Abstract
A number of enzymes, targeting factors and chaperones engage ribosomes to support fundamental steps of nascent protein maturation, including enzymatic processing, membrane targeting and co-translational folding. The method selective ribosome profiling (SeRP) is a new tool for studying the co-translational action of maturation factors that provides proteome-wide information on a factor’s nascent interactome, the onset and duration of binding and mechanisms controlling factor engagement. This method is closely related to proximity-specific ribosome profiling, which has been developed recently to monitor the subcellular localization of translating ribosomes. Both methods can reveal nascent chain-specific co-translational interaction profiles of factors and predominantly differ in the experimental strategy that is employed to selectively purify the desired ribosome subpopulation.
Exemplified for the yeast Hsp70 chaperone Ssb, we provide a detailed SeRP protocol for studying nascent chain interactions that is readily adaptable for the analysis of other co-translationally acting eukaryotic factors. The protocol provides general guidance for experimental design and optimization, as well as detailed instructions for cell growth and harvest, the isolation of (factor-engaged) monosomes, the generation of a cDNA library and data analysis. Experience in biochemistry, RNA handling, and basic programing knowledge is necessary to perform SeRP. Execution of a SeRP experiment takes 8-10 working days and initial data analysis can be completed within 1-2 days. All the software sources used in this protocol are available in the repository webpage: https://github.com/gfkramer/SeRPyeast. This protocol is an extension of the originally developed bacterial SeRP protocol by Becker and colleagues (2013).
Introduction
Efficient production of functional proteins is of vital importance for all living cells. Recent work has demonstrated that all major maturation steps of newly synthesized polypeptides are coupled to protein synthesis, supported by a network of maturation factors that transiently engage the nascent polypeptide chain. Understanding how factor engagement is coordinated during the highly dynamic process of mRNA translation is of fundamental importance for a deeper, mechanistic understanding of the molecular principles governing the final steps of protein synthesis and maturation.
Current models assume that the ubiquitously acting N-terminal processing enzymes are the first interactors of the emerging polypeptide, followed by translocation factors and folding chaperones 1. Recent data further demonstrate that also the assembly of protein complexes can be coupled to protein synthesis in both, pro- and eukaryotes, by one fully synthesized, folded protein complex subunit engaging another subunit of the complex during synthesis 2,3.
Studying in vivo nascent chain interactions has become possible with the development of SeRP, which is an extension of ribosome profiling (RP) that was established by Ingolia and colleagues 4. SeRP was first applied for studying nascent chain interactions of the ribosome-associated bacterial chaperone Trigger factor in Escherichia coli (E. coli) 5,6. Subsequent SeRP studies in bacteria analyzed the signal recognition particle (SRP) mediated targeting of ribosomes to the membrane embedded translocon 7 and the co-translational assembly of the Vibrio harveyi luciferase 2. More recently, we adapted the bacterial SeRP protocol to explore nascent chain interactions in Saccharomyces cerevisiae (S. cerevisiae) 3,8. A significantly modified procedure was used recently to explore the co-translational action of yeast SRP 9.
Analysis of Ssb’s co-translational function by SeRP
The SeRP protocol described here was applied to analyze the co-translational function of the yeast ribosome-associated Hsp70 chaperone Ssb 8 that functions within a network of chaperones to support folding of newly synthesized proteins. A subset of these chaperones named chaperones linked to protein synthesis (CLIPS) engages nascent chains co-translationally 10. Some of the CLIPS also bind the ribosome, among them the hetero-dimeric nascent polypeptide-associated complex (NAC), and the chaperone triad consisting of Ssb and the ribosome-associated complex (RAC) 11,12–14. RAC is composed of the Hsp40 chaperone Zuo1 and the non-canonical Hsp70 Ssz1 and known to stimulate Ssb’s ATPase activity 15. The ablation of either component of the chaperone triad causes highly similar pleiotropic phenotypes, including slow growth, enhanced sensitivity against high salt, cold, and aminoglycosides, massive protein aggregation and defective ribosome biogenesis 16–18. SeRP analyzing yeast Ssb 8 revealed its’ nascent substrates comprising a large subset of cytosolic and nuclear proteins as well as nascent precursors of proteins destined for the endoplasmic reticulum (ER) and mitochondria. Ssb engages many substrates at multiple phases during translation, and binding is triggered by the emergence of a degenerate sequence motif enriched with positively charged and aromatic residues. Studying the importance of other ribosome-associated chaperones for the Ssb function demonstrated that RAC but not NAC is critical for substrate selection and timely Ssb-nascent chain engagement. Comparing SeRP binding profiles with the ribosome densities along open reading frames of wild-type yeast strains revealed that Ssb-bound ribosomes on average translate mRNAs faster than ribosomes not engaged by Ssb. The variation of translation speed is predominantly imposed by features of the mRNA such as codon usage and secondary structure, charge properties of the emerging polypeptide and only to a minor part by the direct action of Ssb.
Applications of the method
Fundamental mechanistic aspects of the co-translational function of many chaperones are currently not understood, including (i) features of nascent chains and the translation process per se that coordinate chaperone recruitment and release, (ii) the interplay of the factors, and (iii) the potential impact of chaperones on protein folding. SeRP can help fill this gap in our knowledge, by revealing nascent substrate pools and the timing of factor engagement with the progress of translation. Correlating interaction profiles with features of the exposed part of nascent chains can reveal functional principles governing factor binding and release. Finally, comparing the interaction profiles of multiple factors can provide a comprehensive view on the cascade of co-translational events.
This advanced protocol is suited to monitor the nascent interactome of any eukaryotic factor and the timing of its nascent chain engagement during protein synthesis. Current studies employing SeRP focused on the analysis of chaperones and targeting factors as well as interactions of protein complex subunits for co-translational complex assembly. However, to reveal a comprehensive view on the cascade of events driving nascent chain maturation, future studies may also include N-terminal processing enzymes, such as methionine aminopeptidases and N-terminal acetyltransferases, as well as factors involved in mRNA quality control that recognize, ubiquitinate and degrade stalled or misfolded nascent chains. Other possible applications of SeRP are the analyses of translation factors acting during protein synthesis, including translation elongation and termination factors. Combined with the protocol of the Preiss laboratory to study scanning 40S ribosomal subunits 19, SeRP may also be suited to explore the function of translation initiation factors that interact with the small subunit, before the 80S ribosome is assembled. Finally, SeRP may also be used for studying the function of the recently discovered hundreds of proteins that constitute the mammalian ribo-interactome 20.
Comparison with other methods
Traditional methods to study interactions of chaperones with translating ribosomes include polysome isolation from cell lysates by sucrose gradient centrifugation followed by the detection of co-migrating factors using immunoblotting. This method has been successfully used for detecting factor – ribosome interactions 21 but is limited by the sensitivity of the immunoblotting. Furthermore, the method cannot reveal length-resolved interaction profiles of specific polypeptides. In vitro interaction studies employing stalled ribosomes exposing nascent chains of defined length can provide length-resolved interaction profiles but are limited to the analysis of factor binding to a small, selected pool of nascent polypeptides under steady state conditions. A global approach to define the co-translational substrate pool of Ssb and NAC in yeast was previously performed by Frydman and co-workers 22,23. Similar to SeRP, the authors purified chaperone-bound ribosome nascent chain complexes (RNC complexes) along with the mRNA that is translated. Comparing the pool of factor-associated mRNAs with the pool of all translated mRNAs, a list of substrates could be defined. However, this approach does not provide length-resolved interaction profiles of nascent chains and is not well suited to identify transient interactions.
Proximity-specific ribosome profiling developed by the Weissman lab allows studying translation at specific subcellular locations 24,25 and can also provide nascent chain length resolved interaction profiles of factors. The major difference to SeRP is that for proximity-specific ribosome profiling one of the ribosomal proteins is Avitagged and the factor explored is fused to a biotin ligase. Cells are grown under biotin-depleted conditions and labelling of ribosomes engaged with (or proximal to) the factor is initiated by a biotin pulse, along with a block of translation elongation using a translation inhibitor. Following the in vivo labelling, cells are lysed and biotinylated ribosomes are affinity purified from the total pool of ribosomes to reveal a selected translatome, very similar to SeRP. Proximity-specific ribosome profiling was used first to identify mRNAs translated near the ER and mitochondria in yeast 24,25. More recently, the method was combined with a rapid, auxin-induced SRP degradation system to identify proteins that rely on SRP for efficient ER targeting in yeast 26.
Experimental design
An outline of SeRP is provided in Figure 1. SeRP is a combination of two ribosome profiling experiments analyzing ribosome-protected mRNA fragments of different ribosome populations derived from the same population of cells. The first RP reveals the mRNA position of all active ribosomes (metagene profile) and provides a representative view on the cellular translatome. Analyzing the read densities for each transcript determines relative expression levels of all genes at the level of translation, while the distribution of footprint densities along transcripts provides position-specific information about the relative translation speed of ribosomes. The second RP, the selected translatome, identifies the sub-population of ribosomes that is engaged by the factor under investigation. This data set provides information on the binding properties of the factor to nascent chains and also encompasses information on the local translation kinetics of ribosomes. Calculating the position-specific ratio of footprint densities along every transcript eliminates read density differences originating from translation speed variation existing in both data sets and reveals the binding profile of the factor to the growing polypeptide. Central to SeRP is the maintenance and selective purification of complexes of translating ribosomes engaged by the factor under investigation that formed in vivo. Therefore, (i) translating ribosomes must be efficiently stalled and stabilized at the time point of cell harvest, (ii) the formation of new complexes after cell lysis must be prevented, and (iii) factor-engaged RNC complexes must be selectively purified. The workflow of SeRP is described in more detail below and summarized in Figure 2.
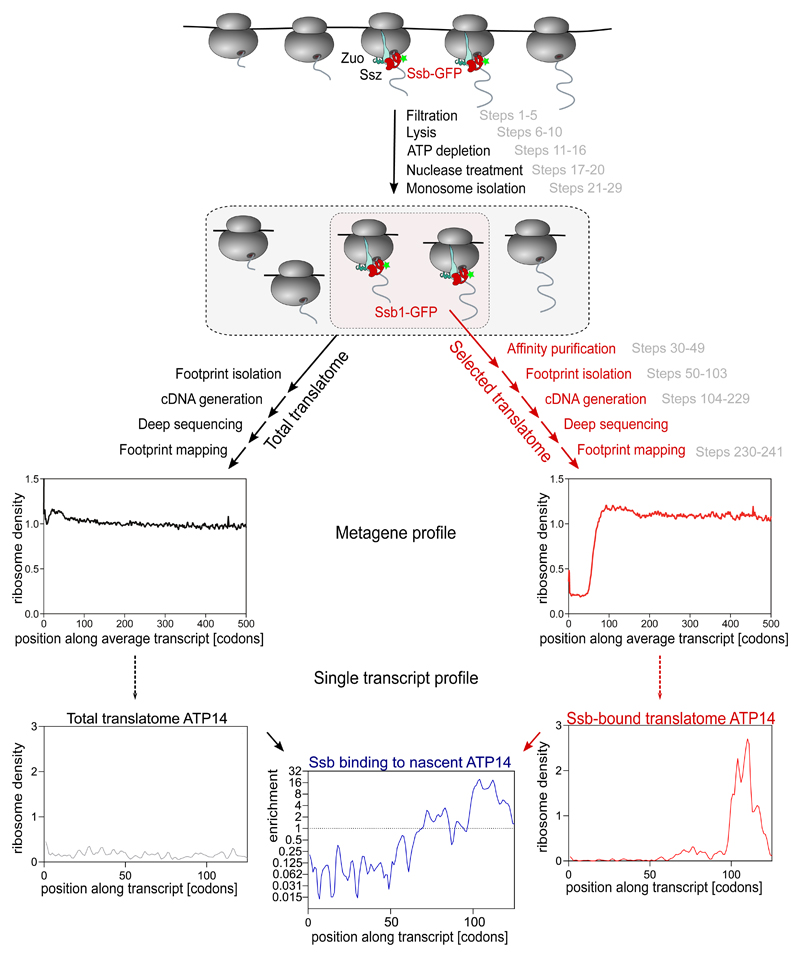
Figure 1. Schematic overview on selective ribosome profiling (SeRP) exemplified for the yeast Hsp70 chaperone Ssb.
Cells are collected by rapid filtration and lysed in frozen state. Lysates are nuclease treated to generate monosomes. Ribosome footprints are isolated and converted to a cDNA library (corresponding procedure steps are shown in grey). Following deep sequencing, footprint reads are mapped to the reference sequence. Metagene analyses, disclose the density of ribosomes averaged over all transcripts. Footprint numbers reveal gene expression levels and the read distribution along transcripts (exemplified for the gene ATP14) provides information on local translation kinetics. SeRP involves parallel processing of two ribosome pools: (i) all ribosomes, revealing the "total translatome" (black) and (ii) the Ssb-bound “selected translatome” (red). Forming the ratio of Ssb-bound translatome and total translatome data reveals length-resolved nascent protein interaction profiles (blue, shown here is the Ssb binding to nascent ATP14).
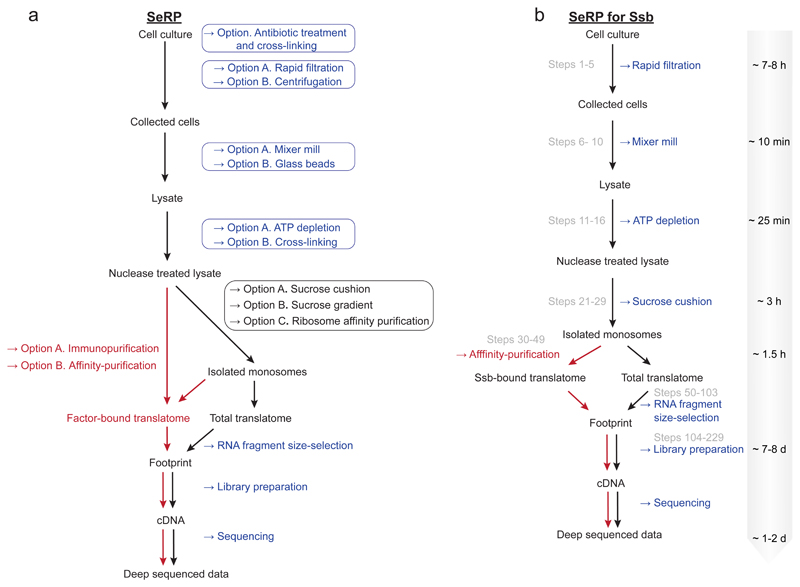
Figure 2. Scheme of the experimental workflow for Selective ribosome profiling (SeRP) in eukaryotic cells.
(a) Experimental procedure starting from cell culture and ending with deep sequencing. The steps to create a total translatome sample or factor-bound (selected) translatome are shown in black or red respectively. Alternative options are indicated in boxes.
(b) SeRP workflow and the corresponding time line applied to study the Ssb-bound translatome (corresponding procedure steps are shown in grey).
General considerations
At the beginning of a SeRP study, a strategy to purify factor-bound ribosomes must be defined. We generally consider two alternative approaches:
(1) Affinity-tagging of the factor under investigation. The E. coli TF interactome was studied by fusing the in vivo biotinylated TEV-Avi-tag to its C-terminus 5 while the analysis of the co-translational assembly of the bacterial heterodimeric luciferase employed an N-terminal CFP/YFP-tag of the subunits LuxA and LuxB 2, which can be selectively purified using a derivate of a lama single chain GFP antibody (GFP-binder) 27. Similarly, the SeRP-based analysis of co-translational interactions of Ssb and the co-translational assembly of yeast protein complexes relied on C-terminal GFP-tagging of the proteins that bind translating ribosomes 3,8 (Figure 2B). Another example for the successful use of affinity-tagging is a study identifying nascent Ssb substrates using microarrays that employed TAP-tagged Ssb 23. Generally, poly-histidine tags should be avoided since we and others observed that this tag can change the ribosome binding properties of factors 28.
(2) Immunopurification (IP) using antibodies that efficiently bind the factor in complex with ribosomes. IPs were successfully used in the past for analyzing the SRP interactome in E. coli 7.
