Abstract
Organic chemistry is ubiquitous in the Solar System, and both Mars and a number of icy satellites of the outer Solar System show substantial promise for having hosted or hosting life. Here, we propose a novel astrobiologically focused instrument suite that could be included as scientific payload in future missions to Mars or the icy moons: the Complex Molecules Detector, or CMOLD. CMOLD is devoted to determining different levels of prebiotic/biotic chemical and structural targets following a chemically general approach (i.e., valid for both terrestrial and nonterrestrial life), as well as their compatibility with terrestrial life. CMOLD is based on a microfluidic block that distributes a liquid suspension sample to three instruments by using complementary technologies: (1) novel microscopic techniques for identifying ultrastructures and cell-like morphologies, (2) Raman spectroscopy for detecting universal intramolecular complexity that leads to biochemical functionality, and (3) bioaffinity-based systems (including antibodies and aptamers as capture probes) for finding life-related and nonlife-related molecular structures. We highlight our current developments to make this type of instruments flight-ready for upcoming Mars missions: the Raman spectrometer included in the science payload of the ESAs Rosalind Franklin rover (Raman Laser Spectrometer instrument) to be launched in 2022, and the biomarker detector that was included as payload in the NASA Icebreaker lander mission proposal (SOLID instrument). CMOLD is a robust solution that builds on the combination of three complementary, existing techniques to cover a wide spectrum of targets in the search for (bio)chemical complexity in the Solar System.
Key Words: Prebiotic chemistry, Mars, Icy moons
1. Introduction: Detecting the Polymers Building (Any Form of) Life
Although a precise definition of life remains elusive (Joyce, 1994; Domagal-Goldman et al., 2016), those focused on the biochemical composition of living beings are gaining consensus. Under this perspective, living entities can be defined as systems out of equilibrium that tend to be complex and highly organized, being endowed with the ability to assimilate matter and energy from the environment and transforming them, thus allowing the growth and reproduction of the organism. Two basic principles should inform the search for life elsewhere: (1) Carbon chemistry in water-based media might be the only viable option for living systems; (2) the combination of compartmentation, a metabolic machinery, and a molecule that is able to be (error-prone) copied and transmitted to the progeny (reproduction) are generic requisites for all forms of life (de la Escosura et al., 2015; Ruiz-Mirazo et al., 2017).
Based on terrestrial life, both amphiphilic molecules and functional biopolymers (such as proteins, nucleic acids, or analogous macromolecules) could be involved in the biochemical processes required to maintain such ternary systems, though alternative and complex enough molecules could perform analogous functions in extraterrestrial environments. Thus, being able to identify (e.g., on Mars or in the subsurface ocean of the icy moons) functional polymers more or less related to those performing biochemical functions on Earth would likely constitute compelling evidence of the existence of present life.
Carbon is ubiquitous in the Universe and exhibits unique capabilities to establish stable covalent bonds with other abundant elements (in particular, H, O, N, P and S), thus leading to the synthesis of millions of organic molecules. Water is also very abundant in the Solar System as well as in interstellar environments, shows optimal performance as a solvent, plays an active role in a number of chemical reactions, offers protection against radiation, and provides a friendly environment for the carbon-based chemistry to develop. The suitability of other alternatives to the carbon chemistry and water solvent as universal ingredients for life has yet to be demonstrated (Cabrol, 2019), and this would probably require geochemical environments with different physicochemical parameters (P, T, salinity, pH) where other solvent-based complex chemistries were possible (Schulze-Makuch and Irwin, 2018). Even in such a case, this chemically different kind of life would emerge as an increased organization level of the molecules supporting it.
Compartmentation is a process that allows concentrating chemicals into a particular niche, separated from the environment through a permanent and semi-permeable structure, which allows a far-from-equilibrium chemistry inside the compartmented system. Examples of membrane-based compartments include vesicles, rods, liposomes, and other cell-like structures that account for chemical organization and system individualization (Monnard and Walde, 2015).
In turn, metabolism is a network of chemical reactions that allow gathering material resources and energy from the environment and transforming them to allow self-maintenance of the whole system. It requires the interconversion of a repertoire of organic molecules, which could be achieved by a complex enough prebiotic chemistry that included catalytic species or mineral surfaces, before the advent of a primitive metabolism already based on enzymatic activities (Peretó, 2005; Ruiz-Mirazo et al., 2014).
Finally, the template-based replication of an information-bearing macromolecule is also required to transmit the “blueprint of the system” to the progeny. Interestingly, the mutations and other kinds of molecular rearrangements introduced during the copying process lead to a certain degree of diversity in the offspring that will ultimately allow the open-ended evolution of the whole system (Szathmary, 2006; Pressman et al., 2015). The self-reproduction of such a compartmented system endowed with metabolism and template replication relies on the combined functionalities of complex polymers and other macromolecules, which must show enough conformational plasticity to allow the noncovalent interactions involved in ligand binding and catalysis (Orgel, 1992; Mansy et al., 2008).
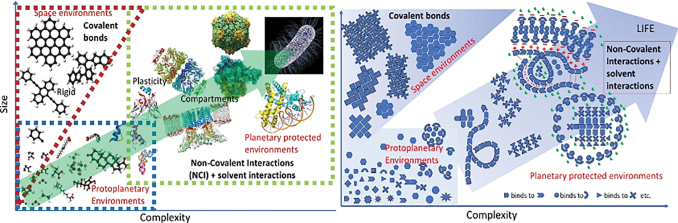
Besides the growing repertoire of low-molecular-weight (MW) compounds detected by astrochemistry in the harsh conditions of the interplanetary and interstellar space, as well as in meteorites and cometary nuclei, only a limited number of complex organic molecules, showing recalcitrant structures (e.g., polycyclic aromatic hydrocarbons [PAH]), have been found (Bernstein et al., 1999; Siebenmorgen and Krügel, 2010; Pizzarello and Shock, 2010; Goesmann et al., 2015; McGuire, 2018). In turn, certain planetary environments may allow carbon chemistry to expand and diversify in such a way that a high number of organic compounds can be formed, differing in molecular size, structure, and/or complexity (Fig. 1).
FIG. 1.
Size versus complexity in the molecular world leading to life. Left: The case for terrestrial life. Red triangle: Harsh space environments only permit simple, highly radiation–resistant molecules, as well as rigid poly-aromatic hydrocarbon structures. Blue rectangle: Mild proto-planetary and unprotected planetary environments favor the formation and stabilization (i.e., the acquisition of a longer half-life) of biomolecules, thus increasing the complexity of the available chemical repertoire. Green rectangle: More protected and stable environments of planets or satellites allow the formation and stabilization of homo- and heteropolymers, whose 3D structure in solution, based on noncovalent interactions, confers them conformational plasticity and functional capabilities. This can allow the systems to undergo self-assembly and self-organization processes, including those leading to the formation of the compartments required for life. Right: The suggested case for a “generic life” involving other biomolecules that might be stable and chemically functional under different physicochemical conditions. Color images are available online.
However, in the mild and wet environment provided by some planets and moons, the chemical complexity of the synthesized molecules is limited by their solubility or even by intramolecular hindrance constraints, as well as by their stability in such water-based media. On the early Earth, a pathway was opened that allowed molecular size and complexity to increase: the oligomerization and eventual subsequent polymerization of a certain kind of monomeric building blocks (Ruiz-Mirazo et al., 2014, and references therein).
Polymers of this kind include, in terrestrial biochemistry, peptides and proteins, nucleic acids (ribonucleic acid [RNA] and deoxyribonucleic acid [DNA]), or oligosaccharides. Polymeric macromolecules open a universe of new chemical and structural possibilities based on noncovalent, supramolecular interactions that include hydrogen bonds, Van der Waals forces, and electrostatic and hydrophobic interactions (Lehn, 1995). On the Earth, polymers such as proteins and RNA add new properties to the chemistry of their monomers: They are flexible, can fold and unfold based on the physicochemical features of the environment (including T, pH, ionic strength, and concentration of divalent cations), can behave as charged polyelectrolytes, can adopt alternative conformations as a function of the intramolecular network of weak interactions available, can interact with other polymers (of either the same or different kind) to form macromolecular assemblies, and can create new microenvironments that allow (bio)chemical reactions to take place by lowering the required activation energy (i.e., they can become catalysts) (Benner, 2017).
Therefore, polymerization of the building blocks achieved by prebiotic chemistry allowed the synthesis of macromolecules endowed with enough conformational plasticity to interact with each other and to form new structures and compartments, thus establishing the biochemical networks that are required for life to originate and evolve.
Specific planetary environments can allow chemical systems to increase in complexity, including molecular size, structure, and functionality of the molecules, as well as the number of interactions established among the individual components (Cockell et al., 2016). The search for extraterrestrial life should be reasonably based on the assumption that its underlying biochemistry may be different from what we know on the Earth. Thus, how to detect signs of life eventually present in favorable environments in the Solar System is still an open question (Cabrol, 2019). It is currently straightforward to detect and recognize terrestrial life (e.g., in extreme environments on our planet), because we already know its main molecular features. Therefore, the identification on other planets or satellites of any low MW biomolecule, biopolymer, or structural component equal to those produced by living beings on the Earth would be considered as evidence consistent with life as we know it.
This might also be the case if cell-like morphologies of carbonaceous composition known in terrestrial microorganisms are detected, though being aware that a number of “abiotic biomorphs” can be chemically formed in the absence of any biological process (Rouillard et al., 2018). However, we would have enormous difficulties to identify other nonterrestrial-like life, even using well-equipped laboratories and trained personnel, if their biochemistry differs from known life. Thus, the key questions become the following: (1) How could we recognize other self-sustaining chemical systems capable of Darwinian evolution if their biochemistry is different from that of terrestrial life?; (2) Is there any generic physicochemical feature or set of features we can detect and measure that could shed light about its biogenicity?; and (3) How difficult is such an ambitious task using only remote exploration on other planets and moons?
Searching for unequivocal signs of life in planetary contexts is a major objective for the space science and astrobiology communities, and thus the main space agencies support this objective in their current programs of planetary exploration through several missions. Regrettably, the exploration of the Solar System so far has involved just one true “search for life” mission: the Viking landers on Mars nearly 50 years ago (Klein et al., 1976). Although some of the upcoming missions to Mars (NASA's Perseverance rover in 2020 and ESA's Rosalind Franklin rover in 2022) or Europa (ESA's JUICE and NASA's Europa Clipper) have the capability to detect molecular biosignatures for past or present life, interpreting these missions' data is not easy and, particularly for the case of extant life (Box 1), a much better job can be done by using (1) an extraction method that is less chemically aggressive than pyrolysis and (2) a more selective analytical technique for molecular identification.
Up to now, the instrumentation devoted to the detection of organics in planetary exploration has been mostly based on the identification of either volatile compounds using gas chromatography/mass spectrometry (GC/MS) or some spectroscopic organic signatures. After the Viking landers' GC/MS, three instruments have focused mainly on the analysis of organic molecules on Mars: NASA's Phoenix Thermal and Evolved Gas Analyzer (TEGA), the current NASA MSL Sample Analysis at Mars (SAM) instrument suite, and the future ESA ExoMars Mars Organics Molecule Analyzer (MOMA) onboard the Rosalind Franklin rover (Table 1).
Table 1.
Different Analytical Methods and Instruments Used (Light Gray) or Proposed (White) for Organic Detection in Planetary Exploration
Our concept scheme (dark gray) is a robust and compact instrument suite that assembles three complementary techniques to cover a wide spectrum of targets.
Ab = antibody; Ap = aptamer; GC-MS = gas chromatography/mass spectrometry; IR = infrared; MOMA = Mars Organics Molecule Analyzer; PAH = polycyclic aromatic hydrocarbon; RLS = Raman Laser Spectrometer; SAM = Sample Analysis at Mars; TEGA = Thermal and Evolved Gas Analyzer.
Box 1.
