Abstract
The aim of this study was to assess pathogen DNA extraction with a new spin column-based method (DNA-XT). DNA from either whole-blood samples spiked with Plasmodium falciparum or Leishmania donovani amastigote culture was extracted with DNA-XT and compared with that produced by a commercial extraction kit (DNeasy®). Eluates from large and small sample volumes were assessed by PCR and spectroscopy. Using a small volume (5 μl) of blood, the DNA-XT and DNeasy methods produced eluates with similar DNA concentrations (0.63 vs 1.06 ng/μl, respectively). The DNA-XT method produced DNA with lower PCR inhibition than DNeasy. The new technique was also twice as fast and required fewer plastics and manipulations but had reduced total recovered DNA compared with DNeasy.
Keywords : blood, diagnostics, DNA extraction, Leishmania, malaria, PCR, Plasmodium
METHOD SUMMARY
DNA-XT, which is designed for small sample volumes, uses a 5-min detergent and enzymatic lysis step to release DNA from cells. Contaminating proteins and lipids are then bound to a matrix within a spin column during a 1-min centrifugation step while DNA passes directly through.
DNA extraction is an essential starting point for methodologies such as PCR, which is used in the laboratory for molecular biology and clinical diagnostics. It was first achieved in 1869 by Friedrich Meischer [1], but a routine laboratory procedure was not developed until 1958 [2]. Today, a variety of methodologies are available (reviewed by Griffiths and Chacon-Cortes [3]); however, commercial spin column purification of DNA from blood and other tissues and fluids is the most common approach used in modern laboratories. The technique, which originated in 1979 [4], uses silica matrices to selectively bind DNA, allowing washing before elution of the purified product.
NanoMal was an EU-funded industrial–academic consortium that was brought together with the aim of developing a simple-to-use, affordable, handheld diagnostic device to detect malaria infection and the drug resistance status of the parasite by identifying associated genetic mutations. The cartridge-based assay was designed to use only a finger prick of blood, with results being obtained approximately 20 min after sampling. The diagnostics platform technology developed by our industrial partner, QuantuMDx Group Ltd (Newcastle upon Tyne, UK), incorporates a novel DNA extraction technique that allows DNA from lysed whole blood (or other sample types) to flow straight through a proprietary material, but cellular debris is retained. The resulting eluate contains DNA that is of a quality that can be used directly in PCR; therefore, the technique removes the need for additional washing and elution steps, which greatly simplifies the extraction procedure. During the NanoMal project, QuantuMDx created a standalone, spin column-based version of this easy-to-use extraction methodology called DNA-XT, which is optimized for human whole blood. Small-volume samples are lysed and placed over a column of the proprietary material, which is housed within a microcentrifuge collection tube. Following centrifugation, the resulting eluate contains purified, high-quality gDNA.
Here, we examine the performance characteristics of DNA-XT in parallel with a widely used commercial DNA extraction spin column kit for the isolation of parasite DNA from small volumes of Plasmodium falciparum-infected erythrocytes spiked into human whole blood (as a model for blood infections) and purified Leishmania donovani amastigotes (as might be required in a laboratory setting). The two primary objectives were to determine the efficiency of gDNA extraction and the quality of the isolated gDNA by undertaking parasite-specific PCR. Secondary objectives were to assess the level of operator input, the time required for extractions and the use of reagents or consumables.
Materials & methods
Parasites
P. falciparum 3D7 parasites were cultured in human red blood cell (RBC) suspensions using RPMI 1640 medium (Sigma-Aldrich, MO, USA; cat. no. R0883-500ML) supplemented with 2-mM l-glutamine, 35-mM HEPES, 0.5% (w/v) AlbuMAX™ I (Thermo Fisher Scientific, MA, USA), 0.2-mM hypoxanthine and 50-μg/ml gentamycin, and were maintained at 37°C under 5% CO2. Parasite growth was followed by microscopic examination of Giemsa-stained thin blood smears and maintained at ≤10% parasitemia, with an approximately 2% hematocrit. Synchronization of early trophozoite stages was achieved by incubating infected RBCs (iRBCs) in 5% (w/v) sorbitol for 10–20 min at room temperature [5]. L. donovani amastigotes were harvested from the spleens of donor mice. Briefly, female RAG1B6 KO mice, infected with L. donovani at least 60 days before, were humanely killed. At necropsy, spleens were dissected and homogenized, and the amastigotes harvested by differential centrifugation [6].
