Abstract
The post-genomic era has seen many advances in our understanding of cancer pathways, yet resistance and tumor heterogeneity necessitate multiple approaches to target even monogenic tumors. Here, we combine phenotypic screening with chemical genetics to identify pre-mRNA endonuclease Cleavage and Polyadenylation Specificity Factor 3 (CPSF3) as the target of JTE-607, a small molecule with previously unknown target. We show that CPSF3 represents a novel synthetic lethal node in a sub-set of acute myeloid leukemia (AML) and Ewing’s sarcoma cancer cell lines. Inhibition of CPSF3 by JTE-607 alters expression of known downstream effectors in AML and Ewing’s sarcoma lines, upregulates apoptosis and causes tumor-selective stasis in mouse xenografts. Mechanistically, it prevents the release of newly synthesized pre-mRNAs, resulting in read-through transcription and the formation of DNA-RNA hybrid R-loop structures. This study implicates pre-mRNA processing, and specifically CPSF3, as a druggable target providing a new avenue to therapeutic intervention in cancer.
Introduction
Phenotypic screening enables interrogation of native targets and pathways in an unbiased, disease-relevant context and has become widely recognized as a key strategy in the discovery of first-in-class drugs.1,2 Large collections of genetically and pharmacologically annotated cancer cell lines can serve as preclinical models and subsequent target identification can provide insights into molecular mechanism, ascribe function to poorly understood proteins, and enable the discovery of novel druggable nodes and pathways.3–5 While knowledge of the biological target is not required for advancing bioactive molecules with compelling phenotypes into clinical studies, an understanding of target and mechanism of action (MoA) greatly enables compound optimization, biomarker identification, efficacy studies and safety monitoring during drug development.
JTE-607 (1) was developed over two decades ago and has progressed to human healthy volunteer clinical studies, even though the mechanism of action and direct target of the compound are not known.6 Pre-clinically, JTE-607 has been shown to inhibit the production of proinflammatory cytokines in rodent and human models of acute injury, septic shock and endotoxemia.6–9 JTE-607 was also shown to cause cell death in AML cell lines in vitro and to prolong survival in a U-937 xenograft mouse model.10,11
We profiled 1 across a defined subset of the Cancer Cell Line Encyclopedia (CCLE) consisting of a panel of 92 genetically characterized human cancer cell lines of diverse lineages, probing for features that may define compound sensitivity and stratify potential patient populations. Amongst lineages sensitive to 1 were AML and Ewing’s sarcoma cell lines, two cancers characterized by aberrant transcription downstream of oncogenic translocations.12–14 To elucidate the mechanism of 1 sensitivity in AML or Ewing’s sarcoma, we set out to identify the target of 1. Through an extensive array of chemical genetics target identification and validation approaches, we determined Cleavage and Polyadenylation Specificity Factor 3 (CPSF3), a core component of the pre-mRNA cleavage and polyadenylation complex, as the single target directly bound by the active derivative of JTE-607. The CPSF complex is composed of at least five proteins (CPSF1, CPSF2, CPSF3, CPSF4 and FIP1L1) that recognizes the canonical nucleotide sequence AAUAAA in a newly synthesized pre-messenger RNA (pre-mRNA). Pre-mRNA is cleaved by CPSF3, the catalytic component of the complex also known as CPSF73, preferentially after a CA sequence motif 10-30 base pairs downstream of an AAUAAA element.15 A poly(A) tail is then added to the newly formed 3’-pre-mRNA end by polynucleotide adenylyltransferase (PAP).
Our data points to a model whereby inhibition of CPSF3 blocks the release of newly synthesized pre-mRNAs, resulting in read-through transcription and the formation of DNA-RNA hybrid R-loop structures. The expression of known downstream effectors in AML and Ewing’s sarcoma is negatively affected in vitro and tumor-selective stasis is observed in vivo. The efforts described here indicate that pre-mRNA processing, and specifically CPSF3, represent a novel druggable node in cancers driven by aberrant transcription. As such, our findings argue that inhibition of pre-mRNA processing, a fundamental cellular function, is not universally limiting and thus opens the possibility for therapeutic intervention in a given oncogenic context.
Results
A subset of cell lines show sensitivity to Compound 1
Profiling compounds across genetically characterized cancer cell lines, such as the CCLE, can inform patient stratification and compound mechanism of action.4,16 To determine if 1 (Fig. 1a) had a lineage-specific viability effect on cancer cells, 92 cell lines of diverse lineages (Supplementary Table 1) were profiled for sensitivity to the compound. The most sensitive cell lines were of AML and Ewing’s sarcoma lineage (Fig. 1b), and included lines harboring genetic translocations of MLL (such as AML lines MV-4-11 (MLL-AF4), ML2 (MLL-AF6) or MOLM-13, MONO-MAC-1, NOMO-1, THP-1 and MONO-MAC-6 (all MLL-AF9)) or EWSR1 (such as Ewing’s lines A-673, SK-ES-1 and MHH-ES-1 (all EWS-FLI)), respectively, a profile that had not previously been commented on.10,11 Functionally, compound 1 treatment of select Ewing’s and AML lines resulted in a shift to a more differentiated phenotype as measured by cell surface marker expression (A-673, Supplementary Fig. 1a; THP-1, Supplementary Fig. 2a), downregulation of known key effectors (A-673, Supplementary Fig. 1b,c; MOLM-13, Supplementary Fig. 2b,c) and increased apoptosis (NOMO-1, Supplementary Fig. 3a,b).17–20 Additionally, compound 1 showed in vivo efficacy and selectivity in human cancer xenograft models when comparing an AML cell line (MOLM-13) to a non-translocated lung adenocarcinoma cell line (A549) in athymic nude mice. Following evaluation of compound plasma stability and pharmacokinetics (Supplementary Fig. 4a-d), treatment of MOLM-13 tumors with 1 resulted in tumor volume stasis in a dose-dependent manner and downregulation of known key effectors (Supplementary Fig. 4e,f), while growth of A549 tumors was not significantly affected despite target engagement as measured by expression of the same set of biomarkers (Supplementary Fig. 4g,h). Taken together, these findings indicate that compound 1 modulates marker gene expression and differentiation status in AML or Ewing’s sarcoma, ultimately exerting a negative effect on cell viability, both in tissue culture and in vivo.
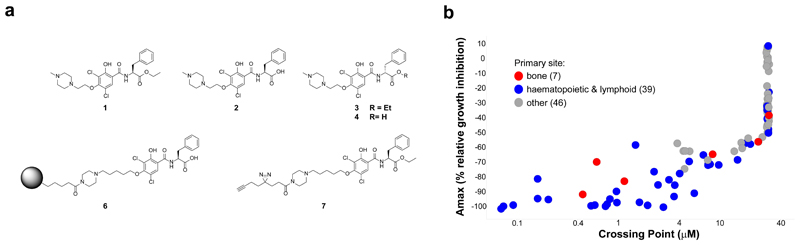
Figure 1. In vitro growth of AML and Ewing’s sarcoma cancer cell lines is sensitive to compound 1.
a, Chemical probes developed for study. b, Cell viability profile of compound 1 against a panel of 92 cancer cell lines, derived via CellTiter Glo (CTG) read-out after 72h of compound treatment in 11-point dose-response from 30 µM to 0.1 nM final concentration. Crossing Point was defined as the concentration at which the fitted curve of compound 1 for a given cell line achieved 50% of the positive control compound MG132. Amax was defined as the maximal inhibition observed with compound 1 for a given cell line over the range of tested concentrations. Cancer cell lines are color-coded by tumor primary site for bone (red, 7 cell lines), hematopoietic and lymphoid (blue, 39 cell lines), or other (grey, 46 cell lines). See Supplementary Table 1 for list of all 92 lines tested detailing primary site, lineage and compound sensitivity.
JTE-607 is a pro-drug and modulates pre-mRNA processing
To determine the cellular efficacy target of 1 we applied a comprehensive chemical genetics strategy focusing on unbiased identification of genes functionally required for compound action as well as physical interactors of the compound.21 The following approaches were conducted in parallel to establish and test target hypotheses.
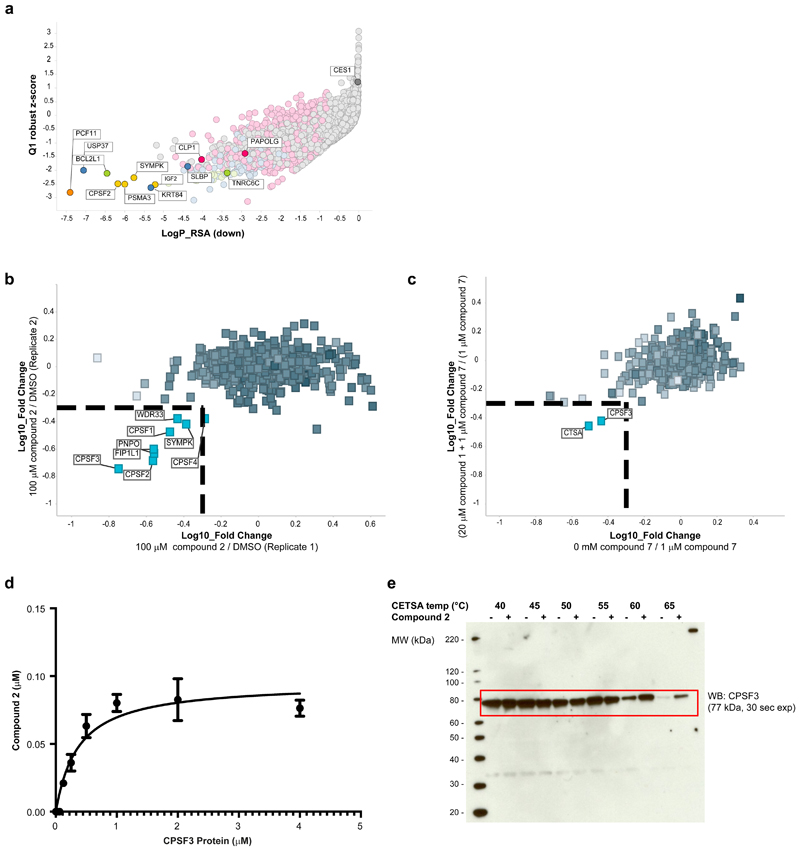
Compound-sensitized siRNA screens enable identification of genes that when knocked down lead to greater (or reduced) sensitivity to compound, thereby informing on pathway nodes affected by the small molecule, if not the direct target itself.22 We conducted two full genome siRNA screens (8 siRNAs/gene, ~19,350 human genes, Supplementary Dataset 1) in A-673 Ewing’s sarcoma cells in the presence of DMSO or an IC10 concentration of compound 1 (0.4 μM). The outcome of each siRNA in the DMSO treated condition were subtracted from that of the compound 1 treated condition before conducting gene level analysis. Using both quartile-based analysis and redundant siRNA analysis (RSA)23 we observed that knockdowns of pre-mRNA processing complex members were significantly enriched as synergy partners for 1 (Fig. 2a, Supplementary Fig. 5a,c,d; Supplementary Dataset 2), including PCF11, SYMPK, CPSF2, SLBP and CLP1 (RSA down p-value < 10-4). GeneMANIA analysis of the top 50 genes selected (ranked by RSA down p-value) highlighted additional genes over-connected to the input genes, including CPSF3, CSTF2, CPSF7, CSTF3 (Supplementary Fig. 5a).24 The data from these synergistic hits thus highlighted pre-RNA processing as a key node modulated by 1.
