Abstract
Fusion proteins involving Nucleoporin 98 (NUP98) are recurrently found in acute myeloid leukemia (AML) and are associated with poor prognosis. Lack of mechanistic insight into NUP98-fusion-dependent oncogenic transformation has so far precluded the development of rational targeted therapies. We reasoned that different NUP98-fusion proteins deregulate a common set of transcriptional targets that might be exploitable for therapy. To decipher transcriptional programs controlled by diverse NUP98-fusion proteins we developed mouse models for regulatable expression of NUP98/NSD1, NUP98/JARID1A and NUP98/DDX10. By integrating chromatin occupancy profiles of NUP98-fusion proteins with transcriptome profiling upon acute fusion protein inactivation in vivo, we defined the core set of direct transcriptional targets of NUP98-fusion proteins. Among those, CDK6 was highly expressed in murine and human AML samples. Loss of CDK6 severely attenuated NUP98-fusion-driven leukemogenesis and NUP98-fusion AML was sensitive to pharmacologic CDK6 inhibition in vitro and in vivo. These findings identify CDK6 as a conserved, critical direct target of NUP98-fusion proteins, proposing CDK4/CDK6 inhibitors as a new rational treatment option for AML patients with NUP98-fusions.
Keywords: Acute Myeloid Leukemia, NUP98-fusion proteins, CDK6, targeted therapy, Patient-derived xenografts
Introduction
Recurrent translocations involving chromosome 11 lead to fusion of the Nucleoporin 98 (NUP98) gene to more than 25 different recipient loci in leukemia.1 While the overall frequency of NUP98-fusions is low, they are significantly overrepresented in pediatric acute myeloid leukemia (AML), where the expression of these fusions defines a clinically and molecularly homogenous group of patients that have a particularly bad prognosis.2–4 Lack of a detailed molecular understanding of the mechanism of action of NUP98-fusion proteins has hampered the development of tailored approaches to efficiently target this leukemia subgroup.
NUP98/NSD1 and NUP98/JARID1A are the most frequent NUP98-fusion proteins in pediatric AML3,4, representing hybrid proteins joining the NUP98-N-terminus with the catalytic domain of the lysine methyltransferase NSD1 (KMT3B) or the lysine demethylase JARID1A (KDM5A), respectively.5,6 In addition, NUP98-fusions with the RNA helicase DDX10, the DNA topoisomerases TOP1, or the adapter protein PSIP1 were identified.7 NUP98 can also be fused to members of the Homeodomain transcription factor family, such as HOXA9, HOXD13 or PMX2.7 Patients with NUP98-fusions often carry additional mutations in NRAS, KRAS, KIT, MYC or FLT3 genes, indicating a potential functional cooperation with NUP98-fusions.1,8 In mouse models, NUP98-fusions recapitulate principal aspects of the human disease, including increased self-renewal of hematopoietic progenitor cells, inhibition of myeloid differentiation and high expression of HOXA genes.9–13
The endogenous NUP98 protein is an integral part of the nuclear pore complex, mediating nucleo-cytoplasmic transport of macromolecules across the nuclear membrane.14 In addition, NUP98 is involved in transcriptional regulation through interactions between the NUP98-N-terminus with the histone-modifying enzymes CBP/p300 and HDAC1.15,16 In hematopoietic cells, NUP98 regulates H3K4me3 levels by recruiting the SET1A histone methyltransferase complex to promoters of highly expressed genes.17 Unlike endogenous NUP98, NUP98-fusion proteins do not localize to the nuclear pore, but are distributed across the nucleus.18,19
Given the involvement of endogenous NUP98 in transcriptional control and the structural and functional diversity of C-terminal fusion partners among NUP98-fusion proteins, it was hypothesized that chromatin targeting of NUP98-fusion proteins depends on the NUP98 N-terminus, while functional properties encoded in or recruited by the C-terminal fusion partners might mediate specific gene regulatory functions that drive leukemogenesis.20 For instance, both NUP98/NSD1 and NUP98/JARID1A fusions contain PHD (Plant Homeodomain) domains, which recognize methylated lysine residues in histone tails.9,10 Mutations of either the PHD domain or the catalytic methyltransferase domain of NUP98/NSD1 abolished its oncogenic potential.9 Similarly, the PHD domain of the NUP98/JARID1A fusion protein was critical for oncogenic transformation.10 This indicates that both chromatin targeting as well as aberrant histone modifications are required for leukemogenesis driven by NUP98/NSD1 and NUP98/JARID1A. Different molecular mechanisms were proposed for other NUP98-fusion proteins. For instance, mutations in the DDX10 helicase domain impaired the transforming potential of a NUP98/DDX10 fusion protein, but the molecular underpinnings of this effect remain unclear.21 Thus, given the structural heterogeneity and the absence of direct DNA-binding domains among non-homeodomain NUP98-fusion proteins, it is possible that they hijack different molecular mechanisms to induce leukemic transformation. Yet, as molecularly different NUP98-fusion proteins can induce leukemia in vivo, different molecular pathways likely converge on a conserved set of target genes that is critical for induction and maintenance of NUP98-fusion AML. Therefore, we reasoned that the identification of actionable gene products among overlapping target gene sets of different NUP98-fusion proteins might provide new insight into developing more efficient therapeutic strategies to combat AML driven by NUP98-fusions.
To identify critical gene targets that are shared by diverse NUP98-fusion proteins, we generated novel mouse models allowing for Doxycycline (Dox)-regulatable expression of NUP98/NSD1, NUP98/JARID1A and NUP98/DDX10. The transcriptome of these model systems recapitulated specific characteristics of gene expression programs of human NUP98-fusion AML. Time-resolved profiling of the transcriptional response upon NUP98-fusion protein withdrawal in vivo combined with ChIP-seq analyses identified a surprisingly small, conserved core of genes whose expression is maintained by all NUP98-fusion proteins. Within this core of direct transcriptional targets, we identified the druggable kinase CDK6, which was highly expressed in murine and human NUP98-fusion AML. CDK6 expression was required for initiation and maintenance of NUP98-fusion-AML. Consistently, murine and patient-derived NUP98-fusion AML cells were sensitive to the CDK4/6 inhibitor Palbociclib, which caused rapid induction of apoptosis and cell cycle arrest in NUP98-fusion-expressing cells in vitro and in vivo. Our study defines the common core transcriptional targets of diverse NUP98-fusion proteins in AML and identifies CDK6 as an actionable critical target in this leukemia subtype.
Methods
Statistics
GraphPad Prism 8.0 (San Diego, CA, USA) was used for statistical analyses. If not stated differently, two-tailed students t-test was used for p-value determination. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Data Sharing Statement
All NGS data are available at GEO (accession ID: GSE134784).
All other Methods are described in the Supplementary Methods section.
Results
Different NUP98-fusion proteins drive common gene expression programs
To investigate shared transcriptional programs of distinct NUP98-fusion proteins in vivo, we focused on the most recurrent NUP98-fusion proteins NUP98/NSD1 and NUP98/JARID1A, as well as on the molecularly distinct NUP98/DDX10 fusion, which lacks an annotated DNA-binding or chromatin-interaction domain.21 We co-transduced murine hematopoietic stem and progenitor cells (HSPC) with retroviral constructs driving tetracycline-dependent expression of GFP and NUP98-fusion proteins, as well as constitutive expression of the tetracycline transactivator protein (tTA), luciferase and activated NrasG12D, as this genetic lesion is commonly found in NUP98-rearranged AML patients1,8 (Fig. 1a). This strategy ensures that only co-infection of both constructs enables NUP98-fusion protein expression.
