Abstract
TLR8 is among the highest-expressed pattern-recognition receptors in the human myeloid compartment, yet its mode of action is poorly understood. TLR8 engages two distinct ligand binding sites to sense RNA degradation products, although it remains unclear how these ligands are formed in cellulo in the context of complex RNA molecule sensing. Here, we identified the lysosomal endoribonuclease RNase T2 as a non-redundant upstream component of TLR8-dependent RNA recognition. RNase T2 activity is required for rendering complex single-stranded, exogenous RNA molecules detectable for TLR8. This is due to RNase T2’s preferential cleavage of single-stranded RNA molecules between purine and uridine residues, which critically contributes to the supply of catabolic uridine and the generation of purine-2ʹ,3ʹ-cyclophosphate-terminated oligoribonucleotides. Thus-generated molecules constitute agonistic ligands for the first and second binding pocket of TLR8. Together, these results establish the identity and origin of the RNA-derived molecular pattern sensed by TLR8.
Introduction
Pattern-recognition receptors (PRRs) of the innate branch of our immune system have evolved to sense the presence of microbe-associated molecular patterns (MAMPs) as non-self. A heterogeneous group of PRRs can detect microbial nucleic acids in different subcellular compartments (Barbalat et al., 2011, Wu and Chen, 2014). Microbe-derived nucleic acids do not always fulfill the criteria of a true MAMP given that they do not necessarily differ in their biochemical structure from host-derived nucleic acids. Here, to ensure discrimination of self versus non-self, additional principles apply (Roers et al., 2016). These principles include the following: the positioning of these PRRs in compartments that are devoid of potential self-ligands (e.g., the endolysosome), the regulation of the abundance of endogenous nucleic acids (e.g., by nucleases), and the modulation of nucleic acid sensors’ thresholds by additional licensing signals (e.g., type I interferons). Among the toll-like-receptor (TLR) family, four TLRs detect nucleic acids in the human system: TLR3 senses long double-stranded RNA (dsRNA), and TLR9 detects CpG-motif-containing DNA molecules, whereas TLR7 and TLR8 sense RNA degradation products. Mice express TLR13 as an additional nucleic-acid-sensing TLR. Interestingly, this TLR seems to respond to single-stranded RNAs (ssRNAs) of a rather specific sequence and conformation, which renders it unique among the other nucleic-acid-sensing TLRs that seem to harbor little sequence specificity (Song et al., 2015).
The role of TLR7 has been extensively studied in the murine system. Here, it has been shown that TLR7 plays a pivotal role in virus recognition and sterile inflammation (Barbalat et al., 2011). Human and murine TLR7 are well expressed in plasmacytoid dendritic cells (pDCs) and B cells, as well as in certain cells of the myeloid lineage. Human TLR8, on the other hand, is not expressed in pDCs or B cells but is highly abundant in cells of the myeloid lineage, including neutrophils. Although the expression profile of murine TLR8 is similar to that of human TLR8, it differs in functionality. TLR7-deficient mouse macrophages display a complete loss of responsiveness toward ssRNA molecules or synthetic agonists that activate human TLR7 or TLR8 (Diebold et al., 2004, Heil et al., 2004). Although studies have reported on the functionality of murine TLR8 under certain conditions, it appears that murine TLR13 acts as a functional homolog of human TLR8 (Krüger et al., 2015, Oldenburg et al., 2012). As such, it has been shown that bacteria and bacterial RNA of various sources are potent activators of hTLR8 and mTLR13, respectively. However, despite these functional commonalities, the modes of recognition between these two TLRs are vastly different (Song et al., 2015, Tanji et al., 2015). With regard to their ligand-sensing capacities, both human TLR7 and TLR8 share a similar mode of action. Their horseshoe-shaped leucine-rich-repeat (LRR) domains form side-to-side homodimers in a rotational symmetry. In this configuration, two distinct sets of ligand-binding locations are available (Tanji et al., 2015, Zhang et al., 2016, Zhang et al., 2018). Two ligand binding pockets, one provided by each protomer, are situated at the apex of the dimerization interface (first binding pocket). For TLR8, this site has been shown to bind uridine molecules, as well as synthetic TLR8 agonists such as TL8-506. Two additional binding pockets, again one from each protomer, are positioned at the concave surface of the LRRs (second binding pocket). This pocket has been shown to bind short oligoribonucleotides, whereas its engagement strongly increases the affinity of the first binding pocket toward uridine molecules. As such, optimal TLR8 agonism is achieved when the first and second binding pockets are occupied. Of note, synthetic agonists and high concentrations of uridine can exert TLR8 agonism in the absence of the engagement of second binding pocket (Tanji et al., 2015). However, to exert agonistic activity, complex RNA ligands require a functional second binding pocket. Despite our detailed structural understanding of the TLR8 ligand-binding domains, it has yet remained elusive how TLR8-agonistic ligands are being formed in the context of sensing complex RNA ligands. In this study, we set out to explore the mechanism by which complex RNA molecules and live pathogens are rendered “visible” to TLR8.
Results
TLR7 and TLR8 Are Functional TLRs in BLaER1 Monocytes
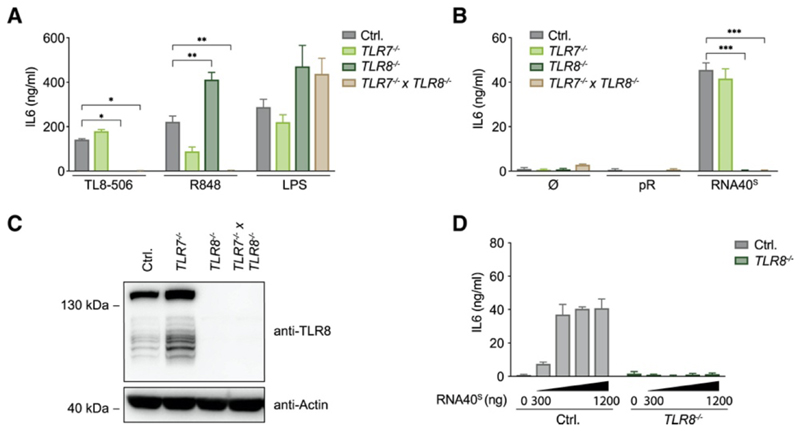
To genetically dissect TLR8 signaling, we turned to the BLaER1 system that we have previously employed to model human monocytes (Gaidt et al., 2017, Gaidt et al., 2016). In these cells, TLR8 expression is strongly upregulated upon transdifferentiation (Figure S1A). We used the well-established RNA40 molecule, which is a 20-mer ssRNA oligonucleotide (ON) derived from the HIV-1 genome (Heil et al., 2004), as a prototypic ligand. We used a variant of RNA40 stabilized by phosphorothioate modifications, designated as RNA40S. This modification renders the inter-nucleotide linkage more nuclease resistant and hence increases the half-life of this ON in biological systems. To deliver RNA into endolysosomal compartments, we used the polycationic polypeptide poly-L-arginine (pR) (Ablasser et al., 2009). As a proxy of pro-inflammatory gene expression, we measured IL-6 production. After stimulation with RNA40S or with small-molecule ligands specific to TLR8 (TL8-506) or to both TLR7 and TLR8 (R848), BLaER1 monocytes produced IL-6 (Figure 1A). As expected, TLR8 −/− BLaER1 monocytes were unresponsive to the TLR8-specific ligand TL8-506, whereas R848-mediated activation was only blunted when both TLR7 and TLR8 were ablated. On the other hand, RNA40S-mediated IL-6 production was completely TLR8 dependent (Figures 1B and 1C). TLR4-dependent stimulation (LPS) was active in all genotypes tested. TLR8 activation was observed when increasing doses of RNA were used and was also seen when RNA40S was transfected with Lipofectamine 2000, albeit at lower activity (Figure 1D and Figure S1B). Studying the phosphodiester version of RNA40 (RNA40O) also showed TLR8-dependent stimulation but with lower potency (Figure S1C). Similar results were obtained when an unrelated phosphodiester ssRNA ON (P20-5M) was studied (Figure S1D). Control BLaER1 monocytes produced IL-6 in response to this ON, whereas TLR8 −/− cells showed no response. As previously noted, a self-complementary version of this ON (P20) did not exert any TLR8-agonistic activity (Ablasser et al., 2009), corroborating the notion that a certain degree of single-strand conformation was required for stimulating this TLR. Altogether, these results establish that BLaER1 monocytes serve as a physiological model for the study of RNA-dependent activation of TLR8.
Figure 1. TLR7 and TLR8 Are Functional TLRs in BLaER1 Monocytes.
(A and B) BLaER1 monocytes of indicated genotypes were stimulated with (A) TL8-506, R848, and LPS or were (B) unstimulated or stimulated with pR and RNA40S. After 14 h, IL-6 release was measured. (C) TLR8 expression in BLaER1 cells of indicated genotypes. (D) Control and TLR8-deficient BLaER1 monocytes were stimulated with increasing amounts of RNA40S. After 14 h, IL-6 was measured. Data are depicted as mean + SEM of three independent experiments (A, B, and D) or one of three representative blots (C). Statistics indicate significance by two-way ANOVA: ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant. See also Figure S1.