Missions to Mars with an Astrobiological Relevance
| The two NASA Vikings, which landed on Mars in 1976, were the two first landers on Mars and they had a clear ambition to detect life. Three experiments were onboard that focused on detecting potential metabolic activities. The Labeled Release experiment tried to detect some metabolism activities in a regolith sample, mixing it with a solution that contained nutrients doped with 14C. The Pyrolitic Release experiment used an atmosphere of CO2 also with14C: After some time, the gases were removed and the sample was volatilized to check whether the 14C had been somehow absorbed by microorganisms. The Gas Exchange experiment analyzed the gases given off by a moistened soil sample in a helium atmosphere. The Labeled Release gave positive results (not conclusive, as some abiotic reactions could have produced similar results). The Pyrolitic Release showed some evidence for organic synthesis that has never been fully explained, whereas Gas Exchange produced O2 when the soil was moistened, which was not considered a biological process. NASA's Phoenix landed close to Mars' north polar cap in 2007, taken with two instruments devoted to perform chemical analysis of soil samples. The TEGA detected gases released during the heating of a sample by mass spectrometry. The MECA allowed the observation of samples by optical and atomic force microscopy, and it determined the pH, conductivity, redox potential, and abundance of minerals such as magnesium and sodium cations or chloride, bromide, and sulfate anions. NASA's Mars Science Laboratory (Curiosity rover) is on Mars since 2012 and is equipped with a GC/MS and a tunable laser spectrometer, with a set of ovens to volatilize soil samples. The SAM has also a nonpyrolysis mode using MTBSTFA derivatization. Their goals are, among others, determining the organic carbon inventory as well as the chemical and isotopic state of lighter elements. The MSL has already detected organic carbon formed in situ in Gale crater, including ion fragments typical of aliphatic and aromatic hydrocarbons as well as chlorobenzene, dichloroalkanes, and thiophenes. SHERLOC will land on Mars in 2021 onboard NASA's Mars 2020. It is a DUV resonance Raman and fluorescence spectrometer that is capable of detecting organics that show natural fluorescence, organic radicals (aromatic and aliphatic), and even some potential biosignatures. ESA's Rosalind Franklin will land on Mars in 2023 with two instruments that are especially relevant in astrobiology. The MOMA is a GC and laser desorption with a linear ion trap MS. It is devoted to the identification of refractory organic substances, as well as chiral and isotopic composition. The MOMA has also a nonpyrolysis mode using TMAH, DMF-DMA, and MTBSTFA derivatization. The RLS has two goals related to life detection: (i) identifying organic compounds, and (ii) identifying mineral products and indicators of biological activity (related to biomineralization processes). |
DUV = deep ultraviolet; GC/MS = gas chromatography/mass spectrometry; MOMA = Mars Organics Molecule Analyzer; RLS = Raman Laser Spectrometer; SAM = Sample Analysis at Mars; TEGA = Thermal and Evolved Gas Analyzer.
2. Strategies for In Situ Detection of Different Levels of (Pre)biotic Chemical and Structural Complexities
Similar to previous missions capable of searching for chemical evidence for life on Mars (Fig. 2), we also propose with Complex Molecules Detector (CMOLD) to explore for chemical biosignatures that combine fundamental biological properties connected to the environment, including chemical disequilibria, distribution of patterns of structurally related compounds, isotopic signatures indicative of the dominance of catalysis in biochemistry, and concentration of chemical monomers that are dictated by their usefulness in the processes of adaptation (e.g., amino acid type, enantiomeric excess of certain monomers, or presence of lipids, among others) (Neveu et al., 2018).
FIG. 2.
List of Mars exploration missions carrying payload instruments focused on organic and chemical detection of (bio)molecules. Pathfinder and MERs are not mentioned, as missions to other planets or moons are not included, because their payload was not intended to detect organics. See additional details in Box 1. Color images are available online.
Within this framework, we propose to focus on the identification and characterization of the eventual (bio)chemical complexity developed on the planet or satellite under study, by using a general chemical approach that also allows the detection of different biomonomers, biopolymers, and cell-like structures that are common to all known terrestrial life. By being capable of detecting these distinctive levels of chemical organization and complexity, it becomes possible to infer a highly advanced and mature prebiotic chemistry or even an active biochemistry operating in a given extraterrestrial life.
The detection of (bio)chemical complexity would be based on the measurable parameters specified in Table 2. Given this novel framework for searching for (chemical evidence for) life, we propose the following three key goals for a hypothetical mission designed to search for universal features of life: (1) searching for signs of higher-order organization and/or compartmentation of chemical systems; (2) searching for complex carbon-based chemistry and signs of potential conformational plasticity of polymeric molecules based on noncovalent interactions; and (3) searching for biomolecules (both low MW compounds and polymers) to compare with those of terrestrial life.
Table 2.
Science Traceability Matrix of Our Proposed Concept Scheme
| Objectives | Measurable | Technique/Instrument | Interpretation | Implications | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Compartmentalization, organization | Morphology | Microscopy | Supramolecular structures. Vesicles, cell-like forms | Y | Terrestrial-like life | Y | CPC-Complex prebiotic chemisry. Potential generic life | Y | Ultra-structure compatible with life | N | Complex prebiotic chemisry. Potential generic life |
| Carbon chemistry and potential metabolism | Vibration of Carbon bonds+Intramolecular noncovalent forces | Raman spectros-copy from 15 to 1800 cm−1 | Prebiotic and/or biotic chemistry Complex molecules with conformational plasticity |
Y | Y | N | Y | ||||
| Biomolecules | Bio-affinity compatible Structure recognition | Biosensor/SOLID | Prebiotic, biochemistry and potential molecular biomarkers | Y | N | N | N | ||||
Implications: Y means positive detection; N, negative. Bold indicates the main conclusion that could be drawn from each set of Y/N results.
CPC = Complex Prebiotic Chemistry.
Such goals can be addressed by identifying several key parameters with specific technologies: (1) Morphologies compatible with potentially compartmented biochemical systems can be recognized by optical microscopy techniques. (2) Covalent bonds between carbon and other elements (such as C-C, C-H, C-N, C-O, C = O) can be detected through their Raman spectroscopic vibration features. In addition, the existence of intramolecular and noncovalent bonds (hydrophobic interactions, hydrogen bonds, Van der Waals forces, and electrostatic interactions), which are characteristic of polymeric molecules, can be also inferred using the same technique, by accessing the low-frequency spectral range. And (3) molecules and chemical structures belonging to either the Complex Prebiotic Chemistry or Complex Bio-Chemistry stages of the transition from chemistry to biology can be identified with the use of bioaffinity-based biosensing platforms (e.g., antibody- and aptamer-based biosensors).
3. Concept of the CMOLD Instrument Suite Design
To fulfill the ambition for in situ detection of different levels of (pre)biotic chemical and structural complexities in selected bodies of the Solar System, we propose an instrument suite based on a microfluidic block that distributes a sample in liquid suspension to the three technologies mentioned earlier and is detailed in the next paragraphs.
3.1. Microscopy
3.1.1. Goals and challenges of microscopy in space exploration
The first and most obvious instrument to be included in the proposed suite is a microscope that is able to resolve evidence for life at sub-micron length scale. The use of microscopy in space exploration requires very fast, robust measurements, ease of use, and contactless systems. So far, only a few microscope instruments have been included in Mars exploration missions: the microscope on the 2003 Mars lander in ESA's Beagle 2 mission, with a resolution of circa 4 μm/pixel with a depth of field of 40 μm (Thomas et al., 2004); the optical microscopy of the MECA instrument on board NASA's 2007 Phoenix lander, with a maximum resolution of 16 μm/pixel and a field of view of 2 × 2 mm (Hecht et al., 2008); and the Close-UP Imager (CLUPI) instrument included on ESA's 2022 Rosalind Franklin Mars rover, which will offer a maximum resolution of 7 μm/pixel at a distance of 10 cm from the target (Josset et al., 2017). Aiming for resolutions below 1 μm, those relevant for the detection of cell-like structures, is therefore already an identified challenge.
The microscope should be able to detect structures that are 0.5 μm or larger, a size chosen based on cell-size distribution of microbial life in Earth's ocean and polar brines (with a diameter ranging from 0.2 to 3 μm). Also, 0.5 μm is the average of Ross Sea bacterial and archaeal sizes (La Ferla et al., 2015), though ultra-small cells have been reported in Antarctic lakes as a result of environmental stress or life cycle-related conditions (Kuhn et al., 2014).
In microscopy techniques, the main goal is to get as much information as possible from the images of the sample, ideally with a very high resolution and contrast sensitivity. Limitations are self-evident, especially for an instrument in space, as performance comes at a cost in complexity and size. Further, just increasing the instrument imaging resolution may not be sufficient, due to the fact that information on chemical composition of the imaged feature, as well as its eventual growth or dynamics, would be necessary for assessing its biological origin. In this context, the main performance requirements for any microscope are to be able to provide information on size, shape, and mobility of the particulates detected. Conventional optical microscopy is, nevertheless, limited in resolution by the wavelength of the light that illuminates the sample. Other microscopy techniques, known as super-resolution microscopy (Stockert and Blázquez-Castro, 2017), can reach resolutions of fractions of the illuminating wavelength, but they represent a huge increase in complexity and require high processing power.
3.1.2. The microscope in CMOLD
The main caveat of microscopy for life detection missions is that morphology alone could not provide definitive criterion for biogenicity of life on another planet not known a priori to be inhabited. CMOLD will cope with this challenge in two ways. First, microscopy in CMOLD will be used in combination with other techniques, such as Raman spectrometry, that will help us to determine the chemical composition and structure of the samples. Nadeau et al. (2018) already pointed out that even the best optical instrument will likely need to be coupled to additional instruments to eliminate ambiguity between the abiotic and biotic origin of relatively complex chemical structures. Second, microscopy in CMOLD will be able to discriminate very efficiently not only between rounded forms and those contained in vesicles but also between static and mobile organisms, thus allowing differentiation between indicators of geological or biological activity.
Our microscope as part of CMOLD will be configured in a fixed focus arrangement that ensures the depth of focus to avoid complex mechanisms and moveable parts to bring the specimen into focus. Digital holographic microscopy (DHM), a new method that digitally implements the holography principle, is particularly suitable for CMOLD. With the current power of computer systems and the rapid development of digital cameras, recorded holograms can be interpreted numerically at video frequencies to simultaneously provide: (1) phase information, which reveals the surface of the object with vertical resolution at nanometric scale along the optical axis; (2) intensity values, as obtained by a conventional optical microscope.
The main advantage of the DHM technique lies in the use of the so-called off-axis configuration, which allows all information to be captured by a single image acquisition, that is, typically in the order of microseconds, and can be monochromatic and with low bit depth. These extremely short acquisition times make DHM systems insensitive to vibrations and allow the acquisition of images with enough quality at a frame rate that makes it possible to observe mobile biological specimens. In addition, the technique proposed does not require a focusing mechanism nor digitally processing onboard, since the reconstruction of the object can be achieved digitally on the Earth by analysis of the interference pattern. Indeed, processing and compressing data onboard will be implemented if needed in case the mission can only achieve a modest data relay rate to the Earth, for example, from a distant jovian or saturnian moon. All these advantages make DHM a very promising technique for CMOLD (Alm et al., 2013).
Another technique that can be implemented and assessed in CMOLD is based on the complementation of the conventional microscope with a digital micro-mirror device (DMD) in the illumination path (Fig. 3, left). Arbitrary patterns can be created on the DMD and projected onto the sample. This patterned illumination can be used to improve lateral and axial resolution over the resolution of a wide-field microscope, as well as to reduce the incoming illumination dose. Different illumination patterns require specific reconstruction strategies and result in an image quality similar to that of confocal or structured illumination microscopy (Dan Dan et al., 2013). Other techniques for illumination patterning, such as diffractive optical elements, can also be considered for CMOLD.
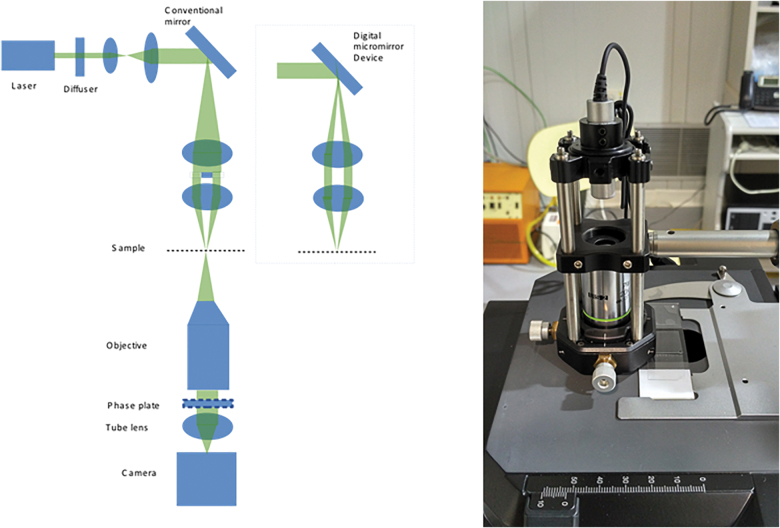
FIG. 3.