Both strategies have specific advantages and limitations. The greatest advantage of IPs is that processes can be analyzed in wild-type cells, preventing the risk of artefacts caused by tagging proteins. Disadvantages are that high-affinity antibodies allowing efficient immunopurifications are not always available for the protein of interest and the specificity of the selected antibody may also bias the outcome of the experiment. Moreover, IPs may create biased data sets, for example if the ribosome-bound factor exists in alternative complexes or conformations that are purified with different efficiency. Finally, for the specific case of Ssb, the use of antibodies would not allow to selectively study the binding properties of both Ssb isoforms separately.
The use of affinity-tags eases the establishment of a robust purification protocol and reduces the risk of IPs to artificially enrich sub-populations of factor-engaged ribosomes. It also allows straightforward control experiments estimating the extent of post-lysis interactions of factors with translating ribosomes (see below). The major disadvantage of factor tagging is the potential negative impact on protein function. Therefore, activity and ribosome binding properties of tagged proteins must be rigorously tested and compared to the native factor in vivo and in vitro. This includes experiments determining the expression level of the tagged variant, the full complementation of a knockout phenotype, as well as experiments exploring whether the binding to vacant and translating ribosomes is affected.
The analysis of Ssb function using SeRP was performed using yeast cells encoding chromosomally tagged Ssb1-GFP or Ssb2-GFP that either lacked or encoded the respective other, untagged Ssb isoform. GFP-tagged strains and deletion strains were constructed according to Janke et al. 2004 29. Each tagged isoform fully complemented all tested phenotypes of ssb1Δ ssb2Δ cells when expressed from its authentic locus and maintained its ribosome binding properties 8. Ssb-bound ribosomes were purified using homemade GFP-binder slurry (Box 1). Alternatively, a commercially available GFP-binder material (e.g. GFP-Trap®, ChromoTek) may be used.
Box 1. GFP-binder purification.
Cell growth and protein expression
-
1
Transform BL21 DE3 Rosetta E. coli cells with the plasmid p2666, which encodes for C-terminal His6−tagged lama single chain GFP antibody (GFP-binder), and grow them on LB plates containing 10 μg/mL chloramphenicol (Cm) and 50 μg/mL kanamycin (Kan).
-
2
Inoculate 50 mL LB medium containing 10 μg/mL Cm and 50 μg/mL Kan with freshly transformed E. coli cells from step 1. Grow the pre-culture overnight shaking at 37 °C.
-
3
Inoculate 2x 1.5 L of LB medium containing 10 μg/mL Cm and 50 μg/mL Kan with the overnight culture from step 2 to a starting OD600 of 0.05.
-
4
Grow the culture shaking at 30 °C to an OD600 of 0.4-0.5.
-
5
Transfer the culture to a 25 °C incubator for ~1 h (culture reaches an OD600 of ~0.8).
-
6
Collect a 1 mL aliquot as uninduced control for an SDS-PAGE analysis. Centrifuge the sample at 16,000g for 1 min at 4 °C, discard the supernatant and resuspend the pellet in 100 µL 2x SDS sample buffer. Incubate the sample for 10 min at 95 °C and keep it for subsequent SDS-PAGE analysis.
-
7
Induce GFP-binder expression with 1 mM freshly prepared IPTG and incubate shaking at 25 °C for 20 h.
-
8
Collect 1 mL aliquot as induced control for an SDS-PAGE analysis and proceed as described in step 6.
-
9
Centrifuge the cultures from step 7 at 4,000g for 10 min at 4 °C and discard the supernatant.
-
10
Transfer cell pellets from the 3 L culture to a 50-mL conical tube using a rubber scraper.
-
11
Flash freeze in liquid nitrogen and store at ࢤ80 °C.
PAUSE POINT Cells can be kept at −80 °C for up to 6 months.
-
12
To test for successful expression of GFP-binder, load ~10 µL (corresponding to an OD600 of 1) of uninduced and induced sample (step 6 and 8) on a 12% SDS gel, run and stain the gel with coomassie solution to check the GFP-binder expression. The induced protein runs at a height of ~15 kDa.
His-tag affinity-purification of GFP-binder
-
13
Resuspend the pellet from 3 L culture (step 10) in 40 mL GFP-binder binding buffer.
-
14
Lyse the cells twice in a French pressure cell press at 1,000 psi.
-
15
Collect 10 µL of the total cell lysate and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C and keep the sample for subsequent SDS-PAGE analysis.
-
16
Centrifuge the total lysate at 20,000g for 20 min at 4 °C to remove unbroken cells and cell debris. Use the supernatant for further purification.
-
17
Collect a 10 µL sample of the clarified cell lysate for SDS-PAGE analysis and proceed as described in step 15.
-
18
Set up a 5-mL HisTrap Crude column for His-tag affinity-purification. Use a 50-mL syringe to equilibrate the column manually with 5 column volumes (CV) of ultrapure water.
-
19
Equilibrate with 25 mL (5 CV) of GFP-binder binding buffer as described in step 18.
CRITICAL STEP Never let the column run dry and never let air bubbles get into the system.
-
20
Load the 30 mL clarified cell lysate (from step 16) onto the equilibrated column. Keep the flow-through.
-
21
Collect a 10 µL sample of the flow-through for SDS-PAGE analysis and proceed as described in step 15.
-
22
Wash the colu mn with 1 x 40 mL (8 CV) of GFP-binder binding buffer.
-
23
Wash the column with 1 x 40 mL (8 CV) GFP-binder wash buffer. Collect the wash fraction in a separate tube.
-
24
Elute the GFP-binder with 1x 35 mL (7 CV) GFP-binder elution buffer. Collect 35 fractions of 1 mL in 1.5 mL tubes. Take 5 µL of each fraction and mix with 100 µL of 1x Bradford solution to check protein concentration.
-
25
Collect 10 µL samples of the elution fractions for SDS-PAGE analysis and proceed as described in step 15.
-
26
Load 4 µL of each sample from steps 17, 21 and 25 on a 10% SDS gel. Run and stain the gel with coomassie solution to check the protein content and purity.
-
27
Clean the column by applying 50 mL (10 CV) of 500 mM Imidazole followed by 50 mL (10 CV) of ultrapure water.
-
28
Add 20 mL (4 CV) of 20% ethanol to the column. The column can be stored for at least 1 year at 4 °C.
-
29
Pool the eluted fractions containing purified GFP-binder and dialyze against 1x PBS buffer (2x 3 L) overnight at 4 °C. Use tubing with a 3.5 kDa cutoff.
-
30
Concentrate the GFP-binder up to 5 mL with a centrifuge filter unit with a 3 kDa cutoff according to the manufacturer’s protocol.
-
31
Centrifuge the concentrated protein at 20,000g for 5 min at 4 °C to remove potential aggregates and transfer the supernatant to a fresh tube.
Size-exclusion chromatography
-
32
Set up HiLoad 16/600 Superdex S75 gelfiltration column in a Åkta purifier.
-
33
Wash column with 150 mL degassed water.
-
34
Equilibrate the column with 150 mL GFP-binder gel filtration running buffer.
-
35
Load the 5 mL of GFP-binder (step 31) on the gel-filtration column with a flow rate of 1 mL/min.
CAUTION Control column pressure and decrease the flow if required.
-
36
Collect 2 mL fractions.
-
37
Collect 10 µL samples of the elution fractions for SDS-PAGE analysis and proceed as described in step 15.
-
38
Load 4 µL of each sample from step 37 on a 10% SDS gel. Run and stain the gel with coomassie solution to check the protein content and purity.
-
39
Combine all fractions from step 36 showing the correctly sized band and no other contaminating bands, measure protein concentration as described in step 24, freeze aliquots in liquid nitrogen and store at −80 °C.
PAUSE POINT Purified GFP-binder can be kept at −80 °C for years.
Coupling of GFP-binder to Sepharose beads
-
40
Couple the GFP-binder (step 39) to NHS-activated Sepharose™ 4 Fast Flow.
-
41
Activate the beads by washing with 10-15 bead volumes (BV) of 1 mM ice-cold HCl.
-
42
Wash the beads three times with 2 BV 1x PBS buffer.
-
43
After each washing step centrifuge at 450g to sediment the beads and remove the supernatant to exchange buffers.
CAUTION Centrifugation at higher forces leads to the collapse of the beads.
-
44
Use 1 mg protein for 1 mL of beads and adjust the GFP-binder concentration with 1x PBS to 1 mg/mL. Mix the protein solution with the beads and incubate at RT for 4 h.
-
45
Remove the solution and add 200 mM glycine solution pH 8.0 at RT for 2 h.
-
46
Wash twice with 2 BV 1x PBS buffer.
-
47
Wash GFP-binder-coupled beads in 0.1 M PBS-Sodium azide and store them in this buffer as a 50% suspension at 4 °C.
Cell growth and harvest
Considering the high sensitivity of RP and SeRP, cells must be cultured using conditions that ensure high reproducibility. This includes tight control of media composition, growth temperature, aeration, number of duplications and culture density at the timepoint of cell harvest. For Ssb-SeRP studies, yeast cultures were inoculated in 200 mL media to an optical density (OD600) of 0.01-0.03 in 1 L flasks, grown under vigorous shaking (120 rpm) and harvested at mid-log phase (OD600 = 0.5-0.6).
Cell harvest is another critical step of the procedure, as it has to preserve both, the position of ribosomes on mRNAs and the engagement of the factor under investigation. Initial protocols used for RP and SeRP in bacteria and eukaryotes often relied on ribosome stalling by antibiotics that were applied to the growth media, specifically chloramphenicol in bacteria and cycloheximide for yeast and higher eukaryotes. Adding these translation elongation inhibitors to growing cultures prior to harvest is necessary if in vivo chemical crosslinking is performed to preserve factor-RNC complexes (Figure 2A). If possible, the treatment of growing cultures with antibiotics should be avoided as it potentially biases the ribosome distribution and impairs the analysis of translation by RP 30,31. In addition, arresting translation in growing cells prior to harvest may change the binding pattern of factors which control binding in response to the speed of translation or the nascent chain folding state. A relatively simple procedure for the harvest of yeast cultures is the rapid filtration using a vacuum manifold. Ideally, the temperature of the medium should not change during filtration, e.g. by using pre-warmed filtration units. Once the filtration is completed, cells must be rapidly (within seconds) scraped of the filter using a scoopula and frozen in liquid nitrogen. Using the described equipment, the harvesting procedure is typically completed within approximately 40 sec.
Cell lysis
An elegant way to preserve the integrity of translating ribosomes and factor-RNC complexes is to lyze frozen cells using a mixer mill (e.g. CryoMill, MM400 (Retsch), TissueLyser (Qiagen)). It is important that the samples remain frozen during the procedure, for example by repeated chilling of the mixer mill jars in liquid nitrogen before and after each pulverization step. Lysis may be performed in the presence of frozen droplets of any buffer of choice to ensure defined conditions during thawing of the frozen lysate. Alternative methods for cell lysate preparation include the glass bead lysis of yeast cells and various protocols for the lysis of adherent or suspended cells grown in culture 32.
Stabilization of factor-RNC complexes and suppression of ex vivo factor-RNC complex formation
To reveal the interaction pattern of a factor within cells, it is critically important that during the course of the experiment (cell lysis, nuclease digestion and purification of factor-RNC complexes), the in vivo established interactions are preserved and the ex vivo formation of new complexes is suppressed. This is a challenging task in particular for factors that rapidly scan translating ribosomes. In some cases, the stability of factor-RNC complexes can be increased enough by performing all steps at low temperature or by using stabilizing buffer conditions (e.g. charge interactions may be stabilized by low ionic strength). Another possibility is to apply chemical crosslinking. In the past, we have successfully used in vivo crosslinking (DSP (Dithiobis (succinimidyl propionate)), formaldehyde) and ex vivo crosslinking (DSP, BS3 (bis(sulfosuccinimidyl)suberate), EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride)), unpublished data and 5,6. For SeRP of Ssb, chaperone-RNC complexes were stabilized by instant ATP depletion in the lysate. The rationale of this strategy is that the ADP-bound form of Hsp70 chaperones stably binds substrates and that substrate release is strongly accelerated by the exchange of ADP with ATP. Thus, the lack of ATP stabilizes Hsp70-substrate complexes. To rapidly deplete ATP, the frozen cell lysate was thawed in buffer containing excess amounts of glucose and hexokinase. An alternative possibility is to deplete ATP using Apyrase 33.
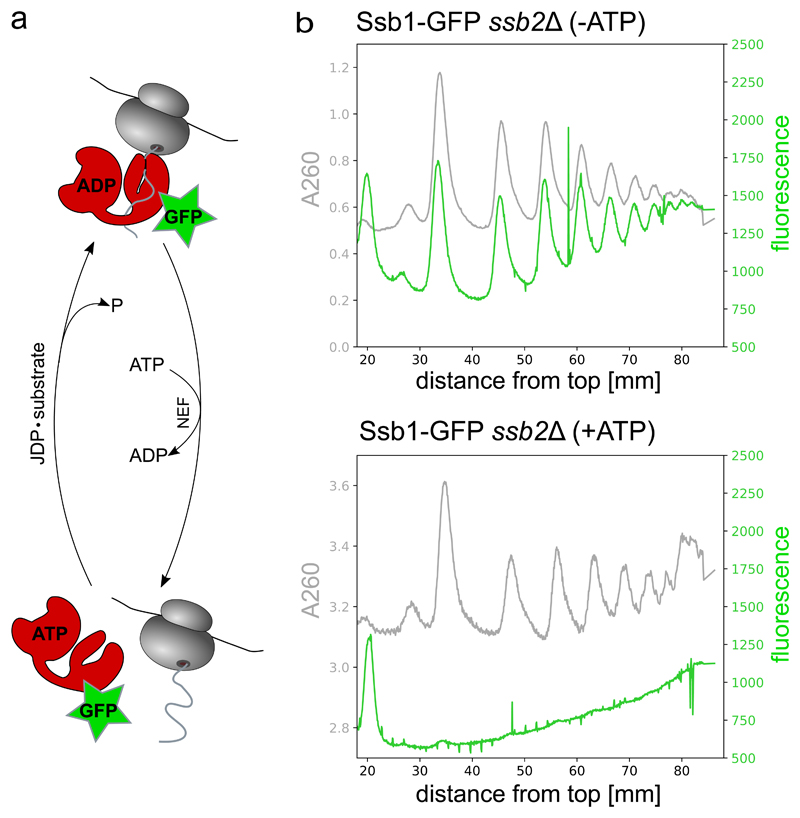
The efficiency of complex stabilization can be tested by a number of different approaches. The simplest approach is to analyze whether complexes remain stable during ribosome purification, e.g. during sucrose gradient centrifugation. For testing the impact of ATP depletion on Ssb-RNC complex stability, we performed sucrose gradient centrifugation of a Ssb1-GFP cell lysate either in the presence of ATP or after ATP depletion and detected the co-migration of Ssb-GFP with ribosomes and polysomes by measuring GFP fluorescence during gradient fractionation (Figure 3 and Box 2). Monitoring Ssb1-GFP fluorescence in sucrose gradients demonstrates the stabilizing effect of ATP depletion. Another possibility to determine complex stability is to analyze fractions of sucrose gradients by western blotting using factor- or tag-specific antibodies (Figure 4). The nascent chain dependence of factor – RNC interactions can be tested using the tRNA analog puromycin. In the absence of cycloheximide, the exposure of RNCs to puromycin and high-salt releases nascent chains. We have analyzed the impact of nascent chains release on the binding of Ssb-GFP to RNCs by performing co-sedimentation analyses followed by western blotting and rRNA detection using a Bioanalyzer Nano chip (Figure S1). Demonstrating the nascent chain-dependent binding of Ssb to RNCs, puromycin treatment interferes with the co-purification of Ssb-GFP and ribosomes.
Figure 3. Detected Ssb-GFP in the sucrose gradient reflects the association of the Hsp70 to ribosomes.