Sample preparation
For fresh whole blood spiked with P. falciparum iRBCs, stock iRBCs with 8–10% parasitemia were diluted in fresh whole blood (Cambridge Biosciences, Cambridge, UK) to attain a parasitemia of 1%. Two additional tenfold dilutions were performed to obtain samples with parasitemias of 0.1 and 0.01%. The parasitemias of the 1 and 0.1% samples were confirmed by microscopy (Supplementary file 1). Nonspiked fresh whole blood was used as a negative control. As a reference, levels of parasitemia of 0.1–0.2% (5000–10,000 parasites per μl of blood) are generally accepted to be the point at which fever begins and a patient becomes symptomatic for P. falciparum malaria [7]. For Leishmania parasite preparations, parasites (amastigotes) were washed twice in RPMI 1640 without serum prior to counting and were used at a concentration of 1 × 107/ml.
Column purification
For iRBC-spiked fresh whole blood, the DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) and DNA-XT™ DNA extraction kit (QuantuMDx) were used to process samples. Samples (iRBC-spiked or nonspiked fresh whole blood) were processed following the manufacturer's instructions (blood protocols). A 10-µl sample volume was used for the DNA-XT kit and a 100-µl sample volume for the DNeasy kit (termed Q100). Notably, only 40 μl of the 80-μl lysis step is passed though DNA-XT columns after initial processing; thus, only 5 μl of the original blood sample passes through the column (Figure 1). In light of this, an additional arm of the study used DNeasy columns with a 5-μl sample volume (termed Q5) to allow a more direct comparison of the two extraction methodologies.
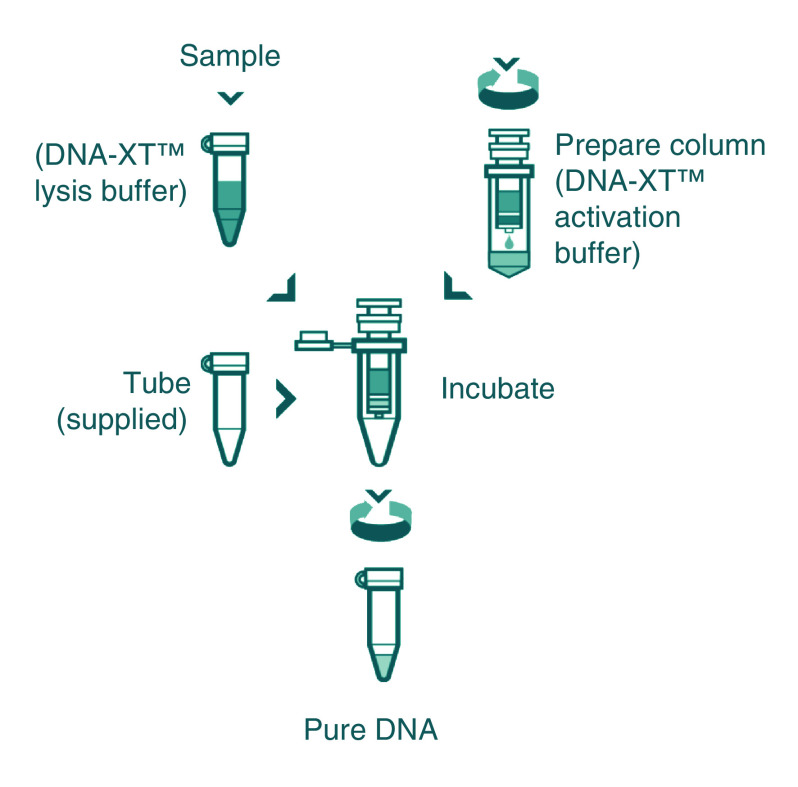
Figure 1. . DNA-XT protocol.
