Abstract
UCP1 and UCP3 are members of the uncoupling protein (UCP) subfamily and are localized in the inner mitochondrial membrane. Whereas UCP1's central role in non-shivering thermogenesis is acknowledged, the function and even tissue expression pattern of UCP3 are still under dispute. Because UCP3 properties regarding transport of protons are qualitatively identical to those of UCP1, its expression in brown adipose tissue (BAT) alongside UCP1 requires justification. In this work, we tested whether any correlation exists between the expression of UCP1 and UCP3 in BAT by quantification of protein amounts in mouse tissues at physiological conditions, in cold-acclimated and UCP1 knockout mice. Quantification using recombinant UCP3 revealed that the UCP3 amount in BAT (0.51 ng/(μg total tissue protein)) was nearly one order of magnitude higher than that in muscles and heart. Cold-acclimated mice showed an approximate three-fold increase in UCP3 abundance in BAT in comparison to mice in thermoneutral conditions. Surprisingly, we found a significant decrease of UCP3 in BAT of UCP1 knockout mice, whereas the protein amount in skeletal and heart muscles remained constant. UCP3 abundance decreased even more in cold-acclimated UCP1 knockout mice. Protein quantification in UCP3 knockout mice revealed no compensatory increase in UCP1 or UCP2 expression. Our results do not support the participation of UCP3 in thermogenesis in the absence of UCP1 in BAT, but clearly demonstrate the correlation in abundance between both proteins. The latter is important for understanding UCP3's function in BAT.
Keywords: Recombinant protein, Anti-UCP3 antibody, UCP3 knockout mice, Cold-acclimated mice, Skeletal muscles, Uncoupling protein 2
1. Introduction
UCP1 and UCP3, proteins of the mitochondrial uncoupling protein subfamily (UCP), are implicated in the pathophysiology of different diseases such as obesity, diabetes mellitus, ischemia, cancer, etc. [1–3]. However, the underlying molecular mechanisms and even the protein distribution among tissues have been controversially discussed for nearly two decades.
UCP3, first described in muscles [4], has 57% and 55% homology to UCP1 in humans and mice, respectively. Both UCP1 and UCP3 were shown to transport protons [5–7]. However, whereas the role of UCP1 in non-shivering thermogenesis is accepted, the function of UCP3 is still under dispute [8–10]. Alternatively, UCP3 was proposed to transport fatty acids, lipid peroxides and pyruvate [11–14]. Results from experiments using knockout or overexpression of UCP3 in murine skeletal muscles imply the protective role of UCP3 against triglyceride accumulation [15,16].
Although the expression of UCP3 at the protein level was reported for muscle [17], heart [18], brown adipose tissue [19], white adipose tissue [20], spleen and thymus [21], the investigation of its function has mostly been focused on how it works in muscles. The generation of UCP1 and UCP3 knockout mice reveals that such mice have no conspicuous phenotype under physiological conditions [22;23]. The cold adaptation of a UCP1 knockout mouse confirmed the function of UCP1 as a mediator of non-shivering thermogenesis [24,25]. Because both UCP1 and UCP3 are localized in BAT, it is possible that the function of one protein can be taken over by the other (sister) uncoupling protein. One of the few existing comparative investigations of UCP1 and UCP3 describes the mRNA interdependence of UCP1 in BAT with UCP3 in skeletal muscles [26]. However, in contrast to UCP1, UCP3 is characterized by a rapid turnover [27] which makes protein expression analysis so important in the investigation of UCP3 function and regulation. It was reported that UCP3 abundance increases in skeletal muscle of cold-acclimated UCP1−/− mice, however, no evidence was found to indicate that it takes over the UCP1 thermogenic function [28].
The understanding of UCP3's expression pattern under different physiological and challenging (cold acclimation) conditions will provide an important indication for revealing its function. Therefore, the goals of the present work were (i) to quantify the expression of UCP3 in mouse tissues using the UCP3 recombinant protein and a novel polyclonal antibody; (ii) to evaluate whether and in which tissues UCP1 and UCP3 may substitute for each other by compensatory increases in protein abundance, using the model of UCP1 and UCP3 knockout mice and (iii) to verify whether there is any interaction between UCP1 and UCP3 at the protein level in general.
2. Materials and methods
2.1. Chemicals
All of the chemicals we used, except for those mentioned below, were acquired by Roche (Austria), Sigma Aldrich (Austria), Biorad (Austria), Amersham Biosciences, and GE Healthcare (Austria).
2.2. Recombinant proteins
Recombinant proteins, murine UCP1 (mUCP1), human UCP2 (hUCP2), mUCP2, mUCP3, mUCP4-eGFP and mUCP5, were used for antibody validation and for quantification as a standard in Western blot analysis. Recombinant production of mUCP1, hUCP2, mUCP4-eGFP and mUCP5 has been previously described [29,30]. Production and reconstitution of mUCP2 and mUCP3 were performed in a similar fashion. In brief, the open reading frames of mUCP2 and mUCP3 were obtained as full length cDNA clones IRAVp968G0921D and IRCKp5014J1815Q (BioCat, Germany). Mouse ucp2 was inserted in the NdeI and EcoRI sites, mouse ucp3 – in the NheI and a BamHI sites of pET24a + (Novagen, Germany), respectively. For protein expression, pET24a + containing mucp2 or mucp3 were transfected into the E. coli expression strain Rosetta (DE3) (Novagen, Germany). Bacteria were grown to OD 600 0.3–0.5 before protein expression was induced by addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After incubation for an additional 2.5 h, bacteria were harvested by centrifugation. A suspended bacteria pellet was lysed using a French press (Constant Systems Limited, UK). Inclusion bodies were collected by centrifugation at 14,000 g. Approximately 3 mg of mUCP2 or mUCP3 were solubilized from inclusion bodies using 2% sarcosyl and 1 mM DTT. 100 mg E. coli polar lipid (Avanti polar lipids, Alabaster, USA), 300 μg Triton X-114, 75 μg octyl-polyoxyethylene and 2 mM GTP were added to the solubilized mUCP2 or mUCP3. Sarcosyl and GTP were removed by dialysis, and the buffer was thereby changed to the assay buffer (50 mM NaSO4, 10 mM Tris, 10 mM MES and 0.6 mM EGTA, pH 7.34). The dialysate was passed through a hydroxyapatite column (Bio-Rad, Germany) to remove decomposed proteins. Non-ionic detergents were eliminated using Bio Beads (Bio-Rad, Germany). Proteoliposomes containing mUCP2 or mUCP3 were stored at −80 °C.