RNase-T2-Deficient Cells Fail to Respond to RNA Oligonucleotides
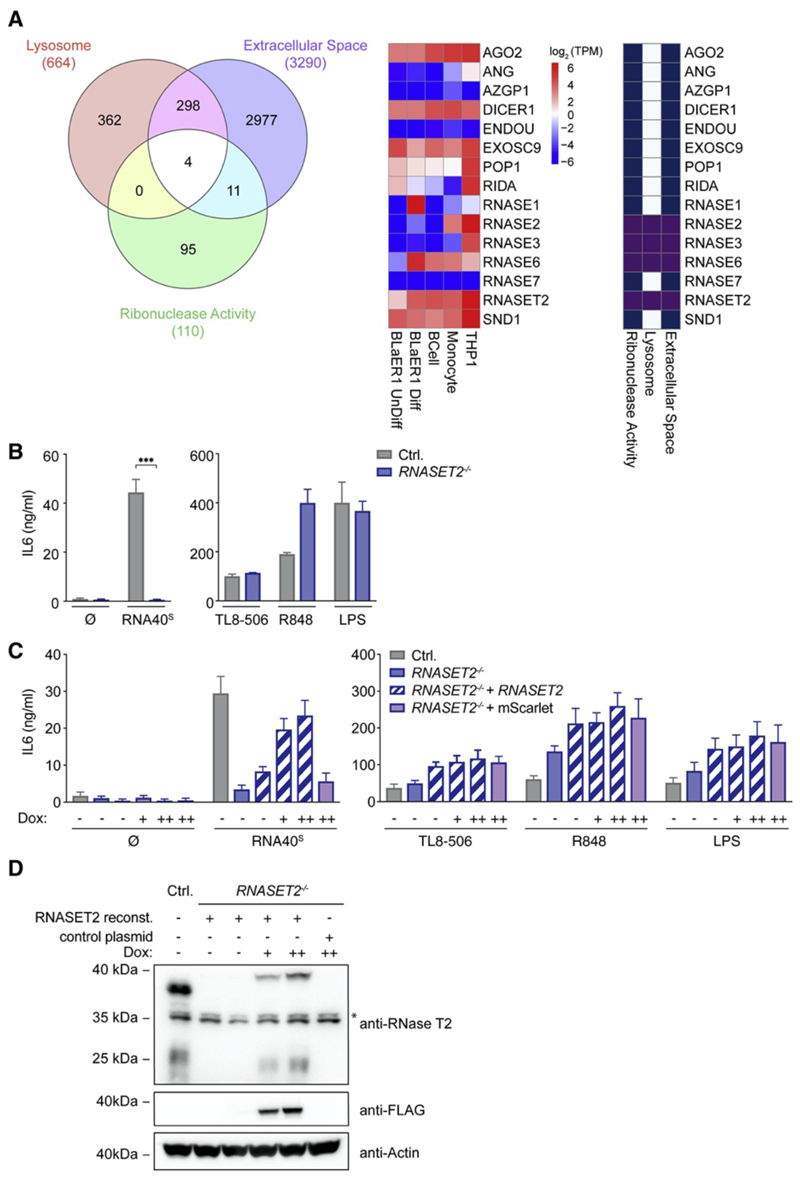
We hypothesized that luminal RNases must function upstream of TLR8 in that they degrade RNA40 into ligands that engage its first and/or second binding pocket. We thus considered annotated enzymes with ribonuclease activity to be located either in the lysosome or in the extracellular space. Prioritizing lysosomal RNases that are well expressed in both primary human monocytes and transdifferentiated BLaER1 monocytes, we first focused on RNASE2, RNASE6, and RNASET2. We additionally included RNASE1 because of its high expression in BLaER1 cells (Figure 2A). Studying knockout cell lines of these RNases indicated that RNASET2 −/− cells, but not RNASE1 −/−, RNASE2 −/−, or RNASE6 −/− cells, displayed a complete loss-of-function phenotype upon RNA40S stimulation (Figure 2B and Figures S2A–S2C). Of note, RNase T2 deficiency had no impact upon TLR4-mediated IL-6 production, and stimulation with synthetic TLR8 agonists was unaffected in the absence of this RNase. This indicates that RNase T2 deficiency has no impact on the functionality of this receptor per se. Analogous results were obtained when we studied human monocytic THP-1 cells deficient for RNase T2 or TLR8 (Figure S2D). Reconstitution of RNase T2 by lentiviral transduction rescued RNASET2 −/− cells in their response toward RNA40S in a dose-dependent fashion (Figures 2C and 2D). Finally, phosphodiester-linked ONs were also completely dependent on RNase T2 in their TLR8-stimulatory activity (Figure S2E). In summary, these results suggest that RNase T2 is non-redundantly required for rendering RNA ONs TLR8 agonistic.
Figure 2. RNase-T2-Deficient Cells Fail to Respond to RNA Oligonucleotides.
(A) Venn diagram of proteins that are, according to their Gene Ontology (GO) terms, annotated as “lysosomal,” “extracellular space,” or “ribonuclease activity” (left). A heatmap shows RNA expression levels of several RNases and their GO-term designation in the indicated cell types (middle and right). (B) IL-6 production in BLaER1 Controls and RNASET2 −/− cells stimulated with RNA40S, TL8-506, R848, and LPS for 14 h. (C) RNASET2 −/− cells were reconstituted with doxycycline-inducible RNase T2 (++, 1 μg/mL; +, 0.5 μg/mL). An inducible mScarlet construct was used as a control. (D) The immunoblot corresponding to the reconstitution experiment is shown. The asterisk indicates unspecific bands. Data are depicted as mean + SEM of three independent experiments or one of three representative blots. Statistics indicate significance by two-way ANOVA: ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant. See also Figure S2.
RNase T2 Cleaves RNA between Purine Bases and Uridine
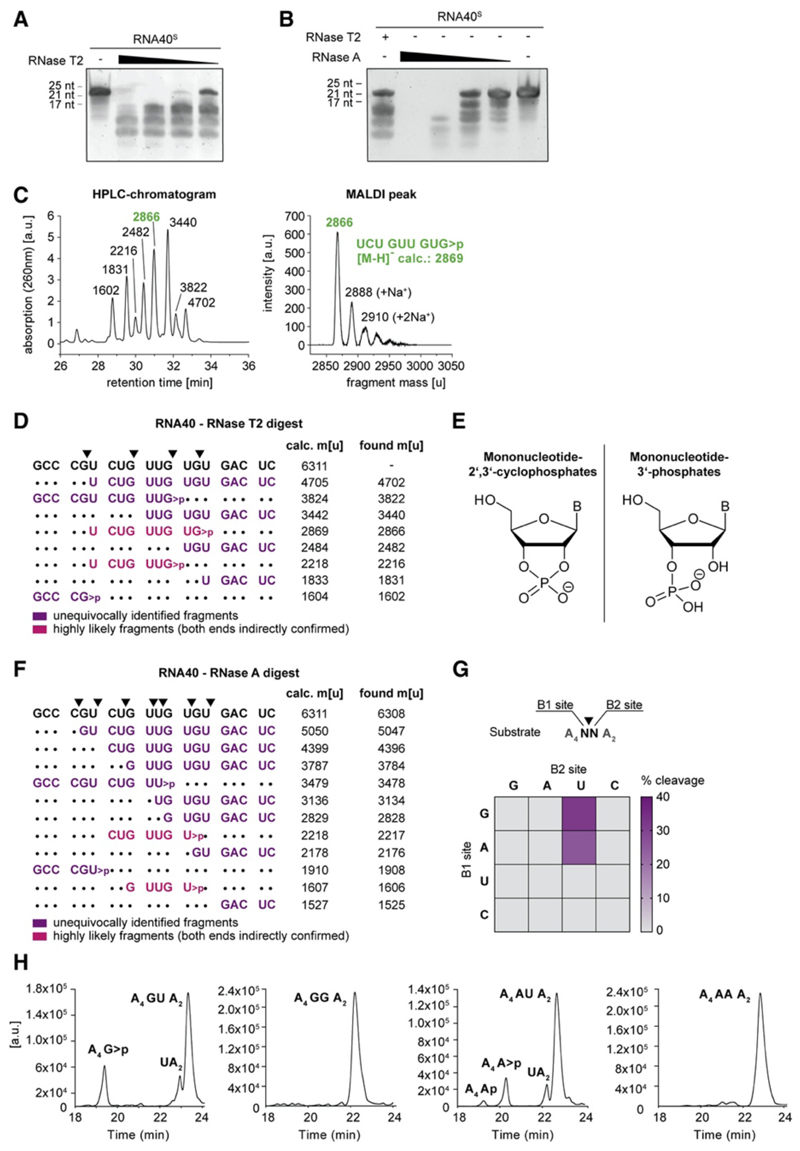
We considered it most likely that RNase T2 degrades RNA into fragments that are rendered agonistic for either the first and/or second binding pocket of TLR8. To characterize the putative degradation pattern of RNase-T2-digested RNA40, we incubated RNA40S by using recombinant RNase T2 at limiting enzyme concentrations. This provided a characteristic cleavage pattern that was different from the one obtained with bovine pancreatic RNase A, which is orthologous to human RNase 1 (Figures 3A and 3B). To further analyze the cleavage pattern of RNase-T2-digested RNA40, we analyzed individual fragments identified by liquid chromatography (LC) by using MALDI-TOF (Figure 3C). Here, we first studied the digestion of RNA40O because RNA40S-derived fragments generated less-defined peaks on LC as a result of the diastereomeric configuration of their phosphorothioate bonds. These analyses revealed at least nine distinct peaks, of which eight peaks could be assigned to distinct masses compatible with endoribonuclease cleavage products of RNA40O. Interestingly, all of the identified cleavage products were consistent with endoribonuclease activity between a guanosine and uridine residue (Figure 3D). Moreover, the majority of the identified fragments displayed a mass that was compatible with a 3ʹ configuration of a 2ʹ,3ʹ-cyclophosphate rather than a 3ʹ phosphate (Figure 3E). Conducting analogous experiments with RNA40S provided comparable results. As such, a similar cleavage pattern was observed with fragments consistent with endoribonuclease activity between guanosine and uridine (GU). Moreover, the mass of the individual fragments indicated the presence of a 2ʹ,3ʹ-cyclophosphate configuration (Figures S3A and S3B). In total, analyzing RNase-T2-digested RNA40 molecules yielded 11 out of 14 possible fragments that were consistent with cleavage between GU (Figure S3C). In line with the well-established notion that RNase A cleaves after pyrimidines, RNA40O digested with RNase A generated fragments that were terminated by a 3ʹ uridine or cytidine (Figure 3F). As observed for RNase T2, the masses of the fragments were consistent with a 2ʹ,3ʹ-cyclophosphate configuration. Because RNA40 provided limited sequence space for systematic exploration of the substrate specificity of RNase T2, we next conducted RNase T2 cleavage assays by using a set of ONs that contained all 16 possible dinucleotide substrates (Figures 3G and 3H). These experiments confirmed that RNase T2 cleaved between GU and furthermore indicated that also AU served as a substrate with comparable efficiency. Of note, treating the self-complementary oligoribonucleotide P20 with RNase T2 did not yield any discernable fragments, whereas its ssRNA counterpart was readily digested (Figure S3D). These results indicate that base-pairing RNA molecules did not serve as substrates for RNase T2, providing a rationale as to why such oligoribonucleotides exerted no TLR8 activity (Figure S1D) (Ablasser et al., 2009). In summary, these results indicate that, under the conditions tested, RNase T2 preferentially cleaves ssRNA between purine and uridine residues and leaves a guanosine or adenosine 2ʹ,3ʹ-cyclophosphate configuration.
Figure 3. RNase T2 Cleaves RNA between Purine Bases and Uridine.
(A and B) Urea gel of RNA40S digested with decreasing RNase T2 (A) or RNase A (B) concentrations. One representative gel of two independent experiments is shown. (C) HPLC chromatogram of RNA40O-derived ON fragments (left; fragment masses determined by MALDI-TOF) and the corresponding MALDI peak of one representative peak (right) (D) RNA40O was digested with RNase T2 and analyzed by HPLC and MALDI-TOF. The most likely assigned fragments based on MALDI-TOF analysis are depicted next to all calculated and found masses. Calculated masses are shown as [M-H]−. (E) Structure of mononucleotide 2ʹ,3ʹ-cyclophosphates (left) and Mononucleotide 3ʹ-phosphates (right) (F) RNA40O was digested in vitro with RNase A and analyzed as above. Masses are shown as [M-H]−. (G) 16 ONs containing all possible dinucleotide combinations (A4NNA2) were analyzed after RNase T2 in vitro digestion. Of note, all ONs with a U at the B1 site were also cut between A and U, whereas cleavage between U and N was not detected. (H) HPLC chromatograms corresponding to the in vitro digests of GU, GG, AU, and AA (G). See also Figure S3.