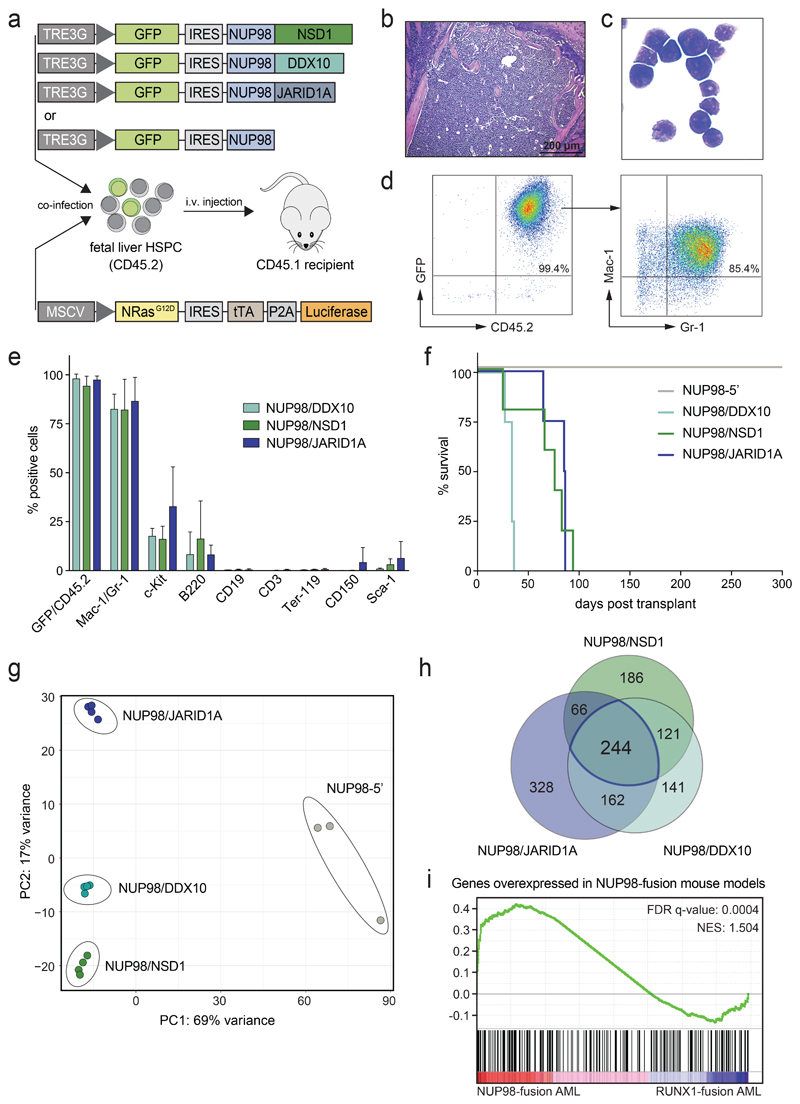
Figure 1. Different NUP98-fusion proteins induce a phenotypically similar AML-like disease in mice.
(a) Schematic outline of the experimental strategy for the generation of transplantation models for Dox-repressible expression of NUP98-fusion proteins. (b-d) Representative bone marrow histology (b), bone marrow cytospin (c) and flow cytometric analysis (d) from a moribund mouse transplanted with NUP98/DDX10-transformed murine HSPC. (e) Relative abundance of indicated cell populations in the bone marrow from moribund mice expressing indicated NUP98-fusion proteins (mean ± s.d. n ≥ 4). (f) Kaplan-Meier survival curves of mice transplanted with murine HSPC expressing indicated NUP98-fusion-proteins (n ≥ 4). (g) Principal component analysis (PCA) of the distances between steady-state gene expression data of NUP98-fusion-protein-driven leukemia cells (n ≥ 3). (h) Venn-diagram illustrating the overlap of overexpressed genes of NUP98-fusion-protein-driven leukemia cells vs. cells expressing NUP98-5’ (>3-fold upregulated, p < 0.01, n ≥ 3). (i) Gene Set Enrichment Analysis (GSEA) indicating conservation of the gene signature found in murine NUP98-fusion-protein-driven leukemia with AML patients harboring NUP98 rearrangements.
Upon transplantation, recipient mice developed an aggressive AML-like disease that was characterized by leukocytosis, anemia, thrombocytopenia, splenomegaly and infiltration of leukemic blasts into bone marrow, spleen and liver22 (Fig. 1b,c and Supplementary Fig. 1a-c). Leukemia cells in bone marrow and spleen expressed high levels of the mature myeloid surface markers Mac-1 and Gr-1 while only a minor fraction of blasts was positive for the progenitor marker c-Kit and the lymphoid marker B220 (Fig. 1d,e and Supplementary Fig. 1d-f). Expression of NUP98/DDX10, NUP98/NSD1 and NUP98/JARID1A led to rapid development of a lethal, phenotypically highly similar disease in vivo (median survival 34-84 days). In contrast, expression of mutated Nras G12D together with the N-terminal NUP98-fusion moiety lacking a fusion partner (referred to as NUP98-5’) did not lead to any disease development within 300 days (Fig. 1f).
To study the transcriptomes associated with these three NUP98-fusion-protein-driven leukemia models we performed RNA-sequencing on ex-vivo-isolated leukemia cells. Global gene expression patterns of leukemia driven by NUP98/DDX10, NUP98/NSD1 and NUP98/JARID1A were closely related and differed significantly from NUP98-5’-expressing bone marrow cells (Fig. 1g). Integration of these data with transcriptomes of purified murine hematopoietic stem and progenitor cell populations23 revealed that global gene expression patterns of NUP98-fusion-driven leukemia cells resembled common myeloid progenitors (CMP) and granulocyte-monocyte progenitors (GMP) (Supplementary Fig. 2a). Expression of 244 genes was significantly increased (>3-fold, p < 0.01) in all three NUP98-fusion-protein-driven leukemia models compared to NUP98-5’-expressing cells (Fig. 1h and Supplementary Table 1). This core set of genes upregulated in NUP98-fusion leukemia was enriched for regulators of normal and aberrant self-renewal, including members of the Hoxa gene family as well as the transcription factor Meis1 24 (Supplementary Fig. 2b). Importantly, the gene expression programs in murine NUP98-fusion-protein-driven AML models recapitulated specific transcriptional signatures observed in AML patients with NUP98-rearrangements, highlighting the clinical relevance of our leukemia models (Fig. 1i and Supplementary Fig. 2c,d).
These data show that expression of distinct NUP98-fusion proteins induce a phenotypically similar, aggressive AML-like disease in vivo that features characteristic transcriptional patterns of human AML with NUP98-fusion proteins.
Maintenance of NUP98-fusion leukemia is dependent on sustained oncogene expression
Next, we aimed to investigate whether expression of NUP98-fusion proteins is required for disease maintenance. Murine leukemia cells driven by NUP98/NSD1, NUP98/JARID1A or NUP98/DDX10 induced a fatal disease with very similar characteristics in secondary recipients (Supplementary Fig. 3a-d). We used these tetracycline-responsive models to induce transcriptional shutdown of NUP98-fusion expression in established leukemias by Dox administration in vivo (Fig. 2a). Bioluminescence imaging confirmed that the disease rapidly progressed in untreated secondary recipient mice (Fig. 2b). In contrast, Dox-induced transcriptional shutdown of NUP98-fusion expression led to disease regression, translating into a significant survival benefit (Fig. 2b,c and Supplementary Fig. 4a,b). The rapid, time-dependent decrease of GFP expression upon Dox treatment indicates that NUP98-fusion protein expression is downregulated with similar kinetics in leukemia cells, as GFP is translationally coupled to NUP98-fusions via an internal ribosome entry site (IRES) (Fig. 2d and Supplementary Fig. 4c). Loss of NUP98-fusion protein expression induced higher levels of the myeloid surface markers Mac-1 and Gr-1 while levels of the progenitor cell marker c-Kit were decreasing within 5 days (Fig. 2e). In line with this, Dox-treated leukemia cells showed strong morphological signs of terminal myeloid differentiation (Fig. 2f). Likewise, in vitro-cultured leukemia cells derived from the bone marrow of our in vivo models remained fully dependent on the expression of NUP98-fusion proteins, as Dox-induced downregulation of fusion protein expression resulted in growth inhibition and induction of apoptosis (Supplementary Fig. 4d-g).