(a) Hsp70 cycle for the example of Ssb-GFP binding to ribosomes. (JDP = J-domain protein, NEF = nucleotide exchange factor)
(b) Polysome profiles recording the co-migration of Ssb-GFP with ribosomes. Ribosome and Ssb-GFP co-migration is analyzed by simultaneous detection of A260 (grey) and GFP fluorescence (green) using the TRIAX™ flow cell (BIOCOMP instruments). Ssb-GFP cell lysate was either thawed in the presence of hexokinase and 0.2% glucose for rapid ATP depletion (-ATP, upper panel) or in the presence of 1 mM ATP (+ATP, lower panel).
Box 2. Sucrose gradient testing Ssb association to polysomes.
-
1
Grow, harvest and lyse cells as described in procedure step 1.-10.
Gradient preparation
-
2
Prepare 50 mL of 2x sucrose gradient buffer, split into two conical 50-mL tubes each containing, either 2.5 g (5% w/v) or 22.5 g (45% w/v) sucrose and fill up to 50 mL with DEPC-water. This buffer amount is sufficient for up to 6 samples.
-
3
Filter 1x sucrose gradient buffers through 0.2 µm filter.
-
4
Mark the open-top polyclear tube according to the metal marker block (half-full mark for short caps).
-
5
Add 5% sucrose solution to the open-top polyclear tube ~2 mm above the mark for short caps.
-
6
Carefully underlay the 45% sucrose solution with a syringe and a needle until reaching again the mark for short caps.
-
7
Close the tubes using short caps and empty the extra solution on top of the caps using a 1,000-μL pipette.
-
8
Place the tubes in a magnetic tube holder, which adheres to the rotary steel plate on the Gradient Master station.
-
9
Generate linear gradients by applying tilted tube rotation in 15 steps with the program: SW40 − short -− Sucr − 05-45% - wv − 15S.
| Gradient preparation steps |
|---|
| start the fractionation station |
| level out the platform |
| press GSTM |
| select rotor: SW40 |
| press rctn |
| select the program |
| press run |
|
|
| Step | Time [min] | Angle [°] | Rotation [rpm] |
|---|---|---|---|
|
| |||
| S01 | 0:09 | 83 | 30 |
| S02 | 0:08 | 83 | 0 |
| S03 | 0:09 | 83 | 30 |
| S04 | 0:08 | 83 | 0 |
| S05 | 0:09 | 83 | 30 |
| S06 | 0:08 | 83 | 0 |
| S07 | 0:09 | 83 | 30 |
| S08 | 0:08 | 83 | 0 |
| S09 | 0:09 | 83 | 30 |
| S10 | 0:08 | 83 | 0 |
| S11 | 0:09 | 83 | 30 |
| S12 | 0:08 | 83 | 0 |
| S13 | 0:01 | 86 | 30 |
| S14 | 4:18 | 90 | 0 |
| S15 | ∞ | 0 | 0 |
|
| |||
| SW40 – short – Sucr – 05-45% – wv – 15S | |||
-
10
After gradient formation, carefully remove the caps and store the gradients at 4 °C until usage (up to 4 h).
Polysome Profiling
-
11
Thaw the cell powder either at 30 °C in a water bath for 2 min (as soon as the powder is melted immediately put the conical tube on ice) or stepwise in a beaker as described in 3.i-ii.
-
12
Transfer the lysate to a 1.5-mL tube.
-
13
Centrifuge at 30,000g for 2 min at 4 °C.
-
14
Transfer the supernatant to a new tube; discard the cell debris pellet.
-
15
Dilute 1 µL of the clarified lysate in 99 µL of DEPC-water. Meassure A260 by NanoDrop to determine the nucleic acid concentration. Keep the sample on ice.
-
16
Load 500 µg RNA sample (up to 400 µL in total) onto each pre-chilled gradient from step 10 and balance the samples with lysis buffer.
CRITICAL STEP Make sure not to mix the lysate with the sucrose gradient to ensure proper separation of ribosomes.
-
17
Centrifuge gradients at 35,000 rpm (220,000g) for 2.5 h at 4 °C in a SW40 rotor.
-
18
Fractionate gradients using a Piston Gradient Fractionator while profiling the Gradient with the Triax™ Flow Cell (FC-2). Therefore, fractionation settings and scanning settings are used as indicated below. Profiling the Gradient with the Triax™ Flow Cell (FC-2) allows obtaining two input wavelengths (UV and VIS scans) in the same gradient, measuring UV at A260 and extinction at 535 nm. Additionally 300 µL fractions may be collected for subsequent SDS-PAGE and western blotting.
|
FRACTIONATION SETTINGS
|
| Volume displaced/mm: 0.143 mL/mm |
| Tubing length: 280 mm |
| Total Dead Volume (= last fraction): 0.294 mL |
| Scan Speed: 0.25 mm/sec |
| Total Distance: 84.4 mm |
| Upper Limit to Slow-down Distance: 37.50 mm |
| Start mode: Meniscus sensing |
| Fail-safe Distance (M): 7 mm |
| Number of fractions: 40 |
| Distance/fraction: 2.11 mm |
| Start Distance: 0.00 mm |
| Volume/fraction: 0.301 mL |
| Data Samples/mm: 12.50 |
|
SCANNING SETTINGS
|
| Scanning mode: UV OD WITH SINGLE |
| FLUORESCENCE SCAN |
| Channel A (LED1) Wavelength: 260 nm |
| Channel B (LED2) Fluor: EGFP |
| Excitation (LED2) Wavelength: 474 nm |
| Excitation filter A: 474/50 nm |
| Emission filter A: 535/50 nm |
| Sensitivity: 0 |
| Integration time: 320 ms |
| Averaging: 1 samples |
| LED1 (260 nm) On Time: 64 ms |
| PD2 (474 nm) On Time: 320 ms |
| Sample A Zero: 792807 |
| Source A Zero: 325159.1 |
| Fluorescence Zero: 50 |
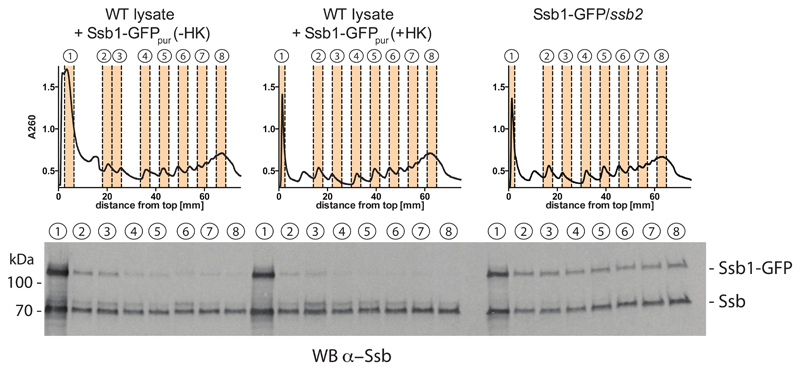
Figure 4. Detected Ssb-RNC complex interactions reflect the in vivo binding properties of Ssb.
Purified Ssb-GFP (Ssb-GFPpur) was added to WT lysates in the absence or presence of hexokinase (HK). Lysates were loaded onto sucrose-gradients, centrifuged and polysome profiles were recorded. Fractions were collected and the indicated fractions were analyzed by western blotting using antibodies targeting Ssb. Demonstrating that Ssb-GFP binds translating ribosomes, a lysate of cells encoding Ssb1-GFP and Ssb2 was analyzed.
The other important feature of a meaningful SeRP experiment is to minimize factor association ex vivo. Complex formation after cell lysis can be reduced by handling samples in the cold, by instant dilution of lysates and by limiting the handling time for purifying factor-RNC complexes. If the factor of interest is tagged, ex vivo formation of factor-RNC complexes can be suppressed by adding excess amounts of purified, untagged factor that competes for binding RNC complexes. We performed two different experiments for testing the prevalence of ex vivo formed Ssb-RNC complexes that may be applied to other SeRP studies. In the first experiment, we thawed frozen cell lysate in buffer containing purified Ssb-GFP in presence or absence of hexokinase. Omitting the nuclease treatment, the lysate was subsequently subjected to sucrose gradient centrifugation and all fractions were tested for the co-migration of Ssb or Ssb-GFP by western blotting (Figure 4). These analyses showed that our experimental conditions (buffer composition, low temperature, dilution, ATP depletion) stabilized Ssb-RNC complexes and largely prevented formation of Ssb-GFP-RNC complexes after cell lysis (Figure 4).
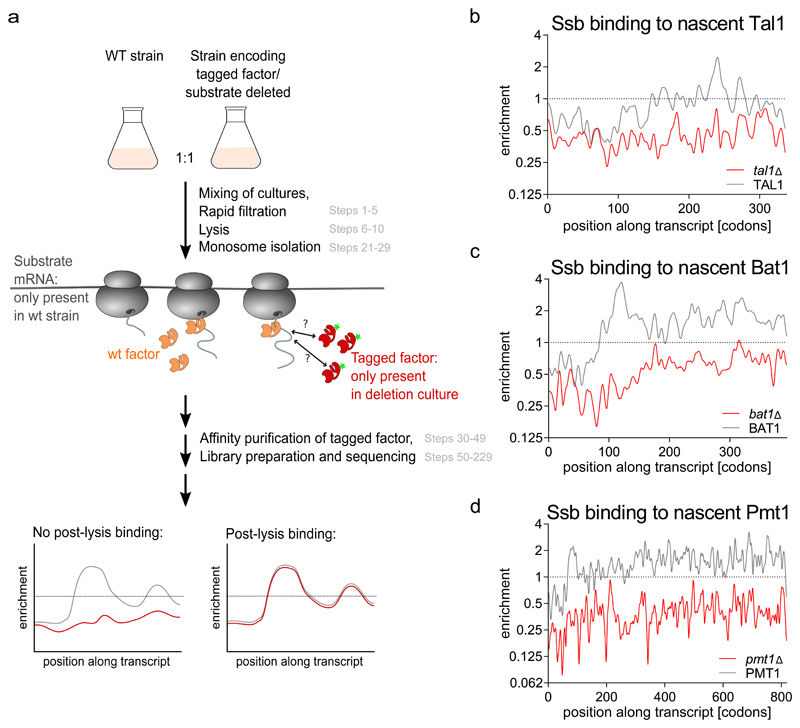
The second experiment tested the extent of ex vivo interactions and can only be performed if the factor of interest is affinity-tagged and once initial SeRP experiments have identified potential substrates. The general strategy is to mix two different strains in a 1:1 ratio directly before harvesting. One strain encodes the tagged factor but lacks genes encoding one or several substrates that were identified in initial experiments. The other strain encodes the untagged factor and the genes encoding these substrates. Mixing both strains followed by SeRP will reveal to what extent the tagged factor engages the nascent substrates that are present only in the mixed lysate. We have performed such Ssb-SeRP studies by independently mixing yeast deletion strains expressing Ssb1-GFP that lacked the Ssb substrates Tal1, Bat1 and Pmt1, respectively, with wild-type cells (Figure 5). Plotting the binding profiles of Ssb1-GFP to substrates that were either expressed in the same (grey) or different (red) cell shows that our SeRP protocol largely suppresses ex vivo formation of Ssb-RNC complexes and specifically detects in vivo nascent chain interactions of Ssb.
Figure 5. Mixing control analyzing the extent of ex vivo interactions in Ssb-SeRP.
(a) Schematic representation of experimental setup (corresponding procedure steps are shown in grey). (b-d) Profiles of Ssb-GFP binding to cytoplasmic protein Tal1 (b), the mitochondrial protein Bat1 (c) or the ER-protein Pmt1 (d). Red curves report on post-lysis binding of Ssb-GFP, black curves on Ssb-GFP binding detected in SeRP.
Nuclease digestion of mRNA
A general step required for RP and SeRP is the nuclease digestion of mRNA parts that are not protected by ribosomes to generate ribosome footprints. Standard RP protocols mostly recommend relatively low concentrations of RNase and long incubation times (30–60 min) at 20–30 °C. However, as discussed before, dissociation and association reactions should be maximally suppressed by reducing both, the temperature and the duration of the nuclease treatment. We recommend to increase nuclease concentrations and to perform RNA digestion at 4 °C for short time (5 min). In the ideal case, nuclease treatment should shift the vast majority of ribosomes from the polysome to the monosome fraction in sucrose gradients. Often a small disome peak remains detectable, which may originate from two neighbouring ribosomes that are too close for the nuclease to efficiently access and cleave the connecting mRNA. The quantity of remaining disomes should be kept at a minimum, to limit the specific loss of footprints derived from ribosomes that are very close and to generate a population of ribosome footprints of similar length. Over-digestion of ribosomes should also be avoided as it enhances the degradation of rRNA and the amount of rRNA fragments that are sequenced. RNaseI is generally used for RP and SeRP of yeast 4,34 but other nucleases such as micrococcal nuclease (MNase) may be used as well 6. Gerashchenko and colleagues 35 provide a detailed discussion of the impact of ribonuclease selection for RP in yeast and mice.
Monosome isolation for total translatome and selected translatome
Once the nuclease treatment is completed, monosomes and factor-engaged RNC complexes must be purified. Different strategies are possible and should be selected based on the specific features of the factor under investigation and the experimental setup used to stabilize factor-RNC complexes. One strategy is to initially isolate ribosomes, either by sucrose cushion centrifugation or by selective purification of monosomes upon sucrose gradient centrifugation. A small fraction of monosomes is subsequently used to determine the total translatome while the larger fraction serves to purify the factor-engaged RNC complexes and determine the selected translatome. An alternative strategy is to divide the lysate into two fractions and use the smaller fraction to isolate monosomes, while the larger fraction is employed to isolate factor-RNC complexes by affinity-purification directly from the nuclease treated lysate (Figure 2A). The advantage of initial ribosome purification is that the selected translatome is isolated from the same pool of purified monosomes that is used to determine the total translatome and that the fraction of the factor not engaged with ribosomes is excluded from the affinity-purification. The disadvantage is that the multi-step purification of factor-RNC complexes takes several hours, increasing the risk for post-lysis artefacts. A potential alternative is to determine both, total and selected translatome, by affinity-purification using lysates from two different strains, one encoding the affinity-tagged factor, and the second strain encoding a tagged ribosomal protein, e.g. the large ribosomal protein Rpl16 23. While this latter possibility has the advantage that both samples are generated by affinity-purification, the major disadvantage of this strategy is that samples from two different strains and cultures must be compared.
Library preparation
Once monosomes are isolated, ribosome footprints must be extracted and converted into a deep sequencing-suited cDNA library, following standard ribosome profiling protocols. A number of different protocols have been published during past years and also commercial kits for library generation are available. In principle, all established protocols can be used, the only specific limitation is that the procedure must allow generating a library from the often very small amounts of ribosome footprints in selected translatome samples. For the sake of completeness, we provide a well-established library preparation protocol that is very similar to the recently updated RP protocol by the Ingolia lab 34.
The first step following monosome preparation is the isolation of ribosome footprints, which includes nucleic acid extraction and subsequent size-selection employing denaturing polyacrylamide gel electrophoresis. We generally select RNA fragments in a size range of 25 to 35 nucleotides. In some cases, alternative size ranges may be selected, for example to detect factor-RNC complexes in alternative ribosomal conformations 36,37. Library generation begins with the 3’-dephosphorylation of RNA fragments using T4 polynucleotide kinase and subsequent determination of concentration and fragment length distribution using an Agilent Bioanalyzer Small RNA chip. Next, the dephosphorylated 3’-ends are ligated to a 5’-adenylated DNA-Linker (3-L1) by the truncated T4 RNA Ligase 2. Linker 3-L1 contains a dideoxyribose at the 3’-end, which prevents a possible circularization of the linker and the concatenation of multiple linker molecules. These ligation products are again purified by denaturing polyacrylamide gel electrophoresis. Subsequently, reverse Linker L(rt) is hybridized to Linker 3-L1 and the RNA fragments are reverse transcribed into cDNA using Superscript III Reverse Transcriptase. Afterwards, RNA molecules are hydrolyzed by high pH and temperature followed by another denaturing polyacrylamide gel electrophoresis to isolate the single-stranded cDNA. Following a circularization with CircLigase II, the single-stranded cDNA is amplified by PCR using Phusion polymerase introducing a barcode for multiplexed sequencing (Table 1). PCR products are gel purified and the size-distribution of the excised product is analyzed employing a High Sensitivity DNA chip. For deep sequencing, DNA concentrations are determined using a Qubit and samples are mixed for multiplexed deep sequencing. A frequent alteration of the protocol is to deplete rRNA fragments during library generation. This may be done subsequent to the initial fragment size selection or after circularized cDNA has been generated, using either customized oligos 32 or commercially available, species-specific rRNA depletion kits (e.g. the ThermoFisher RiboMinus™ Transcriptome Isolation Kit, yeast).