Altered RNA Catabolism In Cellulo in the Absence of RNase T2
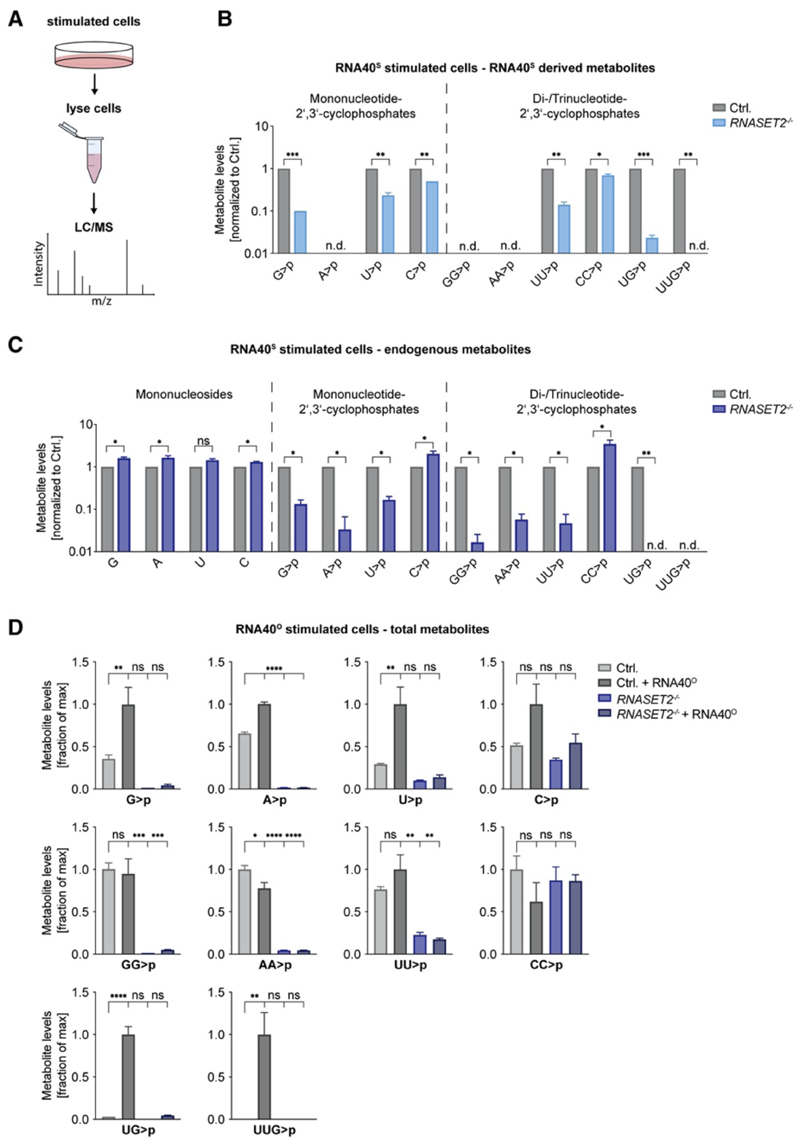
To address how RNase T2 affects the catabolism of RNA substrates in cellulo, we undertook a mass spectrometry approach to determine the abundance of selected metabolites in control and RNASET2 −/− cells (Figure 4A). On the basis of synthetic standards, in vitro digests of RNA40, or unequivocally ascribable masses, we could employ this technology to determine selected metabolites ranging from nucleosides up to certain trinucleotides. We first aimed at identifying RNA40S-derived degradation products by taking advantage of the fact that RNA40S-derived nucleotides are distinguishable by mass from endogenous molecules because the sulfur atom is present as part of the phosphodiester linkage. Analyzing mono- and dinucleotides with a 2ʹ,3ʹ-cyclophosphate moiety showed that the abundance of RNA40S-derived C>p and CC>p was only scarcely affected by RNase T2 deficiency, whereas the levels of G>p were reduced by more than 90% (Figure 4B). Of note, U>p and UU>p levels were also greatly reduced in the absence of RNase T2. RNA40S-derived A>p was undetectable in control and RNase-T2-deficient samples. Moreover, GG>p and AA>p, which are not present in RNA40, were not found in any sample. Intriguingly, peaks assignable to RNA40S-derived UG>p or UUG>p were readily identified in control cells but were only scarcely detectable or undetectable in the lysates of RNase-T2-deficient cells. Analyzing the endogenous pool of nucleotides (non-sulfur-containing nucleotides) in these RNA40-stimulated samples (Figure 4C) or unstimulated samples (Figure S4A) largely mirrored the results obtained with the RNA40S-derived metabolites. Endogenous C>p and CC>p levels were not affected by RNase T2 deficiency, whereas G>p, U>p, and UU>p levels strongly decreased. In addition, endogenous A>p, AA>p, and GG>p levels could now be detected, and these metabolites were also strongly affected by the absence of RNase T2. Of note, mononucleoside levels were not decreased in RNASET2 −/− cells (Figure 4C and Figure S4A). Although these results could provide unequivocal proof of the source of the measured metabolites and also inform on the relative abundance of these metabolites dependent on RNase T2, these experiments cannot provide insight into the relative increase in putative RNA40-derived metabolites upon RNA40 stimulation. To address this question, we conducted stimulation experiments with RNA40O, which gives rise to degradation products that are of the same mass as the endogenous metabolites. These experiments revealed that RNA40O stimulation led to a moderate increase in G>p, A>p, U>p, and C>p levels (2.8-, 1.5-, 3.4-, and 1.9-fold, respectively) and that G>p and A>p levels, and to a lesser extent U>p levels, were RNase T2 dependent (Figure 4D). GG>p, AA>p, UU>p, and CC>p levels were not increased upon RNA40 stimulation, and similarly to the mononucleotides, AA>p, GG>p, and to a lesser extent UU>p levels were RNase T2 dependent. C>p and CC>p levels were unaffected by RNase T2 deficiency. Intriguingly, UG>p levels were induced by 33-fold upon RNA40 stimulation, and UUG>p could only be detected in RNA40-stimulated cells. Both these metabolites were completely RNase T2 dependent. Of note, RNASE2- and RNASE6-deficient cells showed no decrease in RNA40-derived metabolites. A reduction of C>p and CC>p levels was observed only for RNASE1 −/− cells (Figures S4B–S4D). In conclusion, these results indicate that RNase T2 is non-redundantly necessary for generating certain ribonucleotides with a 2ʹ,3ʹ-cyclophosphate moiety of both endogenous and exogenous origins. Most critically, UG>p and UUG>p were completely RNase-T2-dependent metabolites and almost exclusively seen in cells stimulated with exogenous RNA.
Figure 4. RNase T2 Deficiency Leads to Altered RNA Metabolism.
(A) BLaER1 cells were stimulated with RNA40S for 14 h, and then lysates were analyzed by liquid chromatography-mass spectrometry (LC-MS). (B and C) Cell lysates of RNA40S stimulated BLaER1 cells with indicated genotypes were analyzed by LC-MS. RNA40S-derived (B) or endogenous (C) metabolites are shown. Data are normalized to control cells (note logarithmic scale). (D) Cell lysates of RNA40O-stimulated BLaER1 cells with indicated genotypes were analyzed by LC-MS. For each metabolite dataset, the mean of the maximal values was determined, and all data are depicted as a fraction thereof. Data are depicted as mean + SEM of three independent experiments. Statistics indicate significance by a Welch’s unequal variances t test (B and C) or by one-way ANOVA (D). ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant; n.d., not detected. See also Figure S4.
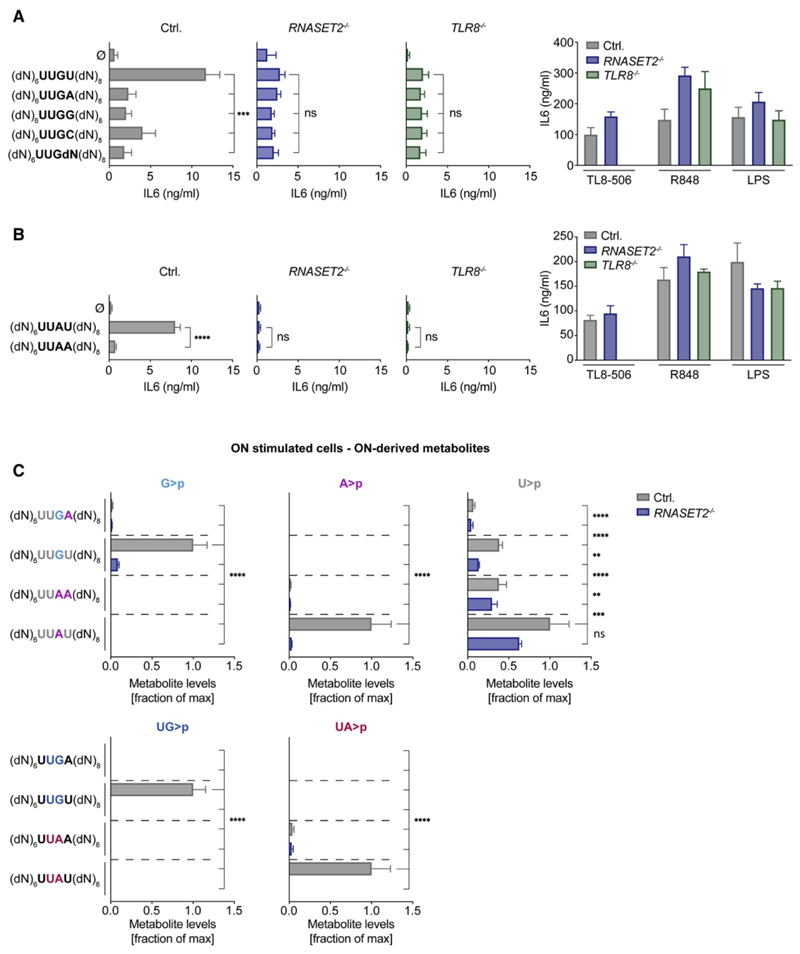
A Minimal Motif for TLR8
The above studies implied that the presence of a GU dinucleotide, which serves as a recognition site for RNase T2, was important for the TLR8 agonistic activity of RNA40. Given that RNA40 contains four such GU dinucleotides, we turned to a more reductionist setting to address this hypothesis. Indeed, previous work showed that a UUGU motif within an otherwise non-stimulatory deoxy ON exerted pro-inflammatory activity in human myeloid cells, consistent with TLR8 stimulation (Forsbach et al., 2008). We hypothesized that the activity of this motif was dictated by the presence of its GU dinucleotide, which serves as a substrate for RNase T2. Indeed, testing the UUGU-containing ON, we observed its activity to be dependent on RNase T2 and TLR8, albeit with lower activity than RNA40 (Figure 5A). Moreover, in line with the substrate specificity of RNase T2, exchanging the terminal uridine in this motif for the other three naturally occurring ribonucleotides, we observed that uridine, but not any other nucleotide following guanosine, was critically required for rendering this motif stimulatory for TLR8 (Figure 5A). Interestingly, in this previous study, AU-rich ONs were also reported to exert pro-inflammatory activity. Again, these observations are in line with the observed substrate specificity of RNase T2 and raise the question as to whether AU-containing oligoribonucleotides exert RNase-T2-dependent TLR8 agonism. Indeed, UUAU stimulated RNase-T2-dependent TLR8, and when the AU motif was permutated to AA (UUAA), no stimulatory activity was observed (Figure 5B). These functional data were reflected by the abundance of respective ON-dependent metabolites in cellulo: only UUGU, but not UUGA-stimulated, cells displayed an increase in G>p and UG>p levels, and this was completely RNase T2 dependent (Figure 5C). At the same time, UUAU, but not UUAA, stimulation led to an RNase-T2-dependent increase in A>p and UA>p levels. Interestingly, U>p levels were disconnected from the stimulatory capacity of these ONs. To this end, the non-stimulatory ON UUAA led to the same levels of U>p as the agonistic ON UUGU, whereas these levels were largely RNase T2 independent. In summary, these data suggest that UURU constitutes a minimal motif for RNase-T2-dependent TLR8 activation by an RNA ON. Moreover, these results indicate that the RNase-T2-dependent release of UR>p fragments highly correlates with TLR8 agonistic activity, although a sole increase in U>p levels in the context of RNA stimulation is not sufficient for TLR8 agonism.