2.3. Animals
UCP1−/− [24], UCP2−/− [31], UCP3−/− [17] and their wild type (wt) controls used in this study were backcrossed for 10 generations on the C57BL/6 background. Alternatively, three months old wt C57BL/6 mice were used for the experiments on isolated mitochondria. The animals were routinely kept in a 12 h:12 h light–dark cycle, were acclimated to 21–23 °C (unless otherwise stated) and had unlimited access to food (R70 Standard Diet, Lactamin) and tap water. In some experiments mice were acclimated to 4 °C or 30 °C for at least 4 weeks. Mice were single caged, cages were enriched with wooden chips, carton tube and paper for mice welfare. All experiments were approved by the Animal Ethics Committee of the North Stockholm region, Sweden.
2.4. Tissue samples preparation
Animals were anaesthetised with a mixture of 79% CO2 and 21% O2 for 1 min and then decapitated. The extracted tissues were immediately shock-frozen in liquid nitrogen for further investigation. We applied two homogenisation protocols for different tissues [30,32]. To homogenize brown (BAT) and inguinal white (WAT, [20]) adipose tissues, we used a Potter homogenizer with a Teflon pestle. BAT and WAT were homogenized in RIPA buffer supplemented with a protease inhibitor (Complete mini). After centrifugation (14,000 g, 15 min, at 4 °C), the supernatant was collected with a syringe, aliquoted and stored at −80 °C. Thymus, spleen, brain and lungs were crushed using a mixer mill (MM200, Retsch, Germany) in lysis buffer (50 mM Tris, 150 mM NaCl, 1% sodium deoxycholate, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, pH 7.4) with a protease inhibitors cocktail and then sonicated. Heart (He), skeletal muscles (SkM) and stomach were first trimmed into smaller pieces with scissors and then treated as described above. After 30 min of incubation on ice, the lysates were centrifuged 10 min at 2,500 g. The supernatants were collected, centrifuged again under same conditions, aliquoted and stored at − 80 °C. The total protein concentration was determined with a Pierce BCA Protein Assay Kit (Thermo Scientific).
2.5. Mitochondrial protein isolation
BAT, WAT, He and SkM mitochondria were used for the UCP3 antibody validation and UCP3 quantification. Extraction of BAT, WAT, He and SkM (skeletal muscles: scapulae muscle, ScM and gastrocnemius muscle, GMsc) and mitochondria isolation were performed according to two different protocols [28] and [30] with a modification as proposed in [33].
2.6. Western blot analysis
Western blot (WB) analysis was performed as described previously [34]. We modified the described protocol by using PVDF membranes (Amersham Hybond-P PVDF Transfer Membrane GE Healthcare). Probes were incubated at 4 °C overnight (or at least for 8 h) with the primary antibody diluted in 2% BSA solution (bovine serum albumin, Sigma Aldrich, Austria) and for 1 h at RT with the secondary antibody diluted in 2.5% milk solution. Detection was performed by luminescence reaction using a secondary antibody against rabbit antibodies linked with horseradish peroxidase (GE Healthcare, Austria) and ECL Western blotting reagent (GE Healthcare, Austria) accomplished with a ChemiDoc-It® 600 Imaging System (UVP, UK). The Launch Vision Works LS software (UVP, UK) was used for quantification. To ensure an accurate comparison of UCP1 and UCP3 amounts in different tissues or under different conditions, we methodically controlled the exposure time to avoid signal saturation using the same software.
To control protein loading, the membranes were stripped with a low pH strip solution (100 mM sodium citrate at pH 2.2) for at least 1 min, washed, blocked for 30 min in block solution and incubated again with antibodies against SDHA (succinate dehydrogenase complex subunit A, Abcam ab14715; dilution 1:5,000), GAPDH (Glyceraldehyde-3-phosphate dehydrogenase, Sigma-Aldrich, G8795; dilution 1:10,000) and NDUFA9 (complex I, Invitrogen, 459100; dilution 1:3,000) diluted in 2% BSA solution for at least 1 h at RT, and subsequently incubated with horseradish peroxidase-linked anti-mouse IgG (GE Healthcare, NA931).
2.7. Statistics
In each experiment 3–5 mice were used per group. All data are presented as mean values ± standard deviation (SD). Mean value for each mouse was built from 2–3 Western blots at the same conditions. Statistical analyses for the comparison of two mice groups were performed using Student's t test. By more than two mice groups one-way ANOVA test was used. *, **, *** indicate significant difference of p < 0.001, p < 0.004 and p < 0.012 respectively. The analysis was carried out using Sigma Plot 12.5 software.
3. Results
3.1. UCP3 antibody generation and validation
The quality of antibodies is crucial for precise assessment of UCP tissue distribution. Because the commercially available antibodies against mUCP3 were not adequately specific enough for our research purposes, we generated a new polyclonal antibody against the peptide sequence located in the third matrix loop of the protein. We used tissue samples from UCP3 knockout mice as a negative and recombinant UCP3 as a positive control to verify the specificity of the novel antibody in WB experiments. Recombinant mUCP1, mUCP2, mUCP4-eGFP, mUCP5 and mUCP3 [30,35] were used to exclude a cross-reactivity with other UCP family members. Figure S1 (Supplementary Materials) demonstrates that the anti-mUCP3 antibody we produced recognized recombinant mUCP3 and UCP3 in skeletal muscle (SkM, Figure S1, A) of wt mice and showed no cross-reactivity with other recombinant UCPs (Figure S1, B).