Table 1. PCR reverse primers.
| Index | PCRr ID | Sequence |
|---|---|---|
| ATCACG | 1 | 5′-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTG-3′ |
| CGATGT | 2 | 5′-CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAGTTCAGACGTGTG-3′ |
| TTAGGC | 3 | 5′-CAAGCAGAAGACGGCATACGAGATGCCTAAGTGACTGGAGTTCAGACGTGTG-3′ |
| TGACCA | 4 | 5′-CAAGCAGAAGACGGCATACGAGATTGGTCAGTGACTGGAGTTCAGACGTGTG-3′ |
| ACAGTG | 5 | 5′-CAAGCAGAAGACGGCATACGAGATCACTGTGTGACTGGAGTTCAGACGTGTG-3′ |
| GCCAAT | 6 | 5′-CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTG-3′ |
| CAGATC | 7 | 5′-CAAGCAGAAGACGGCATACGAGATGATCTGGTGACTGGAGTTCAGACGTGTG-3′ |
| ACTTGA | 8 | 5′-CAAGCAGAAGACGGCATACGAGATTCAAGTGTGACTGGAGTTCAGACGTGTG-3′ |
| GATCAG | 9 | 5′-CAAGCAGAAGACGGCATACGAGATCTGATCGTGACTGGAGTTCAGACGTGTG-3′ |
| TAGCTT | 10 | 5′-CAAGCAGAAGACGGCATACGAGATAAGCTAGTGACTGGAGTTCAGACGTGTG-3′ |
| GGCTAC | 11 | 5′-CAAGCAGAAGACGGCATACGAGATGTAGCCGTGACTGGAGTTCAGACGTGTG-3′ |
| CTTGTA | 12 | 5′-CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAGTTCAGACGTGTG-3′ |
Data Analysis
The sequenced DNA libraries comprising the total translatome and the factor-bound translatome represent a rich data set to study factor engagement properties as well as features of mRNA translation. Initial steps of SeRP data analysis are generally performed using publicly available as well as customized read processing and alignment tools that are also used for RP analysis. A comprehensive list of available tools and detailed discussion of computational analysis strategies is provided by 38,39. We provide SeRP specific tools we have used for identifying Ssb substrates and Ssb binding properties (Figure 6). Additionally, we provide the folder ’demo’ containing a compilation of reduced reference files, a small data set, and the SeRP scripts with preset parameters that can be directly used for testing data analysis (https://github.com/gfkramer/SeRP_yeast).
Figure 6. Data analysis of eukaryotic selective ribosome profiling data.
Initial read processing trims adaptors, removes low quality and noncoding reads, and aligns the filtered reads to the genome of interest. Python scripts to perform subsequent analyses including ribosome A-site assignment, RP quality assessment and Basic SeRP analysis are explained in detail in the text.
Read processing of demultiplexed deep sequencing data sets includes (i) trimming of adaptor derived sequences, (ii) removal of low-quality reads, reads that are either too short or too long, as well as reads of noncoding RNAs, including rRNAs and snoRNAs, and (iii) alignment of the filtered reads to the genome. We provide commands used for demultiplexing and read trimming below (step 230. -232.). Generally used genome alignment tools include Bowtie2 40 and Tophat2 41. The quality of intermediate data sets may be analyzed by e.g. FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), which indicates by green checkmarks whether nucleotide distribution and the number of duplicated or overrepresented sequences are within a normal range and also provides information about the average read length.
We provide a number of python scripts for initial analysis of SeRP data sets, which can be run on standard computers (supplementary script A-G, summary in Table 2). Alternative tools are provided by web-based platforms like RiboGalaxy (https://ribogalaxy.ucc.ie/) 42 and Trips-Viz (https://trips.ucc.ie/)43.
Table 2. Summary of data analysis with supplementary scripts A-G (located at the repository: https://github.com/gfkramer/SeRP_yeast).
| No | Name | Description | Input | Output |
|---|---|---|---|---|
| A | Ribosome_Assignment | General data analysis comprises the assignment of the ribosomal A-site for each footprint. | SAM file from genome alignment, A-site offset (txt file) in case of ‘5-end’ mode | no.of reads at each position along the genome (pkl file) distribution of footprint lengths (txt file) gene expression (txt file) |
| B | 3_nt_Periodicity | The average ribosome density of all included transcripts is visualized to check the periodicity. Similar to a metagene profile but on nt scale. | no.of reads at each position along the genome (pkl file) from A | metagene profile with nt resolution (figure) |
| C | Footprint_Length_Distribution | The calculated footprint length distribution is visualized as dot plot. | distribution of footprint lengths (txt file) from A | distribution of footprint lengths (figure) |
| D | Tota l_Enrichment | The expression of each gene is compared between selective and total data sets, 2 biological replicates required. | gene expression (txt file) from A | total enrichment for each transcript (txt file) |
| E | Transcript_Profiles | An enrichment profile is plotted for each transcript. | no. of reads at each position along the genome (pkl file) from A | transcript profiles (figure) |
| F | Binding_Detection | The enrichment of selective over total translatome is statistically analyzed to identify factor- binding regions. | no. of reads at each position along the genome (pkl file) from A | number, localization and length of each binding region (txt file) |
| G | Metagene_Profiles | The average profile of all included transcripts is plotted for the selective and total data set and the enrichment of both. | no. of reads at each position along the genome (pkl file) from A | metagene profiles (figure) |
Ribosome A-site assignment
Supplementary script A uses genome aligned reads to assign ribosomal A-sites along all transcripts. Two alternative assignment modes, named center weighting and 5’ assignment, are implemented in the provided script. Center weighting, a previously published algorithm, is based on a statistical approximation, in which a center of each footprint is defined as the most likely localization of the ribosomal A-site 5,6,8. The alternative 5’ assignment follows the observation that the 5’-end of RNaseI generated footprints mostly has an offset of 15 nt to the ribosomal A-site 44. Both assignment modes are valid for SeRP data analysis and provide highly similar outcomes concerning the identification of factor-specific binding properties. Offset-based A-site determination provides a better 3 nt periodicity (see below) while center weighting, by smoothing the data set, reduces the number of spikes or gaps within transcript profiles. Application and impact of the different ribosome assignment methods is thoroughly discussed in 38,39. The output ’GeneExpression.txt’ of supplementary script A provides information on the sum of reads per transcript normalized to the total number of reads per sample [RPM] or normalized to the total number of reads per sample and the transcript length [RPKM]. Statistical testing must be performed for each sample set to determine the threshold for reliable footprint density per gene, above which the inter-replicate variation is not a major source of error 4. This threshold must be set in supplementary scripts A, B and G. Genes that do not pass the threshold are labeled ’excluded’ in the output of supplementary script A. Supplementary script A that is provided in the demo folder has a preset threshold and labels transcripts with less than 64 reads as ’excluded’.
RP quality assessment
-
Supplementary scripts B and C analyze the 3 nt periodicity and the footprint length distribution. A pronounced periodicity and a normal Gaussian distribution of the footprint length peaking at around 28-30 nt indicates efficient nuclease treatment of mRNAs.
Basic SeRP analysis tools of total and selected translatome include:
Supplementary script G performs metagene analyses, which reveal the density of ribosomes averaged over all transcripts included in the data set. Comparing metagene profiles of both samples reveals the binding properties of the factor as a function of nascent chain length but does not reveal nascent chain specific factor binding. Metagene profiles can also be limited to specific groups of genes, for example genes that encode proteins of specific cellular compartments. Such analyses require minor modifications of the provided supplementary script G.
Supplementary script D calculates the total enrichment of reads for each transcript by forming the ratio of RPKM values in selected translatomes over total translatomes. Enrichment values can be used to identify factor-bound nascent chains (i.e. substrates). However, transient binding of factors during synthesis of a protein often has negligible effects on the ratio or RPKM values of transcripts and cannot be detected using this tool.
Supplementary script E provides factor specific enrichment profiles on a single transcript level.
-
Supplementary script F identifies mRNAs regions that are enriched in selected translatomes, indicating transient factor-nascent chain interaction. The algorithm computes the ratio of reads at every transcript position and identifies regions of minimal 15 nucleotides of footprint densities enriched over a preselected threshold in the selected over the total translatome. The preset parameters of script F were used for determining binding of Ssb to RNC complexes 8 and must be adjusted for analyzing other factors.
The output of these initial analyses provides a starting point for other, more detailed analyses tailored to the factor under investigation such as the identification of binding motifs within the emerged part of the nascent protein, characterization of binding patterns or gene ontology enrichment analysis, which must be customized for each experiment and are not included in the general data analysis provided here.
Limitations
SeRP is a highly sensitive method to reveal interaction profiles of factors with the nascent proteome at nearresidue resolution. The method has the potential to study factor engagement in vivo, provided that fast and efficient purification conditions can be established which stabilize in vivo factor-RNC complexes and prevent the formation of new complexes in cell lysates. The development of a suitable protocol for the efficient purification of factor-RNC complexes requires a number of optimization and control steps and is often very time consuming. In this regard, affinity-tagging is one attractive possibility to facilitate purification, as it fastens the establishment of the pulldown procedure, but must be tightly controlled to assure that the tagged factor retains its full functionality.
Chemical crosslinking is one universal possibility to stabilize complexes. Since crosslinking can introduce biases it must be carefully optimized and controlled. The risk of creating artefacts is particularly high if the factor is not crosslinked to its potential ribosome binding site but to the nascent chain and if the crosslinker has high amino acid selectivity. Such highly selective crosslinkers may stabilize only the subset of factor-RNC complexes in which a reactive amino acid in the nascent chain is close enough to be crosslinked. Another limitation of SeRP is that the amount of purified factor-RNC complexes is sometimes near or below the limit that is required to produce a sequencable cDNA library. A number of features can contribute to this limitation, for example if the factor is lowly expressed, the number of substrates is limited or if the interactions are too transient to be efficiently stabilized by crosslinking. Some of the commercially available kits for library generation that omit extensive gel purification steps can be used with very small amounts of starting material and may overcome this limitation.
Another limitation of SeRP is that interaction profiles do not provide information on the fraction of a specific nascent chain that is bound by the factor. One potential solution may be to develop a purification procedure that strongly depletes factor-RNC complexes from lysates and to perform a third RP analyzing the unbound fraction. Comparing this RP data set to the total translatome would allow estimating the fraction of any nascent chain population that is factor-engaged at any time during synthesis.
As SeRP is analyzing large populations of monosomes, binding peaks in interaction profiles do not provide direct information on the stability of factor-RNC complexes and the binding kinetics. The only information concerns the maximal life-time of a complex, which can be estimated from the length of a binding peak and the average translation speed. SeRP also cannot directly reveal the binding site of a factor at the ribosome or the nascent chain, but solely indicates the length of the nascent chain that promotes factor binding. This is particularly relevant for factors which do not bind to ribosomes and that may engage any part of the nascent chain that already emerged. Ribosome-bound factors such as Ssb, SRP and NAC likely bind the part of the nascent polypeptide that just emerged from the tunnel, however, such a claim requires supporting evidence. In the case of Ssb, we identified a defined binding motif in the emerging part of nascent chains that selectively recruits Ssb to ribosomes, demonstrating that Ssb binds to nascent chain segments that are close to the tunnel exit.
Finally, the initial nuclease treatment of lysates may limit the use of SeRP. One example is the SeRP analysis of SRP-RNC complex interactions in yeast, which was hampered by the RNaseI sensitivity of the 7S RNA part of yeast SRP 9. The necessity to use RNAses may also exclude a SeRP study of factors that bind to nuclease-sensitive rRNA loops on the ribosomal surface. One potential example is the yeast N-terminal acetyltransferase NatA protein that predominantly binds to ribosomal RNA expansion segments 45. A possibility to overcome this limitation may be to test and employ alternative RNases, for example the micrococcal nuclease (MNase). Another very promising strategy is to use proximity-specific ribosome profiling. Since ribosomes are biotinylated in vivo, the quality of the data cannot be compromised by RNAses that reduce the integrity of factor-RNC complexes.
Reagents
Absolute ethanol (VWR, 20821) CAUTION Ethanol is flammable. Keep away from sources of ignition.
Acid-Phenol-Chloroform Ambion (AM9722) CAUTION Acid-Phenol-Chloroform is corrosive. Acid-Phenol-Chloroform is suspected of causing cancer. Handle with care. Wear gloves.
Ambion™ RNaseI (Thermo Fischer, AM2294)
Aprotinin (Roth, A162.3)
Bacto Tryptone Becton, Dickinson and Company (BD, 211699)
Bacto™Agar Becton, Dickinson and Company (BD, 214030)
Bacto™Peptone Becton, Dickinson and Company (BD, 211820)
Bacto™Yeast extract Becton, Dickinson and Company (BD, 212720)
Bradford protein assay (Bio-Rad, cat. no. 500-0006) CAUTION Bradford reagent is toxic. Handle it with care. Wear gloves.
Bromophenol blue (Chroma,40090)
Chloramphenicol (Sigma, C0378)
Chloroform (Merck, 1.02445) CAUTION Chloroform is irritant and harmful.
Colloidal Coomassie staining solution (Roth, Roti-Blue quick, cat. no. 4829)
cOmplete, EDTA free protease inhibitor tablets Roche Diagnostics GmbH (29384100)
Cycloheximide (Sigma, C0378)
Diethylpyrocarbonate (DEPC, Roth, K028) CAUTION DEPC is carcinogenic. Handle with care. Wear gloves.
DNase I recombinant, RNase-free (Roche, 4716728001)
Glycerol (VWR, 24388.260)
GlycoBlue (c = 15 mg/mL) (Ambion, AM9516)
HCl 1mM (Applichem, A1305) CAUTION Hydrochloric acid is corrosive. Wear gloves and eye protection.
HEPES (Roth, HN78.3)
Hexokinase from Saccharomyces cerevisiae (Sigma, H6380-1.5KU)
Imidazole (Roth, 3899) CAUTION Imidazole is corrosive. Wear gloves and eye protection.
Isopropanol (2-propanol) (Sigma, 33539) CAUTION Isopropanol is flammable. Keep away from sources of ignition.
Isopropyl β-D-1-thiogalactopyranoside (IPTG, Roth, CN08)
Kanamycin (Roth, T832.4)
KCl (Roth, 6781.1)
KH2PO4 (Roth, 3904.1)
KOH (Roth, 6751) CAUTION Potassium hydroxide is corrosive. Wear gloves and eye protection.
Leupeptin (Boehringer, 1017128)
Lysozyme from chicken egg white (Sigma, 62971)
Methanol (VWR, 20847) CAUTION Methanol is toxic and flammable. Handle with care. Wear gloves. Keep away from sources of ignition.
MgCl2 (Roth, KK36.3)
Na2HPO4*2H2O (Roth, T879.3)
NaCl (Roth, 9265)
NHS-Activated Sepharose 4 Fast Flow (GE, Life Sciences, 17090601)
Nonidet P 40 Substitute (NP-40, Sigma, 74385)
Pepstatin A (Roth, 2936.3)
Phenylmethylsulfonylfluoride (PMSF, Roth, 6367) CAUTION PMSF is toxic. Handle it with care. Wear gloves.
Plasmid p2666 encodes for C-terminal His6 –tagged lama single chain GFP antibody (provided upon request)
RNaseI (Ambion™, ThermoFischer, AM2295)
S. cerevisiae strain background: BY4741 (ATCC, 4040002)
SDS pellets (Roth, CN30) CAUTION SDS is carcinogenic. Handle with care. Wear gloves.
-
Sucrose (Sigma, 16104)
CRITICAL the best sucrose gradient profiles were obtained using sucrose puriss. grade (from Sigma)
Superase-In RNase inhibitor (Ambion, AM2696)
Tris (Roth, 4855)
α-D-Glucose monohydrate (Serva, 22720)
β-mercaptoethanol (Roth, 4227) CAUTION β-mercaptoethanol is toxic. Handle with care. Wear gloves.