Figure 2. Chemical genetics and proteomics studies combine to identify mRNA processing and CPSF3 as the target of compound 2.
a, Full genome siRNA plus compound 1 synergy screen in A-673 cells, with cell viability read-out via CellTiter Glo assay. Per siRNA, the differential effect between DMSO-treated versus compound 1-treated cells was calculated. Plotting gene-level redundant siRNA activity (RSA statistical model, one-sided, down) as a measure of significance of effect versus Q1 Z-score as a measure of magnitude of effect identified siRNA-targeted genes whose knock-down sensitized cells to 0.4 µM compound 1 treatment. The siRNA library was tested as one replicate with each gene represented by n=8 siRNAs on average. Top screening hits, including known regulators of mRNA processing, are highlighted and named. b, Chemical proteomics studies using 6 in NOMO-1 cells. Proteins identified as specifically competed are highlighted and named. c, Photo-affinity labeling with PAL probe 7 in A-673 cells. Specifically enriched proteins are highlighted and named. d, Affinity binding measurement of compound 2 to human CPSF3 (Kd = 370 nM) as determined by SEC-TID. Error bars represent standard deviation of the mean (n=2). e, Western blot against CPSF3 following Cellular Thermal Shift Assay (CETSA) with compound 2 (+) or DMSO (-) treatment using NOMO-1 cell lysates (n=1).
In the opposite direction, carboxylesterase 1 (CES1) was revealed to be the top antagonistic hit, i.e. genes which when knocked down confer resistance to compound 1-induced cell death (Supplementary Fig. 5b; RSA up p-value = 10-7). As CES1 protein is known to hydrolyze functional ester groups in small molecules, the strong difference in activity of CES1 knockdown between the DMSO and compound 1 treatments (visualization of the gene-level activity in individual screening conditions shown in Supplementary Fig. 5e,f) suggested that CES1 protein hydrolyzes compound 1 to generate the cellularly active carboxylic acid form (compound 2). In support of such a pro-drug mechanism, mass spectrometry analysis of compound treated A-673 cells confirmed that ester 1 is rapidly taken up and intracellularly converted to the corresponding carboxylic acid 2, while 2 was not taken up as expected due to its polar carboxyl group (Supplementary Fig. 6a). Accordingly, cell lines sensitive to compound 1 were not affected in their viability when treated with compound 2 (Supplementary Fig. 7b). Analogously, ester 3, the enantiomer of compound 1, was intracellularly converted to the corresponding carboxylic acid 4 (albeit at a lower rate, Supplementary Fig. 6a), but it did not negatively affect the viability of sensitive cell lines (Supplementary Fig. 7b). This suggested that an alternate target was responsible for the viability defect, particularly one in which the stereochemical integrity of compound 1 was important for target engagement (vide infra).
Although compound-sensitive cell lines had higher CES1 transcript expression on average (Supplementary Fig. 6b), high CES1 levels were not a prerequisite for compound sensitivity as several lines with high CES1 showed little to no viability defect (Supplementary Table 1). Among translocated AML or Ewing’s sarcoma cell lines, CES1 expression levels varied (Supplementary Fig. 6c), conflating the translocation and CES1 associations to compound sensitivity. Therefore, to assess the effect of intracellular compound 1 ester hydrolysis and release of the corresponding acid on cell viability, we transiently overexpressed CES1 in two leukemia cell lines with low endogenous CES1 levels (Supplementary Fig. 8a), followed by treatment with compound 1 in dose-response (Supplementary Fig. 8b). Ectopic CES1 expression rendered both CMK, a non-translocated AML line, as well as SEM, an MLL-AF4 translocated ALL line, more sensitive to compound 1 in a cell viability assay, shifting IC50 values by approximately a factor of ten in both cases. SEM were generally more sensitive to 1 than CMK, and neither line was as sensitive as MOLM-13, despite comparable CES1 protein expression levels across the three lines as measured by Western blot. Taken together, these results argued that 1 behaves as a pro-drug, entering the cell as the ethyl ester and being rapidly converted by CES1 to the corresponding carboxylic acid, 2, which, in turn, engages its intracellular target or targets.
Compound 2 directly binds CPSF3
To identify the direct target(s) of active species compound 2, we employed a quantitative chemical proteomics approach.25 To enable affinity purification, detailed structure-activity relationship (SAR) studies on compound 1 (Supplementary Fig. 7a) determined a suitable site for incorporation of a linker, which would not impede bioactivity. We assessed a variety of linker surrogates (compound 5 for example, Supplementary Fig. 7b) in cellular assays to ensure that the affinity probe would retain comparable activity and selectivity to parent compound 1, and gain confidence that the linker site was likely solvent exposed. A bioactive linker analog was immobilized on a sepharose resin to generate affinity matrix 6, (Fig. 1a). Affinity enrichment from NOMO-1 whole cell lysate pre-incubated either with DMSO or compound 2 yielded proteins specifically competed (Supplementary Fig. 9), followed by quantitative proteomics analysis by mass spectrometry. Of the 894 proteins identified and quantified across replicates, eight exhibited significant and reproducible concentration-dependent competition in the presence of compound 2 (Fig. 2b, Supplementary Fig. 10, Supplementary Dataset 3). Interestingly, all proteins showing greater than 2-fold competition (except PNPO) belonged to the same protein complex, namely CPSF, suggesting that compound 2 binds to one or more components of this functional protein complex in cell lysate. To determine the protein(s) directly interacting with compound 2, (and to provide evidence of direct cellular engagement), we generated photo-affinity cross-labeling (PAL) probe 7 for photo-crosslinking studies in live cells (Fig. 1a). Compound 7 consists of a photoactivatable diazarine cross-linker (to covalently attach the probe to the binding partner(s) upon UV irradiation) and an alkyne moiety for click chemistry-mediated conjugation of biotin-azide (for affinity capture of the cross-linked binding partner(s) upon cell lysis). Combined probe titration and single dose competition experiments in A-673 intact cells, (pretreating cells with compound 1 prior to treatment with PAL probe 7 and UV irradiation), followed by analysis of the quantitative mass spectrometry data, enabled identification of specific interactions (Fig. 2c, Supplementary Dataset 4).26 The PAL-experiments revealed that while a number of CPSF complex members were identified, CPSF3 was the only CPSF complex member to exhibit appreciable enrichment and competition. Additionally, CTSA showed significant enrichment and competition behavior in the PAL experiment, constituting a potential off-target interaction of 2. However, the convergence of the affinity pulldowns and PAL-based experiments indicated that compound 2 binds directly to CPSF3 in cells.
To further validate CPSF3 as the direct binding partner of compound 2, we purified CPSF3 protein and conducted affinity binding experiments using a size-exclusion chromatography approach known as SEC-TID.27 While compound 1 did not demonstrate any binding, binding between compound 2 and CPSF3 was observed with a dissociation constant (Kd) of 370 nM (Fig. 2d). Furthermore, compound 2 treatment stabilized CPSF3 in a thermal shift assay using cell lysates (CETSA), further supporting direct engagement of compound 2 to CPSF3 (Fig. 2e).
To summarize the target identifications efforts, compound sensitized siRNA screening honed in on the pathway while affinity pulldowns led us to a specific complex. Photoaffinity cross-linking chemical proteomics experiments identified the direct binding partner of the probe molecule, and biochemical and biophysical studies validated the SAR, thus establishing the target.
Compound 2 mimics pre-mRNA binding to CPSF3
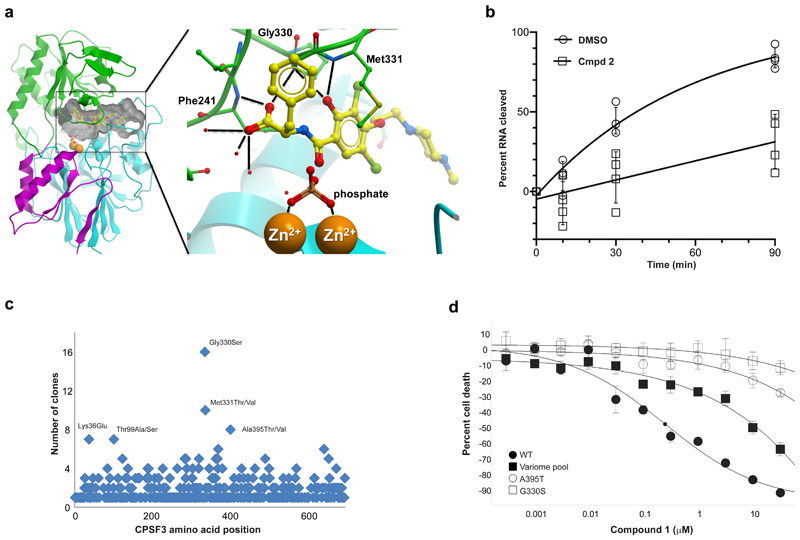
CPSF3 has RNA endonuclease activity and is composed of a metallo-β-lactamase domain and a β-CASP domain. The active site is located at the interface of these two domains and harbors two zinc ions.28 To elucidate how compound 2 modulates pre-mRNA cleavage activity of CPSF3 on a molecular level, we determined the 2.5 Å resolution co-crystal structure of CPSF3 and compound 2 (Fig. 3a and Supplementary Fig. 11). Compound 2 is bound at the large interfacial cavity of CPSF3, positioned approximately 7 Å from the active site zinc ions. The crystallization solution contained ~2 M phosphate and electron density showed a phosphate coordinating both zinc ions, (a sulfate has been observed at the corresponding position in the apo structure of CPSF3).28 In comparison with the apo form of CPSF3, the compound 2 bound structure shows the β-CASP domain rotated by 7.3 degree with respect to the large metallo-β-lactamase domain (Supplementary Fig. 11a, Supplementary Table 2). Compound binding is accompanied by structural rearrangements in loop regions lining the interfacial cavity (Supplementary Fig. 11a,b). These loops have been implicated in gating RNA access to the CPSF3 active site.28 Critical interactions between compound 2 and CPSF3 are contributed by the carboxylate and hydroxyl groups of 2, which form bifurcated hydrogen bonds to the backbone NH groups of Phe241, Gly330 and Met331, respectively (Fig. 3a). Furthermore, the carboxylate contacts three ordered water molecules at the interfacial cavity. We speculate that bound 2 may partially mimic the binding mode of pre-mRNA at the active site of CPSF3: the phosphate molecule and the carboxylate of compound 2 may occupy binding sites for two adjacent phosphodiester groups of a natural pre-mRNA substrate. The corresponding pre-mRNA bases may bind to the two pockets that accommodate the benzyl and dibromo-hydroxyphenyl groups of compound 2 (Supplementary Fig. 11c). This arrangement would position a pre-mRNA phosphodiester bond for nucleophilic attack by a water molecule that is coordinated and polarized by the two zinc ions. Based on the co-crystal structure of 2, we modelled binding of the corresponding (R)-enantiomer 4 and found the benzyl group of 4 clashing with the zinc-coordinating side chain of His73, perhaps explaining why compound 1 was active while the corresponding enantiomer 3 was inactive in the cell panel screens.