3.2. UCP3 distribution in murine tissues at protein level
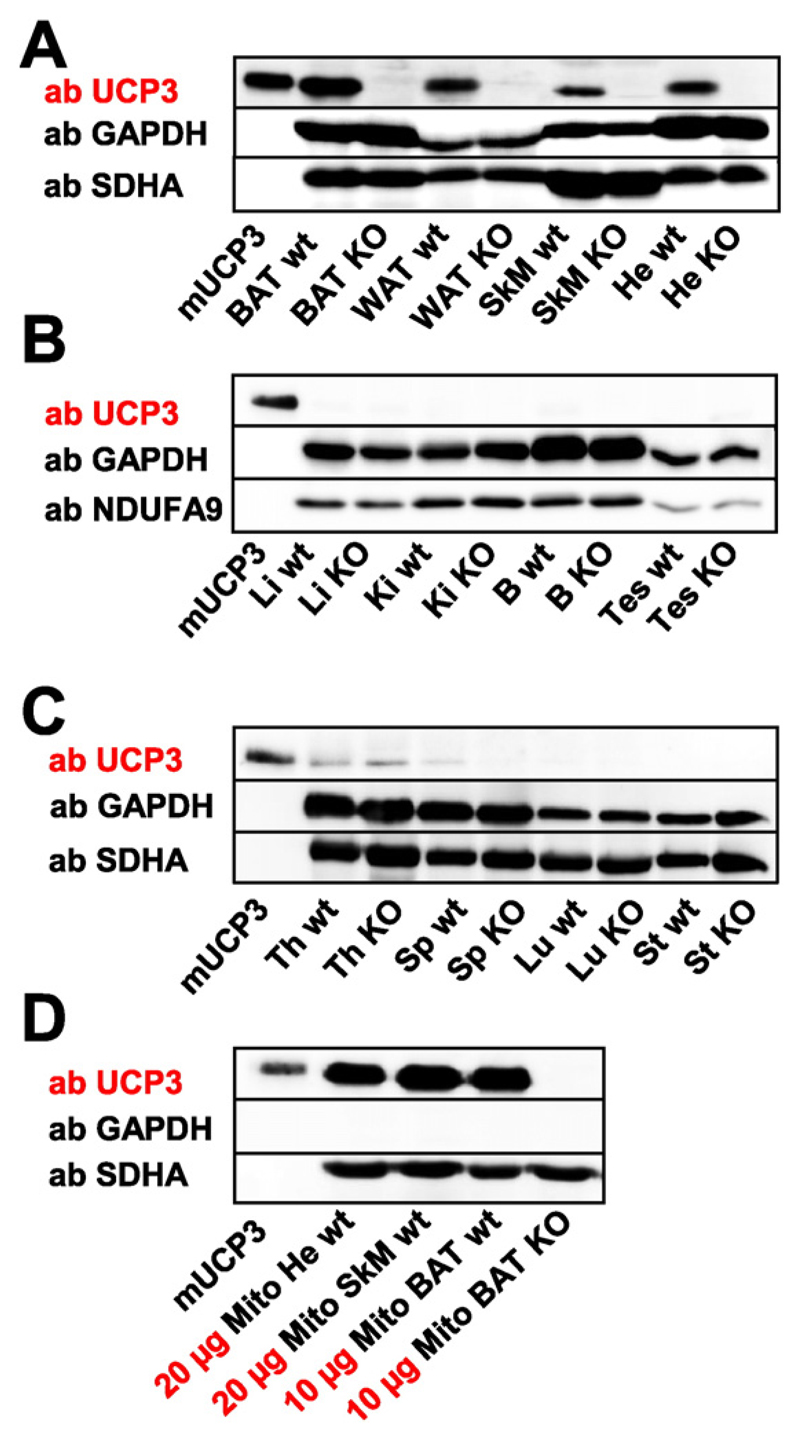
First, we compared the protein distribution in tissues of wt and UCP3 knockout mice under physiological conditions to evaluate the controversial reports about UCP3 tissue localization described in Introduction. Fig. 1A demonstrates that UCP3 is expressed in brown adipose tissue (BAT), inguinal white adipose tissue (WAT), skeletal muscle (SkM) and heart (He) of wt mice, but not in the knockout controls. We did not detect UCP3 in the liver (Li), kidney (Ki), brain (B), testis (Tes), thymus (Th), spleen (Sp), lungs (Lu) and stomach (St) (Fig. 1B–C). This expression pattern generally confirms the previous reports [23,36]. Interestingly, UCP3 expression seems to be different in rats compared with mice; UCP3 was described in the murine thymus and spleen (although in very low abundance), but no UCP3 was found in the rat heart [21,37].
Fig. 1. UCP3 expression patterns in murine tissues under physiological conditions.
(A) UCP3 was detected in skeletal muscle (SkM), heart (He), brown adipose tissue (BAT) and white adipose tissue (WAT) of wild type (wt) mice. Corresponding tissues from UCP3−/− mice were used as a negative control. (B–C) Kidney (Ki), liver (Li), brain (B), testes (Tes), thymus (Th), spleen (Sp), lung (Lu) and stomach (St) show no detectable UCP3 expression. 25 μg of total protein from the tissue were loaded per lane. Five mice per group were tested. (D) UCP3 expression in isolated mitochondria. 20 μg isolated mitochondria from He, SkM and 10 μg isolated mitochondria from BAT were loaded per lane. Recombinant mouse UCP3 (mUCP3, 5 ng) was used as a positive control. Antibodies against GAPDH, SDHA and NDUFA9 were used as a control for protein loading and mitochondria amount. Six mice were tested.
The data imply that UCP3 abundance in BAT is distinctly higher than in SkM. To verify that this difference is not related to the mitochondria amount, we repeated the experiment using isolated mitochondria from heart, muscles and BAT. Fig. 1D shows that the UCP3 amount in mitochondria isolated from BAT is indeed approximately two times higher than in skeletal muscles.