Figure 5. A Minimal Motif for TLR8.
(A and B) BLaER1 cells were stimulated with the indicated DNA-RNA UUG-containing (A) or UUA-containing (B) hybrid ONs or control stimuli. The four bases in the middle are ribonucleotides flanked by random deoxynucleotides (dN). IL-6 was measured after 14 h. (C) BLaER1 cells were stimulated with the indicated DNA-RNA hybrid ONs and analyzed by LC-MS (14 h after stimulation). For all stimulations, 2.4 μg of the respective ON was used. For each metabolite dataset, the mean of the maximal values was determined, and all data are depicted as a fraction thereof. Data are depicted as mean + SEM of three independent experiments. Statistics indicate significance by two-way (A and B) or one-way ANOVA (C): ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant. See also Figure S5.
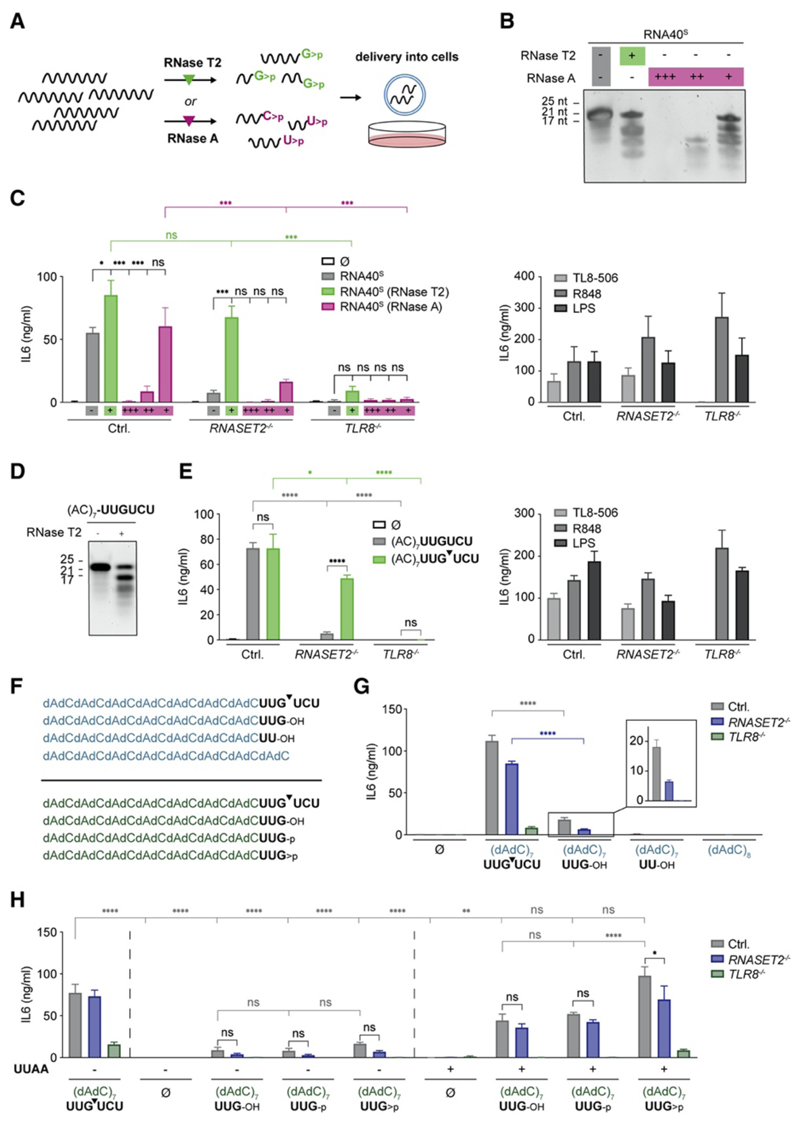
RNase T2 Degradation Products Bypass the Lack of RNase T2 to Exert TLR8 Agonism
We hypothesized that RNase T2 activity acting on exogenous RNA substrates was predominantly required for generating ligands for the second binding pocket of TLR8. This idea was spurred by the notion that previous structural studies had revealed the coordination of a YG or YYG di- or trinucleotide with a 2ʹ,3ʹ-cyclophosphate in the second binding pocket of TLR8 (Figure S5A). The requirement for this (Y)YG>p motif in this binding location—as revealed in these studies—was well in line with the notion that stimulation with TLR8-agonistic ONs resulted in the appearance of such fragments in an RNase-T2-dependent manner (Figure 5C). To address whether we could directly engage TLR8 by providing agonists for the second binding pocket of TLR8, we digested RNA40S by using RNase T2 in vitro by using limiting amounts of enzyme (Figures 6A and 6B) and subsequently delivered the thus-obtained fragments into BLaER1 monocytes. At the same time, we treated RNA40S by using RNase A (Figure 6B), covering a range of three concentrations to encompass both excess (+++) and limiting (+) enzyme amounts. As expected, undigested RNA40 was only active in control cells and non-stimulatory when RNase T2 or TLR8 was absent (Figure 6C, gray bars). However, in vitro RNase-T2-digested RNA40 was active in control cells and also stimulatory in RNASET2 −/− cells. As expected, TLR8 dependency was maintained for these in vitro-digested RNAs (Figure 6C, green bars). Hence, the ex cellulo RNA40 digestion using RNase T2 could bypass the requirement of lysosomal RNase T2. Intriguingly, RNase-A-derived degradation products were unable to trigger TLR8 activation in RNASET2 −/− cells (Figure 6C, magenta bars). Moreover, their activity in control cells was present only when undigested RNA40S was present, as was the case in the undercut preparation (Figure 6C, +). To study this approach with a defined motif, we designed an ON that contained only one UUGU motif in an otherwise inert sequence context: (AC)7UUGUCU. As expected, digestion of this ON by RNase T2 generated a major 17-mer fragment (Figure 6D), and stimulation experiments showed that this ON exerted RNase-T2-dependent TLR8 agonism (Figure 6E). Analogous to the results obtained with RNA40, ex cellulo digestion of this ON bypassed the requirement for endogenous RNase T2 (Figure 6E, green bars). We next wanted to test whether ONs directly terminating with the UUG motif would exert TLR8 agonism on their own. In order to be able to employ high-performance liquid chromatography (HPLC) to purify these ONs, we exchanged the inert AC portion of (AC)7UUGUCU into a non-phosphorothioate deoxynucleotide (dAdC)7 (Figure 6F). Similarly to the previous results, this ON exerted RNase-T2-dependent TLR8 agonism, and ex cellulo digestion bypassed RNase T2 requirement (Figures S6A and S6B). We next compared a version of this ON terminating with UUG with a variant lacking the 3ʹ terminal guanosine (UU). These experiments showed that UUG could exert TLR8 agonism, whereas the UU variant was completely inactive (Figure 6G, insert). However, this activity was greatly reduced in comparison with that of the ex-cellulo-digested RNase T2 substrate UUGUCU (Figure 6G and Figures S6C and S6D) and also partially RNase T2 dependent. We considered that two possible scenarios might be accountable for this difference. On the one hand, the ex-cellulo-digested ON additionally contained the 3ʹ-terminal UCU fragment, which could serve the function of supplying lysosomal uridine, the abundance of which would be dependent on RNase T2 and exogenous RNA delivery (Figures 4 and 5). On the other hand, the 2ʹ,3ʹ-cyclophosphate group of the RNase-T2-digested fragment could be responsible for its higher potency over the here-tested 3ʹhydroxyl-terminated fragment. To address these scenarios, we obtained highly pure 3ʹ OH, 3ʹ phosphate, or 2ʹ,3ʹ-cyclophosphate-terminated (dAdC)8UUG by HPLC purification and tested these fragments in the absence or presence of a “uridine donor” (Figure 6H and Figure S6E). To this end, we co-delivered the previously characterized UUAA ON, which did not exert TLR8 activity but resulted in the release of U>p to the same extent as TLR8-agonistc ONs. These experiments revealed that the three (dAdC)8UUG ONs exerted 10%–20% of the activity of the ex-cellulo-digested substrate on their own (Figure 6H, left). However, co-delivery of UUAA greatly enhanced the activity of the three UUG ONs, now paralleling the activity of the ex-cellulo-digested substrate (Figure 6H, right). The UUAA ON itself exerted no activity at all. Comparing the ONs with the three different UUG 3ʹ termini revealed that the 2ʹ,3ʹ-cyclophosphate moiety increased activity by approximately 2-fold. In summary, these results indicate that the RNase T2 requirement for TLR8 stimulation can be bypassed by the delivery of guanosine-terminated ON fragments through the direct engagement of the second binding pocket of TLR8. Under these conditions, sufficient uridine levels are required so that the engagement of the second binding pocket can trigger TLR8 activation.
Figure 6. RNase T2 Degradation Products Bypass the Lack of RNase T2 to Exert TLR8 Agonism.
(A) RNA40 was digested with either RNase T2 or RNase A, and BLaER1 cells were transfected with thus-obtained degradation products. (B) Urea gel of RNA40S digested with RNase T2 or different concentrations of RNase A. (C) Digested RNA40S from (B) or respective controls were used to stimulate BLaER1 cells. (D) Urea gel of full-length and RNase-T2-digested (AC)7-UUGUCU. (E) BLaER1 cells were stimulated with either digested or undigested (AC)7-UUGUCU and the indicated controls. (F) List of ONs used for the stimulations in (G) (top) and (H) (bottom). Arrow indicates in vitro digestion with RNase T2. (G) ONs depicted in (F) were used to stimulate BLaER1 cells. Control stimuli are shown in Figure S6D. (H) The ONs depicted in (F) were purified by HPLC and then used to stimulate BLaER1 cells. To obtain the (dAdC)7UUG>p cyclophosphate, we digested (dAdC)7-UUGUCU ON by RNase T2 and purified by HPLC. Control stimuli are shown in Figure S6F. In the right panel, all ONs were co-transfected with the DNA-RNA hybrid (dN)6UUAA(dN)8 (1.2 μg), serving as an uridine donor. IL-6 was measured after 14 h. Data are depicted as mean + SEM of three independent experiments or one of three representative gels. Statistics indicate significance by two-way ANOVA: ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant. See also Figure S6.