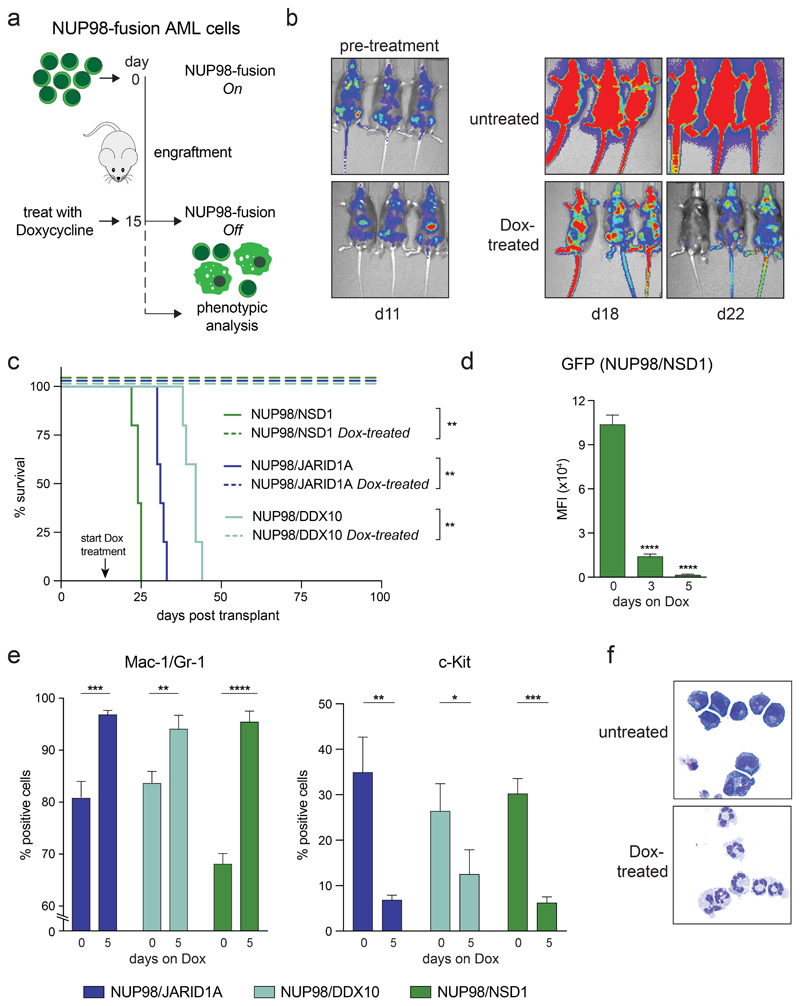
Figure 2. NUP98-fusion-protein-driven AML is highly dependent on sustained oncoprotein expression in vivo .
(a) Schematic illustration of the experimental workflow to induce downregulation of oncogene expression in vivo. 1x106 GFP/CD45.2-positive leukemia cells were transplanted into sublethally irradiated secondary recipients. Animals were treated with Dox (4 mg/ml) after an engraftment period of 15 days. (b) Representative bioluminescence imaging of untreated vs. Dox-treated mice transplanted with NUP98/NSD1-expressing leukemia cells. (c) Kaplan-Meier survival curves of mice transplanted with leukemia cells expressing indicated NUP98-fusion proteins receiving either no treatment or Dox (4 mg/ml) (n ≥ 4). (d-e) Flow cytometric analyses of bone-marrow-derived leukemia at indicated time points after Dox treatment of mice (mean ± s.d. n = 3). (d) Quantification of mean fluorescence intensity (MFI) of GFP. (e) Quantification of the myeloid differentiation markers Mac-1/Gr-1 and the progenitor cell marker c-Kit. (f) Representative cytospin images of untreated vs. Dox-treated (8 days) NUP98/JARID1A-expressing cells in vitro.
Taken together, these data demonstrate that sustained expression of the driver oncogene is required for propagation of NUP98-fusion AML models in vivo and in vitro, and that loss of NUP98-fusion expression results in rapid terminal differentiation of leukemia cells and disease regression.
NUP98-fusion proteins regulate overlapping sets of transcriptional targets
To investigate global effects on gene expression upon transcriptional inactivation of NUP98-fusion expression, we isolated leukemia cells from secondary recipients three and five days after Dox administration and performed RNA-seq analysis (Fig. 3a). Time-resolved analysis of conserved transcriptional kinetics after oncogene withdrawal in all three models revealed four consistent patterns of gene regulation: 2819 genes displayed early and pronounced transcriptional changes three days upon NUP98-fusion shutdown, following a pattern of steady down- or upregulation after oncogene withdrawal (Fig. 3b, left panels). In contrast, changes in the expression of 2847 genes were only observed after five days upon Dox-mediated loss of the driver oncogene (Fig. 3b, right panels). While the first category likely contains the majority of direct gene targets of NUP98-fusion proteins, the latter category might reflect changes in gene expression that are further downstream of oncogene inactivation. Loss of NUP98-fusion proteins affected the differentiation trajectories of NUP98-fusion leukemias in the context of normal hematopoietic lineages. Oncogene shutdown in NUP98-fusion-transformed leukemia cells resulted in global transcriptional reprogramming towards mature monocytes/macrophages or granulocytes (Supplementary Fig. 5a). In line with this, we found gradual upregulation of gene sets characteristic of differentiated immune cells after downregulation of NUP98-fusion proteins, including Tnfa/NFkB signaling, TLR activation and Interleukin signaling (Fig. 3c). These data confirm that leukemia cells lose characteristics of immature hematopoietic progenitors after withdrawal of NUP98-fusion protein expression and initiate differentiation along monocytic and granulocytic lineages.
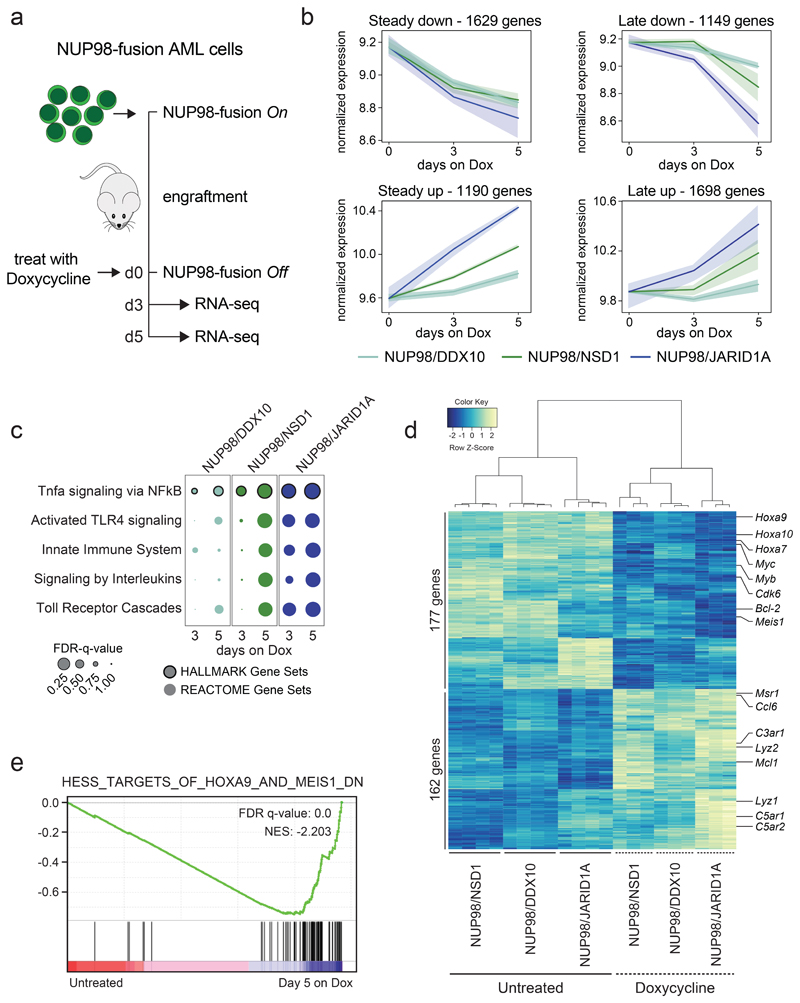
Figure 3. Different NUP98-fusion proteins regulate a common core of transcriptional targets.