3.3. Quantification of UCP3 expression at physiological conditions
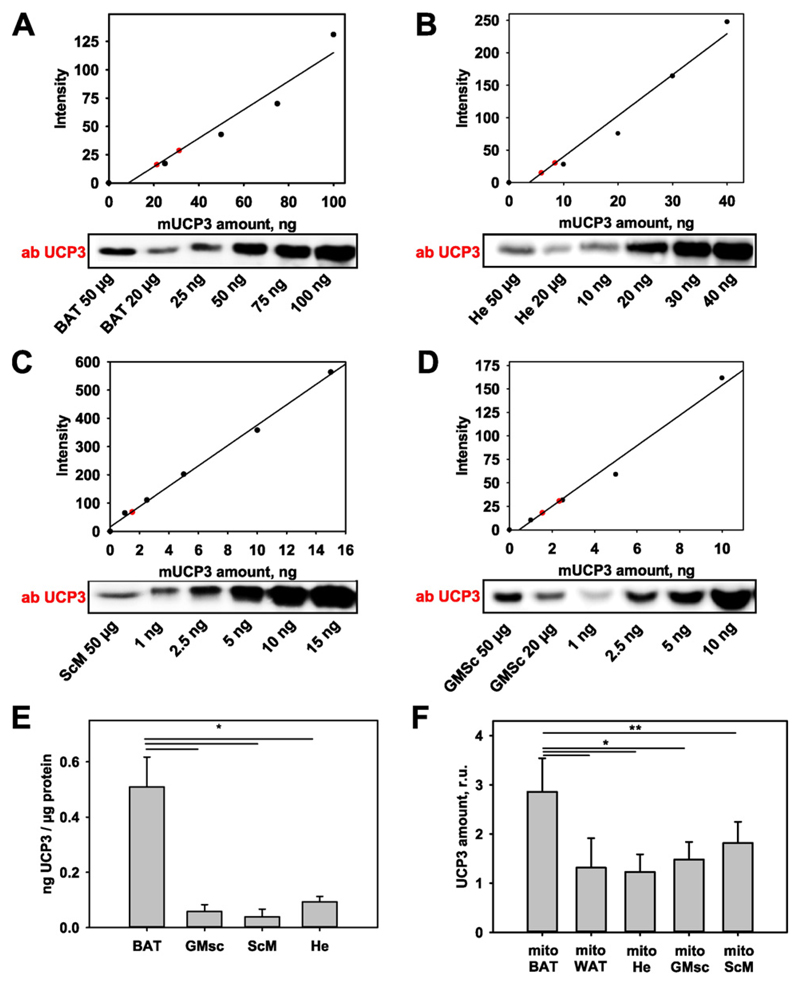
To quantify the differences between the expression of UCP3 in BAT, heart and muscles, we used the recombinant mouse protein (mUCP3). Increasing concentrations of mUCP3 and the investigated tissues (20 and 50 μg of total protein) were gradually loaded onto a WB gel. We plotted the values of WB density against known mUCP3 concentration (Fig. 2A–D) and calculated the UCP3 content per 1 μg total cellular protein (Fig. 2E) to compare UCP3 abundance among different investigated tissues. The UCP3 amount in BAT (0.51 ± 0.11) ng/(μg total protein) was nearly one order of magnitude higher than that in gastrocnemius muscle (0.058 ± 0.024) ng/(μg total protein), scapulae muscle (0.038 ± 0.028) ng/(μg total protein), and heart (0.093 ± 0.02) ng/(μg total protein). We performed the quantification in isolated mitochondria in a similar manner (Fig. 2F). The results correspond with those obtained with tissue. No difference was observed between two diverse types of skeletal muscle tissue, which contradicts the previously reported results [33].
Fig. 2. Protein quantification in tissues expressing UCP3.
(A–D) Representative WBs for UCP3 quantification in BAT (A), He (B), ScM (C) and GMsc (D) in wt mice under physiological conditions. 20 μg and 50 μg of total protein from the investigated tissue (red circles) and different concentrations of recombinant UCP3 (mUCP3) were loaded for quantification. (E–F) Comparison of UCP3 amounts in different tissues (n = 4, E) and in isolated mitochondria (n = 7) of wt mice (F).
3.4. Quantification of UCP3 expression in mice housed at different temperatures
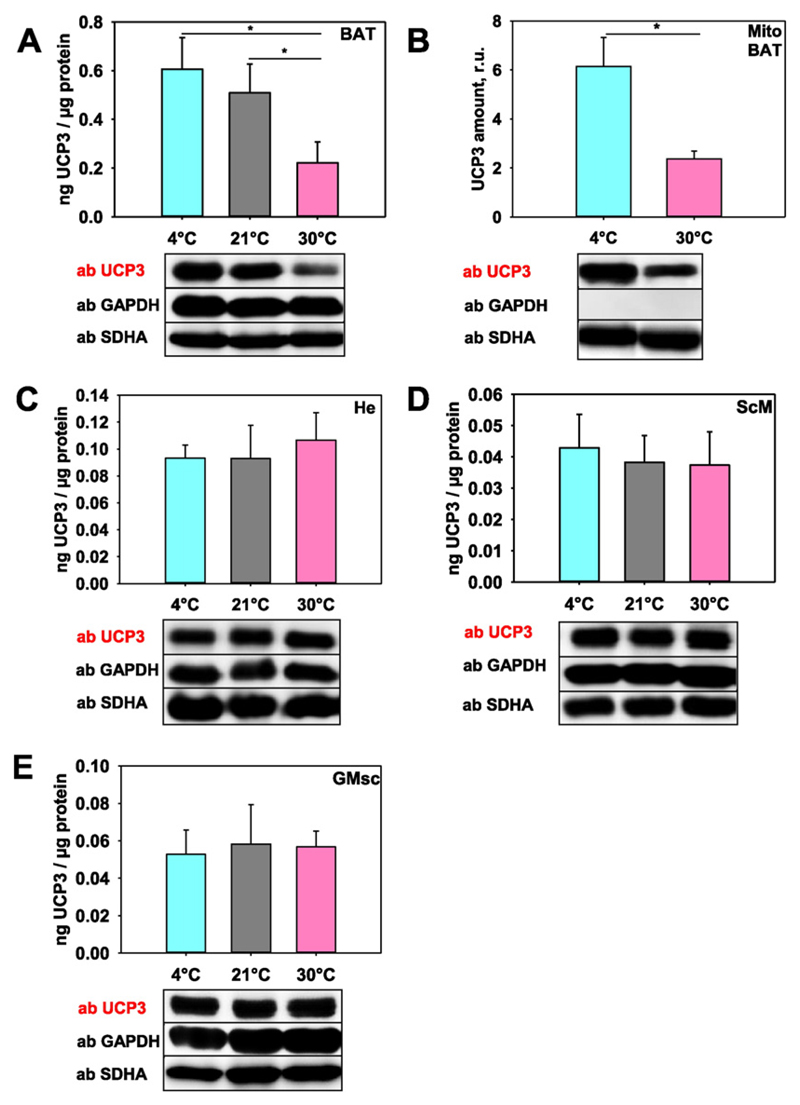
Next, we evaluated whether UCP3 expression is temperature-sensitive akin to UCP1. For this, we measured UCP3 in BAT, ScM, GMsc and He tissue probes taken from mice housed at 4 °C (cold acclimatization), 21 °C (standard housing) and 30 °C (thermoneutral control). Indeed, Fig. 3A shows that cold acclimatization of mice led to a three-fold increase of UCP3 abundance in BAT (0.61 ng/(μg total protein) at 4 °C vs. 0.22 ng/(μg total protein) at 30 °C). To verify that the observed UCP3 increase was not due to an increase in mitochondria number, we performed UCP3 quantification using isolated mitochondria. Fig. 3B confirms that UCP3 abundance is temperature-sensitive and is up-regulated at lower temperatures. Surprisingly, no change in UCP3 abundance was measured in skeletal muscles and heart under similar temperature conditions (Fig. 3C–E).