Left: Baseline wide-field optical microscope with phase-contrast capability. In the dashed box, details on eventual implementation with DMD for the illumination section to project arbitrary patterns on the sample and improve the resolution. Right: Example of a DIHM system consisting of a laser source (central wavelength at 450 nm) whose beam is focused by a microscope objective (20 × ) in a 2 μm diameter pinhole (used for spatial filtering purpose). The sample under test is placed after the pinhole. The laser beam undergoes a phase shift when reflected by the presence of eventual particles within the sample, creating a reference beam. All the beams interfere and create a holographic pattern that is detected by a CMOS detector. This pattern can be processed to detect location, sizes, and number of particles, among other parameters. DMD, digital micro-mirror device. Color images are available online.
The ability of the DMD to work at high speed can also be explored to modulate the illumination light field, by producing optical trapping effects that can be used to further manipulate the particles under observation, fixing them in the focal plane, or rotating them. Besides optimizing the imaging process, such an ability to modulate illumination can also be used to further explore the information carried by the scattered radiation.
The analysis of the mobility of the particles in the sample will be achieved by acquiring several images at reasonable video rates, and searching for differences between those images, therefore discriminating between static (maybe with Brownian motion patterns) and motile components within the sample. The capture of the images at video frequencies can be stored onboard and transferred to the Earth for postprocessing and analysis, based on the reconstruction of the object from the interference pattern by a digital holographic technique. The images can be monochromatic with a low level of digital output.
We suggest that the baseline parameters to be included in the wide-field microscope with phase-contrast capability in CMOLD (Fig. 3, right) should be as follows: (1) a sample illumination device, sharing the same light source as the Raman spectrometer (monochromatic laser excitation Z = 532 nm); (2) fixed-focus infinity-corrected objective; (3) large numerical aperture (NA) of 0.75; (4) resolution: better than 500 nm; (5) field (for a 40 × magnification of view of 250 × 165 μm); and (6) a 2K × 2K charge-coupled device (CCD) or CMOS monochromatic imaging device.
3.2. Raman spectroscopy
3.2.1. Goals and challenges of Raman spectroscopy in space exploration
A Raman spectrometer will fly on a planetary mission for the first time with the Rosalind Franklin (Rull et al., 2017) and Mars 2020 (Beegle and Bhartia, 2016; Wiens et al., 2017) rovers. Such an instrument has been used extensively in the laboratory in recent decades and more recently during field campaigns. A Raman spectrometer working at frequencies from 15 to 1800 cm−1 can detect signatures of organic molecules of different sizes and complexities, including the presence of weak noncovalent bonds (ionic, Van der Waals, hydrophobic, hydrogen bonds) in biopolymers and their aggregates, and in particular the secondary and tertiary structures that account for the functionality of proteins and nucleic acids, with a spectral resolution of 1 cm−1 (Maiti et al., 2004).
Recent advances in microfluidic systems allow miniaturizing and integrating various processes involved in biological analysis and chemical engineering. These advances have led to the development of new systems of lab-on-a-chip devices for analyzing traces of organic substances. In such miniaturized systems, interfacial phenomena have a noticeable effect on the liquid's flow structure and chemical properties due to the large surface-to-volume ratio (Persichetti et al., 2017). Thus, using the Raman scattering from molecules inside micro channel paths, it is possible to analyze the chemical contents of different substances.
One identified challenge when using Raman spectroscopy for looking for signs of life in planetary missions is the masking of biogenic molecules by inorganic ions (e.g., Böttger et al., 2012, see section 4.1). To resolve this problem, a previous dedicated analysis of the region to be studied is a necessary task to be carried out before the analytical instruments arrive at the place of interest. Based on the results obtained predeployment of instruments, the mineral compounds expected to be found and all other related geo-chemical processes in the area will be analyzed.
In addition, decades of exploration in terrestrial analogs, including Raman studies, have demonstrated that, when biogenic traces are found in mineral samples, their proportion is quite low with respect to the main mineral composition. This is why several different spots on the same sample should be analyzed to put the results obtained into context. Finally, to maximize the science return, some dedicated algorithms for improving all spectra obtained with the Raman instrument need to be included, such as fluorescence removal, integration time optimization, and baseline adjustment. All these onboard operation tools will enable the Raman instrument to obtain much cleaner and more interpretable spectra, reducing masking effects or ambiguous results.
3.2.2 Raman spectrometer in CMOLD
A typical Raman spectrometer is composed of three main subsystems: (1) a laser for exciting the sample; (2) two separate optics systems: first, a focusing optics system for concentrating the optical signal at the required irradiance on the sample, and second a collecting optics system for capturing the Raman signal; and (3) a spectrometer to measure the peaks that allow the identification of bands or spectral features of the sample.
3.2.2.1. Laser
The Raman spectrometer to be included in CMOLD will be equipped with a 532 nm pumping laser for exciting the samples. We have developed and validated a laser for space applications (Fig. 4, top left), in which is part of the Raman Laser Spectrometer (RLS) instrument onboard ESA's Rosalind Franklin rover (Rull et al., 2017), as well as the RAX (RAman spectrometer for MMX) instrument for JAXA's MMX (Martian Moons eXplorer) mission. The laser generates a 532 nm monochromatic beam of exciting photons with 0.6–1.2 kW/cm2 irradiance on the sample, limited to 0.3–0.6 kW/cm2 in the RLS instrument to ensure compatibility with other instruments on the rover, particularly MOMA, covering an illuminated total volume of 50 × 50 × 50 μm, which is highly appropriate for microfluidic applications. We selected the 532 nm excitation wavelength, because most of the recent studies and comparative databases in terrestrial conditions use this excitation wavelength (Wang, 2012), which is now included as standard configuration in research and miniaturized equipment.
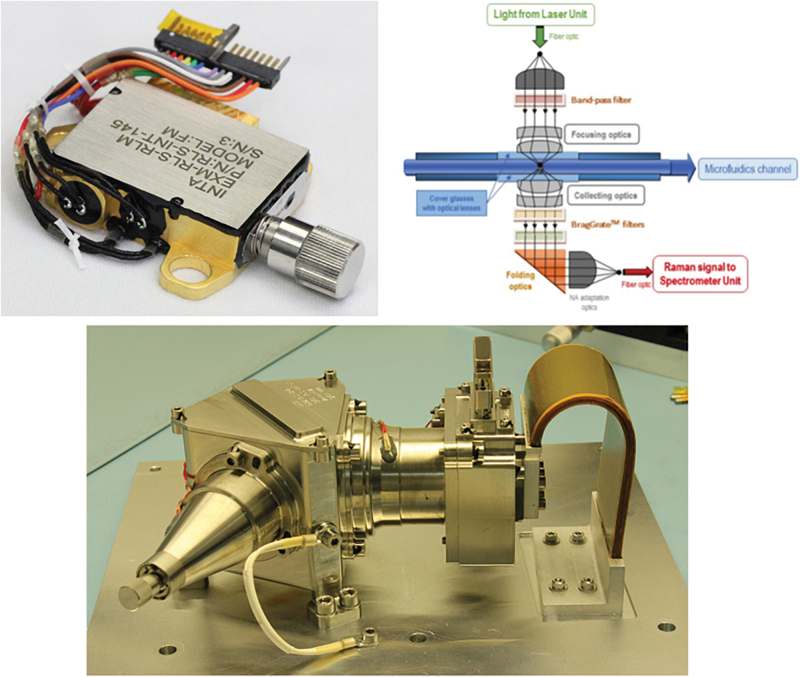
FIG. 4.
Top left: Laser head unit of the Raman spectrometer (based on the RLS instrument onboard ESA's Rosalind Franklin mission). Top right: Optical head of the Raman spectrometer. Bottom: Raman spectrometer designed at INTA-CAB for the ESA's Rosalind Franklin mission. RLS, Raman Laser Spectrometer. Color images are available online.
Some authors have also used ultraviolet (UV) excitation in biochemical applications (Asher, 1993; Sapers et al., 2019), due to the fact that many biomolecules have chromophore groups that absorb the UV wavelength; thus, the limit of detection and quantification can be increased when resonance conditions are fulfilled (Resonant Raman Scattering [RRS]). This technology is also the basis of the SHERLOC instrument (Scanning Habitable Environments with Raman & Luminescence for Organics & Chemicals), included in NASA's Perseverance rover (Beegle and Bhartia, 2016).
This configuration is not yet widespread in terrestrial applications, and the number of relevant studies is still limited. In addition, the interpretation of the RRS spectrum is usually more complex than that of the conventional Raman spectrometer, since the increase in spectral intensity is highly selective, as it occurs only for those molecules that are able to become resonant with the incident excitation. Therefore, although it is possible that very strong Raman signals might be detected for certain molecules using RRS, molecules that are not resonant would not be detected at all.
3.2.2.2. Optics systems
In the proposed instrument suite CMOLD, the laser light is guided to the liquid sample by means of a fiber optics bundle that, in combination with a special focusing optics system (Fig. 4, top right), will concentrate the light directly within the microfluidic channel. This Raman optical head is based on a transmission concept, in which the pumping laser is located in the focal point of a collimated lens that injects the 532 nm beam to the internal flow channel through an aspheric optical system.
The advantage of this strategy is that the Raman scattering of the material that covers the cell can be practically excluded in all cases. The pumping path incorporates a special band-pass filter that allows transmission of only the 532 nm wavelength, while blocking the 1064 nm (first harmonic) and the 808 nm (corresponding to the pumping system) wavelengths. The focusing optics concentrate the beam on the cell containing the sample, thus producing an increase in the irradiance to obtain enough intensity of the Raman signal. Several tests aimed at determining the detection capability of the Raman technique on fluids have been completed with different models of the RLS instrument. These tests not only demonstrated the capability to detect low concentrated perchlorate dissolutions (up to 0.02 M) but also showed the capability to determine the molarity of the measured sample, since the instrument response was linear with concentration (Moral et al., 2019).
The collecting path should be designed considering the refractive index of the microfluidic liquid to get an NA close to 0.85 (immersed lenses). This path is then collimated by a spherical microlens. In this optical system, a minimum of two BragGrate™ filters will be used for getting an ultra-low frequency Raman spectrometer with a minimum range of 15 cm−1, which is appropriate to study 3D features of biological polymers (Ronen et al., 2018). These filters should be tilted nearly 20° to obtain the high rejection factor required to ensure the closest spectral range to the pumping wavelength.
The subsequent optical system (NA adapting optics in Fig. 4, top right) allows adapting the final 0.22 NA required by the optical fiber to transport the spectral flux to the spectrometer, thus diminishing the loss of signal as much as possible. To excite higher volumes of liquid suspension on the microfluidic channel and to get a faster analysis of the sample, it is possible to incorporate some cylindrical lens on both, focusing and collecting systems. The plan is to incorporate anamorphic projection lenses on both optical paths to access larger illuminated areas on the microfluidic channel of the order of 50 × 400 μm (as a target).
Using a fiber optics bundle, the collected light can be directed toward a small spectrometer that collects the Raman peaks of the sample on the detector. The Raman signal is much lower than the Rayleigh-scattered signal onto the CCD detector, because the probability of inelastic collisions is only ∼10−8. In general, the wavenumber shifts are independent of the frequency of the incident light but, to have a good SNR on the electronic chain, the 532 nm wavelength is considered optimal.
3.2.2.3. Spectrometer
A spectrometer is required in CMOLD for recording the Raman spectrum of the potential biomolecules contained in the sample. This technique is well established, and we have developed small spectrometers (already on ESA's Rosalind Franklin rover, ready to be launched in 2022), based on volume holographic grating (WHG) with a spectral resolution of 0.2 nm in the range from 535 to 675 nm wavelength (Fig. 4, bottom) (Rull et al., 2017). The Raman spectrometer to be included in CMOLD will contain a 2048 pixel detector, a built-in 16-bit digitizer, a USB 2.0 interface, and a cooled (the temperature can be easily regulated and maintained by means of a thermoelectric cooler device) CCD for higher dynamic range. This would produce significantly reduced dark counts and the long-term operation stability indicated for low-light level detection and long-term monitoring applications, thus being ideal for in situ planetary exploration.
The spectrometer should be adequate for most Raman applications with spectral configurations from 200 nm to 1050 nm, with resolutions between 0.2 and 4.5 nm, respectively. The advantage of using this kind of spectrometer is that it can be modified to obtain the range from 15 to 3800 cm−1. This configuration would allow recording some additional carbohydrates (C-H stretch) and hydrogen-bonding-based (O-H stretch) features in the potential biopolymers in the analyzed sample. However, such spectral features are less specific than the amide bands.