Sample preparation: 10 μl of blood/sample was added to 70 μl of lysis buffer, and the mixture was incubated at 55°C for 10 min. Column preparation: 350 μl of buffer was added prior to centrifugation (1400 ×g, 3 min), and 40 μl of incubated lysis buffer/sample was added to the prepared column and incubated at room temperature for 3 min prior to centrifugation (1400 ×g, 3 min).
For Leishmania parasite preparations, the same two DNA extraction kits were used to process samples. For the DNA-XT kit, 10 μl of sample was used, following the manufacturer's instructions (blood protocol); for the DNeasy kit, 5 μl of sample was used, again following the manufacturer's instructions (nucleated blood protocol).
DNA quantitation
The volume of the eluate (although technically flow-through in the case of the DNA-XT kit) was noted and the DNA concentration determined by fluorescence using a Qubit™ spectrophotometer (Thermo Fisher Scientific). The two kits have a fivefold difference in final eluate/flow-through volume, with DNeasy recommending eluting in 200 μl and DNA-XT having a flow-through of approximately 40 μl.
PCR
PCR targeting P. falciparum β tubulin (PfBetaT) was performed using Phire™ Hot Start II DNA Polymerase (Thermo Fisher Scientific). Two volumes of template (1 and 10 μl) were used in parallel reactions with primers (PfBetaT F, 5′-TTGGGGTCCTTCCCCTTTATTGTAT-3′; PfBetaT R, 5′-CAAAGGGGCCAGCACGAACACT-3′) at final concentrations of 200 nM. Cycling conditions were 95°C for 30 s, then 35 cycles of 95°C for 30 s, 55°C for 30 s and 68°C for 30 s, followed by 68°C for 10 min.
PCR targeting L. donovani kinetoplast DNA (high copy number) and DNA polymerase I (Pol I; low copy number) was performed using Phusion™ polymerase (Thermo Fisher Scientific). Volumes of 1 and 10 μl L. donovani eluate were used, respectively, with primers for the kinetoplast DNA (LdKinetoplast F2, 5′-CCAATGAAGCCAAGCCAGTG-3′; LdKinetoplast R2, 5′-GGCTGGTTTTAGATGTGGGC-3′) and DNA Pol I (LdDNApol I F, 5′-TGTCGCTTGCAGACCAGATG-3′; LdDNApol I R, 5′-GCATCGCAGGTGTGAGCAC-3′) [8] at final concentrations of 200 nM. Cycling conditions were 98°C for 30 s, then 10 cycles of 10 s at 98°C, 10 s at 69–59°C touchdown and 2 s at 72°C, followed by 20 cycles of 10 s at 98°C, 10 s at 59°C and 2 s at 72°C, which were followed by 72°C for 5 min. PCR products were electrophoresed on 1% (w/v) agarose gels and visualized with UV.
Statistical analysis
For the eluate DNA concentrations, a Student's t-test (unpaired, equal variance) was performed using Prism Mac 6.0h (GraphPad Software, CA, USA).
Results & discussion
Extraction
Process timings
The time to complete extractions from eight samples using each of the kits was recorded. For the DNA-XT kit, the extractions took between 50 and 60 min, compared with 120–130 min for the DNeasy kit.
Manipulations
The number of manipulations required to perform the extractions was also tallied. For an individual sample, the DNA-XT kit required 14 manipulations, whereas the DNeasy kit required 25. A manipulation was considered to be any change of tip or tube/wash column or any action (e.g., centrifugation or incubation step). In terms of plastics usage for a single sample, the DNA-XT kit used five tips and four tubes (including the column), and the DNeasy kit required eight tips and seven tubes.