Fig. 3. Quantification of UCP3 expression at different temperatures.
(A) UCP3 amount in tissue lysates from brown adipose tissue, (C) heart, (D–E) skeletal muscles (ScM, D; GMsc, E) and (B) mitochondria isolated from BAT. Wild type mice were housed at 30 °C, 21 °C and 4 °C. 20 μg of total protein from tissue or 10 μg isolated mitochondria were loaded per lane. The relative UCP3 amount (r.u.) is a ratio between the band intensity of standard (mUCP3, 5 ng) and sample. Five mice were tested at 30 °C and 4 °C and four mice at 21 °C.
3.5. Uncoupling proteins expression patterns in UCP1 or UCP3 — ablated mice
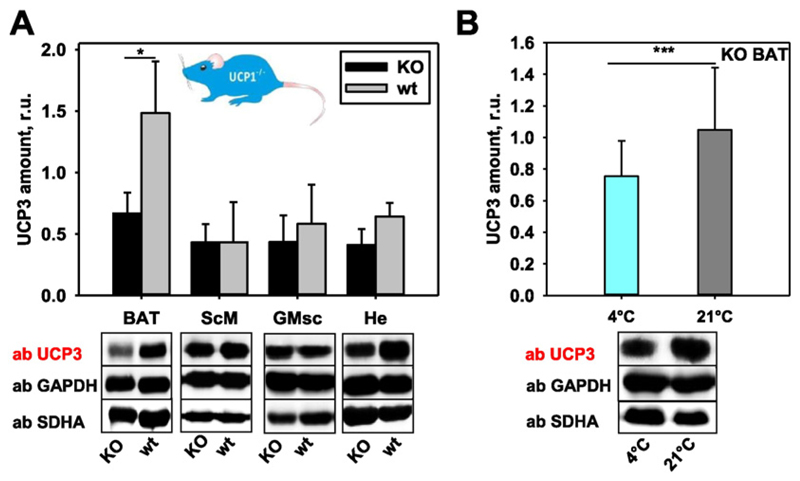
To evaluate whether the lack of UCP1 can be substituted by the up-regulation of UCP3 or vice versa at physiological conditions, we determined the UCP3 expression in several tissues of the UCP1−/− mice (Fig. 4) and UCP1 expression in UCP3−/− mice (Fig. 5). We detected a significantly lower UCP3 expression in BAT of UCP1−/− compared to wt mice, whereas heart and both types of skeletal muscles showed similar UCP3 abundance (Fig. 4A). To test whether the temperature decrease would stimulate UCP3 expression, we compared UCP1−/− mice at 4 °C and 21 °C (Fig. 4B). Surprisingly, UCP1−/− mice acclimated at 4 °C showed the lowest UCP3 amount. Because of high homology between UCP2 and UCP3 (~ 75%), we analysed whether UCP2 is regulated in UCP1−/− mice. The results revealed that UCP2 is not expressed in brown adipose tissue as a compensation for the lack of UCP1, and its abundance in tissues normally expressing UCP2 (spleen and thymus [35]) does not change (Figure S2, A–B, Supplementary Materials).
Fig. 4. Representative Western blots and quantification showing UCP3 expression in UCP1−/− mice.
(A) UCP3 expression in brown adipose tissue, skeletal muscles (ScM, GMsc) and heart of UCP1−/− mice compared to their wild type controls (wt). Data are presented as a mean values from three mice ± SD. (B) UCP3 expression in BAT of UCP1−/− mice housed at different temperatures. The relative amount (r.u.) of UCP3 is a ratio between the band intensity of standard (mUCP3, 5 ng) and sample. 25 μg of total protein from tissues were loaded per lane. The number of tested mice was n = 5 at 4 °C and n = 4 at 21 °C.
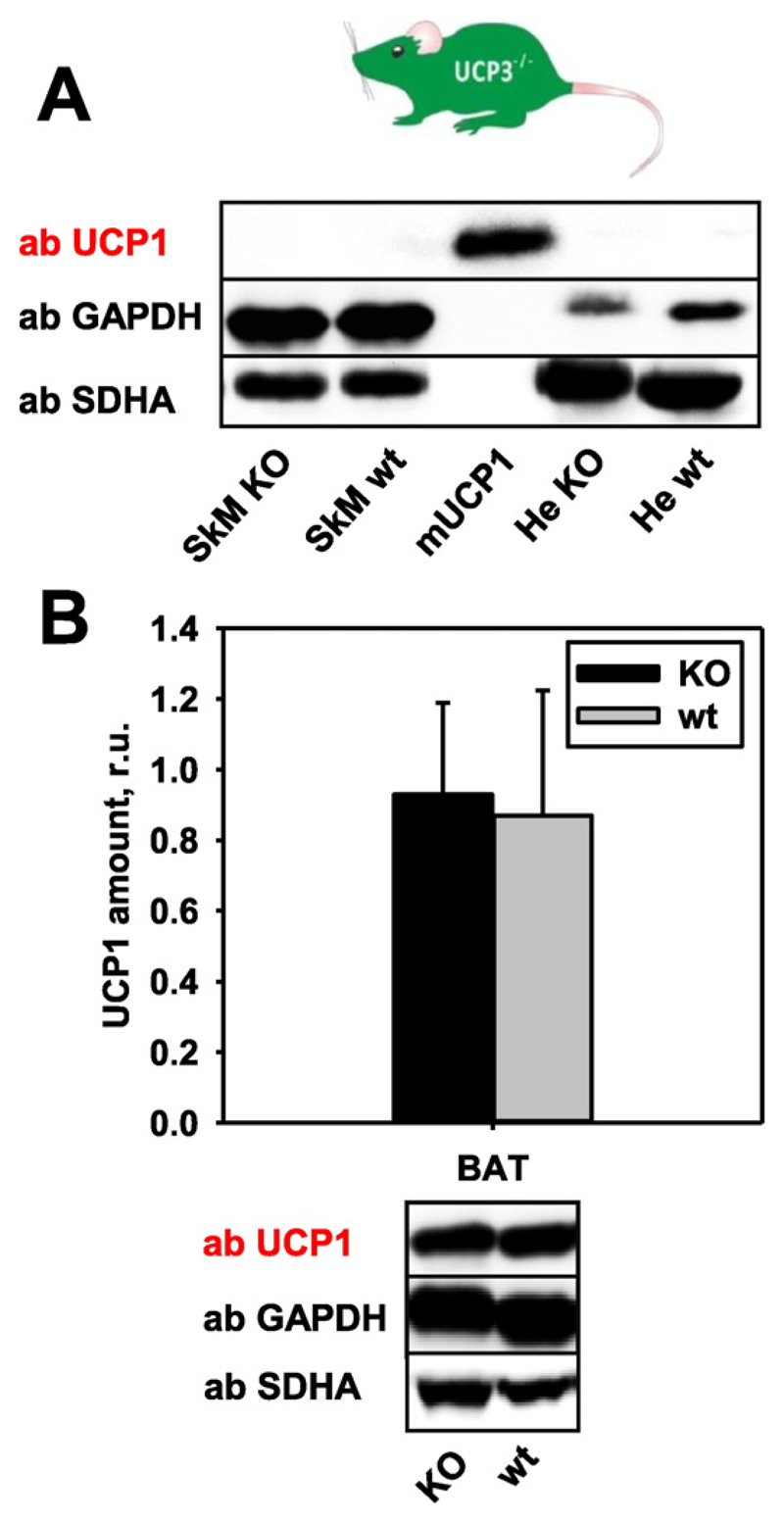
Fig. 5. Representative Western blots and quantification showing UCP1 expression in UCP3−/− mice.