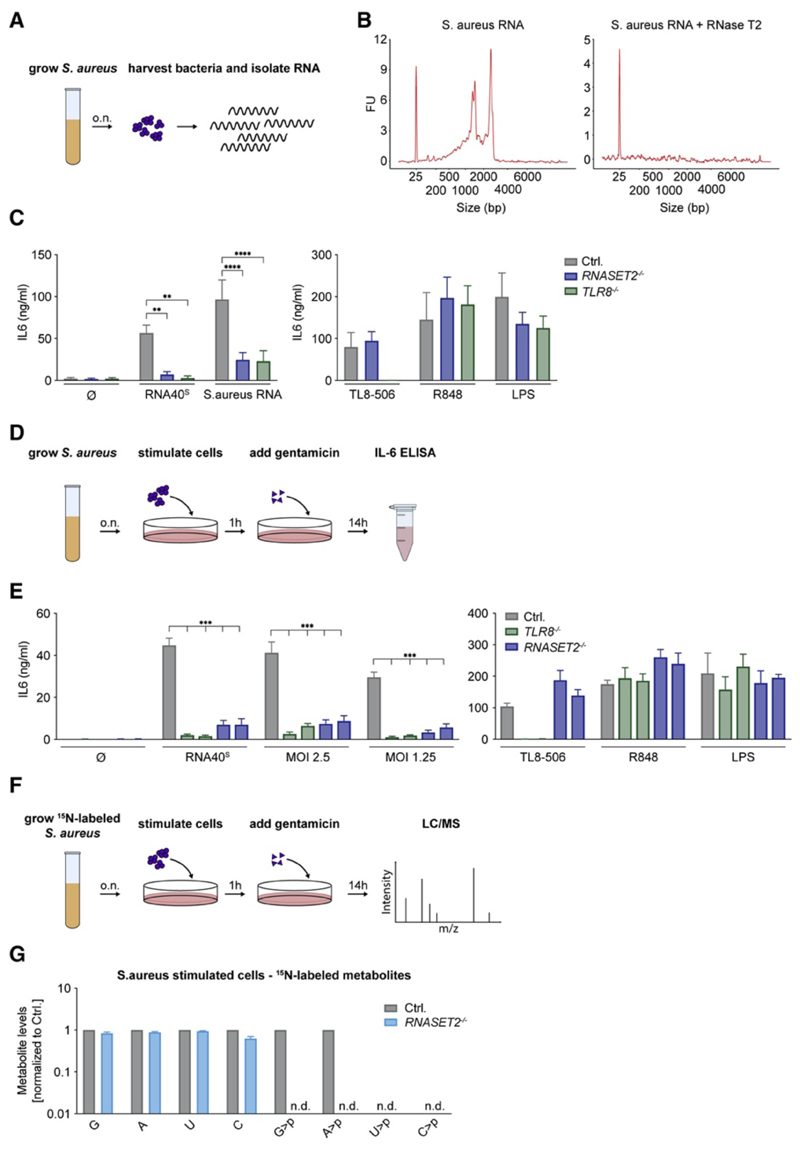
Staphylococcus aureus Detection in Myeloid Cells Depends on RNase T2 Upstream of TLR8
To explore whether RNase T2 also plays a role in the recognition of a microbial pathogen that is sensed by TLR8, we studied the recognition of Staphylococcus aureus. Previous work has established that human myeloid cells largely employ TLR8 to detect S. aureus (Bergstrøm et al., 2015, Krüger et al., 2015). BLaER1 monocytes responded to purified Staphylococcal RNA delivered by pR in an RNase-T2- and TLR8-dependent fashion (Figures 7A–7C). Next, we incubated BLaER1 monocytes with live S. aureus at different MOIs (Figure 7D). Under these conditions, BLaER1 cells displayed a potent IL-6 response upon stimulation with S. aureus, and this response was again largely dependent on RNase T2 and TLR8 (Figure 7E). Furthermore, to address whether RNA from live S. aureus is degraded by RNase T2 in cellulo, we metabolically labeled S. aureus by using stable-isotope-containing medium (15N) and analyzed isotope-labeled metabolites in BLaER1 monocytes upon infection (Figure 7F). Doing so, we could detect S. aureus-derived ribonucleosides in infected cells, yet these metabolites were not affected by RNase T2 deficiency (Figure 7G and Figures S7A and S7B). At the same time, S. aureus-dependent G>p and A>p could be detected in control cells but not in RNASET2 −/− cells (U>p and C>p levels could not be determined). Altogether, these results suggest that the recognition of a complex bacterial RNA molecule, such as the one encountered in the context of S. aureus infection, depends on RNase T2 upstream of TLR8.
Figure 7. S. aureus Detection in Myeloid Cells Depends on RNase T2 Upstream of TLR8.
(A) RNA was isolated from S. aureus to stimulate BLaER1 cells. (B) Bioanalyzer spectrum of undigested or RNase-T2-digested S. aureus RNA. (C) BLaER1 cells stimulated with S. aureus RNA and indicated controls. (D) Schematic view of how live S. aureus was used to stimulate BLaER1 cells at different MOIs. (E) After infection or stimulation with indicated controls, IL-6 was measured. (F) Schematic view of how S. aureus was grown in 15N-labeled medium and used to stimulate BLaER1 cells. (G) S. aureus-derived metabolites were analyzed by LC-MS after infection. Data are normalized to control cells (note logarithmic scale). Data are depicted as mean + SEM of two (G), three (E), or four (C) independent experiments. Statistics indicate significance by two-way ANOVA: ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant; n.d., not detected. See also Figure S7.
Discussion
Despite its central function in antimicrobial immunity and autoimmunity, our mechanistic understanding of the RNA-sensing PRR TLR8 has remained limited. Here, we identified the lysosomal endoribonuclease RNase T2 as a pivotal upstream component for the recognition of RNA molecules by TLR8. Using various model substrates, we found that human RNase T2 cleaves ssRNA molecules at RU, generating adenosine- or guanosine-2ʹ,3ʹ-cyclophosphate-terminated fragments. In line with previous structural studies, such oligoribonucleotides constitute ideal ligands for engaging the second binding pocket of TLR8. However, because of its substrate specificity, RNase T2 also serves to increase catabolic uridine, which engages the first binding pocket of TLR8, another prerequisite for the activation of this PRR. However, this functionality of releasing uridine from exogenous substrate RNA is not sufficient to activate TLR8, and uridine release can also be uncoupled from RNase T2. Interestingly, RNase T2 functions in a largely non-redundant fashion upstream of TLR8. This implies that other lysosomal RNases, of which there are several within the lysosome, cannot produce the 3ʹ termini required for engaging TLR8. In line with this notion, endoribonucleases that are part of the RNase A family cleave 3ʹ of pyrimidines, hence generating incompatible 3ʹ ends for the second binding pocket of TLR8. As such, it appears that hTLR8 has evolved to specifically sense the presence of RNase T2 degradation products within the lysosome.
RNase T2 is part of an endoribonuclease family that consists of RNase A, RNase T1, and RNase T2. Members of these transferase-type endoribonucleases cleave their substrate via a 2ʹ,3ʹ-cyclophosphate intermediate to a 3ʹ-phosphate (MacIntosh, 2011). Apart from their shared mode of action, all of these RNases locate to compartments of the secretory pathway, as such ending up in the lysosome or the extracellular space. Whereas RNase T1 family members are found only in fungi and bacteria, RNase A members exist only in the vertebrate system. Unlike RNase A members, RNase T2 family members are found across all living organisms, excluding the domain Archaea. Indeed, RNase T2 is highly conserved in metazoan, and unlike RNase A family members, of which there are 13 encoded in the human genome, it has not undergone extensive duplication or diversification events. This would argue for an important housekeeping function of RNase T2. One possibility is that RNase T2 plays an important role in ribosome turnover, e.g., in the context of ribophagy. In line with this notion, zebrafish deficient in RNase T2 activity display a marked accumulation of rRNA within lysosomes, although this is mostly confined to the central nervous system (Haud et al., 2011).
Structural studies of RNase T2 have revealed two distinct base-coordinating sites (B1 and B2 sites) located 5ʹ and 3ʹ to the scissile bond (MacIntosh, 2011). Both sites can affect substrate specificity of RNase T2. For example, the B2 site of the bitter gourd RNase T2 ortholog MC1 accommodates uridine and thus dictates its substrate specificity for 5ʹ-NU-3ʹ dinucleotides (Suzuki et al., 2000) (Numata et al., 2003). Human RNase T2 is structurally highly related to MC1, and the base-coordinating amino acid residues in its B2 site are orthologous to the ones of MC1 (Thorn et al., 2012). In line with this notion, all of the here-identified in vitro digestion RNA fragments displayed termini that were the result of a cleavage event upstream of uridine. In addition, we found the B1 site to be specific to purine nucleotides (R), rendering the substrate specificity of RNase T2 to be RU. To further assess the activity and substrate specificity of RNase T2 and RNase A family enzymes in cellulo, we made use of the possibility to measure 2ʹ,3ʹ-cyclophosphate-terminated nucleotides of both exogenous and endogenous origins. Indeed, with currently little known about the fate of lysosomal RNA degradation products, we would consider the analysis of these immediate endonuclease products as an ideal proxy of the catabolic activity of these enzymes. These analyses revealed that RNASET2 −/− cells displayed a marked drop in G>p and A>p and G>p- and A>p-terminated nucleotides of both endogenous and exogenous sources. Given that RNase A family enzymes mainly cleave after pyrimidines, one would expect C>p and U>p levels to be unaffected in the absence of RNase T2. Indeed, C>p levels were largely intact in the absence of RNase T2 yet were affected by deletion of one of the RNase A family enzymes (RNASE1). However, U>p levels were almost decreased as much as the levels of G>p or A>p. Because RNase T2 cleaves upstream of uridine, RNase T2 deficiency is expected to decrease U>p levels by theoretically 50%. Why U>p levels were affected beyond these concentrations is unclear. It is conceivable that RNase T2 also accepts uridine in its B1 site but with lower affinity, thereby masking this activity in our undercutting conditions. At the same time, it is possible that additional factors beyond the dinucleotide substrate specificity of RNase T2 affect its activity in cellulo. Nevertheless, apart from its impact on the abundance of purine-terminated RNA fragments and nucleotides, we can infer from these results that the lysosomal concentration of uridine is markedly decreased in the absence of RNase T2.
By uncovering the role of RNase T2 and its unique substrate specificity upstream of TLR8, our studies shed light on the nature of the MAMP and/or DAMP (damage-associated molecular pattern) that is sensed by TLR8. In fact, RNase T2 affects the availability of ligands for both binding pockets of TLR8. Although these functionalities are interconnected in the context of a physiological RNA ligand being sensed, it is important to consider these steps separately because they differ in quality and relevance.
The second binding pocket of TLR8 has been shown to accommodate guanosine-terminated di- and trinucleotides, whereas the nucleotide(s) preceding guanosine have been found to be pyrimidine, preferably uridine (Tanji et al., 2015). With its unique substrate specificity for RU within ssRNA, RNase T2 generates purine-terminated ribonucleotides that can engage this second binding pocket of TLR8. Indeed, across different stimulatory conditions tested, RNA-derived (U)UR>p fragments were RNase T2 dependent and closely correlated with TLR8 agonistic activity. In fact, our studies suggest that RNase T2 is the only lysosomal endoribonuclease that generates aforementioned 3ʹ termini, thus explaining its non-redundant function in RNA recognition upstream of TLR8. As an additional feature of endoribonuclease cleavage, the thus-generated 3ʹ termini harbor a 2ʹ,3ʹ-cylcophosphate configuration. Although this feature is neither sufficient nor required for TLR8 agonistic activity, it shows enhanced activity compared with that of 3ʹ-OH or 3ʹ-phosphate-terminated fragments.