(a) Schematic illustration of the experimental workflow to investigate NUP98-fusion protein-dependent transcriptional programs. NUP98-fusion-protein-driven leukemia cells were transplanted into secondary recipient mice. Mice were treated with Dox for 3 or 5 days after an initial engraftment phase of 15 days and leukemia cells were sorted from the bone marrow based on GFP/CD45.2 expression and gene expression was analyzed by RNA-seq (n ≥ 3). (b) Representation of dynamics of global gene expression changes after 3 and 5 days of Dox-induced NUP98-fusion-protein-repression. Each line represents the harmonized mean of the median expression of all genes within the cluster of distinct fusion-protein-driven cancer cells. Maximum/Minimum medians are indicated by the colored area. (c) GSEA indicating induction of myeloid differentiation upon fusion protein withdrawal. (d) Heatmap of 339 commonly up- and downregulated genes in all three NUP98-fusion-protein-driven models after 5 days of Dox-mediated fusion-protein repression (≥ 1.5-fold change, p < 0.01). (e) GSEA illustrating the enrichment of HOXA9 / MEIS1-target genes in cells expressing NUP98/NSD1.
The extent of global gene expression changes 5 days upon Dox-mediated oncogene shutdown was significantly different (1110 genes for NUP98/DDX10, 2014 genes for NUP98/NSD1, 3304 genes for NUP98/JARID1A), and only a small set of genes was commonly regulated across different NUP98-fusion models. Among 2340 genes, 162 were jointly upregulated upon transcriptional shutdown of the three NUP98-fusions. Likewise, 177 of 2238 genes were downregulated after oncogene withdrawal in all three models. Thus, the expression of a core set of 339 genes is commonly regulated by NUP98-fusion proteins (Fig. 3d, Supplementary Fig. 5b,c and Supplementary Table 2). Among the 177 genes whose expression was maintained by all NUP98-fusion proteins we found several known regulators of progenitor self-renewal, including Hoxa9, Meis1, Myb and Bcl-2 24,25. (Fig. 3d). Consistently, targets of HOXA9, MEIS1 and E2F transcription factors were downregulated upon NUP98-fusion protein withdrawal (Fig. 3e and Supplementary Fig. 5d).
Altogether, time-resolved mapping of transcriptional responses upon oncogene withdrawal showed that different NUP98-fusion proteins control distinct and complex transcriptional programs that converge on a small set of common gene targets.
Cdk6 is a conserved direct transcriptional target of NUP98-fusion proteins
We next investigated whether NUP98-fusion proteins directly participate in the regulation of these genes. Co-expression of an HA-tagged NUP98/JARID1A variant and Nras G12D in mouse HSPC induced an aggressive AML-like disease in vivo (Supplementary Fig. 6a,b). Leukemic blasts displayed high levels of Mac-1/Gr-1, together with low levels of c-Kit and B220 (Supplementary Fig. 6c,d). As these results show that the N-terminal HA-tag does not interfere with the oncogenic potential of NUP98/JARID1A, we analyzed global chromatin association of the NUP98/JARID1A-fusion protein by ChIP-seq using HA antibodies. ChIP-qPCR confirmed NUP98/JARID1A binding to the Hoxa7 promoter region10 (Supplementary Fig. 6e). Analysis of ChIP-seq data showed that 78% of NUP98/JARID1A-bound sites are located within promoters (2554 regions), while only 14% (439 regions) were found in intronic regions. 4% (128 regions) localized to distal intergenic regions (Fig. 4a and Supplementary Table 3). Within promoter regions, NUP98/JARID1A binding was enriched around annotated transcriptional start sites (TSS) (Fig. 4b,c). We detected NUP98/JARID1A binding in the promoters of genes that are important regulators of NUP98-fusion AML, such as the Hoxa cluster and Meis1 (Fig. 4d).
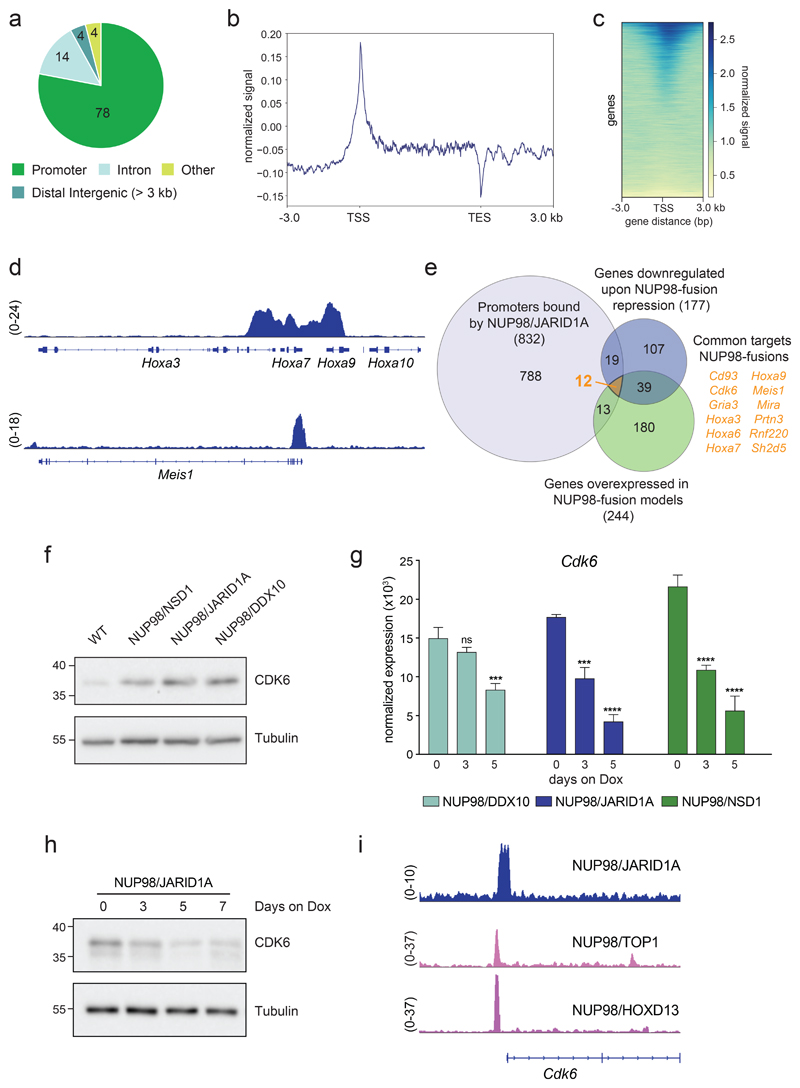
Figure 4. NUP98-fusion proteins regulate the expression and bind the promoter of Cdk6 .
(a) Pie chart of HA-NUP98/JARID1A chromatin binding events (%) in the indicated chromatin contexts. (b) Signal distribution of HA-NUP98/JARID1A normalized to the input signal, between the transcription start sites (TSS) and transcription end sites (TES) +/- 3 kb up- and downstream. (c) Heatmap showing HA-NUP98/JARID1A chromatin binding within +/- 3 kb of annotated TSS. All annotated murine genes are plotted on the y-axis. (d) Representative ChIP-seq tracks showing the binding of HA-NUP98/JARID1A within the Hoxa cluster (top) and the promoter region of Meis1 (bottom). (e) Venn diagram showing the overlap of genes with NUP98/JARID1A-occupied promoter-regions, all downregulated genes upon repression of NUP98-fusion-proteins and all genes that are highly overexpressed in NUP98-fusion-protein-driven mouse models. (f) Western Blot analysis of CDK6 levels in bone-marrow of mice transplanted with leukemia cells expressing different NUP98-fusion proteins, compared to bone-marrow of wild type (WT) mice. (g) Expression kinetics of Cdk6 in ex-vivo-derived leukemia cells from the bone marrow upon Dox-mediated repression of the indicated NUP98-fusion proteins. (mean ± s.d. n = 3). (h) Western Blot analysis of CDK6 levels of in vitro cultured NUP98/JARID1A-driven leukemia cells at indicated time points during Dox-mediated fusion protein repression (0.5 μg/ml). (i) Representative ChIP-seq tracks showing the binding of indicated NUP98-fusion proteins within the promoter region of Cdk6.