(A) No compensatory expression of UCP1 in skeletal muscles, heart and (B) BAT. Data are presented as a mean values from five mice ± SD. 20 μg of total protein from tissues of UCP3−/− or wt mice were loaded per lane. The relative protein amounts of UCP1 were calculated as a ratio between line intensity of standard (mUCP1, 250 ng) and sample.
To test whether the UCP1 expression level is also influenced by the absence of UCP3, we measured it in UCP3−/− mice. UCP1 was neither expressed in SkM nor He (Fig. 5A), and was not up-regulated in BAT (Fig. 5B). Similarly, no compensatory expression of homologous UCP2 was detected in tissues where UCP3 (BAT, SkM or He, Figure S2, C) or UCP2 (Sp and Th, Figure S2, D) are normally expressed.
3.6. Uncoupling proteins expression patterns in UCP2 — ablated mice
Finally, we analysed four UCP2−/− mice to test for the compensatory up-regulation of UCP3 or UCP1. We did not find expression of UCP1 or UCP3 (data not shown) in tissues where UCP2 is normally expressed [35]. BAT in UCP2−/− mice had a similar amount of UCP1 and UCP3 compared to the wt controls (Figure S3, A–B). No compensatory expression of UCP3 was detected in skeletal muscle or heart (Figure S3, B).
4. Discussion
Although one of the first reports about UCP3 described its expression in muscles and BAT [36] more recent morphological and functional studies generally refer to UCP3's predominance in skeletal muscles. The lack of a commercially available, well characterized antibody is still the main obstacle for a reliable comparison of UCP3 abundance in different tissues. For this study we first evaluated a self-designed antibody using positive (recombinant protein) and negative (knockout) controls. The quantitative analysis of UCP3 abundance in skeletal muscles, heart, and brown adipose tissue of mice revealed that BAT has the highest UCP3 amount among the investigated tissues even at physiological conditions. This difference was even more obvious in tissues of cold-acclimated mice. We verified this finding by testing isolated mitochondria to exclude artefacts caused by different amounts of mitochondria in various organs and the adaptive mitochondriogenesis described for cold acclimation [38]. The different expression of rat UCP3 mRNA in BAT in comparison to skeletal muscle after exposure to cold was reported earlier [39]. The higher amounts of expressed protein in BAT and its expression alongside UCP1 may reflect a specific role of UCP3 in this tissue compared to muscles and heart.
Quantitative analysis using recombinant proteins further demonstrated that the abundances of UCP3 and UCP1 in BAT are directly correlated. The functional links between UCP1 and UCP3 were analysed in mice at physiological conditions, in cold-acclimated, UCP1−/− and UCP3−/− mice. Similar to UCP1, UCP3 in BAT was regulated by temperature showing three times higher expression at 4 °C than at 30 °C (thermoneutrality). At first glance, these results would suggest a thermogenic role of UCP3 in BAT as proposed earlier [39]. However, we did not find any compensatory expression of UCP3 in tissues of UCP1−/− mice, in which the ablated UCP1 is normally present. We even detected a significant decrease of UCP3 in BAT of UCP1−/− animals, in contrast to previously reported higher expression of UCP3 in skeletal muscles of cold-acclimated UCP1 knockout mice [28]. This fact rather indicates that UCP3 is not able to take over the thermogenic function of UCP1 in BAT, again supporting the hypothesis about an organ-specific function for UCP3. Moreover, UCP3 abundance in BAT is still approximately 400-fold lower, when compared to UCP1 amounts [35].
The various expression of UCP3 in BAT and muscles may be explained by the recently described differential gene regulation in both tissues [40]. In BAT, the UCP3 transcription is stimulated by an additional intronic enhancer with SP1/3 and PPARγ as core factors for ucp3 gene expression. Also a posttranslational regulation – phosphorylation [41] or glutathionylation [42] – was proposed to influence the uncoupling function of UCP3 and may differ in BAT and skeletal muscles.
We can only speculate about the molecular function of UCP3 and its interconnection with UCP1, as is similar in other groups in this research area to date. The direct correlation between UCP1 and UCP3 amounts in BAT could most likely be explained by the specific UCP3 transport function [43], coherent with the proton transport function of highly abundant UCP1. This idea is supported by other groups that have previously reported on UCP3 exporting fatty acids from mitochondria predominantly during fatty acid oxidation [22]. Bouillaud et al. [44] suggested a UCP3 function in pyruvate and glutamine metabolism as a passive pyruvate transporter. This suggestion would fit to the putative involvement of UCP3 in the regulation of brown fat cell metabolism similar to the function proposed for the highly homologous UCP2 (73%) in rapidly proliferating cells [34,35].
In conclusion, we show that (i) UCP3 is highly abundant in BAT and the sensitivity of the protein expression to temperature is similar to that of UCP1; and (ii) UCP3 expression is significantly decreased when UCP1 is knocked out. Further investigation of the molecular role of UCP3 in BAT is an important task for the future, and may provide new insights into its function.
Supplementary Material
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbabio.2015.10.011.
Acknowledgments
Acknowledgements
This work was partly supported by Austrian Research Fund (FWF, P25357 to EEP). We thank Irina Shabalina for the preparation of brown fat mitochondria. We are grateful to Barbara Cannon, Jan Nedergaard and Irina Shabalina (the Wenner–Gren Institute, Stockholm University, Sweden) for valuable discussion. We thank Quentina Beatty for editorial assistance as a native English speaker.