The incorporation of a second spectrum channel, that is, a second fiber displaced laterally with respect to the central one, can produce a second Raman line to be acquired in the CCD. This approach allows analyzing two regions of the microfluidics system at the same time, if required, or it can be used to compare two Raman spectra with different concentrations of components to select a differential Raman technique. In any case, the classical Stokes solutions will be studied, as they are more intense than those obtained with the anti-Stokes alternatives. Water-based solutions do not significantly affect the Raman signal (though they disturb the infrared spectra), thus the proposed analysis of biomolecules in microfluidic devices can share the spectra database already built for the analysis of solid samples in the RLS instrument onboard the Rosalind Franklin rover.
3.3. Biosensor for biomarker detection
3.3.1. Goals and challenges of biomarker detectors in space exploration
The instrumentation deployed on planetary exploration missions for the detection of organic molecules (only on Mars, so far) has been primarily based on the analysis of heat-extracted volatile compounds (GC/MS) (Klein et al., 1976; Cannon et al., 2012; Ming et al., 2014), a methodology that, due to its limited sensitivity, may be blind to the presence of low levels of organics (Navarro-González et al., 2006; Montgomery et al., 2019). We note, however, that SAM and MOMA have a nonpyrolysis mode, that is, derivatization with MTBSTFA. Regardless, the taphonomy of parent molecules is often lost when using the highly destructive pyrolysis approach. We propose here the alternative strategy of flying a biomarker detector equipped with a bioaffinity-based sensor that is capable of detecting life-related and nonlife-related organic molecules with a wide range of specificities, using liquid suspensions of the powdered samples under study.
The goal of the biomarker detector would be screening for chemical species of different size and complexity, including prebiotic (i.e., nonlife-related) universal organic molecules and others that are comparable (even highly similar) to molecular biomarkers of terrestrial-like life. The most obvious identified challenge of biosensors, particularly immunosensors, is that they are highly specific. However, they may also have a relatively high versatility to recognize structures that are similar and universal for different life-forms, such as the d or l chemical group on the aromatic amino acids (Kassa et al., 2011; Moreno-Paz et al., 2018), or hydrophobic stretches of proteins (regardless the aa sequence). Also, it is possible to produce antibodies (Abs) or aptamers (Aps) to detect xenobiotic (nonlife derived) molecules.
It is true that having relaxed-specificity binding in the biosensing receptors hinders information about the precise molecules they are detecting, but at least the biosensor will infer the type of molecules and their relative size by using a “sandwich-type” assay. By means of combinatorial chemistry and bioinformatics, it is possible to infer the most probable combinations of the chemical structures, comparing them with known prebiotic chemistry and developing specific sensors (Abs or Aps) for their detection.
Our biosensor approach in CMOLD is the best complement to GC/MS and Urey-type instruments. On one hand, although GC/MS narrows its targets through the column resolution and the mass spectra library for peak identification, biosensors select the most universal potential (bio)chemical targets in each planetary environment. On the other hand, the Urey instrument (Table 1) is based on capillary electrophoreses and represents a very powerful fluidic solution for detecting amino acids and other small amino-containing molecules. However, as GC/MS, or even MOMA, its resolution is limited by the size of the compounds that can be detected: The lower the size of the molecule, the lesser the potential (bio)-chemical information on it. That said, Urey would be, indeed, a good complement for biosensing-type systems: The former informs about potential monomeric material, whereas the latter would provide clues about their presence in the form of bio-polymeric forms.
Here, we advocate leveraging the strong SOLID design and testing heritage (Parro et al., 2005, 2008, 2011a, 2011b; Moreno-Paz et al., 2018) to detect nonvolatile organic molecules, as either free monomers and polymers or being part of supra-macromolecular structures (including membranes, vesicles, and nano-micro particles) in liquid suspension. We have developed the SOLID instrument and it is one of the payload instruments in NASA's Icebreaker proposal mission to Mars (McKay et al., 2013). In SOLID, a set of up to 200 specific molecular probes (namely, antibodies and aptamers) can interrogate for the presence of a similar number of compounds (including prebiotic organic molecules, biochemical and microbial markers), in the samples previously extracted and processed.
The latest version of the SOLID instrument (Parro et al., 2011b) has the following specifications: (1) sample volume of the liquid suspension to be analyzed: 100–500 μL; (2) target molecular size: from aromatic amino acids (such as Phe, with a MW of 165 Da) to large and complex polymers (typically, with MW in the range 10–100 kDa), cell debris (<0.1 μm in size), and cells (0.5–1 μm); (3) sensitivity: in the range of 1–10 ppb (ng/mL); (4) reuse: for at least 10 times without relevant loss of performance in a short time slot, which is particularly useful when there is no positive result in one assay or there are only a few positive signals, and then LDChip can be re-used directly without additional treatment; and (5) number of different target molecules simultaneously interrogated: at least 200.
3.3.2. Biomarker detector in CMOLD
Our concept of the biomarker detector to be included in CMOLD consists of four main components, for which we have accumulated an extensive heritage thanks to the design, building, and testing of three prototypes of the SOLID instrument (Fig. 5, top) (Parro et al., 2005, 2008, 2011a, 2011b). The components are: (1) a microarray containing bioaffinity-based sensor elements (antibodies and aptamers); (2) the supporting substrate, which is a wave-guide; (3) an optic package composed of a faceplate, an optical filter, a CCD, and a laser beam or diode laser; and (4) a microfluidic cartridge with chambers, channels, and valves.
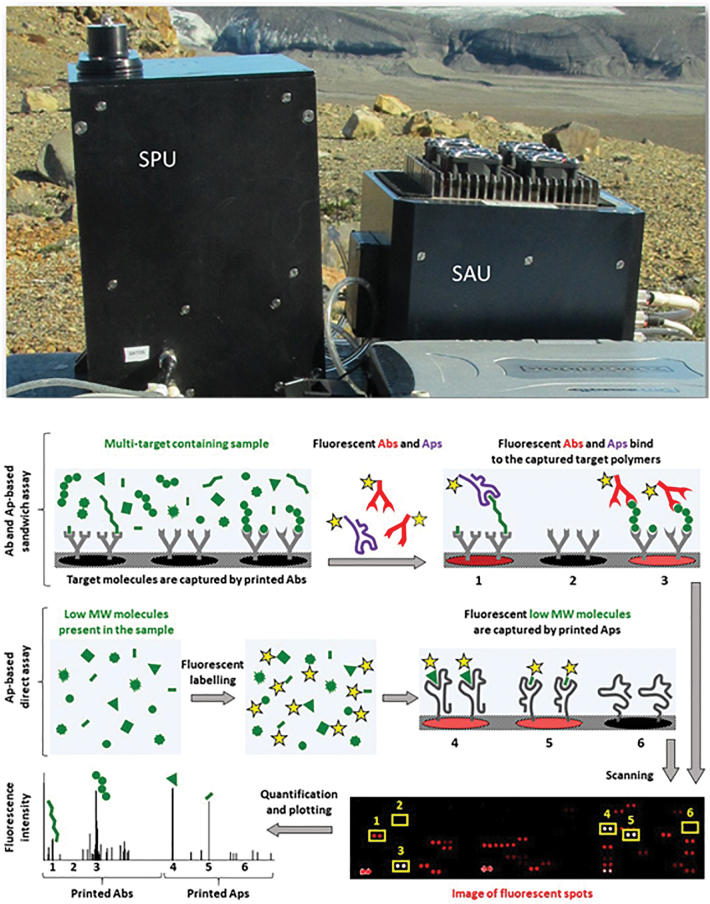
FIG. 5.
Top: SOLID V3.1 showing the single extraction cell SPU and the SAU with LDChip in a field campaign. Bottom: Schematic representation of the FSI protocol, combining the Ab- and Ap-based sandwich assay (top panel) and the Ap-based direct assay (medium panel) to detect the presence of biopolymers and low MW biomolecules, respectively. Spots on the biosensor microarray contain the capturing probes (Abs or Aps, depicted in gray). For analysis in the sandwich format, first a liquid suspension extract of the sample is incubated with the microarray for 1 h at 37°C, and thus the capturing probes printed in the spots bind to the target molecules present in the sample (green). After incubation and washing (to discard the unspecific interactions), the microarray is flooded with fluorescently labeled Abs and/or Aps that specifically bind to epitopes of the polymeric targets already captured by the spotted probes, thus forming a sandwich. In turn, the direct assay relies on the specific interaction of the (previously labeled) low MW biomolecules (including monomers of the interrogated biopolymers) with Aps printed in the corresponding spots of the microarray. Fluorescence is excited with a laser (bottom panel), and spots that contain the detected targets are identified by a bright signal in a CCD image. SI provides the highest level of specificity and sensitivity for the biomolecules used as biomarkers of Earth-like life. Based on test analyses with natural samples, we estimate a precision better than 10% and accuracy better than 15% for the detection of large organic molecules with such a sandwich immunoassay (Rivas et al., 2008), and similar values are expected for the direct assay. Functionally, the limit of detection for each biomolecule corresponds to a fluorescent signal that is two times (2 × background standard deviation) above the background signal. Ab, antibody; Ap, aptamer; CCD, charge-coupled device; FSI, Fluorescence Sandwich Immunoassay; MW, molecular weight; SAU, Sample Analysis Unit; SPU, Sample Processing Unit. Color images are available online.
3.3.2.1. The molecular-affinity-based detection principle
Living beings are characterized by possessing large and conformationally adaptable polymers (mainly proteins and nucleic acids, as well as polysaccharides). They can interact with each other and also with small molecules by means of noncovalent bonds, to form stable, dynamic, and specific complexes. Indeed, this is the basis of the biochemical networks that support life. The affinity and specificity of these interactions is highly variable in nature.
One of the most relevant examples of affinity-based detection in current biology is that of the recognition of antigens (Ags) by antibodies. An Ab or immunoglobulin is a large, Y-shaped protein produced by certain cells of the immune system of some taxonomic classes of animals (including fishes, amphibians, reptiles, birds, and mammals) to neutralize toxins or pathogenic microorganisms by specifically recognizing a unique molecule or epitope of the target, called the Ag. The affinity of Abs for their Ags (measured as their equilibrium dissociation constant, Kd) typically lies in the nanomolar range (from 0.01 to 100 nM). In turn, the Ab-Ag specificity is very variable: Some Abs are specific enough to recognize even each enantiomer (d and l) of a chiral Ag (Hofstetter et al., 2005), whereas others are relatively unspecific and can, for example, bind hydrophobic protein stretches regardless of their amino acid sequence and structure.
In turn, Aps are RNA or single-stranded DNA (ssDNA) oligonucleotides selected in vitro from a large library of synthetic random oligonucleotides, which can bind to their target molecules with affinities and specificities analogous to those of the Ab-Ag pairs (Stoltenburg et al., 2007; Briones, 2015). Aps are produced by using a cell-free, amplification/selection method known as Systematic Evolution of Ligands by EXponential enrichment (SELEX), which was developed in parallel by two groups in 1990 (Tuerk and Gold, 1990; Ellington and Szostak, 1990). A growing number of different Aps have been in vitro selected and are able to recognize a broad range of targets, including low MW organic molecules, amino acids, nucleotides, antibiotics, mono- and oligosaccharides, peptides, proteins, nucleic acids, macromolecular aggregates, viruses, and even whole cells or tissues.
In particular, due to their smaller size in comparison with Abs, Aps show better performance than Abs for the recognition of low MW compounds, without requiring their prior binding to any kind of conjugate molecule (Cho et al., 2009; Kim and Gu, 2014). Also, Aps can be in vitro selected against both toxic targets and low immunogenic compounds, which cannot be recognized by antibodies raised in experimental animals. As nucleic acid chemistry is directly applicable to Aps, they can be chemically modified at one of their 5′ or 3′ ends, thus allowing their straightforward immobilization onto a large variety of solid substrates (including glass, plastics, beads, nanoparticles, activated graphene, or other nanomaterials), and similar assay formats than those used with Abs can be used. As a result of the features mentioned earlier, Aps are being increasingly applied to the fields of biotechnology and biomedicine (Sun and Zu, 2015) and a number of Ap-based biosensors (also called aptasensors) are currently available in different formats, some of which are based on label-free detection methodologies (Cho et al., 2009; Sharma et al., 2017; Moreno, 2019).