Figure 3. Compound 2 binds directly to CPSF3, inhibiting its endonuclease activity and conferring growth inhibitory effects.
a, Co-crystal structure of CPSF3 and 2. Ribbon representation of the CPSF3 metallo-β-lactamase domain in cyan (residues 7 to 208) and magenta (396 to 459), and the β-CASP domain in green (209 to 395). Two zinc ions at the active site (orange spheres) are coordinated by a phosphate. 2 is shown as yellow ball-andstick and the interfacial cavity outlined by a grey surface. Insert highlights polar interactions between 2 and CPSF3. The carboxylate group of 2 forms bifurcated hydrogen bonds to the backbone NH of Phe241 and Gly330, respectively. Three ordered water molecules contact the carboxylate. The hydroxyl group is in hydrogen bonding distance to the backbone NH groups of Gly330 and Met331, respectively. b, In vitro cleavage reaction monitored over time using yeast recombinant 8-subunit core CPF complex, cleavage factors (CFIA and CFIB) and a Cyc1 model RNA substrate in the presence of 1% DMSO or 100 µM compound 2. The data was fitted to an exponential plateau model (solid line), error bars represent standard deviation of the mean (n=4). c, Missense mutation distribution in CPSF3 that desensitize to compound 1 in cell viability assay as identified from variomics studies in A-673. d, Cell viability assessment of A-673 cells expressing in trans either wild-type (WT) CPSF3 encoding cDNA or dominant mutant versions of human CPSF3 as derived from variomics study in response to treatment with compound 1. Small black circle on fitted curve for WT condition indicates IC50 value. Center values represent mean, error bars represent standard deviation (n=3).
Functional validation of compound 2
Previously, recombinant human CPSF3 was shown to have non-specific endonuclease activity in vitro,28 and we were keen to assess if compound 2 could directly inhibit RNA cleavage. In our hands, the enzymatic activity of full-length human CPSF3 (the same protein as used for SEC-TID experiment shown in Fig. 2d) required micromolar concentrations of the enzyme, which prevented direct measurement of inhibition by 2. However, a recent report proposed that active and specific processing of yeast mRNA 3ʹ ends minimally requires an eight-subunit Cleavage and Polyadenylation Factor (CPF) complex.29 Utilizing such a yeast “CPF core” complex (Supplementary Fig. 12a) to monitor in vitro cleavage of a model RNA substrate, we found compound 2 to significantly reduce the accumulation of cleaved RNA substrate over time in comparison to a DMSO-treated control reaction (Fig. 3b). Since compound 2 had not been optimized for binding to yeast CPF, micromolar compound concentrations were required in this assay. Nevertheless, we were able to clearly demonstrate an inhibitory effect on RNA substrate cleavage, distinguishable from that of DMSO, parent ester 1 or inactive enantiomer 4 treatment (Supplementary Fig. 12b), thereby demonstrating compound 2-specific inhibition of CPF endonuclease activity.
To functionally validate CPSF3 as the direct target of compound 2, we next conducted saturating mutagenesis (variomics) of 14 candidate target genes (Supplementary Fig. 13a) identified from siRNA, chemical proteomics and PAL experiments, followed by identification of mutations that confer resistance to compound treatment.30,31 Of the tested genes, CPSF3 showed the most robust resistance in colony formation assays. Resistant clones were sequenced and three mutations were identified that occurred with high frequency (counts >8) and 29 with low frequency (counts >4) (Fig. 3c). Candidate mutants were engineered and transduced into A-673 cells where a shift (by up to 10-fold) in IC50 with compound 1 treatment compared to wild type (WT) was observed in a cell viability assay, assessing either the variome pool or CPSF3 point mutants A395T or G330S, with G330S showing the strongest effect (Fig. 3d). The two most prevalent mutations identified, residues G330 and M331 (Fig. 3c), when mapped onto the co-crystal structure, would sterically clash with compound 2 while all of the low frequency mutant residues would fall immediately around the CPSF3 active site (Supplementary Fig. 13b,c). Accordingly, we found binding of compound 2 to human CPSF3 G330S mutant protein to be strongly reduced as measured by SEC-TID (Supplementary Fig. 13d). Additionally, compound-induced differentiation in A-673 cells was significantly blunted by overexpression of G330S mutant CPSF3 (Supplementary Fig. 13e,f).
CPSF3 inhibition results in mRNA read-through
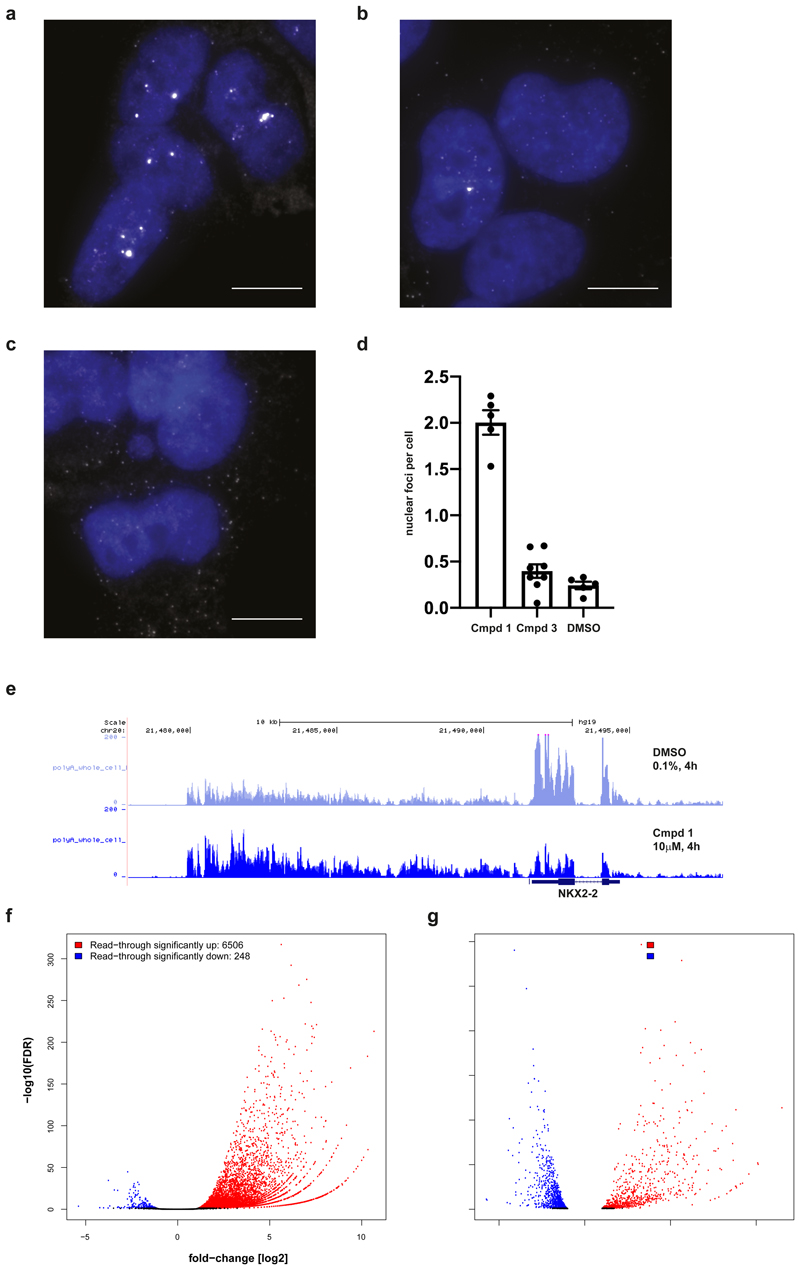
We next monitored pre-mRNA processing to further our mechanistic understanding of compound 1, using NKX2-2, a known downstream effector of EWS-FLI in Ewing’s sarcoma, as a candidate marker. Strikingly, single-molecule fluorescence in situ hybridization (smFISH) against NKX2-2 mRNA in A-673 cells treated with 1 revealed that transcripts become retained in the nucleus and accumulate in bright foci (Fig. 4a). These foci likely correspond to the sites of transcription, which is consistent with loss of CPSF3-mediated mRNA cleavage. In contrast, this effect was not observed following treatment with inactive enantiomer 3 or DMSO (Fig. 4b-d) or when imaging RUNX-3 mRNA as a control (Supplementary Fig. 14a-d). In line with crystallography and SEC-TID binding data, A-673 CPSF3 G330S mutant cells showed fewer foci upon imaging of NKX2-2 mRNA (Supplementary Fig. 15a-d). Furthermore, in agreement with compound-based inhibition of mRNA cleavage, “read-through” transcription of NKX2-2 beyond the gene’s annotated 3ʹ-UTR was found to be consistently upregulated when treated with compound 1 (Fig. 4e), supporting a mechanism of impaired RNA Pol II termination. Compound-induced read-through transcripts of NKX2-2 were not, or only minimally, exported from the nucleus (Supplementary Fig. 16b-d), suggesting a downregulation of cytosolic expression of fully processed NXK2-2 mRNA, in line with reduced NKX2-2 protein in A-673 cells in response to compound 1 treatment (Supplementary Fig. 1c). Globally, we found transcript read-through at about 6500 gene loci in A-673 cells upon treatment with compound 1 (Figure 4f), indicative of CPSF3’s central role in pre-mRNA processing. Transcript read-through resulted in a complex pattern of gene expression changes (Fig. 4g), which did not match or significantly enrich for previously published Ewing’s sarcoma gene signatures.32–34 However, when assessing transcripts downregulated with compound 1, gene sets related to DNA damage (“negative regulation of double-strand break repair” and “resolution of D-loop structures through Holliday junction intermediates”) as well as various RNA modification and metabolism related gene sets were among the most significantly enriched gene ontology (GO) terms (Supplementary Fig. 16a), pointing toward a compound 1 induced DNA damage-based phenotype. In A-673 CPSF3 G330S mutant cells however, compound 1 dependent read-through or changes in gene expression were strongly reduced (Supplementary Fig. 16e,f). Accordingly, at the NKX2-2 locus, read-through was abrogated in CPSF3 G330S mutant cells but not at the RUNX3 locus used as a control, in line with smFISH results (Supplementary Fig. 17a,b).
Figure 4. Compound 1 induces transcript accumulation and RNA Pol II read-through.
a,b,c, Images of A-673 cells that were treated with compound 1 (a), compound 3 (b) or DMSO (c) for 4 h prior to fixation and probed for NKX2-2 mRNAs (white) using FISH probes targeted to the coding sequence of the transcript. Nuclei (blue) were stained with DAPI. Images are representative from three independent experiments performed in duplicate. Scale bar = 10 μm. d, Compound 1 (79 cells) resulted in the formation of bright nuclear foci that contained multiple NKX2-2 transcripts (p-value < 0.0001, unpaired two-tailed t test) while transcripts in cells treated with compound 3 (40 cells) were similar to cells treated with DMSO (59 cells) (p-value=0.15, unpaired two-tailed t test). Data are presented as mean plus/minus standard error of the mean (error bars) for nuclear foci counted in each experiment. e, RNA-seq traces at NKX2-2 locus (hg19, chr20:21,477,893-21,498,177) generated from A-673 cells that were treated with 1 or DMSO for 4 h prior to whole cell lysis and RNA purification. Three biological replicates per condition merged into one trace. f,g, RNA-seq based quantification of (f) read-through expression genome-wide and of (g) changes in global gene expression, both in A-673 cells upon 4 h treatment with 1 versus DMSO, analyzing RNA purified from whole cell extracts of three biological replicates. Normalized read counts were analyzed using a negative binomial generalized log-linear model with two-sided comparisons and false discovery rate controlled using Benjamin & Hochberg multiple comparisons adjustment via edgeR. Significance cut-offs defined as absolute value fold-change [log2] > 1 and adjusted p-val/FDR < 0.05, and marked in blue (down) or red (up).