Abbreviations used in this article
- BAT
brown adipose tissue
- He
heart
- GMsc
gastrocnemius muscle
- mUCP3
recombinant uncoupling protein 3
- RT
room temperature
- ScM
scapulae muscle
- SkM
skeletal muscle
- Sp
spleen
- Th
thymus
- WB
Western blot
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Busiello RA, Savarese S, Lombardi A. Mitochondrial uncoupling proteins and energy metabolism. Front Physiol. 2015;6:36. doi: 10.3389/fphys.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ozcan C, Palmeri M, Horvath TL, Russell KS, Russell RR., III Role of uncoupling protein 3 in ischemia-reperfusion injury, arrhythmias, and preconditioning. Am J Physiol Heart Circ Physiol. 2013 May;304(9):H1192–H1200. doi: 10.1152/ajpheart.00592.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Robbins D, Zhao Y. New aspects of mitochondrial uncoupling proteins (UCPs) and their roles in tumorigenesis. Int J Mol Sci. 2011;12(8):5285–5293. doi: 10.3390/ijms12085285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, et al. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997 May 12;408(1):39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- [5].Echtay KS, Winkler E, Frischmuth K, Klingenberg M. Uncoupling proteins 2 and 3 are highly active H(+) transporters and highly nucleotide sensitive when activated by coenzyme Q (ubiquinone) Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1416–1421. doi: 10.1073/pnas.98.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jaburek M, Garlid KD. Reconstitution of recombinant uncoupling proteins: UCP1, − 2, and − 3 have similar affinities for ATP and are unaffected by coenzyme Q10. J Biol Chem. 2003 Jul 11;278(28):25825–25831. doi: 10.1074/jbc.M302126200. [DOI] [PubMed] [Google Scholar]
- [7].Beck V, Jaburek M, Breen EP, Porter RK, Jezek P, Pohl EE. A new automated technique for the reconstitution of hydrophobic proteins into planar bilayer membranes. Studies of human recombinant uncoupling protein 1. Biochim Biophys Acta. 2006 May;1757(5–6):474–479. doi: 10.1016/j.bbabio.2006.03.006. [DOI] [PubMed] [Google Scholar]
- [8].Nedergaard J, Cannon B. Pros and cons for suggested functions. Exp Physiol. 2003 Jan;88(Pt 1):65–84. doi: 10.1113/eph8802502. [DOI] [PubMed] [Google Scholar]
- [9].Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med. 2011 Jun 24;51(6):1106–1115. doi: 10.1016/j.freeradbiomed.2011.06.022. [DOI] [PubMed] [Google Scholar]
- [10].Toime LJ, Brand MD. Uncoupling protein-3 lowers reactive oxygen species production in isolated mitochondria. Free Radic Biol Med. 2010 Aug 15;49(4):606–611. doi: 10.1016/j.freeradbiomed.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mozo J, Ferry G, Studeny A, Pecqueur C, Rodriguez M, Boutin JA, et al. Expression of UCP3 in CHO cells does not cause uncoupling, but controls mitochondrial activity in the presence of glucose. Biochem J. 2006 Jan 1;393(Pt 1):431–439. doi: 10.1042/BJ20050494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Moore GB, Himms-Hagen J, Harper ME, Clapham JC. Overexpression of UCP-3 in skeletal muscle of mice results in increased expression of mitochondrial thioesterase mRNA. Biochem Biophys Res Commun. 2001 May 18;283(4):785–790. doi: 10.1006/bbrc.2001.4848. [DOI] [PubMed] [Google Scholar]
- [13].Schrauwen P, Hesselink MK. The role of uncoupling protein 3 in fatty acid metabolism: protection against lipotoxicity? Proc Nutr Soc. 2004 May;63(2):287–292. doi: 10.1079/PNS2003336. [DOI] [PubMed] [Google Scholar]
- [14].Lombardi A, Grasso P, Moreno M, de Lange P, Silvestri E, Lanni A, et al. Interrelated influence of superoxides and free fatty acids over mitochondrial uncoupling in skeletal muscle. Biochim Biophys Acta. 2008 Jul;1777(7–8):826–833. doi: 10.1016/j.bbabio.2008.04.019. [DOI] [PubMed] [Google Scholar]
- [15].Costford SR, Chaudhry SN, Crawford SA, Salkhordeh M, Harper ME. Long-term high-fat feeding induces greater fat storage in mice lacking UCP3. Am J Physiol Endocrinol Metab. 2008 Nov;295(5):E1018–E1024. doi: 10.1152/ajpendo.00779.2007. [DOI] [PubMed] [Google Scholar]
- [16].Aguer C, Fiehn O, Seifert EL, Bezaire V, Meissen JK, Daniels A, et al. Muscle uncoupling protein 3 overexpression mimics endurance training and reduces circulating biomarkers of incomplete beta-oxidation. FASEB J. 2013 Oct;27(10):4213–4225. doi: 10.1096/fj.13-234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gong DW, Monemdjou S, Gavrilova O, Leon LR, Marcus-Samuels B, Chou CJ, et al. Lack of obesity and normal response to fasting and thyroid hormone in mice lacking uncoupling protein-3. J Biol Chem. 2000 May 26;275(21):16251–16257. doi: 10.1074/jbc.M910177199. [DOI] [PubMed] [Google Scholar]
- [18].Langdown ML, Smith ND, Sugden MC, Holness MJ. Excessive glucocorticoid exposure during late intrauterine development modulates the expression of cardiac uncoupling proteins in adult hypertensive male offspring. Pflugers Arch. 2001 May;442(2):248–255. doi: 10.1007/s004240100519. [DOI] [PubMed] [Google Scholar]
- [19].Liebig M, von Praun C, Heldmaier G, Klingenspor M. Absence of UCP3 in brown adipose tissue does not impair nonshivering thermogenesis. Physiol Biochem Zool. 2004 Jan;77(1):116–126. doi: 10.1086/381464. [DOI] [PubMed] [Google Scholar]
- [20].Shabalina IG, Petrovic N, de Jong JM, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013 Dec 12;5(5):1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- [21].Kelly OM, Porter RK. Absence of mitochondrial uncoupling protein 3: effect on thymus and spleen in the fed and fasted mice. Biochim Biophys Acta. 2011 Sep;1807(9):1064–1074. doi: 10.1016/j.bbabio.2011.06.002. [DOI] [PubMed] [Google Scholar]
- [22].Himms-Hagen J, Harper ME. Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp Biol Med (Maywood) 2001 Feb;226(2):78–84. doi: 10.1177/153537020122600204. [DOI] [PubMed] [Google Scholar]