Interestingly, a recent report has addressed the resistance of Aps to cosmic rays and other sources of radiation at different energies and doses, using ground-based facilities as well as during the EXPOSE-R2 mission outside the International Space Station (Coussot et al., 2019). The authors found that the functionality of the tested aptamers was not affected after being exposed for 588 days to the outer space in EXPOSE-R2, where they accumulated a radiation dose of 220 mGy, equivalent to that expected during a mission to Mars.
3.3.2.2. Fluorescence Sandwich Immunoassay for detecting biopolymers and large molecular complexes
In a Fluorescence Sandwich Immunoassay (FSI) (Fig. 5, bottom), selected Abs recognize and bind in vitro to the desired target biomarker with high affinity and specificity, similarly to how the immune system of an animal recognizes in vivo the epitopes present in the molecules and cells that are foreign to it (Huang and Honda, 2006). An epitope consists of either a 3D surface feature of a specific biomolecule (conformational epitopes) or a specific sequence of monomers such as amino acids in a peptide or protein (linear epitopes). Using FSI, the presence of a given biomarker showing at least two epitopes (e.g., a relatively complex biomolecule or a biopolymer) can be revealed by means of a two-site noncompetitive (sandwich) immunoassay coupled to fluorescence-based detection, in such a way that the relative abundance of different biomarkers can then be estimated by comparing their corresponding fluorescence intensities (Parro et al., 2005, 2008). Processing the sample at high temperature is not required, thus guaranteeing the integrity and 3D structure of the biomolecules present in it.
The FSI can be performed in microarray format, where fluorescence detection occurs at purposely designed reaction spots, each one containing a different kind of Ab immobilized on it. A blank assay is typically run before sample analysis to obtain a baseline fluorescence signal, and a number of different control reactions can be performed. In the sandwich format (Fig. 5, bottom), the sample (typically, a liquid suspension) is incubated with the microarray, the unspecific reactions are washed away, and a previously designed mixture of soluble and fluorescently marked Abs is added to the reaction chamber.
Thus, the labeled antibodies can bind to certain epitopes present in the target biopolymers that were previously recognized by the immobilized capture antibodies, through similar or different epitopes. As a result, the microarray produces a characteristic immunopattern, that is, a set of positive immunoreactions to different compounds, thus indicating the presence of multiple biomarker epitopes in a sample. We have developed a collection of more than 300 polyclonal and monoclonal Abs that are ready to be implemented in planetary exploration missions (Parro et al., 2011b). This allows adapting the library of Abs to specific mission objectives (e.g., search for subsets of biomarkers on Mars or in icy moons), with minimal impact on hardware.
We have also been able to detect certain low MW compounds (such as hopane) by using Abs in a competitive approach. For that, the reactive spots on the microarray are first coated with conjugate molecules, that is, large molecules carrying the small target compound to be detected (e.g., bovine serum albumin [BSA] bound to hopane). The microarray is flooded with fluorescent Abs, which specifically bind to such conjugates. An image of the microarray is obtained after exciting the fluorescent marker with a laser: Such an image represents 100% of positive Ab-Ag coupling and is used as a baseline.
Next, the sample to be analyzed (in our example, containing hopane) is mixed with fluorescent Abs on the microarray. Both free hopane and the BSA-hopane conjugates compete for the fluorescent Abs, and the former inhibits the Ab binding to the conjugate spots. Then, a second image of the microarray is obtained after exciting fluorescence: The signal intensity retrieved from the corresponding spot will be inversely proportional to the amount of free hopane in the interrogated sample. Based on test analyses with natural samples, we estimated a precision to within 15% and accuracy to within 10% for the detection of the tested low MW molecules, showing that a 10–15% fluorescence loss is required to achieve a lower limit of detection for specific compounds (Fernandez-Calvo et al., 2006; Blanco et al., 2013).
We have been using FSI for many years, both in the laboratory and in multiple field campaigns, and the sensitivities of detection obtained range from 0.1 to 10 ppm for biochemical compounds and 103 to 104 cells or spores per milliliter (Fernandez-Calvo et al., 2006; Rivas et al., 2008; Parro et al., 2011a; Blanco et al., 2013; Lezcano et al., 2019; García-Descalzo et al., 2019; Sánchez-García et al., 2020).
3.3.2.3. Direct and competitive aptamer-based assay for detecting low MW compounds and small epitope-containing polymers
We have developed Aps to specifically bind different kind of biomolecules, including structured nucleic acids (Sanchez-Luque et al., 2014), proteins (Moreno et al., 2019), and low MW compounds such as amino acids and antibiotics. We have also used some of our protein-specific ssDNA aptamers as bioaffinity probes to develop graphene-based aptasensors (Bueno et al., 2019). In parallel, we have accumulated experience in the use of nucleic acid-based microarrays for the characterization of fluorescence-labeled, structured RNAs (Fernández et al., 2011; Romero-López et al., 2014) as well as for mutation screening in bacteria (Garrido et al., 2006) and viruses (Martín et al., 2016).
In our CMOLD concept, Ap-based biosensing will be used to detect biomonomers and other low MW compounds, including those that only have one available epitope, by means of either direct or competitive/inhibitory assays. Direct detection of low MW biomarkers requires the fluorescent labeling of the sample to be analyzed, so that the measured fluorescent signal will be directly proportional to the amount of biomarker in such a sample (Fig. 5, bottom).
In turn, in competitive assays the basic scheme will be as follows: (1) Fluorescently labeled biomarkers (previously known and available as control samples) react with their corresponding Aps immobilized onto the biosensor surface, and the basal fluorescence signal is measured at each spot; (2) an unlabeled sample is incubated with the biosensor; (3) the competition between unlabeled and labeled biomarkers for their recognition domains in Aps results in the displacement of a fraction of the labeled biomarkers initially bound to the Aps; (4) the fluorescence signal detected at each spot of the biosensor will, thus, be directly proportional to the amount of the corresponding target molecule in the sample. Therefore, Ap-based, competitive biosensing is the preferred way to detect biomarkers present at low concentrations in the interrogated sample, as it will likely be the case when analyzing liquid suspensions collected on Mars or on icy moons by rovers and/or landers.
In addition, fluorescently labeled aptamers can also be used instead of (or together with) fluorescent Abs in FSI to reveal the presence of an epitope in a biomolecule, once another (similar or different) epitope has reacted with the specific capture antibody (Fig. 5, bottom). Therefore, tunable combinations of Abs and Aps, both capture probes and labeled, can be tested in our biosensor format.
A different and more complex Ap-based approach with potential applicability in astrobiology has been recently published, which relies on mixing a large, randomly generated library of folded Aps (of unknown sequences, and whose affinity and specificity has not been previously tested) with a sample, assuming that complex biological materials (such as a cell membrane, highly complex biomolecules, and even unknown nonterran biosignatures) will be able to be recognized by a higher number of Aps, whereas inorganic substrates will barely interact with any aptamer in the mixture.
The proposed method would require the bound Aps to be recovered from their target, polymerase chain reaction-amplified, and finally sequenced (or, alternatively, fluorescently labeled and hybridized to an oligonucleotide microarray) in situ. This would reveal a “fingerprint” of Ap sequences corresponding to the detected biomarker (Johnson et al., 2018). Although the interest and novelty of such an Ap-based, agnostic approach for life detection is evident, its intrinsic methodological complexity is much higher than that of the one proposed here, as the aptasensor included in CMOLD is focused on the identification of known, Earth-based biomolecules (both biomonomers and biopolymers) once the detection of any kind of biopolymers (both terran and nonterran) in the collected sample has been performed by Raman spectroscopy, as detailed in Section 3.2.
3.3.2.4. Specific molecules and degradation products that SOLID can detect
Potential molecular targets for CMOLD may be those selected to represent meteoritic input, fossil organic matter, and extant (living, recently dead) organic matter, as reviewed in the work of Parnell et al. (2007), with a special focus on the potential polymeric material they can form (mainly peptides). The current list of compounds that can be detected with SOLID-LDChip comprises many of these: PAHs and derivatives such as bezo[a]pyrene and pentachlorophenol, aromatic d/l amino acids, or xenobiotics (Moreno-Paz et al., 2018); irradiated proteins, peptides, and other molecules (Blanco et al., 2018); and a variety of polymeric biomarkers from exopolysaccharide, lipopolysaccharide, humic substances, lipoteichoic acids, and peptidoglycans from bacterial cell walls, proteins from primitive metabolism, nucleic acids, and their derivatives (oxidized as 8-oxo-guanine, or thymidine dimers formed after irradiating nucleic acids with UV radiation), aromatic compounds, or short peptides (see tables from Fernandez-Calvo et al., 2006; Rivas et al., 2008; Parro et al., 2011b, 2018).
Similarly, we have developed specific aptamers to bind structured nucleic acids (Sanchez-Luque et al., 2014), proteins (Moreno et al., 2019), as well as some amino acids, antibiotics, and peptides (not shown). We have also tested the performance of aptamers as capture probes in different kinds of biosensors (Bueno et al., 2019). In a hypothetical mission, the detection of several of these structures, either with SOLID or with CMOLD, will inform about the existence of a complex chemistry that could be compatible with a carbon-based biology.
4. Building the CMOLD Instrument Suite
Design criteria aiming that in the future our instrument suite could be part of a planetary exploration payload include: (1) must be able to concentrate liquid sample and handle low liquid volumes to allow instrument miniaturization; (2) the number of samples to be analyzed by CMOLD is highly dependent on the mission and the system designed to get them, with a minimum number of two; (3) must minimize size, mass, and power consumption; (4) must be robust enough to survive a standard space environment, with special care in the design to ensure the instrument survival in an extreme radiation environment, particularly in outer Solar System's moons; and (5) modularity to simplify integration and tests.
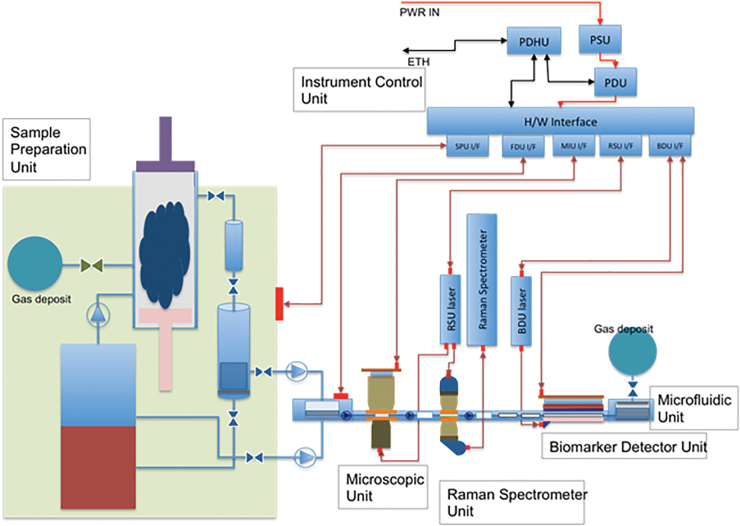
To work with the microscope, the Raman spectrometer, and the biomarker detector, any concept for instrument design envisaged to be part of a planetary exploration payload must also include three additional modules. (1) A Sample Processing Unit (SPU), which receives the soil or powdered rock (e.g., from the surface or subsurface of Mars) or ice (e.g., from the surface of Europa or Enceladus) sample and processes it properly. It must be able to prepare and concentrate the liquid suspension sample (or the melted ice) and to handle small volumes, thus allowing further instrument miniaturization. The number of samples to be analyzed by CMOLD is highly dependent on the mission concept and the system designed to collect them. (2) A Fluidic Distribution Unit (FDU), which takes the sample from the SPU and pumps it through the different sensors, controlling the flow with the help of a set of valves, until reaching the waste deposit. (3) An Instrument Control Unit with the proper hardware and software specifications.