Equipment
2100 Bioanalyzer Instruments (Agilent Technologies, G2939A)
ÄKTApurifier 10 (GE Healthcare, 28406264)
Centrifuge filter units (Sartorius, Vivaspin6, cutoff 3 kDa, VS2001)
Centrifuge for conical tubes (Heraeus / Thermo Scientific, Multifuge 3SR Plus, 75004371)
Centrifuges (Sorvall, Discovery M120 SE)
Conical tubes, 15 mL (Sarstedt, 62.554.512)
Conical tubes, 50 mL (Sarstedt, 62.547.254)
Criterion™ TGX™ Precast Gels 10% and 12% (BIO-RAD, 5671034, 5671045)
Dialysis tubing (Fisherbrand, 3.5 kDa cutoff, 21-152-10)
Filtering equipment: glass filter holder with glass funnel (1 L), vacuum base and cap, stainless steel screen, gasket and spring clamp (Millipore, 90 mm, XX1009020); ground joint flask 1 L (Millipore, XX1504705)
Filtropur, 0.2 µm (Sarstedt, 831822.101)
French Pressure Cell Press (SIM-AMINCO)
Gel filtration column HiLoad™ 16/600 Superdex™ 75 pg column L × I.D. 60 cm × 16 mm (GE, Healthcare, 28-9893-33).
Gradient station (BIO-COMP, 153)
High Sensitivity DNA chip (Agilent Technologies, 5067-4626)
HisTrap FF crude columns, 5 mL column volume (GE, 17-5286-01)
Incubator (Kuhner, Climo-Shaker ISF1-X)
Magnetic stir bar (Roth, PK74.1)
Magnetic stirrer (Heidolph MR Hei-Mix L, P/N 505-00000-00)
Mixer mill (Retsch, MM400, 20.745.0001)
Nanodrop spectrophotometer (Thermo Scientific, NanoDrop2000, ND2000)
Non-stick, RNase-free tubes (Ambion, AM12450)
Open-top polyclear tubes (Seton, 7031)
Overhead roller (Neolab, Intelli-Mixer, cat. no. 7-0045)
Polycarbonate centrifuge tubes (Beckman Coulter, 343778)
Protran nitrocellulose transfer membrane, 0.45 µm (Amersham, 1000002)
Reaction tubes, 1.5 mL (Sarstedt, 72.690.001)
Rotor Type SW 40 Ti (Beckman, 331302)
Rotor Type TLA120 (Beckman, 357656)
Rubber Scraper (Durawear, 5922)
Safe-lock microcentrifuge tubes 1.5 mL (Eppendorf, T9661-1000EA)
Scoopula (Fisher Scientific, 14-357Q)
Serological pipette, 10 mL (Greiner Bio-One, Cellstar, 607107)
Stainless steel grinding balls (Retsch, 12 mm, 05.368.0037, and 25 mm, 05.368.0105)
Stainless steel jars (Retsch, 10 mL, 01.462.0236, and 50 mL, 01.462.0216)
Syringe filters, 0.22 µm (Sarstedt, 83.1826.001)
Syringe, 50 mL (BD, 300865)
Tabletop centrifuge (Eppendorf, Centrifuge 5424, 5424 000.614)
Tabletop centrifuge, refrigerated (Eppendorf, 5417R, 5407 000.317)
Thermomixer comfort (Eppendorf, 5355)
TriaxTM Flow Cell UV/Fluorescence Gradient Profiling (BIO-COMP, FC-2)
Ultracentrifuge (Sorvall WX90, 46901)
UV/Vis Spectro-photometer (Amersham Bioscience, Novaspec Plus, 80-2117-50)
Vacuum pump unit, (Vacuubrand, F162936 0044F)
Python3 (http://www.python.org/download/)
Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
Reagent Setup
CRITICAL STEP Dissolve all chemicals in ultrapure water in the indicated concentrations, unless noted otherwise. Adjust the pH if necessary.
70% ethanol. Absolute ethanol 70% (v/v) and water 30% (v/v). Store at room temperature (RT, 20-25 °C). The solution is stable for at least three months when not opened.
Aprotinin 10 mg/mL. Store at −80 °C. The solution is stable for at least three months.
Chloramphenicol 10 mg/mL in 70% (v/v) ethanol. Store at −20 °C. The solution is stable for at least three months.
Cycloheximide 10 mg/mL in ethanol. Store at −20 °C. The solution is stable for at least three months.
DEPC-water. Add 1 mL of DEPC to 1 L of ultrapure water, shake vigorously for 1 min, and incubate overnight under the hood with open lid, autoclave the next day to inactivate DEPC. Store at RT. The solution is stable for years.
GFP-binder binding buffer. 1x PBS pH 8.0, 0.5 M NaCl, 20 mM Imidazole, 10 μg/mL Lysozyme, DNaseI 50 mg/mL, 1 mM PMSF, 40 μg/mL Bestatin, 20 μg/mL Leupeptin, 20 μg/mL Aprotinin.
GFP-binder elution buffer. 1x PBS pH 8.0, 0.5 M NaCl, 200 mM Imidazole, 10 μg/mL Lysozyme, DNaseI 50 mg/mL, 1 mM PMSF, 40 μg/mL Bestatin, 20 μg/mL Leupeptin, 20 μg/mL Aprotinin.
GFP-binder gel filtration running buffer. 1x PBS (sterile filtered and degassed)
GFP-binder wash buffer. 1x PBS pH 8.0, 0.5 M NaCl, 50 mM Imidazole, 10 μg/mL Lysozyme, DNaseI 50 mg/mL, 1 mM PMSF, 40 μg/mL Bestatin, 20 μg/mL Leupeptin, 20 μg/mL Aprotinin
Glucose 40% (w/v). Sterile filter. Store at RT. The solution is stable for at least six months.
Glycine solution 200 mM, pH 8.0
HEPES 1 M, adjust pH to 7.0, 7.5 and 8.0 with KOH. Sterile filter. Store at RT. The solution is stable for at least six months.
Hexokinase buffer. 20 mM Tris pH 8.0, 40 mM NaPO4 pH 7.0, 0.2% glucose
Imidazole 1 M, adjust pH to 7.5 with NaOH. Sterile filter. Store at RT in the dark. The solution is stable for at least three months.
IPTG 1 M. Sterile filter. Store at −20 °C. The solution is stable for at least one year.
Kanamycin 50 mg/mL. Sterile filter. Store at −20 °C. The solution is stable for at least one year.
KCl 1 M. Autoclave or sterile filter. Store at. The solution is stable for years.
KOH 10 N. Store at RT. The solution is stable for years.
LB medium. For 1 L weigh 10 g of NaCl, 5 g of Bacto Yeast Extract, and 10 g of Bacto Tryptone. Dissolve in 1 L of deionized water. Autoclave. Store at RT. Medium is stable for at least three months.
Leupeptin 5 mg/mL. Store at °80 °C. The solution is stable for at least three months.
Lysis buffer. 20 mM HEPES pH 7.5, 140 mM KCl, 10 mM MgCl2, 0.1 mg/mL Cycloheximide, 0.1% NP-40 (v/v), 1 mM PMSF, 1x Roche Inhibitor Cocktail, 0.02 U/μl DNaseI in DEPC-water. Always prepare fresh and keep on ice until use.
MgCl2 1 M. Autoclave or sterile filter. Store at RT. The solution is stable for years.
NaCl 5 M. Autoclave or sterile filter. Store at RT. The solution is stable for years.
NaOH 10 N. Store at RT. The solution is stable for years.
NP-40 20% (v/v). Sterile filter. Store at RT. The solution is stable for at least three months.
PBS buffer. 10 mM Na2HPO4, 1.8 KH2PO4, 2.7 mM KCl, 137 mM NaCl. Adjust the pH to 7.4 with HCl. Store at RT. The solution is stable for years.
Pepstatin A 1 mg/mL. Weigh 10 mg of pepstatin A, dissolve in 10 mL of methanol. Store at −20 °C. The solution is stable for at least three months.
PMSF 100 mM. Dissolve 174 mg of PMSF in 10 mL of isopropanol. Store at −20 °C. The solution is stable for at least three months.
SDS sample buffer 2x. 250 mM Tris pH 7.0, 6% (w/v) SDS, 6% (v/v) β-mercaptoethanol, 20% (v/v) glycerol, 0.02% (w/v) bromphenole blue. Store at 4 °C. Add freshly every time β-mercaptoethanol. The solution is stable for years.
SDS 20%. Weigh 20 g of SDS, dissolve in 100 mL of hot (approximately 70 °C) ultrapure water. Autoclave. Store at RT. The solution is stable for years.
Sucrose cushion buffer 1x. 20 mM HEPES pH 7.5, 140 mM KCl, 10 mM MgCl2, 0.1 mg/mL Cycloheximide, 1x Roche Inhibitor Cocktail, 25% (w/v) sucrose in DEPC-water. Always prepare fresh, filter before use and keep on ice until use.
Sucrose gradient buffer 2x. 40 mM HEPES pH 7.5, 280 mM KCl, 20 mM MgCl2, 0.2 mg/mL Cycloheximide, 2x Roche Inhibitor Cocktail in DEPC-water. Always prepare fresh and keep on ice until use. Add desired sucrose concentration and fill up to 1x gradient buffer; filter before use.
Tris 1 M, adjust pH to 7.0, 7.5 and 8.0 with HCl. Autoclave or sterile filter. Store at RT. The solution is stable for at least one year.
Wash buffer I. 20 mM HEPES pH 7.5, 140 mM KCl, 10 mM MgCl2, 0.1 mg/mL Cycloheximide, 0.1% (v/v) NP-40, 1x Roche Inhibitor Cocktail in DEPC-water. Always prepare fresh and keep on ice until use.
Wash buffer II. 20 mM HEPES pH 7.5, 140 mM KCl, 10 mM MgCl2, 0.1 mg/mL Cycloheximide, 0.01% (v/v) NP-40, 1x Roche Inhibitor Cocktail, 10% (v/v) glycerol in DEPC-water. Always prepare fresh and keep on ice until use.
YPD medium. 1% (w/v) yeast extract and 2% (w/v) bacto peptone. Dissolve both in deionized water, fill up to 95% of the final volume, autoclave and add the remaining 5 %vol sterile 40% (w/v) glucose solution. Store at RT. Medium is stable for at least three months.
Procedure
The following protocol describes SeRP for GFP-tagged Ssb as outlined in Figure 2B.
Cell growth and harvest TIMING 7-8 h
-
1
Inoculate a 1 L flask containing 200 mL of YPD medium with a fresh overnight culture to an OD600 of 0.01 to 0.03 (allowing at least 4 cell-doublings before harvest).
-
2
Grow culture at 30 °C with shaking at 120 rpm to an OD600 of 0.5 to 0.6.
-
3
Prewarm the 90-mm glass filtration system and the scoopula; prepare the nitrocellulose membrane (0.45 μm).
-
4
Filter one flask at a time, using the scoopula, rapidly scrape the cells of the membrane and immediately freeze in liquid nitrogen.
CRITICAL STEP The filtration step and in particular the scraping and freezing of filtered cells has to be accomplished in a short time period (40 sec).
-
5
Remove the frozen cells from the scoopula and collect them in a 50-mL conical tube filled with liquid nitrogen. Pierce the lid with a needle and invert to spray out the remaining liquid nitrogen.
PAUSE POINT Cells can be kept at −80 °C for up to 6 months.
Cell lysis TIMING 10 min
Once the cells have been harvested and frozen, perform lysis using a mixer mill.
Prepare lysis buffer: Fill a 50-mL conical tube with liquid nitrogen and drip 600 µL of lysis buffer into liquid nitrogen. Remove excess liquid nitrogen and store at −80 °C until use.
-
6
Chill 10-mL jars and 12-mm grinding balls in liquid nitrogen.
-
7
Add frozen cells (derived from 200 mL cell culture) and 600 µL of frozen lysis buffer to one jar.
-
8
Mixer mill at 30 Hz for 2 min (always use both jars to balance the mixer mill. If only one jar is needed, leave the second jar empty).
-
9
Chill jars in liquid nitrogen.
-
10
Scrape out pulverized cells into a 50-mL conical tube. Store at −80 °C.
CRITICAL STEP Make sure the lysate stays frozen all the time.
PAUSE POINT Lysate can be stored at −80 °C for up to four weeks.
Thawing and ex vivo ATP depletion TIMING 10 min
-
11
Prepare a 5-mL glass beaker on a magnetic stirrer at RT with 100 µL of hexokinase buffer and 100 units of hexokinase enzyme.
-
12
Stepwise thaw the frozen powder (derived from 200 mL cell culture) by adding it into the beaker while stirring with a magnetic flea to immediately deplete ATP.
-
13
Transfer the thawed lysate into a 1.5-mL non-stick tube.
CRITICAL STEP Henceforth use RNase-free reagents and materials. Wear gloves during all RNA preparation steps.
-
14
Centrifuge the tubes at 30,000g for 2 min at 4 °C in a tabletop centrifuge.
-
15
Transfer the supernatant into a fresh 1.5-mL non-stick tube.
-
16
Collect 10 µL of clarified lysate and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C and keep the sample for subsequent SDS-PAGE and western blotting.
Nuclease digestion TIMING 15 min
-
17
Dilute 1 µL of clarified lysate (from step 15.) in 99 µL of DEPC-water. Measure A260 by NanoDrop to determine the nucleic acid concentration. Compare absorbance to a blank containing a 1:100 dilution of lysis buffer.
-
18
Calculate the nucleic acid concentration of clarified lysate considering that 1 A260 (blank-corrected, multiplied by dilution factor) unit corresponds to a nucleic acid concentration of 40 μg/mL.
-
19
Digest the lysate by adding 60 U per 1 A260 of RNaseI and incubate for 5 min at 4 °C in an overhead roller.
-
20
Add 10 µL of SuperaseIn to stop the nuclease digestion and put tubes on ice. Proceed to the next step immediately.
Monosome isolation TIMING 3 h
-
21
Load 400 µL aliquots of the digested lysate onto 800 µL ice-cold sucrose cushions in 1.5-mL polycarbonate centrifuge tubes.
CRITICAL STEP Make sure not to mix the lysate with the sucrose cushion to ensure proper separation of molecules.
-
22
Pellet ribosomal particles by centrifugation at 75,000 rpm (245,000g) for 1.5 h at 4 °C in a TLA120 rotor.
-
23
Quickly remove the supernatant by aspiration and overlay the pellet with 300 µL wash buffer I.
-
24
Resuspend pelleted ribosomes by shaking at 4 °C for about 30-60 min.
-
25
Resuspend residual pellet by pipetting and transfer to a fresh non-stick tube.
-
26
Combine all resuspended ribosomal pellets that belong to the same sample, and measure the nucleic acid concentration of 1:100 dilutions using the NanoDrop.
-
27
Collect 10 µL of the resuspended ribosomal pellet and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C and keep the sample for subsequent SDS-PAGE and western blotting.
-
28
Transfer 100-200 µg of total RNA to a fresh non-stick tube for analyzing the total translatome, fill up the volume to 700 µL using 10 mM Tris pH 7.0 and freeze in liquid nitrogen.
Use these isolated ribosomes for the subsequent RNA extraction and library preparation, they can be stored at −80 °C for up to 7 days.
-
29
Use the rest of the resuspended pellet for Ssb-RNC complex purification revealing the Ssb-bound translatome.
Ssb-GFP affinity-purification TIMING 1.5 h
-
30
Equilibrate GFP-binder slurry before use: Transfer 500 µL of GFP-binder slurry into a 1.5 mL non-stick tube.
-
31
Centrifuge the GFP-binder slurry at 450g for 1 min and remove the supernatant.
CAUTION Centrifugation at higher forces leads to collapse of the beads. Avoid exerting shearing forces.
-
32
Add 1 mL of wash buffer I to the pelleted beads and resuspend gently using a 1,000-μL pipette.
-
33
Repeat equilibration steps (31. and 32.) two more times.
-
34
Centrifuge the GFP-binder slurry at 450g for 1 min and remove the supernatant.