As promoter binding by regulatory proteins often controls the expression of the corresponding gene, we reasoned that any direct transcriptional target gene of NUP98-fusion proteins would need to fulfill the following criteria: (i) It is overexpressed in NUP98-fusion AML compared to normal HSPC; (ii) its expression is downregulated upon shutdown of fusion protein expression; and (iii) it exhibits NUP98-fusion protein binding in its promoter. Using this stringent approach, only 12 candidate genes classified as common direct transcriptional targets of NUP98-fusion proteins, including several genes of the Hoxa cluster and Meis1 (Fig. 4e). However, the cyclin-dependent kinase 6 (Cdk6) gene particularly stood out. CDK6 was shown to be a critical target in MLL-fusion-expressing AML26 and is highly overexpressed in different AML subtypes (Supplementary Fig. 6f). Cdk6 was among the highest overexpressed genes in all three murine NUP98-fusion-protein-driven AML models (Fig. 4f and Supplementary Fig. 6g) and its expression was rapidly downregulated upon loss of the driver fusion protein (Fig. 4g,h). Cdk6 promoter binding was not restricted to NUP98/JARID1A, as re-analysis of published ChIP-seq datasets revealed that NUP98/TOP1 and NUP98/HOXD13 fusion proteins were also associated with the same genomic region19 (Fig. 4i).
These data show that orthogonal integration of RNA-seq and ChIP-seq datasets efficiently identifies direct transcriptional targets of NUP98-fusion proteins and highlight Cdk6 as a regulated, direct target gene of multiple NUP98-fusion proteins.
Loss of Cdk6 impairs NUP98-fusion-protein-driven leukemogenesis
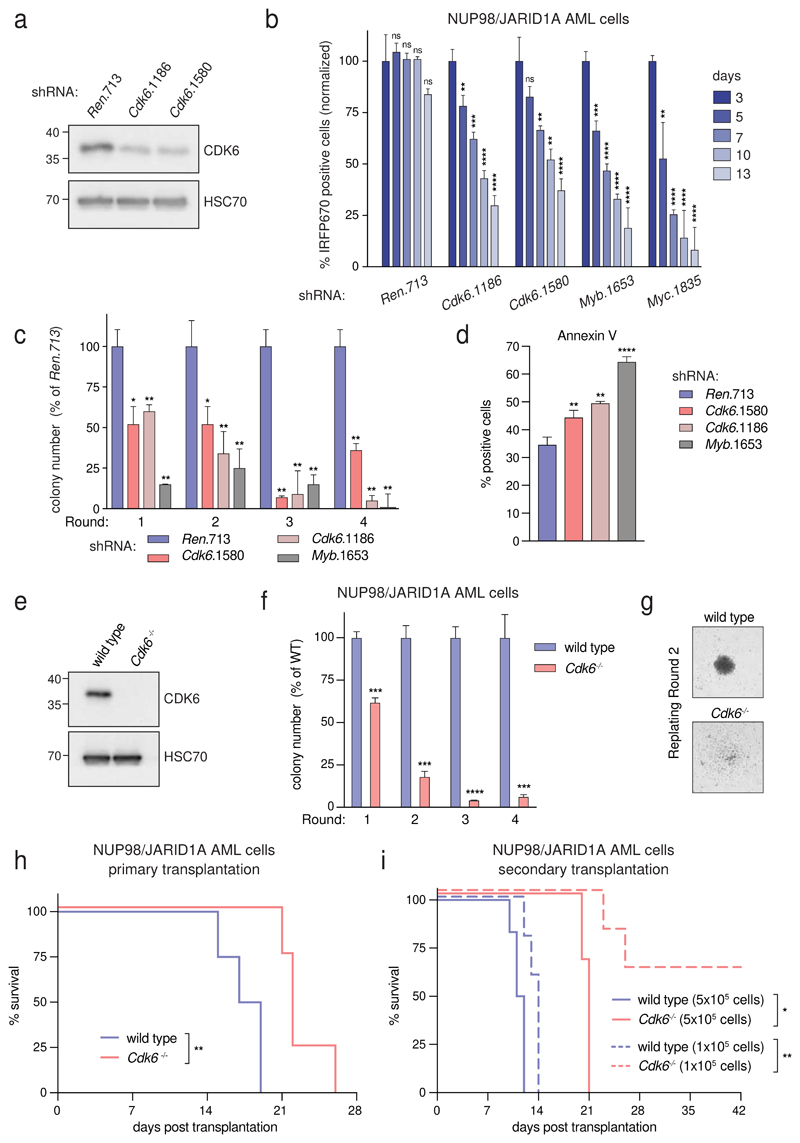
To investigate whether Cdk6 expression is required for development and maintenance of NUP98-fusion driven AML we re-engineered our transplantation-based leukemia models to combine constitutive NUP98-fusion expression with the presence of the reverse tet-transactivator (rtTA3) to be able to modulate Cdk6 expression by inducible RNAi27 (Supplementary Fig. 7a). Mice transplanted with cells expressing these vectors developed an aggressive AML-like disease that showed similar features as our other NUP98-fusion-protein-driven in vivo models (Supplementary Fig. 7b-d). NUP98/JARID1A-driven leukemia cells were transduced with lentiviral vectors enabling Dox-inducible expression of Cdk6-targeting shRNAs (Fig. 5a and Supplementary Fig. 8a). shRNA-mediated downregulation of Cdk6 caused a severe proliferative disadvantage of NUP98/JARID1A-driven cells in competitive growth assays in vitro and induced a reduction of actively cycling leukemia cells (Fig. 5b and Supplementary Fig. 8b). Cdk6 knockdown strongly impaired the colony formation capacity of NUP98/JARID1A-expressing AML cells in semisolid media and was associated with increased apoptosis and elevated levels of the myeloid maturation markers Mac-1 and Gr-1 and lower expression of the progenitor marker c-Kit (Fig. 5c,d and Supplementary Fig. 8c).
Figure 5. NUP98-fusion-protein-driven leukemia is dependent on CDK6 expression.
(a) Western Blot analysis of CDK6 levels in NUP98/JARID1A-expressing AML cells after 4 days of Dox-mediated Cdk6 shRNA induction. (b) Results of the competitive proliferation assay shown as the percentage of IRFP670-positive murine NUP98/JARID1A-driven leukemia cells expressing indicated shRNAs in the presence of Dox (0.5 μg/ml) over 13 days (mean ± s.d. n = 3). The non-targeting shRNA (shRen.713) is used as neutral control, shRNAs targeting essential genes in hematopoietic cells Myb (shMyb.1653) or Myc (shMyc.1835) as positive controls. (c) Colony formation assay of G418-selected murine NUP98/JARID1A-driven leukemia cells expressing indicated shRNAs in the presence of Dox (0.5 μg/ml) over 4 rounds of replating. Colony numbers were normalized to cells expressing shRen.713 (mean ± s.d. n = 3). (d) Quantification of AnnexinV-positive cells of murine NUP98/JARID1A-driven leukemia cells expressing indicated shRNAs after two rounds of replating (mean ± s.d. n = 3). (e) Western Blot analysis of CDK6 levels in wild type vs. Cdk6 -/- fetal liver cells. (f) Colony formation assay of bone marrow-derived leukemia cells of mice transplanted with NUP98/NSD1-transformed Cdk6 -/- or wild type fetal liver cells. Colony numbers were normalized to wild type NUP98/NSD1 leukemia cells (mean ± s.d. n = 3). (g) Representative image of colonies from wild type or Cdk6-/- NUP98/NSD1 leukemia cells. (h) Kaplan-Meier survival curves of mice transplanted with wild type vs. Cdk6 -/- fetal liver cells transformed with NUP98/NSD1 (n = 4). (i) Kaplan-Meier survival curves of secondary transplantations of bone-marrow-derived wild type or Cdk6 -/- leukemia cells at indicated concentrations into sublethally irradiated (4.5 Gray) recipient mice (5x105 transplanted cells: n ≥ 3, 1x105 transplanted cells: n = 5).