- [23].Vidal-Puig AJ, Grujic D, Zhang CY, Hagen T, Boss O, Ido Y, et al. Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem. 2000 May 26;275(21):16258–16266. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]
- [24].Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997 May 1;387(6628):90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- [25].Golozoubova V, Hohtola E, Matthias A, Jacobsson A, Cannon B, Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 2001 Sep;15(11):2048–2050. doi: 10.1096/fj.00-0536fje. [DOI] [PubMed] [Google Scholar]
- [26].de Queiroz KB, Rodovalho GV, Guimaraes JB, de Lima DC, Coimbra CC, Evangelista EA, et al. Endurance training blocks uncoupling protein 1 up-regulation in brown adipose tissue while increasing uncoupling protein 3 in the muscle tissue of rats fed with a high-sugar diet. Nutr Res. 2012 Sep;32(9):709–717. doi: 10.1016/j.nutres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- [27].Azzu V, Mookerjee SA, Brand MD. Rapid turnover of mitochondrial uncoupling protein 3. Biochem J. 2010 Feb 15;426(1):13–17. doi: 10.1042/BJ20091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shabalina IG, Hoeks J, Kramarova TV, Schrauwen P, Cannon B, Nedergaard J. Cold tolerance of UCP1-ablated mice: a skeletal muscle mitochondria switch toward lipid oxidation with marked UCP3 up-regulation not associated with increased basal, fatty acid- or ROS-induced uncoupling or enhanced GDP effects. Biochim Biophys Acta. 2010 Jun;1797(6–7):968–980. doi: 10.1016/j.bbabio.2010.02.033. [DOI] [PubMed] [Google Scholar]
- [29].Rupprecht A, Sokolenko EA, Beck V, Ninnemann O, Jaburek M, Trimbuch T, et al. Role of the transmembrane potential in the membrane proton leak. Biophys J. 2010 Apr 21;98(8):1503–1511. doi: 10.1016/j.bpj.2009.12.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Smorodchenko A, Rupprecht A, Sarilova I, Ninnemann O, Brauer AU, Franke K, et al. Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochim Biophys Acta. 2009 Oct;1788(10):2309–2319. doi: 10.1016/j.bbamem.2009.07.018. [DOI] [PubMed] [Google Scholar]
- [31].Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000 Dec;26(4):435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- [32].Kramarova TV, Shabalina IG, Andersson U, Westerberg R, Carlberg I, Houstek J, et al. Mitochondrial ATP synthase levels in brown adipose tissue are governed by the c-Fo subunit P1 isoform. FASEB J. 2008 Jan;22(1):55–63. doi: 10.1096/fj.07-8581com. [DOI] [PubMed] [Google Scholar]
- [33].Jimenez M, Yvon C, Lehr L, Leger B, Keller P, Russell A, et al. Expression of uncoupling protein-3 in subsarcolemmal and intermyofibrillar mitochondria of various mouse muscle types and its modulation by fasting. Eur J Biochem. 2002 Jun;269(12):2878–2884. doi: 10.1046/j.1432-1033.2002.02953.x. [DOI] [PubMed] [Google Scholar]
- [34].Rupprecht A, Sittner D, Smorodchenko A, Hilse KE, Goyn J, Moldzio R, et al. Uncoupling protein 2 and 4 expression pattern during stem cell differentiation provides new insight into their putative function. PLoS One. 2014;9(2):e88474. doi: 10.1371/journal.pone.0088474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rupprecht A, Brauer AU, Smorodchenko A, Goyn J, Hilse KE, Shabalina IG, et al. Quantification of uncoupling protein 2 reveals its main expression in immune cells and selective up-regulation during T-cell proliferation. PLoS One. 2012;7(8):e41406. doi: 10.1371/journal.pone.0041406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun. 1997 Jun 9;235(1):79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- [37].Cunningham O, McElligott AM, Carroll AM, Breen E, Reguenga C, Oliveira ME, et al. Selective detection of UCP 3 expression in skeletal muscle: effect of thyroid status and temperature acclimation. Biochim Biophys Acta. 2003 Jul 10;1604(3):170–179. doi: 10.1016/s0005-2728(03)00057-4. [DOI] [PubMed] [Google Scholar]
- [38].Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004 Jan;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- [39].Larkin S, Mull E, Miao W, Pittner R, Albrandt K, Moore C, et al. Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem Biophys Res Commun. 1997 Nov 7;240(1):222–227. doi: 10.1006/bbrc.1997.7636. [DOI] [PubMed] [Google Scholar]
- [40].Hoffmann C, Zimmermann A, Hinney A, Volckmar AL, Jarrett HW, Fromme T, et al. A novel SP1/SP3 dependent intronic enhancer governing transcription of the UCP3 gene in brown adipocytes. PLoS One. 2013;8(12):e83426. doi: 10.1371/journal.pone.0083426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kelly OM, McNamara YM, Manzke LH, Meegan MJ, Porter RK. The preservation of in vivo phosphorylated and activated uncoupling protein 3 (UCP3) in isolated skeletal muscle mitochondria following administration of 3,4-methylenedioxymethamphetamine (MDMA aka ecstasy) to rats/mice. Mitochondrion. 2012 Jan;12(1):110–119. doi: 10.1016/j.mito.2011.03.011. [DOI] [PubMed] [Google Scholar]
- [42].Mailloux RJ, Seifert EL, Bouillaud F, Aguer C, Collins S, Harper ME. Glutathionylation acts as a control switch for uncoupling proteins UCP2 and UCP3. J Biol Chem. 2011 Jun 17;286(24):21865–21875. doi: 10.1074/jbc.M111.240242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Goglia F, Skulachev VP. A function for novel uncoupling proteins: antioxidant defense of mitochondrial matrix by translocating fatty acid peroxides from the inner to the outer membrane leaflet. FASEB J. 2003 Sep;17(12):1585–1591. doi: 10.1096/fj.03-0159hyp. [DOI] [PubMed] [Google Scholar]
- [44].Criscuolo F, Mozo J, Hurtaud C, Nubel T, Bouillaud F. UCP2, UCP3, avUCP, what do they do when proton transport is not stimulated? Possible relevance to pyruvate and glutamine metabolism. Biochim Biophys Acta. 2006 Sep;1757(9–10):1284–1291. doi: 10.1016/j.bbabio.2006.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.