4.1. Sample Processing Unit
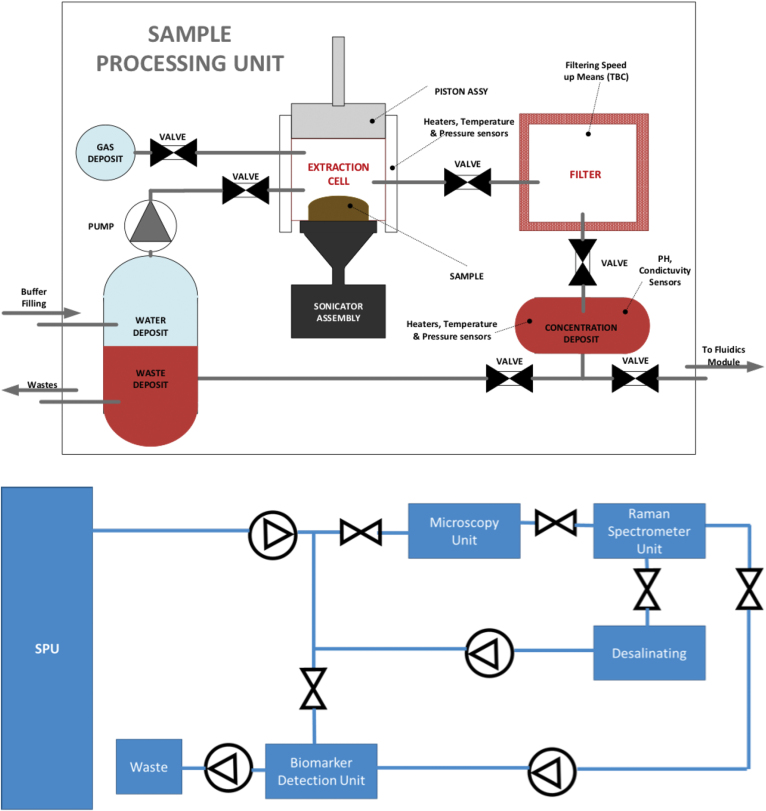
After other external equipment in the spacecraft collects the sample (drill or scoop) and inputs the sample into the extraction cell, the SPU (Fig. 6, top) receives a solid sample and processes it properly to deliver a concentrated sample to the FDU (see Section 4.2). The main functionalities for the SPU will be: (1) storing the sample (solid, liquid, permafrost) and isolating it from the external environment; (2) dilution of the sample in deionized and sterile water (temperature above 0°C); (3) homogenization of the sample by ultrasonication at 40 kHz, as the vibrations generate pressure waves in the dissolution of the processed sample causing cavitation within the dissolution, homogenizing the sample (by selecting the amplitude of vibrations, the intensity of cavitation can be adjusted, and therefore the degree of homogeneity of the mixture, covering a range that goes from soft homogenization, at low amplitudes, up to the breakdown of cells at high amplitude levels); (4) filtering the homogenized sample; (5) concentration of the filtered sample (with temperatures below 50°C) by evaporation/sublimation, sending the residual gases to a waste deposit; (6) measurement of the physicochemical parameters (pH, conductivity); and (7) delivery of the concentrated sample to the FDU module for downstream analysis.
FIG. 6.
Top: SPU functional diagram. SPU receives a solid or ice sample and processes it properly. In the end, it leaves a concentrated sample that is absorbed by the FDU Bottom: FDU block diagram. Once the FDU receives the sample from the SPU, it pumps it throughout the different sensors controlling the flow with the help of a set of valves. The sample ends in the waste deposit. FDU, Fluidic Distribution Unit. Color images are available online.
4.2. Fluidic Distribution Unit
The FDU (Fig. 6, bottom) takes the sample from the SPU and pumps it throughout the different sensors, controlling the flow with the help of a set of valves, distributing the sample through the different analytical systems in CMOLD. Due to the small volume of sample available after the treatment in the SPU, a microfluidic system is the best option to circulate the sample from one analysis system to the next. The microfluidic system must contain a series of micro-pumps and micro-valves, allowing the precise control and distribution of the fluid.
Depending on the sampling site and the type of sample, the liquid extract from the SPU might contain high salt or metal concentrations. Under these conditions, the bioaffinity interaction with antibodies or aptamers may be severely affected due to the high ionic strength. The CMOLD will use low-organic retention desalting cartridges or filters so that the potential biological polymers can move through and specifically bind to the capturing probes in the biomarker chip.
4.3. Additional elements
Five additional aspects to be considered when building CMOLD are: (1) All the modules need to be thermally controlled at a working temperature at which water solutions and suspensions remain in liquid state and the biochemical interactions required in the biosensor are allowed (i.e., typically in the range of 20–37°C); (2) the module must also be sealed to avoid any liquid leakage, a very critical point to comply with planetary protection restrictions of the mission (Fairén et al., 2019); (3) space design criteria, requiring minimization of the instrument size and mass, as well as its power consumption, also taking into account the required robustness to survive the space conditions; (4) rational design to ensure the instrument survival under an extreme radiation environment, which is particularly relevant in the exploration of outer Solar System moons; and (5) modularity, to simplify the required integration and tests during the construction and assembly (Fig. 7).
FIG. 7.
Block diagram of our instrument concept, including the complete instrument suite. Color images are available online.
In addition, our proposed suite is not meant to be the entire payload of a life-seeking mission to Mars or icy moons. It will be integrated with other instruments in the mission payload, including a suite of surface cameras to image the lander's surroundings to understand where the mission is, to choose targets to collect samples from, and to image the samples before processing them.
5. Applications of CMOLD to Astrobiological Missions in the Solar System
Studies on selected Mars (Fairén et al., 2010) and Europa (Lorenz et al., 2011) analogues on Earth show that, generally, in extreme environments we should expect chemolithotrophic metabolisms and a very low biomass of cells, which are in most cases very small in size (Table 3). This is so because the stability and functionality of the biomolecules of most organisms (and, thus, their viability) are strongly affected by the extreme physicochemical parameters such as the acidity and redox state. Therefore, the sensitivity of the proposed instruments must be high enough to guarantee the detection of a limited number of cell-like morphologies as well as a low concentration of biomolecules. In any case, additional work in terrestrial analogues deploying CMOLD is critical to better understand what morphological and biochemical diversity could be expected in other planets and moons of the Solar System.
Table 3.
Cell Content in Different Earth Analog Environments Assayed
| Analogue to | Environment | Number of cells (bacteria) | Organic concentration estimation (ppb)a |
|---|---|---|---|
| Europa | Lake Vostok | 120 cell/mL | 0.18 |
| Lake Vida | 444.000 cell/mL | 666 | |
| Deep Ocean | 65.000 cell/mL | 97.5 | |
| Mars | Artic permafrost sediments ≥300 m Antarctic permafrost sediments (20m) Atacama desert Bottom sediments of lakes (upper layer) |
102–108cfu/gdw 101–104 cell/mL 103–106 cell/mL 102–106 cell/mL |
3 × 10−4–0.3 3 × 10−1–330 3 × 10−2–330 |
Cell mass 10−12 g; organic content: 30% of cell mass = 3 × 10−13 g.
cfu = colony-forming units; dw = dry weight.
5.1. Mars
Solid rocks and powdered material are dominant on the martian surface. Upcoming missions to Mars plan to explore lacustrine and depositional paleo-environments, where a water cycle was likely active leaching sedimentary or volcanic deposits (Vago et al., 2017; Williford et al., 2018; Loizeau et al., 2019), because Mars is believed to have been much more of an Earth-like planet between 4.5 and 3.5 Gyr ago, with an active surface hydrosphere that included glaciers, rivers, ice-covered lakes, deltas, and maybe even a hemispheric ocean (Fairén, 2010), making Mars the only known planetary body, other than Earth, where liquid water appears to have played a key role in its early surface evolution.
Due to the preferred Noachian to Hesperian age for the materials (Fairén et al., 2010), they could be either buried underground or exhumed to the surface, where they would be exposed to high radiation doses and extreme temperature changes. The mineral composition of the interesting regions includes the presence of phyllosilicates, oxides, and several types of salts (mainly sulfates, chlorides, and perchlorates). The interaction of some of these minerals, particularly perchlorates, with the organic material determines their stability and preservation on Mars in the long term (Kounaves et al., 2014; Archer et al., 2019), and to obtain reliable results with CMOLD aimed at in situ analyzing different levels of prebiotic/biotic chemical and structural complexities.
5.2. Icy moons. Examples: Europa, Enceladus, and Titan
The approach to icy moon analog samples is different than that for Mars. Europa, Enceladus, and Titan are icy moons that deserve consideration because of their astrobiological interest.
It has been described, from previous missions to the Jupiter system, that the surface composition of Europa is dominated by water ice plus other dark albedo materials, identified as hydrated salts (Carlson et al., 1999; McCord et al., 1999; Dalton et al., 2005; Trumbo et al., 2019). These salts are associated with tectonic and cryovolcanic features, suggesting that they have a subsurface origin linked to aqueous reservoirs. Endogenic materials are strongly affected by the surface radiation environment (Dalton et al., 2013; Hand and Carlson, 2015). Moreover, some volatiles are detected (e.g., CO2, H2S, SO2) as ice phases mixed with water ice, either trapped as inclusions between crystals or forming hydrate clathrates (Prieto-Ballesteros et al., 2005).
There are three terrestrial analogs that are considered suitable places to understand the geochemistry and potential habitability of Europa: aphotic systems, deep cold brines, and subglacial liquid–water environments (Painter et al., 1982; Murray et al., 2012; Priscu and Hand, 2012). Additional laboratory experiments and field work are required to better anticipate what we can find in Europa with an instrument suite such as CMOLD. This will allow determination of different levels of prebiotic/biotic chemical and structural complexities, using aqueous solutions that resemble the melted endogenous materials, though enriched in varying concentrations of sulfates, sulfuric acid, and chlorides, and also doped with different concentrations of the target biomolecules to be detected (e.g., antibiotics, peptides, proteins, nucleic acids, lipids, or whole cells).
The plume activity detected to be emerging from outer Solar System moons, such as Europa, Enceladus, or Triton, facilitates access to materials sourced from deep aqueous layers. This process seems to be relatively continuous at present in Enceladus. The main composition of the plumes, which has been investigated by the Cassini INMS, shows the presence of water, organic molecules, and particles of silica and salts (Hansen et al., 2019). The possibility of collecting particles (perhaps of organic nature) from the plume with different spacecraft devices has been already suggested (McKay et al., 2014; Lunine, 2017). This would enable an investigation of the (bio)chemical complexity of the ejecting material with the CMOLD suite adapted to a spacecraft platform.
In the case of Titan, a rich repertoire of organic prebiotic molecules is expected in the atmosphere, in the dunes, and in the liquid surface bodies (Imanaka, 2019). The CMOLD instruments may help to evaluate how organics evolve in environments with solvents different than liquid water, and potentially to detect traits of life unlike from those on Earth.
6. Conclusions
We have outlined here a novel suite of first-order in situ analytical instruments and sensors that would allow determining different levels of prebiotic/biotic chemical and structural complexities in planets and satellites of the Solar System, as well as their (dis)similarity with the biochemistry of terrestrial life. This CMOLD is a unique concept that aims at addressing one of the main scientific challenges in planetary exploration, namely, to establish a minimal set of criteria and observations to identify life beyond the Earth.
The concept for a future astrobiological mission is most timely, because all the instrumentation devoted toward seeking organics in planetary exploration so far has been mostly based on the detection of volatile compounds by GC/MS, or on the identification of some spectroscopic organic signatures. This has been performed by the Viking Landers GC/MS, the Phoenix TEGA instrument, and the current MSL SAM, and it will also be the task for the Rosalind Franklin's MOMA instrument. However, the extraction of organic molecules into a liquid suspension and the detection of nonvolatile complex compounds with powerful analytical techniques are only now being considered by way of derivatization with SAM and MOMA instrumentation.
We have proposed here a step forward by targeting not only low MW biomolecules but also biopolymers and other macromolecules, supra-macromolecular complexes, as well as vesicular and cell-like morphologies by using three complementary astrobiological tools: (1) an optical microscope, to resolve microscale evidence for life; (2) a Raman spectrometer, to detect noncovalent bonds and atomic composition, resolve 3D secondary and tertiary polymeric structures, and identify other signatures of organic molecules of different sizes and complexities, not restricted to the known biochemistry operating in our planet; and (3) a biomarker detector equipped with a biosensor containing multiple bioaffinity probes (antibodies and aptamers) for up to 200 life-related and nonlife-related chemical compounds, able to identify the nature and structure of the molecules detected, at least the part of the molecule (epitope) that has been captured by the receptor molecule (Ab or Ap), following the lock-and-key principle.
Our concept is a robust and compact solution that covers a wide spectrum of targets, building on technologies and capabilities developed in the framework of recent space science exploration missions. We have developed state-of-the-art versions of both the proposed Raman spectrometer and the biomarker detector: The RLS instrument is included in the payload of the 2022 ESA Rosalind Franklin Mars rover, whereas the biomarker detector (SOLID instrument) is part of the payload on the NASA Icebreaker mission proposal for a Mars lander. CMOLD and the associated mission concept outlined here would be in line with the already scheduled or planned planetary exploration missions to Mars (Mars 2020 and ExoMars 2022) as well as to Jupiter's and Saturn's icy moons (Europa, Enceladus, Titan), which clearly sets the objective of developing agnostic approaches to search for signs of life in the Solar System.