Eluate characteristics
P. falciparum
All three column extraction arms (Q100, Q5 and DNA-XT) had eluates that contained DNA, as measured by fluorescence spectrometry (Table 1). Given that the theoretical amount of human DNA in each sample far exceeds that of any parasite DNA, samples spiked with different levels of iRBCs were analyzed both alone and together when comparing yields. Unsurprisingly, in the Q100 arm there was a greater yield of DNA than in the Q5 arm. This difference was 21-fold (mean ± SEM, 4420 ± 340 vs 205 ± 11 ng; n = 12), which is in line with the different volumes of blood processed in the Q100 and Q5 arms (100 and 5 μl, respectively). Assessing the Q5 and DNA-XT arms where the same amount of sample was passed through the respective column, similar concentrations of DNA were obtained (mean ± SEM, 0.63 ± 0.04 vs 1.06 ± 0.06 ng/μl, respectively; n = 12), with the Q5 arm performing significantly better (p = 0.013), albeit by 1.7-fold (only a single batch of each column type was tested here). Notably, the volume of eluate produced when using each manufacturer's protocol is different: ∼200 μl for Q100 and Q5 and ∼40 μl for DNA-XT. Thus, the total DNA yield from the Q5 column was higher than that of the DNA-XT column (7.8-fold).
Table 1. . Eluate characteristics for Plasmodium falciparum-spiked whole blood-derived samples.
| Column | Parasitemia | Eluate volume (mean ± SEM, μl) | [DNA] (mean ± SEM, ng/μl) | Total DNA yield (mean ± SEM, ng) | n |
|---|---|---|---|---|---|
| Q100 | Overall | 197 ± 1.4 | 22.4 ± 1.6 | 4420 ± 340 | 12 |
| 1% | 22.1 ± 1.5 | 4320 ± 290 | 3 | ||
| 0.1% | 21.4 ± 2.2 | 4260 ± 500 | 3 | ||
| 0.01% | 24.5 ± 0.5 | 4880 ± 170 | 3 | ||
| 0% | 21.5 ± 1.1 | 4210 ± 210 | 3 | ||
| Q5 | Overall | 194 ± 0.4 | 1.06 ± 0.06 | 205 ± 11 | 12 |
| 1% | 1.06 ± 0.02 | 204 ± 4.5 | 3 | ||
| 0.1% | 1.09 ± 0.12 | 211 ± 23 | 3 | ||
| 0.01% | 1.08 ± 0.03 | 209 ± 5.6 | 3 | ||
| 0% | 1.02 ± 0.05 | 197 ± 10 | 3 | ||
| DNA-XT | Overall | 42 ± 0.4 | 0.63 ± 0.04 | 26.3 ± 1.7 | 12 |
| 1% | 0.65 ± 0.03 | 28.4 ± 1.1 | 3 | ||
| 0.1% | 0.59 ± 0.05 | 24.8 ± 2.3 | 3 | ||
| 0.01% | 0.68 ± 0.05 | 28.9 ± 2.6 | 3 | ||
| 0% | 0.55 ± 0.07 | 22.8 ± 3.7 | 3 |
L. donovani
For the L. donovani extractions, only the Q5 and DNA-XT protocols were used (i.e., those that use 5 μl of sample). Both Q5 and DNA-XT had eluates that contained detectable, albeit low, levels of DNA, with the exception of one replicate of the Q5 arm (Table 2). Again, similar concentrations of DNA were obtained in the Q5 and DNA-XT arms (mean ± SEM, 0.06 ± 0.002 vs 0.17 ± 0.01 ng/μl, respectively; n = 2 or 3), with the DNA-XT arm performing slightly better by 2.8-fold. Due to the different eluate volumes, the total DNA yield from the Q5 column was slightly higher than that of the DNA-XT column (1.5-fold).
Table 2. . Eluate characteristics for purified Leishmania donovani amastigote samples.
| Column | Eluate volume (μl) | [DNA] (ng/μl) | Total DNA yield (ng) | |
|---|---|---|---|---|
| Q5 | Mean ± SEM | 193 ± 1.2 | 0.06 ± 0.002 | 11 ± 0.4 |
| A | 191 | Too low | nd | |
| B | 195 | 0.06 | 11 | |
| C | 193 | 0.05 | 10 | |
| DNA-XT | Mean ± SEM | 42 ± 0.3 | 0.17 ± 0.01 | 7.3 ± 0.2 |
| A | 42 | 0.16 | 6.6 | |
| B | 42 | 0.11 | 7.2 | |
| C | 43 | 0.19 | 8.1 |
nd: Not determined.