-
35
Mix the equilibrated GFP-binder beads with the resuspended ribosome pellet from step 29.
-
36
Incubate the mixture in an overhead roller for 30 min at 4 °C.
-
37
Centrifuge the mixture at 450g for 1 min at 4 °C, transfer the supernatant into a fresh tube.
-
38
Collect 10 µL of the unbound fraction and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C and keep the sample for subsequent SDS-PAGE and western blotting.
-
39
Resuspend the pelleted beads from step 37. in 1 mL wash buffer I and incubate in an overhead roller at 4 °C for 20 min.
-
40
Pellet the beads as described in step 34. and transfer the supernatant to a fresh non-stick tube.
-
41
Collect 10 µL of the wash fraction and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C and keep the sample for subsequent SDS-PAGE and western blotting.
-
42
Repeat washing steps (39. – 40.) three more times but incubate for 5 min instead.
-
43
Resuspend the pelleted beads in 1 mL wash buffer II and incubate in an overhead roller at 4 °C for 20 min.
-
44
Pellet the beads as described in step 34. and remove the supernatant.
-
45
Resuspend the pelleted beads in 1 mL wash buffer II and incubate in an overhead roller at 4 °C for 5 min.
-
46
Collect 50 µL of the beads and add the same amount of 2x SDS sample buffer. Incubate for 5 min at 95 °C, centrifuge at 450g and keep the supernatant for subsequent SDS-PAGE and western blotting.
-
47
Pellet the beads as described in step 34. and remove the supernatant.
CRITICAL STEP Use fresh non-stick tubes every second wash step.
-
48
Load 10 µL of each sample from steps 16. (Lysate), 27. (Pellet), 38. (Unbound), 41. (Wash) and 46. (Bound) on a 12% SDS gel. Run SDS-PAGE and perform western blotting with an Ssb specific antibody to check efficiency of the AP (compare Figure S1).
-
49
Take the rest of the beads (step 47.) and fill up the volume to 700 µL using 10 mM Tris pH 7.0 and freeze in liquid nitrogen.
Use these isolated ribosomes for the subsequent RNA extraction and library preparation.
PAUSE POINT Beads can be stored at −80 °C for up to 7 days.
RNA extraction and precipitation TIMING 2.5 h
WORK UNDER THE HOOD – Acid phenol-chloroform EVAPORATES QUICKLY and is corrosive. Acid-Phenol-Chloroform is suspected of causing cancer. Handle with care. Wear gloves.
-
50
Pre-warm 750 µL acid phenol-chloroform per sample to 65 °C in 1.5-mL safe-lock non-stick tubes.
-
51
Thaw samples from step 28. and step 49. on ice.
-
52
Add 40 µL 20% SDS to each RNA sample and invert 2 times. The sample turns immediately white, caused by protein precipitation.
-
53
Add 750 µL pre-warmed acid phenol-chloroform.
-
54
Incubate at 65 °C for 5 min shaking at 1,400 rpm in a Thermomixer.
CAUTION Make sure that tubes do not open by using safe-lock tubes or lid clamps.
-
55
Chill samples on ice for 5 min.
-
56
Centrifuge for 2 min at 20,000g. Transfer top, aqueous layer to fresh safe-lock non-stick tube (~700 µL can be recovered).
-
57
Add 700 µL acid phenol-chloroform.
-
58
Incubate at RT for 5 min with occasional vortexing.
-
59
Centrifuge for 2 min at 20,000g. Transfer top, aqueous layer to a fresh safe-lock non-stick tube (~650-700 µL can be recovered).
-
60
Add 600 µL chloroform to the aqueous sample and mix by vortexing.
-
61
Centrifuge for 1 min at 20,000g. Transfer top, aqueous layer to a fresh non-stick tube (~650 µL can be recovered; safe-lock tubes are not required anymore).
-
62
Precipitate nucleic acids by adding 78 µL 3 M sodium acetate and 2 µL GlycoBlue (vortex before use), inverting the tube and adding 750 µL isopropanol.
CRITICAL STEP For precipitation always add reagents in the indicated order.
-
63
Mix by vortexing.
-
64
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
65
Centrifuge at 20,000g for 60 min at 4 °C.
-
66
Remove the supernatant by pipetting.
-
67
Wash the pellet with 750 µL ice-cold 70% ethanol and invert the tube several times.
-
68
Centrifuge at 20,000g for 5 min at 4 °C and remove the supernatant.
-
69
Pulse spin to collect remaining ethanol and aspirate remaining liquids by pipetting.
-
70
Dry the pellet for 2 min at 55 °C with the lid open.
-
71
Resuspend each sample in 21 µL 10 mM Tris pH 7.0 by pipetting and vortexing.
-
72
Spin down and transfer sample to fresh non-stick tube.
-
73
Prepare a 1:5 dilution of the resuspended RNA sample and load 1 µL of the dilution on a Bioanalyzer Nano chip according to the manufacturer’s protocol to measure the co-purified RNA in the bound fractions of the APs (Figure S1).
-
PAUSE POINT Extracted RNA can be stored at −80 °C for months.
TROUBLESHOOTING
-
Library Preparation for Next generation sequencing TIMING 7-8 d
REAGENT Library Preparation for Next generation sequencing
ATP (Adenosine 5′-Triphosphate, 10 mM) (NEB P0756S)
20% SDS Ambion (AM, 9820)
3′-phosphorylated 28 nucleotide RNA control oligonucleotide (IDT, RNasefree HPLC purification): 5′_AUGUACACGGAGUCGACCCGCAACGCGA/3Phos/_3′
Bioanalyzer High Sensitivity DNA kit (Agilent, 5067-4626)
Bioanalyzer Small RNA kit (Agilent, 5067-1548)
Chloroform (Merck, 1.02445) CAUTION Chloroform is irritant and harmful. Wear protection. Work only under the hood.
CircLigase™ II ssDNA Ligase (100 U/μL) (Epicentre, CL9025K) supplied with CircLigase™ II 10x reaction buffer, betaine (5 M), MnCl2 (50 mM), CircLigase™ ssDNA Control Oligo (2 pmol/μL) and sterile water.
Deoxynucleotide (dNTP) Solution Set (NEB, N0446S)
Dimethylsulfoxide (DMSO, Sigma, 276855)
DNA Ladder 10 bp O’RangeRuler (Thermo Scientific, SM1313)
DNA loading dye 6x (Thermo, R0631)
Ethylenediaminetetraacetic acid (EDTA, di-sodium-di-hydrate, Roth, 8043)
Glycoblue (Ambion, AM9516)
HF Phusion polymerase (NEB, M0530L), supplied with 5x Phusion HF reaction buffer.
Murine RNase Inhibitor (NEB, M0314L)
NaOH (Roth, P031) CAUTION Sodium hydroxide is corrosive. Wear gloves and eye protection.
Novex™ TBE-Urea sample buffer (2x) (Invitrogen, LC6876)
Polyethylene glycol (PEG) 8000 (Roth, 0263.2)
Qubit™ dsDNA HS Assay Kit (ThermoFisher, Q32851)
Sodium acetate (3M) pH 5.5 (Ambion, AM9740)
Sodium chloride (5M) (Invitrogen, AM97606)
Sodium dodecyl sulfate 20% (w/v) (SDS, Ambion, AM9820) CAUTION SDS is irritating to eyes and skin. Wear protection.
Sodium hydroxide (1N), (Sigma, S2770)
Superscript III Reverse Transcriptase (Invitrogen, 18080-044), supplied with 5x FS buffer and 100 mM DTT
SYBR gold (Invitrogen, S11494)
T4 polynucleotide kinase (NEB, M0201L), supplied with 10x T4 polynucleotide kinase reaction buffer.
T4 RNA Ligase 2, truncated (NEB, M0242L), supplied 10x T4 RNA Ligase reaction buffer.
TB polyacrylamide gel 8% (Invitrogen, Novex, EC6215BOX)
TBE-Urea polyacrylamide gel 10% (Invitrogen, Novex, EC68752BOX)
TBE-Urea polyacrylamide gel 15% (Invitrogen, Novex, EC6885BOX)
Tris (1M), pH 7.0 (Ambion, AM9851)
Tris (1M), pH 8.0 (Ambion,AM9856)
UltraPure 10x TBE buffer (Invitrogen, 15581-044)
Water DEPC-treated and sterile filtered (Sigma, 95284)
Linker 3-L1 with 5′ adenylation and 3′ dideoxy-Cytidine, unique molecular identifiers (’NN…’) (IDT, RNase-free HPLC purification): 5′-/5rApp/NNNNNATCGTAGATCGGAAGAGCACACGTCTGAA/3ddC/-3′
Linker reverse transcription L(rt) with 5′ phosphorylated, unique molecular identifiers (’NN…’) (IDT, RNase-free HPLC purification): 5′-/5Phos/NNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG/iSp18/GTGACTGGAGTTCAGACGTGTGCTC-3′
PCR forward primer PCRf: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC-3′
-
PCR reverse primers PCRr:
EQUIPMENT Library Preparation for Next generation sequencing
Bioanalyzer 2100 (Agilent, G2943CA)
Electrophoresis power supply (Consort, EV231)
Filter-tips 10 µL, 200 µL, and 1,000 µL (Steinbrenner, 976-010XL, 976-020, 976-200 and 976-1000)
Gel breaker tubes, DNase- and RNase-free (IST Engineering Inc, 3388-100)
Gel chamber (Invitrogen, XCell SureLock Mini-Cell, EI0001)
PCR reaction vials 0.2 mL (Roth, Rotilabo, PC75.1)
Qubit 3 Fluorometer, (ThermoFisher, Q33216)
Qubit™ Assay Tubes (ThermoFisher, Q32856)
Scalpel (Braun, Surgical Disposable Scalpel, 5518016)
Spin-X cellulose acetate columns, 2 mL, 0.45 µm pore size (Fisher, 07-200-387)
Thermocycler (Biometra, TPersonal 48, 050-551)
Transilluminator Bluelight (Herolab UVT-22-LED, NN24)
REAGENT SETUP Library Preparation for Next generation sequencing
CRITICAL STEP Dissolve all chemicals in RNase-free water in the indicated concentrations, unless noted otherwise. Adjust pH if indicated.
Self-made DEPC-water is used for steps prior to acid phenol extraction and for preparation of 1x TBE running buffer, commercial DEPC-water for all reagents used after footprint extraction.
EDTA to 0.5 M, adjust pH to 8.0 with NaOH. Autoclave or sterile filter. Store at RT. The solution is stable for years.
RNA and DNA oligonucleotides. Briefly centrifuge the vials at 500g for 30 sec at 4 °C before opening. Dissolve all oligonucleotides to 100 μM. Aliquot to 20-μL aliquots. Dilute the RNA control oligonucleotide further to 20 μM and the linker L(rt) to 25 μM. Store at −20 °C. Oligonucleotides are stable for years.
Ethanol 80% (v/v). Store at RT. The solution is stable for years.
PEG 8000 50% (w/v). Store at RT. The solution is stable for up to six weeks.
cDNA Library preparation of ribosome footprints
CRITICAL STEP Use RNase-free reagents and materials and wear gloves, while handling RNA-samples.
RNA Quantification by Nanodrop TIMING 5 min
-
74
Dilute 1 µL of extracted nucleic acid in 9 µL of DEPC-water. Measure A260 by Nanodrop.
-
75
Dilute 50 µg of acid phenol extracted RNA (from step 72.) into 20 µL of 10 mM Tris pH 7.0. (The suggested amount increases the probability of success however, we generated cDNA libraries using 0.5 µg of acid phenol extracted RNA as input material.)
Gel-purification of ribosome protected footprints TIMING 4.5 h
-
76
Set up a 15% TBE-Urea polyacrylamide gel in 1x TBE running buffer, wash the wells with running buffer and pre-run it for 1 h at 200 V (gel needs to be warm when samples are loaded).
-
77
Add 20 µL 2x TBE-Urea sample buffer to each sample (from step 75.).
-
78
Prepare the control oligo: Dilute 1 µL control oligo in 9 µL 10 mM Tris pH 7.0 and add 10 µL 2x TBE-Urea sample buffer.
-
79
Thaw the 10 bp O’RangeRuler DNA ladder.
-
80
Denature all samples and the control oligo at 80 °C for 2 min, return to ice - do NOT heat the ladder.
-
81
Load each sample on two adjacent lanes and keep an empty well between different samples. Run the gel for 65 min at 200 V.
-
82
Dilute 6 µL SYBR gold in 60 mL 1x TBE buffer and stain gel for 10-20 min while shaking (use light-protected boxes).
-
83
Prepare labeled gel breaker tubes in labeled 1.5-mL non-stick tubes and a sterile scalpel. Cut a foil for the gel.
-
84
Visualize the stained nucleic acids on a Transilluminator and take a picture before and after cutting.
-
85
Excise desired fragment (optimal: 25-35 nt) with sterile scalpel and place the excised gel slice in the prepared gel breaker tube. Avoid cross contamination of the samples.
-
86
Centrifuge at 20,000g for 5 min at 4 °C.
-
87
Collect and transfer remaining gel pieces from the gel breaker tube to the non-stick tube.
TROUBLESHOOTING
-
88
Add 500 µL 10 mM Tris pH 7.0.
-
89
Incubate at 70 °C for 10 min shaking at 1,400 rpm in a Thermomixer.
-
90
Transfer the gel slurry to a Spin-X cellulose acetate column using a 1,000-μL pipette tip.
TROUBLESHOOTING
-
91
Centrifuge at 20,000g for 3 min at 4 °C.
-
92
Transfer the flow-through to a fresh non-stick tube and place on ice.
-
93
Precipitate nucleic acids by adding 55 µL 3 M sodium acetate and 2 µL GlycoBlue (vortex before use), inverting the tube and adding 550 µL isopropanol.
CRITICAL STEP Add reagents in the indicated order.
-
94
Mix by vortexing.
-
95
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
96
Centrifuge at 20,000g for 60 min at 4 °C.
-
97
Remove the supernatant by pipetting.
-
98
Wash the pellet with 750 µL ice-cold 70% ethanol and invert the tube several times.
-
99
Centrifuge at 20,000g for 5 min at 4 °C and remove the supernatant.
-
100
Pulse spin to collect remaining ethanol and aspirate remaining liquids by pipetting.
-
101
Dry the pellet for 2 min at 55 °C with the lid open.
-
102
Resuspend each sample in 15 µL 10 mM Tris pH 7.0 by pipetting and vortexing.
-
103
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT RNA can be stored at −80 °C for months.
Dephosphorylation TIMING 5 h
-
104
Add 2 µL 10x T4 polynucleotide kinase buffer without ATP, 1 µL murine RNase Inhibitor to each sample and mix well by pipetting. (Prepare a master mix of aforementioned reagents if working with multiple samples to reduce variability.)
-
105
Add 2 µL T4 polynucleotide kinase 2.
-
106
Mix thoroughly by pipetting.
-
107
Incubate at 37 °C for 2 h.
-
108
Inactivate the enzyme by incubation at 75 °C for 10 min and pulse spin sample.
-
109
Add 500 µL 10 mM Tris pH 7.0.
-
110
Precipitate nucleic acids by adding 55 µL 3 M sodium acetate, 2 µL GlycoBlue and 550 µL isopropanol.
-
111
Mix by vortexing.
-
112
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
113
Pellet as described in step 96. −101.
-
114
Resuspend pellet in 11 µL 10 mM Tris pH 7.0.
-
115
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT Dephosphorylated RNA can be stored at −80 °C for months.
Quantification by Bioanalyzer TIMING 45 min
-
116
Prepare a 1:5 dilution of the resuspended RNA sample (1 µL sample in 4 µL DEPC-water).
-
117
Run a Bioanalyzer Small RNA chip according to the manufacturer’s protocol.
Expected RNA fragment size: 28-30 nt.
Linker Ligation at 3’ end TIMING 5 h
-
118
Dilute 5 pmol (= 92.4 ng of 28 nt fragment) of the samples from step 114. to 10 µL with 10 mM Tris pH 7.0.
TROUBLESHOOTING
-
119
Denature all samples at 80 °C for 2 min and return to ice.