To investigate the role of CDK6 in leukemia initiation we co-transduced wild type or Cdk6 -/- HSPC with retroviral constructs for the expression of NUP98/NSD1 and Nras G12D (Fig. 5e and Supplementary Fig. 8d). Cdk6 deficiency strongly abolished the serial re-plating capacity of NUP98/JARID1A-expressing AML cells and caused loss of blast-like colony morphology (Fig. 5f,g). In vivo, disease latency was significantly increased in primary recipients of NUP98/NSD1-transformed Cdk6 -/- cells as compared to mice receiving wild type AML cells (Fig. 5h). The reduced leukemogenic potential of NUP98/NSD1-transformed Cdk6 -/- cells was even more pronounced in secondary transplantations despite similar levels of engraftment between NUP98/NSD1-transformed wild type and Cdk6 -/- cells (Fig. 5i and Supplementary Fig. 8e-h).
Taken together, these data demonstrate that CDK6 is required for both the establishment and the maintenance of NUP98-fusion-protein-driven AML.
Pharmacologic CDK4/CDK6-inhibition efficiently targets NUP98-fusion AML
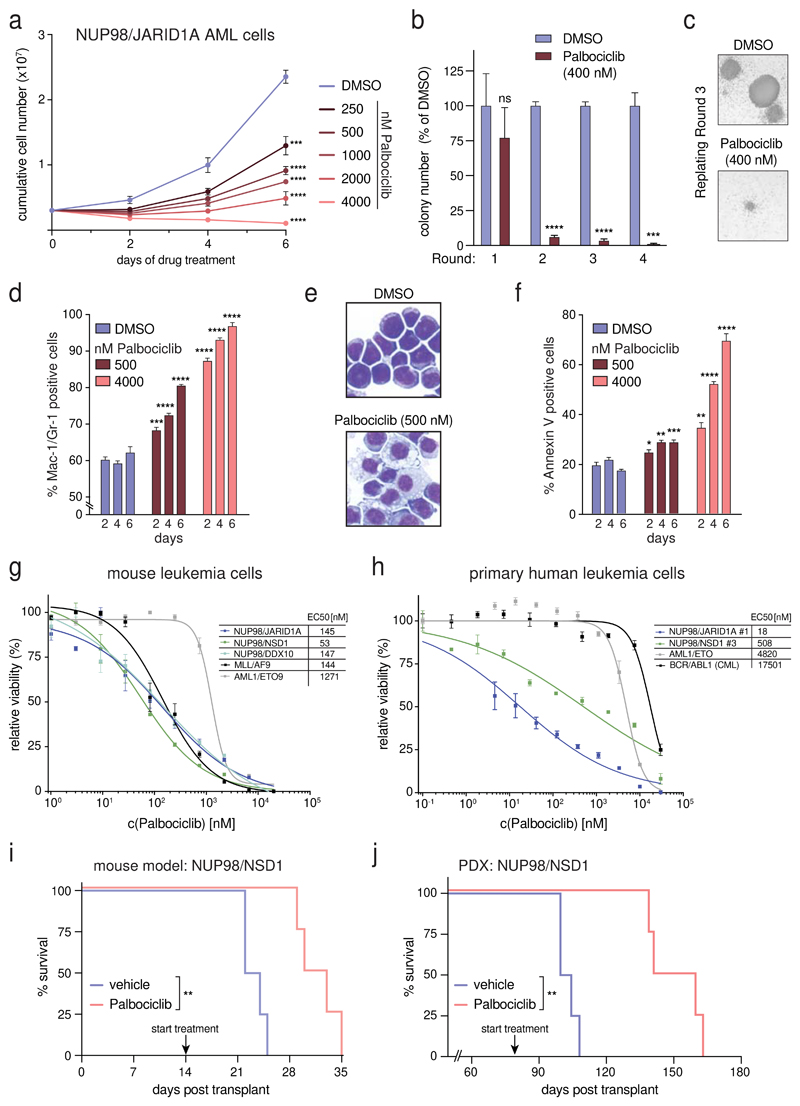
As NUP98-fusion AML is dependent on CDK6 expression, we reasoned that inhibition of CDK6 kinase activity might represent an attractive targeting strategy in this AML subtype. The small-molecule inhibitor Palbociclib inhibits CDK4/CDK6 and is approved for breast cancer therapy. We found that Palbociclib exerted significant dose- and time-dependent anti-proliferative effects on murine AML cells driven by NUP98/JARID1A and NUP98/NSD1 (Fig. 6a and Supplementary Fig. 9a). CDK4/CDK6 inhibition strongly impaired the re-plating capacity of NUP98/JARID1A-driven AML cells (Fig. 6b,c). Palbociclib led to a reduction in actively proliferating cells (Supplementary Fig. 9b) and induced myeloid differentiation of NUP98/JARID1A-driven AML blasts, as measured by increased levels of Mac-1/Gr-1, lower levels of c-Kit and loss of progenitor morphology (Fig. 6d,e and Supplementary Fig. 9c). Finally, Palbociclib treatment caused a dose- and time-dependent increase in apoptosis (Fig. 6f).
Figure 6. NUP98-fusion-protein-driven leukemia is highly sensitive to pharmacological CDK4/CDK6 inhibition.
(a) Proliferation curves of murine NUP98/JARID1A-expressing leukemia cells in the presence of indicated concentrations of Palbociclib. (mean ± s.d. n = 3). (b) Colony formation assay of murine NUP98/JARID1A-expressing leukemia cells in the presence of DMSO or Palbociclib. Colony numbers were normalized to DMSO (mean ± s.d. n = 3). (c) Representative image of colonies in the presence of DMSO or Palbociclib after three rounds of replating. (d) Flow cytometric analyses of Mac-1/Gr-1 surface marker expression in NUP98/JARID1A-expressing AML cells treated with indicated concentrations of Palbociclib (mean ± s.d. n = 3). (e) Representative Cytospin images illustrating the morphology of NUP98/JARID1A-expressing AML cells 7 days after DMSO or Palbociclib treatment. (f) Flow cytometric analyses of AnnexinV expression in NUP98/JARID1A-expressing leukemia cells treated with indicated concentrations of Palbociclib (mean ± s.d. n = 3). (g,h) Dose-response curves of Palbociclib-mediated growth inhibition of murine leukemia cells (g) and primary patient cells (h) driven by different fusion proteins (mean ± s.e.m. n = 3). (i) Kaplan-Meier survival curve of C57BL/6 Ly5.1 recipients transplanted with murine NUP98/NSD1-driven leukemia cells after vehicle or Palbociclib treatment (50mg/kg, n = 4). (j) Kaplan-Meier survival curves of NSG mice transplanted with NUP98/NSD1-PDX AML cells after vehicle or Palbociclib treatment (50mg/kg, n = 4).