The first binding pocket of TLR8 binds uridine. With its strict specificity for uridine in the B2 position, RNase T2 also critically contributes to catabolism-associated uridine levels and thus the activation of TLR8. To this end, lack of RNase T2 resulted in approximately 4-fold reduced U>p levels under steady-state conditions, and after stimulation an approximately 15-fold difference was observed. In that regard, upon delivery of a sole second binding pocket agonist (UUG), the TLR8 response of control and RNase-T2-deficient cells was much reduced in comparison with that of cells that were stimulated with an ex-cellulo-digested ON (UUG↓UCU). However, this could be reverted by co-delivery of a non-stimulatory ON (UUAA) to increase uridine levels to concentrations that paralleled the uridine concentrations of TLR8-agonistic ONs. Under these conditions, the stimulatory capacity of the UUG-terminated ONs was strongly increased to the level of the ex cellulo RNase-T2-digested precursor. From these experiments we can conclude that uridine released from exogenous RNA sources strongly contributes to TLR8 activation under these conditions. Nevertheless, it is important to note that despite its critical role in TLR8 activation, catabolic uridine is not sufficient in the context of a complex RNA molecule’s being sensed, and RNase T2 is not its sole source. As such, the stimulatory activity of an ON did not correlate with the abundance of ON-derived uridine levels, and an increase in uridine levels could be achieved in the absence of RNase T2.
In summary, RNase T2 activity is required for the TLR8-dependent recognition of complex RNA molecules by carrying out two independent functions that are both dictated by its substrate specificity. On the one hand, by regulating catabolism-derived uridine levels, RNase T2 critically contributes to the engagement of the first binding pocket. On the other hand, RNase T2 non-redundantly generates agonistic ligands for the second binding pocket of TLR8, which allosterically controls the affinity for uridine of the first binding pocket. Of note, this regulatory step is critically required in the context of a natural RNA substrate being sensed.
Loss-of-function mutations within the human RNASET2 gene result in a cystic leukoencephalopathy, which resembles congenital cytomegalovirus infection in clinical and neuroradiological features (Henneke et al., 2009). It is tempting to speculate that this RNASET2-related leukodystrophy shares the same pathomechanism with Aicardi-Goutières syndrome (AGS) (Rice et al., 2017). In AGS, defects in nucleic acid metabolism result in the inadvertent engagement of nucleic-acid-sensing PRRs and thus the initiation of autoinflammation (Crow and Manel, 2015). At first sight, this association seems counterintuitive given that our data imply a pro-inflammatory role of RNase T2 upstream of TLR8. However, the fact that different RNA-sensing PRRs harbor different ligand specificities could explain this scenario. It is conceivable that RNase T2 deficiency results in the accumulation of undigested RNA in the lysosome and leads to its cytosolic translocation and engagement of cytosolic RNA sensors. Interestingly, a similar scenario has been described for the lysosomal endonuclease DNase II. On the one hand, DNase II activity is required for degrading complex DNA molecules within the lysosome into fragments so that TLR9 can be engaged (Chan et al., 2015). On the other hand, lack of DNase II results in the accumulation of lysosomal DNA, which subsequently leads to its translocation and activation of the cGAS-STING pathway (Ahn et al., 2012, Gao et al., 2015). As such, RNase T2 activity could require a tight balance between its pro-immunogenic role upstream of TLR8 and its potentially anti-inflammatory role as an RNA-degrading enzyme. In that respect, the activity of these lysosomal nucleases appears to be well protected against potential pathogen-encoded, counter regulatory mechanisms in that perturbation of their activities would result in the initiation of antimicrobial defense mechanisms.
Star Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Direct-Blot HRP anti-FLAG tag | BioLegend | Cat#637311 |
| Anti-RNase T2 | Sigma-Aldrich | Cat#HPA029013 |
| Anti-rabbit IgG HRP linked | Cell Signaling Technology | Cat#7074 |
| Anti-TLR8 (D3Z6J) | Cell Signaling Technology | Cat#11886S |
| Bacterial Strains | ||
| S. aureus subsp. aureus | ATCC | ATCC 6538 |
| Chemicals | ||
| Agilent RNA 6000 Nano Kit | Agilent | Cat#5067-1511 |
| Ammonium persulfate | Sigma-Aldrich | Cat#A3678 |
| Blood agar plate | OXOID | Cat#PB5039A |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891 |
| EDTA (0.5M) | Thermo Fisher | Cat#15575020 |
| GeneJuice® Transfection Reagent | Merck Chemicals GmbH | Cat#70967 |
| Gentamicin | Thermo Fisher | Cat#15750060 |
| HisTrap™ | GE Healthcare | Cat#17-5247-01 |
| ISOGRO®-15N Powder-Growth Medium | Sigma-Aldrich | Cat#606871 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668019 |
| LPS-EB Ultrapure | InvivoGen | Cat#tlrl-3pelps |
| nitrocellulose membrane (0.45μm) | GE Healthcare | Cat#10600002 |
| Pam3CSK4 | Invivogen | Cat#tlrl-pms |
| Penecilin-Streptomycin | Life Technologies | Cat#15140122 |
| Phenol/Chloroform/Isoamylalkohol | Roth | Cat#A156.3 |
| Phorbol 12-myristate 13-acetate (PMA) | Enzo Life Sciences | Cat#BML-PE160-0005 |
| poly-L-arginine | Sigma-Aldrich | Cat#P7762 |
| Recombinant Human IL-3 | MPI of Biochemistry, Munich | N/A |
| Recombinant Human INF-γ | PeproTech | Cat#300-02 |
| Recombinant Human M-CSF | MPI of Biochemistry, Munich | N/A |
| R848 SequaGel Concentrate |
InvivoGen national diagnostics |
Cat# tlrl-r848 Cat#EC8301L |
| SequaGel Buffer | national diagnostics | Cat#EC835200ML |
| SequaGel Diluent | national diagnostics | Cat# EC8401L |
| Superdex-200 16/600 | GE Healthcare | Cat#28989335 |
| SYBR Gold Nucleic Acid Gel Stain | Thermo Fisher Scientific | Cat#S11494 |
| TL8-506 | InvivoGen | Cat# tlrl-tl8506 |
| TEMED | Roth | Cat#2367.3 |
| UltraPure 0.5M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| ß-Estradiol | Sigma-Aldrich | Cat#E8875-250MG |
| Critical Commercial Assays | ||
| Human IL-6 ELISA Set | BD Biosciences | Cat#555220 |
| MiSeq Reagent Kit v2, 300 Cycles | Illumina | Cat#MS-102-2002 |
| Deposited Data | ||
| BLaER1 RNA Seq data | This paper | GEO: GSE138913 |
| Experimental Models: Cell Lines | ||
| BLaER1 human b-cell to monocyte trans-differentiation cell line | (Rapino et al., 2013) | N/A |
| HEK293T | (Gaidt et al., 2017) | N/A |
| THP-1 | DSMZ | Cat#ACC 16 |
| Oligonucleotides | ||
| RNA40S (rG*rC*rC*rC*rG*rU*rC*rU*rG*rU*rU*rG*rU*rG*rU* rG*rA*rC*rU*rC) |
Miltenyi | 130-104-429 |
| RNA40 (rGrCrCrCrGrUrCrUrGrUrUrGrUrGrUrGrArCrUrC) |
IDT | N/A |
| P20 (rUrUrGrArArGrGrArCrArUrGrUrCrCrUrUrCrArA) | IDT | N/A |
| P20-5M (rUrGrUrCrCrUrGrArCrArUrGrUrCrCrUrUrCrArA) |
IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*G*U*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*G*A*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*G*G*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*G*C*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*G*dN*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*A*U*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| dN*dN*dN*dN*dN*dN*U*U*A*A*dN*dN*dN*dN*dN*dN*dN*dN | IDT | N/A |
| rArArArGrGrArArA | IDT | N/A |
| rArArArGrArArArA | IDT | N/A |
| rArArArGrUrArArA | IDT | N/A |
| rArArArGrCrArArA | IDT | N/A |
| rArArArArGrArArA | IDT | N/A |
| rArArArArArArArA | IDT | N/A |
| rArArArArUrArArA | IDT | N/A |
| rArArArArCrArArA | IDT | N/A |
| rArArArUrGrArArA | IDT | N/A |
| rArArArUrArArArA | IDT | N/A |
| rArArArUrUrArArA | IDT | N/A |
| rArArArUrCrArArA | IDT | N/A |
| rArArArCrGrArArA | IDT | N/A |
| rArArArCrArArArA | IDT | N/A |
| rArArArCrUrArArA | IDT | N/A |
| rArArArCrCrArArA | IDT | N/A |
| rA*rC*rA*rC*rA*rC*rA*rC*rA*rC*rA*rC*rA*rC*rU*rU*rG*rU*rC*rU | IDT | N/A |
| dAdCdAdCdAdCdAdCdAdCdAdCdAdCdAdC | IDT | N/A |
| dAdCdAdCdAdCdAdCdAdCdAdCdAdC*rU*rU*rG*rU*rC*rU | IDT | N/A |
| dAdCdAdCdAdCdAdCdAdCdAdCdAdC*rU*rU*rG | IDT | N/A |
| dAdCdAdCdAdCdAdCdAdCdAdCdAdC*rU*rU*rG-3‘-phosphate | IDT | N/A |
| dAdCdAdCdAdCdAdCdAdCdAdCdAdC*rU*rU | IDT | N/A |
| Recombinant DNA | ||
| pcDNA3.1_RNase T2_PreScission_6xHis | This study | N/A |
| pLI_hu_RNase T2_FLAG_Puro | This study | N/A |
| pLI_mScarlet _Puro | This study | N/A |
| CMV-mCherry-Cas9 | (Schmid-Burgk et al., 2014) | N/A |
| pLK0.1-gRNA-CMV-GFP | (Schmid-Burgk et al., 2014) | N/A |
| pCAS9-mCherry -gRNA | (Schmid-Burgk et al., 2016) | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad | N/A |
| Outknocker | (Schmid-Burgk et al., 2014) | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Veit Hornung (hornung@genzentrum.lmu.de).
Experimental Model and Subject details
Cell culture
BLaER1 cells and THP-1 were cultured in RPMI medium 1640, supplemented with 10% (v/v) FCS, L-glutamine and 100 U/mL Penicillin-Streptomycin. HEK293T cells were cultured in DMEM (same supplements as RPMI). BLaER1 cells were trans-differentiated for 5-6 day in medium containing 10 ng/mL IL-3, CSF1 (MPI of Biochemistry, Munich) and 100 nM β-estradiol. BLaER1 cells with a CASP4 −/− genetic background were used as control cells (Ctrl.) throughout the whole study. In the course of these studies, we serendipitously identified that BLaER1 cells express transcripts of SMRV (Squirrel monkey retrovirus) and subsequent experiments confirmed that BLaER1 cells harbor the SMRV proviral genome. Testing early passages of BLaER1 cells by Dr. Thomas Graf (Thomas.Graf@crg.eu) confirmed that the parental BLaER1 cell line (Rapino et al., 2013) is positive for SMRV. Of note, extensive characterization of BLaER1 monocytes in comparison to other human myeloid cells has not provided any indication that SMRV positivity would impact on the functionality of these cells as myeloid cells.
Method Details
Bioanalyzer
S. aureus total RNA analysis was performed using the Agilent Bioanalyzer device. The RNA 6000 Nano kit (#5067-1511) was used according to the supplier’s protocol.