When profiling global transcript changes in the NOMO-1 AML cell line in response to treatment with compound 1, read-through transcription was observed at thousands of loci (Supplementary Fig. 18a), resulting in wide-spread changes in gene expression in either direction (Supplementary Fig. 18b). Analogous to our observation in A-673 Ewing’s sarcoma cells, genes downregulated in NOMO-1 cells did not significantly enrich for published MLL- or MYB-dependent AML signatures.17,35–37 Instead, gene sets “leukocyte differentiation” and “cytokine production” were the two most enriched gene ontology terms in line with our biomarker results and published data,6 respectively (Supplementary Fig. 18c). Interestingly, the gene set “regulation of response to DNA damage stimulus” was among the top ten enriched gene ontology terms, pointing to possible mechanistic overlaps in compound-induced effects between AML and Ewing’s sarcoma cells. Non-translocated, CES1-high lung adenocarcinoma A549 cells also showed comparable levels of transcript read-through and misregulation of gene expression in response to compound 1 (Supplementary Fig. 18d,e) with “double-strand break repair via homologous recombination” as the second most enriched gene ontology term (Supplementary Fig. 18f), indicative of CPSF3 target engagement yet without the detrimental effect on cell viability as seen with A-673 or NOMO-1 cells.
Compound 1 causes accumulation of nuclear R-loops
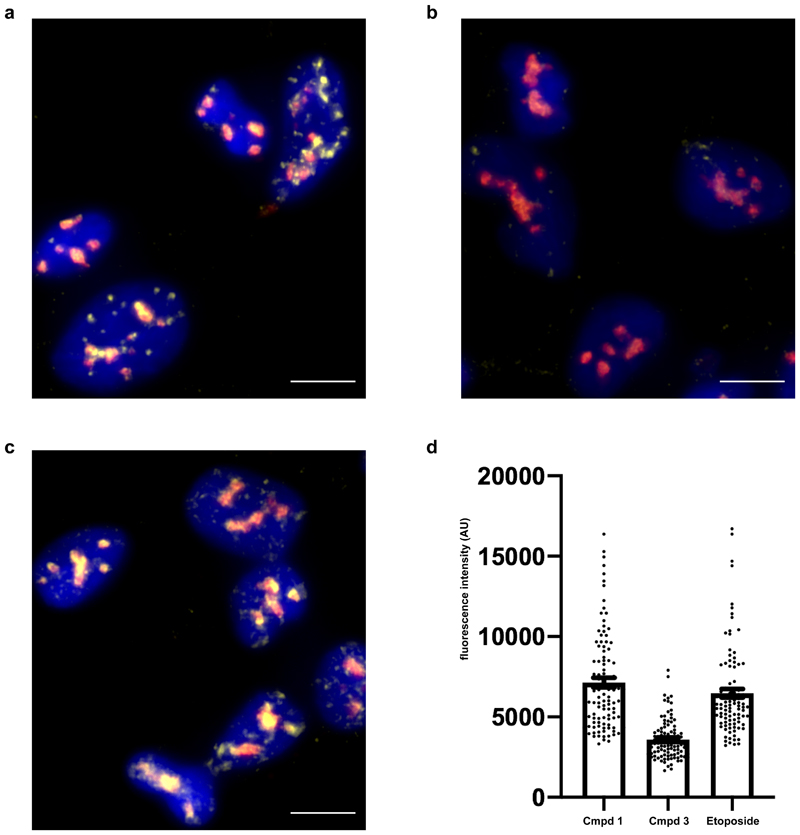
In line with our observed enrichment for DNA double-strand break repair associated genes among those downregulated with compound 1 in A-673 cells, Ewing’s sarcoma cells were recently shown to display elevated levels of three-stranded DNA-RNA hybrid structures called R-loops, which cause replication stress, DNA damage and ultimately cell death if not resolved.38 Therefore, we hypothesized that the lack of pre-mRNA cleavage and upregulation of read-through transcription observed in A-673 cells upon treatment with compound 1 may result in a further increase in R-loops, similar to the effects observed for mRNA cleavage and polyadenylation mutants.39 Staining against DNA-RNA hybrids in A-673 cells revealed that compound 1 did indeed strongly upregulate the formation of R-loops while treatment with the inactive enantiomer compound 3 did not. In fact, compound 1 induced R-loop formation to a level similar to that observed with etoposide, a genotoxin known to promote R-loop formation, (Fig. 5a-d), whose AML and Ewing’s sarcoma cell line sensitivity profile was similar to the one observed with compound 1 (Supplementary Fig. 19).
Figure 5. Compound 1 results in accumulation of nuclear R-loops.
a,b,c, Images of A-673 cells treated with compound 1 (a), compound 3(b) or etoposide (c) for 20 h prior to fixation and staining for R-loops (DNA-RNA hybrids, yellow), nucleolin (red) and DAPI (blue). Images are representative from two independent experiments performed in duplicate. Scale bar = 10 μm. d, Quantification of nucleoplasmic R-loop staining. Etoposide (100 cells) and compound 1 (100 cells) treatment result in increased R-loop staining compared to compound 3 (100 cells) (p-value <0.001, unpaired two-tailed t test). Data are presented as mean plus/minus standard error of the mean (error bars) for R-loop staining combined from all experiments.
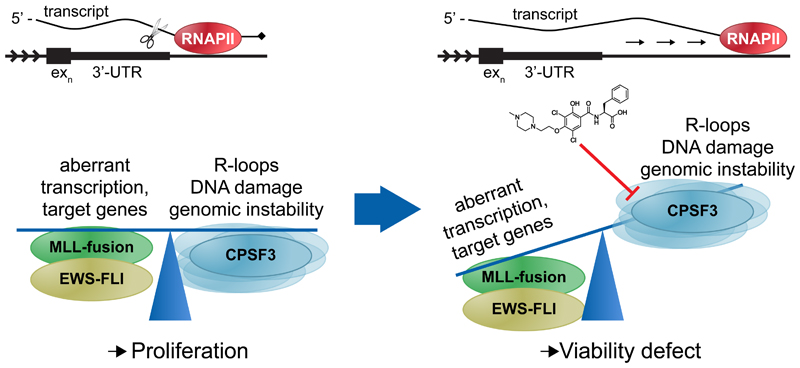
Thus, in summary, the results presented here point to a mechanism in which modulation of CPSF3 by JTE-607 prevents mRNA cleavage in conjunction with increased read-through transcription. This negatively affects the expression of key effectors such as NKX2-2 specifically as well as DNA damage response pathways globally, in turn impairing the cells’ ability to resolve the observed increase in R-loop formation, ultimately resulting in a cell viability defect (Fig. 6).
Figure 6. Proposed model for compound 2 mechanism of action.
Cancers such as EWSFLI translocated Ewing’s sarcoma or MLL-translocated AML have selected for a balance between oncogenic fusion protein-driven aberrant transcription and its associated genomic instability that allows for proliferation. Under such conditions, the mRNA processing and termination machinery, specifically one of its core components, the cleavage and polyadenylation specificity factor 3 (CPSF3), represents a previously undescribed novel synthetic lethal node. Compound 2 inhibition of CPSF3-mediated mRNA cleavage results in RNA Pol II read-through beyond the 3’-UTR and transcript accumulation. As a result, gene expression is perturbed, including downregulation of genes involved in DNA double-strand break repair, while R-loop formation is increased to levels that the cell can no longer buffer or balance, ultimately leading to a cell viability defect.
Discussion
JTE-607 (1) was first described as an inhibitor or blocker of cytokine release almost 20 years ago6 and has since been tested in various preclinical disease models as well as in healthy human volunteer studies,6–9 albeit without knowledge of its protein target. Similarly, while sensitivity of AML cells to compound 1 had been reported previously,10,11 the underlying subtype selectivity and mechanism of action remained unknown.
In this study we identify CPSF3, a core component of the pre-mRNA cleavage and polyadenylation complex, as the target of compound 2. Our work underscores the power of unbiased phenotypic screening coupled with a confluence of orthogonal approaches towards target identification, functional validation and deciphering of MoA. Correlating compound sensitivity with large-scale genomic data, we found AML and Ewing’s sarcoma cell lines enriched among those lines most sensitive to compound 1 and embarked on further experiments to better understand the target of the compound as well as its MoA. As such, 1 acts as a pro-drug which when converted by CES1 to its active form, compound 2, causes cellular viability defects in sensitive cancer cell lines in vitro as well as selective tumor stasis in vivo. Given such a pro-drug mechanism and its dependence on esterase, CES1 expression levels acted as a confounding factor when stratifying cell lines for compound sensitivity. However, going forward, our identification of CPSF3 as the efficacy target and presentation of a compound 2/CPSF3 co-crystal structure should enable future studies to circumvent this issue by screening for direct binders of the target protein, enabling the development of LMW inhibitors of CPSF3 that do not require a pro-drug motif. During the revision of this article a paper appeared which independently confirmed that compound 1 interacts with CPSF3.40
Since we did not observe a pan-toxic phenotype in our cell panel, nor have any adverse effects been reported in in vivo studies of compound 1 in both rodents and humans,7 it suggests that selective LMW targeting of CPSF3 could offer a therapeutic window.
While pharmacological modulation of a process as central to cellular function as mRNA processing may have been expected to be detrimental to cell viability in a non-selective manner, our data shows that CPSF3 is in fact not universally limiting in a given context. Importantly, as Cpsf3-null mice are embryonic lethal (http://www.mousephenotype.org/data/genes/MGI:1859328#section-associations) and genetic loss of function of CPSF3 across cancer cell line panels is consistently growth-inhibitory (https://depmap.org/portal/gene/CPSF3?tab=dependency), our findings highlight a difference between pharmacological modulation and genetic loss of function, thereby underscoring the utility of using small molecule-mediated phenotypic screening approaches to discover novel biology.
Based on the cumulative evidence presented in this study, inhibition of mRNA cleavage and concomitant transcript read-through emerge as a previously unrecognized synthetic lethality in cancers such as Ewing’s sarcoma or AML. Specifically, inhibition of CPSF3 represents a novel druggable node whose modulation tips the balance between aberrant transcription characterizing these cancers and the genomic instability resulting from it towards misregulated gene expression, DNA damage and downstream cellular viability defects.
Our work extends findings from recent studies that mechanistically linked EWS-FLI to pre-mRNA splicing, and reported Ewing’s sarcoma to be vulnerable to the induction of replication stress or inhibition of DNA damage repair pathways.38,41–45 We demonstrate Ewing’s sarcoma to be dependent on pre-mRNA processing via the pharmacological targeting of CPSF3, thereby offering a new paradigm for therapeutic intervention. While we focused our mechanistic studies on Ewing’s sarcoma cells, we speculate that a similar mechanism underlies the observed sensitivity of AML cells. That is, aberrant transcription sensitizes the cell to interference with CPSF3 mRNA processing. In support of our mechanistic model, a recent study described loss of CPSF complex members or associated RNA-binding protein WDR33 (most of which, including WDR33, were hits in our chemical proteomics experiments), to sensitize cancer cells to replication stress induced by nucleotide depletion or inhibition of the replication checkpoint kinase ATR, leading to RNA Pol II read-through and R-loop formation.46
Targeting of RNA biology with LMW therapeutics has recently emerged as a viable intervention point for diverse pathologies. Besides pharmacological inhibition of transcription and translation, and along with recent findings for the role of small molecule modulation of splicing,47,48 this work illustrates the impact of small molecules on mRNA processing in disease. In addition to targeting CPSF3 as a new therapeutic paradigm in cancer as described here, CPSF3 has recently been suggested as an anti-malarial target49 and modulation of CPSF3 by JTE-607 most likely underlies its previously described effects on cytokine production and immune modulation.6,40 As such, the probe molecule and target identification presented here should help drive what will undoubtedly be a rapidly growing area of scientific investigation as it is becoming increasingly clear that all steps of the gene expression pathway may be druggable.