Parasite DNA quality
P. falciparum
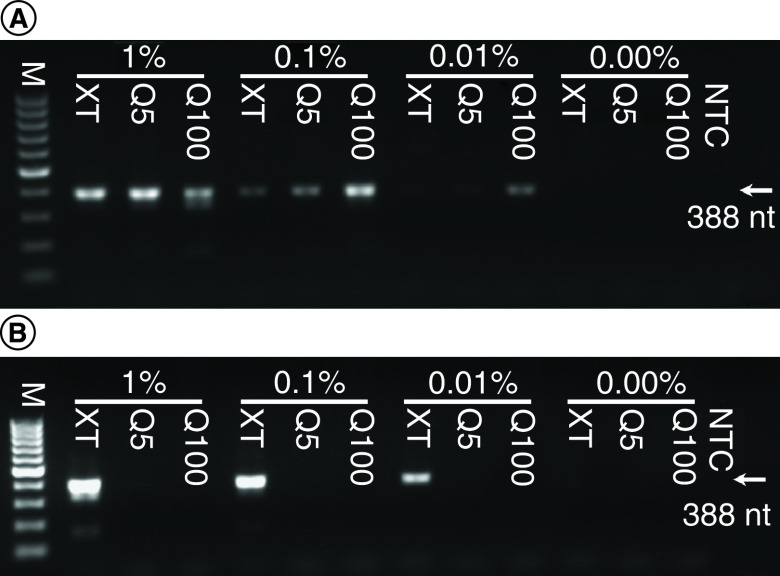
While the DNA extracted in these experiments reflects human DNA predominantly, parasite DNA was collected from spiked samples. Its presence and quality in eluates were determined by PCR detection of PfBetaT. Two different volumes of eluate were used, a low volume (1 μl) and a high volume (10 μl). In most cases, amplicon bands of the appropriate size (388 bp) could be detected in samples in which parasite DNA should be present (Figure 2). When using the low volume of eluate, the strongest bands were produced from the Q100 eluates. This was to be expected, as the Q100 eluates contain the highest total DNA concentrations (Table 1). Bands were also visible, though fainter, in lanes using Q5 and DNA-XT samples, even at the lowest (0.01%) parasitemia used. However, when a greater (tenfold) volume of eluate was used, only the DNA-XT-derived samples resulted in positive amplicon bands in a parasitemia-dependent manner. This suggests an apparent inhibition of PCR when using high volumes of the Q5 and Q100 samples. The findings presented in Figure 2 are supported by data from three independent repeats of the experiment (with duplicate technical repeats within each). The number of positive bands in each (see Supplementary file 2 for PCR gel images not present in the main text) are presented in Table 3.
Figure 2. . Plasmodium falciparum DNA from spiked whole blood determined by PfBeta tubulin PCR.
(A) Low-volume eluate template, 1 μl. (B) High-volume eluate template, 10 μl. Percentage denotes the parasitemia after spiking.
M: 100-bp marker; NTC: No template control.
Table 3. . PCR positive outcomes for Plasmodium falciparum-spiked whole blood-derived samples.
| Human whole blood | 1% parasitemia (pos/total) | 0.1% parasitemia (pos/total) | 0.01% parasitemia (pos/total) | 0% unspiked (pos/total) |
|---|---|---|---|---|
| High-volume template Q5 | 2/6 | 2/6 | 2/6 | 0/6 |
| High-volume template Q100 | 0/6 | 1/6 | 0/6 | 0/6 |
| High-volume template DNA-XT | 6/6 | 6/6 | 6/6 | 0/6 |
| Low-volume template Q100 | 6/6 | 6/6 | 5/6 | 0/6 |
| Low-volume template Q5 | 5/6 | 6/6 | 6/6 | 0/6 |
| Low-volume template DNA-XT | 6/6 | 6/6 | 5/6 | 0/6 |
pos: Gel band positive.