Cell stimulation
BLaER1 cells were stimulated 5-6 days after trans-differentiation with ORN RNA40, which has a phosphorothioate backbone (RNA40S), or with the un-stabilized RNA40 (RNA40O). If not otherwise indicated, RNA40 was complexed in a 1:1 ratio with poly-L-arginine (0.6 μg per condition). For conditions involving ex cellulo digests (Figure 6), 1.2 μg per condition were used for all ONs to account for the cleavage activity of RNase T2. RNA and poly-L-arginine were separately incubated for 5 min at RT with Opti-MEM (GIBCO®) (25 μl per 96-well each). Afterward the two reagents were mixed and incubated another 20 min at RT. The medium was changed with 100 μl RPMI per 96-well prior to adding 50 μl transfection mix. The transfection of RNA40 with Lipofectamine 2000 (LF) was performed according to the supplier’s protocol. Furthermore, cells were stimulated with either 200 ng/mL LPS, 1 μg/mL R848 or 100 ng/mL TL8-506. To stimulate cells with degradation products, ONs were digested with either RNase T2 (self-purified) or bovine pancreatic RNase A (Invitrogen, Maxiprep Kit, K210017). The RNA was mixed with IDTE buffer pH 8 in a 1:1 ratio and after adding the enzyme incubated for 20 min at 37°C. The digested RNA was transfected according to the protocol above. All stimulations were carried out for 14-16 h at 37°C.
For infections with S. aureus subsp. aureus (ATCC® 6538) bacteria were grown overnight on a blood agar plate at 37°C. The next day bacteria were resuspended in PBS, centrifuged 5 min at 4000 g and resuspended in RPMI medium (without antibiotics). Cells were stimulated with 100 μl per 96-well at the indicated MOI for 1 h at 37°C. Afterward the medium was topped up with 50 μl fresh RPMI medium containing gentamicin, leading to a final concentration of 50 μg/mL. Cells were incubated for another 14 h at 37°C. Stimulation with 15N labeled S.aureus was performed the same way as with unlabeled bacteria.
THP-1 cells were differentiated with 100 ng/mL PMA for 16 h. Afterward cells were washed twice with PBS and seeded into new dishes. THP-1 cells were stimulated after a resting phase of 3 days and a 6 h incubation with IFN-γ. For stimulation, RNA40 (1200 ng/ 96-well) was complexed in a 1:1 ratio with poly-L-arginine. Therefore, the RNA and poly-L-arginine were separately incubated for 5 min at RT with Opti-MEM (GIBCO®) (25 μl per 96-well each). Afterward the two reagents were mixed and incubated another 20 min at RT. The medium was changed with 100 μl RPMI per 96-well prior to adding 50 μl transfection mix. As control, THP-1 cells were stimulated with either 0.33 μg/mL Pam3CSK4, 2 μg/mL R848 or 200 ng/mL TL8-506. All stimulations were carried out for 14-16 h at 37°C.
CRISPR/Cas9 mediated knockout-cell line generation
Gene deficient BLaER1 cells were generated using a CRISPR/Cas9 based knockout workflow as previously described (Schmid-Burgk et al., 2014). Briefly, sgRNAs (18 or 20-mer) targeting an early coding exon were designed for each gene. BLaER1 cells were electroporated with one plasmid expressing sgRNA and one expressing mCherry-Cas9 (pLK0.1-gRNA-CMV-GFP, CMV-mCherry-Cas9), using a Biorad GenePulser device. mCherry positive cells were sorted and cloned by limiting dilution. After identifying monoclones, cells were replated and grown to genotype them using deep sequencing (Illumina’s Miseq-platform). For every knockout several clones containing all-allelic frameshift mutations were picked and used for experiments.
Immunoblotting
Immunoblotting of whole cell lysate was performed as follows: Cells were detached from the dish using PBS-EDTA (diluted 1:250 in PBS), pooled and centrifuged 5 min at 500 g. The pellet was lysed in DISC buffer and again centrifuged 10 min at 16.000 g to get rid of the nuclei. The supernatant was mixed with 6x Lämmli buffer and denatured for 5 min at 85°C. After separation by tris-glycine denaturing SDS-PAGE, proteins were blotted onto 0.45 mm nitrocellulose membranes, blocked in 5% milk and incubated with indicated primary and corresponding secondary antibodies. Chemiluminescent signals were recorded with a CCD-camera and respective images contrast-enhanced in a linear fashion.
Kits
hIL-6 ELISA were performed according to the supplier’s protocol.
Lentiviral expression
RNase T2 was amplified from cDNA derived of BLaER1 cell lysate and cloned into a doxycycline inducible (dox-on) plasmid using conventional restriction enzyme cloning (pLIX_RNase T2_3xFLAG_Puro). As control a pLIX_mScarlet_Puro plasmid was used. BLaER1 cells of indicated genotype were transduced and selected by using puromycin. The polyclonal cell population was then used for further experiments.
LC/MS
3x106 BLaER1 cells were lysed for 10 min on ice in 750 μl of a 1:1 mixture of water and acetonitrile. Cells were spun down at 21.000 g for 10 min and the supernatant was flash frozen. The samples were freeze-dried overnight and the fluffy white content was redissolved in 300 μL of milliQ-water. Each sample was centrifuged at 21.000 g and 4°C for 30 min, and the supernatant was transferred to a new tube. Prior to MS-measurement, 10 μL of a 0.3 μM solution of 2-(D3-methyl)guanosine in milliQ-water (D3-m2G); synthesized in the Carell group (Globisch et al., 2011) was added as internal standard to 90 μL of the sample. The mixture was vortexed for 60 s. The LC-HESI-MS analysis of the samples, containing the soluble pool of the cells and 3 pmol of D3-m2G as internal standard, was performed on a Dionex Ultimate 3000 HPLC system coupled to a Thermo Fisher LTQ Orbitrap XL mass spectrometer. For comparability, the injection volume was always 90 μL (of 300 μL total sample volume) per technical replicate. Nucleosides / nucleotides were separated on an Interchim Uptisphere120-3HDO C18 column whose temperature was maintained at 30°C. Elution buffers were buffer A (2 mM NH4HCOO in H2O; pH 5.5) and buffer B (2 mM NH4HCOO in H2O/MeCN 20/80 v/v; pH 5.5) with a flow rate of 0.15 mL/min. The gradient was as follows: 0→10 min, 0% B; 10→15 min, 0→0.1% B; 15→50 min, 0.1→5% B; 50→90 min, 5→100%. The chromatogram was recorded at 260 nm with a Dionex Ultimate 3000 Diode Array Detector, and the chromatographic eluent was directly injected into the ion source of the mass spectrometer without prior splitting. Ions were scanned in the positive polarity mode over a full-scan range of m/z 225-2000 with a resolution of 60,000. Parameters of the mass spectrometer were tuned with a freshly mixed solution of inosine (5 μM) in buffer A and set as follows: Capillary temperature 275.00°C; source voltage 4.80 kV; capillary voltage 0.00 V; tube lens voltage 45.00 V. The ion chromatograms of the compounds of interest were extracted from the total ion current (TIC) chromatogram and the areas under the curves were integrated. The resulting integrals were divided by the corresponding integral obtained for the internal standard D3-m2G improving the accuracy with respect to sample comparability. Of note, due to the fact that the introduction of the sulfur atoms created diastereomeric configurations of RNA40S-derived fragments, several liquid chromatography peaks were detected for these molecules. For our analyses, we quantified the most abundant peak. In case of the cell extracts derived from S. aureus-infected cultures, fully 15N-labeled compounds were by far more abundant than the respective compounds lacking one or more of these heavy-atom labels. In these experiments, we therefore only considered the fully labeled compounds for MS-based quantification.
For studying the substrate specificity of RNase T2, the same HPLC-ESI-MS-setup was used (a Dionex Ultimate 3000 HPLC system coupled to a Thermo Fisher LTQ Orbitrap XL mass spectrometer). While the elution buffers A (2 mM NH4HCOO in H2O; pH 5.5) and B (2 mM NH4HCOO in H2O/MeCN 20/80 v/v; pH 5.5) were the same as before, the flow rate (0.2 mL/min) and the gradient were optimized for these experiments. Here, the gradient was as follows: 0→5 min, 0% B; 5→20 min, 0→5% B; 20→30 min, 5→100% B. The chromatogram was again recorded at 260 nm with a Dionex Ultimate 3000 Diode Array Detector, and the UV trace was used to calculate the percentage of strand cleavage. To this end, the sum of the UV-peak integrals of the cleavage products was divided by the sum of both the UV-peak integrals of the cleavage products and of the full strand. The MS-data generated in parallel was used to identify and verify the nature of all compounds. Please note that the oligomers were bearing multiple charges, so that the observed m/z-values were still within the MS full-scan range of m/z 225-2000.
HPLC and MALDI
1200 ng RNA was digested with RNase T2 (5-50 pg/μl) or RNase A (5-50 pg/μl) at 37°C for 20 min. The reaction was performed in a total volume of 10 μl and filled up with water to 300 μl after digesting. The sample was mixed with an equal volume of Roti®-Phenol/Chloroform/Isoamylalcohol and flash frozen. The following steps were performed at room temperature. After thawing, the samples were centrifuged at 21.000 g for 3 min and the aqueous (upper) phase was transferred to a new tube. To remove traces of phenol, one volume-equivalent of chloroform was added to the aqueous phase and the mixture was vortexed for 30 s. After centrifugation at 21.000 g for 3 min, the aqueous (upper) phase was again transferred to a new tube. This chloroform extraction was repeated once and the remaining aqueous phase was subsequently freeze-dried overnight. The samples were redissolved in 60 μL of milliQ-water and centrifuged at 21.000 g and 4°C for 30 min. Of the supernatant, 55 μL were transferred to a new vial and analyzed by HPLC with an injection volume of 50 μL. HPLC-analyses were performed on a Waters e2695 Separations Module. The ONs were separated on an EC 250/4 Nucleodur 100-3 C18ec column whose temperature was maintained at room temperature, and the chromatogram was recorded at 260 nm by a Waters 2489 UV/Visible Detector. Elution buffers were buffer C (0.1 M NEt3/HOAc in H2O); and buffer D (0.1 M NEt3/HOAc in H2O/MeCN 20/80 v/v) with a flow rate of 0.5 mL/min. The gradient was as follows: 0→45 min, 0→25% D; 45→47 min, 25→100% D. The collected fractions were freeze-dried overnight, redissolved in 10 μL of milliQ-water, and desalted for 3 h using a MF-Millipore Membrane Filter with 0.025 μm pore size. The subsequent MALDI-MS-analysis was performed on an autoflex II system from Bruker Daltonics in the negative polarity mode with 1.5 μL of HPA-matrix (3-hydroxypicolinic acid) per 1.5 μL of the desalted sample. All MALDI experiments were repeated two times; oligomer masses were calculated as [M-H]− of the most abundant isotopologue.
15N labeling of S. aureus
To obtain S. aureus with 15N labeled RNA, ISOGRO-15N growth medium was used. It was dissolved in water to a final concertation of 0.5 g/ 50 mL and supplemented as follows: K2HPO4 (10 mM), KH2PO4 (10 mM), MgSO4 (10 mM) and CaCl2 (0.1 mM). 5 mL of the growth medium were inoculated with S. aureus subsp. aureus (ATCC® 6538) and grown for 16 h. After 8 min centrifugation at 5000 g the bacteria were washed twice with PBS and used for stimulation.