Online Methods
Preparation of compounds 1-7 is outlined in Supplementary Note 1 and proton and carbon NMR spectra for compounds 2, 3, 4, 5 and 7 are provided in Supplementary Note 2
Compound detection by mass spectrometer
Cells were plated at a density of 2 x 106 cells per well of a 6 cm dish and treated with 10 µM compound 1, 2, 3, 4, or DMSO in biological triplicate for 4 h. Cell-free media was plated in dishes without cells and 10 µM compound was added for 4 h. After 4 h, supernatant media was removed from cultured cells, centrifuged to remove any detached cells and transferred to a new tube. Adherent cells were washed three times with cold PBS before being scraped off the dish and centrifuged to remove all PBS. Cell pellets, cell media supernatant, and cell-free media were stored at -80°C. For downstream analysis, all samples were thawed at room temperature. Cell pellets were treated with 200 µL of acetonitrile, capped and vortexed for 5 min. Per sample of supernatant media, 50 µl were transferred to a well plate, treated with 450 µL of acetonitrile, then vortexed for 5 min. All samples were centrifuged for 5 min at 4000 rpm at 4°C. After centrifugation, 100 µL of each sample were transferred to a new well plate and diluted with 100 µL of water. For 1:10 dilutions of cell pellets, 20 µL of acetonitrile extract was transferred to a well plate and diluted with 180 µL of water:acetonitrile (50:50, v:v). All samples were then heat-sealed, vortexed, and stored at 4°C prior to analysis. Calibration curves were prepared individually for each standard and were made up in a water:acetonitrile (50:50, v:v), covering a range of 10 pM to 10000 nM. Samples were analyzed using a 2 min gradient from 5-95% B utilizing an Agilent 1290 Infinity II UPLC equipped with a Sciex 6500+ triple quad mass spectrometer. Mobile phase A is water with 0.1% formic acid, mobile phase B is acetonitrile with 0.1% formic acid. The system was equipped with at Waters HSS T3, 2.1x50 mm, 1.8 µm column, the flow rate was set to 1 mL/min, with an 80/20 (to MS) split at the mass spectrometer. Sample injections were 5 µL. Compound esters were monitored using 524.3>127.2, CE = 45, and compound acids were monitored via 496.3>127.3, CE = 45. Source conditions were optimized for the compounds.
Plasma stability assay
Plasma stability assays were conducted with final concentrations of 99% plasma, 1% DMSO, and 5 μM compound. Incubations occurred in EDTA-treated plasma (BioreclamationIVT) from five species – C57BL/6 mouse, Sprague Dawley rat, Beagle dog, Cynomolgus monkey, and human. Time points up to 4 h were collected and quenched on ice with 6X volume of acetonitrile containing an internal standard, 100 ng mL-1 glyburide. Samples were vortexed then centrifuged to precipitate protein. A 6:5 ratio of water:supernatant were mixed for injection on a Sciex 4000 with a TSI source in positive mode with a source temperature, spray voltage, gas 1, gas 2, CAD gas, and curtain gas settings of 500, 5500, 60, 60, 10, and 30, respectively. Transitions for 1, 2, and glyburide were 524.2 > 84.1, 496.4 > 127.1, and 494.2 > 168.8, respectively, with declustering potentials of 70, 100, and 70 V, respectively, and collision energies of 45, 65, and 40 V, respectively. An Agilent liquid chromatography system used a ACE C18 30x2.0 mm and 3 μm column (Advanced Chromatography Technologies Ltd.) with solvents A and B containing 0.1% formic acid in water (A) and acetonitrile (B) at a flow rate of 0.7 mL min-1 with a gradient profile of 2% B from 0 to 0.5 min, an increase from 2% to 98% B from 0.5 to 2 min, 98% B from 2 to 2.5 min, a decrease from 98% to 2% B from 2.5 to 2.6 min, and 2% B from 2.6 to 3 min.
CES1 overexpression experiment
Homo sapiens carboxylesterase 1 (CES1), transcript variant 1 (NM_001025195.1) was Gateway LR cloned (ThermoFisher Scientific 11791020) from pDONR221 (ThermoFisher Scientific 12536017) into pLNCX2 Retrovival Vector (Takara 631503) modified by adding Gateway cassette into the MCS. Retrovirus was generated in HEK293T (RRID:CVCL_0063) by transient transfection of pLNCX2-CES1 with pLP-GAG-Pol and pLP-VSVG. CMK (RRID:CVCL_0216) and SEM (RRID:CVCL_0095) cells were infected with virus at an MOI of 1 and cells were selected with Neomycin for 7-10 days. For western blot cells were treated with 10uM of 1 or 3 for 4 h and prepared for western blot as detailed below. For cell viability cells were treated with an 8 pt. 3.16-fold dilution of 1 with a top concentration of 30 μM and experiment was carried out as detailed below.
Cell viability experiments
Cell viability experiments against the 92 cell line panel (Supplementary Table 1), were performed using an ultra-high throughput screening system (GNF Systems). Each cell line was harvested and suspended to a concentration of 50,000 cells mL-1 in the appropriate medium (RPMI or DMEM, supplemented with 10% Fetal Bovine Serum). Cells (250 cells/well, 5 µL/well) were then dispensed into Greiner white, solid-bottom, TC-treated, 1536-well assay plates (Greiner 789173-A) and incubated for 10 hours at 37°C (95% humidity, 5% CO2). Compound (15 nL/well; 11 pt., 3.16-fold dilutions, 30 µM to 0.1 nM final concentration) and controls (15 nL/well; MG-132 and DMSO) were then added to the assay plates in duplicate using an Echo acoustic liquid dispenser (Labcyte). Assay plates were incubated 3 days at 37°C (95% humidity, 5% CO2) before addition of CellTiter Glo (4 µL/well, Promega). Assay plates were incubated for 15 min at room temperature and then luminescence was measured using a ViewLux uHTS Microplate Imager (PerkinElmer). Reported IC50 values were calculated as described.16
In vivo efficacy studies
All animal studies were carried out in accordance with federal, state, local and institutional guidelines governing the use of laboratory animals in research, and were approved by the Novartis Institutes for BioMedical Research Institutional Animal Care and Use Committee. PK studies were conducted using male B6 mice (C57BL/6NCrl, Charles River) at 8 weeks of age. For efficacy experiments, 2x106 cells were implanted subcutaneously with 50% Matrigel, in 8 female athymic nude mice (Hsd:Athymic Nude-Foxn1nu, Harlan) per treatment group at age 8 weeks. Dosing began when tumor volume reached 150-200 mm3 (8 days post-implant for Molm-13, 21 days post-implant for A549). Compound 1 was formulated in 20% 2-hydoxyproply-β-cyclodextrin at 20 mg mL-1, and delivered twice per day by subcutaneous injection. Tumor volume was determined by caliper measurements obtained in two dimensions and calculated as width2 x length 2-1. Error bars represent standard error of the mean, and were calculated using GraphPad Prism software. Viable tumor tissue was preserved in RNALater (Sigma), and total RNA was isolated by standard methods using a QIAsymphony instrument (Qiagen). TaqMan assays were performed with optimized probes (Applied Biosystems), using PPIA (cyclophilin A) as an internal standard in multi-plexed format. Error bars represent standard error of the mean, and were calculated using GraphPad Prism software.
FACS
For cell surface antibody staining, 1x 106 either A-673 (ATCC CRL-1598) Ewing’s sarcoma cells, A-673 cells with FLI1 shRNA1917, or THP-1 (ATCC TIB-202) AML cells were plated, incubated overnight (37°C, 5% CO2, 95% relative humidity), and treated with compound 1 (1μM), DMSO (0.1%) or doxycycline (100 ng mL-1) the following day for 72 h (THP-1), 96 h (A-673), or 120 h (A-673 FLI1 shRNA1917). Cells were washed into FACS staining buffer (PBS, with 1% BSA) and stained with either PE-conjugated CD73 (BioLegend 344004), CD44 (BioLegend 338808), CD54 (BioLegend 353106), or CD11b (BioLegend 301306). Samples were processed on a LSR Fortessa cell analyzer (BD Biosciences). Data was analyzed using FlowJo V10 (FLOWJO, LLC) by gaiting cells based on SSC-A/FSC-A for live cells and FSC-W/FSC-A for single cells.
For cell cycle and Annexin V staining, 5x 105 NOMO-1 cells were plated, incubated overnight (37°C, 5% CO2, 95% relative humidity), and treated the following day with either compound 1 (1 or 10 μM) or DMSO (0.1%) for 1.5, 4, or 24 h. Cells were lifted, washed with PBS and each sample’s volume was split, half for Annexin V staining and half for cell cycle analysis. To assess apoptosis levels, cells were stained with Annexin V Dead Cell Apoptosis Kit (Thermo Fisher Scientific) according to manufacturer’s instructions. To assess cell cycle status, cells were permeabilized in 70% ethanol at -20°C for 15 min, followed by PBS washes and staining with BD Pharmingen PI/RNase Staining Buffer (BD Biosciences). Samples were processed on a LSR Fortessa cell analyzer (BD Biosciences) and data was analyzed using FlowJo V10 by gaiting cells based on SSC-A/FSC-A for live cells and FSC-W/FSC-A for single cells. For Annexin V analysis, PI-A/FITC-A scatter plots were generated with quadrant parameters based on DMSO-treated controls. For cell cycle analysis, cell count/PI-A histograms were generated and analyzed using the FlowJo cell cycle analysis module.
Western blot
To assess protein levels by western blot, 5x 106 cells (A-673, Molm-13, Nomo-1, or HEL92.1.7) were plated, incubated overnight (37°C, 5% CO2, 95% relative humidity), and treated with compound 1 (1 or 10μM), compound 3 (10μM), or DMSO (0.1%) for 4 h. Cells were washed with PBS and lysed in RIPA buffer (Boston Bioproducts) containing protease inhibitor (Roche Complete Mini) and phosphatase inhibitor (Roche Phos-Stop). Protein was quantitated using a Lowry assay kit (BioRad) and a SpectraMax plate reader (Molecular Devices). All samples were normalized to 30μg total protein in NuPage LDS Sample Buffer (Thermo Fisher Scientific) with NuPage Sample Reducing Agent (Thermo Fisher Scientific) and heated to 70°C for 10 min. Samples were run on a NuPage Novex 4-12% Bis-Tris gel (Thermo Fisher Scientific), followed by transfer to nitrocellulose membrane (Thermo Fisher Scientific). Blots were probed with either anti-FLI1 antibody (Abcam ab124791, dilution 1:1000), anti-NKX2.2 antibody (Abcam ab187375, dilution 1:1000), anti-MYC antibody (Cell Signaling Technologies 13987, dilution 1:1000), anti-MYB antibody (Cell Signaling Technologies 12319, dilution 1:1000), anti-MEK1/2 (Cell Signaling 8727, dilution 1:1000), anti-Vimentin (Cell Signaling 5741, dilution 1:1000), anti-CPSF3 (Abgent AT1610a, dilution 1:1000), anti-CES1 (Abcam ab68190, dilution 1:1000), or anti β-actin antibody (Sigma A5441, dilution 1:10,000) in 3% non-fat dry milk in TBST (TBS, 0.1% Tween-20) at 4°C overnight (see Supplementary Table 3 for list of antibodies used in this study). Blots were washed in TBST, followed by 1 h room temperature incubation in either anti-mouse HRP (GE Amersham) or anti-rabbit HRP (GE Amersham) secondary antibody, followed by washed in TBST. Blots were developed using SuperSignal West Dura Chemiluminescent substrate ECL (Thermo Fisher Scientific) on film.