L. donovani
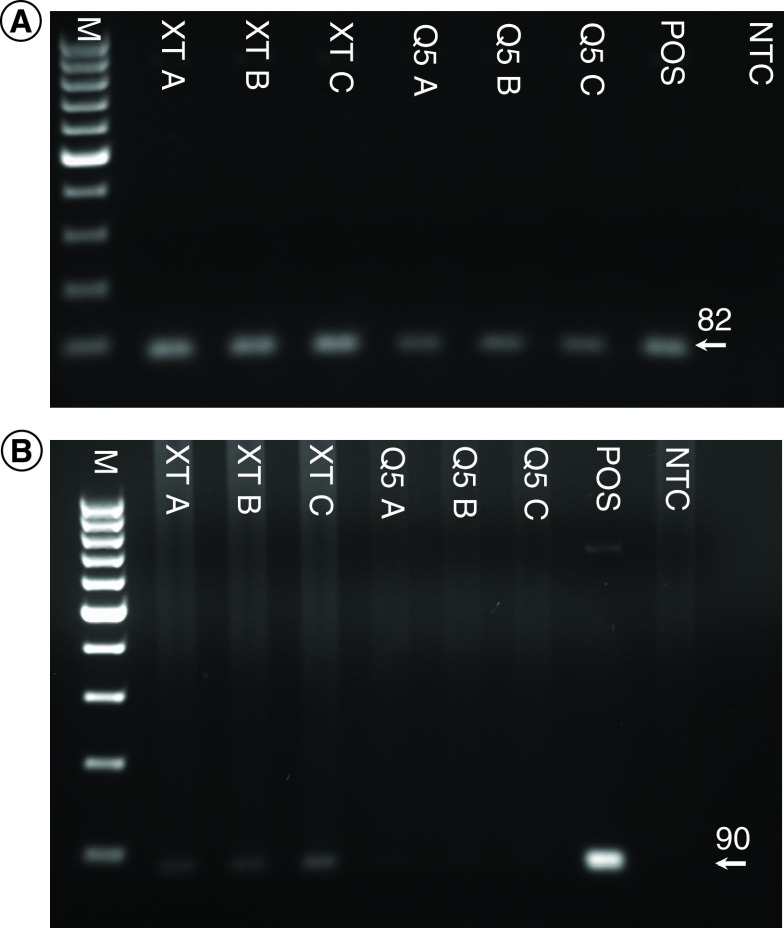
DNA was successfully amplified from low-volume (1 μl) eluates of purified L. donovani amastigotes derived from Q5 and DNA-XT extraction technologies when targeting the kinetoplast, a high copy number DNA sequence (Figure 3A). Targeting a single copy number gene, Leishmania spp. DNA Pol I, a PCR amplicon, was obtained with a larger volume (10 μl) of DNA-XT-derived eluate, but not with Q5-derived eluate (Figure 3B).
Figure 3. . Leishmania donovani DNA extracted from purified amastigotes and determined by PCR targeting the kinetoplast sequence.
(A) 1 μl of eluate from each manufacturer's column was used as a template. (B) 10 μl of eluate from each manufacturer's column was used as a template. Eluates from three separate columns for each manufacturer were used.
M: 100-bp marker; NTC: No template control; POS: Positive control.
Here, a simple process for the extraction of DNA from small, parasite-containing culture samples was assessed against a standard commercial, spin column-based extraction process that is widely used in research and diagnostic laboratories. A considerable difference between the two techniques is that DNA is retained on the Qiagen DNeasy mini spin column prior to elution, but the QuantuMDx DNA-XT column retains contaminants and only the DNA is eluted in one centrifugation step. The advantages of this process, as demonstrated in this study, are time savings, reduced plastics use, reduced reagent use (including removing the necessity to use guanidinium chloride) and a reduction in PCR-inhibiting contaminants. However, total yields of DNA are much lower, generally, when using the DNA-XT protocol.