Abbreviations Used
- Ab
antibody
- Ag
antigen
- Aps
aptamers
- BSA
bovine serum albumin
- CCD
charge-coupled device
- cfu
colony-forming units
- CMOLD
Complex Molecules Detector
- CPC
Complex Prebiotic Chemistry
- DHM
digital holographic microscopy
- DMD
digital micro-mirror device
- DNA
deoxyribonucleic acid
- DUV
deep ultraviolet
- dw
dry weight
- FDU
Fluidic Distribution Unit
- FSI
Fluorescence Sandwich Immunoassay
- GC/MS
gas chromatography/mass spectrometry
- IR
infrared
- MMX
Martian Moons eXplorer
- MOMA
Mars Organics Molecule Analyzer
- MW
molecular weight
- NA
numerical aperture
- PAH
polycyclic aromatic hydrocarbons
- RAX
RAman spectrometer for MMX
- RLS
Raman Laser Spectrometer
- RNA
ribonucleic acid
- RRS
Resonant Raman Scattering
- SAM
Sample Analysis at Mars
- SELEX
Systematic Evolution of Ligands by EXponential enrichment
- SPU
Sample Processing Unit
- ssDNA
single-stranded DNA
- TEGA
Thermal and Evolved Gas Analyzer
- UV
ultraviolet
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work has been funded by the project “MarsFirstWater,” European Research Council Consolidator grant number 818602 to Alberto G. Fairén; the Spanish Ministry of Economy and Competitiveness (MINECO) and EU FEDER program projects no. ESP2015-69540-R, RTI2018-094368-B-I00, BIO2016-79618-R, and ESP2017-89053-C2-1-P; and the Spanish Agencia Estatal de Investigación (AEI) project number MDM-2017-0737–Unidad de Excelencia “María de Maeztu.”
References
- Alm K, El-Schich Z, Falck Miniotis M, et al. (2013) Cells and holograms: holograms and digital holographic microscopy as a tool to study the morphology of living cells. In Holography: basic principles and contemporary applications, edited by Mihaylova E, Intech., Rijeka, Croatia, pp 335–351 [Google Scholar]
- Archer PD, Ming DW, Sutter B, et al. (2019) Perchlorate on Mars—overview and implications. In 9th Mars Conference, Pasadena, California [Google Scholar]
- Asher SA. (1993) UV resonance Raman Spectroscopy for analytical, physical, and biophysical chemistry. Anal Chem 65:59A–66A [DOI] [PubMed] [Google Scholar]
- Aubrey AD, Chalmers JH, Bada JF, et al. (2008) The Urey instrument: an advanced in situ organic and oxidant detector for Mars exploration. Astrobiology 8:583–595 [DOI] [PubMed] [Google Scholar]
- Beegle L and Bhartia R (2016) SHERLOC: an investigation for Mars 2020. In EGU General Assembly Conference Abstracts, vol. 18, Vienna, Austria, EGU2016-11215 [Google Scholar]
- Benner SA. (2017) Detecting Darwinism from molecules in the Enceladus Plumes, Jupiter's Moons, and other planetary water lagoons. Astrobiology 17:840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein MP, Sandford SA, Allamandola LJ, et al. (1999) UV Irradiation of polycyclic aromatic hydrocarbons in ices: production of alcohols, quinones, and ethers. Science 283:1135–1138 [DOI] [PubMed] [Google Scholar]
- Blanco Y, Rivas LA, Ruiz-Bermejo M, et al. (2013) Immunological detection of metilic acid in the Atacama desert: implication for organics detection on Mars. Icarus 224:326–333 [Google Scholar]
- Blanco Y, de Diego-Castilla G, Viúdez-Moreiras D, et al. (2018) Effects of gamma and electron radiations on the structural integrity of organic molecules and macromolecular biomarkers measured by microarray immunoassays and their astrobiological implications. Astrobiology 18:1497–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger U, de Vera JP, Fritz J, et al. (2012) Optimizing the detection of carotene in cyanobacteria in a martian regolith analogue with a Raman spectrometer for the ExoMars mission. Planet Space Sci 60:356–362 [Google Scholar]
- Briones C. (2015) Aptamer. In Encyclopedia of Astrobiology, 2nd ed., edited by Gargaud M and Irvine WM, Springer, Heidelberg, pp. 112–113 [Google Scholar]
- Bueno R, Marciello M, Moreno M, et al. (2019) Versatile graphene-based platform for robust nanobiohybrid interfaces. ACS Omega 4:3287–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol N. (2019) Beyond the Galileo experiment. Nat Astronomy 3:585–587 [Google Scholar]
- Cannon KM, Sutter B, Ming DW, et al. (2012) Perchlorate induced low temperature carbonate decomposition in the Mars Phoenix Thermal and Evolved Gas Analyzer (TEGA). Geophys Res Lett 39 L13023 https://agupubs.onlinelibrary.wiley.com/doi/pdf/10.1029/2012GL051952 [Google Scholar]
- Carlson RW, Johnson RE, and Anderson MS (1999) Sulfuric acid on Europa and the radiolytic sulfur cycle. Science 286:97. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Lee J-W, and Ellington AD (2009) Applications of aptamers as sensors. Annu Rev Anal Chem 2:241–264 [DOI] [PubMed] [Google Scholar]
- Cockell C, Bush T, Bryce C, et al. (2016) Habitability: a review. Astrobiology 16:89–117 [DOI] [PubMed] [Google Scholar]
- Coussot G, Le Postollec A, Incerti S, et al. (2019) Photochemistry on the Space Station-aptamer resistance to space conditions: particles exposure from irradiation facilities and real exposure outside the International Space Station. Astrobiology 19:1063–1074 [DOI] [PubMed] [Google Scholar]
- Dalton JB, Prieto-Ballesteros O, Kargel JS, et al. (2005) Spectral comparison of heavily hydrated salts with disrupted terrains on Europa. Icarus 177:472–490 [Google Scholar]
- Dalton JB, Cassidy T, Parnicas C, et al. (2013) Exogenic controls on sulfuric acid hydrate production at the surface of Europa. Planet Space Sci 77:45–63 [Google Scholar]
- Dan Dan ML, Yao B, Wang W, et al. (2013) DMD-based LED-illumination super-resolution and optical sectioning microscopy. Sci Rep 3:1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Escosura A, Briones C, and Ruiz-Mirazo K (2015) The systems perspective at the crossroads between chemistry and biology. J Theor Biol 381:11–22 [DOI] [PubMed] [Google Scholar]
- Domagal-Goldman SD, Wright KE, Adamala K, et al. (2016) The Astrobiology Primer v2.0. Astrobiology 16:561–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD and Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818. [DOI] [PubMed] [Google Scholar]
- Evans-Nguyen T, Becker L, Doroshenko V, et al. (2008) Development of a low power, high mass range mass spectrometer for Mars surface analysis. Int J Mass Spectrom 278:170–177 [Google Scholar]
- Fairén AG. (2010) A cold and wet Mars. Icarus 208:165–175 [Google Scholar]
- Fairén AG, Davila AF, Lim D, et al. (2010) Astrobiology through the ages of Mars. Astrobiology 10:821–843 [DOI] [PubMed] [Google Scholar]
- Fairén AG, Schulze-Makuch D, Parro V, et al. (2019) Planetary Protection and the astrobiological exploration of Mars: proactive steps in moving forward. Adv Space Res 63:1491–1497 [Google Scholar]
- Fernández N, Fernández-Miragall O, Ramajo J, et al. (2011) Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res 39:8572–8585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Näke C, Rivas LA, et al. (2006) A multi-array competitive immunoassay for the detection of broadrange molecular size organic compounds relevant for astrobiology. Planet Space Sci 54:1612–1621 [Google Scholar]
- García-Descalzo L, Parro V, García-Villadangos M, et al. (2019) Microbial markers profile in anaerobic Mars analogue environments using the LDChip (Life Detector Chip) antibody microarray core of the SOLID (Signs of Life Detector) platform. Microorganisms 7:E365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido P, Blanco M, Moreno-Paz M, et al. (2006) STEC-EPEC oligonucleotide microarray: a new tool for typing genetic variants of the LEE pathogenicity island of human and animal Shiga toxin-producing E. coli (STEC) and enteropathogenic E. coli (EPEC) strains. Clin Chem 52:192–201 [DOI] [PubMed] [Google Scholar]
- Goesmann F, Rosenbauer H, Bredehoft J, et al. (2015) Organic compounds on comet 67P/Churyumov-Gerasimenko revealed by COSAC mass spectrometry. Science 349:aab0689. [DOI] [PubMed] [Google Scholar]
- Goesmann F, Brinckerhoff WB, Raulin F, et al. (2017) The Mars Organic Molecule Analyzer (MOMA) instrument: characterization of organic material in martian sediments. Astrobiology 17:655–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand KP and Carlson RW (2015) Europa's surface color suggests an ocean rich with sodium chloride. Geophys Res Lett 42:3174–3178 [Google Scholar]
- Hansen CJ, Esposito LW, Colwell JE, et al. (2019) The composition and structure of Enceladus' plume from the complete set of Cassini UVIS occultation observations. Icarus 344:113461 [Google Scholar]
- Hecht MH, Marshall J, Pike WT, et al. (2008) Microscopy capabilities of the microscopy, electrochemistry, and conductivity analyzer. J Geophys Res Planets 113 E00A22 https://agupubs.onlinelibrary.wiley.com/doi/pdf/10.1029/2008JE003077 [Google Scholar]
- Hofstetter H, Cary JR, Eleniste PP, et al. (2005) New developments in the production and use of stereoselective antibodies. Chirality 17(Suppl 1):S9–S18 [DOI] [PubMed] [Google Scholar]
- Huang J and Honda W (2006) CED: a conformational epitope database. BMC Immunol 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka H. (2019) Astrobiology on Titan: geophysics to organic chemistry. In Astrobiology, edited by Yamagishi A, Takegawa T, and Usui T, Springer, Singapore, pp. 409–418 [Google Scholar]
- Johnson SS, Anslyn EV, Graham HV, et al. (2018) Fingerprinting non-terran biosignatures. Astrobiology 18:915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset JL, Westall F, Hofmann BA, et al. (2017) The Close-Up Imager onboard the ESA ExoMars Rover: objectives, description, operations, and science validation activities. Astrobiology 17:595–611 [DOI] [PubMed] [Google Scholar]
- Joyce GF. (1994) ‘Foreword.’ In Origins of Life: The Central Concepts, edited by Deamer DW and Fleischaker GR, Jones and Bartlett, Boston, pp. xi–xii [Google Scholar]
- Kassa T, Undesser LP, Hofstetter H, et al. (2011) Antibody-based multiplex analysis of structurally closely related chiral molecules. Analyst 136:1113–1115 [DOI] [PubMed] [Google Scholar]
- Kim YS and Gu B (2014) Advances in aptamer screening and small molecule aptasensors. Adv Biochem Eng Biotechnol 140:29–67 [DOI] [PubMed] [Google Scholar]
- Klein HP, Lederberg J, Rich A, et al. (1976) The Viking Mission search for life on Mars. Nature 262:24 [Google Scholar]
- Kounaves SP, Chaniotakis NA, Chevrier VF, et al. (2014) Identification of the perchlorate parent salts at the Phoenix Mars landing site and possible implications. Icarus 232:226–231 [Google Scholar]
- Kuhn E, Ichimura AS, Peng V, et al. (2014) Brine assemblages of ultrasmall microbial cells within the ice cover of Lake Vida, Antarctica. Appl Environ Microbiol 80:3687–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ferla R, Maimone G, Lo Giudice A, et al. (2015) Cell size and other phenotypic traits of prokaryotic cells in pelagic areas of the Ross Sea (Antarctica). Hydrobiologia 761:181–194 [Google Scholar]
- Lehn JM. (1995) Supramolecular Chemistry: Concepts and Perspectives. Wiley−VCH, New York [Google Scholar]
- Lezcano MÁ, Moreno-Paz M, Carrizo D, et al. (2019) Biomarker profiling of microbial mats in the geothermal band of Cerro Caliente, Deception Island (Antarctica): life at the edge of heat and cold. Astrobiology 19:1490–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizeau D, Balme MR, Bibring JP, et al. (2019) ExoMars 2020 surface mission: choosing a landing site. In Lunar and Planetary Science Conference, #2378, Lunar and Planetary Institute [Google Scholar]
- Lorenz RD, Gleeson D, Prieto-Ballesteros, et al. (2011) Analog environments for a Europa lander mission. Adv Space Res 48:689–696 [Google Scholar]
- Lunine JI. (2017) Ocean worlds exploration. Acta Astronautica 131:123–130 [Google Scholar]
- Mahaffy PR, Webster CR, Cabane M, et al. (2012) The sample analysis at Mars investigation and instrument suite. Space Sci Rev 170:401–478 [Google Scholar]
