128/AEM.71.8.4241-4247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riedel C.U., Monk I.R., Casey P.G., Morrissey D., O'Sullivan G.C., Tangney M. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl Environ Microbiol. 2007;73(9):3091–3094. [Google Scholar]
- 43.Churchill R.L., Lee H., Hall J.C. Rapid purification of recombinant listeriolysin O (LLO) from Escherichia coli. J Ind Microbiol Biotechnol. 2005;32(8):355–363. doi: 10.1007/s10295-005-0002-2. [DOI] [PubMed] [Google Scholar]
- 44.Bermudez-Humaran L.G., Cortes-Perez N.G., Le Loir Y., Alcocer-Gonzalez J.M., Tamez-Guerra R.S., de Oca-Luna R.M. An inducible surface presentation system improves cellular immunity against human papillomavirus type 16 E7 antigen in mice after nasal administration with recombinant lactococci. J Med Microbiol. 2004;53(Pt 5):427–433. doi: 10.1099/jmm.0.05472-0. [DOI] [PubMed] [Google Scholar]
- 45.Bermudez-Humaran L.G., Langella P., Commissaire J., Gilbert S., Le Loir Y., L’Haridon R. Controlled intra- or extracellular production of staphylococcal nuclease and ovine omega interferon in Lactococcus lactis. FEMS Microbiol Lett. 2003;224(2):307–313. doi: 10.1016/S0378-1097(03)00475-0. [DOI] [PubMed] [Google Scholar]
- 46.Steidler L., Hans W., Schotte L., Neirynck S., Obermeier F., Falk W. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289(5483):1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 47.Drouault S., Corthier G., Ehrlich S.D., Renault P. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl Environ Microbiol. 1999;65(11):4881–4886. doi: 10.1128/aem.65.11.4881-4886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyoshi A., Bermudez-Humaran L.G., Ribeiro L.A., Le Loir Y., Oliveira S.C., Langella P. Heterologous expression of Brucella abortus GroEL heat-shock protein in Lactococcus lactis. Microb Cell Fact. 2006;5:14. doi: 10.1186/1475-2859-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Portnoy D.A., Tweten R.K., Kehoe M., Bielecki J. Capacity of listeriolysin O, streptolysin O, and perfringolysin O to mediate growth of Bacillus subtilis within mammalian cells. Infect Immun. 1992;60(7):2710–2717. doi: 10.1128/iai.60.7.2710-2717.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goetz M., Bubert A., Wang G., Chico-Calero I., Vazquez-Boland J.A., Beck M. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc Natl Acad Sci USA. 2001;98(21):12221–12226. doi: 10.1073/pnas.211106398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joseph B., Przybilla K., Stuhler C., Schauer K., Slaghuis J., Fuchs T.M. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188(2):556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Decatur A.L., Portnoy D.A. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science. 2000;290(5493):992–995. doi: 10.1126/science.290.5493.992. [DOI] [PubMed] [Google Scholar]
- 53.Jin B., Wang R.Y., Qiu Q., Sugauchi F., Grandinetti T., Alter H.J. Induction of potent cellular immune response in mice by hepatitis C virus NS3 protein with double-stranded RNA. Immunology. 2007;122(1):15–27. doi: 10.1111/j.1365-2567.2007.02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson K., Chamberlain L.M., Lopez M.C., Rush C.M., Marcotte H., Le Page R.W. Mucosal and cellular immune responses elicited by recombinant Lactococcus lactis strains expressing tetanus toxin fragment C. Infect Immun. 2004;72(5):2753–2761. doi: 10.1128/IAI.72.5.2753-2761.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Igwe E.I., Geginat G., Russmann H. Concomitant cytosolic delivery of two immunodominant listerial antigens by Salmonella enterica serovar typhimurium confers superior protection against murine listeriosis. Infect Immun. 2002;70(12):7114–7119. doi: 10.1128/IAI.70.12.7114-7119.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimoto H., Nomura M., Kobayashi M., Mizumachi K., Okamoto T. Survival of lactococci during passage through mouse digestive tract. Can J Microbiol. 2003;49(11):707–711. doi: 10.1139/w03-092. [DOI] [PubMed] [Google Scholar]
- 57.Lee M.H., Roussel Y., Wilks M., Tabaqchali S. Expression of Helicobacter pylori urease subunit B gene in Lactococcus lactis MG1363 and its use as a vaccine delivery system against H. pylori infection in mice. Vaccine. 2001;19(28/29):3927–3935. doi: 10.1016/s0264-410x(01)00119-0. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z.H., Jiang P.H., Li N.J., Shi M., Huang W. Oral vaccination of mice against rodent malaria with recombinant Lactococcus lactis expressing MSP-1(19) World J Gastroenterol. 2005;11(44):6975–6980. doi: 10.3748/wjg.v11.i44.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vesa T., Pochart P., Marteau P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment Pharmacol Ther. 2000;14(6):823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 60.Bahey-El-Din M., Griffin B.T., Gahan C.G.M. Attack and counter-attack: targeted immunomodulation using bacterial virulence factors. In: Sleator R., Hill C., editors. Patho-Biotechnology. Landes Bioscience; Austin, TX: 2008. pp. 163–172. [Google Scholar]
- 61.Darji A., Chakraborty T., Wehland J., Weiss S. Listeriolysin generates a route for the presentation of exogenous antigens by major histocompatibility complex class I. Eur J Immunol. 1995;25(10):2967–2971. doi: 10.1002/eji.1830251038. [DOI] [PubMed] [Google Scholar]
- 62.Higgins D.E., Shastri N., Portnoy D.A. Delivery of protein to the cytosol of macrophages using Escherichia coli K-12. Mol Microbiol. 1999;31(6):1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 63.Radford K.J., Higgins D.E., Pasquini S., Cheadle E.J., Carta L., Jackson A.M. A recombinant E. coli vaccine to promote MHC class I-dependent antigen presentation: application to cancer immunotherapy. Gene Ther. 2002;9(21):1455–1463. doi: 10.1038/sj.gt.3301812. [DOI] [PubMed] [Google Scholar]
- 64.Catic A., Dietrich G., Gentschev I., Goebel W., Kaufmann S.H., Hess J. Introduction of protein or DNA delivered via recombinant Salmonella typhimurium into the major histocompatibility complex class I presentation pathway of macrophages. Microbes Infect. 1999;1(2):113–121. doi: 10.1016/s1286-4579(99)80001-x. [DOI] [PubMed] [Google Scholar]
- 65.Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F. Comparative genomics of Listeria species. Science. 2001;294(5543):849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 66.Gasson M.J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]