qPCR
To assess mRNA expression levels by quantitative real-time PCR, 2x 106 cells (A-673, A-673 FLI1 shRNA1917, or Molm-13) were plated, incubated overnight (37°C, 5% CO2, 95% relative humidity), and treated with compound 1 (as indicated), DMSO (0.1%) or doxycycline (100 ng ml-1) the following day for 2, 4, 8, or 120 h. Cells were washed with PBS and lysed in RLT buffer (Qiagen), followed by RNA isolation using RNeasy Mini Kit (Qiagen). cDNA was prepared using SuperScript III First-Strand cDNA Synthesis System (Thermo Fisher Scientific). qPCR was performed using the following TaqMan assays (Applied Biosystems) with TaqMan Fast Advanced Mastermix (Thermo Fisher Scientific): EWSR1-FLI1 (Hs03024497_ft), NKX2-2 (Hs00159616_m1), MLL-AF9 (Hs03296416_ft), MYB (Hs00920556_m1), MYC (Hs00905030_m1), CEBPa (Hs00269972_s1), EGR1 (Hs00152928_m1), GFI1 (Hs00382207_m1), IL-8 (Hs00174103_m1), MPO (Hs00924296_m1), and PPIA endogenous control (Hs99999904_m1). Relative RNA quantities were determined against a standard curve generated from pooled cDNA using a ViiA-7 qPCR instrument (Applied Biosystems) and normalized to endogenous control per sample and to DMSO control per time point.
siRNA screening procedure
The siRNA screening library consisted of ~150,000 individual siRNAs (composed of 4 Qiagen unmodified siRNAs and 4 Dharmacon On-Target-Plus siRNAs for ~19,350 human genes, Supplementary Dataset 1). The siRNAs were suspended in water at 2 aM and 60 nL of each was dispensed into a well of white TC treated 1536-well assay plates (Corning, Cat#789173), sealed, and stored at -20°C as assay-ready library stamps. A-673 cells were reverse-transfected by first resuspending siRNAs in 3 CL transfection buffer (Lipofectamine RNAiMAX (Thermo Fisher Scientific) 1:200 dilution in Opti-MEM) for 20 minutes. A-673 cells (250 cells/well) in RPMI with 20% Fetal Bovine Serum were dispensed (3 /L/well) into the siRNA plates using a GNF bottle valve liquid dispenser, resulting in a final siRNA concentration of 20 nM. Two parallel screens were conducted, one screen was performed with treatment of 0.4 µM compound 1 and a second set with DMSO treatment only (0.4% final concentration). The cells, siRNA, and compound (or DMSO) were incubated for 72 hours at 37°C and 5% CO2. Cell viability was determined using the CellTiterGlo reagent (Promega; 3 .L per well), using a Perkin Elmer Viewlux plate reader.
siRNA screening data analysis
The CellTiterGlo activity measurement (Promega), aquired on a Perkin Elmer Envision plate reader, was normalized by plate using the median measurement of 32 negative control siRNA (Qiagen AllStars Negative Control siRNA SI03650318) transfected wells (neutral control, NC) and the median measurement of 32 PLK1 positive control siRNA (Qiagen Hs_PLK1_6 FlexiTube siRNA Cat#SI02223837 siRNA) transfected wells (active control, AC) per plate as follows: Nx = (-100)*(x-NC)/(ACNC) where x is the measurement for siRNAx. The activity measurement corrected for the minimum value of N was log2 transformed. The log2-transformed value for each siRNA in the individual conditions (DMSO or 1 at 0.4 µM) or in the differential (log2-transformed value in DMSO condition subtracted from low concentration 1 condition) was then robust zscore-normalized (robust z-score = (log2_value – Median_log2_values)/(1.4826*MAD_log2_values)), where MAD is the median absolute deviation. The activity at gene-level was determined and graphed using a combination of 3 calculations. (1) The RSA (redundant siRNA activity23) statistic was performed without setting the upper or lower limits for each of the assay directions (up and down); (2) the quartiles Q1 and Q3, representing the 25th percentile and 75th percentile activity value, respectively, out of the number of siRNAs values for each gene; (3) the z_count_down and z_count_up values representing the number of siRNAs for a gene passing a threshold of robust z-score less than or equal to -2.0 or more than or equal to +2.0, respectively. See Supplementary Tables 8 for gene-level activity (“differential”, “cmpd1”, “DMSO”), including a column “num_sirna_tested” per gene, with on average 8 siRNAs tested per gene.
The screen results were graphed as the Quartile (Q1 or Q3) of corresponding RSA statistic (log10 p-value) with Spotfire 6.5. Color was set as the z_count viewed.
Gene set enrichment analysis. The top 50 genes based on RSAdown_log10 p-value ranking were selected and used as input gene list. GeneMANIA24 (version and content available at http://genemania.org/data/archive/2013/10/15) extended extended the gene list to over connected neighbor interacting genes based on selected interaction datasets. Gene Ontology (GO) terms enrichments passing a FDR p-value of 0.05 are reported.
Chemical proteomics experiments
Preparation of cell lysate, affinity matrix synthesis, lysate-based enrichment, sample processing, and data acquisition were performed as previously described.25 In brief, NOMO-1 lysates were treated either DMSO, 1 µM 2, 10 µM 2, 100 µM 2 for 1 h at 4°C. Following preincubation with competition compounds, each lysate was incubated with compound 17 conjugated to sepharose beads (2 µmol ml-1; 35 µl of packed bead volume per treatment condition) for 4 h at 4°C. Experiments were performed in duplicate.
Photoaffinity labeling experiments
In brief, 10 million cells (either NOMO-1 or A-673) were added to individual 15 cm2 plates and allowed to recover overnight at 37°C with 5% CO2. The following day, each plate was washed with 3x with PBS and incubated in Opti-MEM Reduced Serum Media with 0% FBS. DMSO was added to 5 plates, while the remaining plate was treated with 20 µM 1 for 1 h at 37°C with 5% CO2. DMSO-treated cells were then treated with 0, 0.01, 0.1, 1 or 10 µM 7, while 1-treated cells were treated with 1 µM 7 for 1 h at 37°C with 5% CO2. Irradiation, cell lysis, enrichment, sample processing and data acquisition were performed as previously described.26
Quantitative Proteomics by 2D nanoLC-MS/MS
The TMT-labeled sample was reconstituted in 0.1 % TFA/2% acetonitrile for offline high-pH reversed phase separation (RP10) using a Dionex UltiMate™ 3000 HPLC system with fraction collection operating Chromeleon v. 6.8 (Thermo). Separation was acheived on a Waters Xbridge™ C18 3.5 µm 2.1 x 150 mm HPLC column (Waters) using a flow rate of 250 µL/min and the following ternary gradient and conditions: Mobile phase A=Water (HPLC grade), Mobile phase B=Acetonitrile (HPLC grade), and Mobile phase C=200 mM ammonium acetate, pH 10. Mobile phase C is held at 10% throughout gradient. Starting conditions are 89% A and 1% mobile phase B, ramping mobile phase B to 50% over 65 minutes, and returning to 1% B from 65-75 minutes. Fractions were collected in U-bottom 96 well plate at 1 min/well. Fractions from 3-min intervals were combined, dried, and reconstituted in 0.1 % TFA/2% acetonitrile for nLC-MS/MS comprising 16 fractions. The fractions were analyzed by top 12 data-dependent nLC-MS/MS using a LTQ Orbitrap Elite (Thermo) coupled to an Easy-nLC 1000 nanoflow liquid chromatograph operating at 275 nL/min using custom made nanoESI interface source. MS1 Data was acquired m/z 350-1400 using 60,000 resolution with a target value of 1 x 106, and a maximum injection time of 150 ms. HCD MS2 was collected using collision energy of 40% at a resolution of 17,500 (@ 400 m/z) and a target value was 5 x 104 with an isolation window of 1.5 Da. MS2 maximum injection time of 100 ms, with dynamic exclusion set for 60 s with a repeat count of 1.
Data Processing and Analysis
Xcalibur (Thermo) Rawfiles were converted to Mascot-compatible mgf-files as well as subsequent mzXML, pepXML, protXML files using the Trans-proteomic pipeline (TPP) modules (Institute for Systems Biology). Mgf-files were searched against human fasta database using Mascot v 2.5.1 (Matrix Science) with peptide mass tolerance set to 10 ppm, fragment tolerance set to 0.1 Da and trypsin cleavage specificity (cleavage at K, R except if followed by P) allowing for 2 missed cleavages. Carbamidomethylation of cysteine was set as fixed modification, and methionine oxidation, and TMT-modification of N-termini and lysine residues were set as variable modifications. Peptide and protein data validation was performed using PeptideProphet and ProteinProphet modules in TPP, deriving a complete dataset from combined Mascot search results using the decoy database function, nonparametric model, and the standard peptide probability threshold of 0.05. TPP output (Interact) files containing peptide and protein information and corresponding TMT reporter ion peak intensity information for each peptide–to-spectrum match (as parsed from the Mascot result files) were uploaded to Oracle using in-house software. For each peptide sequence and modification state, reporter ion signal intensities from all spectral matches were summed for each reporter ion type to account for lower precision of reporter ion ratios at low absolute signal intensities. Only peptide-to-spectrum matches that are unique assignments to a given identified protein within the total dataset were considered for protein quantitation. High confidence protein identifications were reported based on protein probability cut-off values based on a false positive prediction of <1% by ProteinProphet. For each protein in the high confidence dataset, individual peptide fold changes were calculated by normalizing each channel to the control channel. Protein fold changes over control were derived as median peptide fold change per protein.
CPSF3 protein expression and purification
A DNA construct of human CPSF3 (based on pdb entry 2I7T) with an N-terminal His-tag and thrombin cleavage site was cloned into a customized vector. Protein expression in E. coli NiCo21(DE3) was induced with 100 μM IPTG at 20°C. CPSF3 was purified by Ni sepharose affinity and size exclusion chromatography on Superdex200 (GE Healthcare) equilibrated with 20 mM HEPES pH 7.5, 150mM NaCl, 5% glycerol and 1 mM DTT. Peak fractions were pooled, concentrated to 10 mg/mL and used for binding assays and crystallization.
SEC-TID
Size-exclusion chromatography for target identification (SEC-TID) was performed as previously described.27
Cellular Thermal Shift Assay (CETSA)
NOMO-1 lysates utilized in the chemical proteomics experiments were used for cellular thermal shift experiments. Lysates were thawed on ice and spun at 20,000g rcf for 20 min at 4°C to remove any precipitate protein resulting from freeze thaw cycle. Lysates were diluted to 2 mg mL-1 in buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 5% glycerol and treated with DMSO or 100 µM 2 at 25°C for 1 h. Treated lysates were then aliquot to individual wells of a 96-well PCR plate; 6x100 µL aliquots of DMSO-treated lysates and 6x100 µl aliquots of 2-treated lysates. Lysates were heated between 40 and 65°C for 3 min, and cooled to 25°C for 1 min. Lysates were then transferred to individual 0.5 ml tubes and spun at 20,000g rcf for 20 min at 4°C. The resulting supernatants were transferred to individual 0.5 ml tubes and CPSF3 levels were assessed by western blot.