Next we compared the Palbociclib sensitivity of NUP98/JARID1A-, NUP98/NSD1- and NUP98/DDX10-driven AML samples to leukemia cells driven by the fusion proteins MLL/AF9 and AML1/ETO9a, which were both shown to be sensitive to CDK4/CDK6 inhibition.26,28 All three murine NUP98-rearranged cell lines were very sensitive to Palbociclib treatment (EC50 values ranging from 53-147 nM). MLL/AF9-driven cells showed comparable inhibitor sensitivity (EC50: 144 nM), while AML1/ETO9a-expressing AML tolerated higher concentrations (EC50: 1271 nM) (Fig. 6g). To validate these findings in human AML, we examined six distinct primary, patient-derived leukemia samples expressing different NUP98-fusion proteins (Supplementary Table 4) for their Palbociclib sensitivity. Also in NUP98/NSD1-expressing primary human AML cells, CDK4/CDK6 inhibition induced cell cycle arrest and apoptosis (Supplementary Fig. 9d,e). Four of six NUP98-rearranged patient samples were highly sensitive to CDK4/CDK6 inhibition (EC50: 18-508 nM), while two samples tolerated slightly higher doses (EC50: 2300-2400 nM). However, all six NUP98-fusion AML samples were more sensitive than a primary AML sample expressing AML1/ETO9a (EC50: 4820 nM) and a BCR/ABL1-expressing CML sample (EC50: 17501 nM) (Fig. 6h and Supplementary Fig. 9f). To investigate whether Palbociclib displays anti-leukemic activity in vivo that can be harnessed to efficiently combat NUP98-fusion-driven AML, we transplanted NUP98/NSD1-driven leukemia cells into recipient mice and initiated Palbociclib treatment as soon as leukemia cells were detectable by bioluminescence imaging (Supplementary Fig. 9g). CDK4/CDK6 inhibition delayed leukemia progression, caused increased myeloid differentiation of leukemic blasts and led to a significant survival benefit in vivo (Fig. 6i, Supplementary Fig. 9g-i).
Finally, we employed a patient-derived xenograft model of NUP98/NSD1-rearranged AML (Supplementary Fig. 9j,k). Palbociclib monotherapy was initiated 80 days after transplantation. Strikingly, Palbociclib treatment caused a highly significant prolongation of survival compared to the vehicle-treated control cohort (median survival 152 vs 103 days) (Fig. 6j).
In summary, these data show that NUP98-rearranged AML is sensitive towards CDK4/CDK6 inhibition in vivo and in vitro. Therefore, we propose Palbociclib may be a new rational treatment option for patients suffering from this disease.
Discussion
Targeting critical effectors of oncogenes represents an attractive strategy in cancer therapy, particularly when the opportunity to inhibit the driver oncoprotein itself is limited. NUP98-fusion-protein-driven AML is a devastating disease with very poor prognosis. No convincing targeting approach has been developed for this leukemia subtype. We established novel mouse models for controllable expression of three distinct NUP98-fusion proteins and used RNA-seq and ChIP-seq to identify conserved transcriptional targets. These studies revealed CDK6 as a highly expressed, directly regulated target of NUP98-fusion proteins and demonstrate that genetic and pharmacologic interference with CDK6 results in cell cycle arrest, myeloid differentiation and apoptosis in vitro and in vivo. We thus propose CDK4/6 inhibition as a rational strategy to target AML with NUP98-fusions.
The cyclin-dependent kinase CDK6 is highly expressed in AML patient samples and represents a promising target in MLL-fusion expressing- and FLT3-ITD-positive leukemia.26,29 In addition, a recent report identified deregulation of cell cycle control via aberrant regulation of cyclin D2/CDK6 as a critical feature of RUNX1/ETO-driven AML.28 The canonical role of the serine/threonine kinase CDK6 and its homolog CDK4 is to regulate cell cycle progression through association with D-type cyclins.30 As CDK6 but not CDK4 is amplified in human hematopoietic malignancies, it was hypothesized that CDK6 exerts distinct functions in addition to cell cycle control. In fact, CDK6 acts as a transcriptional regulator in cancer cells. While the expression of genes important for proliferation, survival, and cytokine production depend on its kinase activity, CDK6-dependent regulation of angiogenesis and stem cell functions are kinase-independent.29,31–34 As NUP98-fusion-protein-driven leukemia was sensitive to treatment with the CDK4/CDK6 inhibitor Palbociclib, it is likely that targeting the kinase-dependent role of CDK6 is of particular importance within the context of NUP98-fusion AML. Loss of RB1 expression or the acquisition of TP53 mutations cause resistance to CDK4/6 inhibition.34,35 Thus, future efforts should focus on the development of specific CDK6 inhibitors and on the identification of targets that synergize with CDK6 inhibition to maximize therapy response.
In our study, structurally distinct NUP98-fusion proteins induce oncogenic transformation in mouse models. Although we cannot exclude that different NUP98-fusion proteins employ unrelated molecular pathways to induce leukemia, we show that all NUP98-fusions share common sets of target genes that are critical for their oncogenicity. As several partner genes among NUP98-fusion proteins lack an annotated DNA-binding- or chromatin-interaction domain, it is likely that chromatin targeting by the amino terminus of NUP98, which is shared between all distinct fusion partners, is essential for leukemogenesis. Fusion proteins exert their oncogenic activity in the context of large protein complexes.36–39 NUP98-fusion proteins interact with the MLL1/NSL complexes, and MLL1 function is critical for NUP98-fusion-dependent leukemogenesis.19,40 It is thus appealing to speculate that the NUP98-moiety is required for chromatin targeting, while the C-terminal fusion partner exerts critical functions in gene control during leukemogenesis. This concept may also explain how fusions without annotated DNA-binding capacity, such as NUP98/DDX10 still result in aberrant expression of defined genes.
The two most common NUP98-fusion proteins, NUP98/NSD1 and NUP98/JARID1A, but also the molecularly distinct NUP98/DDX10-fusion protein share several highly overexpressed target genes in addition to Cdk6. A common core signature of 244 genes whose expression is high in our NUP98-fusion AML mouse models was strongly enriched in patients harboring NUP98-rearrangements. This signature contained many genes with critical roles in normal and malignant hematopoiesis. For instance, genes of the Hoxa cluster and Meis1 are critical factors in murine and human cells transformed by NUP98-fusion proteins and AML patients harboring NUP98 gene rearrangements.2,9,11 Among the 12 shared direct targets of NUP98-fusion-potein-driven AML there are several interesting candidates with potential relevance for leukemia biology. For instance, the C-type lectin CD93 is highly expressed in leukemia-initiating subpopulations of MLL-rearranged AML cells and was proposed to regulate self-renewal by downregulating the tumor suppressor gene CDKN2B.41 Moreover, CD93 expression has been shown to discriminate leukemia cells in the monitoring of minimal-residual-disease (MRD).42 Screening for CD93 could thus be useful in the detection of MRD among AML patients with NUP98-fusion proteins. Further, the RING domain E3 ubiquitin ligase RNF220 was shown to stabilize b-catenin, resulting in increased Wnt signaling43, an essential pathway in AML cells.44
NUP98-fusions also regulate a number of genes whose expression is not particularly high in our murine AML-like model systems but whose functions are still pivotal in leukemia biology, such as the transcription factors Myc and Myb.45 Interestingly, CDK6 was shown to be a target gene of MYB, indicating that CDK6 expression could be regulated at multiple levels downstream of NUP98-fusion proteins46. Furthermore, the promoter of the anti-apoptotic Bcl-2 gene is bound by NUP98/JARID1A in the steady state and Bcl-2 expression was rapidly downregulated upon oncoprotein shut-down. BCL2 inhibition by the small molecule Venetoclax has recently been approved for AML therapy in a combinatorial treatment regimen.47 Combined inhibition of CDK6 and BCL2 might thus be an attractive approach in the therapy of AML patients with NUP98-rearrangements, as both genes are direct targets of NUP98-fusion proteins.
In summary, our findings show that combining advanced modeling of NUP98-fusion-dependent leukemogenesis in mice with integrated transcriptomic analyses provides valuable insight into shared fusion-protein driven transcriptional circuitries. The identification of CDK6 as a direct target in NUP98-fusion-protein-driven AML has potential clinical implications, as CDK6 inhibition represents a rational therapeutic intervention strategy for this subgroup of leukemia patients with particularly poor prognosis.
Supplementary Material
Key Points.
NUP98-fusion proteins directly regulate leukemia-associated gene expression programs in AML.