RNA precipitation
Following HPLC purification, sodium acetate precipitation of the obtained RNA fragments or oligomers was performed. A 3 M sodium acetate solution was added to the RNA sample resulting in a final concertation of 0.3 M. 4 volumes of 100% ethanol were added, and the mixture was incubated for 1 h at −20°C. Afterward, the sample was centrifuged for 30 min at full speed and the pellet washed twice with 80% ethanol. After air drying the pellet, it was dissolved in RNase free water.
Protein purification
Human WT RNase T2 (encoding residues 1-256) was amplified from cDNA derived from BLaER1 cell lysate and cloned into either the piggyBac vector system Li et al. (2013) or a pcDNA3.1 vector using conventional restriction enzyme cloning. Both vectors were used to produce RNase T2 to over 90% purity determined by Coomassie stained protein gels and mass spectrometry. Activity was comparable between both sources.
The pcDNA3.1_RNase T2_PreScission_6xHis plasmid was transfected into HEK293T cells. The supernatant of six 10 cm dishes were harvested and clarified (centrifugation at 500 g for 5 min, 4°C). The supernatant was filtered and loaded onto a 1 mL HisTrap. The column was washed using 20 column volumes (CV) of wash buffer (20 mM K2HPO4, pH 7.5, 0.5 M NaCl, 20 mM Imidazole), the protein was eluted using a linear gradient of buffer A (wash buffer) and buffer B (1 M imidazole) over 10 CV. RNase T2 containing fractions were subjected to size exclusion chromatography (SEC) via a Superdex-200 16/600 in SEC buffer (200 mM NaAc pH 6.0, 50 mM NaCl). Fractions containing RNase T2 were pooled, concentrated and flash frozen for storage at −80°C.
The piggyBac_RNase T2_PreScission_6xHis plasmid was used to generate stable HEK293T/17 SF cells. Cells were grown to a density of 1 × 106 cells/mL in 1 L and treated with doxycycline (1 μg/mL). After 7 days, supernatant containing secreted RNase T2 was harvested. 1 L of harvested supernatant was concentrated using a Sartocon Slice 200, 10 kDa MW cutoff (Sartorius) in PBS. Concentrated protein was subjected to size exclusion chromatography in SEC buffer (200 mM NaAc, 50 mM NaCl, pH 6.0) using a HiLoad 26/60 Superdex 200 (GE healthcare). Fractions containing RNase T2 were concentrated using a 10 kDa cut-off, Amino Ultra 15 concentrator (Amicon), to a concentration of 11 mg/mL and flash frozen in liquid nitrogen.
RNA-Seq
Undifferentiated as well as differentiated BLaER1 cells were lysed in Trizol (2x106 / 1 ml) and total RNA was purified using the Zymo Direct-zol RNA Miniprep. Total RNA was used to generate stranded RNA sequencing libraries using the Encore Complete RNA-Seq library system of NuGEN. The libraries were sequenced on an Illumina HiSeq1500 device. The reads were aligned to the human reference genome (Ensemble genome version 91) using STAR (Dobin et al., 2013). The transcripts were quantified using RSEM (Li and Dewey, 2011) and TPM (Transcripts Per Kilobase Million) values are shown as gene expression. TPM values below 2-6 or above 26 were trimmed to allow for data visualization (Figure 2A). All RNA-seq datasets were generated in this study, except the THP-1 one which is derived from a public database (GEO: GSE62171).
Urea gel
Urea gel was casted according to the supplier’s protocol using SequaGel Concentrate, SequaGel Diluent and SequaGel Buffer. The gel was run at 250 V for 70 min and stained using SYBR Gold. Afterward the gel was imaged.
Quantification and Statistical Analysis
If not otherwise indicated, statistical significance was determined by two-way ANOVA, and a post hoc test using Dunnett’s or Tukey’s correction for multiple testing. The exact number of replicates (n) is indicated within figure legends. All statistical analysis, except the analysis of the normalized LC/MS data, was performed using GraphPad Prism 8. **** p ≤ 0.0001; *** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05, ns = not significant. If multiple comparisons are depicted withone comparison bar, the major tick of the comparison bar indicates the reference data to which the statements regarding the level of significance are made. LC/MS data was normalized by dividing the values of RNASET2 −/− cells by the values of Ctrl. cells. These values were then transformed into a Log2 scale and a Welch’s unequal variances t test was performed. Undetectable values were replaced with 1/10 of the lowest non-zero value of the corresponding dataset. The analysis was performed using R.
Data and Code Availability
BLaER1 RNA-Seq data can be accessed at Gene Expression Omnibus (GEO: GSE138913) or at Sequence Read Archive (SRA: SRP225808).
Supplementary Material
Highlights.
Lysosomal RNase T2 activity functions upstream of TLR8
RNase T2 cleaves ssRNA between purines and uridine (U)
This releases U and purine-2ʹ,3ʹ-cyclophosphate-terminated RNAs activating TLR8
Staphylococcus aureus is sensed in a manner dependent on RNase T2 and TLR8
Acknowledgments
We kindly thank Larissa Hansbauer, Andreas Wegerer, and Romy Böttcher for great technical support; LAFUGA for deep sequencing; the BioSysM FACS Core Facility for cell sorting; and the BioSysM Automation Facility for liquid handling (all Gene Center, LMU). We kindly thank Sabine Suppmann from the Biochemistry Core Facility (Max Planck Institute of Biochemistry) for protein expression and purification, Andreas Pichlmair (Technical University of Munich) for the pLIX plasmid, and Christine Josenhans (Max von Pettenkofer-Institute, LMU) for providing us with S. aureus subsp. aureus. A.L. is supported by the Else Kröner-Fresenius-Stiftung. This work was supported by grants from the European Research Council (ERC) (ERC-2016-ADG – project 741912 EPiR) and by the Deutsche Forschungsgemeinschaft (DFG) via the programs SFB 1309 (project 325871075) and SFB 1361 (project 393547839) to T.C. Moreover, support was provided by grants from the ERC (ERC-2014-CoG – project 647858 GENESIS) and the DFG TRR 237 (project 404446805) to V.H.
Footnotes
Author Contributions
Conceptualization, W.G., M.W., M.G., T.C., and V.H.; Formal Analysis, Y.C.; Investigation, W.G., M.W., M.G., C.S., and A.L.; Writing, V.H. with input from all authors; Resources, T.C. and V.H.; Funding Acquisition, T.C. and V.H.; Supervision, T.C. and V.H.
Declaration of Interests
The authors declare no competing interests
References
- Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, et al. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J Immunol. 2009;182:6824–6833. doi: 10.4049/jimmunol.0803001. [DOI] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, Flo TH, Mollnes TE, Espevik T, Stenvik J. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-beta Production via a TAK1-IKKbeta-IRF5 Signaling Pathway. J Immunol. 2015;195:1100–1111. doi: 10.4049/jimmunol.1403176. [DOI] [PubMed] [Google Scholar]
- Chan MP, Onji M, Fukui R, Kawane K, Shibata T, Saitoh S, Ohto U, Shimizu T, Barber GN, Miyake K. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun. 2015;6 doi: 10.1038/ncomms6853. 5853. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, e Sousa C Reis. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, O'Duill F, Schmid-Burgk JL, Hoss F, Buhmann R, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 2017;171:1110–1124 e1118. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AA, Cooper MA, Graf T, Hornung V. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A. 2015;112:E5699–5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Pearson D, Hienzsch A, Bruckl T, Wagner M, Thoma I, Thumbs P, Reiter V, Kneuttinger AC, Muller M, et al. Systems-based analysis of modified tRNA bases. Angew Chem Int Ed Engl. 2011;50:9739–9742. doi: 10.1002/anie.201103229. [DOI] [PubMed] [Google Scholar]
- Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gartner J, Alia A, et al. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci U S A. 2011;108:1099–1103. doi: 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Henneke M, Diekmann S, Ohlenbusch A, Kaiser J, Engelbrecht V, Kohlschutter A, Kratzner R, Madruga-Garrido M, Mayer M, Opitz L, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- Kruger A, Oldenburg M, Chebrolu C, Beisser D, Kolter J, Sigmund AM, Steinmann J, Schafer S, Hochrein H, Rahmann S, et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 2015;16:1656–1663. doi: 10.15252/embr.201540861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC. In: RNase T2 Family: Enzymatic Properties, Functional Diversity, and Evolution of Ancient Ribonucleases. Ribonucleases AW Nicholson., editor. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. pp. 89–114. [Google Scholar]
- Numata T, Suzuki A, Kakuta Y, Kimura K, Yao M, Tanaka I, Yoshida Y, Ueda T, Kimura M. Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity. Biochemistry. 2003;42:5270–5278. doi: 10.1021/bi034103g. [DOI] [PubMed] [Google Scholar]
- Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- Rapino F, Robles EF, Richter-Larrea JA, Kallin EM, Martinez-Climent JA, Graf T. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013;3:1153–1163. doi: 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Rice GI, Melki I, Fremond ML, Briggs TA, Rodero MP, Kitabayashi N, Oojageer A, Bader-Meunier B, Belot A, Bodemer C, et al. Assessment of Type I Interferon Signaling in Pediatric Inflammatory Disease. J Clin Immunol. 2017;37:123–132. doi: 10.1007/s10875-016-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Hiller B, Hornung V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity. 2016;44:739–754. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 2014;24:1719–1723. doi: 10.1101/gr.176701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Schmid-Burgk JL, Ebert TS, Gaidt MM, Hornung V. Designer Nuclease-Mediated Generation of Knockout THP1 Cells. Methods Mol Biol. 2016;1338:261–272. doi: 10.1007/978-1-4939-2932-0_19. [DOI] [PubMed] [Google Scholar]
- Song W, Wang J, Han Z, Zhang Y, Zhang H, Wang W, Chang J, Xia B, Fan S, Zhang D, et al. Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13. Nat Struct Mol Biol. 2015;22:782–787. doi: 10.1038/nsmb.3080. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yao M, Tanaka I, Numata T, Kikukawa S, Yamasaki N, Kimura M. Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity. Biochem Biophys Res Commun. 2000;275:572–576. doi: 10.1006/bbrc.2000.3318. [DOI] [PubMed] [Google Scholar]
- Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, Miyake K, Shimizu T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat Struct Mol Biol. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- Thorn A, Steinfeld R, Ziegenbein M, Grapp M, Hsiao HH, Urlaub H, Sheldrick GM, Gartner J, Kratzner R. Structure and activity of the only human RNase T2. Nucleic Acids Res. 2012;40:8733–8742. doi: 10.1093/nar/gks614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, Tanji H, Isobe T, Uchiyama S, Miyake K, et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity. 2016;45:737–748. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ohto U, Shibata T, Taoka M, Yamauchi Y, Sato R, Shukla NM, David SA, Isobe T, Miyake K, et al. Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands. Cell Rep. 2018;25:3371–3381 e3375. doi: 10.1016/j.celrep.2018.11.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BLaER1 RNA-Seq data can be accessed at Gene Expression Omnibus (GEO: GSE138913) or at Sequence Read Archive (SRA: SRP225808).