The DNA-XT methodology is similar to a previously described DNA isolation method that uses a polyaniline-containing silica sorbent [9]. In terms of ease of use, the DNA-XT methodology is also comparable with DNA extraction protocols that use Chelex® 100 (Bio-Rad Laboratories, CA, USA). Both of these alternative DNA extraction techniques have been used for forensics applications and perform well with samples that have low DNA concentrations [10–12]. In this study, DNA-XT was used to extract DNA from whole blood spiked with P. falciparum-infected RBCs, simulating a clinical diagnostic sample, and purified L. donovani amastigotes, as might be used in a research laboratory. In both cases, it was possible to amplify parasite DNA, although the majority of DNA in whole-blood extracts would be expected to be human DNA. Furthermore, with comparable input volumes, the DNA-XT technology compared favorably with the Qiagen extraction kit. Thus, DNA-XT has potential uses within laboratories where DNA extraction is required from small sample volumes.
The technology behind DNA-XT was designed for microfluidic-based, point-of-care, molecular diagnostics platforms, where cell lysis and DNA extraction occur within a cassette to provide DNA for microfluidic PCR and subsequent amplicon sensing. In the case of malaria diagnosis at point-of-care (and other diseases caused by blood-borne pathogens), capillary sampling from finger or heel pricks are the preferred options to generate test samples for parasite detection. Capillary sampling can be safely performed by staff with basic training and is often quicker and less stressful when compared with venepuncture [13]. Capillary sampling has also been used extensively for epidemiological studies of malaria with the use of blood spot collection on filter paper [14–16].
Future perspective
Because DNA-XT has been demonstrated here to produce high-quality parasite gDNA (seemingly free of the types of PCR inhibitors that frequently contaminate whole-blood extracts [17–19]), this technology may also be useful for extracting blood from dried blood spots, as has been demonstrated for other DNA extraction methodologies [20]. Further applications for high-quality DNA extraction include genotyping for therapeutic diagnostics (e.g., parasite drug resistance/susceptibility) and epidemiology for monitoring asymptomatic individuals [21,22] and those with submicroscopic parasitemia within a population [23]. These could be particularly useful in potential eradication programs [24–26]. Future work for diagnostic applications for the DNA-XT technology should include patient samples and a larger sample size to field-test this new technology. In addition, a more accurate method of DNA quantification should be used in these proof-of-concept studies, namely quantitative PCR.
Supplementary Material
Footnotes
Author contributions
Experimentation was designed by D Clark, C Moore, S Croft, J Tyson, S Whitehouse, J O’Halloran, S Krishna and H Staines and was undertaken by D Clark, C Moore, M Flanagan, K Van Bocxlaer, E-T Piperaki and V Yardley. The manuscript was prepared by D Clark, C Moore, V Yardley, S Krishna and H Staines. All authors had the opportunity to read and approve the manuscript.
Financial & competing interests disclosure
We gratefully acknowledge support for this work from the European Community's Seventh Framework Programme (FP7/2007-2013); NanoMal under grant agreement number 304948 to S Krishna, H Staines, S Whitehouse and J O’Halloran; and MRC Confidence in Concept award number MC_PC_14111 (2014–2015) to S Croft, H Staines and S Krishna. H Staines is supported by the Wellcome Trust Institutional Strategic Support Fund (204809/Z/16/Z) awarded to St. George's University of London. The funders had no role in the design of the study, data collection, analysis, or preparation of the manuscript. M Flanagan, J Tyson, S Whitehouse and J O’Halloran work for QuantuMDx, the company that developed DNA-XT. S Krishna is a paid advisor and chairs the infectious diseases advisory board for QuantuMDx. S Krishna and H Staines are both shareholders in QuantuMDx. S Krishna and H Staines are in receipt of funds from QuantuMDx to develop diagnostic technologies and assays (that have supported D Clark and C Moore). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
All experiments involving animals were conducted at the London School of Hygiene & Tropical Medicine under license (PPL 70/8427) according to UK Home Office Regulations, Animals (Scientific Procedures) Act 1986 and European Directive 2010/63/EU.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