CDK6 expression is under direct transcriptional control of NUP98-fusions and NUP98-fusion AML is particularly sensitive to CDK6 inhibition.
Acknowledgements
We thank all members of the Grebien and Zuber laboratories for stimulating discussions, in particular Mareike Roth. Julian Jude is acknowledged for providing help with molecular cloning, Michaela Fellner for excellent technical assistance and Matthias Muhar for providing murine AML1/ETO9a-driven cells. We thank Gerald Schmauss and Marietta Weninger for expert FACS sorting, Manuela Telsnig for animal care and the Molecular Biology Services at the Vienna Biocenter (VBC). Irina Sadovnik, Rene Gaupmann, Gabriele Stefanzl, Barbara Peter, Daniela Berger and Karin Nebral are acknowledged for excellent technical assistance. R.I. and A.P. are indebted to Jihène Benlagha and Jean-Michel Cayuela from the Saint-Louis Hospital Cell Bank. Next Generation Sequencing was performed at the VBCF NGS Unit (www.viennabiocenter.org/facilities). This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement n° 636855 to F.G. and n° 336860 to J.Z.). The Austrian Science Fund (FWF) supported J.Z. with the SFB grant F4710, P.V. with SFB F4704-B20, R.M. and H.T.T.P with grants SFB F4707 and SFB-F06105. J.S. and L.S. are recipients of a DOC Fellowship of the Austrian Academy of Sciences at the University of Veterinary Medicine and the Ludwig Boltzmann Institute for Cancer Research, respectively. Research at the IMP is generously supported by Boehringer Ingelheim.
Footnotes
Conflict of interest: COI declared - see note
COI notes: P.V. has received research grants and/or honoraria from Pfizer, Celgene and Novartis. No potential conflicts of interests are disclosed by other authors.
Preprint server: No;
Agreement to Share Publication-Related Data and Data Sharing Statement: All RNA-seq and ChIp-seq Data are uploaded to GEO (Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE134784
Clinical trial registration information (if any):
Data Sharing Statement
All NGS data are available at GEO (accession ID: GSE134784).
References
- 1.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247–57. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollink IHIM, van den Heuvel-Eibrink MM, Arentsen-Peters STCJM, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645–56. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 3.Struski S, Lagarde S, Bories P, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31(April):565–572. doi: 10.1038/leu.2016.267. [DOI] [PubMed] [Google Scholar]
- 4.Bolouri H, Farrar JE, Triche T, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaju RJ, Fidler C, Haas OA, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98(4):1264–1268. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij JDE, Hollink IHIM, Arentsen-Peters STCJM, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27(12):2280–8. doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 7.Romana SP, Radford-Weiss I, Ben Abdelali R, et al. NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogénétique Hématologique. Leukemia. 2006;20(4):696–706. doi: 10.1038/sj.leu.2404130. [DOI] [PubMed] [Google Scholar]
- 8.Lavallée V-P, Lemieux S, Boucher G, et al. Identification of MYC mutations in acute myeloid leukemias with NUP98-NSD1 translocations. Leukemia. 2016;30:1621–1624. doi: 10.1038/leu.2016.19. [DOI] [PubMed] [Google Scholar]
- 9.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 10.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough SM, Lee F, Yang F, et al. NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function. Cancer Discov. 2014;4(5):564–577. doi: 10.1158/2159-8290.CD-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanasopoulou A, Tzankov A, Schwaller J. Potent co-operation between the NUP98-NSD1 fusion and the FLT3-ITD mutation in acute myeloid leukemia induction. Haematologica. 2014;99(9):1465–71. doi: 10.3324/haematol.2013.100917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dash AB, Williams IR, Kutok JL, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99(11):7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franks TM, Hetzer MW. The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol. 2013;23(3):112–117. doi: 10.1016/j.tcb.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasper LH, Brindle PK, Schnabel CA, et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–76. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai XT, Gu BW, Yin T, et al. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006;66(9):4584–4590. doi: 10.1158/0008-5472.CAN-05-3101. [DOI] [PubMed] [Google Scholar]
- 17.Franks TM, McCloskey A, Shokirev M, et al. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 2017;31(22):2222–2234. doi: 10.1101/gad.306753.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahrenkrog B, Martinelli V, Nilles N, et al. Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS One. 2016;11(3):e0152321. doi: 10.1371/journal.pone.0152321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Valerio DG, Eisold ME, et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell. 2016;30(6):863–878. doi: 10.1016/j.ccell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franks TM, McCloskey A, Shokirev M, et al. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells - online version. Genes Dev. 2017;31(22):2222–2234. doi: 10.1101/gad.306753.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yassin ER, Abdul-Nabi aM, Takeda A, Yaseen NR. Effects of the NUP98-DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia. 2010;24(5):1001–11. doi: 10.1038/leu.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):246–258. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 23.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Chromatin state dynamics during blood formation. Science (80-.) 2014;345(6199):943–950. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27(5):1000–8. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 25.Konopleva M, Contractor R, Tsao T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10(5):375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Placke T, Faber K, Nonami A, et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood. 2014;124(1):13–23. doi: 10.1182/blood-2014-02-558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber J, McJunkin K, Fellmann C, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29(1):79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Soria N, McKenzie L, Draper J, et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell. 2018;34(4):626–642. doi: 10.1016/j.ccell.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uras IZ, Walter GJ, Scheicher R, et al. Palbociclib treatment of FLT3-ITD + AML cells uncovers a kinase-dependent transcriptional regulation of FLT3 and PIM1 by CDK6. Blood. 2016;127(23):2890–2902. doi: 10.1182/blood-2015-11-683581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Nicolay BN, Chick JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546(7658):426–430. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schleicher R, Hoelbl-Kovacic A, Bellutti F, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125(1):90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24(2):167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellutti F, Tigan AS, Nebenfuehr S, et al. CDK6 antagonizes P53-induced responses during tumorigenesis. Cancer Discov. 2018;8(7):884–897. doi: 10.1158/2159-8290.CD-17-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condorelli R, Spring L, O’Shaughnessy J, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29(3):640–645. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 36.Brehme M, Hantschel O, Colinge J, et al. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci U S A. 2009;106(18):7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skucha A, Ebner J, Schmöllerl J, et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulay G, Sandoval GJ, Riggi N, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017;171(1):163–178. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride MJ, Pulice JL, Beird HC, et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell. 2018;33(6):1128–1141. doi: 10.1016/j.ccell.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima Y, Yumoto M, Katsumoto T, Kitabayashi I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia. 2017;31(10):2200–2210. doi: 10.1038/leu.2017.62. [DOI] [PubMed] [Google Scholar]
- 41.Iwasaki M, Liedtke M, Gentles AJ, Cleary ML. CD93 Marks a Non-Quiescent Human Leukemia Stem Cell Population and Is Required for Development of MLL-Rearranged Acute Myeloid Leukemia. Cell Stem Cell. 2015;17(4):412–421. doi: 10.1016/j.stem.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coustan-Smith E, Song G, Shurtleff S, et al. Universal monitoring of minimal residual disease in acute myeloid leukemia. JCI Insight. 2018;3(9):e98561. doi: 10.1172/jci.insight.98561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma P, Yang X, Kong Q, et al. The Ubiquitin Ligase RNF220 Enhances Canonical Wnt Signaling through USP7-Mediated Deubiquitination of β-Catenin. Mol Cell Biol. 2014;34(23):4355–4366. doi: 10.1128/MCB.00731-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Krivtsov AV, Sinha AU, et al. The wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science (80-.) 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pattabiraman DR, Gonda TJ. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 46.Zhong X, Prinz A, Steger J, et al. HoxA9 transforms murine myeloid cells by a feedback loop driving expression of key oncogenes and cell cycle control genes. Blood Adv. 2018;2(22):3137–3148. doi: 10.1182/bloodadvances.2018025866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konopleva M, Letai A. BCL-2 inhibition in AML: an unexpected bonus? Blood. 2018;132(10):1007–1013. doi: 10.1182/blood-2018-03-828269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.