Purification of Core CPF
Recombinant 8-subunit core CPF was purified from insect cells as previously described.29
Cleavage Assays
Cleavage assays were carried out as previously described29with some modifications. Briefly, reactions were reconstituted with 50 nM core CPF, 300 nM CFIA and 300 nM CFIB, 3 mM DTT, 1X polyA buffer (5 mM HEPES pH 8.0, 150 mM KOAc, 2 mM MgOAc, 0.05 mM EDTA, 2% PEG 6000) and 1 U/µl RiboLock (Thermo Fisher, EO0381). Reactions were pre-incubated on ice with 1% DMSO or 100 µM compound 2, 1 or 4 for 10 min, and transferred to 30°C where the reaction time course was started by adding 50 nM Cyc1 RNA. Reactions were sampled at t = 0, 10, 30 and 90 min by taking 15 µL aliquots of the reaction, adding them to 4 µL stop buffer (1X polyA buffer, 130 mM EDTA, 5% SDS, 12 mg/ml proteinase K) and incubating them at 37°C for at least 10 min. 20 µl of 2X RNA loading buffer (1 M NaCl, 1 mM EDTA, 78% formamide, 0.01% bromophenol blue, 0.005% xylene cynol) was added and incubated at 70°C for 5 min before running the reaction on prewarmed (30W for 30 min) 6% TBE-Urea PAGE. Gels were stained with SYBR Green II for 15 min in 1X TBE, and visualized on a BioRad ChemiDoc. Gel images were background subtracted and contrast adjusted using ImageJ50 and substrate bands quantified by integration. Percentage of cleaved substrate (% RNA cleaved) was calculated by (1) measuring the intensity of the substrate band at t = 10, 30 or 90 and normalizing it to t = 0 within the corresponding time course, (2) subtracting that ratio from 1 and (3) multiplying by 100.
CPSF3 crystallization and structure determination
CPSF3 in 20 mM Hepes pH 7.5, 150 mM NaCl, 5% Glycerol, 1 mM DTT was supplemented with 2 to a final concentration of 0.5 mM, concentrated to 10.5 mg mL-1 and crystallized in sitting drops at 293 K. Crystals grew in conditions containing 100 mM Tris/HCl pH 7.5, 200 mM NaCl, 0.4 M NaH2PO4 and 1.47 M K2HPO4. For cryoprotection, crystals were incubated in reservoir solution supplemented with 20% glycerol (v/v) before freezing in liquid nitrogen. X-ray data was collected on a Pilatus 6M detector at the Advanced Photon Source beamline 17-ID at a wavelength of 1.0 Å. Reflections were indexed, integrated, and scaled using autoPROC.51The space group was determined to P43212 with two CPSF3 molecules in the asymmetric unit. The structure was determined by molecular replacement in PHASER52 using the two domains of apo CPSF3 (pdb code 2i7t, residues 9 to 207// 396 to 459 and 208 to 396, respectively) as individual search models. Fo-Fc difference electron density at the active site indicated the presence of 2 (Supplementary Figure 11d). The compound was placed into the density in coot53 followed by iterative rounds of model building and refinement in BUSTER.54 The final model with residues 7 to 459 was refined to an R-factor of 0.165 (Rfree=0.221), and has good geometry without residues in the disallowed regions of the Ramachandran plot. Data collection and refinement statistics are shown in Supplementary Table 2.
Variomics
The Variomics lentiviral expression vector pXP1510 was modified from pINDUCER2155 by replacing the existing protein expression units with a synthesized expression cassette that uses the EF1a promoter to drive expression of a transcript encoding both the mutant protein and the Neomycin resistance marker separated by an IRES element. Gateway cloning was used to create a wildtype CPSF3 (NP_057291.1) pXP1510 template. Error-prone PCR55 using the primers XPO1920 (5’GAGCCTACCTAGACTCAGC3’) and XPO2007 (5’CTGCTTCCTTCACGACATTC3’) followed by cloning of NotI-AscI-digested PCR products into pXP1510 yielded a library of CPSF3 mutants with an average of 4 mutations per clone (confirmed using Sanger sequencing 48 clones with XPO1920 and XPO2007). Variomics library DNA was packaged in lentivirus by transfecting into HEK293T cells using TransIT-293 transfection reagent (Mirus Bio) with Cellecta Packaging vector mix (Cellecta). Virus was generated over the course of 3 d and harvested through 0.45 μM cellulose acetate filters (Corning). A-673 cells were infected with virus by plating 1.5x 105 cells in 2 mL of media, incubating overnight (37°C, 5% CO2, 95% relative humidity), then adding 100 μL of viral supernatant with 5 μg mL-1 polybrene (Millipore) and incubated for 24 h. Cells were selected with 0.5 μg mL-1 Geneticin (Thermo Fisher Scientific) for 7 d, or until non-transduced cells were dead. Genomic DNA was prepared using PureLink Genomic DNA kit (Thermo Fisher Scientific). The primers XPO1920 and XPO2007 were used with Kapa HiFi (Kapa Biosystems) to amplify CPSF3 mutants from genomic DNA extracted from cells that survived selection. PCR products were digested with NotI-AscI and cloned into pXP1510. The CPSF3 mutation profile was determined by Sanger sequencing 294 resulting clones with XPO1920 and XPO2007 and compiling the observed mutations.
Colony formation assay
Colony formation assays were performed by plating 2 x 105 cells per 6 well and treating with 50, 25, 12.5, 6.25, 3.13, and 1.56 μM of compound for 7 d, followed by a media change and re-application of compound for additional 7 d incubation. To quantify colony formation, cells were washed with PBS and stained with crystal violet (4% PFA, 0.1% crystal violet, in PBS) for 20 min, followed by PBS washes. For cell viability curves, 250 cells per well were seeded in 1536-well plates and incubated overnight (37°C, 5% CO2, 95% relative humidity). The following day compound was added using an Echo 555 Acoustic Dispenser (Labcyte), followed by 72 h incubation. Viability was assayed using CellTiterGlow reagent (Promega). Viability curves were NC1 normalized to DMSO (0) and 8 μM MG132 (-100) treatments.
RNA FISH
A-673 cells were cultured on poly-lysine coated #1 coverslips (Neuvitro) in DMEM (Life Technologies) supplemented with 10% FBS (Sigma) and 1% Pen Strep (Life Technologies). Cells were treated with either DMSO, 10 μM compound 1, or 10 μM compound 3 for 4 h before fixation with 4% paraformaldehyde (Electron Microscopy Sciences) in PBSM (1X PBS with 5mM MgCl2) for 20 min at room temperature. Cells were washed two times with PBSM and permeabilized with 0.5% (v/v) Triton X-100 (Sigma) in PBSM for 5 min at room temperature. After permeabilization, cells were washed two times with PBSM and then incubated with pre-hybridization solution (2X SCC, 10% (v/v) formamide (Sigma)) for 5 min at room temperature. Cells were then hybridized with 250nM of NKX2-2 or RUNX3 FISH probes fluorescently labeled with Quasar 570 (Biosearch Technologies) in 2X SSC, 10% (v/v) formamide (Life Technologies), 10% (w/v) dextran sulfate (Sigma) for 16 h at 37°C. Following hybridization, cells were washed two times with pre-hybridization solution warmed to 37°C for 30 min. Cells were washed three times with PBSM for 5 min at room temperature before counterstaining DNA with DAPI (0.5 mg l-1) and mounted on slides using ProLong gold reagent (Life Technologies). For sequences of FISH probes, see Supplementary Table 4.
RNA FISH image acquisition
RNA FISH slides were imaged on a Zeiss Axioimager Z1 microscope using a 100x 1.4NA Plan-APOCHROMAT oil immersion objective (Zeiss) and AxioCam 506 mono camera (Zeiss). An X-cite 120 (EXFO) light source was used for illumination with filter sets for DAPI and Cy3 (Zeiss). Images were acquired with Zen software (Zeiss) as z-stacks (5 μm range in 0.24 μm steps).
RNA FISH image analysis
Three-dimensional image stacks were maximally projected along the z-axis to a single two-dimensional plane for analysis of RNA particles using ImageJ.49 The position and intensity of RNA particles were automatically detected as diffraction-limited fluorescence signals using custom software written in Matlab (Mathworks).56 Nuclei were automatically segmented on the maximum intensity projection of the DAPI z-stack (ImageJ). Nuclear foci were identified as spots that were three times the average intensity of a single mRNA and also located within the nucleus.
R-loop staining
A-673 cells were cultured on poly-lysine coated #1 coverslips (Neuvitro) in DMEM (Life Technologies) supplemented with 10% FBS (Sigma) and 1% Pen Strep (Life Technologies). Cells were treated with either etoposide (10 μM), compound 1 (10 μM) or compound 3 (10 μM) for 20 h. The cytoplasm was extracted using 0.1% Triton-X100 in 1X PBS for 30 s before fixation with 4% paraformaldehyde (Electron Microscopy Sciences) in 1X PBS for 10 min at room temperature. Cells were washed two times with 1X PBS and permeabilized with 0.5% (v/v) Triton X-100 (Sigma) in 1X PBS for 5 min at room temperature. After permeabilization, cells were washed two times with 1X PBS and then blocked with 1X PBS, 0.1% Tween20, 1% BSA and 4% goat serum for 1 h. Cells were incubated with primary antibody against R-loops (mouse monoclonal anti-DNA-RNA Hybrid [S9.6] antibody, dilution 1:200, Kerafast ENH001, generated against a ΦX174 bacteriophage-derived synthetic DNA-RNA antigen) and nucleolin (dilution 1:1000, Abcam ab22758) overnight at 4°C. The following day, cells were washed two times with 1X PBS, 0.1% Tween20, 1% BSA and 4% goat serum for 5 min and then incubated with fluorescent secondary antibodies (Alexa 568 goat anti-mouse (dilution 1:1000, ThermoFisher Scientific A11031) and Alexa 647 goat anti-rabbit (dilution 1:1000, ThermoFisher Scientific A21245)) for 30 min at room temperature in the dark. Cells were then washed two times with 1X PBS before counterstaining DNA with DAPI (0.5 mg l-1) and mounted on slides using ProLong gold reagent (Life Technologies).
R-loop image acquisition
R-loop slides were imaged on a Zeiss Axioimager Z1 microscope using a 100x 1.4NA Plan-APOCHROMAT oil immersion objective (Zeiss) and AxioCam 506 mono camera (Zeiss). An X-cite 120 (EXFO) light source was used for illumination with filter sets for DAPI, Cy3 and Cy5 (Zeiss). Images were acquired with Zen software (Zeiss) as z-stacks (5μm range in 0.24μm steps).
R-loop image analysis
Three-dimensional image stacks were maximally projected along the z-axis to a single two-dimensional plane for analysis of R-loops using ImageJ.50 Nuclei were automatically segmented on the maximum intensity projection of the DAPI z-stack (ImageJ). The nucleolus was masked using the nucleolin signal and only the resulting nucleoplasmic R-loop staining was quantified.
RNA sequencing
Global transcript levels and RNA Pol II read-through in A-673 cells, A-673 CPSF3 G330S mutant cells, NOMO-1 cells or A549 cells were measured using RNA sequencing (RNA-seq) technology. Cells were treated with compound 1 at 10 μM or with 0.1% DMSO for 4 h prior to RNA purification from whole cell extracts or cytoplasmic extracts, as indicated. Cytoplasmic extracts were prepared using a Cell Fractionation kit (Cell Signaling Technology). Each experimental condition was performed in triplicate and RNA was isolated using the RNeasy Plus Mini kit (Qiagen) and polyadenylated mRNA was enriched using oligo-dT capture. The amount of RNA was quantified using the Qubit RNA HS Assay Kit (Invitrogen) and inspected using the Agilent RNA 6000 Nano Kit (Agilent Technologies). RNA libraries were prepared using the Illumina TruSeq Stranded mRNA Sample Preparation Kit and sequenced using the Illumina HiSeq2500 platform following the manufacturer’s protocol. Samples were sequenced in paired-end mode to a length of 2x76 base-pairs. Images from the instrument were processed using the manufacturer’s software to generate FASTQ sequence files. Read quality was assessed by running FastQC on the FASTQ files. Sequencing reads showed excellent quality, with a mean Phred score higher than 30 for all base positions. The raw RNA-sequencing reads are available in the NCBI Short Read Archive under accession number SRP158650.