- 1.Dahm R. Friedrich Miescher and the discovery of DNA. Dev. Biol. 278(2), 274–288 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Meselson M, Stahl FW. The replication of DNA in Escherichia coli. Proc. Natl Acad. Sci. USA 44(7), 671–682 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths L, Chacon-Cortes D. Methods for extracting genomic DNA from whole blood samples: current perspectives. J. Biorepository Sci. Appl. Med. 2, 1 (2014). [Google Scholar]
- 4.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc. Natl Acad. Sci. USA 76(2), 615–619 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65(3), 418 (1979). [PubMed] [Google Scholar]
- 6.Croft SL, Yardley V. Animal models of visceral leishmaniasis. : Handbook of Animal Models of Infection. Zak O, Sande MA (). Academic Press, CA, USA, 783–787 (1999). [Google Scholar]
- 7.Hanscheid T. Diagnosis of malaria: a review of alternatives to conventional microscopy. Clin. Lab. Haematol. 21(4), 235–245 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Bretagne S, Durand R, Olivi M. et al. Real-time PCR as a new tool for quantifying Leishmania infantum in liver in infected mice. Clin. Diagn. Lab. Immunol. 8(4), 828–831 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapustin DV, Yagudaeva EY, Zavada LL. et al. A composite polyaniline-containing silica sorbent for DNA isolation. Russ. J. Bioorganic Chem. 29(3), 281–285 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Kopka J, Leder M, Jaureguiberry SM, Brem G, Boselli GO. New optimized DNA extraction protocol for fingerprints deposited on a special self-adhesive security seal and other latent samples used for human identification. J. Forensic Sci. 56(5), 1235–1240 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Matai AS, Sijen T. Improving a rapid DNA profiling service regularly applied in forensic casework. Forensic Sci. Int. Genet. Suppl. Ser. 4(1), e284–e285 (2013). [Google Scholar]
- 12.Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 54(3), 506–513 (2013). [PubMed] [Google Scholar]
- 13.WHO. WHO Guidelines on Drawing Blood: Best Practices in Phlebotomy. World Health Organization, Geneva, Switzerland: (2017). [PubMed] [Google Scholar]
- 14.Bereczky S, Mårtensson A, Gil JP, Färnert A. Short report: rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 72(3), 249–251 (2005). [PubMed] [Google Scholar]
- 15.Corran PH, Cook J, Lynch C. et al. Dried blood spots as a source of anti-malarial antibodies for epidemiological studies. Malar. J. 7(1), 195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiang MS, Lin M, Dokomajilar C. et al. PCR-based pooling of dried blood spots for detection of malaria parasites: optimization and application to a cohort of Ugandan children. J. Clin. Microbiol. 48(10), 3539–3543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 39(2), 362–372 (1994). [PubMed] [Google Scholar]
- 18.Al-Soud WA, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39(2), 485–493 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Soud WA, Jönsson LJ, Râdström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38(1), 345–350 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strøm GEA, Tellevik MG, Hanevik K, Langeland N, Blomberg B. Comparison of four methods for extracting DNA from dried blood on filter paper for PCR targeting the mitochondrial Plasmodium genome. Trans. R. Soc. Trop. Med. Hyg. 108(8), 488–494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.le Fichoux Y, Quaranta JF, Aufeuvre JP. et al. Occurrence of Leishmania infantum parasitemia in asymptomatic blood donors living in an area of endemicity in southern France. J. Clin. Microbiol. 37(6), 1953–1957 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riera C, Fisa R, Udina M, Gállego M, Portus M. Detection of Leishmania infantum cryptic infection in asymptomatic blood donors living in an endemic area (Eivissa, Balearic Islands, Spain) by different diagnostic methods. Trans. R. Soc. Trop. Med. Hyg. 98(2), 102–110 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Lin JT, Saunders DL, Meshnick SR. The role of submicroscopic parasitemia in malaria transmission: what is the evidence? Trends Parasitol. 30(4), 183–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silal SP, Little F, Barnes KI, White LJ. Hitting a moving target: a model for malaria elimination in the presence of population movement. PLoS ONE 10(12), e0144990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White LJ, Maude RJ, Pongtavornpinyo W. et al. The role of simple mathematical models in malaria elimination strategy design. Malar. J. 8(1), 212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tietje K, Hawkins K, Clerk C. et al. The essential role of infection-detection technologies for malaria elimination and eradication. Trends Parasitol. 30(5), 259–266 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.