-
120
Add 16 µL 50% sterile filtered PEG MW 8000, 4 µL DMSO, 4 µL 10x T4 RNA Ligase 2 buffer, 2 µL murine RNase Inhibitor, 0.1 µL 10 mM adenylated linker 3-L1 and 2.9 µL DEPC-water to each sample. (Prepare a master mix of aforementioned reagents if working with multiple samples to reduce variability.)
-
121
Add 1 µL T4 RNA Ligase 2.
-
122
Mix thoroughly by pipetting.
-
123
Incubate at 23 °C for 2 h.
-
124
Add 500 µL 10 mM Tris pH 7.0.
-
125
Precipitate nucleic acids by adding 55 µL 3 M sodium acetate, 2 µL GlycoBlue and 550 µL isopropanol.
-
126
Mix by vortexing.
-
127
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
128
Pellet as described in step 96. − 101.
-
129
Resuspend pellet in 6 µL 10 mM Tris pH 7.0.
-
130
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT Ligated RNA can be stored at −80 °C for months.
Gel-purification of 3’ linked footprints TIMING 5.5 h
-
131
Set up a 10% TBE-Urea polyacrylamide gel in 500 mL 1x TBE buffer and pre-run for 1 h at 200 V (gel needs to be warm when samples are loaded).
-
132
Add 6 µL 2x TBE-Urea sample buffer to each sample.
-
133
Prepare the control oligo: Dilute 1 µL control oligo in 9 µL 10 mM Tris pH 7.0 and add 10 µL 2x TBE-Urea sample buffer.
-
134
Thaw the 10 bp O’RangeRuler DNA ladder.
-
135
Denature all samples and the control oligo at 80 °C for 2 min, return to ice - do NOT heat the ladder.
-
136
Load each sample on one lane and keep an empty well between different samples. Run the gel for 50 min at 200 V.
-
137
Dilute 6 µL SYBR gold in 60 mL 1x TBE buffer (for each gel) and stain gel with SYBR gold for 10-20 min while shaking (use light-protected boxes).
-
138
Prepare labeled gel breaker tubes in labeled 1.5-mL non-stick tubes and a sterile scalpel. Cut a foil for the gel.
-
139
Photograph gels.
-
140
Excise desired fragment with sterile scalpel and place the excised gel slice in the prepared gel breaker tube. Avoid cross contamination of the samples.
Expected RNA band size: 64 - 66 nt (28 - 30 nt footprint + 36 nt 3-L1).
TROUBLESHOOTING
-
141
Photograph gels.
-
142
Spin at 20,000g for 5 min at 4 °C.
-
143
Collect and transfer remaining gel pieces from the gel breaker tube to the non-stick tube.
-
144
Add 500 µL 10 mM Tris pH 7.0.
-
145
Incubate at 70 °C for 10 min shaking at 1,400 rpm in a Thermomixer.
-
146
Transfer the gel slurry to a Spin-X cellulose acetate column using a 1,000-μL pipette tip.
-
147
Spin at 20,000g for 3 min at 4 °C.
-
148
Transfer the flow-through to a fresh non-stick tube.
-
149
Precipitate nucleic acids by adding 55 µL 3 M sodium acetate, 2 µL GlycoBlue and 550 µL isopropanol.
-
150
Mix by vortexing.
-
151
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
152
Pellet as described in step 96. − 101.
-
153
Resuspend pellet in 10 µL 10 mM Tris pH 7.0.
-
154
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT Ligated RNA can be stored at −80 °C for months.
Reverse transcription of 3’ linked footprints to ssDNA TIMING 6 h
-
155
Add 1 µL 10 mM dNTPs, 0.5 µL 25 μM Linker L(rt) (reverse transcript) and 2 µL DEPC-water to each sample. (Prepare a master mix of aforementioned reagents if working with multiple samples to reduce variability.)
-
156
Mix sample by vortexing and pulse spin.
-
157
Incubate samples at 65 °C for 5 min to denature nucleic acids.
-
158
Return samples to ice.
-
159
Add 4 µL 5x FSB buffer, 1 µL murine RNase Inhibitor, 1 µL 0.1 M DTT to each sample. (Prepare a master mix of aforementioned reagents if working with multiple samples to reduce variability.)
-
160
Mix sample by vortexing and pulse spin.
-
161
Add 1 µL Superscript III to each sample.
-
162
Mix thoroughly by pipetting.
-
163
Incubate at 50 °C for 30 min.
-
164
Add 2.3 µL 1 N NaOH to hydrolyze RNA and to quench the reverse transcription.
-
165
Incubate at 95 °C for 15 min (the sample turns pink in color).
-
166
Set up a 10% TBE-Urea polyacrylamide gel in 500 mL 1x TBE buffer and pre-run for 1 h at 200 V (gel needs to be warm when samples are loaded).
-
167
Add 23 µL 2x TBE-Urea sample buffer to each sample.
-
168
Thaw the 10 bp O’RangeRuler DNA ladder.
-
169
Denature all samples and the control oligo at 80 °C for 2 min, return to ice - do NOT heat the ladder.
-
170
Load each sample on two adjacent lanes and keep an empty well between different samples. Run the gel for 70 min at 200 V.
-
171
Dilute 6 µL SYBR gold in 60 mL 1x TBE buffer and stain gel for 10-20 min while shaking (use light- protected boxes).
-
172
Prepare labeled gel breaker tubes in labeled non-stick tubes and a sterile scalpel. Cut a foil for the gel.
-
173
Visualize the stained nucleic acids and take a picture before and after cutting.
-
174
Excise desired fragment with sterile scalpel and place the excised gel slice in the prepared gel breaker tube. Avoid cross contamination of the samples.
Expected DNA band size: 104 nt
-
175
Centrifuge at 20,000g for 5 min.
-
176
Collect and transfer remaining gel pieces from the gel breaker tube to the non-stick tube.
-
177
Add 500 µL 10 mM Tris pH 8.0.
-
178
Incubate at 70 °C for 10 min shaking at 1,400 rpm in a Thermomixer.
-
179
Transfer the gel slurry to a Spin-X cellulose acetate column using a 1,000-μL pipette tip.
-
180
Spin at 20,000g for 3 min at 4 °C.
-
181
Transfer the flow-through to a fresh non-stick tube and place on ice.
-
182
Precipitate nucleic acids by adding 32 µL 5 M NaCl, 1 µL 0.5 M EDTA, 2 µL GlycoBlue and 550 µL isopropanol.
-
183
Mix by vortexing.
-
184
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
185
Pellet as described in step 96. − 101.
-
186
Resuspend in 15 µL 10 mM Tris pH 8.0.
-
187
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT DNA can be stored at −80 °C for months.
Circularization of ssDNA TIMING 2.5 h
-
188
Add 2 µL 10x CircLigase buffer, 1 µL 1 mM ATP, 1 µL 50 mM MnCl2 to each sample. (Prepare a master mix of aforementioned reagents if working with multiple samples to reduce variability.)
-
189
Add 1 µL CircLigase II to each sample.
-
190
Incubate at 60 °C for 1 h.
-
191
Add additional 1 µL CircLigase II to each sample.
-
192
Incubate at 60 °C for 1 h.
-
193
Incubate at 80 °C for 10 min to inactivate the enzyme.
-
194
Return to ice.
-
195
Directly go to the PCR amplification or store at −80 °C.
PAUSE POINT Circularized DNA can be kept at −80 °C for years.
PCR amplification TIMING 4 h
-
196Prepare the following PCR reagents for each sample:
DEPC-H2O 62.6 μL 5x HF buffer 16.7 μL 10 mM dNTPs 1.7 μL 100 μM PCR fwd primer 0.4 μL HF Phusion 0.8 μL -
197
Add 82.2 µL master mix (MM) to a fresh non-stick tube for each sample.
-
198
Add 5 µL of circularized DNA to each tube with MM.
-
199
Keep the rest of circularized DNA at −80 °C.
-
200
Add 1 µL 20 μM barcode primer to each sample.
-
201
Mix by vortexing.
-
202
Aliquot 17 µL PCR mix to 4 separate PCR strips.
-
203PCR reaction:
Step number Time Temp Repeats 1 30 s 98 °C 1 2 10 s 98 °C 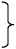 7-16
7-163 10 s 60 °C 4 5 s 72 °C -
204
Remove strips after cycles 9/10/11/12 as a first attempt. The cycle numbers need to be adjusted according to the amount of circularized DNA in each reaction.
-
205
Add 3.5 µL 6x DNA loading dye to each 17 µL reaction.
-
206
Thaw the 10 bp O’RangeRuler DNA ladder.
-
207
Set up a 8% TBE-Urea polyacrylamide gel in 500 mL 1x TBE buffer and wash wells with running buffer.
-
208
Load the four different cycle numbers of each sample on four adjacent lanes. Run the gel for 55 min at 180 V.
-
209
Dilute 6 µL SYBR gold in 60 mL 1x TBE buffer and stain the gel for 10-20 min while shaking (use light-protected boxes).
-
210
Prepare labeled gel breaker tubes in labeled non-stick tubes and a sterile scalpel. Cut a foil for the gel.
-
211
Visualize the stained nucleic acids and take a picture before and after cutting.
-
212
Excise desired fragment with sterile scalpel and place the excised gel slice in the prepared gel breaker tube. Avoid cross contamination of the samples.
Expected DNA band size: 162 bp
-
213
Centrifuge at 20,000g for 5 min.
-
214
Collect and transfer remaining gel pieces from the gel breaker tube to the non-stick tube.
-
215
Add 500 µL 10 mM Tris pH 8.0.
-
216
Incubate at 70 °C for 10 min shaking at 1,400 rpm in a Thermomixer.
-
217
Transfer the gel slurry to a Spin-X cellulose acetate column using a 1,000-μL pipette tip.
-
218
Centrifuge at 20,000g for 3 min.
-
219
Transfer the flow-through to a fresh non-stick tube and place on ice.
-
220
Precipitate nucleic acids by adding 32 µL 5 M NaCl, 1 µL 0.5 M EDTA, 2 µL GlycoBlue and 550 µL isopropanol.
-
221
Mix by vortexing.
-
222
Chill samples at −80 °C for at least 1 h.
PAUSE POINT The precipitation can be kept at −80 °C overnight or longer.
-
223
Pellet as described in step 96. - 101.
-
224
Resuspend in 11 µL 10 mM Tris pH 8.0.
-
225
Spin down and transfer sample to fresh non-stick tube.
PAUSE POINT The dsDNA can be kept at −80 °C for years.
Quantify size distribution by Bioanalyzer
-
226
Make a 1:5 dilution with 1 µL sample and 4 µL DEPC-water.
-
227
Run a Bioanalyzer high sensitivity DNA chip according to the manufacturers recommendations. Expected sample size: 162 bp +/- 5 bp.
TROUBLESHOOTING
Determine DNA concentration by Qubit
-
228
Run a dsDNA high sensitivity test with the Qubit according to the manufacturers recommendations.
-
229
Multiplex and sequence samples according to the Illumina recommendations.
Data analysis - Execution TIMING 1-2 d
The generated cDNA libraries are sequenced on an Illumina NextSeq system according to manufacturer’s protocol and data processing commands are provided below. However the cDNA library generated as described in step 74.-229. is compatible with other sequencing systems as the HiSeq technology as well.
-
230
Process the obtained reads using bcl2fastq2: filter reads based on sequencing quality and demultiplex, i.e. sort reads according to index sequence to the corresponding sample. This step requires a reference file containing sample names and corresponding indices localized in the input folder. Please note that setting the number of threats by −p, −r and −w depends on the used machine and can drastically impact the run time.
bcl2fastq --no-lane-splitting -R /path/to/input -o /path/to/output/
-
231
Combine same samples from different runs, rename and unzip given fastq.gz files with OS specific programs. This step can be performed before or after adaptor trimming.
-
232
Trim adaptor sequence from 3’-end of each read. Identified adaptor sequences that match 6 or more nucleotides of the given sequence and low quality ends (phred quality <20) are removed. Additionally, each read is flanked by 2 and 5 nt unique molecular identifiers at the 5’- and 3’-end, respectively 34, which are removed. Processed reads are written to the output file only after successful trimming and if the final length is between 20 nt and 45 nt.
cutadapt -u 2 -a ATCGTAGATCGGAAGAGCACACGTCTGAA -nextseq-trim=20 --discard-untrimmed -m20 -M45 -O6 -o <filename_output_1>.fastq <filename_input>.fastq cutadapt -u -5 -o <filename_output_2>.fastq <filename_output_1>.fastq
-
233
Remove reads that correspond to noncoding elements e.g. rRNA, snoRNA, exploiting Bowtie2. For the sake of rapid data analysis we prefer using Bowtie2, a program that does not consider splicing. A fasta file (or a list of fasta files) containing the given elements must be transformed into a set of 6 reference files before first alignment:
bowtie2-build <noncoding_RNA_file>.fa <noncoding_RNA>A file containing all noncoding elements for S. cerevisiae can be derived from SGD. Align all trimmed reads (from step 232.) to the generated reference files containing noncoding elements. Use default settings for alignment (end-to-end alignment mode, 20 nt seed length, 0 mismatches) and dismiss aligned reads if not required for further analyses. Continue with non-aligned reads.
bowtie2-un<filename_output>.fastq<noncoding_RNA> <filename_input>.fastq -S /dev/null
-
234
Align noncoding depleted reads (from step 233.) to the organism’s genome. For this purpose 6 reference files are required and can be generated based on the genome containing fasta file, which for yeast can be downloaded from SGD:
bowtie2-build <genome_file>.fa <genome>
To align footprints to eukaryotic genomes a splicing-aware aligner must be used. We generally use the splicing-aware aligner Tophat2, which by default uses Bowtie2 algorithms for read mapping and aligns reads with 2 mismatches or less. For yeast data sets, the minimal and maximal intron lengths are set to 40 nt and 2000 nt. These parameters need to be adjusted for analyzing RP data sets of other organisms. Only reads are considered that span the intron in either direction by at least 4 nt. To save time and computer capacity, the genome position of yeast introns is provided in a gff file and the identification of novel introns, insertions and deletions is disabled.
tophat2 -a4 -i40 -I2000 -g1 --max-insertion-length 0 --max-deletion-length 0 -G <known_introns>.gff --no-novel-juncs --no-convert-bam -o <output_file> <genome> <input_file>.fastq
After aligning the reads to the genome, further data processing is performed using additional Python scripts, as explained below. This includes assignment of ribosome positions (supplementary script A), periodicity of footprint-derived read distribution (supplementary script B), analysis of read lengths (supplementary script C), gene expression per sample (supplementary script A) and in comparison between total translatome and selected translatome samples (supplementary script D), ribosome distribution along specific (supplementary script E) or average transcripts (supplementary script G) and identification of factor-bound nascent chain regions (supplementary script F) (provided at: https://github.com/gfkramer/SeRP_yeast). For each script, the specific usage and positional and optional arguments can be seen by calling the −h option. Reference files (yeast_transcripts.pkl, yeast_introns.pkl, yeast_sequence.pkl, yeast_tRNA.pkl) required to run the supplementary python scripts contain information about the genome sequence, gene positions, tRNA positions and intron positions for yeast. These files are provided and must be organized in the subfolder references_yeast, which itself is localized in the same folder as the scripts. In addition, we provide a script (build_references.py) to independently generate these reference files, for yeast as well as other organisms. The script generates these files based on publicly available fasta files enclosing the genome or transcriptome sequence and a table that contains positional information for each transcript.
-
235
Assignment of the ribosomal position along transcripts is performed based on the SAM file, generated during genome alignment (step 234.). Use this file, containing information about start position and length of each read, as input file (input_name; with file ending) for supplementary script A. The output_name (without file ending) is the new name of this sample and will be used as input name (sample_name) in all subsequent scripts. If no new name should be given, use the input_name but without file ending. Optionally, set the input_path as the location of this SAM file and the output_path as the folder in which all generated files are stored − the output folder must exist. Otherwise the current working directory will be used by default as both, input and output path. Additionally, the mode of ribosomal A-site assignment (’center’ or default: ’5-end’), the minimal and maximal footprint length (default: 23-40) and the read threshold for included genes must be given. Center weighting (’center’) means that the likelihood of 1 (=100%) gets equally distributed to all positions of a read at which the A-site can reside (=center). This region is defined as the remaining (central) positions of a read after removal of 11 nt from both 5’- and 3’-end. The alternative A-site assignment according to the 5’-end of each footprint (’5-end’) complies with reports, that identified the most accurate A-site prediction after RNaseI treatment 15 nt downstream of the footprint’s 5’-end 44. Accordingly, the default setting for all footprint lengths is a 15 nt offset. More accurate A- and P-site determination may be achieved by inferring a read length specific offset, using one of several tools available 46,47. These length-specific offset values can be individually adjusted in the file Asite_offset.txt.