RNA-seq data processing
Reads were aligned to the February 2009 (hg19) human genome assembly from UCSC.57 Alignments were performed using STAR v.2.5.0a with the following settings to allow reporting of one randomly chosen alignment per multi-mapping read: “— outFilterMultimapMax 20 –outMultimapperOrder Random --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMmultNmax 1 --outSAMtype BAM SortedByCoordinate”.58 Aligned and sorted reads were indexed using SAMtools v.1.3.1.59
Differential expression and read-through analysis
For differential mRNA expression analysis, reads were counted over all exons from the hg19 UCSC Genes annotation track using the qCount function from the QuasR package (v.1.14.0). Exons were collapsed to yield one value per gene. Genes with a mean count of at least 16 reads across all samples were used as an input for differential expression analysis using edgeR (v. 3.22.3, with R v. 3.5.0). A threshold test was performed using the glmTreat function to detect genes with a change in expression of at least 2-fold in either direction (log2 fold-change cutoff of 1, adjusted p-value < 0.05) upon compound treatment. In order to identify and analyze read-through, gene bodies were first defined as the distance between the start of the 5’-most annotated exon and the end of the 3’-most annotated exon, then extended by 20 bp in both directions. Downstream regions were then defined as the distance between the 3’ end of each gene and the 5’ end of the next gene on the same strand, or up to 10 kb downstream of the 3’ end if no downstream gene occurred within that distance. Reads overlapping all downstream regions were quantified in a strand-specific manner. Downstream regions with > 16 counts in at least 3 samples were then used as input for differential expression analysis using edgeR. As for genes, a thresholded test was performed using the glmTreat function. Genes whose downstream regions showed at least 4-fold up-regulation upon compound treatment (log2 fold-change cutoff of 2, adjusted p-value < 0.05) were considered to show read-through. Pathway and process enrichment analysis followed by hierarchical clustering was performed using Metascape with the default settings. The following ontologies were included: KEGG Pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways and CORUM. Cutoffs for significantly enriched terms were p-value < 0.01, minimum count of 3, and enrichment of observed / expected counts > 1.5. For bar plots, the single most statistically significant term per cluster is shown. Further methodological details can be found at www.metascape.org.
Supplementary Material
Acknowledgements
The authors wish to thank Dale Porter, Ralph Tiedt, Jeremy Baryza, and Greg Rice for helpful discussions through the course of this research study. This work was supported by the Novartis Research Foundation (J.A.C.), the SNF-NCCR RNA & Disease network (J.A.C.) and the Medical Research Council (MRC) grant MC_U105192715 (L.A.P.).
Footnotes
Author contributions
R.E.J.B., F.L., and N.T.R conceptualized research, interpreted data, and wrote the manuscript; S.C. conducted and interpreted biomarker, variomics, and mechanism of action studies; A.F. synthesized all compounds; W.A.W. conducted crystallography experiments and solved structures; M.H. conducted the siRNA synergy experiment; M.S. and G.M. conducted and interpreted CPSF3 biochemical binding studies; J.T., S.C., and F.A.A. prepared RNA samples; J.K. and W.C. conducted RNA sequencing experiments; S.H.C., S.G., S.S. and G.R. interpreted RNA-seq results; S.B., J.M., J.T., and M.S. conducted and interpreted chemical proteomics experiments; G.M. and S.C. conducted and interpreted FACS experiments; Y.W. and J.J. contributed to in silico target predictions; F.S. analyzed siRNA experiments; H.M. conducted cell line cytotoxicity studies; G.B. and E.G. conducted and interpreted in vivo xenograft experiments; A.C. and K.X. conducted variomics cloning experiments; J.S.R.-H. designed variomics cloning strategy; M.Z. and M.S. conducted mass spectrometry experiments; C.S. and E.T.W. conducted in vivo compound PK experiments; J.H.W., C.A.-R., M.J. and J.A.C. conducted and interpreted FISH and R-loop experiments; J.B.R.M. and L.A.P. conducted and interpreted in vitro cleavage experiments; J.A.C. and J.T. helped write the manuscript.
Competing Financial Interests Statement
All authors (except otherwise noted) were employees of Novartis Institutes for BioMedical Research at the time of their involvement in this study and may hold stock in Novartis. Correspondence should be addressed to R.E.J.B. (rohan.beckwith@novartis.com).
Data Availability
The datasets generated during and/or analyzed during the current study are included in this published article (and its supplementary information files). The raw RNA-sequencing reads are available in the NCBI Sequence Read Archive under accession number SRP158650. X-ray structure data has been deposited to wwPDB with pdb code 6M8Q. All other relevant data are available from the corresponding author on reasonable request.
References
- 1.Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507–19. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 2.Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber SL. Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg. Med. Chem. 1998;6:1127–1152. doi: 10.1016/s0968-0896(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 4.Carson C, et al. Englerin A agonizes the TRPC4/C5 cation channels to inhibit tumor cell line proliferation. PLoS One. 2015;10:e0127498. doi: 10.1371/journal.pone.0127498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothman DM, et al. Metabolic Enzyme Sulfotransferase 1A1 Is the Trigger for N-Benzyl Indole Carbinol Tumor Growth Suppression. Chem. Biol. 2015;22:1228–1237. doi: 10.1016/j.chembiol.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Kakutani M, Takeuchi K, Waga I, Iwamura H, Wakitani K. JTE-607, a novel inflammatory cytokine synthesis inhibitor without immunosuppression, protects from endotoxin shock in mice. Inflamm. Res. 1999;48:461–468. doi: 10.1007/s000110050487. [DOI] [PubMed] [Google Scholar]
- 7.Borozdenkova S, et al. Effects of a cytokine inhibitor, JTE-607, on the response to endotoxin in healthy human volunteers. Int. Immunopharmacol. 2011;11:1837–1843. doi: 10.1016/j.intimp.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Ryugo M, et al. Pharmacologic preconditioning of JTE-607, a novel cytokine inhibitor, attenuates ischemia-reperfusion injury in the myocardium. J. Thorac. Cardiovasc. Surg. 2004;127:1723–1727. doi: 10.1016/j.jtcvs.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Jian MY, Koizumi T, Tsushima K, Kubo K. JTE-607, a cytokine release blocker, attenuates acid aspiration-induced lung injury in rats. Eur. J. Pharmacol. 2004;488:231–238. doi: 10.1016/j.ejphar.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Uesato N, Fukui K, Maruhashi J, Tojo A, Tajima N. JTE-607, a multiple cytokine production inhibitor, ameliorates disease in a SCID mouse xenograft acute myeloid leukemia model. Exp. Hematol. 2006;34:1385–1392. doi: 10.1016/j.exphem.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Tajima N, et al. JTE-607, a multiple cytokine production inhibitor, induces apoptosis accompanied by an increase in p21waf1/cip1 in acute myelogenous leukemia cells. Cancer Sci. 2010;101:774–781. doi: 10.1111/j.1349-7006.2009.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li BE, Ernst P. Two decades of leukemia oncoprotein epistasis: The MLL1 paradigm for epigenetic deregulation in leukemia. Exp. Hematol. 2014;42:995–1012. doi: 10.1016/j.exphem.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev. Pathol. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat. Rev. Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danckwardt S, Hentze MW, Kulozik AE. 3’ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somervaille TCP, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith R, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Tirode F, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 21.Schirle M, Jenkins JL. Identifying compound efficacy targets in phenotypic drug discovery. Drug Discov. Today. 2016;21:82–89. doi: 10.1016/j.drudis.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 22.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 23.König R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 24.Zuberi K, et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gower CM, et al. Conversion of a Single Polypharmacological Agent into Selective Bivalent Inhibitors of Intracellular Kinase Activity. ACS Chem. Biol. 2016;11:121–131. doi: 10.1021/acschembio.5b00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas JR, et al. A Photoaffinity Labeling-Based Chemoproteomics Strategy for Unbiased Target Deconvolution of Small Molecule Drug Candidates. In: Lazar IM, Kontoyianni M, Lazar AC, editors. Proteomics for Drug Discovery: Methods and Protocols. Springer New York; New York: 2017. pp. 1–18. [DOI] [PubMed] [Google Scholar]
- 27.Salcius M, et al. SEC-TID: A label-free method for small-molecule target identification. J. Biomol. Screen. 2014;19:917–927. doi: 10.1177/1087057114522691. [DOI] [PubMed] [Google Scholar]
- 28.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3’-endprocessing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill CH, et al. Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex. Mol. Cell. 2019;73:1217–1231. doi: 10.1016/j.molcel.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bill A, et al. Variomics screen identifies the re-entrant loop of the calcium-activated chloride channel ANO1 that facilitates channel activation. J. Biol. Chem. 2015;290:889–903. doi: 10.1074/jbc.M114.618140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Z, et al. A Functional Variomics Tool for Discovering Drug-Resistance Genes and Drug Targets. Cell Rep. 2013;3:577–585. doi: 10.1016/j.celrep.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grohar PJ, et al. Functional Genomic Screening Reveals Splicing of the EWS-FLI1 Fusion Transcript as a Vulnerability in Ewing Sarcoma. Cell Rep. 2016;14:598–610. doi: 10.1016/j.celrep.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riggi N, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechansims to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomazou EM, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion proteins EWS-FLI1. Cell Rep. 2015;10:1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerry J, et al. MLL-AF4 Spreading Identifies Binding Sites that Are Distinct from Super-Enhancers and that Govern Sensitivity to DOT1L Inhibition in Leukemia. Cell Rep. 2017;18:482–495. doi: 10.1016/j.celrep.2016.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorthi A, et al. EWS-FLI1 increases transcription to cause R-Loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555:387–391. doi: 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakegawa J, Sakane N, Suzuki K, Yoshida T. JTE-607, a multiple cytokine production inhibitor, targets CPSF3 and inhibits pre-mRNA processing. Biochem. Biophys. Res. Commun. 2019;518:32–37. doi: 10.1016/j.bbrc.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Iniguez AB, et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer Cell. 2018;33:202–216. doi: 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidaleo M, et al. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget. 2015;6:31740–31757. doi: 10.18632/oncotarget.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selvanathan SP, et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA. 2015;112:E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubbury SJ, Boutz PL, Sharp PA. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature. 2018;564:141–145. doi: 10.1038/s41586-018-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teloni F, et al. Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability. Mol. Cell. 2019;73:670–683. doi: 10.1016/j.molcel.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 48.Palacino J, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 49.Sonoiki E, et al. A potent antimalarial benzoxaborole targets a Plasmodium falciparum cleavage and polyadenylation specificity factor homologue. Nat. Commun. 2017;8:1–11. doi: 10.1038/ncomms14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, et al. Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angew. Chemie Int. Ed. Engl. 2013;52:8551–8556. doi: 10.1002/anie.201300683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are included in this published article (and its supplementary information files). The raw RNA-sequencing reads are available in the NCBI Sequence Read Archive under accession number SRP158650. X-ray structure data has been deposited to wwPDB with pdb code 6M8Q. All other relevant data are available from the corresponding author on reasonable request.