- Maiti NC, Apetri MM, Zagorski MG, et al. (2004) Raman spectroscopic characterization of secondary structure in natively unfolded proteins: alpha synuclein. J Am Chem Soc 126:2399–2408 [DOI] [PubMed] [Google Scholar]
- Mansy SS, Schrum JP, Krishnamurthy M, et al. (2008) Template-directed synthesis of a genetic polymer in a model protocell. Nature 454:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V, Perales C, Fernández-Algar M, et al. (2016) An efficient microarray-based genotyping platform for the identification of drug-resistance mutations in majority and minority subpopulations of HIV-1 quasispecies. PLoS One 11:e0166902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord TB, Hansen GB, Matson DL, et al. (1999) Hydrated salt minerals on Europa's surface from the Galileo near-infrared mapping spectrometer (NIMS) investigation. J Geophys Res Planets 104:11827–11851 [Google Scholar]
- McGuire BA. (2018) 2018 Census of interstellar, circumstellar, extragalactic, protoplanetary disk, and exoplanetary molecules. Astrophys J Supplement Series 239:17–65 [Google Scholar]
- McKay CP, Stoker CR, Glass BJ, et al. (2013) The Icebreaker life mission to mars: a search for biomolecular evidence for life. Astrobiology 13:334–353 [DOI] [PubMed] [Google Scholar]
- McKay CP, Anbar AD, Porco C, et al. (2014) Follow the plume: the habitability of Enceladus. Astrobiology 14:352–355 [DOI] [PubMed] [Google Scholar]
- Ming DW, Archer PD Jr., Glavin DP, et al. (2014) Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale Crater, Mars. Science 343:388. [DOI] [PubMed] [Google Scholar]
- Monnard P-A and Walde P (2015) Current ideas about prebiological compartmentalization. Life (Basel) 5:1239–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery W, Jaramillo EA, Royle SH, et al. (2019) Effects of oxygen-containing salts on the detection of organic biomarkers on Mars and in terrestrial analog soils. Astrobiology 19:711–721 [DOI] [PubMed] [Google Scholar]
- Moral AG, Rull F, Maurice S, et al. (2019) Design, development, and scientific performance of the Raman Laser Spectrometer EQM on the 2020 ExoMars (ESA) Mission. J Raman Spectrosc. doi: 10.1002/jrs.5711 Special Issue Review: https://onlinelibrary.wiley.com/doi/abs/10.1002/jrs.5711 [DOI]
- Moreno M. (2019) Sensors/Aptasensors. In Encyclopedia of Analytical Science, 3rd ed., edited by Worsfold P, Townshend A, Poole C.F., and Miró M., Elsevier, Amsterdam, The Netherlands, pp. 150–153 [Google Scholar]
- Moreno-Paz M, Gómez-Cifuentes A, Ruiz-Bermejo M, et al. (2018) Detecting nonvolatile life- and nonlife-derived organics in a carbonaceous chondrite analogue with a new multiplex immunoassay and its relevance for planetary exploration. Astrobiology 18:1041–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M, Fernández-Algar M, Fernández-Chamorro J, et al. (2019) A combined ELONA-(RT)qPCR approach for characterizing DNA and RNA aptamers selected against PCBP-2. Molecules 24:1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Kenig F, Fritsen CH, et al. (2012) Microbial life at -13°C in the brine of an ice-sealed Antarctic lake. Proc Natl Acad Sci USA 109:20626–20631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JL, Bedrossian M, and Lindensmith CA, 2018. Imaging technologies and strategies for detection of extant extraterrestrial microorganisms. Adv Phys 3:1424032 [Google Scholar]
- Navarro-González R, Navarro KF, De La Rosa J, et al. (2006) The limitations on organic detection in Mars-like soils by thermal volatilization–gas chromatography–MS and their implications for the Viking results. Proc Natl Acad Sci USA 103:16089–16094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu M, Hays LE, Voytek MA, et al. (2018) The ladder of life detection. Astrobiology 18:1375–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann HB, Atreya SK, Bauer SJ, et al. (2005) The abundances of constituents of Titan's atmosphere from the GCMS instrument on the Huygens probe. Nature 438:779. [DOI] [PubMed] [Google Scholar]
- Novotny M, Hayes JM, Bruner F, et al. (1975) Gas chromatographic column for the Viking 1975 molecular analysis experiment. Science 189:215–216 [DOI] [PubMed] [Google Scholar]
- Orgel LE. (1992) Molecular replication. Nature 358:203–209 [DOI] [PubMed] [Google Scholar]
- Painter PC, Mosher LE, Rhoads C (1982) Low-frequency modes in the Raman spectra of proteins. Biopolymers 21:1469–1472 [DOI] [PubMed] [Google Scholar]
- Parnell J, Cullen D, Sims MR, et al. (2007) Searching for life on Mars: selection of molecular targets for ESA's aurora ExoMars mission. Astrobiology 7:578–604 [DOI] [PubMed] [Google Scholar]
- Parro V, Rodríguez-Manfredi JA, Briones C, et al. (2005) Instrument development for biomarker search on Mars: acidophile, iron powered chemolithoautotrophic communities as terrestrial model system. Planet Space Sci 53:729–737 [Google Scholar]
- Parro V, Fernández-Calvo P, Rodríguez Manfredi JA, et al. (2008) SOLID2: an Antibody Array-Based Life-Detector Instrument in a Mars Drilling Simulation Experiment (MARTE). Astrobiology 8:987–999 [DOI] [PubMed] [Google Scholar]
- Parro V, de Diego-Castilla G, Moreno-Paz M, et al. (2011a) A microbial oasis in the hypersaline Atacama subsurface discovered by a life detector chip: implications for the search for life on Mars. Astrobiology 11:969–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parro V, de Diego-Castilla G, Rodríguez-Manfredi JA, et al. (2011b) SOLID3: a multiplex antibody microarray-based optical sensor instrument for in situ life detection in planetary exploration. Astrobiology 11:15–28 [DOI] [PubMed] [Google Scholar]
- Parro V, Blanco Y, Puente-Sánchez F, et al. (2018) Biomarkers and metabolic patterns in the sediments of evolving glacial lakes as a proxy for planetary lake exploration. Astrobiology 18:586–606 [DOI] [PubMed] [Google Scholar]
- Peretó J. (2005) Controversies on the origin of life. Int Microbiol 8:23–31 [PubMed] [Google Scholar]
- Persichetti G, Grimaldi IA, Testa G, et al. (2017) Multifunctional optofluidic lab-on-chip platform for Raman and fluorescence spectroscopic microfluidic analysis. Lab Chip 17:2631–2639 [DOI] [PubMed] [Google Scholar]
- Pizzarello S and Shock E (2010) The organic composition of carbonaceous meteorites: the evolutionary story ahead of biochemistry. Perspect Biol 2:a002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressman A, Blanco C, and Chen IA (2015) The RNA world as a model system to study the origin of life. Curr Biol 25:R953–R963 [DOI] [PubMed] [Google Scholar]
- Prieto-Ballesteros O, Kargel JS, Fernández-Sampedro M, et al. (2005) Evaluation of the possible presence of clathrate hydrates in Europa's icy shell or seafloor. Icarus 177:491–505 [Google Scholar]
- Priscu JC and Hand KP (2012) Microbial habitability of icy worlds. Microbe 7:167–172 [Google Scholar]
- Pullan D, Westall F, Hofmann BA, et al. (2008) Identification of morphological biosignatures in martian analogue field specimens using in situ planetary instrumentation. Astrobiology 9:119–156 [DOI] [PubMed] [Google Scholar]
- Rivas LA, García-Villadangos M, Moreno-Paz M, et al. (2008) A 200-antibody microarray biochip for environmental monitoring: searching for universal microbial biomarkers through immunoprofiling. Anal Chem 80:7970–7979 [DOI] [PubMed] [Google Scholar]
- Romero-López C, Barroso-delJesus A, García-Sacristán A, et al. (2014) End-to end cross-talk within the hepatitis C virus genome mediates the conformational switch of the 3’X-tail region. Nucleic Acids Res 42:567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen M, Kalanoor BS, Orend Z, et al. (2018) Characterization of peptides self-assembly by low frequency Raman spectroscopy. RSC Adv 8:16161–16170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard J, García-Ruiz JM, Gong J, et al. (2018) A morphogram for silica-witherite biomorphs and its application to microfossil identification in the early earth rock record. Geobiology 16:279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Mirazo K, Briones C, and de la Escosura A (2014) Prebiotic systems chemistry: new perspectives for the origins of life. Chem Rev 114:285–366 [DOI] [PubMed] [Google Scholar]
- Ruiz-Mirazo K, Briones C, and de la Escosura A (2017) Chemical roots of biological evolution: the origins of life as a process of development of autonomous functional systems. Open Biol 7:170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rull F, Maurice S, Diaz E, et al. (2017) The Raman Laser Spectrometer for the ExoMars Rover Mission to Mars. Astrobiology 17:627–654 [Google Scholar]
- Sánchez-García L, Fernández-Martínez MA, Moreno-Paz M, et al. (2020) Simulating Mars drilling mission for searching for life: ground-truthing lipids and other complex microbial biomarkers in the iron-sulfur rich Río Tinto analog. Astrobiology [Epub ahead of print]. doi: 10.1089/ast.2019.2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Luque FJ, Stich M, Manrubia S, et al. (2014) Eficient HIV-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimization strategies. Sci Rep 4:6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapers HM, Hollis JR, Bhartia R, et al. (2019) The cell and the sum of its parts: patterns of complexity in biosignatures as revealed by deep UV Raman Spectroscopy. Front Microbiol 10:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Makuch D and L Irwin (2018) Life in the Universe: Expectations and Constraints, 3rd ed., Springer Nature Switzerland, Springer-Verlag [Google Scholar]
- Sharma TK, Bruno JG, and Dhiman A (2017) ABCs of DNA aptamer and related assay development. Biotechnol Adv 35:275–301 [DOI] [PubMed] [Google Scholar]
- Siebenmorgen R and Krügel E (2010) The destruction and survival of polycyclic aromatic hydrocarbons in the disks of T Tauri stars. A&A 511:A6 [Google Scholar]
- Stockert JC and Blázquez-Castro A (2017) “Super-resolution Microscopy”. In Fluorescence Microscopy in Life Sciences, edited by Stockert JC and Blázquez-Castro A, Bentham Science Publishers, Sharjah, UAE, pp. 687–711 [Google Scholar]
- Stoltenburg R, Reinemann C, and Strehlitz B (2007) SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24:381–403 [DOI] [PubMed] [Google Scholar]
- Sun H and Zu Y (2015) A highlight of recent advances in aptamer technology and its application. Molecules 20:11959–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary E. (2006) The origin of replicators and reproducers. Philos Trans R Soc Lond B Biol Sci 361:1761–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N, Lüthi BS, Hviid SF, et al. (2004) The microscope for Beagle 2. Planet Space Sci 52:853–866 [Google Scholar]
- Trumbo SK, Brown ME, and Hand KP (2019) Sodium chloride on the surface of Europa. Sci Adv 5:eaaw7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C and Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510 [DOI] [PubMed] [Google Scholar]
- Vago JL, Westall F, Coates A, et al. (2017) Habitability on early Mars and the search for biosignatures with the ExoMars rover. Astrobiology 17:471–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. (2012) In situ laser Raman spectroscopy for Mars Sample Return Mission. In 43rd Lunar and Planetary Science Conference; March19-23, 2012; Abstract 2149, Houston, TX, USA [Google Scholar]
- Wiens RC, Maurice S, and Rull F (2017) The SuperCam remote sensing instrument suite for the Mars 2020 rover mission: a preview. Spectroscopy 32:50–55 [Google Scholar]
- Williford KH, Farley KA, Stack KM, et al. (2018) The NASA Mars 2020 rover mission and the search for extraterrestrial life. In From Habitability to Life on Mars, edited by Cabrol NA and Grin EA, Amsterdam, Netherlands, Elsevier, pp. 275–308 [Google Scholar]