The generated output files include a python dictionary (<sample>_Reads.pkl with each genomic position and the corresponding sum of footprints at this position, raw and normalized) and three text files containing information on the footprint length distribution (<sample>_FootprintLength.txt), the number of reads per chromosome, the number of all reads (<sample>_TotalReads.txt) and the gene expression (<sample>_GeneExpression.txt).
TROUBLESHOOTING
These files serve as input files for the subsequent scripts. Setting the text option to yes (default: no) generates an additional output file (<sample>_Reads.txt) containing the dictionary content in readable text format.
python Supplementary_script_A.py [-h] [-i INPUT_PATH] [-o OUTPUT_PATH] [-m MODE] [-l LIMITS] [-t THRESHOLD] [-x TEXT] input_name output_name
-
236
Before continuing with more detailed data analyses, perform quality control steps of each generated and assigned (selective) ribosome profiling data set. Use supplementary script B and provide the output folder and output sample name from step 235. as input_path (default: current working directory) and sample_name, and the same threshold as in supplementary script A. The algorithm uses the dictionary (<sample>_Reads.pkl) as input and calculates the 3 nt periodicity based on all ribosomal footprints that are located within genes. The output figure (<sample>_3ntPeriodicity.pdf/png) shows the periodicity along an average transcript, which is usually more pronounced using 5’ assignment than using center weighting and furthermore depends on the nuclease treatment and allowed read lengths. The axis start and stop can be set by limits (default: -10 to 50 nt).
python Supplementary_script_B.py [-h] [-i INPUT_PATH] [-t THRESHOLD] [-l LIMITS] sample_name
-
237
Check the read length distribution using supplementary script C and the output folder and output sample name from step 235. as the input_path (default: current working directory) and sample_name, respectively. The script displays all given ribosome footprints from step 235., independently of the given length limits in step 235. in the output files <sample>_FootprintLengthDistribution.pdf/png. The length distribution depends on the organism studied, the nuclease treatment and the size selection during library preparation. An ideal length distribution is between 27 nt and 32 nt.
python Supplementary_script_C.py [-h] [-i INPUT_PATH] sample_name
-
238
Total enrichment calculation computes separately the ratio of summed up reads per transcript of the selected translatome data set over the total translatome data set. The value reveals whether a transcript is enriched in the selected translatome and can reveal factor-bound nascent chains. For this analysis, provide the output folder from step 235. as input_path (default: current working directory) and the sample names from two total (file_total_1, file_total_2) and two selected translatome data sets (file_selec_1, file_selec_2) to the supplementary script D. For this and all subsequent analyses, data of two biological replicates from each SeRP experiment are required. Set an additional name for the experiment (e.g. SeRP_experiment1) as output_name. The script uses the gene expression files (introduced in step 235.) to calculate the required ratio for each transcript and provides the output as text file (<output_name>_TE.txt).
python Supplementary_script_D.py [-h] [-i INPUT_PATH] file_total_1 file_total_2 file_selec_1 file_selec_2 output_name
-
239
Generate enrichment profiles for each transcript separately with supplementary script E. This script uses the same input as supplementary script D (step 238.) and additionally generates an output folder called <output_name>_TranscriptProfiles, which contains two image files for each gene (<output_name>_transcript name.pdf/png). Transcript profiles allow a manual inspection of the ribosome enrichment (selected vs. total translatome) along each transcript separately and in a position resolved manner. For these graphs, the number of selected translatome footprints is divided by the number of total translatome footprints for each single position along each transcript. Data smoothing and filtration of spikes and other technical artifacts is possible to increase analyzability and interpretability but not per se included. Transcripts whose ratio cannot be calculated due to missing reads in total or selective translatome data sets are omitted from this analysis.
python Supplementary_script_E.py [-h] [-i INPUT_PATH] file_total_1 file_total_2 file_selec_1 file_selec_2 output_name
-
240
Metagene profiles show the ribosomal enrichment as in step 239., and additionally distribution of selected translatome footprints and total translatome footprints separately, but averaged between all transcripts which are ’included’. Supplementary script G uses the dictionaries with position-resolved read numbers from step 235. (<sample>_Reads.pkl) of four different samples, two total and two selected translatome data sets. To this end, set the sample_name (used in step 235.) of four samples as file_total_1, file_total_2, file_selec_1, file_selec_2, their localization as input_path (default: current working directory) and an output_name to label the output files <output_name>_metagene.pdf/png. The algorithm weights all transcripts the same, independent of expression level, by normalizing to the number of reads per transcript and excludes transcripts with a total number of reads below the threshold.
python Supplementary_script_G.py [-h] [-i INPUT_PATH] [-t THRESHOLD] file_total_1 file_total_2 file_selec_1 file_selec_2 output_name
-
241
To identify factor-bound nascent chain regions, run supplementary script F with four samples as file_total_1, file_total_2, file_selec_1, file_selec_2 and their localization as input_path (default: current working directory). Provide the sample names given in step 235. and additionally define an output_name. The script scans individual transcript profiles for the reproducible presence of at least 15 nt long stretches that have an enrichment of at least 1.5-fold (= ratio selected vs. total) at every position to be classified as strong binders. Transcripts that are not classified as strong binders are scanned for at least 15 nt long stretches that show an enrichment of at least 3-fold above the binding background (= the first 90 nt where no nascent protein dependent factor binding can occur) and are thereby classified as binders. The width of the interaction and other parameters fit for Ssb binding but need to be adjusted for other factors. Both, strong binders and binders, are given in the output text files (<output_name>_StrongBinders.txt, <output_name>_Binders.txt) together with the number of bound regions, their respective start sites and lengths.
python Supplementary_script_F.py [-h] [-i INPUT_PATH] [--peakwidth] [--minRPKM] [--background] [--minHeightBinders] [--min HeightStrongBinders] [--threshold_Binders] [--threshold_strongBinders] [--threshold_average] [--minCorrelation] file_total_1 file_total_2 file_selec_1 file_selec_2 output_name
Troubleshooting.
| Step | Problem | Possible reason | Possible solution |
| 73 | Inefficient co-purification of RNC complexes upon affinity purification. | Factor-RNC complexes are not stabilized enough to sustain the purification process. | Fast ATP depletion (for Ssb) or chemical crosslinking. In vitro ATP depletion or crosslinking can be accelerated by the stepwise thawing of small frozen lysate amounts in the stabilizing buffer (that provides ATP depleting conditions or contains chemical crosslinker). Add new powder only after the previously added powder is completely melted. |
| 73 | Factor-independent binding of ribosomes during affinity purification in the control (high background). | Inefficient washing condition. | High background binding may be reduced by applying harsher washing conditions, either by changing the composition of washing buffer (e.g. by increasing the salt or detergent concentrations), or by washing more intensely (more or longer washing steps). Another possibility is to change the kind of beads used for the purification. |
| 87 | Gel pieces remain in the gel breaker tube after centrifugation. | The gel pieces are too big. | Take a sterile 200-µL filter-tip and smash gel-pieces, centrifuge again. |
| 90 | The 1,000 µL filter-tip clogs while transferring the broken gel pieces | The gel pieces produced during the gel breaker tube centrifugation are too big. | Cut the pipet tip to widen the tip opening. |
| 118 | Less than 5 pmol of RNA are available for the linker ligation reaction | Small amount of RNC- factor complexes used for acid phenol extraction of nucleic acids | If only small amounts of extracted RNA fragments are available, the amount of linker 3-L1 and Linker L(rt) should be reduced to maintain the proper stoichiometry of reactants. We have successfully generated libraries starting from 0.5 pmol of dephosphorylated RNA by using 10-times less linker 3-L1 and Linker L(rt). |
| 140 | Inefficient ligation reaction | The ligation reaction (and other enzymatic reactions) may be inhibited by elevated salt concentration. | The amount of co-precipitating salt can be reduced by precipitating nucleic acids over night at −20 °C instead of precipitating one hour or more at −80 °C. |
| 227 | Limited amount of cDNA product obtained by PCR | Small amount of PCR template. | Increase the number of PCR cycles. Be aware that as little cycles as possible should be used to limit the PCR bias and to avoid the accumulation of unspecific PCR-products. |
| 235 | Elevated expression of stress genes | Cells were exposed to stress (e.g. nutrient starvation) during growth or cell harvest. | Cell harvest by filtration should be done using prewarmed equipment and as fast as possible. Cells must be scraped and frozen immediately once the medium has passed the membrane. |
TIMING
Step 1.-5., cell growth and harvest: ~ 7-8 h
Step 6.-10., cell lysis: ~ 10 min
Step 11.-20., thawing of the lysate and nuclease digestion: ~ 25 min
Step 21.-29., monosome isolation: ~ 3 h including resuspension of ribosomes
Step 30.-49., Ssb-GFP affinity-purification: ~ 1.5 h
Step 50.-73., RNA extraction: ~ 2.5 h
Step 74.-229., cDNA library preparation and deep sequencing: 7-8 d
Step 230.-241., initial data analysis: 1-2 d
Box 1, GFP-binder purification: ~ 4 d
Box 2, Sucrose gradient testing SSB association to polysomes ~ 6 h
Anticipated Results
200 mL yeast culture harvested by filtration at an OD600 of ~0.5 and lysed by mixer milling together with 600 µL frozen buffer droplets typically yields about 0.7 mL lysate (nucleic acid concentration of 3-5 mg/mL). The ribosomal pellet of an ultracentrifugation contains ~3 mg nucleic acid. About 75-100 µg nucleic acid is used for revealing the total translatome. Phenol extraction of 100 µg input generally recovers ~60 µg of nucleic acid. Affinity-purification of Ssb-RNC complexes using 500 µL GFP-binder slurry reveals about 30-50 µg nucleic acid (measured after phenol extraction). Up to 50 µg are loaded on a polyacrylamide gel for size-selection. About 200-250 ng of RNA fragments (size range 25-35 nucleotides) are typically recovered by gel extraction (determined using a Small RNA Bioanalyzer chip), corresponding to ~15 pmol of 30 nt RNA fragments (calculated based on an average molecular weight of 340 g/mol RNA nucleotide). Ideally, about 5-10 pmol RNA fragments should be used for subsequent steps. The terminal PCR step should yield about 0.4-0.7 pmol DNA fragment (length 162 bp, determined using the DNA HS Bioanalyzer chip).
Good read coverage for determining factor interactomes starts from about 10 million genome-aligned reads. Approximately 95-99% of all reads pass the filters applied during trimming. If no rRNA depletion was performed, about 55-85% of all reads correspond to rRNA sequences. About 95% of the remaining reads align to protein coding sequences.
Supplementary Material
Acknowledgement
We thank L. Eismann and other members of the Bukau laboratory for valuable comments on the manuscript. This work was supported by the ERC (advanced grant 743118), and the DFG (SFB1036 and FOR1805).
Footnotes
Data and code availability
Scripts provided in this protocol and a demo data set are available in the repository under the GNU General Public License: https://github.com/gfkramer/SeRP yeast
The data sets analyzed during the current protocol are available in repository with the identifier GEO: GSE93830 (primary Ssb data set) and GEO: GSE123166 (rebinding control experiments).
Author contributions
G.K., and B.B. designed the study. K.D. and D.M. performed experiments. K.D., D.M. and C.G. set up the protocol for general ribosome profiling in yeast. K.D., G.K., and B.B. established the protocol for selective ribosome profiling. U.F. and K.D. generated the python scripts, and performed data analysis. C.G., D.M. and G.K. wrote the manuscript.
Competing financial interests.
The authors declare no competing financial interests.
References
- 1.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nature structural & molecular biology. 2009;16:589–597. doi: 10.1038/nsmb.1614. nsmb.1614 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Shieh YW, et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 3.Shiber A, et al. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018;561:268–272. doi: 10.1038/s41586-018-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. 1168978 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nature protocols. 2013;8:2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schibich D, et al. Global profiling of SRP interaction with nascent polypeptides. Nature. 2016;536:219–223. doi: 10.1038/nature19070. [DOI] [PubMed] [Google Scholar]
- 8.Doring K, et al. Profiling Ssb-Nascent Chain Interactions Reveals Principles of Hsp70-Assisted Folding. Cell. 2017;170:298–311 e220. doi: 10.1016/j.cell.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartron JW, Hunt KC, Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016 doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 11.Kramer G, Shiber A, Bukau B. Mechanisms of Cotranslational Maturation of Newly Synthesized Proteins. Annual review of biochemistry. 2018 doi: 10.1146/annurev-biochem-013118-111717. [DOI] [PubMed] [Google Scholar]
- 12.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends in biochemical sciences. 2012 doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 14.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Molecular cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 16.Craig EA, Jacobsen K. Mutations in cognate genes of Saccharomyces cerevisiae hsp70 result in reduced growth rates at low temperatures. Molecular and cellular biology. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 KDa heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 18.Koplin A, et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. The Journal of cell biology. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- 20.Simsek D, et al. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell. 2017;169:1051–+. doi: 10.1016/j.cell.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. The Journal of biological chemistry. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- 22.del Alamo M, et al. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS biology. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willmund F, et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346 doi: 10.1126/science.1257521. 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa EA, Subramanian K, Nunnari J, Weissman JS. Defining the physiological role of SRP in protein- targeting efficiency and specificity. Science. 2018;359:689–692. doi: 10.1126/science.aar3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. doi:M700342-MCP200 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Pech M, Spreter T, Beckmann R, Beatrix B. Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. The Journal of biological chemistry. 2010;285:19679–19687. doi: 10.1074/jbc.M109.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 30.Marks J, et al. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:12150–12155. doi: 10.1073/pnas.1613055113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teter SA, et al. Polypeptide flux through bacterial Hsp70: DnaK cooperates with Trigger Factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 34.McGlincy NJ, Ingolia NT. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerashchenko MV, Gladyshev VN. Ribonuclease selection for ribosome profiling. Nucleic acids research. 2017;45:e6. doi: 10.1093/nar/gkw822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diament A, et al. The extent of ribosome queuing in budding yeast. PLoS computational biology. 2018;14:e1005951. doi: 10.1371/journal.pcbi.1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. Clarifying the Translational Pausing Landscape in Bacteria by Ribosome Profiling. Cell reports. 2016;14:686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Wang Y, Xie Z. Computational resources for ribosome profiling: from database to Web server and software. Brief Bioinform. 2019;20:144–155. doi: 10.1093/bib/bbx093. [DOI] [PubMed] [Google Scholar]
- 39.Calviello L, Ohler U. Beyond Read-Counts: Ribo-seq Data Analysis to Understand the Functions of the Transcriptome. Trends Genet. 2017;33:728–744. doi: 10.1016/j.tig.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michel AM, et al. RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 2016;13:316–319. doi: 10.1080/15476286.2016.1141862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiniry SJ, O’Connor PBF, Michel AM, Baranov PV. Trips-Viz: a transcriptome browser for exploring Ribo-Seq data. Nucleic acids research. 2019;47:D847–D852. doi: 10.1093/nar/gky842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martens AT, Taylor J, Hilser VJ. Ribosome A and P sites revealed by length analysis of ribosome profiling data. Nucleic Acids Res. 2015;43:3680–3687. doi: 10.1093/nar/gkv200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knorr AG, et al. Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nature structural & molecular biology. 2019;26:35–39. doi: 10.1038/s41594-018-0165-y. [DOI] [PubMed] [Google Scholar]
- 46.Dunn JG, Weissman JS. Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC genomics. 2016;17:958. doi: 10.1186/s12864-016-3278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malone B, et al. Bayesian prediction of RNA translation from ribosome profiling. Nucleic acids research. 2017;45:2960–2972. doi: 10.1093/nar/gkw1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.